Abstract
Phenotypic profiling by high throughput microscopy, including Cell Painting, has become a leading tool for screening large sets of perturbations in cellular models. To efficiently analyze this big data, available open-source software requires computational resources usually not available to most laboratories. In addition, the cell-to-cell variation of responses within a population, while collected and analyzed, is usually averaged and unused. We introduce SPACe (Swift Phenotypic Analysis of Cells), an open-source platform for analysis of single-cell image-based morphological profiles produced by Cell Painting. We highlight several advantages of SPACe, including processing speed, accuracy in mechanism of action recognition, reproducibility across biological replicates, applicability to multiple models, sensitivity to variable cell-to-cell responses, and biological interpretability to explain image-based features. We illustrate SPACe in a defined screening campaign of cell metabolism small-molecule inhibitors tested in seven cell lines to highlight the importance of analyzing perturbations across models.
Subject terms: High-throughput screening, Software
Phenotypic profiling by high-throughput imaging can aid in the screening of perturbations in cell models, but most studies often overlook cell-to-cell variation of responses within samples/populations. Here, the authors present SPACe, an easy-to-deploy, open-source platform for analysis of single-cell image-based morphological profiles produced by Cell Painting.
Introduction
Measuring biological complexity of physiological and pathological states from single cells to whole organisms is the basis for developing models and analytical methods that result in new knowledge moving toward new interventions. Arguably, one of the most successful attempts in measuring cell states, characterizing biological pathways, and testing thousands of small-molecules for drug development has been the application of Cell Painting (CP) or its multiple variants1–7. CP combines several fluorescent dyes to illuminate cellular structures, allowing for an inexpensive, high content (HC), high-throughput (HT) microscopy-based assay that is scalable to whole genome knock-out/overexpression and large small-molecule inhibitor libraries8–12. Information extraction from CP images involves two main steps: firstly, identifying regions of interest (e.g., cellular substructures) and extracting relevant features, typically done using open-source (e.g., CellProfiler13,14) or commercial software; secondly, performing feature reduction and representation1,15–17 to facilitate further downstream analyses such as clustering and classification. Due to its biological and analytical relevance, single-cell data in HT imaging-based campaigns are now widely used both for data quality control and for hit identification associated with various treatments based upon clear phenotypic differences6,18–23. Nonetheless, while CP approaches have entered the mainstream for phenotypic screening, there is still a very active research effort to enhance robustness, processing speed, and sensitivity to best capture cell population heterogeneity. As AI-driven strategies have become more prevalent for high-dimensional and large-scale data analysis, integration of such strategies into CP and its variants has stimulated a major interest due to the promise of higher accuracy, faster processing times and the potential for fusing multimodal data18,19. However, with the continual advancement of HT microscopes and laboratory automation, and resultant large datasets, a key roadblock appears to be how to analyze data efficiently, in a timely manner, and the availability of massive computational resources needed to carry out such analyses.
To address these outstanding challenges, we developed an open-source, Python-based, easy-to-deploy, single-cell image analysis platform named SPACe (Swift Phenotypic Analysis of Cells), that tackles object segmentation, quality control, feature extraction and analysis of large image datasets collected from HC/HT imaging campaigns while requiring significantly less computational resources with respect to existing methods. In fact, SPACe can process large datasets commonly used in HT imaging-based campaigns approximately ten times faster than CellProfiler using a standard personal computer, without performance loss in downstream analysis (e.g., mechanism of action – MoA - recognition accuracy).
The SPACe platform includes additional properties that were designed to ensure reproducibility and the ability to improve interpretability of downstream analysis of extracted features. Specifically, SPACe includes a state-of-the-art approach for cell segmentation, the ability to segment multiple subcellular compartments and the application of a directional Earth Mover’s Distance (signed EMD) to quantify differences in single-cell feature distributions. We recall that most of existing cell screening analytical platforms, while extracting information from individual cells, ultimately utilize only per-well or per treatment central tendency values like mean or median for downstream analysis. A notable exception is the work from Pearson et al.18, that demonstrated the potential advantage of interrogating single-cell information by analyzing the statistical distribution of data within cellular populations. While the traditional per-well average approach has proven to be successful and sufficient for hit calling from single endpoint assays in large-scale screening campaigns, it ignores the inherent phenotypic heterogeneity in a cell population and the fact that many biological responses do not follow a normal distribution6,19,23.
While the SPACe platform is applicable to virtually any screening campaign using various instruments, we focus here on analyzing images captured with JUMP Consortium-vetted high-throughput widefield and spinning disk confocal microscopes.
Results
Development of a single-cell-based image analysis pipeline – SPACe
The traditional way of analyzing CP images relies on either the open-source CellProfiler1,13 or commercial software (i.e., Columbus and others). For the larger datasets typically associated with screening, it is recommended to run these solutions on distributed computing resources (i.e., CPU clusters), Cloud computing, or powerful workstations. However, many labs around the world considering using CP may not have access to such resources. To overcome this limitation, we developed SPACe (Fig. 1A, B), an open-source, user-friendly Python-based analysis pipeline that can efficiently analyze CP images using a standard desktop PC equipped with a consumer grade graphics processing unit (GPU). SPACe was measured to be approximately 10× faster than the open-source CellProfiler pipeline (Fig. 1C), and is provided to the community on GitHub (https://github.com/dlabate/SPACe) as a downloadable version for local installation, customization, and possible linkage to additional user-defined modules. We also include a Google Colab version of the code (https://github.com/dlabate/SPACe/blob/main/SPACe_colab.ipynb), designed for testing the software under different hyperparameter settings.
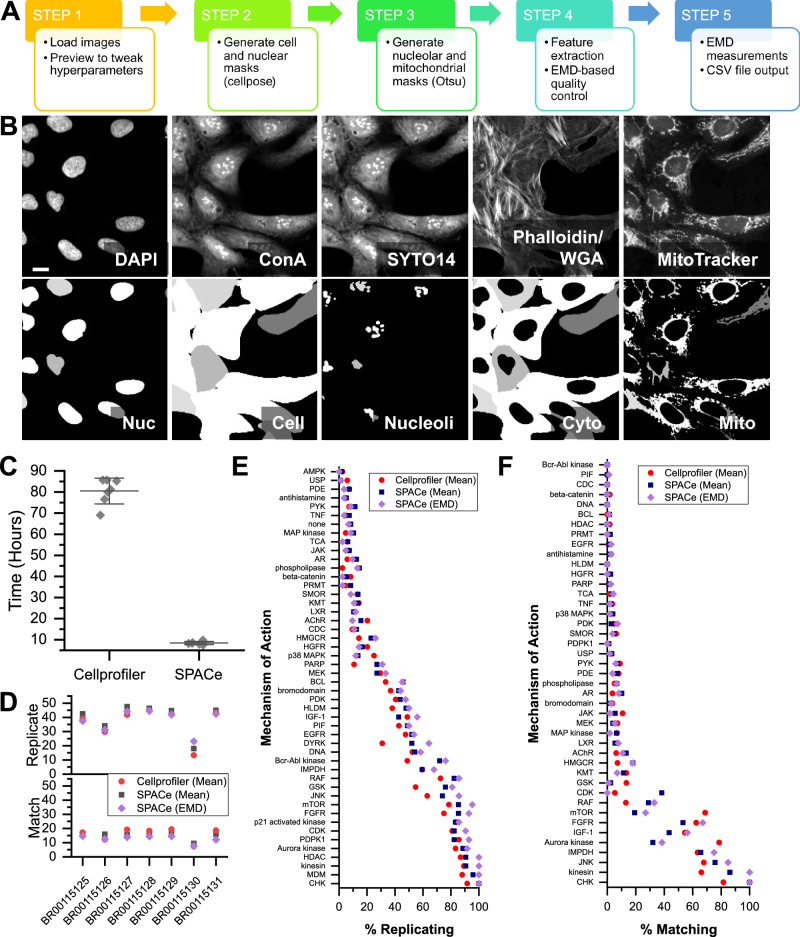
Fig. 1. SPACe workflow and performance on JUMP MOA reference datasets.
A A graphical depiction of the steps included in the SPACe pipeline. B Example of four SPACe segmented objects (out of thousands of images) in CP-stained U-2 OS cells, please note that the fifth compartment (cytoplasm) is obtained by subtraction of the nuclear mask from the cell mask. Scale bar: 5 µm. C Processing time for JUMP MoA reference datasets using either CellProfiler or SPACe, each symbol represents running time for a plate of the reference dataset, with also the mean ± standard deviation overlayed. D Assessment of percent replicating (top) and percent matching (bottom) based on population mean (CellProfiler, SPACe) or EMD (SPACe only). Assessment of analysis feature percent replicating (E) or percent matching (F) by the mechanisms of actions present in the JUMP reference datasets. Source data are provided as a Source Data file.
As part of streamlining the CP approach, we decided to not include a module for illumination correction due to the fact that this can be performed as a preprocessing step following established methods2,24,25. We have often found that instrument-associated image correction is sufficient to remove illumination errors. In the case of experiments performed in this study, we relied upon the Yokogawa CV8000 software to correct non-uniform illumination, pixel misalignment between cameras, and fluorescence channel crosstalk.
