Abstract
The carboxy terminus-encoding portion of the gag gene of Mason-Pfizer monkey virus (M-PMV), the prototype immunosuppressive primate type D retrovirus, encodes a 36-amino-acid, proline-rich protein domain that, in the mature virion, becomes the p4 capsid protein. The p4 domain has no known role in M-PMV replication. We found that two mutants with premature termination codons that remove half or all of the p4 domain produced lower levels of stable Gag protein and of self-assembled capsids. Interestingly, yeast two-hybrid screening revealed that p4 specifically interacted with TCP-1γ, a subunit of the chaperonin TRiC (TCP-1 ring complex). TRiC is a cytosolic chaperonin that is known to be involved in both folding and subunit assembly of a variety of cellular proteins. TCP-1γ also associated with high specificity with the M-PMV pp24/16-p12 domain and human immunodeficiency virus p6. Moreover, in cells, Gag polyprotein associated with the TRiC chaperonin complex and this association depended on ATP hydrolysis. In the p4 truncation mutants, the Gag-TRiC association was significantly reduced. These results strongly suggest that cytosolic chaperonin TRiC is involved in Gag folding and/or capsid assembly. We propose that TRiC associates transiently with nascent M-PMV Gag molecules to assist in their folding. Consequently, properly folded Gag molecules carry out the intermolecular interactions involved in self-assembly of the immature capsid.
The infectious virus particle of the Mason-Pfizer monkey virus (M-PMV) contains at least six capsid proteins: p10 (MA; matrix), pp24/16, p12, p27 (CA; capsid), p14 (NC; nucleocapsid), and p4 (3, 46). As with other retroviruses, these capsid proteins are produced by proteolytic cleavage, during or shortly after budding, of the gag gene-encoded precursor polyprotein (Gag polyprotein). Gag polyproteins are synthesized in M-PMV-infected cells along with two other Gag-related polyproteins (Gag-Pro and Gag-Pro-Pol, encoded by the gag-pro gene and the gag-pro-pol gene, respectively). The three Gag-containing polyproteins are then assembled within the cytoplasm into an immature capsid and transported to the plasma membrane, where budding occurs. In the past two decades, extensive molecular studies of M-PMV and other retroviruses have examined the biological roles of the capsid proteins during retroviral infection. In addition to their roles as processed components of mature virions, capsid proteins are critical as constituents of the Gag precursor for the multiple events of protein folding, transport, and assembly in the final stages of retrovirus replication (reviewed in references 12 and 44).
The capsid proteins of MA, CA, and NC, although they show very little conservation in amino acid sequences among different retroviruses, are located in the same relative positions on the Gag precursor and have some shared functions (50). However, the carboxy-terminal domain is highly diversified. In Rous sarcoma virus (RSV), the virus protease is found at the carboxy terminus of the Gag polyprotein (2), whereas in murine leukemia virus no additional protein is encoded 3′ of the NC coding sequence (4). By contrast, this region of M-PMV yields a small protein, p4. The p4 protein is composed of 36 amino acids, of which approximately 22% are proline (46). Interestingly, a small, proline-rich protein, p6, is also found at the equivalent position in the Gag polyprotein of human immunodeficiency virus type 1 (HIV-1). Mutagenic studies on this 6-kDa protein have suggested that p6 is involved in efficient virus release (14) and in direct interaction with regulatory protein Vpr for virion incorporation (1). Furthermore, Parent et al. showed that, during the late stages of budding, HIV p6 could functionally replace RSV p2b, a PPPY motif-containing protein of Gag (33). Because there is no primary sequence homology between these two proteins, it was speculated that a host factor(s) might be recruited in a sequence-independent manner through the proline-rich domain of these proteins to mediate retroviral budding. In contrast to HIV p6, M-PMV p4 has no function identified as yet.
To understand the biological roles of p4 in M-PMV replication, we made two p4 truncation mutants, Mp4L17 and Mp4G1, which have a carboxy-terminal 20-amino-acid deletion and a complete deletion of p4, respectively. We found that the carboxy-terminal proline-rich domain of M-PMV Gag appears to play a role in both stabilizing the molecule and facilitating capsid assembly. Furthermore, yeast two-hybrid screening revealed that this domain interacts with TCP-1γ, a subunit of TRiC. TRiC is a chaperonin that is involved in the folding of numerous cellular proteins including actins and tubulins (9, 13, 25, 47, 51). TRiC also participates in the assembly of a functional complex of the von Hippel-Lindau (VHL) tumor suppressor protein with its partner proteins (10). Thus, our findings suggest that the TRiC chaperonin complex assists nascent M-PMV Gag molecules to fold into a stable structure, thereby allowing the intermolecular interactions of capsid assembly to occur.
MATERIALS AND METHODS
DNAs.
Two M-PMV mutants, Mp4G1 and Mp4L17, with premature termination codons within the p4 coding region were generated by oligonucleotide-directed mutagenesis on single-stranded M13.SBGAG DNA, which contains the 3′ half of the gag gene, as previously described (54). After mutagenesis, the mutated fragments were recloned into M-PMV expression vector pSHRM15 (39) to replace the wild-type fragment. The presence of the mutations was confirmed by dideoxy sequencing of the double-stranded DNA (45).
To construct bait plasmids for the yeast two-hybrid screen, the entire coding sequence for each domain of various retroviral Gag polyproteins used in this report was amplified from an infectious proviral genome by PCR and then was inserted into LexA DNA binding domain plasmid pEG202 (Origin Technologies, Inc.). The prey plasmids were constructed by cloning HeLa cDNAs (kindly provided by R. Finley, Wayne State University School of Medicine) into hemagglutinin epitope-tagged B42 transcription activation domain plasmid pJG4-5.
To generate glutathione S-transferase (GST) fusion constructs for in vitro binding assays, the DNA fragments encoding various M-PMV Gag domains were amplified by PCR and ligated in frame into the pGEX-5X-1 vector (Promega). The full-length cDNA of the human TCP-1γ coding region was generated from a HeLa cDNA library by PCR and then was cloned into prokaryotic expression vector pET21a (Novagen), to express T7 epitope-tagged TCP-1γ in bacteria. It was also engineered into Myc epitope-tagging vector pcDNA3.1/myc (Invitrogen) for expression in mammalian cells. All constructs were verified by DNA sequencing.
Radiolabeling and immunoprecipitation of virus proteins.
