Abstract
Background
Hyperactive Hedgehog (Hh) signaling initiates and drives the progression of various tumors. Despite the clinical success of Hh inhibitors targeting Smoothened (SMO), drug resistance, often stemming from SMO mutations, remains a formidable obstacle in cancer therapy. Here, we investigated the potential of imperatorin (IMP), a Chinese herbal medicine, to overcome drug resistance and revealed the potential mechanisms.
Methods
The effect of IMP on Hh signaling pathway was evaluated via Quantitative reverse transcription-polymerase chain reaction, Dual-luciferase reporter assay and Western blot. Meanwhile, we tested its ani-proliferative potential on Hh-driven tumor cells. Loss/gain-of-function, network pharmacology analysis, RNA-sequence analysis and molecular docking were performed to investigate the potential mechanisms of IMP-mediated functions. Furthermore, we established a subcutaneous Hh-driven medulloblastoma xenograft model using the DAOY cell line and examined the in vivo therapeutic efficacy of IMP.
Results
We identified IMP as a novel Hh inhibitor capable of overcoming drug-resistance caused by SMO mutants by inhibiting downstream transcription factor GLI1. IMP suppressed the proliferation of Hh-dependent cancer cells along with Hh activity inhibition. Mechanistically, IMP attenuated the phosphorylation of signal transducer and activator of transcription 3 (STAT3) and its interaction with GLI1 promoter, consequently blocking GLI1 transcription and the target gene expressions. Molecular docking analysis revealed the favorable binding affinity between IMP and STAT3. Importantly, IMP application effectively inhibited the growth of medulloblastoma in vivo, accompanied by the downregulation of GLI1 and phosphorylated STAT3.
Conclusion
Our findings revealed IMP as an innovative approach to combat the drug resistance of SMO inhibitors in Hh-driven tumors, highlighting the crucial role of STAT3 as a transcriptional regulator in Hh signaling.
Keywords: Hedgehog, resistance, GLI1, imperatorin, STAT3, medulloblastoma
Introduction
The Hedgehog (Hh) signaling pathway orchestrates critical processes in embryonic patterning, tissue homeostasis, and regeneration.1,2 However, aberrantly activated Hh signaling is implicated in various aspects of tumor development and progression, including tumor proliferation, differentiation, angiogenesis, metastasis, tumor resistance and immunity.3–6 Therefore, the inhibition of hyperactive Hh represents an enticing therapeutic strategy in cancers.2,7–9
In mammals, the canonical Hh signaling is triggered by secreted Hh ligands (Sonic Hedgehog (Shh), Desert Hedgehog (Dhh), and Indian Hedgehog (Ihh)), which directly bind to 12-pass transmembrane receptor patched 1 (PTCH1),10,11 enabling the ciliary translocation and activation of smoothened (SMO), a 7-pass cell surface protein.10,12–14 Once activated, SMO activates the downstream glioma-associated oncogene (GLI) transcription factors (GLI1, GLI2, and GLI3) to translocate to the nucleus, where GLIs transactivate the expression of Hh target genes, such as Gli1, Ptch1 and Myc. GLI1 also functions as an amplifier of the Hh signals through a positive feedback loop mechanism, promoting the transcription of GLI2.15 In the absence of Hh ligand, PTCH suppresses SMO activity and downstream signals. This finely tuned regulatory mechanism ensures precise control of Hh pathway activity in response to extracellular cues.
Dysregulated Hh signaling activity acts as a significant player in tumor development.3–6,16 Activation mutations in key Hh pathway components, such as PTCH, SMO, and GLI1 are pivotal in fueling tumor initiation and progression, notably observed in medulloblastoma (MB), basal cell carcinoma (BCC), and rhabdomyosarcoma.17–19 Almost all sporadic BCC and more than 30% cases of MB are driven by Hh mutational activation. In addition to the tumor cells, hyperactive Hh pathway activity has also been reported in tumor stromal cells, wherein nonneoplastic cells in tumor microenvironment receive Hh ligands from tumor cells and subsequently secrete mediators, such as IL-6, MMP and TGF-β to further facilitate tumor growth.20–23 Such mechanism is widely reported in numerous tumors, such as pancreatic ductal adenocarcinoma, bladder cancer and cholangiocarcinoma.20–23 This intricate interplay underscores the urgency in unraveling the regulatory mechanisms of Hh signaling and developing targeted antagonists to counteract its pro-tumor effects.
SMO is the predominant pharmacological target of the Hh signaling pathway. Currently, three inhibitors against SMO are on the market, namely vismodegib (GDC-0449, GDC),24 sonidegib25 and glasdegib,26 for tumor therapy. However, the emerging resistances pose significant treatment challenges. The resistance of SMO inhibitors could derive from SMO point mutations, loss-of-function mutations in SUFU, which negatively regulate GLIs, or GLI1 and GLI2 amplifications.27–29 In addition, GLI expression can also be induced through non-classical pathways or other bypass pathways in multiple tumors.30–33 Our group has revealed that prostaglandin E2 (PGE2)-JNK signaling axis promoted GLI2 expression by inhibiting GLI2 ubiquitination degradation in a SMO-independent manner in colorectal cancer.34 Unfortunately, there are no drugs that can overcome the resistance of SMO inhibitors in clinics, prompting the development of alternative strategies.
Inhibition of GLI has currently emerged as a promising therapeutic strategy, with the potential to break the resistance of SMO antagonists.35 Meanwhile, as discussed above, multiple tumors harbor hyperactivated GLI activity in a SMO-independent mechanism, as exemplified by noncanonical activation of GLI1 in prostate, breast lung, and colon cancers.32,36–38 Hence, inhibitors against GLI would display wider therapeutic spectrum. Arsenic trioxide that targets both GLI1 and GLI2 has shown therapeutic efficacy on MB and drug refractory MB xenograft in mice,39–41 and has entered clinical studies, however, adverse side effects and modest overall activity limit its clinic use.42
Imperatorin (IMP) is a natural product isolated from angelica dahurica and possesses diverse pharmacological activities that have garnered significant attention, particularly for its anti-tumor effects.43–45 It has been reported that IMP overcame chemotherapy resistance by inducing MCL-1 degradation in liver cancer and inhibited tumor metastasis by CREB1-TGF-β 2/ERK signaling axis in esophageal squamous cell carcinoma.43,46 However, it is still unknown whether IMP can suppress Hh signaling pathway and the tumors with hyperactive Hh. In this study, we revealed that IMP inhibited Hh signal at the level of GLI and overcame the resistance of SMO inhibitors. Mechanistically, IMP suppressed GLI1 transcription via STAT3. Additionally, IMP effectively delayed the growth of MB xenografts in vivo. These results have significant implications for the development of alternative strategies to overcome the resistance of current SMO inhibitors and provide novel insights into Hh pathway modulation.
Materials and Methods
Cells and Cell Culture
Light II was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). NIH3T3, DAOY and 293T cells were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Light II cells stably expressing 8 × 3 Gli1-firefly luciferase and TK-Renilla reporters were cultured in DMEM supplemented with 10% FBS (Gibco, USA), 0.4 mg/mL G418 (Selleck Chemicals, USA) and Zeocin 0.15 mg/mL (Gibco, USA) as previously described.47 NIH3T3, DAOY and 293T cells were cultured in DMEM supplemented with 10% FBS. All cells were grown at 37 °C under 5% CO2 in a humidified incubator.
Conditioned Medium Preparation
The Shh was collected as previously reported.47 Briefly, 293T cells were plated in 10 cm dishes in 10 mL of DMEM containing 10% FBS with a density of 2.1 × 106 cells/dish. After 24 hours, the culture medium was switched to 10 mL of DMEM containing 0.5% FBS, and the cells were transfected with ShhN plasmid (#37680, Addgene, USA), the active form of Shh,47 using Lipofectamine 3000 (#L3000008, Invitrogen, USA) according to the manufacturer’s protocol. Eight hours after transfection, the cells were washed with PBS and fresh medium supplemented with 10% FBS was added for an additional 16 hours. Subsequently, cells were incubated in 10 mL of 0.5% FBS medium for 48 hours and the cell supernatant was collected. The cell supernatant was referred to as Shh, which was either used for subsequent experiments or stored at −80 °C immediately.
