Abstract
The Autographa californica multiple nucleopolyhedrovirus (AcMNPV) GP64 protein is an essential virion protein that is involved in both receptor binding and membrane fusion during viral entry. Genetic studies have shown that GP64-null viruses are unable to move from cell to cell and this results from a defect in the assembly and production of budded virions (BV). To further examine requirements for virion budding, we asked whether a GP64-null baculovirus, vAc64−, could be pseudotyped by introducing a heterologous viral envelope protein (vesicular stomatitis virus G protein [VSV-G]) into its membrane and whether the resulting virus was infectious. To address this question, we generated a stably transfected insect Sf9 cell line (Sf9VSV-G) that inducibly expresses the VSV-G protein upon infection with AcMNPV Sf9VSV-G and Sf9 cells were infected with vAc64−, and cells were monitored for infection and for movement of infection from cell to cell. vAc64− formed plaques on Sf9VSV-G cells but not on Sf9 cells, and plaques formed on Sf9VSV-G cells were observed only after prolonged intervals. Passage and amplification of vAc64− on Sf9VSV-G cells resulted in pseudotyped virus particles that contained the VSV-G protein. Cell-to-cell propagation of vAc64− in the G-expressing cells was delayed in comparison to wild-type (wt) AcMNPV, and growth curves showed that pseudotyped vAc64− was generated at titers of approximately 106 to 107 infectious units (IU)/ml, compared with titers of approximately 108 IU/ml for wt AcMNPV. Propagation and amplification of pseudotyped vAc64− virions in Sf9VSV-G cells suggests that the VSV-G protein may either possess the signals necessary for baculovirus BV assembly and budding at the cell surface or may otherwise facilitate production of infectious baculovirus virions. The functional complementation of GP64-null viruses by VSV-G protein was further demonstrated by identification of a vAc64−-derived virus that had acquired the G gene through recombination with Sf9VSV-G cellular DNA. GP64-null viruses expressing the VSV-G gene were capable of productive infection, replication, and propagation in Sf9 cells.
Baculoviruses constitute a family of viruses that are pathogenic to certain insect species but do not appear to productively infect other invertebrates or vertebrates. Baculoviruses such as the Autographa californica multiple nucleopolyhedrovirus (AcMNPV) have been developed as biological control agents and as protein expression vectors. AcMNPV also serves as the primary model system for studies of baculovirus gene regulation and structure. AcMNPV has a large double-stranded DNA genome (134 kbp) and produces two virion phenotypes during the infection cycle (27). One virion phenotype, the occlusion-derived virions (ODV), is adapted for survival in the environment and propagation of infection from animal to animal, through oral transmission and infection of the midgut epithelial cells. In contrast, the other virion phenotype, the budded virions (BV), is adapted for propagation of infection from cell to cell throughout the animal, after infection is established by ODV in the midgut (9, 20, 21, 29). BV efficiently enter many cell types in the infected animal, including most notably hemocytes, tracheal epithelial cells, and fat body cells (6–8). The infection cycle is completed when ODV are assembled (enveloped) and occluded within occlusion bodies in the nuclei of infected cells. Occlusion bodies are then released by cell lysis.
Because BV are generated only after successful infection of the midgut epithelial cells, BV appear to have adopted a strategy of promiscuous infection of many insect cell types. Studies of baculovirus BV entry into mammalian cell lines and cultured primary cells show that in culture, BV from AcMNPV can enter primary rat hepatocytes as well as a number of human cell lines (4, 15, 41) although baculoviruses do not productively replicate there. When a reporter gene driven by a mammalian promoter is inserted into the AcMNPV genome, expression can be readily detected in many mammalian cell types. In contrast, gene expression could not be detected from a reporter under the control of the baculovirus polyhedrin promoter (4). Thus, baculovirus BV enter mammalian cells and appear to selectively express only genes under the control of mammalian promoters. Such studies suggested that baculovirus BV may be an effective vehicle for gene delivery to mammalian cells, perhaps as gene theraphy agents (1, 15, 16 38, 41). Indeed, several features of baculoviruses are highly desirable for the development of baculoviruses as potential vectors for gene therapy. These include the capacity of the baculovirus genome to accommodate very large insertions of foreign DNA, the inability of the virus to replicate within mammalian cells, and the apparent absence of expression of most baculovirus genes. Other studies (5) have shown that baculoviruses incorporating selectable markers (such as the neomycin phosphotransferase II gene) under a mammalian regulatory context, can be used to generate stably transformed mammalian cell lines.
During virion entry, the AcMNPV GP64 protein is involved in binding of virions to host cells (13). GP64 also mediates low-pH-triggered membrane fusion during entry by endocytosis (2, 22–24, 28, 34, 35). Genetic studies with GP64-null viruses (containing a gp64 knockout) showed that GP64 is also necessary for efficient virion budding from the cell surface (29, 30). Interestingly, GP64 proteins containing C-terminal truncations that removed portions or all of the GP64 cytoplasmic tail domain (CTD) did not show the same severity of the defect in budding as the complete GP64 deletion. This suggests that the CTD is not required for efficient budding and that some other feature of GP64 is important for virion assembly and budding. In certain retrovirus and rhabdovirus systems, heterologous envelope proteins can complement the absence of the endogenous envelope protein. Virions carrying a heterologous envelope protein are referred to as “pseudotyped” viruses. Pseudotyped virions have been used for applications such as gene therapy but also serve as valuable tools for dissecting the functions necessary for assembly of mature virions and budding at the cell surface. Thus, to better understand the requirements for baculovirus budding, we asked whether a heterologous viral envelope glycoprotein might complement the deletion of the gp64 gene from the AcMNPV genome. In a previous study, it was shown that when the vesicular stomatitis virus G protein (VSV-G) was expressed from a recombinant AcMNPV baculovirus, the presence of VSV-G in BV appeared to enhance infectivity in mammalian cells (1). In that study, BV presumably contained both VSV-G and GP64. In the present study, we asked whether VSV-G was capable of complementing both virion budding and infectivity in the context of a GP64-null virus, vAc64−. To examine this question, we first generated and characterized an Sf9-derived cell line that inducibly expressed the VSV-G protein upon infection with AcMNPV. The cell line, Sf9VSV-G, was then infected with vAc64−, and cells were monitored for movement of infection from cell to cell. Using this procedure, we generated pseudotyped virions that contain the VSV-G protein and were able to propagate infection from cell to cell in VSV-G-expressing cells, but not in Sf9 cells. Although cell-to-cell propagation of the GP64-null virus was delayed in comparison to wild-type (wt) AcMNPV propagation in the VSV-G-expressing cells, growth curves showed that pseudotyped virions were generated at titers of approximately 106 to 107 infections units (IU)/ml, compared with titers of approximately 108 for wt AcMNPV. In this study we demonstrate that several functions of GP64 can be replaced by the VSV-G protein, and we provide the first example of functionally pseudotyped baculovirus virions.
MATERIALS AND METHODS
Construction of plasmid pSM8141-VSV-G.
The VSV-G gene (Indiana serotype) was isolated from plasmid VSVG-BP95NOTSV (kindly provided by F. Boyce) as a 1,692-bp BamHI fragment containing the entire VSV-G open reading frame (ORF). The VSV-G gene was inserted into the unique BamHI site of a dual expression p10 locus transfer plasmid (kindly provided by D. H. L. Bishop), between a polyhedrin gene promoter and a simian virus 40 terminator, to create plasmid pSM8135. The presence and orientation of the VSV-G gene was confirmed by sequencing using primers located in the polyhedrin promoter. As a marker for expression in infected insect cells, a BglII-BamHI fragment containing a polyhedrin gene promoter and beta-glucuronidase (GUS) reporter gene was inserted into the BglII site of plasmid pSM8135 to create plasmid pSM8141-VSV-G (see Fig. 1).
