Abstract
Klebsiella pneumoniae is a potent human pathogen and a prevalent ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species). Considerably, K. pneumoniae becomes a major clinical problem due to numerous AMR genes [extended-spectrum β-lactamase, plasmid-mediated AmpC, carbapenemases, tigecycline resistance, and New Delhi Metallo-β-lactamase-1 (NDM-1)] and can hydrolyze the majority of β-lactam antibiotics. Hence, targeting NDM-1 could be an effective approach to eradicate K. pneumoniae pathogenesis. A plethora of reports suggests that the plant compounds possess an anti-microbial activity and their utilization could be a promising strategy to develop novel antibiotics. Our study utilized the hydromethanolic leaves extract of Acorus calamus L. (AC) to target NDM-1 containing K. pneumonia using an in silico approach. At first, we determined the phytochemical composition of AC using GC-HRMS. Further, the phytoconstituents were screened against the NDM-1 (PDB ID: 3ZR9) of K. pneumoniae through molecular docking studies. Our results revealed the compounds from AC such as (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one (−9.5 kcal/mol), 4,4,5,8-tetramethyl-2,3-dihydrochromen-2-ol (−6.6 kcal/mol), 5-chloro-2-(2,4-dichloro phenoxy)phenol (−6.0 kcal/mol), [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate (−5.7 kcal/mol), 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol (−5.6 kcal/mol), and (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid (−5.6 kcal/mol) possess substantial docking scores against NDM-1. Therefore, our study concludes that phytochemicals of AC may inhibit NDM-1-mediated resistance in K. pneumoniae and could be an alternative therapeutic strategy for targeting NDM-1-containing K. pneumoniae.
Keywords: Klebsiella pneumoniae, MDR, NDM-1, β-lactam, Acorus calamus L
1. Introduction
K. pneumoniae is a pathogen that causes pneumonia, severe infections, and human sepsis [1]. It is a ubiquitous, gram-negative, encapsulated, non-motile, rod-shaped, lactose fermenting, and facultative anaerobic bacterium that belongs to the Enterobacteriaceae family [2,3]. It is a resident and transient flora of the gastrointestinal tract and is also found in sewage, soil, industrial effluents, drinking water, polluted surface water, and vegetation [4]. K. pneumoniae is responsible for hospital-acquired infections (HAI) in the urinary tract, bloodstream, and neonates and also causes pyogenic liver abscesses and meningitis [5,6].
Infections of K. pneumoniae are generally eradicated by utilizing various antibiotic classes such as cephalosporins, aminoglycosides, fluoroquinolones, and carbapenems. However, the misutilization of these antibiotics for more than seven decades resulted in the development of antimicrobial resistance (AMR) which could be the major challenge for effective therapeutics against pathogens [7]. AMR is an outcome of any one or combination of complex mechanisms such as inactivating drugs, activating efflux signaling, limiting drug uptake, and modifying drug targets [8]. Furthermore, continuous exposure to antibiotics leads to AMR phenotypes in microbes [9,10]. The increasing incidences of MDR (multidrug-resistant) and XDR (extremely drug-resistant) pathogens of the Enterobacteriaceae family have become an alarming. Additionally, these infections account for higher rates of mortality in patients [11]. In the current scenario, multi and extreme drug-resistant K. pneumoniae variants belong amidst the deadly ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) [12]. These ESKAPE pathogens adopt AMR owing to horizontal gene transfer directed by the plasmids or mobile gene elements [13].
Numerous β-lactam antibiotics are known to target K. pneumoniae due to their broad-spectrum activity, higher efficiency, and lower toxicity. However, due to the overuse of these antibiotics, K. pneumoniae develops diverse drug-resistance mechanisms to β-lactam antibiotics [14]. Several β-lactam resistance genes such as extended-spectrum β-lactamase gene, plasmid-mediated AmpC genes, carbapenemases, tigecycline resistance genes, and blaNDM gene have been found in K. pneumoniae [15]. blaNDM gene encodes for all the variants of New Delhi Metallo-β-lactamases. Among all NDM variants NDM-1, NDM-4, NDM-5, and NDM-7 have been found dominant in Indian sub-continent [16]. The blaNDM-1 gene containing K. pneumoniae has recently been reported as a ‘superbug’ [17]. NDM-1 belongs to B1 β-lactamases and can hydrolyze most β-lactam antibiotics. It spreads quickly between the same or different species [18]. The structure of NDM-1 is typical to metallo-hydrolase/oxidoreductase superfamily with their distinct αβ/βα sandwich fold. NDM-1 has a narrow active site groove consisting of divalent zinc ions surrounded by a flexible hairpin loop structure. Both the zinc ions stabilize the enzyme-substrate complex (NDM-1-Antibiotic complex) and promote hydrolysis. The process of hydrolysis leads to the cleavage of the C-N bond of core β-lactam ring [19]. Likewise, NDM-1 hydrolyses almost all β-lactam antibiotics. Major antibiotics include carbapenem and cephalosporins regarded as last-resort antibiotics to prevent the infection of K. pneumoniae [20,21]. The NDM-1-containing K. pneumoniae is the major organism responsible for the higher mortality rate [22]. The risk of death for carbapenem-resistant K. pneumoniae patients was approximately three times higher than that of carbapenem-sensitive K. pneumoniae patients [23]. Currently, no inhibitors have been reported for clinical use against NDM-1-mediated infection caused by K. pneumoniae [18]. Thus, extensive research to identify potent metallo-β-lactamases inhibitors is the need of an hour. In line, the deprived discovery rate of novel antibiotics and active molecules is compelling for developing alternative therapeutic strategies. Utilization of phytochemicals as an antimicrobial agent could be a promising approach in the face of the incompetence of available drugs [24]. Plants produce secondary metabolites such as phenolics, alkaloids, flavonoids, terpenoids, glycosides, and sulfur-containing compounds possessing significant pharmacological activity and are presumed to combat the AMR crisis [25].
Indian plants have been studied for multifarious therapeutic applications including antimicrobial activities. Moreover, the World Health Organization (WHO) has published guidelines in support of the utilization of plants and their parts for therapeutic regime design [26]. In recent years, many Indian plants have been under study for their antimicrobial potential and drug discovery. In our study, we have investigated the antimicrobial potential of Acorus calamus L. (AC) (also known as sweet flag or “calamus”) leaves extract. AC is classified under the Acorales order and Acoraceae family and is known for its therapeutic applications [27,28]. AC leaves and rhizomes are reported to treat cough, fever, digestive disorder, asthma, and bronchitis [29]. It contains 1,2,4-trimethoxy-5-[(E)-prop-1-enyl]benzene and 1,2,4-trimethoxy-5-[(Z)-prop-1-enyl]benzene which have been reported for anti-microbial, insecticidal, anti-cancer, anti-ulcer, immunosuppressive, anti-inflammatory, anti-diabetic, anti-hepatotoxic and antioxidant activities [[30], [31], [32], [33]].
In our study, we identified the phytochemical compounds present in AC through GC-HRMS and analyzed their interaction with NDM-1 by molecular docking studies, which eventually decide the potency of the compounds of AC to inhibit NDM-1.
2. Materials and methods
2.1. Collection of plant material
A. calamus L. leaves were collected from Anand, Gujarat, India, spanning from March to September. The entire plant (Voucher specimen ACRBS01) was deposited at the herbarium of Department of Biosciences at Sardar Patel University [33].
2.2. Plant extract preparation
The collected leaves were rinsed with water, air-dried under shade, ground into powder, sieved, and stored in a container at 4 °C. The mixture of plant powder (10 g) and 80 % methanol (100 ml) was prepared and incubated for 24 h at room temperature. Thereafter, the extract was filtered using Whatmann filter paper and spun at 2500 rpm for 20 min. The supernatant was collected and stored at 4 °C. Percentage yield was calculated by formula (B/A) x 100, B represents the weight of powder after extraction and A represents the weight of powder before extraction [34].
2.3. Phytochemical analysis
Hydromethanolic extract of AC was subjected to various phytochemical analyses to assess the content of alkaloids, glycosides, flavonoids, phenols, saponins, lignins, sterols, anthraquinones, and tannins in leaves using standard procedures [[35], [36], [37]].
