Abstract
Background
Congenital heart disease (CHD) is a heterogeneous collection of structural abnormalities of the heart or great vessels that are present at birth. These birth defects are one of the leading causes of infant mortality and morbidity worldwide. The etiology and pathogenesis of CHD are unclear and largely considered to be multifactorial in nature. Since the chromosomal profile of CHD has not been analyzed in a large sample size, we aimed to summarize the clinical features, cytogenetics findings, and pregnancy outcomes of CHD to provide a clinical reference for prenatal diagnosis.
Methods
Among 21,152 pregnant women, 471 (2.23%) showed fetal CHD on cordocentesis or amniocentesis. The number of cases showing simple CHD, simple CHD with concomitant extracardiac structural abnormalities, complex CHD, and complex CHD with concomitant extracardiac structural abnormalities was 128, 124, 89, and 130, respectively. For prenatal genetic diagnosis, karyotyping was performed with single-nucleotide polymorphism array(SNP-array)-based chromosomal microarrays, fluorescence in situ hybridization (FISH), copy number variation sequencing (CNV-seq), and BACs-on-Beads™ (BoBs) analyses. The results of ultrasonography examinations, genetic analyses, and pregnancy outcomes were recorded in detail.
Results
Ventricular septal defects (VSDs) were observed in 245 (52.02%) cases of fetal CHD. Among the 471 cases of CHD, 258 (54.78%) showed other ultrasound abnormalities. The most common ultrasound abnormalities were abnormalities of the central nervous system. The 471 cases included 93 (19.75%) cases showing chromosomal abnormalities, and the incidence of these abnormalities increased with advanced maternal age or the presence of other ultrasound abnormalities. In eight cases, karyotype analysis showed normal results while SNP-array or CNV-seq results were abnormal. Among the 453 cases that were followed up, 166 (36.64%) involved pregnancy termination, 273 (60.26%) involved live births, 7 (1.55%) involved fetal death in utero, and 7 (1.55%) involved neonatal death after birth.
Conclusions
Fetuses with CHD showed higher rates of chromosomal abnormalities. In cases diagnosed with fetal CHD during fetal ultrasonic examination, the mothers should undergo a careful and comprehensive fetal ultrasound scan as well as prenatal genetic testing, including karyotype analysis and SNP-array or CNV-sequencing. The prognosis for simple fetal CHD is good, while the prognosis for complex fetal CHD and extracardiac anomalies is poor.
Keywords: Congenital heart defect, Copy number variation sequencing, Single-nucleotide polymorphism-based chromosomal microarrays, Prenatal diagnosis, Chromosome abnormality
Background
The term “congenital heart disease” (CHD) refers to a heterogeneous collection of structural abnormalities of the heart or great vessels that are present at birth. These birth defects are observed in more than 1 million live births worldwide and are responsible for 10% of stillbirths. The incidence of CHD in live births is approximately 5%-8%, making it the most frequently occurring birth defect [1, 2]. Due to its high morbidity, disability, and lethality, CHD poses a major threat to the health of mothers and infants. The pregnancy outcomes of fetuses with CHD are influenced by a variety of factors, including the type and severity of CHD and the presence of chromosomal abnormalities, which are the most important abnormalities associated with such cases.
The existing understanding of the etiology and pathogenesis of fetal CHD is incomplete. CHD is considered to be a condition influenced by complex genetic traits, with genetic and environmental factors playing major roles in disease development. Increased awareness of prenatal screening among pregnant women and advancements in testing methods have increased the detection rate of fetal CHD. In clinical settings, chromosomal karyotyping used to be the most common method for diagnosing chromosomal disorders prenatally, but this method has a long turnaround time and yields large segments, which can complicate timely or comprehensive diagnosis of chromosomal abnormalities. Advancements in molecular genetics technology have resulted in a constant influx of new prenatal diagnostic techniques and other new technologies, which can provide more accurate prenatal diagnoses for fetuses with normal karyotypes and abnormal ultrasound findings. To further elucidate the pathogenesis and prenatal diagnosis of CHD, the clinical data of 471 patients with CHD were summarized in this study.
Methods
During the period from January 2009 to August 2022, a total of 21,152 pregnant women underwent prenatal genetic diagnosis with amniocentesis or cordocentesis under ultrasound guidance at Hebei General Hospital. Cordocentesis or amniocentesis was performed in 471 pregnant women with fetal CHD. The diagnostic criteria for fetal CHD were based on the National Health Ministry’s birth defect map and the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD-10). Simple CHD refers to heart disease with a single defect, which generally does not cause hemodynamic changes. Complex CHD refers to one or more malformations of the intracardiac and/or macrovascular structure, which cause severe hemodynamic alterations.
On the basis of the complexity of the intracardiac malformations and the presence of concomitant extracardiac abnormalities, fetuses with CHD were classified into four groups: Group A, simple CHD without any concomitant extracardiac abnormalities; Group B, simple CHD combined with extracardiac abnormalities; Group C, complex CHD without concomitant extracardiac abnormalities; and Group D, complex CHD with concomitant extracardiac abnormalities. Groups A and B were categorized as simple fetal CHDs, while groups C and D were categorized as complex fetal CHDs.
The results of intracardiac and extracardiac ultrasonography examinations and genetic analysis of the investigated cases were also tabulated. Fetal ultrasonography using a 5-MHz transducer (IU22, Philips; VOLUSON E8, General Electric) was performed and evaluated by a team consisting of an obstetrician, an experienced sonographer, and a pediatric cardiologist specializing in the use of fetal echocardiography to detect fetal CHD. The prenatal ultrasound analysis was performed using the nine-segment sequential method, which covered nine sections: transverse section of the upper abdomen, four-chamber heart section, left ventricular outflow tract section, right ventricular outflow tract section, three-vascular tracheal section, bilateral subclavian-artery section, long-axis section of the superior and inferior vena cava, long-axis section of the aortic arch, and long-axis section of the ductus arteriosus. All samples underwent karyotyping, while only 91, 26, 37, and 13 samples underwent SNP-array, CNV-seq, BoBs, and FISH analyses.