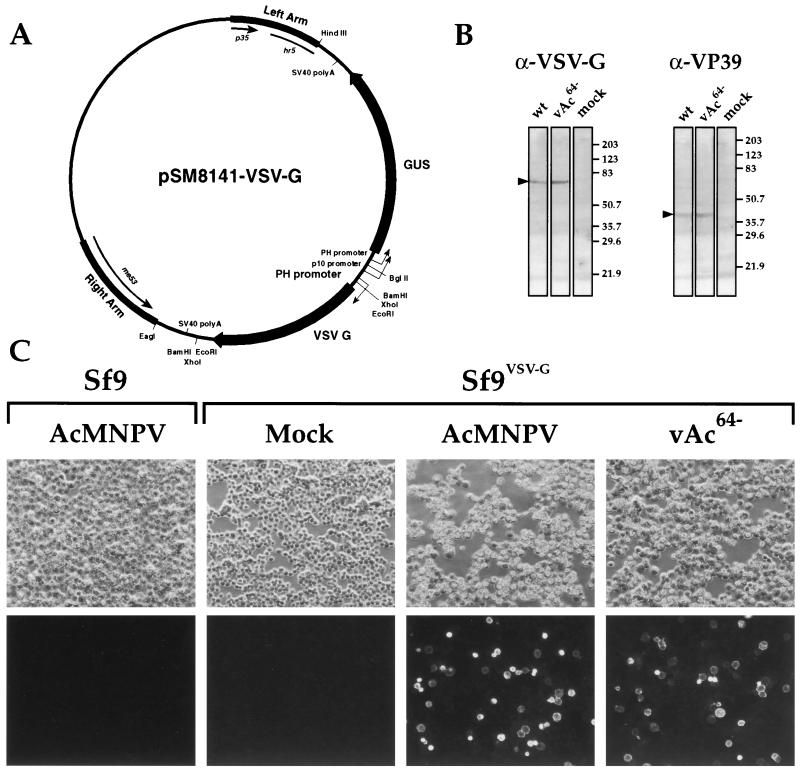
FIG. 1.
Construction and analysis of cell line Sf9VSV-G. (A) Plasmid pSM8141-VSV-G contains a VSV-G gene under the control of an AcMNPV polyhedrin (PH) promoter, and a GUS gene under the control of PH and p10 promoters from AcMNPV. Each gene cassette is terminated by a simian virus 40 (SV40) poly(A) cleavage and addition site. The two genes are flanked by left- and right-arm sequences from the AcMNPV p35-hr5 region and the me53 region, respectively. Plasmid pSM8141-VSV-G was transfected into Sf9 cells to generate a stably transfected cell line (Sf9VSV-G). (B) Western blot analysis of VSV-G protein induction in Sf9VSV-G cells infected with wt AcMNPV (wt) or a GP64-null virus (vAc64−). Sf9VSV-G cells were infected at an MOI of 1, harvested at 46 hp i, and then examined for VSV-G protein expression and VP39 protein expression, using MAbs (see Materials and Methods). The specificity of each MAb is indicated above each group of blots (α-VSV-G or α-VP39), and positions and molecular weights (in thousands) of protein size markers are indicated on the right. The positions of VSV-G and VP39 are indicated by an arrowhead on the left of each group of blots. (C) Immunofluorescent detection of VSV-G protein. SF9 or Sf9VSV-G cells (upper labels) were infected with wt AcMNPV or vAc64− (lower labels) at an MOI of 10 and then fixed at 40 hpi and immunostained with an anti-VSV-G MAb (P5D4) and goat anti-mouse IgG fluorescein isothiocyanate conjugate. Cells were examined and photographed by epifluorescence microscopy.
Cell line generation and propagation.
To generate cells expressing VSV-G protein, Sf9 cells adapted to serum-free medium (ESF921; Expression Systems LLC) were plated in T75 flasks (7.5 × 106 cells per flask), and flasks were transfected with either (i) 2 μg of pSM8141-VSV-G plus 1 μg of pIE-neo (Novagen), (ii) 2 μg of pSM8141-VSV-G alone, or (iii) no DNA (mock transfected). After transfection, cells were incubated in ESF921 medium for 24 h and then resuspended, diluted 1:4, and replated in T75 flasks. ESF921 medium was replaced with ESF921 containing G418 (1 mg/ml). After 2 weeks, cells were monitored to confirm that all mock-transfected cells were dead. Small cell colonies that had grown from cells transfected with pSM8141-VSV-G plus pIE-neo were selected as single, well-isolated colonies and were picked using sterile micropipettor tips, transferred to individual wells of a 24-well dish, and cultured in ESF921 plus 5% fetal bovine serum (FBS). A cell line derived from one colony was selected and named Sf9VSV-G.
Cell lines Sf9, Sf9op1D (34), and Sf9VSV-G were propagated at 27°C in TNMFH medium containing 10% FBS (31). The wt AcMNPV virus used for these studies was AcMNPV strain E2, and the construction of the GP64-null virus, vAc64−, was described previously (30). Infectious vAc64− was generated in Sf9Op1D cells by infecting cells at a multiplicity of infection (MOI) of 1, which was followed by harvest of virus at approximately 3 days postinfection. The titer of vAc64− was determined on Sf9Op1D cells. Virus stocks of vAc64− were monitored for the presence of rescued virus containing the Orgyia pseudotsugata MNPV (OpMNPV) gp64 gene, by infecting Sf9 cells at a low MOI (approximately 10−2 to 10−4) followed by prolonged incubation and observation.
SDS-polyacrylamide gel electrophoresis and Western blot analysis.
Samples were prepared for western blot analysis in the following manner. Cell extracts from infected or uninfected cells were lysed in 1x Laemmli buffer (125 mM Tris, 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue, pH 6.8) and heated to 100°C for 5 min prior to electrophoresis. Virions of wt AcMNPV or pseudotyped GvAc64− BV were prepared from tissue culture supernatants by centrifugation at 80,000 × g for 75 min at 4°C through a 25% sucrose cushion in phosphate-buffered saline (PBS) (31) and subsequent resuspension of the pellet in 1 × Laemmli buffer. Samples were heated to 100°C for 5 min and subjected to SDS–10% polyacrylamide gel electrophoresis. Approximately 2.6 × 104 cells or 8 × 106 virions were electrophoresed in each lane. Gels were blotted onto Immobilon-P filters (Millipore) and incubated with the following primary monoclonal antibodies (MAbs): AcV5, an anti-GP64 MAb (17); MAb P10, an anti-VP39 MAb (a gift from JaRue Manning); or P5D4, an anti-VSV-G MAb (Sigma). The MAbs above were diluted 1:100, 1:1,000, and 1:100,000, respectively, in TBST (10 mM Tris [pH 8], 150 mM NaCl, 0.05% Tween 20) with 0.02% sodium azide. After washing, blots were incubated with a secondary antibody consisting of a goat anti-mouse immunoglobulin G (IgG)-alkaline phosphatase conjugate (Promega) at a dilution of 1:10,000. Western blots were processed as described earlier (2).
Immunofluorescence microscopy.
For immunofluorescence staining, 105 Sf9 or Sf9VSV-G cells were plated per well on two-well slides (Nunc Inc.), the cells were allowed to attach for 1 h, and then they were mock infected or infected with AcMNPV or vAc64− (MOI = 10) for 1 h. At 40 h post infection (hpi), cells were washed three times with 1 ml of PBS (pH 6.4) and then fixed in 100% methanol at −20°C for 10 min. Cells were then air dried 10 min and rehydrated in 300 μl of buffer A (5% filtered FBS, 0.1% saponin, 1X PBS) for 10 min. Cells were then incubated with anti-VSV-G antibody (P5D4, mouse ascites fluid; Sigma) (diluted 1:10,000 in buffer A) for 45 min at room temperature. After three washes with 300 μl of buffer A (10 min per wash), cells were incubated with a 1:100 dilution of goat anti-mouse IgG fluorescein isothiocyanate conjugate (Sigma) for 30 min at room temperature. Cells were washed four times with buffer A and then sealed in GelMount (Biomedia, Inc.) and viewed on an Olympus IX70 epifluorescence microscope.
Plaque assays, growth curves and TCID50 assays.
