Abstract
Microbial biosurfactants have garnered significant interest from industry due to their lower toxicity, biodegradability, activity at lower concentrations and higher resistance compared to synthetic surfactants. The deep‐sea Rhodococcus sp. I2R has been identified as a producer of glycolipid biosurfactants, specifically succinoyl trehalolipids, which exhibit antiviral activity. However, genome mining of this bacterium has revealed a still unexplored repertoire of biosurfactants. The microbial genome was found to host five non‐ribosomal peptide synthetase (NRPS) gene clusters containing starter condensation domains that direct lipopeptide biosynthesis. Genomics and mass spectrometry (MS)‐based metabolomics enabled the linking of two NRPS gene clusters to the corresponding lipopeptide families, leading to the identification of 20 new cyclolipopeptides, designated as rhodoheptins, and 33 new glycolipopeptides, designated as rhodamides. An integrated in silico gene cluster and high‐resolution MS/MS data analysis allowed us to elucidate the planar structure, inference of stereochemistry and reconstruction of the biosynthesis of rhodoheptins and rhodamides. Rhodoheptins are cyclic heptapeptides where the N‐terminus is bonded to a β‐hydroxy fatty acid forming a macrolactone ring with the C‐terminal amino acid residue. Rhodamides are linear 14‐mer glycolipopeptides with a serine‐ and alanine‐rich peptide backbone, featuring a distinctive pattern of acetylation, glycosylation and succinylation. These molecules exhibited biosurfactant activity in the oil‐spreading assay and showed moderate antiproliferative effects against human A375 melanoma cells.
Combining genome mining and mass spectrometry‐based metabolomics disclosed the deep‐sea Rhodococcus sp. I2R as a source of two families of novel lipopeptide biosurfactants with antiproliferative activity against human A375 melanoma cells.
INTRODUCTION
Surfactants are amphiphilic molecules with hydrophilic heads and hydrophobic tails, conferring these compounds the natural tendency to form self‐aggregates and lower water surface tension. Due to their various applications, such as detergents in household and industrial cleaning products, or as vehicles for active ingredients in pharmaceutical formulations, surfactants are extremely important from an industrial perspective. Currently, exhaustible fossil fuels are the starting material for surfactant production (Nagtode et al., 2023). Therefore, in response to increasing industrial demand, finding alternative and renewable sources for surfactants presents both economic and ecological imperatives. Microbial biosurfactants have raised significant attention as being less toxic, biodegradable, active at lower concentrations and more resistant to extreme conditions as compared to synthetic ones (Jahan et al., 2020). Moreover, biosurfactants are often endowed with pharmacological properties, such as antimicrobial, antitumor, antiviral and anti‐inflammatory activities (Buonocore et al., 2023; Giugliano et al., 2023; Ceresa et al., 2023; Pilz et al., 2023; Subramaniam et al., 2020).
Bacteria usually secrete biosurfactants to emulsify hydrophobic substrates and improve nutrient availability in harsh conditions. Moreover, biosurfactants exert antibacterial functions to gain an advantage over competitors and mediate physiological processes such as quorum sensing, biofilm formation, cell attachment/dissociation to surfaces and swarming motility (Dias & Nitschke, 2023).
Among microbial surfactants, lipopeptides (LPs) are low‐molecular‐weight metabolites, which have emerged as promising anticancer and antimicrobial agents. For instance, pseudofactin II triggers apoptosis in melanoma A375 cells as a consequence of plasma membrane permeabilization (Janek et al., 2013). Iturin A prompts apoptotic cell death in breast cancer via inhibition of the Akt pathway (Dey et al., 2015). Surfactin enhances a ROS/JNK‐mediated mitochondrial/caspase pathway to kill MCF‐7 cancer cells, but also disrupts cell membrane and protein synthesis in pathogenic bacteria (Cao et al., 2010; Chen et al., 2022). Remarkably, the LP daptomycin is a membrane‐active antibiotic currently used in clinical settings (Huang, 2020).
Biosynthetically, LPs are assembled primarily through the sequential condensation of proteinogenic and non‐proteinogenic amino acids by large multimodular enzymes, known as non‐ribosomal peptide synthetases (NRPSs). Only recently were LPs of ribosomal origin, such as lipolanthines, discovered (Wiebach et al., 2018).
Typically, each NRPS module contains a condensation (C) domain, an adenylation (A) domain, and a peptidyl carrier protein (PCP) domain, adding a single amino acid to the peptidyl backbone. The specific fatty acyl chains anchored to the peptide core in LPs form hydrophobic tails that strongly influence LP bioactivity. Fatty acyl chains are incorporated into the peptidyl backbone during the first step of LP biosynthesis through the N‐acylation of the α‐amino group of the first amino acid, a process known as lipoinitiation.
Bacteria use different lipoinitiation strategies to build up LPs (Figure 1) (Chooi & Tang, 2010).
FIGURE 1.
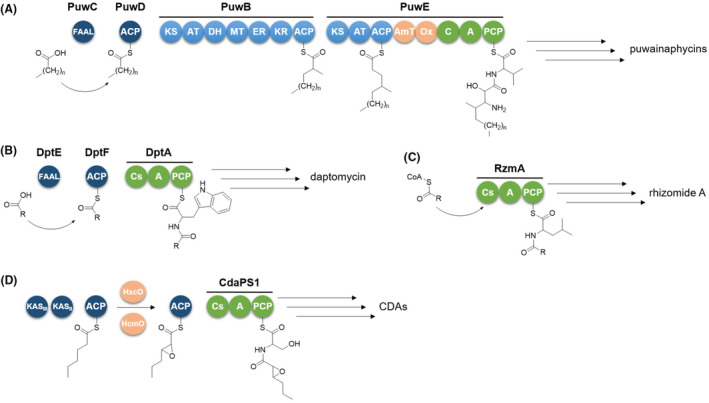
Lipoinitiation strategies employed in the biosynthesis of puwainaphycins (A), daptomycin (B), rhizomide A (C), and calcium‐dependent antibiotics (CDAs) (D). FAAL, fatty acyl‐AMP ligase; ACP, acyl carrier protein; KS, ketosynthase; AT, acyltransferase; DH, dehydratase; MT, methyltransferase; ER, enoylreductase; KR, ketoreductase; AmT, aminotransferase; Ox, monooxygenase; C, condensation domain; A, adenylation domain; PCP, peptidyl carrier protein; Cs, starter condensation domain; KASII, beta‐ketoacyl‐acyl carrier protein synthase II; KASIII, beta‐ketoacyl‐acyl carrier protein synthase III; HxcO and HcmO, FAD‐dependent oxidases.
Within the biosynthetic machinery of puwainaphycins (Mareš et al., 2014), a hybrid polyketide synthase and non‐ribosomal synthetase (PKS/NRPS) system incorporates the lipid chain (Figure 1A). The fatty acyl‐AMP ligase (FAAL) PuwC selects and activates the fatty acid, which is then transferred to the acyl carrier protein (ACP) PuwD. The fatty acid is subsequently elongated, aminated and hydroxylated by two adjacent PKS modules (PuwB and PuwE). Finally, the modified lipid chain is incorporated by the first NRPS module, priming the synthesis of the peptide backbone.
In most cases, lipoinitiation is directed by the starter condensation domain (Cs) located in the initial NRPS module, which catalyses formation of an amide bond between the fatty acyl chain and the first amino acid. Cs domains can be promptly detected through bioinformatics, as being phylogenetically distinguishable from other C‐domain subtypes (Rausch et al., 2007). In ramoplanin and daptomycin biosynthesis (Hoertz et al., 2012; Wittmann et al., 2008), FAAL and ACP directly activate and transfer the starter fatty acyl unit to the Cs of the initial NRPS module (Figure 1B). In case of the calcium‐dependent antibiotics (CDAs), a dedicated fab operon within the biosynthetic gene cluster (BGC) synthesizes 2,3‐epoxyhexanoic acid, which is tethered to an ACP and transferred to the Cs domain to be channelled into the NRPS assembly line (Hojati et al., 2002) (Figure 1D). The fatty acyl unit can be also delivered to the Cs domain as a free acyl‐CoA derivative, without FAAL and ACP‐mediation, as in the case of SrfAA surfactin, RzmA rhizomide and HolA holrhizin synthetases (Kraas et al., 2010; Zhong et al., 2021) (Figure 1C).
Following the One Strain Many Compounds (OSMAC) approach (Pan et al., 2019) with 22 different culture media, the marine isolate Rhodococcus sp. I2R (actinomycetota) was identified as a producer of more than 30 novel glycolipid biosurfactants, specifically succinoyl trehalolipids, which exhibit potent antiviral effects against herpes simplex virus and human coronaviruses, likely through a detergent‐like mechanism (Palma Esposito et al., 2021). However, genome mining of Rhodococcus sp. I2R (from now on R. I2R) highlighted a number of cryptic BGCs, some of which are predicted to encode new surface‐active natural products. Given the potential of this strain, we have taken an interest in further exploring its biosurfactant repertoire using a metabologenomic approach.
Herein, we report on the structure and biosynthesis of two LP families from R. I2R. Mining the bacterial genome for Cs domain‐containing NRPS revealed four putative LP biosynthetic operons. Linking genomics with mass spectrometry (MS)‐based metabolomics enabled us to connect two NRPS pathways to the corresponding LP families, leading to the identification of 20 new cyclic lipoheptapeptides, designated as rhodoheptins, and 33 new glycolipopeptides, designated as rhodamides. The integrated in silico genome analysis and comprehensive investigation of high‐resolution MS/MS data from individual LPs proved valuable in elucidating the planar structure, inferring stereochemistry and reconstructing the biosynthetic route of rhodoheptins and rhodamides, as they are produced in quantities too small for isolation and characterization by NMR. This study represents a step forward in exploring the biotechnological potential of the Rhodococcus genus.
RESULTS AND DISCUSSION
Identification of non‐ribosomal peptide lipopeptide gene clusters
The draft genome of R. I2R (accession: JAHUTG000000000) consists of 72 contigs, with an overall size of 5.29 Mb and GC content of 64.01%. A preliminary antiSMASH analysis (Blin et al., 2023) revealed several BGCs split across multiple contigs (Palma Esposito et al., 2021). To improve assembly continuity, the 72 contigs were joined into 36 scaffolds using the multi‐draft‐based scaffolder MEDUSA (http://150.217.159.17/medusa, accessed on 07/12/2023) (Bosi et al., 2015). Aiming to identify genes encoding the biosynthesis of LPs, de novo assembled scaffolds were mined for the presence of NRPS modules containing a Cs domain. Five novel intact multimodular NRPS systems with Cs domains were detected using antiSMASH. Four NRPS were predicted to catalyse LP biosynthesis, one of them being a hydroxamate siderophore synthetase. The last one was predicted to assemble a hydroxamate‐catechol hybrid siderophore, where the Cs domain is expected to incorporate a 2,3‐dihydroxybenzoic acid moiety. As modular NRPS biosynthesis usually follows the collinearity rule, and functions of catalytic domains within each module can be fairly inferred in silico (Della Sala et al., 2020), the putative structures and/or chemical substructures of the encoded LPs could be predicted from gene cluster sequences. Mass spectrometry data from R. I2R extracts were analysed for metabolites matching the chemical features predicted. This ‘feature‐based matching approach’ enabled us to link two of the five NRPS gene clusters, designated as rhp and rmd, to their corresponding LP families, namely rhodoheptins and rhodamides respectively. These LPs were detected via molecular networking in the R. I2R extract and structurally characterized using liquid chromatography coupled with high‐resolution tandem mass spectrometry (LC‐HRMS2) (see Section 2.4). Rhodoheptins were shown to be cyclic lipoheptapeptides while rhodamides were identified as linear glycolipopeptides.
