Abstract
Non-small cell lung cancer (NSCLC) is histologically classified into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LSCC). However, some tumors are histologically ambiguous and other pathophysiological features or microenvironmental factors may be more prominent. Here we report integrative multiomics analyses using data for 229 patients from a Korean NSCLC cohort and 462 patients from previous multiomics studies. Histological examination reveals five molecular subtypes, one of which is a NSCLC subtype with PI3K-Akt pathway upregulation, showing a high proportion of metastasis and poor survival outcomes regardless of any specific NSCLC histology. Proliferative subtypes are present in LUAD and LSCC, which show strong associations with whole genome doubling (WGD) events. Comprehensive characterization of the immune microenvironment reveals various immune cell compositions and neoantigen loads across molecular subtypes, which predicting different prognoses. Immunological subtypes exhibit a hot tumor-enriched state and a higher efficacy of adjuvant therapy.
Subject terms: Non-small-cell lung cancer, Cancer genomics, Proteomics, Immunogenetics
Subtyping of non-small cell lung cancer can be challenging based on pathology. Here, the authors utilise multi-omics analysis of 229 patients to identify further subtypes, and altered immune composition between subtypes.
Introduction
Lung cancer is a major health concern worldwide, accounting for 18% of global cancer-related deaths1. Surgical resection, with or without postoperative adjuvant therapy, is currently the first-line treatment for locally advanced or early-stage lung cancers2. In advanced cases, genomics-based targeted therapies and immunotherapies using immune checkpoint inhibitors provide promising treatment options3, often in addition to chemotherapy. Although the mortality rates associated with lung cancer are continuously decreasing with appropriate surgical and medical treatment4, the 5-year survival rates for localized, regional, and metastatic non-small cell lung cancer (NSCLC) remain unsatisfactory (64%, 37%, and 8%, respectively), mainly due to recurrence after treatment5. Therefore, tailoring treatments based on patient stratification according to molecular characteristics is of increasing interest to improve patient survival.
High-throughput omics approaches have facilitated the classification of NSCLC molecular subtypes and candidate molecular targets as well as prognostic or predictive biomarkers for lung cancer treatment6. Initially, The Cancer Genome Atlas Research Network identified the molecular pathways of lung cancer and classified lung squamous cell carcinoma (LSCC) into classical, basal, secretory, and primitive subtypes7, and lung adenocarcinoma (LUAD) into terminal respiratory unit, proximal inflammatory, and proximal proliferative subtypes8. Recently, proteomic and proteogenomic approaches9–14 have been introduced to identify novel subtypes and their druggable targets.
Although these efforts have advanced our understanding of cancer biology, the clinical implementation of these findings in high-throughput approaches remains challenging15,16. First, many proteogenomic studies have focused on a single subtype of NSCLC (such as LUAD or LSCC), but the approach excludes cases in which the pathological classification is ambiguous and/or discrepant17,18, which eventually leads to the formation of study cohort that may not be representative of real NSCLC population. Second, the sensitive prediction of postoperative survival beyond those achieved using conventional prognostic factors is a prerequisite for personalized surgical oncology treatment. Further characterization of causative biomarkers associated with prognosis may identify crucial treatment targets19, and requires a multiomics-based analysis of well-annotated clinical data. Finally, most multiomics data have been generated from resected lung cancer tissues, and adjuvant chemotherapy after surgery is the primary treatment for locally advanced lung cancer. Adjuvant chemotherapy largely relies on platinum doublet and platinum-based chemotherapeutic regimens20 in addition to a cytotoxic agent (e.g., pemetrexed, gemcitabine, vinorelbine, or paclitaxel). Although this approach has improved patient survival21, few studies22 have investigated the molecular basis of the response to postoperative adjuvant treatment, and consensus biomarkers for predicting treatment efficacy have not yet been identified.
In this study, we conducted a comprehensive multiomics analysis to define the molecular subtypes of NSCLC using a Korean NSCLC discovery cohort of 229 patients and a replication cohort of 462 NSCLC patients from previous multiomics studies. Our aim was to expand the scope of histological subtyping and identify molecular subtypes of NSCLC with potential prognostic and therapeutic implications. We further characterized the subtypes by integrating a large-scale single-cell RNA sequencing dataset23 of NSCLC and evaluated their cellular specificity and histopathological relevance. In addition, our study included an extensive histological review of patient data for tumor-infiltrating lymphocytes (TILs), identified potential neoantigens and cryptic peptides characterizing the immune microenvironment, and noted the varying efficacy of adjuvant therapies between subtypes. Overall, our study represents a considerable advance in the field of NSCLC research and has important implications for precision medicine and personalized therapies.
Results
Identification of subtypes in NSCLC patients by multiomics
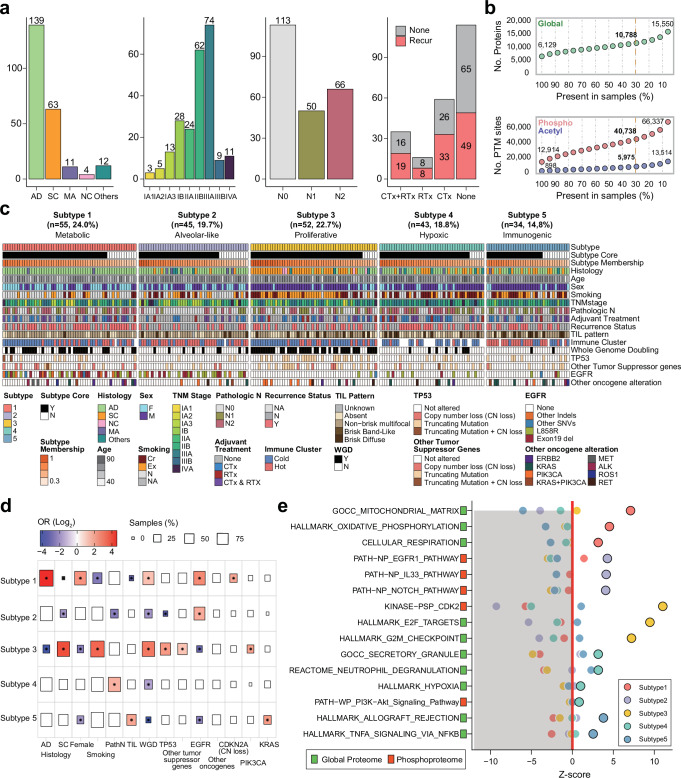
The multiomics analysis utilized a retrospective cohort of 229 Korean patients diagnosed with NSCLC at Asan Medical Center in Seoul, Korea (Supplementary Data 1a). Tumor samples with matched normal tissues or blood samples were collected via surgery between 2010 and 2019. Patient demographics, disease parameters (including histology and tumor-node-metastasis [TNM] staging), survival, and treatment response indicated good coverage of disease severity and patient population. Histologically, the tumor samples included 139 adenocarcinomas (LUADs, 61%), 63 squamous cell lung carcinomas (LSCCs, 27%), and 27 tumors of other types (12%) (Fig. 1a, Supplementary Data 1a). LUAD cases were almost equally distributed between the sexes (66 males and 74 females), whereas LSCC cases were found mostly among males (97%, n = 61), which was previously shown in other Korean LSCC cohort24. Self-reported smoking status indicated that 61% of the patients had a smoking history, with a higher prevalence of smoking in males and patients with LSCC. The TNM-based stages ranged from IA1 to IVA, with approximately 40% of the patients having late-stage disease (IIIA, n = 75; IIIB, n = 9; IVA, n = 11; Fig. 1a). Approximately half of the patients had lymph node metastases (51%, n = 116) at the time of pathological diagnosis after surgery. Adjuvant therapy, including chemotherapy (CTx) or radiation therapy (RTx), was administered to 48% of the patients (110/229 patients; 59 CTx, 16 RTx, and 35 CTx and RTx) according to the NCCN guidelines. Tumor recurrence was observed in 54% of the patients who received adjuvant therapy, and the recurrence rate was similar across treatments (CTx, 56%, 33/59; RTx, 50%, 8/16; CTx and RTx, 54%, 19/35) (Fig. 1a) and histological diagnoses (53%, AD, 72/137; 44%, SC, 27/62) (Supplementary Fig. 1a).
Fig. 1. Identification of multiomics subtypes in Korean NSCLC patients.
a Summary of clinical information for the Korean NSCLC patient cohort. Bar plots show NSCLC histology, tumor stages, pathologic N status, and tumor recurrence status with adjuvant treatment. b Numbers of quantified proteins and PTM sites identified in global proteomic, phosphoproteomic, and acetylproteomic analyses. The number of features quantified at <30% missing values across 229 samples is represented by the yellow line. c Overview of NMF clustering. Other tumor suppressor genes consisted of CDKN2A, STK11, KEAP1, RB1, PPP2R1A, and SMARCA4. Other oncogene alterations consisted of frameshift deletions; in-frame deletion/insertion and missense mutations in KRAS, ERBB2, and PIK3CA; exon skipping in MET; and gene fusion in ALK, ROS1, and RET. Copy number loss was defined as homozygous deletion (absolute copy number <0.5). d Enrichment of the five identified NMF clusters for clinical variables. Tests indicating statistical significance (P < 0.05, two-sided Fisher’s exact test) are colored according to the odds ratio (OR). Box size indicates the proportion of the cohort characterized by a given clinical variable. e Pathway enrichment analyses of the five subtypes using GSVA and PTM-SEA. Pathways with statistical significance (FDR < 0.05, permutation) and positive enrichment scores (z-score) are represented by dots.
We generated genomic, transcriptomic, proteomic, phosphoproteomic, and acetylproteomic datasets from the samples. A genomic dataset was generated by whole-exome sequencing (WES) of 228 normal adjacent tissue (NAT)-matched tumors and one tumor-only sample with a read depth sufficient for variant discovery (tumor: ~300X, NAT: ~100X). We observed 33,301 somatic small mutations that contained single nucleotide variants (SNVs) and indels (on average, 145 per sample), 470,836 copy-number alterations encompassing amplification, gain, heterozygous deletion, and homozygous deletion (on average, 2056 per sample) and a 2.7 tumor mutation burden (TMB) score in 229 tumors. For transcriptomic analysis, we performed bulk RNA-seq for 205 tumors and 85 matched NATs for deep coverage (approximately 120 M reads per sample), enabling gene expression quantification and alternative splicing isoform discovery. We acquired 60,688 transcripts and selected 20,088 transcripts, based on low-count genes across samples, for subsequent analysis. For proteomic analysis using tandem mass tag (TMT)-based isobaric labeling, proteomic data were collected from 229 tumor samples and 26 matched NATs. A total of 10,788 proteins, 40,738 phosphosites, and 5975 acetylation sites were observed in at least 30% of the samples and quantified as a log2 ratio to the common reference (CR) sample (Fig. 1b).
For multiomics analysis, we integrated proteomic, phosphoproteomic, and acetylproteomic data and conducted non-negative matrix factorization (NMF) clustering to identify multiomics subtypes in the 229 NSCLC samples. We identified five multiomics subtypes: metabolic (Subtype 1), alveolar-like (Subtype 2), proliferative (Subtype 3), hypoxic (Subtype 4), and immunogenic (Subtype 5) (Fig. 1c), characterized based on genetic mutations, clinical phenotypes, and molecular pathways (Fig. 1d, e, Supplementary Data 1b–d).
Subtype 1 was composed mainly of LUAD females (64%, 35/55) with EGFR and TP53 mutations, as well as a high frequency of whole genome doubling (WGD) events (i.e., phenomena in which more than half of the chromosomes are gained; Fig. 1c), suggesting a chromosomally unstable co-driven subtype. Significant enrichment of CDKN2A copy number loss in Subtype 1 (OR: 3.63, P = 2.35 × 10−2, Fisher’s exact test) also supports the observation (Fig. 1d). Subtype 2 mainly comprised patients with LUAD (71%, 32/45) with EGFR mutations (49%, 22/45) and without WGD events (Fig. 1c), suggesting a chromosomally stable oncogene-driven subtype. These samples showed a significantly lower frequency of TP53 mutations (6%; odds ratio [OR]: 0.04, P = 3.1 × 10−9, Fisher’s exact test; Fig. 1d) and much lower tumor mutational burden (TMB) than samples representing other subtypes (Subtype 2 = 0.9 variants per Mb, others = 3.2 variants per Mb; P = 3.6 × 10−10, Wilcoxon ranked sum test) (Supplementary Fig. 1b, c). Despite the upregulation of the EGFR pathway in both subtypes, molecular pathways were specifically enriched for each cluster. Subtype 1 exhibited significant upregulation of proteins involved in oxidative phosphorylation (adjusted P = 1.7 × 10−6, Wilcoxon rank-sum test), mitochondrial matrix (adjusted P = 8.1 × 10−9, Wilcoxon rank-sum test), and cellular respiration (adjusted P = 4.2 × 10−5, Wilcoxon rank-sum test), indicating an association with metabolic pathways. Conversely, Subtype 2 was characterized by a significant upregulation of phosphorylation in the IL-33 (adjusted P = 1.8 × 10−15, Wilcoxon rank-sum test) and Notch pathways (adjusted P = 2.4 × 10−16, Wilcoxon rank-sum test) (Fig. 1e), implicating these molecular features in early tumorigenesis and early-stage disease.
Subtype 3 was significantly associated with WGD events (OR: 13.5, P = 4.6 × 10−10, Fisher’s exact test), TP53 (OR: 13.5, P = 4.6 × 10−10, Fisher’s exact test) and PIK3CA mutations (OR: 3.89, P = 3.16 × 10−2, Fisher’s exact test), and was more prevalent in patients with LSCC (65%, 34/52 patients), in males (83%, 43/52 patients), and smokers (85%, 44/52 patients) (Fig. 1d). Subtype 3 exhibited a highly proliferative phenotype, as evidenced by significant enrichment in cell cycle-related pathways, including E2F/MYC target (adjusted P = 4.7 × 10−22, 1.3 × 10−20, Wilcoxon rank-sum test), G2M checkpoint (adjusted P = 2.4 × 10−23, Wilcoxon rank-sum test), and cyclin-dependent kinase (CDK) target pathways (adjusted P = 5.3 × 10−24, Wilcoxon rank-sum test) (Fig. 1e). Thus, this subtype can be defined as a chromosomally unstable tumor suppressor-deficient proliferative subtype.
Subtype 4 was not associated with any specific histological type of NSCLC but was significantly enriched for metastasis (OR: 3.0, P = 5.6 × 10−3, Fisher’s exact test). We found that phosphorylated sites in this subtype were upregulated in hypoxia (adjusted P = 2.3 × 10−4, Wilcoxon rank-sum test), PI3K-Akt (adjusted P = 4.7 × 10−3, Wilcoxon rank-sum test), and neutrophil degranulation (adjusted P = 1.7 × 10−4, Wilcoxon rank-sum test) pathways, and proteins were also enriched for neutrophil degranulation (adjusted P = 4.6 × 10−6, Wilcoxon rank-sum test), suggesting a potential role in the promotion of tumor migration, invasion, and metabolism in tumor metastasis. Therefore, Subtype 4 can be considered as a chromosomally stable mesenchymal subtype.
Subtype 5 showed a significantly elevated proportion of tumor-infiltrating lymphocyte (TIL)-associated patterns (OR: 3.6, P = 1.2 × 10−2, Fisher’s exact test; Fig. 1d) and enrichment of immune-related pathways, such as TNFα signaling via NF-κB (adjusted P = 1.1 × 10−6, Wilcoxon rank-sum test, Fig. 1e), suggesting that this subtype was a high-immune and chromosomally stable tumor-suppressor-driven inflammatory subtype. KRAS mutation was significantly enriched in Subtype 5 (OR: 4.51, P = 3.53 × 10−2, Fisher’s exact test) (Supplementary Fig. 1d), with four cases presenting concurrent TP53 mutation. STK11 and KEAP1 mutations were each identified only in 1 case, although the trends were not statistically significant.