Dual-Luciferase Reporter Assay
Light II cells were seeded at a density of 8000 cells/well into 96-well plates. After attachment, culture medium was changed to starvation medium (0.5% FBS), and the cells were treated with various compounds in the presence or absence of Hh agonists (Shh or SAG) for 36 h. Then, the activities of firefly and Renilla luciferase were monitored using a luminometer according to the Dual-Luciferase Reporter Assay kit protocol (#E1960, Promega, USA). The firefly luciferase signal was normalized by TK-Renilla luciferase signal. The IC50 values were obtained via a nonlinear regression analysis at least triplicates.
Cell Viability Analysis
Cells were seeded into 96-well plates at the density of 5000 cells/well. Once the cell confluence reached approximately 70%, cells were treated with IMP as indicated for 24, 48 or 72 hours. Subsequently, the CCK-8 reagent (#C0037, Beyotime, China) was added into cells, and optical density (OD) values at 450 nm were examined using a Tecan plate reader after 2 hours of incubation.
RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (#15596026, Invitrogen, USA) as the manufacturer’s instructions. The Reverse Transcription Kit (#R323-01, Vazyme, China) was employed to synthesize complementary DNA (cDNA). Subsequently, qRT-PCR was performed using SYBR Green (#Q711-02, Vazyme, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene. The relative gene expression was calculated using 2−ΔΔCt method. The primer sequences were as follows: mouse Gli1 Forward: 5’-GCAGTGGGTAACATGAGTGTCT-3’, Reverse: 5’-AGGCACTAGAGTTGAGGAATTGT-3’; mouse Ptch1 Forward: 5’-AAAGAACTGCGGCAAGTTTTTG-3’, Reverse: 5’- CTTCTCCTATCTTCTGACGGGT-3’; mouse GAPDH Forward: 5’- CATGTTTGTGATGGGTGTGA-3’, Reverse: 5’- AATGCCAAAGTTGTCATGGA-3’; human GLI1 Forward: 5’- GTTCACATGCGCAGACACACT-3’, Reverse: 5’-TTCGAGGCGTGAGTATGACTTC; human PTCH1 Forward: 5’- CCACAGAAGCGCTCCTACA-3’, Reverse: 5’- CTGTAATTTCGCCCCTTCC-3’; human GAPDH Forward: 5’-GGCAAATTCCATGGCACCG-3’, Reverse: 5’- ATGACGAACATGGGGGCATC-3’.
Lenti-Virus Infection
shRNA lentiviral construct against SUFU was obtained from Santa Cruz (Santa Cruz, USA). To generate SUFU lentiviruses, 293T cells were transfected with SUFU shRNA, envelop (VSV-G) and packaging (pDelta 8.9) plasmids using Lipofectamine 3000 (#L3000015, Invitrogen, USA). After 48 hours of transfection, the cell supernatant was collected after centrifugation, which was either used immediately or stored at −80 °C. For virus infection, Light II cells were incubated with SUFU lentivirus for 8 hours. Subsequently, the cell culture was switched to normal culture medium and cultured for 40 hours, followed by selection with puromycin for 72 hours. The SUFU knockdown efficacy was examined using Western blot analysis.
Western Blot
Total protein was extracted using RIPA buffer (#P0013K, Beyotime, China) containing PMSF (#ST506, Beyotime, China) and cocktail inhibitors (P8340, Sigma, USA). We used BCA assay kit (#P0010, Beyotime, China) to quantify the protein contents. About 5–10 μg of protein was separated with SDS-PAGE gel and transferred to the polyvinylidene fluoride (PVDF, Pall, USA) membrane. Subsequently, it was blocked with 5% of skimmed milk for 45 minutes at room temperature. Thereafter, the membrane was treated with primary antibodies at 4° C overnight. After washing, the membrane was treated with corresponding secondary antibodies for 45 minutes. Finally, the protein bands were examined using ECL UltraPlus kit (#WBULP, Millipore, USA). The following antibodies were used: SUFU (#26118, CST, USA), GLI1 (#2643, CST, USA), STAT3 (#9139S, CST, USA), P-STAT3 (#9145S, CST, USA), Flag (#14793, CST, USA), β-Actin (#20536-1-AP, proteintech, USA), GAPDH (#60004-1-ig, proteintech, USA).
Molecular Docking
The cocrystal structure of STAT3 was derived from the Protein Data Bank (1BGl). The chemical structure of IMP was retrieved from PubChem database. Prior to docking, AutoDock Tools were applied to prepare the receptor protein and ligand, including the removal of excess structures and water molecules. Molecular docking was performed using AutoDock v1.5.7 software.48 PyMOL software was used to analyze the docking results and output the picture.
Network Pharmacology Analysis
We built the chemical structural formula of IMP via the pubChem database. The Swiss Target Prediction (http://www.swisstargetprediction.ch) and SEA were employed to predict IMP targets. GeneCards (https://www.genecards.org/) and OMIM (https://www.omim.org/) were employed to predict the disease targets of MB. Venny platform (https://bioinfogp.cnb.csic.es/tools/venny/) was used to obtain the intersected genes between IMP and MB. The search tool for retrieval of interacting genes (STRING) (https://string-db.org/) database was used to obtain protein–protein interaction (PPI) network by choosing the highest confidence of 0.9.49 Cytoscape3.9.2 was used to calculate the PPI node degree.50 Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed with the R package.
Xenograft Tumor Models
All in vivo studies were approved by Institutional Animal Care and Use Committee of Shanghai University and performed following China Animal Welfare Legislation. Ethical Approval Number: No.2019–0020. Six-week-old female nude mice were purchased from Shanghai Model Organisms (Shanghai, China). 5×106 DAOY cells in PBS were mixed with Matrigel (1:1) and immediately injected into the right flank of nude mice. When the average tumor volumes reached 50 mm3, mice were separated into two groups randomly (n = 5 mice per group) and orally administrated either with vehicle or IMP (50 mg per kg body weight) once daily for 38 days consecutively. At the end of the experiment, tumor samples were collected for subsequent examinations. Tumor volume was measured with calipers once every three days and calculated as follows: tumor volume=1/2 (length × width2).
Morphometric Analysis and Immuno-Staining
Tumor samples were subjected to 4% paraformaldehyde (PFA), dehydrated and then embedded into paraffin wax for sectioning. For the hematoxylin and eosin (H&E) staining, the sections were stained with H&E as reported procedures.34 For Ki67 staining, the sections were treated with Ki67 (#GB111499, 1:400, Servicebio, China) overnight at 4°C, following by detection with DAB kit. Image J software was used to count the Ki67 positive cells.
Statistical Analysis
Data were shown as mean ±SD from at least 3 independent experiments. Two-way ANOVA was used for comparing tumor growth curves. Student’s t test was used for comparisons between two different groups. P < 0.05 was considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001; # P > 0.05).
