Abstract
Skeletal muscle activation using optogenetics has emerged as a promising technique for inducing noninvasive muscle contraction and assessing muscle function both in vivo and in vitro. Transgenic mice overexpressing the optogenetic fusion protein, Channelrhodopsin 2‐EYFP (ChR2‐EYFP) in skeletal muscle are widely used; however, overexpression of fluorescent proteins can negatively impact the functionality of activable tissues. In this study, we characterized the contractile properties of ChR2‐EYFP skeletal muscle and introduced the ChR2‐only mouse model that expresses light‐responsive ChR2 without the fluorescent EYFP in their skeletal muscles. We found a significant reduction in the contractile ability of ChR2‐EYFP muscles compared with ChR2‐only and WT mice, observed under both electrical and optogenetic stimulation paradigms. Bulk RNAseq identified the downregulation of genes associated with transmembrane transport and metabolism in ChR2‐EYFP muscle, while the ChR2‐only muscle did not demonstrate any notable deviations from WT muscle. The RNAseq results were further corroborated by a reduced protein‐level expression of ion channel‐related HCN2 in ChR2‐EYFP muscles and gluconeogenesis‐modulating FBP2 in both ChR2‐EYFP and ChR2‐only muscles. Overall, this study reveals an intrinsic skeletal dysfunction in the widely used ChR2‐EYFP mice model and underscores the importance of considering alternative optogenetic models, such as the ChR2‐only, for future research in skeletal muscle optogenetics.
Keywords: Channelrhodopsin‐2, function, optogenetics, skeletal muscle, structure
Optogenetic tools like Channelrhodpsin‐2 (ChR2) are useful for inducing skeletal muscle contraction using blue light. However, the prevalence of fluorescent proteins like EYFP may cause functional issues. Here, Lamia et al. showed that the contractile properties of ChR2‐EYFP skeletal muscle had aggregated EYFP in sarcolemma, decreased contractile force production, and smaller fibers compared with ChR2‐only and WT mice. This study introduced an alternative optogenetic model (ChR2‐only) for more physiological studies using optogenetics than the commonly used ChR2‐EYFP.
Abbreviations
- ACTN3
Alpha‐actinin‐3
- ChR2
Channelrhodopsin‐2
- CSA
Cross sectional area
- EDL
Extensor digitorum longus
- EYFP
Enhanced yellow fluorescent protein
- FBP2
Fructose‐bisphosphatase 2
- GFP
Green fluorescent protein
- HCN2
Potassium/sodium hyperpolarization‐activated cyclic nucleotide‐gated ion channel 2
- LED
Light emitting diode
- PCSA
Physiological cross sectional area
- TIMP
Tissue inhibitor of metalloproteinases 2
1. INTRODUCTION
The ability to control skeletal muscle activation is essential for assessing muscle function both in vivo and in vitro. In a controlled laboratory environment, skeletal muscle activation is predominantly assessed using either nerve‐mediated or direct electrical stimulation. While electrical stimulation techniques are well‐established and repeatable, significant technical challenges exist, including the iatrogenic effects of needle insertions and the inability to activate denervated muscles, which is common in peripheral nerve injuries (e.g., brachial plexus avulsion and spinal cord injuries) and neuromuscular disorders (e.g., muscular dystrophies and amyotrophic lateral sclerosis). 1 , 2 , 3 Furthermore, the use of established electrical stimulation techniques presents significant challenges when applied to neonatal mice and preclinical models used for studying muscle development and pathology. Smaller muscles in neonatal mice and less resilient muscles in pathological mouse models can exacerbate iatrogenic issues due to repeated needle insertions. Consequently, traditional electrical stimulation methods might compromise the integrity of muscle structure, affect perfusion, and induce inflammation, thereby diminishing the physiological relevance of experimental outcomes.
To overcome these challenges, skeletal muscle activation with optogenetic stimuli (i.e., optogenetics) has emerged as a noninvasive technique to induce muscle contraction. 4 , 5 , 6 , 7 , 8 Optogenetic techniques are well established in neurobiological research due to high spatio‐temporal resolution. 9 , 10 Optogenetic stimulation can elicit muscle contraction via depolarization of muscle fibers that express light‐sensitive opsins, such as Channelrhodopsin‐2 (ChR2) protein. 4 , 5 ChR2 is a nonselective transmembrane cation channel derived from the green microalgae Chlamydomonas reinhardtii. 9 When exposed to blue light (455 nm), the transmembrane ChR2 opens to allow nonselective cation flow for muscle fiber depolarization and contraction. 10 , 11 Thus, optogenetic contractions can be elicited independent of motoneuron excitation, leading to muscle stimulation even when muscle is denervated. Direct optogenetic stimulation of excitable muscle fibers have been used to induce contraction noninvasively via transdermal light exposure 1 , 4 , 5 , 6 , 7 and serve as an alternative to electrical stimulation. 1 Optogenetic stimulation has primarily been conducted using transgenic mice expressing ChR2 (H134R)‐enhanced yellow fluorescent protein (EYFP) fusion (commonly known as Ai32) under the Sim1 and chicken‐β‐actin promoters. 4 , 5 In these mice, skeletal muscle localizes ChR2‐EYFP within the sarcolemma and t‐tubules, as well as within voltage‐gated sodium channels of the sarcolemma. 4 Our group has previously used the established Ai32 transgenic strain to achieve optogenetic control of skeletal muscle using the Acta1 promoter, and we showed that ChR2‐EYFP was expressed only in the skeletal muscle fibers and not in the nerves. 6
While the EYFP present in the commonly used Ai32 mouse line is useful for detecting Cre recombination within tissues, fluorescent reporters such as EYFP and green fluorescent protein (GFP) have been associated with cytotoxicity. 12 , 13 , 14 , 15 Moreover, Agbulut et al. demonstrated that cytosolic GFP in muscle impairs actin‐myosin head binding, leading to contractile dysfunction. 16 High levels of ChR2‐EYFP expression have also been shown to alter the electrophysiological properties and morphology of neurons. 17 , 18 , 19 Furthermore, ChR2‐EYFP‐mediated immunogenicity has been reported to cause motor neuron death and muscle atrophy. 20 We have previously shown that the doxycycline‐inducible Acta1‐tetO‐rtTA‐Cre; Ai32 mouse model exhibited aggregation of EYFP in skeletal muscle. 6 We were concerned that this clustering of EYFP had the potential to influence the contractile and electrophysiological function of muscle. To address this concern, we deleted the EYFP gene from the ChR2‐EYFP mice using CRISPR/Cas9. To test if and how EYFP influences skeletal muscle contractility, we compared contractile properties using electrical stimulation. We also evaluated the effect of EYFP on light‐induced contractile force in vitro with collimated blue light. In addition, we determined how the presence of ChR2/EYFP affects muscle ultrastructure and fiber size. To obtain a mechanistic view behind the contractile dysfunction, we used RNA sequencing (RNAseq) to identify the transcriptional responses of naïve muscle, as well as following daily bouts of optogenetic stimulation in these two ChR2‐expressing optogenetic strains.
2. MATERIALS AND METHODS
The Unit for Laboratory Animal Medicine at the University of Michigan approved all animal procedures. The study design is shown in Figure 1.
FIGURE 1.
Study design: schematic of experiments with mouse strains and number of mice utilized. Mice with muscle‐specific expression of optogenetic ChR2‐EYFP fusion or ChR2‐only protein (referred to as ChR2‐EYFP or ChR2‐only strains, respectively) and their wild‐type (WT) littermates were used in various experiments. Young adult (2–3‐month‐old) mice were used in microscopy, muscle contractility with both optogenetic and gold standard nerve or electrical stimulation, and fiber cross‐sectional area measurement. Muscles from young (5‐week‐old) mice were harvested for protein measurements. For RNA sequencing, right triceps surae muscles of young (4‐week‐old) ChR2‐EYFP and ChR2‐only mice were stimulated with blue light (455 nm) to induce optogenetic muscle contraction. After 5 daily bouts of 20 min stimulation, stimulated and age‐matched WT mice were euthanized to harvest the triceps surae for sequencing. The superscripts ‘u’ or ‘v’ indicate same mice were used across different experiments. The number of mice used in each experiment are presented as WT, ChR2‐EYFP, and ChR2‐only, respectively.
2.1. Animal models
All animals were kept on a 12‐hour light/dark cycle under standard housing conditions with ad libitum access to water and chow throughout the experiment. Mice were genotyped by PCR (Transnetyx, TN, USA). Wild‐type (WT) littermates were used as controls. Acta1Cre;ChR2‐EYFP mice were generated as described previously. 6 Briefly, tetracycline‐inducible Acta1Cre 21 male mice were bred with Ai32fl/fl (Jax ID: 024109, C57BL6/J background) female mice to generate Acta1Cre;Ai32fl/fl mice, referred to as ChR2‐EYFP onward. Cre recombination was induced via doxycycline (tetracycline) chow (Inotiv, 200 Doxycycline) at mating and continued till pups were weaned. The ChR2‐only mouse line was generated by deleting the EYFP‐WPRE DNA fragment from the ChR2‐EYFP mice (Jax Stock No. 024109) using the CRISPR/Cas9 technology (Figure S1). 22 Briefly, two single‐guide RNAs (sgRNA), one targeting the junction between ChR2 and EYFP (AGTACCCGCGGCCGCCACCA) and the other one targeting the end of WPRE (GGCCCTAGGCGGTATCGATG in reverse orientation), were generated using ThermoFisher's in vitro transcription service. These two sgRNAs (20 ng/μL each) were co‐microinjected with Cas9 mRNA (50 ng/μL, TriLink BioTechnologies) into the cytoplasm of zygotes collected from ChR2‐EYFP mouse mating pairs. A single‐strand oligonucleotide donor (GAGGTCGAGACGCTGGTGGAGGACGAGGCCGAGGCTGGCGCAGTACCCGCGGCCGCCACCGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCGATACCGCCTAGGGCCTCGACTGTGCCTTCTAGTTGCCAGCCATCTGTTGTTTGCCCCT) was also included in the injection mix (100 ng/μL) for mediating precise deletion as well as adding a V5 tag (sequence in bold) to the C‐terminus of the ChR2 transgene. Injected embryos were cultured overnight in KSOM medium (Millipore Sigma) in a 37°C incubator with 6% CO2. In the next day, embryos that had reached the two‐cell stage of development were implanted into the oviducts of pseudo‐pregnant surrogate mothers (CD1 strain from Charles River Laboratory). Offspring born to the foster mothers were genotyped using PCR followed by Sanger sequencing. Founder mice with the desired deletion V5 tagging were expanded to establish the ChR2‐only mouse line. Acta1Cre;ChR2‐onlyfl/fl mice, referred to as ChR2‐only onward were generated similarly as ChR2‐EYFP mice. For light and electron microscopy, constitutive CKCre;Ai32fl/fl mice were also used and were generated in Glancy laboratory.