A NSCLC subtype associated with poor prognosis and frequent metastasis
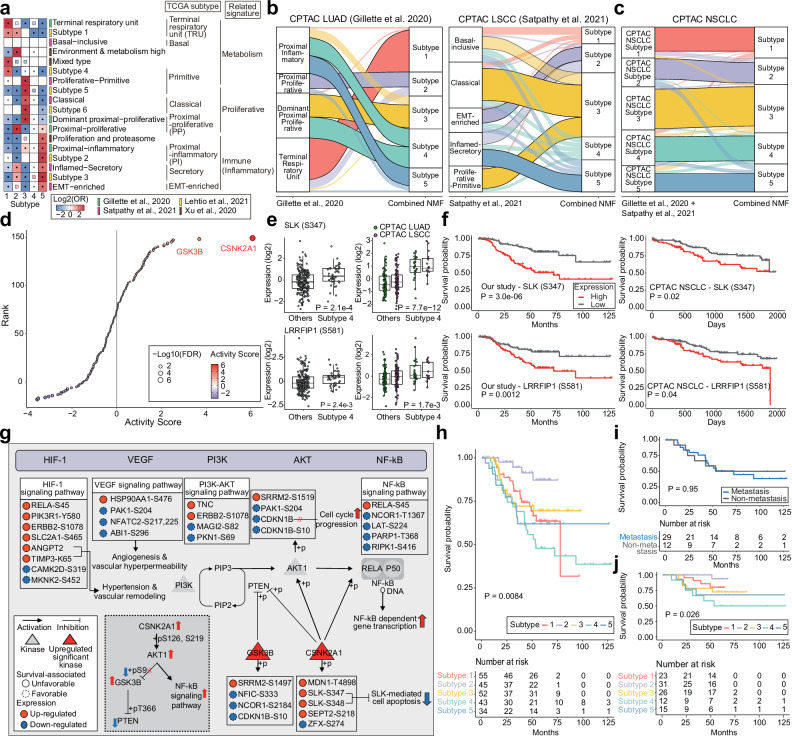
To replicate our subtype classification, we utilized multiomics or proteomics data from 462 patients with NSCLC obtained from previous studies, including two LUAD studies by Gillette et al.10 (n = 110) and Xu et al.11 (n = 103), an LSCC study by Satpathy et al.13 (n = 108), and an NSCLC study by Lehtio et al.14 (n = 141). We compared the top features of the NMF subtypes between our cohort and those of other studies and found significant overlaps (FDR < 0.01, Fisher’s exact test): the terminal respiratory unit subtype of adenocarcinoma (Subtype 1), inflammatory subtypes (Subtypes 2 and 5), and proliferative subtypes of LUAD and LSCC (Subtype 3) (Fig. 2a and Supplementary Data 2a). In contrast, Subtype 4 was distinct, showing enrichment for phosphorylation features associated with the EMT-enriched (LSCC)13 subtype and acetylation features from the inflammatory subtypes of LUAD and LSCC (Supplementary Fig. 2a, b). We also performed a combined NMF analysis for a total of 447 patients with NSCLC by integrating proteome, phosphoproteome, and acetylome datasets from our study with those from previous studies10,13 (hereafter called the “combined CPTAC dataset”). We identified five subtypes, referred to as “Combined NMF” (Fig. 2b). The combined NMF showed a highly consistent pattern with the feature overlap analysis (Fig. 2a and Supplementary Data 2a) and confirmed that the four subtypes (Subtype 1, 2, 3, 5) showed a consistency with the previously identified NMF subtypes (Fig. 2b, Supplementary Fig. 2c and Supplementary Data 2b). Subtype 4 did not cluster with any single subtype of LUAD or LSCC in the combined NMF analysis (Fig. 2b). A comparison without the Korean cohort (“CPTAC NSCLC”), constituting 51% (229/447) of the Combined NMF, showed 77% of patients maintained their subtype classification when including the Korean cohort (Fig. 2c). Additionally, Subtype 4 consistently comprised 18.8% (43/229) of the Korean NSCLC cohort and 18.3% (40/218) of the combined CPTAC dataset without histological types. Collectively, our findings suggest that Subtype 4 represents a NSCLC subtype that requires further clinical and molecular characterization.
Fig. 2. Novel NSCLC subtype associated with poor survival.
a Overlap of subtype features between the five NMF subtypes in this study and subtypes identified in previous NSCLC multiomics studies. Protein enrichment is in the heatmap. Full rectangle and asterisk indicate significant overlaps (Two-sided fisher’s exact test adjusted P ≤ 0.05, Benjamini-Hochberg adjustment), faint rectangle indicates overlaps which pass only nominal P value (Two-sided fisher’s exact test P ≤ 0.05, two-sided fisher’s exact test adjusted P > 0.05), and blank indicates overlaps which is not significant (two-sided fisher’s exact test P > 0.05). b, c Reclassification of samples from previously defined multiomics subtypes10,13 according to our combined NMF subtypes. The statistical significance of the relationship is visually represented by the clarity and transparency of the lines (Supplementary Fig. 2c). d Subtype 4-specific kinase activity scores estimated from phosphoproteomic data and the kinase-substrate network database (PHONEMeS). The colors of the points represent the estimated kinase activity scores. The sizes of the points represent the -log10(FDR) of the kinase activity estimates. There were two significantly upregulated kinases: CSNK2A1 and GSK3B (FDR < 0.05). e Expression of poor prognosis markers containing phosphorylated sites on SLK (S347) and LRRFIP1 (S581) is shown for our study (Subtype 4, n = 43; others, n = 186) and CPTAC (LUAD (Subtype 4, n = 26; others, n = 84), LSCC (Subtype 4, n = 15; others, n = 93)). Wilcoxon rank-sum test was performed to test the differences in expression. The color of the dots in the right panel represents the study type in CPTAC. For box-plots, middle line, median: box edges, 25th and 75th percentiles; whiskers, most extreme points that do not exceed ±1.5 × IQR. f Cancer-specific overall survival length according to the expression of poor prognosis markers in our study and the CPTAC dataset (integrated with LUAD and LSCC). The p-value was calculated with the log-rank test. g Intracellular signaling pathways underlying poor prognosis in Subtype 4. The blue box represents the main signaling pathways, including the HIF-1, VEGF, PI3K-AKT, and NF-κB signaling pathways. The red triangular nodes are kinases identified as significantly upregulated in Subtype 4. The colors of the points represent the log2FC values obtained through differential expression (DE) analysis of Subtype 4 and the other subtypes. The border style of the point indicates the prognostic direction of the feature. h Cancer-specific overall survival length between our subtypes indicating significant changes in survival probability (y-axis) over time (x-axis). i Survival curves for patients with (n = 35) and without metastasis (n = 8) in Subtype 4 (n = 43) and ( j) patients without metastasis in each subtype (n = 91). The p-value was calculated with the log-rank test (h–j).
To examine the molecular characteristics of Subtype 4, we extracted distinct features (proteins, phosphorylation, or acetylation) of the subtypes in our NMF clustering analysis (Supplementary Data 1e). We found that the majority of NMF features present in Subtype 4 were phosphorylated sites (96%, 178/186, Supplementary Data 1e), indicating that phospho-kinase interactions are a major signature. Therefore, we investigated the kinase activity of this subtype using phosphoproteomic data. We found significant enrichment of two kinases, CSNK2A1 (FDR = 2.3 × 10−7) and GSK3B (FDR = 1.9 × 10−2), which are known to phosphorylate various proteins in the PI3K-AKT signaling pathway, in Subtype 4 compared to other subtypes (Fig. 2d, Supplementary Fig. 2d and Supplementary Data 2c). Upon evaluating the relationship between the activity and expression levels of significant kinases (P < 0.05), we observed a moderate correlation (Supplementary Fig. 2e). Survival analysis based on feature expression (Supplementary Data 2d) showed that most of the unfavorable prognostic factors were phosphorylated sites differentially expressed in Subtype 4 (91%, 104/114) (Supplementary Fig. 2f). Notably, STE20-like serine/threonine-protein kinase at serine 347 (SLK (S347)), a protein phosphorylated by CSNK2A1, was significantly upregulated in Subtype 4 (adjusted P = 8.0 × 10−3, Benjamini–Hochberg adjustment, Supplementary Data 3a) and associated with unfavorable prognostic features (P = 3.0 × 10−6, log-rank test) (Fig. 2e, f). In the combined CPTAC dataset, we also found increased phosphorylation of SLK (S347) in Subtype 4 and poor survival outcomes (Fig. 2e, f, Supplementary Fig. 2g and Supplementary Data 2e). SLK mediates apoptosis downstream of the ErbB2 and PI3K pathways25 and is activated in a CSNK2A1-dependent manner26. Recent studies have reported that high SLK expression is associated with reduced overall survival in HER2-positive patients27 and in glioma28. Using both our cohort and the CPTAC cohort to check the ROC curve, we found that SLK (S347) is an effective marker for distinguishing Subtype 4 not only in our cohort but also more effectively in the CPTAC cohort (Supplementary Fig. 2h). Collectively, these results suggest that SLK participates in tumor progression and could be a key marker specific to Subtype 4. Furthermore, Subtype 4 showed a significant upregulation of leucine-rich repeat flightless-interacting protein 1 at serine 581 (LRRFIP1 (S581)) phosphorylation (adjusted P = 1.3 × 10−2, Benjamini–Hochberg adjustment, Supplementary Data 3a), which was correlated with unfavorable prognostic features (P = 1.2 × 10−3, log-rank test) (Fig. 2e, f). Similarly, in the combined CPTAC dataset, elevated phosphorylation of LRRFIP1 (S581) was observed in Subtype 4, which coincided with poor survival outcomes (Fig. 2e, f, Supplementary Fig. 2g and Supplementary Data 2e). Notably, LRRFIP1 stimulates the epithelial–mesenchymal transition (EMT) pathway by modulating the Wnt/β-catenin signaling pathway29. A recent study30 demonstrated that high LRRFIP1 expression was associated with reduced overall survival in glioma. These findings collectively suggest that LRRFIP1 may also contribute to cellular invasion and metastasis and could serve as a key marker specific to Subtype 4.
Subtype 4 included numerous prognostic features in the HIF-1 and PI3K-AKT signaling pathways (Fig. 2g and Supplementary Fig. 2i). Among these unfavorable features, the significantly upregulated ANGPT2 protein is known to increase in a hypoxic environment, which can promote the release of pro-angiogenic cytokines, such as VEGF, through HIF-1 accumulation. We also observed increased acetylation of TIMP3 at K65, which is presumably involved in the ANGPT2-induced hypertensive response. In contrast, CDKN1B, which was significantly downregulated (adjusted P = 7.1 × 10−11, two-sided t-test, Supplementary Data 3a), was a favorable prognostic feature in our cohort (P = 1.3 × 10−4, log-rank test, Supplementary Data 2d) and the combined CPTAC dataset (P = 1.6 × 10−2, log-rank test). Given that the loss of CDKN1B increases the risk of cancer metastasis31, the reduced expression of CDKN1B implies an advanced phenotype of Subtype 4.
We evaluated the clinical significance of Subtype 4 according to the survival rates of patients with this subtype. Subtype 4 in the Korean NSCLC cohort was associated with a significantly poorer survival rate compared to that in the other subtypes (P = 8.4 × 10−3, log-rank test, Fig. 2h), possibly influenced by stage distribution. Since Subtype 4 showed the highest rate of metastasis among the five subtypes, metastasis may have an important impact on the poor survival outcomes. Interestingly, there was no variation in the survival rates of patients with and without metastasis in Subtype 4 (P = 9.5 × 10−1, log-rank test, Fig. 2i), suggesting that metastasis may not be the sole mechanism leading to poorer survival. When comparing survival rates among non-metastatic patients of the five subtypes, we observed a significant disadvantage for patients with Subtype 4 (P = 2.6× 10−2, log-rank test, Fig. 2j), indicating that other biological factors may contribute to the poor prognosis of this subtype. In contrast, we did not find any significant difference in overall survival between Subtype 4 and the other subtypes in the combined CPTAC cohort (P = 7.9 × 10−1, log-rank test, Supplementary Fig. 2j), indicating the effect of differences in ethnicity and treatment records.
Cellular landscape of the five subtypes of NSCLC
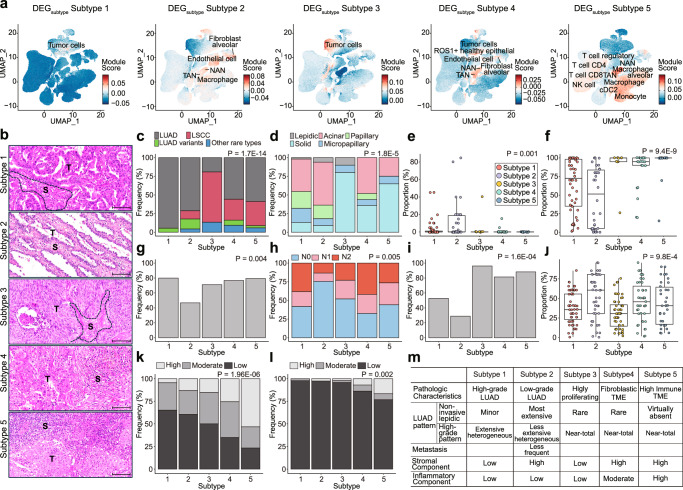
Exploring the tumor microenvironment is crucial for understanding the mechanisms underlying cancer progression and for developing effective therapeutic strategies to target not only cancer cells but also the surrounding microenvironment. We assessed the tumor microenvironment of the five NSCLC subtypes based on cell type specificity through a comparative analysis of subtype-specific genes with a range of cell type-specific genes using an integrated single-cell RNA (scRNA) sequencing dataset23 of NSCLC patients. First, we computed the differentially expressed genes (DEGs) for each subtype compared to the NAT samples (Supplementary Data 3b). As expected, we found that the DEG sets of all five subtypes were significantly upregulated in tumor cell types (FDR < 0.001, permutation; Supplementary Fig. 3a and Supplementary Data 3c), highlighting the overall degree of tumorigenicity of the subtypes.
Next, we identified DEGs for each subtype by comparing all subtypes (DEGsubtype, e.g., Subtype 1 vs. other subtypes) (Supplementary Data 3d). In Subtypes 1 and 3, the DEGsubtype was enriched for cell types related to tumors (FDR < 0.01, permutation): Subtype 1 corresponding to LUAD and LUAD mitotic cell types and Subtype 3 corresponding to LSCC and LSCC mitotic cell types as defined23, suggesting actively proliferating tumor cell components (Fig. 3a, Supplementary Fig. 3b and Supplementary Data 3e). In contrast, the DEGsubtype sets of Subtypes 2, 4, and 5 were enriched in neutrophils. While a high proportion of NAT-associated neutrophils (NANs) was present in these subtypes (Supplementary Data 3e), the tumor-associated neutrophil (TAN) cell types were enriched in Subtypes 4 and 5, with the largest proportion in Subtype 4. Furthermore, the DEGsubtype set of Subtype 2 appeared to be enriched for alveolar-type fibroblasts and endothelial cells, implying that these tumors have stromal components similar to those of the normal alveolar interstitium (Supplementary Data 3e). Most immune-related cell types were enriched in the DEGsubtype set of Subtype 5 (Supplementary Data 3e), reflecting a high proportion of TIL patterns.
Fig. 3. Landscape of cell type-specific subtype characteristics.
a UMAP plot of single-cell types specific to Subtypes 1 to 5, using each DEGsubtype. The color of each point represents the module score of each cell; the more relevant the module is to the cell type, the higher the score and the redder the color. UMAP information was obtained from the original study23. b Representative histologic images of the subtypes. The tumor cell (T) and stromal (S) components are separately labeled. Note the irregularly fused tumor glands in subtype 1 tumor compared to those in subtype 2 tumors composed of small, uniform tumor cells lying within the elastic stroma similar to the normal alveolar wall. Dense stromal inflammatory cell infiltration in Subtype 5 tumors. Scale bars for b = 100 μm. c Proportions of samples with different pathologic diagnoses within each subtype. LUAD was predominant in Subtypes 1 and 2, whereas LSCC was predominant in Subtype 3. d–f Histologic patterns of LUADs in each subtype. The predominant patterns of Subtype 1 and 2 LUADs were most commonly acinar or papillary but were quite heterogeneous (d). The proportion of the lepidic pattern, considered to indicate noninvasive LUAD, was enriched mostly in Subtype 2 LUADs, suggesting that Subtype 2 is most like early LUAD (Subtype 1, n = 55; Subtype 2, n = 34, Subtype 3, n = 10; Subtype 4, n = 26; Subtype 5, n = 20) (e). Consistently, the proportion of high-grade histologic patterns (including solid, micropapillary, cribriform, and complex glandular patterns) was lowest in Subtype 2 LUADs. The high-grade histologic pattern was more extensive in Subtype 1 than in Subtype 2, but these subtypes were remarkably heterogeneous compared to Subtype 3–5 LUADs, which were mostly composed of high-grade histologic patterns (Subtype 1, n = 55; Subtype 2, n = 34, Subtype 3, n = 10; Subtype 4, n = 24; Subtype 5, n = 20) (f). For box-plots, middle line, median: box edges, 25th and 75th percentiles; whiskers, most extreme points that do not exceed ±1.5 × IQR. g–I Lymphovascular invasion (g), lymph node metastasis (h), and tumor necrosis (i) were less common in Subtype 2 tumors, which also implies that Subtype 2 tumors are in a clinically early, nonprogressed stage. j Microscopically, the stromal component was more extensive in Subtype 2, 4, and 5 tumors (Subtype 1, n = 55; Subtype 2, n = 43, Subtype 3, n = 52; Subtype 4, n = 43; Subtype 5, n = 34). For box-plots, middle line, median: box edges, 25th and 75th percentiles; whiskers, most extreme points that do not exceed ±1.5 × IQR. k–l Tumor-infiltrating lymphocytes (k) and stromal neutrophilic infiltration (l) were most extensive in Subtype 5 tumors. The p-value was calculated using the chi-square test (c, d, g–I, k, and l) the Kruskal-Wallis test (e, f, and j). m Summary of the histopathologic characteristics of the NSCLC subtypes.
We correlated these findings with the histopathological features of the subtypes (Fig. 3b–m). Subtypes 1 and 2 were predominantly well-to-moderately differentiated adenocarcinomas, in which acinar and papillary patterns were predominant (Fig. 3c, d). Comparisons of the histological patterns for LUAD cases showed that the lepidic pattern, reminiscent of the normal alveolar structure of the lung parenchyma and a non-invasive component of lung adenocarcinoma32, was more extensive in Subtype 2 (Fig. 3e), confirming the DEGsubtype findings. In contrast, Subtype 1 had a higher proportion of high-grade histologic patterns, including solid, micropapillary, cribriform, and complex glandular patterns33 compared to those in Subtype 2 (Fig. 3f). Consistent with these observations, Subtype 2 tumors exhibited less frequent lymphovascular invasion (Fig. 3g) and lymph node metastasis (Fig. 3h). Subtype 3 mainly consisted of squamous cell carcinomas and a subset of solid-predominant adenocarcinomas (Fig. 3c, d). Subtype 3 tumors had enlarged pleomorphic nuclei with high mitotic activity and frequent tumor necrosis (Fig. 3i), which was consistent with the DEGsubtype analysis. In contrast to Subtypes 4 and 5, the stromal components of Subtype 3 tumors were relatively scarce (Fig. 3b–j). Subtype 4 tumors had moderate-to-large amounts of desmoplastic stromal components with variable amounts of inflammatory cells, whereas Subtype 5 tumors exhibited high tumor infiltration by immune cell components in approximately half of the cases (Fig. 3k). Likewise, in the DEGsubtype analysis, tumors of Subtypes 4 and 5 were more likely to be accompanied by moderate-to-high stromal neutrophilic infiltration, but the proportion of such tumors was not high (Fig. 3l). Collectively, we obtained representative histological images of the five subtypes that reflected their histopathological characteristics and clinical relevance.