COS-1 cells were transiently transfected with either wild-type or mutant proviral DNAs of M-PMV (5 μg/35-mm-diameter plate) by a modified calcium phosphate precipitation method (5). At 48 h after transfection, cells were pulse-labeled for 20 min with [3H]leucine (0.8 mCi/ml, 157 Ci/mmol; DuPont Co.) and chased for various periods in complete growth medium (35). Then, cells were lysed in lysis buffer A (50 mM Tris [pH 7.5], 1% Triton X-100, 1% sodium deoxycholate, 0.15 M NaCl), and cell-associated viral proteins were immunoprecipitated with rabbit anti-p27 CA antiserum. Radiolabeled, extracellular virus particles were pelleted from the culture medium of the pulse-chase-labeled cells by centrifugation for 15 min at 80,000 rpm in a Beckman TLA 100 rotor at 4°C. The virus pellet was suspended in lysis buffer A supplemented with 0.1% sodium dodecyl sulfate (SDS). Virion-associated viral proteins were then immunoprecipitated with goat anti-M-PMV antiserum (Division of Cancer Cause and Prevention, National Cancer Institute). The immunoprecipitated viral proteins were separated with a 10% resolving gel by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Fractionation of Gag polyproteins.
Gag polyproteins were fractionated into free and capsid-associated forms as previously described (39). HOS (human osteosarcoma cells; ATCC CRL-1543) cell lines containing integrated wild-type or mutant proviral DNAs of M-PMV were established by transfection with proviral DNAs linearized with Fsp 1 and subsequent selection in medium containing 350 μg of hygromycin B (GIBCO BRL)/ml as described previously (37). Cells were washed twice with TNE buffer (10 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 1 mM EDTA) and lysed with Triton X-100 lysis buffer (0.25 M sucrose, 1% Triton X-100, 10 mM Tris-HCl [pH 7.5], 0.14 M NaCl, 1 mM EDTA, 10 μg of DNase I/ml) for 1 h at room temperature. After removal of nuclei from the lysates by centrifugation for 5 min in a microcentrifuge at 4°C, capsids were pelleted through a 20% sucrose cushion by centrifugation at 80,000 rpm for 15 min in a Beckman TLA 120.2 rotor at 4°C. Viral proteins in supernatant and pellet fractions were separately immunoprecipitated with rabbit anti-Gag antiserum, separated by SDS-PAGE, and detected by Western blot assay with rabbit anti-Gag antibodies followed by peroxidase-conjugated goat anti-rabbit antibodies using an enhanced chemiluminescence detection system (ECL; Amersham). Quantitation of bands was performed using the Gel-document system (Bio-Rad).
Yeast two-hybrid screening and in vitro binding assay.
To identify the host cellular proteins that interact with M-PMV p4, a LexA-based, two-hybrid screening assay was carried out according to the manufacturer's instructions (Origin Technologies, Inc.). Briefly, bait plasmids containing coding sequences of the p4 domain were cotransformed with a LacZ reporter plasmid (pSH18-34) in the EGY48 strain (auxotrophic to histidine, leucine, tryptophan, and uracil). The yeast was then transformed on a large scale with prey plasmids of a HeLa cDNA library. Double transformants were obtained by plating on minimal selective medium lacking histidine, uracil, and tryptophan and further selected by growth on medium lacking leucine. The positive colonies were cultured in yeast extract-peptone-dextrose rich medium, and library plasmids were rescued and partially sequenced.
The in vitro binding assay was performed to determine direct interactions between an M-PMV Gag domain and a TCP-1γ subunit protein as described previously with some modifications (41). In brief, GST or fusions of GST proteins with various M-PMV Gag domains in DH5α or BL21 (DE3) cells were induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 3 h and subsequently purified by binding to glutathione-Sepharose (Amersham Pharmacia Biotech Inc.). The Sepharose beads were then incubated with 0.3 μg of unfractionated whole lysates of BL21 cells expressing a T7-tagged TCP-1γ protein/ml for 1 h at 4°C in binding buffer (25 mM HEPES [pH 7.4], 25 mM NaCl, 2.5 mM CaCl2, 1 mM MgCl2, 0.1% Triton X-100, 0.1% bovine serum albumin [BSA], 1 mM phenylmethylsulfonyl fluoride, and 1 μg of aprotinin, 1 μg of leupeptin, and 0.1 μg of pepstatin A/ml). After extensive washing in the same binding buffer lacking BSA, beads were resuspended in SDS sample buffer and boiled for 5 min, and the bound proteins were resolved by 10%–SDS PAGE. After electrophoresis, proteins were transferred to nitrocellulose and incubated with alkaline phosphatase-conjugated T7 antibody (Novagen). The presence of the expected GST or GST fusion proteins was checked by Coomassie brilliant blue staining.
Cell fractionation and coimmunoprecipitation of TCP-1γ with M-PMV Gag polyprotein.
M-PMV Gag polyprotein associated with chaperonin TRiC was detected by a combined experiment of cell fractionation and coimmunoprecipitation. 293T cells were transiently cotransfected with a proviral M-PMV DNA and a Myc-tagged TCP-1γ-expressing plasmid by using DMRIE-C reagent (GIBCO BRL). At 60 h after transfection, the cells were washed once with cold phosphate-buffered saline (pH 7.4) and lysed on ice for 10 min in 0.5% NP buffer (0.5% Nonidet P-40, 20 mM Tris [pH 7.4], 0.15 M NaCl, 5 mM EDTA, 1mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 1 μg of aprotinin, 1 μg of leupeptin, and 0.1 μg of pepstatin A/ml). Nuclei and cell debris were removed by centrifugation for 10 min at 10,000 rpm in a microcentrifuge at 4°C. Postnuclear supernatants were layered onto a prechilled continuous 5-to-40% (wt/vol) linear sucrose gradient, which was then centrifuged for 183 min at 50,000 rpm in a Beckman SW55Ti rotor at 4°C as described previously (27). Fractions of 400 μl each were collected from the bottom, and the Myc-tagged TCP-1γ protein with its associated proteins were coimmunoprecipitated with a mouse monoclonal anti-Myc antibody (Invitrogen) and protein G-agarose (Boehringer Mannheim). Proteins in the immune complex were separated by 8%–SDS PAGE and were analyzed by Western blot assay with a mouse monoclonal anti-Myc antibody, a rat monoclonal anti-TCP-1α antibody (StressGen), and a purified rabbit anti-Gag antibody.
RESULTS
Synthesis and processing of wild-type and p4-truncated viral proteins.
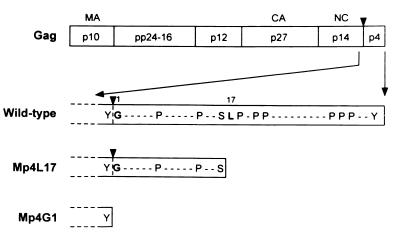
To understand the role of p4 in virus assembly and replication, we generated two p4-truncated M-PMV mutants (Fig. 1). The first truncation mutant, Mp4G1, was constructed to completely delete the p4-coding sequences of the gag gene by changing the first codon of p4 from GGG (glycine) to TAG (stop). The second truncation mutant, Mp4L17, was created to delete the carboxy-terminal half of the p4 protein, including proline clusters PEPP and PPP at positions 18 to 21 and 31 to 33, respectively. In this mutant, the 17th codon was changed from TTA (leucine) to TAA (stop). Since ribosomal frameshifting for the pro reading frame in M-PMV occurs upstream of the p4 coding sequences (16, 46), the two-nucleotide substitutions in Mp4G1 result in a change of codon TGG (tryptophan) to TTA (leucine) in the pro reading frame. Thus, translation is expected to produce gag-pro readthrough products (Gag-Pro polyprotein) with a single amino acid substitution. However, the mutation in Mp4L17, changing from CTT (leucine) to CTA (leucine) in the pro reading frame, should bring no amino acid substitutions in the Gag-Pro precursor.