2.2. Light and electron microscopy
Extensor digitorum longus (EDL) muscles from adult (>2‐months old) WT, ChR2‐EYFP, and ChR2‐only male mice were flash‐frozen for histology following standard protocols (n = 1/group). Tissues were sectioned at 10 μm thickness longitudinally or transversely and were imaged in a confocal or widefield microscope. For transmission electron microscopy (TEM), EDL muscles from adult WT, ChR2‐EYFP, and ChR2‐only male mice were fixed with glutaraldehyde and osmium tetroxide and were embedded in plastic (n = 1/group). Ultrathin sections were produced to visualize the ultrastructure in JEOL‐1400 transmission electron microscope. To visualize muscle cell membrane, in vivo microscopy of the Tibialis anterior muscle of ChR2‐only mice was performed as described previously. 23 Briefly, the tibialis anterior muscle fibers were exposed by removal of the overlying skin and fascia and superfused with Tyrodes buffer (pH 7.1 at 37°C) containing HEPES (10 mM), NaCl (137 mM), KCl (4.5 mM), MgSO4 (0.5 mM), KPO4 (0.5 mM), glucose (10 mM), CaCl2 (1.8 mM) and Di‐8‐ANEPPS (1 μM). Di‐8‐ANEPPS fluorescence was imaged using an upright Leica SP8 multiphoton microscope with resonant scanning and a Nikon 25× (1.1 NA) water‐immersion objective using 900 nm excitation light and emission collected from 605 to 680 nm.
2.3. In situ contractility of gastrocnemius muscle using nerve stimulation
Experiment was conducted as detailed in Larkin et al. 2011. 24 Briefly, 3‐month‐old WT, ChR2‐EYFP, and ChR2‐only male (n = 4, 3, and 5, respectively, per group) mice were anesthetized using 2,2,2‐Tribromoethanol suspended in 2‐methyl‐2‐butanol and sterile saline. 0.3–0.5 mL anesthetic was injected intraperitoneally as frequently as needed to maintain anesthesia. The mouse under anesthesia was placed on a heated platform to maintain optimal body temperature throughout the procedure. The gastrocnemius muscle was carefully separated from the surrounding musculature without damaging the blood supplies and nerves. A suture was tied around the calcaneal tendon and calcaneus bone and then the calcaneus was dissected. The knee and foot were tied to fixed posts to limit motion and maintain isometric condition. The calcaneal tendon was tied to a dual mode lever system (6650LR, Cambridge Technology). The sciatic nerve under the hamstring muscle was exposed and a bipolar platinum wire electrode was placed upon it. Heated saline (37°C) was dripped continuously on the muscle to prevent it from drying. The gastrocnemius muscle was activated by stimulating the nerve with electrical pulses to induce contraction. Muscle contraction was induced using 0.2 ms electrical stimulation pulses. The applied voltage was incrementally increased until the maximum isometric twitch force was achieved. After identifying the voltage that produced maximum force, the voltage was kept constant, and the muscle length was adjusted to find the optimal length (L o) that produced the maximum twitch force. Trains of electrical pulses were then applied to produce isometric tetanic contractions. Forces at frequencies ranging from 40 to 220 Hz were recorded. We normalized the maximum isometric tetanic force with respect to the physiological cross‐sectional area (PCSA) of the muscle using Equation (1).
| (1) |
where m = muscle mass, ρ = density (1.06 g/cm3), and L = muscle fiber length. To obtain L, L o was multiplied by 0.45 (L to L o ratio).
2.4. In vitro contractility of EDL and soleus muscles
2.4.1. Electrical stimulation
Experiment was conducted as detailed in Brooks and Faulkner 1988. 25 Briefly, EDL muscles were dissected from 3‐month‐old WT, ChR2‐EYFP, and ChR2‐only male mice (n = 5, 3, and 5, respectively, per group) under anesthesia. The isolated EDL muscle was placed in buffered mammalian Ringer solution bath to maintain physiological condition. Distal and proximal tendons were tied with sutures to a fixed post and force transducer (BG‐50, Kulite semiconductor), respectively. Two platinum electrodes were placed on either side of the muscle parallel to its long axis. An electrical field was applied between the electrodes and the muscle was tested following the same protocol as the in situ test described before. Time to peak twitch force and half relaxation times were also recorded. Soleus muscles were tested similarly following this same method (n = 3, 3, and 4, respectively, per group, all male except two in the ChR2‐EYFP group). Maximum isometric tetanic forces were normalized with respect to respective muscles' PCSA using Equation (1). To obtain L, L o was multiplied by 0.44 or 0.71 for EDL or soleus muscle, respectively.
2.4.2. Optogenetic stimulation
With the muscle remaining in the same experimental chamber as was used for electrical stimulation, we optogenetically stimulated EDL (n = 3 and 4, respectively, per group) and soleus (n = 3 and 4, respectively, per group, all male except two in the ChR2‐EYFP group) muscles following electrical stimulation. A blue LED (455 nm wavelength, Thorlabs) was placed directly above the muscle at an irradiance of 140 mW/cm2. We measured muscle peak twitch force with incremental light pulse duration ranging from 1 to 100 ms. The ability of the muscle to sustain tetanic contraction with optogenetic stimulation was also tested by varying LED pulse durations and frequency over 300 ms duration.
2.5. Muscle fiber cross‐sectional area (CSA) measurements
The fiber cross‐sectional area (CSA) was determined using the contralateral unstimulated EDL muscles from the mice utilized in the contractility experiments (n = 3 per group). The EDL muscle was flash‐frozen following standard histology protocols. And 10 μm thick transverse sections were obtained from the mid belly region and were stained with Alexa 647 fluorophore conjugated wheat germ agglutinin (WGA). Sections were imaged in fluorescence microscope (BioTek Lionheart FX with Cy5 cube) and fiber CSA were determined with FIJI particle analysis.
2.6. RNA sequencing of muscles
2.6.1. In vivo muscle stimulation
1‐month‐old ChR2‐EYFP and ChR2‐only male mice (n = 3 per genotype) were subjected to isometric unilateral optogenetic muscle stimulation for 5 days, as described previously. 7 Briefly, with the mouse in prone position, right limb was placed in an in vivo muscle stimulation apparatus (Aurora Scientific, ON, Canada), with the foot resting against a flat plate. The limb was clamped at the knee to stabilize the joint for isometric triceps surae contractions. A custom‐built setup was used to shine controlled light pulses on the tricep surae, 6 which consisted of a blue‐light LED (455 nm, 900 mW, Thorlabs) and a driver (DC2200, Thorlabs) for pulse modulation. Muscle was stimulated at 10 Hz frequency (70 ms on‐/30 ms off‐time; 10 repetitions/second) for 20 minutes for 5 days. The left limbs served as unstimulated internal controls. After each daily session, mice were returned to cages to resume regular activity.
2.6.2. RNA isolation and sequencing
Mice were euthanized following institutional guidelines 4 h after their last stimulation session. Age‐matched WT mice were euthanized at the 5‐day time points. From stimulated mice, tricep surae muscles of both limbs were rapidly dissected, flash‐frozen in liquid nitrogen, and stored at −80°C until RNA isolation. From WT mice only the right tricep surae muscles were harvested. Samples were pulverized in TRIzol (Ambion, Austin, TX, USA) using Precellys tissue pulverizer. Total RNA was extracted using Invitrogen PureLink RNA Mini Kit with on‐column DNA digestion (Ambion). RNA was eluted in 50 μL RNase‐free water. RNA integrity number (RIN) was measured using Agilent bioanalyzer. A total of 15 muscle samples were sequenced in two different batches. Next‐generation sequencing was performed on poly‐A mRNA libraries as described before. 7
2.7. Computational estimation of light penetrance during optogenetic stimulation
We utilized a Monte Carlo (MC) photon transport model to quantify light penetration depth in muscle tissue for our optogenetics experiments. 26 , 27 MC simulations are physics‐based, stochastic models of photon propagation through a turbid biological medium. Individual photons are launched into a simulated tissue with user‐specified optical properties (scattering coefficient [μ s (cm−1)], absorption coefficient, [μ a (cm−1)], scattering anisotropy factor [g]). Photon movement and local energy deposition is tracked until the photon has either lost all its original energy or exited the tissue volume (Figure S6). This process is repeated for many photons (~106–108) such that average distribution of the light propagation path within the tissue can be reliably estimated. By tracking the locally deposited power of each photon during the simulation, the normalized fluence rate, or NFR (cm−2) can be output for each voxel within the tissue cuboid. This conceptually represents the fraction of irradiance that was incident on that voxel during the simulation per Watt of incident light.