Proteogenomic features underlying whole-genome doubling in NSCLC subtypes
The proliferative subtype is common in NSCLC and is mainly characterized by the upregulation of cell cycle pathways, including E2F target, G2M checkpoint, and the MYC targets v1 and v2. Previous proteogenomic study10 reported a proliferative subtype of LUAD, the dominant proximal-proliferative cluster, which accounts for 27% of patients with LUAD (30/110 samples; Supplementary Fig. 4a). Similarly, a proliferative subtype, called the proliferative-primitive cluster, affects 16% of patients with LSCC (17/108 samples, Supplementary Fig. 4b). When considering the classical subtype, which also exhibits upregulation of cell cycle pathways, proliferative subtypes collectively constituted approximately 44% of patients with LSCC13 (47/108 samples).
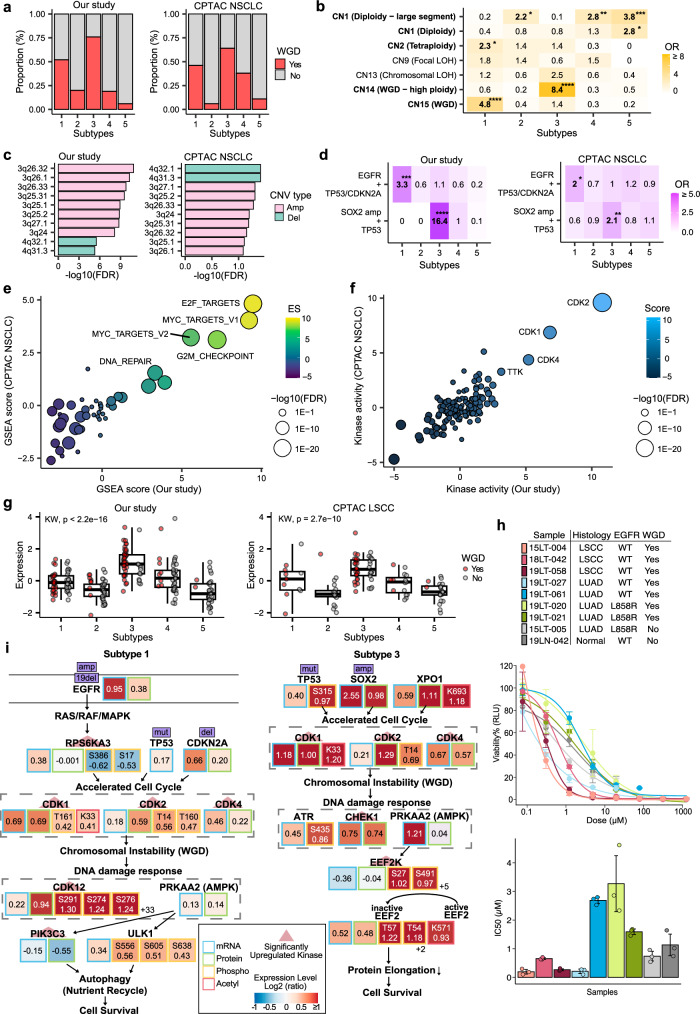
Our multiomics analysis identified Subtype 3 as a proliferative subtype characterized by a WGD event. In Subtype 3, 75% of the patients (39/52) exhibited WGD (Fig. 4a and Supplementary Data 4a, c). Copy-number signature analysis revealed a significant association between Subtype 3 and the CN14 signature (OR: 8.4, Fisher’s exact test; Fig. 4b and Supplementary Fig. 4d), which is indicative of high ploidy (absolute copy numbers 3 to 8), relatively large segment size ( > 40 Mb), and whole-chromosome or chromosome arm-scale losses prior to a single genome-doubling event. In particular, Subtype 3 showed amplification on chromosome 3q and significant enrichment for the co-occurrence of SOX2 amplification and TP53 mutations compared to the other subtypes (OR: 16.4, Fisher’s exact test; Fig. 4c, d and Supplementary Data 4e, f). Furthermore, Subtype 3 in the combined NMF analysis, which integrated the CPTAC LUAD and LSCC datasets also showed a high frequency of WGD (64%, 41/64; Fig. 4a and Supplementary Data 4b, d) and enrichment of the CN10 (28%, 18/64) and CN16 signatures (17%, 11/64), which indicated focal and chromosomal losses before single- and twice-genome doubling, respectively (Supplementary Fig. 4c and e). Subtype 3 exhibited a greater TMB compared to that in the other subtypes in both our (P = 5.7 × 10−11, Wilcoxon rank-sum test) and previous studies (P = 6.9 × 10−6, Wilcoxon rank-sum test). Although LSCC is the most common histology (67%, 35/52 in Korean NSCLC and 66%, 42/64 in CPTAC NSCLC), Subtype 3 is an NSCLC tumor characterized by the presence of WGD events due to copy number alterations and cell cycle pathway enrichment.
Fig. 4. Proteogenomic features underlying whole-genome doubling (WGD) in NSCLC subtypes.
a Barplot showing WGD fraction in each multiomics subtype from our study and CPTAC NSCLC patients. b Overlap of copy number signatures for the five multiomics subtypes, with the colors indicating the odds ratio from one-sided Fisher’s exact test. The COSMIC v3 signature and etiology for each signature are indicated on the y-axis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-sided Fisher’s exact test). c Top 10 most significantly enriched copy number variations (CNVs) in Subtype 3 (FDR < 0.1). The y-axis indicates the cytoband and the x-axis shows the log10-scaled FDR from linear regressions comparing Subtype 3 and other samples. d Enriched co-mutations in each subtype in both our and the CPTAC cohorts. EGFR mutation, missense mutation, in-frame deletion, frameshift deletion, and amplifications were counted. For TP53 and CDKN2A, amplifications were excluded, and for SOX2, only amplifications were counted. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-sided Fisher’s exact test). e Protein-level gene set enrichment analysis (GSEA) revealing upregulated and downregulated pathways in Subtype 3 of both cohorts. The x- and y-axis are enrichment scores (ES) from the current study and CPTAC NSCLC data, respectively. Labeled pathways are the top-5 upregulated pathways in Subtype 3. The Molecular Signatures Database (MSigDB) hallmark gene set v7.4 was used for GSEA. f Subtype 3-specific kinase activity scores. The sizes of the points indicate -log10(FDR) from kinase activity estimation. Significantly up- and downregulated kinases are labeled (FDR < 0.05). g Elevated protein expression of XPO1 in Subtype 3 is shown for our study (Subtype 1, n = 55; Subtype 2, n = 45; Subtype 3, n = 52; Subtype 4, n = 43; Subtype 5, n = 34) and CPTAC LSCC (Subtype 1, n = 11; Subtype 2, n = 19; Subtype 3, n = 42; Subtype 4, n = 15; Subtype 5, n = 21). Kruskal-Wallis test was performed to test the differences in expression. WGD status is marked by red dots and the y-axis shows log2 protein expression levels. For box-plots, middle line, median; box edges, 25th and 75th percentiles; whiskers, most extreme points that do not exceed ± 1.5 × IQR. h Sample information (top), drug response curve (middle), and IC50 (bottom) for selinexor (XPO1 inhibitor) for lung organoids highlighting a higher sensitivity in WGD-positive LSCC organoids. Three technical replicates were tested in each organoid sample. For IC50 barplot, dots indicate each replicate, and error bars indicate average ± 1 standard deviation. i WGD-related pathway underlying Subtype 1 LUAD tumors and Subtype 3 LSCC tumors. Significantly upregulated kinases are highlighted with red triangles (FDR < 0.05) and mutations are shown in purple boxes. Kinase activity scores are estimated from phosphoprotein expression. The log2 fold changes from DE analyses are indicated by the color in each box. For the phosphoproteome, only features with FDR less than 0.1 are displayed.
Consistent with previous studies, Subtype 3 showed upregulation of proteins, PTMs, and kinases in cell cycle pathways (Fig. 4e, f, Supplementary Fig. 4f, g and Supplementary Data 3a, 4g–j). The key proteins and PTMs involved in this subtype included SRSF1 (proteins S199 and K179), SRSF2 (T25 and S26), and XPO1 (protein K693). SRSF1 and SRSF2, both proto-oncogenes, modulate the splicing patterns of tumor suppressor genes, kinases, and kinase receptors into their oncogenic isoforms, thereby promoting cell cycle activity34. XPO1 was upregulated at both the protein and acetylated protein levels in this subtype (Fig. 4g). Overexpression of XPO1 has been observed in many malignancies35 and its inhibition could potentially reduce cell proliferation and promote cell cycle arrest in proliferative subtypes. To assess this hypothesis, we selected tumor organoids from the organoid biobank of SG Medical, Inc. (Seoul, Korea) generated from WGD-positive LUAD and LSCC patients and WGD-negative controls. After treating the organoids with selinexor, a targeted XPO1 inhibitor, we found higher drug sensitivity in WGD-positive LSCC organoids than in other samples (Fig. 4h and Supplementary Data 4k). This finding suggests that targeting XPO1 with selinexor may be a promising therapeutic approach for patients with LSCC and WGD tumors, which warrants further investigation into its potential clinical applications.
To a lesser extent, Subtype 1 also included patients with WGD (51%, 28/55 patients; Fig. 4a). Subtype 1 was significantly enriched in the CN15 signature (OR: 4.8, Fisher’s exact test; Fig. 4b), evenly distributed ploidy (absolute copy number from 2 to more than 9), and a segment size of 1 to 10 Mb. This subtype was predominantly found in patients with LUAD and was associated with co-occurring mutations in EGFR and tumor suppressor genes (TP53 or CDKN2A) (OR: 5.3, Fisher’s exact test; Fig. 4d), implying that subtype 1 is LUAD-prevalent and WGD is activated through LUAD-specific tumor evolution events. In the combined NMF analysis, we validated the proportion of WGD-positive patients (46%, 19/41 patients; Fig. 4a) and the enrichment of the WGD-related signature CN16 in Subtype 1 from previous LUAD and LSCC studies (34%, 14/41 patients; Supplementary Fig. 4c).
WGD characterizes two major subtypes of NSCLC but seems to harbor different driver genes and underlying pathways for each subtype (Fig. 4i and Supplementary Data 4l). Subtype 1 was characterized by a high rate of in-frame deletion and copy number gain in EGFR, with a high mutational burden on tumor suppressor genes. Significantly upregulated kinases in Subtype 1 included RPS6KA3, which mediates cell growth signaling initiated by EGFR activation, CDK1, CDK2, and CDK4, indicating an accelerated cell cycle (FDR < 0.1) (Supplementary Fig. 4h and Supplementary Data 4m). Additionally, CDK12 and PIK3C3, which are related to DNA repair and autophagy, respectively, were significantly upregulated (FDR < 0.1). Since increased cell proliferation induces hypoxia and nutrient depletion36, these kinases may be activated in response to WGD to promote the cell’s adaptation to the nutrient-deprived environment and stabilize the genome, ultimately leading to cell survival. Similarly, together with CDK1, CDK2, and CDK4, kinases related to the DNA damage and nutrient deprivation responses were activated in Subtype 3 LSCC samples (Supplementary Fig. 4i and Supplementary Data 4m). However, the putative driver mutations of WGD in Subtype 3 are TP53 mutations and copy number amplification on chromosome 3q, where many cell cycle genes reside, including SOX2, ATR, STAG1, GSK3B, TFDP2, and MCM2. Since LUSC tumors tend to have copy number gain in the 3q arm37, Subtype 3 may have benefited from a 3q gain after WGD, thus having a higher WGD fraction than Subtype 1 samples. In addition, unlike Subtype 1, CHEK1 and EEF2K play important roles after undergoing WGD. EEF2K is known to inactivate EEF2 by phosphorylation under dietary restriction, which in turn contributes to cell survival38. Overall, these results show that Subtypes 1 and 3 represent proliferative subtypes that have undergone WGD in LUAD and LSCC tumors, respectively, and that selinexor may be an effective treatment for patients with Subtype 3 will requires further validation study.
Heterogeneous immune landscapes in NSCLC
Understanding the tumor immune microenvironment (TIME) is crucial in the molecular characterization of cancer subtypes39,40. To profile the TIME in patients with NSCLC, two clustering analyses based on cell type41 and pathways42 were performed, with enrichment scores inferred from curated gene signatures. We identified three major immune clusters, namely, hot-tumor-enriched (HTE), cold-tumor-enriched (CTE), and NAT-enriched, across 205 tumors and 85 NATs (Fig. 5a and Supplementary Data 5a). HTE and CTE immune clusters overlapped considerably between the cell type- and pathway-based clusters (Supplementary Fig. 5a and Supplementary Data 5b). HTE tumors were mainly enriched in CD8+ and CD4 + T cells, regulatory T (Treg) cells, B cells, natural killer (NK) cells, neutrophils, dendritic cells (DCs), monocytes, and macrophages at the cell-type level, compared to CTE tumors. In the pathway-based clustering results, the HTE cluster showed a greater activation of immune-related pathways than the CTE cluster. Because the cell-type enrichment score was inferred based on curated gene signatures, we confirmed the cell-type specificity of the immune clusters using scRNA-seq data from multiple NSCLC studies23. From the top-300 DEGs in the HTE (DEGHTE) and CTE (DEGCTE) clusters, we investigated the enrichment of each DEG in scRNA-seq data. The DEGHTE set was enriched in CD8+ and CD4 + T cells, regulatory T cells, B cells, NK cells, neutrophils, DCs, monocytes, and macrophages, whereas the DEGCTE set was primarily enriched in epithelial cells, including LUAD and LSCC tumor cells (Fig. 5b, top, and Supplementary Data 5c). Additionally, the DEGs of each cell type inferred from the gene signatures were enriched in the corresponding cell types in the scRNA-seq data (Supplementary Fig. 5b). The cell-type enrichment score was also highly correlated with the level of TILs (Fig. 5b, bottom, and Supplementary Data 5d). These results indicate that the single-cell and immunohistochemical analyses achieved good immune clustering based on the cell type score.
Fig. 5. Landscapes of immune clusters and cell types across NSCLC subtypes and cohorts.
a Immune subtyping based on cell type and pathway enrichment scores. Cell type-based clustering was performed with 205 tumor and 85 normal adjacent to the tumor (NAT) samples, and pathway-based clustering was performed using only tumor samples. The tumor-infiltrating lymphocyte (TIL) pattern, clinical histology (diagnostics, DX), multiomics subtype, tumor stage, and tissue information are described. IC, immune cluster. b (top) DEGHTE and DEGCTE were used to generate the UMAP plot of scRNA-seq data. A two-sided t-test was conducted to assess the statistical significance of the differences in gene expression. The color of each point represents the module score of each cell; higher scores are shown in red. UMAP information was obtained from multiple NSCLC studies23. (bottom) The correlations of 10 cell types and immune clusters with the pattern of TILs were analyzed. The sizes and colors of the circles indicate the statistical significance and correlation coefficient of the correlations, respectively. The horizontal black dotted line indicates P = 0.05. c Hazard ratios for overall survival (OS, left) and relapse-free survival (RFS, right) related to various cell types in the cell type-based immune cluster. A hazard ratio lower than zero (blue box) indicates that the hot-tumor-enriched (HTE) status or a high cell type score was associated with prolonged survival. Error bars (gray lines) represent mean ± 95% confidence interval (CI). Red text indicates statistical significance in the survival analysis by the log-rank Mantel‒Cox test (n = 174). d Correlations of the RNA expression, protein expression, and protein activity of 10 immunomodulators with immune cluster status for our cohort as well as other lung cancer multiomics cohorts10,13. Correlation coefficients and p-values were obtained from a generalized linear model (GLM). e Correlations between the expression or activity of immunomodulators and the status of driver mutations in our cohort and the Satpathy and Gillette cohorts. The top associations between immunomodulators and known driver genes are described. f The left boxplot shows the RNA and protein expression of SLAMF7 in samples (n = 205) with wild-type or mutant SMARCA4 (n = 205); the right boxplot shows the RNA and protein expression of SLAMF7 in HTE and cold-tumor-enriched (CTE) samples (n = 174). The two-sided t-test was performed to test the differences in expression. The box represents the 25th and 75th percentiles, the central mark denotes the median, and the whiskers extend to the most extreme points within ±1.5 × IQR. g (left) Box (top) and balloon (bottom) plots showing the mean expression of marker genes of HTE and 10 cell types across the multiomics subtypes (n = 174). The marker genes were defined as the top−300 and −30 most overexpressed genes in HTE samples and highly cell type-enriched samples, respectively. (right) The bar (top) and box (middle and bottom) plots show the mutation frequency of SMARCA4 and RNA/protein expression of SLAMF7 across multiomics subtypes (n = 174), respectively. The Kruskal‒Wallis test was performed to assess the differences among the multiomics subtypes. The box represents the 25th and 75th percentiles, the central mark denotes the median, and the whiskers extend to the most extreme points within ±1.5 × IQR.