Results
IMP Antagonizes Hh Signaling
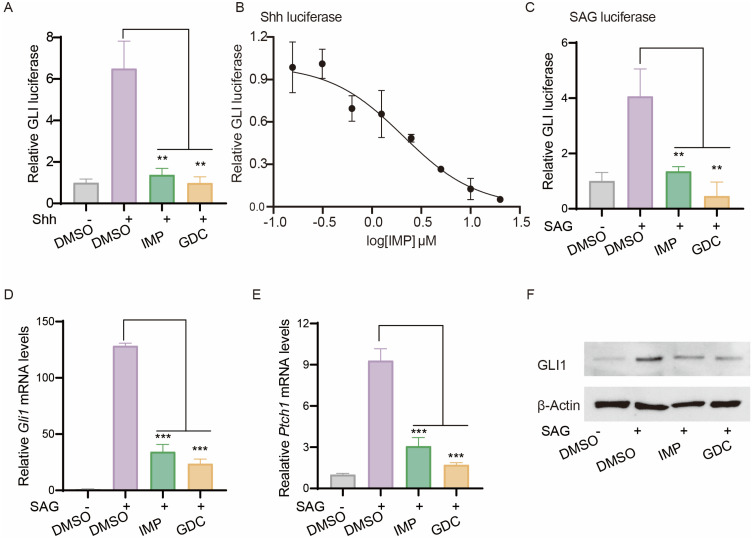
We examined the effect of IMP on Hh signaling pathway using the classic Light II cell model, a NIH3T3 reporter cell line stably expressing GLI-firefly luciferase that mirrors GLI dependent transcriptional activity, as well as TK-Renilla luciferase that functions as an internal control.34,47 Treatment of Light II cells with Shh conditional medium (referred as Shh) induced GLI-luciferase activity, which was significantly inhibited by IMP and GDC (Figure 1A). IMP exhibited a dose-dependent inhibition of Shh-induced pathway activity, with an IC50 value around 2 μM (Figure 1B). Moreover, we found that, in line with GDC, IMP effectively suppressed pathway activity induced by SAG, an agonist of SMO,51 as demonstrated by decreased GLI-luciferase (Figure 1C), as well as downregulated levels of Hh target genes Gli1 and Ptch1 (Figure 1D and E), and GLI1 protein (Figure 1F) in Light II cells. Notably, IMP treatment at concentrations up to 20 μM did not significantly affect the viability of Light II cells (Supplementary Figure S1), ruling out the cytotoxicity as a factor to dampen Hh signaling. Altogether, these findings confirm the inhibitory effect of IMP on Hh signaling pathway.
Figure 1.
IMP antagonizes Hh signaling. (A) GLI-luciferase reporter activity in Light II cells treated with Shh in the presence of IMP (10 μM), GDC (1μM) or DMSO for 36 h. (B) Dose-response inhibition of GLI-luciferase activity by IMP in Light II cells. (C) GLI-luciferase reporter activity of Light II cells treated with SAG (100 nM) in the presence of IMP (10 μM), GDC (1μM) or DMSO for 36 h. (D and E) qRT-PCR analysis of Gli1 and Ptch1 mRNA expressions in Light II cells treated with SAG (100 nM) in the presence of IMP (10 μM), GDC (1μM) or DMSO for 24 h. (F) Western blot analysis of GLI1 expression in Light II cells treated with SAG in the presence of IMP (10 μM), GDC (1μM) or DMSO for 24 h. The β-Actin was shown as a loading control. All error bars represent mean ± s.d. Experiments were repeated at least three times. Statistical significance was calculated using Student t-test, (**P <0.01, ***P <0.001).
IMP Suppresses Cell Proliferation of Hh-Driven Medulloblastoma
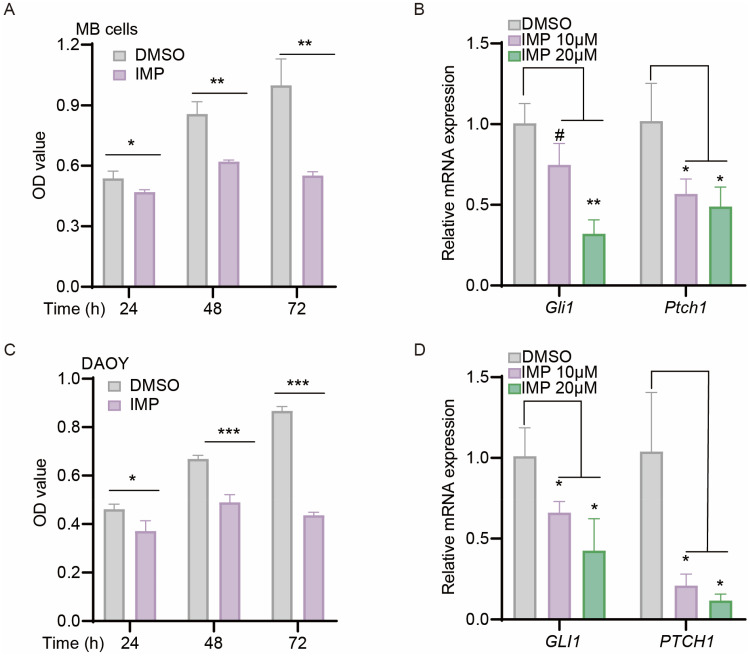
Since hyperactive Hh signal is a critical driver in cell proliferation and tumorigenesis, we hypothesized that the observed inhibitory effect on pathway could translate into an inhibition on cell biology. Accordingly, we examined the effects of IMP on Hh-driven Shh subtype MB cells using primary mouse MB cells isolated from autochthonous MB arising in Ptch+/−; Trp53−/− mice,47 and human DAOY cell lines.52 As expected, IMP treatment inhibited the cell proliferation, and decreased the expressions of mouse Gli1 and Ptch1 mRNA as well as human GLI1 and PTCH1 mRNA in MB cells and DAOY, respectively (Figure 2A-D), indicating that IMP has the ability to suppress MB proliferation by inhibiting Hh signaling pathway.
Figure 2.
IMP inhibits cell viability and Hh pathway in Hh-driven medulloblastoma. (A and C) CCK-8 analysis of primary Ptch+/−; Trp53−/− mutated MB cells (A), human DAOY cells (C) administrated with IMP (20 μM) or DMSO at indicated time. (B and D) qRT-PCR analysis of Gli1 and Ptch1 mRNA expressions in primary Ptch+/−; Trp53−/− mutated MB cells (B), and GLI1 and PTCH1 mRNA in human DAOY cells (D) treated with IMP (10 μM, 20μM) or DMSO for 24 h. All error bars represent mean ± s.d. All experiments were repeated at least three times. Statistical significance was calculated using Student t test, #P>0.05, *P <0.05, **P <0.01, ***P <0.001.
IMP Inhibits Hh Signal at the Level of GLI
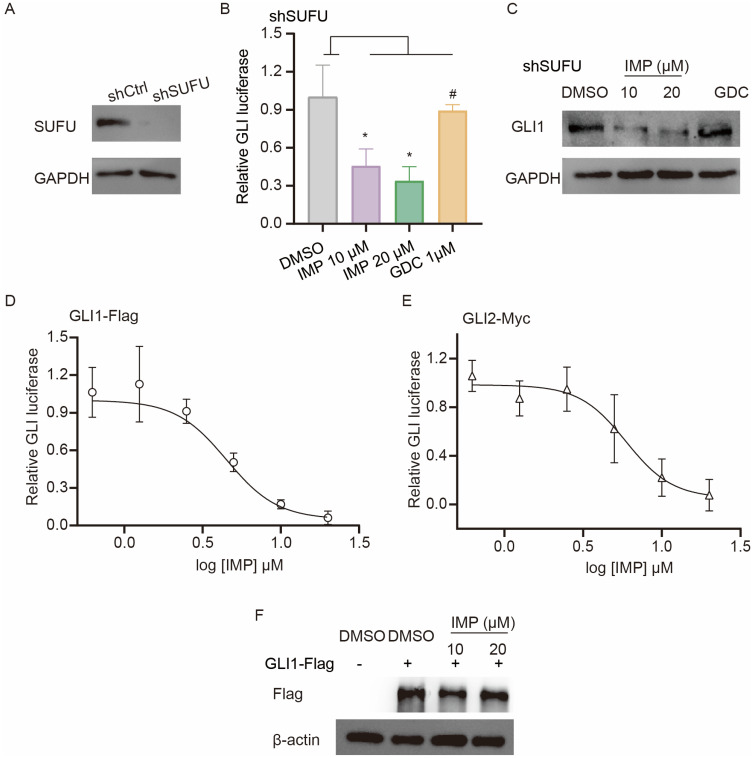
We next sought to investigate the molecular mechanisms of how IMP inhibited Hh signal. SUFU is a negative regulator of GLI.53 Therefore, we first established a SUFU knockdown cell line (shSUFU) using Light II cells to address the contribution of SUFU to IMP-mediated Hh regulation (Figure 3A). As reported, shSUFU cells exhibited increased Hh transcriptional activity in comparison with negative control (shCtrl), as shown by augmented GLI-luciferase reporter activity (Supplementary Figure S2A), and this effect was not influenced by 1 μM GDC (Figure 3B), although GDC showed significant inhibition at this concentration in response to SAG or Shh-induced GLI transcription activity (Figure 1). In contrast, IMP displayed remarkable inhibition on GLI-luciferase activity and GLI1 protein expression in shSUFU cells (Figure 3B and C), hinting that IMP likely targeted downstream of SUFU.