Plaque assays were performed in six-well plates, as previously described (31). Sf9, Sf9Op1D, and Sf9VSV-G cells were plated at 1.5 × 106 cells/well and, after a 1-h attachment period, were infected with vAc64− at several dilutions. Cells were monitored for infection and plaque formation over an 18-day period. At 10 or 18 days, each well was overlaid with neutral red (50 μg/ml) in 1% agarose. Growth curves were carried out by a modification of a previously described protocol (30). Sf9 cells were infected with AcMNPV, and Sf9Op1D and Sf9VSV-G cells were infected with vAc64− at an MOI of 5. After an initial 1-h infection period, cells were washed three times with TNMFH and supernatants were collected at the indicated time points. Data from each time point represent accumulated infectivity from infection through the indicated time. The titers of all supernatants were determined by 50% tissue culture infective dose (TCID50) assay on Sf9Op1D cells (31).
PCR analysis.
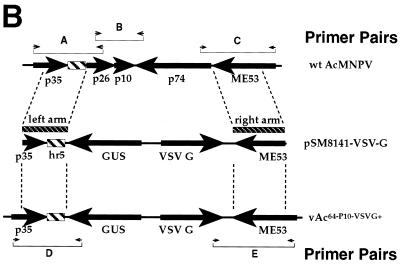
For preliminary PCR analysis, oligonucleotide primers specific to regions within the VSV-G, gp64, p35, or vp39 ORF were synthesized. Primer pairs were composed of oligonucleotides with the following nucleotide sequences: primer pair VSV-G, 5′-TCCGATCCTTCACTCCATCTG-3′ and 5′-TAGCTGAGATCCACTGGAGAG-3′; primer pair gp64, 5′-GTTGTTATTGGCTACAAGGGC-3′ and 5′-TGAGTAGAGCGTGGCGTTGAGC-3′; primer pair p35, 5′-CAGAATTCATGTGTGTAATTTTTCCGGTAG-3′ and 5′-AATGCTCTAGATTATTTAATTGTGTTTAATATTAC-3′; primer pair vp39, 5′-CGGGATCCAATGGCGCTAGTGCCCGTGGGTATGG-3′ and 5′-CGGGATCCGCGACGGCTATTCCTCCACCTGCTTC-3′. PCR amplification mixtures contained a 0.3 μM concentration of each primer; 0.3 mM (each) dATP, dGTP, and dTTP; 50 mM KCl; 10 mM Tris-HCl, pH 8.3; 1.5 mM MgCl; 0.1% Triton X-100; and 0.4 U of Taq DNA polymerase (Eppendorf) in a final volume of 20 μl. Reactions were subjected to 94°C for 3 min, followed by 3 cycles at an annealing temperature of 52°C and then 27 cycles at an annealing temp of 53°C where each cycle consists of denaturation at 94°C for 30 s, annealing at the prescribed temperature (above) for 40 s, and extension at 72°C for 1.5 min. The final extension was held at 72°C for 10 min. Products were electrophoresed on 1% agarose gels and stained with ethidium bromide.
To amplify portions of the p10 locus from the genomes of GP64-null and pseudotyped GP64-null viruses, we used primer pairs that would amplify (i) fragments from only the intact wt p10 locus (see Fig. 6B, primer pairs A, B, and C) or (ii) portions of the VSV-G ORF and p10 locus if the VSV-G gene were integrated at the predicted site (see Fig. 6B, primer pairs D and E). Each 50-μl reaction mixture contained a 0.3 μM concentration of each primer, a 0.3 mM concentration of each deoxynucleoside triphosphate, 10 mM KCl, 10 mM (NH4)2S04, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, and 0.6 U of Vent DNA polymerase (New England Biolabs). Amplification reactions were held at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 53°C for 45 s, and 72°C for 5 min. Finally, reactions were held at 72°C for 5 min. Primer pairs consisted of oligonucleotides with the following nucleotide sequences: primer pair A, 5′-TGCGTGTTGAAGCCGGGATTTG-3′ and 5′-GTCCCGACAGCTGGGACGCCT-3′; Primer pair B, 5′-CGAATGGCTGTTACCGGTGACG-3′ and 5′-CTCGCTATACACTCGCATGGAG-3′; primer pair C, 5′-CGATGCATATGTATGGCATACC-3′ and 5′-GAGTTTGGGAACAAGTTTGAAGG-3′; primer pair D, 5′-TGCGTGTTGAAGCCGGGATTTG-3′ and 5′-GTGAAGAGTATCAGTGTGCATG-3′; primer pair E, 5′-GTAGAAGGTTGGTTCAGTAGTTG-3′ and 5′-GAGTTTGGGAACAAGTTTGAAGG-3′.
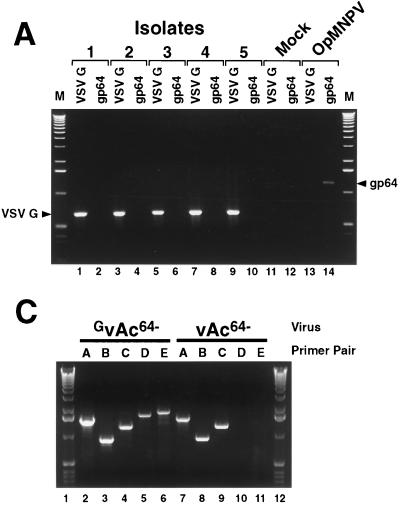
FIG. 6.
PCR analysis of virus preparations of GvAc64−. (A) PCR analysis of five GvAc64− virus preparations. Gene-specific primer pairs were used to examine DNAs from cell lysates of Sf9 cells infected with five GvAc64− virus preparations. DNA preparations were examined for the presence of the OpMNPV gp64 gene or the VSV-G gene. Isolation of individual preparations is described in the text and preparations are indicated as 1 to 5. The gene-specific primer pairs are indicated above individual lanes (VSV-G or gp64). (B) Strategy for PCR analysis of the p10 locus of AcMNPV in wt, vAc64−, and GvAc64− virus preparations. Large arrows show locations of AcMNPV ORFs in the p10 region (top line) or the predicted positions of the GUS and VSV-G ORFs integrated into the p10 region (bottom line). Small arrowheads show the locations of PCR primers on the wt AcMNPV genome (primer pairs A through C) and a predicted recombinant GvAc64− genome (vAc64−P10−VSVG+) (primer pairs D and E). Dashed lines indicate locations of the “arm” regions (potential recombination regions) present in the pSM8141-VSV-G plasmid. (C) PCR analysis of the p10 locus and the potential integration site of the VSV-G gene in vAc64− and GvAc64− virus preparations. DNAs from infected Sf9 cell lysates were used as templates for PCR analysis. Primer pairs specific for the wt AcMNPV p10 region in vAc64− (primer pairs A, B, and C) were compared with primer pairs specific for the predicted integration of VSV-G and GUS into the p10 region in viruses GvAc64− (primer pairs D and E).
Electron microscopy.
For transmission electron microscopy, virions were purified from cell culture supernatants and then fixed, embedded, sectioned, and stained. For each virus preparation (wt AcMNPV or GvAc64−), 33 ml of infected cell culture supernatants (representing 3 × 109 or 1 × 109 IU, respectively) was pelleted by centrifugation at 80,000 × g for 75 min at 4°C through a 25% sucrose cushion. The resulting virus pellet was fixed in 2.5% glutaraldehyde in 100 mM cacodylate buffer (pH 7.2) and then postfixed in 1.5% osmium tetroxide overnight at 4°C. After fixing, virions were dehydrated through a graded series of ethanol washes and embedded in Spurr's embedding medium (42). Ultrathin sections were stained by incubation for 5 to 30 min in 2% uranyl acetate in H2O, washed three times in distilled H2O (dH2O), stained for 5 min in Reynolds lead citrate, and washed five times in dH2O. Sections were examined at magnifications of ×15,000 and ×70,000 at 80 or 100 kV on a Phillips 201 transmission electron microscope.
