Abstract
Background
Suppression Subtractive Hybridization PCR (SSH PCR) is a sophisticated cDNA subtraction method to enrich and isolate differentially expressed genes. Despite its popularity, the method has not been thoroughly studied for its practical efficacy and potential limitations.
Results
To determine the factors that influence the efficacy of SSH PCR, a theoretical model, under the assumption that cDNA hybridization follows the ideal second kinetic order, is proposed. The theoretical model suggests that the critical factor influencing the efficacy of SSH PCR is the concentration ratio (R) of a target gene between two cDNA preparations. It preferentially enriches "all or nothing" differentially expressed genes, of which R is infinite, and strongly favors the genes with large R. The theoretical predictions were validated by our experiments. In addition, the experiments revealed some practical limitations that are not obvious from the theoretical model. For effective enrichment of differentially expressed genes, it requires fractional concentration of a target gene to be more than 0.01% and concentration ratio to be more than 5 folds between two cDNA preparations.
Conclusion
Our research demonstrated theoretical and practical limitations of SSH PCR, which could be useful for its experimental design and interpretation.
Background
Alterations in gene expression are associated with a large spectrum of biological and pathological process [1]. The identification of differentially expressed genes often leads to greater insight into the molecular mechanisms underlying disease progression or biological development. To facilitate the discovery of differentially expressed genes, a variety of methods have been developed in recent years including Differential Display PCR [2], RNA fingerprinting [3], SAGE [4], Real-time Quantitative PCR (TaqMan) [5-7], Subtractive Suppression Hybridization PCR (SSH) [8], and hybridization to gene arrays of various formats [9,10]. Although each method has advantages and drawbacks, the general methodology for identification of differentially expressed genes has progressed from labor-intensive procedures, such as polyacrylamide gel-based differential display, to automatic high throughput methods such as hybridization-based gene arrays. Commercial gene arrays, which contain probes bound to small glass plates or chips representing many genes and ESTs, provide simultaneous measurement of gene abundance and have greatly accelerated the search for differentially expressed genes. However, such arrays and associated equipment are expensive and beyond the access of most academic laboratories. Commercial arrays also suffer by being restricted to available gene sequences to serve as templates for probe design. They generally only cover human and the most common model organisms. Thus, to identify novel genes or to study other organisms such as agricultural crops and live stocks, it is still necessary to utilize additional methods beyond such gene chips and arrays.
Subtractive hybridization is an attractive method for enriching differentially expressed genes. This method was first used by Bautz and Reilly to purify phage T4 mRNA in the mid-1960's [11]. Pure subtractive methodologies are of limited use due to the need for a large quantity of mRNA to drive hybridization to completion as well as the difficulty in cloning the tiny amount of cDNA remaining after hybridization. The method was greatly improved when Duguid and Dinauer adapted generic linkers to cDNA [12] allowing the selective PCR amplification of tester cDNA between hybridization cycles. Diatchenko et al . further introduced the technique of Suppression Subtractive Hybridization PCR (SSH PCR) in which differentially expressed genes could be normalized and enriched over 1000-fold in single round of hybridization [8]. The recent commercialization of an SSH PCR kit by Clontech (CLONTECH Laboratories, Palo Alto, CA, USA) has lead to its increasing popularity in biological research laboratories [13-17].
Despite the popularity of SSH PCR, this complicated method has not been thoroughly studied for its practical efficacy and potential limitations. In this work, we have proposed a theoretical model of SSH PCR based on the assumption that cDNA hybridization follows the ideal second kinetic order. We further tested the theoretical predictions by several SSH experiments.