Analysis of the rhp biosynthetic gene cluster
The putative rhp gene is 26,760 bp long and encodes for a heptamodular NRPS, which is expected to assemble a lipoheptapeptide molecule (Figure 2, Table S1). AntiSMASH analysis of rhp allowed for the rough prediction of the peptide backbone of rhodoheptins by integrating the A domain specificity conferring code identified by Stachelhaus et al. and the NRPyS library (Blin et al., 2023) (Table 1). Rhodoheptin biosynthesis starts with the loading module which selects and loads an l‐Leu residue undergoing N‐acylation catalysed by the Cs. A putative N‐acyl transferase (orf5, Table S1) is located upstream of the NRPS gene and presumably recruits and loads the acyl group to the loading module. Based on the subsequent MS‐based structure elucidation of rhodoheptins (Section 2.5), the Cs domain recognizes saturated and monounsaturated β‐hydroxy fatty acids ranging from C16 to C24. The N‐acylated Leu residue is subsequently transferred to the first elongation module featuring an LCL‐A‐PCP domain architecture and catalysing the condensation with an l‐Ser residue (LCL is a C domain catalysing condensation of two L amino acids). The following elongation module (LCL‐A‐PCP‐E) extends the N‐acylated leucylseryl intermediate with a d‐Leu or allo‐Ile residue, generated by the epimerization domain (E) from the corresponding l‐epimer. Accordingly, module 4 starts with a d CL forming a peptide bond between the d‐Leu/allo‐Ile and the fourth unknown l‐monomer. The growing peptide chain is forwarded sequentially to the following three extension modules, each adding one amino acid, predicted to be threonine, valine, and X. Specificity‐pocket structures predicted for the Rhp_A4 and Rhp_A7 domains did not resolve to a single building block, likely due to either a relaxed substrate selectivity or an unprecedented binding site signature. The antiSMASH algorithm refined the prediction of Rhp_A4 selectivity for branched aliphatic amino acids, while it resulted in ambiguous matches for Rhp_A7. In light of these findings, Rhp_A4 was hypothesized to select l‐Leu/Ile, consistent with the peptide sequence deduced by MS/MS analysis of rhodoheptins (Section 2.5). On the other hand, structural analysis of the rhodoheptin variants demonstrated that Rhp_A7 possesses substrate promiscuity, being able to activate l‐Leu/Ile, l‐Phe and l‐MetO (Section 2.5). Module 5 displays the same organization as module 3 (LCL‐A‐PCP‐E), accounting for the incorporation of d‐Thr at position 5 of the peptide moiety. As expected, module 6 has a d CL‐A‐PCP domain architecture and is involved in the condensation between d‐Thr and l‐Val. Finally, the last module terminates with a canonical thioesterase/cyclase domain, which may catalyse the LP release either through hydrolysis or cyclization. In a BlastP search against thioesterases (TEs) from the ThYme database (https://thyme.engr.unr.edu/v2.0/, accessed on 20/12/2023), TEs belonging to the TE16 family were identified as the closest homologues to RhP_TE (E value = 1e‐98 ≤ × ≤ 2e‐38 for the first 500 hits). TE16 enzymes have α/β‐hydrolase folds and in several PKSs and NRPs may use a hydroxyl group from the substrate chain for lactonization, thus yielding macrocyclic products (Cantu et al., 2010). This is consistent with the structure of rhodoheptins, where the β‐hydroxyl group of the N‐terminal fatty acid forms an ester bond with the last amino acid incorporated during biosynthesis (Figure 2), similar to the process observed in surfactin biosynthesis (Kraas et al., 2010).
FIGURE 2.
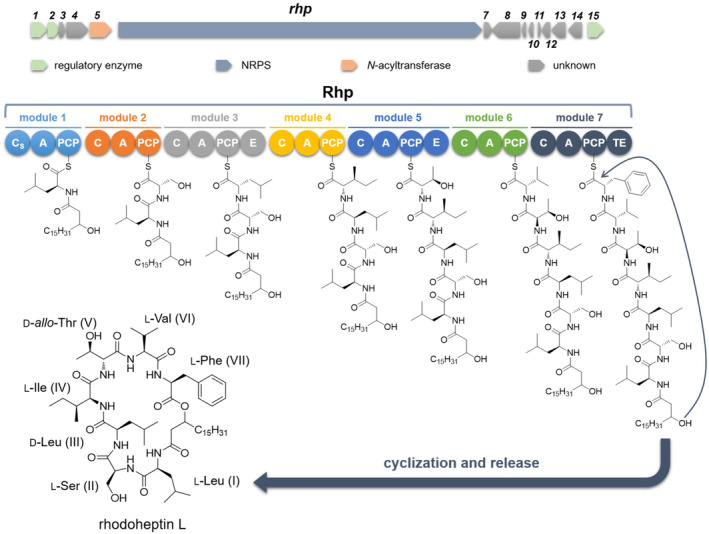
Putative biosynthesis of rhodoheptins. C, condensation domain; A, adenylation domain; PCP, peptidyl‐carrier protein; E, epimerase; Cs, starter condensation domain; TE, thioesterase.
TABLE 1.
Binding pocket signatures of adenylation domains from the Rhp synthetase.
| AntiSMASH prediction | Binding pocket signatures (position) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | ||
| Rhp_A1 | Leu | D | A | L | F | V | G | A | V | F | K |
| Rhp_A2 | Ser | D | V | W | H | F | S | L | V | D | K |
| Rhp_A3 | Leu/Ile | D | A | L | F | A | G | A | I | F | K |
| Rhp_A4 | Unknown | D | A | L | F | V | G | A | V | F | K |
| Rhp_A5 | Thr | D | F | W | N | I | G | M | V | H | K |
| Rhp_A6 | Val | D | A | L | F | V | G | G | I | M | K |
| Rhp_A7 | Unknown | D | A | Y | F | A | G | G | I | Q | K |
As reported by Della Sala et al. (2020).
Analysis of the rmd biosynthetic gene cluster
The putative rmd operon is 47,562 bp long and consists of 14 NRPS modules (Figure 3, Table S2). In addition, the putative biosynthetic genes, rmdE and rmdF, encode for two distinct acyl‐CoA synthetases, which are predicted to cooperate with the NRPS machinery to assemble rhodamides. RmdE and RmdF showed 50% and 33% identity, respectively, with YngI (ABS74201.1) and VrtB (ADI24927.1) fatty acyl‐CoA ligases in a BlastP search against the MIBiG database (Terlouw et al., 2023). Therefore, they are expected to catalyse formation of CoA‐activated fatty acids to fuel the rmd biosynthetic route. NRPS lipopeptide synthetase assembly lines very often require availability of acyl‐CoA thioesters to be initiated, as in the case of surfactin biosynthesis, where four different fatty acyl‐CoA ligases—LcfA, YhfL, YhfT, and YngI—are involved in providing the activated pool of fatty acid starter units (Kraas et al., 2010). Based on the subsequent MS‐based structural characterization of rhodamides (Section 2.6), RmdE and RmdF catalyse thioester formation with CoA from saturated and monounsaturated C16, C20, C22, and C24 fatty acids. The RmdA_Cs domain catalyses the transfer of CoA‐activated fatty acids from its donor site to the PCP‐bound amino acid, namely glycine. The N‐acylated glycyl intermediate is elongated by modules 2–14 in an assembly‐line fashion as amino acid residues are added sequentially to yield the LP backbone. Although antiSMASH analysis revealed the modular architecture of the rmd gene cluster, as well as the domain organization of each NRPS module, substrate selectivity of the A‐domain‐binding pocket could be predicted only for 6 out of 14 modules, as indicated in Table 2. RmdA_A3, RmdA_A6 and RmdB_A9 activate and select serine residues while RmdA_A7, RmdB_A10 and RmdD_A13 show selectivity for Thr, Ala, and Leu, respectively. Building block prediction based on the adenylation domain specificity was consistent with the structure of rhodamides, except for RmdA_A3. This is not surprising considering that according to the model proposed by Rausch et al. (2005), the 8 Ǻ signature sequence extracted from the RmdA_A3 active site shares only 65% identity with Ser‐selective A domains. Notably, modules 5 and 12 of the rhodamide synthetase apparently lack a functional A domain. Prior examples reported the complementation of adenylation activity of NRPS modules lacking their own A domain with upstream/downstream A domains from other modules (Du et al., 2000; Felnagle et al., 2007; Magarvey et al., 2006; Thomas et al., 2003; Zhang et al., 2020). According to this biosynthetic model, which specifies serine specificity, RmdA_A6 and/or RmdB_A9 should deliver Ser to modules 5 and 12 to match the chemical structures of rhodamides. However, it cannot be excluded that the two A‐less modules may result from contig/scaffold misassembly caused by either low‐depth sequencing data or the presence of highly similar A‐domain sequences within the BGC.
FIGURE 3.
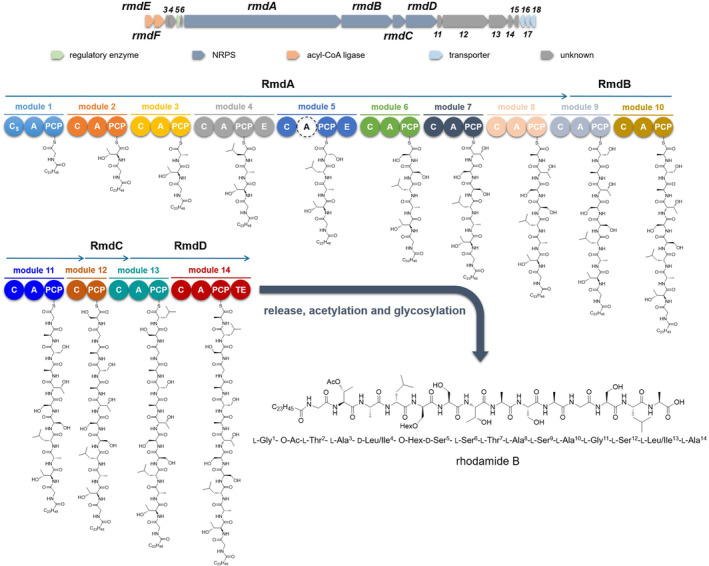
Putative biosynthesis of rhodamides. C, condensation domain; A, adenylation domain; PCP, peptidyl carrier protein; E, epimerase; Cs, starter condensation domain; TE, thioesterase; Hex, hexosyl; Ac, acetyl. The A domain labelled with the dashed circle is a putative incomplete or inactive A domain.
TABLE 2.
Binding pocket signatures of adenylation domains from the Rmd synthetase.
| AntiSMASH prediction | Binding pocket signatures (position) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | ||
| RmdA_A1 | Unknown | D | V | W | D | F | I | L | V | S | K |
| RmdA_A2 | Unknown | D | P | L | I | S | G | A | I | V | K |
| RmdA_A3 | Ser | D | V | W | S | L | A | L | V | H | K |
| RmdA_A4 | Unknown | D | P | L | I | S | G | G | I | I | K |
| RmdA_A6 | Ser | D | V | W | H | F | S | L | V | D | K |
| RmdA_A7 | Thr | D | F | W | N | V | G | M | V | H | K |
| RmdA_A8 | Unknown | D | L | W | H | L | I | L | V | S | K |
| RmdB_A9 | Ser | D | V | W | H | F | S | L | V | D | K |
| RmdB_A10 | Ala | D | V | W | S | Q | A | L | V | H | K |
| RmdB_A11 | Unknown | D | V | W | D | F | I | L | V | S | K |
| RmdD_A13 | Leu | D | A | L | F | V | G | A | V | V | K |
| RmdD_A14 | Unknown | D | L | W | H | L | I | V | V | S | K |
As reported by Della Sala et al. (2020).
