Abstract
A virulent double-stranded DNA bacteriophage, ΦK1-5, has been isolated and found to be capable of infecting Escherichia coli strains that possess either the K1 or the K5 polysaccharide capsule. Electron micrographs show that the virion consists of a small icosohedral head with short tail spikes, similar to members of the Podoviridae family. DNA sequence analysis of the region encoding the tail fiber protein showed two open reading frames encoding previously characterized hydrolytic phage tail fiber proteins. The first is the K5 lyase protein gene of ΦK5, which allows this phage to specifically infect K5 E. coli strains. A second open reading frame encodes a protein almost identical in amino acid sequence to the N-acetylneuraminidase (endosialidase) protein of ΦK1E, which allows this phage to specifically infect K1 strains of E. coli. We provide experimental evidence that mature phage particles contain both tail fiber proteins, and mutational analysis indicates that each protein can be independently inactivated. A comparison of the tail gene regions of ΦK5, ΦK1E, and ΦK1-5 shows that the genes are arranged in a modular or cassette configuration and suggests that this family of phages can broaden host range by horizontal gene transfer.
Escherichia coli capsular polysaccharides (K antigens) have often been associated with increased virulence (17). The K1 antigen in particular increases the invasiveness of E. coli, and these strains are often involved in cases of meningitis and septicemia (32). These polysaccharide coats also act as recognition sites for bacteriophages, which often carry tail spikes that contain polysaccharide depolymerization activities. Several K1-specific phages have been described (10), one of which, ΦK1E, was found to possess N-acetylneuraminidase (endosialidase) as a part of the tail fiber protein (37). This enzyme catalyzes the cleavage of α-2,8-linked poly-N-acetylneuraminic acid carbohydrate polymer of the K1 capsule. It has been suggested that the tail fiber protein is involved in both adsorption to the cell surface and penetration into the cell by enzymatically degrading the polysaccharide capsule. The ΦK1E endosialidase gene has been cloned and sequenced (20). A similar gene has been cloned and sequenced from ΦK1F (29).
ΦK5 is a related bacteriophage specific for E. coli strains that display the K5 antigen, a polymer consisting of a repeating structure of 4-linked α-N-acetylglucosamine and β-glucuronic acid (N-acetyl heparosin). In this case, ΦK5 encodes a tail-associated K5 specific lyase protein that is also responsible for attachment to the cell surface and degradation of the K5 polysaccharide capsule (12, 14). Phage specific for other E. coli polysaccharide antigens, including K3, K7, K12, K13, and K20 (26, 27), have also been found; all probably possess specific polysaccharide depolymerization activities as part of the phage particle.
Both ΦK5 and ΦK1E have a Salmonella phage SP6-like promoter upstream of their tail proteins as well as a region of homology which is just downstream of the lyase gene of ΦK5 and just upstream of the endosialidase gene of ΦK1E (6). The sequences upstream of the tail gene promoters in ΦK1E and ΦK5 are highly similar as well. ΦK5, ΦK1E, and SP6 share a common morphology and life cycle, suggesting that they may be closely related.
ΦK1-5 is a morphologically similar phage that we recently isolated from the Montgomery County (Maryland) sewage treatment plant using a K5 strain of E. coli as a host. In this study, we analyzed the host range of ΦK1-5 and found that it can successfully infect and grow on either K1 or K5 strains. DNA sequence analysis of the tail fiber genes revealed that it encodes both a K5 lyase protein similar to that of ΦK5 and an endosialidase protein similar to that of ΦK1E. The arrangement of these genes suggests that phage host range can be broadened or changed in nature by the acquisition of new tail genes by recombination.
MATERIALS AND METHODS
Isolation of K1-5.
ΦK1-5 was isolated from raw sewage by the plaque technique. Briefly, a 1-liter sample of sewage was centrifuged at 6,000 rpm in a GSA rotor to remove solid matter and was then passed through a 0.45-μm-pore-size nitrocellulose filter (Nalgene); 100 μl of filtrate was added to 200 μl of an overnight culture of E. coli ATCC 23506 (K5) grown in Luria-Bertani (LB) medium. Then 3 ml of melted tempered top agar (5 g/liter in LB) was added, and the mix was plated onto an LB agar plate and incubated at 37°C overnight. The following day, plaques were picked and replaqued three times to ensure pure culture. Final plaque isolates were stored as an agar plug from a Pasteur pipette deposited in 1 ml of SM buffer (10 mM MgSO4, 100 mM NaCl, 0.01% gelatin, 50 mM Tris [pH 7.5]).
Host range was initially screened by spotting 10 μl of SM buffer containing a plaque plug onto a lawn of an appropriate strain. Host ranges of interesting phage isolates were further confirmed by the plaque assay. All phage titrations were done by the plaque assay technique.
Large-scale purification.
