Abstract
Background
The clinically high comorbidity between polycystic ovary syndrome (PCOS) and breast cancer (BC) has been extensively reported. However, limited knowledge exists regarding their shared genetic basis and underlying mechanisms.
Method
Leveraging summary statistics from the largest genome-wide association studies (GWASs) to date, we conducted a comprehensive genome-wide cross-trait analysis of PCOS and BC. A variety of genetic statistical methods were employed to uncover potential shared genetic causes.
Results
Our analysis revealed genetic overlap between the three trait pairs. After partitioning the genome into 2,495 independent regions, we identified two loci, chr8: 75,011,700–76,295,483 and chr17: 6,305,079–7,264,458, with significant localized genetic correlations. Pleiotropic analysis under a composite null hypothesis identified 1,183 significant pleiotropic single nucleotide polymorphisms (SNPs) across three trait pairs. FUMA mapped 26 pleiotropic loci, with regions 16q12.2 and 6q25.1 duplicated across all three trait pairs, while COLOC detected three loci with colocalization evidence. Gene-based analysis identified 23 unique candidate pleiotropic genes, including the FTO shared by all trait pairs, as well as SER1, RALB, and others in two trait pairs. Pathway enrichment analysis further highlighted key biological pathways, primarily involving the significant biological pathways were the metabolism of regulation of autophagy, regulation of cellular catabolic process, and positive regulation of catabolic process. Latent Heritable Confounder Mendelian randomization (LHC-MR) supported a positive causal relationship between PCOS and both BCALL and ERPBC but not with ERNBC.
Conclusion
In conclusion, our genome-wide cross-trait analysis identified a shared genetic basis between PCOS and BC, specific identical genetic mechanisms and causality between PCOS and various BC subtypes, which could better explains the genetics of the co-morbidity of PCOS and ERPBC rather than PCOS and ERNBC. These findings provide new insights into the biological mechanisms underlying the co-morbidity of these two complex diseases, which have important implications for clinical disease intervention, treatment, and improved prognosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13058-024-01923-5.
Keywords: Polycystic ovary syndrome, Breast cancer, GWAS, Shared genetic
Introduction
Breast cancer (BC) remains a significant global health challenge, ranking as the second most lethal malignancy in women, surpassed only by lung cancer [1]. Lymphatic tract metastasis is the most common mode of spread of BC, and more than half of the patients will be found to have axillary lymph node involvement on examination [2]. The median survival time after metastasis of this aggressive cancer is so short that patients live less than 24 months merely. Polycystic ovary syndrome (PCOS), a common multi-system disorder, affects 5 to 8% of women of reproductive age [3, 4]. A population-based case–control study indicated that premenopausal women with PCOS have nearly triple the risk of developing BC compared to those without PCOS [5]. This increased risk may stem from the substantial overlap in risk factors between BC and PCOS. Endogenous hormone levels, which have been strongly linked to BC risk in previous prospective cohort studies, play a crucial role in this association [6]. Additionally, elevated androgen levels resulting from PCOS can directly stimulate the synthesis of epidermal growth factor (EGF) by breast apical cells, thereby activating the ErbB family of receptors. Simultaneously, serum androgen levels also promote the growth of a specific subset of tumors characterized as ER-negative/AR-positive apocrine tumors, thus establishing their comorbidity [7, 8]. Furthermore, key clinical manifestations of PCOS (such as elevated androgen levels, later onset of menarche, and delayed first childbirth) are also recognized risk factors for BC [9–11]. The considerable heritability of PCOS, estimated at up to 70%, and the variable heritability among BC subtypes, ranging from 35 to 80%, suggest a genetic architecture for their comorbidity [12, 13]. Given the high hereditary nature of both PCOS and BC, the high prevalence of comorbidities among patients may indeed be attributed to a shared genetic mechanism.
PCOS and BC are both recognized as polygenic diseases, their development attributable to the cumulative effects of multiple genes, and such diseases are often more affected by environmental factors. To date, major genome-wide association studies (GWAS) have identified at least 17 loci associated with PCOS, while advances in GWAS have unveiled over 180 genetic variants (single nucleotide polymorphisms, SNPs) linked to BC [14]. Notably, a Mendelian randomization (MR) study investigating BC risk in PCOS patients found an association between PCOS and an increased risk of ERPBC, but not with ERNBC. This study identified three SNPs (rs10739076, rs13164856, and rs11031005) among the PCOS-associated genetic variants significantly associated with an increased risk of ERPBC [15]. Further, a MR analysis by Wen et al. explored the causal relationship between PCOS and more specifically classified subtypes of BC, revealing an association with increased risk for luminal A-like, luminal B/HER2-negative-like, and luminal B-like BC subtypes [16]. These MR studies underscore the concept of vertical pleiotropy as one type of genetic mechanism, wherein a SNP influences one trait, which in turn influences another. In recent years, the use of horizontal pleiotropy as the other type of genetic mechanism to explain genetic associations between different diseases has become a research hotspot; for example, a recent study published in JAMA identified shared genetic determinants between gastrointestinal and psychiatric disorders by exploring horizontal pleiotropy between the two diseases. However, there has been a lack of systematic studies to date on horizontal pleiotropy between PCOS and BC, where SNPs impact both conditions through independent pathways. Therefore, our study undertook a comprehensive genome-wide cross-trait analysis to explore pleiotropic genetic variants and loci, aiming to elucidate the shared genetic mechanism between PCOS and BC.
In this genome-wide association study, we leveraged the latest large-scale GWAS summary statistics and applied multiple genetic statistical methods to explore pleiotropy at various levels, including SNP, gene, and biological pathway. Our goal was to elucidate the shared genetic architecture and underlying mechanisms between PCOS and BC. Initially, we quantified the genetic associations of the two diseases at both the genome-wide and regional levels, estimating the genetic overlap. We identified pleiotropic SNPs and annotated them as pleiotropic loci and further conducted positional mapping to further investigate potential pleiotropic genes. In addition, we expanded our analysis to downstream pleiotropic genes and performed tissue-specific enrichment analysis to better understand the biological pathways linking PCOS and BC. Furthermore, we employed MR analysis methods to investigate potential causal relationships between the two diseases, focusing on vertical pleiotropy. This comprehensive approach aims to construct refined genetic linkage, enhancing our understanding of the causes of comorbidity Ultimately, our findings aim to contribute valuable insights for future therapeutic strategies and risk prediction models.
Method
GWAS data sets for PCOS and BC
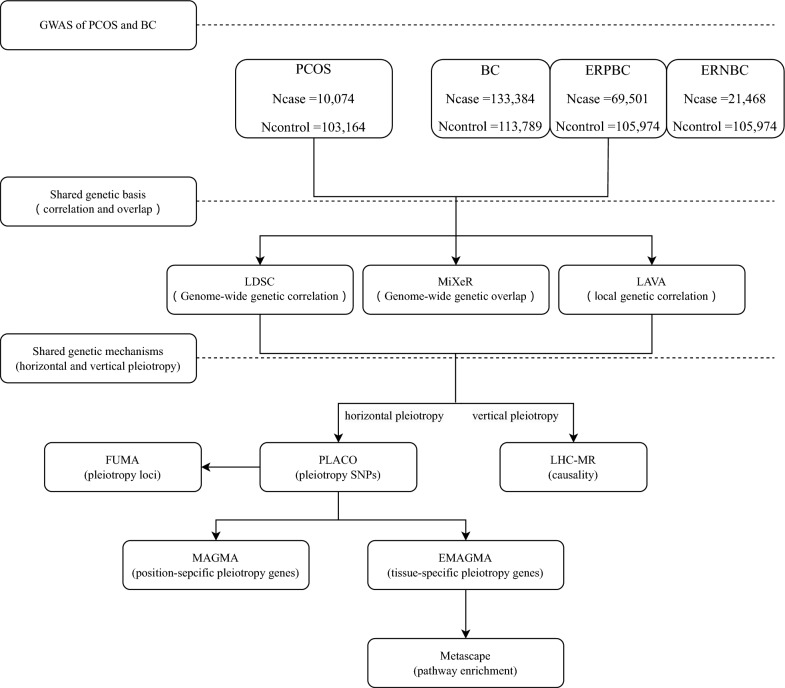
Figure 1 outlines the workflow for our study. Summary statistics were retrieved from publicly available GWASs conducted for PCOS and BC. The latest GWAS of PCOS was performed by Day F et al. in 2018, which combined 10,074 PCOS-affected women and 103,164 female control subjects (all of European ancestry). Diagnosis of PCOS was made based on the National Institutes of Health (NIH) criteria or Rotterdam criteria (Zawadzki et al., 1992; ESHRE and Group, 2004) or by self-report. The NIH criteria require androgen excess and ovulatory dysfunction, whereas polycystic ovarian morphology is included in the Rotterdam criteria. In addition, estimates of each variable collected from summary statistics of contributing studies were meta-analyzed using fixed-effects, and inverse-weighted variance meta-analysis using GWAMA or METAL [17]. For BC overall, the most recent, also the largest GWAS, was performed by Zhang et al. in 2020, meta-analyzing data from 82 participating studies of the Breast Cancer Association Consortium (BCAC) and 11 other BC genetic studies. This GWAS combined 133,384 BC-affected women and 113,789 female control subjects (all of European ancestry) [13]. In this meta-analysis, standard and novel approaches were used to better account for potential tumor heterogeneity, specifically by estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 status and tumor grade. For subtype-specific BC, we retrieved summary statistics from the largest published GWAS on estrogen receptor ER + and ER– BC performed by Michailidou et al. in 2017. This GWAS meta-analyzed data from BCAC and DRIVE, combining 122,977 BC-affected women (of which 69,501 were ER + cases and 21,468 were ER–) and 105,974 female control subjects (all of European ancestry) [18]. Details of the dataset used are available in Table 1. To reduce population stratification bias we obtained Europe-only summary statistics and conducted strict quality controls: (i) compared with the reference of the 1000 Genomes Project v3 Europes (1000G) constructed from the hg19 genome and deleted SNPs that did not match alleles; (ii) excluded sex chromosomes to ensure that all SNPs were restricted to autosomes; (iii) deleted SNPs that duplicate or missing rsIDs appeared in the GWAS summary data; (iv) filtered SNPs with minor allele frequency (MAF) < 0.01. After we unified and filtered the SNPs shared between the GWAS summary statistics for PCOS and three BCs, there were a total of 7,270,318 SNPs.