Results
Theoretical model of SSH PCR
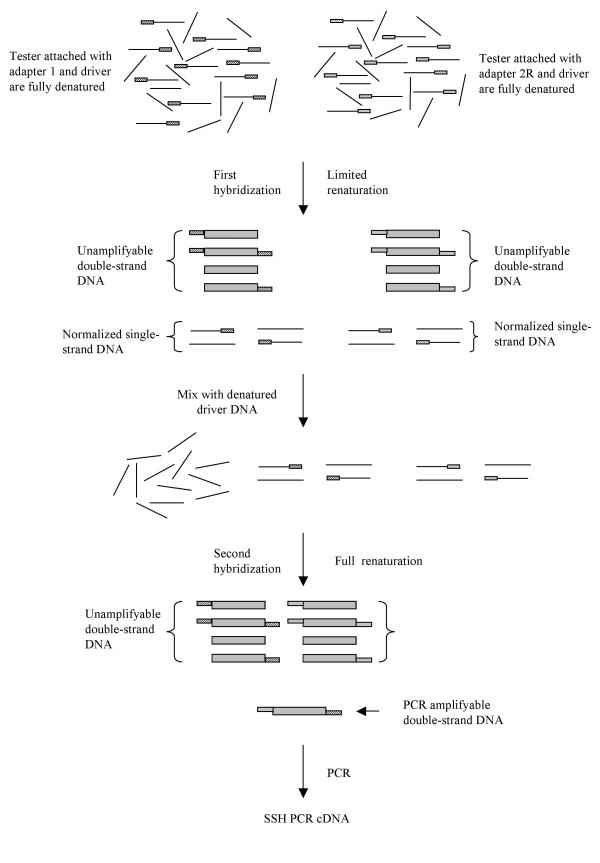
The strategy of SSH PCR to enrich differentially expressed gene is depicted in Figure 1. The procedure consists primarily of two substrative hybridizations and a single PCR amplification. In the first hybridization step, tester cDNA fitted with adapter 1 or 2R is mixed with a large excess of driver cDNA and denatured separately. They are then subjected to limited renaturation, also separately. Because the renaturation process, which is random collision of complementary strands, obeys the ideal second-order kinetics, the rate of the reaction can be described by Equation 1 [18,19]:
Figure 1.
Scheme of SSH PCR
![]()
where C is molar concentration of a single-strand target gene, t is time and k is the rate constant.
Equation 1 can be integrated and solved yielding Equation 2:
![]()
where C0 is the starting concentration of the single-strand DNA, and Ct is the concentration of the remaining single-strand DNA at time t. When C0kt>>1, Equation 2 simplifies to Equation 3:
![]()
Equation 3 implies that when hybridization time is long enough, or when C0kt>>1, the concentration of remaining single-strand DNA is determined mainly by its hybridization rate constant k and hybridization time t, and is independent of its starting concentration C0. This is the basis of normalization in the first hybridization reaction.
Because single-strand cDNAs consist of both tester cDNAs, which are fitted with adapter, and driver cDNAs, which are not fitted with adapters, and if we further assume that DNA with and without adapter have the same hybridization kinetics or to say simply that adapter will not interfere with DNA hybridization, then the concentration of the PCR amplifiable cDNA (those with adapters) can be calculated from Equation 4:
![]()
where Ct' is the concentration of a target single-strand cDNA with adapter, N is the ratio of the driver to tester in the first hybridization, and the R is the concentration ratio of the target cDNA in tester to that in driver.
In the first hybridization none of the double-strand cDNA can be amplified by PCR because it either lacks adapter sequences for binding of PCR primer(s) or PCR is suppressed by a so-called "panhandle" structure that is formed by long complementary sequences of 5' and 3' ends of adapters [21]. Therefore, only the single-strand cDNAs containing adapters are of consequence in the second hybridization.
In the second hybridization, the single-strand cDNAs from the first hybridization are mixed with new denatured driver cDNAs to form double-strand cDNAs. The second hybridization is carried out over a longer time period to ensure that all cDNAs become double-stranded. This reaction can be described by Equation 5:
![]()
where A and B are a single-strand cDNA with its complementary strand respectively. A' and A" are strands fitted with adapter 1 and 2R respectively. B' and B" are fitted with adapter 1 and 2R respectively. In the second hybridization, only the double-strand cDNAs with two different adapters at each end (A'B" and A"B') can be amplified by PCR. The amount of product (A'B"+A"B') available for amplification can determined by Equation 6:
![]()
Given that A = B = MC0/R, where M is the ratio of driver to tester in the second hybridization and R is the concentration ratio of a target cDNA of tester to driver and given Equation 4 the following hold true: A' = B' = A" = B" = Ct' = Ct/(1 + N/R). Thus the concentration of target double-strand cDNA with hetero-adapters can be calculated by Equation 7:
![]()
where Ct is the concentration of remaining single-strand cDNA after the first hybridization, N is the ratio of driver to tester in the first hybridization (30 in our experiments), M is the ratio of driver to tester in the second hybridization (5 in our experiments), and the R is the concentration ratio of the target cDNA in tester to that in driver.
