Abstract
We have used multiple sequencing approaches to sequence the genome of a volunteer from Saudi Arabia. We use the resulting data to generate a de novo assembly of the genome, and use different computational approaches to refine the assembly. As a consequence, we provide a contiguous assembly of the complete genome of an individual from Saudi Arabia for all chromosomes except chromosome Y, and label this assembly KSA001. We transferred genome annotations from reference genomes to fully annotate KSA001, and we make all primary sequencing data, the assembly, and the genome annotations freely available in public databases using the FAIR data principles. KSA001 is the first telomere-to-telomere-assembled genome from a Saudi individual that is freely available for any purpose.
Subject terms: Genome informatics, Genomics
Background & Summary
The first complete, or almost complete, sequence of a human genome was made available in 2020 and published in 20221, based on the functionally haploid cell line CHM13. Since then, several human genome assemblies were published2,3, including the diploid genome sequences of 47 individuals4. The availability of these genomes is driven both by advances in sequencing technology which make it possible to sequence more accurate and longer reads, and advances in assembly and read mapping algorithms5–8 which can efficiently assemble and map sequence reads obtained from different sequencing technologies and of different quality, as well as assemble complex regions of genomes.
The availability of cheap, fast, and accurate sequencing technologies now enables sequencing of multiple genomes from diverse populations in order to understand their genetic variability. For each population, it also becomes possible to develop computational resources and databases that capture their diversity. Creating population-specific computational resources can serve as the foundation for bioinformatics workflows and thereby improve the accuracy of genomic analyses within a population as well as when comparing results between multiple different populations9.
Reference genomes in particular are a foundation of workflows that analyze genomic data, and it is well-known that current reference genomes exhibit population bias10 that may affect analyses built on them9. Specifically, population-specific structural variants will likely not be included in reference genomes that are not derived from the same population, and, consequently, sequencing reads may not be aligned accurately when the corresponding genomic region is missing from the reference genome; this can affect the success of identifying disease-causing variants or pre-disposing variants for common diseases within the population.
In the Middle East, the Qatari genome project11 and the United Arab Emirates population genome project12 aimed to address this challenge by producing a population-specific reference genome that includes as major alleles the most frequent ones identified within their respective populations. However, many populations in the Middle East have historically been organized in tribal structures where marriages occur predominantly within a tribe13 leading to several populations that were isolated for a period of time and therefore exhibit different genetic structure14. Additionally, the tribal structure also resulted in a relatively high prevalence of consanguineous marriages and consequently homozygosity and Mendelian diseases15. No single reference genome can fully capture the genetic diversity found in different populations within the Middle East.
Although there are many genetic studies of ancient and current populations of the Middle East16–19, only little genomic data from the Middle East is publicly available, or data that is available can only be used under prohibitive licenses, not be shared publicly, or not use commercially. However, public availability and permissive licences are crucial for resources that need to be shared and utilized broadly, in particular for reference genomes. To further ensure broad usability, resources that underlie common analyses and workflows should be Findable, Accessible, Interoperable, and Reusable (FAIR)20.
We sequenced the genome of a female volunteer from Saudi Arabia who consented to make her genomic data public and freely available. We used three different sequencing strategies based on long and short reads. The first draft genome was created based on a de novo genome assembly using these reads. The assembly was further refined using the CHM13 genome1 resulting in the most complete publicly available genome sequence of a Saudi individual so far. We use a variety of tools to fully annotate this genome, either transferring information from public databases or using methods from bioinformatics to identify functional elements (genes, regulatory regions) using the genome sequence directly. Our sequencing, assembly, and annotation effort resulted in a reference-quality personal genome from a Saudi individual which we label KSA001.
The assembled genome sequence of KSA001, the primary sequencing data, and the workflows used to construct the genome sequence are freely available on https://github.com/bio-ontology-research-group/KSA001. The sequence reads and assembly are further available in public sequence databases, and the genome is available in standard formats, making this the first telomere-to-telomere-assembled genome from the Arabian peninsula. We provide genome annotations for KSA001 and make a variant calling workflow using KSA001 freely available as well to enable its immediate use in molecular genetics studies.
We make this genome publicly available following the FAIR principles20. The genome is findable as it is deposited in repositories containing genome assemblies, and accessible through common protocols and data download utilities. It is also interoperable as we use standard formats used across bioinformatics, and it is reusable as we make the primary data available as well as a description of the workflow that led to the assembled genome. The quality of the assembly and the set of genome annotations we make available, but in particular its free availability, allows KSA001 to be shared and used as a reference genome for genomic studies in Saudi Arabia and the Middle East. We demonstrated that KSA001 can be used for variant calling and therefore can contribute to the success of diagnostic or prognostic genomic methods.
Methods
Recruitment
One volunteer provided the sample for KSA001, and the volunteer’s parents also donated samples. The donors provided informed consent for the collection of blood, DNA sequencing, and for making all data public. The criteria for selecting the donor were Saudi nationality with tribal origin, over 18 years old, and the ability to provide informed consent.
The study and recruitment process were advertised in the research center at King Fahad Medical City between 2022 and 2023 and were overseen by Dr. Malak Abedalthagafi. This process included detailed bilingual consent (Arabic and English) and extensive counseling. The donors voluntarily agreed to donate their anonymous samples, with written informed consent obtained from all participants, allowing the data and analyses from the genome sequencing to be made public. The parents of the individual designated as KSA001 agreed to be part of the study and were provided with detailed consent forms and counseling.