2.4. Quantification of total flavonoids
The total flavonoid content of AC was determined by the method described earlier [38]. Flavonoids were measured using 200 μl of AC extract mixed with 5 % NaNO2, 10 % AlCl3 6H2O, and 1 M NaOH at 510 nm. The Quercetin (25–500 mg/ml) was used as a standard.
2.5. Determination of antioxidant activities
2.5.1. Superoxide scavenging activity
Superoxide activity of AC was determined as described earlier [33]. 2 ml NBT (25 μM), 2 ml NADH (39 μM), and a sample solution of extract (10–500 μg/ml) in distilled water should be added to a Tris-HCl buffer (6 ml, 8 mM, pH 8.0). 2 ml of PMS solution (0.5 μM) was added to the mixture to begin the reaction. After incubating the reaction mixture at 25 °C for 5 min, the absorbance was measured at 560 nm and compared to the control. The control group was ascorbic acid. The reaction mixture's lower absorbance indicated that it was better at scavenging superoxide anion.
2.5.2. Hydrogen peroxide radical scavenging activity
The Hydrogen peroxide radical scavenging activity of AC was determined as described earlier [39]. The extract's capacity to scavenge hydrogen peroxide (H2O2) was determined. In phosphate buffer at pH 7.4, a 40 mM hydrogen peroxide solution was prepared. A UV–visible spectrophotometer was used to determine hydrogen peroxide level by measuring absorbance at 230 nm. The extracts (10–500 μg/ml) in distilled water were added to a hydrogen peroxide solution after 10 min, and the absorbance at 230 nm was compared to a blank solution containing just phosphate buffer and no hydrogen peroxide. Ascorbic acid was used as a standard.
2.6. Compounds identification by GC-HRMS
The Photochemical compounds of AC were identified by GC-HRMS analysis at SAIF, IIT-Bombay, India. The 20:80 split injection mode was used in the Agilent GC-MS system for better separation. The NIST Library was used to identify the chemicals after they had been separated using the traditional procedure using the HP5 capillary column.
2.7. In silico molecular docking studies
The flexible docking was performed by taking NDM-1 as a target and phytoconstituents of A. calamus as ligands. The three-dimensional structure of the NDM-1 (PDB ID: 3ZR9) was retrieved from Protein Data Bank (https://www.rcsb.org/) and modified by removing water molecules and addition of polar hydrogen atoms followed by the addition of Kollaman charge using AutoDockTools (Version 1.5.7). The modified NDM-1 structure was saved in.pdbqt format and used for docking studies. The native structures of the ligands were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in SDF format (.sdf) which was subsequently converted into the PDB format in PyMOL (Version 2.5). Thereafter, the ligands were prepared for molecular docking study in AutoDockTools (Version 1.5.7) and saved in.pdbqt format. Further, the structure of NDM-1 and respective ligands (.pdbqt format) was retrieved in the AutoDockTools (Version 1.5.7). After that, a grid box was prepared using a grid map of 40 × 40 × 40 Å points with a spacing of 0.375 Å. The values of the x center, y center, and z center of the grid box were 26.887, 10.935, and 17.058 Å respectively. The energy range and exhaustiveness value were 4 and 8 respectively. 31 ligands (23 compounds identified from hydromethanolic extract of AC leaves and 8 standard antibiotics which were used as controls) were docked with enzyme NDM-1 by using AutoDock 4.2.
AutoDock 4.2 and Graphical user interface AutoDockTools has proven to be an effective tool for accurately predicting the binding energy and conformation of protein-ligand complexes. The algorithm used for docking is Lamarckian Genetic Algorithm (LGA) and empirical free energy scoring function. This algorithm provides reproducible docking results for ligands with approximately 10 flexible bonds and predicted energy. For ligand sampling, a stochastic algorithm was used. We used multiple parameters of Lamarckian GA such as 31 independent runs with a population size of 150, a maximum number of 25 × 105 energy evaluations, the maximum number of generations simulated was 27000, an individual is guaranteed to survive into the next generation, mutation rate, and crossover rate were 0.02 and 0.80 respectively, 10 generations were taken into consideration when deciding the threshold for the worst individual in the current population. AutoDock 4.2 is effective for systems with approximately 10 torsional degrees of freedom as systems with more torsional flexibility may not give reliable results. All the conformations are evaluated based on scoring function in the form of binding energy required for ligand-protein interactions. The process of determining a particular pose into NDM-1 was based on the number of favourable intermolecular interactions such as hydrogen bonds, van der Waals interaction, and hydrophobic interaction. Conformations with the lowest binding energies are selected for further analysis. The docking pose having the lowest ΔG-score was best fitted with the receptor and used further for analysis of Ligand-receptor interactions [[40], [41], [42]]. In our study, we used Discovery Studio 2021 Client (Version 21.1) for the visualization and analysis of the ligand-receptor complex.
2.8. In silico ADMET study
For in silico studies, the chemical structure was drawn using ChemDraw Ultra 8.0. Structured activity metrics, Absorption, Distribution, Metabolism, Excretion, Toxicity, (ADMET) features, and drug-likeness (Lipinski's rule of five, and Jorgenson's rule of 3) are important determinants of drug development attrition that were evaluated using pkCSM (https://biosig.lab.uq.edu.au/pkcsm/) [33,[43], [44], [45]]. Early optimization of these aspects throughout the drug development process may reduce the likelihood of clinical trial failure [[46], [47], [48]].
3. Results and discussion
3.1. Total yield of plant crude extract
The hydromethanolic extract of AC was prepared by cold maceration method as it offers a superior percentage yield and prevents the loss of heat-sensitive compounds [28]. Solvent selection adhered to the principles of similarity and inter-miscibility. Extraction yields are optimized when the polarity of the solvent closely matches that of the solute. Alcohols are proven to be surpassingly effective solvents for phytochemical analysis [49]. Hydromethanol, with its gentle nature, high boiling point, and other favourable qualities, stands out as an excellent solvent for extracting secondary plant metabolites. During hydroalcoholic solvent extraction, AC demonstrated a 5.08 % yield. Flavonoids, alkaloids, phenolic, tannins, saponins, terpenoids, cardiac glycosides, phlobatannins, and carbohydrates were detected in the extract, but there was an absence of anthraquinones, steroids, and anthocyanides.
3.2. Quantification of total flavonoids
Flavonoids play a pivotal role in determining the flavor, scent, and color of fruits, flowers, and seeds. Beyond their sensory contributions, flavonoids also serve as attractants for mammals, birds, and insects. Moreover, they exhibit inhibitory effects on enzymes involved in the generation of reactive oxygen species (ROS), including cyclooxygenase, lipoxygenase, monooxygenase, and xanthine oxidase. Additionally, flavonoids contribute to the regulation of singlet oxygen production [50]. Here, we estimated the flavonoid content in the crude extract of AC and found that the flavonoid concentration was 170 ± 0.008 mg/ml.
3.3. Antioxidant activities
3.3.1. Superoxide scavenging activity
Superoxide anions can cause direct or indirect damage to biomolecules in various pathologies and aging, by generating H2O2, −OH, peroxynitrite, or singlet oxygen. It has been implicated in direct lipid peroxidation [51]. However, the majority of plant compounds possess antioxidant activity, which is beneficial for human health. The superoxide scavenging activity of AC leaves is detailed in Table 1. Notably, the superoxide scavenging capacity of AC exhibited a significant increase with concentration. Given their pronounced inhibitory effects on superoxide anion generation, AC emerges as a potent antioxidant. These findings strongly suggest that AC functions as a robust scavenger of superoxide radicals.
Table 1.