FIG. 1.
Schematic representation of M-PMV mutants. The arrangement of the structural proteins within the Gag polyprotein is schematically presented (top). The partial amino acid sequences of the M-PMV p4 proteins are shown below, using single-letter amino acid codes. Prolines and carboxy-terminal residues are displayed, and other residues are indicated by dashed lines. Numerals indicate residue numbers. In Mp4L17, the 17th codon (TTA) was changed to a stop codon (TAA). In Mp4G1, the first codon (GGG) was changed to a stop codon (TAG). Arrowhead, cleavage site between the p14 NC protein and the p4 protein.
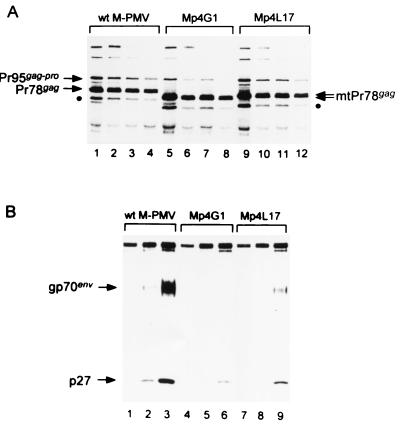
In order to determine whether the Gag proteins of these p4-truncated mutants are synthesized and assembled normally into mature virions, we performed a pulse-chase experiment (Fig. 2). COS-1 cells transiently transfected with either wild-type or mutant proviral DNAs were pulse-labeled for 20 min with [3H]leucine and chased for 0.5, 1, and 2 h. Both cell-associated (Fig. 2A) and released virion-associated proteins (Fig. 2B) were analyzed. Two major Gag-related precursor proteins, Pr78gag and Pr95gag-pro, were efficiently synthesized by all cells during the pulse-labeling (Fig. 2A, lanes 1, 5, and 9). Truncated Gag polyproteins (mtPr78gag) of Mp4G1 and Mp4L17 were smaller by an amount consistent with their deletions (lanes 5 and 9). As expected, the Gag-Pro polyproteins (Pr95gag-pro) were all the same size, while the internal initiation products of the gag gene (36) were smaller in accordance with the sizes of the deletions (Fig. 2A).
FIG. 2.
Immunoprecipitation of intracellular and extracellular viral proteins. To examine the biosynthesis and turnover of Gag (Pr78gag) and Gag-related precursor polyproteins (Pr95gag-pro), wild-type and mutant M-PMV proviral DNAs were transfected into COS-1 cells. (A) At 48 h after transfection, cells were pulse-labeled for 20 min with [3H]leucine (lanes 1, 5, and 9) and chased for 0.5 (lanes 2, 6, and 10), 1 (lanes 3, 7, and 11), and 2 h (lanes 4, 8, and 12). Cell-associated Gag-specific viral polyproteins were immunoprecipitated with rabbit anti-p27 CA antibody. An internal initiation product of the gag gene, Pr68gag, is marked (●). (B) Extracellular virions were pelleted from the culture medium of pulse-labeled cells after 0.5- (lanes 1, 4, and 7), 1- (lanes 2, 5, and 8), and 2-h (lanes 3, 6, and 9) chases and then immunoprecipitated with a goat anti-M-PMV antibody.
Late in the budding process or shortly thereafter, the Gag protein is proteolytically cleaved into capsid proteins (46). We observed this processing as a decrease in the amount of cell-associated, pulse-labeled Gag precursors during the chase period (Fig. 2A). Simultaneously, virion-associated p27 and mature envelope glycoprotein gp70env appeared in the culture medium (Fig. 2B, lanes 1 to 3). Approximately half of the newly synthesized Gag polyproteins appeared to be processed into mature proteins and released as virions during the 2-h chase, consistent with our previous results (38). In cells with either mutant virus, much fewer of these two virion-associated proteins were detected (Fig. 2B, lanes 4 to 9), even though a significant proportion of the radiolabeled mutant Gag proteins were lost during the chase period (Fig. 2A, lanes 5 to 8 and 9 to 12): after a 2-h chase about 55 to 60% of the pulse-labeled Gag molecules were detected in cells. Thus, both p4-truncated mutant Gag polyproteins were efficiently synthesized, but were turned over (with an approximate 2-h half-life) without being efficiently released as virions into the culture medium. Since the truncation of p4 triggered a degradation of the entire Gag polyprotein, these results suggest that the proline-rich domain of the Gag carboxy terminus is crucial for stabilizing the molecule.
We also determined the spread of these mutant viruses through the HeLa cells by measuring reverse transcriptase (RT) activity in culture fluids (38). There was a rapid increase in RT activity 6 to 12 days after infection with wild-type virus; in contrast, Mp4L17 mutant viruses showed a much slower rate of RT activity increase and the levels remained low. In mutant Mp4G1-infected cells RT activity was just above that detected with the uninfected cells (data not shown). These results coincided with the impaired virion release from these mutant virus-infected cells (Fig. 2B).
Fractionation of Gag polyprotein.
Since the newly synthesized Gag proteins are assembled into capsids with a half-life of approximately 45 min (37), it was of interest to determine whether the p4-truncated Gag proteins were assembled into a capsid prior to degradation. Completely assembled intracytoplasmic immature capsids (ICAP) of M-PMV are stable and pelletable under mild non-ionic-detergent conditions, while mature capsids and unassembled capsid precursors (either at the plasma membrane or within the cytoplasm) are soluble (36–38). Exploiting these different properties of the capsids, we carried out a Gag protein fractionation experiment to determine the extent of capsid formation with the mutant Gag polyproteins. Since Gag polyproteins have been observed to form pelletable aggregates when they were overexpressed in COS-1 cells (36), we established HOS cell lines with low-level expression of integrated wild-type or mutant proviral genomes.
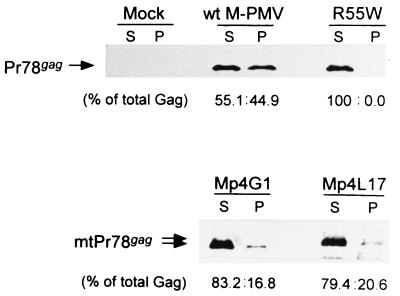
After lysis of transfected HOS cells in Triton X-100 lysis buffer, Gag polyproteins were fractionated by sedimentation into soluble (free-protein) and pelleted (capsid-associated) forms (Fig. 3). Gag molecules in each fraction were then analyzed by immunoprecipitation and immunoblotting with rabbit anti-Gag antiserum. As observed previously (36), a significant proportion (40 to 45% of total Gag) of cell-associated wild-type M-PMV Gag proteins were found in the pelleted fraction. The R55W mutant, which is defective in ICAP assembly, produced no pelleted Gag. Interestingly, in the Mp4G1 and Mp4L17 mutant viruses, approximately 15 to 20% of the total cell-associated Gag polyproteins were incorporated into stable, pelletable particles within the cytoplasm.