For statistical analyses, we used the Statistical Package for Social Sciences Version 26.0 (SPSS, IBM, Chicago, IL, USA). Quantitative data were expressed as mean ± standard deviation or median (range) using the Kruskal–Wallis H test, while qualitative data were expressed as frequency (n) and percentage (%). Comparisons between groups were performed using the Chi-squared test (P = 0.05), The P value was corrected using Bonferroni correction (P, = 0.05/4 × (4–1)/2 = 0.0083). P, values were calculated using a two-sided test with 0.0083 as a statistically significant level (P, < 0.0083).
Results
A total of 471 pregnant women with fetal CHD underwent invasive sampling procedures, including 169 amniocenteses and 302 cordocenteses. The mean maternal age was 28.73 ± 4.80 years (range, 18–45 years), The mean maternal ages in groups A, B, C, and D were 27.73 ± 3.56 years, 30.40 ± 5.98 years, 27.97 ± 3.48 years, and 28.63 ± 4.99 years, respectively; the age data of the four groups did not conform to a normal distribution, and the differences were significant when evaluated using the Kruskal–Wallis H test (H = 15.118, P = 0.002). The median gestational age at ultrasound examination was 26.91 ± 1.96 weeks (range, 20–28 weeks).
A total of 23 CHDs were identified in this study, and the top 10 CHDs were, in order, ventricular septal defects (VSDs; 245 cases, 52.02%), right aortic arch (13 cases, 2.76%), tetralogy of Fallot (11 cases, 2.34%), Ebstein anomaly (9 cases, 1.91%), hypoplastic left heart syndrome (9 cases, 1.91%), endocardial cushion defects (5 cases, 1.06%), coronary sinus malformation (5 cases, 1.06%), cardiac tumor (4 cases, 0.85%), hypoplastic right heart syndrome (4 cases, 0.85%), and right and left pulmonary artery crossing (4 cases, 0.85%). The top 10 CHDs accounted for 65.60% of the cases. Among the 471 cases of CHDs, 254 (53.93%) showed abnormalities on ultrasound examinations. Central nervous system (CNS) ultrasound abnormalities accounted for 32.28% of the fetal CHD abnormalities. Among the non-CNS abnormalities, placenta and umbilical cord abnormalities accounted for 23.23% of the cases, followed by polyhydramnios and digestive abnormalities. Some patients also showed concomitant pathological ultrasound findings such as pyelectasis and abnormal limb development (Table 1). (Because this study mainly focused on the four groups of fetal congenital heart disease [A, B, C, and D], the cases were not subdivided on the basis of the type of ventricular septal defect (muscular or perimembranous. Nevertheless, the cytogenetics of muscular and perimembranous ventricular septal defects require further investigation).
Table 1.
Fetal CHD presenting with other ultrasound abnormalities
| Ultrasound abnormalities | Total | Rate/(%) |
|---|---|---|
| Central Nervous System | 82 | 32.28 |
| Ventriculomegaly | 23 | 9.06 |
| Choroid plexus cyst | 45 | 17.72 |
| Abnormal cavum septum pellucidum | 10 | 3.94 |
| Blake’s pouch cyst | 3 | 1.18 |
| Meningocele | 1 | 0.39 |
| Digestive system | 38 | 14.96 |
| Digestive tract obstruction | 20 | 7.87 |
| Small or undetected stomach vesicles | 11 | 4.33 |
| Gallbladder enlargement | 7 | 2.76 |
| Extremity | 17 | 6.69 |
| Abnormal posture | 15 | 5.90 |
| Shortness | 1 | 0.39 |
| Polydactyly | 1 | 0.39 |
| Abnormalities of the renal system | 29 | 11.42 |
| Hydronephrosis | 23 | 9.05 |
| Enlarged kidney | 6 | 2.36 |
| Face & neck | 6 | 2.36 |
| Neck cyst | 4 | 1.57 |
| Nasal bone dysplasia | 2 | 0.78 |
| Other | ||
| Single umbilical artery | 46 | 18.11 |
| Polyhydramnios | 39 | 15.35 |
| Nuchal translucency thickening | 4 | 1.57 |
| Intrauterine growth restriction | 2 | 0.78 |
| Umbilical hernia | 2 | 0.78 |
| Visceral version | 1 | 0.39 |
| Umbilical cord cyst | 13 | 5.12 |
| Herniation of the diaphragm | 1 | 0.39 |
Cases were counted twice if the same fetus showed ≥2 abnormal ultrasound indices; Ventriculomegaly: atrial width of the lateral ventricles was ≥10 mm; Choroid plexus cyst: bilateral or unilateral choroidal cysts greater than 1.0 cm in diameter from the choroid; Abnormal cavum septum pellucidum: A narrow cavum septum pellucidum (CSP) was defined as a CSP smaller by 2 standard deviations (SDs) than the mean width according to gestational age (width < (mean normal width - 2SD)). A wide CSP was defined as a CSP larger by 2 SDs than the mean width according to gestational age (width > (mean width + 2SD)). In addition, the CSP was considered to have an abnormal shape if its anteroposterior diameter was smaller than its width. If the measured value was greater than two times the SD of the reference range for the corresponding gestational age, then the gallbladder was identified as being enlarged
Chromosomal abnormalities were detected in 93 (19.75%) of the 471 cases of fetal CHD in this study. Groups A (simple fetal CHD), B (simple fetal CHD with concomitant extracardiac structural abnormalities), C (complex fetal CHD), and D (complex fetal CHD with concomitant extracardiac structural abnormalities) showed 11 (8.59%), 34 (27.42%), 9 (10.11%), and 39 (30%) cases of chromosomal abnormalities, respectively. In comparisons of groups A and B, B and C, and C and D, the differences were statistically significant. The remaining comparisons did not show significant differences (P > 0.083), as reported in Table 2.