To analyze the prognostic value of the immune landscape in patients with NSCLC, each immune cell enrichment score, along with the immune cluster, was tested against overall and relapse-free patient survival rates (Fig. 5c, Supplementary Fig. 5c and Supplementary Data 5e). HTE status was the most favorable factor for patient survival, and high enrichment of CD8+ and CD4 + T cells, B cells, and NK cells were positively correlated with a good prognosis, in contrast to the enrichment of epithelial cells or neutrophils, which had negative correlations. None of the immune-related pathway-based enrichment scores were significantly associated with patient survival, although most showed a positive association with prolonged survival (Supplementary Fig. 5d). HTE tumors with high enrichment of CD8+ and CD4 + T cells and immune-related pathways are known to be associated with favorable prognoses43. Additionally, Previous studies10,13 reported that Tregs were enriched in HTE tumors, but had an immunosuppressive effect and were associated with poor prognosis. We found a concordant trend of Treg enrichment in HTE tumors (Fig. 5a, b); patients with Treg-enriched HTE had worse survival rates than those with low Treg levels (Supplementary Fig. 5e and Supplementary Data 5f).
Further analysis of the immune clusters was performed based on their cell-type composition, which showed significant correlations with patient survival. We performed a pathway enrichment analysis comparing the HTE and CTE immune clusters using multiomics features (Supplementary Fig. 5f and Supplementary Data 5g). A set of immune-related and EMT signaling pathways was enriched in HTE tumors, whereas cell cycle-related pathways and glycolysis were enriched in CTE tumors. Specifically, some metabolic pathways and oxidative phosphorylation were enriched in CTE tumors at the phosphoprotein level but were enriched in HTE tumors at the acetylated-protein level. Many of these results are consistent with the findings of previous lung cancer multiomics studies10,13.
The differences between HTE and CTE immune clusters may be better elucidated by examining their respective regulators. To identify putative regulators of patient immune landscapes, we analyzed RNA and protein expression data and inferred protein regulatory activity according to the systematical influence of a protein in the transcription of relevant targets44. We performed a regression analysis of gene expression or activity and enrichment scores for cell type or immune clusters. We found that the majority of immunomodulators, which mainly comprised cancer cell ligands and immune cell receptors, were positively correlated with HTE tumors and most immune cells, but not epithelial cells45 (Supplementary Fig. 5g and Supplementary Data 5h). Notably, the expression and activity of VEGF-A were negatively correlated with HTE tumors, which contrasts the patterns observed with other immunomodulators. VEGF-A was negatively associated with TILs and has potential implications for cancer risk46,47. Specifically, 10 immunomodulators showed significant correlations with immune cluster status, with a consistent correlation direction not only in our cohort, but also in other lung cancer multiomics studies10,13 (Fig. 5d and Supplementary Data 5i).
We profiled the associations between the immunomodulators and known driver mutations to determine the putative mechanism(s) underlying changes in immunomodulators. Because Subtype 5 is immunogenic, we only considered the most activated or repressed immunomodulators to identify the potential regulatory mechanisms that distinguish it from the other subtypes. Ultimately, the three immunomodulators showed significant correlations with mutations in one of the eight driver genes, with consistent patterns observed in other multiomics NSCLC cohorts10,13, although these correlations were not statistically significant (Fig. 5e and Supplementary Data 5j). Among these immunomodulators, the SMARCA4 mutation was positively correlated with the expression of SLAMF7 at both the RNA and protein levels, and positively correlated with HTE status in both our study cohort and the independent cohort10 (Fig. 5f, Supplementary Fig. 5h, and Supplementary Data 5k). Based on these results, SMARCA4 is a potential regulator of the immunomodulator SLAMF7, which is associated with HTE status.
We further analyzed the distribution of the immune landscape among the multiomics subtypes (Fig. 5g, left, and Supplementary Data 5l). The expression of HTE feature genes (DEGHTE) was highest in Subtype 5, which had immunogenic characteristics. This subtype also showed the highest activation of CD8+ and CD4 + T cells, Treg cells, B cells, NK cells, neutrophils, and macrophage marker genes. Neutrophils and Tregs, which are associated with a poor prognosis in all patients and specifically in THE patients, respectively (Fig. 5c and Supplementary Fig. 5e), were also found to be enriched in Subtype 5. This enrichment may be linked to the intermediate survival characteristics observed in this subtype, despite Subtype 5 exhibiting the strongest THE tumor signal. Moreover, the highest frequencies of SMARCA4 mutations and SLAMF7 overexpression were observed in Subtype 5 (Fig. 5g, right). We confirmed that the distribution of cell types within the immune cluster and their correlation with the multiomics subtype were consistent in the integrated cohort combining our cohort with those of recent multiomics studies10,13 (Supplementary Fig. 5i and Supplementary Data 5m). Our results showed that HTE tumors were positively associated with better prognosis in patients with NSCLC. These tumors were enriched in immunogenic Subtype 5, which was also positively associated with mutations in the putative regulator SMARCA4, which targets SLAMF7, an immunomodulator.
Multiomics profiling of neoantigens and immune clusters
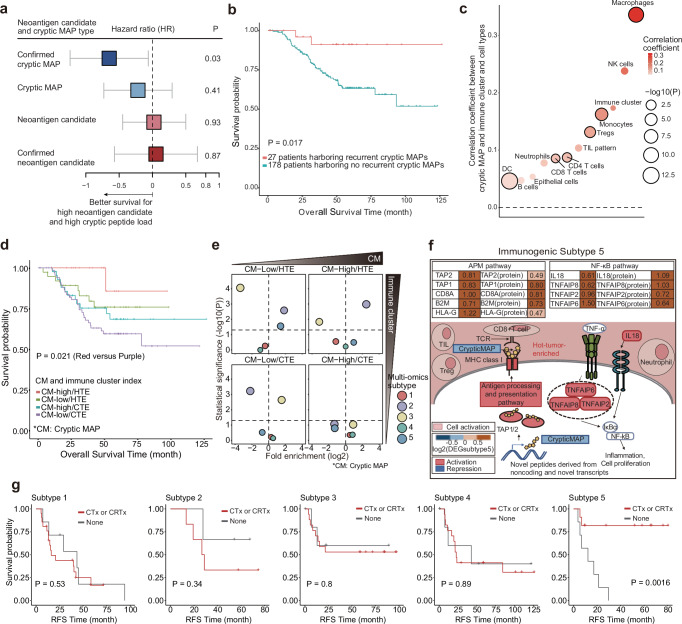
Neoantigens are tumor-specific antigens generated by tumor cells and are key factors affecting the immune landscape of cancer patients48. Therefore, we predicted neoantigens in our NSCLC cohort using a multiomics dataset. Neoantigens derived from somatic mutations in the coding regions were predicted using WES data and confirmed by MS analysis to determine whether they were expressed as peptides of the “confirmed neoantigen candidates.” Furthermore, we identified peptides derived from non-coding and non-annotated transcripts that were distinct from the canonical neoantigens. Among them, we selected “cryptic MHC class I-associated peptides (MAPs),” which were described as noncanonical peptides predicted to bind to MHC class I molecules in a previous study49–51. We identified “confirmed cryptic MAPs” based on a previously determined expression threshold52. We inferred 85,430 neoantigen candidates and 775 cryptic MAPs (Supplementary Fig. 6a) and annotated the origin of the cryptic MAPs based on the matched transcripts (Supplementary Fig. 6b and Supplementary Data 6a). Non-annotated isoforms, pseudogenes, and untranslated regions (UTRs) accounted for 90.97% of the sources of cryptic MAPs, which is consistent with previous studies53–58.
We tested the associations of the neoantigen candidates, confirmed neoantigen candidates, cryptic MAPs, and confirmed cryptic MAPs with patient survival (Fig. 6a and Supplementary Data 6b). Interestingly, only confirmed cryptic MAPs showed a strong positive correlation with improved survival, although the number of cryptic MAPs was notably low across patients (Supplementary Fig. 6c and Supplementary Data 6b). We also found 12 confirmed cryptic MAPs in more than three patients, called them recurrent cryptic MAPs, some of which were derived from the same gene of origin (Supplementary Fig. 6d and Supplementary Data 6c). Furthermore, the presence of recurrent cryptic MAPs was significantly correlated with prolonged survival (Fig. 6b and Supplementary Data 6b), indicating the prognostic value of cryptic MAPs in patients with NSCLC.
Fig. 6. Clinical relevance of neoantigens and cryptic MAPs, and their associations with multiomics subtypes.
a Survival estimated according to the type of neoantigen and cryptic MAP. A lower hazard ratio (blue box) indicates that a high load of neoantigens or cryptic MAPs is associated with prolonged overall survival, and a high hazard ratio (red box) indicates the opposite. Hazard ratios for individual trials and overall effects are given with 95% CIs. Log (HR) values and their corresponding 95% confidence intervals (CIs) are depicted in grey. b Kaplan‒Meier curve showing the survival of two groups of patients (n = 204) according to whether they did (blue line) or did not have (red line) recurrent cryptic MAPs. The p-value was derived by comparing the curves with the log-rank Mantel‒Cox test. c Correlations between the number of cryptic MAPs and enrichment scores of 10 cell types and the immune cluster. Correlation coefficients were calculated by a linear regression model with the covariates of sample batches and histological diagnosis. The size of the dots indicates the degree of the -log10-scaled p-value, and the color of the dots represents the strength of the correlation coefficient. The bold-lined dots indicate statistical significance. d Kaplan‒Meier curve showing the survival patterns of four groups of patients (n = 174) stratified by cryptic MAP load and immune cluster. The p-value was obtained by comparing curves between the two groups with the largest difference in the log-rank Mantel‒Cox test. e Enrichment analysis of the four groups described in Fig. 4d for the multiomics subtypes. The x- and y-axis indicate enrichment and statistical significance calculated using a two-sided Fisher’s exact test with the Benjamini‒Hochberg adjustment, respectively. The size of each dot indicates the level of significance. f Features of patients with multiomics Subtype 5 disease who had a low cryptic MAP load with an HTE status, activated APM, and activated NF-κB pathway. g Kaplan–Meier curve of recurrence-free survival according to treatment status (chemotherapy [CTx] or chemoradiation therapy [CRTx]) in patients categorized by multiomics subtype. The p-value was derived by comparing the curves with the log-rank Mantel‒Cox test.
We also investigated the association between cryptic MAPs, immune clusters, and cell-type enrichment. Cryptic MAPs were positively correlated with most immune cell types, including HTE status, although some correlations were weak or insignificant (Fig. 6c and Supplementary Data 6b). In our cohort, patient prognosis was evaluated after stratification according to immune cluster criteria and cryptic MAP load. Patients with high cryptic MAP load and HTE status showed the longest survival, whereas those with low cryptic MAP load and CTE status showed the worst survival (Fig. 6d and Supplementary Data 6b).
We determined the association between the integrated immune landscape and multiomics subtypes by evaluating the distributions of the multiomics subtypes based on the combined status of the cryptic MAP load and immune clusters. Multiomics Subtype 5 was most enriched in the low cryptic MAP load and HTE groups (Fig. 6e and Supplementary Data 6d). Interestingly, a similar survival trend and multiomics subtype distribution were obtained when the immune cluster was replaced with antigen processing, presentation machinery (APM), and TIL pattern (Supplementary Fig. 6e–g and Supplementary Data 6e, f), which was proposed as a critical factor for evaluating the immune-activating potential with neoantigens14. Notably, patients with Subtype 5, who showed moderate survival, had a low cryptic MAP load with HTE and activated APM. This is consistent with the large proportion of TILs observed in single-cell and histopathological analyses (Fig. 3a–k). Patients with Subtype 5 also showed high activity of the TNF-α-pathway via NF-κB in addition to APM. Consequently, the moderate survival associated with this subtype appeared to be linked to a low load of cryptic MAPs and high enrichment of immunosuppressive Tregs and neutrophils despite having an active immune landscape with HTE status (Fig. 6f and Supplementary Data 1a). Remarkably, the use of adjuvant chemotherapy or chemoradiation therapy substantially enhanced the survival of patients with Subtype 5 tumors (Fig. 6g). In contrast, no substantial improvement in survival was observed in patients with other subtypes who underwent adjuvant chemotherapy or chemoradiation therapy. Subtype 5 also demonstrated the most favorable prognosis compared to other subtypes in the overall population treated with adjuvant chemotherapy, despite the marginal statistical significance (data not shown). This underscores that subtype 5 could be associated with a clinical benefit from adjuvant chemotherapy. These results imply that understanding patient prognosis and multiomics subtypes requires a multifactorial consideration of the TIME. Furthermore, the distribution of HLA alleles before and after the binding prediction was consistent with previous reports on the Korean population or lung cancer studies59–62, thereby corroborating the reliability of our results (Supplementary Fig. 6h–j).
Discussion
In this study, we conducted a comprehensive multiomics analysis of 229 patients from a Korean NSCLC cohort and identified five NSCLC subtypes enriched for WGD, oncogenes, metastasis, and the immune microenvironment. The phosphoproteome dataset was the most informative for subtype identification, contributing 80% (911 out of 1,134) of the features, while global proteome and acetylome data also played crucial roles in decoding signaling pathways across the identified subtypes. By extending our analysis to integrate multiomics data from 462 patients with NSCLC, we validated our subtype classification, confirming its alignment with previously established multiomics subtypes. Utilizing an extensive single-institute clinical dataset allowed us to delineate detailed histopathological beyond histologic subtype and clinical relevance of our multiomics subtypes, such as prevalent alterations of targetable oncogenic drivers, histologic grade of LUAD, metastatic potential, and tumor immune response, and finally complement previous findings on the underlying biology of NSCLC.
We identified a NSCLC subtype, Subtype 4, which was associated with a high frequency of metastasis and poor outcomes, independent of the NSCLC histological type. This subtype contained a large proportion of desmoplastic stromal components, suggesting that predominant tumor invasion and metastasis contribute to its aggressive behavior. Increased phosphorylation in hypoxia and activation of the PI3K-Akt signaling pathway were key characteristics of this subtype. In particular, the upregulation of SLK and LRRFIP1 phosphorylation predicted poor survival outcomes and has been consistently observed in other NSCLC multiomics studies. Further research on these events may identify targets for therapeutic intervention and enhance our understanding of poor outcomes in NSCLC.
Characterizing NSCLC according to multiomics subtypes has important implications for personalized treatment strategies. Our study revealed that Subtype 3, characterized by high levels of chromosomal instability and XPO1 expression, represents a highly proliferative NSCLC subtype with co-occurring TP53 and cell cycle gene amplification. Patient-derived organoid experiments indicated that selinexor, a targeted inhibitor, could be effective against Subtype 3 with WGD. In Subtype 1, WGD only occurred in a limited number of patients with LUAD (51%, 28/55 patients) and was enriched for co-occurring mutations in EGFR and tumor suppressor genes. However, XPO1 expression was not remarkably increased and selinexor did not show strong efficacy in this subtype, suggesting potential subtype-specific differences in XPO1 expression and therapeutic response to XPO1 inhibition. These findings suggest that WGD events are associated with tumor evolution via genomic alterations specific to the subtype, even in WGD-predominant subtype7,63.
We demonstrated that Subtype 5, enriched in the inflammatory tumor microenvironment, exhibited extraordinary improvements in survival with conventional adjuvant chemotherapy. This finding implies that the selection of postoperative adjuvant treatment for NSCLC could be helpful only for the case with high tumor immune response. Additionally, patients within this subtype could potentially benefit from immune checkpoint inhibitor therapy. Identifying suitable patients for coventional adjuvant chemotherapy could be facilitated by various biomarkers indicating a positive tumor immune response, including PD-L1 immunostaining, T-cell marker presence, and the histological nature of the tumor immune microenvironment. In contrast, adjuvant chemotherapy did not significantly alter the survival of node-positive patients with Subtypes 1 or 2, which are enriched in driver oncogene alterations. Given the minimal survival improvement with cytotoxic chemotherapy in EGFR-mutant LUAD observed in prior trials64,65, targeted therapies aimed at driver mutations may offer better outcomes for these patients.
Multiomics-based profiling has the potential to enhance our understanding of the TIME. The HTE signature exhibited significant activation in Subtype 2, but was repressed in Subtype 4, which is inconsistent with our histopathological observations. This highlights the value of multiomics profiling as a complementary tool to histopathological examination in the functional characterization of the tumor microenvironment. We identified SLAMF7 as an immunomodulator significantly associated with HTE and SMARCA4 mutations in Subtype 5. Given the efficacy of immune checkpoint inhibitors in NSCLC patients with SMARCA4 mutations66, SLAMF7, and SMARCA4 are promising molecular targets for enhancing cancer immunotherapy. Our multiomics analyses also revealed cryptic MAPs derived from non-coding and non-annotated transcripts, which were subsequently confirmed using proteomics. Unlike neoantigen candidates, cryptic MAPs were significantly correlated with favorable survival and immune cell enrichment. Thus, experimental validation of MHC-binding prediction using immunopeptidomics for recurrent cryptic MAPs could facilitate the development of a cancer vaccine.
Our study provides a comprehensive profile of the multiomics subtypes of NSCLC, but there are several limitations to consider. First, we used exome sequencing to infer whole-genome doubling, which may be less accurate than whole-genome sequencing. Ethnic differences may also be present in certain subtypes, particularly in Subtype 1 for EGFR female patients, although some subtypes were consistently found in both our cohort and the CPTAC cohorts. Additionally, the confirmation of cryptic MAPs by proteomics is limited owing to the experimental availability of normal samples. Further studies using larger normal sample cohorts are needed to confirm these findings. A prospective cohort study is also needed to test the effectiveness of drugs, such as selinexor, in the various subtypes. Finally, more comprehensive studies are needed to understand the effectiveness of selinexor in treating the WGD subtype.