DNA extraction
DNA was extracted using two methods. Ultra high molecular weight DNA (uHMW) was isolated from fresh blood samples using New England Biolabs (NEB) Monarch High Molecular Weight (HMW) DNA isolation kit following manufacturer’s protocol (New England Biolabs, UK) with modification, agitation was set at 700 rpm during the lysis step. DNA was kept at 4 °C until library preparation for long-read sequencing using the PacBio and Oxford nanopore sequencing platforms.
For short read sequencing of KSA001, we isolated donor DNA using the DNeasy Blood & Tissue kit (Qiagen) following the manufacturer’s instructions. DNA was kept at −20 °C until library preparation.
For the parents of KSA001, we extracted genomic DNA from blood samples of the parents of KSA001 using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Per the manufacturer’s instructions, we placed 200 μL of blood in a 1.5 mL microcentrifuge tube and mixed it with 20 μL of proteinase K and 200 μL of Buffer AL. We vortexed the sample for 15 seconds and incubated it at 56 °C for 10 minutes to allow complete lysis. We followed the consequent steps in the protocol, which consisted of DNA binding to the silica membrane and washing to remove residual contaminants and proteins. Finally, we eluted the DNA, where we placed the DNeasy Mini spin column in a clean 1.5 mL microcentrifuge tube and pipetted 200 μL of Buffer AE directly onto the membrane. After incubating at room temperature for 1 minute, we centrifuged the column at 6,000 x g for 1 minute.
Sequencing library preparation and sequencing
We used three platforms for sequencing the extracted DNA: Illumina NovaSeq 6000 (Illumina, San Diego, USA), PacBio Sequel II (Pacific Biosciences, Menlo Park, USA), and Oxford Nanopore PromethION (Oxford Nanopore Technologies, Oxford, UK).
We used 100 ng of genomic DNA (gDNA) as an input to construct a whole genome library to be sequenced using the NovaSeq 6000 platform. The DNA was mechanically sheared with Covaris (Covaris, Woburn, USA) and converted to a sequence-ready library using the TruSeq DNA Nano Library Kit (Illumina, San Diego, USA). Subsequently, the library was quantified using Qubit high-sensitivity dsDNA Assays (model Q33230, ThermoFisher Scientific, Waltham, USA), and the quality control was performed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA).
The KSA001 sample was sequenced on two lanes of an SP flowcell with a read length of 2 × 150 bp in paired-end format, which generated 311 GB of data, resulting in an estimated 100x coverage. For the parents of KSA001, we sequenced both libraries on a NovaSeq 6000 system (Illumina, USA) using an SP flowcell (2 × 150 cycles), with each sample sequenced on a single lane. The genome coverage was approximately 23× and 29× for maternal and paternal samples, respectively.
7.5 μg of high molecular weight gDNA was sheared with Megaruptor 3 (Diagenode, Denville, USA) to the size range of 15-20 kb. SMRTbell was prepared with HiFi Express Template prep kit 2.0 (102-088-900), and size-selected with the PippinHT System (Sage Science HTP0001). Finally, SMRTbell QC was assessed with Qubit dsDNA High Sensitivity (model Q33230; ThermoFisher Scientific, Waltham, USA) and FEMTO Pulse (Inc. P-0003-0817; Agilent Technologies, Santa Clara, USA). Sequencing of SMRTbell was set up on PacBio Sequel II system with Sequel II Binding kit 2.2 (101-894-200), Sequel II Sequencing Kit 2.0 (101-820-200), and SMRTcell 8M Tray (101-389-001), according to conditions specified in SMRTlink with 30 hour movie times, 2 hour pre-extension time, and adaptive loading mode.
For Nanopore sequencing, two libraries were prepared using the Ultra-Long Sequencing Kit (SQK-ULK001) and its recommended protocol from Oxford Nanopore Technologies (Oxford Nanopore Technologies, Oxford, UK). 30 μg of uHMW gDNA was used as input for each library. A PromethION flowcell (FLO-PRO002) was primed and prepared according to the same protocol and 75 μl of a sequencing library was loaded. 24 hours after the first sequencing run, a nuclease flush and priming step were performed according to the protocol, and an additional 75 μl of library was loaded before starting another sequencing run. This process was repeated until each library was loaded three times. Basecalling was performed using Guppy v5.1.13 with the super-accurate basecalling model. The entire process is repeated with the second prepared library and another PromethION flowcell.
We employed the Dovetail Omni-C protocol (Dovetail Genomics, USA) to construct Hi-C libraries, a widely adopted method for interrogating chromatin interactions on a genome-wide scale. We initiated the Omni-C Proximity Ligation Assay protocol using 1 ml of fresh whole human blood collected in EDTA-coated tubes as an anticoagulant. The sample was processed to reach a cell density of 1 million cells/mL as an input that then crosslinked using formaldehyde. This process preserves the native chromatin structure, allowing us to work with genomic DNA in its original nuclear context. After cell lysis, we captured chromatin using Chromatin Capture Beads and fragmented it through in situ nuclease digestion. We then performed proximity ligation using an Intra-Aggregate Ligation Enzyme Mix, resulting in chimeric DNA molecules that represent the original three-dimensional chromatin interactions. Following DNA purification and size selection, we quantified the purified DNA using Qubit fluorometry and used 150 ng as input for library preparation. This DNA represents crosslinked, proximity-ligated genomic DNA fragments. The library was generated through end repair, adapter ligation, and index PCR steps. Finally, we subjected the library to a ligation capture step using the Dovetail™ Primer Set for Illumina and amplified it by PCR to generate the final Omni-C library. This approach allowed us to capture and analyze genome-wide chromatin interactions from our blood samples, providing insights into the three-dimensional organization of the genome.