FIG. 3.
ICAP formation. To determine whether the p4-truncated Gag polyproteins can be assembled into capsids, ICAPs were prepared from lysates of HOS cells expressing wild-type M-PMV, the Mp4L17 and Mp4G1 truncation mutants, and a negative-control R55W mutant that is defective in ICAP assembly (36). Soluble (S) and capsid-associated pelletable (P) Gag polyproteins were fractionated and then immunoprecipitated with rabbit anti-Gag antiserum and visualized by Western blot assay with rabbit anti-Gag antibodies. The amount of Gag polyprotein in each band was quantified, and the ratios to the total amount of Gag protein (% of total Gag) were determined.
Together with the results in Fig. 2A showing that mutant Gag proteins were turned over, but with a much longer half-life than that for capsid assembly, these results strongly suggested that p4-truncated Gag molecules assemble capsids with reduced efficiency (approximately 30 to 50% of that for wild-type Gag molecules). However, it was not clear whether these mutant Gag molecules assembled no capsids or assembled mutant capsids that were readily disrupted under the experimental conditions. To examine this further, we used electron microscopy to study COS-1 cells transfected with each mutant genome. No large intracytoplasmic accumulations of assembling or assembled capsids were observed in the mutant virus-expressing cells (data not shown). Collectively, these results led us to conclude that the rate-limiting step of these mutant Gag polyproteins in the late stage of retrovirus replication appears to be capsid assembly. Thus, this proline-rich domain of the Gag molecule at the carboxy terminus may be required not only to stabilize the molecules but also to facilitate the process of capsid assembly.
Previously we have shown that Gag molecules of the P43L M-PMV mutant could assemble capsids, even though they were very unstable and were turned over rapidly with a half-life of 1 h (38). More interestingly, electron-microscopic studies on thin sections of these mutant virus-infected cells revealed intracytoplasmic inclusions of partially assembled capsids with very few complete ones. In contrast, the p4-truncated Gag molecules had a longer half-life (2 h; Fig. 2A) and they did not contain these structures. These observations strengthened our hypothesis that the proline-rich sequences at the carboxy terminus of the M-PMV Gag protein may play a role in intracytoplasmic capsid assembly.
Specific interactions between p4 and the TCP-1γ subunit of chaperonin TRiC.
Proline-rich motifs and domains in many cellular and viral proteins are thought to be involved in protein-protein and protein-DNA interactions (7, 11, 18, 42). To determine whether the M-PMV p4 domain interacts with specific host cellular proteins to facilitate the capsid assembly process, we carried out yeast two-hybrid screening against a HeLa cDNA library. Among several candidates with positive signals, the most abundant were cDNAs encoding the human TCP-1γ subunit of cytoplasmic chaperonin TRiC (TCP-1 ring complex).
To confirm the specificity of the interaction of p4 with TCP-1γ, we retransformed the prey plasmid coding for B42 fused to full-length protein TCP-1γ into yeast containing bait plasmids that express proteins resulting from the fusion of LexA with various domains of a retrovirus Gag polyprotein (Table 1). Expression of LexA-Gag domain fusion proteins as well as a hemagglutin-tagged full-length TCP-1γ protein under the control of the GAL promoter was confirmed (data not shown). The reporter assays of leucine prototrophy and β-galactosidase activity revealed that the interaction of TCP-1γ with M-PMV p4 was specific, since no activation of either reporter gene was detected with the LexA DNA binding domain per se (Table 1; for Mp4G1, no p4 domain was fused to LexA because of the termination codon substitution for the first codon in the p4 gene). The p4 truncation mutant, Mp4L17, failed to activate reporter genes, suggesting that p4, and particularly the proline clusters in the carboxy-terminal half of p4, are crucial for binding the TCP-1γ protein.
TABLE 1.
Interaction between TCP-1γ and various domains of retroviral Gag polyprotein
| Gag domain | Growth on Leu− platea | β-Galactosidase activity (liquid assay)b | In vitro interactionc |
|---|---|---|---|
| M-PMV p4 | ++ | 3.293 ± 0.409 | + |
| Mp4L17 | − | 0.299 ± 0.033 | − |
| Mp4G1 | − | 0.156 ± 0.041 | − |
| M-PMV MA | − | 0.327 ± 0.015 | − |
| M-PMV pp24/16-p12 | ++ | 3.677 ± 0.237 | ND |
| M-PMV CA | + | 1.893 ± 0.142 | ND |
| M-PMV NC | − | 0.632 ± 0.081 | − |
| HIV p6 | ++ | 4.485 ± 0.374 | ND |
Leu2 reporter gene activity was defined by the time required for positive colonies to reach 0.5 mm in diameter. ++, <3 days; +, 3 to 6 days; −, no detection after 9 days.
The activation of the lacZ reporter gene was determined by β-galactosidase activity as described previously (30). The β-galactosidase units (U) were calculated as follows: U = (1,000 × OD at 420 nm)/(reaction time [minutes] × OD at 600 nm × concentration factor), where OD is the optical density. Data are the means and standard errors from four independent transformants.
T7 epitope-tagged TCP-1γ binding to GST fused with the indicated Gag domain was examined in in vitro binding assays as described in Materials and Methods. +, positive result of test; −, negative result of test; ND, not determined.
We also tested other retroviral proteins with this TCP-1γ-expressing prey plasmid (Table 1). Interestingly, the HIV-1 p6 protein, a carboxy-terminal Gag domain with a high proline content similar to that of M-PMV p4, was also found to interact with TCP-1γ. In addition, the M-PMV pp24/16-p12 construct had a strong interaction with TCP-1γ protein. This region of M-PMV Gag contains both a p12 protein and a phosphorylated pp24/16 protein with a proline-rich motif that was identified as a late-budding domain (53). In contrast, the other M-PMV Gag domains showed no interaction (MA and NC proteins) or a very weak interaction (CA protein).