To perform these simulations, an open‐source, MATLAB‐based MC model (“MC Matlab”) was used. A one‐ or two‐layer model was generated to describe both the in vitro (muscle only) and in vivo (skin + skeletal muscle) experiments, respectively. The following optical properties were extracted from relevant literature: scattering coefficient, μ s = 200 and 90 cm−1, absorption coefficient, μ a = 6 and 1 cm−1, scattering anisotropy factor, g = 0.75 and 0.9, for skin and muscle, respectively. 28 , 29 , 30 Refractive indices were assumed 1.4 for both tissues. The light source was described as an infinite plane wave at 455 nm with normal incidence on the surface of the tissue. For each simulation, a 3D volume of 100 × 100 × 300 bins was generated for a total tissue volume of .35 × .35 × .35 cm (voxel size of .035 × .035 × .012 mm; length, width, height, respectively). A total of 108 photons were launched per simulation. All cuboid boundaries were set as non‐reflective to best mimic native conditions and not forcibly and erroneously “trap” light within the simulated sub volume of the muscle. As such photons could escape from the lateral edges of the volume and not be “reflected,” “transmitted,” or “dead” within the tissue volume.
2.8. Western blotting
Following euthanasia, gastrocnemius and soleus muscles from 5‐week‐old female and male mice (n = 3 per group) were dissected, flash‐frozen in liquid nitrogen, and stored in −80°C until protein extraction. Briefly, 20–30 mg of tissue was lysed in radio immunoprecipitation assay (RIPA) buffer supplemented with protease and phosphatase inhibitors. Following lysis, the suspension was centrifuged, and the resulting supernatant was collected for blotting. Protein concentration was measured with Pierce BCA protein assay (Thermo Scientific, #23225). For ChR2 blotting, 10 μg gastrocnemius muscle protein diluted in loading buffer was reduced with dithiothreitol and was loaded onto 8–16% tris glycine gel. Following running, proteins were transferred to polyvinylidene difluoride (PVDF) membrane. The membrane with transferred protein was stained with Ponceau S stain (Sigma‐Aldrich #P7170) to determine the quality of transfer and protein load. The membrane was incubated with anti‐ChR2 primary antibody (Table 1). A separate membrane was developed with soleus muscle protein similarly. Other targets, TIMP1, FBP2, ACTN3, and HCN2 were selected from the sequencing result based on their differential expression intensity. For these targets, 15 μg gastrocnemius muscle protein was denatured and reduced before loading onto the gel and primary antibodies were used at dilution ratios mentioned in Table 1. The membranes were then incubated with horseradish peroxidase‐conjugated secondary antibody (Table 1). Protein bands were developed with enhanced chemiluminescent substrate (SuperSignal™ West Pico PLUS Chemiluminescent Substrate, #34577) and were imaged with ChemiDoc MP imaging system (Bio‐Rad). All blots included a well with protein isolated from the gastrocnemius muscle of a 6‐month‐old C57BL/6J (B6J) female mouse. Band intensity for immunoblots were normalized with respect to representative bands from total protein staining (Ponceau S) 31 , 32 or to intensity of the control B6J band. Images were analyzed with FIJI Gel plugin.
TABLE 1.
Western blot antibody information.
| Primary antibody dilution | Secondary antibody dilution |
|---|---|
| ChR2 (Progen #651180) 1:150 | Goat anti‐mouse (ProteinTech #SA00001‐2) 1:6000 |
| TIMP1 (ProteinTech, #16644‐1‐AP) 1:1500 | Goat anti‐rabbit (ProteinTech #SA00001‐2) 1:6000 |
| FBP2 (ProteinTech, #25192‐1‐AP) 1:1000 | Goat anti‐rabbit (ProteinTech #SA00001‐2) 1:6000 |
| ACTN3 (ProteinTech, #24378‐1‐AP) 1:1000 | Goat anti‐rabbit (ProteinTech #SA00001‐2) 1:6000 |
| HCN2 (ProteinTech, #55245‐1‐AP) 1:1000 | Goat anti‐rabbit (ProteinTech #SA00001‐2) 1:6000 |
2.9. Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 9 or higher, La Jolla, USA) or in RStudio (version 4.1.0).
2.9.1. Contractility, muscle fiber CSA, and western blot measurements
Body masses, muscle masses, maximum specific tetanic forces, and muscle fiber CSAs were analyzed with one‐way ANOVA corrected for multiple testing (Tukey's post hoc) across genotypes. Tetanic forces were compared using two‐way repeated measures ANOVA to determine significance across genotypes and stimulation frequencies. Significance was set at p < .05. For western blots, band intensities (calculated as adjusted volumes in ImageLab software) were compared using one‐way ANOVA with Tukey's or Sidak's correction for multiple comparisons.
2.9.2. Differential expression of genes (DEG)
We used DESeq2 in R/Bioconductor 33 to determine the DEGs. Count data were organized in two different count matrices: matrix 1: WT, ChR2‐EYFP unstimulated, and ChR2‐only unstimulated; and matrix 2: ChR2‐EYFP unstimulated, ChR2‐EYFP stimulated, ChR2‐only unstimulated, and ChR2‐only stimulated. In matrix 1, we analyzed ChR2‐EYFP and ChR2‐only unstimulated samples with WT as baseline controlling for batch variability. In matrix 2, ChR2‐EYFP and ChR2‐only stimulated samples were analyzed with their respective unstimulated samples as baseline controlling for batch, genotype, and animal variability. Principal component analysis (PCA) was performed with the top 500 most variable genes across samples in a matrix to visualize sample clustering. For analyses in matrix 1, significance was tested against log2(FoldChange) = .15, while in matrix 2 significance was tested against .32. p‐values were adjusted for multiple testing using the Benjamini and Hochberg method, and differential expression significance was set at p‐adjusted (p‐adj) <.05.
2.9.3. Enrichment analysis
To determine enriched biological processes, the list of up and downregulated DEGs was input separately into the database for annotation, visualization, and integrated discovery (DAVID). 34 , 35 A threshold of false‐discovery rate (FDR) <0.1 was applied to identify significantly enriched gene ontology biological processes.
3. RESULTS
3.1. High ChR2‐EYFP expression and sarcolemma clustering of EYFP was observed in ChR2‐EYFP muscles
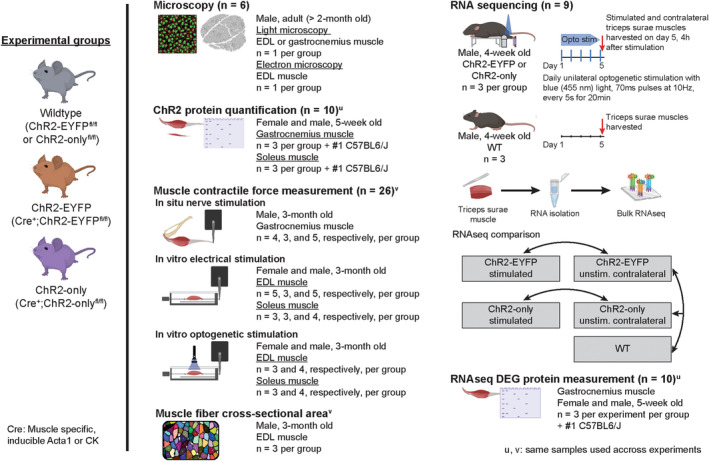
To investigate the effects of ChR2‐EYFP overexpression in skeletal muscle, we generated a transgenic mouse line to express ChR2 in skeletal muscle with EYFP fluorescent reporter. 6 , 7 Doxycycline‐treated, ACTA1‐rtTA;tetO‐Cre mice were bred with ChR2‐EYFPfl/fl mice (known as Ai32 mice) to generate Acta1Cre;ChR2‐EYFPfl/fl mice, referred to as ChR2‐EYFP mice onward. We observed potentially abnormal expression of ChR2‐EYFP fusion protein in the gastrocnemius muscle sarcolemma of 2‐month‐old ChR2‐EYFP mouse (Figure 2). To determine if this phenomenon was independent of Cre expression, we visualized EYFP expression in other muscle‐specific Cre‐lines, such as CKCre;ChR2‐EYFPfl/fl (Figure 2), Myf6Cre;ChR2‐EYFPfl/fl, and Sim1Cre;ChR2‐EYFPfl/fl (Figure S2). We observed overexpression of ChR2‐EYFP protein in the sarcolemma in all strains. Next, we bred doxycycline‐treated, ACTA1‐rtTA;tetO‐Cre mice with ChR2‐onlyfl/fl mice (developed in Glancy lab, see Methods for details) to generate Acta1Cre;ChR2‐onlyfl/fl mice, referred to as ChR2‐only mice onward. ChR2‐only muscle, when stained with the cell membrane dye Di‐8‐ANEPPS, demonstrated regular muscle structure (Figure 2). Using transmission electron microscopy (TEM), we identified an accumulation of vacuoles in the ChR2‐EYFP EDL muscle along the sarcolemma (Figure S2) which were not seen in wild‐type (WT) or ChR2‐only muscles.
FIGURE 2.
Abnormal ChR2‐EYFP clustering was observed in EYFP‐positive optogenetic mouse skeletal muscle. Confocal imaging demonstrated ChR2‐EYFP overexpression in the longitudinal plane of EDL muscle (CKCre;ChR2‐EYFPfl/fl muscle) and the transverse plane of gastrocnemius muscle (Acta1Cre;ChR2‐EYFPfl/fl muscle). Normal muscle membranes were visible in Acta1Cre;ChR2‐onlyfl/fl mice when stained with Di8ANEPPS. Scale bar: 100 μm.