Assembly
We used PacBio HiFi reads and the ONT ultra-long reads together with Hi-C or reads from the parents to resolve the haplotypes. We applied Hifiasm v0.19.0-r53421 and Verkko (commit 508efb)22 in Hi-C and Trio modes to generate four haplotype-resolved assemblies. Table 1 shows basic statistics of the sequencing data. We evaluated the quality of the assemblies using Merqury23 with k-mer size of 21. Table 2 provides statistics on each of our assemblies. We use the Verkko’s Trio assembly as our primary diploid assembly because it resulted in highest sequence length of the shortest contig at 50% (N50) and Quality Value (QV) score. Trio mode assemblies also enable us to determine the maternal and paternal haplotypes.
Table 1.
Sequencing reads statistics.
| Sequencing Technology | Number of Reads | Avg. Read length | GC(%) | Coverage |
|---|---|---|---|---|
| Illumina | 2,061,737,744 | 151 | 41.3 | 100x |
| PacBio | 8,994,317 | 17,657 | 40.6 | 51x |
| Oxford Nanopore | 1,863,431 | 51,076 | 40.3 | 30x |
| Hi-C | 729,360,320 | 151 | 45.6 | 35x |
| Illumina (Parent 1) | 477,854,908 | 151 | 40.1 | 23x |
| Illumina (Parent 2) | 602,505,048 | 151 | 40.6 | 29x |
Table 2.
Assembly statistics and quality.
| Tool | Maternal | Paternal | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Scaffolds | Gaps | N50 | Length | QV | Scaffolds | Gaps | N50 | Length | QV | |
| Hifiasm - HiC | 728 | 0 | 104.145 MB | 3062.204 MB | 66.0647 | 626 | 0 | 105.218 MB | 3027.886 MB | 66.4196 |
| Hifiasm - Trio | 494 | 0 | 106.330 MB | 3036.153 MB | 65.5969 | 701 | 0 | 99.993 MB | 3068.265 MB | 66.8000 |
| Verkko - HiC | 45 | 11 | 136.125 MB | 3031.626 MB | 65.2061 | 75 | 11 | 135.417 MB | 3008.205 MB | 65.6895 |
| Verkko - Trio | 92 | 18 | 145.092 MB | 3026.057 MB | 67.2079 | 173 | 14 | 136.125 MB | 3029.708 MB | 63.6325 |
Refinement of assembly
After the initial assembly process, we aligned the contigs to CHM13 using minimap224 and manually inspected the gaps that were not linked in previous steps. We found that most of them are in centromeres which contain highly repetitive regions and are generally difficult to align and assemble25,26. Using the alignments, we placed the assembly scaffolds into chromosomes. We found that 18 chromosomes completely align to one scaffold in our assembly and chromosomes 13, 15, 19, 21, X map to 2 or 3 scaffolds with gaps around highly repetitive centromeric regions. After this step, maternal and paternal haplotypes had 25 and 23 gaps, respectively.
We performed an additional assembly using the ultra-long ONT reads only with the Flye assembler (2.9.1)27 and used MaSuRCA Samba (v4.1.0)28 to close the gaps using the ONT based assembly. After this step, the maternal haplotype had 18 gaps and the paternal haplotype had 17 gaps left. We also used the Flye assembly to derive a circular contig with two copies of the mitochondrial genome. Furthermore, we polished the mitochondrial genome from the ONT-based assembly using NextPolish (v1.4.1)29 guided by quality-filtered Illumina reads.
We generated a haploid assembly by merging both haplotypes and selecting the best chromosome based on its QV score. We computed QV scores using Merqury23 with k-mer size of 21 from both Illumina short reads and PacBio HiFi reads. Then, we selected chromosomes with higher QV scores for our haploid KSA001 assembly. We used the BioPython library version 1.8130 to generate the haploid assembly using custom code that is available on the project’s code repository. In the haploid KSA001 genome, 15 chromosomes (containing 2,098,976,917 bp) and the mitochondrial genome (16,567 bp) were derived from the maternal haplotype, and 7 chromosomes (containing 959,396,833 bp) were derived from the paternal haplotype. The haploid KSA001 has 12 gaps in total in chromosomes 7 (2 gaps), 9 (1 gap), 13 (2 gaps), 15 (2 gaps), 21 (1 gap), 22 (1 gap), and X (3 gaps). We compared to CHM13 and found that all the gaps are around the centromeric regions with highly repetitive genome sequence.
Genome annotation
We used the Liftoff tool (v1.6.1)31 to transfer gene annotations from CHM13 (v2.0) and GRCh38 (v40). We lifted over UCSC GENCODEv35 CAT/Liftoff v2 gene annotations from CHM13 and UCSC KnownGene, UCSC RefSeq and NCBI RefSeq from GRCh38.
To use KSA001 in downstream applications, we generated chain files between KSA001 (v1.0.0) and both CHM13 (v2.0) and GRCh38 (v40) using minimap2 (v.2.26)24 together with ChainTools32, rustybam33 and paf2chain34. Furthermore, we used BCFtools liftover35 to transfer the dbSNP (v156) and gnomAD annotation from GRCh38 to KSA001.