Fig. 1.
Study design. Summary statistics were retrieved from publicly available GWASs conducted for PCOS and BC. The genetic basis between the two diseases was explored through multiple analytical methods. Genome-wide genetic correlations were estimated using LDSC, while genome-wide genetic overlap was assessed through MiXeR. Additionally, local genetic correlations were examined using LAVA. Together, these analyses provide a comprehensive view of the shared genetic architecture between the two conditions. The shared genetic mechanism of the two diseases was analyzed, and the horizontal effect was studied at four levels. PLACO was employed to identify pleiotropic SNPs, which were subsequently annotated to genomic loci using FUMA. Location-specific pleiotropic genes were identified through MAGMA, while eMAGMA was used to uncover tissue-specific pleiotropic genes at the gene level. Lastly, Metascape was applied to perform functional enrichment analysis on the significant genes identified by eMAGMA, providing insights into the biological processes and pathways involved. We applied LHC-MR to evaluate the pairwise causal associations between PCOS and the BC, primarily elucidating the contributions from vertical pleiotropy
Table 1.
Overview of PCOS and BC included in this study
| Diseases | Abbreviations | PMID | Data source | Year | Population | N | Cases | Controls | Reference genome |
|---|---|---|---|---|---|---|---|---|---|
| breast cancer | BC | 32,424,353 | https://bcac.ccge.medschl.cam.ac.uk/bcacdata/oncoarray/oncoarray-and-combined-summary-result/ | 2020 | European | 247,173 | 133,384 | 113,789 | GRCh39 (hg19) |
| ER + breast cancer | ERPBC | 29,059,683 | ieu-a-1127 | 2017 | European | 175,475 | 69,501 | 105,974 | GRCh39 (hg19) |
| ER- breast cancer | ERNBC | 29,059,683 | ieu-a-1128 | 2017 | European | 127,442 | 21,468 | 105,974 | GRCh39 (hg19) |
| polycystic ovarian syndrome | PCOS | 30,566,500 | https://www.repository.cam.ac.uk/handle/1810/289950 | 2018 | European | 113,238 | 10,074 | 103,164 | GRCh39 (hg19) |
Overview of PCOS and BC, abbreviations as used throughout the manuscript, associated PubMed ID, Data source and Year of publication, the sample size, population and reference genome on which summary statistics are based, and the number of SNPs included in the original summary statistics, before we applied filtering
Genome-wide correlation between PCOS and BC
We first used cross-trait linkage disequilibrium score regression (LDSC) [19] to evaluate the heritability of PCOS and BC and their genetic correlation. LDSC is a newly developed SNP heritability estimation method that can estimate genetic correlation through GWAS summary data instead of a single level of genotype data. This approach minimizes the effects of confounding factors and population stratification and evaluates genetic correlations without bias due to sample overlap. First, SNP-based heritability (h2, representing the SNP fraction of phenotypic variation explained by common genetic variations under study) of PCOS and BC was estimated using univariate LDSC. Then, bivariate LDSC was used to estimate genetic correlations (rg) between PCOS and BC. The z-score for each variant in trait one is multiplied by the z-score for each variant in trait 2. LDSC builds a weighted linear model by regressing the product of the Z statistic on the LD score. We utilized the pre-computed LD scores of European ancestry derived from the third phase of the 1000 Genomes project provided by the LDSC developers as a reference and excluded the MHC region. The Bonferroni-corrected significant threshold was set at P < 1.67E-02 (0.05/3).
To determine tissues linked to PCOS and BC, the linkage disequilibrium score for the specific expression of genes (LDSC-SEG) [20] analysis using tissue gene expression data was performed. LDSC-SEG is a computational method that uses hierarchical LD score regression to identify phenotypically relevant tissues, allowing discernment of which tissues or cell types are most associated with a specific disease or health phenotype. Genotype‐Tissue Expression (GTEx) project served as the genome annotation during the analysis. We utilized the 49 tissues and used P < 3.40E-04 (0.05/49/3) as the threshold to find some traits with significant tissue type-specific enrichment and continue to conduct subsequent tissue-specific pleiotropic studies research.
Polygenic overlap analysis using bivariate causal mixture model (MiXeR)
LDSC rg as a genome-wide summary measure, returns a total measure of the correlation of all SNP effect sizes, it does not differentiate genetic overlap with a mixture of concordant and discordant effects from an absence of genetic overlap, returning an estimate close to 0 in both scenarios. MiXeR can quantify the number of shared causal variants even when the effects are mixed it can detect genetic overlap that LDSC may miss, offering a more nuanced understanding of shared genetic architecture. To quantify the number of shared ‘causal’ variants and identify genetic overlap between trait pairs, we applied the statistical tool MiXeR software (https://github.com/precimed/mixer) which is the complement of genetic correlation, can be more comprehensive understanding of the genetic relationship between the phenotype [21]. In our study, 1000 Genomes Project v3 was used as a reference panel for European samples, and the MHC region was excluded (Frei et al., 2019) due to the intricate Chained disequilibrium structure. MiXeR provides univariate estimates for each trait of interest regarding the number of loci affecting the trait and provides a bivariate analysis model for determining additive genetic effects [22]. Univariate mixed models can estimate the number of disease-affecting SNPs (i.e., SNPs associated with diseases other than LD). MiXeR uses maximum likelihood estimation to estimate the polygenicity and discoverability of a given trait. We also used a bivariate Gaussian mixture model to determine additive genetic effects as a mixture of 4 bivariate Gaussian components, representing SNPs that do not affect either phenotype, the only SNPs that affect the primary phenotype, the only SNPs that affect the minor phenotype SNPs, and SNPs that affect both phenotypes. After fitting the model parameters, the Dice coefficient (i.e., the proportion of shared variants to the total number of variants) was also calculated. The overall measure of multigene overlap was quantified via the Dice coefficient in the range zero to one. The results were presented in the form of Venn diagrams, and the difference between the Akaike Information Criterion (AIC) and the reference model was used to evaluate the model fit, that is, the ability of the MiXeR model to predict the actual GWAS data. We chose the "infinitesimal model" as the reference model of the univariate MiXeR. The most suitable fit model was compared in terms of the overlap in maximum and minimum possible values, with a positive AIC difference considered evidence of a distinction between the best-fitting MiXeR estimate and the reference model. In addition, logarithmic likelihood plots are generated to visually understand the analysis process. We also used conditional Q-Q plots to visually assess the shared genetic background between two phenotypes.
Calculating local genetic correlations and emphasizing the relevance of mixed effects
In addition to exploring genome-wide genetic correlations, the Local Analysis of Variance Annotation (LAVA) method was employed to resolve issues of opposing directional confounding within specific genomic regions. This approach provided a clearer determination of whether a shared genetic correlation exists between PCOS and BC in independent regions of the genome. LAVA, an integrated framework for local rg analysis that, in addition to testing the standard bivariate local rgs between two phenotypes, can further emphasize the relevance of mixed effects [23]. In this analysis, we used the LD reference panel based on 1000 Genomes phase 3 genotype data for European samples, and the genome is divided into 2495 partitioned semi-independent genomic regions, with an average size of 1 Mb. To study regions with apparent genetic relatedness, we estimated local genetic correlations using univariate LAVA to test for local genetic signals within each trait in 2,495 regions (P < 1.00E-04 as threshold). Bivariate tests were then performed on loci and traits with significant univariate genetic signals. Using the Bonferroni method, we corrected the P-values for local genetic correlations for multiple testing (P < 0.05/no. of bivariate tests = 0.05/182 = 2.75E-04).