Antioxidant activity of plant crude leaves extract of Acorus calamus L. by Superoxide scavenging activity.
| Concentration (μg/ml) | Inhibitory activity (%) |
|
|---|---|---|
| Ascorbic acid | Acorus calamus L. | |
| 300 | 90.59 ± 0.24 | 94.10 ± 1.28 |
| 150 | 51.71 ± 0.55 | 55.47 ± 0.93 |
| 75 | 40.30 ± 0.24 | 28.29 ± 1.32 |
| 37.5 | 30.79 ± 0.39 | 25.01 ± 1.52 |
| 18.75 | 25.83 ± 0.41 | 22.85 ± 0.84 |
| 9.37 | 19.75 ± 0.09 | 18.92 ± 1.15 |
| IC50 | 145.03 ± 0.55 | 135.20 ± 1.33 |
3.3.2. H2O2 radical scavenging activity
As indicated in Table 2, AC demonstrated concentration-dependent hydrogen peroxide decomposition activity. Hydrogen peroxide, being a mild oxidizing substance, can directly inactivate only a limited number of enzymes by oxidizing essential thiol (-SH) groups. Nevertheless, hydrogen peroxide exhibits a high rate of penetration through cell membranes, and once inside the cell, it is prone to interacting with Fe2+ and possibly Cu2+ ions, leading to the generation of hydroxyl radicals and potentially causing various negative effects [52]. Consequently, cells biologically benefit from regulating the accumulation of hydrogen peroxide. The antioxidant and free radical-scavenging properties of AC may play a role in the breakdown of H2O2, contributing to cellular homeostasis.
Table 2.
Antioxidant activity of plant crude leaves extract of Acorus calamus L. by Hydrogen Peroxide scavenging activity.
| Concentration (μg/ml) | Inhibitory activity (%) |
|
|---|---|---|
| Ascorbic acid | Acorus calamus L. | |
| 250 | 89.71 ± 1.74 | 96.19 ± 0.87 |
| 125 | 47.42 ± 3.71 | 61.52 ± 1.18 |
| 62.5 | 29.71 ± 2.28 | 37.14 ± 1.14 |
| 31.25 | 26.28 ± 2.37 | 29.14 ± 0.57 |
| 15.62 | 13.14 ± 2.00 | 21.90 ± 1.18 |
| 7.8 | 4.00 ± 2.16 | 14.67 ± 0.87 |
| IC50 | 131.80 ± 3.71 | 102.09 ± 1.30 |
3.4. Determination of potent bioactive compounds of AC
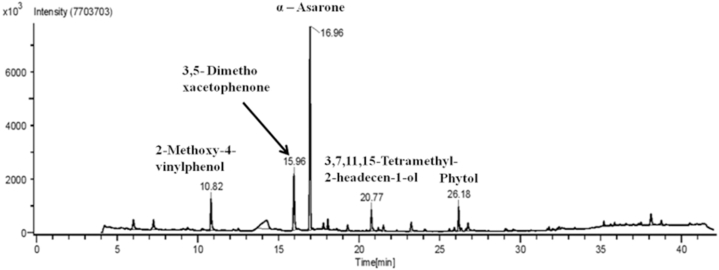
The phytoconstituents present in the hydromethanolic extract of AC were determined by GC-HRMS and we found multiple compounds (Fig. 1) (Table 3).
Fig. 1.
GC-HRMS profile of Acorus calamus L. extract.
Table 3.
Bioactive compounds identified from hydromethanolic extract of Acorus calamus L. leaves.
| Sr. No. | Compound Name | RT min | Content (%) | Base m/z |
|---|---|---|---|---|
| 1 | N-methyloctadcylamine | 4.51 | 17.7 | 44 |
| 2 | 2,5-Dimehyl-4-hydroxy-3[2H]-fuanone | 5.57 | 19.6 | 43 |
| 3 | 3-hydroxy-2-methylpyran-4-one | 6.01 | 15.54 | 126 |
| 4 | 3,5-dihydroxy-6-methyl-2,3-dihydropyran-4-one | 7.25 | 94.8 | 43 |
| 5 | [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate | 9.37 | 37.0 | 43 |
| 6 | 4-ethenyl-2-methoxyphenol | 10.82 | 60.3 | 139 |
| 7 | 1-(3,5-dimethoxyphenyl)ethenone | 15.97 | 22.8 | 165 |
| 8 | (+-)-[2-carboxy-cis-propenyl]-2,2-dimethyl cyclopropane-cis-1-carboxylic acid | 16.23 | 30.1 | 153 |
| 9 | 1,2,4-trimethoxy-5-[(E)-prop-1-enyl]benzene | 16.96 | 65 | 208 |
| 10 | 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one | 17.13 | 32.6 | 190 |
| 11 | 4,4,5,8-tetramethyl-2,3-dihydrochromen-2-ol | 17.79 | 41.4 | 173 |
| 12 | 1,2,4-trimethoxy-5-[(Z)-prop-1-enyl]benzene | 18.07 | 36.1 | 208 |
| 13 | 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol | 19.30 | 35.6 | 137 |
| 14 | (E)-3,7,11,15-tetramethylhexadec-2-en-1-ol | 20.77 | 30.4 | 81 |
| 15 | n-Hexadecanoic acid | 23.24 | 79 | 60 |
| 16 | hexadecan-1-ol | 25.61 | 4.53 | 55 |
| 17 | 10,12-Hexadecadien-1-ol | 25.92 | 23.8 | 67 |
| 18 | (E,7R,11R)-3,7,11,15-tetramethylhexadec-2-en-1-ol | 26.17 | 68.4 | 71 |
| 19 | 5-chloro-2-(2,4-dichlorophenoxy)phenol | 26.34 | 92.8 | 288 |
| 20 | octadec-17-ynoic acid | 26.64 | 6.29 | 55 |
| 21 | (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid | 26.76 | 20.6 | 79 |
| 22 | 2-methylundecanal | 29.08 | 8.82 | 58 |
| 23 | Spiro[Andros-5-ene-17-1′-cycobutan]-2′-one, 3-hydroxy | 32.42 | 39 | 41 |
| 24 | 3-(3,4-dimethoxyphenyl)prop-2-enoic acid | 35.19 | 17 | 208 |
| 25 | (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid | 35.62 | 14.6 | 208 |
| 26 | 11- trans- Octadecenoic acid, trimethylsilyl ester | 35.86 | 9.95 | 73 |
| 27 | (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one | 36.57 | 51.4 | 314 |
| 28 | Pyridine-3-carbonirile,2-[2-[3,4-dihydoxyphenyl]-2-oxoethylthio]-4-methoxymethyl-6-methyl | 38.11 | 26.3 | 137 |
| 29 | Cis-13-Octadecenoic acid, trimethylsilyl ester | 38.76 | 10.3 | 73 |
| 30 | 1- Monolinoleoylglcerol trimeylsilyl ether | 40.53 | 23.6 | 73 |
3.5. Screening and identification of phytochemicals of AC targeting NDM-1
Molecular docking is an efficient and reliable tool to reveal the binding stability of protein and ligand complexes. Hydrogen bonds and hydrophobic interaction majorly govern the binding stability of complexes [53]. The lowest binding energy represents the least energy required for the enzymatic reactions in the case of inhibition constant. The minimum energy represents a higher number of hydrogen bonds and hydrophobic interactions [54]. To recognize the most favourable pose, each pose is evaluated based on compatibility with the target in terms of shape and properties such as electrostatics and generates a corresponding dock score. The docking score is the main parameter that shows the favourable binding of the ligand to the target NDM-1. Our study revealed that Cefepime and Tigecycline, widely used antibacterial drugs have the highest docking score among all standard antibiotics, which were taken as controls. However, (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethyl spiro [5-oxapentacyclo [10.8.0.02,9.04,8.013,18] icos-1(12)-ene-6,2′-oxane]-11-one has a higher docking score (−9.5 kcal/mol) than Cefepime and Tigecycline. 1-(3,5-dimethoxyphenyl)ethanone, 3-(3,4-dimethoxyphenyl)prop-2-enoic acid, 4-ethenyl-2-methoxyphenol, 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one, (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid, 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol, [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate, 5-chloro-2-(2,4-dichloro phenoxy)phenol and 4,4,5,8-tetramethyl-2,3-dihydrochromen-2-ol are considered to be the key molecules owing to the docking score range between −5.1 kcal/mol to −6.6 kcal/mol which are almost at par to the Cefepime and Tigecycline. Molecules like (E)-3,7,11,15-tetramethylhexadec-2-en-1-ol, 3-hydroxy-2-methylpyran-4-one, n-hexadecadien-1-ol, 1,2,4-trimethoxy-5-[(E)-prop-1-enyl]benzene, (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid, 1,2,4-trimethoxy-5-[(Z)-prop-1-enyl]benzene, (E,7R,11R)-3,7,11,15-tetramethylhexadec-2-en-1-ol and 3,5-dihydroxy-6-methyl-2,3-dihydropyran-4-one are the moderately active molecules with docking score in a range of −4.5 kcal/mol to −5.0 kcal/mol. Except for the hexadecan-1-ol, 10,12-Hexadecadien-1-ol, 2-methylundecanal, N-methyloctadylamine, and octadec-17-ynoic acid, all the molecules in the present study have shown favourable binding with the target, which has been expressed through the higher docking scores.