RESULTS
A previous study (30) showed that the AcMNPV GP64 protein was necessary for efficient assembly and budding of virions from the surface of infected cells. In addition to its role in virion budding, GP64 also is necessary for virion entry and is involved in virion binding to host cells and low-pH-triggered membrane fusion during entry by endocytosis (2, 13, 22, 23, 28, 29, 34, 43). To begin to examine the requirements for virion assembly, budding, and infectivity, we asked whether the VSV-G protein was capable of substituting for the AcMNPV GP64 protein. To address this question, we generated a cell line that expresses the VSV-G protein, then infected that cell line with a GP64-null virus, vAc64−, and asked whether infectious virus progeny were generated.
Expression of VSV-G in Sf9VSV-G cells.
Because it was previously reported that expression of VSV-G is toxic in some cell lines (32), we used a strategy in which expression of VSV-G in insect Sf9 cells was dependent on infection with AcMNPV. A plasmid (pSM8141-VSV-G) containing the VSV-G gene under the control of an AcMNPV polyhedrin gene promoter (Fig. 1A) was constructed and cotransfected into Sf9 cells with the plasmid pIE1-neo, which contains the Escherichia coli neomycin phosphotransferase II gene under the control of the AcMNPV ie1 promoter. G418 was used to select and clone a cell line that was named Sf9VSV-G. To determine whether the VSV-G gene was stably inserted into the cell line and whether expression of G was inducible by infection with AcMNPV, we examined induced (infected) and uninduced (mock-infected) Sf9VSV-G cells by Western blot analysis and immunofluorescence microscopy (Fig. 1 B and C). Figure 1B shows a comparison of VSV-G expression in infected and mock-infected Sf9VSV-G cells. VSV-G protein was not detected in mock-infected cells but was detected in Sf9VSV-G cells infected with either wt AcMNPV or vAc64−. An antiserum directed against the AcMNPV major capsid protein (VP39) was used as an internal control to confirm infection (Fig. 1B [α-VP39]). Examination of VSV-G expression by immunofluorescence microscopy showed that VSV-G was detected from AcMNPV- or vAc64−-infected Sf9VSV-G cells but not from mock-infected Sf9VSV-G cells or Sf9 cells infected with wt AcMNPV (Fig. 1C). Immunofluorescent staining of infected cells was consistent with VSV-G presence at the periphery of infected cells, suggesting that G was likely transported to the surface of these cells. Thus, infection of cell line Sf9VSV-G with wt AcMNPV or vAc64− results in the induction of VSV-G protein expression, and G appears to be present at the surface of these cells.
Propagation and amplification of vAc64− in Sf9VSV-G cells.
To determine if the presence of VSV-G protein was sufficient to facilitate the production of infectious baculovirus in the absence of GP64, a GP64-null virus (vAc64−) containing no gp64 gene was used to infect Sf9VSV-G cells. Infected cells were examined for the capacity to propagate the GP64-null virus infection. In the experiments shown in Fig. 2A and B, three cell lines were infected with virus vAc64−. The cell lines included Sf9Op1D, an Sf9-derived cell line expressing the OpMNPV GP64 protein that was previously shown to complement vAc64− (29, 30); Sf9, a cell line that does not support propagation of vAc64−; and Sf9VSV-G, a cell line that inducibly expresses VSV-G protein. The GP64-null virus used for these experiments was propagated in Sf9Op1D cells as described previously (30). Therefore, the virus inoculum contained the OpMNPV GP64 protein in the virion envelope but no gp64 gene in the viral genome. Each cell line was infected with vAc64−, and plaque formation was examined over an extended time period. As expected, vAc64− infection of Sf9Op1D cells resulted in cell-to-cell movement of infection and abundant formation of plaques, as OpMNPV GP64 is known to effectively complement the AcMNPV gp64 deletion (Fig. 2A and B [Sf9Op1D]). In contrast, vAc64− infection of Sf9 cells resulted in an initial infection of single cells but the virus failed to propagate from cell to cell and did not form plaques (Fig. 2A and B [Sf9]). In vAc64−-infected Sf9VSV-G cells, we initially observed single infected cells, and plaques were not clearly visible at 5 to 7 days postinfection (dpi). However, upon further incubation, plaques were detected in Sf9VSV-G cells by 10 dpi and had expanded significantly by 16 to 18 dpi (Fig. 2A and B [Sf9VSV-G]). These observations suggested that the VSV-G protein was capable of complementing the defect in the GP64-null virus, vAc64−, but virus propagation appeared to be delayed. A similar delay was not observed when wt AcMNPV was used to infect Sf9VSV-G cells (data not shown), indicating that the Sf9VSV-G cells were not responsible for the delay. The formation of plaques by vAc64− in Sf9VSV-G cells suggested that the defects in both virion exit from the initial infected cell, and entry into neighboring cells, were complemented.
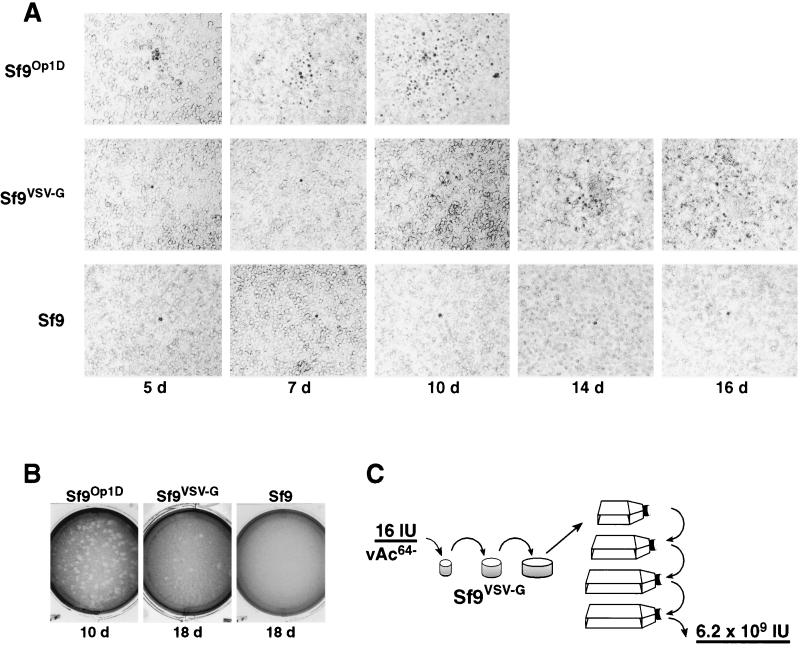
FIG. 2.
GP64-null virus propagation in Sf9VSV-G cells. (A) A monolayer of cells (Sf9Op1D, Sf9VSV-G, or Sf9) was infected with vAc64− at an MOI of approximately 6 × 10−5, and infected cells were identified at the indicated intervals (5, 7, 10, 14, or 16 days). Infected cells were identified by the presence of occlusion bodies, and these infected cells appear as dark cells against the background of lighter cells. (B) Plaque formation in vAc64− infected Sf9Op1D, Sf9VSV-G, and Sf9 cell monolayers was examined after 10, 18, and 18 days, respectively. (C) Schematic of vAc64− propagation in Sf9VSV-G cells. Sf9VSV-G cells (7.2 × 105 cells) were infected with 16 IU of vAc64−, and cells and supernatants were passaged until all cells appeared to be infected (6 to 7 passages). Titration of the supernatant resulted in a final virus titer of 6.2 × 109 IU. Control cells (Sf9 and Sf9Op1D) were also infected in parallel (see Results).
To confirm that the GP64-null virus could be propagated and amplified in Sf9VSV-G cells, we performed the following experiment. Sf9, Sf9Op1D, or Sf9VSV-G cells were infected with vAc64− at an MOI of 2.2 × 10−5 (16 IU per 7.2 × 105 cells), and cells were incubated at 27°C until the cells were 90% confluent. The medium and cells from each well were then transferred into successively larger wells and then to T flasks. At each step, cells were transferred when they reached approximately 90% confluency (Fig. 2C). Passage of vAc64− in Sf9Op1D cells in this manner resulted in a rapid propagation of infection such that cell growth was arrested and all cells were infected after the third passage. This was expected since the OpMNPV GP64 protein expressed by the Sf9Op1D cells complements the absence of GP64 in virus vAc64−. Attempted passage of vAc64− in Sf9 cells in this manner resulted in no spread of infection. Although vAc64− appeared to propagate slowly in Sf9VSV-G cells, continued passage resulted in increasing numbers of infected cells until most cells were infected at passage six or seven. Supernatants were then harvested, and the titers of viruses were determined on Sf9Op1D cells, which are sensitive indicators of infection by the GP64-null virus. We measured 6.2 × 109 IU from the vAc64− virus passaged in Sf9VSV-G cells in the manner described above. Thus, the vAc64− virus was amplified approximately 3.9 × 108-fold in Sf9VSV-G cells in this experiment. From this point on, we will refer to the vAc64− that was amplified in Sf9VSV-G cells as pseudotyped GP64-null virus, or GvAc64−.