Pleiotropy insights to causal inference between PCOS and BC
We used LHC-MR to investigate the possible bidirectional effect that exists between PCOS and BC, resolving their causal relationship (i.e. vertical pleiotropy). LHC-MR is a new method that extends standard two-sample MR by modelling potential heritable confounders that influence exposure and outcome characteristics. This method appropriately uses genome-wide genetic markers to estimate bidirectional causal effects, direct heritability, and confounding effects while accounting for sample overlap. The standard error of each parameter estimated using LHC-MR was obtained by implementing a block jackknife procedure, in which the SNP effects were divided into blocks, and the maximum likelihood estimate(MLE) value was calculated again in a block-leaving manner. The estimated variance can then be calculated based on the results of various MLE optimizations [24]. LHC-MR framework has multiple pathways through which SNPs can affect traits and allows for null effects, making it more precise and efficient than many MR methods (i.e., MR egger, weighted median, inverse variance weighted, simple mode, and weighted mode). Its features make LHC-MR suitable for our purpose because we assume that the trait pairs have a common cause and that traits mutually affect each other. When Paxy < 0.05/3, Payx > 0.05 represents the one-way causal relationship between PCOS and BC; when Payx < 0.05/3, Paxy > 0.05 represents the one-way causal relationship between BC and PCOS, both positive and negative pass Strict correction represents bidirectional causation.
Identification of pleiotropic SNPs of PCOS and BC
While the aforementioned methods suggest genetic sharing between PCOS and BC, they do not specifically address shared genetic variation at the SNP level. To assess pleiotropy (specifically horizontal pleiotropy) influencing genetic variation and to further elucidate the genetic architecture of these complex traits at the SNP level, we conducted a pleiotropy analysis using the composite null hypothesis (PLACO) approach [25]. PLACO was applied to conduct a genome-wide search for SNPs influencing the pleiotropy of both PCOS and BC. It is a new statistical approach to detecting pleiotropic loci between two traits by considering potential composite null hypotheses. This method allows correlation in summary statistics between studies that may arise due to the sharing of controls between disease traits. We calculated this using the PLACO test statistic (Z1*Z2), where Z1 and Z2 are the observed Z-scores for two traits in a given genetic variant. This method examines one SNP at a time with two sets of Z-statistics as input and proceeds by dividing the composite null hypothesis of pleiotropy into the following scenarios: (i) H00: the SNP is not associated with either of the two disorders. (ii) H10: The SNP is associated with the first disorder but not the second. (iii) H01: The gene is not associated with the first disorder but the second. (iv) H11: the SNP is related to both disorders. Moreover, SNPs with very high Z2 (> 100) values were removed because spurious pleiotropic signals could be caused. For genome-wide significant SNPs, we define PPLACO < 5.00E − 08.
Identifying genetic shared loci using mapping
Functional Mapping and Annotation (FUMA) can be used to discover independent genomic loci, identify pleiotropic loci in PCOS and BC, and functionally annotate pleiotropic SNPs identified by PLACO. It is helpful to better elucidate the genetic mechanism of PCOS and BC. FUMA is a platform that built a series of SNP functional annotation approaches and then by chromosome, base-pair position, reference and alternate alleles to multiple publicly available databases to annotate candidate SNPs for functional consequences on gene functions. All LD information is calculated from the 1000 Genomes Phase 3 published reference panel. Among them, the LD region is defined as 500kbp upstream and downstream of lead SNP (1Mbp in total). We first identified independently significant SNPs with genome-wide significant p-values (P < 5.00E-08) and r2 < 0.6. If the independently significant SNPs are independent of each other and r2 < 0.1, we define their subset as independent leading SNPs. Positional mapping in FUMA was performed using the ANNOVAR annotations to specify the maximum distance between SNPs and genes and using Combined Annotation Dependent Depletion (CADD) scores to predict the functional consequences of SNPs on genes. The CADD scores predict how deleterious the effect of an SNP is likely to be for a protein structure or function, with higher scores referring to higher deleteriousness. 12.37 could be viewed as the threshold for a deleterious score [26]. Independent significant SNPs and SNPs in LD with them were annotated for consequences on gene function using ANNOVAR, Regulome DB score, and 15-core chromatin state predicted. Among them, Greater evidence for a variant to be located in a functional region is shown by lower scores in the RegulomeDB, and a score of 7 indicates that there is no data about the function of a certain SNP.
Moreover, we conducted a FUMA analysis to conduct gene mapping to understand better the genetic mechanisms underlying PCOS and BC. One of the functions of FUMA, SNP2GENE, is used to annotate the biological functions of SNPs and map SNPs to genes. Using the SNP2GENE function, we performed both positional mapping (maximum distance 10 kb) and eQTL mapping (cis-eQTL, i.e., up to 1 Mb) using GTEx v8. The reference panel for extracting chromosomes and locations was the human genome hg19, using only protein-coding genes from the genetic map.
Colocalization analysis
We conducted a colocalization analysis of potential pleiotropic loci identified by FUMA to identify shared causal variants across pairwise traits within each locus. In short, COLOC analysis is based on five hypotheses: H0, not related to any trait; H1, related to trait 1, but not related to trait 2; H2, related to trait 2, but not related to trait 1; H3, traits 1 and 2 are both related to two independent SNPs; H4, traits 1 and 2 are both related to a shared SNP (Trait 1 is PCOS, trait 2 is BCALL, ERPBC, ERNBC). We focus mainly on the last hypothesis, which quantifies the support dimension via the posterior probability PPH4. Colocalization is represented if the posterior PPH4 of a model with shared causal variables is 0.70 [27] or higher. Furthermore, the SNP with the largest PPH4 is a candidate causal variant.
Variant annotation and MAGMA gene-based analysis
Based on the PLACO results and single-trait GWAS, we attempted to use multimarker genomic annotation analysis (MAGMA) to identify important genes associated with PCOS or BC and potential pleiotropic genes associated with both diseases. MAGMA is a gene-based association approach using only summary statistics [28]. It aggregated the joint associations of multiple SNPs within whole gene areas with PCOS or BC, considering the LD between SNPs. MAGMA computed the statistics results more efficiently with multiple linear regression in a gene while accounting for the LD Build 37 (GRCh37/hg19) and included 17,636 autosomal protein-coding genes. Phase 3 of the 1000 Genomes Project was exploited as a reference panel for calculating the LD matrix, where LD is defined as 10 kb. The Bonferroni-corrected significant threshold was set at P < 9.45E-07 (0.05/17,636/3).
Tissue-specific pleiotropic genes
MAGMA may not accurately identify functionally related genes, as it assigns trait-related SNPs to the nearest gene within a 10 kb range, potentially overlooking regulatory effects on gene expression (eQTLs) that are located 1 Mb upstream and downstream of the gene promoters they influence. However, their remote regulatory roles can significantly influence gene expression by interacting with enhancers, promoters, and other regulatory elements. This allows eQTLs to play crucial roles in biological pathways, providing insight into gene function and disease mechanisms that are not captured by proximity alone. Therefore, to more accurately study the biological mechanism of pleiotropy genes, we used eQTL genome annotation multi-marker analysis (eMAGMA) to analyze the gene association between PCOS and BC, thereby more precisely reflecting the functional relationship between SNPs and cis-eQTLs in specific tissues [29]. E-MAGMA modifies the MAGMA method by integrating eQTL information from the GTEx project, providing better statistical performance in gene-based association analysis and taking tissue specificity into account. This method could conduct eQTL-informed gene-based tests by assigning SNPs to tissue-specific genes so that they provide more biologically meaningful and interpretable results. We use GTEx v8-based annotation files that are available on the eMAGMA website. Tissue-specific gene analysis was performed based on the significantly enriched tissues in the LDSC-SEG results that were thought to be associated with PCOS and BC and on the established consensus biological knowledge. Specifically, E-MAGMA analysis included 6098 Breast Mammary Tissue-specific genes, 2719 Cells EBV-transformed lymphocyte-specific genes, 2872 Ovary-specific genes, 1698 Uterus-specific genes, 1702 Vagina-specific genes and 7931 Whole Blood-specific genes. We performed Bonferroni correction for each tissue and selected important genes with a P value less than 0.05/ no. of gene sets tested/no. of trait pairs [such as P Whole Blood < 2.10E-06 (0.05/7931/3), P Uterus < 9.80E-06 (0.05/1698/3)].