If we make some simple approximations by a. ignoring the cDNAs that cannot be amplified by PCR, which is logical considering the exponential amplification by PCR which results in unamplified cDNA comprising only a tiny portion of the total final cDNA, b. ignoring differences in PCR efficiency between amplifiable cDNAs, which is reasonable considering that all cDNAs have identical adapters, then Equation 7 gives the relative amount of all cDNAs after SSH PCR.
Thus, several predictions can be directly made by Equation 7. 1. when R = ∞, meaning that the target cDNA is an 'all or nothing' differentially expressed cDNA due to its presence only in tester and not in driver cDNA, then A'B" + A"B' = Ct = 1/kt (Equation 4), then every 'all or nothing' differentially expressed cDNA will be enriched to a fixed level irrespective of its starting concentration; 2. when R is a small number (<10 for example), meaning the target is a ratio differentially expressed cDNA present both in tester and driver cDNA but at different concentrations, then C0>>Ct and N>>R. Equation 7 can therefore be simplified to:
![]()
Equation 8 demonstrates that the enrichment of a ratio differentially expressed gene is proportional to the cube of R, implying that the greater the expression ratio is between a cDNA in driver vs. tester the more likely it is to be detected by SSH PCR.
Experimental Test of SSH PCR
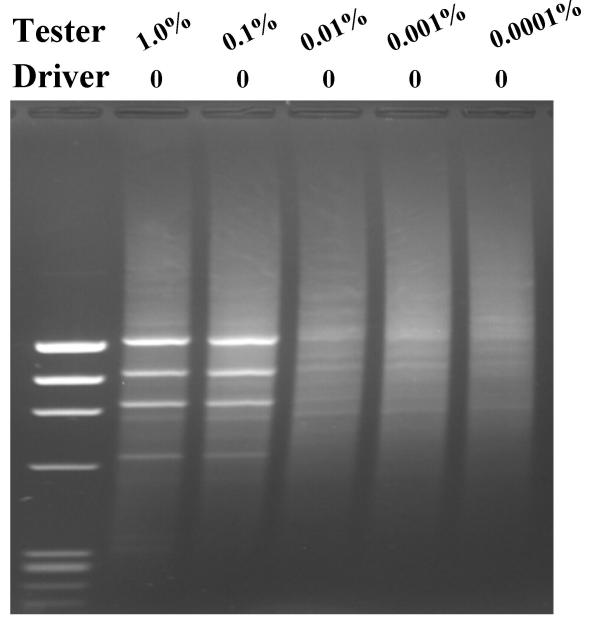
To experimentally test the two predictions of the theoretical model we designed a series of experiments. First, we tested SSH PCR for enrichment 'all or nothing' differentially expressed genes. We prepared a series of tester cDNAs by artificially adding φx174 DNA to fibroblast cDNA to simulate differentially expressed genes and extracted the tester cDNAs by using fibroblast cDNA as driver. The results (Fig 2) demonstrated that 'differentially expressed' φx174 DNA can be enriched to clearly visible bands when its fractional concentration is more than 0.01% of tester cDNA (Fig 2. lanes 2,3). When the starting fractional concentrations of φx174 were 1.0% and 0.1% respectively in the tester cDNA preparations, the SSH PCR φx174 bands were of similar intensity after SSH PCR as shown in lanes 2 and lane 3 (Fig 2), indicating their enrichment to the same level. This is in consistent with the theoretical prediction. Fig 2 also revealed a practical limitation of SSH PCR not obvious from the theoretical model. When φx174 DNA is less than 0.01% of tester cDNA, no clearly visible bands of φx174 are apparent after agarose gel electrophoresis (lane 4,5,6), indicating that most of SSH PCR cDNAs are not the 'differentially expressed' target φx174, but are predominantly randomly amplified fibroblast cDNAs.
Figure 2.