Table 2.
Results of chromosome analyses of 471 fetal CHD cases
| Groups | Total | Abnormalresults | Frequency ofChromosomalabnormalities(%) |
|---|---|---|---|
| simple fetal CHD | 128 | 11 | 8.59% |
| simple fetal CHD with concomitant extracardiac structural abnormalities | 124 | 34* | 27.42% |
| complex fetal CHD | 89 | 9# | 10.11% |
| complex fetal CHD with concomitant extracardiac structural abnormalities | 130 | 39% | 30.00% |
*p = 0.00<0.0083 vs. simple fetal CHD; # p = 0.002 <0.0083 vs. simple CHD with concomitant extracardiac structural abnormalities;% p = 0.00 < 0.0083 vs. complex fetal CHD. The diagnostic criteria for fetal CHD were based on the National Health Ministry’s birth defect map and the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD-10). Simple CHD refers to heart disease with a single defect, which generally does not cause hemodynamic changes. Complex CHD refers to one or more malformations of the intracardiac and/or macrovascular structure, which cause severe hemodynamic alterations
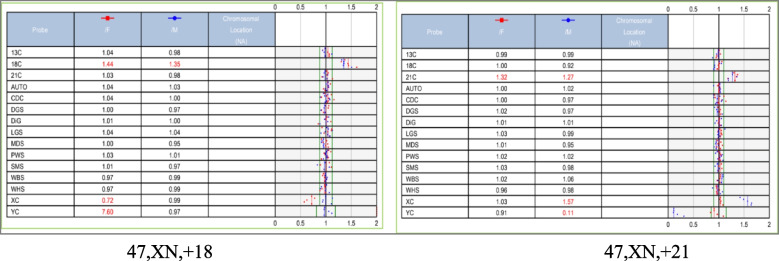
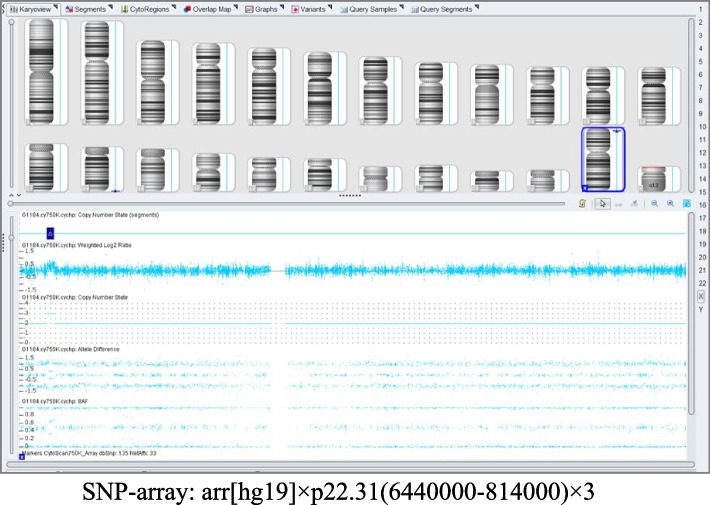
The karyotyping of the 471 fetuses revealed 85 abnormalities. This study identified 10 cases of structural chromosome abnormalities and 75 cases of aneuploidies, including 38 cases of trisomy 18, 26 cases of risomy 21, and three cases of trisomy 13. Four cases showed sex chromosome abnormalities, including two cases of 45,X, one case of 47,XYY, and one case of 47,XXX. One case showed a 48,XXX, + 18 sex chromosome and three cases showed chromosome chimerism: 47,XXY/46,XY, 45,X/46,XX, and 46,XY/46,XX. More details are provided in Table 3, and a part of the chromosomal karyotype is shown in Fig. 1. BoBs, FISH, CNV-seq, and SNP-array examinations were performed in 37, 13, 26, and 91 fetuses, respectively. Some of the BoBs results are plotted in Figs. 2 and one of the SNP-array results is plotted in Fig. 3. While the results of all BoBs and FISH examinations were consistent with the results of the karyotype analyses, only 87 of the 91 SNP-array examinations showed results consistent with the results of the karyotype analyses. In the remaining four cases, karyotype results were normal, but chromosomal SNP-array results indicated the presence of microdeletions in three fetuses and microduplications in one fetus (Table 4). Among of the 26 fetuses that underwent CNV-seq examination, 22 showed results consistent with the results of karyotype analysis. Among the remaining four fetuses, karyotype analyses indicated normal results, but chromosomal CNV-seq results indicated the presence of microdeletions (Table 5). SNP-array and CNV-seq analyses increased the detection rate of chromosomal abnormalities by 8.60%. The results of FISH, BoBs, SNP-array, and CNV-seq analyses are detailed in Table 6.
Table 3.
Results of chromosomal karyotyping analysis of fetal CHD
| Chromosome abnormalities | Total [n(1)(%)] | A [n(2)(%)] | B [n(3)(%)] | C [n(4)(%)] | D [n(5) (%)] |
|---|---|---|---|---|---|
| Numerical abnormalities | 75(15.92) | 9(7.03) | 30(24.19) | 5(5.62) | 31(23.85) |
| Trisomy-18 | 38(8.07) | 2(1.56) | 17(13.71) | 0 | 19(14.62) |
| Trisomy-21 | 26(5.52) | 4(3.13) | 11(8.87) | 2(2.25) | 9(6.92) |
| Trisomy-13 | 3(0.64) | 0 | 0 | 1(1.12) | 2(1.54) |
| Abnormal number of sex chromosomes | 4(0.85) | 2(1.56) | 0 | 1(1.12) | 1(0.76) |
| Abnormal numbers of sex chromosomes with autosomal staining. | 1(0.21) | 1(0.78) | 0 | 0 | 0 |
| Chromosome chimerism | 3(0.63) | 0 | 2(1.61) | 1(1.12) | 0 |
| Structural abnormalities | 10(2.12) | 0 | 4(3.23) | 2(2.25) | 4(3.08) |
| Total | 85(18.05) | 9(7.03) | 34(2.74) | 7(7.87) | 35(26.92) |
(1) total of 471; (2) total of 128; (3) total of 124; (4) total of 89; (5) total of 130. Group A, simple fetal CHD; Group B, simple fetal CHD with concomitant extracardiac structural abnormalities; Group C, complex fetal CHD; and Group D, complex fetal CHD with concomitant extracardiac structural abnormalities
Fig. 1.