Two canonical epimerization domains have been detected in modules 4 and 5, indicating conversion of l‐Ser to the d‐configuration at positions 4 and 5 of the peptide backbone. The last NRPS module contains a terminal thioesterase domain belonging to the TE16 family mediating the LP release through hydrolysis, thus yielding a linear product as revealed by MS/MS analysis of rhodamide congeners (Section 2.6).
To achieve the end‐product, the LP backbone undergoes (a) side‐chain glycosylation at Ser‐5 and/or Thr‐7 and/or Ser‐12 and (b) side‐chain acetylation at Thr/Dab‐2 and/or Thr‐4. Since suitable tailoring enzymes could not be bioinformatically detected within the relevant modules, trans‐acting glycosyl‐ and acetyl‐transferases are likely to cooperate with the Rhd synthetase to yield the final product. Two putative glycosyltransferases (WP_230557595.1 and WP_230555296.1) belonging to the glycosyltransferase family A are located adjacent to the Rmd NRPS and could O‐glycosylate the rhodamide peptide backbone. On the other hand, no acetyl‐transferases were identified within the rmd gene cluster. More broadly, it cannot be excluded that glycosyl‐ and acetyl‐transferases located at different loci in the genome of R. I2R may catalyse these tailoring steps in rhodamide biosynthesis, as reported for the glycosylation of macrolactin and bacillaene (Qin et al., 2014). In addition, the glycosyl moiety in most rhodamide variants undergoes O‐succinylation. R. I2R has been already reported to assemble succinic saccharide esters, and a probably responsible acyltransferase (WP_230555977) has been identified in the bacterial genome (Palma Esposito et al., 2021).
Tandem mass spectrometry molecular networking analysis of R. I2R culture
Our previous OSMAC screening (22 culture conditions) failed to trigger LP production in R. I2R (Palma Esposito et al., 2021). In this work, we tried to enhance LP biosynthesis in ASG medium by hexadecane and iron supplementation, as previously described (de Oliveira Schmidt et al., 2021; Peng et al., 2008). After cultivation, biomass of R. I2R and exhausted culture broth were harvested separately and extracted using MeOH and AcOEt, respectively. Then, crude extracts were combined and fractionated by C18 reversed‐phase (RP) chromatography, thus yielding five fractions F1‐F5, eluted using different mixtures of H2O and MeOH (100% H2O, 50% MeOH, 90% MeOH, 100% MeOH, and 100% MeOH supplemented with 0.1% TFA).
To map a metabolic profile of R. I2R, each fraction was analysed by untargeted LC‐HRMS2. MS2 data from all fractions were used to generate a global molecular network using the feature‐based molecular networking (FBMN) tool (Nothias et al., 2020), which enables clustering of similar molecules based on MS/MS fragmentation similarity (Figure S1). Molecules are subsequently visualized in cytoscape as nodes, connected by edges, with the thickness of each edge corresponding to the similarity of the MS/MS spectra. Linking the molecular networking analysis with the manual detection of specific substructures in MS2 data led to the identification of two large molecular clusters, which matched the predictions of the new rhp and rmd BGCs, respectively. The rhodoheptins and rhodamides were shown to be mainly located in the F5 fraction eluted with 100% MeOH plus 0.1% TFA. Therefore, following the workflow described above, a molecular network of the F5 fraction was created to assess its chemical composition (Figure 4). Besides the presence of rhodoheptins (red nodes) and rhodamides (orange nodes), F5 also contained a minor molecular group composed of succinoyl trehalolipids (blue nodes).
FIGURE 4.
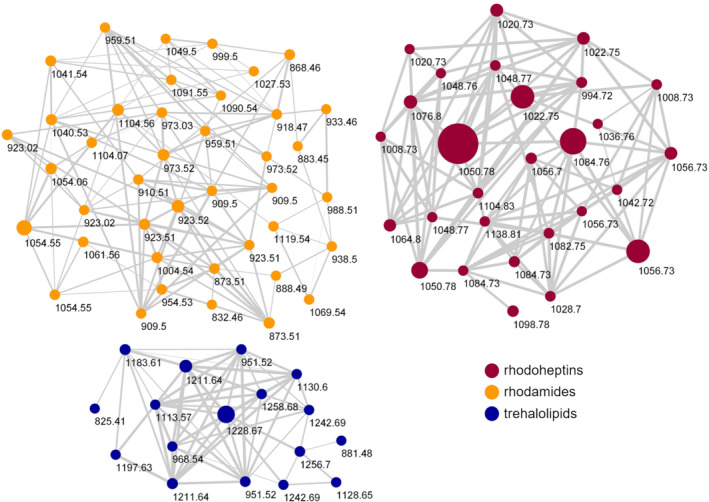
Molecular network of the F5 fraction obtained by reversed‐phase chromatography of the organic extract of Rhodococcus sp. I2R. Nodes are coloured according to biosurfactant classes and their size is related to metabolite amounts. Edge thickness reflects cosine scores which reveal the MS/MS spectra similarity.
To search for LPs encoded by the rhp and rmd BGCs, LC‐HRMS2 data were imported into the MZmine platform. The data were mined for precursor ions that generated fragment ions and/or neutral losses diagnostic of chemical substructures inferred by genome mining. Specifically, rhodoheptins were identified by looking for precursor ions showing neutral losses of 313.2002 amu (C15H27N3O4) and/or 200.1161 amu (C9H16N2O3) corresponding to the Leu‐Ser‐Ile/Leu and Thr‐Val motifs, respectively, consistent with the prediction of the rhp BGC (Table 1, Figure S2). Additionally, rhodamides were detected by searching for precursor ions that generated the fragment ions at m/z 159.0764 (C6H11N2O3 +) and 189.0870 (C7H13N2O4 +) corresponding to the Ser‐Ala and Thr‐Ser motifs, respectively, as inferred from the prediction of the rmd gene cluster (Table 2, Figure S3).
Structure elucidation of rhodoheptins by mass spectrometry
For structural characterization of rhodoheptins, F5 was analysed by LC‐HRMS2 in the positive ion detection mode on a Q Exactive Focus Orbitrap mass spectrometer. Precursor ions included in the rhodoheptin network (red nodes, Figure 4) were selected to be fragmented in the higher‐energy collision dissociation cell (HCD). The high‐resolution (HR) masses of the pseudo‐molecular ions [M + H]+ indicated the molecular formulas reported in Table 3 (mass accuracy ≤1.4 ppm).
TABLE 3.
Structures of rhodoheptins A‐U (1–21) from Rhodococcus sp. I2R.
| Compound | [M + H]+ | m/z | R t (min) | R (FA) a | Aa‐5 | Aa‐6 | Aa‐7 |
|---|---|---|---|---|---|---|---|
| Rhodoheptin A (1) | C52H96N7O11 | 994.7152 | 22.3 | C13H27 (C16:0;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin B (2) | C53H98N7O11 | 1008.7305 | 26.0 | C14H29 (C17:0;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin C (3) | C53H98N7O11 | 1008.7311 | 27.3 | C15H31 (C18:0;O) | d‐Ser | l‐Val | l‐Leu/Ile |
| Rhodoheptin D (4) | C54H98N7O11 | 1020.7308 | 23.2 | C15H29 (C18:1;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin E (5) | C54H100N7O11 | 1022.7470 | 30.1 | C15H31 (C18:0;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin F (6) | C55H94N7O11 | 1028.7001 | 21.4 | C13H27 (C16:0;O) | d‐allo‐Thr | l‐Val | l‐Phe |
| Rhodoheptin G (7) | C56H96N7O11 | 1042.7154 | 26.6 | C15H31 (C18:0;O) | d‐Ser | l‐Val | l‐Phe |
| Rhodoheptin H (8) | C56H102N7O11 | 1048.7637 | 29.8 | C17H33 (C20:1;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin I (9) | C56H104N7O11 | 1050.7787 | 19.1 | C17H35 (C20:0;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin J (10) | C57H96N7O11 | 1054.7154 | 22.3 | C15H29 (C18:1;O) | d‐allo‐Thr | l‐Val | l‐Phe |
| Rhodoheptin K (11) | C53H98N7O12S | 1056.6981 | 21.8 | C15H31 (C18:0;O) | d‐allo‐Thr | l‐Val | l‐MetO |
| Rhodoheptin L (12) | C57H98N7O11 | 1056.7317 | 28.8 | C15H31 (C18:0;O) | d‐allo‐Thr | l‐Val | l‐Phe |
| Rhodoheptin M (13) | C57H106N7O11 | 1064.7940 | 22.1 | C18H37 (C21:0;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin N (14) | C58H100N7O11 | 1070.7470 | 30.4 | C15H31 (C18:0;O) | d‐allo‐Thr | l‐Ile | l‐Phe |
| Rhodoheptin O (15) | C58H106N7O11 | 1076.7948 | 14.7 | C19H37 (C22:1;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin P (16) | C59H100N7O11 | 1082.7472 | 28.6 | C17H33 (C20:1;O) | d‐allo‐Thr | l‐Val | l‐Phe |
| Rhodoheptin Q (17) | C55H102N7O12S | 1084.7290 | 25.8 | C17H35 (C20:0;O) | d‐allo‐Thr | l‐Val | l‐MetO |
| Rhodoheptin R (18) b | C59H102N7O11 | 1084.7633 | 15.3 | C17H35 (C20:0;O) | d‐allo‐Thr | l‐Val | l‐Phe |
| Rhodoheptin S (19) | C60H104N7O11 | 1098.7781 | 18.9 | C18H37 (C21:0;O) | d‐allo‐Thr | l‐Val | l‐Phe |
| Rhodoheptin T (20) | C60H110N7O11 | 1104.8262 | 34.8 | C21H41 (C24:1;O) | d‐allo‐Thr | l‐Val | l‐Leu/Ile |
| Rhodoheptin U (21) | C63H108N7O11 | 1138.8105 | 32.3 | C21H41 (C24:1;O) | d‐allo‐Thr | l‐Val | l‐Phe |
Abbreviations: aa, amino acid; FA, fatty acid; MetO, methionine sulfoxide; R t, retention time.
Fatty acids have been reported in parenthesis using the LIPID MAPS shorthand notation (Liebisch et al., 2020). Fatty acyl chains are indicated as C:N;O, where C is the number of carbon atoms, N is the number of double bond equivalents and O is the number of additional oxygen atoms linked to the hydrocarbon chain.
Frankfater et al. (2020).