Pathway enrichment analysis
We performed functional enrichment analysis by Metascape using significant genes in eMAGMA to find enriched pathways and related functional annotations of target genes. Metascape is a database that utilizes more than 40 independent knowledge bases to collect data and combines functional enrichment, interactome analysis, gene annotation and membership search [30]. It presents the results in the form of high-quality graphs and concise explanations. The GO database can be used to analyze the identified important genes in biological processes (BP), cellular components (CC), etc., to understand biological mechanisms better. The Gene Ontology (GO) resource (http://geneontology.org/) is a bioinformatics tool that provides a framework and set of concepts for describing the function of gene products in all organisms. GO terms of biological functions associated with setting the cut-off P value as 0.01(the default settings of the Metascape) were considered significantly enriched.
Result
Estimated overall genetic correlation between PCOS and BC
The results of univariate LDSC showed that the LDSC SNP-based heritability (h2SNP) of PCOS was 11.69% (SE = 0.0214). Among three breast cancer traits, the heritabilities of BC, ERNBC, and ERPBC were 12.51% (SE = 0.0092), 7.30% (SE = 0.0065), and 13.30% (SE = 0.0114), respectively. It can be seen that the heritability of the three types of BC ranges from 7.30% to 13.30%, among which ERPBC has the strongest heritability and ERNBC has the lowest heritability. To study genetic correlations between pairs of traits, we performed bivariate LDSC. The P values between PCOS and BCALL (P = 8.71E-01), PCOS and ERNBC (P = 8.71E-01), PCOS and ERPBC (P = 8.26E-01) are all greater than 1.67E-02 (0.05/3) (Supplementary Table 1). The P values of the three trait pairs are not significant, indicating that there is no genetic correlation between them. However, genome-wide genetic correlations are unable to distinguish mixtures of congruent and discordant genetic effects from the absence of genetic overlap, potentially underestimating the shared genetic basis of PCOS and BC. Further analyses were implemented to explore the specific genetic mechanisms.
Polygenic overlap analysis using MiXeR
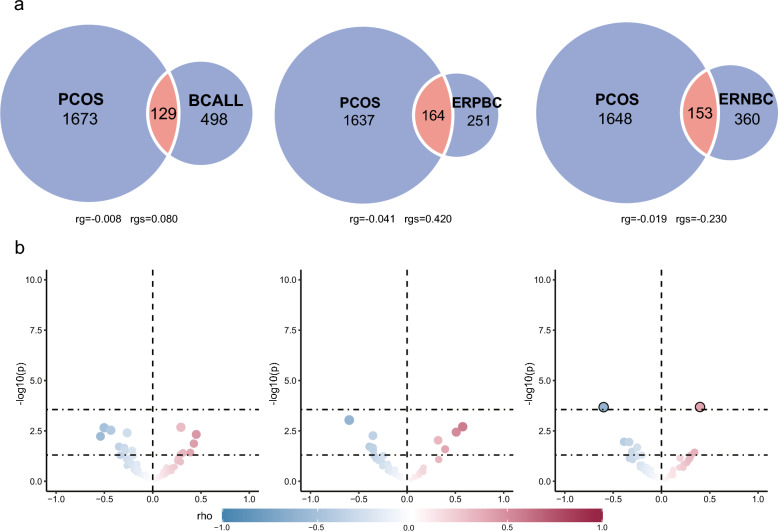
Although no genetic correlation existed between trait pairs estimated by LDSC, MiXeR was then applied to elucidate genetic overlap between PCOS and BC, irrespective of the direction of their effects. Univariate MiXeR found that 627 variants [standard error (SE) = 116] are associated with BCALL, 416 variants (SE = 88) are shared with ERPBC and 514 variants (SE = 60) are shared with ERNBC(Supplementary Table 2). These reflected their different genetic architectures and also showed that BCALL has the highest polygenicity among the three breast cancer traits; its SNPs can annotate more genes, while ERPBC has the lowest polygenicity. Bivariate MiXeR analysis revealed polygenic overlaps of PCOS-influencing variants with each breast cancer. As shown in the Venn diagram (Fig. 2a), the estimated number of shared 'causal' variants between PCOS and BCALL was 129 (SE = 142), with 1673 (690) unique PCOS variants and 498 (199) unique BCALL variants. The Dice coefficient was 0.104 for variants shared between PCOS and BCALL(Supplementary Table 2). MiXeR estimated 153 (130) shared 'causal' variants between PCOS and ERNBC, with 1648 (712) unique PCOS variants and 360 (135) unique ERNBC variants. Moreover, 164 (101) shared 'causal' variants between PCOS and ERPBC, with 1637 (693) unique PCOS variants and 251 (138) unique ERPBC variants. The Dice coefficient was 0.132 for variants shared between PCOS and ERNBC. The proportion of shared 'causal' variants with concordant effects for PCOS-BCALL was 0.530(SE = 0.080), 0.420(SE = 0.110) for PCOS-ERNBC and 0.640(SE = 0.080) for PCOS-ERPBC. Moreover, there was a positive genetic correlation between PCOS and BCALL(rg = 0.004, rho = 0.080) and ERPBC(rg = 0.066, rho = 0.420), while a negative genetic correlation exists between PCOS and ERNBC(rg = −0.026, rho = −0.230). The results of AIC differences are shown in a log-likelihood plot based on the modelling quantity of causal variables. The minimum value of the X-axis of the image represents the minimum value of causal variables, while the maximum value of the X-axis represents the maximum value of causal variables. In comparing the possible maximum overlap in model fitting among the three traits (PCOS-BCALL, PCOS-ERNBC, and PCOS-ERPBC) with AIC values of 13.52, 3.30, and 7.92 respectively, and the minimum overlap with AIC values of 4.76, 0.85, and 5.60 respectively, positive AIC differences were supported in all cases(Additional file 2). However, the result of small genetic overlap was still obtained, which may be due to the low polygenicity of these phenotypes, resulting in the inconsistent causality of several SNPs with a high single driving force in the region confounding the genetic correlation. Based on the above analysis, we have revealed no genetic correlation despite polygenic overlaps in PCOS and three types of breast cancer, which may be due to the existence of “mixed effects”.
Fig. 2.
Local genetic correlations, and genetic overlap of the polycystic ovary syndrome and breast cancer. (a)Volcano plots of LAVA local genetic correlation coefficients (rho, y-axis) against -log10 (p-values) for each pairwise analysis per locus. Larger dots with black circles represent significantly correlated loci after the Bonferroni correction. Note that the Volcano plots were plotted at p-values truncated by 1.00E-04 for better visualisation. (b) MiXeR Venn diagrams show the number of shared and disorder-specific ‘causal’ variants in thousands for each pair of disorders
Calculating local genetic correlations and emphasising the relevance of mixed effects
For local rg analysis, we used LAVA (Fig. 2b, Supplementary Tables 3–4). Among 2495 areas, regions with a P-value < 1.00E-04 were considered to be significant. The results indicate that 1553 regions are significant, with the highest count associated with PCOS at 514, whereas ERNBC had the fewest with 289. The regions obtained by ERPBC and BCALL are 338 and 412, respectively. Moreover, A total of 182 bivariate tests were conducted to explore the relationship between PCOS and three types of breast cancer diseases (BCALL, ERNBC, and ERPBC) in the context of bivariate LAVA analysis. Local genetic correlations LAVA analysis of loci shared between PCOS and BC revealed 30 significant local genetic correlations (P < 0.05). When p is less than 0.05, among the 14 sites connecting PCOS and BCALL, 8 exhibit negative rho values, while the remaining six show positive values. Similarly, out of the nine sites linking PCOS and ERNBC, 5 demonstrate negative rho values, with the remaining 4 displaying positive values. Notably, the number of positive and negative correlation areas is relatively balanced. The mixture of positive and negative regions leads to a lack of genome-wide significance, potentially explaining our prior observations of genetic overlap without significant genome-wide genetic correlation. Further, two loci (chr8: 75,011,700–76,295,483, chr17: 6,305,079–7,264,458) through Bonferroni correction (P < 0.05/182 = 2.75E-04) with significant local heritability were found in PCOS and ERPBC (p values respectively: 2.06E-04, 2.09E-04). One of the loci, situated on chromosome 8, shows a positive correlation, whereas the second locus exhibits a negative correlation.
Estimating bidirectional causal effects
We used LHC-MR to test for bidirectional causality between trait pairs using GWAS summary statistics (Fig. 3, Supplementary Table 5). The study results showed that LHC-MR did not provide any evidence of a causal relationship effect of PCOS on ERNBC (P = 4.67E-01) and no evidence for the reverse causal effect (P = 5.93E-02). However, LHC-MR revealed that corresponding to PCOS on BCALL showed a potential positive causal effect of PCOS on BCALL (P = 1.07E-03). We found no evidence for the reverse causal effect (P = 2.08E-01). At the same time, It showed a potential positive causal effect of PCOS on ERPBC (P = 2.28E-03) and found no evidence for the reverse causal effect (P = 2.05E-01). In essence, the presence of PCOS exacerbates the likelihood of BCALL, particularly the emergence of ERPBC, while showing nearly no correlation with ERNBC incidence.