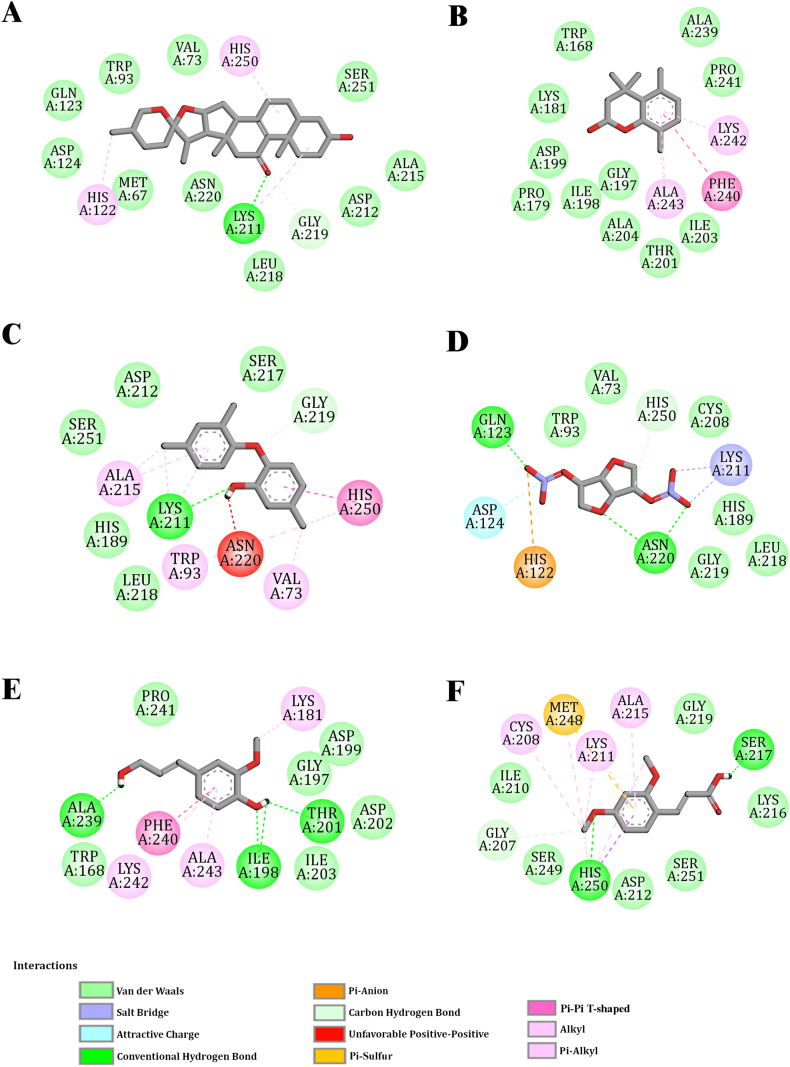
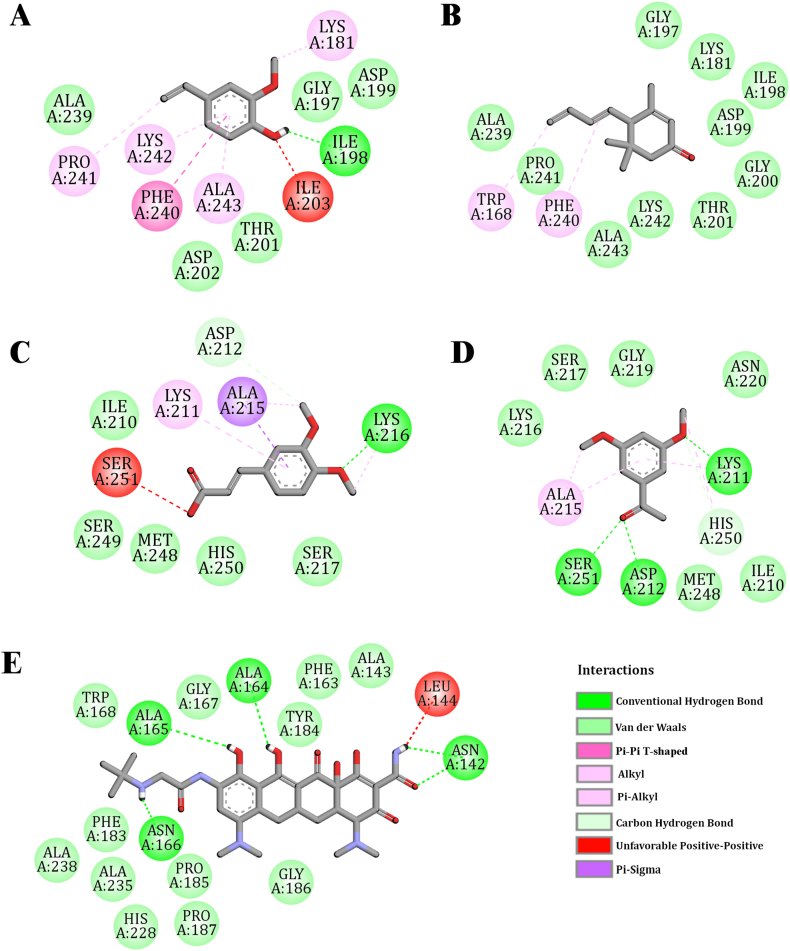
For most ligands, the proposed binding mode is consistent with that of the seven amino acid residues viz. LYS A:211, GLY A:197, GLY A:219, ASP A:199, ALA A:243, LYS A:242, and PHE A:240, and could be the key interaction points of the target. The key amino acid residues involved in the interaction with the target through the different bonds are shown in Table 4 & Supplementary Table 1. The graphical binding modes of all molecules and standard drugs are shown in Fig. 2, Fig. 3 & Supplementary Figs. 1–3. In a previous report, Molecular modeling suggests that LYS A:211 is one of the most imperative amino acid residues for conventional hydrogen bonding and Pi-Alkyl bonding interaction. In the present study, this residue forms conventional hydrogen and Pi-Alkyl bonds with diverse molecules including the (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one, (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid, 3-(3,4-dimethoxyphenyl)prop-2-enoic acid, 5-chloro-2-(2,4-dichlorophenoxy)phenol, 1,2,4-trimethoxy-5-[(Z)-prop-1-enyl]benzene, 1,2,4-trimethoxy-5-[(E)-prop-1-enyl]benzene, 1-(3,5-dimethoxyphenyl)ethanone and five of the standard antibiotics such as Meropenem, Penicillin, Cefotaxime, Piperacillin, Levofloxacin and Cefepime. Pi-Alkyl bonding amino acid residues are LYS A:242, ALA A:243, and PHE A:240. These amino acid residues are involved in the binding of N-methyloctadylamine, 3-hydroxy-2-methylpyran-4-one, 3,5-dihydroxy-6-methyl-2,3-dihydropyran-4-one, 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one, (E)-3,7,11,15-tetramethylhexadec-2-en-1-ol, hexadecan-1-ol and 2-methylundecanal molecules with the target. GLY A:219 forms Carbon hydrogen bonding with the (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one, Levofloxacin, Cefotaxime, Piperacillin, and Cefepime. GLY A:219 also forms van der Waals bond with the Meropenem, Penicillin, [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate, 1-(3,5-dimethoxyphenyl)ethanone, 1,2,4-trimethoxy-5-[(Z)-prop-1-enyl]benzene, 1,2,4-trimethoxy-5-[(E)-prop-1-enyl]benzene, 5-chloro-2-(2,4-dichlorophenoxy)phenol, and (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid. Additionally, N-methyloctadylamine, 3-hydroxy-2-methylpyran-4-one, 3,5-dihydroxy-6-methyl-2,3-dihydropyran-4-one, 4-ethenyl-2-methoxyphenol, 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one, 4,4,5,8 Tetramethylchroman-2-ol, 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol, (E)-3,7,11,15-tetramethylhexadec-2-en-1-ol, n-Hexadecadien-1-ol, hexadecan-1-ol, 10,12- Hexadecadien-1-ol, (E,7R,11R)-3,7,11,15-tetramethylhexadec-2-en-1-ol, (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid and 2-methylundecanal form van der Waals bond with the GLY A:197 and ASP A:199.