Ethical approval
This work was approved by the Institutional Review Board (IRB) at the Faculty of Medicine, King Fahad Medical City (KFMC) under approval number 22-037, and by the Institutional Bioethics Committee (IBEC) at King Abdullah University of Science and Technology (KAUST) under approval number 22IBEC023. The approval covers the recruitment of one Saudi individual with at least three generations of tribal roots in Saudi Arabia to provide a blood sample. The approved protocol specifies that this sample will be sequenced using multiple sequencing technologies, processed with bioinformatics tools, and published without access restrictions for use in bioinformatics workflows. Additionally, the approval includes the recruitment of 10 more Saudi individuals to provide blood samples for sequencing, with their data also made freely available.
The consent forms were available in both English and Arabic. Participants were recruited by Dr. Malak Abedalthagafi, who is the Principal Investigator (PI) for the approved IRB protocol at King Fahad Medical City. The consent for volunteer KSA001 was co-signed by two witnesses, including the IRB chairperson. Dr. Abedalthagafi personally obtained consent from the volunteers, who participated without any payment or compensation and were recruited from research clinic care. The consent was signed in the presence of two witnesses. There is no known professional or personal relationship between the authors and the participants, except for Dr. Malak Abedalthagafi, who ensured that participants were fully aware of the risks and were competent to understand them. As part of the consent process, participants underwent genetic counseling, as required by the IRB protocol.
None of the other authors were involved in the ethical approval processes at KFMC or KAUST. Robert Hoehndorf was a member of the IBEC at KAUST at the time of approval but did not participate in the review or discussion of the research protocol, and was not present during its discussion.
Data Records
Data, supporting information, and a description of the assembly workflow are available at https://github.com/bio-ontology-research-group/KSA001. Sequencing reads are available on the Sequence Read Archive under accession numbers SRR2192783636, SRR2192783537, SRR2192783438, SRR2192783339, SRR2912251940, and SRR2909248741, and collected under the project SRP402943 (NCBI BioProject PRJNA891101). Assembled genomes for maternal and paternal haplotypes are available at NCBI Datasets Genomes under accession numbers GCA_037177635.142 and GCA_037177555.143, respectively.
Technical Validation
Comparison to CHM13 and GRCh38
We first annotated the KSA001 genome by transferring genome features from CHM13 to KSA001 using the Liftoff (v1.6.1) tool31. Liftoff successfully mapped 62,351 genes out of 63,494 genes of all types (98%, Table 3) to KSA001. When we further considered overlapping genome feature annotations, 1,072 additional genes were mapped. By default, Liftoff considers a gene as mapped when it satisfies at least 50% of alignment coverage and sequence identity. Most annotated genes were mapped with sequence identity higher than 99%. Table 4 provides the summary of mapped genes by the sequence identity and Table 5 provides a summary of all mapped genome features. Out of 19,969 protein coding genes, the Proline Rich 32 (PRR32, ENTREZ:100130613) gene is completely missing from KSA001 in comparison to genes in CHM13.
Table 3.
LiftOff annotation statistics.
| Genes | Transcripts | ||||
|---|---|---|---|---|---|
| Total | Protein coding | Non-coding | Total number | Protein coding | |
| CHM13 | 63,494 | 19,969 | 43,525 | 234,903 | 156,412 |
| KSA001 | 63,421 | 19,968 | 43,453 | 233,494 | 155,918 |
Table 4.
LiftOff annotation statistics. Number of mapped genes from CHM13 grouped by sequence identity.
| Genes | 100% | ≥99% | ≥95% | ≥90% | ≥75% | Total |
|---|---|---|---|---|---|---|
| All | 22,593 | 59,936 | 62,560 | 62,933 | 63,257 | 63,421 |
| Protein coding | 1,581 | 18,484 | 19,591 | 19,769 | 19,906 | 19,968 |
| Non-coding | 21,012 | 41,452 | 42,969 | 43,164 | 43,351 | 43,453 |
Table 5.
Gene annotation summary.
| Gene category | Gene Biotype | CHM13 | KSA001 | Count of unmapped |
|---|---|---|---|---|
| Protein-coding | protein coding | 19,969 | 19,968 | 1 |
| Non-coding RNA | lncRNA | 17,482 | 17,482 | 0 |
| miRNA | 2,045 | 2,223 | 20 | |
| misc RNA | 2,224 | 2,221 | 3 | |
| Mt rRNA | 3 | 3 | 0 | |
| Mt tRNA | 29 | 29 | 0 | |
| ribozyme | 8 | 8 | 0 | |
| rRNA | 1,007 | 1,007 | 0 | |
| rRNA pseudogene | 506 | 503 | 3 | |
| scaRNA | 48 | 48 | 0 | |
| scRNA | 2 | 2 | 0 | |
| snoRNA | 945 | 944 | 1 | |
| snRNA | 1,886 | 1,883 | 3 | |
| sRNA | 5 | 5 | 0 | |
| TEC | 1,341 | 1,341 | 0 | |
| vault RNA | 1 | 1 | 0 | |
| Pseudogenes | pseudogene | 18 | 15 | 3 |
| polymorphic pseudogene | 50 | 50 | 0 | |
| processed pseudogene | 10,769 | 10,764 | 5 | |
| transcribed processed pseudogene | 551 | 550 | 1 | |
| transcribed unitary pseudogene | 138 | 137 | 1 | |
| transcribed unprocessed pseudogene | 941 | 941 | 0 | |
| translated processed pseudogene | 2 | 2 | 0 | |
| translated unprocessed pseudogene | 1 | 1 | 0 | |
| unitary pseudogene | 98 | 98 | 0 | |
| unprocessed pseudogene | 2,725 | 2,723 | 2 | |
| Immunoglobulin/T-cell receptor | IG C gene | 15 | 15 | 0 |
| IG D gene | 10 | 0 | 10 | |
| IG C pseudogene | 9 | 10 | 0 | |
| IG J gene | 18 | 8 | 10 | |
| IG J pseudogene | 3 | 3 | 0 | |
| IG pseudogene | 1 | 1 | 0 | |
| IG V gene | 148 | 148 | 0 | |
| IG V pseudogene | 216 | 214 | 2 | |
| TR C gene | 7 | 7 | 0 | |
| TR J gene | 80 | 73 | 7 | |
| TR J pseudogene | 4 | 4 | 0 | |
| TR V gene | 108 | 108 | 0 | |
| TR V pseudogene | 33 | 33 | 0 | |
| Unknown | StringTie | 48 | 48 | 0 |
| total | 63,494 | 63,421 | 73 |
Counts of CHM13 genes (without chrY) mapped by Liftoff to the KSA001 genome assembly.