Schematic of partial karyotypes
Fig. 2.
BOBs outcome Plot
Fig. 3.
SNP outcome Plot
Table 4.
Fetal CHD with normal karyotype and abnormal SNP-array results
| Age (years) | Gestation (week) | Ultrasound Findings | Area | Size | Type | Nature |
|---|---|---|---|---|---|---|
| 25 | 26+2 | Abnormal intraabdominal circumference of the umbilical vein; ductus venosus absent; enlarged right heart; widening of the inferior vena cava; intrahepatic strong echoes; enhanced intestinal wall echoes. | Chromosome 15q11.2 | 512.3Kb | Loss | Uncertain significance |
| 33 | 29+6 | Vascular Ring | Chromosome 22q11.21 | 2.88Mb | Loss | Uncertain significance |
| 24 | 30 | Increased right heart proportion; large tricuspid valve regurgitation; Mild mitral regurgitation;dextroposition | Chromosome1p36.33p36.32 | 2.387Mb1.972Mb | LossLoss | Pathogenic |
| 24 | 24 | Congenital complex cardiovascular abnormalities in the fetus (double right ventricular outlet; VSD; right aortic arch; enlarged right heart proportion) fetal hemivertebral body; right polycystic dysplastic kidney; Single Umbilical Artery; | Chromosome Xp22.31 | 1.695Mb | Duplication | Uncertain significance |
Table 5.
Fetal CHD with normal karyotype and abnormal CNV-seq
| Age (years) | Gestation (week) | Ultrasound Findings | Area | Size | CNVType | Nature |
|---|---|---|---|---|---|---|
| 19 | 27 | VSD; malproportion of main and pulmonary arteries; tricuspid mitral regurgitation | Chromosome1q21.1 | 0.38Mb | LOH | Uncertain significance |
| 29 | 28 | Bilateral aortic arch in the fetus; perpetual left superior cavity. | Chromosome16 p13.11 | 1.24Mb | Loss | Pathogenic |
| 30 | 22 | cysticlymphatic malformation; Widening of the coronary venous sinus; perpetual left superior cavity | Chromosome14q24.1q24.2 | 2.64Mb | Loss | Uncertain significance |
| 24 | 26+6 | Fetal dysplasia of the nasal bones; dextroposition | Chromosome21q11.2 | 2.34Mb | LOH | Pathogenic |
Table 6.
Results of chromosome microarray analyses of fetal CHD
| Chromosome microarray technology | SNP-array (n=91) | CNV-seq (n=26) | BoBs (n=37) | FISH (n=13) | ||||
|---|---|---|---|---|---|---|---|---|
| normal | abnormal | normal | abnormal | normal | abnormal | normal | abnormal | |
| A | 33(36.26) | 1(1.10) | 9(34.62) | 1(3.85) | 9 (24.32 ) | 0 | 1(7.69) | 1(7.69) |
| B | 15(16.48) | 2(2.20) | 3(11.54) | 1(3.85) | 10 (27.03 ) | 3(8.11) | 1(7.69) | 2(15.38) |
| C | 13(14.29) | 1(1.10) | 4(15.38) | 1(3.85) | 5(13.51) | 0 | 0 | 0 |
| D | 16(17.58) | 10(10.99) | 5(19.23) | 2(7.69) | 8(21.62) | 2(5.41) | 5(38.46) | 3(23.08) |
| Total | 77(84.62) | 14(15.38) | 21(80.77) | 5(19.23) | 32(86.49) | 5(13.51) | 7(53.85) | 6(46.15) |
Among the 471 pregnant women, 418 were aged < 35 years, of whom 77 (18.42%) had chromosomal abnormalities. Fifty-three women were aged ≥ 35 years, of whom 16 (30.19%) had chromosomal abnormalities. Older pregnant women showed a significantly higher incidence of chromosomal abnormalities than younger pregnant women (P < 0.05).
Among the 471 cases of fetal CHD, 453 (96.18%) were followed-up. Fetal CHD resulted in termination of 166 pregnancies (36.64%); among the pregnancies that were not terminated, seven (1.55%) ended in intrauterine death (Table 7), 280 (61.81%) were delivered as live births; and seven (1.54%) involved neonatal death after birth (Table 8). The rate of pregnancy terminations progressively increased from group A to D and was 12.20%, 37.07%,38.20%, and 59.20%, respectively, in these four groups (P < 0.0083for all differences). As shown in Table 9, the survival rates of fetuses in cases of continued pregnancy in groups A, B, C, and D were 96.30%, 98.63%, 90.91%, and 92.16%, respectively (Table 9). Thus, the survival rate of continued pregnancies was the highest in cases showing simple fetal CHD with extracardiac abnormalities. Tables 9 show that the pregnancy termination rate and continued pregnancy outcomes differed significantly among the four groups (χ2 = 59.164, P = 0.000; χ2 = 59.936, P = 0.000). Further two-by-two comparisons were performed using Bonferroni's method, and the corrected P’ = 0.0083, which indicated no statistically significant differences in the outcomes (The rate of pregnancy terminations and the survival rates of fetuses in cases of continued pregnancy) between groups B and C (χ2 = 0.028, P = 0.868; χ2 = 2.646, P = 0.104), while the differences between the remaining groups were statistically significant (P < 0.083).