Phages were prepared by the cesium chloride density gradient method. One liter of an appropriate host was grown to an optical density at 600 nm of between 0.4 and 0.6 at 37°C with shaking at 200 rpm in LB broth. Phage were added at a multiplicity of infection (MOI) of 1 phage to 100 bacteria, and the culture was allowed to incubate until the optical density reached a minimum for 30 min. After 10 ml of chloroform was added, the mixture was allowed to shake for 10 min and then centrifuged for 20 min at 6,000 rpm in a GSA rotor to remove cellular debris. The supernatant was collected, and 1/10 volume of 5 M NaCl and 1/10 (wt/vol) of polyethylene glycol was added to precipitate the phage; this preparation was held at 4°C overnight. The phage were then pelleted by centrifugation at 6,000 rpm in a GSA rotor at 4°C. The pellet was resuspended in phosphate-buffered saline, and CsCl was added to a density of 1.5 g/ml. The sample was spun in Ti80 (Beckman) rotor at 34,000 rpm overnight. The phage band was extracted with a syringe and was dialyzed against phosphate-buffered saline (pH 7.4).
DNA isolation and sequencing.
DNA was isolated from CsCl-purified phage by phenol-chloroform extraction. The phage DNA was used directly as a template for DNA sequencing, which was carried out by Commonwealth Biotechnologies, Richmond, Va. Both strands were sequenced. DNA database searches were done by BLAST (1), and sequence alignments were performed with the Wisconsin Package (9).
Mutagenesis.
Cesium-purified phage were mutagenized with UV light using a model TM 36 chromatovue transilluminator (UVP, Inc.). Phage were typically exposed for 10 to 20 sec, which reduced viability 1,000-fold. The mutagenized phage were then amplified on ATCC 23503 or ATCC 23506 and subjected to selection and amplification as described in Results. Phage were also mutagenized by incubation with 400 mM hydroxylamine until the phage titer was reduced 100-fold. They were then plated on a double lawn of ATCC 23503 and ATCC 23506. Turbid plaques were picked, replaqued for isolation, and tested for growth against a collection of K1 and K5 E. coli strains.
Nucleotide sequence accession numbers.
GenBank accession numbers are as follows: ORFP of ΦK5, AF322018; tail gene region of ΦK1-5, AF322019; and region downstream of the endosialidase gene of ΦK1E, AF322020.
RESULTS
Isolation and characterization of ΦK1-5.
ΦK1-5 was isolated using E. coli ATCC 23506 (K5) as a host (see Materials and Methods). Electron micrographs show that ΦK1-5 is morphologically similar to members of the Podoviridae family, which includes coliphages T7 and T3, and Salmonella phages SP6 and P22. The phage particle consists of an icosohedral head about 60 nm in diameter with a small tuft of short tail fibers (Fig. 1). ΦK1-5 is highly lytic. When phage were added to a logarithmic culture of a susceptible host at an MOI of 1:1, lysis occurs in 15 to 20 min. Burst size was determined by a one-step growth curve and found to be to be about 110. ΦK1-5 plaques are clear and large, about 4.0 to 5.0 mm in diameter, with a halo of about 12.0 to 15.0 mm in diameter on LB agar plates. The plaques reached a limit in size after 24 h. In contrast, T7 plaques can continue to grow for several days (38). DNA was isolated from cesium chloride density gradient-purified phage by phenol extraction. Digestion of the DNA with several restriction enzymes indicated that is double stranded, with an estimated size of 40 kb.
FIG. 1.
Electron micrograph of ΦK1-5 negatively stained with phosphotungstic acid at a magnification of ×115,500. Morphologically this phage can be classified in the Podoviridae family which includes T7 and SP6.
Extended host range of ΦK1-5.
The host range of ΦK1-5 was compared to that of ΦK1E (K1 antigen specific) and ΦK5 (K5 antigen specific). E. coli strains ATCC 23506 and ATCC 23508 possess the K5 polysaccharide capsule, and strains ATCC 23503 and ATCC 23511 possess the K1 capsule. We also tested a set of K5 strains collected by Ian Roberts from the University of Manchester and a set of K1 isolates collected by Richard Silver from the University of Rochester (Table 1). ΦK1E, ΦK5, and ΦK1-5 also failed to grow on ATCC strains 23502 (K4), 23504 (K8), 23505 (K9), 23507 (K10), 23509 (K11), 23510 (K14), 23515 (K17), 23516 (K20), 23517 (K13), 23518 (K18), 19110 (K7), 19138 (K2), and 31616 (K35).
TABLE 1.