We further compare KSA001 with CHM13 and GRCh38 in two ways. First, we perform variant calling of short reads derived from sequencing KSA001, using CHM13 and GRCh38 as reference genomes, and report the number of variants in KSA001 when aligned against both the reference genomes (Table 6). Consistent with previously reported results9, we observe a substantially lower number of variants called when using CHM13 as a reference compared to using GRCh38.
Table 6.
Variant call statistics for KSA001 Illumina reads aligned to GRCh38 and CHM13.
| Total Number of variants | SNPs | Indels | |
|---|---|---|---|
| KSA001 aligned to GRCh38 | 5,337,778 | 4,317,999 | 1,115,246 |
| KSA001 aligned to CHM13 | 4,888,875 | 3,934,939 | 1,036,797 |
Comparison between numbers of genes and transcripts in CHM13 and KSA001.
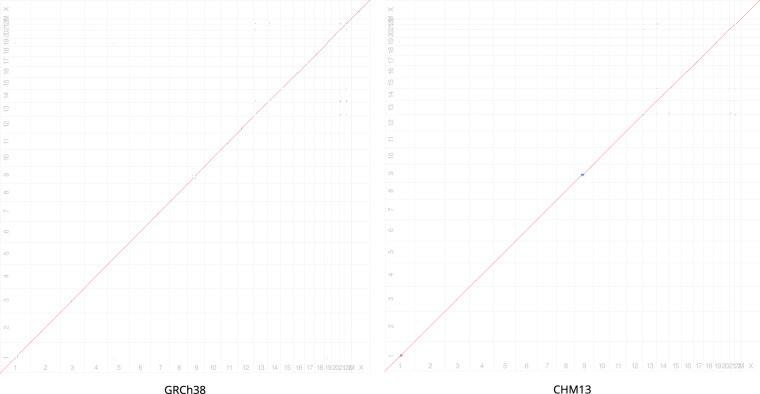
Second, we aligned KSA001 to CHM13 and GRCh38 using Minimap2 (v2.24-r1122) and used the call command within paftools.js to call variants. Table 7 provides summary statistics of the variants. Figure 1 depicts a Harr plot of the assemblies (Harr plots were generated using minidot from miniasm, v0.3-r17944).
Table 7.
Comparison of KSA001 with CHM13 and GRCh38 based on assembly-to-assembly alignment.
| SNPs/Indels | SVs | ||||
|---|---|---|---|---|---|
| SNPs | Insertion | Deletion | Insertion | Deletion | |
| Alignment of KSA001 and CHM13 | 4,012,344 | 379,878 | 358,464 | 13,541 | 13,467 |
| Alignment of KSA001 and GRCh38 | 3,463,231 | 373,606 | 353,703 | 7,539 | 12,523 |
SNPs: Single nucleotide polymorphisms, Indels: insertion or deletion, SV: Structural Variants.
Fig. 1.
Harr plot of KSA001 aligned with GRCh38 (left) and CHM13 (right).
Table 8 shows the completeness of the chromosomes and the number of basepairs in each chromosome of KSA001 in comparison to the CHM13 v2.0 assembly. Overall, the numbers of base-pairs in each chromosome of KSA001 are comparable to CHM13. Also, we report base-level quality (QV) scores for the KSA001 assembly for each chromosome and the entire assembly. QV score represents a log-scaled probability of error for the consensus base calls. Maternal and paternal haplotypes of KSA001 achieve a quality of 68.79 and 64.43 with some of the chromosomes reaching a QV score of almost 80. The merged haploid genome has a QV score of 72.97.
Table 8.
Completeness of chromosomes of KSA001 compared to CHM13 and QV scores.