The interaction between p4 and TCP-1γ was confirmed by in vitro binding assays (Table 1). Partially purified p4- and truncated p4 (Mp417)-GST fusion proteins were bound to glutathione-Sepharose beads and mixed with equal amounts of bacterial lysates containing T7-tagged TCP-1γ proteins. The amount of T7-tagged TCP-1γ protein bound to the beads was examined by Western blotting. The GST-p4 fusion protein precipitated the T7-tagged TCP-1γ protein, while the p4 mutant fusion protein did not. As a negative control, GST alone (Table 1; Mp4G1) did not bind the tagged TCP-1γ protein. GST-M-PMV MA and GST-M-PMV NC were included in these assays to confirm that no T7-tagged TCP-1γ was precipitated by these fusion proteins. These results distinctly suggested that the proline-rich domain at the carboxy terminus of the retrovirus Gag molecule interacts with the TCP-1γ subunit protein of the TRiC chaperonin complex. In M-PMV Gag, there appeared to be multiple interactions between Gag and TCP-1γ, through at least two different domains in p4 and pp24/16-p12.
The in vivo interaction of M-MPV p4 with TCP-1γ.
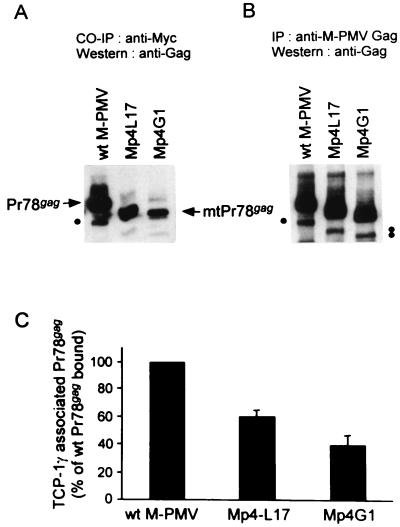
The association of p4 with TCP-1γ was further examined in cells. A coimmunoprecipitation experiment was carried out with 293T cells transiently cotransfected with wild-type or p4-truncated proviral DNAs and a Myc-tagged TCP-1γ-expressing plasmid. Precipitation with an anti-Myc antibody brought down wild-type Gag polyprotein Pr78gag bound to TCP-1γ (Fig. 4A). The internal initiation product of Gag, Pr68gag (as shown in Fig. 2A) can also be seen (Fig. 4A). As a negative control, no Gag was found in the immune precipitates of antibody and protein G-agarose when cells were transfected only with M-PMV DNAs (data not shown). These results clearly demonstrated that M-PMV Gag polyproteins interact with TCP-1γ in cells.
FIG. 4.
Coimmunoprecipitation of wild-type and p4-truncated Gag polyproteins with the TCP-1γ subunit. (A) The cellular interactions between M-PMV Gag polyprotein and TCP-1γ protein were examined by a coimmunoprecipitation (CO-IP) experiment. 293T cells transiently cotransfected with wild-type or p4 mutant (Mp4L17 or Mp4G1) proviral DNA and Myc-tagged TCP-1γ-expressing plasmid DNA were lysed in 1% NP-40-containing TNE buffer. TCP-1γ–Myc and its associated proteins were coimmunoprecipitated with mouse monoclonal anti-Myc antibody for 2 h at 4°C and then separated by 8%–SDS PAGE. TCP-1γ-Myc-bound Gag polyproteins were detected by Western blot assay with rabbit anti-Gag antibody. Dots, internal initiation products (Pr68gag). (B) To measure the total amount of Gag polyprotein in each sample, equal numbers of cotransfected 293T cells were lysed in lysis buffer A supplemented with 0.1% SDS and total Gag polyproteins were immunoprecipitated (IP) with rabbit anti-Gag antibody. Gag polyproteins were detected by Western blot assay with the same antibody. (C) The amount of Gag polyprotein in each band of panel A was quantified. The amount of TCP-1γ–Myc-bound Gag protein was normalized by that of total Gag (B) and then plotted as a percentage. The amount bound by wild-type (wt) M-PMV is arbitrarily presented as 100%. The data are the means and standard errors from three separate experiments.
In contrast, much less Gag was coprecipitated with TCP-1γ for mutants Mp4L17 and Mp4G1 (Fig. 4A). We also used an anti-Gag antibody to precipitate these lysates to verify that total Gag expression with these mutants was similar to or higher than that with wild-type virus (Fig. 4B). After normalizing for total Gag protein, we quantified the Gag association with TCP-1γ from Fig. 4A. The Mp4L17 and Mp4G1 mutants had approximately 40 and 60% less Gag association with TCP-1γ, respectively, than wild-type M-PMV (Fig. 4C). Comparable amounts of Myc-tagged TCP-1γ protein were detected in all immune complexes when the same membrane was reprobed with the anti-Myc antibody (data not shown). These results agree with those obtained from the yeast two-hybrid system and in vitro binding assays presented in Table 1: p4 and pp24/p16-p12 proteins in M-PMV Gag are able to associate with the TCP-1γ subunit protein of chaperonin TRiC. The interaction of p4-truncated mutant Gag with TCP-1γ was of lower affinity and appeared to be mediated through other domain(s), yet to be identified, including one in the pp24/p16 and p12 proteins.
Association of M-PMV Gag polyprotein with the TRiC cytoplasmic chaperonin complex.
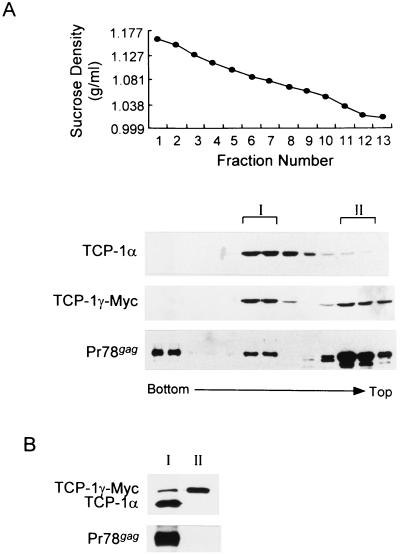
Having shown that the M-PMV Gag polyprotein interacts with TCP-1γ, we wanted to find whether Gag is associated with the whole TRiC chaperonin complex, not just with free TCP-1γ subunits. 293T cells were transiently cotransfected with M-PMV proviral DNA pSHRM15 and a Myc-tagged TCP-1γ-expressing plasmid. The intact TRiC chaperonin complex has a relative molecular mass of 800 to 950 kDa and is composed of eight different but homologous subunits (22, 24, 26). When we performed a 5-to-40% (wt/vol) sucrose gradient, anti-TCP-1α detected the TRiC complex at 19 to 23% sucrose (fractions 6 to 8), equivalent to a density of 1.076 to 1.094 g/ml (Fig. 5A). This result is consistent with densities previously observed (22, 27). Specific antibodies against Gag and Myc-tagged TCP-1γ detected proteins at two distinct regions of the gradient, fractions 6 to 8 and 11 to 13 (Fig. 5A). In addition, Gag could be seen in fractions 1 and 2 with a higher density, which appears to reflect assembled capsids normally recovered from the bottom of the gradient, with a density of 1.20 g/ml (23, 43). Proteins in the later fractions (11 to 13), being at the top of the gradient, were expected to be free molecules.
FIG. 5.