After loading images (Step 1), the SPACe pipeline automatically defines nuclear (“Nucleus”) and cell (“Cell”) segmentation boundaries using the popular AI-based Cellpose package and its pretrained generalist model(s)26,27 (Step 2). Although Cellpose can automatically determine segmentation hyperparameters on a per image basis, for more consistent and faster performance, users should indicate the expected nuclear and cell diameters in terms of number of pixels, which is often cell model specific. To facilitate this task in SPACe, we implemented a preview function that allows the user to test selected fields of view (FOVs) making sure that the segmentation is accurate before analyzing the entire dataset. Following nuclear and cell regions identification, an adaptive Otsu & MaxEntropy thresholding routine is applied to identify nucleoli (“Nucleoli”) and mitochondria (“Mito”, Step 3). A separate cytoplasmic region (“Cyto”) is defined by subtracting each nuclear region from each cell region. An example of segmented objects is shown in Fig. 1B.
SPACe extracts more than 400 curated features from each object mask including intensity, morphology, and textural measurements (Step 4) that are described in Supplementary Table 1. Again, experienced users can alter/add/subtract the set of extracted features as needed. As we describe in detail below (Supplementary Figs. 2 and 3), we demonstrate that the number of features extracted by SPACe is sufficient to capture phenotypic changes in Mechanism of Action (MoA) datasets produced by the JUMP consortium.
A version of our previously published19 quality control (QC) pipeline for high-throughput single-cell data was added in SPACe (Step 4); this step leverages the analysis of the empirical probability distribution of single-cell features in control samples (i.e., DMSO treated) to identify and discard outlier wells and to generate a reference distribution for each feature, defined as the median empirical distribution of all remaining DMSO wells. This reference distribution is then used to quantify the effect of perturbations based on the Earth Mover’s Distance (EMD), a metric that quantifies the dissimilarity between probability distributions and has been shown to work well for phenotypic screens18. Additional details, including data normalization, can be found in the “Methods” section. Here we adopt a directional variant of the EMD (signed EMD), where we assign a positive sign to the EMD if the median has increased with respect to DMSO, and a negative sign if the median has decreased. The SPACe pipeline ultimately provides users with the saved object masks, single-cell raw data, and a CSV file containing the signed EMD values plus the well-averaged mean and median values for each feature (“distance maps” – Step 5); all of which can be further analyzed using preferred software packages as suggested in several publications1,15–17. Filtering wells with a low number of detected objects can also be used as an additional QC metric to avoid analyzing the distribution of single-cell data from wells with an insufficient number of data points. In our previous study19, albeit not using CP, we estimated that a minimum of ∼1000 cells are needed to properly reconstruct a faithful distribution from single-cell data.
Comparing SPACe with CellProfiler using JUMP Consortium reference datasets
As one of the goals of SPACe is to be useful to a large community of researchers with potentially limited computational resources, we compared its performance to CellProfiler using a highly diverse CP experiment by downloading seven reference datasets provided by the JUMP consortium. These JUMP MoA datasets (BR00115125-31) contain 90 unique treatments with 47 annotated mechanisms of action (MoA) along with negative control DMSO wells1. Using a standard PC, of the type available to most research labs (Intel i7 13700 CPU, NVIDIA 3070 GPU, 32 GB memory), the average processing time per plate was almost 10× lower using SPACE (8.5 ± 0.5 h) compared to CellProfiler running the pipeline recommended by the JUMP consortium (80.2 ± 5.3 h) (Fig. 1C). The primary contribution to this difference is the extraction of a reduced, curated set of features within SPACe and its implementation of the Pyradiomics library to extract texture features using GPU acceleration. As a result of this, the primary contributor to processing time for SPACe is the ROI segmentation of using the pretrained cNN models in Cellpose, whereas for CellProfiler it is the extraction of texture features. When using the percent replicating and percent matching1 calculations that define the correlation of extracted features between replicate wells and those with different treatments, but matching annotated MoAs, there was no significant difference between SPACe and CellProfiler generated outputs (Fig. 1D), with the largest determinant in performance being the source dataset. However, when percent replicating was ranked for each dataset, SPACe mean well-values ranked significantly better (Supplementary Fig. 1A, B). In percent matching, both SPACe and CellProfiler mean well-values ranked significantly better than SPACe EMD values (Supplementary Fig. 1C-D). When examined by individual MoAs, the trend in percent replicating and percent matching were similar between analysis methods (Fig. 1E, F). Although no difference was statistically significant, there were examples (e.g., GSK, Bcr-Abl kinase, Aurora kinase, mTOR) where one method appeared to outperform the other, likely reflecting differences in the underlying segmentation and texture feature extraction approaches used.
One of the key differences between CellProfiler and SPACe is the reduced number of features extracted (∼4000 versus ∼400). To understand the uniqueness of the features collected by each method, Spearman correlation between features across all samples in the JUMP reference datasets were examined (Supplementary Fig. 2A–C). The relative frequency of features with correlation values between 0.2 and 0.8 were similar for each method, however, SPACe-extracted feature sets contain a greater proportion of highly correlated features (Spearman correlation >0.8) than the CP feature set (Supplementary Fig. 2E). SPACe mean and EMD feature sets contain a higher proportion (24% and 32%, respectively) of ‘unique’ features (defined as a feature with no correlation) compared to the CellProfiler feature set (16%), despite the absolute number of unique features being lower (Supplementary Fig. 2D). This suggests that although SPACe collects a smaller feature set, the feature set contains sufficient diversity to recapitulate the CellProfiler generated results from the reference datasets, similar to other published work that reduced the CellProfiler feature set to a little over 60028.
A concern with collecting a smaller number of features is the potential inability to accurately capture phenotypes associated with various MoAs. To address this concern, we generated five random forest (RF) models for each JUMP reference dataset for each of the CellProfiler well mean, SPACe well mean, and SPACe EMD feature sets. Each RF model was trained with half of the treatment replicates, randomly selected for each model replicate. Model performance was assessed by the ability to correctly predict the MoA of treatment replicates not selected for training. Across all samples, the feature set used to train the model did not make a significant difference in the accuracy of MoA prediction (Supplementary Fig. 3A). When examined by individual MoA, RF models trained with the different feature sets showed similar trend in prediction accuracy (Supplementary Fig. 3B). RF models trained with SPACe (either well mean or EMD) features were noted to better predict several MoAs, particularly the DYRK (significant, p < 0.05), MEK, LXR, PRMT, and beta-catenin signaling pathways. When examined by prediction accuracy rank for each MoA, RF models trained with SPACe EMD values ranked significantly better (Supplementary Fig. 3C). When a confusion matrix was examined for each set of RF models (Supplementary Fig. 3D–F), all incorrect MoA predictions shared a similar pattern, which was to predict a sample as inactive vs. active (i.e., “none” category in the graphs). Taken together, these results indicate that the reduced feature set collected by SPACe, especially the EMD-based feature set, can capture induced phenotypes as well as, if not slightly better, than the feature sets generated using the JUMP consortium CellProfiler pipeline.
