Abstract
Purpose
The pathogenesis of head and neck squamous cell carcinoma (HNSCC) was complex and the overall survival was not satisfying. It was urgent to uncover novel molecules that play vital role in HNSCC for disease monitoring and drug development.
Methods
Distinguished expression of FCGBP mRNA in HNSCC was analyzed by TCGA-HNSC and three GEO datasets, the relationship between FCGBP and clinical stage and survival was analyzed by GEPIA 2, the immune infiltration pattern analysis was conducted by TIMER 2.0, pathways affected by FCGBP was conducted by GSEA and GO/KEGG. In vitro experiments (including qRT-PCR, siRNA transfection, CCK8, transwell assay and flow cytometry) were conducted to confirm bioinformatic analysis.
Results
FCGBP was down-regulated in tumor samples compared with normal tissues at both mRNA and protein levels, and positively correlated with survival in HNSCC. Genes co-expressed with FCGBP were mainly enriched in immune-related biological processes and pathways. GSEA indicated that FCGBP was associated with activated immune reaction and inhibiting well-known pro-tumor pathways. GSE41613 validated FCGBP as an independent prognostic marker for HNSCC and FCGBP was down-regulated in HNSCC cell lines by qRT-PCR. Migration and invasion of SCC9 and CAL27 were enhanced by FCGBP-targeting siRNAs, the ratio of cytotoxic T lymphocytes were down-regulated while the ratio of myeloid-derived suppressor cells were increased by FCGBP-targeting siRNAs.
Conclusion
FCGBP was a tumor suppressor gene and was an independent prognostic marker for better survival. The underlying mechanism may be that FCGBP inhibited tumor migration and invasion and activated immune response against tumor cells.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-024-01607-8.
Keywords: HNSCC, FCGBP, Immune infiltration, Prognostic marker, Tumor suppressor gene
Introduction
Head and neck squamous cell carcinoma (HNSCC) refers to a malignancy originating from mucosal epithelial tissues of oral cavity, pharynx, larynx and paranasal sinuses, which accounts for the vast majority of head and neck tumors [1]. The incidence of HNSCC ranked sixth among all cancers worldwide with 890,000 new cases and 450,000 deaths in 2018 [2], alcohol and tobacco consumption, environmental pollutants and microorganism infection are common risk factors for HNSCC [1]. The treatments for HNSCC are multimodal, including surgery, chemotherapy and radiotherapy, despite the progress of surgery and drug development, the 5-year overall survival (OS) rate of human papilloma virus (HPV)-negative HNSCC patients across all stages remains at 40–50% with exclusive of Epstein-Barr virus (EBV)-related nasopharyngeal, and the 5-year OS get much worse when referring to recurrent or metastatic (R/M) HNSCC patients with the median value was only 10–13 months [3]. In recent years, immunotherapy has gradually emerged as a promising strategy for HNSCC patient, especially for immune checkpoint inhibitors (ICI). Nivolumab and pembrolizumab were approved for treatment of R/M HNSCC patients by FDA in 2016, and pembrolizumab for the first-line treatment for inoperable R/M HNSCC patients in 2019, however, the outcome was not thrilling as anticipated initially. It was shown that only 18% R/M HNSCC patients benefited from ICI treatments in phase 1/2 KEYNOTE-012 [4]. Therefore, it is urgent to unveil new molecular mechanism attributing to initiation, tumorigenesis and progression of HNSCC.
TCGA (The cancer genome atlas) database and TCGA based data analysis platform (such as GEPIA 2, Ualcan, cBioPortal, and so on) provide abundant biological information of various tumors, such as transcriptome, proteome and methylomics data, which it is convenient for researches to reveal potential molecules that may play important roles in occurrence or progression of tumors. By analyzing TCGA-HNSC data set, we found that the IgG Fc-binding protein (FCGBP) was found to be down-regulated expressed in HNSCC samples compared with normal tissues and correlated with better survival at the same time, therefore, FCGBP was speculated to function as a tumor suppressor gene in HNSCC.
Physiologically, FCGBP is the major component of mucus which predominantly secreted by goblet cells of intestine and form a net-like structure covering inner mucus layer to prevent bacterial invasion [4, 5]. FCGBP is named on account of its capability to binding immunoglobulin G (IgG) and proposed to protect mucosa as it binds another mucus component trefoil factor 3 (TFF3) and cross-links mucus [6]. However, in recent years, some studies shown that FCGBP also played a role in a variety of tumors, especially in colorectal cancer (CRC) [7, 8]. There were several studies indicated that FCGBP gene expression was down-regulated in HNSCC samples and was a favorable maker for better overall survival in HNSCC based on TCGA and Gene Expression Omnibus (GEO) datasets [9–11], however, those GEO datasets was not specialized to HNSCC, but also included lung adenocarcinoma cell line and the size number of samples was not abundant, meanwhile, those studies were lack of experiments for further validation. Therefore, our study was conducted by utilizing TCGA and other well-known public databases and validated by in vitro experiments.