Fig. 3.
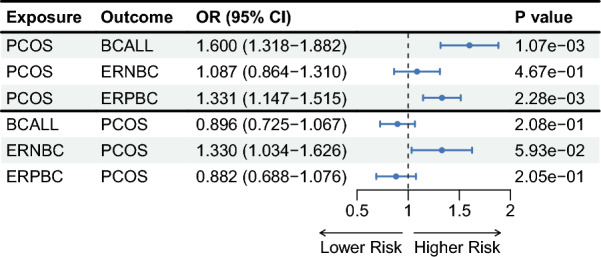
The forest plot for associations between pairs of PCOS and BC in bidirectional MR analysis. The estimates presented in the forest plot were obtained using the LHC-MR method. A positive association is indicated by the odd ratio (OR > 1), while a negative association is indicated by OR < 1. The results of forward causality from LHC-MR method are in the up panel, and the results of reverse causality from the LHC-MR method are in the down panel
Identification of pleiotropic loci of PCOS and BC
We applied PLACO to identify 1,183 SNPs with potential pleiotropic effects associated with PCOS and breast cancer, including 491 were identified in the PCOS and BCALL trait pairs, 224 in the PCOS and ERNBC trait pairs, and 468 in the PCOS and ERPBC trait pairs. FUMA mapped 26 genetic risk loci involving 15 unique chromosomal regions, with several pleiotropic regions, such as 16q12.2 and 6q25.1 (mapping positional: CASC10, SKIDA1, MLLT10 and DNAJC1) (Fig. 4, Supplementary Tables 6–7), were found to be duplicated across all three pairs of traits (Table 2). Additional multiple effector regions, such as 2q14.2, overlapped in PCOS-BCALL and PCOS-ERNBC trait pairs, suggesting that the loci have a wide range of pleiotropic effects. Previous studies have shown that high expression of the RALB gene on chromosome 2q14.2 is significantly associated with reduced survival in breast cancer patients with metastatic progression[31].
Fig. 4.
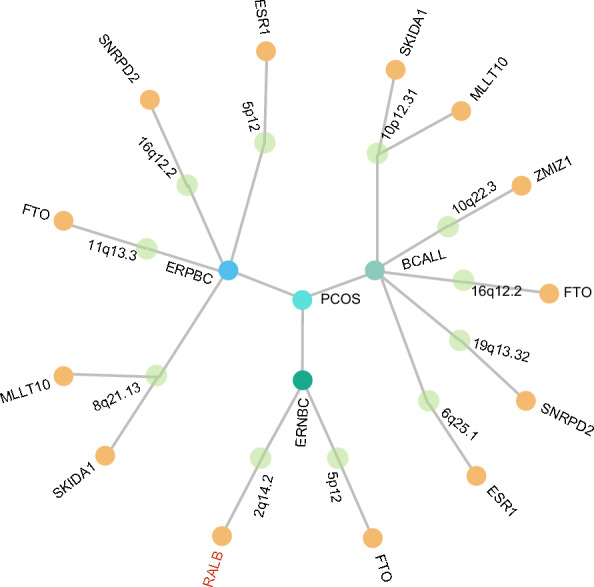
The Overall Landscape of the Pleiotropic Associations Across polycystic ovary syndrome and breast cancer. A circular dendrogram included polycystic ovary syndrome(PCOS) at the centre point and three types of breast cancer(including breast cancer all[BCALL], estrogen receptor-positive breast cancer[ERPBC], estrogen receptor-negative breast cancer[ERNBC]) in the inner circle, resulting in 3 trait pairs. A total of 11 pleiotropic loci were identified across 3 trait pairs (second circle). A total of 13 significant pleiotropic genes were further identified by MAGMA and the genes with the tissue specificity identified by E-MAGMA were also highlighted (red font annotation)
Table 2.
26 pleiotropic genomic loci identified by FUMA using PLACO results
| Trait pair | Top SNP | Locus boundary | Region | Nearest gene | PP.H3 | PP.H4 | Best causal SNP | SNP.PP.H4 |
|---|---|---|---|---|---|---|---|---|
| PCOS-BCALL | rs10423928 | 19:46,179,043–46202172 | 19q13.32 | GIPR | 2.53E-01 | 2.28E-02 | rs4849879 | 1.40E-01 |
| PCOS-BCALL | rs11012730 | 10:21,766,969–22,275,118 | 10p12.31 | SKIDA1 | 1.99E-01 | 1.63E-02 | rs552647 | 1.24E-01 |
| PCOS-BCALL | rs147872430 | 5:44,992,980–45933347 | 5p12 | NA | 4.53E-02 | 2.14E-02 | rs930395 | 7.44E-01 |
| PCOS-BCALL | rs17630711 | 3:27,421,024–27553889 | 3p24.1 | SLC4A7 | 2.91E-01 | 2.43E-02 | rs60954078 | 2.13E-01 |
| PCOS-BCALL | rs2926589 | 8:76,362,101–76689287 | 8q21.13 | HNF4G | 1.93E-01 | 5.37E-02 | rs72658071 | 4.03E-01 |
| PCOS-BCALL | rs45577136 | 16:52,566,220–52630972 | 16q12.1 | TOX3 | 8.82E-02 | 1.22E-01 | rs12251016 | 2.07E-01 |
| PCOS-BCALL | rs4980554 | 11:69,281,227–69,285,786 | 11q13.3 | NA | 2.00E-01 | 1.35E-02 | rs719338 | 2.76E-01 |
| PCOS-BCALL | rs61874140 | 10:123,435,982–123,435,982 | 10q26.13 | NA | 1.33E-01 | 1.31E-02 | rs2981579 | 8.35E-01 |
| PCOS-BCALL | rs73949122 | 2:121,153,284–121,180,803 | 2q14.2 | NA | 1.39E-01 | 2.37E-02 | rs78540526 | 1.00E + 00 |
| PCOS-BCALL | rs753271 | 10:80,853,575–80869471 | 10q22.3 | ZMIZ1 | 2.41E-01 | 1.97E-02 | rs4784227 | 9.55E-01 |
| PCOS-BCALL | rs8050136 | 16:53,797,908–53845487 | 16q12.2 | FTO | 1.83E-01 | 8.12E-01 | rs62048402 | 2.03E-01 |
| PCOS-BCALL | rs851984 | 6:151,970,639–152052215 | 6q25.1 | ESR1 | 7.87E-02 | 6.43E-01 | rs10423928 | 2.69E-01 |
| PCOS-ERNBC | rs1121980 | 16:53,797,908–53845487 | 16q12.2 | FTO | 2.37E-01 | 7.79E-02 | rs4528762 | 9.99E-01 |
| PCOS-ERNBC | rs1293960 | 6:151,970,639–151970639 | 6q25.1 | NA | 2.83E-01 | 2.43E-02 | rs9397437 | 2.28E-01 |
| PCOS-ERNBC | rs4849857 | 2:120,986,708–121070811 | 2q14.2 | NA | 1.60E-01 | 8.35E-01 | rs55872725 | 9.14E-02 |
| PCOS-ERPBC | rs10423928 | 19:46,179,043–46202172 | 19q13.32 | GIPR | 1.66E-01 | 1.70E-02 | rs490706 | 6.21E-02 |
| PCOS-ERPBC | rs11012730 | 10:21,766,969–22,275,118 | 10p12.31 | SKIDA1 | 1.42E-01 | 3.64E-02 | rs985261 | 3.97E-02 |
| PCOS-ERPBC | rs11706019 | 3:27,421,024–27553889 | 3p24.1 | SLC4A7 | 2.91E-01 | 2.43E-02 | rs60954078 | 3.34E-01 |
| PCOS-ERPBC | rs12380632 | 9:110,891,494–110941095 | 9q31.2 | LOC105376214 | 1.30E-01 | 1.41E-02 | rs7862747 | 2.54E-01 |
| PCOS-ERPBC | rs13131992 | 4:175,819,086–175870676 | 4q34.1 | ADAM29 | 8.71E-02 | 1.32E-01 | rs10828248 | 2.73E-01 |
| PCOS-ERPBC | rs45577136 | 16:52,566,220–52630972 | 16q12.1 | TOX3 | 1.33E-01 | 1.31E-02 | rs2981579 | 7.34E-01 |
| PCOS-ERPBC | rs4980554 | 11:69,281,227–69,285,786 | 11q13.3 | NA | 1.39E-01 | 2.37E-02 | rs78540526 | 1.00E + 00 |
| PCOS-ERPBC | rs61874140 | 10:123,435,982–123,435,982 | 10q26.13 | NA | 2.41E-01 | 1.94E-02 | rs4784227 | 9.93E-01 |
| PCOS-ERPBC | rs715537 | 22:28,625,685–29,257,046 | 22q12.1 | NA | 1.45E-01 | 8.50E-01 | rs62048402 | 1.29E-01 |
| PCOS-ERPBC | rs8050136 | 16:53,797,908–53845487 | 16q12.2 | FTO | 1.05E-01 | 5.24E-01 | rs10423928 | 3.40E-01 |
| PCOS-ERPBC | rs851984 | 6:152,008,780–152024985 | 6q25.1 | ESR1 | 1.84E-01 | 4.62E-02 | rs62237573 | 1.00E + 00 |
PCOS, polycystic ovary syndrome; BC, breast cancer; ERNBC, estrogen receptor-negative breast cancer; ERPBC, estrogen receptor-positive breast cancer; Best causal, the candidate causal single-nucleotide polymorphism (SNP); PP.H3, posterior probability of H3; PP.H4, posterior probability of H4; SNP.PP.H4, posterior probability of the best causal polymorphism. Locus boundary of each pleiotropic genomic risk locus was denoted as “chromosome: start–end” defined by FUMA for the corresponding trait pair. The top SNP in this locus was also identified as a candidate causal SNP
The result of ANNOVAR, which annotates recent gene and gene expression consequences for each SNP, shows that among all variants, 8 SNPs (30.8%) were located in intronic regions, and 3 (11.5%) were located in intergenic regions. We also calculated a CADD score to indicate whether each variant would have a deleterious effect. The only SNP with a CADD score greater than 12.37 is located at 9q31.2. 4 SNPs had an RDB score of 4a, indicating slightly stronger evidence. While 7 SNPs had a score of 7, suggesting that the support for a regulatory potential is the least.