Mass fragmentation spectra of rhodoheptins suggested a cyclic LP structure (Figure 5), as being characterized by several b‐ and y‐type ion series arising from random ring opening at different amide bonds and at the ester bond. Particularly, most rhodoheptin variants first undergo α‐ε bond cleavages, thus yielding five different linear fragments (Figure 6 and Figures S5–S23). Then, sequential N‐ and C‐terminal cleavage of these linear fragments resulted in five series of ions, which allowed for the assignment of the amino acid sequence of rhodoheptins. In addition, the linear fragment β (Figure 6) displays neutral loss of a β‐hydroxy fatty acyl moiety as ketene, which is then linked to Leu1 through an amide bond. This is consistent with the genome mining prediction results, thereby indicating an LP structure. The β‐hydroxy function of the fatty acyl group forms an ester bond with the carboxylic group of aa‐7, as inferred from the generation of (a) the y 6 and y 5 ions during fragmentation of the δ linear fragment and (b) the y 5, y 4 and y 4 # ions during fragmentation of the ε linear fragment (Figure 6). As a result, the structure of rhodoheptins was determined as indicated in Table 3. Rhodoheptins have the common tetrapeptide motif Leu1‐Ser2‐Leu3‐Ile4, while structural diversification results from amino acid substitutions in positions 5, 6, and 7 and/or fatty acyl degree of unsaturation and length. Thus, either Thr or Ser (the latter only in rhodoheptins C (3) and G (7)) is incorporated into position 5, whereas Val or Ile (the latter only in rhodoheptin N (14)) may occupy position 6. However, the highest variation is due to exchange of amino acids in position 7 where Leu/Ile, Phe or MetO may be present. Incorporation of MetO in rhodoheptins K (11) and Q (17) is corroborated by the presence of fragment ions arising from the neutral loss of methanesulfenic acid (CH3SOH, 63.9983 Da), which is indicative of the sulfoxide group in side‐chain of MetO (Figures S14 and S20). Saturated and monounsaturated β‐hydroxy fatty acids ranging from C16 to C24 are the fatty‐acyl moieties embedded in the peptide ring of rhodoheptins.
FIGURE 5.
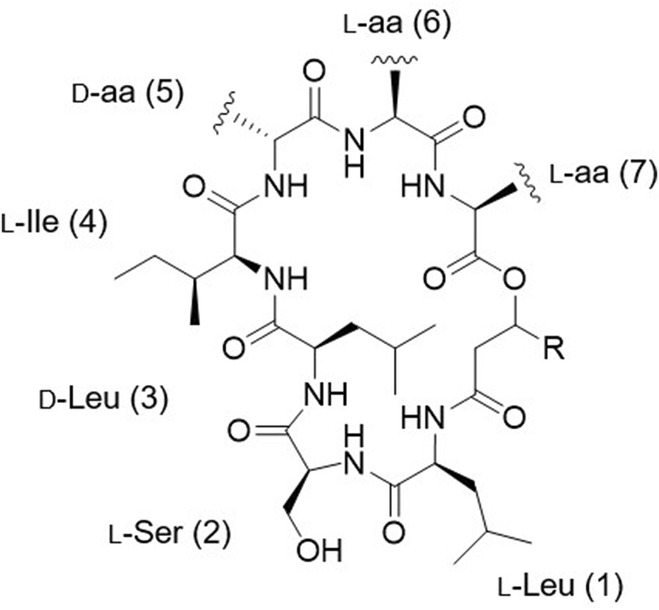
General structure of rhodoheptins.
FIGURE 6.
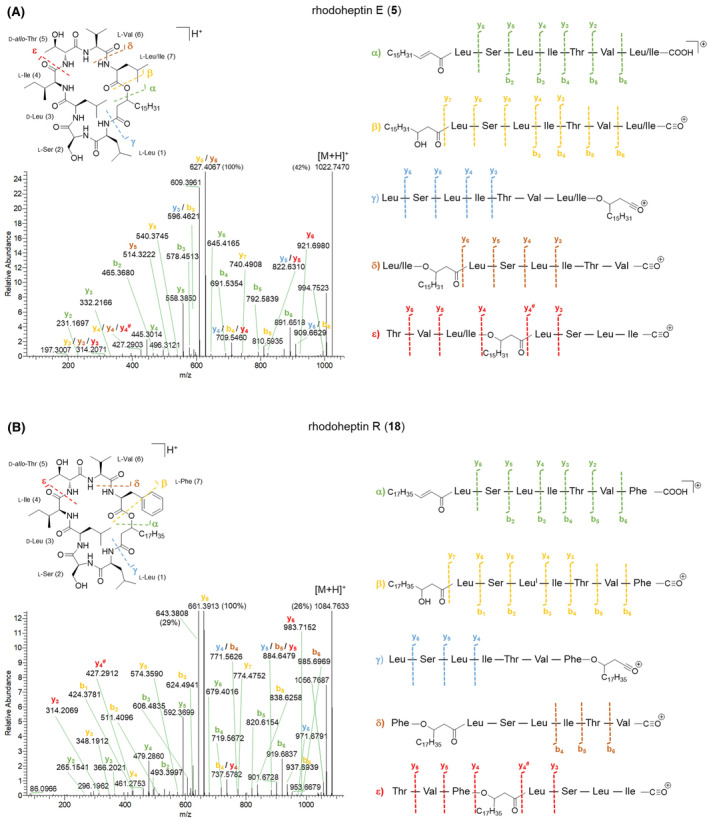
HR‐MS2 spectra of the [M + H]+ pseudo‐molecular ions of rhodoheptin E (5) (A) and rhodoheptin R (18) (B) and their proposed fragmentation pathways.
Finally, the structures of 21 rhodoheptins have been characterized, 20 of them being new compounds. Based on the similarity of the MS/MS spectrum, only one congener, that is, rhodoheptin R (18), was already reported from Rhodococcus equi and Rhodococcus opacus (Frankfater et al., 2020), which were shown to produce a family of cyclic and linear LPs closely related to rhodoheptins. By analogy with the structure of rhodoheptin R (18) and the other peptidolipid variants elucidated by MS, NMR spectroscopy and GC/MS by Frankfater and co‐workers (Frankfater et al., 2020), we tentatively distinguished between the isomeric Ile and Leu residues at positions 1, 3, 4, and 6 in rhodoheptins, as indicated in Table 3. Notably, Leu1 also correlates with prediction of the adenylation domain selectivity (Table 1). However, it was not possible to recognize by analogy the Ile/Leu7 moieties in rhodoheptins A‐E (1–5), H‐I (8–9), M (13), O (15) and T (20), as such a residue is not present in any peptidolipid variants from R. equi and R. opacus, where the last amino acid is always Phe.
Rhodoheptins are composed of mixed d‐ and l‐amino acids. The absolute configuration of the seven amino acids was assigned through bioinformatic prediction. As the rhodoheptin synthetase includes two epimerization domains within modules 3 and 5 (Figure 2), respectively, the presence of d‐Leu3 and d‐ allo‐Thr5/d‐Ser5 was inferred, while the remaining amino acids were assumed to possess the l configuration. Nevertheless, the absolute configuration of the β‐carbon of the β‐hydroxy fatty acids in rhodoheptins remains unsolved, as not predictable by gene cluster analysis.
Structure elucidation of rhodamides by mass spectrometry
After reversed‐phase liquid chromatography, MS/MS spectra of [M + 2H]2+ ions of rhodamides (orange nodes, Figure 4) were acquired at lower and higher normalized collision energies (NCE), that is, 15 and 30, to obtain both larger and smaller fragment ions for de novo sequencing of the peptide backbone. The high‐resolution ESI mass spectrum of [M + 2H]2+ ions of rhodamides was indicative of the molecular formulas reported in Table 4 (mass accuracy ≤3 ppm). Rhodamides were shown to be linear glycosylated and acetylated peptidolipids consisting of a tetradecapeptide linked to a saturated or monounsaturated fatty acid via an amide bond with the N‐terminal amino acid residue (Figure 7).
TABLE 4.
Structures of rhodamides A‐Z and A1‐D1 (22–54) from Rhodococcus sp. I2R.
| Compound | [M + 2H]2+ | m/z | R t (min) | R1 (FA) a | R2 | R3 | R4 | l‐aa‐2 | d‐aa‐4 |
|---|---|---|---|---|---|---|---|---|---|
| Rhodamide A (22) | C76H132N14O31 | 868.4610 | 5.9 | C15H31 (C16:0) | Succ‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide B (23) | C80H142N14O28 | 873.5066 | 20.3 | C23H45 (C24:1) | Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Isorhodamide B (24) | C80H142N14O28 | 873.5066 | 21.6 | C23H45 (C24:1) | H | Hex | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide C (25) | C76H130N14O33 | 883.4483 | 5.2 | C15H31 (C16:0) | Succ‐Hex | H | H | O‐Ac‐Thr | O‐Ac‐Thr |
| Rhodamide D (26) | C80H140N14O30 | 888.4929 | 18.4 | C23H45 (C24:1) | Hex | H | H | O‐Ac‐Thr | O‐Ac‐Thr |
| Rhodamide E (27) | C82H142N14O31 | 909.4992 | 14.4 | C21H41 (C22:1) | Succ‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Isorhodamide E (28) | C82H142N14O31 | 909.4992 | 15.2 | C21H41 (C22:1) | H | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide F (29) | C82H144N14O31 | 910.5064 | 21.0 | C21H43 (C22:0) | Succ‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide G (30) | C80H136N14O34 | 918.4683 | 6.1 | C15H31 (C16:0) | Disucc‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide H (31) | C84H147N15O30 | 923.0219 | 18.2 | C23H45 (C24:1) | Succ‐Hex | H | H | N‐γ‐Ac‐Dab | Leu/Ile |
| Rhodamide I (32) | C84H146N14O31 | 923.5143 | 20.0 | C23H45 (C24:1) | Succ‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Isorhodamide I (33) | C84H146N14O31 | 923.5143 | 21.4 | C23H45 (C24:1) | H | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide J (34) | C80H134N14O36 | 933.4566 | 5.4 | C15H31 (C16:0) | Disucc‐Hex | H | H | O‐Ac‐Thr | O‐Ac‐Thr |
| Rhodamide K (35) | C84H144N14O33 | 938.5007 | 18.4 | C23H45 (C24:1) | Succ‐Hex | H | H | O‐Ac‐Thr | O‐Ac‐Thr |
| Rhodamide L (36) | C86H152N14O33 | 954.5326 | 18.6 | C23H45 (C24:1) | Hex | Hex | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide M (37) | C86H146N14O34 | 959.5073 | 14.4 | C21H41 (C22:1) | Disucc‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide N (38) | C88H151N15O33 | 973.0306 | 17.9 | C23H45 (C24:1) | Disucc‐Hex | H | H | N‐γ‐Ac‐Dab | Leu/Ile |
| Rhodamide O (39) | C88H150N14O34 | 973.5222 | 19.7 | C23H45 (C24:1) | Disucc‐Hex | H | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide P (40) | C88H148N14O36 | 988.5094 | 18.2 | C23H45 (C24:1) | H | Disucc‐Hex | H | O‐Ac‐Thr | O‐Ac‐Thr |
| Rhodamide Q (41) | C86H146N14O39 | 999.4961 | 5.2 | C15H31 (C16:0) | Succ‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide R (42) | C90H156N14O36 | 1004.5404 | 18.4 | C23H45 (C24:1) | Succ‐Hex | Hex | H | O‐Ac‐Thr | Leu/Ile |
| Rhodamide S (43) | C90H154N14O39 | 1027.5263 | 13.1 | C19H39 (C20:0) | Succ‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide T (44) | C92H156N14O39 | 1040.5341 | 13.1 | C21H41 (C22:1) | Succ‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide U (45) | C92H158N14O39 | 1041.5411 | 19.2 | C21H43 (C22:0) | Succ‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide V (46) | C90H150N14O42 | 1049.5042 | 5.4 | C15H31 (C16:0) | Disucc‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide W (47) | C94H161N15O38 | 1054.0575 | 16.5 | C23H45 (C24:1) | Succ‐Hex | H | Succ‐Hex | N‐γ‐Ac‐Dab | Leu/Ile |
| Rhodamide X (48) | C94H160N14O39 | 1054.5492 | 18.3 | C23H45 (C24:1) | Succ‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide Y (49) | C94H158N14O41 | 1069.5369 | 16.9 | C23H45 (C24:1) | Succ‐Hex | H | Succ‐Hex | O‐Ac‐Thr | O‐Ac‐Thr |
| Rhodamide Z (50) | C96H160N14O42 | 1090.5419 | 13.2 | C21H41 (C22:1) | Disucc‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide A1 (51) | C96H162N14O42 | 1091.5485 | 18.8 | C21H43 (C22:0) | Disucc‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide B1 (52) | C98H165N15O41 | 1104.0656 | 16.3 | C23H45 (C24:1) | Disucc‐Hex | H | Succ‐Hex | N‐γ‐Ac‐Dab | Leu/Ile |
| Rhodamide C1 (53) | C98H164N14O42 | 1104.5573 | 18.0 | C23H45 (C24:1) | Disucc‐Hex | H | Succ‐Hex | O‐Ac‐Thr | Leu/Ile |
| Rhodamide D1 (54) | C98H162N14O44 | 1119.5451 | 16.8 | C23H45 (C24:1) | Disucc‐Hex | H | Succ‐Hex | O‐Ac‐Thr | O‐Ac‐Thr |
Abbreviations: aa, amino acid; Dab, diaminobutyric acid; Disucc, disuccinyl; FA, fatty acid; Hex, hexosyl; R t, retention time; Succ, succinoyl.