Host ranges of phages ΦK1E, ΦK5, ΦK1-5, ΦK1-5(K1−), and ΦK1-5(K5−)a
| E. coli strain | K antigen | ΦK1-5 | ΦK1E | ΦK5 | ΦK1-5(K1−) | ΦK1-5(K5−) |
|---|---|---|---|---|---|---|
| ATCC 23503 | K1 | + | + | − | − | + |
| ATCC 23511 | K1 | + | + | − | − | + |
| RS164 | K1 | + | + | − | − | + |
| RS166 | K1 | + | + | − | − | + |
| RS167 | K1 | + | + | − | − | + |
| RS168 | K1 | + | + | − | − | + |
| RS176 | K1 | + | + | − | − | + |
| RS179 | K1 | + | + | − | − | + |
| RS180 | K1 | + | + | − | − | + |
| RS188 | K1 | + | + | − | − | + |
| RS203 | K1 | + | + | − | − | + |
| RS215 | K1 | + | + | − | − | + |
| RS218 | K1 | + | + | − | − | + |
| RS228 | K1 | + | + | − | − | + |
| ATCC 23506 | K5 | + | − | + | + | − |
| ATCC 23508 | K5 | + | − | + | + | − |
| 20026 | K5 | + | − | + | + | − |
| 21195 | K5 | + | − | + | + | − |
| 21386 | K5 | + | − | + | + | − |
| 21786 | K5 | + | − | + | + | − |
| 21795 | K5 | + | − | + | + | − |
| 21831 | K5 | + | − | + | + | − |
| 21832 | K5 | + | − | + | + | − |
| 21834 | K5 | + | − | + | + | − |
| 21835 | K5 | + | − | + | + | − |
Plaque assays were done to determine host ranges of the phages against a collection of K1 and K5 strains of E. coli. ΦK1E grows only on K1 strains, ΦK5 grows only on ΦK5 strains, and ΦK1-5 grows on both. ΦK1-5(K1−) and ΦK1-5(K5−) are mutants of ΦK1-5 defective in growth on K1 and K5 strains, respectively.
Because of the promoter sequence similarity between ΦK1-5 and SP6, we tested if ΦK1-5 could grow on Salmonella serovar enterica Typhimurium strain LT2 (the host for SP6) and if SP6 could grow on any of the E. coli isolates sensitive to ΦK1-5. SP6 did not grow on any of the E. coli strains, and likewise ΦK1-5 did not grow on Salmonella serovar Typhimurium.
ΦK1-5 encodes two tail genes.
ΦK1E and ΦK5 share a region of sequence similarity upstream of the tail proteins (including the SP6-like promoter [3]). Since ΦK1-5 had structural, biological, and host similarities to these two phages, we speculated that all three may be closely related and share this upstream sequence similarity. We designed a primer based on the sequence of this region in ΦK1E and ΦK5 to determine the sequence downstream of the promoter. When ΦK1-5 DNA was used as a template, the primer did hybridize, and we were able to generate sequence. We continued sequencing downstream by primer walking. The sequence immediately downstream of the promoter was very similar to that of ΦK5 and encoded an open reading frame with a high degree of similarity (>92% amino acid identity) to that of ΦK5 tail protein. Continued sequencing downstream revealed a second open reading frame that is nearly identical (>97% amino acid identity) to the endosialidase protein of ΦK1E. A region of 85 bp lies between the termination codon of the lyase gene and the start codon of the endosialidase gene. This region is also present in ΦK5, immediately following the K5 lyase gene, and also in ΦK1E, immediately upstream of the endosialidase gene and immediately downstream of a 111-amino-acid open reading frame (ORFL [6]). No recognizable promoter was found in this region, but there are two strong regions of symmetry, which may act as a Rho-independent transcriptional terminator. Sequence was determined 598 bp downstream of the termination codon of the endosialidase gene, at which point the end of the DNA molecule was reached. No open reading frames were found in this area.
The sequence 500 bp upstream of the K5 lyase gene in ΦK1-5 was also determined. Like in the other phages, an SP6-like promoter is present and is probably required for transcription of the tail genes. The upstream sequence shares a high degree (>90%) of identity to that of the analogous region in ΦK5 and ΦK1E.
We also sequenced downstream of the endosialidase gene of ΦK1E; 718 bp downstream from the endosialidase termination codon we reached the end of the DNA molecule. There is little sequence similarity between this region and the analogous region in ΦK1-5.
Each ΦK1-5 virion contains both tail proteins.