Co-localization analysis further identified three signals with PPH4 greater than 0.7 among the 26 potential pleiotropic sites, among which the two SNPs were identified as candidate co-causal variants. One signal between PCOS and BC traits resides on chromosome 16q12.2 (PPH4 = 0.81), while another is situated on chromosome 2q14.2 (PPH4 = 0.84), where the association is between PCOS and ERNBC. The third signal is identified within a region on chromosome 22q12.1 (PPH4 = 0.85) in the PCOS-ERPBC trait pairs.
Variant annotation and MAGMA gene- and gene-set-based analysis
MAGMA association analysis was performed on the colocalized potential pleiotropic loci to identify genes related to PCOS, ERNBC and ERPBC. We identified a total of 23 genes, with 10 exhibiting pleiotropic effects in the PCOS-BCALL trait pair, four pleiotropic genes in the PCOS-ERNBC trait pair, and nine pleiotropic genes found between PCOS and ERPBC. The FTO gene was present in all three trait pairs, while ESR1, FGFR2, SNRPD2, MLLT10, ZMIZ1, SKIDA1 and CASC10 were found in both the PCOS-BCALL and PCOS-ERPBC trait pairs (Supplementary Tables 9–10). The ESR1 gene, which was duplicated in both trait pairs, was shown in previous studies to be a key factor in BC susceptibility. For PCOS, estrogen promotes cAMP synthesis and triggers the activation of phosphoinositide 3-kinase and extracellular signal-regulated kinase so that ESR1 is in the development of comorbidities. In addition, the FTO gene, a crucial gene among 23 genes, is positioned at 16q12.2, 5p12, and 11q13.3 across three distinct trait pairs. FTO is overexpressed in breast cancer cells, which affects the energy metabolism of the cells [32]. Furthermore, currently, studies indicate that FTO is a crucial component of m6A modification; it regulates cancer stem cell function and promotes the growth, self-renewal, and metastasis of cancer cells [33]. Meanwhile, FTO polymorphisms exacerbate susceptibility to PCOS by influencing the function of neighbouring genes, such as IRX5 and IRX3. MAGMA analysis identified a total of 23 pleiotropic genes, of which 2 unique genes (SPRY4, CASC10) are novel for BC, and all genes are new to PCOS. Thirteen genes (SKIDA1, MLLT10, ZMIZ1, FTO, SNRPD2, ESR1, FTO, RALB, SKIDA1, MLLT10, FTO, SNRPD2 and ESR1) identified by MAGMA passed the verification of FUMA position mapping (Supplementary Table 8).
Candidate tissue-specific pleiotropic genes were identified by eQTL mapping
LDSC‐SEG analyses using GTEx were carried out to identify tissues linked to PCOS and BC. According to P < 1.02E-03 (0.05/49)(Supplementary Table 12), significant genetic enrichment of BCALL-related SNP was found in breast mammary tissue, followed by Uterus and Vagina. Taking all these into consideration, we selected six tissues(breast mammary tissue, uterus, ovary, vagina, cells EBV-transformed lymphocytes and whole blood) to continue the subsequent study on tissue-specific pleiotropy.
We used the E-MAGMA method to map pleiotropic SNPs to genes based on tissue-specific information from the six tissues we chose. Utilizing E-MAGMA analysis, a result unveiled a total of 4 pleiotropic genes linked with PCOS, while 431 pleiotropic genes were associated with BCALL (Supplementary Tables 13–14). Furthermore, the analysis identified 68 pleiotropic genes correlated with ERNBC, along with 284 pleiotropic genes associated with ERPBC. In six different tissues, a total of 20 genes were identified to be associated with three trait pairs, with PCOS-BCALL involving 9 genes, PCOS-ERPBC involving 8 genes, and PCOS-ERNBC associated with 3 genes. Among these findings, the GDI2 gene stands out, being highly enriched in the PCOS-ERPBC trait pair across four tissues: Cells EBV-transformed lymphocytes, ovary, uterus, and whole blood demonstrating its significant tissue specificity within this trait pair and its profound biological relevance. Similarly, the CYBRD1 gene is noteworthy for its enrichment not only in the PCOS-ERPBC trait pair within the ovary, uterus, and vagina tissues but also in the PCOS-BCALL trait pair in the ovary and uterus tissues. These results collectively highlight the critical histological value of GDI2 and CYBRD1 genes in the ovary and uterus tissues for the comorbidity mechanisms of PCOS-BCALL and PCOS-ERPBC, further revealing their essential roles in the underlying biological mechanisms and providing important insights and directions for future research. Most importantly, we took the intersection of the location-based gene obtained by MAGMA and the pleiotropic gene obtained by eMAGMA and finally identified a gene: RalB (Supplementary Table 8). In previous research, automated quantification of RalA/B expression levels by immunohistochemistry in primary tumors of breast cancer patients unravelled overexpression of both proteins in tumors from patients with metastasis [31]. In addition, RalB has previously been considered a poor prognostic factor in breast cancer patients.
Pathway enrichment analysis
We then employed the Metascape website to conduct GO enrichment analysis on the significant genes identified by e-MAGMA analysis to identify several represented biological processes (Supplementary Table 11). The 18 pathways shown by GO enrichment analysis all belong to the biological process (BP) category. It revealed biological processes containing negative regulation of protein localization, small GTPase-mediated signal transduction, negative regulation of cellular component organization, response to metal ion, regulation of autophagy, regulation of cellular catabolic process, positive regulation of, catabolic process, response to nutrient levels, regulation of secretion by cell process, etc. Remarkably, the significant biological pathways were the metabolism of regulation of autophagy, regulation of cellular catabolic process, and positive regulation of catabolic process, as they are pathways enriched by RALB, the only one gene overlapped by MAGMA and e-MAGMA analysis. Previous studies have shown that metabolic disorders are closely related to breast cancer incidence and outcome [34]. It is also a major cause of PCOS, as it is a widespread endocrine and metabolic disorder. The ovarian autophagy mechanism is one of the pathogenesis of PCOS.[35] In terms of cancer initiation, autophagy is considered tumor suppressive due to its cytoprotective role. This is evidenced by an increased autophagy-related gene signature in normal mammary glands, which is lost during breast cancer progression [36]. In addition, autophagy regulation is also a type of metabolic regulation, which, together with the regulation of cellular catabolic processes and the positive regulatory pathways of catabolic processes, further supports the common genetic mechanism of PCOS and BC in metabolism.
Discussion
To our knowledge, this investigation represents the first and most extensive cross-trait analysis to date, utilizing the most comprehensive GWAS data available for PCOS and BC, including ERNBC, ERPBC, and BCALL subtypes. This study systematically examines the shared genetic foundations and mechanisms between these two complex diseases. Although genome-wide genetic associations between the three pairs of traits were considered weak, our analysis revealed substantial evidence of genetic overlap. Remarkably, through the division of the genome into multiple smaller regions, our regional genetic correlation analysis identified significant heritable gene regions with opposite correlation directions between PCOS and ERPBC. Further cross-trait meta-analyses illuminated multiple shared loci, with comprehensive analyses elucidating the biological pathways involved. Ultimately, MR analysis substantiated a positive causal relationship between PCOS and ERPBC, while not establishing any causal relationship with ERNBC. This pioneering study not only sheds light on the intricate genetic interplay between PCOS and various subtypes of BC but also paves the way for future research to delve deeper into these complex relationships, offering potential insights for therapeutic strategies and risk assessment.