Table 7.
Findings in cases involving intrauterine death
| Case | Ultrasound findings | Chromosomal karyotyping findings | Chromosome microarray findings | Outcome | Autopsy result |
|---|---|---|---|---|---|
| Case 1 | Vacancy | Normal | SNP-array: arr[hg19]1p36.33p36.32(849466-3236685)×1;arr[hg19]1p36.32p36.31(4536675-6328248)×1 | Cesarean delivery because of fetal distress at 34 weeks of gestation (birth weight, 2050 g); echocardiogram findings showed atrial defect; the infant died due to fetal asphyxia despite receiving resuscitation. | - |
| Case 2 | Widening of the lateral ventricles; loss of hyaline septum; VSD; small amount of tricuspid regurgitation | Normal | - | Cesarean delivery of one male infant after a full-term pregnancy (birth weight, 2000 g); the infant died due to fetal asphyxia despite receiving resuscitation. | Structural malformations of the heart and brain |
| Case 3 | VSD | Normal | SNP-array: normal findings | Female infant born at 36 weeks' gestation due to premature rupture of membranes (birth weight, 2500 g); the infant died due to congenital asphyxia despite receiving resuscitation. | - |
| Case 4 | Complex fetal CHD | Normal | BOBs: normal findings | Cesarean delivery of a female infant after a full-term pregnancy; the infant died of fetal asphyxia. | - |
| Case 5 | VSD; nasal bone dysplasia | Normal | SNP-array: normal findings | Male stillbirth due to HELLP syndrome. | - |
| Case 6 | VSD | Normal | - | Delivery of a female infant after a full-term pregnancy; the infant died due to fetal asphyxia despite receiving resuscitation. | - |
| Case 7 | Complex fetal CHD; ectopic kidney; single umbilical artery | Normal | - | Cesarean delivery of a female infant at 36+2 weeks of gestation; the infant died due to fetal asphyxia despite receiving resuscitation. | - |
Table 8.
Findings in cases involving neonatal death
| Case | Ultrasound findings | Chromosomal karyotyping findings | Chromosome microarray technology | Outcome | Autopsy result |
|---|---|---|---|---|---|
| Case 1 | VSD | Normal | - | A female infant was hospitalized for neonatal asphyxia after birth, and auxiliary tests showed ventricular defects and abnormal lung and bronchial development. She died after surviving for more than 1 month. | - |
| Case 2 | VSD | Normal | SNP-array:1seq[hg19]1q21.1(145380000-145760000)×1 | A male infant born at full term (birth weight, 2750 g) died of asphyxia after surviving for more than 1 month. | - |
| Case 3 | VSD; aortic constriction; choroid plexus cyst | Normal | - | A male infant (birth weight, 4000 g) was delivered by cesarean section after a full-term pregnancy and died of survived for 7 months after an echocardiogram indicated complex cardiac malformations. | - |
| Case 4 | VSD; left subclavian artery isolation; Right aortic arch | Normal | CNV-seq: normal | A female infant born at full term showed ultrasound evidence of complex congenital heart disease and esophageal dysplasia and died after surviving for more than a month. | -。 |
| Case 5 | Complex fetal CHD | Normal | - | A male infant was delivered preterm due to perinatal cardiomyopathy (birth weight, 2,000 g). The echocardiogram suggested complex congenital heart disease, and the infant died after surviving for 7 days. | - |
| Case 6 | Complex fetal CHD | Normal | - | A postnatal echocardiogram indicated cardiac disease and showed atrial agenesis, aortic duct failure, and pulmonary hypertension (primary type). The infant survived for 4 months and died without undergoing an autopsy. | - |
| Case 7 | VSD; excessive amniotic fluid | 46,XY/46,XX | SNP-array: arr(12)x2-3,(X)x1-2,(Y)x0-1 | A male infant was delivered by cesarean section at 35 weeks' gestation due to fetal distress. The infant showed muscle weakness, poor feeding, and respiratory distress and died 25 days after birth. | - |
Table 9.
Pregnancy outcomes by group
| Groups | Effective tracking | Termination | Intrauterine death | Dying | Survival | Continued pregnancy survival |
|---|---|---|---|---|---|---|
| A | 123 | 15(12.20%) | 1 | 3 | 104(84.55%) | 96.30%(104/108) |
| B | 116 | 43(37.07%)* | 1 | 0 | 72(62.07%)* | 98.63%(72/73) |
| C | 89 | 34(38.20%)* | 2 | 3 | 50(56.18%)* | 90.91%(50/55) |
| D | 125 | 74(59.20%)*#% | 3 | 1 | 47(37.60%)*#% | 92.16 %(47/51) |
*P’(*PBAT=0.000;*PCAT=0.000; *PDAT=0.000; *PBAS=0.000; *PCAS=0.000; *PDAS=0.000) < 0.0083 vs Group A; #P’(#PDBT=0.001;#PDBS=0.000)< 0.0083 vs Group B; %P’(%PDCT=0.002;%PDCS=0.007) < 0.0083 vs Group C
Discussion
CHDs are the most common congenital malformations [2, 3], and their prevalence varies widely in studies around the world; the best estimate is 8/1000 live births [1]. In this study, the prognosis for simple fetal CHD was good, while that for complex fetal CHD and extracardiac anomalies was poor. Fetuses with CHD showed higher rates of chromosomal abnormalities. Despite extensive advances in medical care, CHD remains a major cause of infant morbidity and mortality. An accurate prenatal diagnosis of the CHD subtype is essential for better clinical decision-making, including recommendations for termination of pregnancy, postnatal management, and early referrals to pediatric cardiology and cardiovascular surgery centers [4].