Fatty acids have been reported in parenthesis using the LIPID MAPS shorthand notation (Liebisch et al., 2020). Fatty acyl chains are indicated as C:N;O, where C is the number of carbon atoms, N is the number of double bond equivalents and O is the number of additional oxygen atoms linked to the hydrocarbon chain.
FIGURE 7.
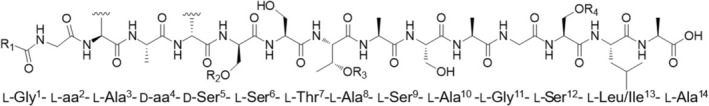
General structure of rhodamides.
The product ion spectra of [M + 2H]2+ ions of rhodamides at NCE 15 displayed y 12 ‐y 2 and b 12 ‐b 1 ions together with b‐type internal fragments arising from fragmentation of the y 10 ion, the latter being useful to identify the last two amino acids as Ile/Leu13 and Ala14 (Figures 8 and Figure S24–S26). The observed fragmentation pattern at NCE 15 allowed for the assignment of the peptide sequence as shown in Table 4, however, it was necessary to increase collision energy (NCE 30) to identify the structure of the b 1 fragment ion, which features in all rhodamide isoforms a Gly residue loss to yield the acylium ion corresponding to the fatty acid starter unit (Figure 8B). With this regard, rhodamides were shown to possess C16:0, C20:0 C22:0, C22:1 and C24:1 fatty acyl chains (Table 4).
FIGURE 8.
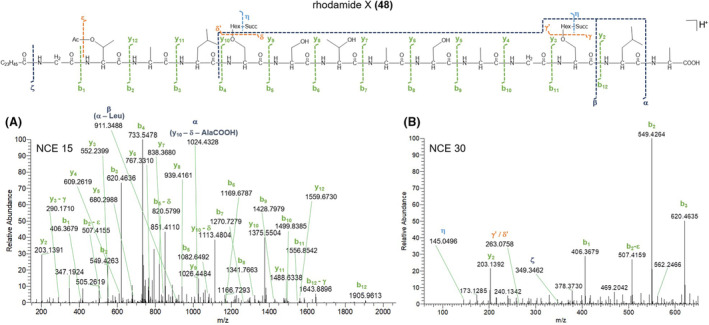
HR‐MS2 spectra of the [M + 2H]2+ pseudo‐molecular ion of rhodamide X (48) acquired at NCE 15 (A) and 30 (B) and its proposed fragmentation pathway. Succ, succinoyl; Hex, hexosyl; Ac, acetyl.
As a result, the peptide backbone of rhodamides was determined as Gly1‐aa2‐Ala3‐aa4‐Ser5‐Ser6‐Thr7‐Ala8‐Ser9‐Ala10‐Gly11‐Ser12‐Leu/Ile13‐Ala14, where the different congeners contain (a) O‐acetylthreonine (O‐Ac‐Thr) or N‐γ‐acetyl‐2,4‐diaminobutyric acid (N‐γ‐Ac‐Dab) and (b) Leu/Ile or O‐Ac‐Thr at the variable amino acid positions 2 and 4, respectively. Particularly, the presence of O‐Ac‐Thr or N‐γ‐Ac‐Dab at positions 2 and/or 4 was inferred from the loss of the acetyl group as ketene (C2H2O, 42.0106 Da) from b 2 and/or b 4 fragment ions, followed by the loss of a Thr or Dab residue (Figures 8, and Figure S24–S26). An in‐depth investigation of the MS/MS spectra led to the identification and position assignment of the substitution patterns of rhodamides as detailed in Table 4. Rhodamides bear up to two pendant hexosyl groups, which may undergo or not mono‐ or di‐succinylation. Indeed, the MS/MS spectra of rhodamides displayed low‐abundance fragment ions at m/z 163.0601 (C6H11O5 +), m/z 145.0495 (C6H9O4 +) and/or m/z 263.0761 (C10H15O8 +) and/or m/z 363.0922 (C14H19O11 +), as well as the corresponding neutral losses, which were indicative of the dehydrated forms of the hexoside (Hex), succinoyl‐hexoside (Succ‐Hex) and disuccinoyl‐hexoside (Disucc‐Hex) moieties (Figures 8B and Figure S24B–S26B). Indeed, fragment ions at m/z 263.0761 and m/z 363.0922 undergo further fragmentation resulting in the neutral loss of one molecule of succinic acid (C4H6O4, 118.0260 amu) and sequential losses of succinic acid and succinic anhydride (C4H4O3, 100.0160 amu), respectively, thereby suggesting one or two succinate units ester linked to the hexosyl group. The product ion spectra of rhodamides bearing the Hex/Succ‐Hex/Disucc‐Hex on Ser5 side‐chain hydroxy function exhibit glycosylated b 12 ‐b 5 ion fragments (besides the corresponding non‐glycosylated counterparts) but only non‐glycosylated b 4 ‐b 1 ions, thus allowing for the unambiguous assignment of the substituent position (Figure S24). Also, rhodamide variants with a pendant glycosyl moiety on Thr7 or Ser12 side‐chain hydroxy group display glycosylated y 12 ‐y 8 or y 12 ‐y 3 ions (together with the corresponding non‐glycosylated fragments) but only the non‐glycosylated y 7 ‐y 2 or y 2 ions (Figure S25 and S26). The simultaneous presence of glycosylated y 12 ‐y 8 /y 12 ‐y 3 and b 12 ‐b 5 ion series in MS/MS spectra was indicative of rhodamide congeners featuring substituents on both the Thr7/Ser12 and Ser5 side‐chains (Figure 8).
The genomics‐driven absolute configuration assignment of rhodamides indicated the presence of d‐Leu/Ile4, d‐ allo‐O‐Ac‐Thr4 and d‐Ser5, as the rhodamide synthetase features two epimerization domains within modules 4 and 5, respectively (Figure 3). Consequently, the remaining amino acids were assumed to possess the l configuration.
Finally, the structures of 33 glycolipopeptides have been reported here for the first time. Interestingly, a group of cell wall glycolipopeptides related to rhodamides has been isolated from R. erytropolis 134 and differs from rhodamides in the amino acid composition, the substitution pattern and the N‐terminal fatty acid moiety, including mycolic acids besides normal fatty acids (Koronelli, 1988).
In addition, while glycolipopeptides from R. erytropolis 134 and peptidolipids from R. equi and R. opacus have been described as cell wall components, rhodamides and rhodoheptins have been detected both in the cell pellet and the supernatant of R. I2R. Therefore, it can be argued that the bacterium may secrete these biosurfactants to emulsify and intake hexadecane from the growth medium (see Section 4.2), as widely reported for Corynebacteriaceae (Koronelli, 1988).
Bioactivity evaluation of rhodoheptins and rhodamides
The F5 fraction (including rhodoheptins, rhodamides, and trehalolipids) exhibited biosurfactant activity in the oil‐spreading assay, causing oil displacement and forming a clear zone in the oil layer (Figure S4) as for other surface active agents of microbial origin (Giugliano et al., 2023; Buonocore et al., 2020). Therefore, the identification of rhodoheptins and rhodamides further expands the biosurfactant repertoire of R. I2R, previously recognized solely as a source of glycolipid biosurfactants (Palma Esposito et al., 2021). An increasing body of evidence suggests that many biosurfactants may interfere with different steps of cancer development and bacterial infections (Wang et al., 2024). Consequently, we assessed potential antimicrobial and antiproliferative effects of LPs from R. I2R.
F5 did not show any activity in antibacterial assays against Staphylococcus aureus ATCC 6538, Listeria monocytogenes MB 677, E. coli ATCC 10536, and Pseudomonas aeruginosa PA01.
Moreover, F5 and a purified rhodoheptin‐enriched fraction were evaluated for antiproliferative activity against human A375 melanoma cells using the MTT assay (Figure 9). The purified rhodoheptin‐enriched fraction was obtained following the procedure described in Section 4.2 and its composition was assessed using LC‐HRMS2 (Figure S27 and Table S4), thus confirming the presence of all the rhodoheptin variants listed in Table 3. F5 induced approximately 50% cancer cell death after a 48‐hour treatment at the highest concentration (500 μg/mL). In contrast, the purified rhodoheptin‐enriched fraction elicited weaker yet significant growth inhibition (33%) in A375 cells at the same concentration. The synergestic effects of rhodoheptins, trehalolipids and presumably rhodamides may explain the increased reduction in cell viability, considering that trehalolipids from R. I2R have been already shown to induce cytotoxicity against PC3 prostate cancer cells (Palma Esposito et al., 2021).
FIGURE 9.
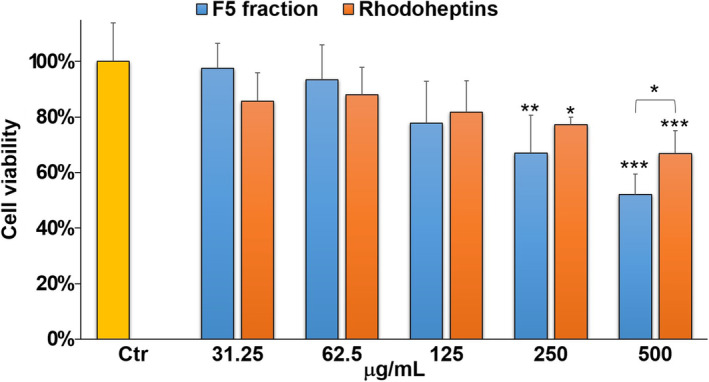
Cell viability of A375 cells exposed to 0.25% DMSO vehicle (Ctr) and different concentrations of F5 and rhodoheptin mixture (31.25, 62.5, 125, 250 or 500 μg/mL). Data have been normalized to the relevant control (Ctr) and are presented as mean ± SD; n = 4. Where not indicated, statistical significances are referred to as control (Ctr). *p < 0.05; **p < 0.01; ***p < 0.001.