Testing selected reference chemicals with the pipeline and their reproducibility
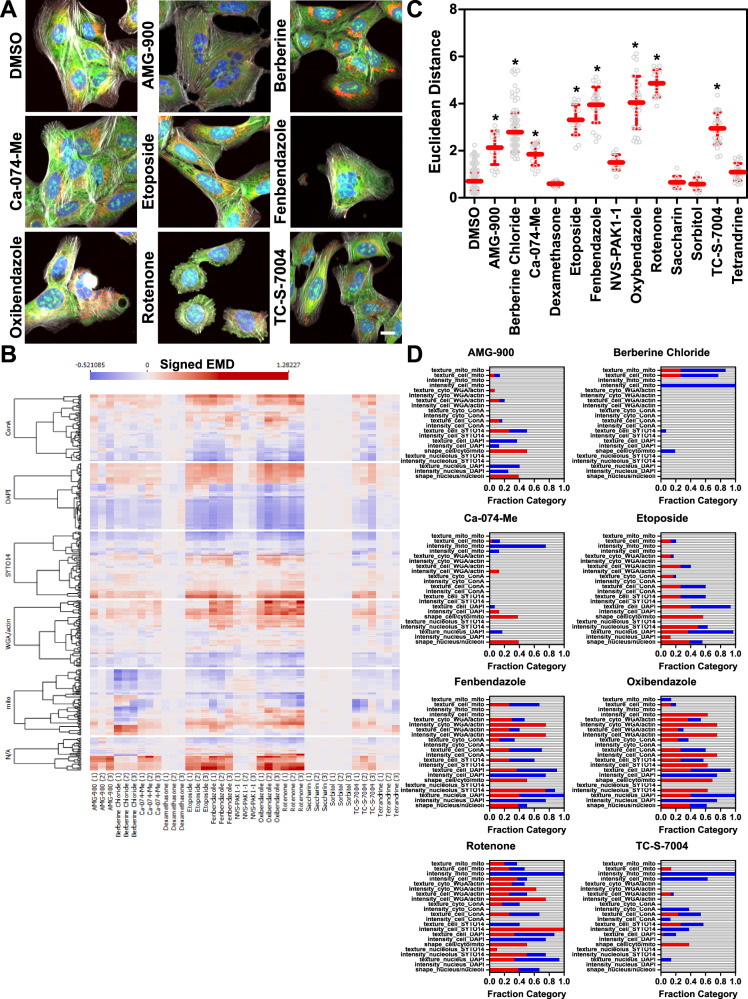
We used the standard JUMP consortium U-2 OS osteosarcoma cell line1–3 to investigate reproducibility and interpretability of the results obtained with SPACe. We chose a selection of potential reference chemicals (Table 1) that have been previously shown in publications or by the JUMP consortium to alter the phenotype of U-2 OS cells3,29–31, including saccharin and sorbitol as negative controls. Cells were treated for 24 h ahead of the CP protocol, which was performed either manually or robotically following published reports1–3 and imaged using a Yokogawa CV8000 high-throughput spinning disk confocal using the established conditions32. Figure 2A shows representative color images of the cells treated with the indicated compounds. In Fig. 2B the signed EMD from the median DMSO control (which has distance = 0) for each of the >400 features, from three independent biological replicates (labeled 1,2,3 in parentheses below the treatment names), is shown as a heatmap for the “reference compound set” with each of the fluorescence channels separately highlighted. The quantification of all the data in Fig. 2B is shown in Fig. 2C, where we used Euclidean distance between the treatment fingerprint (i.e., represented by changes in all >400 features) for each well (represented as a circle in the graph), from the median of DMSO wells of each biological replicate. Overall, in terms of Euclidean distance, we observed good concordance between the three independent biological experiments, and a compound with Euclidean Distance >2 was considered active as it was fully separated from the DMSO control wells and was significant by non-parametric ANOVA, a parameter that we used for the rest of the study. As expected, the negative controls (saccharin and sorbitol) had no effect across the feature space. Of the tested reference compounds, we also failed to detect significant and/or reproducible effects of tetrandrine, dexamethasone and NVS-PAK1-1. Berberine chloride caused a redistribution of well-resolved mitochondria, which was evident from the images (Fig. 2A31) and was readily measured by changes exclusively in the MitoTracker channel (Fig. 2B, D). To aid with interpretability of the results (Fig. 2D), we subdivided the features in categories (based on cell compartment and fluorescence channel) and then represented changed features (defined as EMD > 0.15 or <−0.15 in at least two out of three biological replicates) as a stacked bar graph (blue features are reduced and red features are increased as compared to DMSO). AMG-900 and Etoposide showed changes in the nuclear and nucleolar compartments, which are compatible with their known mechanism of action (aurora kinase and topoisomerase inhibitors, respectively). Rotenone affected multiple compartments, including mitochondria. Similarly, fenbendazole and oxibendazole, two anti-parasitic drugs, showed changes across all compartments, including visual evidence of polynucleation, as previously described3. In the case of Ca-074-Me, we did not observe the increase in ConA staining intensity that was previously reported3, but the most reproducible changes were measured at the level of nuclear and cellular size and shape. TC-S-7004, a DYRK1A/1B inhibitor30,33, also had a fingerprint that was significantly different from DMSO, with changes in the SYTO14 and MitoTracker channels. It is important to note that at the concentration used, the compounds could have mixed mechanisms of action through off-target and secondary effect(s), which can be sorted out in follow-up experiments (i.e., concentration-response, time-response and target knock-down experiments).
Table 1.
List of potential reference chemicals tested
| Chemical Name | CAS # | Reported Phenotype (3,29,31,34) | Source | Notes |
|---|---|---|---|---|
| AMG-900 | 945595-80-2 | JUMP positive control | Selleckchem | Aurora kinase inhibitor |
| Berberine Chloride | 633-65-8 | Redistribution of mitochondria | Sigma | Target unclear |
| Ca-074-Me | 147859-80-1 | Bright ConA staining | Sigma | Cathepsin B inhibitor |
| Dexamethasone | 50-02-2 | JUMP positive control | Tocris | GR agonist |
| Etoposide | 33419-42-0 | Large nucleoli | Sigma | Topoisomerase II inhibitor |
| Fenbendazole | 43210-67-9 | Multi-nucleated cells | Sigma | Anti-helmintic |
| NVS-PAK1-1 | 1783816-74-9 | JUMP positive control | Tocris | PAK1 inhibitor |
| Oxibendazole | 20559-55-1 | Multi-nucleated cells | Sigma | Anti-helmintic |
| Rotenone | 83-79-4 | Mitochondrial fission | Sigma | Complex I inhibitor |
| Saccharin | 81-07-2 | Negative control | Sigma | Artificial sweetener |
| Sorbitol | 50-70-4 | Negative control | Sigma | Sugar alcohol |
| TC-S-7004 | 1386979-55-0 | JUMP positive control | Tocris | DYRK1A/1B inhibitor |
| Tetrandrine | 518-34-3 | Abundant WGA | Sigma | Calcium channel blocker |
Fig. 2. Testing a set of potential reference compounds with SPACe.
U-2 OS were treated for 24 h with the indicated compounds at 10 µM ahead of the CP protocol. Representative 20× color images from the same experiment out of three independent biological replicates are shown in (A). Scale bar is 10 µm. B Heatmap showing the signed EMD (Earth Mover’s Distance) from DMSO controls of the indicated compounds from three independent biological replicates (labeled in parentheses below the name of the compounds as 1,2,3). Each channel is separated to show the changes by fluorescent dye. N/A represents shape features and ratios (i.e., nuclear size/cell size). C Euclidean distance is used as a measure to compare the features fingerprint of each compound from the median DMSO control. This is measured for each well (represented as a hollow circle in the graph) treated with the indicated compound in three independent biological replicates, shown together in the graph. Overlayed in red the mean ± standard deviation is represented. * is p < 0.05 using non-parametric ANOVA (Kruskal–Wallis) compared to DMSO group. D Stacked bar graphs representing the fraction of changed features (in blue if they are reduced or red if they are increased, gray means no change) in the defined groups for each treatment. The chosen threshold for significance was EMD distance <−0.15 or >0.15 (see “Methods” section for description). Source data are provided as a Source Data file.
Reproducibility and interpretability: berberine chloride as a case study
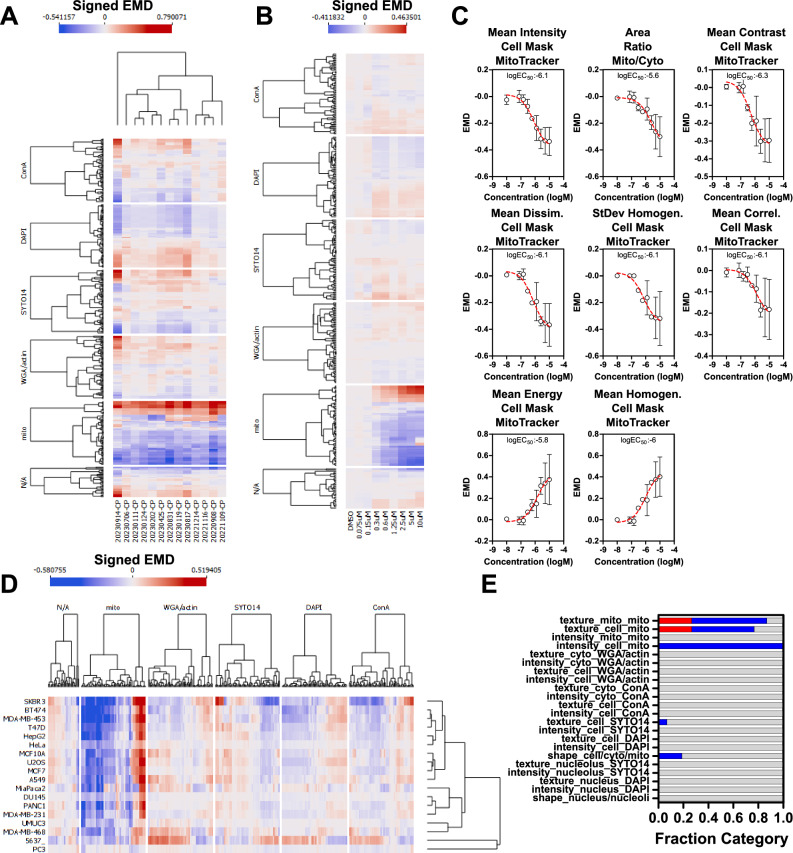
To further analyze the quality of SPACe pipeline outputs, we first focused on berberine chloride as it showed a unique phenotype that is immediately evident from visual inspection of the images (e.g., redistribution and size reduction of mitochondria). In Fig. 3A, the analysis of thirteen independent biological replicate experiments confirmed high reproducibility of the berberine-induced, visible phenotype in terms of the mitochondria features changing from the DMSO control. Only one experiment was somewhat an outlier with additional cellular compartments being more affected; however, the top mitochondrial features remained similarly altered. We performed berberine chloride concentration-response experiments (four biological replicates) to verify if the top changing mitochondrial features were indeed responding in a concentration-dependent manner. Figure 3B shows the berberine chloride concentration-response experiments in a heatmap format, confirming that most responding features indeed have a concentration dependency, an indication of their biological specificity likely linked to the mechanism of action of any compound. We next selected eight features (intensity, texture, plus the ratio between mitochondrial mask and cell mask areas, Fig. 3C), that exhibit clear concentration dependency, with very similar EC50s, indicating they are likely related to the berberine chloride mechanism of action, which remains ill-defined34,35.