Enrichment of 'all or nothing' differentially expressed φx174 DNA HaeIII by SSH PCR. Various amount of φx174 HaeIII DNA was added in tester. None was added in driver. SSH PCR was carried out as described in Materials and Methods. Lane 1, φx174 HaeIII DNA marker; lane 2, 1.0% of φx174 DNA HaeIII in tester; lane 3, 0.1% of φx174 DNA HaeIII in tester; lane 4, 0.01% of φx174 HaeIII DNA in tester; lane 5, 0.001% of φx174 HaeIII DNA in tester; lane 5, 0.0001% of φx174 DNA HaeIII in tester.
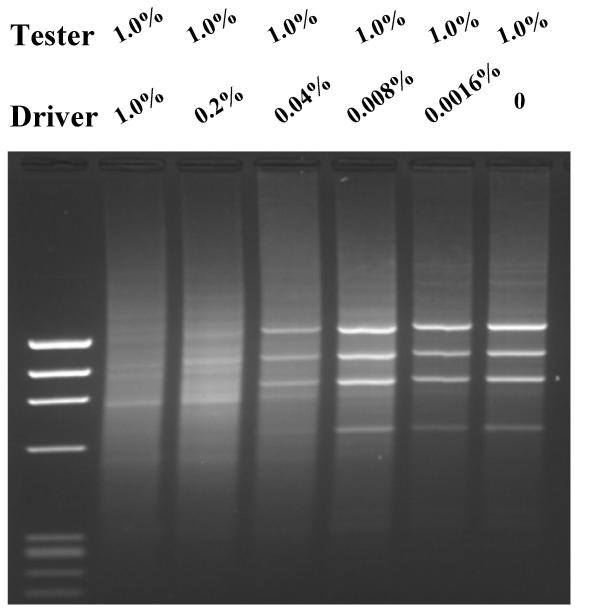
We also tested SSH PCR for efficacy in enriching ratio differentially expressed genes. We prepared a series of tester and driver cDNAs by adding different amounts of φx174 DNA to fibroblast cDNA. In the first series, the tester cDNA contained a fixed amount (1.0%) φx174 DNA added to fibroblast cDNA while a series of driver cDNAs were made by adding φx174 DNA ranging from 1% to 0% to fibroblast cDNA. Then we enriched the 'differentially expressed' φx174 DNA by SSH PCR. The results (Fig 3) demonstrated that 'differentially expressed' φx174 DNA can be enriched to clearly visible bands only when it is 5-fold or more concentrated in tester compared to driver cDNAs (lane 4, 5, 6 and 7). When the differentially expressed cDNA is less than 5-fold concentrated in the tester, no distinguishable φx174 DNA bands were seen (lane 2 and 3), suggesting that the "differentially expressed' φx174 DNA was not enriched enough by SSH PCR and that the resulting SSH library consists mainly of randomly amplified fibroblast cDNAs.
Figure 3.
Enrichment of ratio differentially expressed φx174 DNA HaeIII by SSH PCR. Fixed amount of 1.0% φx174 HaeIII DNA was added in tester and various amount of φx174 HaeIII DNA in driver, which resulted in a serial five folds of dilutions. SSH PCR was carried out as described in Materials and Methods. Lane 1, φx174 HaeIII DNA marker; lane 2, 1.0% of φx174 HaeIII DNA in driver; lane 3, 0.2% of φx174 HaeIII DNA in driver; lane 4, 0.04% of φx174 HaeIII DNA in driver; lane 5, 0.008% of φx174 HaeIII DNA in driver; lane 6, 0.0016% of φx174 HaeIII DNA in driver in driver; lane 7, no φx174 HaeIII DNA in driver driver.