Figure 3.
IMP inhibits Hh signal at the level of GLI. (A) Western blot analysis of SUFU expression in wild-type (shCtrl) and SUFU-knockdown (shSUFU) Light II cells. The GAPDH was shown as a loading control. (B) GLI-luciferase reporter activity in shSUFU Light II cells treated with IMP (10 μM, 20 μM) or GDC (1μM) for 36 h. (C) Western blot analysis of GLI1 expression in shSUFU-Light II cells treated with IMP (10 μM, 20 μM) or GDC (1μM) for 24 h. The GAPDH was shown as a loading control. (D and E) Dose-response inhibition of GLI-luciferase activity by IMP in Light II cells with overexpression of GLI1-Flag (D) or GLI2-Myc (E). (F) Western blot analysis of Flag expression in Light II cells with overexpression of GLI1-Flag. The β-actin was shown as a loading control. All experiments were repeated at least three times. Statistical significance was calculated using Student t test, #P>0.05, *P <0.05.
Given that GLI transcription factors are downstream effectors of SUFU, we therefore probed IMP-mediated effect on pathway activity induced by overexpression of Flag-tagged GLI1 (GLI1-Flag) or full-length Myc-tagged GLI2 (GLI2-Myc). We found that such cells displayed activated GLI reporter activity, and IMP suppressed this activity to a considerably lower basal level (Figure 3D and E). Consistently, the overexpression of full-length GLI2-Myc or GLI2 detaN, a truncated active form of GLI2,54 induced Gli1 mRNA expression, which was remarkably blocked by IMP, but not GDC (Supplementary Figure S2B and C). These findings suggested that IMP inhibited Hh signal at the level of GLI. GLI1 and GLI2 are highly homologous; thus, most reported GLI1 inhibitors also target GLI2.54–57 Moreover, given the importance of GLI1 in tumorigenesis and tumor progression,58 we then mainly focus on the effect of IMP on GLI1. Interestingly, we did not observe any inhibition in exogenous Flag-GLI1 protein expression driven by CMV promoter (Figure 3F), whereas downregulated endogenous GLI1 protein was observed in IMP-treated cells (Figure 1F). Therefore, we reasoned that the anti-Hh effect of IMP may be associated with GLI1 promoter, thus negatively modulating GLI1 transcription.
Exploring IMP Targets Through Network Pharmacology
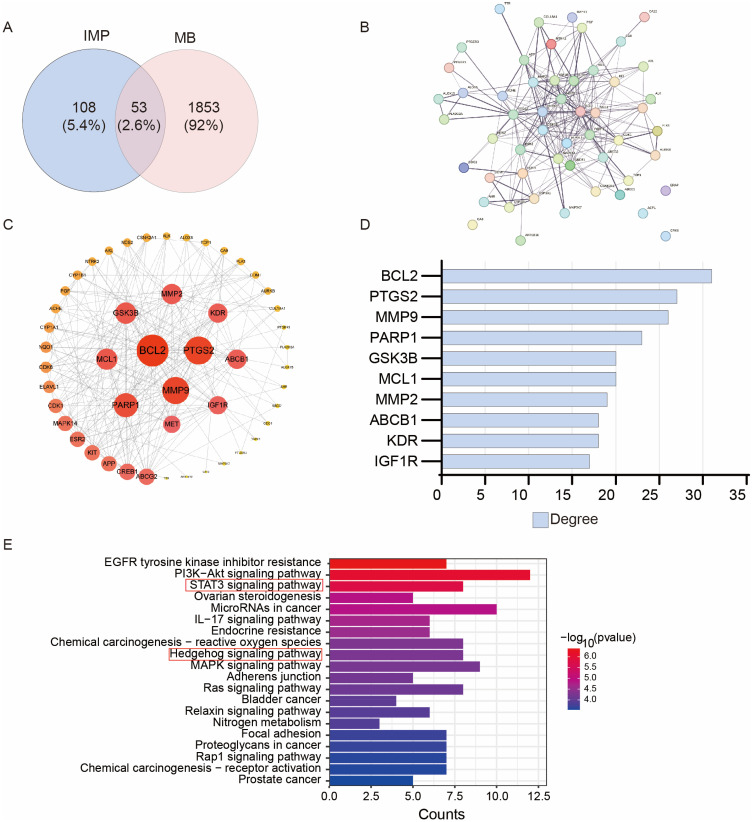
The above findings suggested that IMP indirectly inhibits GLI by modulating its transcription. Therefore, we next investigated the possible targets of IMP via network pharmacology analysis. In this case, we selected MB as the disease modality. Swiss Target Prediction and SEA database prediction identified 161 possible targets of IMP. GeneCards and OMIM databases generated 1906 possible targets of MB. Furthermore, Venny analysis produced 53 intersected targets between IMP and MB, accounting for 2.6% (Figure 4A). In order to develop the connections among these genes (PPI networks), STRING was used (Figure 4B). Subsequently, cytoscape 3.9.2 analysis was used to visualize corresponding node degree, resulting in top 10 key genes in the network (Figure 4C and D). Among those, the BCL2, MMP2, MMP9, MCL1 are the target genes of both STAT3 and GLI.59,60 Similarly, KEGG analysis demonstrated that STAT3 and Hh signaling pathways were enriched in the intersection targets (Figure 4E). These findings demonstrate that the STAT3 pathway is highly associated with IMP functions on Hh-dependent tumors.
Figure 4.
Network pharmacology investigation of IMP targets. (A) Analysis of the targets related to IMP and MB. (B) PPI network based on the intersected proteins. (C and D) Cytoscape analysis the node degree of PPI network. (D) The degree top 10 genes in Cytoscape analysis. (E) KEGG analysis of the intersected genes.