We addressed the question of whether ΦK1-5 particles contain both tail fiber proteins, or if two populations of particles (one containing the K5 lyase and the other containing the endosialidase) were produced after infection. We made a phage preparation using ATCC 23506 (K5) as a host and determined its titer on ATCC 23506 (K5) and ATCC 23503 (K1) (Table 2). A sample of the phage was then incubated with ATCC 23506 for 5 min, which is long enough for phage to attach and possibly inject the DNA but not long enough for production of new phage particles. The MOI was 1 phage particle to 100. The mixture was then rapidly filtered. Phage particles that had attached to the cells would be eliminated from the filtrate. The filtrate was then titered on both K1 and K5 strains. If the phage preparation was initially a mixture of two populations, then only those displaying the K5 lyase would attach and be eliminated. The remaining phage would be mainly those that contained the K1-specific endosialidase, and therefore the titer would be higher on the K1 E. coli strains than on the K5 strain. On the other hand, if each of the phage particles contained both tail proteins, titers of the phage remaining in the filtrate would be the same on the two strains; i.e., levels of the K5 lyase-containing phages would not be selectively reduced. We found the latter to be the case and concluded that each virion has both the K1 endosialidase and the K5 lyase. Similar results were seen in the converse experiment in which the 5-min incubation was performed with the K1 E. coli strain (Table 2). As controls we performed the experiments with both ΦK1E and ΦK5, using both strains for the incubation. ΦK1E titers were reduced 99% by preincubation with the K1 strain but not with the K5 strain, and ΦK5 titers were similarly reduced after preincubation with the K5 strain but not with the K1 strain.
TABLE 2.
Preincubation experiments to show that all ΦK1-5 particles contain both tail proteinsa
| Phage | Titer |
|---|---|
| ΦK1-5 | |
| Preincubated with 23506 | 5.2 × 108 |
| Titered on 23506 after preincubation | 3.1 × 106 |
| Titered on 23503 after preincubation | 3.7 × 106 |
| Preincubated with 23503 | 5.2 × 108 |
| Titered on 23506 after preincubation | 6.8 × 106 |
| Titered on 23503 after preincubation | 6.4 × 106 |
| ΦK5 | |
| Preincubated with 23506 | 4.0 × 108 |
| Titered on 23506 after preincubation | 8.5 × 105 |
| Preincubated with 23503 | 4.0 × 108 |
| Titered on 23506 after preincubation | 3.3 × 108 |
| ΦK1E | |
| Preincubated with 23506 | 7.7 × 108 |
| Titered on 23503 after preincubation | 6.8 × 108 |
| Preincubated with 23503 | 7.7 × 108 |
| Titered on 23503 after preincubation | 3.0 × 106 |
Preincubation of ΦK1-5 with either ATCC 23503 (K1) or ATCC 23506 (K5) for 5 min results in roughly 100-fold loss of phage particles when titered on either strain. If the phage preparation consisted of two populations that each contained just one tail protein, then titers after the incubation would be reduced only on the strain that was used in the preincubation. ΦK5 titers are reduced by preincubation with a permissive host (23506) but not with a nonpermissive host (23503). ΦK1E titers are also reduced only by preincubation on a permissive host (23503) and not a nonpermissive host (23506).
ΦK1-5 mutants defective in growth on either K1 or K5 E. coli.
A mixed lawn of K1 and K5 strains of E. coli was used to screen for ΦK1-5 mutants defective in growth on one or the other host strains. ΦK1-5 forms clear plaques on a mixed lawn of K1 and K5 E. coli; mutants in either tail would result in turbid plaques due to growth of the nonpermissive host. Phage were treated with the mutagen hydroxylamine and plated on a double lawn. Turbid plaques were identified, picked, and purified by multiple plaque isolations on the double lawn. These were then screened by separately testing for growth on each strain. Of eight isolates purified, three were unable to plaque on the K5 strain but could plaque on the K1 strain. One of these, ΦK1-5(K5−), was screened for growth against the entire host collection and found to be unable to replicate on any of the K5 strains but able to grow on all of the K1 strains (Table 1). Five of the isolates could still replicate on both K1 and K5 strains but gave a turbid plaque morphology on the K5 strains.
None of the mutants isolated in this way were defective in growth on K1 strains, so we devised a selection/amplification scheme to enrich for those that can replicate on K5 but not K1 hosts. Mutagenized phage were amplified on a K5 strain, filtered to remove bacterial debris, and then used to infect a logarithmically growing K1 strain for 5 min. This mixture was rapidly filtered before phage burst could occur. Phage able to grow on the K1 strain would attach to the cells and be eliminated from the filtrate. We then reamplified the sample on the K5 strain and repeated the cycle eight times. This strongly selects for phage that can replicate on K5 hosts but not K1 hosts. Titers of the filtrate were 200-fold higher on the K5 strain than on the K1 strain. Several were picked and purified by multiple rounds of single-plaque isolation. One isolate, ΦK1-5(K1−), was further characterized and found to be unable to grow on any of the K1 strains (Table 1).
DNA sequence of a putative ΦK5 tail gene.
Clarke et al. described a partial sequence of an open reading frame (ORFP) in ΦK5 immediately downstream of the 85-base region common to the three phages (6). We continued sequencing downstream and found that the complete open reading frame is 523 amino acids. A BLAST search revealed a small region of sequence similarity with the N-acetylglucosamine-permease IIABC component near the N terminus. It has no significant sequence similarity with any other entry in the database or any of the tail proteins described here. Sequence was determined an additional 163 bases downstream, at which point the end of the DNA molecule was reached.