Rhodoheptin and rhodamide‐related lipopeptides from other Rhodococcus strains
As R. equi and R. opacus are able to synthesize cyclic and linear LPs structurally related to rhodoheptins, genomes of both species (NCBI accession numbers NZ_CP027793.1 and NZ_CP080954.1) were mined in silico for the relevant BGCs. Unfortunately, the genome of R. erytropolis 134 is not publicly available and, therefore, it was not possible to look for the BGC encoding for the biosynthesis of the rhodamide‐related glycolipopeptides described by Koronelli. (1988). Nevertheless, an antiSMASH analysis of several R. erytropolis strains (CCM2595, JCM 2895, X5, CERE8 and D310) disclosed the presence of multimodular NRPS putatively involved in the assemblage of serine‐ and threonine‐containing LPs resembling rhodamide architecture.
As expected, R. equi and R. opacus were found to host the heptamodular NRPS gene cluster, specifically WP_106851421.1 and WP_025433610.1, respectively, featuring the same organization as rhp and showing collinearity with the peptidolipid family described by Frankfater and co‐workers (Figure S28). These findings further support the link between the BGCs and the encoded molecules, thus shedding light on a biosynthetic route conserved across different Rhodococcus strains. Basically, rhodoheptins from R. I2R differ from peptidolipid variants of R. equi and R. opacus as a result of (a) the relaxed substrate selectivity of the adenylation domain present in the last module, which allows for the accommodation of Leu/Ile and MetO, beyond Phe, and (b) the fatty acyl moiety specificity of the Cs domain, which selects C16 and C17, beyond C18‐C24 β‐hydroxy fatty acids.
In the most general terms, the LP profile of R. I2R is more similar to that of R. equi as being exclusively characterized by cyclic LPs. Notably, detection of a putative esterase‐encoding gene (WP_005249251.1) flanking the NRPS cluster solely in R. opacus could explain why this strain is able to produce also linear LPs, likely arising from the corresponding cyclic variants after hydrolytic cleavage of the lactone ring. As for the rhodoheptin synthetase, the heptamodular NRPS from R. equi contains two E domains within the modules 3 and 5, thus unveiling the conserved d absolute configuration of aa‐3 and ‐5 (Figure S28). On the other hand, the heptamodular NRPS from R. opacus bears two E domains within modules 1 and 6, thus accounting for a different stereochemistry of the entire LP family in this strain (Figure S28).
To determine if the rhodoheptin BGC is widespread in Rhodococcus species, a cblaster analysis (Gilchrist et al., 2021) was performed. All genomic data from Rhodococcus available on NCBI were used. We detected a total of 2224 clusters distributed across 1936 genomic scaffolds originating from 856 different organisms. Among these, 322 unique species were identified to have at least 60% of the genes present in the query cluster (Figure S29). For the rhodamide BGC, cblaster detected 2499 clusters across 2150 genomic scaffolds from 860 organisms within the NCBI Rhodococcus data. Of these, 478 unique species were found to contain at least 60% of the genes present in the query cluster (Figure S30). This indicates a wide distribution of the rhodoheptin and rhodamide biosynthetic pathways across a diverse set of Rhodococcus species.
CONCLUSIONS
The genome and metabolome analysis of R. I2R enabled the structure and biosynthetic pathway discovery of two LP families with biosurfactant and moderate antiproliferative activity. These families would have remained cryptic if a genomic bottom‐up approach had not been applied. Indeed, genome mining guided the selection of tailored growth media to unlock LP biosynthesis and supported detection and stereo‐structural elucidation of rhodoheptins and rhodamides as well. So far, few studies report about LPs from Rhodococcus spp. (Chiba et al., 1999; Habib et al., 2020; Peng et al., 2008), as the most prolific strains belong to Bacillus and Paenibacillus species, Pseudomonas spp., Cyanobacteria and Actinomycetota, mainly represented by the Streptomyces genus (Carolin et al., 2021; Cock & Cheesman, 2023; Li et al., 2021; Zhang et al., 2023). Therefore, our results represent a step forward in the investigation of the underexplored Rhodococcus genus as a biotechnological resource. Nevertheless, as widely documented for biosurfactants (Hashemi et al., 2024), the low microbial productivity remained a bottleneck which hampered isolation and NMR structural characterization of rhodoheptins and rhodamides. Notably, R. I2R was shown to possess multiple biosynthetic routes for biosurfactant production, thus making this bacterium a factory of diverse surface‐active natural products, i.e., saccharide succinic esters (Palma Esposito et al., 2021), LPs and glycolipopeptides. As R. I2R has been isolated from deep‐sea sediments, the bacterium has likely maintained this synergy as a result of the environmental pressure (e.g. light and nutrient shortage, high pressure, and salinity), which might have imposed R. I2R to synthesize molecules such as biosurfactants, for complex carbon source degradation, nutrient retrieval and chemical defence. Due to the novelty of the chemical structures and the availability of the BGCs of rhodoheptins and rhodamides, our future work aims at the large‐scale production of these potentially industrially relevant biosurfactants through heterologous expression.
EXPERIMENTAL PROCEDURES
Bioinformatics
The draft genome of R. I2R (accession: JAHUTG000000000) consisting of 72 contigs was assembled into 36 scaffolds employing the software MeDuSa (Multi‐Draft based Scaffolder) (Bosi et al., 2015) and selecting the genomes of Rhodococcus sp. P1Y (NZ_CP032762.1) and Rhodococcus sp. BP‐261 (NZ_JABUCO000000000.1) as references to guide contigs ordering and orientating, as previously described (Palma Esposito et al., 2023). BGCs encoding the biosynthesis of LPs were identified by using the genome mining tool antiSMASH 7.0 (Blin et al., 2023). The rhodoheptin and rhodamide gene clusters were deposited in GenBank under accession numbers PP729154 and PP729155, respectively. Cblaster (Gilchrist, et al. 2021) was run using the search module (default settings), against the NCBI Database (limited to Rhodococcus genomes). To visualize the similarity of the clusters, clinker was used.
Cultivation, extraction and fractionation
The marine bacterium R. I2R was isolated on agar plates containing mineral salt medium (MSM) supplemented with 1.0 mM of phenanthrene from deep‐sea sediments collected in the Southern Tyrrhenian Sea, as previously reported (Palma Esposito et al., 2021). For LP production, R. I2R was grown at 28°C under shaking (200 rpm) for 6 days in 200 mL of ASG medium (casamino acids 3 g/L; MgSO4 × 7 H2O 12 g/L; KCl 0.75 g/L; NaCl 15 g/L; CaCl2 3 g/L; NH4Cl 1 g/L; NaHCO3 0.17 g/L) supplemented with 0.5% (v/v) hexadecane and 0.5 μM FeCl3. Hexadecane and iron were added as potential hydrophobic and hydrophilic inducers, respectively, to stimulate LP synthesis (de Oliveira Schmidt et al., 2021; Peng et al., 2008; Solyanikova & Golovleva, 2019). After the 6 days of growth, biomass of R. I2R and exhausted broth were harvested separately and extracted with MeOH (3 × 100 mL) and AcOEt (2 × 200 mL) respectively. The combined MeOH and AcOEt extracts (1.734 g) were purified by reversed‐phase chromatography using a C18 SPE column eluted with different mixtures of H2O and MeOH to obtain five fractions: F1, 100% H2O (88.9 mg); F2, 50% MeOH (32.7 mg); F3, 90% MeOH (51,8 mg); F4, 100% MeOH (20.4 mg); F5, 100% MeOH supplemented with 0.1% TFA (6.5 mg). To obtain the rhodoheptin‐enriched fraction, crude bacterial extract was separated over a C18 SPE column using the above‐mentioned gradient elution and finally flushed with CHCl3, to collect an additional fraction (0.5 mg) containing only residual rhodoheptins previously retained by the stationary phase.
Liquid chromatography–high‐resolution tandem mass spectrometry (LC‐HRMS2 )
The five SPE fractions were suspended in CH3OH at a concentration of 1 mg/mL and analysed by untargeted LC‐HRMS2 on a Thermo Scientific Q Exactive Focus Orbitrap mass spectrometer coupled to a Thermo Ultimate 3000 HPLC System equipped with an Hypersil C18 column (100 × 4.6 mm, 3 μm). The RP18 column was maintained at 25°C and eluted at 400 μL/min with H2O and CH3CN, both supplemented with 0.1% formic acid, setting the following gradient program: 70% CH3CN 10 min (equilibration), 70% → 75% CH3CN over 3 min, 75% → 85% CH3CN over 5 min, 85% → 100% CH3CN over 22 min, 100% CH3CN 11 min. MS spectra were recorded in the positive ion detection mode and HESI source parameters were set as follows: a sheath gas flow rate of 32 units N2, an auxiliary gas flow rate of 15 units N2, a spray voltage of 4.8 kV, a capillary temperature of 285°C, an S‐lens RF level of 55 and an auxiliary gas heater temperature of 150°C. MS survey scans (800–1400 m/z) were acquired at a resolution of 70,000 and an AGC target of 1e6. HRMS2 spectra were acquired in the DDA mode at a resolution of 70,000 and an AGC target of 5e4, setting three MS2 events after each full MS scan. HRMS2 scans were obtained with HCD fragmentation, using an isolation width of 2.0 m/z, a normalized collision energy of 15 and 30 units and an automated injection time.
Feature‐based molecular networking (FBMN)
To chart a metabolic overview of R. I2R, the molecular network of the F1‐F5 fractions from R. I2R was generated with the FBMN workflow (Nothias et al., 2020; Teta et al., 2016) on GNPS (https://gnps.ucsd.edu), as previously reported (Teta et al., 2021). MS raw data were initially processed by MZmine2 (Pluskal et al., 2010) using the parameters reported in the Supplementary Material (Table S3). Briefly, after chromatogram building and deconvolution, peaks from the SPE fractions and blank sample (i.e. methanol used for fraction suspension) were aligned to subtract the background spectrum and eliminate interference. In addition, [M + Na–H], [M + K–H], [M + Mg − 2H], [M + NH3], [M‐Na + NH4], [M + 1, 13C] adducts and peaks without associated MS2 spectra were filtered out. The results were exported as mgf file to GNPS for FBMN analysis. For FBMN analysis, the precursor ion mass tolerance was set to 0.02 Da and the MS2 fragment ion tolerance to 0.05 Da. A molecular network was then created where edges were filtered to have a cosine score above 0.7 and more than three matched peaks. The molecular network (Figure S1) was visualized using the Cytoscape software (Shannon et al., 2003) and can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=1a8c0d5b8c0a42aaaa1a8b5520f5764c. Following the same workflow as described above, a molecular network of the F5 fraction containing rhodoheptins and rhodamides was created (Figure 4) and is available at the following link https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4a962a758d304461a855f935f0295109.
Oil‐spreading assay
The oil‐spreading assay was performed following the protocol previously reported (Giugliano et al., 2023). In brief, 60 μL of exhaust motor oil was added to 25 mL of distillate water in a Petri dish (Ø 9 cm) to create a thin oil layer. Then, F1‐F5 fractions were dissolved in DMSO at 50 mg/mL concentration and 1 μL of each fraction was poured into the centre of the oil layer. The presence of biosurfactants was indicated by oil displacement and formation of a clear zone in the oil layer.
