Abstract
Background
Co-occurring mutations in pairs of genes can pinpoint clinically relevant subgroups of cancer. Most colorectal cancers (CRCs) are microsatellite stable (MSS) and have few frequent mutations. Large patient cohorts and broad genomic coverage are needed for comprehensive co-mutation profiling.
Methods
Co-mutations were identified in a population-based Swedish cohort analyzed by whole-genome sequencing (n=819 stage I-IV MSS CRCs). Prognostic value was further evaluated in a publicly available dataset of clinically sequenced metastatic CRCs (MSK-IMPACT; n=934 MSS). Multivariable Cox proportional hazards analyses with clinicopathological parameters were performed for locoregional (stage I-III) and metastatic (stage IV and recurrent) cancers separately.
Results
Prevalent co-mutations were detected in 23 unique gene pairs, 20 of which included APC, TP53, KRAS and/or PIK3CA. Several co-mutations involving APC were associated with good overall survival in locoregional CRC, including APC-TCF7L2 (multivariable HR: 0.49, 95% CI 0.27-0.89). This co-mutation was prognostic also in metastatic cancers (multivariable HR: 0.49 and 0.37, 95% CI: 0.24-0.98 and 0.17-0.82 in the Swedish and MSK cohorts, respectively). APC-SOX9 co-mutations were mutually exclusive with APC-TCF7L2, and the co-mutations combined had stronger prognostic associations than APC alone in both metastatic cohorts. BRAF p.V600E-RNF43 co-mutations were associated with poor overall and recurrence-free survival in locoregional CRC (multivariable HR: 4.13 and 3.2, 95% CI: 1.78-9.54 and 1.53-8.04, respectively).
Conclusions
We report a genome-wide evaluation of co-occurring mutations in MSS CRCs, and suggest that co-mutations can improve the prognostic stratification compared to single mutations alone.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02173-x.
Keywords: CRC, Locoregional, Metastatic, MSS, Survival, Prognosis, Co-mutations
Introduction
Most colorectal cancers (CRCs) are microsatellite stable (MSS) and have a moderate mutation rate compared to other solid malignancies. Five to seven driver alterations have been estimated to be sufficient for cancer development [1]. Few CRC-critical mutations have clear prognostic implications, with the notable exception of the poor survival associated with BRAF p.V600E, and potentially with mutations of KRAS/NRAS (RAS) [2]. Mutations with lower prevalence can also have important prognostic impact, as illustrated with the pathogenic POLE exonuclease mutations found in approximately 1% of tumors [3]. Over the past 5 years there has been increased focus on subgroups of CRCs defined by co-occurring mutations of gene pairs. In particular, co-mutations of RAS and TP53 are associated with poor survival after liver resection of metastatic CRCs (mCRCs) [4]. Furthermore, co-mutations of BRAF p.V600E and RNF43 have been associated with improved benefit from BRAF-targeted combination therapy of mCRCs [5]. Most co-mutation studies have focused on genes typically covered by targeted sequencing panels and analyzed in a clinical setting. Broader genomic coverage and larger patient cohorts are needed for more systematic evaluation of co-occurring mutations and their prognostic implications.