3.2. ChR2 protein expression was not significantly different between ChR2‐EYFP and ChR2‐only muscles
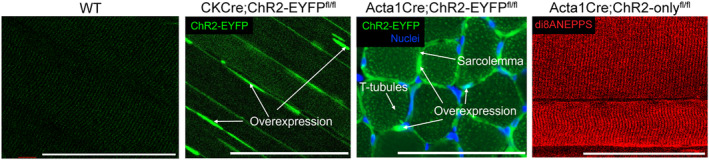
We measured ChR2 protein levels in the muscles of the 5‐week‐old ChR2‐EYFP and ChR2‐only mice to determine if ChR2 expression is different between the strains. ChR2 protein was identified at two different molecular weights in ChR2‐EYFP and ChR2‐only groups (55 and 35 kDa, respectively) (Figure 3). The ChR2‐EYFP gastrocnemius muscles had a slight, but not statistically significant, increase in ChR2 protein expression compared with the ChR2‐only gastrocnemius muscles (Figure 3A). No difference in ChR2 expression was observed in the soleus muscle (Figure 3B). The larger molecular weight for ChR2‐EYFP was expected given the fusion with EYFP (~25 kDa).
FIGURE 3.
ChR2‐EYFP gastrocnemius muscle had moderate, but not significant, elevation of ChR2 protein compared with ChR2‐only gastrocnemius muscle, and no difference was observed in the soleus muscle. ChR2 western blot and quantification of (A) gastrocnemius and (B) soleus muscle. n = 3 mice per group. Data were compared using one‐way analysis of variance (ANOVA) with Tukey's correction for multiple comparisons. Error bars denote means ± SD. WT group included n = 1 ChR2‐EYFPfl/fl and n = 2 ChR2‐onlyfl/fl mice (all cre‐negative).
3.3. The presence of ChR2‐EYFP fusion protein led to force deficits in fast‐twitch muscle
We found that the 3‐month‐old ChR2‐EYFP mice had lower body mass than age‐matched ChR2‐only mice (Figure 4A). To understand if and how the ChR2‐EYFP fusion protein was influencing the growth and function of skeletal muscle, we evaluated the individual weights and force generation potential of muscles with a battery of in situ and in vitro tests. The mass of gastrocnemius muscles from ChR2‐EYFP mice were significantly lower than age‐ and sex‐matched WT or ChR2‐only strains (Figure 4B). Isometric tetanic force of ChR2‐EYFP gastrocnemius muscles were also significantly reduced compared with WT and ChR2‐only mice at all test frequencies except for 40 Hz (Figure 4C). To account for differences in mouse and muscle size, we normalized the maximum tetanic force with respect to muscle's physiological cross‐sectional area (PCSA) to obtain the maximum specific tetanic force (Equation 1). However, the normalized force remained significantly lower in the ChR2‐EYFP gastrocnemius muscle compared with WT and ChR2‐only muscles (Figure 4D), suggesting the functional deficit was not scaled to animal/muscle size alone.
FIGURE 4.
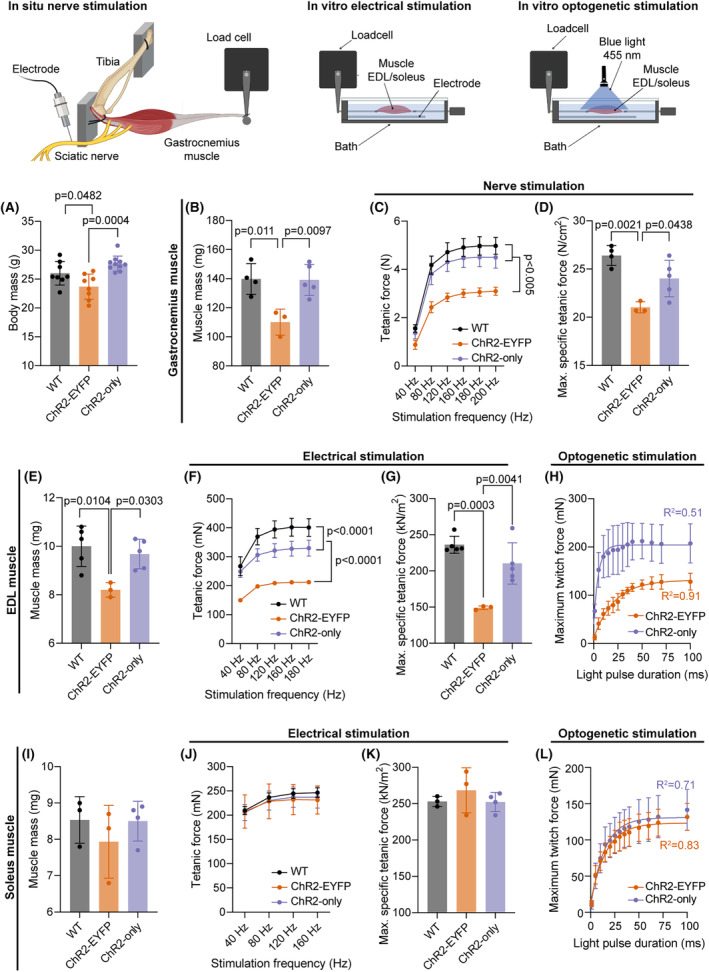
The presence of EYFP with ChR2 compromised the electrical and optogenetic contractile properties of fast‐twitch skeletal muscle compared with ChR2 only and WT muscle. (A) Body mass of all mice used in the experiment (N = 26; n ≥ 8 per genotype; WT group included n = 6 ChR2‐EYFPfl/fl and n = 2 ChR2‐onlyfl/fl mice, all cre‐negative). (B, E, I) Gastrocnemius and EDL, but not soleus muscle masses, were significantly different in ChR2‐EYFP mice compared with WT and ChR2‐only mice. (C, F, J) Tetanic forces of the gastrocnemius and EDL, but not soleus muscles, were significantly reduced in ChR2‐EYFP mice compared with WT and ChR2‐only at incremental electrical stimulation frequencies. (D, G, K) Maximum specific tetanic forces of the gastrocnemius, EDL, and soleus muscles, respectively; and (H, L) Light induced twitch forces of EDL and soleus muscle, respectively, measured at light pulse durations ranging from 1 to 100 ms. For (A), (B), (D), (E), (G), (I), and (K), data were compared using one‐way ANOVA with Tukey's correction for multiple comparisons. For (C), (F), and (J), data were compared using repeated measures two‐way ANOVA with Tukey's correction. For (H) and (L), data points from the average maximum twitch within strains were fitted into one‐phase decay model. Error bars denote means ± SD. EDL, extensor digitorum longus. Each dot represents a biological replicate (individual mouse).
Next, we evaluated direct muscle contractility in vitro to assess muscle function independent of motor neuron innervation. The EDL, a muscle composed primarily of fast‐twitch, glycolytic fibers, also weighed significantly less in ChR2‐EYFP mice compared with EDLs of WT mice (Figure 4E). Tetanic forces were significantly different between the three groups (Figure 4F). The normalized maximum specific tetanic force of the EDL muscle was significantly lower in ChR2‐EYFP EDLs compared with WT and ChR2‐only EDLs, however, no difference was observed in ChR2‐only and WT muscles (Tukey's multiple comparisons adjusted p = .14, Figure 4G). This deficit in direct muscle contractility in the ChR2‐EYFP EDLs was further tested in our optogenetic strains using blue‐light activation. ChR2‐only EDL muscles produced higher twitch forces than ChR2‐EYFP muscle at all pulse duration (Figure 4H). We modeled the force response to pulse duration using Akike information criteria (AIC) using one phase decay (R 2 = .51 and .91, respectively, for ChR2‐only and ChR2‐EYFP) and measured higher contractile forces at smaller pulses in ChR2‐only EDLs compared with ChR2‐EYFP EDLs. Specifically, ChR2‐only EDL muscles twitch force plateaued at shorter pulse durations (i.e., 30 ms duration) compared with ChR2‐EYFP EDL muscles (100 ms). Similarly, ChR2‐only EDLs generated higher forces with partially fused tetanus at 50 Hz, 10 ms pulse (Figure S3), which was not achieved in ChR2‐EYFP EDLs. However, it is also possible that an increase in light irradiance could lead to increased optogenetic‐induced contractile force. 4
To determine if this force deficit was dependent on fiber types, we measured the contractile properties of the soleus muscle, which comprise a substantial population of slow‐twitch, oxidative fibers, using in vitro contractility experiments. Contrary to what we found in fast‐twitch muscles, we did not observe any differences in muscle mass (Figure 4I), isometric tetanic force (Figure 4J), or maximum specific tetanic force (Figure 4K), suggesting that the contributions of EYFP in contractility in muscle‐specific optogenetic strains may be fiber‐type‐dependent. The optogenetic twitch forces in soleus muscle were also similar in both strains at all pulse durations and they plateaued at similar pulse durations (70 ms, Figure 4L, AIC‐validated one phase decay, R 2 > .7 for both strains). Soleus muscles from ChR2‐EYFP and ChR2‐only generated similar forces with partially fused tetanus at 50 Hz, 10 ms pulse (Figure S3).
To investigate if changes in calcium handling mechanism of muscle were influenced by EYFP expression, we recorded the twitch times of EDL and soleus muscles during in vitro experiments. We observed a small but significant increase in the half‐relaxation times of EDL and soleus muscles (Figure S4) suggesting potential alterations in calcium machineries in muscle fibers. ChR2‐only gastrocnemius, EDL and soleus muscles demonstrated a similar PCSA compared with their WT counterparts (Figure S5). ChR2‐EYFP‐expressing EDL and gastrocnemius muscles had the smallest PCSA, however, soleus muscle area was similar to WT and ChR2‐only muscles.
3.4. ChR2‐EYFP EDL muscle fibers were smaller compared with WT and ChR2‐only fibers
Because we observed a significant decrease in ChR2‐EYFP EDL muscle mass compared with WT and lower specific force production of ChR2‐EYFP muscles, we next determined the fiber area distribution of EDL muscles in each strain at 3‐month age (Figure 5A). Muscle fiber area of both optogenetic strains were smaller compared with WT muscle, with ChR2‐EYFP displaying a significantly reduced average fiber size (Figure 5B). While WT muscle demonstrated a uniform distribution of fiber CSA, ChR2‐EYFP muscles had higher percentage of smaller fibers (Figure 5C).