Methods
Human subjects with investigation of clinicopathologic features
A total of 229 samples, self-reported as Korean ethnicity, were histologically defined as NSCLC and selected for this study. Tumors and normal tissues adjacent to the tumor (NAT) were harvested under the Clinical Proteomic Tumor Analysis Consortium (CPTAC) guidelines from Asan Medical Center, Korea, with the approval of the Institutional Review Board of Asan Medical Center (Approval number: 2019-1210). In brief, we retrieved the 408 NSCLC cases whose cold ischemic time of fresh frozen tumor and NAT tissue sample was less than 15 minutes from the NSCLC cases of the bioresource center of Asan Medical Center deposited from January 2010 to March 2019. To investigate the impact of proteogenomic findings on post-operative therapy and metastasis, we preferentially selected 137 NSCLC patients showing lymph node metastasis at the pathologic examination of resection specimens. We further included 113 cases with absent lymph node metastasis at the time of surgical resection. With the further exclusion of 21 cases showing inadequate nucleic acid quality metrics, 229 patients were finally enrolled in the study. The clinical information of the enrolled patients, including age, gender, smoking history, adjuvant therapy, presence of post-operative recurrence, and treatment histories, was reviewed and documented by the thoracic oncologists (W.J.J, C.M.C., and J.C.L.). All the histopathologic slides of the resection specimens removed from the enrolled patients were independently reviewed by two thoracic pathologists (H.S.H. and S.J.J.) and documented for pathologic findings such as pathologic diagnosis, grade, tumor size, pleural invasion, histologic pattern (for non-mucinous adenocarcinoma, lymphovascular invasion, spreading through alveolar space, and lymph node metastasis. The cases with discrepancy between the independent assessments were discussed in consensus meetings. Based on these findings, all enrolled patients were re-staged according to the 8th lung cancer TNM stage67. In addition, pattern of tumor-infiltrating lymphocytes (TIL) was reviewed by the pathologists and classified according to the classification by Saltz et al.68.
Whole exome sequencing
Genomic DNA was isolated from a FFPE (formalin-fixed paraffin embedded) tumor and normal samples (NATs or blood buffy coat69 if NATs are not available). We generated whole exome sequencing (WES) libraries using SureSelect V6-Post (Agilent, CA, USA). Pooled libraries were run on the Illumina NovaSeq to obtain an average of 300x depth per tumor library and 100x depth per NAT and blood buffy coat library. The raw Illumina BCL (base calls) binary files were converted into Fastq files using the Illumina package bcl2fastq. Sequencing read data was checked for quality and adaptor/overrepresented sequence (ORS) by FastQC (v0.11.9). There was no ORS that could be a quality problem, and since the adapter sequence was less than 1% in all samples, the additional trimming process was skipped. Fastq files were aligned to the human reference GENCODE GRCh38.p13 v32 primary assembly genome by BWA (v0.7.17). Among the three algorithms of BWA, the latest BWA-MEM was selected as it is suitable for Illumina sequences ranging from 70 bp to 1 mbp with fast and accurate performance. After alignment, we sorted the BWA process output by coordinates via Picard SortSam and checked duplicate reads using Picard MarkDuplicates (v2.23.1). Two types of duplicated reads, optical duplicates (incorrectly detected as multiple clusters by the optical sensor of the sequencing machine) and library duplicates (created during PCR library preparation), were annotated for downstream analyses. For all tools requiring interval files, we used V6_S07604514_hs_hg38_S07604514_Covered.bed provided by Agilent. Because of systematic errors from the sequencing machine, base quality scores were recalibrated by the GATK4 BQSR algorithm (v4.1.8.0). The algorithm calculates the average Phred score by assuming the mismatch base is an error by matching known variants with the data base. We used the known variants VCF from dbSNP (broad_hg38_v0_Homo_sapiens_assembly38.dbsnp138.vcf, broad_hg38_v0_Homo_sapiens_assembly38.known_indels.vcf.gz) and the Mills and 1000 G project (broad_hg38_v0_Mills_and_1000G_gold_standard.indels.hg38.vcf.gz) from the GATK4 bucket (https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0/). Then, the empirical Phred score is calculated using the average Phred score and then recalibration is performed based on the empirical Phred score.
Variant calling
We called somatic variants from the recalibrated BAMs using GATK4 Mutect2 (v4.1.8.0). In the Mutect2 pipeline, somatic-hg38_1000g_pon.hg38.vcf.gz data was used as panel-of-normals and somatic-hg38_af-only-gnomad.hg38.vcf.gz data was used for germline variants removal. We performed tumor with paired normal mode to exclude germline variants. As one tumor sample had no matching normal sample, the tumor only mode was performed. After calling somatic variants from GATK4 Mutect2, a read support reference for the well-known variant sites was created from the tumor recalibrated BAMs using GATK4 GetPileupSummaries. The tool requires a common somatic mutation sites VCF, so we used the somatic-hg38_small_exac_common_3.hg38.vcf.gz file created from the gnomAD resource from the GATK4 bucket (https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0/). The GetPileupSummaries table was used in the GATK4 CalculateContamination to calculate the fraction of reads deriving from cross-sample contamination. The calculated cross-sample contamination data were saved as a table for each sample, and tumor segmentation data by minor allele fraction were additionally collected. The raw output of Mutect2 was converted into filtered VCF using cross-sample contamination table and tumor segmentation data by GATK4 FilterMutectCalls. The VCFs generated from GATK4 FilterMutectCalls were converted to annotated VCF and MAF files via GATK4 funcotator. We used GATK4 4.2.0.0 version solely for the annotation step to use the latest annotation data source for GATK4 funcotator. The latest pre-packaged data sources (funcotator_dataSources.v1.7.20200521 s) was downloaded from the GATK4 bucket (gs://broad-public-datasets/funcotator/). All annotations were performed in CANONICAL transcript-selection-mode after choosing a custom transcript list. Finally, the annotated MAF files were merged into one using maftools v2.8.0570.
Germline variants were called from the recalibrated BAMs using GATK4 HaplotypeCaller (v4.1.8.0). The reference confidence scores were confirmed in GVCF mode using the option -ERC GVCF, which was a reference model emitted with condensed non-variant blocks. For variant calls, -G StandardAnnotation and -G AS_StandardAnnotation annotation options were applied. All single sample GVCFs from GATK4 HaplotypeCaller were imported into GenomicsDB by GATK4 GenomicsDBImport (v4.1.8.0) for joint genotyping. Joint genotyping was performed using GATK4 GenotypeGVCFs (v4.1.8.0) from the constructed GenomicsDB. The produced joint VCF was recalibrated in variant quality by two steps. The first step was the recalibration of the SNPs. After calculating the exome specific recalibration score through GATK4 VariantRecalibrator (v.4.1.8.0), it was applied to the joint VCF using GATK4 ApplyVQSR (v.4.1.8.0). We used the training sets from the HapMap project (broad_hg38_v0_hapmap_3.3.hg38.vcf.gz), Mills and 1000 G project (broad_hg38_v0_1000G_omni2.5.hg38.vcf.gz, broad_hg38_v0_1000G_phase1.snps.high_confidence.hg38.vcf.gz) and dbSNP (broad_hg38_v0_Homo_sapiens_assembly38.dbsnp138.vcf) from GATK4 bucket (https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0/). The second step was the recalibration of indels. The same tools in the first step were used and the training sets from Mills and 1000 Genome Project (broad_hg38_v0_Mills_and_1000G_gold_standard.indels.hg38.vcf.gz) and the dbSNP genotyping calls (broad_hg38_v0_Homo_sapiens_assembly38.dbsnp138.vcf). The recalibrated joint VCF was converted to annotated VCF via the GATK4 funcotator. The 4.2.0.0 version of GATK4 was again used at the annotation step to utilize the latest annotation data source for GATK4 funcotator. The latest pre-packaged data sources (funcotator_dataSources.v1.7.20200521 g) was downloaded from the GATK4 bucket (gs://broad-public-datasets/funcotator/). All annotations were performed in CANONICAL transcript-selection-mode.
Identification of copy number alterations
DNA somatic copy number variations (CNVs) were detected using CNVkit v0.9.871. We labeled the copy number status with the following criteria: genes with an absolute copy number greater than or equal to 4 were labeled as “amplification”, and genes greater than or equal to 2.5 and less than 4 were labeled as “gain”. Similarly, genes with an absolute copy number from 0.5 to 1.5 were labeled “heterozygous deletion”, and those less than 0.5 were labeled “homozygous deletion”.
Copy number signature analysis and WGD detection
For copy number signature analysis and WGD detection, we used FACETS v0.16.072 to identify allele-specific copy number information. Preprocessed paired tumor-normal BAM files and a VCF file of common and germline polymorphic sites downloaded from https://www.ncbi.nlm.nih.gov/variation/docs/human_variation_vcf/ were used as the input for FACETS. Two samples were dropped out due to quality problems; therefore, 228 samples in total were used for copy number signature analysis and WGD detection. We defined samples as ‘WGD-positive’ if greater than 50% of their autosomal genome had an absolute copy number greater than or equal to two.
Mutational signature analysis was performed using the COSMIC signature database v373 and R package Sigminer v2.1.574, as previously described75. Briefly, three copy number features, including total copy number, segment length, and loss-of-heterozygosity state were extracted and classified into 48 components that categorize the continuous values regarding the value range and biological significance. To decide the number of signature groups (or a factorization rank), non-negative matrix factorization (NMF) was performed using a tumor-by-component matrix with 50 runs checking 2 to 12 ranks. Based on the cophenetic score plot, we determined to use rank seven for copy number variants. Each signature was nominated by the COSMIC signature of the highest cosine similarity. Then, by performing hierarchical clustering, we assigned the samples to one of the signatures based on the consensus matrix.
RNA-seq data generation and preprocessing
Total mRNA-seq libraries were prepared using Library-TruSeq Stranded Total RNA with Ribo-Zero H/M/R_Gold. Pooled libraries were run on the Illumina NovaSeq6000 to generate an average of 200 million reads per library with 100-bp pair-end. The raw Illumina BCL binary files were converted into Fastq files using the Illumina package bcl2fastq. The sequenced read data was checked for quality and adaptor/overrepresented sequence (ORS) by FastQC (v0.11.9). There was no ORS that could be a quality problem, and since the adapter sequence was less than 1% in all samples, the additional trimming process was skipped. Fastq files were aligned to the human reference GENCODE GRCh38.p13 v32 primary assembly genome using STAR (v2.7.3a). After alignment, we sorted the output by coordinates via Picard SortSam and checked duplicate reads using Picard MarkDuplicates (v2.23.1). The reads that contain Ns in the cigar string were split using GATK4 SplitNCigarReads. Regarding systematic errors from the sequencing machine, base quality scores were recalibrated by the GATK4 BQSR algorithm (v4.1.8.0). The aligned BAMs from STAR were sorted by coordinates using Samtools Sort (v1.10). The Salmon algorithm (v1.4.0) was applied to quantify transcript-level expression, and raw read counts were produced. The output files (quant.sf) produced by Salmon, quantified transcript-level estimates, were imported to R using tximport and converted to a gene-level expression matrix. Then, we estimated the size factors using the “median ratio method” with the “estimateSizeFactors” function supported by DESeq2 R packages. With estimated size factors, we normalized the gene-level read count matrix using the “counts” function in DESeq2 R packages76, which divides the read counts by the size factors.
Isoform expression analysis
Isoform scale expression was quantified from transcriptome data using a pipeline including StringTie77. STAR-aligned BAM without going through GATK4 SplitNCigarReads in RNA preprocessing was used. Transcripts of each sample were assembled using StringTie v2.1.778. The first analyzed outputs were combined into a single assembly using merge command of StringTie (stringtie –merge), creating a single merged gtf containing the transcript of the entire sample. After that, a secondary analysis was conducted to calculate the expression in isoform level of each sample based on the merged transcript assembly using the StringTie Ballgown command (stringtie -eB). The novel isoform in the StringTie outputs were compared and annotated using gffcompare v0.11.679. Processed isoform gtfs are made into a single count matrix using predDE.py, a python script provided by StringTie.
The abundance or exon usage change between normal and cancer tissue was analyzed and visualized using IsoformSwitchAnalyzeR pipeline. IsoformSwitchAnalyzeR v1.17.480 pipeline includes DRIMseq81 to find isoform switch by condition and extract the sequence of mRNA or amino acid produced from the isoform. We analyzed predicted isoforms or ORF lists with additional tools as CPAT82, PFAM83,84, SignalP85 and IUPred2A86.
Fusion gene analysis
To detect the fusion genes, RNA-seq fastq was aligned, and fusion calling and filtering were performed. First, RNA-seq reads were mapped to the GENCODE GRCh38.p13 v32 primary assembly genome using STAR aligner v2.7.3a87. Unlike previous STAR mapping, ‘–chimOutType WithinBAM’ was included in the STAR output to include chimeric read to be suitable for use in arriba fusion calling. To increase sensitivity, the parameters were adjusted as recommended by the author as follows:
--chimSegmentMin 10
--chimOutType WithinBAM SoftClip
--chimJunctionOverhangMin 10
--chimScoreMin 1
--chimScoreDropMax 30
--chimScoreJunctionNonGTAG 0
--chimScoreSeparation 1
--alignSJstitchMismatchNmax 5 -1 5 5
--chimSegmentReadGapMax 3
We found fusion gene by applying Arriba v1.2.088 to the STAR mapped BAM file. Mismatches were discovered by comparing chimeric reads with reference genome assembly fasta and annotation gtf. Additionally, fusion genes had bad quality of frequently found healthy tissues were excluded from the analysis using the hg38 fusion blacklist provided by Arriba. Arriba outputs were labeled with sample name and tissue and concatenated with one table.
From the Arriba output, fusion genes with confidence of “low” was removed. At this time, the fusion genes appearing in normal tissues were collected and used as a blacklist for tumor tissue fusion genes. If the fusion genes found in tumor tissue were included in this blacklist, they were excluded from the analysis. Furthermore, fusion genes including known cancer-related genes (ALK, ROS1, RET, and PTK2)89 were annotated as “known fusion” and fusion genes found across several tumor samples as “recurrent fusion”. They were used for downstream analysis. After filtering, we visualized the structure and protein domain of fusion genes using the R script (”draw_fusion.R”) provided by arriba.
Protein extraction and tryptic digestion
For in-depth proteomic experiments, fifty milligrams of cryopulverized human NSCLC and NAT samples were homogenized in lysis buffer at a ratio of about 300 μL lysis buffer for every tissue. The lysis buffer consisted of 5% SDS, 50 mM TEAB (pH 7.55, Thermo Fisher Scientific, USA, 90114), protease inhibitor cocktail (1:100; Thermo Fisher Scientific, USA, 78430), 1 EA of PhosSTOP (Roche, Swiss, 4906845001), and 20 μM PUGNAc (Sigma-Aldrich, USA, A7229). Tissue lysis was performed with a couple of probe sonications using the Digital Sonifier SFX 550(Branson, USA). Sonication time was set at 30 s with a cycle of on-time 5 s and an interval of 3 s at 28%. Lysates were centrifuged at 14,500 g at 4 °C for 5 min, followed by measuring protein concentration using the BCA assay (Thermo Fisher Scientific, USA, 23225), and kept at -80 °C until used for analysis.
Protein lysates were reduced at 95 °C for 10 minutes with 20 mM 1,4-dithiothreitol (Roche, Swiss, 10708984001), followed by alkylation with 40 mM iodoacetamide (Sigma-Aldrich, USA, I6125) in the dark at RT for 30 minutes. 12% phosphoric acid (Sigma-Aldrich, USA, 695017) of 1/10 of the sample volume and 7 multiple volumes of S-trap binding buffer (100 mM TEAB in 90% MeOH) were added sequentially for formation of colloidal status. The solution was loaded on an S-Trap™ spin column (Protifi, USA) and centrifuged to 4000 g for 30 s. The captured protein was washed three times at 4000 g for 30 s with 400 μL of binding buffer. For tryptic digestion, 125 μL of digestion buffer (50 mM TEAB) containing Pierce Trypsin/Lys-C Mix (Thermo Fisher Scientific, USA, A41007) (w/w ratio of 1:25) was added and incubated at 37 °C for 6 h. The digested sample was sequentially centrifuged with 80 μL of digestion buffer, 80 μL of 0.2% formic acid (Honeywell, USA, 94318) at 1000 g for 60 s, and 80 μL of 0.2% formic acid in 50% acetonitrile (ACN) at 4000 g for 60 s. Eluted peptides were dried and stored at −80 °C until the next process. Samples were desalted using a C18 spin column (Harvard Apparatus, USA) and dried. A205 application of NanoDrop One (Thermo Fisher Scientific, USA) was employed for peptide concentration measurements. A baseline was established using 1 µL of HPLC water, and dried samples were each diluted with HPLC water and measured at 1 µL. For the common reference sample, 10 μg aliquots were taken from the sample being analyzed, pooled together, and used for each TMT batch.
Construction of the common reference pool
In-depth proteomic analyses for this study were organized as TMTpro 16 plex experiments. For comparative quantification between all samples across experiments, a common reference (CR) sample was involved at the 134 N channel in each 16-plex. The CR is a protein mixture that contains all samples analyzed in the TMT experiments. All subsequent procedures, including digestion, were performed along with other individual samples to minimize variation.