| Chromosome | CHM13 | KSA001.mat | QV.mat | KSA001.pat | QV.pat | KSA001 | QV |
|---|---|---|---|---|---|---|---|
| chr1 | 248,387,328 | 246,893,779 | 72.66 | 248,515,191 | 68.87 | 249,555,359 | 72.66 |
| chr2 | 242,696,752 | 242,046,071 | 67.91 | 242,632,705 | 75.97 | 243,650,403 | 75.97 |
| chr3 | 201,105,948 | 201,462,031 | 75.93 | 200,338,375 | 77.26 | 200,901,366 | 77.26 |
| chr4 | 193,574,945 | 191,676,519 | 76.81 | 191,977,661 | 76.28 | 193,119,457 | 76.81 |
| chr5 | 182,045,439 | 181,678,762 | 70.83 | 182,455,576 | 58.06 | 182,340,321 | 70.83 |
| chr6 | 172,126,628 | 171,636,591 | 77.00 | 171,747,260 | 73.48 | 172,212,606 | 77.00 |
| chr7 | 160,567,428 | 160,688,708 | 74.99 | 160,686,623 | 78.21 | 161,491,582 | 78.21 |
| chr8 | 146,259,331 | 145,082,495 | 71.28 | 145,751,733 | 78.73 | 146,283,325 | 78.73 |
| chr9 | 150,617,247 | 142,392,075 | 69.80 | 137,639,207 | 59.85 | 144,206,915 | 69.80 |
| chr10 | 134,758,134 | 137,124,436 | 79.28 | 136,124,862 | 75.99 | 137,487,260 | 79.28 |
| chr11 | 135,127,769 | 134,887,184 | 79.08 | 135,515,747 | 67.33 | 135,526,054 | 79.08 |
| chr12 | 133,324,548 | 133,634,782 | 75.29 | 133,706,131 | 61.20 | 137,511,954 | 75.29 |
| chr13 | 113,566,686 | 113,178,117 | 70.10 | 109,813,855 | 60.32 | 114,155,372 | 70.10 |
| chr14 | 101,161,492 | 98,200,010 | 71.78 | 104,568,253 | 75.09 | 104,854,739 | 75.09 |
| chr15 | 99,753,195 | 95,263,998 | 62.42 | 97,348,991 | 72.54 | 100,665,885 | 72.54 |
| chr16 | 96,330,374 | 91,348,093 | 77.78 | 84,634,195 | 74.86 | 91,858,333 | 77.78 |
| chr17 | 84,276,897 | 83,888,424 | 73.11 | 84,257,549 | 76.14 | 84,644,139 | 76.14 |
| chr18 | 80,542,538 | 80,057,793 | 69.88 | 79,675,856 | 55.70 | 80,394,818 | 69.88 |
| chr19 | 61,707,364 | 63,671,480 | 63.93 | 64,545,147 | 65.98 | 66,377,225 | 65.98 |
| chr20 | 66,210,255 | 66,487,749 | 76.83 | 66,499,802 | 79.99 | 66,944,161 | 79.99 |
| chr21 | 45,090,682 | 43,459,253 | 75.13 | 42,689,246 | 75.21 | 43,720,397 | 75.21 |
| chr22 | 51,324,926 | 42,716,621 | 65.58 | 49,396,945 | 66.70 | 49,931,329 | 66.70 |
| chrX | 154,259,566 | 154,358,968 | 68.79 | 153,509,181 | 65.26 | 154,680,412 | 68.79 |
| chrM | 16,569 | 16,567 | 49.39 | 0 | 0.00 | 16,567 | 49.39 |
| chrY | 62,460,029 | 0 | - | 0 | - | 0 | - |
| Total | 3,117,292,070 | 3,021,850,506 | 68.79 | 3,024,030,091 | 64.43 | 3,062,529,979 | 72.97 |
Pathogenic variants in KSA001
We aligned KSA001 to GRCh38 and called the variants using minimap224 assembly-to-assembly alignment strategy. We screened the ClinVar45 database and found that KSA001 is carrying four variants (ClinVar accessions VCV000360644.10, VCV000298183.17, VCV000802557.8, and VCV000293285.5) that are reported in the database as “Conflicting classifications of pathogenicity” and none of “Pathogenic” or “Likely pathogenic” variants.
Using KSA001 in variant calling workflows
Saudi Arabia, and the Middle East and North Africa (MENA) region, have a higher burden of genetic diseases due to consanguinity46, the accurate calling of rare genomic variants is particularly important for the diagnosis of Mendelian disorders which are more prevalent in the region. Identifying functionally important structural variants (SVs) is more challenging as population-specific SVs are not included in reference genomes. We have identified thousands of SNPs, Indels, and SVs with a size reaching up to 102,752 bp in KSA001 compared to CHM13 (Table 7). While a single genome is not a sufficient representation of genetic variation within a population, KSA001 will likely contain several common variants from the Saudi population; including these variants in a reference used for variant calling has the potential to provide more accurate alignments of sequencing reads, and can make variant calling more efficient and specific to disease-causing variants.
The majority of genomic samples, in particular in a clinic, are still processed using short-read sequencing methods. To test whether KSA001 can be used for and potentially improve variant calling in Saudi individuals, we utilized a public genome sequence from a Saudi individual (SRR2700225647) sequenced on an Illumina NovaSeq 6000.
We aligned the Illumina reads to KSA001, GRCh38, and CHM13 using BWA-MEM (v0.7.17)48. We then sorted and indexed the alignment files using Picard (v2.20.4)49, and marked duplicate reads using the same tool. To reduce systematic errors in base quality scores caused by sequencers, we performed base quality score recalibration (BQSR) using GATK (v4.1.2.0)49 by first building a covariation model and then applying it to adjust the quality scores based on the model. For variant calling, we used HaplotypeCaller within GATK to call SNPs and small indels through a local de novo assembly of haplotypes in specific regions. As part of this process, we also generated a number of common resources that transfer information from CHM13 to KSA001. Table 9 shows a summary of the number of variants called on the Saudi individual we are using. We identify fewer average number of variants when using KSA001 and CHM13 compared to using GRCh38, indicating that KSA001 captures major alleles in the Saudi population better than GRCh38; the number of variants identified using KSA001 as reference is similar to (although slightly more than) the number of variants identified using CHM13.
Table 9.