In this study, VSD was the most common type of fetal CHD. The ten most common CHDs accounted for 65.60% of the cases of CHD. The frequencies of these abnormalities in the present study were slightly different from those reported in previous studies. Because of the focus on comprehensive sonographic evaluation rather than on detecting a single deformity, we observed more cases of complex multi-system abnormalities in this study. Complex multi-system fetal anomalies can be rapidly diagnosed by systematic and specialized ultrasound examinations. Nevertheless, since our hospital provides prenatal diagnosis and consultation as a provincial referral center, most cases involved complex CHD, which may have influenced the statistical distribution, and the deeper reasons are subject to further and more in-depth study.
This study showed extracardiac abnormalities in 254 (53.93%) cases of fetal CHD were, of which neurologic ultrasound anomalies were the most common, followed by abnormalities in the placenta and umbilical cord (24.01%) and abnormalities in the amniotic fluid (15.35%). In one previous study, extracardiac anomalies were observed in 66% of fetuses with CHD, and these primarily affected the central nervous system (31%), the genitourinary system (26%), and the gastrointestinal system (24%) [5]. Other studies have reported that CHD with concomitant extracardiac defects is associated with fetal growth restriction, amniotic fluid anomalies, neurologic malformations, and urinary tract anomalies [6–9]. The findings of these studies and our study indicate a high incidence of fetuses showing congenital heart defects with concomitant extracardiac structural anomalies; while the types and proportions of fetuses with extracardiac structural anomalies were not entirely the same across studies, the findings still implied an increased incidence of amniotic fluid anomalies and neurological abnormalities. Thus, to avoid clinical underdiagnosis, pregnant women should undergo fetal echocardiography as soon as ultrasonography indicates neurological abnormalities or abnormalities in the fetal amniotic fluid.
The overall rate of chromosomal abnormalities in fetuses with CHD was higher in this study. Chromosomal abnormalities were detected more frequently in complex fetal CHD than in simple fetal CHD, but the difference was not statistically significant. However, the detection rate of chromosomal abnormalities differed significantly between the group showing simple fetal CHD with concomitant extracardiac abnormalities and the group showing complex fetal CHD. Thus, fetal chromosomal abnormalities do not significantly correlate with fetal CHD complexity. Analyzing the data, we found that fetuses showing cardiac structural abnormalities with concomitant extracardiac abnormalities had a significantly higher detection rate of chromosomal abnormalities than those without extracardiac abnormalities. Other studies have also reported similar results [10]. The presence of even a small segmental variant in one chromosome may affect multiple neighboring genes, influencing the development of multiple fetal systems [11]. Therefore, to avoid the birth of infants with defects, prompt prenatal genetic diagnosis is essential in cases showing fetal CHD with concomitant extracardiac abnormalities on ultrasonography.
Genetic and environmental factors are known to contribute to the etiology of fetal CHD, and chromosome number and structural abnormalities are known to play a role in 10%-15% of CHD cases [12]. For decades, traditional karyotyping has been the gold standard for prenatal diagnosis. The first documented genetic cause of CHD was also chromosomal aneuploidy [13]. One of the most common causes of fetal CHD is trisomy 21, followed by trisomy 18 [14]. Common CHDs in cases showing trisomy 21 include endocardial cushion defects, VSDs, atrial septal defects, and aortic arch anomalies. VSD, patent ductus arteriosus, atrial septal defects, and aortic constriction are common clinical manifestations of trisomy 18 [15]. The present study identified 75 cases of abnormal chromosome number, with an abnormality percentage of 15.92%; among these, trisomy 18 was the most common, accounting for 44.71% of all abnormal karyotypes and 40.86% of all abnormal genetic factors. According to the literature [15], the correlation between trisomy 18 and VSDs is high, and the present study showed a high percentage of VSDs. Therefore, the high incidence of trisomy 18 in the patient population in present study may be attributable to the high incidence of VSDs. Our study population also included eight cases (8.60%) of sex chromosome abnormalities. Two of them involved Turner syndrome, two showed sex chromosome triploids, one showed 48,XXX, + 18, and three were chimeric. Due to their specific structure and gene content, sex chromosomes are more prone to recombination errors than autosomes [16]. The clinical phenotypes of sex chromosomes are correlated with their chimeric ratios, which may indicate early sexual anomaly inactivation. The present study, however, did not investigate this issue due to the small sample size. Future studies with larger sample sizes should aim to address this issue. Fetal CHD is also associated with chromosomal structural abnormalities, such as recombination and chimerism. Such abnormalities were observed in 10.75% of the cases in this study. Fetal CHD is primarily caused by chromosome number abnormalities. Therefore, timely prenatal diagnosis should be performed if screening or ultrasound findings indicate chromosomal abnormalities.
Chromosomal karyotyping is the most classical prenatal diagnosis method. However, its long detection period and low resolution make it unsuitable for genetic counseling. Advancements in chromosome microarray technology have allowed the detection of microdeletions or microduplications of genomic segments [17] as well as chromosomal imbalance rearrangements, uniparental diploidy, and triploidy with this technique. Moreover, these techniques do not require cell culture and have a shorter detection cycle [18]. The wide range of phenotypes of CHD can be attributed to the presence of many structural abnormalities of the heart and genetic variants that contribute to this disease. These include the 22 q11.2 deletion syndrome, 1p36 deletion syndrome, Williams–Beuren syndrome (7q11.23 deletion syndrome), and WHS syndrome (4P syndrome) [19, 20]. The development of chromosome microarrays has increased the CHD detection rate by 5% [12, 21, 22], highlighting the importance of promoting advancements in chromosome microarray technology. These microdeletions and microduplications were not observed in the cases reported in the present study. In this study, eight fetuses showing normal karyotypes and abnormal SNP-array or CNV-seq results were identified using chromosomal microarray technology, increasing the detection rate of fetal CHD by 8.60%. CNV-seq or SNP-array technologies are suitable for first-line testing in most cases and have been shown to be synergistically effective. SNP-array techniques evaluate the genetic markers formed at the genomic level by DNA sequence polymorphisms caused as a result of variations in individual nucleotides. On the other hand, CNV-seq, which involves the detection of CNVs by low-level whole-genome sequencing, evaluates large segments of CNVs at the whole-genome level. Both CNV-seq and high-density SNP-array techniques can achieve 100% detection of CNVs that are known to be pathogenic. However, CNV-seq performs better than medium-density SNP-array techniques. Both techniques can detect chimeras, with CNV-seq showing a lower limit of detection (20%) than SNP-array techniques (30%). However, only SNP-array techniques can detect uniparental diploid (UPD) and heterozygous omission (LOH), [23]. However, these techniques cannot detect balanced chromosomal rearrangements, and karyotyping is required if such rearrangements are suspected. For detection of single-gene defects associated with CHD, whole-exome testing may be considered if the results of SNP-array or CNV-seq analyses are negative.