Fig. 3. Berberine chloride as an example of a reference compound with a clear, interpretable phenotype.
A Feature response reproducibility. Thirteen independent experiments were conducted with U-2 OS cells treated with berberine chloride (10 µM) for 24 h, features were extracted with SPACe and signed Earth Mover’s Distance (signed EMD) is represented as a heatmap. B Signed EMD heatmap showing all the extracted features after a berberine chloride concentration-response (75 nM to 10 µM) at the 24 h time point. C Average ± standard deviation of the top, non-redundant eight features changing after berberine chloride treatment, with EC50 indicated, from the experiments in (A). D Eighteen cell lines were treated with berberine chloride 10 µM for 24 h and the signed EMD of all features is represented as a heatmap. E Stacked bar graph showing a berberine chloride “consensus fingerprint” of the changing features across cell lines. Increased features are in red, decreased in blue, and unchanged in gray. N/A represents shape features and ratios (i.e., nuclear size/cell size). Source data are provided as a Source Data file.
To determine if the response to berberine chloride was universal so that it could be employed as a true reference compound for all CP experiments, we treated 18 human cell lines for 24 h (Fig. 3D). 14 out of 18 cell lines showed a strong mitochondrial phenotype, while three more had a partial responsiveness to berberine chloride, reinforcing the assumption that the mechanism of action of this chemical is largely universal and visually affects primarily mitochondria. Moreover, it is interesting to point out that berberine was non-toxic in all the cell lines tested. PC-3, a prostate cancer cell line, was the only clear outlier for unknown reasons, with bladder cell lines UM-UC-3 and 5637 also having a muted and a more diverse fingerprint. The consistent response allowed us to extract a “berberine chloride consensus fingerprint” that is visible as aggregate results in a stacked bar plot subdivided by feature classes and cellular compartments (Fig. 3E). A list of the selected features that constitute such a fingerprint is available in Supplementary Table 2. Collectively, the fingerprint signature allowed us to add interpretability to this treatment as the selected features can be visually linked to the images. For example, the intensity of the MitoTracker channel in the cell compartment is reduced as is the ratio of mitochondria-to-cell area, while several distinct mitochondrial texture features are changing both within the cell and mitochondria compartments.
Cell Painting in breast cancer cell lines: analysis of a small panel of chemicals reveals both cell-type specific and broad effects
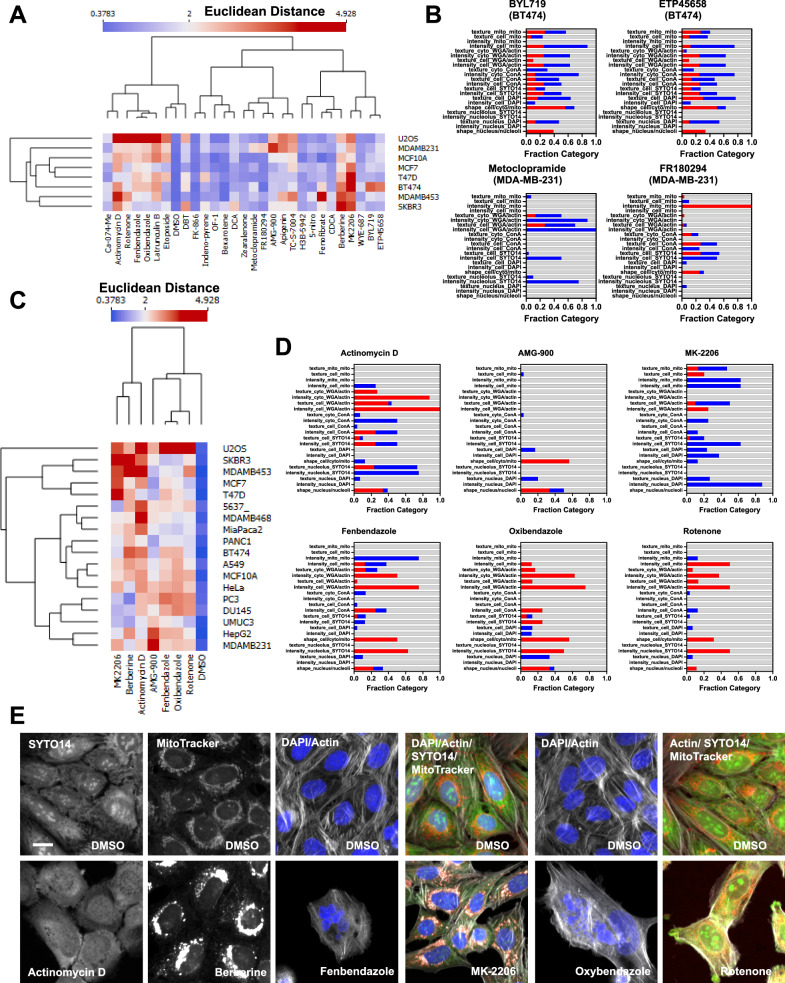
To further explore the potential of SPACe for analysis of cell lines outside the canonical U-2 OS and A549 models often used in CP, we tested a set of breast cancer models representing different tumor subtypes (luminal, Her2, and triple negative), by treating them for 24 h with a small set of 28 diverse chemicals, including those deemed active in Fig. 2C. Figure 4A shows the Euclidean distance of each compound from DMSO across multiple experiments and cell lines, represented as a heatmap. Overall, 19 out of 28 compounds were found to be active (Euclidean distance >2, 68%) in at least one cell line; however, only 8 out of 28 (29%) were active in at least five cell lines. These included the Akt inhibitor MK-2206, berberine chloride, fenbendazole, oxibendazole, rotenone, TC-S-7004, actinomycin D, and latrunculin B; all of which have been used as references before or have established strong responses and mechanism of action. Interestingly, we had no compound that showed specificity for U-2 OS, however, a few were only active in one breast cancer cell line, including BYL-719 (PI3K inhibitor, BT-474), DCA – deoxycholic acid (bile acid, SK-BR-3), ETP45658 (PI3K inhibitor, BT-474), FR180294 (ERK1/2 inhibitor, MDA-MB-231), and metoclopramide (dopamine receptor antagonist, MDA-MB-231). A more detailed inspection of the phenotypic changes revealed that the two PI3K inhibitors had a very similar overall profile in BT-474, indicating a likely class (and perhaps cell line) specific phenotypic readout (Fig. 4B). In contrast, the two MDA-MB-231 specific compounds modulated multiple compartments and channels, being nucleolus and actin for metoclopramide, mitochondria and concanavalin A/SYTO14 for FR180294.
Fig. 4. Testing SPACe on a set of breast cancer models.
A Cell lines were treated with 28 chemicals for 24 h and then analyzed with SPACe. The heatmap shows the Euclidean distance of each compound from the DMSO wells of each cell line. B Stacked bar graphs showing the interpretability profile of four cell line specific hit chemicals with increased features in red, decreased in blue, and unchanged in gray. C Eighteen cell lines were treated with the indicated compounds for 24 h and then Euclidean distance was calculated and represented as a heatmap. D Stacked bar graphs showing tentative “consensus fingerprints” of the changing features across a minimum of five cell lines, with increased features in red, decreased in blue, and unchanged in gray. E Selected images (zoom in from 20×/1.0 images) from one out of three independent biological replicates to showcase specific phenotypic changes caused by the indicated compounds in highlighted channels/compartments. Scale bar: 10 µm. Source data are provided as a Source Data file.
We then selected seven of the best responders and tested them across all 18 cell lines (Fig. 4C) to determine their universality and attempt to identify their response fingerprint, akin to what we showed for berberine chloride in Fig. 3E. Overall, all compounds elicited a response in at least 10 cell lines, but none affected all 18, indicating that it is unlikely to identify compounds that would act in a universal manner and can be used as controls across all experimental models. This complicates the analysis, prediction, and MoA interpretation for compounds when based uniquely on phenotypic screening in a single-cell model. We attempted to add interpretability by selecting only features that were changing in at least 5 cell lines, as very few to none significantly changed across all models, hoping to mitigate the off-target effects that can be seen at µM concentrations. Figure 4D shows the interpretation using stacked bar graphs for all seven compounds, except for berberine chloride that was already presented in Fig. 3E. Actinomycin D showed major changes in the nucleolus (Fig. 4E), compatible with its activity as a general inhibitor of RNA polymerases and gene transcription, and actin cytoskeleton, which can represent signs of toxicity, even though we were still able to collect information from more than 1000 cells/experiment. For AMG-900, only a few features were consistent across cell lines, and these revolved around cell and nuclear size/shape and DAPI texture features, which is compatible with its known mechanism of action in mitosis as an Aurora kinase inhibitor. MK-2206 is a specific Akt inhibitor that can be an autophagy activator36 and the main features that appear to be linked to this phenotype include reduction in nuclear DAPI signal and changes in texture features for SYTO14, MitoTracker, and WGA/phalloidin that can be visually interpreted as the formation of autophagolysosomes in the cytoplasm (Fig. 4E).