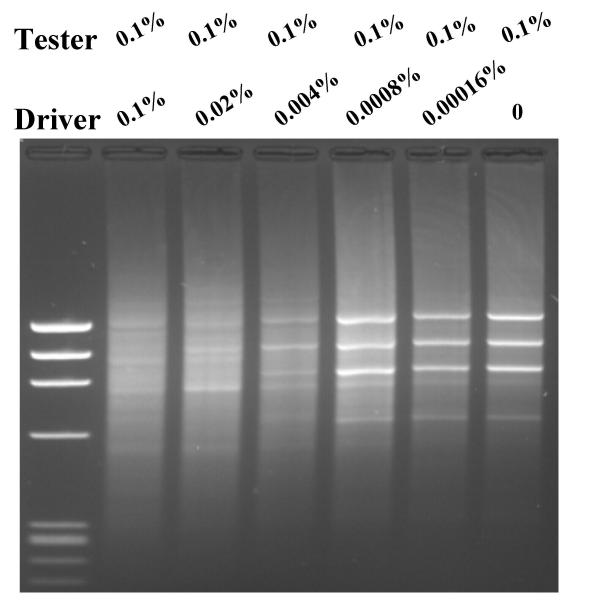
To further examine the role of the concentration ratio R and the effect of target abundance on efficiency of SSH PCR, we made a second series of tester and driver cDNA for SSH PCR. Tester cDNA contained 0.1% φx174 DNA, one-tenth the amount in the previous experiment added to fibroblast cDNA and the series of driver cDNAs was also reduced by 10-fold driver ranging from 0.1% to 0% φx174 DNA in fibroblast cDNA. Thus, the absolute amount of 'differentially expressed' φx174 DNA is one tenth of the amount in the previous experiment, however, the corresponding concentration ratios are identical. We again enriched the 'differentially expressed' φx174 DNA using SSH PCR and the results are shown in Fig 4. The results of this experiment were almost identical to the previous experiment in that the dependence on concentration ratio for effective enrichment was similar requiring more than five-fold more φx174 DNA in tester than in driver (lane 3, 4, 5 and 6). The results in Fig 3 and Fig 4 together demonstrate that effective enrichment by SSH PCR is highly dependent on concentration ratio of the differentially expressed gene. Enrichment is far more effective for genes that are highly differentially expressed. These results are consistent with the theoretical prediction described in Equation 8.
Figure 4.
Enrichment of ratio differentially expressed φx174 DNA HaeIII by SSH PCR. Fixed amount of 0.1% φx174 HaeIII DNA was added in tester and various amount of φx174 HaeIII DNA in driver, which resulted in serial five folds of dilutions. SSH PCR was carried out as described in Materials and Methods. Lane 1, φx174 HaeIII DNA marker; lane 2, 0.1% of φx174 HaeIII DNA in driver, lane 3, 0.02% of φx174 HaeIII DNA in driver; lane 4, 0.004% of φx174 HaeIII DNA in driver; lane 5, 0.0008% of φx174 HaeIII DNA in driver; lane 6, 0.00016% of φx174 HaeIII DNA in driver in driver; lane 7, no φx174 HaeIII DNA in driver.
Discussion
We presented a theoretical model to describe SSH PCR based on the well-established second order kinetic of DNA hybridization [18,19]. Recent kinetic modeling and computer simulation of subtractive hybridization based on the similar principles have shown that they agree well with existing experimental data [20,22]. Our mathematical calculations described in Equation 7 and 8 reveal the relative importance of factors such as concentration ratio (R) and target abundance for any specific cDNA to be present in an SSH PCR library. When R→∞, that is when differentially expressed genes are 'all or nothing', they are effectively enriched to a fixed concentration of 1/kt. When R is a small number, enrichment is proportional to R3, favoring highly differentially expressed genes. Our experiments confirmed the theoretical prediction that the primary factor influencing enrichment is the concentration ratio R and not the absolute difference. This was supported by the similar enrichment of 1.0% and 0.1% φx174 DNA shown in Fig 3 and 4. On the contrary side, SSH PCR cannot exclude all non-differentially expressed gene from a library. This was demonstrated the evenly distributed DNA surrounding the φx174 DNA bands which are evidently derived from 'non-differentially' expressed fibroblast cDNA. Contrary to the theoretical prediction, however, our SSH PCR experiment failed to enrich φx174 DNA when less than 0.01% (Fig 2 lane 4, 5 and 6). A possible explanation is that target cDNA less than 0.01% is too low to drive hybridization to completion in the second hybridization. Because formation of double-stranded cDNA is required for PCR amplification in SSH PCR, the result will be low representation of the rare target cDNA in the SSH PCR library even if it is of the 'all or nothing' differentially expressed cDNAs.
Practical factors, such as PCR amplification efficiency, have not been taken into our theoretical consideration. As note before, the PCR amplification efficiency is sequence-dependent, which may result in fortuitous over-representation or under-representation of certain sequences in SSH PCR library. The factors may change the outcomes of SSH PCR experiments serendipitously. They, however, don't constitute the basis for SSH PCR to enrich differentially expressed genes. For simplicity, they are not included in our theoretical consideration.