A family history of CHD, maternal disease (diabetes, collagen vascular disease), teratogenic exposure, viral infections (rubella virus, etc.), advanced maternal age, twin pregnancies, maternal obesity, maternal alcohol or drug use history, and in vitro fertilization all increase the risk of fetal CHD [24, 25]. As an independent risk factor, age is of considerable clinical significance. Pregnancies involving older women were significantly more likely to show chromosomal abnormalities than those involving younger women in this study. Thus, timely prenatal diagnosis in patients with CHD and advanced maternal age is essential to avoid serious birth defects in clinical practice.
The rates of complex and simple fetal CHD differed significantly in this study, and the rate of fetal CHD with concomitant extracardiac structural abnormalities was greater than that of fetal CHD without extracardiac anomalies. When choosing whether to terminate a pregnancy, the complexity of cardiac malformations as well as the presence of extracardiac anomalies are important factors worth consideration. A complex malformation cannot be surgically corrected and is associated with high morbidity and mortality rates, which may be an important factor in the family’s decision to terminate pregnancy. The survival rate in complex fetal CHD was significantly lower than that in simple fetal CHD in this study, consistent with the literature [26], suggesting that the severity of fetal malformation and the presence of concomitant extracardiac anomalies have a significant impact on the prognosis of the fetus. In this study, the survival rate for continuing pregnancies in Group B was significantly higher than those in Group A and Group D, which in turn were significantly higher than that in Group C. Thus, the survival rate for continuing pregnancies showing fetal CHD with concomitant extracardiac anomalies was significantly higher than that for pregnancies without fetal CHD with concomitant extracardiac structural anomalies. This may be attributed to the higher detection rate of chromosomal anomalies in this study as well as the timely termination of pregnancies with chromosomal abnormalities. Thus, fetal CHDs with timely prenatal diagnosis may have a higher survival rate, and timely prenatal diagnosis of fetal CHD can improve the survival rate of continuing pregnancy in CHD patients. Further evidence is required to confirm the importance of prenatal diagnosis in reducing birth defects and improving eugenics.
Conclusions
Fetal VSD was the most common type of fetal CHD. Other ultrasound abnormalities were commonly associated with fetal CHD, of which CNS abnormalities are the most common. Chromosomal abnormalities were common in fetuses with CHD, and trisomy 18 was the most common chromosomal abnormality. An improved detection rate for chromosomal abnormalities in fetal CHD was achieved by combining karyotype analyses with SNP-array or CNV-seq analysis. Chromosomal abnormalities are more likely to be detected in older pregnancies with CHD. Simple fetal CHD has a good prognosis, but complex CHD and combined extracardiac anomalies have a poor prognosis.
Acknowledgements
We would like to thank all the patients and family members participating in this work for their cooperation and patience. We also thank clinical geneticists and certified genetic counselors of Hebei General Hospital for their support.
Abbreviations
- BoBs
BACs on Beads™
- CHD
Congenital heart disease
- CNS
Central nervous system
- CNV
Copy number variation
- FISH
Fluorescence in situ hybridization
- SNP
Single-nucleotide polymorphism
- VSD
Ventricular septal defects
Authors’ contributions
Data interpretation: Yulu Quan, Yan Luo and Tao Wang. Methodology: Yulu Quan, and Juan li. Statistical analysis: Yulu Quan. Writing original draft: Yulu Quan and Yali Li. Writing review and editing: Pingping Zhang and Yali Li. All authors reviewed the drafts of the manuscript and approved the final version of the manuscript.
Funding
This work was supported by the Clinical Medical Personnel Training Programs subsidized by the Chinese government [grant number 2021040], the High-level Talent Funding Project of Hebei Province, China [grant number C20231018] and Key R&D Science and Technology Projects in Hebei Province [grant number22377792D].
Data availability
Data will be made available on request.
Declarations
Ethics approval and consent to participate
This study was approved by the Hebei General Hospital Ethics Committee and conducted in accordance with the ethical standards set forth by the 1964 Helsinki declaration and its later amendments. Informed consent was obtained from all participants.
Consent for publication
Individual informed consent was obtained from all subjects. Individual consent was specifically gathered from each participant, ensuring their understanding and willingness to participate. As our study does not include any details, images or videos about an individual person, consent for publication is not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pingping Zhang, Email: jumping.zpp@yeah.net.
Yali Li, Email: li_y_li@sina.com.