Materials and methods
Exploration of key genes both affect survival and expressed distinguishly between tumor and normal tissues in HNSCC
Firstly, the list of differentially expressed genes (DEGs) between tumor and normal tissues in HNSCC was downloaded from GEPIA 2 website (http://gepia2.cancer-pku.cn/), |Log2FC| cutoff was 1 and q-value cutoff was 0.01, list of DEGs was shown in Table S1. Then, the list of top 100 genes which affect patient survival in HNSCC was also downloaded from GEPIA 2, genes affecting both OS and disease free survival (DFS) was analyzed to get survival predictors as more as possible, cutoff to define expression level (high/low) was set as median value and repeated analysis by quartile value, gene list was shown in Table S2. By analyzing those two lists, several genes were found to affect survival and be expressed differently between tumor and normal tissues in HNSCC, among those genes, FCGBP was the only gene both affect OS and DFS and then was selected for further investigation.
Analyzing the relationship between FCGBP mRNA expression and clinical stage
On GEPIA 2 website, Expression Analysis part, expression DIY subpart, Stage Plot module, we use major stage as classification, and Log2(TPM + 1) for log-scale, we get violin plots of FCGBP mRNA expression pattern among different clinical stages in different tumors.
Survival analysis
On GEPIA 2 website, Expression Analysis part, Survival analysis subpart, first, we analyzed OS, and Group Cutoff was set as median, if P > 0.05, we changed Group Cutoff as Quartile and repeated analysis, FCGBP was defined as no prognostic marker only when both P > 0.05, it went the same way when analyzing DFS.
Immune infiltration analysis
Immune infiltration analysis was performed by TIMER 2.0. On Immune part, Gene module, FCGBP was input in Gene Expression and then immune cell types were selected, “Purity Adjustment” option were selected and partial Spearman’s correlation was adopted to conduct this analysis, P value and Rho value across all tumors were gained and displayed in heatmap, and then we select TCGA-HNSC cohort for scatter diagram display.
GSEA (Gene Set Enrichment Analysis) and Gene Ontology (GO)/ Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
Pathways affected by FCGBP was investigated by GSEA 3.0 software. The transcriptome data of HNSCC were downloaded from TCGA (https://portal.gdc.cancer.gov/), in Cases part, TCGA-HNSC was select for Project, in Files part, HTSeq-FPKM was selected for Workflow type, 502 tumor samples with 44 adjacent normal tissues were obtained for further study. Single gene mRNA expression of FCGBP in tumor samples were extracted from transcriptome data by using Limma 3.13 package for R 4.1.1 software, and then tumor samples were divided into two groups (high/low group) according to the median value of FCGBP mRNA expression level, and then conduct pathways enrichment analysis of two groups by GSEA 3.0 software, we choose c2.cp.kegg.v7.4.symbols.gmt as the reference gene set to perform GSEA.
Moreover, genes correlated with FCGBP were also investigated for GO/KEGG analysis, co-expression gene list was downloaded from cBioPortal database to profile genes that were co-expressed with FCGBP. Those 478 co-expression genes with the absolute value of Spearman’s correlation was more than 0.3 were further analyzed by GO/KEGG (detailed list of genes was shown in Table S3).
Validation by GEO dataset
Microarray raw data of three GEO datasets (GSE37991, GSE29330, GSE30784) was downloaded to validate FCGBP mRNA expression in tumor and normal tissues, one GEO dataset (GSE41613) was utilized to validate the relationship between FCGBP mRNA and survival and validate FCGBP correlated genes.
Cell culture
HNSCC cell lines (SCC9 and CAL27) were kindly donated by Professor Wu Lihong (School and Hospital of Stomatology, Guangdong Engineering Research Center of Oral Restoration and Reconstruction, Guangzhou Medical University), and those cells have STR identification reports. Human oral keratinocytes (HOK) were kindly donated by Professor Liao Guiqing (Hospital of Stomatology, Guanghua School of Stomatology, Sun Yat-sen University). SCC9 were cultured in DMEM/F12 1:1 (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, BI, Israel) and 400 ng/ml hydrocortisone (Solarbio, Beijing, China), CAL27 and HOK were cultured in DMEM with high glucose (Gibco, USA) supplemented with 10% FBS. All types of cells were cultured at atmosphere with 37 °C, 95% humidity and 5% carbon dioxide.
Quantitative real-time PCR (qRT-PCR)
Rnase free principle was followed in this whole process. Firstly, total RNA was extracted by using RNAzol agent (AG, China) and then quantified using NanoDrop 2000. Then RNA was reverse transcripted into cDNA by using Evo M-MLV RT Premix for qPCR (AG, China). Finally, real-time PCR was carried out in Agilent system (Agilent, USA) by utilizing Hieff UNICON qPCR SYBR Green Master Mix (Yeasen Biotech, China). The amplification conditions set as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s, and then the procedure for melting curve were set as follows: 95 °C for 5 s and 65 °C for 1 min, maintained at 97 °C continuously. The GAPDH were used as internal control. The relative mRNA level among samples was calculated using the 2−ΔΔCT formula. Each sample had 3 duplicates and this experiment was repeated independently by 3 times. The primers for FCGBP and GAPDH were listed as follows:
FCGBP
Forward primer: 5′-GCCAAGGCTGAGATGATAGGC-3′
Reverse primer: 5′- CCTGCACAGAGATGGCAT
GAPDH
Forward primer: 5′-GGAGCGAGATCCCTC CAAAAT-3′
Reverse primer: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Knockdown of FCGBP
To knockdown FCGBP expression, siRNA transfection was performed in HNSCC cell lines (SCC9 and CAL27). The siRNA sequences specifically targeting FCGBP were designed and synthesized by RiboBio (China). The reagent used for siRNA transfection was Lipofectamine 3000 (Thermo Fisher Scientific, USA). The siRNA transfection procedure was carried out by following the manufacture’s instructions.