FIGURE 5.
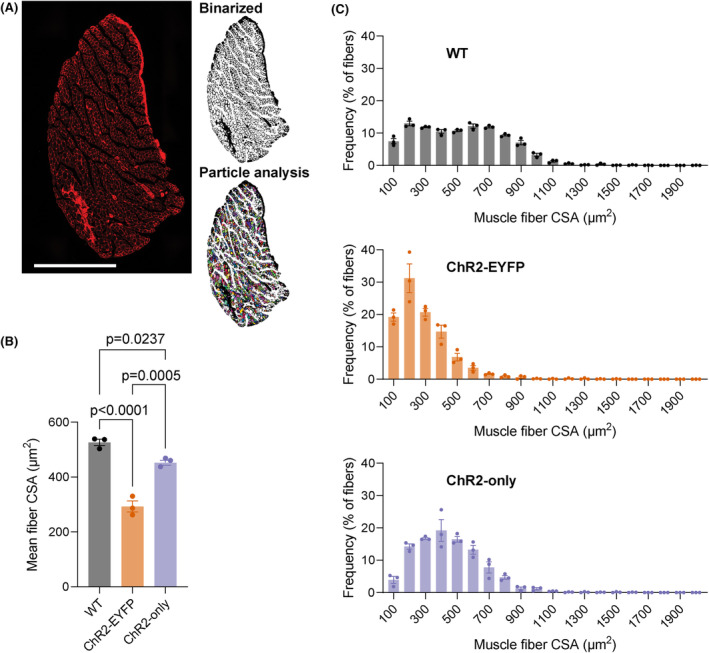
The presence of EYFP in ChR2‐expressing EDL muscle led to smaller fiber size. (A) WGA‐stained EDL muscle section. Images were binarized before particle analysis. (B) Mean muscle fiber area; and (C) fiber size frequency distribution. WGA, wheat germ agglutinin; CSA, cross‐sectional area. Scale bar = 500 μm. n = 3 per group; WT group included n = 1 ChR2‐EYFPfl/fl and n = 2 ChR2‐onlyfl/fl mice (all cre‐negative). Error bars denote mean ± SEM. For (B), data were compared using one‐way ANOVA with Tukey's multiple correction.
3.5. Skin acts as a barrier to light penetration depth for in vivo optogenetic stimulation of muscle
To estimate how deep our illumination conditions penetrate the skeletal muscle during in vivo optogenetics experiments, we utilized a Monte Carlo (MC) photon transport model. Qualitatively, the Monte Carlo simulations of steady‐state illumination within the optogenetically simulated tissues reflect expected behavior of photon transport in a turbid biological media (Figure S6). In both simulated scenarios, normalized fluence rate (NFR) peaked around the mean free path of the tissue. The mean free path represented the path length a photon will travel on average before changing its direction due to scattering and is equivalent to 1/μs. In the case of the in vitro muscle only results, this is the μs of the muscle, but in the in vivo condition, this is the μs of the most superficial layer, the skin. The photon NFR monotonically decreases deep to this peak in both conditions due to progressive photon energy loss and deposition associated with scattering and absorption events. In the in vivo case, the rate of decay of the decreasing NFR changes at the boundary between the skin and muscle due to the change in underlying optical properties of the medium.
For the single‐layer in vitro muscle only condition, 23.13% of incident photon energy was absorbed within the simulated tissue cuboid boundaries and 37.6% was absorbed for the two‐layer in vivo skin‐muscle condition (Table 2). For the in vivo case, we considered only the total percentage of light that was deposited in the muscle and not the skin. A large percentage of light was reflected in both cases (19.07% muscle only, 30.4% skin‐muscle). Despite the highly scattering nature of these tissues, some light (2.94% muscle only, 1.18% skin‐muscle) was able to be transmitted through the entire tissue volume. As the x and y edges of the cuboid were simulated as non‐reflecting boundaries, any remaining light escaped out the volume from the lateral edges of the cuboid (54.86% muscle only, 30.81% muscle‐skin). When considering how much of the incident light penetrated the muscle specifically, this was higher in the muscle only case (23.12%) than the muscle‐skin case (8.32%) (Table S1). This is due to the increased reflection caused by the skin layer, as well as the fluence that remained in the skin layer itself (4.78%).
TABLE 2.
Percentages of total light absorbed within volume or lost at each boundary.
| % Light absorbed within boundaries | % Light reflected | % Light lost to edges | % Light transmitted | |
|---|---|---|---|---|
| in vitro | 23.13 | 19.07 | 54.86 | 2.94 |
| In vivo | 37.6 | 30.40 | 30.81 | 1.18 |
In the in vitro experiments, an incident irradiance of 140 mW/cm2 was used. Therefore, for the cuboid size used in the MC simulations, this would result in an average irradiance of 32.32 mW/cm2. Therefore, for the light pulses ranging from 1 to 100 ms duration used in these studies, this is equivalent to a 32.32 μJ/cm2 to 3.23 mJ/cm2 light dose delivered to the muscle volume, respectively. For the in vivo experiments, an incident irradiance of 150 mW/cm2 was used, resulting in 12.48 mW/cm2 average muscle irradiance. This results in a dose of 873.6 μJ/cm2 delivered per 70 ms pulse.
3.6. Unstimulated ChR2‐EYFP muscle were transcriptionally unique compared with ChR2‐only and WT muscle
To evaluate if the muscles from the ChR2‐EYFP and ChR2‐only optogenetic strains respond differently to daily repeated optogenetic stimulation, we unilaterally stimulated the triceps surae muscles of 4‐week‐old mice from these two strains for 5 days (Figure 1). In total, we ran four RNAseq comparisons as shown in Table S2. Transcriptomes of WT and unstimulated ChR2‐only muscles were in general similar, as shown by the unsupervised clustering of WT and unstimulated ChR2‐only samples in the PCA (Figure 6A). We identified 1283 genes that were differentially expressed in the ChR2‐EYFP and 410 in the ChR2‐only muscle compared with WT, respectively (Table S2). Among the 632 and 251 downregulated genes in ChR2‐EYFP and ChR2‐only muscles, respectively, 142 were common in both strains. We identified 44 overlapping upregulated genes between ChR2‐EYFP and ChR2‐only muscles. We used the Database for annotation, visualization, and integrated discovery (DAVID) and found that downregulated genes in ChR2‐EYFP muscle transcriptome enriched metabolism, phosphorylation, transmembrane transport, and muscle contraction biological processes (Figure 6B). Upregulated genes in ChR2‐EYFP muscle were associated with inflammatory response and extracellular matrix (ECM) organization (Figure 6C). To understand the magnitude of differential expression between genotypes at baseline (unstimulated), we plotted the log2(FoldChanges; FC) of genes associated with several processes, such as muscle contraction, gluconeogenesis, collagen accumulation and degradation, and transmembrane (ion channel) transport (Figure 6D–G, respectively). We observed downregulation in fast‐twitch muscle contraction‐related genes, such as fMybpc2, Mylpf, Tnni2, and Tnnt3, which may explain the loss of force in fast‐twitch EDL muscle 36 , 37 , 38 , 39 but not in the slow‐twitch soleus muscle. In ChR2‐EYFP muscle, we observed a downregulation of transmembrane transport genes that encode the voltage‐gated potassium and sodium ion channels (Hcn2, Kcnf1, Kcnc3, Kcnb1, etc.) compared with WT muscle. While ECM degradation‐associated genes, such as Mmps, Ctss, and Ctsk were mostly upregulated in the ChR2‐EYFP muscle, MMP inhibitor Timp1 expression was also upregulated. Genes modulating gluconeogenesis, such as Fbp2, Pgam2, and Gpd1 were downregulated in ChR2‐EYFP muscle; however, they were unchanged in the ChR2‐only muscle. To assess if reduced gene expression was translated to protein level, we measured expression of several targets: fructose‐1,6‐bisphosphatase 2 (FBP2), actinin alpha 3 (ACTN3), tissue inhibitor of metalloproteinases 1 (TIMP1), and hyperpolarization‐activated cyclic nucleotide‐gated potassium channel 2 (HCN2) in 5‐week‐old gastrocnemius muscles. Both ChR2‐EYFP and ChR2‐only muscles showed decreased FBP2 protein expression compared with WT (Figure 7B). HCN2 protein was significantly reduced at the protein level in ChR2‐EYFP gastrocnemius muscle compared with ChR2‐only muscle (Figure 7A). However, ACTN3 and TIMP1 protein expression did not match the transcription levels (Figures 7C, and S7). No biological processes were significantly enriched by the ChR2‐only DEGs compared with the WT muscle transcriptome (false‐discovery rate (FDR) <0.1).
FIGURE 6.