Pseudotyped virus growth curve.
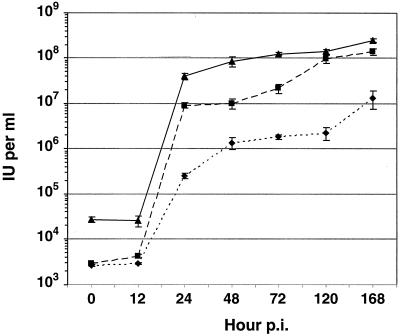
In our initial analysis of vAc64− virus propagation in Sf9VSV-G cells, we observed that plaque formation was significantly delayed compared with plaque formation by the same virus in Sf9Op1D cells. This could result from a delay in the infection cycle, low virus yields, lowered infectivity of the pseudotyped virus, or some combination of these factors. To examine the kinetics of virion production in vAc64−-infected Sf9VSV-G cells, we generated a one-step growth curve of infectious virus production and compared that curve to similar curves generated from wt AcMNPV-infected Sf9 cells and vAc64−-infected Sf9Op1D cells. Because the infectivity of virions carrying VSV-G protein may differ from those carrying GP64 and because the observed propagation of viruses in G-expressing cells was delayed, the titers of all virus samples collected from growth curve experiments were determined simultaneously by TCID50 on Sf9Op1D cells. The one step growth curves are compared in Fig. 3. Each cell line (Sf9, Sf9VSV-G, or Sf9Op1D) was infected at an MOI of 5 with either wt AcMNPV or vAc64−, and supernatants were harvested at the indicated times postinfection. The temporal kinetics of the growth curves of all viruses were similar (Fig. 3), although peak virion production of the G pseudotyped vAc64− appeared to lag behind that of wt-AcMNPV-infected Sf9 cells and vAc64−-infected Sf9Op1D cells. For the two control infections (AcMNPV-infected Sf9 cells or vAc64−-infected Sf9Op1D cells), titers of ≥107 IU/ml were observed by 24 to 48 hpi. In contrast, vAc64−-infected Sf9VSV-G cells produced titers in the range of 105 IU/ml at 24 h and approximately 106 IU/ml by 48 h. Titers of the pseudotyped virus increased to 107 IU/ml at 168 hpi. AcMNPV-infected Sf9 cells and vAc64−-infected Sf9Op1D cells generated titers of approximately 108 IU/ml by 72 to 120 hpi. Thus, while the kinetics of virus production were generally similar, the production of infectious virus particles that were pseudotyped with VSV-G protein lagged slightly behind that of wt AcMNPV. Final yields of infectious pseudotyped virus were reduced by at least 1 log and therefore represented approximately 10% of the final yield from wt AcMNPV.
FIG. 3.
One-step growth curves. Growth curves are plotted for the GP64-null virus (vAc64−) in cells expressing VSV-G (Sf9VSV-G) (⧫) or OpMNPV GP64 (Sf9Op1D) (■). For comparison, a growth curve of wt AcMNPV infected Sf9 cells was generated in parallel and is also plotted (▴). Cells were infected and supernatants were collected at the indicated times postinfection, and virus yields were determined as TCID50 values on Sf9Op1D cells. Each data point represents three individual infections, and error bars represent standard error.
Western blot analysis of infected cells.
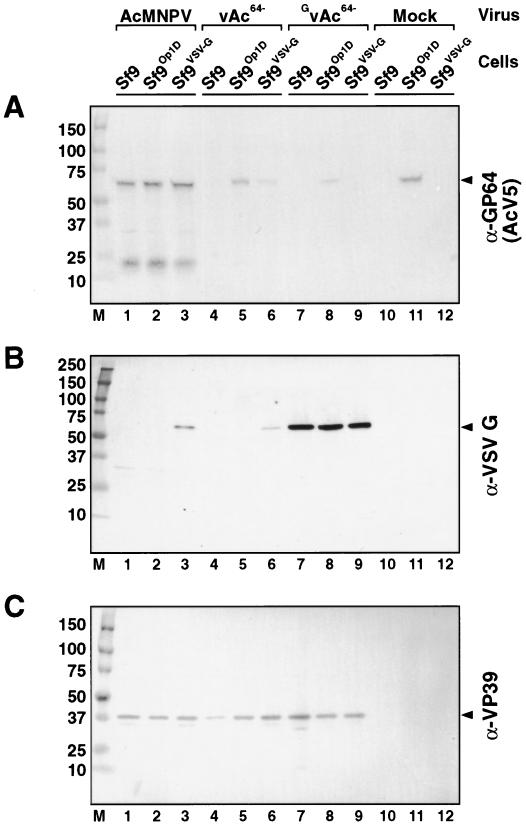
To confirm that the amplified virus (GvAc64−) did not result from contamination with wt AcMNPV or a GP64-null virus that had acquired the OpMNPV gp64 gene during prior propagation in Sf9Op1D cells, we used Western blot analysis to examine cells infected with either wt AcMNPV, vAc64−, or GvAc64− (Fig. 4). The GP64 protein was detected from cells infected with wt AcMNPV and from virus infections in Sf9Op1D cells, which constitutively express OpMNPV GP64 (Fig. 4A, lanes 1 to 3, 5, 8, and 11). In addition, a weak GP64 signal was frequently observed from cells infected with vAc64−. Because vAc64− was previously passaged in Sf9Op1D cells and carries wt OpMNPV GP64 in the envelope, low levels of GP64 detected from these samples can result from GP64 carried in with the inoculum virus (Fig. 4A, lanes 4 and 6). However, GP64 was not detected from Sf9 or Sf9VSV-G cells infected with GvAc64− (Fig. 4A, lanes 7 and 9). VSV-G was detected in all infected Sf9VSV-G cells as expected (Fig. 4B, lanes 3, 6, and 9). Interestingly, a strong VSV-G signal was detected in extracts from all cells infected with the pseudotyped virus, GvAc64− (Fig. 4B, lanes 7 to 9). One possible explanation for this result was that some of the GvAc64− virus may have acquired the VSV-G gene during passage through the Sf9VSV-G cells and thus was expressing G protein from the virus genome. This possibility was addressed in detail in subsequent experiments. In the present experiments, we found that GP64 was not detected in Sf9 or Sf9VSV-G cells infected with the G-pseudotyped virus, GvAc64−. Thus, the observed propagation of vAc64− in Sf9VSV-G cells was not due to contamination with a virus expressing GP64. These data confirm that the GP64-null virus (vAc64−) can be propagated in G-expressing cells in the absence of GP64.
FIG. 4.
Western blot analysis of GP64, VSV-G, and VP39 proteins in infected cell lysates. Cells were infected with viruses at an MOI of 1, and lysates were harvested at 75 hpi. The viruses and cells used for each infection are indicated above panel A. Western blots were treated with either anti-GP64 MAb AcV5 (A), anti-VSV-G MAb P5D4 (B), or anti-VP39 MAb P10 (C).
Biochemical and genetic analysis of GvAc64− BV.