IMP Inhibits GLI1 Transcription Through STAT3
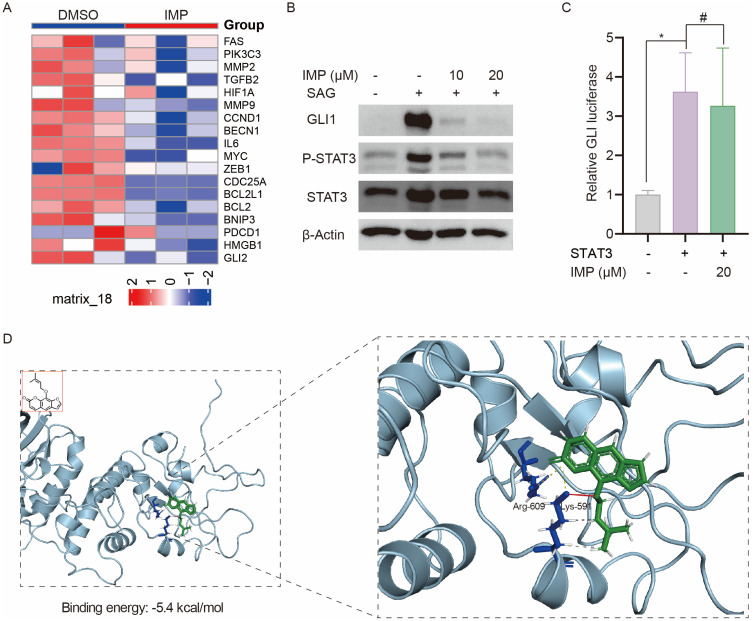
STAT3 is a transcription factor that regulates the transcription of multiple oncogenic genes.61 Since IMP inhibited the transcription of GLI1, we sought to investigate whether STAT3 serves as a key mediator of IMP’s inhibitory effect on GLI1. 143B is an osteosarcoma cell line with hyperactive Hh and STAT3.62,63 Analysis of the GEO database (GSE180321)64 revealed that IMP treatment resulted in decreased Hh and STAT3 activity in these cells, as evidenced by reduced GLI2 expression and downregulation of multiple STAT3 target genes (Figure 5A). In response to external stimuli, STAT3 is phosphorylated and activated to exert its functions.61 Interestingly, Western blot analysis showed that IMP remarkably inhibited GLI1 and the phosphorylation of STAT3 in Light II cells (Figure 5B). In addition, IMP inhibited SUFU knockdown induced GLI-luciferase activity; however, this effect was reversed by STAT3 overexpression in shSUFU cells (Figure 5C), indicative of the critical role of STAT3 in the regulation of GLI1 transcription by IMP. Previous studies have shown that STAT3 directly binds and activates GLI1 promoter in fibrocytes and chronic lymphocytic leukemia cells.65,66 Indeed, using JASPAR, we identified multiple putative-binding sites for STAT3 on GLI1 proximal promoter (Supplementary Figure S3A). In our study, we also observed STAT3 activation with IL-6 promoted GLI-luciferase activity and this effect was suppressed by IMP in DAOY cells (Supplementary Figure S3B). These data suggest that STAT3 is a potential transcriptional factor of GLI1, and IMP inhibits GLI1 transcription and expression via STAT3.
Figure 5.
IMP inhibits GLI1 transcription through STAT3. (A) Heatmap of GLI2 and STAT3 target genes in 143B cells treated with or without IMP from GEO database (GSE180321). (B) Western blot analysis of GLI1, P-STAT3 and STAT3 expressions in Light II cells treated with SAG (100 nM) with or without IMP (10 μM, 20 μM) treatment for 24 h. The β-actin was shown as a loading control. (C) GLI-luciferase activity in shSUFU-Light II cells with or without STAT3 overexpression (100 ng) in the presence or absence of IMP (20 μM). (D) Molecular docking of the binding model between IMP and STAT3 (PDB: 1BG1). All experiments were repeated at least three times. Statistical significance was calculated using Student t test, #P>0.05, *P <0.05.
Next, we probed the possibility of IMP directly targeting STAT3 via bioinformatics analysis. Molecular docking was employed and revealed that the binding energy between IMP and STAT3 was −5.4 kcal/mol (Figure 5D). Given that the binding energy below −5 kcal/mol denotes strong binding affinity, the observed results indicated that IMP was able to bind with STAT3. In addition, similar to previous reports,67 salt bridges were formed between IMP and residue Arg-609, Lys-591 on STAT3 at distances of 3.57 Å and 3.98 Å, respectively. In particular, Lys-591 displayed the hydrogen bond and hydrophobic interactions with IMP, which provided strong van der Waals forces (Figure 5D). Altogether, these findings demonstrate that IMP has the potential to directly bind with STAT3 and thus inhibit the transcription of GLI1.
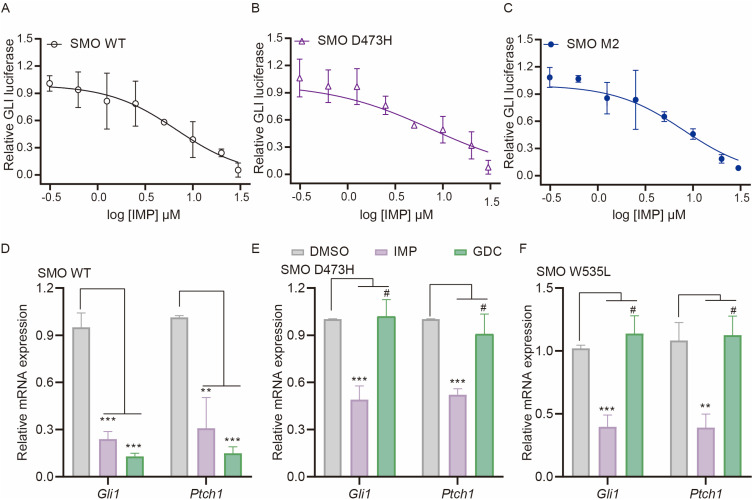
IMP Inhibits Drug-Resistant SMO Mutants
Having revealed that IMP inhibited Hh signaling by targeting at the level of GLI, we investigated whether IMP could overcome the resistance of SMO inhibitors induced by SMO mutations. We examined its effect on Hh activity induced by SMO D473H and SMO M2 (also known as SMO W535L), both of which contribute to the resistance to GDC in clinic.28,68,69 As expected, IMP inhibited SMO D473H and SMO M2-induced GLI-luciferase in a dose-dependent manner with the IC50 value around 6–8 μM, which was comparable to the potency against SMO WT and Shh-induced activity (Figures 6A-C and 1B). In this case, the decrease in Gli1 and Ptch1 gene expressions was also observed after IMP treatment, whereas no inhibition was shown after GDC treatment (Figure 6D-F). These findings suggest that IMP has the potential to overcome the drug-resistance caused by SMO mutations.
Figure 6.
IMP inhibits drug-resistant SMO mutants. (A-C) GLI-luciferase reporter activity in Light II cells with overexpression of SMO WT (A), SMO D473H (B) or SMO W535L (C) in the presence of IMP as indicated concentrations for 36 h. (D-F) qRT-PCR analysis of Gli1 and Ptch1 mRNA expressions in Light II cells with overexpression of SMO WT (D), SMO D473H (E) or SMO W535L (F) in the presence of IMP (20 μM) or GDC (1μM) for 36 h. All error bars represent mean ± s.d. All experiments were repeated at least three times. Statistical significance was calculated using Student t test, #P>0.05, **P <0.01, ***P <0.001.
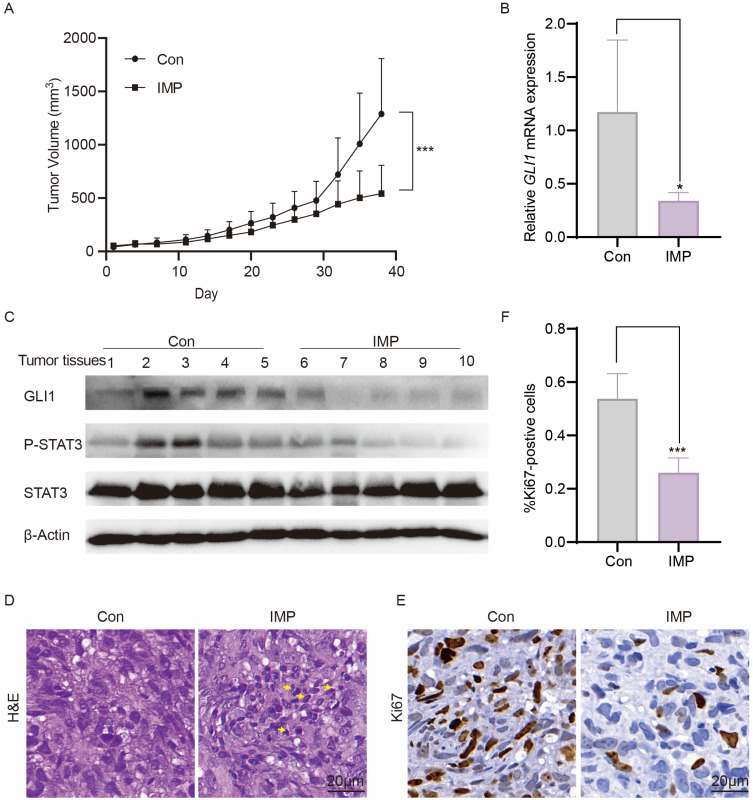
IMP Impairs the Growth of Shh-Medulloblastoma in vivo
The therapeutic efficacy of IMP was evaluated in vivo using xenograft model DAOY, a widely used human Shh-MB model for Hh drug screening.52,70 We injected DAOY cells subcutaneously into the flank of nude mice. The mice were orally administrated with either IMP (50 mg/kg) or vehicle control (Con) once a day when the tumor volumes reached around 50 mm3. We observed an obvious delay of tumor growth after IMP treatment (Figure 7A). In line with the inhibitory effect on GLI1 transcription and STAT3 phosphorylation in vitro, IMP also diminished the expression of GLI1 mRNA, as well as the levels of GLI1 and phosphorylated STAT3 proteins in tumor samples (Figure 7B and C). Furthermore, the IMP treatment increased necrosis (yellow arrowhead) as shown by H&E staining (Figure 7D). Ki67 staining revealed a reduction in Ki67-positive proliferating cells in IMP-treated tumors compared to control (Figure 7E and F). Of importance, no body weight loss or death was observed (Supplementary Figure S4), indicative of no overt toxicity. Altogether, these in vivo observations demonstrate an ability of IMP against cancer growth by inhibiting Hh signaling pathway.