Coimmunoprecipitation of M-PMV Gag polyprotein with the chaperonin TRiC. Cell fractionation and coimmunoprecipitation were carried out with 293T cells transiently cotransfected with M-PMV proviral DNA pSHRM15 and a Myc-tagged TCP-1γ-expressing plasmid. (A) The cells were lysed in 0.5% NP buffer and then fractionated through a continuous 5-to-40% (wt/vol) linear sucrose gradient. The fractions were collected from bottom to top and measured for sucrose density (top) Aliquots of each fraction were analyzed by Western blot assay with rat monoclonal anti-TCP-1α (TCP-1α), mouse monoclonal anti-Myc (TCP-1γ–Myc), and rabbit anti-Gag (Pr78gag). The densities of fractions 6 and 7 (1.076 and 1.086 g/ml, respectively) are near the expected density of the TRiC complex. (B) Peaks I and II (pooled fractions 6 and 7 and 11 and 12, respectively) were diluted with 0.5% NP buffer and then used to coimmunoprecipitate TCP-1γ–Myc and its associated proteins with a mouse monoclonal anti-Myc antibody. Proteins in the immune complex were analyzed as described above for TCP-1γ–Myc, TCP-1α, and Gag by Western blot assay.
For further analysis, we pooled fractions 6 and 7 (peak I; density of 1.076 to 1.086 g/ml) and fractions 11 and 12 (peak II). These fractions were then used to coimmunoprecipitate TCP-1γ–Myc as well as its associated proteins. A Myc-specific monoclonal antibody precipitated endogenous TCP-1α, TCP-1γ, and Gag from fractions 6 and 7 (Fig. 5B, lane I). By contrast, neither Gag nor TCP-1α was detected in fractions 11 and 12, even though substantial amounts of TCP-1γ were precipitated (Fig. 5B, lane II). These results clearly demonstrated that M-PMV Gag polyproteins associate with cytoplasmic chaperonin TRiC in virus-infected cells, presumably through the TCP-1γ subunit.
It is noteworthy that significant steady-state levels of Myc-tagged TCP-1γ were found with the complex, suggesting that the exogenous, overexpressed TCP-1γ molecules were folded correctly and complexed with other subunits of TRiC. Furthermore, we found that TCP-1α-specific antibodies coimmunoprecipitated both Gag and TCP-1γ, along with TCP-1α, from fractions 6 and 7 (data not shown). Thus, these findings raised the possibilities that the interaction with TRiC allows nascent Gag molecules to be properly folded into a native structure and/or assembled into a capsid.
ATP requirement for M-PMV Gag release from TRiC.
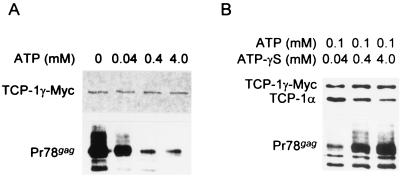
TRiC, like other chaperones, assists in protein folding by ATP-dependent cycles of release and rebinding (13, 17, 28, 31). Indeed, substrates, such as tubulin, actin, and firefly luciferase, are released from the substrate-TRiC complex upon incubation with ATP, but not with nonhydrolyzable analogues (13, 31, 52). Since the data described above suggested that TRiC mediates M-PMV Gag folding and/or assembly, we investigated whether Gag proteins dissociate from TRiC in an ATP-dependent fashion.
Aliquots of 293T cell lysates expressing M-PMV viral and TCP-1γ–Myc proteins were treated with 0.04, 0.4, and 4 mM ATP on ice for 1 h in the presence of 2 mM MgCl2 prior to coimmunoprecipitation with a Myc-specific antibody. In the presence of ATP, even as little as 0.04 mM, much less Gag associated with TCP-1γ (Fig. 6A). In addition, the effect of ATP activity on Gag release was concentration dependent with nearly maximal release around 0.1 to 0.4 mM. These observations were confirmed by competition assays with nonnonhydrolyzable ATP analog ATP-γ-S (Fig. 6B). When increasing amounts of ATP-γ-S were added to the reaction mixture with 0.1 mM ATP, release was inhibited in a concentration-dependent manner, while there were no changes in the amount of TCP-1α associated with the complex (Fig. 6B). Therefore, we can conclude that M-PMV Gag, like other TRiC-associated proteins, depends on ATP hydrolysis for its release from the chaperone.
FIG. 6.
Effects of ATP and ATP-γ-S on the M-PMV Gag association with the chaperonin TRiC. 293T cells transiently cotransfected with an M-PMV proviral DNA and a TCP-1γ–Myc-expressing plasmid were lysed in 1% NP-40-containing TNE buffer. (A) Cell lysates were incubated with 2 mM MgCl2 and various concentrations of ATP on ice for 1 h prior to coimmunoprecipitation with mouse monoclonal anti-Myc antibody. TCP-1γ–Myc and Gag proteins were detected by Western blot assay with mouse anti-Myc and rabbit anti-Gag antibodies, respectively. The sample with no ATP was sham treated with H2O. (B) To confirm the effect of ATP on dissociation of Gag proteins from TRiC, competition assays were carried out with nonhydrolyzable ATP analog ATP-γ-S. Cell lysates were treated for 1 h on ice with various concentrations of ATP-γ-S in the presence of 0.1 mM ATP and 2 mM MgCl2. After coimmunoprecipitation with an anti-Myc antibody, TCP-1γ–Myc, TCP-1α, and Gag proteins were detected as described above with mouse anti-Myc, rat anti-TCP-1α, and rabbit anti-Gag antibodies, respectively.
DISCUSSION
In cells, newly synthesized proteins fold into a native structure, a state of minimum potential energy, by poorly defined mechanisms, in which molecular chaperones play a fundamental role (8, 15, 21). In addition, molecular chaperones are believed to assist in intracellular targeting and complex assembly of folded native proteins (29). Two major families of molecular chaperones, the 70-kDa heat shock protein cognate (Hsc70) and the chaperonin TCP-1 ring complex (TRiC), mediate such functions in the eukaryotic cell cytosol. Hsc70 appears to be predominant, interacting with the majority of cytosolic nascent polypeptides over 20 kDa, whereas TRiC has been found to interact with a small subset of newly synthesized proteins of 30 and 60 kDa, about 9 to 15% of all cytosolic proteins (49). To date, only a few proteins have been identified as TRiC folding substrates, including actin, tubulin (13), Gα-transducin (9), myosin II (47), and the VHL tumor suppressor protein (10). Here we describe the ATP-dependent interaction of the gag gene product of type D retroviruses with cytosolic chaperonin TRiC, with the likelihood that the Gag polyprotein is a TRiC folding substrate.