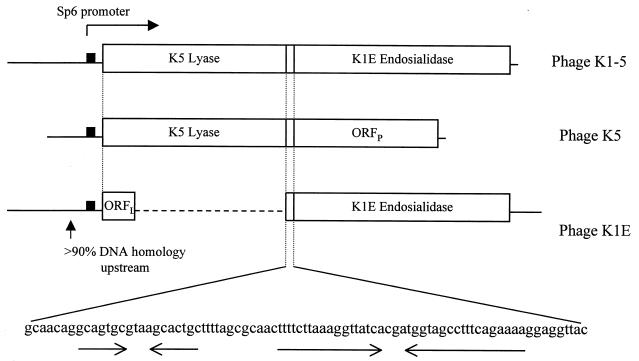
Figure 2 compares the regions encoding tail proteins in all three phages. ΦK1-5 has a K5 lyase protein in the same position as that of ΦK5. ΦK1E has a 111-amino-acid open reading frame (ORFL) of unknown function in this position. Immediately downstream, all three phages have an intergenic region of 85 bases that has two dyad axis of symmetry. Immediately downstream of this region ΦK1-5 encodes its endosialidase protein, which is in the analogous position as the ΦK1E endosialidase. ΦK5 encodes a 523-amino-acid open reading frame (ORFP) in this position. The three phages share sequence similarity upstream of the tail genes. No sequence similarity was noted downstream, and in all three phages the DNA molecule ends downstream.
FIG. 2.
Comparison of the coding regions of the tail proteins of ΦK1-5, ΦK5, and ΦK1E. All three phages share sequence similarity in the upstream region (which contains an SP6 promoter) as well as an 85-base intergenic region. Just downstream of the promoter, ΦK1-5 and ΦK5 encode a lyase protein and ΦK1E encodes ORFL. Immediately following the termination codons of the lyases or ORFL is the intergenic region that contains a potential hairpin structure, the first of which could be a Rho-independent transcription terminator. Immediately following this, ΦK1-5 and ΦK1E encode an endosialidase where ΦK5 encodes ORFP. None of the three phages have any coding regions downstream, and the DNA molecule ends in all three cases. No homology exists in this terminal region.
DISCUSSION
We isolated a bacteriophage, ΦK1-5, that is able to infect and grow on either K1 or K5 strains of E. coli. It appears that its ability to replicate on these strains is due to the fact that it encodes two different hydrolytic tail fiber proteins. One is an endosialidase protein, almost identical to a similar protein from ΦK1E, that allows it to attach to and degrade the K1 polysaccharide capsule. The other is almost identical to a lyase protein that has been shown to allow ΦK5 attach to and degrade the K5 polysaccharide capsule. This is the first example of a phage that has a dual host specificity based on having two different tail fiber proteins. All three of these phages share sequence similarity upstream of the region encoding the tail proteins, and all have an SP6-like promoter that probably drives transcription of the tail gene(s). In ΦK1-5 and ΦK5, the first gene downstream of this promoter is the K5 lyase protein. ΦK1E does not encode this protein and instead has a 111-amino-acid open reading frame (ORFL) of unknown function. Immediately downstream of the K5 lyase proteins of ΦK1-5 and ΦK5, and downstream of ORFL in ΦK1E is an 85-base region of similarity between all three phages. This region contains two strong symmetrical elements that may be involved in transcription termination. Further downstream, phages ΦK1-5 and ΦK1E encode the endosialidase gene. ΦK5 does not encode this gene but instead encodes the 523-amino-acid ORFP. The morphologies and life cycles of these three phages are all very similar, and they all have similar SP6-like promoter sequences (and therefore probably SP6-like RNA polymerases). We believe that these phages are all very closely related and differ mainly in the tail fiber proteins.