Transwell assay
Cells which had been transfected by FCGBP-targeting siRNA for 48 h were then digested for migration and invasion detection. Transwell migration and invasion assays were performed as previously described [12].
Proliferation detection by CCK8
After transfected with siRNA for 48 h, CAL27 and SCC9 with good growth status were digested and then seeded into 96 well culture plates at a cell density of 4000 cells per well. Five duplicates were set in each group, on the 1st, 2nd and 3rd day cultivation, CCK8 solution was diluted in DMEM in a ratio of 1:9 and then added and incubated for another 2 h. The absorbance value at 450 nm was measured on an microplate reader, and the cell growth curve was plotted using the average absorbance value.
Isolation of peripheral blood mononuclear cells (PBMCs)
PMBCs were isolated from peripheral venous blood donated by healthy volunteers who had signed informed consent. The procedures were carried out by following the manufacture’s instructions. Human whole blood mononuclear cell isolation reagent (Tianjin Haoyang Biotech, China) was used in this study.
Phenotype detection of PBMCs by flow cytometry
The culture medium of CAL27 and SCC9 cells was changed into fresh RPMI 1640 complete medium after transfected with FCGBP siRNA for 6 h, then the cell culture supernatant after transfected for 72 h was collected and centrifuged (2000 RPM, 10 min, 4 °C) to remove cells and cell debris (termed as conditioned media). Different groups of conditioned media were mixed with RPMI 1640 complete medium in a ratio at 1:1 and culturing PBMC for another 48 h. Then PBMCs were collected and washed twice with PBS, then stained with Ghost Dye 510 at room temperature at dark for 30 min (1 μl/ml), after washed twice with PBS, the primary antibodies were added and incubated with PBMCs for 30 min (4 °C, at dark). For the detection of Granzyme B, cells were fixed and permeabilized for 30 min and then the antibody was added and incubated overnight (4 °C, at dark). After washed twice with PBS, the proportion of cytotoxic T lymphocytes (CTLs, CD3+CD8+Granzyme B+), B cells (CD19+), and myeloid-derived suppressor cells (MDSCs, CD33+CD11b+HLA-DR−) in PBMCs of each group were detected by flow cytometry.
Statistic analysis
All the in vitro experiments were repeated three times independently. SPSS 20.0 software (SPSS, USA) was used for statistical analysis. Graphs were produced in GraphPad Prism 8 (GraphPad Software, USA) and One-Way ANOVA was used for comparison between multiple groups, and Student’s t-test for comparison between two groups. P < 0.05 was considered statistically significant.
Results
Key genes both affect survival and expressed differently between tumor and normal tissues in HNSCC
By comparing DEG-List and Survival-List, we found that there were several genes that both affected survival and expressed differently between tumor and normal tissues in HNSCC, which was shown in Table S4. Those genes were speculated to play certain roles in tumorigenesis or progression of HNSCC, among them, FCGBP were selected as the key gene for further investigation as it affected both OS and DFS.
FCGBP expression level and correlation with clinical stage and survival in HNSCC
GEPIA 2 revealed that FCGBP mNRA was expressed differently between tumor and normal tissues in diverse tumors, further indicated that FCGBP may play a vital role in tumors. In HNSCC, FCGBP were down-regulated in HPV-negative HNSCC compared with HPV-positive HNSCC, which were in consistent with previous study based on GEO datasets [9] (Fig. 1a). Furthermore, FCGBP protein level were significantly down-regulated in HNSCC tumors compared with normal tissues by analyzing Ualcan database (Fig. 1b). Down-regulated expression of FCGBP mRNA in tumor tissues was further validated by three GEO datasets (GSE37991, GSE29330, GSE30784) (Fig. 1c). FCGBP mRNA level was tended to negatively correlate with clinical stage in HNSCC although with no statistical significance (Fig. 1d). Survival analysis revealed that patients in higher FCGBP expression group had more favorable OS and DFS in HNSCC (Fig. 1e).
Fig. 1.
FCGBP expression level and correlation with clinical stage and survival in HNSCC. a Differential FCGBP mRNA expression in tumor and normal tissues by pan-cancer analysis downloaded from TIMER 2.0, FCGBP level was lower in tumor tissues and HPV− tumor tissues in HNSCC. b FCGBP protein expression in HNSCC samples and normal tissues based on Ualcan database. c Differential expression of FCGBP mRNA between tumor and normal tissues of HNSCC based on 3 GEO datasets (GSE37991, GSE29330 and GSE30784). d FCGBP mRNA level was negatively correlated with clinical stage in HNSCC, although there was no statistical significance. e the detailed information of Kaplan–Meier analysis, for OS, the cut-off value was set as median of FCGBP mRNA expression, while for DFS, the cut-off was set as quartile. d and e were analyzed by GEPIA 2. * means P < 0.05, ** means P < 0.01, *** means P < 0.001, **** means P < 0.0001
Low FCGBP mRNA level was related with a suppressive immune infiltration pattern
By analyzing TIMER 2.0 database, FCGBP mRNA level was positively correlated with effective immune cells participating in tumor killing, like B cells (Fig. 2a), CD8+ T cells (Fig. 2b), dendritic cells (Fig. 2c), monocytes (Fig. 2d) and neutrophiles (Fig. 2e), whereas negatively correlated with myeloid-derived suppressor cells (MDSCs) (Fig. 2f) which functioned as immune suppressive cells. Therefore, FCGBP was a marker for effective immune cell infiltration which was in accordance with the above results showing that FCGBP functioned as a tumor suppressor gene in HNSCC.