Results and discussion
Patient cohorts and prognostic gene mutations
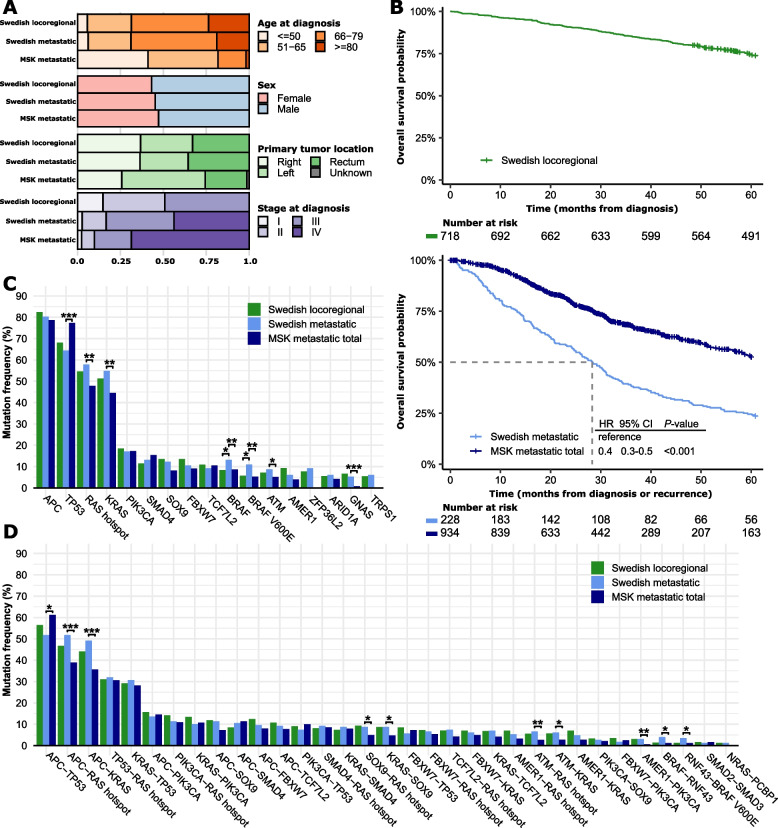
Co-mutation discovery focusing on nonsynonymous single nucleotide variants (SNVs) and small insertions and deletions (indels) was performed in a Swedish population-based series of stage I-IV MSS CRCs analyzed by whole-genome sequencing (n = 819) [6]. Survival analyses were performed separately for locoregional (n = 719 stages I-III) and metastatic cancers (n = 228; Fig. 1A and Supplementary Table 1) using overall survival (OS) as the primary endpoint, and recurrence-free survival (RFS) as secondary endpoint for locoregional cancers. The metastatic cohort included patients diagnosed with synchronous metastases (n = 100 stage IV, 44%) and recurrent cases from the locoregional cohort (n = 128). Additional prognostic analyses were performed in a publicly available single-hospital series of MSS mCRCs sequenced with the MSK-IMPACT gene panel (n = 934, 69% diagnosed with stage IV; Fig. 1A and Supplementary Table 1) [7]. The MSK metastatic cohort had younger patient age, less frequent right-sided primary tumors, and better OS than the Swedish metastatic cohort (Fig. 1A-B and Supplementary Table 1). Clinicopathological parameters with prognostic associations were included with the mutations in multivariable survival models (Supplementary Fig. 1). Metastasectomy status was a strong prognostic factor in both metastatic cohorts (Supplementary Fig. 1) and was included for stratified analyses of the MSK cohort (Supplementary Fig. 2A-B and Supplementary Table 2). The Swedish metastatic cohort was not sufficiently sized for stratified analyses, but there was no difference in the frequency of metastasectomy between the two cohorts (Supplementary Table 1).
Fig. 1.