Phages that are specific for K antigens are believed to have the hydrolytic activity associated with the tail proteins, which are part of the mature virion. This has been directly shown for ΦK1F by immunoelectron microscopy (29). In the case of ΦK1-5, we have experimental evidence that both the endosialidase and the lyase are part of each phage particle, and we have shown that the ability to grow on either of the host types can be deleted by mutagenesis. This raises the question of how the tail proteins are attached to the capsid. The ΦK1F endosialidase protein has an N-terminal region that is not part of the ΦK1E protein. This N-terminal region has some sequence similarity to the N terminus of the T7 tail protein, which is thought to be involved in attachment (29). Since neither the endosialidase nor the K5 lyase has this region, or any other region similar to any other tail protein (or with each other), it is impossible to predict what part of the protein serves this function. Morphologically, ΦK1E, ΦK5, and ΦK1-5 are similar to Salmonella phage P22. The tail protein of P22 has been extensively studied and is also a hydrolytic protein involved in degradation of the Salmonella serovar Typhimurium O antigen. This protein is a homotrimer with six copies per phage (30). The gp17 tail fiber of T7 is also a trimer with six copies of the trimer per phage particle (33). The endosialidase of ΦK1E is also a trimer (20), but it has yet to be shown that there are six copies of the trimer per phage particle. Bacteriophage 63D is another newly characterized sialidase-containing phage in which it has has been shown by electron microscopy that the sialidase is present with six copies per particle (21). This phage is quite different morphologically from ΦK1E, ΦK5, and ΦK1-5 and has a long tail similar to that of bacteriophage lambda, with the sialidase located at the end of the tail. Six copies of a trimeric tail protein appears to be a general structural motif. Assuming that the endosialidase and K5 lyase are also arranged in six copies per virion, it is interesting to speculate how the two tail proteins are arranged on the head structure of ΦK1E. They may be arranged in an alternating fashion where there are three copies of each (Fig. 3a). In the case of P22, there is evidence that only three copies of the tail are needed for infection (16), suggesting that this model is theoretically possible. The fact that there are no sequence similarities between the two tail proteins argues against this model, since one might predict a common motif within the tail proteins that is required to attach to similar regions of the head structure. An alternative model is that there may be six copies of each tail protein, one attached to the other (Fig. 3b). It seems that phage mutants unable to grow on K1 strains are rarer that those defective in growth on K5. It is possible that the endosialidase plays a more important structural role than the K5 lyase, supporting the second model.
FIG. 3.
Two possible models for the arrangement of the tails proteins on the phage capsid. (a) There are three copies of each tail forming a hexamer. (b) There are six copies of each tail. One is attached to the head and is part of the core of the tail; the other is then attached to the first tail protein, in effect making a longer tail fiber with two different enzymatic activities.
ΦK1E encodes a 111-amino-acid ORFL located in the analogous position as the K5 lyase gene in ΦK1-5. The function of this ORF is unknown, but the first nine amino acids are identical to those of the K5 lyase protein. Clarke et al. pointed this out and speculated that ΦK5 could be the progenitor ΦK1E, in which ΦK5 acquired the endosialidase gene by a horizontal event and then lost the lyase gene (6). In that scenario, ΦK1-5 would be the intermediate. However, it is difficult to predict the order of events that may have occured in this two-gene system. For instance, ΦK5 could have arisen from ΦK1-5 by a replacement of the endosialidase with ORFP, making ΦK1-5 the progenitor of both ΦK5 and ΦK1E.
ΦK5 is able to also grow on K95 strains of E. coli (28). Since ORFP is in a position analogous to that of the endosialidase of ΦK1-5, we speculate that it may also be a tail protein and could be responsible for growth on K95 strains. Future work will be needed to determine if this is in fact a tail fiber protein. Another K antigen-specific phage, ΦK20, is also able to lyse two different types of E. coli hosts, those that possess the K5 antigen and those that possess the K20 polysaccharide (26). It is quite possible that ΦK20 carries a K5 lyase protein similar to the ΦK5/ΦK1-5 protein along with a yet unidentified K20-specific hydrolytic tail fiber protein. Phages that are specific to each of the capsular antigens K3, K7, K12, and K13 of E. coli have also been isolated (27). Presumably these phages have corresponding K-specific hydrolytic tail proteins. It may be possible to find phages that have double specificities with other combinations of K antigens, and we may find that the occurrence of multispecificity phages is common.
The theory of modular evolution of phages is well established (2, 34). The basic idea is that phage genomes have evolved by interchanging different modules that may consist of single genes or clusters of related genes. Recent evidence for modular evolution has been shown for the Salmonella phages P22, ES18, and L (31), for the T-even/pseudo T-even phages (22), for Streptococcus thermophilus phages (8, 25), for Lactococcus phage sk1 (5), and for E. coli lambdoid phages (4, 15, 19). Evidence that tail proteins have evolved by horizontal transfer also exists. Phages P1, P2, and Mu (phages that are unrelated but have similar host ranges) exhibit sequence similarities at the carboxyl ends of the tail fiber proteins (13). It is this portion of the protein that is responsible for host recognition; the amino-terminal portions of the genes are not similar presumably because they are required for attachment to different structures on the phage particles. The T-even family of phages also show a high level of variability in the tail fiber adhesion genes (gene 37 of T4), allowing for changes in host range (24, 35, 36). In this case, duplications and rearrangements of regions within the tail fiber genes as well as possible recombination between tail fiber genes of different phages seem to mediate changes in host specificity. Another study showed that it quite likely that a T4 ancestor may have picked up the tail fiber assembly protein and the side tail fiber from a lambdoid phage (23). It has also been shown that partially fibered T4 particles can be complemented by addition of tail T6 fibers (7).