Fig. 2.
Low FCGBP mRNA level was related with a suppressive immune infiltration pattern. Immune infiltration analysis showed that FCGBP expression was positively correlated with immune cells which play a dominant role in anti-tumor immune response and negatively correlated with suppressive immune cells MDSCs, a–f showed B cells, CD8+T cells, dendritic cells, monocytes, neutrophils and MDSCs respectively
GSEA of pathways affected by FCGBP and GO/KEGG analysis of FCGBP co-expression genes in HNSCC
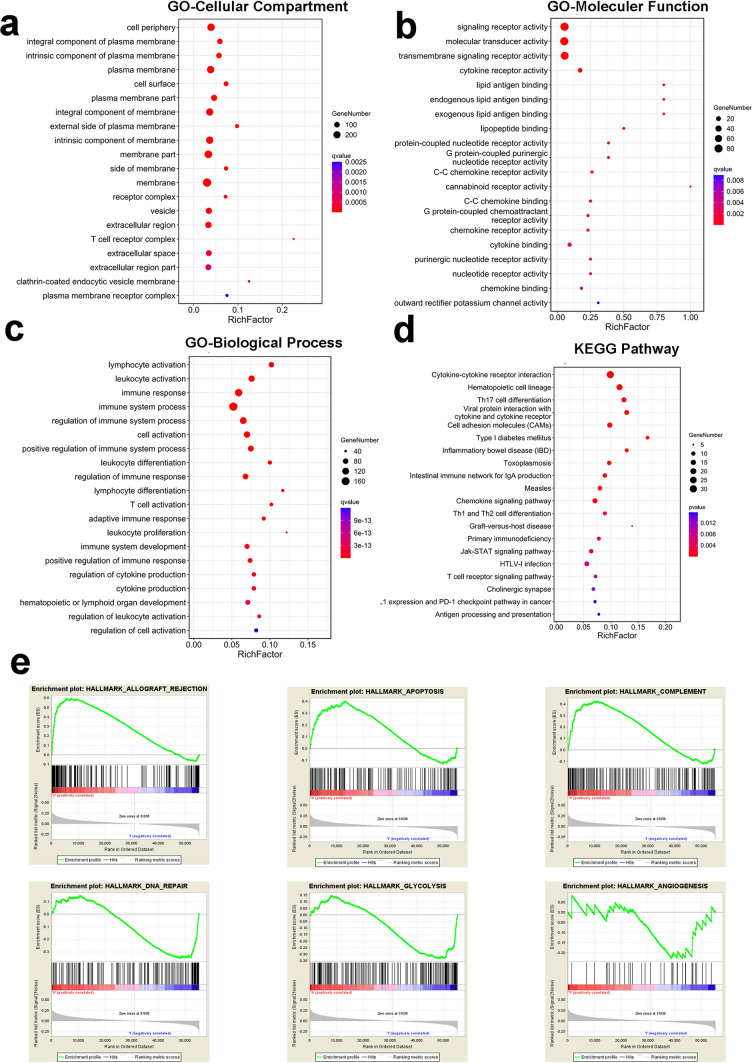
The results of GO/KEGG analysis of FCGBP co-expression genes shown that these genes were mainly located in plasma membrane and cell periphery (Fig. 3a), enriched in signaling receptor activity, molecular transducer activity, transmembrane signaling receptor activity and cytokine receptor activity (Fig. 3b), enriched in immune-related biological process, such as lymphocyte activation, leukocyte activation, immune response, immune system process and so on (Fig. 3c), KEGG analysis showed these genes were enriched in cytokine-cytokine receptor interaction, hematopoietic cell lineage, Th17 cell differentiation and so on (Fig. 3d).
Fig. 3.
Pathways affected by FCGBP in HNSCC by GO/KEGG analysis and GSEA. GO and KEGG analysis were conducted for genes co-expressed with FCGBP by Omicshare website (https://www.omicshare.com/tools/). a GO-Cellular compartment analysis showed these genes were mainly located in plasma membrane and cell periphery, b GO-Molecular function showed these genes were enriched in signaling receptor activity, molecular transducer activity, transmembrane signaling receptor activity and cytokine receptor activity, c GO-Biological process showed these genes were enriched in immune-related biological process, such as lymphocyte activation, leukocyte activation, immune response, immune system process, d KEGG analysis showed these genes were enriched in cytokine-cytokine receptor interaction, hematopoietic cell lineage, Th17 cell differentiation and so on. e GSEA showed allograft rejection, apoptosis and complement were activated in the group with high FCGBP expression, while DNA repair, glycolysis and angiogenesis were activated in the group with low FCGBP expression
By GSEA, it showed that pathways associated with immune reaction (Hallmark_Allograft rejection, Hallmark_Complement) were activated in group with high FCGBP expression, Hallmark_Apoptosis were also activated in this group. Meanwhile, pathways associated with tumor initiation and progression (including Hallmark_DNA repair, Hallmark_Glycolysis, Hallmark_Angiogenesis) were activated in group with low FCGBP expression, which further indicated that FCGBP might function as a tumor suppressor gene in HNSCC (Fig. 3e).