Comparison of clinicopathological parameters and mutations among cohorts. A Clinicopathological characteristics of the Swedish locoregional (n = 719; 1 patient with missing survival information), Swedish metastatic (n = 228; 128 patients with metachronous metastases overlap with the locoregional cohort), and MSK (all patients, n = 934) cohorts. B Kaplan–Meier plots of overall survival in each cohort. C-D Bar plots of the frequency of single (C) and co-occurring (D) gene mutations in each cohort. Fisher exact analyses were performed between the cohorts and statistically significant differences are indicated by asterisks (***p < 0.001, **p < 0.01, and *p < 0.05)
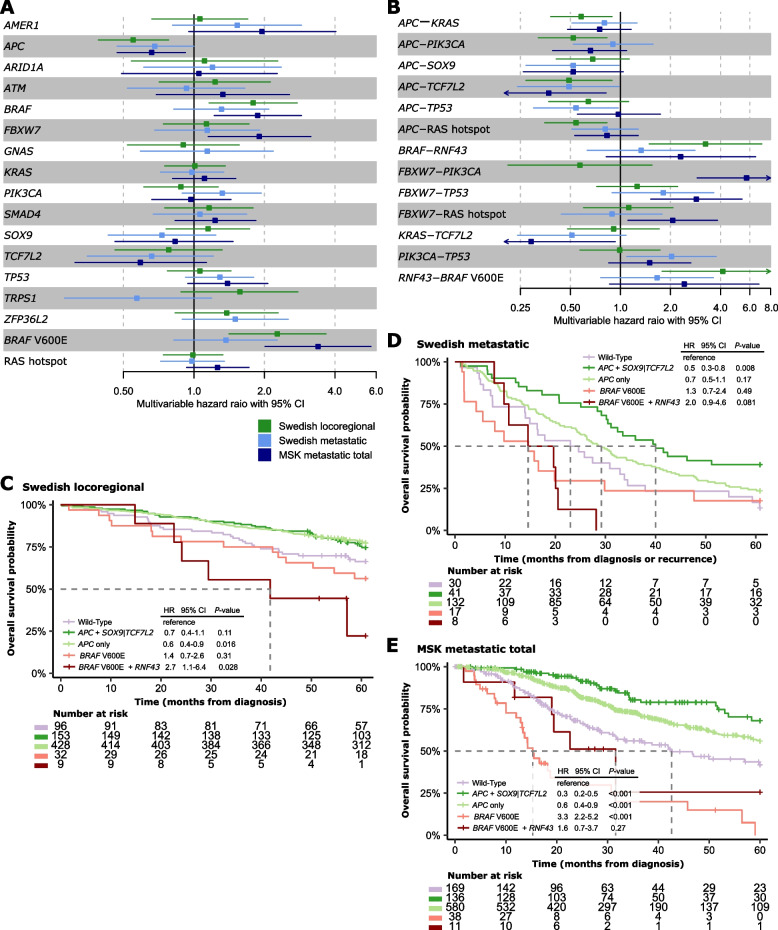
Among the most frequently mutated genes in the full Swedish cohort (n = 15 genes with SNVs or indels in ≥ 5% of tumors), only BRAF had a different mutation frequency between the locoregional and metastatic/recurrent cancers (BRAF: 8% vs 13%, p = 0.04; BRAF p.V600E: 6% vs 11%, p = 0.01; Fig. 1C). The Swedish metastatic cohort had more frequent ATM, BRAF, GNAS, and KRAS (including RAS hotspot) mutations and less frequent TP53 mutations than the MSK cohort. APC mutations were associated with good OS and BRAF p.V600E mutations with a poor OS among both locoregional and metastatic cancers (Fig. 2A and Supplementary Table 3). Results were similar for RFS as endpoint in the locoregional cohort (Supplementary Table 3). In a stratified analysis of the MSK cohort, several genes had prognostic associations in either the metastasectomy group (poor prognosis with FBXW7 mutations) or the no-metastasectomy group (poor prognosis with ATM and TP53 mutations and good prognosis with SOX9 mutations; Supplementary Fig. 2D). Among these, only FBXW7 mutations had prognostic associations in the total MSK cohort, and none were prognostic in the Swedish metastatic cohort.
Fig. 2.
Survival analyses of single mutations and co-occurring mutations in each cohort. A-B Forest plots for multivariable Cox proportional hazards models of overall survival according to each of the most frequent (A) single mutations and (B) co-mutations in each cohort. The reference group is cancers without mutations in the respective genes. Only co-mutations with significant prognostic associations (p < 0.05) in at least one cohort are shown. C-E Kaplan–Meier plots of overall survival according to selected co-mutations in each cohort. The wild-type group represents patients without mutations in any of the genes considered, and was used as reference group in statistical comparisons. Hazard ratios are from Cox proportional hazard analyses, and p-values from Wald’s tests. One patient in the Swedish locoregional cohort had missing survival information
The frequency of copy number amplifications (5 or more additional copies) and homozygous deletions (complete loss) of each of the 15 genes was lower than 6% in each cohort (Supplementary Fig. 3A-B). Incorporation of these copy number alterations had little impact on the prognostic associations of each gene, with the exception that mutation or deletion of SMAD4 was associated with a poor OS in the MSK cohort (Supplementary Table 4).