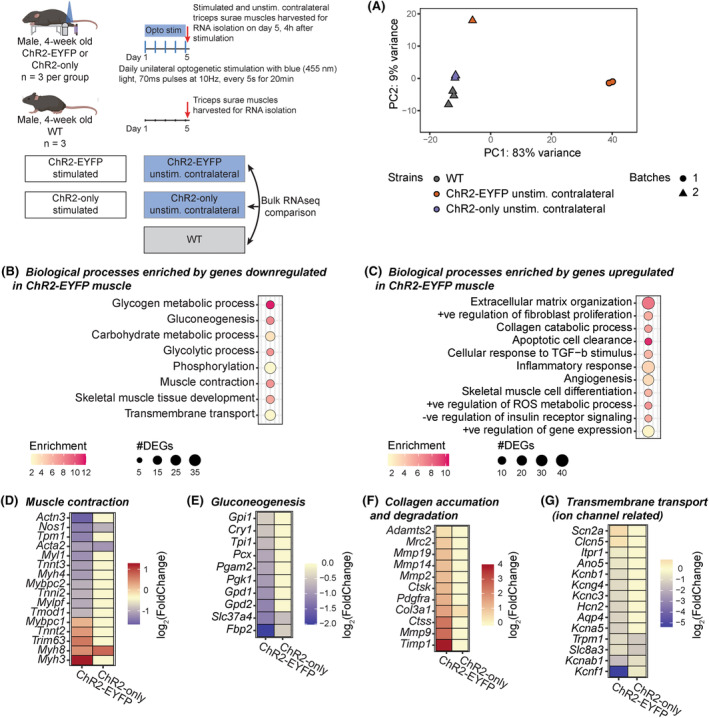
RNA sequencing revealed differential gene expression between WT, unstimulated ChR2‐EYFP, and unstimulated ChR2‐only triceps surae (i.e., gastrocnemius and soleus) muscles. (A) Principal component analysis of gene expression shows clustering of WT (all cre‐negative ChR2‐onlyfl/fl), ChR2‐EYFP, and ChR2‐only muscles. Shapes indicate different sequencing batches. Biological processes enriched in ChR2‐EYFP skeletal muscle compared with WT indicated (B) downregulation of metabolism, muscle contraction, and transmembrane transport‐related genes and (C) upregulation of inflammatory response and ECM related genes. (D–G) Gene expressions associated with enriched muscle contraction, gluconeogenesis, collagen catabolism biological processes, and transmembrane transport, respectively. n = 3 per group.
FIGURE 7.
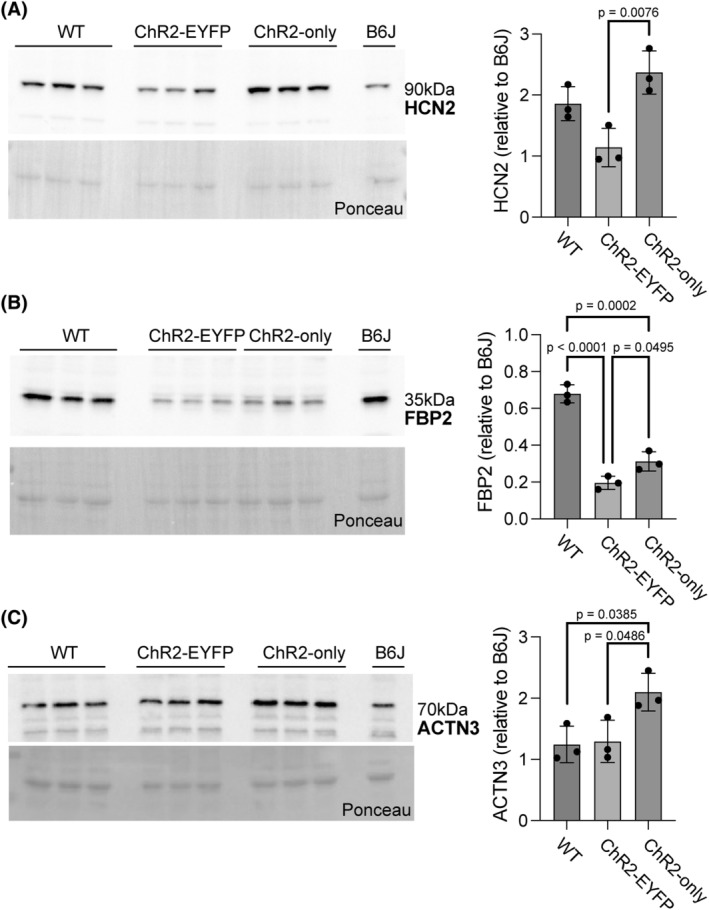
Presence of EYFP led to reduced HCN2 and FBP2 protein levels in gastrocnemius skeletal muscle. Significantly reduced protein‐level expressions of (A) HCN2 in the ChR2‐EYFP muscle and (B) FBP2 in both ChR2‐EYFP and ChR2‐only muscles were observed, compared with ChR2‐only and WT muscles, respectively. (C) ACTN3 protein expression was significantly higher in ChR2‐only muscle compared with WT and ChR2‐EYFP muscles. Error bars denote means ± SD. WT group included n = 1 ChR2‐EYFPfl/fl and n = 2 ChR2‐onlyfl/fl mice (all cre‐negative).
3.7. Presence of EYFP with ChR2 led to reduced transcriptional sensitivity to optogenetic stimulation compared with ChR2‐only muscles
To evaluate if and how muscles from ChR2‐EYFP and ChR2‐only optogenetic strains differentially respond to daily bouts of repeated optogenetic stimulation, we compared the transcriptomes of stimulated and contralateral unstimulated muscles within each strain. Optogenetic stimulation of ChR2‐only muscle resulted in a high number of DEGs compared with the unstimulated side (5698 DEGs), whereas only 267 DEGs were identified when comparing stimulated ChR2‐EYFP muscle to their respective unstimulated side (Table S2). In both strains, we observed an upregulation of inflammatory response‐related genes with daily optogenetic stimulation as identified by biological processes enrichment analysis (Figure 8C,E). In addition to inflammatory response, we also found that ECM organization, collagen organization, and response to mechanical stimulus were enriched by upregulated genes in both strains. Although we observed a small number of biological processes enriched by the downregulated DEGs in stimulated ChR2‐EYFP, these all had high enrichment score (Figure 8B). The downregulated genes in the stimulated ChR2‐only genes were primarily associated with mitochondria and metabolism (Figure 8D).
FIGURE 8.

Repeated bouts of optogenetic stimulation led to a more robust transcriptional response in ChR2‐only muscle compared with ChR2‐EYFP muscle. (A) Principal component analysis of gene expression. Shapes indicate different sequencing batches. Biological processes obtained from genes (B) downregulated and (C) upregulated in ChR2‐EYFP stimulated muscle and (D) downregulated and (E) upregulated in ChR2‐only stimulated muscle compared with unstimulated contralateral muscle. n = 3 per group.
4. DISCUSSION
In this study, we characterized the functional response of optogenetic skeletal muscle with or without the expression of a commonly fused fluorescent reporter, EYFP. Previous studies have explored if and how the overexpression of ChR2‐EYFP fusion influences the electrophysiological responses of neuron and HEK293 cells. 17 , 18 Yet, the role of ChR2‐EYFP overexpression in muscle functionality is largely unknown, except for a few studies on the effect of GFP and YFP accumulation in cardiac, neural, and muscle tissues. 12 , 15 , 16 We developed a new mouse strain by replacing the fluorescent EYFP of light responsive Ai32 with nonfluorescent v5 epitope and expressed this construct in skeletal muscle. Similar to our established ChR2‐EYFP mouse model (Acta1‐rtTA‐tetO‐Cre;Ai32), 6 , 7 we leveraged this new ChR2‐only model to induce optogenetic muscle contraction. We also showed that the ChR2‐EYFP fusion protein accumulated in the sarcolemma of the gastrocnemius and EDL muscles. Expression of ChR2‐EYFP led to significant impairments of muscle function, assessed using electrical and optogenetic contractility experiments. With the new ChR2‐only mouse model, we overcame this issue and returned muscle contractility using electrical stimulation to that of WT mice. These new findings suggest that ChR2‐only muscles are physiologically more similar to the WT muscles than the ChR2‐EYFP muscles. Green fluorescent protein (GFP) and YFP have been implicated in causing damage in various tissues, including skeletal and cardiac muscles and neurons. 12 However, if EYFP or ChR2 alone are responsible for functional deficit need further studies. Although not significant, the new ChR2‐only mouse model had moderately reduced ChR2 expression in the gastrocnemius muscle compared with ChR2‐EYFP muscle which could have subtly contribute to differences in muscle function. We expect, but did not directly investigate in this study, if translational efficiency of ChR2 was impaired in ChR2‐only muscle, given that the woodchuck hepatis virus posttranscription regulatory element (WPRE) was removed in addition to EYFP. 40 , 41 Any subtle differences in ChR2 protein may influence skeletal muscle function and should be investigated in future.
Unlike humans, limb muscles in mice are primarily composed of fast‐twitch, glycolytic fibers, except for the soleus muscles which consists of around 80% oxidative (type I and IIA) fibers. 42 Different types of muscles (oxidative and glycolytic) have varied responses to genetic manipulation and stimuli. 43 In this study, we accounted for different types of muscles, and although we observed significant force and muscle mass deficit in EDL and gastrocnemius muscle, we did not find these same differences in soleus muscles. This differential response may be explained by muscle types, that is, the ChR2‐EYFP overexpression may influence contractility of glycolytic fibers more than oxidative fibers. In line with this differential response based on fiber type, we observed downregulation of fast‐twitch muscle‐related genes in ChR2‐EYFP muscles, however, further studies are needed to better understand this phenomenon.
