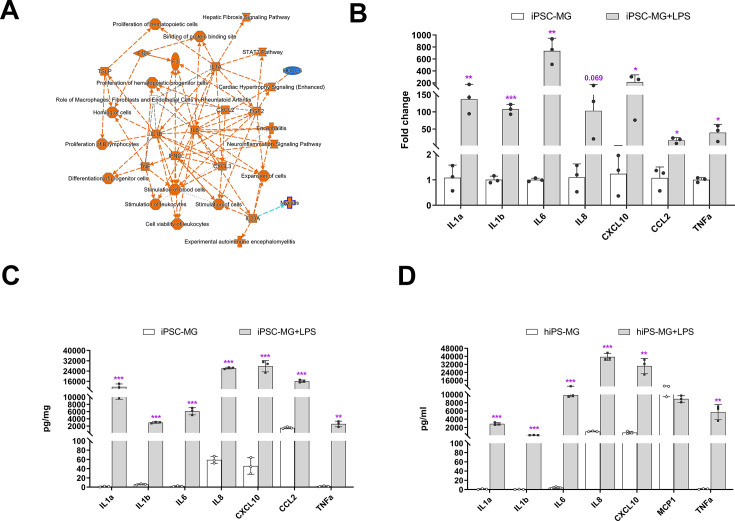
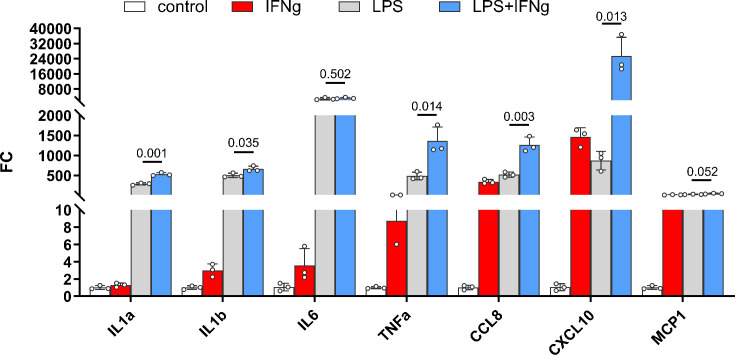
Figure 3. Inflammation responses of human-induced pluripotent stem cell (hiPSC)-derived microglial cells following lipopolysaccharide (LPS) stimulation.
(A) Ingenuity Pathway Analysis (IPA) showed different gene expression (fold change >twofold, p < 0.05) between LPS-treated and control hiPSC-derived microglial cells, demonstrating activation of core pathways involving IL6, IL1A, IL1B, and IFNG. (B) Assessment of mRNA expression of selected genes for inflammatory cytokines using quantitative reverse transcription-PCR (qRT-PCR; Oligonucleotide primers are provided in Supplementary 4) demonstrated increased expression following LPS (0.1 µg/ml) stimulation for 6 hr (3-6 replicates). These changes corresponded to increases in the protein expression levels of inflammatory cytokines following 24 hr of LPS stimulation as measured with a Multiplex kit (Millipore) in cell lysate (C) and conditioned media (D). The data in (C) and (D) are presented as means ± SEM (3-6 replicates). *p < 0.05, **p < 0.01, ***p < 0.001.