Cell viability assay
Cell viability was assessed by the MTT assay as previously described (Villanova et al., 2022). A375 melanoma cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, penicillin–streptomycin (100 U/mL) and 2 mM L‐glutamine, at 37°C under a 5% CO2 humidified atmosphere. A375 cells were seeded at a cell density of 6000 cells/well in a final volume of 100 μL per well. Approximately 24 h after seeding, medium was removed and cells were treated with medium containing DMSO vehicle, F5 fraction and rhodoheptin mixture at the indicated concentrations for 48 h. Samples were dissolved in DMSO to prepare 200 mg/mL stock solutions and then diluted in RPMI medium for antiproliferative assays. In all experiments, the final concentration of DMSO did not exceed 0.25%, as it was not toxic to A375 cancer cells. After 48 h treatment, 0.5 mg/mL MTT was added to each well. Then, cells were incubated for 3 h at 37°C in a 5% CO2 humidified atmosphere. After incubation, medium was discarded and formazan salts generated by living cells were dissolved in 100 μL isopropanol at room temperature for 60 min under shaking. To assess cell viability, sample absorbance was measured at 570 nm using an Infinite M1000Pro (TECAN, Männedorf, Switzerland) plate reader.
Data from MTT assay are reported as the mean ± standard deviation (SD) of four independent experiments. Data statistical analysis was performed by using the GraphPad Prism Software version 5. One‐way analysis of variance (ANOVA) and Student's t‐test were used to compare means between groups. Dunnett test was applied as a post hoc test in ANOVA for multiple comparisons with the control group. Differences were considered statistically significant if p < 0.05.
Antibacterial assays
Antibacterial activity was evaluated following the broth microdilution method, as previously reported (Giugliano et al., 2023). Briefly, after reaching the log‐phase, Staphylococcus aureus ATCC 6538, E. coli ATCC 10536, and Pseudomonas aeruginosa PA01 were diluted in MH (Mueller Hinton) medium and dispensed into a microtitre plate to have a titre of 5 × 104 CFU per well in a final volume of 200 μL. Unlike the other pathogens, Listeria monocytogenes MB 677 was diluted in TSB medium supplemented with 0.6% yeast extract. Pathogens were treated with 2% DMSO vehicle (control) and the F5 fraction at different concentrations (1.0–0.500–0.250–0.125–0.062 mg/mL) for 24 h at 37°C. Antibacterial activity was assessed by comparing the optical density (OD) values for the treated strain with the control. OD was measured at 600 nm by using a Tecan plate reader (Tecan, Männedorf, Switzerland).
AUTHOR CONTRIBUTIONS
Costanza Ragozzino: Data curation; methodology; investigation; validation. Fortunato Palma Esposito: Investigation; methodology; data curation; writing – review and editing; conceptualization. Carmine Buonocore: Investigation; writing – review and editing; visualization. Pietro Tedesco: Visualization; writing – review and editing. Daniela Coppola: Visualization; writing – review and editing. Davide Paccagnella: Investigation; formal analysis; data curation; writing – review and editing. Nadine Ziemert: Writing – review and editing; visualization; validation; formal analysis; supervision. Gerardo Della Sala: Conceptualization; investigation; methodology; supervision; writing – review and editing; writing – original draft; validation; formal analysis; data curation. Donatella de Pascale: Conceptualization; funding acquisition; writing – review and editing; visualization; resources; project administration; supervision.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
Data S1:
ACKNOWLEDGEMENTS
This research was funded by H2020‐FNR‐11‐2020: SECRETED—Grant agreement: 101000794.
Ragozzino, C. , Palma Esposito, F. , Buonocore, C. , Tedesco, P. , Coppola, D. , Paccagnella, D. et al. (2024) Integrated genome and metabolome mining unveiled structure and biosynthesis of novel lipopeptides from a deep‐sea Rhodococcus . Microbial Biotechnology, 17, e70011. Available from: 10.1111/1751-7915.70011
Costanza Ragozzino and Fortunato Palma Esposito shared first co‐authorship.
Gerardo Della Sala and Donatella de Pascale shared last co‐authorship.
Contributor Information
Gerardo Della Sala, Email: gerardo.dellasala@szn.it.
Donatella de de Pascale, Email: donatella.depascale@szn.it.
DATA AVAILABILITY STATEMENT
The molecular networks and mass specrometry data can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=1a8c0d5b8c0a42aaaa1a8b5520f5764c and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4a962a758d304461a855f935f0295109. The rhodoheptin and rhodamide gene clusters were deposited in GenBank under accession numbers PP729154 and PP729155, respectively.
REFERENCES
- Blin, K. , Shaw, S. , Augustijn, H.E. , Reitz, Z.L. , Biermann, F. , Alanjary, M. et al. (2023) antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Research, 51(W1), W46–W50. Available from: 10.1093/nar/gkad344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi, E. , Donati, B. , Galardini, M. , Brunetti, S. , Sagot, M.F. , Lió, P. et al. (2015) MeDuSa: a multi‐draft based scaffolder. Bioinformatics (Oxford, England), 31, 2443–2451. Available from: 10.1093/bioinformatics/btv171 [DOI] [PubMed] [Google Scholar]
- Buonocore, C. , Giugliano, R. , Della Sala, G. , Palma Esposito, F. , Tedesco, P. , Folliero, V. et al. (2023) Evaluation of antimicrobial properties and potential applications of pseudomonas gessardii M15 Rhamnolipids towards multiresistant Staphylococcus aureus . Pharmaceutics, 15(2), 700. Available from: 10.3390/pharmaceutics15020700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore, C. , Tedesco, P. , Vitale, G.A. , Esposito, F.P. , Giugliano, R. , Monti, M.C. et al. (2020) Characterization of a new mixture of mono‐Rhamnolipids produced by pseudomonas gessardii isolated from Edmonson point (Antarctica). Marine Drugs, 18(5), 269. Available from: 10.3390/md18050269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu, D.C. , Chen, Y. & Reilly, P.J. (2010) Thioesterases: a new perspective based on their primary and tertiary structures. Protein Science: A Publication of the Protein Society, 19(7), 1281–1295. Available from: 10.1002/pro.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X.H. , Wang, A.H. , Wang, C.L. , Mao, D.Z. , Lu, M.F. , Cui, Y.Q. et al. (2010) Surfactin induces apoptosis in human breast cancer MCF‐7 cells through a ROS/JNK‐mediated mitochondrial/caspase pathway. Chemico‐Biological Interactions, 183(3), 357–362. Available from: 10.1016/j.cbi.2009.11.027 [DOI] [PubMed] [Google Scholar]
- Carolin, C.F. , Kumar, P.S. & Ngueagni, P.T. (2021) A review on new aspects of lipopeptide biosurfactant: types, production, properties and its application in the bioremediation process. Journal of Hazardous Materials, 407, 124827. Available from: 10.1016/j.jhazmat.2020.124827 [DOI] [PubMed] [Google Scholar]
- Ceresa, C. , Fracchia, L. , Sansotera, A.C. , De Rienzo, M.A.D. & Banat, I.M. (2023) Harnessing the potential of biosurfactants for biomedical and pharmaceutical applications. Pharmaceutics, 15(8), 2156. Available from: 10.3390/pharmaceutics15082156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Lu, Y. , Shan, M. , Zhao, H. , Lu, Z. & Lu, Y. (2022) A mini‐review: mechanism of antimicrobial action and application of surfactin. World Journal of Microbiology and Biotechnology, 38(8), 143. Available from: 10.1007/s11274-022-03323-3 [DOI] [PubMed] [Google Scholar]
- Chiba, H. , Agematu, H. , Sakai, K. , Dobashi, K. & Yoshioka, T. (1999) Rhodopeptins, novel cyclic tetrapeptides with antifungal activities from Rhodococcus sp. III. Synthetic study of rhodopeptins. The Journal of Antibiotics, 52(8), 710–720. Available from: 10.7164/antibiotics.52.710 [DOI] [PubMed] [Google Scholar]
- Chooi, Y.H. & Tang, Y. (2010) Adding the lipo to lipopeptides: do more with less. Chemistry and Biology, 17(8), 791–793. Available from: 10.1016/j.chembiol.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Cock, I.E. & Cheesman, M.J. (2023) A review of the antimicrobial properties of cyanobacterial natural products. Molecules (Basel, Switzerland), 28(20), 7127. Available from: 10.3390/molecules28207127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Schmidt, V.K. , de Souza Carvalho, J. , de Oliveira, D. & de Andrade, C.J. (2021) Biosurfactant inducers for enhanced production of surfactin and rhamnolipids: an overview. World Journal of Microbiology and Biotechnology, 37(2), 21. Available from: 10.1007/s11274-020-02970-8 [DOI] [PubMed] [Google Scholar]
- Della Sala, G. , Mangoni, A. , Costantino, V. & Teta, R. (2020) Identification of the biosynthetic gene cluster of Thermoactinoamides and discovery of new congeners by integrated genome mining and MS‐based molecular networking. Frontiers in Chemistry, 8, 397. Available from: 10.3389/fchem.2020.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, G. , Bharti, R. , Dhanarajan, G. , Das, S. , Dey, K.K. , Kumar, B.N. et al. (2015) Marine lipopeptide Iturin a inhibits Akt mediated GSK3β and FoxO3a signaling and triggers apoptosis in breast cancer. Scientific Reports, 5, 10316. Available from: 10.1038/srep10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, M.A.M. & Nitschke, M. (2023) Bacterial‐derived surfactants: an update on general aspects and forthcoming applications. Brazilian Journal of Microbiology, 54(1), 103–123. Available from: 10.1007/s42770-023-00905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L. , Sánchez, C. , Chen, M. , Edwards, D.J. & Shen, B. (2000) The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chemistry and Biology, 7(8), 623–642. Available from: 10.1016/s1074-5521(00)00011-9 [DOI] [PubMed] [Google Scholar]
- Esposito, F.P. , Vecchiato, V. , Buonocore, C. , Tedesco, P. , Noble, B. , Basnett, P. et al. (2023) Enhanced production of biobased, biodegradable, poly(3‐hydroxybutyrate) using an unexplored marine bacterium Pseudohalocynthiibacter aestuariivivens, isolated from highly polluted coastal environment. Bioresource Technology, 368, 128287. Available from: 10.1016/j.biortech.2022.128287 [DOI] [PubMed] [Google Scholar]
- Felnagle, E.A. , Rondon, M.R. , Berti, A.D. , Crosby, H.A. & Thomas, M.G. (2007) Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin. Applied and Environmental Microbiology, 73(13), 4162–4170. Available from: 10.1128/AEM.00485-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfater, C. , Henson, W.R. , Juenger‐Leif, A. , Foston, M. , Moon, T.S. , Turk, J. et al. (2020) Structural determination of a new Peptidolipid family from Rhodococcus opacus and the pathogen Rhodococcus equi by multiple stage mass spectrometry. Journal of the American Society for Mass Spectrometry, 31(3), 611–623. Available from: 10.1021/jasms.9b00059 [DOI] [PubMed] [Google Scholar]