Table 4.
Molecular docking scores and intermolecular bonding residues with compounds of Acorus calamus L. leaves and standard drug.
| Sr. No. | Compound name | Docking Score (kcal/mol) | Conventional hydrogen | Van der Waals | Pi interaction | Carbon Hydrogen |
|---|---|---|---|---|---|---|
| C1 | (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one | −9.5 | LYS A:211 | VAL A:73 TRP A:93 GLN A:123 ASP A:124 MET A:67 ASN A:220 LEU A:218 ASP A:212 ALA A:215 SER A:251 |
Pi-Alkyl: HIS A:250 HIS A:122 |
GLY A:219 |
| C2 | 4,4,5,8-tetramethyl-2,3-dihydrochromen-2-ol | −6.6 | TRP A:168 LYS A:181 ASP A:199 PRO A:179 ILE A:198 GLY A:197 ALA A:204 THR A:201 ILE A:203 PRO A:241 ALA A:239 |
Pi-Pi T-Shaped: PHE A:240 Pi-Alkyl: ALA A:243 LYS A:242 |
||
| C3 | 5-chloro-2-(2,4-dichlorophenoxy)phenol | −6.0 | LYS A:211 | SER A:217 ASP A:212 SER A:251 LEU A:218 HIS A:189 GLY A:219 |
Pi-Alkyl: ALA A:215 TRP A:93 VAL A:73 Pi-Pi Stacked: HIS A:250 |
|
| C4 | [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate | −5.7 | GLN A:123 ASN A:220 |
TRP A:93 VAL A:73 CYS A:208 HIS A:189 GLY A:219 LEU A:218 |
Pi-Anion: HIS A:122 |
|
| C5 | 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol | −5.6 | ALA A:239 ILE A:198 THR A:201 |
PRO A:241 TRP A:168 ILE A:203 ASP A:202 GLY A:197 ASP A:199 |
Pi-Pi T-Shaped: PHE A:240 Pi-Alkyl: ALA A:243 LYS A:242 LYS A:181 |
|
| C6 | (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid | −5.6 | HIS A:250 SER A:217 |
GLY A:219 ILE A:210 SER A:249 ASP A:212 SER A:251 LYS A:216 |
Pi-Alkyl: ALA A:215 LYS A:211 CYS A:208 Pi-Sulfur: MET A:248 Pi-Sigma: HIS A:250 |
GLY A:207 |
| C7 | 4-ethenyl-2-methoxyphenol | −5.5 | ILE A:198 | ALA A:239 ASP A:202 THR A:201 GLY A:197 ASP A:199 |
Pi-Pi T-Shaped: PHE A:240 Pi-Alkyl: LYS A:242 PRO A:241 ALA A:243 LYS A:181 |
|
| C8 | 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one | −5.5 | GLY A:197 LYS A:181 ILE A:198 ASP A:199 GLY A:200 THR A:201 LYS A:242 ALA A:243 PRO A:241 ALA A:239 |
Pi-Alkyl: TRP A:168 PHE A:240 |
||
| C9 | 3-(3,4-dimethoxyphenyl)prop-2-enoic acid | −5.2 | LYS A:216 | ILE A:210 SER A:249 MET A:248 HIS A:250 SER A:217 |
Pi-Alkyl: LYS A:211 Pi-Sigma: ALA A:215 |
|
| C10 | 1-(3,5-dimethoxyphenyl)ethanone | −5.1 | SER A:251 ASP A:212 LYS A:211 |
ASN A:220 GLY A:219 SER A:217 LYS A:216 MET A:248 ILE A:210 |
Pi-Alkyl: ALA A:215 HIS A:250 LYS A:211 |
HIS A:250 |
| S1 | Tigecycline | −7.6 | ALA A:164 ALA A:165 ASN A:166 ASN A:142 |
TRP A:168 PHE A:183 ALA A:238 ALA A:235 PRO A:185 HIS A:228 PRO A:187 GLY A:186 GLY A:167 TYR A:184 PHE A:163 ALA A:143 |
∗C1-C10 represents the compounds identified from Acorus Calamus L. extract. S1 represents Standard Drug (Tigecycline).
Fig. 2.
Schematic 2D diagram of the interaction between NDM-1 (PDB ID: 3ZR9)
(A) (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo [10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one (B) 4,4,5,8-tetramethyl-2,3-dihydrochromen-2-ol, (C) 5-chloro-2-(2,4-dichlorophenoxy)phenol (D) [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate (E) 4-(3-hydroxyprop-1-enyl)-2-methoxypheno (F) (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid.
Fig. 3.
Schematic 2D diagram of the interaction between NDM-1 (PDB ID: 3ZR9)
(A) 4-ethenyl-2-methoxyphenol (B) 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one (C) 3-(3,4-dimethoxyphenyl)prop-2-enoic acid (D) 1-(3,5-dimethoxyphenyl)ethanone (E) Tigecycline.
In the present study, molecular docking revealed the formation of hydrogen and hydrophobic bonds with key residues of the NDM-1 active site. The residues HIS:122, HIS:189, CYS:208, LYS:211, ASN:220, and HIS:250 of NDM-1 are reported as conserved amino acids in metallo-β-lactamase and play an important role in substrate recognition [55,56]. Our study also revealed that (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one interact with key active site amino acid LYS A:211, ASN A:220, HIS A:250, and HIS A:122 either by hydrogen bond or hydrophobic bond and most of the other molecules having stable interaction with LYS A:211 and HIS A:250 revealed by respective docking score (Table-4).
Meropenem and NDM-1 (PDB ID: 4EYL) complex was stabilized by six hydrogen bonds (HIS:120, HIS:189, LYS:211, ASN:220, HIS:250) and one hydrophobic interaction with TRP:93. Other residues such as VAL:73, TRP:93, HIS:122, GLU:123, ASP:124 and CYS:208 also gave stability to meropenem at the active site of NDM-1 [57]. Our investigation also reveals that meropenem and NDM-1 (PDB-ID: 3ZR9) complex was stabilized by 2 hydrogen bonds (ASN A:220 and LYS A:211) and hydrophobic bonds such as ALA A:215, LYS A:216, VAL A:73, ASP A:124, GLN A:123, TRP A:93, HIS A:122, MET A:67, HIS A:189, LEU A:218, CYS A:208, GLY A:219, HIS A:250 and SER A:251. Further, the binding frequency of standard antibiotics (Cefepime, Carbapenem, Levofloxacin, Piperacillin, Cefotaxime, Penicillin, and Tigecycline) towards NDM-1 was also monitored by analysis of hydrogen as well as hydrophobic bonds through visualizing the protein-drug complexes as in Fig. 3 & Supplementary Fig. 3. The present study has revealed the potent inhibitors of NDM-1 from AC through virtual screening. Among these inhibitors, (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro [5-oxapentacyclo [10.8.0.02,9.04,8.013,18] icos-1(12)-ene-6,2′-oxane]-11-one showed the highest affinity/stability with NDM-1 and its active site residues as the Meropenem (Table 4 & Supplementary Table 1). Moreover, 5-chloro-2-(2,4-dichloro phenoxy)phenol, [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate, (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid, 3-(3,4-dimethoxyphenyl)prop-2-enoic acid, 1-(3,5-dimethoxyphenyl)ethanone, 1,2,4-trimethoxy-5-[(E)-prop-1-enyl]benzene and 1,2,4-trimethoxy-5-[(Z)-prop-1-enyl]benzene interact with some of the active site residues of NDM-1 as Meropenem-NDM-1 (Table 4 & Supplementary Table 1). Due to the continuous evolution of MBL variants, conventional drugs and inhibitors are becoming ineffective against K. pneumoniae [58,59]. This warrants the need to discover potential inhibitors for NDM-1. In this study, we identified some potent inhibitors of NDM-1 from phytoconstituents of the medicinal plant source. Phytocompounds from A. calamus interact with the key to amino acids NDM-1 active site and hence could lead to a promising therapeutic strategy to inhibit NDM-1 mediated resistance in K. pneumoniae.