Utilizing LDSC analysis, we observed a minimal genetic correlation between PCOS and BC at the whole-genome level. Notably, despite a very weak genome-wide genetic correlation, bivariate MiXeR analysis employed to quantify the genetic overlap between PCOS and BC revealed a certain degree of shared genetic risk the genetic correlation between them. This suggests potential extensive confounding effects among trait pairs. Continuing our investigation with the LAVA analytical method, we delved into local genetic correlations among the three trait pairs. Post Bonferroni correction, we identified two shared regions exhibiting significant yet opposite genetic correlations between PCOS and ERPBC, with effect sizes closely mirroring each other, indicative of a counterbalancing effect. The nine shared regions identified in PCOS and ERNBC also matched this cancelling effect, suggesting that LAVA has a pattern of confounding effect directions masked by estimates of genome-wide genetic correlations, further supporting our previous inferences. In summary, our analysis uncovers a shared genetic foundation between PCOS and BC, hinting at the presence of significant pleiotropic effects influencing both conditions, despite the lack of clear evidence at the genome-wide level. Univariate MiXeR analysis was used to estimate the polygenicity of PCOS and BC. Although these findings are still in the clinical stage to be confirmed, polygenotypes may be useful markers of clinical-level heterogeneity.
Based on the substantial genetic overlap between BC and PCOS, we further explored the shared genetic mechanisms underlying these disorders at the multiplicity level (both vertical multiplicity and horizontal multiplicity), and we first explored the causal relationship between them at the vertical multiplicity level using LHC-MR analysis. The results of the analysis confirmed the presence of positive causality exclusively between PCOS and both ERPBC and all BC while identifying no positive causal relationship with ERNBC. These findings align with previous MR studies, which also reported a positive association between PCOS and ERPBC [15]. Our findings are also consistent with other studies wherein genetically predicted PCOS was associated with estrogen receptor (ER) positive rather than ER-negative breast cancer [16]. Our LHC-MR analyses are based on larger sample sizes while being able to effectively mitigate a variety of problems such as sample overlap, bidirectional causality, and the inability to predict confounding effects, which provides a guarantee of the reliability of the conclusions. Our findings do not support a causal relationship between PCOS and ERNBC, suggesting that vertical pleiotropy alone may not fully account for the comorbidity of these conditions. Thus, in the next study, multiple analytical approaches were used to probe the shared genetic mechanisms of the two diseases at the level of horizontal pleiotropy.
Initially, the PLACO analysis method was employed to identify pleiotropic genes at the variant level in order to validate the genetic mechanism of co-morbid risk. It is worth noting that PLACO identified rs1121980 and rs8050136 were localized to locus 16q12.2, while risk locus 16q12.2 was detected in all three trait pairs and the gene FTO, which is associated with adiposity and obesity, was located at this risk locus [37]. The FTO gene (associated with fat mass and obesity) promotes proliferation, enrichment, and metastasis of BC cells by mediating m6A demethylation of BNIP3 within the pro-apoptotic protein family, and the stability of this effect is negatively regulated via a YTHDF2-independent mechanism [33, 38]. In PCOS patients, FTO polymorphisms ultimately mediate increased susceptibility to PCOS by modulating the activity of nearby genes, such as IRX5 and IRX3, with effects on fat mass, BMI, and risk of obesity. Interestingly enough, even after adjusting for BMI, effects on PCOS manifestation caused by FTO mutation have not been completely eliminated, and the potential mechanism has not been studied yet, which needs to be further explored [39].
Subsequent to our analyses, we conducted a MAGMA association analysis to identify genes based on position linked with PCOS and BC, and we employed eMAGMA to pinpoint tissue-specific pleiotropic genes. The RalB gene was identified as an overlapping gene of MAGMA and eMAGMA, and is thought to play an important role downstream of Ras oncoproteins. Specifically, RalB promotes cell invasion and accelerates cancer progression through two cytokines, RGL 1 and RGL 2, as well as by regulating a group of proteins called exocyst that are present in cells. A set of clinical data investigating RalB levels in breast cancer patients showed a significant positive relationship between BC disease progression and elevated RalB levels [40], while Shima Ghoroghi et al. identified RalB as an important factor in the poor prognosis of BC patients [31]. However, the mechanism of the genetic role of the RalB gene in PCOS patients has not been systematically elaborated by research, and more samples and clinical studies as well as experimental designs are needed to fill this gap in the future. This analysis reiterated the pleiotropic nature of the pleiotropic loci targeted gene FTO across all three trait pairs, supporting its role in promoting comorbidity, as previously discussed. Moreover, we observed that the ESR1 gene recurred in the trait pairs PCOS-BCALL and PCOS-ERPBC. The ESR1 gene, encoding estrogen receptor-α (ER-α), emerges as a critical factor for sensitivity in BC [41], stimulating cell proliferation through the induction of MYC and cyclin D1 expression [42]. Additionally, mutations in ESR1 enhance drug resistance in this type of BC by altering protein structure and recruiting coactivators, resulting in reduced sensitivity to endocrine therapy (ET) [43, 44]. In the context of PCOS, estrogen is known to facilitate cAMP production and activate phosphoinositide 3-kinase (PI3K) and extracellular signal-regulated kinase (ERK), further implicating ESR1 in the development of comorbidities. Apart from FTO, no additional overlapping pleiotropic genes were identified in PCOS-ERNBC.
Based on the eMAGMA analysis, RALB has been identified as a pleiotropic gene associated with both PCOS and BC. Pathway enrichment via Metascape highlights its involvement in regulating cellular catabolic processes, positive regulation of catabolic processes, and autophagy, all crucial for understanding its pleiotropic roles. The regulation of cellular catabolism is key for maintaining energy homeostasis, which is vital in metabolic diseases like PCOS, and in cancer progression, where altered catabolism supports rapid tumor growth.[45] In BC, tumor cells often exhibit upregulated autophagy to survive metabolic stress and nutrient deprivation [46], a process similarly observed in PCOS. For instance, heightened autophagy in ovarian cells can disrupt follicular development in PCOS [47], while in BC, autophagy aids cancer cell survival during chemotherapy [48]. Positive regulation of catabolism enhances macromolecule breakdown, reducing oxidative stress and creating an environment conducive to both tumor progression and metabolic imbalance [49]. These shared pathways suggest a common mechanistic link in RALB-mediated processes, providing further insights into the genetic architecture underlying both conditions. Understanding these biological mechanisms offers deeper insight into how PCOS and BC share catabolism-related genetic mechanisms.
Based on the information from the genetic data, our study identified multiple risk loci shared by both PCOS-ERPBC and PCOS-BCALL, providing a basis for the existence of a pleiotropic effect that is difficult to ignore. The common genetic mechanism of PCOS-ERPBC and PCOS-BCALL observed in multi-pleiotropic analyses at all levels further supports a possible common aetiology, although there is no evidence of a significant genome-wide genetic association. Although PCOS and ERNBC trait pairs have a certain genetic basis, we only found that this trait pair overlaps FTO with the other two trait pairs at the SNP level, and this overlap has not been found at the level of genes and biological pathways. This suggests that a high genetic association can explain PCOS and ERPBC comorbidity, but not PCOS and ERNBC comorbidity.
We recognize that this study has some limitations. First, given the statistical power of this study and to avoid bias due to population stratification, the genetic data we chose were derived only from common variants of European ancestry, which limits the generalizability of our findings to a wide range of ethnic populations. Second, this study only explored the shared genetic mechanisms between BCALL, ERNBC, ERPBC, and PCOS, while other BC subtypes based on the expression of other hormone receptors, such as human epidermal growth factor receptor 2 (HER2), were not investigated, and future BC-specific typing cross-sectional studies based on a larger sample size are necessary to further broaden our research results. Third, while the genes associated with PCOS and BC were identified in this study, longitudinal studies and more experimental data are needed in the future to unravel the underlying biological mechanisms.
In conclusion, our research elucidates the intricate genetic connections underpinning these two prevalent female conditions. Through demonstrating genetic correlations, identifying potential pleiotropic loci, and suggesting causal links, our study identified the genetic mechanism of the shared gene between PCOS and BC, and shared pathways suggest a common mechanistic link in RALB-mediated processes, providing insights into the genetic basis underlying both conditions. This comprehensive analysis opens new avenues for exploring the complex interplay between genetic factors and offers advanced insights into the converging pathways that may underlie these diseases.