Figure 7.
IMP impairs the growth of Shh-medulloblastoma in vivo. (A) The tumor volume of DAOY allografts over the time course of treatments with vehicle control (Con) or IMP (50 mg/kg) once daily (n=5). (B) qRT-PCR analysis of GLI1 expression in treated DAOY tumors (n=5). (C) Western blot analysis of GLI1, P-STAT3, and STAT3 expressions of treated DAOY tumor tissues. (D) Representative images of H&E staining of treated DAOY tumor tissues (n=5). (E) Representative images of Ki67 staining of treated DAOY tumor tissues (n=5). (F) Quantification of Ki67 positive cells in tumor tissues from E. All error bars represent mean ± s.d. Statistical significance was calculated using Student t-test or two-way ANOVA, *P <0.05, ***P <0.001.
Discussion
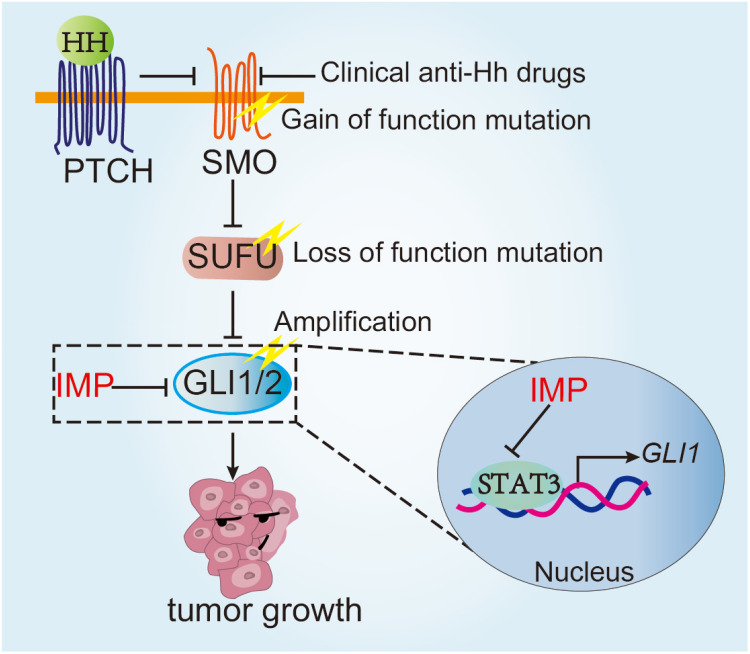
Abnormal activation of Hh signaling pathway has been shown to drive tumor initiation and progression, and preclinical and clinical studies provide strong evidence supporting it as a promising treatment target in a wide range of tumors. There are multiple reported Hh pathway inhibitors, and several have moved into clinic. However, clinically available treatment approaches, mainly targeting SMO, are challenged by the primary and acquired resistance as well as side effects, thus calling for an alternative strategy. We and other studies have revealed that targeting downstream GLI represents a superior strategy against multiple resistant mechanisms, such as mutations of SMO.71,72 In this study, we have shown that IMP, a natural product, inhibited GLI1 transcription by acting at GLI1 promoter via STAT3. IMP significantly inhibited Hh-driven tumor models in vitro and in vivo. Of importance, IMP possessed the ability to circumvent various resistance mechanisms of clinically available anti-Hh drugs, including SMO mutations, loss of SUFU and GLI2 amplifications, thus providing preclinical support for future clinical trials (Figure 8).
Figure 8.
Schematic illustration of the function and mechanism of IMP in overcoming the resistance of SMO inhibitors. IMP inhibited GLI1 transcription by acting at its promoter via STAT3, thereby circumventing various resistance mechanisms of clinical available anti-Hh drugs, including SMO mutations, loss of SUFU and GLI2 amplifications.
IMP presents in multiple traditional Chinese medicine with high safety, extensive sources and low price.73 The anticancer properties of IMP have attracted great attention in recent years. IMP has been shown to induce MCL-1 degradation to inhibit drug-resistance liver cancer.46 A more recent study showed that IMP improved chemosensitivity by downregulating P-glycoprotein expression in tumors.74 However, previous studies mainly focused on its application in tumor chemoresistance. We here provided the first evidence of IMP against Hh-driven tumors and overcoming the resistance of clinic available SMO inhibitors. It is also worth noting that IMP has distinct structures with reported Hh inhibitors, thus providing a novel tool for chemistry optimization.
Here, we demonstrated that IMP acted through STAT3 to inhibit the Hh signaling. Our findings revealed that IMP treatment robustly suppressed the transcription of GLI1, a core transcriptional factor of Hh pathway. Network pharmacology and RNA-seq analyses linked IMP targets with STAT3, prompting us to investigate whether STAT3 was the key player in IMP-mediated Hh inhibition. Indeed, STAT3 overexpression mitigated the inhibitory effect of IMP on Hh pathway (Figure 5C) and molecular docking showed the possible interactions between IMP and STAT3 via salt bridges, hydrogen bond and hydrophobic interactions. However, the precise bonding mechanisms of IMP to STAT3 and the specific sites involved warrant further investigation.
STAT3 has been reported to activate GLI1 expression in chronic lymphocytic leukemia cells and fibrocytes, and we thus evaluated whether the interaction between IMP and STAT3 could inhibit GLI1 transcription in MB cell. Multiple putative STAT3-binding sites were identified on the promoter region of GLI1 gene via JASPAR. Dual-luciferase reporter assay revealed that STAT3 activation induced the GLI1 transcription, whereas IMP attenuated this effect (Figure S3). However, the biological and cellular effects of this regulation remain to be determined in future studies. Of importance, multiple STAT3 inhibitors have entered into clinical trials, and data presented in this work might encourage further studies to explore the potential of STAT3 inhibition alone or in combination with the treatment of SMO constitutive activation-driven tumors, such as MB and BCC.
Conclusion
Taken together, our study provides compelling evidence demonstrating that IMP effectively suppresses aberrant Hh pathway by inhibiting GLI1 and overcomes the resistance of current SMO inhibitors. Our data elucidate that IMP exerts its inhibitory effects on GLI1 transcription by targeting and repressing STAT3. Furthermore, our findings highlight the potential of STAT3 as both a therapeutic target in Hh-dependent tumors. Therefore, our study provides novel therapeutic approaches for patients with Hh-dependent tumors, particularly those who are resistant to current SMO inhibitors.