This study shows that the carboxy-terminal proline-rich protein, p4, of M-PMV Gag polyprotein interacts specifically in vitro and in vivo with TCP-1γ, a subunit of TRiC, thereby mediating the interaction between Gag polyprotein and TRiC (Table 1 and Fig. 4). When the p4 domain was partially or completely deleted through premature termination mutations, the interaction between the p4-truncated Gag and TCP-1γ was impaired: there was a 40 to 60% drop in the amount of TCP-1γ-associated mutant Gag compared to that with wild-type Gag (Fig. 4). The pp24/16-p12 domain on Gag was also identified in vitro as an additional TCP-1γ binding site, suggesting that multiple bindings of M-PMV Gag to TRiC may be required for stable interaction between the two molecules. Of note, capsid assembly by these mutants occurred at similarly reduced kinetics: immature capsids were assembled in the cytoplasm inefficiently, with a 50 to 70% drop in the relative ratio of particle-associated Gag to total Gag (Fig. 3). Together, these data suggest that the M-PMV Gag-associated TRiC chaperonin complex probably promotes the process of capsid assembly.
Retroviral capsid assembly requires a series of events including synthesis and modification of the Gag polyprotein, folding into a stable conformation, transport to the site of assembly, and multimerization to form a complex of 2,000 to 3,000 molecules. These processes depend on host cell proteins and machinery. In particular, the folding of Gag to an assembly-competent form is critical in the late phase of the retrovirus life cycle, capsid assembly being dependent on the intermolecular interactions between Gag molecules. Our previous studies of M-PMV mutants with in-frame deletions (37) or proline-to-leucine substitution mutations (38) within MA agree well with this notion. Mutant Gag molecules appeared to be folded into an unfavorable conformation, exhibiting rapid turnover with a half-life of less than 1 h. The unfavorable conformation interfered with the assembly process such that very few capsids were completed. It should be noted that these characteristics of stability-assembly-defective mutants differ from those seen with assembly-defective ones with single-amino-acid substitutions in the major homology region of M-PMC CA (48). Typically, the latter mutants express stable Gag molecules with a half-life of 4 h but with no visibly assembled or assembling capsids, suggesting that these mutant Gag proteins fold into a favorable ternary conformation but without the ability to assemble a capsid. Consequently, no virus particles are released. Interestingly, the phenotype observed with the p4-truncated mutants we present here is somewhat different from both defective phenotypes. The p4-truncated Gag molecules were turned over with intermediate kinetics (half-life of 2 h; Fig. 2 A) and assembled into capsids with a 50%-reduced efficiency compared to that of the wild type (Fig. 3); those that were assembled were able to be transported to the plasma membrane and released (Fig. 2B). Thus, p4, most likely through the TRiC interaction, appears to play a critical role in the processes whereby Gag acquires stable conformations and assembly competency. We argue that TRiC assists type D retrovirus Gag folding into a native structure, which confers stability on the molecule. The impaired interaction with TRiC may render intermediate stability to the p4 mutant Gag, which was then subject to degradation mechanisms, resulting in inefficient capsid assembly.
Feldman et al. (10) demonstrated that, in addition to a role in chaperoning monomeric protein folding, TRiC assists in the assembly of a functional VBC complex by mediating incorporation of tumor suppressor protein VHL into a complex with its partner proteins, elongin B and elongin C. Such involvement in protein multimerization was also implicated in assembly of hepatitis B virus capsid by showing that TRiC or a related chaperonin associates with assembly intermediates but not with either the initial unassembled virus core proteins or the mature capsid (29). Thus, it remains possible that TRiC mediates capsid assembly as well as Gag folding in M-PMV. The reduced capsid assembly observed with the p4 truncation mutants might reflect a mere defect in this additional role of TRiC. Although this is a formal possibility, we favor the interpretation that TRiC assists in Gag folding, not assembly, because we have shown that the purified recombinant M-PMV Gag molecules can assemble capsid-like structures in vitro (23).
The p4 domain of M-PMV Gag is proline rich, with the proline residues clustered at the carboxy half of the domain. This proline-rich domain appears to be necessary for Gag interaction with TCP-1γ. In many proteins, cellular and viral proline-rich motifs have been shown to be involved in multiprotein interactions, as was found in the vesicle-associated proteins and the SH3 domain binding proteins. Mutational studies on various retroviral Gag polyproteins have also suggested that conserved proline-rich motifs serve as docking sites for the cellular protein(s) to mediate the budding process (14, 19, 33, 34, 53). In addition, HIV-1 Gag association with molecular chaperone cyclophilin A (11) is mediated by a proline-rich segment within CA. More interestingly, HIV-1 p6, analogous to M-PMV p4, exhibits a strong interaction with TCP-1γ in the yeast two-hybrid system (Table 1). However, no sequence homology between the p4 and p6 proteins can be seen except for their unusually high proline content. The core TRiC binding domain of β-tubulin was defined through mutagenesis and proteolytic analyses; it spans amino acids 150 to 350 and contains many proline and hydrophobic residues (6, 40). There are no conserved sequences among the core domains of β-tubulin, VHL (10), and β-actin (20). Thus, our data support the speculation that TRiC may interact with its folding substrates in a sequence-independent manner through structural motifs contributed by proline residues.
In summary, the data presented here suggest a mechanism by which the gag gene products of a type D retrovirus acquire an assembly-competent folded conformation. Although the details of the mechanism involved in this folding process have yet to be investigated, our findings imply that the TRiC chaperonin complex assists in the folding of newly synthesized Gag polyproteins to a native structure. The TRiC-Gag interaction can be achieved through the association between the p4 and pp24/16-p12 domains of Gag and the TRiC subunit protein, TCP-1γ. This interaction is transient, with release of Gag from the chaperonin complex in an ATP hydrolysis-dependent reaction, as seen in TRiC-mediated folding of firefly luciferase and tubulin (13, 32). Insufficient interaction between these molecules, shown with the p4-truncated mutants, appears to impair the TRiC folding process, which likely results in incorrect folding reactions that induce Gag protein degradation. Consequently, capsid assembly and virus release are inefficient. Many molecular chaperones, such as cyclophilin A, hsp27, hsp70, and hsp78, have been shown to interact with HIV Gag (11). It seems likely, then, that several cellular chaperones function in tandem to promote productive folding of retroviral Gag to a native form.
ACKNOWLEDGMENTS
We thank R. Finley for the HeLa cDNA library. We are grateful to members of our laboratory for discussion during the project and to J. Macke for substantive editing of the manuscript.
This work was supported by grant B-98015 to S.S.R. from the Samsung Biomedical Research Institute and by grant R39 CA27834 to E.H. from the National Cancer Institute.