The attenuated muscle force generation in ChR2‐EYFP mice that we observed may be in part explained by either decreased calcium handling in muscle cells limited by ChR2‐EYFP fusion protein or by structural damage caused by excessive accumulation of EYFP. In our mouse models, ChR2‐EYFP co‐localized at the sarcolemma and T‐tubules. 4 ChR2‐EYFP colocalization may hinder the functionality of voltage‐gated dihydropyridine (DHP) calcium channels, which could negatively impact the calcium transients, thereby reducing contractile force. We saw a small but significant increase in the half relaxation time of the ChR2‐EYFP EDL muscle compared with WT muscle, which may suggest that calcium handling during excitation‐contraction coupling was slightly affected by ChR2‐EYFP. However, additional tests such as calcium transient analysis are required to confirm this. The attenuated muscle force generation in ChR2‐EYFP mice that we observed may also be attributed to potential alterations in the native electrophysiology of muscle fibers caused by ChR2‐EYFP overexpression. Elevated ChR2‐EYFP can modify cell's electrophysiological properties (capacitance, thresholds, and currents), 17 , 18 which can also decrease conduction velocity of action potentials and inadvertently limit muscle's force generation capacity and cause contractile dysfunction. 12 , 16 Additionally, we observed a downregulation in ion channel‐associated genes in addition to a reduction in HCN2 protein levels in ChR2‐EYFP muscle. Hyperpolarization‐activated cyclic nucleotide‐gated (HCN) channels activate in response to hyperpolarization and generate an inward cation current that regulates resting membrane potential. 44 Thus, it is possible that the decrease of HCN2 with ChR2‐EYFP overexpression may be impairing the regular electrophysiological activities of muscle fibers. Meng et al. demonstrated that expression of ChR2‐EYFP in neurons caused slight hyperpolarization of HCN half‐activation voltages, 17 indicating the potential of ChR2‐EYFP overexpression to regulate cell's intrinsic properties. Moreover, EYFP, a 25 kDa molecule fused to the C‐terminus of the 35 kDa ChR2 protein can potentially alter its native conformation, impacting ChR2's function and possibly leading to ion imbalance. While studies using ChR2‐YFP demonstrated that the fusion protein retained functionality in terms of ion conductance and optogenetic control, they did not directly compare ChR2‐YFP to untagged ChR2 to assess potential structural alterations. 10 , 11 Notably, fusion of GFP on C‐terminus significantly modifies Connexin 43's gating mechanisms and electrophysiological behavior, likely due to steric hindrance or interference with intermolecular interactions. 45 , 46 , 47 This disruption has been observed to alter conductance states and voltage sensitivity, which may contribute to dysfunctions like ion imbalances in cells. Similar changes in ChR2 with EYFP conjugation is possible, ultimately leading to the contractile and metabolic dysfunction we have observed. Furthermore, the structural damage caused by excessive EYFP clustering in the ChR2‐EYFP muscle could also be responsible for these force deficits. Membranous whorls, similar to what we observed in TEM images, are commonly found in muscles exposed to myotoxins such as vincristine, which can lead to muscle weakness and are hypothesized as a response to the necessity of managing toxic effects. 48 Thus, it is possible that the functional deficit of muscle is a result of the macro‐morphological alterations in muscle cell and tissue structure.
The biological processes enriched by genes downregulated in ChR2‐EYFP muscle compared with WT suggest impaired glycolytic metabolism, which may lead to reduced contractile force of EDL and gastrocnemius muscles. This hypothesis is further supported by the downregulation of FBP2 protein which regulates glycogenesis. 49 Many of the downregulated DEGs in 3‐month‐old ChR2‐EYFP muscle overlapped with those associated with aging (e.g., from SarcoAtlas, 7 , 50 ), suggesting a severe metabolic dysfunction in the presence of ChR2‐EYFP fusion protein. Thus, in addition to functional impairments, metabolic dysfunction in ChR2‐EYFP muscle may further compromise the physiological relevance of experimental findings obtained using this optogenetic model. Furthermore, we observed downregulation in fast‐twitch fiber contraction related genes which may explain the loss of force in fast‐twitch EDL muscle 36 , 37 , 38 but not in the slow‐twitch soleus muscle. The strong and significant downregulation of Actn3 is consistent with the functional and structural changes we have observed, as mice lacking Actn3 have been reported to have reduced muscle strength, mass, and a shift of fast‐twitch fibers toward slow‐twitch fibers. 51 , 52 The slight increase in half‐relaxation time in ChR2‐EYFP muscle may also indicate a slowing fiber phenotype, however, additional tests such as single fiber contractility and calcium transient analysis are required to confirm this. We also observed a downregulation of transmembrane transport genes encoding the voltage‐gated potassium and sodium ion channels, which may indicate changes in intracellular and transmembrane ion transport mechanisms. Additional contributors of functional impairments could be explained by increased inflammatory responses in the ChR2‐EYFP muscle. 53 The ECM in muscle plays a critical role in contractile force transmission, thus the upregulation of ECM and collagen catabolism‐related genes may influence muscle's force transmission efficiency. 54 , 55
Our in vivo 10 Hz optogenetic stimulation protocol generated similar ankle torque as 150 Hz electrical stimulation protocol. 6 However, ankle torque decayed as the muscle gets fatigued and retained at only ~50% of the initial torque by the end of the 20 min stimulation protocol. Generally, an exercise protocol is considered either endurance or resistance type depending on the amount force or torque produced and number of contractions elicited. Skeletal muscle secretes proinflammatory myokines under mechanical loading, 56 and biological processes indicative of inflammatory and immune responses were upregulated with optogenetic stimulation in both of our optogenetic strains, as previously reported in other studies on exercise. 57 , 58 However, the ChR2‐only muscle was more sensitive than the ChR2‐EYFP muscle as the same stimulation protocol yielded a higher number of DEGs in the ChR2‐only muscle. We observed downregulation of genes related to metabolism and mitochondrial regulation in the stimulated ChR2‐EYFP muscle; however, this phenomenon is not uncommon in resistance exercise. 59 Viggars et al. demonstrated that with daily electrical stimulation in rats, mitochondrial genes were downregulated in rat skeletal muscle until training period reached 30 days. 58 Thus, our optogenetic muscle stimulation protocol may be considered resistance exercise as it produces high torque with fewer contractions. 60 , 61
This study holds great significance in the field of musculoskeletal bioengineering as we demonstrated the potential adverse effects of fluorescent Cre reporters on muscle physiology. Cre reporter strains are popular in biomedical research for a variety of tasks, such as cell lineage tracing, validating recombination, tracking cellular processes, and monitoring expression in living tissues and cultured cells. However, our findings revealed significant dysfunctions in skeletal muscle expressing ChR2 with EYFP (i.e., Ai32). Upon EYFP removal (ChR2‐only), we observed substantial improvement in muscle contractility and physiological response to stimulation. This work has notable implications for the field, as it shows the potential to improve ChR2 delivery and efficacy in skeletal muscle thus strengthening the translational potential of optogenetics for regulating muscle function.
AUTHOR CONTRIBUTIONS
Syeda N. Lamia, Brian Glancy, Susan V. Brooks, and Megan L. Killian designed research. Syeda N. Lamia, Carol S. Davis, Peter C. D. Macpherson, Leanne Iannucci, Elahe Ganji, Desmond Harden, and Megan L. Killian analyzed data. Syeda N. Lamia, Carol S. Davis, T. Brad Willingham, Yingfan Zhang, Chengyu Liu, Leanne Iannucci, Elahe Ganji, Desmond Harden, Iman Bhattacharya, Adam C. Abraham, and Megan L. Killian performed research. Syeda N. Lamia and Megan L. Killian wrote the paper. Brian Glancy and Leanne Iannucci contributed new reagents or analytic tools.
DISCLOSURES
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
Supporting information
Data S1.
ACKNOWLEDGMENTS
National Science Foundation (CAREER 144448 to MLK); National Institutes of Health (R01AR079367 to MLK, ACA, SVB); Michigan Integrative Musculoskeletal Health Center P30AR069620 to MLK, SVB, CSD; 1ZIAHL006221 to BG; K12HD073945 and R03HD094594 to MLK; University of Michigan Rackham Graduate Student Research Grant (to SL); thanks to Robert Balaban at the National Institutes of Health and the NHLBI Electron Microscopy Core.
Lamia SN, Davis CS, Macpherson PCD, et al. Overexpression of enhanced yellow fluorescent protein fused with Channelrhodopsin‐2 causes contractile dysfunction in skeletal muscle. The FASEB Journal. 2024;38:e70185. doi: 10.1096/fj.202401664RR
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Dryad at doi: 10.5061/dryad.mpg4f4r8h or available in the Results and/or Supporting Information of this article.