Variant calling statistics for the Saudi donor (SRR27002256), using Illumina reads aligned to KSA001, CHM13, and GRCh38.
| Total Number of variants | SNPs | Indels | Homozygous variants | Heterozygous variants | |
|---|---|---|---|---|---|
| KSA001 | 4,632,600 | 3,756,816 | 875,784 | 1,413,245 | 2,552,866 |
| CHM13 | 4,525,713 | 3,671,251 | 854,462 | 1,306,039 | 2,562,750 |
| GRCh38 | 4,928,855 | 4,012,232 | 916,623 | 1,595,018 | 2,594,096 |
Furthermore, we repeated the variant calling workflow using Illumina reads of the mother (SRR2912251940) and father (SRR2905592250) of KSA001 (Table 10). We identify considerably fewer variants of all types using KSA001 as a reference compared to CHM13 and GRCh38, with the exception of heterozygous variants using reads from the father. We also find fewer variants when aligning reads from the mother compared to the father which is consistent with the fact that approximately 2/3 of the haploid KSA001 genome is derived from the maternal haplotype.
Table 10.
Variant calling statistics for the mother (SRR29122519) and father (SRR29055922) of KSA001, using Illumina reads aligned to KSA001, CHM13, and GRCh38.
| Sample | Reference | Variants | SNPs | Indels | Homozygous | Heterozygous |
|---|---|---|---|---|---|---|
| Mother | KSA001 | 3,844,863 | 3,137,864 | 706,999 | 575,836 | 2,718,732 |
| CHM13 | 4,609,105 | 3,776,510 | 832,595 | 1,204,406 | 2,751,851 | |
| GRCh38 | 4,979,364 | 4,089,298 | 890,066 | 1,474,714 | 2,757,196 | |
| Father | KSA001 | 4,446,695 | 3,615,255 | 831,440 | 816,030 | 2,733,448 |
| CHM13 | 4,647,609 | 3,762,777 | 884,832 | 1,281,940 | 2,693,290 | |
| GRCh38 | 5,012,366 | 4,075,934 | 936,432 | 1,525,662 | 2,726,041 |
Additionally, we performed variant calling against the same three reference genomes using short reads from individuals of different ethnicities: an East Asian Han Chinese female (SRR129555451), a European British male (SRR129102652), an American Peruvian male (SRR129542653), and a Middle Eastern Bedouin male (ERR75783154) (Table 11). In terms of KSA001 as a reference, we find an overall lower number of variants when aligning reads of the Han Chinese individual, also the only female compared. We also find that using CHM13 as a reference resulted in the lowest number of variants across the different ethnicities, with the exception of the Peruvian individual, which had slightly less variants when aligned to GRCh38.
Table 11.
Variant calling statistics for individuals of varying ethnicities using Illumina reads aligned to KSA001, CHM13, and GRCh38.
| Sample | Reference | Variants | SNPs | Indels | Homozygous | Heterozygous |
|---|---|---|---|---|---|---|
| Han Chinese | KSA001 | 4,659,795 | 3,891,211 | 768,584 | 1,591,064 | 2,323,911 |
| CHM13 | 4,464,929 | 3,729,342 | 735,587 | 1,478,926 | 2,305,843 | |
| GRCh38 | 4,824,298 | 4,043,873 | 780,425 | 1,709,181 | 2,341,850 | |
| British | KSA001 | 4,989,584 | 4,155,236 | 834,348 | 1,469,535 | 2,523,526 |
| CHM13 | 4,438,021 | 3,685,963 | 752,058 | 1,300,352 | 2,451,308 | |
| GRCh38 | 4,868,354 | 4,053,247 | 815,107 | 1,608,939 | 2,477,185 | |
| Peruvian | KSA001 | 5,235,296 | 4,458,182 | 777,114 | 1,682,902 | 2,775,561 |
| CHM13 | 5,213,319 | 4,441,326 | 771,993 | 1,690,479 | 2,770,716 | |
| GRCh38 | 5,213,056 | 4,443,981 | 769,075 | 1,669,264 | 2,773,262 | |
| Bedouin | KSA001 | 5,057,761 | 4,320,790 | 826,971 | 1,464,537 | 2,642,526 |
| CHM13 | 4,560,734 | 3,802,390 | 758,344 | 1,343,928 | 2,568,971 | |
| GRCh38 | 4,945,775 | 4,132,974 | 812,801 | 1,604,459 | 2,605,295 |
The individuals are an East Asian Han Chinese (SRR1295554), a European British (SRR1291026), an American Peruvian (SRR1295426), and a Middle Eastern Bedouin (ERR757831).
Acknowledgements
This work has been supported by funding from King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) under Award No. REI/1/4789-01-01, URF/1/4675-01-01, REI/1/4938-01-01, REI/1/5235-01-01, and URF/1/4697-01-01; by funding from King Abdullah University of Science and Technology (KAUST) - KAUST Center of Excellence for Smart Health (KCSH) under award number 5932; and by funding from King Abdullah University of Science and Technology (KAUST) - Center of Excellence for Generative AI under award number 5940. MSA is supported by King Salman Center for disability research grant R-20190016. We acknowledge support from the KAUST Supercomputing Laboratory, and support from the KAUST Bioscience Core Laboratory.
Author contributions
M.K.: draft assembly and refinement, analysis; R.T.: variant calling, analysis; Y.L.: variant calling, technical validation; H.A.A.: genome annotation, inversions; M.Alarawi: DNA extraction; MAbdelhakim: sample processing, DNA extraction, library preparation; H.A.: library preparation; E.A.: sample processing, DNA extraction; D.A.: DNA extraction; A.Alth.: variant calling, analysis; A.Ang.: PacBio and ONT protocol optimization and sequencing; S.B.: draft assembly and refinement; P.D.: PacBio and ONT protocol optimization and sequencing, supervision; C.P.: PacBio and ONT protocol optimization and sequencing; A.P.: PacBio and ONT protocol optimization and sequencing; A.R.R.: PacBio and ONT protocol optimization and sequencing; CAEH: supervision; N.C.: supervision; M.S.A.: conception, sample processing, supervision, funding acquisition; R.H.: conception, supervision, funding acquisition.