Co-mutations and prognostic associations
Co-mutations were defined as gene pairs with co-occurring SNVs or indels in at least 10% of tumors with mutation of each gene. A total of 33 co-mutations with prevalence above 5% were identified in the full Swedish cohort, of which 23 involved unique gene pairs (not counting hotspot mutations of RAS or BRAF p.V600E separately; Fig. 1D). The frequency of co-mutations corresponded largely with the frequency of the individually mutated genes, and only 3 co-mutations did not involve APC, TP53, KRAS and/or PIK3CA (BRAF-RNF43, NRAS-PCBP1, SMAD2-SMAD3). The frequency of co-mutations was similar between locoregional and metastatic cancers in the Swedish cohort, as well as between the Swedish metastatic and MSK metastatic cohorts, with the exception that the MSK cohort had more frequent APC-TP53 co-mutations and less frequent co-mutations of RAS hotspots with APC, SOX9 and ATM, as well as BRAF p.V600E-RNF43 and AMER1-PIK3CA.
Co-mutations previously reported to be prognostic in CRC were evaluated specifically, but neither RAS-TP53 [4], APC-PIK3CA [8] nor KRAS-PIK3CA [9] were associated with poor survival in any of the cohorts (Supplementary Table 5). This inconsistency might be related to more heterogeneous patient populations in our study, considering that the previous studies included patients with resectable liver metastases only [4, 8]. Furthermore, RNF43 mutations have been proposed to be associated with improved survival after BRAF-targeted combination therapy of BRAF p.V600E mCRCs [5]. In our study, BRAF p.V600E-RNF43 co-mutations were associated with poor OS and RFS among patients with locoregional cancer (multivariable HR: 4.13 and 3.2, 95% CI: 1.78–9.54 and 1.53–8.04, respectively; Fig. 2B), and had prognostic value also in univariable models of OS in both metastatic cohorts (Supplementary Table 5). However, there was no added prognostic effect of the co-mutation compared to BRAF p.V600E alone (Supplementary Fig. 4), consistent with the lack of a prognostic value of RNF43 in BRAF p.V600E mCRCs not receiving targeted treatment [5]. In the Swedish locoregional cohort, patients with the co-mutation had a numerically shorter median OS and RFS than patients with BRAF p.V600E alone (Fig. 2C and Supplementary Fig. 4A-B), suggesting a higher prognostic effect of the co-mutation in locoregional compared to metastatic MSS CRCs. However, the survival difference was not statistically significant, and the small sample size of the mutation subgroups precluded conclusions of added prognostic value. The lack of a locoregional validation cohort is a limitation of this study.
Consistent with the prognostic value of APC mutations alone, co-mutations of APC with KRAS, PIK3CA, TCF7L2 and RAS hotspots were associated with good OS among patients with locoregional cancer (Fig. 2B). The prognostic association was consistent among mCRCs for APC-TCF7L2 (multivariable HR: 0.49 and 0.37, 95% CI: 0.24–0.98 and 0.17–0.82 in the Swedish and MSK cohorts, respectively; Fig. 2B), but limited to patients not treated by metastasectomy in the MSK cohort (Supplementary Fig. 2E). Similarly, APC-SOX9 co-mutations were also associated with a good prognosis among mCRCs, and limited to patients not treated by metastasectomy in the MSK cohort (multivariable HR: 0.52 and 0.52, 95% CI: 0.27–0.98 and 0.26–1.04 in the Swedish and total MSK cohorts, respectively; Supplementary Fig. 2E). There was no difference in the frequency of APC-TCF7L2 co-mutations according to metastasectomy status in the MSK cohort, but APC-SOX9 co-mutations were less frequent in the cancers not treated by metastasectomy (Supplementary Fig. 2C).