Our results have a significant bearing on the use SSH PCR application and the interpretation of experimental results. Because SSH PCR favors highly differentially expressed genes, the primary application of SSH PCR should be to detect dramatic alteration of gene expression, such as comparison of gene expression after viral infection or gene expression profiling of two different tissues. In profiling gene expression differences in diseased vs. normal tissues or over an experimental time course where small changes in gene expression are more likely to be physiologically relevant, SSH PCR would be highly ineffective in profiling gene expression changes. In such situations, differential screening of very large SSH PCR libraries can potentially compensate but at high costs in time and labor. In addition, for effective enrichment by SSH PCR the target mRNA must be at least 0.1% of the total mRNA, thus low abundance genes such as transcription factors, cytokines, and receptors which are key regulators of many pathological processes would not be detected by this method.
Care must be also be taken in the interpretation of SSH PCR results. The presence of many non-differentially expressed genes in an SSH PCR library may not result from experimental error but maybe due to the absence of significantly differentially expressed genes between the chosen driver and tester samples. The failure of a SSH PCR library to include a known differentially expressed mRNA may also not be a result of experimental error. From Equation 8, a differentially expressed cDNA is only R3-fold enriched in a SSH PCR library as compared with an unsubtracted cDNA library. Thus it should not be surprising that a small SSH PCR library does not contain a known differentially expressed gene.
Conclusions
Our theoretical model suggests that effective enrichment of a target gene by SSH PCR is determined by its concentration ratio (R) between tester and driver. The enrichment is far more efficient for differentially expressed genes with a large value for R. Our experiments validate the theoretical predictions that enrichment by SSH is greatly influenced by concentration ratio R. They also revealed practical limitations: for effective enrichment of 'all or nothing' differentially expressed genes, the fractional concentration of a target gene needs be more than 0.01%. For effective enrichment of ratio differentially expressed genes, the concentration ratio needs to be more than 5-fold.
Materials and Methods
Total RNAs were isolated from primary cell cultures of human fibroblast using the RNeasy Mini kit (Qiagen, Chatsworth, CA, USA). cDNAs were synthesized and amplified from the total RNA with the SMART PCR cDNA Synthesis kit (Clontech, Palo Alto, CA, USA). The cDNAs were purified by the QIAquick PCR Purification kit (Qiagen, Chatsworth, CA, USA). The purified cDNAs were digested by RsaI and repurified by the QIAquick PCR procedure. The digested cDNAs were suspended at a concentration 360 ng/μl and used directly for SSH PCR.
Defined amounts of HaeIII-digested φx174 DNA to human fibroblast cDNA to simulate differentially expressed genes in tester cDNAs. Human fibroblast cDNAs were used as the driver. SSH PCR methods were those described in PCR-Select cDNA Subtraction kit (Clontech, Palo Alto, CA, USA). The appearance of φx174 HaeIII bands following agarose gel electrophoresis of SSH PCR products in ethidium bromide-stained gels was taken as an indicator of enrichment. In short, various amounts of HaeIII digested phage φx174 DNA were added to the RsaI digested cDNAs to simulate differentially expressed genes. Tester cDNAs were fitted with either adapter 1 or adapter 2R by T4 DNA ligase. In the first SSH PCR hybridization, 18 ng of tester cDNAs fitted with either adapter 1 or 2R were mixed with 540 ng of driver cDNA and hybridization buffer in a volume of 5 μl. They were denatured and allowed to undergo 8 hr of limited renaturation at 68°C separately. In the second SSH PCR hybridization, 360 ng of freshly denatured driver DNA and the two reactions of the first hybridization were mixed in a volume of 14 μl and allowed to undergo 20 hr of hybridization at 68°C. The subtracted tester cDNA was then diluted with 235 μl of dilution buffer. 1 μl of the diluted subtracted cDNA was amplified by PCR in 25 μl of reaction mixture containing: 1× PCR reaction buffer, 200 μM dNTP, 400 nM PCR primer 1 and 1× Advantage cDNA Polymerase Mix. The PCR was performed on a MJ Research PTC 200 thermocycler with program: 75°C 5 min, 94°C 25 sec, 27 cycles of 94°C 10 sec, 66°C 30 sec, 72°C 1.5 min. The PCR products were diluted 10 times with H2O. 1 μl of the diluted PCR products was amplified again by nested PCR in 25 μl of reaction mixture containing: 1× PCR reaction buffer, 200 μM dNTP, 400 nM Nested PCR primer 1, 400 nM Nested PCR primer 2R and 1× Advantage cDNA Polymerase Mix. The PCR was performed on a MJ Research PTC 200 thermocycler with 12 cycles of 94°C 10 sec, 68°C 30 sec, 72°C 1.5 min. The nested PCR products were separated electrophoretically on 2% agarose gels. The agarose gels were stained with ethidium bromide and pictures were taken under UV illumination at 254 nm.