References
- 1.Van Der Linde D, Konings EEM, Slager MA, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–7. 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–900. 10.1016/s0735-1097(02)01886-7. (2002). [DOI] [PubMed] [Google Scholar]
- 3.Ozkutlu S, Akça T, Kafali G, et al. The results of fetal echocardiography in a tertiary center and comparison of low- and high-risk pregnancies for fetal congenital heart defects. Anadolu Kardiyol Derg. 2010;10(3):263–9. 10.5152/akd.2010.068. [DOI] [PubMed] [Google Scholar]
- 4.Bernier PL, Stefanescu A, Samoukovic G, et al. The challenge of congenital heart disease worldwide: epidemiologic and demographic facts. Semin Thoracic Cardiovasc Surg Pediatr Cardiac Surg Ann. 2010;13(1):26–34. 10.1053/j.pcsu.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Tennstedt C, Chaoui R, Körner H, et al. Spectrum of congenital heart defects and extracardiac malformations associated with chromosomal abnormalities: results of a seven year necropsy study. Heart. 1999;82(1):34–9. 10.1136/hrt.82.1.34. (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krampl E, Chalubinski K, Schatten C et al. Does acute hypoxia cause fetal arterial blood flow redistribution? Ultrasound Obstet Gynecol: Off J Int Soc Ultrasound Obstet Gynecol. 2001;18(2):175–177. 10.1046/j.1469-0705.2001.00501.x. [DOI] [PubMed]
- 7.Wilson RD, Chitayat D, McGillivray BC. Fetal ultrasound abnormalities: correlation with fetal karyotype, autopsy findings, and postnatal outcome–five-year prospective study. American J Med Genet. 1992;44(5):586–90. 10.1002/ajmg.1320440511. [DOI] [PubMed] [Google Scholar]
- 8.Van Nisselrooij AEL, Jansen FAR, Geloven NA, et al. Impact of extracardiac pathology on head growth in fetuses with congenital heart defect. Ultrasound Obstet Gynecol. 2020;55(2):217–25. 10.1002/uog.20260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oral O, Toprak MHH, Uysal F, et al. The frequency of asymptomatic urinary system abnormalities in children detected with cineurography imaging during angiocardiography. Cardiol Young. 2019;29(2):119–22. 10.1017/s1047951118001828. (2019). [DOI] [PubMed] [Google Scholar]
- 10.Mustafa HJ, Jacobs KM, Tessier KM, et al. Chromosomal microarray analysis in the investigation of prenatally diagnosed congenital heart disease. Am J Obstet Gynecol MFM. 2020;2(1):100078. 10.1016/j.ajogmf.2019.100078. [DOI] [PubMed] [Google Scholar]
- 11.Trevisan P, Zen TD, Rosa RFM, et al. Chromosomal abnormalities in patients with congenital heart disease. Arq Bras Cardiol. 2013;101(6):495–501. 10.5935/abc.20130204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng J, Picker J, Zheng ZJ, et al. Chromosome microarray testing for patients with congenital heart defects reveals novel disease causing loci and high diagnostic yield. BMC Genomics. 2014;15(1):1127. 10.1186/1471-2164-15-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman RJ, Rasmussen SA, Botto LD, et al. The contribution of chromosomal abnormalities to congenital heart defects: a population-based study. Pediatr Cardiol. 2011;32:1147–57. 10.1007/s00246-011-0034-5. [DOI] [PubMed] [Google Scholar]
- 14.Cheng P J, Liu C M, Chueh H Y, et al. First-trimester nuchal translucency measurement and echocardiography at 16 to 18 weeks of gestation in prenatal detection for trisomy 18. Prenat Diagn. 2003;23(3):248–251. 10.1002/pd.581. [DOI] [PubMed] [Google Scholar]
- 15.Musewe NN, Alexander DJ, Teshima I, et al. Echocardiographic evaluation of the spectrum of cardiac anomalies associated with trisomy 13 and trisomy 18. J American College Cardiol. 1990;15(3):673–7. 10.1016/0735-1097(90)90644-5. [DOI] [PubMed] [Google Scholar]
- 16.Sun YM, Zhang PP, Zhang N, et al. Cytogenetic analysis of 3387 umbilical cord blood in pregnant women at high risk for chromosomal abnormalities. Mol Cytogenet. 2020;13:2. 10.1186/s13039-020-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annual review of genomics and human genetics. 2006;2006(7):407–42. 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 18.Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109(2):201–12. 10.1016/j.fertnstert.2018.01.005. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan M, Deng L, Yang Y, et al. Intrauterine phenotype features of fetuses with Williams-Beuren syndrome and literature review. Annals of human genetics. 2020;84(2):169–76. 10.1111/ahg.12360. [DOI] [PubMed] [Google Scholar]
- 20.Gao L, Zhang J, Han X, et al. A rare cardiac phenotype of dextrocardia observed in a fetus with 1p36 deletion syndrome and a balanced translocation: a prenatal case report. Mol Cytogenet. 2020;13(1):48. 10.1186/s13039-020-00514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. American journal of human genetics. 2010;86(5):749–64. 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochstenbach R, Buizer-Voskamp JE, Vorstman JA, et al. Genome arrays for the detection of copy number variations in idiopathic mental retardation, idiopathic generalized epilepsy and neuropsychiatric disorders: lessons for diagnostic workflow and research. Cytogenet Genome Res. 2011;135(3–4):174–202. 10.1159/000332928. [DOI] [PubMed] [Google Scholar]
- 23.Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–601. 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins K J, Correa A, Feinstein J A, et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics Circulation. 2007;115(23):2995–3014. 10.1161/circulationaha.106.183216. [DOI] [PubMed]
- 25.Crino JP, Finberg HJ, Frieden F, Kuller J, Odibo A, Robichaux A, Bohm-Velez M, Pretorius DH, Sheth S, Angtuaco TL, Hamper UM. AIUM practice guideline for the performance of obstetric ultrasound examinations. J Ultrasound Med. 2013;32(6):1083–1101. 10.7863/ultra.32.6.1083. [DOI] [PubMed]
- 26.Qiu X, Weng ZJ, Liu M, et al. Prenatal diagnosis and pregnancy outcomes of 1492 fetuses with congenital heart disease: role of multidisciplinary-joint consultation in prenatal diagnosis. Sci Rep. 2020;10(1):7564. 10.1038/s41598-020-64591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.