TMTpro 16-plex labeling
Peptides, 250 μg per sample (based on peptide level quantification with NanoDrop One), were labeled with 16-plex TMT reagents (Thermo Fisher Scientific, USA, A44520) according to the manufacturer’s protocol. For each peptide aliquot of an individual sample, 1 mg of labeling reagent was used. Dissolve each sample in 50 μL of 100 mM TEAB (pH 8.5), add 20 μL of labeling reagent dissolved in anhydrous acetonitrile, and incubate for 1 h with shaking. 1.8 μg of labeled peptides from each channel were taken out and subjected to LC-MS/MS analysis to confirm labeling efficiency before pooling. Label efficiency criteria were set as having a minimum of 95% fully labeled MS/MS spectra in each sample. To quench the reaction, 5 μL of 5% hydroxylamine was added and incubated for 15 minutes. Samples were pooled in each 16-plex experiment and sequentially desalted with Sep-Pak C18 3 cc Vac Cartridge (Waters, USA) and dried.
Mid pH reverse-phase liquid chromatography fractionation
To minimize sample complexity, samples were fractionated by mid-pH reversed phase (RP) separation using Shimadzu LC20A (Shimadzu, Japan) with an analytical column (XBridge Peptide BEH C18 Column; 300 Å, 5 µm, 4.6 × 250 mm, Waters, USA) and a guard column (SecurityGuard cartridge C18, 4 ×3.0 mm, Phenomenex, USA). We performed the peptide fractionation at a flow rate of 0.5 mL/min. Mobile phases A and B were 10 mM TEAB and 10 mM TEAB in 90% ACN, and the sample was dissolved in 0.1% formic acid in 90 μL. The LC gradient of mobile phase B was 5% in 8 min, 40% in 65 min, 44% in 69 min, 60% from 74 min to 88 min, and 5% in 90 min. We collected a total of 96 fractions from 8 min, pooling them into 24 non-consecutive fractions every 0.91 min. We then concatenated 95% of each fraction into 12 non-consecutive fractions for post-translational modifications (PTMs) analysis.
Phosphopeptide enrichment
For the phosphopeptide enrichment, immobilized metal affinity chromatography (IMAC) was used. 300 μL of Ni-NTA agarose bead slurry (QIAGEN, Germany, 30410) was resuspended in the tube. The slurry was spun down for 1 minute, and the supernatant was removed. The bead was washed three times with 1 mL of HPLC water and then incubated with 1.2 mL of 100 mM EDTA (Sigma-Aldrich, USA, E7889) for 30 minutes by end-over-end turning at RT. After three washes with 1 mL of HPLC water, the beads were incubated with 1.2 mL of 10 mM FeCl3 for 30 minutes with end-over-end turning at room temperature. 12 fractions for PTM analysis are reconstituted in 113 μL of 0.1% TFA in 50% ACN, and then 337 μL of 0.1% TFA in 95% ACN is added sequentially. Beads were resuspended in 140 μL of 1:1:1 ACN:MeOH:0.01% acetic acid solution and 10 μL of beads were aliquoted into 12 tubes for each fraction. We resuspended the beads in the sample solution to bind the phosphopeptides, and then gently mixed them for 30 minutes at room temperature. The supernatant was collected separately for the next acetyl peptide enrichment step. Beads coupled with phosphopeptides were dissolved in 180 μL of 0.1% TFA in 80% ACN and desalted using a C18 stage-tip to elute for LC-MS/MS.
Acetylpeptide enrichment
For the acetylpeptide enrichment, the PTMScan® Acetyl-Lysine Motif [Ac-K] Kit (Cell Signaling Technology, USA, 13416) was used. IMAC eluents were concatenated into four fractions and dried. Peptides were dissolved in 1.4 mL of 1 x IAP buffer, in which 10 x IAP buffer (5.78 g MOPS-NaOH, 1.461 NaCl, 0.8 g dibasic sodium phosphate, and 0.08 g monobasic sodium phosphate) were adjusted to pH with acetic acid ( ~ 1.4 to 1.6 mL). Agarose beads bound to the acetyl-lysine motif antibody were pre-washed a total of 4 times using 1 x IAP buffer under ice and split into 2 tubes with equal volume. The peptides were transferred to a tube and incubated at 4 °C for 3 hours with end-over-end turning. Each tube was centrifuged (2,000 g, 30 sec, 4 °C), and the supernatant was separated, followed by washing the beads twice with 1 mL of ice-cold PBS and three times with chilled HPLC water. 100 μL of 0.15% TFA was added to beads coupled with acetyl peptides and then incubated at RT for 10 minutes, gently mixed every 2-3 minutes, and then centrifuged to elute. We desalted the eluted acetyl peptides using a C18 stage-tip (IMAC procedure) and dried them after a total of two elution steps.
LC-MS/MS for proteomics analyses
For global proteomic analyses, the Ultimate 3000 RSLC nano system (Thermo Fisher Scientific, USA) coupled with the Q Exactive HF-X hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, USA) was used. Trap column (Acclaim™ PepMap™ 100 C18 LC Column, C18, 100 um x 2 cm, 5 μm, Thermo Fisher Scientific, USA) and analytical column (EASY-Spray™ LC Columns, C18, 75 um x 50 cm, 2 μm, Thermo Fisher Scientific, USA), which were heated to 50 °C to prevent over-pressuring, were equipped with UHPLC separation. Mobile phase flow rate was 0.3 μL/min, and solvents A and B were 97% water, 3% ACN, 0.1% formic acid, and 90% ACN, 0.1% formic acid, respectively. The gradient profile of solvent B was 2% in 12 min, 8% in 16 min, 25% in 140 min, 35% in 150 min, 85% from 155 min to 163 min, and 2% from 165 min to 185 min. Data-dependent acquisition was performed at a spray voltage of 1.5 kV. MS1 spectra were measured with a resolution of 120,000 and a mass range of 350 to 1500 m/z. The AGC target of 3e6, maximum injection time of 50 ms, and isolation window of 0.7 m/z were set. Top 15 most abundant precursors per cycle were selected to trigger MS/MS. MS2 spectra were measured with a resolution of 45,000, 1e5 of the AGC target, 120 ms of maximum injection time, 34% of collision energy, and 110 of the fixed first mass (m/z). The precursor charge state was 2-5, the intensity threshold was 1e4 and the dynamic exclusion time was 45 s.
For PTM analysis, the Ultimate 3000 RSLC nano system (Thermo Fisher Scientific, USA) coupled with the Exploris 480 orbitrap mass spectrometer (Thermo Fisher Scientific, USA) was used. Trap column (Acclaim™ PepMap™ 100 C18 LC Column, C18, 100 um x 2 cm, 5 μm, Thermo Fisher Scientific, USA) and analytical column (EASY-Spray™ LC Columns, C18, 75 um x 50 cm, 2 μm, Thermo Fisher Scientific, USA) were equipped with UHPLC separation. The mobile phase flow rate was 0.3 μL/min, and solvents A and B were 0.1 % formic acid in water and 0.1 % formic acid in ACN, respectively. The LC gradient of solvent B was 2% in 14 min, 4% in 17 min, 16% in 120 min, 25% in 145 min, 85% from 150 min to 158 min, and 4% from 160 min to 185 min. Data-dependent acquisition was performed at a spray voltage of 2.1 kV and the cycle time was set to 3 sec. MS1 spectra were measured with a resolution of 120,000 and a mass range of 350 to 1500 m/z. The normalized AGC target (%) was 300, and the maximum injection time was 50 ms. MS2 spectra were measured with a resolution of 45,000, 7.5 e4 of AGC target, 120 ms of maximum injection time, 38% of collision energy, and 0.7 m/z of isolation window. Peptides were selected for a 2-6 precursor charge state with an intensity threshold of 1e4. Peptides that triggered MS/MS scans were dynamically excluded from further MS/MS scans for 45 sec, with a +/- 10 ppm mass tolerance.
The database search strategy
In addition to canonical peptides, to reliably identify variant, post-translational modified, and novel peptides in global proteomics, we used a multi-stage search strategy: 1) the identification of canonical peptides (primary database search), 2) the identification of variant and modified peptides by considering 2355 modifications in Unimod, and 3) the identification of novel peptides. For each stage, the FDR was calculated separately using a target-decoy strategy, and identifications were obtained at 1% FDR. Only unidentified spectra from the previous stage were subjected to the subsequent stage. Details of each stage are described in the following sections: 1) “The Primary Database Search”, 2) “Identification of Variant and Modified Peptides”, and 3) “Identification of Novel Peptides”.
The primary database search
All MS raw files were analyzed using Proteome Discoverer v2.4 with the SequestHT search engine90 against the UniProt human protein database v2021.01 (97,795 entries), combined with 179 common contaminant proteins. The search parameters were set to 10 ppm for precursor mass tolerance, 0.02 Da for fragment mass tolerance, fully-tryptic enzyme specificity allowing for up to 2 missed cleavages, carbamidomethylation ( + 57.021 Da) on Cys residues and TMT modification ( + 304.207 Da) on the peptide N terminus and Lys residues for static modifications, and oxidation ( + 15.995 Da) on Met residues, Met-loss (-131.040 Da) on protein N-terminal Met residues and deamidation ( + 0.984 Da) on Asn and Gln residues for dynamic modifications. The minimum length of a peptide was set at six residues.
For phosphoproteome and acetylproteome, the search parameters additionally included phosphorylation ( + 79.966 Da) on Ser, Thr, and Tyr residues and acetylation ( + 42.016 Da) on the protein N terminus and Lys residues for dynamic modifications. The TMT modifications were set as dynamic modifications for acetylproteome, to prevent co-assignment of TMT and acetylation. All peptide spectrum matches (PSMs) were subsequently rescored by Percolator91 and validated at an estimated false discovery rate (FDR) of 1%. For phosphosite and acetylsite localization, ptmRS92 was used, and modified sites with ptmRS probability greater than 0.95 were regarded as confident.
Identification of variant and modified peptides
Using CustomProDB93, each patient’s germline and somatic mutations were used to generate variant protein sequences. The variant protein sequences were merged for each batch and combined with 16,130 UniProt human proteins identified from the primary database search and 179 common contaminant proteins to generate combined customized databases.
All MS raw files were converted to MGF files using msconvert v3.0.1, and the precursor m/z values were replaced with the recalibrated values by Proteome Discoverer in the primary database search (this recalibrated MS2 spectra were used in all subsequent searches). The unidentified MS2 spectra from the primary database search were searched against the combined customized databases using MODplus v1.0294. The search parameters were set to 10 ppm for precursor mass tolerance, 0.01 Da for-fragment mass tolerance, semi-tryptic enzyme specificity allowing for up to 2 missed cleavages, -1 to +2 for 13 C isotope error, carbamidomethylation ( + 57.021 Da) on Cys residues and TMT modification ( + 304.207 Da) on the peptide N terminus and Lys residues for static modifications. All modifications in Unimod database v2020.10 (2355 entries) including all amino acid substitutions were considered for dynamic modification (multiply modified peptides were allowed within the modified mass range of -150 to +350 Da). All PSMs were validated at an estimated FDR of 1% using MODplus FDR toolkit.
To identify neoantigen candidates, the proteomics-supported somatic mutations were filtered by examining the intensity of TMT reporter ion matched to the patient for whom the somatic mutation was called (the corresponding reporter ion intensity must account for at least 20% of the total intensity of all reporter ions). The immunogenicity of filtered somatic mutations was also evaluated by predicting binding affinity to human leukocyte antigen (HLA)-I molecules.
Identification of novel peptides
All unidentified MS2 spectra from the primary and subsequent variant database searches were analyzed to identify novel peptides originating from pseudogenes, long noncoding RNAs (lncRNAs), untranslated regions (UTRs), and novel isoforms (including fusion genes). For pseudogenes, lncRNAs, and UTRs whose RNA transcripts’ FPKMs are greater than 0, the RNA sequences were selected from the GENCODE transcript fasta files v32, and their three frame translations were generated for each patient, except for UTRs, whose translations were generated by the UTR sequence database construction method55, which assumes that alternative cognate and near-cognate start codons and translational readthrough in the stop codons can result in abnormal translation. These noncoding peptide sequences were merged for each batch and matched to MS2 spectra by MS-GF+ v2021.0995 with the following search parameters: 10 ppm for precursor mass tolerance, tryptic enzyme specificity allowing up to 2 missed cleavages (also permitting non-enzymatic terminals), no 13 C isotope error allowed, carbamidomethylation ( + 57.021 Da) on Cys residues and TMT modification ( + 304.207 Da) on the peptide N terminus and Lys residues for static modifications, oxidation ( + 15.995 Da) on Met residues and deamidation ( + 0.984 Da) on Asn and Gln residues for dynamic modifications, and TMT protocol. The minimum length of a peptide was set at 8 residues. All PSMs were validated at an estimated FDR of 1%.
The novel transcripts, including fusion genes, were predicted by StringTie v2.1.7 (class code =, c, k, m, n, and j)78,96 and Arriba v1.2.088, respectively, and their three frame translations were generated to compose the novel protein isoform sequences for each patient. The novel isoform sequences were merged for each batch and matched to MS2 spectra using Comet v2021.02.097 with the following parameters: 10 ppm for precursor mass tolerance, semi-tryptic enzyme specificity allowing up to two missed cleavages, no isotope error allowed, carbamidomethylation ( + 57.021 Da) on Cys residues and TMT modification ( + 304.207 Da) on the peptide N terminus and Lys residues for static modifications, and oxidation ( + 15.995 Da) on Met residues and deamidation ( + 0.984 Da) on Asn and Gln residues for dynamic modifications. The minimum length of a peptide was set at 8 residues. All PSMs were subsequently rescored by Percolator and validated at an estimated FDR of 1%.
We rejected PSMs conflicting with both MS-GF+ and Comet (i.e., identical spectra but different peptides assigned). The remaining novel peptides were searched using BLAST98 and filtered out if there were peptide sequence matches in the Uniprot (v2022.03, 226,999 entries), RefSeq (v2022.09, 130,184 entries) and/or Gencode (v41, 110,224 entries) human protein sequences, allowing no more than a single amino acid substitution. We also removed the frameshift peptides to concentrate solely on novel peptides derived from noncoding regions and novel isoforms. Finally, we used peptides supported by at least one RNA-Seq read and a quantifiable PSM to retrieve unique peptides at the gene level. In total, we obtained and used 1045 novel sequences for further analysis.
Quantification of Protein, Phosphosite and Acetylsite
Peptide abundances were normalized to have the same total amount of peptide abundances between TMT channel values in the same batch by Proteome Discoverer v2.4 with the SequestHT search engine90 against the UniProt human protein database v2021.01 (97,795 entries), combined with 179 common contaminant proteins. The co-isolation threshold 50% and the average reporter S/N threshold 10% were used to filter out quantification values of low quality. For global proteome quantification, proteins with at least one unique and/or razor peptides were exported, and normalization between batches was achieved by dividing each TMT channel value by that of the common reference (CR) channel in the same batch. The log2 ratio value was used for the subsequent quantification analysis.
The quantification of phosphosites and acetylsites was performed using the same method: the sum of peptide abundances containing each modification site was calculated to represent modification site abundance, and each TMT channel value was normalized using the same method as the global proteome data. After applying two-component normalization of TMT ratios for each proteomics dataset99, the log2 ratio value was used for the subsequent quantification analysis.
We selected features that have less than 30% missing values across all tumors for the global proteomics, phosphoproteomics, and acetylproteomics dataset using preprocessed normalized log2 ratio values. After then, we performed k-nearest neighbor imputation (k = 5) for missing values using the impute R package (10.18129/B9.bioc.impute) for the subsequent quantification analysis.
NMF clustering analysis for multiomics subtypes
To identify patient subtypes, we performed non-negative matrix factorization (NMF) analysis on global proteins and PTM sites as instructed in previous CPTAC studies10,13. We concatenated preprocessed imputated global proteomics, phosphoproteomics, and acetylproteomics dataset into a single matrix. Then, we excluded features with the lowest standard deviation (bottom 5th percentile), followed by scale and z-score transformation. Using the NMF R-package100, we performed the NMF analysis for factorization ranks from 2 to 10 using the standard NMF algorithm from Brunet et al.101 and 2000 max iterations for convergence and repeated 50 times to compute clustering statistics. We determined the optimal factorization rank k, representing the number of clusters, from the rank with the maximal cophenetic correlation coefficient value and its drastic decrease. With the optimal factorization rank k, we sought to robust clusters from the NMF analysis using 200 runs and 5000 max iterations. From decomposed NMF matrices, we assigned samples to NMF clusters corresponding to the optimal factorization rank k and obtained features specific to each cluster according to the row-wise feature score102. We defined a “core sample” for those having a cluster membership score ≥ 0.5 as described previously10,13.
Combined NMF and Comparison of NMF subtypes with previous NSCLC subtypes
To assess the concurrence between our subtypes and lung cancer subtypes defined through recent multi-omics studies, we conducted NMF analysis by integrating our study cohort with previous CPTAC study cohort, which contains LUAD10 and LSCC13. Prior to running NMF, each study was subjected to pre-processing using the methodology outlined in the “NMF clustering analysis for multiomics subtypes” section. Features pertaining to proteomics, phosphoproteomics, and acetylproteomics datasets were examined, and only those identified as commonly present in all three studies were selected and concatenated into a single matrix for NMF clustering. Using the NMF R-package, we sought to robust cluster from the NMF analysis using 200 runs and 5000 max iterations with rank 5. We analyzed the subtypes derived from NMF decomposition matrices for each individual study within the study cohort samples of LUAD and LSCC and compared them with the subtypes identified in the combined NMF results.