We have evidence that ΦK1-5, ΦK5, and ΦK1E can rapidly extend or change host specificity by acquiring a second completely different tail protein or perhaps replace an existing protein with another. It appears that little if any sequence changes within the tail proteins are required for function on the new phage, and it appears that the functions of the tail proteins are independent. The mechanisms for exchange can be explained simply by recombination and or deletion. All of these phages have a common sequence upstream of the tail proteins and a common sequence between the two tail proteins. While the latter sequence is only 85 bases, it still may be enough for homologous recombination to occur. The 85-base intergenic region contains a putative Rho-independent terminator. It has been shown that such sequences can effectively terminate transcription by SP6 and T3 RNA polymerases (18). Juhala et al. describe potential mobile genetic element “morons” that appear to be common in lambdoid phages (19). Moron genes are characteristically flanked by a sigma 70 promoter and Rho-independent terminator and thought to be expressed from the prophage. The K5 lyase gene and OrfL are flanked by an SP6 promoter and a Rho-independent terminator. Although these phages are not known to enter a prophage state and it appears that the genes are transcribed by the phage promoter, the modular structure is interestingly similar to that of morons.
Double host specificity clearly has an evolutionary advantage in an environment with a mixture of bacteria types. However, if a phage finds itself among a single population of bacteria, there would be a growth advantage to having one tail and therefore a smaller genome. It is quite possible that ΦK1E arose in this way, the ancestor being a ΦK1-5-like phage that lost a tail by deletion. It is also interesting to speculate how these phages acquired hydrolytic tail proteins. Recombination with other multiple-tailed phages is very likely. If two phages, each with two tail genes one of which is common between them infect the same cell, a third completely different phage possessing the two other host specific tail proteins could arise. Enzymatic tail proteins may also arise when a phage acquires a similar cellular gene.
ACKNOWLEDGMENTS
We thank Ian Roberts and Richard Silver for kindly supplying strains and phages and Kunio Nagashima for assistance with electron microscopy.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Botstein D. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci. 1980;354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown J E, Klement J F, McAllister W T. Sequences of three promoters for the bacteriophage SP6 RNA polymerase. Nucleic Acids Res. 1986;14:3521–3526. doi: 10.1093/nar/14.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell A, Botstein D. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 365–380. [Google Scholar]
- 5.Chandry P S, Moore S C, Boyce J D, Davidson B E, Hillier A J. Analysis of the DNA sequence, gene expression, origin of replication, and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. doi: 10.1046/j.1365-2958.1997.5491926.x. [DOI] [PubMed] [Google Scholar]
- 6.Clarke B R, Esumah F, Roberts I S. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J Bacteriol. 2000;182:3761–3766. doi: 10.1128/jb.182.13.3761-3766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford J T, Goldberg E B. The function of the tail fibers in triggering baseplate expansion of bacteriophage T4. J Mol Biol. 1980;139:679–690. doi: 10.1016/0022-2836(80)90054-6. [DOI] [PubMed] [Google Scholar]
- 8.Desiere F, Lucchini S, Brussow H. Evolution of Streptococcus thermophilus bacteriophage genomes by modular exchanges followed by point mutations and small deletions and insertions. Virology. 1998;241:345–356. doi: 10.1006/viro.1997.8959. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross R J, Cheasty T, Rowe B. Isolation of bacteriophages specific for the K1 polysaccharide antigen of E. coli. J Clin Microbiol. 1977;6:548–550. doi: 10.1128/jcm.6.6.548-550.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta D S, Jann B, Schmidt G, Golecki J R, Ørskov I, Ørskov F, Jann K. Coliphage K5, specific for E. coli exhibiting the capsular K5 antigen. FEMS Microbiol Lett. 1982;14:75–78. [Google Scholar]
- 12.Gupta D S, Jann B, Jann K. Enzymatic degradation of the capsular K5-antigen of E. coli by coliphage K5. FEMS Microbiol Lett. 1983;16:13–17. [Google Scholar]
- 13.Haggaård-Ljungquist E, Halling C, Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of the tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992;174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanfling P, Shashkov A S, Jann B, Jann K. Analysis of the enzymatic cleavage (beta elimination) of the capsular K5 polysaccharide of Escherichia coli by the K5-specific coliphage: a reexamination. J Bacteriol. 1996;178:4747–4750. doi: 10.1128/jb.178.15.4747-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrix R W, Smith M C M, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Israel V. A model for the adsorption of phage P22 to Salmonella typhimurium. J Gen Virol. 1978;40:669–673. doi: 10.1099/0022-1317-40-3-669. [DOI] [PubMed] [Google Scholar]