Funding Statement
This work was funded by the National Natural Science Foundation of China (grant number 82204450 and 82272278), Science and Technology Commission of Shanghai Municipality (grant number 23ZR1409100), Shanghai Municipal Health Commission (grant number 202140069), Young Clinical Scientist Training Program at Shanghai Medical College of Fudan University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Dr Juan Wang reports a patent pending “Imperatorin in the preparation of drugs that inhibit the Hedgehog signaling pathway (202311098277.8)”. The authors declare no other conflicts of interest in this work.
References
- 1.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- 2.Jing J, Wu Z, Wang J, et al. Hedgehog signaling in tissue homeostasis, cancers, and targeted therapies. Signal Transduct Target Ther. 2023;8(1):315. doi: 10.1038/s41392-023-01559-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grund-Gröschke S, Stockmaier G, Aberger F. Hedgehog/GLI signaling in tumor immunity - new therapeutic opportunities and clinical implications. Cell Commun Signal. 2019;17(1):172. doi: 10.1186/s12964-019-0459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J. Hedgehog signaling mechanism and role in cancer. Semin Cancer Biol. 2022;85:107–122. doi: 10.1016/j.semcancer.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raleigh DR, Reiter JF. Misactivation of Hedgehog signaling causes inherited and sporadic cancers. J Clin Invest. 2019;129(2):465–475. doi: 10.1172/jci120850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Cui B, Li X, et al. The emerging roles of Hedgehog signaling in tumor immune microenvironment. Front Oncol. 2023;13:1171418. doi: 10.3389/fonc.2023.1171418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes JE, Gutzmer R, Kieran MW, Solomon JA. Hedgehog signaling inhibitors in solid and hematological cancers. Cancer Treat Rev. 2019;76:41–50. doi: 10.1016/j.ctrv.2019.04.005 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen NM, Cho J. Hedgehog pathway inhibitors as targeted cancer therapy and strategies to overcome drug resistance. Int J Mol Sci. 2022;23(3). doi: 10.3390/ijms23031733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bariwal J, Kumar V, Dong Y, Mahato RI. Design of Hedgehog pathway inhibitors for cancer treatment. Med Res Rev. 2019;39(3):1137–1204. doi: 10.1002/med.21555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong X, Qian H, Cao P, et al. Structural basis for the recognition of sonic hedgehog by human patched1. Science. 2018;361(6402). doi: 10.1126/science.aas8935 [DOI] [PubMed] [Google Scholar]
- 11.Rudolf AF, Kinnebrew M, Kowatsch C, et al. The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism. Nat Chem Biol. 2019;15(10):975–982. doi: 10.1038/s41589-019-0370-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Bulkley DP, Xin Y, et al. Structural basis for cholesterol transport-like activity of the hedgehog receptor patched. Cell. 2018;175(5):1352–1364.e1314. doi: 10.1016/j.cell.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi C, Di Minin G, Vercellino I, Wutz A, Korkhov VM. Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci Adv. 2019;5(9):eaaw6490. doi: 10.1126/sciadv.aaw6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–376. doi: 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- 15.Regl G, Neill GW, Eichberger T, et al. Human GLI2 and GLI1 are part of a positive feedback mechanism in basal cell carcinoma. Oncogene. 2002;21(36):5529–5539. doi: 10.1038/sj.onc.1205748 [DOI] [PubMed] [Google Scholar]
- 16.Tan Y, Hu Y, Xiao Q, et al. Silencing of brain-expressed X-linked 2 (BEX2) promotes colorectal cancer metastasis through the Hedgehog signaling pathway. Int J Biol Sci. 2020;16(2):228–238. doi: 10.7150/ijbs.38431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsuka A, Levesque MP, Dummer R, Kabashima K. Hedgehog signaling in basal cell carcinoma. J Dermatol Sci. 2015;78(2):95–100. doi: 10.1016/j.jdermsci.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 18.Menyhárt O, Győrffy B. Molecular stratifications, biomarker candidates and new therapeutic options in current medulloblastoma treatment approaches. Cancer Metastasis Rev. 2020;39(1):211–233. doi: 10.1007/s10555-020-09854-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellei B, Caputo S, Carbone A, et al. The role of dermal fibroblasts in nevoid basal cell carcinoma syndrome patients: an overview. Int J Mol Sci. 2020;21(3). doi: 10.3390/ijms21030720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steele NG, Biffi G, Kemp SB, et al. Inhibition of hedgehog signaling alters fibroblast composition in pancreatic cancer. Clin Cancer Res. 2021;27(7):2023–2037. doi: 10.1158/1078-0432.Ccr-20-3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Li H, Li Z, et al. SHH/GLI2-TGF-β1 feedback loop between cancer cells and tumor-associated macrophages maintains epithelial-mesenchymal transition and endoplasmic reticulum homeostasis in cholangiocarcinoma. Pharmacol Res. 2023;187:106564. doi: 10.1016/j.phrs.2022.106564 [DOI] [PubMed] [Google Scholar]
- 22.Hasselluhn MC, Decker-Farrell AR, Vlahos L, et al. Tumor explants elucidate a cascade of paracrine SHH, WNT, and VEGF signals driving pancreatic cancer angiosuppression. Cancer Discov. 2024;14(2):348–361. doi: 10.1158/2159-8290.Cd-23-0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petty AJ, Dai R, Lapalombella R, et al. Hedgehog-induced PD-L1 on tumor-associated macrophages is critical for suppression of tumor-infiltrating CD8+ T cell function. JCI Insight. 2021;6(6). doi: 10.1172/jci.insight.146707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dlugosz A, Agrawal S, Kirkpatrick P. Vismodegib. Nat Rev Drug Discov. 2012;11(6):437–438. doi: 10.1038/nrd3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Migden MR, Guminski A, Gutzmer R, et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind Phase 2 trial. Lancet Oncol. 2015;16(6):716–728. doi: 10.1016/s1470-2045(15)70100-2 [DOI] [PubMed] [Google Scholar]
- 26.Hoy SM. Glasdegib: first Global Approval. Drugs. 2019;79(2):207–213. doi: 10.1007/s40265-018-1047-7 [DOI] [PubMed] [Google Scholar]
- 27.Yauch RL, Dijkgraaf GJ, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574. doi: 10.1126/science.1179386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharpe HJ, Pau G, Dijkgraaf GJ, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):327–341. doi: 10.1016/j.ccell.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ocasio JK, Babcock B, Malawsky D, et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat Commun. 2019;10(1):5829. doi: 10.1038/s41467-019-13657-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitson RJ, Lee A, Urman NM, et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat Med. 2018;24(3):271–281. doi: 10.1038/nm.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang YA, Chen YF, Bao Y, et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote chemoresistance in colorectal cancer. Proc Natl Acad Sci. 2018;115(26):E5990–e5999. doi: 10.1073/pnas.1801348115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neelakantan D, Zhou H, Oliphant MUJ, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8(1):15773. doi: 10.1038/ncomms15773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nolan-Stevaux O, Lau J, Truitt ML, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23(1):24–36. doi: 10.1101/gad.1753809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Wang J, Liu Y, et al. PGE2-JNK signaling axis non-canonically promotes Gli activation by protecting Gli2 from ubiquitin-proteasomal degradation. Cell Death Dis. 2021;12(7):707. doi: 10.1038/s41419-021-03995-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dusek CO, Hadden MK. Targeting the GLI family of transcription factors for the development of anti-cancer drugs. Expert Opin Drug Discov. 2021;16(3):289–302. doi: 10.1080/17460441.2021.1832078 [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Ingram L, Tolosa EJ, et al. Gli transcription factors mediate the oncogenic transformation of prostate basal cells induced by a kras-androgen receptor axis. J Biol Chem. 2016;291(49):25749–25760. doi: 10.1074/jbc.M116.753129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Po A, Silvano M, Miele E, et al. Noncanonical GLI1 signaling promotes stemness features and in vivo growth in lung adenocarcinoma. Oncogene. 