REFERENCES
- 1.Bachand F, Yao X J, Hrimech M, Rougeau N, Cohen É. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 Gag precursor. J Biol Chem. 1999;274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 2.Bennett R P, Rhee S, Craven R C, Hunter E, Wills J W. Amino acids encoded downstream of gag are not required by Rous sarcoma virus protease during Gag-mediated assembly. J Virol. 1991;65:272–280. doi: 10.1128/jvi.65.1.272-280.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradac J, Hunter E. Polypeptides of Mason-Pfizer monkey virus. I. Synthesis and processing of the gag-gene products. Virology. 1984;138:260–275. doi: 10.1016/0042-6822(84)90350-7. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay S K, Sengupta D N, Fredrickson T N, Morse H C, Hartley J W. Characteristics and contributions of defective, ecotropic, and mink cell focus-inducing viruses involved in a retrovirus-induced immunodeficiency syndrome of mice. J Virol. 1991;65:4232–4241. doi: 10.1128/jvi.65.8.4232-4241.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrzynski J K, Sternlicht M L, Farr G W, Sternlicht H. Newly-synthesized β-tubulin demonstrates domain-specific interactions with the cytosolic chaperonin. Biochemistry. 1996;35:15870–15882. doi: 10.1021/bi961114j. [DOI] [PubMed] [Google Scholar]
- 7.Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle M C, Golsteyn R M. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 8.Ellis J. Proteins as molecular chaperones. Nature. 1987;328:378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- 9.Farr G W, Scharl E C, Schumacher R J, Sondek S, Horwich A L. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 10.Feldman D E, Thulasiraman V, Ferreyra R G, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 11.Franke E K, Yuan H E, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 12.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 13.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall J S, Tempst P, Hartl F U. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 16.Hatfield D L, Levin J G, Rein A, Oroszlan S. Translational suppression in retroviral gene expression. Adv Virus Res. 1992;41:193–239. doi: 10.1016/S0065-3527(08)60037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrick J P, Hartl F U. The role of molecular chaperones in protein folding. FASEB J. 1995;9:1559–1569. doi: 10.1096/fasebj.9.15.8529835. [DOI] [PubMed] [Google Scholar]
- 18.Houchens C R, Montigny W, Zeltser L, Dailey L, Gilbert J M, Heintz N H. The dhfr oriβ-binding protein RIP60 contains 15 zinc fingers: DNA binding and looping by the central three fingers and an associated proline-rich region. Nucleic Acids Res. 2000;28:570–581. doi: 10.1093/nar/28.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes G M, Willison K R. Individual subunits of the eukaryotic cytosolic chaperonin mediate interactions with binding sites located on subdomains of β-actin. J Biol Chem. 2000;275:18985–18994. doi: 10.1074/jbc.M910297199. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J L, Craig E A. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 22.Joly E C, Tremblay E, Tanguay R M, Wu Y, Bibor-Hardy V. TRiC-P5, a novel TCP1-related protein, is localized in the cytoplasm and in the nuclear matrix. J Cell Sci. 1994;107:2851–2859. doi: 10.1242/jcs.107.10.2851. [DOI] [PubMed] [Google Scholar]
- 23.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota H, Hynes G, Willison K. The chaperonin containing t-complex polypeptide 1 (TCP-1). Multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. Eur J Biochem. 1995;230:3–16. doi: 10.1111/j.1432-1033.1995.tb20527.x. [DOI] [PubMed] [Google Scholar]
- 25.Leroux M R, Hartl F U. Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol. 2000;10:R260–R264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- 26.Lewis S A, Tian G, Vainberg I E, Cowan N J. Chaperonin-mediated folding of actin and tubulin. J Cell Biol. 1996;132:1–4. doi: 10.1083/jcb.132.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis V A, Hynes G M, Zheng D, Saibil H, Willison K. T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature. 1992;358:249–252. doi: 10.1038/358249a0. [DOI] [PubMed] [Google Scholar]
- 28.Li W Z, Lin P, Frydman J, Boal T R, Cardillo T S, Richard L M, Toth D, Lichtman M A, Hartl F U, Sherman F. Tcp20, a subunit of the eukaryotic TRiC chaperonin from humans and yeast. J Biol Chem. 1994;269:18616–18622. [PubMed] [Google Scholar]
- 29.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Welsh M J. Identification of a site of Hsp27 binding with Hsp27 and αB-crystallin as indicated by the yeast two-hybrid system. Biochem Biophys Res Commun. 1999;255:256–261. doi: 10.1006/bbrc.1999.0174. [DOI] [PubMed] [Google Scholar]
- 31.Marco S, Carrascosa J L, Valpuesta J M. Reversible interaction of β-actin along the channel of the TCP-1 cytoplasmic chaperonin. Biophys J. 1994;67:364–368. doi: 10.1016/S0006-3495(94)80489-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nimmesgern E, Hartl F U. ATP-dependent protein refolding activity in reticulocyte lysate. Evidence for the participation of different chaperone components. FEBS Lett. 1993;331:25–30. doi: 10.1016/0014-5793(93)80290-b. [DOI] [PubMed] [Google Scholar]
- 33.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 37.Rhee S S, Hunter E. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J Virol. 1990;64:4383–4389. doi: 10.1128/jvi.64.9.4383-4389.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee S S, Hunter E. Amino acid substitutions within the matrix protein of type D retroviruses affect assembly, transport and membrane association of a capsid. EMBO J. 1991;10:535–546. doi: 10.1002/j.1460-2075.1991.tb07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee S S, Hui H X, Hunter E. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J Virol. 1990;64:3844–3852. doi: 10.1128/jvi.64.8.3844-3852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rommelaere H, De Neve M, Melki R, Vandekerckhove J, Ampe C. The cytosolic class II chaperonin CCT recognizes delineated hydrophobic sequences in its target proteins. Biochemistry. 1999;38:3246–3257. doi: 10.1021/bi9815905. [DOI] [PubMed] [Google Scholar]
- 41.Rossi F, Evstafieva A, Pedrali-Noy G, Gallina A, Milanesi G. HsN3 proteasomal subunit as a target for human immunodeficiency virus type 1 Nef protein. Virology. 1997;237:33–45. doi: 10.1006/viro.1997.8752. [DOI] [PubMed] [Google Scholar]
- 42.Roth J, Koch P, Contente A, Dobbelstein M. Tumor-derived mutations within the DNA-binding domain of p53 that phenotypically resemble the deletion of the proline-rich domain. Oncogene. 2000;19:1834–1842. doi: 10.1038/sj.onc.1203500. [DOI] [PubMed] [Google Scholar]
- 43.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakalian M, Hunter E. Molecular events in the assembly of retrovirus particles. Adv Exp Med Biol. 1998;440:329–339. doi: 10.1007/978-1-4615-5331-1_43. [DOI] [PubMed] [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 47.Srikakulam R, Winkelmann D A. Myosin II folding is mediated by a molecular chaperonin. J Biol Chem. 1999;274:27265–27273. doi: 10.1074/jbc.274.38.27265. [DOI] [PubMed] [Google Scholar]
- 48.Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thulasiraman V, Yang C F, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 51.Won K A, Schumacher R J, Farr G W, Horwich A L, Reed S I. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yaffe M B, Farr G W, Miklos D, Horwich A L, Sternlicht M L, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- 53.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984;3:479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]