FCGBP was an independent prognostic marker for HNSCC by analyzing GEO dataset and validation of its mRNA level by in vitro experiments
By analyzing GSE41613 dataset, FCGBP was an independent prognostic marker for HNSCC and negatively correlated with patients survival, which was analyzed by uni-variate cox, multivariate cox and Kaplan–Meier methods (Fig. 4a–c).
Fig. 4.
FCGBP was an independent prognostic marker for HNSCC. Prognostic value of FCGBP were investigated by using GSE41613 dataset. a FCGBP was a significant protective factor for HNSCC prognosis, while stage was a risk factor by univariate cox analysis. b FCGBP was an independent prognostic marker for HNSCC by multivariate cox analysis. c Survival analysis validation by GSE41613 dataset. d FCGBP mRNA level were significantly down-regulated in HNSCC cell lines compared with HOK by qRT-PCR. *** means P < 0.001, **** means P < 0.0001
Furthermore, FCGBP mRNA level was confirmed to be down-regulated in HNSCC cell lines compared with HOK (Fig. 4d), which further verified FCGBP expression results based on TCGA and GEO databases.
Knockdown of FCGBP enhanced migration and invasion of SCC9 and CAL27 cell lines
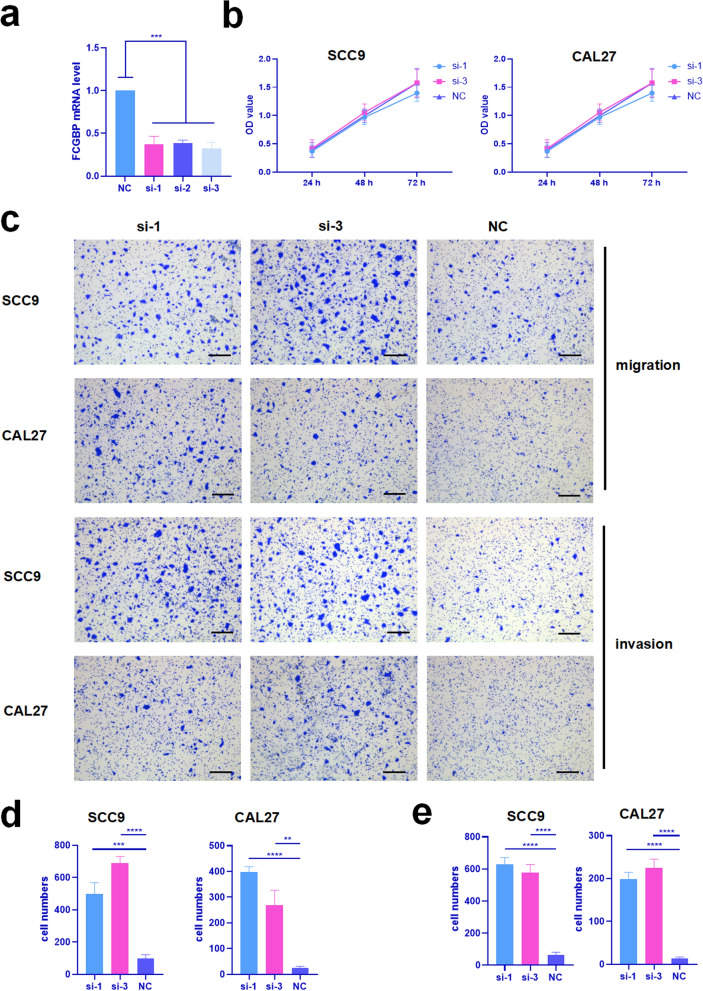
CCK8 assay and transwell assay were utilized to explore the function of FCGBP in SCC9 and CAL27 cell lines. Firstly, siRNA targeting FCGBP were employed to knockdown FCGBP gene expression, the knockdown efficiency on mRNA validated by qRT-PCR (Fig. 5a). It demonstrated that knockdown FCGBP had no influence on proliferation of SCC9 and CAL27 cell lines compared with negative control groups (Fig. 5b). However, it was obvious that knockdown FCGBP enhanced migration and invasion of both SCC9 and CAL27 cell lines significantly (Fig. 5c–e).
Fig. 5.
Knockdown of FCGBP enhanced migration and invasion of SCC9 and CAL27 cell lines. a Validation of knockdown efficiency in SCC9 by qRT-PCR, si-1 and si-3 were selected for the following experiments. b Proliferation of SCC9 and CAL27 after knockdown of FCGBP were detected by CCK8. c Migration and invasion of SCC9 and CAL27 after knockdown of FCGBP were detected by transwell assay, all scale lables represented 50 μm. d Statistic analysis of migration. e Statistic analysis of invasion. ** means P < 0.01, *** means P < 0.001, **** means P < 0.0001
Knockdown of FCGBP promoted an immunosuppressive phenotype formation of PBMCs
After cultured with conditioned medium collected from SCC9 and CAL27 cell lines which were transfected with FCGBP-targeting siRNAs, the PBMCs exerted an immunosuppressive phenotype. The flow cytometry results showed that the proportion of Granzyme B+CD8+T cells (CTLs) were significantly reduced in siRNA groups (Fig. 6a). Meanwhile, the proportion of immunosuppressive cells MDSCs were significantly increased in siRNA groups (Fig. 6b). However, the proportion of B cells were not significantly influenced by FCGBP-targeting siRNAs (Fig. 6c), which was not in line with the results analyzed by TIMER 2.0.
Fig. 6.