Several co-mutations involving FBXW7 had poor-prognostic associations among mCRCs in the MSK cohort, but were either too rare for analysis or not prognostic in the Swedish metastatic cohort (Supplementary Table 5). Co-mutations of FBXW7 and PIK3CA were prognostic both among patients treated and not treated by metastasectomy, while co-mutations of FBXW7 and TP53 or RAS hotspots had significant prognostic associations only in the no-metastasectomy and metastasectomy cohorts, respectively (Supplementary Fig. 2E). Incorporation of gene amplifications and homozygous deletions (in addition to SNVs and indels) had little impact on the prognostic associations of co-mutated gene pairs (Supplementary Table 6). However, co-occurrence of mutation or deletion of SMAD4 with RAS hotspot mutations was associated with a poor survival in the MSK metastasectomy cohort (multivariable HR 4.0, 95% CI 1.7–9.4; Supplementary Fig. 3C-D).
Co-mutations can enhance the prognostic effect
Co-mutations involving APC had prognostic value beyond the effect of the individual genes in the metastatic cohorts (Fig. 2C-E). Co-mutations of APC with TCF7L2 or SOX9 were mutually exclusive, and APC-TCF7L2/SOX9 mutations combined were found in 18% and 15% of mCRCs in the Swedish and MSK cohorts, respectively (p = 0.22 by Fisher exact test). These patients had a better OS than patients with APC mutations alone (Swedish metastatic cohort HR: 0.65, 95% CI: 0.42–1, p = 0.05, and MSK cohort HR: 0.60, 95% CI: 0.39–0.91, p = 0.016). There were no differences in clinicopathological features between patients with co-mutations and APC mutations alone, but co-mutated cancers had a lower frequency of TP53 mutations (Supplementary Tables 7–9). The consistent prognostic value of the co-mutation in two metastatic cohorts with substantial differences in clinicopathological features support robustness. Both SOX9 and TCF7L2 are transcription factors involved in the WNT signaling pathway. This pathway is activated in most MSS CRCs due to inactivating mutations of APC and failure to sequester the transcriptional co-activator β-catenin. Active β-catenin binds to TCF7L2 and activates the transcription of WNT target genes, including SOX9 [10]. Overexpression of SOX9 inhibits the β-catenin-TCF7L2 complex to reduce WNT pathway activity [11]. SOX9 expression was higher in tumors with APC-SOX9 co-mutations in the Swedish cohort (Supplementary Fig. 5), and the positive prognostic effect of APC-TCF7L2/SOX9 co-mutations suggests that disruption of the balance between WNT pathway activation (due to loss of APC activity) and its modulation by TCF7L2 or SOX9 might partially reduce the oncogenic effects, leading to less aggressive cancers.
Neither APC mutations nor any of the co-mutations involving APC were prognostic in the MSK metastasectomy cohort separately (Supplementary Fig. 6A-B). However, the high OS rate and distribution of clinicopathological features suggest that this patient cohort is not population-representative (Supplementary Fig. 2A and Supplementary Table 2), and prognostic data should be interpreted with care. Nonetheless, co-mutations of FBXW7 with PIK3CA and/or RAS hotspots identified a small subgroup of patients in this cohort (4%) who had a poor survival, also compared to patients with PIK3CA and/or RAS hotspot mutations alone, supporting the potential of co-mutations to improve the prognostic stratification (Supplementary Fig. 6C-D). There were no differences in clinicopathological features or other mutations between patients with co-mutations and the individually mutated genes (Supplementary Table 10). It has also previously been reported that FBXW7 mutations predominantly occur alongside with KRAS mutations in advanced CRC [12], and both FBXW7 and RAS mutations have been associated with worse survival after liver resection for mCRC [13]. FBXW7 mutations can cause increased signaling in the EGFR pathway, similarly to RAS mutations [14], and the poor-prognostic effect of the co-mutations might therefore reflect enhanced oncogenic activity of the EGFR pathway.