Code availability
Source code and additional resources are available at https://github.com/bio-ontology-research-group/KSA001.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Malak S. Abedalthagafi, Robert Hoehndorf.
Contributor Information
Malak S. Abedalthagafi, Email: malak.althgafi@emory.edu
Robert Hoehndorf, Email: robert.hoehndorf@kaust.edu.sa.
References
- 1.Nurk, S. et al. The complete sequence of a human genome. Science376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimin, A. V. et al. A reference-quality, fully annotated genome from a Puerto Rican individual. Genetics220, iyab227 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang, C. et al. The complete and fully-phased diploid genome of a male han chinese. Cell Research33, 745–761 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao, Wen-Wei et al. A draft human pangenome reference. Nature617, 312–324 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mc Cartney, A. M. et al. Chasing perfection: validation and polishing strategies for telomere-to-telomere genome assemblies. Nature Methods19, 687–695 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain, C., Rhie, A., Hansen, N. F., Koren, S. & Phillippy, A. M. Long-read mapping to repetitive reference sequences using Winnowmap2. Nature Methods19, 705–710 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vollger, M. R. et al. Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. Annals of Human Genetics84, 125–140, 10.1111/ahg.12364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurk, S. et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Research30, 1291–1305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aganezov, S. et al. A complete reference genome improves analysis of human genetic variation. Science376, eabl3533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paten, B., Novak, A. M., Eizenga, J. M. & Garrison, E. Genome graphs and the evolution of genome inference. Genome Research27, 665–676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fakhro, K. A. et al. The Qatar genome: a population-specific tool for precision medicine in the Middle East. Human Genome Variation3, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daw Elbait, G., Henschel, A., Tay, G. K. & Al Safar, H. S. A Population-Specific Major Allele Reference Genome From The United Arab Emirates Population. Frontiers in Genetics12, 660428 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakoush, O., Bredan, A. & Denic, S. KIN AND NON-KIN MARRIAGES AND FAMILY STRUCTURE IN A RICH TRIBAL SOCIETY. Journal of Biosocial Science48, 797–805 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Mineta, K., Goto, K., Gojobori, T. & Alkuraya, F. S. Population structure of indigenous inhabitants of Arabia. PLOS Genetics17, e1009210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alkuraya, F. S. Genetics and genomic medicine in Saudi Arabia. Molecular Genetics & Genomic Medicine2, 369–378, 10.1002/mgg3.97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John, SumiElsa et al. Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Scientific Reports8, 16583 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott, E. M. et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nature Genetics48, 1071–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rautiainen, M. et al. Telomere-to-telomere assembly of diploid chromosomes with verkko. Nature Biotechnology41, 1474–1482 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biology21, 245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics37, 4572–4574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden, K. E. Human centromere genomics: now it’s personal. Chromosome Research20, 621–633 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Altemose, N. et al. Complete genomic and epigenetic maps of human centromeres. Science376, eabl4178 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Zimin, A. V. & Salzberg, S. L. The SAMBA tool uses long reads to improve the contiguity of genome assemblies. PLOS Computational Biology18, e1009860 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, J., Fan, J., Sun, Z. & Liu, S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics36, 2253–2255 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Cock, PeterJ. A. et al. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. Bioinformatics37, 1639–1643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, N.-C. and Hansen, N. F. milkschen/chaintools: v0.1 (2022).
- 33.Vollger, M. R. mrvollger/rustybam: v0.1.29 (2022).
- 34.Guarracino, A. Andreaguarracino/paf2chain: v0.1.0 (2023).
- 35.Genovese, G. et al. Bcftools/liftover: an accurate and comprehensive tool to convert genetic variants across genome assemblies. Bioinformatics40, btae038 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927836 (2022).
- 37.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927835 (2022).
- 38.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927834 (2022).
- 39.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927833 (2022).
- 40.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR29122519 (2022).
- 41.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR29092487 (2022).
- 42.NCBI genomes, https://identifiers.org/assembly:GCA_037177635.1 (2024).
- 43.NCBI genomes, https://identifiers.org/assembly:GCA_037177555.1 (2024).
- 44.Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics32, 2103–2110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landrum, M. J. et al. Clinvar: improvements to accessing data. Nucleic Acids Research48, D835–D844 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott, E. M. et al. Characterization of greater middle eastern genetic variation for enhanced disease gene discovery. Nature genetics48, 1071–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR27002256 (2022).
- 48.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, May arXiv:1303.3997 [q-bio] (2013).
- 49.O’Connor, B.D. and van der Auwera, G. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. O’Reilly Media, Incorporated (2020).
- 50.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR29055922 (2022).
- 51.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR1295554 (2022).
- 52.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR1291026 (2022).
- 53.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR1295426 (2022).
- 54.NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:ERR757831 (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927836 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927835 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927834 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR21927833 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR29122519 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR29092487 (2022).
- NCBI genomes, https://identifiers.org/assembly:GCA_037177635.1 (2024).
- NCBI genomes, https://identifiers.org/assembly:GCA_037177555.1 (2024).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR27002256 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR29055922 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR1295554 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR1291026 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRR1295426 (2022).
- NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:ERR757831 (2022).
Data Availability Statement
Source code and additional resources are available at https://github.com/bio-ontology-research-group/KSA001.