Contributor Information
Wan Ji, Email: jiwanaustin@aol.com.
Matthew B Wright, Email: matthew.wright@roche.com.
Li Cai, Email: licai2000@yahoo.com.
Angel Flament, Email: angel.flament@roche.com.
Klaus Lindpaintner, Email: klaus.lindpaintner@roche.com.
References
- Lewin B. Gene V. Oxford, UK: Oxford University Press. 1994.
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Welsh J, Chada K, Dalal SS, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Bassler HA, Flood SJ, Livak KJ, Marmaro J, Knorr R, Batt CA. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SP. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1006/jmbi.1998.2134. [DOI] [PubMed] [Google Scholar]
- Bautz EK, Reilly E. Gene-specific messenger RNA: isolation by the deletion method. Science. 1966;151:328–330. doi: 10.1126/science.151.3708.328. [DOI] [PubMed] [Google Scholar]
- Duguid JR, Dinauer MC. Library subtraction of in vitro cDNA libraries to identify differentially expressed genes in scrapie infection. Nucleic Acids Res. 1990;18:2789–2792. doi: 10.1093/nar/18.9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang L, Shao ZM, Beatty P, Sartippour M, Lane TF, Barsky SH, Livingston E, Nguyen M. Identification of a Novel Endothelial-Derived Gene EG-1. Biochem Biophys Res Commun. 2002;290:602–12. doi: 10.1006/bbrc.2001.6119. [DOI] [PubMed] [Google Scholar]
- Bahn SC, Bae MS, Park YB, Oh SI, Jeung JU, Bae JM, Chung YS, Shin JS. Molecular cloning and characterization of a novel low temperature-induced gene, blti2, from barley (Hordeum vulgare L.)(1). Biochim Biophys Acta. 2001;1522:134–137. doi: 10.1016/S0167-4781(01)00317-7. [DOI] [PubMed] [Google Scholar]
- Stassar MJ, Devitt G, Brosius M, Rinnab L, Prang J, Schradin T, Simon J, Petersen S, Kopp-Schneider A, Zoller M. Identification of human renal cell carcinoma associated genes by suppression subtractive hybridization. Br J Cancer. 2001;85:1372–1382. doi: 10.1054/bjoc.2001.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Rossman TG. Genes upregulated in lead-resistant glioma cells reveal possible targets for lead-induced developmental neurotoxicity. Toxicol Sci. 2001;64:90–99. doi: 10.1093/toxsci/64.1.90. [DOI] [PubMed] [Google Scholar]
- Uchijima M, Raz E, Carson DA, Nagata T, Koide Y. Identification of immunostimulatory DNA-induced genes by suppression subtractive hybridization. Biochem Biophys Res Commun. 2001;286:688–691. doi: 10.1006/bbrc.2001.5453. [DOI] [PubMed] [Google Scholar]
- Wetmur JG. Hybridization and renaturation kinetics of nucleic acids. Annu Rev Biophys Bioeng. 1976;5:337–361. doi: 10.1146/annurev.bb.05.060176.002005. [DOI] [PubMed] [Google Scholar]
- Wetmur JG, Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968;31:349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Milner JJ, Cecchini E, Dominy PJ. A kinetic model for subtractive hybridization. Nucleic Acids Res. 1995;23:176–187. doi: 10.1093/nar/23.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho TJ, Park SS. A simulation of subtractive hybridization. Nucleic Acids Res. 1998;26:1440–1448. doi: 10.1093/nar/26.6.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]