To validate the robustness of our subtype classification in a recent NSCLC study10,11,13,14 comprising 462 patients, we conducted a comparative analysis of subtype features across multiomics datasets included in each individual study. To identify the features associated with the subtypes of each study, we performed re-clustering of NMF features using the omics datasets and parameters employed in prior studies and selected features utilizing the “Max” method102. Gillette study was conducted with RNA, global-, phospho- and acetyl-proteomics datasets for NMF, and used 4 ranks, 200 runs and 5000 max iterations. Satpathy study was conducted with CNV, RNA, global-, phospho-, and acetyl-proteomics dataset for NMF and used 5 ranks, 500 runs, and 5000 max iterations. Also, Lehtio study was conducted with only global proteomics dataset for NMF, and used 6 ranks, 100 runs, and 5000 max iterations. For Xu’s (2020) study, the up-regulated protein list for each subtype as presented was utilized as a subtype-specific features. To confirm a statistically significant relationship, an odds ratio and p value were calculated by conducting a Fisher’s exact test in R between each study cluster’s features.
Gene set enrichment analysis
To obtain characteristic analysis information corresponding to NMF subtype based on the expression data of multi-omics of tumor samples, we performed PTM signature Enrichment Analysis (PTM-SEA103) for phosphoprotoemics dataset and R package GSVA104 for global-, phospho- and acetyl protoemic datasets.
For GSVA, pathways from hallmark, KEGG, Reactome, GO and Wikipathway databases which were downloaded from MSigDB were considered and we used only pathways which contains more than 200 genes and lower than 1000 genes in each pathway.
database: “h.all.v2023.1.Hs.symbols.gmt.txt”, “c2.cp.kegg.v2023.1.Hs.symbols.gmt.txt”, “c2.cp.reactome.v2023.1.Hs.symbols.gmt.txt”, “c2.cp.wikipathways.v2023.1.Hs.symbols.gmt.txt”, “c5.go.v2023.1.Hs.symbols.gmt.txt”
For PTM-SEA, flank amino acid sequence (+/- 7 aa) was added to PTM data as primary identifier and used. We computed normalized enrichment scores (NES) of gene sets. We used the implementation which contains PTM-SEA available on GitHub (https://github.com/broadinstitue/ssGSEA2.0) using the command interface R-script (ssgsea-cli.R) using the following parameters:
database:”ptm.sig.db.all.flanking.human.v1.9.0.gmt”
sample. norm. Type: “rank”
output. score. Type = “NES”
nperm = 100
For identifying NMF subtype feature pathway, we select Hallmark pathway gene sets (MSigDB), which summarize specific cancer-related biological states or processes for ssGSEA and PTM signatures database (PTMsigDB) containing a list of modification site-specific signatures.
Identification of differentially enriched copy number variations
Gene-level copy numbers detected by CNVkit were converted to cytoband-level by calculating the mean copy numbers in each cytoband. Then we used a linear regression model to identify differentially enriched copy number variations in each NMF Subtype. To control the effect of the pathological diagnosis classification (DX), we set DX as a covariate.
β0 and β1 indicate the intercept and the log2FC value, respectively. Cytobands with an adjusted p-value (calculated using Benjamini–Hochberg method) less than 0.05 were selected as significant CNVs.
Identification of differentially expressed genes (DEGs)
To identify differentially expressed genes between conditions, we used “DESeq” function provided by DESeq2 R packages76. Quantified transcript-level estimates from Salmon were imported to R using “tximport” and were converted to a gene-level expression matrix. With this gene-level expression matrix, DESeqDataSet was generated using “DESeqDataSetFromTximport” function. Then, differential expression analysis based on the negative binomial distribution was performed using “DESeq” function. Notably, we conducted the DEG analysis using the formula below.
In this model, Group represent the conditions under comparison. DX is the name of the pathological diagnosis classification, including LUAD or LSCC. Batch information for RNA (rna.batch) was also used as a covariate in DEG analysis to remove batch effects from DEG analysis. The adjusted p-value was calculated using the Benjamini-Hochberg method, and an adjusted p-value less than 0.05 was used as a criterion for differentially expressed genes.
Identification of differentially expressed proteins, phosphorylation, and acetylation (DEP)
We identified differentially expressed proteins, phosphorylation, and acetylation using linear regression models as below.
yi is a vector of dependent variables that represents the expression values of each proteomic feature. Group is a vector of independent variables that represents the state we are trying to compare. DX is the name of the pathological diagnosis classification, and Batch is a TMT channel number that represents batch information. β0 is intercept, β1 is beta coefficient for Group variables, and represents the log2FC value. The linear regression’s calculated p-value tests the null hypothesis that the coefficient is equal to zero, indicating no difference between groups. The adjusted p-value was calculated using Benjamini–Hochberg method, and an adjusted p-value less than 0.05 was used as a criterion for differentially expressed proteins, phosphorylation, and acetylation.
Survival analysis
To compare the survival probability across our five NMF subtypes, we measured overall survival (OS) length (Supplementary Data 1a), the time from the date of tumor resection surgery to the time of NSCLC-induced death, for 229 patients. With NSCLC-death, we performed a survival analysis based on Kaplan–Meier estimation. In the Kaplan–Meier estimation model, NSCLC-death was used as an indicator of the ending time of the OS length. It is labeled as a right-censored observation in the case of patients with no deaths, which causes NSCLC at the end of the study. For patients who have died but whose cause of death is not NSCLC, we can use partial information that they survived beyond a certain point, but the exact date of NSCLC-death is uncertain.
The Kaplan-Meier estimation was used to measure survival curves for each subtype. The Kaplan-Meier survival curve is defined as the probability of surviving for each length of time after tumor resection surgery while considering time at many small intervals. For each time interval, survival probability is calculated as the number of patients who survived divided by the number of patients at risk. Patients who have died, dropped out, or moved out are not counted as “patients at risk”. The total probability of survival until that time interval is a cumulative probability, calculated based on the law of multiplication of probability by multiplying all the probabilities of survival at all time intervals until a certain point105. We used the log-rank test to statistically test the null hypothesis that there is no difference between the survival curves of each subtype. For the log-rank test, the total number of observed events in each subtype, i.e., O1 and O2 was used, and the expected number of events in each group, i.e., E1 and E2, was calculated. The total number of expected events is calculated as the sum of the expected number of events at the time of each event in any of the subtype, bringing all subtypes together. The expected number of events at the time of each event is the result of multiplying the total number of patients surviving at the time of events in all subtypes by the risk of events at time105. Log-rank test statistic is calculated as below.
Since there are five subtypes in total, the pooled p value is calculated and shown in Fig. 2g. We perform pairwise comparison between Subtypes to decide which Subtypes represent poor prognosis subtype of NSCLC and adjusted p-value was calculated using the Benjamini–Hochberg procedure.
To identify the molecular features that result in survival probability differences, we also performed feature-wise survival analyses comparing survival curves between group of patients with top 50% expression and bottom 50% expression for each protein and PTM features based on Kaplan-Meier estimation. The prognostic direction, which shows whether the prognosis is favorable or unfavorable, was determined by the sign of coefficient from Cox proportional-hazard (CPH) model not from Kaplan-Meier model. Since in many cases, the survival probability does not reach 0.5, it was difficult to compare the median survival time that can be used as an indicator of prognostic direction in the Kaplan–Meier model. CPH model was used for univariate analysis and categorical analysis. h(t) is hazard function determined by the univariate, high- or low expression group. h0 is called the baseline hazard. It corresponds to the value of the hazard if x1 is equal to zero. The coefficient b1 measure the impact of univariate and tells us prognostic direction by sign. x1 is categorical variable, low- or high expression group. When sign of the coefficient b1 for high expression group is positive, its feature classified as unfavorable prognostic features and vice versa.
For significance, a log-rank test was used, and the nominal p-value < 0.001 was used as a criterion for survival-associated features. The results of the feature-wise survival analysis are listed in Supplementary Data 2d. We selected the top 1% of features by statistical significance for survival-associated features.
Kinase activity estimation based on phosphoproteomic data
To understand the Subtype-specific kinase activities, we used decoupleR R package to infer kinase activity from the results of differentially expressed phosphorylation (DEPP) using prior knowledge network, such as databases for kinase/phosphatase and substrate interactions106. We used prior knowledge network provided by PHONEMeS107. Also, we set minimum size of regulons to 5, and provided 1000 deregulated phosphorylation (each 500 phosphorylation for up-, down-regulated). There are a variety of methods to infer kinase activities and we used multivariate linear model (MLM)-based method as below:
where the dependent variable Y represents the t-statistic measurements of phosphorylation from the results of the differentially expressed phosphorylation analysis, and the independent variable X is the connectivity matrix representing the associations with kinases. Xij equals 1 when phosphosite i is a known substrate of the kinase j, 0 otherwise. Ψ represents the normally distributed error of the fit and β represents the scores of the kinase activity108. In decoupleR packages in R, we used “run_mlm” function to estimate kinase activity. P-value was obtained from a multivariate linear model and the adjusted p-value was calculated using the Benjamini–Hochberg adjustment.
Single-cell specific subtype distribution
To assess the tumor microenvironment in five NSCLC subtypes by comparing subtype-specific genes with a diverse set of cell type-specific genes, using the dataset ‘local_extended.rds’ obtained from https://luca.icbi.at. Subtype-specific gene sets were generated based on the results of differential gene expression analysis (DEGs), including only genes with significantly increased fold change and a significant adjusted p-value (<0.05).
For the tumor versus tumor comparison, we considered only the cell type-specific genes originating from ‘tumor_primary’ cells. Conversely, for the tumor versus normal adjacent tissue (NAT) comparison, we used a subset of cell type-specific genes from both ‘tumor_primary’ and ‘normal_adjacent’ cells.
Clustering analysis for immune microenvironment
We used transcriptome data of NSCLC patients to define the subtypes of tumor immune microenvironment. Two clustering approaches were used, which were based on the enrichment score of cell types41 and immune-related pathways42. The cell type-based clustering was performed with 205 tumors and 85 normal samples. The enrichment scores of 64 cell types were inferred by Xcell41, which performed gene set enrichment analysis based on the curated gene signatures for each cell type. The inferred cell enrichment scores were then normalized as z-score and consensus clustering was performed using R package CancerSubtypes109. After the consensus clustering, the partitioning around medoids (PAM) algorithm was used to divide the three clusters. We first assigned a NAT-enriched cluster among the three clusters when it was mainly matched to normal samples. After that, the immune score calculated by Xcell was used to determine the HTE and CTE immune clusters, so that the cluster with the higher immune score was defined as having an HTE tumor. We also performed pathway-based clustering with 205 tumor samples. For that, GSVA was performed across the patients based on seven curated immune-related pathways. The enrichment scores of the pathways were normalized as z-scores, and k-means clustering was performed based on the z-scores to identify two immune clusters. The cluster having a higher pathway enrichment score was defined as HTE tumor.
Survival and regression analysis with immune cluster and cell types
The enrichment score of cell type and the status of the immune cluster across the patients were tested for their correlation with a set of clinical and molecular features. The pattern of TILs was examined by regression analysis using MASS package and GLM package of R. Overall survival and relapse-free survival were tested for their correlations with cell types and immune clusters by Cox proportional hazards regression analysis using CoxPHFitter package of Python. All analyses were performed with clinical histology and sample batches as covariates.
Identifying putative regulators associated with immune landscape
To identify putative regulators associated with immune landscape, we first inferred protein activity using transcriptome data of the patients. For the protein activity, we obtained regulon networks of lung cancer from ARACNe package110 of R and calculated protein activity of each patient using viper tool44 that inferred how each protein regulates its target genes based the regulon network. The positive value of protein activity indicates that the protein positively regulates its target genes to be overexpressed, and the negative value indicates vice versa. We then calculated the correlation between the protein activity of immunomodulators and the enrichment score of cells, or the status of the immune cluster. The RNA and protein expression of the same immunomodulators was also analyzed for their associations with the immune landscapes. To corroborate the putative regulation of the immune landscape, we further analyzed whether a specific driver mutation was involved in the association between immunomodulators and immune cells or immune clusters. The curated driver mutations were derived from Oncovar111, DriverDBv3112, Intogen113, and mutation catalogue from Martínez-Jiménez et al.114.
Immune landscape analysis with the integrated cohort
To test the reproducibility of our analysis about the immune landscape across NSCLC patients, we used two independent NSCLC cohorts of Satpathy et al.13 and Gillette et al.10. The z-score normalization was performed for the enrichment scores of 64 cell types in each cohort that had 202 (Satpathy et al.), 211 (Gillette et al.), and 290 (our cohort) patients. The normalized enrichment scores of cell types were then integrated into a single cohort and performed consensus clustering. PAM algorithm was used to make three clusters and they were mapped to NAT, HTE, and CTE, respectively, according to the method used in our original cohort.
Neoantigen and cryptic peptide prediction
We predicted different types of neoantigens and cryptic peptides according to their origin and validation (Supplementary Fig. 6a). Firstly, canonical neoantigens were predicted from the mutated peptides that had a length between 9mer and 12mer amino acids including somatic mutations. The binding affinity of the mutated peptides was predicted using MHCflurry115 and NetMHCpan116 for the patient HLAs that were identified by OptiType117. Only the mutated peptides to be predicted as binding to the patient HLAs were defined as the potential neoantigen. Among the neoantigen candidates, we further defined “confirmed neoantigens” when they had evidence of MS experiments (see the section “Identification of variant and modified peptides”). The remaining two types were defined by non-canonical peptides that originated from unconventional translation of pseudogenes, lncRNAs, UTRs, or novel isoform transcripts (Refer to section “Identification of novel peptides”). We identified novel isoform transcripts that were expressed only in tumor samples when they matched normal samples. The mean expression of normal samples was used when the tumor sample did not match normal. We treated a degree of FPKM below 1 as indicating no expression. Only the novel peptides that occurred in more than two tumor samples and not in at least one normal sample were predicted as “cryptic peptides” using the same methodology for neoantigen candidates in the prediction of binding to HLAs. We further determined “confirmed cryptic peptide” when the cryptic peptides also showed substantial expression (FPKM > = 1) in the tumor samples as described in the previous study118.
Analysis of multi-omics subtype distribution according to the cryptic peptide load and immune landscape status
We analyzed the distribution of the patients of each multi-omics subtype across the status of cryptic peptide load and immune landscape. Two features were chosen as the surrogates of the immune landscape, which were the status of the immune cluster (HTE/CTE) and the antigen processing and presentation machinery (APM). The patients were first separated into two groups that had a high and low number of cryptic peptides. They were also divided into two other groups: HTE and CTE, or high APM and low APM. The status of APM was determined based on the pathway enrichment score of “Antigen_Processing_and_Presentation” in KEGG pathway119. We created four categories based on the status of cryptic peptide load and immune landscape, and the degree of over-representation for each multi-omics subtype was calculated by a two-sided Fisher’s exact test.
Patient-derived lung tumoroid culture
Patient-derived lung tumoroid culture and drug screening were performed by SG Medical, Inc. (Seoul, Korea) and conducted as previously described120. Briefly, tissues were kept in a cold Hank’s balanced salt solution (HBSS) with antibiotics (Gibco, OK, USA) and on ice after dissection. Samples were washed three times with cold HBSS with antibiotics and sectioned with sterile blades. Sectioned samples were incubated with 0.001% DNase (Sigma-Aldrich, MO, USA), 1 mg/ml collagenase (Roche, IN, USA), 200 U/ml penicillin, and 200 mg/ml streptomycin in DMEM/F12 medium (Gibco, OK, USA) at 37 °C for 1 h with intermittent agitation. After incubation, the suspensions were repeatedly triturated by pipetting and passed through 40-μm cell strainers (BD Falcon, CA, USA). The strained cells were centrifuged at 112 × g for 3 min, and the pellet was resuspended in lung tumoroid culture media (DMEM/F12 supplemented with 20 ng/ml of bFGF (Invitrogen, CA, USA), 50 ng/ml human EGF (Invitrogen), N2 (Invitrogen), B27 (Invitrogen), 10 μM ROCK inhibitor (Enzo Life Sciences, NY, USA), and 1% penicillin/streptomycin (Gibco, OK, USA). The suspension was mixed with matrigel and seeded onto 6-well plates. Culture media were replaced every 4 days.
Drug screening
Lung tumoroids cultured were harvested and dissociated using TrypLE Express (Gibco, OK, USA). The dissociated lung tumoroids were diluted in a lung tumoroid culture media-matrigel mixture and seeded onto 384-well plates (250 cells per well). After lung tumoroid generation, 8 concentrations of Selinexor (Selleckchem, TX, USA) and vehicles (DMSO) as a negative control were added in triplicate. After 6 days, quantification of cell viability was done by adding 10 μl of CellTiter-Glo 3D (Promega) to each well according to the manufacturer’s instructions on a Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, MA, USA). The determination of IC50 values was conducted using GraphPad Prism.
CPTAC NSCLC data download and preprocessing
Clinical data, MAF file, and gene-level CNV data are downloaded from the supplementary tables provided by previous studies10,13. Segment-level CNV data is downloaded from the GDC data portal (https://portal.gdc.cancer.gov), and global proteome, phosphoproteome, acetylproteome, and clinical data, including survival information, were downloaded from LinkedOmics (http://www.linkedomics.org). For global, phospho-, and acetyl-proteomics data, we performed the imputation in the same way as in our cohort. Briefly, features with more than 30% missing values across all tumors were discarded, and k-NN imputation using k = 5 was performed. We conducted all downstream analyses using these data in the same manner as we did with our cohort.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank the patients who participated in this research, without whose contributions the current study would be impossible. This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2019M3E5D3071921) and the Korea University Grant (to J.Y.A.). E.H.C., G.H.L., C.H., and S.Y.K. received a scholarship from the BK21 FOUR education program. This work was done under the auspices of a Memorandum of Understanding between the Kyung Hee University (KHU) & Daegu Gyeongbuk Institute of Science and Technology (DGIST) and the U.S. National Cancer Institute’s International Cancer Proteogenome Consortium (ICPC). ICPC encourages international cooperation among institutions and nations in proteogenomic cancer research in which proteogenomic datasets are made available to the public. This work was also done in collaboration with the U.S. National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC).