2017;36(32):4641–4652. doi: 10.1038/onc.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazumdar T, DeVecchio J, Shi T, et al. Hedgehog signaling drives cellular survival in human colon carcinoma cells. Cancer Res. 2011;71(3):1092–1102. doi: 10.1158/0008-5472.Can-10-2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beauchamp EM, Ringer L, Bulut G, et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest. 2011;121(1):148–160. doi: 10.1172/jci42874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Lee JJ, Kim J, Gardner D, Beachy PA. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc Natl Acad Sci. 2010;107(30):13432–13437. doi: 10.1073/pnas.1006822107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Aftab BT, Tang JY, et al. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell. 2013;23(1):23–34. doi: 10.1016/j.ccr.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ally MS, Ransohoff K, Sarin K, et al. Effects of combined treatment with arsenic trioxide and itraconazole in patients with refractory metastatic basal cell carcinoma. JAMA Dermatol. 2016;152(4):452–456. doi: 10.1001/jamadermatol.2015.5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu WW, Huang ZH, Liao L, et al. Direct targeting of CREB1 with imperatorin inhibits TGFβ2-ERK signaling to suppress esophageal cancer metastasis. Adv Sci. 2020;7(16):2000925. doi: 10.1002/advs.202000925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang N, Wang J, Zhang Y, et al. Imperatorin ameliorates mast cell-mediated allergic airway inflammation by inhibiting MRGPRX2 and CamKII/ERK signaling pathway. Biochem Pharmacol. 2021;184:114401. doi: 10.1016/j.bcp.2020.114401 [DOI] [PubMed] [Google Scholar]
- 45.Niu X, Yang Y, Yu J, et al. Panlongqi tablet suppresses adjuvant-induced rheumatoid arthritis by inhibiting the inflammatory response in vivo and in vitro. J Ethnopharmacol. 2023;308:116250. doi: 10.1016/j.jep.2023.116250 [DOI] [PubMed] [Google Scholar]
- 46.Li X, Zeng X, Sun J, et al. Imperatorin induces Mcl-1 degradation to cooperatively trigger Bax translocation and Bak activation to suppress drug-resistant human hepatoma. Cancer Lett. 2014;348(1–2):146–155. doi: 10.1016/j.canlet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Zhang Y, Huang WJ, et al. ABT-199 inhibits Hedgehog pathway by acting as a competitive inhibitor of oxysterol, rather as a BH3 mimetic. Acta Pharmacol Sin. 2021;42(6):1005–1013. doi: 10.1038/s41401-020-00504-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forli S, Huey R, Pique ME, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–919. doi: 10.1038/nprot.2016.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci. 2002;99(22):14071–14076. doi: 10.1073/pnas.182542899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Magno L, Manni S, Di Pastena F, et al. Phenformin inhibits hedgehog-dependent tumor growth through a complex I-independent redox/corepressor module. Cell Rep. 2020;30(6):1735–1752.e1737. doi: 10.1016/j.celrep.2020.01.024 [DOI] [PubMed] [Google Scholar]
- 53.Han Y, Shi Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci. 2015;112(20):6383–6388. doi: 10.1073/pnas.1421628112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Y, Gholamin S, Schubert S, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20(7):732–740. doi: 10.1038/nm.3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lauth M, Bergström A, Shimokawa T, Toftgård R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci. 2007;104(20):8455–8460. doi: 10.1073/pnas.0609699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo J, Liu F, Sun X, et al. Inhibition of the FACT complex targets aberrant hedgehog signaling and overcomes resistance to smoothened antagonists. Cancer Res. 2021;81(11):3105–3120. doi: 10.1158/0008-5472.Can-20-3186 [DOI] [PubMed] [Google Scholar]
- 57.Liu F, Jiang W, Sui Y, et al. CDK7 inhibition suppresses aberrant hedgehog pathway and overcomes resistance to smoothened antagonists. Proc Natl Acad Sci. 2019;116(26):12986–12995. doi: 10.1073/pnas.1815780116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doheny D, Manore S G, Wong G L, Lo H W, Hedgehog Signaling and Truncated GLI1 in Cancer. Cells. 2020;9(9). doi: 10.3390/cells9092114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu X, An L, Fan G, et al. EGFR signaling promotes nuclear translocation of plasma membrane protein TSPAN8 to enhance tumor progression via STAT3-mediated transcription. Cell Res. 2022;32(4):359–374. doi: 10.1038/s41422-022-00628-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Beachy PA. Cellular and molecular mechanisms of Hedgehog signalling. Nat Rev Mol Cell Biol. 2023;24(9):668–687. doi: 10.1038/s41580-023-00591-1 [DOI] [PubMed] [Google Scholar]
- 61.Hu Y, Dong Z, Liu K. Unraveling the complexity of STAT3 in cancer: molecular understanding and drug discovery. J Exp Clin Cancer Res. 2024;43(1):23. doi: 10.1186/s13046-024-02949-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang JH, Xu Y, Lin FY. The inhibition of microRNA-326 by SP1/HDAC1 contributes to proliferation and metastasis of osteosarcoma through promoting SMO expression. J Cell Mol Med. 2020;24(18):10876–10888. doi: 10.1111/jcmm.15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuo D, Zhou Z, Wang H, et al. Alternol, a natural compound, exerts an anti-tumour effect on osteosarcoma by modulating of STAT3 and ROS/MAPK signalling pathways. J Cell Mol Med. 2017;21(2):208–221. doi: 10.1111/jcmm.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv M, Xu Q, Zhang B, et al. Imperatorin induces autophagy and G0/G1 phase arrest via PTEN-PI3K-AKT-mTOR/p21 signaling pathway in human osteosarcoma cells in vitro and in vivo. Cancer Cell Int. 2021;21(1):689. doi: 10.1186/s12935-021-02397-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manshouri T, Veletic I, Li P, et al. GLI1 activates pro-fibrotic pathways in myelofibrosis fibrocytes. Cell Death Dis. 2022;13(5):481. doi: 10.1038/s41419-022-04932-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rozovski U, Harris DM, Li P, et al. STAT3 induces the expression of GLI1 in chronic lymphocytic leukemia cells. Oncotarget. 2021;12(5):401–411. doi: 10.18632/oncotarget.27884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen L, Xu W, Yang Q, et al. Imperatorin alleviates cancer cachexia and prevents muscle wasting via directly inhibiting STAT3. Pharmacol Res. 2020;158:104871. doi: 10.1016/j.phrs.2020.104871 [DOI] [PubMed] [Google Scholar]
- 68.Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71(15):5057–5061. doi: 10.1158/0008-5472.Can-11-0923 [DOI] [PubMed] [Google Scholar]
- 69.Atwood SX, Sarin KY, Whitson RJ, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):342–353. doi: 10.1016/j.ccell.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bufalieri F, Infante P, Bernardi F, et al. ERAP1 promotes Hedgehog-dependent tumorigenesis by controlling USP47-mediated degradation of βTrCP. Nat Commun. 2019;10(1):3304. doi: 10.1038/s41467-019-11093-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Zhang Y, Li Y, et al. Development of hedgehog pathway inhibitors by epigenetically targeting GLI through BET bromodomain for the treatment of medulloblastoma. Acta Pharm Sin B. 2021;11(2):488–504. doi: 10.1016/j.apsb.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang W, Liu H, Tan W, Wang J. ABT-737 suppresses aberrant Hedgehog pathway and overcomes resistance to smoothened antagonists by blocking Gli. Med Oncol. 2022;39(12):188. doi: 10.1007/s12032-022-01794-w [DOI] [PubMed] [Google Scholar]
- 73.Kozioł E, Skalicka-Woźniak K. Imperatorin-pharmacological meaning and analytical clues: profound investigation. Phytochem Rev. 2016;15(4):627–649. doi: 10.1007/s11101-016-9456-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao ZG, Tang T, Guan XJ, et al. Improvement of transmembrane transport mechanism study of imperatorin on P-glycoprotein-mediated drug transport. Molecules. 2016;21(12):1606. doi: 10.3390/molecules21121606 [DOI] [PMC free article] [PubMed] [Google Scholar]