Fenbendazole and oxibendazole are anti-parasitic drugs that have been shown to act through multiple mechanisms including microtubule destabilization, G2/M arrest, and apoptosis37,38. Both drugs produced complex CP profiles modifying all the compartments in various ways, most notably higher SYTO14 intensity in the nucleolus and reduced MitoTracker signal in mitochondria. These observations highlight the utility of including these subcellular compartment masks in SPACe. Inspecting the images clearly shows that both compounds cause multi-nucleated cells and dying cells, confirming their main mechanism of action (e.g., cell division and cell death, Fig. 4E). Finally, rotenone, a natural isoflavone, is a strong inhibitor of mitochondrial complex I, which is reflected by the alterations observed in selected MitoTracker features. Interestingly, rotenone appears to cause broader changes also affecting the nucleolus and actin cytoskeleton, likely due to off-target effects.
Cellular metabolism screening library
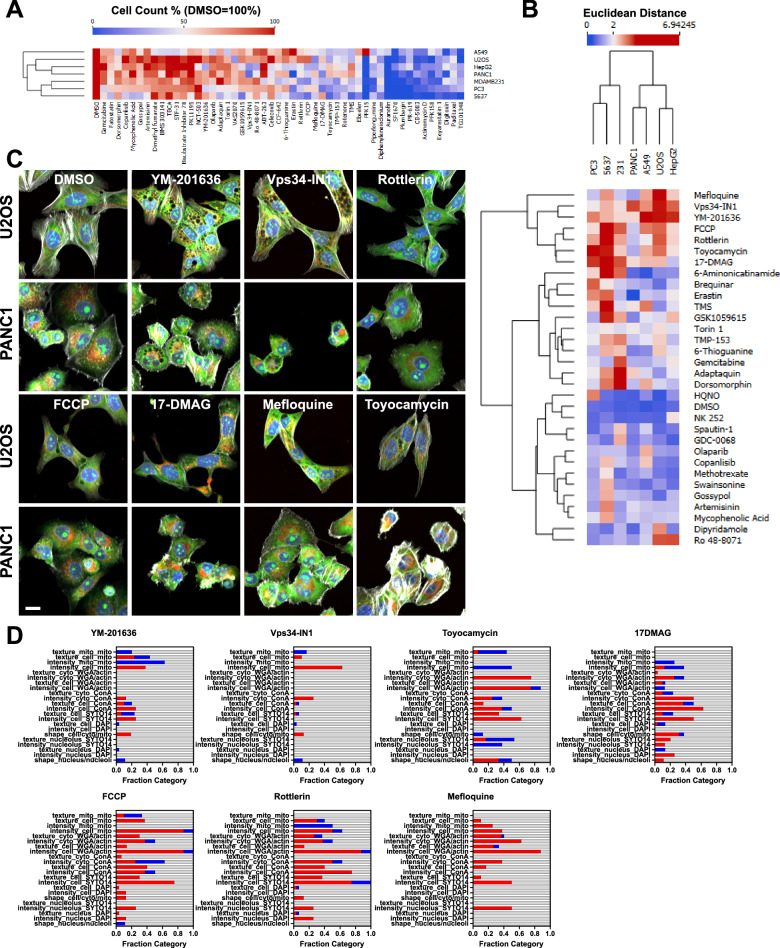
To expand our understanding of the universe of phenotypic changes across models, we treated seven cancer cell lines with different origins (U-2 OS - bone, Hep G2 - liver, 5637 - bladder, PANC-1 - pancreas, PC-3 - prostate, MDA-MB-231 - breast, and A549 - lung) in duplicate plates with the Cayman Chemical Cellular Metabolism Screening Library containing 160 small-molecule modulators of diverse targets and metabolic pathways. First, we analyzed the responses to the library by identifying toxic compounds. In Fig. 5A, treatments that caused a reduction in cell number by >50% in at least one cell line, as compared to DMSO control, are shown in a heatmap format. This is a relevant step as recent work from Dahlin et al., identified a set of compounds that interfere with CP screening through cell injury39. A note here is that the screening was performed solely in U-2 OS cells and will need to be validated in other models. Overall, about a third of the library showed some toxicity in at least one cell line. Ten compounds were the most toxic across all models (auranofin, SF1670, plumbagin, PR-619, CB-5083, PFK158, eeyarestatin-1, digitoxin, paclitaxel, and TG101348) and should be tested at lower concentrations to measure changes at non-toxic levels, as some of them show potentially very interesting phenotypes in the surviving cells. For example, PR-619 and eeyarestatin-1, in U-2 OS and across cell lines, respectively, show cytoplasmic vacuolization and redistribution of mitochondria; plumbagin, in PANC-1 cells, also affects mitochondria plus on cell shape/size; while TG101348, in PANC-1 and Hep G2, causes large changes in the WGA/actin, MitoTracker and some morphological features (Supplementary Fig. 5). Examples of compounds that showed some cell-type selective toxicity were gemcitabine (PC-3, nucleoside analog), copanlisib (5637, PI3K), mycophenolic acid (5637, inosine-5′-monophosphate dehydrogenase), and NCT-503 (Hep G2, PHGDH). Of note, when considering the most toxic compounds we found, only paclitaxel and plumbagin are in the cellular injury list39, together with gemcitabine, which was cell line specific.
Fig. 5. Screening a cellular metabolism modulators library with SPACe analysis.
A Analysis of the library compounds induced toxicity represented as heatmap where cell count was normalized to DMSO, set as 1; the compounds represented caused a cell loss of >50% in at least one cell line. B Heatmap and hierarchical clustering of hits (Euclidean distance >2 in at least one cell line) from the screen after SPACe analysis. C 20× zoomed in images from screen run number 1 of the indicated hits shown in U-2 OS and PANC-1 cells. Scale bar: 10 µm. D Interpretability stacked bar graphs for the compounds shown in (C) with increased features in red, decreased in blue, and unchanged in gray. Source data are provided as a Source Data file.
In Fig. 5B we present the 31 hit compounds from the library screen in a heatmap, defined as treatments with Euclidean distance >2 cutoff in at least one cell line, and in both replicate plates, after filtering out the abovementioned toxic chemicals. Compounds with discordant replicates or with obvious imaging artifacts were also excluded after manual inspection. To improve accuracy and interpretability, it is important to note that every multiwell plate was run with several internal controls (DMSO, actinomycin D, MK-2206, and berberine chloride). Additionally, actinomycin D and rotenone were present as components of the library itself and served as additional quality control treatments as active compounds, and as such were excluded from Fig. 5B.
Perhaps interestingly, only seven compounds in the screen showed effects in a single-cell line: HQNO (PC-3), mycophenolic acid (5637), olaparib (A549), GDC-0068 (MDA-MB-231), dipyridamole (U-2 OS), spautin-1 (MDA-MB-231), and NK 252 (Hep G2); further validating the importance of screening across a wide range of cellular models. Seven compounds showed various phenotypes across four or more cell lines, despite having a somewhat reduced cell number in specific models (top cluster in Fig. 5B). These were the PIKfyve inhibitor YM-201636, the VPS34 inhibitor Vps34-IN1, the mitochondrial uncouplers FCCP and rottlerin, the IRE1 inhibitor toyocamycin, the Hsp90 inhibitor 17-DMAG, and the antimalarial mefloquine. Visual representations of the phenotypes induced by these compounds are shown for U-2 OS and PANC-1 cells in Fig. 5C, while their interpretability plots are shown in Fig. 5D. In the case of all these compounds, it was much harder to identify a common fingerprint between cell lines, which was especially true for Vps34-IN1, where only 25 features were common between three out of seven cell lines. However, in individual cell lines, visually, YM-201636 and Vps34-IN1 responses looked reasonably similar (U-2 OS cells are shown in Fig. 5C), and indeed we found a set of features that matched these two treatments; for example, increased intensity of the concanavalin A and MitoTracker in the cell mask, reduction in nuclear perimeter and various changes of MitoTracker textural features. Also, the two mitochondrial uncouplers, rottlerin and FCCP, had several matching features in the mitochondrial compartment (e.g., increase intensity and mitochondria/cell area ratio).
Discussion
Over the last few years, the leading approaches to HT genetic and chemical screens have shifted from classical single-endpoint cell free assays to unbiased multi-endpoint imaging approaches that are based on phenotypic profiling of intact cells. The success of the latter approach stemmed from the development of CP protocols that allow for economical and fast characterization of a cell state by illuminating specific cellular components. This strategy, coupled with automated image analysis and machine learning, has been proven to be very effective in measuring phenotypic changes upon a large variety of perturbations, including small-molecules, knock-down and overexpression.