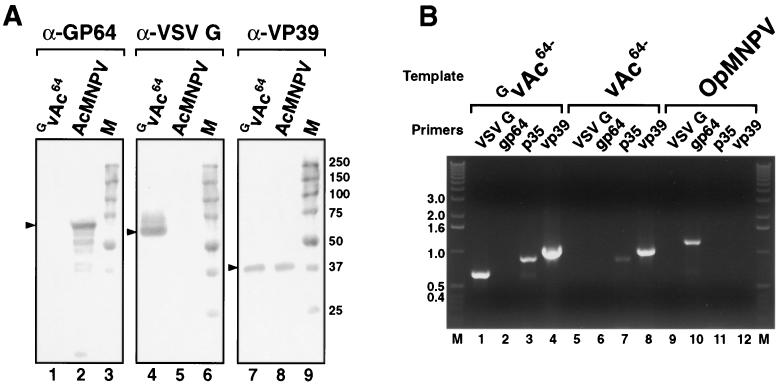
Virus particles pseudotyped with VSV-G protein were examined biochemically for the presence of the G protein. GvAc64− virions were prepared from supernatants of vAc64− infected Sf9VSV-G cells after multiple passages in Sf9VSV-G cells and then examined by Western blot analysis. As a comparison, wt AcMNPV virions were examined in parallel. Preparations of GvAc64− virions contained substantial quantities of G, but GP64 was not detected (Fig. 5A). As expected, G protein was not detected in wt AcMNPV preparations produced in Sf9 cells. The major capsid protein, VP39, was detected in both wt AcMNPV, and GvAc64− preparations.
FIG. 5.
Western blot and PCR analysis of virion preparations of wt AcMNPV, vAc64−, and GvAc64−. (A) Western blot analysis of GP64, VSV-G, and VP39 proteins in wt AcMNPV and GvAc64− virion preparations. Virus preparation GvAc64− represents the GP64-null virus (vAc64−) passaged in VSV-G expressing cells (Sf9VSV-G). Specificities of MAbs are indicated at the top (α-GP64, α-VSV-G, and α-VP39), and virion preparations are indicated above each lane (GvAc64− and AcMNPV). Marker lanes (M) are also indicated, and sizes of protein molecular weight standards (in thousands) are indicated on the right. (B) PCR analysis of viral DNAs isolated from purified virions of GvAc64− and vAc64−. Template viral DNA is indicated at the top, and gene specificities of primer pairs are indicated above individual lanes. Primer pairs were specific for VSV-G, OpMNPV gp64, AcMNPV p35, and AcMNPV vp39 genes. Lane M contains DNA size markers, and sizes (in kilobase pairs) are indicated on the left.
Because VSV-G was detected at relatively high levels in cells infected with GvAc64− (Fig. 4B) and VSV-G was also abundant in virion preparations (Fig. 5A), it was possible that the VSV-G gene had been acquired by the vAc64− virus through homologous recombination. We therefore asked whether the VSV-G gene could be identified in DNA isolated from GvAc64− virions prepared after passage in Sf9VSV-G cells (Fig. 5B). DNA prepared from virions of GvAc64−, vAc64− (passaged in Sf9Op1D cells), or OpMNPV were used with a series of oligonucleotide primers to amplify portions of the VSV-G ORF, the OpMNPV gp64 ORF, and the AcMNPV p35 and vp39 ORFs. Primers specific for portions of the AcMNPV p35 and vp39 ORFs were included as positive controls for AcMNPV genes, and the OpMNPV gp64 ORF-specific primers were included as a control to confirm that viruses had not acquired the gp64 gene during propagation in Sf9Op1D cells. As expected, p35- and vp39-specific primers amplified the expected PCR products (900 and 1,044 bp, respectively) from GvAc64− and vAc64− DNAs but not from OpMNPV DNA (Fig. 5B). The OpMNPV gp64-specific primers amplified the appropriate (1,259-bp) fragment from only the OpMNPV DNA, indicating that the OpMNPV GP64 gene was not detected in GvAc64− and vAc64− DNAs. Using VSV-G-specific primers, a 673-bp fragment was amplified from GvAc64− virion DNA, but not from vAc64− virion DNA. These data suggested that GvAc64− virions had acquired the VSV-G gene during propagation in the Sf9VSV-G cells.
If the GvAc64− virus had acquired the G gene, this virus should no longer have required the Sf9VSV-G cells for propagation. To determine whether acquisition of the G gene would permit the GvAc64− virus to propagate independently of the Sf9VSV-G cell line, we infected Sf9 cells with a GvAc64− virus preparation that was passaged in Sf9VSV-G cells. This resulted in a spreading infection, and the virus was passaged twice in Sf9 cells (5 days per passage); then, several isolates were generated by limiting dilutions in Sf9 cells. G-specific primer pairs were used to examine DNA from infected cell lysates by PCR. Figure 6A shows that the VSV-G gene was present in five isolates generated in this manner, suggesting that each contained viruses with a copy of the VSV-G gene. Although these viruses were able to propagate in Sf9 cells, each isolate was negative for the OpMNPV gp64 gene (Fig. 6A), indicating that virus propagation in Sf9 cells was not due to contamination with a virus that had acquired the OpMNPV gp64 gene from Sf9Op1D cells.
Because the plasmid used to generate the Sf9VSV-G cell line was derived from a transfer vector plasmid that contained sequences flanking the AcMNPV p10 locus, we reasoned that acquisition of VSV-G by the vAc64− virus would most likely occur by homologous recombination at the p10 locus. We therefore used a PCR strategy to examine the p10 locus of viruses passaged first in Sf9VSV-G cells, then in Sf9 cells. Figure 6B shows the PCR strategy and Fig. 6C shows the results from one isolate compared with that from the parental vAc64− virus. As expected, the appropriate PCR products were identified from the parental virus, vAc64−, when primer pairs specific for the wt AcMNPV p10 locus were used (Fig. 6C, lanes 7 to 9). In addition, no PCR products were detected from the vAc64− template when we used primers specific for the predicted VSV-G insertion in the p10 locus (Fig. 6C, lanes 10 to 11). Interestingly, DNAs from GvAc64− that was passaged first through the Sf9VSV-G cells and then through Sf9 cells were positive for both sets of primers (Fig. 6B and C, primers A-C and D-E). This indicates that genotypes containing (i) a wt p10 locus and (ii) a VSV-G insertion in the p10 locus were both present in the preparation. Other similarly derived isolates also showed the same result, suggesting that these virus preparations likely contained mixtures of parental viruses (vAc64−) and recombinant viruses in which the G gene was inserted at the p10 locus. We might speculate that because these recombinant viruses abundantly express G protein they may serve as helper viruses for the defective vAc64− viruses containing no GP64 envelope protein. In summary, we found that the VSV-G protein was sufficient to complement the defect in the vAc64− (GP64-null) virus when G was provided by the cell line Sf9VSV-G. In addition, we detected integration of the VSV-G gene in the p10 locus of vAc64− in several virus preparations and found that these viruses were capable of independent propagation in SF9 cells.
GvAc64− virion morphology.
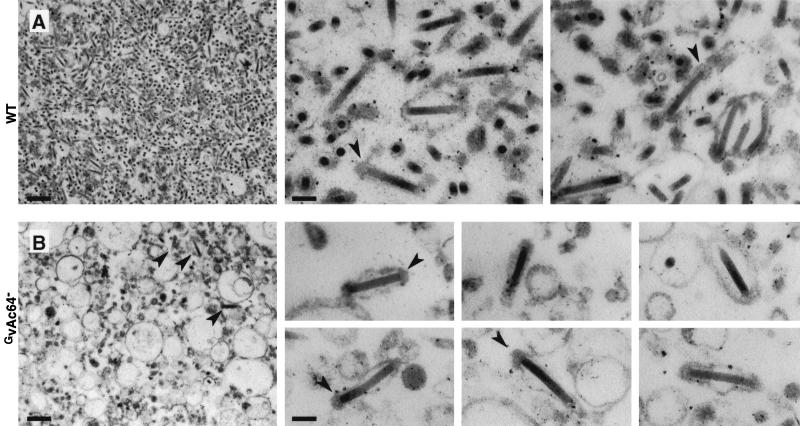
To determine if virions generated in the presence of the VSV-G protein and in the absence of GP64 were altered in morphology, we used transmission electron microscopy to compare GvAc64− virions with those from wt AcMNPV (Fig. 7). An obvious initial difference between preparations of wt AcMNPV and GvAc64− was the presence of numerous vesicles of various sizes (ranging from approximately 120 nm to 1 μm in diameter) in the GvAc64− preparation. Such vesicles may result from vesicle budding mediated by expression of the G protein. Vesiculation has been previously reported in mammalian cells expressing VSV-G protein (37, 44). Infectious virus titers were lower in the GvAc64− preparation, and virus particles were less numerous than in wt AcMNPV preparations. However, GvAc64− virions were clearly visible (Fig. 7B, left panel). wt AcMNPV BV consist of enveloped rod-shaped nucleocapsids. The envelope is typically composed of an apparently loosely adhering (lipid bilayer) membrane with a thickened or dense region in the membrane, near one end of the rod-shaped nucleocapsid (Fig. 7A, right panels). These characteristics were also typical of BV from the GvAc64− preparation (Fig. 7B, right panels). We did not typically observe nucleocapsids within enlarged or distended envelopes, and nucleocapsids within larger vesicles were not observed. Thus, although GvAc64− virions were less abundant, GvAc64− virions appeared to be similar in morphology to those from wt AcMNPV, and they were not morphologically distinguishable.