Author contributions
K.P.K., M.S.K., S.J.J., H.S.H., E.P., K.K., and J.Y.A. designed and directed the integrated proteogenomic analysis. H.S.H., W.J., C.M.C., J.C.L., H.R.K., K.G.K., H.S.A. and J.Y.Y. collected, characterized, and processed the tumor samples. K.P.K. and M.S.K. designed the proteomic experiments. K.J.S., K.C.C., and W.J.A. performed global proteome and phosphoproteome profiling experiments. E.P., S.N., and S.C. unified database searches and analyzed the proteomic data. J.Y.A., E.H.C., G.H.L., Y.J.H., C.H., S.Y.K., S.H.C., K.K., S.B.S., J.S.P., D.Y.Y., E.P., S.N., S.C., H.S.H. and K.J.S. performed integrated analyses with genomic and proteomic data. J.Y.A., E.H.C., K.K., E.P., S.N., S.C., H.S.H., S.J.J., K.P.K. and K.J.S. wrote the manuscript and J.Y.A., K.K., E.P., S.N., S.C., H.S.H., S.J.J. and K.P.K. supervised. All authors have read and approved the final version of the manuscript for publication.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Genomic and transcriptomic raw data generated in this study are available in EGA under restricted access under the study ID EGAS50000000592 and Dataset ID EGAD50000000844. Data will be made available following a data access agreement, and there are no restrictions on who will be granted access. Requests will be assessed by the data access committee, and further information can be requested from Dr Kwang Pyo Kim (kimkp@khu.ac.kr). Additionally, these data were also available in the Korean Nucleotide Archive (KoNA, https://kobic.re.kr/kona) with the accession ID KAP210028. Raw mass spectrometry-based global, phosphoproteome, and acetylome data were deposited in the ProteomeXchange Consortium (accession number: PXD053969, PXD053921, PXD053903) via the jPOST partner repository (accession number: JPST003210, JPST003211, JPST003212)121.
All histologic details and sample annotations can be accessed from Supplementary Data 1a. Processed and normalized gene expression data file is provided in Supplementary Data 7 and the proteomic data files are provided in K-BDS with the accession IDs KAP240387, KAP240391, and KAP240392.
Code availability
No custom code was used or developed for the analyses presented in this study. Standard workflows and open-source R packages and software were used (Methods). The codes used for the analyses included in our manuscript were uploaded to GitHub repository with instructions for users: https://github.com/joonan-lab/PDIAMOND-NSCLC.
Competing interests
Kwang Pyo Kim is the CEO of NioBiopharmaceuticals, Inc. Se Jin Jang is the chief technology officer of SG Medical, Inc. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kyu Jin Song, Seunghyuk Choi, Kwoneel Kim, Hee Sang Hwang.
These authors jointly supervised this work: Seungjin Na, Se Jin Jang, Joon-Yong An, Kwang Pyo Kim.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54434-4.
References
- 1.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Arriagada, R. et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J. Clin. Oncol.28, 35–42 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Yang, C. Y., Yang, J. C. & Yang, P. C. Precision management of advanced non-small cell lung cancer. Annu Rev. Med71, 117–136 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin.71, 7–33 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Submission., N. C. I. S. S. D. N. https://seer.cancer.gov/data-software/documentation/seerstat/nov2020/.
- 6.Heo, Y. J., Hwa, C., Lee, G. H., Park, J. M. & An, J. Y. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes. Mol. Cells44, 433–443 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature489, 519-525 10.1038/nature11404 (2012). [DOI] [PMC free article] [PubMed]
- 8.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature511, 543-550 10.1038/nature13385 (2014). [DOI] [PMC free article] [PubMed]
- 9.Stewart, P. A. et al. Proteogenomic landscape of squamous cell lung cancer. Nat. Commun.10, 3578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillette, M. A. et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell182, 200–225.e235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, J. Y. et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell182, 245–261.e217 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y. J. et al. Proteogenomics of non-smoking lung cancer in east asia delineates molecular signatures of pathogenesis and progression. Cell182, 226–244.e217 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Satpathy, S. et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell184, 4348–4371.e4340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehtio, J. et al. Proteogenomics of non-small cell lung cancer reveals molecular subtypes associated with specific therapeutic targets and immune evasion mechanisms. Nat. Cancer2, 1224–1242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demirel, H. C., Arici, M. K. & Tuncbag, N. Computational approaches leveraging integrated connections of multi-omic data toward clinical applications. Mol. Omics18, 7–18 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Tarazona, S., Arzalluz-Luque, A. Conesa, A. Undisclosed, unmet and neglected challenges in multi-omics studies. Nat. Comput. Sci.10.1038/s43588-021-00086-z (2021). [DOI] [PubMed]
- 17.Stang, A. et al. Diagnostic agreement in the histopathological evaluation of lung cancer tissue in a population-based case-control study. Lung Cancer52, 29–36 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Grilley-Olson, J. E. et al. Validation of interobserver agreement in lung cancer assessment: hematoxylin-eosin diagnostic reproducibility for non-small cell lung cancer: the 2004 World Health Organization classification and therapeutically relevant subsets. Arch. Pathol. Lab Med137, 32–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, L. et al. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat. Commun.5, 5469 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Ardizzoni, A. et al. Cisplatin- versus carboplatin-based chemotherapy in first-line treatment of advanced non-small-cell lung cancer: an individual patient data meta-analysis. J. Natl Cancer Inst.99, 847–857 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Arriagada, R. et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N. Engl. J. Med350, 351–360 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Wallerek, S. & Sorensen, J. B. Biomarkers for efficacy of adjuvant chemotherapy following complete resection in NSCLC stages I-IIIA. Eur. Respir. Rev.24, 340–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salcher, S. et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell40, 1503–1520.e1508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, Y. et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J. Clin. Oncol.32, 121–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roovers, K. et al. The Ste20-like kinase SLK is required for ErbB2-driven breast cancer cell motility. Oncogene28, 2839–2848 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Chaar, Z., O’Reilly, P., Gelman, I. & Sabourin, L. A. v-Src-dependent down-regulation of the Ste20-like kinase SLK by casein kinase II. J. Biol. Chem.281, 28193–28199 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Al-Zahrani, K. N. et al. Loss of the Ste20-like kinase induces a basal/stem-like phenotype in HER2-positive breast cancers. Oncogene39, 4592–4602 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Wang, K., Hong, R. L., Lu, J. B. & Wang, D. L. Ste20-like kinase is upregulated in glioma and induces glioma invasion. Neoplasma65, 185–191 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Douchi, D. et al. Silencing of LRRFIP1 reverses the epithelial-mesenchymal transition via inhibition of the Wnt/beta-catenin signaling pathway. Cancer Lett.365, 132–140 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Ma, W. et al. LRRFIP1, an epigenetically regulated gene, is a prognostic biomarker and predicts malignant phenotypes of glioma. CNS Neurosci. Ther.28, 873–883 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faisal, F. A. et al. CDKN1B deletions are associated with metastasis in african american men with clinically localized, surgically treated prostate cancer. Clin. Cancer Res26, 2595–2602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Travis, W. D. et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol.10, 1243–1260 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Moreira, A. L. et al. A grading system for invasive pulmonary adenocarcinoma: a proposal from the international association for the study of lung cancer pathology committee. J. Thorac. Oncol.15, 1599–1610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karni, R. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol.14, 185–193 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, A. Y. & Liu, H. The past, present, and future of CRM1/XPO1 inhibitors. Stem Cell Investig.6, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muz, B., de la Puente, P., Azab, F. & Azab, A. K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl.)3, 83–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu, Z. W., Bi, J. H., Gazdar, A. F. & Song, K. Genome-wide copy number variation pattern analysis and a classification signature for non-small cell lung cancer. Genes Chromosomes Cancer56, 559–569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leprivier, G. et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell153, 1064–1079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tekpli, X. et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat. Commun.10, 5499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfannstiel, C. et al. The tumor immune microenvironment drives a prognostic relevance that correlates with bladder cancer subtypes. Cancer Immunol. Res7, 923–938 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol.18, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu, Y., Sun, H., Zhang, H. & Wang, X. An immunogram for an individualized assessment of the antitumor immune response in patients with hepatocellular carcinoma. Front Oncol.10, 1189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruni, D., Angell, H. K. & Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer20, 662–680 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Alvarez, M. J. et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet48, 838–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorsson, V. et al. The immune landscape of cancer. Immunity48, 812–830.e814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao, X. et al. Immune landscape of invasive ductal carcinoma tumor microenvironment identifies a prognostic and immunotherapeutically relevant gene signature. Front Oncol.9, 903 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, L. et al. Immune landscape of colorectal cancer tumor microenvironment from different primary tumor location. Front Immunol.9, 1578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Efremova, M., Finotello, F., Rieder, D. & Trajanoski, Z. Neoantigens generated by individual mutations and their role in cancer immunity and immunotherapy. Front Immunol.8, 1679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laumont, C. M. et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat. Commun.7, 10238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erhard, F., Dolken, L., Schilling, B. & Schlosser, A. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol. Res8, 1018–1026 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Starck, S. R. & Shastri, N. Nowhere to hide: unconventional translation yields cryptic peptides for immune surveillance. Immunol. Rev.272, 8–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nejo, T. et al. Reduced neoantigen expression revealed by longitudinal multiomics as a possible immune evasion mechanism in glioma. Cancer Immunol. Res7, 1148–1161 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Kim, M. S. et al. A draft map of the human proteome. Nature509, 575–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, Y. et al. Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat. Commun.9, 903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi, S. et al. Proteogenomic approach to UTR peptide identification. J. Proteome Res19, 212–220 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Cao, X. et al. Comparative proteomic profiling of unannotated microproteins and alternative proteins in human cell lines. J. Proteome Res19, 3418–3426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hari, P. S. et al. Proteogenomic analysis of breast cancer transcriptomic and proteomic data, using de novo transcript assembly: genome-wide identification of novel peptides and clinical implications. Mol. Cell Proteom.21, 100220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, H. et al. Integration of mass spectrometry and RNA-Seq data to confirm human ab initio predicted genes and lncRNAs. Proteomics14, 2760–2768 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Choe, W. et al. Identification of 8-Digit HLA-A, -B, -C, and -DRB1 Allele and Haplotype Frequencies in Koreans Using the One Lambda AllType Next-Generation Sequencing Kit. Ann. Lab Med41, 310–317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee, K. W., Oh, D. H., Lee, C. & Yang, S. Y. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens65, 437–447 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol.38, 199–209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang, T. et al. Heterogeneity of neoantigen landscape between primary lesions and their matched metastases in lung cancer. Transl. Lung Cancer Res9, 246–256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nahar, R. et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat. Commun.9, 216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Isaka, T. et al. Efficacy of platinum-based adjuvant chemotherapy on prognosis of pathological stage ii/iii lung adenocarcinoma based on egfr mutation status: a propensity score matching analysis. Mol. Diagn. Ther.23, 657–665 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Kawaguchi, Y. et al. Epidermal growth factor receptor mutation subtype has differential effects on adjuvant chemotherapy for resected adenocarcinoma pathological stages II-III. Oncol. Lett.18, 6451–6458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, H., Shen, J., Liu, J., Fang, W. & Zhang, L. Efficacy of immune checkpoint inhibitors in SMARCA4-Mutant NSCLC. J. Thorac. Oncol.15, e133–e136 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Amin, M. B. et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin.67, 93–99 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Saltz, J. et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep.23, 181–193.e187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.ML, T. Clinical Hematology: Theory and Procedures, enhanced 6th edition. (Jones & Bartlett Learning (Burlington), 2017).
- 70.Mayakonda, A., Lin, D. C., Assenov, Y., Plass, C. & Koeffler, H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res28, 1747–1756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol.12, e1004873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tate, J. G. et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S. et al. Copy number signature analysis tool and its application in prostate cancer reveals distinct mutational processes and clinical outcomes. PLoS Genet17, e1009557 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steele, C. D. et al. Signatures of copy number alterations in human cancer. Nature606, 984–991 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc.11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol.33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pertea, G. & Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Res910.12688/f1000research.23297.2 (2020). [DOI] [PMC free article] [PubMed]
- 80.Vitting-Seerup, K. & Sandelin, A. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics35, 4469–4471 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Nowicka, M. & Robinson, M. D. DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Res5, 1356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, L. et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res41, e74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res44, D279–D285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Potter, S. C. et al. HMMER web server: 2018 update. Nucleic Acids Res46, W200–W204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol.37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Meszaros, B., Erdos, G. & Dosztanyi, Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uhrig, S. et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res31, 448–460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Creason, A. et al. A community challenge to evaluate RNA-seq, fusion detection, and isoform quantification methods for cancer discovery. Cell Syst.12, 827–838.e825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom.5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 91.Kall, L., Canterbury, J. D., Weston, J., Noble, W. S. & MacCoss, M. J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods4, 923–925 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Taus, T. et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res10, 5354–5362 (2011). [DOI] [PubMed] [Google Scholar]
- 93.Wang, X. & Zhang, B. customProDB: an R package to generate customized protein databases from RNA-Seq data for proteomics search. Bioinformatics29, 3235–3237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Na, S., Kim, J. & Paek, E. MODplus: robust and unrestrictive identification of post-translational modifications using mass spectrometry. Anal. Chem.91, 11324–11333 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Kim, S. & Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun.5, 5277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park, C. N. Mathematical models in quantitative assessment of carcinogenic risk. Regul. Toxicol. Pharm.9, 236–243 (1989). [DOI] [PubMed] [Google Scholar]
- 97.Eng, J. K., Jahan, T. A. & Hoopmann, M. R. Comet: an open-source MS/MS sequence database search tool. Proteomics13, 22–24 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 99.Krug, K. et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell183, 1436–1456.e1431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinforma.11, 367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brunet, J. P., Tamayo, P., Golub, T. R. & Mesirov, J. P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl Acad. Sci. USA101, 4164–4169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carmona-Saez, P., Pascual-Marqui, R. D., Tirado, F., Carazo, J. M. & Pascual-Montano, A. Biclustering of gene expression data by Non-smooth Non-negative Matrix Factorization. BMC Bioinforma.7, 78 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krug, K. et al. A curated resource for phosphosite-specific signature analysis. Mol. Cell Proteom.18, 576–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma.14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goel, M. K., Khanna, P. & Kishore, J. Understanding survival analysis: Kaplan-Meier estimate. Int J. Ayurveda Res1, 274–278 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Badia, I. M. P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform Adv.2, vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gjerga, E., Dugourd, A., Tobalina, L., Sousa, A. & Saez-Rodriguez, J. PHONEMeS: efficient modeling of signaling networks derived from large-scale mass spectrometry data. J. Proteome Res20, 2138–2144 (2021). [DOI] [PubMed] [Google Scholar]
- 108.Hernandez-Armenta, C., Ochoa, D., Goncalves, E., Saez-Rodriguez, J. & Beltrao, P. Benchmarking substrate-based kinase activity inference using phosphoproteomic data. Bioinformatics33, 1845–1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu, T. et al. CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation and visualization. Bioinformatics33, 3131–3133 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Lachmann, A., Giorgi, F. M., Lopez, G. & Califano, A. ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics32, 2233–2235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang, T. et al. OncoVar: an integrated database and analysis platform for oncogenic driver variants in cancers. Nucleic Acids Res49, D1289–D1301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu, S. H. et al. DriverDBv3: a multi-omics database for cancer driver gene research. Nucleic Acids Res48, D863–D870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gundem, G. et al. IntOGen: integration and data mining of multidimensional oncogenomic data. Nat. Methods7, 92–93 (2010). [DOI] [PubMed] [Google Scholar]
- 114.Martinez-Jimenez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer20, 555–572 (2020). [DOI] [PubMed] [Google Scholar]
- 115.O’Donnell, T. J., Rubinsteyn, A. & Laserson, U. MHCflurry 2.0: improved pan-allele prediction of MHC Class I-presented peptides by incorporating antigen processing. Cell Syst.11, 42–48.e47 (2020). [DOI] [PubMed] [Google Scholar]
- 116.Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res48, W449–W454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Szolek, A. et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics30, 3310–3316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xiang, R. et al. Increased expression of peptides from non-coding genes in cancer proteomics datasets suggests potential tumor neoantigens. Commun. Biol.4, 496 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res49, D545–D551 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim, M. et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun.10, 3991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Okuda, S. et al. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res45, D1107–D1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Genomic and transcriptomic raw data generated in this study are available in EGA under restricted access under the study ID EGAS50000000592 and Dataset ID EGAD50000000844. Data will be made available following a data access agreement, and there are no restrictions on who will be granted access. Requests will be assessed by the data access committee, and further information can be requested from Dr Kwang Pyo Kim (kimkp@khu.ac.kr). Additionally, these data were also available in the Korean Nucleotide Archive (KoNA, https://kobic.re.kr/kona) with the accession ID KAP210028. Raw mass spectrometry-based global, phosphoproteome, and acetylome data were deposited in the ProteomeXchange Consortium (accession number: PXD053969, PXD053921, PXD053903) via the jPOST partner repository (accession number: JPST003210, JPST003211, JPST003212)121.
All histologic details and sample annotations can be accessed from Supplementary Data 1a. Processed and normalized gene expression data file is provided in Supplementary Data 7 and the proteomic data files are provided in K-BDS with the accession IDs KAP240387, KAP240391, and KAP240392.
No custom code was used or developed for the analyses presented in this study. Standard workflows and open-source R packages and software were used (Methods). The codes used for the analyses included in our manuscript were uploaded to GitHub repository with instructions for users: https://github.com/joonan-lab/PDIAMOND-NSCLC.