3.6. Evaluation of pharmacokinetic parameters of compounds by in silico ADMET study
All 10 compounds having good docking scores (>−5.0 kcal/mol) follow Lipnski's rule of five (RO5). They all have a molecular weight of less than 500 Da, rotational bond numbers, hydrogen bond acceptors of less than 10, and hydrogen bond donors of less than 5. All compounds except [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b] furan-6-yl] nitrate showed log P values of - 0.4 to + 5.6. Polar surface area for all compounds less than 140 Å2 except (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro [5-oxapentacyclo [10.8.0.02,9.04,8.013,18] icos-1(12)-ene-6,2′-oxane]-11-one. RO5 is important for drug-likeness properties and for oral administration. Further, the compounds were analyzed for their ADMET properties. All the analyzed molecules represented good water solubility (i.e., greater than 1.7 Log mol/L) and elevated Caco-2 permeability. Interestingly, [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate showed poor Caco-2 permeability. Good water solubility leads to good intestinal absorption of the drug. (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one, 4,4,5,8- Tetramethylchroma n-2-ol, 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol, (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid, 4-ethenyl-2-methoxyphenol, 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one, 3-(3,4-dimethoxyphenyl)prop-2-enoic acid, 1-(3,5-dimethoxyphenyl)ethanone showed more than 93 % intestinal absorption while 5-chloro-2-(2,4-dichlorophenoxy)phenol showed 87.45, and [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate showed 74.73 %. (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one, [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate, 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol, (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid, 3-(3,4-dimethoxyphenyl)prop-2-enoic acid, showed relatively less skin permeability > -2.5. (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid, 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one, 3-(3,4-dimethoxyphenyl)prop-2-enoic acid, 1-(3,5-dimethoxyphenyl)ethanone can readily cross the blood-brain barrier, while others (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one, 4,4,5,8-Tetramethylchroma n-2-ol, 5-chloro-2-(2,4-dichlorophenoxy)phenol, [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate, 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol, 4-ethenyl-2-methoxyphenol) are poorly distributed to the brain. All molecules are unable to penetrate the central nervous system except 5-chloro-2-(2,4-dichlorophenoxy)phenol. None of the molecules inhibited the cytochrome P450, an important detoxification enzyme associated with metabolism in the human liver. Moreover, none of the studied compounds showed hepatotoxicity indicating no possibility of drug-induced liver injury to the user (Table 5) [33,44,46,47,60,61]. Collectively, ADMET analysis revealed the drug-likeliness of our compound, and the phytochemicals of A. calamus L. exhibited no apparent toxicity.
Table 5.
Evaluation of pharmacokinetic parameters.
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Properties: | ||||||||||
| Molecular weight | 428.613 | 206.285 | 289.545 | 236.136 | 180.203 | 208.213 | 150.177 | 190.286 | 208.213 | 180.203 |
| LogP | 5.037 | 2.681 | 5.144 | −1.062 | 1.406 | 1.801 | 2.043 | 3.290 | 1.801 | 1.906 |
| Number of rotatable bonds | 0 | 0 | 2 | 4 | 3 | 4 | 2 | 2 | 4 | 3 |
| Number of acceptors | 4 | 2 | 2 | 8 | 3 | 3 | 2 | 1 | 3 | 3 |
| Number of donors | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 1 | 0 |
| Surface area | 186.686 | 90.888 | 113.306 | 88.221 | 76.904 | 87.749 | 65.744 | 86.205 | 87.749 | 77.280 |
| ADMET Parameters: | ||||||||||
| Water solubility (Log mol/L) | −5.385 | −2.712 | −5.013 | −2.370 | −1.748 | −2.984 | −1.958 | −3.651 | −2.984 | −1.949 |
| Caco2 Permeability (log Papp in 106 cm/s) | 1.361 | 1.819 | 1.848 | 0.218 | 1.227 | 1.225 | 1.499 | 1.345 | 1.225 | 1.445 |
| % intestinal absorption | 97.046 | 94.062 | 87.455 | 74.734 | 91.734 | 94.767 | 91.965 | 96.688 | 94.767 | 96.413 |
| Skin Permeability (log Kp) | −3.853 | −2.092 | −2.268 | −2.585 | −2.848 | −2.712 | −2.262 | −1.671 | −2.712 | −2.307 |
| BBB permeability (Log BB) | −0.341 | 0.205 | 0.159 | −1.243 | −0.182 | 0.502 | 0.289 | 0.620 | 0.502 | 0.421 |
| CNS permeability | −2.801 | −2.253 | −1.533 | −3.131 | −2.569 | −2.518 | −2.042 | −2.163 | −2.518 | −2.192 |
| CYP2C19 inhibitor | No | No | Yes | No | No | No | No | No | No | No |
| CYP2C9 inhibitor | No | No | Yes | No | No | No | No | No | No | No |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | No | No | No | No | No | No |
| Total Clearance (Log ml/min/kg) | 0.256 | 0.908 | 0.058 | 0.618 | 0.233 | 0.833 | 0.233 | 0.304 | 0.719 | 0.723 |
| hERG I inhibitor | No | No | No | No | No | No | No | No | No | No |
| Hepatotoxicity | No | No | No | No | No | No | No | No | No | No |
| Skin Sensitisation | No | Yes | No | No | No | No | Yes | Yes | No | Yes |
∗C1: (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one; C2: 4,4,5,8-tetramethyl-2,3-dihydrochromen-2-ol; C3: 5-chloro-2-(2,4-dichlorophenoxy)phenol; C4: [(3S,3aS,6R,6aS)-3-nitrooxy-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] nitrate; C5: 4-(3-hydroxyprop-1-enyl)-2-methoxyphenol; C6: (E)-3-(2,4-dimethoxyphenyl)prop-2-enoic acid; C7: 4-ethenyl-2-methoxyphenol; C8: 4-[(1E)-buta-1,3-dienyl]-3,5,5-trimethylcyclohex-2-en-1-one; C9: 3-(3,4-dimethoxyphenyl)prop-2-enoic acid; C10: 1-(3,5-dimethoxyphenyl)ethanone.
In current study, we have evaluated the inhibitory potential of AC phytoconstituents against NDM-1 solely by in silico approach and it could be enriched with in vitro studies and hence opens opportunities for other researchers as well. Similarly, other MBL classes can also be targeted with the set of phytocompounds of AC.
4. Conclusion
After determining the phytochemical composition of AC as well as in silico analysis of the interaction between the phytoconstituents and enzyme NDM-1, we conclude that the compound (2R,4S,6R,7S,8R,9S,13S)-16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo [10.8.0.02,9.04,8.013,18] icos-1(12)-ene-6,2′-oxane]-11-one strongly binds with the NDM-1 with lower binding energy (−9.5 kcal/mol) by interacting with key active site amino acid residues involve in substrate recognition such as LYS A:211, ASN A:220, HIS A:250, and HIS A:122 suggesting its inhibitory activity. Besides, it could be investigated against other Metallo β-lactamase (MBL) classes responsible for MDR and XDR. This phytocompound might be considered for further in vitro experiments to develop safe and effective NDM-1 inhibitors to target NDM-1-containing K. pneumoniae. Further, the limited in vitro and pre-clinical studies on plant-derived NDM-1 inhibitors could stall the design of optimal treatment parameters for NDM-1-containing K. pneumoniae infections.
CRediT authorship contribution statement
Janki V. Rojmala: Writing – original draft, Investigation, Formal analysis, Data curation. Anjali B. Thakkar: Writing – original draft, Formal analysis, Data curation. Dhruti Joshi: Formal analysis, Data curation. Bhargav N. Waghela: Writing – review & editing, Supervision, Data curation. Parth Thakor: Writing – review & editing, Supervision, Methodology, Formal analysis, Conceptualization.