FIG. 7.
Electron micrographs of wt AcMNPV (A) and GvAc64− (B) virion preparations. Arrows indicate virions containing thickened regions near the termini. Bars on the panels at far left represent 500 nm, and bars on the rightmost panels represent 100 nm. Thin sections of virion preparations were stained with uranyl acetate and lead citrate and examined on a Philips 201 transmission electron microscope.
DISCUSSION
In previous studies, the roles of the GP64 protein in virion attachment, membrane fusion, and budding were examined. GP64 is involved in virion binding (13) and mediates low-pH-triggered membrane fusion during virion entry (2, 23, 43). A GP64 knockout virus was used to show that GP64 is also necessary for efficient virion assembly and budding during virion exit from the infected cell (29, 30). Although the absence of GP64 resulted in an approximately 98% reduction in virion budding, deletion of the CTD resulted in only an approximately 50% reduction in budding efficiency, suggesting that other portions of the GP64 protein play important roles in budding. In other enveloped viruses, the role of the major envelope protein in virion budding is highly variable. For example, retroviruses such as human immunodeficiency virus type 1 or Rous sarcoma virus do not require the envelope protein (Env) for virion budding, although virions generated in the absence of Env are not infectious. In contrast, envelope proteins from influenza viruses are believed to encode important functions necessary for efficient virion budding and envelope proteins also influence virion morphology. These important functions are thought to be redundant in the hemagglutinin (HA) and neuraminidase proteins of influenza A virus (19). Rhabdoviruses such as VSV and rabies virus require the major envelope protein (G protein) for efficient budding. In the absence of G, budding of VSV or rabies virus virions is reduced by approximately 97% (26, 40). Heterologous proteins substituted for G can partially complement virion budding in VSV and rabies virus (25, 40), and recent studies suggest that important signals necessary for efficient budding reside in nonspecific signals in the CTD (39). Efficient budding of VSV in the absence of intact G protein can be reconstituted by providing only a stem region containing the membrane-proximal 12 amino acids of the G protein ectodomain, combined with the transmembrane domain and CTD (36). The small stem region appears to be a functional budding domain necessary to promote efficient budding of VSV in the absence of the majority of the G protein.
One hypothesis to explain the synergistic roles of various proteins in the budding process is the push-pull model (26), in which the push represents the role of matrix and perhaps other proteins on the inner surface of the plasma membrane and the pull represents the role of the membrane proteins within and on the exterior of the membrane. Budding may be accomplished by the concerted or synergistic effects of the two components. While a very low level of budding may be observed in the absence of one component, efficient budding would require the activities of both components. As with VSV and rabies rhabdoviruses lacking G protein, very low levels of apparent budding were previously detected from the AcMNPV baculovirus in the absence of the GP64 protein. Thus, while GP64 appears to catalyze efficient virion assembly and budding at the cell surface, a low level of budding may occur in the absence of GP64 (31). Although the VSV-G protein was able to complement the deletion of GP64 in AcMNPV, the detection of infectious virions was 1 to 10% of that detected from wt AcMNPV that contains GP64 (Fig. 3), suggesting that the level of compatibility in these interactions may not be optimal. Factors that may effect complementation by VSV-G in this system include the timing of VSV-G expression (VSV-G was expressed by the very late polyhedrin promoter), the efficiency of VSV-G interactions during virion assembly and budding, and/or effects of VSV-G on infectivity of the pseudotyped virions.
In a previous study, VSV-G protein was expressed in a baculovirus in the presence of wt GP64. In that study, virions with an altered morphology were reported and these altered virions were described as containing an oval envelope and sometimes containing tail-like projections (1). Examination of virions generated from the G-pseudotyped GP64-null virus generated in this study showed clearly that virions were similar in morphology to wt AcMNPV virions. Nucleocapsids were not observed within vesicles nor within virions that appeared as oval-shaped particles with tail-like structures as reported earlier. A possible explanation for the observed differences is that interactions between GP64 and VSV-G proteins within the membrane might cause the previously reported aberrant virion morphology.
In the present study, we asked whether the VSV-G protein could complement a deletion of GP64. Expression of VSV-G from a cell line was sufficient to complement the production of infectious virus particles that could be passaged in G-expressing cells. Although the levels of virions generated were substantially lower than those generated by wt AcMNPV, the success of complementation was underscored by the observation that homologous recombination between the GP64-null virus (vAc64−) and the VSV-G construct in Sf9VSV-G cells resulted in viruses that expressed the VSV-G protein and were able to propagate infection in Sf9 cells. The acquisition of G by the GP64-null AcMNPV virus poses interesting questions regarding the evolution of this virus family. GP64 is highly conserved among a number of baculoviruses (such as AcMNPV and OpMNPV) that are relatively closely related, yet a number of more distantly related baculoviruses possess an unrelated envelope protein that appears to serve as a functional homolog of GP64. The major BV envelope proteins from two of these more distantly related viruses, Spodoptera exigua MNPV (Se8) and Lymantria dispar MNPV (Ld130), are both envelope fusion proteins (18, 33) and thus serve at least one of the important functions of GP64. However, these proteins and homologs from Xestia c-nigrum granulovirus (XcGV) and Plutella xylostella granulovirus (P×GV) (11, 12) show a higher degree of divergence than that observed among GP64 proteins identified to date. It has therefore been proposed that GP64 may represent a more recent acquisition of an envelope glycoprotein in the Baculoviridae (18, 33). Several orthomyxoviruses contain an envelope protein, GP75, that is phylogenetically related to the baculovirus GP64 protein. The GP75 proteins have been identified from only a small subset of the orthomyxoviruses, and GP75 is not related to the HA proteins found in other orthomyxoviruses. Therefore, it is possible that the GP75 protein was also recently acquired by a member of the orthomyxovirus family. Alternatively, GP75 may represent an ancestral orthomyxovirus envelope gene that was subsequently replaced by HA in some orthomyxoviruses. In the current study, we show that in the presence of selection pressure, a heterologous envelope protein can be acquired through recombination with the host cell genome. Thus, the presence of GP64 in baculoviruses may represent the acquisition of a new envelope protein by either recombination with the host cell genome or during a mixed infection.
In a previous study, a baculovirus expressing the VSV-G protein was reported to have an enhanced ability to transduce mammalian cells (1). In that study, G was expressed in the presence of wt GP64, presumably generating virions with a mosaic of GP64 and G protein in the envelopes. G protein did not appear to interfere with the infectivity of the virus on insect cells but enhanced infectivity on mammalian cells. A potential problem with the utilization of baculovirus virions (BV) containing GP64 for mammalian cell transduction in vivo is the rapid detection of GP64 and inactivation of the virus by the complement system (10, 14). Baculoviruses pseudotyped with VSV-G may be more resistant to inactivation by complement than viruses containing GP64. Therefore, GP64-null viruses carrying the VSV-G protein could provide benefits for use of baculoviruses in vivo in mammalian gene therapy strategies.