Knockdown of FCGBP promoted an immunosuppressive phenotype formation of PBMCs. The immune phenotype of PBMCs after cultured by conditioned media from SCC9 and CAL27 which FCGBP had been knockdown were detected by flow cytometry. a The proportion of CTL/T cells in PBMCs. b The proportion of MDSCs/live cells in PBMCs. c The proportion of B cells/live cells in PBMCs. * means P < 0.05, ** means P < 0.01
Discussion
Tumors can be divided into cold (non-inflamed) tumors or hot (inflamed) tumors according to the level of inflammatory cytokines and immune cell infiltration in tumor microenvironment (TME) [13], it was reported that different immune phenotype led to different prognosis and responses to immunotherapy. Patients with a hot tumor were often more sensitive to immunotherapy in the high T cell infiltration situation [14], while patients with a low ICI score had a more favorable survival [15]. Multiple molecules participated in constitution of tumor immune phenotype in HNSCC, Zhang’s research constructed an immune-related gene signature based on 17 genes to predict patients prognosis [16], in addition, other molecules were also unearthed to contribute to tumor immune microenvironment formation and associated with clinical outcome of HNSCC patients [17, 18]. Immune checkpoint inhibitors exerted powerful effect in certain patients of HNSCC, however, only less than 20% patients benefited from this therapy [19], laying stress on the importance of uncovering other molecules participating in tumor immune microenvironment formation.
This study revealed that FCGBP functioned as an tumor suppressor gene in HNSCC. The possible mechanism in our study based on two aspects. On the one hand, FCGBP inhibited cell migration and invasion intrinsically, on the other hand, FCGBP secreted by tumor cells communicated with immune cells in TME and had an influence on immune cells thus facilitating killing tumor cells.
In the first place, FCGBP was documented to play a role in cancer initiation, progression and prognosis in several types of tumors. In gallbladder adenocarcinoma cells, FCGBP was down-regulated during TGF-β1 induced epithelial mesenchymal transition (EMT) process and patients with high FCGBP expression had a better prognosis [20]. In CRC, FCGBP expression was decreased in colorectal adenoma and CRC compared with normal colorectal tissues, moreover, FCGBP protein was expressed decreasingly with the increase of clinical stage and FCGBP expression was linked with worse prognosis [21]. In other studies focused on HNSCC, FCGBP over-expression inhibited cell proliferation and EMT in FaDu and CAL27 cells, similarly, FCGBP expression was also down-regulated by TGF-β1 [9]. Therefore, FCGBP inhibited tumor cell metastasis possibly by mediating TGF-β1 induced epithelial mesenchymal transition (EMT).
Second and most important, FCGBP was indicated to participate in immune reaction both in TME and non-tumor sites. The complex formed by the combination of FCGBP and TFF3 was reported to be involved in numerous mucosal innate immunity, such as oral cavity [22], respiratory [23], intestinal [24] and vagina [25]. With referring to malignancies, there are completely different reports on the relationship between FCGBP and immune response. For example, in gliomas, bioinformatics analysis revealed a positive correlation between FCGBP and immunosuppressive cells, such as regulatory T cells (Tregs) and Tumor associated macrophages (TAM) [26] and immunosuppressive molecules PD-L1, HAVCR2 and TGFB1 [27]. Besides, FCGBP facilitated M2 macrophage polarization in ovarian cancer. But in most tumors, FCGBP has been reported to be positively correlated with cells or immune molecules that functioned as immunostimulators, for example, there was positive relationship between FCGBP expression level and the extent of infiltrating immune cells, such as B cells and dendritic cells in CRC [21].
In HNSCC, other researchers revealed that FCGBP positively correlated with tumor‐infiltrating CD4+ T cells, CD8+ T cells, dendritic cells, macrophages, NK cells and plasma cells [28]. In another study, FCGBP was indicated positively associated with infiltrating rates of B cells, CD8+ T lymphocytes, T helper follicular cells and mast cells in TME [10], which was strikingly similar to our results. FCGBP was revealed as a potential tumor suppressor gene in HNSCC and negatively correlated with paclitaxel resistance and the PART1/AC007728.2/LINC00885/hsa-miR-877-5p/FCGBP axis was speculated to regulated FCGBP expression [28]. In addition, FCGBP was identified as an independent biomarker for HNSCC prognosis [11], FCGBP expression was upregulated by HPV infection while inhibited by TGF-β, and was correlated to the prognosis of HNSCC patients [9].
In our study, FCGBP was significantly down-regulated in tumor samples compared with normal tissues based on TCGA database and GEO database, which was further validated by qRT-PCR in vitro. The results in our study was in accordance with its expression in gallbladder cancer [29], prostate cancer [20], as for clinical significance, FCGBP was negatively correlated with survival and tended to negatively correlate with clinical stage, which was also reported in gallbladder cancer [30] and osteosarcoma [31]. Those results revealed that FCGBP functioned as a tumor suppressor gene in HNSCC and affected tumor development and clinical outcome. Our study further indicated that FCGBP participated in tumor immune microenvironment formation in HNSCC. Based on TIMER 2.0 analysis, FCGBP mRNA level was positively correlated with B cells, CD8+ T cells, dendritic cells, monocytes and neutrophils while negatively correlated with MDSC, indicating FCGBP promoted an immune-hot microenvironment formation and was in favor of powerful immune response, which was further indicated by GSEA and GO/KEGG analysis in this study.