Data availability statement
Data will be provided upon request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are thankful to Bapubhai Desaibhai Patel Institute of Paramedical Sciences (BDIPS), Charotar University of Science and Technology, Changa, Atmiya University, Rajkot, and Post Graduate Department of Biosciences, Satellite Campus, Sardar Patel University, Vallabh Vidyanagar, Gujarat, India for providing infrastructure facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40211.
Contributor Information
Bhargav N. Waghela, Email: wbhargav@gmail.com.
Parth Thakor, Email: parth7218@gmail.com, parththakor.cips@charusat.ac.in.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Chang D., Sharma L., Dela Cruz C.S., Zhang D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.750662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James Rray C.G. McGraw-Hill Professional; Med/Tech: 2004. Sherris Medical Microbiology: an Introduction to Infectious Diseases. [Google Scholar]
- 3.Chang R.K., Miller M., Shahin K., Batac F., Field C.L., Duignan P., Struve C., Byrne B.A., Murray M.J., Greenwald K. Genetics and pathology associated with Klebsiella pneumoniae and Klebsiella spp. isolates from North American Pacific coastal marine mammals. Vet. Microbiol. 2022;265 doi: 10.1016/j.vetmic.2021.109307. [DOI] [PubMed] [Google Scholar]
- 4.Bagley S.T. Habitat association of Klebsiella species. Infect. Control Hosp. Epidemiol. 1985;6(2):52–58. doi: 10.1017/s0195941700062603. [DOI] [PubMed] [Google Scholar]
- 5.Jarvis W.R., Munn V.P., Highsmith A.K., Culver D.H., Hughes J.M. The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infect. Control Hosp. Epidemiol. 1985;6(2):68–74. doi: 10.1017/s0195941700062639. [DOI] [PubMed] [Google Scholar]
- 6.Siu L.K., Yeh K.-M., Lin J.-C., Fung C.-P., Chang F.-Y. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 2012;12(11):881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 7.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 8.Reygaert W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS microbiology. 2018;4(3):482. doi: 10.3934/microbiol.2018.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies Jdavies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottery M.J., Pitchford J.W., Friman V.-P. Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J. 2021;15(4):939–948. doi: 10.1038/s41396-020-00832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alkofide H., Alhammad A.M., Alruwaili A., Aldemerdash A., Almangour T.A., Alsuwayegh A., Almoqbel D., Albati A., Alsaud A., Enani M. Multidrug-resistant and extensively drug-resistant enterobacteriaceae: prevalence, treatments, and outcomes–a retrospective cohort study. Infect. Drug Resist. 2020:4653–4662. doi: 10.2147/IDR.S283488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulani M.S., Kamble E.E., Kumkar S.N., Tawre M.S., Pardesi K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyres K.L., Holt K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018;45:131–139. doi: 10.1016/j.mib.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Galani I., Karaiskos I., Giamarellou H. Multidrug-resistant Klebsiella pneumoniae: mechanisms of resistance including updated data for novel β-lactam-β-lactamase inhibitor combinations. Expert Rev. Anti-infect. Ther. 2021;19(11):1457–1468. doi: 10.1080/14787210.2021.1924674. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Kumar S., Zhang L., Wu H., Wu H. Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae. Open Med. 2023;18(1) doi: 10.1515/med-2023-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan A.U., Maryam L., Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17:1–12. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Ren R., Peng C., Ji Y., Liu S., Wang F. Genomic characterisation of a multidrug-resistant Klebsiella pneumoniae co-harbouring blaNDM-1, blaKPC-2, and tet (A) isolated from the bloodstream infections of patients. Int. J. Antimicrob. Agents. 2024;64(3) doi: 10.1016/j.ijantimicag.2024.107290. [DOI] [PubMed] [Google Scholar]
- 18.Linciano P., Cendron L., Gianquinto E., Spyrakis F., Tondi D. Ten years with New Delhi metallo-β-lactamase-1 (NDM-1): from structural insights to inhibitor design. ACS Infect. Dis. 2018;5(1):9–34. doi: 10.1021/acsinfecdis.8b00247. [DOI] [PubMed] [Google Scholar]
- 19.Wang T., Xu K., Zhao L., Tong R., Xiong L., Shi J. Recent research and development of NDM-1 inhibitors. Eur. J. Med. Chem. 2021;223 doi: 10.1016/j.ejmech.2021.113667. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira R.L., Da Silva B.C., Rezende G.S., Nakamura-Silva R., Pitondo-Silva A., Campanini E.B., Brito M.C., da Silva E.M., Freire CCdM., Cunha AFd. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front. Microbiol. 2019;9:3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalasseril S.G., Krishnan R., Vattiringal R.K., Paul R., Mathew P., Pillai D. Detection of New Delhi metallo-β-lactamase 1 and cephalosporin resistance genes among carbapenem-resistant Enterobacteriaceae in water bodies adjacent to hospitals in India. Curr. Microbiol. 2020;77:2886–2895. doi: 10.1007/s00284-020-02107-y. [DOI] [PubMed] [Google Scholar]
- 22.Jamal W.Y., Albert M.J., Rotimi V.O. High prevalence of New Delhi metallo-β-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0152638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S., Yang Z., Sun L., Wang Z., Sun L., Xu J., Zeng L., Sun T. Clinical observation and prognostic analysis of patients with Klebsiella pneumoniae bloodstream infection. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.577244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorlenko C.L., Kiselev H.Y., Budanova E.V., Zamyatnin Jr AA., Ikryannikova L.N. Plant secondary metabolites in the battle of drugs and drug-resistant bacteria: new heroes or worse clones of antibiotics? Antibiotics. 2020;9(4):170. doi: 10.3390/antibiotics9040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maran S., Yeo W.W.Y., Lim S.-H.E., Lai K.-S. Plant secondary metabolites for tackling antimicrobial resistance: a pharmacological perspective. Antimicrobial Resistance: Underlying Mechanisms and Therapeutic Approaches. 2022:153–173. [Google Scholar]
- 26.Organization W.H. World Health Organization; 2000. General Guidelines for Methodologies on Research and Evaluation of Traditional Medicine. [Google Scholar]
- 27.Singh R., Sharma P.K., Malviya R. Pharmacological properties and ayurvedic value of Indian buch plant (Acorus calamus): a short review. Adv. Biol. Res. 2011;5(3):145–154. [Google Scholar]
- 28.Thakkar A.B., Sargara P., Subramanian R.B., Thakkar V.R., Thakor P. Induction of apoptosis in lung carcinoma cells (A549) by hydromethanolic extract of Acorus calamus L. Process Biochem. 2022;123:1–10. [Google Scholar]
- 29.Balakumbahan R., Rajamani K., Kumanan K. Acorus calamus: an overview. J. Med. Plants Res. 2010;4(25):2740–2745. [Google Scholar]
- 30.Rajput S.B., Tonge M.B., Karuppayil S.M. An overview on traditional uses and pharmacological profile of Acorus calamus Linn.(Sweet flag) and other Acorus species. Phytomedicine. 2014;21(3):268–276. doi: 10.1016/j.phymed.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 31.Archana S.N., Pant A., Prakash O. Acorus calamus (Sweet flag): a medicinally important plant. Int J Inst Pharm Life Sci. 2015;5:232–240. [Google Scholar]
- 32.Shahzad N., Alam A., Sultana N., Asma K. Phytochemical and pharmacological studies of vaj turki (Acorus calamus Linn.) & Unani description—a review. Int. J. Pharma Sci. Res. 2015;6:1241–1244. [Google Scholar]