- Gilchrist, C.L.M. , Booth, T.J. , van Wersch, B. , van Grieken, L. , Medema, M.H. & Chooi, Y.H. (2021) Cblaster: a remote search tool for rapid identification and visualization of homologous gene clusters. Bioinformatics Advances, 1, vbab016. Available from: 10.1093/bioadv/vbab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giugliano, R. , Della Sala, G. , Buonocore, C. , Zannella, C. , Tedesco, P. , Palma Esposito, F. et al. (2023) New Imidazolium alkaloids with broad Spectrum of action from the marine bacterium Shewanella aquimarina . Pharmaceutics, 15(8), 2139. Available from: 10.3390/pharmaceutics15082139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib, S. , Ahmad, S.A. , Wan Johari, W.L. , Abd Shukor, M.Y. , Alias, S.A. , Smykla, J. et al. (2020) Production of Lipopeptide biosurfactant by a hydrocarbon‐degrading Antarctic Rhodococcus . International Journal of Molecular Sciences, 21(17), 6138. Available from: 10.3390/ijms21176138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi, S.Z. , Fooladi, J. , Vahidinasab, M. , Hubel, P. , Pfannstiel, J. , Pillai, E. et al. (2024) Toward effects of hydrophobicity on biosurfactant production by Bacillus subtilis isolates from crude‐oil‐exposed environments. Applied Microbiology, 4, 215–236. Available from: 10.3390/applmicrobiol4010015 [DOI] [Google Scholar]
- Hoertz, A.J. , Hamburger, J.B. , Gooden, D.M. , Bednar, M.M. & McCafferty, D.G. (2012) Studies on the biosynthesis of the lipodepsipeptide antibiotic Ramoplanin A2. Bioorganic and Medicinal Chemistry, 20(2), 859–865. Available from: 10.1016/j.bmc.2011.11.062 [DOI] [PubMed] [Google Scholar]
- Hojati, Z. , Milne, C. , Harvey, B. , Gordon, L. , Borg, M. , Flett, F. et al. (2002) Structure, biosynthetic origin, and engineered biosynthesis of calcium‐dependent antibiotics from Streptomyces coelicolor. Chemistry and Biology, 9(11), 1175–1187. Available from: 10.1016/s1074-5521(02)00252-1 [DOI] [PubMed] [Google Scholar]
- Huang, H.W. (2020) DAPTOMYCIN, its membrane‐active mechanism vs. that of other antimicrobial peptides. Biochimica et Biophysica Acta, Biomembranes, 1862(10), 183395. Available from: 10.1016/j.bbamem.2020.183395 [DOI] [PubMed] [Google Scholar]
- Jahan, R. , Bodratti, A.M. , Tsianou, M. & Alexandridis, P. (2020) Biosurfactants, natural alternatives to synthetic surfactants: physicochemical properties and applications. Advances in Colloid and Interface Science, 275, 102061. Available from: 10.1016/j.cis.2019.102061 [DOI] [PubMed] [Google Scholar]
- Janek, T. , Krasowska, A. , Radwańska, A. & Łukaszewicz, M. (2013) Lipopeptide biosurfactant pseudofactin II induced apoptosis of melanoma a 375 cells by specific interaction with the plasma membrane. PLoS One, 8(3), e57991. Available from: 10.1371/journal.pone.0057991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronelli, T.V. (1988) Investigation of the lipids of saprophytic mycobacteria in the U.S.S.R. Journal of Chromatography, 440, 479–486. Available from: 10.1016/s0021-9673(00)94553-9 [DOI] [PubMed] [Google Scholar]
- Kraas, F.I. , Helmetag, V. , Wittmann, M. , Strieker, M. & Marahiel, M.A. (2010) Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chemistry and Biology, 17(8), 872–880. Available from: 10.1016/j.chembiol.2010.06.015 [DOI] [PubMed] [Google Scholar]
- Li, J.Y. , Wang, L. , Liu, Y.F. , Zhou, L. , Gang, H.Z. , Liu, J.F. et al. (2021) Microbial Lipopeptide‐producing strains and their metabolic roles under anaerobic conditions. Microorganisms, 9(10), 2030. Available from: 10.3390/microorganisms9102030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch, G. , Fahy, E. , Aoki, J. , Dennis, E.A. , Durand, T. , Ejsing, C.S. et al. (2020) Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS‐derived lipid structures. Journal of Lipid Research, 61(12), 1539–1555. Available from: 10.1194/jlr.S120001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarvey, N.A. , Haltli, B. , He, M. , Greenstein, M. & Hucul, J.A. (2006) Biosynthetic pathway for mannopeptimycins, lipoglycopeptide antibiotics active against drug‐resistant gram‐positive pathogens. Antimicrobial Agents and Chemotherapy, 50(6), 2167–2177. Available from: 10.1128/AAC.01545-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareš, J. , Hájek, J. , Urajová, P. , Kopecký, J. & Hrouzek, P. (2014) A hybrid non‐ribosomal peptide/polyketide synthetase containing fatty‐acyl ligase (FAAL) synthesizes the β‐amino fatty acid lipopeptides puwainaphycins in the cyanobacterium Cylindrospermum alatosporum. PLoS One, 9(11), e111904. Available from: 10.1371/journal.pone.0111904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagtode, V.S. , Cardoza, C. , Yasin, H.K.A. , Mali, S.N. , Tambe, S.M. , Roy, P. et al. (2023) Green surfactants (biosurfactants): a petroleum‐free substitute for sustainability‐comparison, applications, market, and future prospects. ACS Omega, 8(13), 11674–11699. Available from: 10.1021/acsomega.3c00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias, L.F. , Petras, D. , Schmid, R. , Dührkop, K. , Rainer, J. , Sarvepalli, A. et al. (2020) Feature‐based molecular networking in the GNPS analysis environment. Nature Methods, 17(9), 905–908. Available from: 10.1038/s41592-020-0933-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma Esposito, F. , Giugliano, R. , Della Sala, G. , Vitale, G.A. , Buonocore, C. , Ausuri, J. et al. (2021) Combining OSMAC approach and untargeted metabolomics for the identification of new glycolipids with potent antiviral activity produced by a marine Rhodococcus . International Journal of Molecular Sciences, 22(16), 9055. Available from: 10.3390/ijms22169055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, R. , Bai, X. , Chen, J. , Zhang, H. & Wang, H. (2019) Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: a literature review. Frontiers in Microbiology, 10, 294. Available from: 10.3389/fmicb.2019.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, F. , Wang, Y. , Sun, F. , Liu, Z. , Lai, Q. & Shao, Z. (2008) A novel lipopeptide produced by a Pacific Ocean deep‐sea bacterium, Rhodococcus sp. TW53. Journal of Applied Microbiology, 105(3), 698–705. Available from: 10.1111/j.1365-2672.2008.03816.x [DOI] [PubMed] [Google Scholar]
- Pilz, M. , Cavelius, P. , Qoura, F. , Awad, D. & Brück, T. (2023) Lipopeptides development in cosmetics and pharmaceutical applications: a comprehensive review. Biotechnology Advances, 67, 108210. Available from: 10.1016/j.biotechadv.2023.108210 [DOI] [PubMed] [Google Scholar]
- Pluskal, T. , Castillo, S. , Villar‐Briones, A. & Oresic, M. (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry‐based molecular profile data. BMC Bioinformatics, 11, 395. Available from: 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, W. , Liu, Y. , Ren, P. , Zhang, J. , Li, H. , Tian, L. et al. (2014) Uncovering a glycosyltransferase provides insights into the glycosylation step during macrolactin and bacillaene biosynthesis. Chembiochem: A European Journal of Chemical Biology, 15(18), 2747–2753. Available from: 10.1002/cbic.201402384 [DOI] [PubMed] [Google Scholar]
- Rausch, C. , Hoof, I. , Weber, T. , Wohlleben, W. & Huson, D.H. (2007) Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evolutionary Biology, 7, 78. Available from: 10.1186/1471-2148-7-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch, C. , Weber, T. , Kohlbacher, O. , Wohlleben, W. & Huson, D.H. (2005) Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Research, 33(18), 5799–5808. Available from: 10.1093/nar/gki885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N.S. , Wang, J.T. , Ramage, D. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. Available from: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyanikova, I.P. & Golovleva, L.A. (2019) Hexadecane and hexadecane‐degrading bacteria: mechanisms of interaction. Microbiology, 88, 15–26. Available from: 10.1134/S0026261718060152 [DOI] [Google Scholar]
- Subramaniam, M.D. , Venkatesan, D. , Iyer, M. , Subbarayan, S. , Govindasami, V. , Roy, A. et al. (2020) Biosurfactants and anti‐inflammatory activity: a potential new approach towards COVID‐19. Current Opinion in Environmental Science & Health, 17, 72–81. Available from: 10.1016/j.coesh.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlouw, B.R. , Blin, K. , Navarro‐Muñoz, J.C. , Avalon, N.E. , Chevrette, M.G. , Egbert, S. et al. (2023) MIBiG 3.0: a community‐driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Research, 51(D1), D603–D610. Available from: 10.1093/nar/gkac1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta, R. , Della Sala, G. , Mangoni, A. , Lega, M. & Costantino, V. (2016) Tracing cyanobacterial blooms to assess the impact of wastewaters discharges on coastal areas and lakes. International Journal of Sustainable Development and Planning, 11, 804–811. Available from: 10.2495/SDP-V11-N5-804-811 [DOI] [Google Scholar]
- Teta, R. , Sala, G.D. , Esposito, G. , Stornaiuolo, M. , Scarpato, S. , Casazza, M. et al. (2021) Monitoring cyanobacterial blooms during the COVID‐19 pandemic in Campania, Italy: the case of Lake Avernus. Toxins, 13(7), 471. Available from: 10.3390/toxins13070471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M.G. , Chan, Y.A. & Ozanick, S.G. (2003) Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrobial Agents and Chemotherapy, 47(9), 2823–2830. Available from: 10.1128/AAC.47.9.2823-2830.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanova, V. , Galasso, C. , Vitale, G.A. , Della Sala, G. , Engelbrektsson, J. , Strömberg, N. et al. (2022) Mixotrophy in a local strain of Nannochloropsis granulata for renewable high‐value biomass production on the west coast of Sweden. Marine Drugs, 20(7), 424. Available from: 10.3390/md20070424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , An, J. , Cao, T. , Guo, M. & Han, F. (2024) Application of biosurfactants in medical sciences. Molecules (Basel, Switzerland), 29(11), 2606. Available from: 10.3390/molecules29112606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebach, V. , Mainz, A. , Siegert, M.J. , Jungmann, N.A. , Lesquame, G. , Tirat, S. et al. (2018) The anti‐staphylococcal lipolanthines are ribosomally synthesized lipopeptides. Nature Chemical Biology, 14(7), 652–654. Available from: 10.1038/s41589-018-0068-6 [DOI] [PubMed] [Google Scholar]
- Wittmann, M. , Linne, U. , Pohlmann, V. & Marahiel, M.A. (2008) Role of DptE and DptF in the lipidation reaction of daptomycin. The FEBS Journal, 275(21), 5343–5354. Available from: 10.1111/j.1742-4658.2008.06664.x [DOI] [PubMed] [Google Scholar]
- Zhang, J.J. , Tang, X. , Huan, T. , Ross, A.C. & Moore, B.S. (2020) Pass‐back chain extension expands multimodular assembly line biosynthesis. Nature Chemical Biology, 16(1), 42–49. Available from: 10.1038/s41589-019-0385-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Chen, Y. , Zhu, J. , Lu, Q. , Cryle, M.J. , Zhang, Y. et al. (2023) Structural diversity, biosynthesis, and biological functions of lipopeptides from Streptomyces . Natural Product Reports, 40(3), 557–594. Available from: 10.1039/d2np00044j [DOI] [PubMed] [Google Scholar]
- Zhong, L. , Diao, X. , Zhang, N. , Li, F. , Zhou, H. , Chen, H. et al. (2021) Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis. Nature Communications, 12(1), 296. Available from: 10.1038/s41467-020-20548-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1:
Data Availability Statement
The molecular networks and mass specrometry data can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=1a8c0d5b8c0a42aaaa1a8b5520f5764c and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4a962a758d304461a855f935f0295109. The rhodoheptin and rhodamide gene clusters were deposited in GenBank under accession numbers PP729154 and PP729155, respectively.