A major limitation to a wider adoption and deployment of CP-like phenotypic screening is the significant computational resources required to run current image analysis solutions such as CellProfiler and other commercial applications. To address this challenge, here we introduce SPACe, an open-source, lightweight, Python-based CP analysis workflow that differs from most current analysis tools in several important ways. First, SPACe integrates the use of Cellpose pretrained AI-based nuclear and cell segmentation models to achieve highly accurate object segmentation accelerated by widely available GPU-based processing; moreover, we segment two additional cell compartments, i.e., nucleoli and mitochondria, to provide more specific biological information and improve interpretability of downstream analysis. Second, SPACe collects a carefully selected set of ∼400 image-based features as compared to the ∼4000 features collected by CellProfiler, reducing redundancy and data management loads; as we have shown, this implementation choice did not reduce the downstream analysis performance while making data exploration more efficient. Third, SPACe includes the calculation of directional EMD for feature analysis, as EMD values have been shown to be superior18 in capturing diverse responses in a heterogeneous cell population and can be used for both quality control and hit calling19,21–23, while providing the canonical per-well statistics of other platforms. The comparison between mean and EMD that we detail in Supplementary Fig. 1 suggests that central tendency metrics (mean values) are more effective at capturing the relevant features for MoA classification than distribution-based metrics (EMD values) when all features contribute equally, as is the case for ‘Percent Matching’. One possible explanation is that central tendency metrics can more robustly summarize the overall characteristics of a well, making them less sensitive to variations and noise present at the single-cell level/measurements. In the RF analysis, the ability to predict MoA accurately might benefit more from the detailed distribution information captured by EMD feature sets within SPACe. EMD indeed provides a more sensitive measure of the differences between feature sets, which allows the RF model to efficiently capture phenotypic variations resulting in more accurate MoA prediction. In addition, in contrast to ‘Percent Matching’, the weight/contribution of each feature to the prediction can differ in RF models. Therefore, the difference in the relative performance of EMD-based features in ‘Percent Matching’ and RF model outcomes suggest there is likely a subset of EMD-based features that better capture the MoA phenotype than any subset of mean-based features.
Due to these design properties, the SPACe pipeline can analyze the large imaging data generated by typical CP phenotypic screens very efficiently using the computational resources of a standard PC while maintaining sufficient morphological sensitivity and specificity to train predictive machine learning models for treatment targeted MoAs that perform as well, or better, than predictive models derived from much larger feature sets. This confirms the competitivity of our feature selection process given that MoA prediction is known to be a very challenging task15,40,41. However, SPACe does not replicate all feature types extracted by the CellProfiler pipeline used by the JUMP consortium. In particular, SPACe extracts limited features that capture the spatial relationship between cells contained in the sample. Therefore, in more complex samples such as 3D organoids or tissue, we would recommend modifying the feature set within SPACe to include this information.
The ability of SPACe to efficiently capture single-cell morphological features also explains the potential of this pipeline for providing biological interpretation of image-based fingerprints. In this study, we applied SPACe to define phenotypic fingerprints of common reference compounds, small targeted chemical sets, and a larger chemical library targeting cellular metabolism in U-2 OS cells and then expanding up to 18 different cell models. We demonstrated that very often, potent well-defined reference compounds do elicit a phenotypic response across most (but never all) cell lines. However, we found that changes in the underlying features are rarely the same, making it very challenging to generate “universal fingerprints” that could be used to establish universal reference compounds. In larger screens, most chemicals elicit cell-type and feature specific effects; with some cell lines being overall more responsive (i.e., U-2-OS, MDA-MB-231, and 5637). This is not unexpected due to each cell model representing a unique set of activated cellular signaling pathways upon which the chemical perturbation may alter. For laboratories initiating large screening campaigns, we would suggest testing a few cell lines of interest, plus U-2-OS as a gold standard, with a small number of control chemical and/or chemicals of interest, including the proposed “nuisance informer set” to help hit prioritization and triaging39. This is one reason why it is difficult to train a single prediction model to identify and predict MoA-specific phenotypes that are accurate across multiple cell models. To obviate to this problem, plates containing small-molecules with well-annotated MoAs can be used, as indeed it has been proven useful in the past31,39. However, exceptions exist, the most prominent being berberine chloride which affects only the mitochondrial compartment across almost all models we tested. This finding allowed us to extensively test and conclude that berberine chloride-induced phenotypic changes and the SPACe-extracted features are indeed highly reproducible across multiple biological replicates performed months apart, and across multiple cell models. Further work is needed to determine if this phenomenon is specific to berberine chloride and the mitochondrial compartment, or if other compartment-specific perturbations could be identified.
In conclusion, SPACe offers an open-source, user-friendly and efficient platform for the analysis of single-cells HT and HC phenotypic screenings. Due to its lightweight implementation, we expect that this computational software will be particularly beneficial to the large community of researchers who are interested in exploring CP analysis but do not have access to large computational resources, e.g., multi-core computing clusters, required to run current software solutions (e.g., CellProfiler) to process CP-generated feature data.
Methods
Cell culture and treatments
All cell lines were obtained directly from ATCC or from the BCM cell culture core (Department of Molecular and Cellular Biology). 5637 cells were grown in RPMI 1640 + 10% FBS; PC-3 and MDA-MB-468 in DME/F12 + 10% FBS, Hep G2 in DME/F12 + 10% FBS + 1.6% L-glutamine + 1% NEAA, and MCF10A in DME/F12 + 5% horse serum + 20 ng/ml EGF + 0.5 µg/ml hydrocortisone + 100 ng/ml cholera toxin + 10 µg/ml insulin. All other cell lines were grown in DMEM HG PR-free + 10% FBS + L-glutamine + Na pyruvate + penicillin/streptomycin.
Cells were routinely checked for mycoplasma contamination by DAPI staining and high magnification imaging42. Cells were plated in 384 well optical bottom plates (PerkinElmer cat# 6057302) at a density of 2000–3000 cells/well and allowed to settle at room temperature for 30 min prior to being placed in the incubator at 37 °C/5% CO2. After 24 h, without changing media, cells were then treated with the indicated compounds at indicated concentrations for an additional 24 h. All chemicals were reconstituted in DMSO at a stock concentration of 20 mM, which is 2000× of the highest concentration tested, unless otherwise specified. Information on reference chemicals is in Table 1, the other indicated compounds were acquired from: BioVision (WYE-687), Cayman Chemicals (actinomycin D, apigenin, MK-2206), Enzo Life Sciences (BYL-719), Selleckchem (FR 180204, OF-1, H3B-5942), Sigma (bexarotene, CDCA, DCA, DBT, fenofibrate, latrunculin B, 5-Nitro-2-(3-phenylpropylamino) benzoic acid, indeno[1,2,3 cd]pyrene, zearalenone, metochlopramide), and Tocris (ETP45658, FK866). The basic protocol requires plating cells in 20 µl of media, treatments are then added on top in 20 µl of media (final dilution of the chemicals is 1000×).
The cellular metabolism screening library (Cayman Chemical, cat #33705, batch #0609421) contains 160 known modulators of metabolic pathways. The library was arrayed in the test plates using a Labcyte Echo 550 acoustic liquid handler at the Texas A&M Institute of Bioscience and Technology, together with DMSO, berberine chloride, actinomycin D, and MK-2206, which were used as reference treatments (negative and positive). A few untreated wells were also included to confirm the control DMSO concentration was non-toxic and did not affect extracted features. Every compound in the library was used at 10 µM for 24 h of treatment before following the Cell Painting protocol described below.
Cell Painting (CP) processing and imaging
Following the JUMP consortium CP protocol1, multiwell plates were processed either manually or robotically (Beckman Coulter Biomek i5). As a reminder, the CP dyes include: DAPI/Hoechst 33342 (DNA/nucleus), Concanavalin A -AF488 (ER), SYTO14 (RNA, nucleolus), Phalloidin-AF568 (F-actin), WGA-AF555 (cytoplasm, Golgi), and MitoTracker DeepRed (mitochondria). In practice, 20 µl of MitoTracker (3000×) was added to live cells for 30 min and incubated at 37 °C. 20 µl of fixative (16% EM grade formaldehyde in PBS – final concentration in the well is 4%) was then added for 15 min at room temperature before proceeding with application of the fluorescent dyes. Slight changes were needed for robotic processing, specifically, the treatment media was removed, 20 µl of MitoTracker (1000×) was added and, after 30 min, 20 µl of fixative (8% EM grade formaldehyde in PBS, final concentration is 4%) was added.