- 33.Thakkar A.B., Subramanian R., Thakkar S.S., Thakkar V.R., Thakor P. Isolation, identification, and characterization of α-asarone, from hydromethanolic leaf extract of Acorus calamus L. and its apoptosis-inducing mechanism in A549 cells. J. Biomol. Struct. Dyn. 2023:1–21. doi: 10.1080/07391102.2023.2227712. [DOI] [PubMed] [Google Scholar]
- 34.Thakor P., Mehta J.B., Patel R.R., Patel D.D., Subramanian R.B., Thakkar V.R. Extraction and purification of phytol from Abutilon indicum: cytotoxic and apoptotic activity. RSC Adv. 2016;6(54):48336–48345. [Google Scholar]
- 35.Edeoga H.O., Okwu D., Mbaebie B. Phytochemical constituents of some Nigerian medicinal plants. Afr. J. Biotechnol. 2005;4(7):685–688. [Google Scholar]
- 36.Veerachari UBopaiah A. Phytochemical investigation of the ethanol, methanol and ethyl acetate leaf extracts of six Cassia species. Int. J. Pharma Bio Sci. 2012;3(2):260–270. [Google Scholar]
- 37.Nag S., Paul A., Dutta R. Phytochemical analysis of methanolic extracts of leaves of some medicinal plants. International Journal of Scientific and Research Publications. 2013;3(4):1–5. [Google Scholar]
- 38.Park Y.-S., Jung S.-T., Kang S.-G., Heo B.G., Arancibia-Avila P., Toledo F., Drzewiecki J., Namiesnik J., Gorinstein S. Antioxidants and proteins in ethylene-treated kiwifruits. Food Chem. 2008;107(2):640–648. [Google Scholar]
- 39.Nabavi S., Ebrahimzadeh M., Nabavi S., Hamidinia A., Bekhradnia A. Determination of antioxidant activity, phenol and flavonoids content of Parrotia persica Mey. Pharmacologyonline. 2008;2(9):560–567. [Google Scholar]
- 40.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosconati S., Forli S., Perryman A.L., Harris R., Goodsell D.S., Olson A.J. Virtual screening with AutoDock: theory and practice. Expet Opin. Drug Discov. 2010;5(6):597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forli SOlson AJ. A force field with discrete displaceable waters and desolvation entropy for hydrated ligand docking. J. Med. Chem. 2012;55(2):623–638. doi: 10.1021/jm2005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namera D.L., Thakkar S.S., Thakor P., Bhoya U., Shah A. Arylidene analogues as selective COX-2 inhibitors: synthesis, characterization, in silico and in vitro studies. J. Biomol. Struct. Dyn. 2021;39(18):7150–7159. doi: 10.1080/07391102.2020.1806109. [DOI] [PubMed] [Google Scholar]
- 44.Thakkar S.S., Shelat F., Thakor P. Magical bullets from an indigenous Indian medicinal plant Tinospora cordifolia: an in silico approach for the antidote of SARS-CoV-2. Egyptian Journal of Petroleum. 2021;30(1):53–66. [Google Scholar]
- 45.Socha B.N., Pandya S.B., Patel U.H., Patel R., Bhatt B.S., Bhakhar S., Vekariya N., Valand J., Thakor P., Thakkar A.B. 1-D MOF [Ag2 (C10H10N3O3S) 2 (C4H8N) 2] n: photocatalytic treatment, crystallographic evaluation, ADMET parameters, CT-DNA and anticancer activity. J. Biomol. Struct. Dyn. 2023:1–16. doi: 10.1080/07391102.2023.2236732. [DOI] [PubMed] [Google Scholar]
- 46.Thakkar S.S., Thakor P., Doshi H., Ray A. 1, 2, 4-Triazole and 1, 3, 4-oxadiazole analogues: synthesis, MO studies, in silico molecular docking studies, antimalarial as DHFR inhibitor and antimicrobial activities. Bioorg. Med. Chem. 2017;25(15):4064–4075. doi: 10.1016/j.bmc.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 47.Thakor P., Subramanian R.B., Thakkar S.S., Ray A., Thakkar V.R. Phytol induces ROS mediated apoptosis by induction of caspase 9 and 3 through activation of TRAIL, FAS and TNF receptors and inhibits tumor progression factor Glucose 6 phosphate dehydrogenase in lung carcinoma cell line (A549) Biomed. Pharmacother. 2017;92:491–500. doi: 10.1016/j.biopha.2017.05.066. [DOI] [PubMed] [Google Scholar]
- 48.Thakor P., Subramanian R.B., Thakkar S.S., Thakkar V.R. Drug Development for Cancer and Diabetes. Apple Academic Press; 2020. Cytotoxic, apoptosis inducing effects and anti-cancerous drug candidature of jasmonates; pp. 117–127. [Google Scholar]
- 49.Zhang Q.-W., Lin L.-G., Ye W.-C. Techniques for extraction and isolation of natural products: a comprehensive review. Chin. Med. 2018;13:1–26. doi: 10.1186/s13020-018-0177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higdon J.V., Frei B. Tea catechins and polyphenols: health effects, metabolism, and antioxidant functions. 2003 doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- 51.Kumar N., Biswas S., Mathew A.E., Varghese S., Mathew J.E., Nandakumar K., Aranjani J.M., Lobo R. Pro-apoptotic and cytotoxic effects of enriched fraction of Elytranthe parasitica (L.) Danser against HepG2 Hepatocellular carcinoma. BMC Compl. Alternative Med. 2016;16:1–11. doi: 10.1186/s12906-016-1395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siddique N.A., Mujeeb M., Najmi A.K., Akram M. Evaluation of antioxidant activity, quantitative estimation of phenols and flavonoids in different parts of Aegle marmelos. Afr. J. Plant Sci. 2010;4(1):1–5. [Google Scholar]
- 53.Vaidyanathan R., Sreedevi S.M., Ravichandran K., Vinod S.M., Krishnan Y.H., Babu L.K., Parthiban P.S., Basker L., Perumal T., Rajaraman V. Molecular docking approach on the binding stability of derivatives of phenolic acids (DPAs) with Human Serum Albumin (HSA): hydrogen-bonding versus hydrophobic interactions or combined influences? JCIS Open. 2023;12 [Google Scholar]
- 54.Chetia H., Sharma D.K., Sarma R., Verma A. An in silico approach to discover potential inhibitors against multi-drug resistant bacteria producing New-Delhi metallo-β-Lactamase 1 (NDM-1) enzyme. Int. J. Pharm. Pharmaceut. Sci. 2014;6(4):299–303. [Google Scholar]
- 55.Guo Y., Wang J., Niu G., Shui W., Sun Y., Zhou H., Zhang Y., Yang C., Lou Z., Rao Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein & cell. 2011;2(5):384–394. doi: 10.1007/s13238-011-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang gHao Q. Crystal structure of NDM‐1 reveals a common β‐lactam hydrolysis mechanism. Faseb. J. 2011;25(8):2574–2582. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- 57.Rehman M.T., AlAjmi M.F., Hussain A., Rather G.M., Khan M.A. High-throughput virtual screening, molecular dynamics simulation, and enzyme kinetics identified ZINC84525623 as a potential inhibitor of NDM-1. Int. J. Mol. Sci. 2019;20(4):819. doi: 10.3390/ijms20040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farhat N., Khan A.U. Evolving trends of New Delhi Metallo-betalactamse (NDM) variants: a threat to antimicrobial resistance. Infect. Genet. Evol. 2020;86 doi: 10.1016/j.meegid.2020.104588. [DOI] [PubMed] [Google Scholar]
- 59.Mojica M.F., Rossi M.-A., Vila A.J., Bonomo R.A. The urgent need for metallo-β-lactamase inhibitors: an unattended global threat. Lancet Infect. Dis. 2022;22(1):e28–e34. doi: 10.1016/S1473-3099(20)30868-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thakkar S.S., Thakor P., Ray A., Doshi H., Thakkar V.R. Benzothiazole analogues: synthesis, characterization, MO calculations with PM6 and DFT, in silico studies and in vitro antimalarial as DHFR inhibitors and antimicrobial activities. Bioorg. Med. Chem. 2017;25(20):5396–5406. doi: 10.1016/j.bmc.2017.07.057. [DOI] [PubMed] [Google Scholar]
- 61.Thakkar A.B., Subramanian R.B., Thakkar S.S., Thakkar V.R., Thakor P. Biochanin A–A G6PD inhibitor: in silico and in vitro studies in non-small cell lung cancer cells (A549) Toxicol. Vitro. 2024;96 doi: 10.1016/j.tiv.2024.105785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be provided upon request.