In the present study we expressed VSV-G in the absence of GP64 and found that G complemented virion infectivity and possibly virion budding, although the efficiency of infectious virion production appears to be low. This represents the first example of pseudotyping baculovirus virions in the absence of the baculovirus GP64 protein. Such pseudotyped baculovirus virions may be useful for developing baculoviruses for potential gene therapy applications. It will be of interest to determine if these pseudotyped baculovirus particles are capable of enhanced infection of mammalian cells, as was demonstrated in the presence of GP64 (1). Our experiments show that G-pseudotyped GP64-null AcMNPV does not propagate in insect Sf9 cells as efficiently as wt AcMNPV, since plaque formation was substantially delayed in comparison to wt AcMNPV, or GP64-null AcMNPV complemented with OpMNPV GP64. However, it is not yet clear whether this delay results from decreased virion yields or from reduced infectivity of G-pseudotyped virions to Sf9 cells. Further studies will be necessary to address these issues.
ACKNOWLEDGMENTS
We thank F. M. Boyce for providing plasmid VSVG-BP95NOTSV, D. H. L. Bishop for providing the p10 locus transfer vector, and J. Manning and P. Faulkner for providing MAbs P10 and AcV5, respectively.
This work was supported by National Institutes of Health grant AI33657 and Boyce Thompson Institute project 1255–17.
REFERENCES
- 1.Barsoum J, Brown R, McKee M, Boyce F M. Efficient transduction of mammalian cells by a recombinant baculovirus having the vesicular stomatitis virus G glycoprotein. Hum Gene Ther. 1997;8:2011–2018. doi: 10.1089/hum.1997.8.17-2011. [DOI] [PubMed] [Google Scholar]
- 2.Blissard G W, Wenz J R. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH dependent membrane fusion. J Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 4.Boyce F M, Bucher N L R. Baculovirus-mediated gene transfer into mammalian cells. Proc Natl Acad Sci USA. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condreay J P, Witherspoon S M, Clay W C, Kost T A. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc Natl Acad Sci USA. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelhard E K, Kam-Morgan L N W, Washburn J O, Volkman L E. The insect tracheal system: a conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc Natl Acad Sci USA. 1994;91:3224–3227. doi: 10.1073/pnas.91.8.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federici B A. Baculovirus pathogenesis. New York, N.Y: L. K. Miller Plenum Press; 1997. pp. 33–60. [Google Scholar]
- 8.Flipsen J T M, Martens J W M, Van-Oers M M, Vlak J M, Van-Lent J W M. Passage of Autographa californica nuclear polyhedrosis virus through the midgut epithelium of Spodoptera exigua larvae. Virology. 1995;208:328–335. doi: 10.1006/viro.1995.1156. [DOI] [PubMed] [Google Scholar]
- 9.Granados R R, Lawler K A. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology. 1981;108:297–308. doi: 10.1016/0042-6822(81)90438-4. [DOI] [PubMed] [Google Scholar]
- 10.Gronowski A M, Hilbert D M, Sheehan K C, Garotta G, Schreiber R D. Baculovirus stimulates antiviral effects in mammalian cells. J Virol. 1999;73:9944–9951. doi: 10.1128/jvi.73.12.9944-9951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto Y, Hayakawa T, Ueno Y, Fujita T, Sano Y, Matsumoto K. Sequence analysis of the Plutella xylostella granulovirus genome. Virology. 2000;275:358–372. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa T, Ko R, Okano K, Seong S I, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 13.Hefferon K, Oomens A, Monsma S, Finnerty C, Blissard G. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology. 1999;258:455–468. doi: 10.1006/viro.1999.9758. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann C, Huser A, Lehnert W, Strauss M. Protection of baculovirus-vectors against complement-mediated inactivation by recombinant soluble complement receptor type 1. Biol Chem. 1999;380:393–395. doi: 10.1515/BC.1999.052. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci USA. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann C, Strauss M. Baculovirus-mediated gene transfer in the presence of human serum or blood facilitated by inhibition of the complement system. Gene Ther. 1998;5:531–536. doi: 10.1038/sj.gt.3300607. [DOI] [PubMed] [Google Scholar]
- 17.Hohmann A W, Faulkner P. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology. 1983;125:432–444. doi: 10.1016/0042-6822(83)90214-3. [DOI] [PubMed] [Google Scholar]
- 18.IJkel W F J, Westenberg M, Goldbach R W, Blissard G W, Vlak J M, Zuidema D. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology. 2000;275:30–41. doi: 10.1006/viro.2000.0483. [DOI] [PubMed] [Google Scholar]
- 19.Jin H, Leser G P, Zhang J, Lamb R A. The influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keddie B A, Aponte G W, Volkman L E. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science. 1989;243:1728–1730. doi: 10.1126/science.2648574. [DOI] [PubMed] [Google Scholar]
- 21.Keddie B A, Volkman L E. Infectivity difference between the two phenotypes of Autographa californica nuclear polyhedrosis virus: importance of the 64K envelope glycoprotein. J Gen Virol. 1985;66:1195–1200. [Google Scholar]
- 22.Kingsley D H, Behbahani A, Rashtian A, Blissard G W, Zimmerberg J. A discrete stage of baculovirus GP64-mediated membrane fusion. Mol Biol Cell. 1999;10:4191–4200. doi: 10.1091/mbc.10.12.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leikina E, Onaran H O, Zimmerberg J. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 1992;304:221–224. doi: 10.1016/0014-5793(92)80623-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markovic I, Pulyaeva H, Sokoloff A, Chernomordik L V. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J Cell Biol. 1998;143:1155–1166. doi: 10.1083/jcb.143.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mebatsion T, Finke S, Weiland F, Conzelmann K. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 26.Mebatsion T, Konig M, Conzelmann K K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 27.Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. [Google Scholar]
- 28.Monsma S A, Blissard G W. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J Virol. 1995;69:2583–2595. doi: 10.1128/jvi.69.4.2583-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monsma S A, Oomens A G P, Blissard G W. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol. 1996;70:4607–4616. doi: 10.1128/jvi.70.7.4607-4616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oomens A G P, Blissard G W. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology. 1999;254:297–314. doi: 10.1006/viro.1998.9523. [DOI] [PubMed] [Google Scholar]
- 31.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors, a laboratory manual. W. H. New York, N.Y: Freeman and Co.; 1992. [Google Scholar]
- 32.Ory D S, Neugeboren B A, Mulligan R C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson M N, Groten C, Rohrmann G F. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J Virol. 2000;74:6126–6131. doi: 10.1128/jvi.74.13.6126-6131.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plonsky I, Cho M S, Oomens A G P, Blissard G W, Zimmerberg J. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology. 1999;253:65–76. doi: 10.1006/viro.1998.9493. [DOI] [PubMed] [Google Scholar]
- 35.Plonsky I, Zimmerberg J. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J Cell Biol. 1996;135:1831–1839. doi: 10.1083/jcb.135.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robison C S, Whitt M A. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J Virol. 2000;74:2239–2246. doi: 10.1128/jvi.74.5.2239-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolls M M, Webster P, Balba N H, Rose J K. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 38.Sandig V, Hofmann C, Steinert S, Jennings G, Schlag P, Strauss M. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum Gene Ther. 1996;7:1937–1945. doi: 10.1089/hum.1996.7.16-1937. [DOI] [PubMed] [Google Scholar]
- 39.Schnell M J, Buonocore L, Boritz E, Ghosh H P, Chernish R, Rose J K. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 41.Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I, Miyamura T, Matsuura Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J Gen Virol. 1997;78:2657–2664. doi: 10.1099/0022-1317-78-10-2657. [DOI] [PubMed] [Google Scholar]
- 42.Spurr A R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 43.Volkman L E, Goldsmith P A. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology. 1985;143:185–195. doi: 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 44.Whitt M A, Chong L, Rose J K. Glycoprotein cytoplasmic domain sequences required for rescue of a vesicular stomatitis virus glycoprotein mutant. J Virol. 1989;63:3569–3578. doi: 10.1128/jvi.63.9.3569-3578.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]