Plates were imaged inside a three-day window on a Yokogawa CV8000 high-throughput spinning disk confocal microscope with sequential imaging of the five CP channels and appropriate laser/emission filter combinations as described in refs. 1,32. Specifically DAPI was excited with 405 nm laser and collected with a 445/45 filter, ConA with 488 nm laser and collected with a 525/50 filter, SYTO14 with a 488 nm laser and collected with 600/37 filter, actin/WGA with a 561 nm laser and collected with 600/37 filter, and, mitotracker was excited with 647 nm laser and collected with 676/29 filter. Imaging was performed with a 20×W/1.0 objective, and a short z-stack (3× z planes, 1 µm apart) was captured to correct for uneven plates. Images were first processed using Yokogawa software to correct non-uniform illumination, pixel misalignment between cameras and fluorescence channel crosstalk. Max intensity projections were saved as 16 bit TIFFS for automated image analysis. A minimum of nine fields of view were collected from each well in an experimental campaign. With these parameters, a single 384 well plate could be imaged in 3–5 h depending on exposure time and number of FOVs with a file size of ∼120 Gb. For campaigns with 4 or more plates, imaging was automated using a BioAssemblyBot 400 robot (Advanced Solutions), a fully integrated plate-loading solution synchronized with the Yokogawa CV8000.
SPACe image analysis pipeline details
For downloading and installing SPACe, the follow this link has all the instructions: https://github.com/dlabate/SPACe. Note, for the correct deployment of SPACe, the plate map must be properly formatted (see example downloadable at the GitHub link called platemap_template.xlsx inside the SPACe folder). SPACe was tested on images captured from a Yokogawa CV8000 HT spinning disk confocal, an ImageXpress Micro XLS widefield HT microscope and a Yokogawa CQ1 spinning disk confocal. SPACe includes the following steps (Fig. 1A):
Step 1 – load images and select hyperparameters (preview)
After loading the images, the preview function allows users to optimize the hyperparameters linked to identifying the nuclear and cell mask. A Google Colab notebook (https://github.com/dlabate/SPACe/blob/main/SPACe_colab.ipynb) is available for users to preview sample images and set best hyperparameters for segmentation using Cellpose. Hyperparameters include minimum diameter size for nucleus and cytoplasm (default parameters: 100 pixels for both), minimum number of nucleus, cytoplasm, mitochondria, and nucleoli (default parameters: 600, 700, 40, 200, respectively), minimum and maximum of nucleoli size (default parameters 0.005 and 0.3 control lower and upper threshold). Image intensities are rescaled before segmentation to ensure uniform processing across all image channels. Despite the high precision achieved by Cellpose segmentation, it is a good practice for users to use the SPACe output masks to inspect the segmentation performance in samples in which a dramatic change in extracted profile is detected, especially those with large changes in nuclear or cell area. Treatment-specific changes in cell morphology or dye labeling quality may result in systemic Cellpose segmentation errors not present in samples used for initial set-up and preview and may indicate a need for further Cellpose parameter optimization.
Step 2-3 – segmentation
This step detects each cell and, within each detected cell, identifies cytoplasm, nuclei, nucleoli, and mitochondria. It also ensures that each cellular subcompartment is assigned to the corresponding cell with the same label. We apply the generalist learning-based segmentation algorithm Cellpose v2.226,27 to the DAPI and ConA channels to generate nuclear, cell, and cytoplasm masks. Cellpose requires a user-defined hyperparameter that estimates expected cell and nuclear diameters in pixels, that should be optimized in Step 1 using the preview function. Following this segmentation step, we apply label matching to ensure that each segmented nucleus and cytoplasm is assigned to the corresponding cell with the same label. This routine corrects potential errors introduced by Cellpose that might mistakenly detect multiple nuclei per cytoplasm, or a cytoplasm without nucleus. Next, we apply another custom-designed segmentation routine based on Otsu and MaxEntropy thresholding to segment nucleoli (using the SYTO14 channel, within the nucleus mask) and mitochondria (using the MitoTracker channel, within the cytoplasmic mask), followed by another label matching step.
Step 4 - feature extraction and quality control
This step computes single-cell features (shape, intensity, and texture) using the five masks described above (cell, cytoplasm, nuclear, nucleolar, and mitochondria) and images from the five acquired channels. We selected ∼400 features that are widely used while paying special attention to features that are biologically interpretable. A complete list is included in Supplementary Table 1. In SPACe, the texture features are based on the well-established Pyrodiomics library43. SPACe calculates texture features across all distances and angles in the GLCM based on objects that are rescaled to 20 × 20 pixels and image intensities rescaled to 8 grayscale levels based on minimum and maximum object intensity. For each texture feature (contrast, dissimilarity, homogeneity, energy, correlation), SPACe generates a set of values corresponding to the various distance-angle combinations. SPACe then computes statistical descriptors—percentiles, mean, standard deviation (SD), and mean absolute deviation (MAD)—from these sets of values for each texture feature. The output is a vector that includes these statistical descriptors for each texture feature, essentially forming a 4D array where each dimension represents a texture feature and its corresponding statistical descriptors.
The QC routine is designed to establish a reliable ground truth for single-cell distributions in control samples (e.g., DMSO). The idea stems from our prior publication19 that demonstrated the value of distribution analysis as a quality control step for high-throughput microscopy assays and subsequent single-cell analyses. The QC step establishes a reference distribution for the DMSO negative control wells (eliminating outliers because of low object count or aberrant phenotypic profile). The reference distribution is defined as the median of the DMSO distribution in each experiment calculated from the distributions of each of the DMSO wells. The same QC step can also be applied to each set of replicate treatment wells, if appropriate, to discard outlier wells (i.e., wells with missing/low number of cells, artifacts, no or super response). Please note that the lower threshold of cell numbers to discard wells can be altered by the user, which might be useful under conditions of cytostatic compounds when the initial starting cell count is low or in situations where compounds might change cell adhesion.
Step 5 – Directional Earth Mover’s Distance (signed EMD)
For each well and feature, this step computes the EMD to the measured reference distribution. Before computing the EMD, features were normalized independently per plate by removing the top and bottom 2% and standardized in the interval [−1.1] using the Python command robustscale in scikit-learn. A sign (plus or minus) is next assigned to the EMD to indicate the direction of the response as compared to the reference distribution, plus sign indicates that the median value has increased with respect to DMSO and negative sign to indicate that it has decreased. The reference distribution is the median DMSO distribution calculated from the DMSO distributions across all DMSO wells in each experiment.
SPACe output
The results are available in a set of folders that contain intermediate steps (i.e., masks from each compartment – Step 2 & Step 3 folders), single-cell data (Step 4), and distance maps (Step 5). The distance map.csv file contains the signed EMD values for each analyzed well plus per-well mean and median values.
Analysis of JUMP MoA reference datasets
Seven JUMP MoA reference datasets (BR00115125-31) were downloaded from the JUMP Cell Painting gallery (https://registry.opendata.aws/cellpainting-gallery/). For analysis time calculations, all datasets were processed using CellProfiler version 4 (http://www.cellprofiler.org) and SPACe using a desktop PC equipped with a 16-core Intel i7 13700 CPU, NVIDIA 3070 GPU, and 32 GB memory. CellProfiler analysis utilized the segmentation and feature extraction pipelines provided by the JUMP consortium (https://github.com/broadinstitute/imaging-platform-pipelines/tree/master/JUMP_production). The illumination correction and quality control pipelines were excluded since equivalent operations are not present in SPACe.
Calculation of percent replicating and percent matching
Percent replicating and percent matching was evaluated as previously described1. In practice, single-cell data was aggregated at the per-well level by calculating the mean or EMD (SPACe only) value for each feature. All features were then normalized based upon plate Z-score. Using normalized values, we generated a null distribution of 20,000 random non-matching pair-wise correlations (Spearman) for each dataset. Using a threshold based on the 95% percentile of the null distribution, the percentage of replicating or matching well pairs with a correlation above this threshold was determined to calculate the percent replicating and percent matching values for each dataset and/or MoA. All calculations were completed using Biovia Pipeline Pilot (version 18.0) software.
Generation of random forest (RF) MOA prediction models
Data for model generation consisted of mean- or EMD-aggregated and plate Z-score normalized well-values generated using either CellProfiler’s CPA pipeline or SPACe from six JUMP MOA reference datasets. No data cleaning or handling of missing values was required. For each dataset, a total of 5 replicate RF models were generated and tested for a total of 30 models for each analysis method. For each model, replicate treated wells were evenly and randomly assigned for training or testing purposes. A RF model to predict MOAs was initialized with the following hyperparameters: 500 total trees, maximum depth of 50 trees, no minimum samples per leaf, and maximum features per tree set at the square root of total features. The RF model was trained using treatment replicate samples assigned to the training group. Each decision tree in the forest was constructed using the Bagging method, short for Bootstrap Aggregation. For each tree, a bootstrap sample of the original data is taken, and this sample is used to grow the tree. For model development, bootstrap sample is a dataset of the same size as the original one, but in which the same data record can be included multiple times. Excluding duplicates, a bootstrap sample on average contains about 63% of the original data records. Data excluded from the sample are called out-of-bag (OOB) data and used in model validation during tree construction. At each split in a tree, a random subset of features was considered for splitting. The Gini impurity criterion was used to measure the quality of splits. The trees were grown until they reached maximum depth or when the minimum number of samples per leaf (=1) was met. The trained RF model was then used to make MoA predictions of samples assigned to the testing group. Model performance was evaluated by the percent of correct predictions (accuracy) and a confusion matrix was generated. Models were generated using Pipeline Pilot and the R RF library.