Conclusion
We report genome-wide profiling of co-occurring mutations in MSS CRCs, and suggest that co-mutations can improve the prognostic stratification compared to single mutations alone. In particular, co-mutations of APC with TCF7L2 or SOX9 may identify a subgroup of metastatic cancers with favorable prognosis.
Supplementary Information
Abbreviations
- CI
Confidence interval
- CRC
Colorectal cancer
- HR
Hazard ratio
- mCRC
Metastatic colorectal cancer
- MSS
Microsatellite stable
- OS
Overall survival
- RFS
Recurrence free survival
Authors’ contributions
Conceptualization: LN, RAL, AS; Data acquisition and analysis: LN, AS; Data interpretation: all authors; Supervision: RAL, AS; Writing-original draft: LN, AS; Writing-review & editing: All authors reviewed the manuscript.
Funding
This study was funded by grants from the Research Council of Norway (project no. 287899 to AS), the South-Eastern Norway Regional Health Authority (project no. 2021025 funding JMS’s Ph.D. fellowship, project no. 2023101 to AS, and project no. 2021058 and 2024108 to RAL), the Norwegian Cancer Society (project no. 246954–2022 to AS and project no. 223319-2021to RAL), the Swedish Cancer Society (CAN 2018/772 and 21 1719 Pj to TS, and 22 2054 Pj 01H to BG), and the Uppsala Cancer Foundation to BG.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was conducted using publicly available data from two previously published studies: Nunes et al., Nature 2024 [6] and Yaeger et al., Cancer Cell 2018 [7]. Ethical approvals were obtained from the respective institutional review boards, and all participants provided informed consent, as described in the original publications. No new data was collected specifically for this study, and no additional ethical approval was required. All analyses complied with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17:11–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1:207–16. [DOI] [PubMed] [Google Scholar]
- 4.Chun YS, Passot G, Yamashita S, Nusrat M, Katsonis P, Loree JM, et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg. 2019;269:917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elez E, Ros J, Fernández J, Villacampa G, Moreno-Cárdenas AB, Arenillas C, et al. RNF43 mutations predict response to anti-BRAF/EGFR combinatory therapies in BRAFV600E metastatic colorectal cancer. Nat Med. 2022;28:2162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunes L, Li F, Wu M, Luo T, Hammarström K, Torell E, et al. Prognostic genome and transcriptome signatures in colorectal cancers. Nature. 2024;633:137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33:125-136.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita S, Chun Y-S, Kopetz SE, Maru D, Conrad C, Aloia TA, et al. APC and PIK3CA Mutational Cooperativity Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colorectal Liver Metastases. Ann Surg. 2020;272:1080–5. [DOI] [PubMed] [Google Scholar]
- 9.Luo Q, Chen D, Fan X, Fu X, Ma T, Chen D. KRAS and PIK3CA bi-mutations predict a poor prognosis in colorectal cancer patients: A single-site report. Transl Oncol. 2020;13: 100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of β-Catenin-Tcf Signaling in Colon Cancer by Mutations in β-Catenin or APC. Science. 1997;275:1787–90. [DOI] [PubMed] [Google Scholar]
- 11.Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jardim DL, Wheler JJ, Hess K, Tsimberidou AM, Zinner R, Janku F, et al. FBXW7 mutations in patients with advanced cancers: clinical and molecular characteristics and outcomes with mTOR inhibitors. PLoS ONE. 2014;9: e89388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y, Newhook TE, Tran Cao HS, Tzeng CWD, Chun YS, Aloia TA, et al. Alteration of FBXW7 is associated with worse survival in patients undergoing resection of colorectal liver metastases. J Gastrointest Surg. 2021;25:186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boretto M, Geurts MH, Gandhi S, Ma Z, Staliarova N, Celotti M, et al. Epidermal growth factor receptor (EGFR) is a target of the tumor-suppressor E3 ligase FBXW7. Proc Natl Acad Sci. 2024;121: e2309902121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.