REFERENCES
- 1. Gundelach LA, Hüser MA, Beutner D, Ruther P, Bruegmann T. Towards the clinical translation of optogenetic skeletal muscle stimulation. Pflugers Arch. 2020;472:527‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peckham PH, Knutson JS. Functional electrical stimulation for neuromuscular applications. Annu Rev Biomed Eng. 2005;7:327‐360. [DOI] [PubMed] [Google Scholar]
- 3. Thomas CK, Zaidner EY, Calancie B, Broton JG, Bigland‐Ritchie BR. Muscle weakness, paralysis, and atrophy after human cervical spinal cord injury. Exp Neurol. 1997;148:414‐423. [DOI] [PubMed] [Google Scholar]
- 4. Magown P, Shettar B, Zhang Y, Rafuse VF. Direct optical activation of skeletal muscle fibres efficiently controls muscle contraction and attenuates denervation atrophy. Nat Commun. 2015;6:8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruegmann T, van Bremen T, Vogt CC, Send T, Fleischmann BK, Sasse P. Optogenetic control of contractile function in skeletal muscle. Nat Commun. 2015;6:7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ganji E, Chan CS, Ward CW, Killian ML. Optogenetic activation of muscle contraction in vivo. Connect Tissue Res. 2021;62:15‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ganji E, Lamia SN, Stepanovich M, et al. Optogenetic‐induced muscle loading leads to mechanical adaptation of the Achilles tendon enthesis in mice. Sci Adv. 2023;9:eadf4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rousseau E, Raman R, Tamir T, et al. Actuated tissue engineered muscle grafts restore functional mobility after volumetric muscle loss. Biomaterials. 2023;302:122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagel G, Szellas T, Huhn W, et al. Channelrhodopsin‐2, a directly light‐gated cation‐selective membrane channel. Proc Natl Acad Sci. 2003;100:13940‐13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin‐2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279‐2284. [DOI] [PubMed] [Google Scholar]
- 11. Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond‐timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263‐1268. [DOI] [PubMed] [Google Scholar]
- 12. Lipták N, Bősze Z, Hiripi L. GFP transgenic animals in biomedical research: a review of potential disadvantages. Physiol Res. 2019;68:525‐530. [DOI] [PubMed] [Google Scholar]
- 13. Ansari AM, Ahmed AK, Matsangos AE, et al. Cellular GFP toxicity and immunogenicity: potential confounders in in vivo cell tracking experiments. Stem Cell Rev and Rep. 2016;12:553‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang W‐Y, Aramburu J, Douglas PS, Izumo S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat Med. 2000;6:482‐483. [DOI] [PubMed] [Google Scholar]
- 15. Bridge KE, Berg N, Adalbert R, et al. Late onset distal axonal swelling in YFP‐H transgenic mice. Neurobiol Aging. 2009;30:309‐321. [DOI] [PubMed] [Google Scholar]
- 16. Agbulut O, Huet A, Niederländer N, Puceat M, Menasché P, Coirault C. Green fluorescent protein impairs actin‐myosin interactions by binding to the actin‐binding site of myosin. J Biol Chem. 2007;282:10465‐10471. [DOI] [PubMed] [Google Scholar]
- 17. Meng X, Murali S, Cheng Y‐F, et al. Increasing the expression level of ChR2 enhances the optogenetic excitability of cochlear neurons. J Neurophysiol. 2019;122:1962‐1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zimmermann D, Zhou A, Kiesel M, et al. Effects on capacitance by overexpression of membrane proteins. Biochem Biophys Res Commun. 2008;369:1022‐1026. [DOI] [PubMed] [Google Scholar]
- 19. Miyashita T, Shao YR, Chung J, Pourzia O, Feldman DE. Long‐term channelrhodopsin‐2 (ChR2) expression can induce abnormal axonal morphology and targeting in cerebral cortex. Front Neural Circuits. 2013;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maimon BE, Diaz M, Revol ECM, et al. Optogenetic peripheral nerve immunogenicity. Sci Rep. 2018;8:14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao P, Monks DA. A tetracycline‐inducible and skeletal muscle‐specific Cre recombinase transgenic mouse. Devel Neurobio. 2009;69:401‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H, Yang H, Shivalila CS, et al. One‐step generation of mice carrying mutations in multiple genes by CRISPR/Cas‐mediated genome engineering. Cell. 2013;153:910‐918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willingham TB, Zhang Y, Andreoni A, Knutson JR, Lee D, Glancy B. MitoRACE: evaluating mitochondrial function in vivo and in single cells with subcellular resolution using multiphoton NADH autofluorescence. J Physiol. 2019;597:5411‐5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larkin LM, Davis CS, Sims‐Robinson C, et al. Skeletal muscle weakness due to deficiency of CuZn‐superoxide dismutase is associated with loss of functional innervation. Am J Physiol‐Regul Integr Compar Physiol. 2011;301:R1400‐R1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brooks SV, Faulkner JA. Contractile properties of skeletal muscles from young, adult and aged mice. J Physiol. 1988;404:71‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L, Jacques SL, Zheng L. MCML—Monte Carlo modeling of light transport in multi‐layered tissues. Comput Methods Prog Biomed. 1995;47:131‐146. [DOI] [PubMed] [Google Scholar]
- 27. Marti D, Aasbjerg RN, Andersen PE, Hansen AK. MCmatlab: an open‐source, user‐friendly, MATLAB‐integrated three‐dimensional Monte Carlo light transport solver with heat diffusion and tissue damage. J Biomed Opt. 2018;23:1‐6. [DOI] [PubMed] [Google Scholar]
- 28. Bashkatov AN, Genina EA, Tuchin VV. Optical properties of skin, subcutaneous, and muscle tissues: a review. J Innov Opt Health Sci. 2011;4:9‐38. [Google Scholar]
- 29. Jacques SL. Optical properties of biological tissues: a review. Phys Med Biol. 2013;58:R37‐R61. [DOI] [PubMed] [Google Scholar]
- 30. Sabino CP, Deana AM, Yoshimura TM, et al. The optical properties of mouse skin in the visible and near infrared spectral regions. J Photochem Photobiol B Biol. 2016;160:72‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brooks HL, Lindsey ML. Guidelines for authors and reviewers on antibody use in physiology studies. Am J Physiol Heart Circ Physiol. 2018;314:H724‐H732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fosang AJ, Colbran RJ. Transparency is the key to quality. J Biol Chem. 2015;290:29692‐29694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44‐57. [DOI] [PubMed] [Google Scholar]
- 35. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li A, Nelson SR, Rahmanseresht S, et al. Skeletal MyBP‐C isoforms tune the molecular contractility of divergent skeletal muscle systems. Proc Natl Acad Sci USA. 2019;116:21882‐21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y, Szczesna‐Cordary D, Craig R, et al. Fast skeletal muscle regulatory light chain is required for fast and slow skeletal muscle development. FASEB J. 2007;21:2205‐2214. [DOI] [PubMed] [Google Scholar]
- 38. Ju Y, Li J, Xie C, et al. Troponin T3 expression in skeletal and smooth muscle is required for growth and postnatal survival: characterization of Tnnt3 tm2a(KOMP)Wtsi mice. Genesis. 2013;51:667‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song T, McNamara JW, Ma W, et al. Fast skeletal myosin‐binding protein‐C regulates fast skeletal muscle contraction. Proc Natl Acad Sci USA. 2021;118:e2003596118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Madisen L, Mao T, Koch H, et al. A toolbox of Cre‐dependent optogenetic transgenic mice for light‐induced activation and silencing. Nat Neurosci. 2012;15:793‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burkholder TJ, Fingado B, Baron S, Lieber RL. Relationship between muscle fiber types and sizes and muscle architectural properties in the mouse hindlimb. J Morphol. 1994;221:177‐190. [DOI] [PubMed] [Google Scholar]
- 43. Talbot J, Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. WIREs Dev Biol. 2016;5:518‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ludwig A. Absence epilepsy and sinus dysrhythmia in mice lacking the pacemaker channel HCN2. EMBO J. 2003;22:216‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bukauskas FF, Jordan K, Bukauskiene A, et al. Clustering of connexin 43–enhanced green fluorescent protein gap junction channels and functional coupling in living cells. Proc Natl Acad Sci USA. 2000;97:2556‐2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bukauskas FF, Bukauskiene A, Bennett MVL, Verselis VK. Gating properties of gap junction channels assembled from Connexin43 and Connexin43 fused with green fluorescent protein. Biophys J. 2001;81:137‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carnarius C, Kreir M, Krick M, et al. Green fluorescent protein changes the conductance of Connexin 43 (Cx43) Hemichannels reconstituted in planar lipid bilayers. J Biol Chem. 2012;287:2877‐2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khan MA. Effects of myotoxins on skeletal muscle fibers. Prog Neurobiol. 1995;46:541‐560. [DOI] [PubMed] [Google Scholar]
- 49. Park H‐J, Jang HR, Park S‐Y, Kim Y‐B, Lee H‐Y, Choi CS. The essential role of fructose‐1,6‐bisphosphatase 2 enzyme in thermal homeostasis upon cold stress. Exp Mol Med. 2020;52:485‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Börsch A, Ham DJ, Mittal N, et al. Molecular and phenotypic analysis of rodent models reveals conserved and species‐specific modulators of human sarcopenia. Commun Biol. 2021;4:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. MacArthur DG, Seto JT, Chan S, et al. An Actn3 knockout mouse provides mechanistic insights into the association between ‐actinin‐3 deficiency and human athletic performance. Hum Mol Genet. 2008;17:1076‐1086. [DOI] [PubMed] [Google Scholar]
- 52. Seto JT, Roeszler KN, Meehan LR, et al. ACTN3 genotype influences skeletal muscle mass regulation and response to dexamethasone. Sci Adv. 2021;7:eabg0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dalle S, Rossmeislova L, Koppo K. The role of inflammation in age‐related sarcopenia. Front Physiol. 2017;8:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Street SF. Lateral transmission of tension in frog myofibers: a myofibrillar network and transverse cytoskeletal connections are possible transmitters. J Cell Physiol. 1983;114:346‐364. [DOI] [PubMed] [Google Scholar]
- 55. Csapo R, Gumpenberger M, Wessner B. Skeletal muscle extracellular matrix—what do we know about its composition, regulation, and physiological roles? A narrative review. Front Physiol. 2020;11:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scheele C, Nielsen S, Pedersen BK. ROS and myokines promote muscle adaptation to exercise. Trends Endocrinol Metab. 2009;20:95‐99. [DOI] [PubMed] [Google Scholar]
- 57. Fernandez‐Gonzalo R, Willis CRG, Etheridge T, Deane CS. RNA‐sequencing muscle plasticity to resistance exercise training and disuse in youth and older age. Phys Ther. 2022;2:164‐179. [Google Scholar]
- 58. Viggars MR, Sutherland H, Lanmüller H, Schmoll M, Bijak M, Jarvis JC. Adaptation of the transcriptional response to resistance exercise over 4 weeks of daily training. FASEB J. 2023;37:e22686. [DOI] [PubMed] [Google Scholar]
- 59. Pillon NJ, Gabriel BM, Dollet L, et al. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat Commun. 2020;11:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahoney DJ, Tarnopolsky MA. Understanding skeletal muscle adaptation to exercise training in humans: contributions from microarray studies. Phys Med Rehabil Clin N Am. 2005;16:859‐873. [DOI] [PubMed] [Google Scholar]
- 61. Ashida Y, Himori K, Tatebayashi D, Yamada R, Ogasawara R, Yamada T. Effects of contraction mode and stimulation frequency on electrical stimulation‐induced skeletal muscle hypertrophy. J Appl Physiol. 2018;124:341‐348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at doi: 10.5061/dryad.mpg4f4r8h or available in the Results and/or Supporting Information of this article.