- 17.Jann K, Jann B. Polysacharide antigens of E. coli. Rev Infect Dis. 1987;9:S517–S526. doi: 10.1093/clinids/9.supplement_5.s517. [DOI] [PubMed] [Google Scholar]
- 18.Jeng S T, Lay S H, Lai H M. Transcription termination by bacteriophage T3 and SP6 RNA polymerases at Rho-independent terminators. Can J Microbiol. 1997;43:1147–1156. doi: 10.1139/m97-163. [DOI] [PubMed] [Google Scholar]
- 19.Juhala R J, Ford M E, Duda R L, Youlton A, Hatfull G F, Hendrix R W. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 20.Long G S, Bryant J M, Taylor P W, Luzio J P. Complete nucleotide sequence of the gene encoding bacteriophage E endosialidase: implications for K1E endosialidase structure and function. Biochem J. 1995;309:543–550. doi: 10.1042/bj3090543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machida Y, Miyake K, Hattori K, Yamamoto S, Kawase M, Iijima S. Structure and function of a novel coliphage-associated sialidase. FEMS Microbiol Lett. 2000;182:333–337. doi: 10.1111/j.1574-6968.2000.tb08917.x. [DOI] [PubMed] [Google Scholar]
- 22.Monod C, Repoila M, Kutateladze F, Tetart F, Krisch H M. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J Mol Biol. 1997;267:237–249. doi: 10.1006/jmbi.1996.0867. [DOI] [PubMed] [Google Scholar]
- 23.Montag D, Schwarz H, Henning U. A component of the side tail fiber of Escherichia coli bacteriophage λ can functionally replace the receptor-recognizing part of a long tail fiber protein of unrelated bacteriophage T4. J Bacteriol. 1989;171:4378–4384. doi: 10.1128/jb.171.8.4378-4384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montag D, Hashemolhosseini S, Henning U. Receptor recognizing proteins of T-even type bacteriophages. The receptor recognizing area of proteins 37 of phages T4 and TuIa and TuIb. J Mol Biol. 1990;216:327–334. doi: 10.1016/S0022-2836(05)80324-9. [DOI] [PubMed] [Google Scholar]
- 25.Neve H, Zenz K I, Desiere F, Koch A, Heller K J, Brussow H. Comparison of the lysogeny modules from the temperate Streptococcus thermophilus bacteriophages TP-J34 and Sfi21: implications for the modular theory of phage evolution. Virology. 1998;241:61–72. doi: 10.1006/viro.1997.8960. [DOI] [PubMed] [Google Scholar]
- 26.Nimmich W, Schmidt G, Krallmann-Wenzel U. Two different E. coli capsular polysaccharide depolymerases each associated with one of the coliphage ΦK5 and ΦK20. FEMS Microbiol Lett. 1991;82:137–142. doi: 10.1016/0378-1097(91)90322-2. [DOI] [PubMed] [Google Scholar]
- 27.Nimmich W, Krallman-Wenzel U, Muller B, Schmidt G. Isolation and characterization of bacteriophages specific for capsular antigens K3, K7, K12, and K13 of E. coli. Int J Med Microbiol Virol Parasitol Infect Dis. 1992;276:213–220. doi: 10.1016/s0934-8840(11)80008-3. [DOI] [PubMed] [Google Scholar]
- 28.Nimmich W. Detection of E. coli K95 strains by bacteriophages. J Clin Microbiol. 1994;32:2843–2845. doi: 10.1128/jcm.32.11.2843-2845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petter J G, Vimr E R. Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J Bacteriol. 1993;175:4354–4363. doi: 10.1128/jb.175.14.4354-4363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seckler R. Folding and function of repetitive structure in the homotrimeric phage P22 tailspike protein. J Struct Biol. 1998;122:216–222. doi: 10.1006/jsbi.1998.3974. [DOI] [PubMed] [Google Scholar]
- 31.Schicklmaier P, Schmeiger H. Sequence comparison of the genes for immunity, DNA replication, and cell lysis of the P22-related Salmonella phages ES18 and L. Gene. 1997;195:93–100. doi: 10.1016/s0378-1119(97)00182-0. [DOI] [PubMed] [Google Scholar]
- 32.Silver R P, Vimr E R. The bacteria. 11. Molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press, Inc.; 1990. Polysialic acid capsule of E. coli K1; pp. 39–60. [Google Scholar]
- 33.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S, Hainfield J F, Studier W F. Molecular substructure of a viral receptor-recognition protein. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 34.Szybalski W, Szybalski E H. Viruses, evolution and cancer. New York, N.Y: Academic Press; 1974. pp. 563–582. [Google Scholar]
- 35.Tetart F, Repoila F, Monod C, Krisch H M. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesion. J Mol Biol. 1996;258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- 36.Tetart F, Desplats C, Krisch H M. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesion specificity. J Mol Biol. 1998;282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- 37.Tomlinson S, Taylor P W. Neuraminidase associated with coliphage E that specifically depolymerizes the Escherichia coli K1 capsular polysaccharide. J Virol. 1985;55:374–378. doi: 10.1128/jvi.55.2.374-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin J. Evolution of bacteriophage T7 in a growing plaque. J Bacteriol. 1993;175:1272–1277. doi: 10.1128/jb.175.5.1272-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]