It was worth mentioning that the function of FCGBP in immune infiltration in HNSCC had been explored by bioinformatic analysis previously but lack of laboratory evidence. Our research overcame this deficiency through in vitro experiments. The conditioned media containing different concentrations of FCGBP secreted by SCC9 and CAL27 after knockdown of FCGBP expression indeed affected immune-phenotype formation of PBMCs. The conditioned media with low concentration of FCGBP significantly decreased the ratio of CTLs, which were the main force of tumor killing cells in adaptive immunity [32], meanwhile, it significantly increased the proportion of MDSCs, which suppressed T cell function through a number of mechanisms involving arginase 1, NO synthase and reactive oxygen species thus establishing an immunosuppressive network in diverse tumors [33]. But the ratio of B cells was not significantly influenced by conditioned media, which was not in consistent with analysis by TIMER 2.0 database.
In addition to be involved in phenotype change of immune cells, FCGBP was demonstrated to be acted as an oncogene intrinsically by enhancing cell migration and invasion of SCC9 and CAL27 while not affecting proliferation of tumor cells, which was partly inconsistent with previous reports in oral squamous cell carcinoma (OSCC), which was a major type of HNSCC. The possible explanation may be that the FCGBP expression was knockdown in our study while it was over expressed in Liu’s study [34].
The limitation of this study was that most of the results were based on bioinformatic analysis, and the in vitro experiments are relatively simple. After interfering with the expression of FCGBP in HNSCC cell lines, the biological behavior of tumor cells was only detected for cell proliferation, invasion, and migration, and lack of further underlying mechanism exploration. The biggest limitation of this research was the lack of in vivo experimental evidence to support the conclusions of this study.
However, this study did suggest a new molecule that may play an important role in HNSCC tumorigenesis and development, and the underlying mechanism may be that FCGBP can promote immune reaction and immune-hot microenvironment formation. Moreover, FCGBP maybe a diagnostic and prognostic marker in HNSCC due to FCGBP was secreted to extracellular matrix, to the best of our knowledge, saliva and serum were convenient sources of liquid biopsy to early diagnose and monitor tumor progression, especially for OSCC. Therefore, the study focused on the level of FCGBP in patient’s saliva and serum tended to be a meaningful and promising research we would carried out in the near future.
In brief, our study revealed that FCGBP played as a tumor suppressor gene in HNSCC possibly by inhibiting tumor migration and invasion and promoting immune reaction.
Conclusion
FCGBP was a tumor suppressor gene and was an independent prognostic marker for better survival. The underlying mechanism may be that FCGBP inhibited tumor migration and invasion and activated immune response against tumor cells.
Supplementary Information
Acknowledgements
Thanks to all the healthy volunteers for donating peripheral venous blood, thanks to the nurses in Department of Oral and Maxillofacial Surgery, School and Hospital of Stomatology for helping to extract peripheral venous blood from healthy volunteers, and thanks to Guangdong Engineering Research Center of Oral Restoration and Reconstruction & Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou Medical University for providing various laboratory instruments. Finally, thanks to Special Fund for High-Level University Construction Talents of Guangzhou Medical University (Guangzhou, Guangdong Province, China. B185006003006) for supporting the study.
Author contributions
All authors contributed to the study conception and design. Investigation and manuscript preparation were conducted by Lijuan Zeng. Material preparation, data collection and analysis were performed by Jun Zeng, Jianfeng He and Yongqi Li. Visualization was performed by Chengwei Li and Zhiyan Lin. Language editing was performed by Guangwei Chen and Huilin Wu. Conceptualization, supervision and reviewing manuscript were performed by Libin Zhou. The first draft of the manuscript was written by Lijuan Zeng and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Special Fund for High-Level University Construction Talents of Guangzhou Medical University (Guangzhou, Guangdong Province, China: B185006003006).
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by The Medical Ethics Committee of Affiliated Stomatology Hospital of Guangzhou Medical University (JCYJ2024027).