Statistical analysis
All experiments were performed a minimum of three times (biological replicates) with a minimum of 4 wells/treatment (technical replicates), except for the Cell Metabolism screen where only two wells/treatment were used. In screening campaigns, 32 DMSO wells are used, while in other cases a minimum of 8 wells are used.
To compare fingerprints, Euclidean distance was measured between signed EMDs of all features in the treatment wells and the median DMSO control wells. Euclidean distance is a standard method used to measure the distance between two points in a multi-dimensional space and has been employed from gene expression analysis to image-based morphological profiling2,5,16,44,45. Groups were compared with non-parametric Kruskal–Wallis test. Heatmaps and clustering were generated using Orange Data Mining v.3.36, graphs were made in GraphPad.
Interpretation plots
The stacked bar graphs were prepared as follows: features were grouped into classes based on compartment and channel, then, for each treatment, features with EMD > 0.15 and EMD < −0.15 were assigned to the class of “changing features” (the threshold was based on the reproducibility analysis of berberine chloride treatment as shown in Fig. 3). Number of changing features were then transformed into a fraction by dividing over the total number of features (hence, sum = 1) and labeled as up (EMD > 0.15, red in the figures), down (EMD < −0.15, blue in the figures), or unchanging features (gray in the figures).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Software development, experimental approaches and imaging for this project was supported by the Center for Advanced Microscopy and Image Informatics (CAMII, CPRIT RP170719 (to MAM)) and the Integrated Microscopy Core at Baylor College of Medicine (funding from NIH (DK56338, CA125123, ES030285, S10OD030414 (to MAM)), and CPRIT (RR200043)).
Author contributions
F.S., A.R.T., C.D.C., M.G.M., E.A.M., and M.J.B. performed experiments; F.S., P.K.S., M.M., K.S., A.T.S., and C.D.C. analyzed experiments, P.K.S., M.M., K.S., and C.D.S. developed the software platform, F.S., P.K.S., D.L., and M.A.M. developed the original ideas and supervised experimental and analytical work; F.S., A.T.S., P.K.S., K.S., M.M., D.L., and M.A.M. wrote and edited the manuscript.
Peer review
Peer review information
Nature Communications thanks Jayme Dahlin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw data generated for the figures in this study are provided in the Source Data file. A set of images to run the code is provided within the GitHub repository linked to the SPACe pipeline. Additional images can be requested directly to the corresponding author. Source data are provided with this paper.
Code availability
The code is available in GitHub (https://github.com/dlabate/SPACe) and was deposited in Zenodo (10.5281/zenodo.13821484).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fabio Stossi, Email: fstossi@gmail.com.
Michael A. Mancini, Email: mancini@bcm.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54264-4.
References
- 1.Cimini, B. A. et al. Optimizing the Cell Painting assay for image-based profiling. Nat. Protoc.18, 1981–2013 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray, M. A. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc.11, 1757–1774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gustafsdottir, S. M. et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS ONE8, e80999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loo, L. H., Wu, L. F. & Altschuler, S. J. Image-based multivariate profiling of drug responses from single cells. Nat. Methods4, 445–453 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Perlman, Z. E. et al. Multidimensional drug profiling by automated microscopy. Science306, 1194–1198 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Stossi, F. et al. High throughput microscopy and single cell phenotypic image-based analysis in toxicology and drug discovery. Biochem. Pharmacol.216, 115770 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Laber, S. et al. Discovering cellular programs of intrinsic and extrinsic drivers of metabolic traits using LipocyteProfiler. Cell Genom.3, 100346 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray, M. A. et al. A dataset of images and morphological profiles of 30 000 small-molecule treatments using the Cell Painting assay. GigaScience6, 1–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caicedo, J. C. et al. Cell Painting predicts impact of lung cancer variants. Mol. Biol. Cell33, ar49 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekaran, S. N. et al. Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations. Nat. Methods21, 1114–1121 (2024). [DOI] [PMC free article] [PubMed]
- 11.Haghighi, M., Caicedo, J. C., Cimini, B. A., Carpenter, A. E. & Singh, S. High-dimensional gene expression and morphology profiles of cells across 28,000 genetic and chemical perturbations. Nat. Methods19, 1550–1557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohban, M. H. et al. Systematic morphological profiling of human gene and allele function via Cell Painting. eLife6, e24060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinform.22, 433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol.16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Fossa, F. et al. Interpreting image-based profiles using similarity clustering and single-cell visualization. Curr. Protoc.3, e713 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caicedo, J. C. et al. Data-analysis strategies for image-based cell profiling. Nat. Methods14, 849–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D. & Carpenter, A. E. Image-based profiling for drug discovery: due for a machine-learning upgrade? Nat. Rev. Drug Discov.20, 145–159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson, Y. E. et al. A statistical framework for high-content phenotypic profiling using cellular feature distributions. Commun. Biol.5, 1409 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stossi, F. et al. Quality control for single cell imaging analytics using endocrine disruptor-induced changes in estrogen receptor expression. Environ. Health Perspect.130, 27008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stossi, F. et al. Single-cell distribution analysis of AR levels by high-throughput microscopy in cell models: application for testing endocrine-disrupting chemicals. SLAS Discov.25, 684–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gough, A., Shun, T. Y., Lansing Taylor, D. & Schurdak, M. A metric and workflow for quality control in the analysis of heterogeneity in phenotypic profiles and screens. Methods96, 12–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gough, A. H. et al. Identifying and quantifying heterogeneity in high content analysis: application of heterogeneity indices to drug discovery. PLoS ONE9, e102678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gough, A. et al. Biologically relevant heterogeneity: metrics and practical insights. SLAS Discov.22, 213–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray, M. A., Fraser, A. N., Hasaka, T. P. & Carpenter, A. E. Workflow and metrics for image quality control in large-scale high-content screens. J. Biomol. Screen17, 266–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh, S., Bray, M. A., Jones, T. R. & Carpenter, A. E. Pipeline for illumination correction of images for high-throughput microscopy. J. Microsc.256, 231–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods19, 1634–1641 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cross-Zamirski, J. O. et al. Label-free prediction of cell painting from brightfield images. Sci. Rep.12, 10001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyffeler, J. et al. Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol. Appl. Pharmacol.389, 114876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.JUMP-Cell Painting Consortium [Internet] https://jump-cellpainting.broadinstitute.org/results (2023).
- 31.Willis, C., Nyffeler, J. & Harrill, J. Phenotypic profiling of reference chemicals across biologically diverse cell types using the cell painting assay. SLAS Discov.25, 755–769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tromans-Coia, C. et al. Assessing the performance of the Cell Painting assay across different imaging systems. Cytom. Part J Int. Soc. Anal. Cytol.103, 915–926 (2023). [DOI] [PMC free article] [PubMed]
- 33.Blake, D. R. et al. Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer. Sci. Signal.12, eaav7259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ai, X. et al. Berberine: a review of its pharmacokinetics properties and therapeutic potentials in diverse vascular diseases. Front. Pharmacol.12, 762654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu, C. et al. Identification of berberine as a novel drug for the treatment of multiple myeloma via targeting UHRF1. BMC Biol.18, 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simioni, C. et al. The AKT inhibitor MK-2206 is cytotoxic in hepatocarcinoma cells displaying hyperphosphorylated AKT-1 and synergizes with conventional chemotherapy. Oncotarget4, 1496–1506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dogra, N., Kumar, A. & Mukhopadhyay, T. Fenbendazole acts as a moderate microtubule destabilizing agent and causes cancer cell death by modulating multiple cellular pathways. Sci. Rep.8, 11926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, D., Lee, J. H. & Yoon, S. P. Anti-cancer effects of fenbendazole on 5-fluorouracil-resistant colorectal cancer cells. Korean J. Physiol. Pharmacol.26, 377–387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahlin, J. L. et al. Reference compounds for characterizing cellular injury in high-content cellular morphology assays. Nat. Commun.14, 1364 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng, Y., Mitchison, T. J., Bender, A., Young, D. W. & Tallarico, J. A. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat. Rev. Drug Discov.8, 567–578 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Biswas, S. High content analysis across signaling modulation treatments for subcellular target identification reveals heterogeneity in cellular response. Front Cell Dev. Biol.8, 594750 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung, H. et al. Detection and treatment of mycoplasma contamination in cultured cells. Chang Gung Med. J.26, 250–258 (2003). [PubMed] [Google Scholar]
- 43.van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res.77, e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woehrmann, M. H. et al. Large-scale cytological profiling for functional analysis of bioactive compounds. Mol. Biosyst.9, 2604–2617 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Trapotsi, M. A. et al. Cell morphological profiling enables high-throughput screening for PROteolysis TArgeting Chimera (PROTAC) phenotypic signature. ACS Chem. Biol.17, 1733–1744 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data generated for the figures in this study are provided in the Source Data file. A set of images to run the code is provided within the GitHub repository linked to the SPACe pipeline. Additional images can be requested directly to the corresponding author. Source data are provided with this paper.
The code is available in GitHub (https://github.com/dlabate/SPACe) and was deposited in Zenodo (10.5281/zenodo.13821484).