Consent for publication
All healthy volunteers have understood and signed the informed consent form for donating peripheral venous blood to isolate PBMCs.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6(1):92. 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–53. 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Cohen EEW, Bell RB, Bifulco CB, et al. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J Immunother Cancer. 2019;7(1):184. 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–65. 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 5.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105(39):15064–9. 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma R, Jing C, Zhang Y, et al. The somatic mutation landscape of Chinese colorectal cancer. J Cancer. 2020;11(5):1038–46. 10.7150/jca.37017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrencrona E, van der Post S, Gallego P, et al. The IgGFc-binding protein FCGBP is secreted with all GDPH sequences cleaved but maintained by interfragment disulfide bonds. J Biol Chem. 2021;297(1): 100871. 10.1016/j.jbc.2021.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Shi J, Zhou Y, Liu T, Zhan F, Zhang K, Liu N. Integrating proteomics and transcriptomics for the identification of potential targets in early colorectal cancer. Int J Oncol. 2019;55(2):439–50. 10.3892/ijo.2019.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Liu Y, Liu H, Zhang Q, Song H, Tang J, Fu J, Wang X, et al. FcGBP was upregulated by HPV infection and correlated to longer survival time of HNSCC patients. Oncotarget. 2017;8(49):86503–14. 10.18632/oncotarget.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YH, Yang YF, Shiue YL. Multi-omics analyses to identify FCGBP as a potential predictor in head and neck squamous cell carcinoma. Diagnostics (Basel). 2022;12(5):1178. 10.3390/diagnostics12051178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi LH, Wu ATH, Hsiao M, Li YJ. A transcriptomic analysis of head and neck squamous cell carcinomas for prognostic indications. J Pers Med. 2021;11(8):782. 10.3390/jpm11080782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Huang S, Zeng L, et al. Necroptosis in head and neck squamous cell carcinoma: characterization of clinicopathological relevance and in vitro cell model. Cell Death Dis. 2020;11(5):391. 10.1038/s41419-020-2538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Guan C, Shang X, et al. A bioinformatic analysis: the overexpression and clinical significance of FCGBP in ovarian cancer. Aging. 2021;13(5):7416–29. 10.18632/aging.202601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan Q, Zhang H, Zheng J, Zhang L. Turning cold into hot: firing up the tumor microenvironment. Trends Cancer. 2020;6(7):605–18. 10.1016/j.trecan.2020.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18(3):197–218. 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Zhang W, Zhang J. Comprehensive analysis of immune cell infiltration and significant genes in head and neck squamous cell carcinoma. Oral Oncol. 2022;126: 105755. 10.1016/j.oraloncology.2022.105755. [DOI] [PubMed] [Google Scholar]
- 17.de Medeiros MC, Liu M, Banerjee R, Bellile E, D’Silva NJ, Rossa C Jr. Galanin mediates tumor-induced immunosuppression in head and neck squamous cell carcinoma. Cell Oncol (Dordr). 2022;45(2):241–56. 10.1007/s13402-021-00631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiang W, Dai Y, Xing X, Sun X. Identification of a metabolic reprogramming-related signature associated with prognosis and immune microenvironment of head and neck squamous cell carcinoma by in silico analysis. Cancer Med. 2022;11(16):3168–81. 10.1002/cam4.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forster MD, Devlin MJ. Immune Checkpoint Inhibition in Head and Neck Cancer. Front Oncol. 2018;8:310. 10.3389/fonc.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong L, Wen Y, Miao X, Yang Z. NT5E and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal transition (EMT) are associated with tumor progression and survival of patients with gallbladder cancer. Cell Tissue Res. 2014;355(2):365–74. 10.1007/s00441-013-1752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang Q, Shen A, Liu L, et al. Prognostic and immunological roles of Fc fragment of IgG binding protein in colorectal cancer. Oncol Lett. 2021;22(1):526. 10.3892/ol.2021.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houben T, Harder S, Schlüter H, Kalbacher H, Hoffmann W. Different forms of TFF3 in the human saliva: heterodimerization with IgG Fc binding protein (FCGBP). Int J Mol Sci. 2019;20(20):5000. 10.3390/ijms20205000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weste J, Houben T, Harder S, et al. Different molecular forms of TFF3 in the human respiratory tract: heterodimerization with IgG Fc binding protein (FCGBP) and proteolytic cleavage in bronchial secretions. Int J Mol Sci. 2022;23(23):15359. 10.3390/ijms232315359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albert TK, Laubinger W, Müller S, et al. Human intestinal TFF3 forms disulfide-linked heteromers with the mucus-associated FCGBP protein and is released by hydrogen sulfide. J Proteome Res. 2010;9(6):3108–17. 10.1021/pr100020c. [DOI] [PubMed] [Google Scholar]
- 25.Laskou A, Znalesniak EB, Harder S, et al. Different forms of TFF3 in the human endocervix, including a complex with IgG Fc binding protein (FCGBP), and further aspects of the cervico-vaginal innate immune barrier. Int J Mol Sci. 2024;25(4):2287. 10.3390/ijms25042287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang JJ, Zhang Y, Chen Q, et al. A novel prognostic marker and therapeutic target associated with glioma progression in a tumor immune microenvironment. J Inflamm Res. 2023;16:895–916. 10.2147/JIR.S398775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan T, Tian D, Chen J, et al. FCGBP is a prognostic biomarker and associated with immune infiltration in glioma. Front Oncol. 2022;11: 769033. 10.3389/fonc.2021.769033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Q, Lin F, Huang Z, et al. Non-coding RNA-related FCGBP downregulation in head and neck squamous cell carcinoma: a novel biomarker for predicting paclitaxel resistance and immunosuppressive microenvironment. Sci Rep. 2024;14(1):4426. 10.1038/s41598-024-55210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi K, Ogata H, Morikawa M, et al. Distribution and partial characterisation of IgG Fc binding protein in various mucin producing cells and body fluids. Gut. 2002;51(2):169–76. 10.1136/gut.51.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazi MH, He M, Cheville JC, Young CY. Downregulation of IgG Fc binding protein (Fc gammaBP) in prostate cancer. Cancer Biol Ther. 2008;7(1):70–5. 10.4161/cbt.7.1.5131. [DOI] [PubMed] [Google Scholar]
- 31.Dong S, Huo H, Mao Y, Li X, Dong L. A risk score model for the prediction of osteosarcoma metastasis. FEBS Open Bio. 2019;9(3):519–26. 10.1002/2211-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi Y, Ueyama A, Funaki S, et al. In situ analysis of CCR8+ regulatory T cells in lung cancer: suppression of GzmB+ CD8+ T cells and prognostic marker implications. BMC Cancer. 2024;24(1):627. 10.1186/s12885-024-12363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–68. 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen T, Zhang J, Wang Y, Liu B. FCGBP is a promising prognostic biomarker and correlates with immunotherapy efficacy in oral squamous cell carcinoma. J Immunol Res. 2022;2022:8443392. 10.1155/2022/8443392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.