Supplementary Information
Acknowledgements
We would like to thank the authors of all the GWAS who made their summary statistics available for the benefit of this study, including the following: the Breast Cancer Association Consortium (BCAC) and IEU OpenGWAS without whom this effort would not be possible. We also thank Day, Felix (https://orcid.org/0000-0003-3789-7651) who provide polycystic ovarian syndrome data.
Author contributions
H.J. conceptualized, supervised this project, and wrote the manuscript. K.B., M.C., Q.Z., and T.Y. performed the main analyses and wrote the manuscript. W.X. and W.M. performed the statistical analysis and assisted with the interpretation of results. All authors discussed the results and commented on the paper. All authors reviewed the manuscript.
Funding
This study was supported by the Research Project Supported by the Shanxi Scholarship Council of China (2021-157), Key Laboratory of Cellular Physiology, Ministry of Education, Shanxi Medical University (No. CPOF202310), the Youth Project of Shanxi Basic Research Program (No. 202303021222339), and Central Guidance for Local Science and Technology Development Funds Project (No. YDZJSX2024D068). The funder had no role in the design, implementation, analysis, interpretation of the data, approval of the manuscript, and decision to submit the manuscript for publication.
Availability of data and materials
No datasets were generated or analysed during the current study. The datasets supporting the conclusions of this article are included within the article and its additional files.
Declarations
Ethics approval and consent to participate
All cited genome-wide association studies were public, epigenome-wide association studies, and summary-level and individual-level data had been approved by a relevant review board.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kaixin Bi and Miaoran Chen have contributed equally to this work.
References
- 1.Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, Jemal A, Siegel RL. Breast cancer statistics, 2022. CA Cancer J Clin. 2022;72(6):524–41. 10.3322/caac.21754. [DOI] [PubMed] [Google Scholar]
- 2.Belaid A, Kanoun S, Kallel A, Ghorbel I, Azoury F, Heymann S, Pichenot C, Verstraet R, Marsiglia H, Bourgier C. Breast cancer with axillary lymph node involvement. Cancer Radiother. 2010;14(Suppl 1):S136-146. 10.1016/s1278-3218(10)70017-2. [DOI] [PubMed] [Google Scholar]
- 3.Livadas S, Diamanti-Kandarakis E. Polycystic ovary syndrome: definitions, phenotypes and diagnostic approach. Front Horm Res. 2013;40:1–21. 10.1159/000341673. [DOI] [PubMed] [Google Scholar]
- 4.Pasquali R, Gambineri A. Glucose intolerance states in women with the polycystic ovary syndrome. J Endocrinol Invest. 2013;36(8):648–53. 10.1007/bf03346757. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Mersereau JE, Khankari N, Bradshaw PT, McCullough LE, Cleveland R, Shantakumar S, Teitelbuam SL, Neugut AI, Senie RT, et al. Polycystic ovarian syndrome (PCOS), related symptoms/sequelae, and breast cancer risk in a population-based case-control study. Cancer Causes Control. 2016;27(3):403–14. 10.1007/s10552-016-0716-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Key T, Appleby P, Barnes I, Reeves G. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94(8):606–16. 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 7.Berrino F, Pasanisi P, Bellati C, Venturelli E, Krogh V, Mastroianni A, Berselli E, Muti P, Secreto G. Serum testosterone levels and breast cancer recurrence. Int J Cancer. 2005;113(3):499–502. 10.1002/ijc.20582. [DOI] [PubMed] [Google Scholar]
- 8.Secreto G, Girombelli A, Krogh V. Androgen excess in breast cancer development: implications for prevention and treatment. Endocr Relat Cancer. 2019;26(2):R81-r94. 10.1530/erc-18-0429. [DOI] [PubMed] [Google Scholar]
- 9.Collaborative Group on Hormonal Factors in Breast Cancer. Breast cancer and hormonal contraceptives: collaborative reanalysis of individual data on 53 297 women with breast cancer and 100 239 women without breast cancer from 54 epidemiological studies. Lancet. 1996;347(9017):1713–27. 10.1016/s0140-6736(96)90806-5. [DOI] [PubMed] [Google Scholar]
- 10.Anderson KN, Schwab RB, Martinez ME. Reproductive risk factors and breast cancer subtypes: a review of the literature. Breast Cancer Res Treat. 2014;144(1):1–10. 10.1007/s10549-014-2852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunter MJ, Hoover DR, Yu H, Wassertheil-Smoller S, Rohan TE, Manson JE, Li J, Ho GY, Xue X, Anderson GL, et al. Insulin, insulin-like growth factor-I, and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 2009;101(1):48–60. 10.1093/jnci/djn415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mykhalchenko K, Lizneva D, Trofimova T, Walker W, Suturina L, Diamond MP, Azziz R. Genetics of polycystic ovary syndrome. Expert Rev Mol Diagn. 2017;17(7):723–33. 10.1080/14737159.2017.1340833. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Qi G, Jiang X, O’Mara TA, Zhao N, Bolla MK, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet. 2020;52(6):572–81. 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavala VA, Serrano-Gomez SJ, Dutil J, Fejerman L. Genetic epidemiology of breast cancer in latin America. Genes (Basel). 2019. 10.3390/genes10020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen Y, Wu X, Peng H, Li C, Jiang Y, Su Z, Liang H, Liu J, He J, Liang W. Breast cancer risk in patients with polycystic ovary syndrome: a Mendelian randomization analysis. Breast Cancer Res Treat. 2021;185(3):799–806. 10.1007/s10549-020-05973-z. [DOI] [PubMed] [Google Scholar]
- 16.Zhu T, Cui J, Goodarzi MO. Polycystic ovary syndrome and breast cancer subtypes: a Mendelian randomization study. Am J Obstet Gynecol. 2021;225(1):99–101. 10.1016/j.ajog.2021.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, Kraft P, Lin N, Huang H, Broer L, et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018;14(12): e1007813. 10.1371/journal.pgen.1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, Lemaçon A, Soucy P, Glubb D, Rostamianfar A, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–4. 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh PR, Lareau C, Shoresh N, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50(4):621–9. 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindley G, Frei O, Shadrin AA, Cheng W, O’Connell KS, Icick R, Parker N, Bahrami S, Karadag N, Roelfs D, et al. Charting the landscape of genetic overlap between mental disorders and related traits beyond genetic correlation. Am J Psychiatry. 2022;179(11):833–43. 10.1176/appi.ajp.21101051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connell KS, Frei O, Bahrami S, Smeland OB, Bettella F, Cheng W, Chu Y, Hindley G, Lin A, Shadrin A, et al. Characterizing the genetic overlap between psychiatric disorders and sleep-related phenotypes. Biol Psychiatry. 2021;90(9):621–31. 10.1016/j.biopsych.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Fominykh V, Shadrin AA, Jaholkowski PP, Bahrami S, Athanasiu L, Wightman DP, Uffelmann E, Posthuma D, Selbæk G, Dale AM, et al. Shared genetic loci between Alzheimer’s disease and multiple sclerosis: crossroads between neurodegeneration and immune system. Neurobiol Dis. 2023;183:106174. 10.1016/j.nbd.2023.106174. [DOI] [PubMed] [Google Scholar]
- 24.Darrous L, Mounier N, Kutalik Z. Simultaneous estimation of bi-directional causal effects and heritable confounding from GWAS summary statistics. Nat Commun. 2021;12(1):7274. 10.1038/s41467-021-26970-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray D, Chatterjee N. A powerful method for pleiotropic analysis under composite null hypothesis identifies novel shared loci between Type 2 Diabetes and Prostate Cancer. PLoS Genet. 2020;16(12): e1009218. 10.1371/journal.pgen.1009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Liu P, Zhang Z, Ye Y, Yi S, Fan C, Zhao W, Liu J. Genetic overlap and causality between COVID-19 and multi-site chronic pain: the importance of immunity. Front Immunol. 2024;15:1277720. 10.3389/fimmu.2024.1277720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4): e1004219. 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerring ZF, Mina-Vargas A, Gamazon ER, Derks EM. E-MAGMA: an eQTL-informed method to identify risk genes using genome-wide association study summary statistics. Bioinformatics. 2021;37(16):2245–9. 10.1093/bioinformatics/btab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghoroghi S, Mary B, Larnicol A, Asokan N, Klein A, Osmani N, Busnelli I, Delalande F, Paul N, Halary S, et al. Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. Elife. 2021. 10.7554/eLife.61539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Chen H, Heine J, Lindstrom S, Turman C, Warner ET, Winham SJ, Vachon CM, Tamimi RM, Kraft P, et al. A genome-wide association study of mammographic texture variation. Breast Cancer Res. 2022;24(1):76. 10.1186/s13058-022-01570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci. 2022. 10.3390/ijms23073800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cazzaniga M, Bonanni B. Relationship between metabolic disorders and breast cancer incidence and outcomes. Is there a preventive and therapeutic role for berberine? Anticancer Res. 2018;38(8):4393–402. 10.21873/anticanres.12741. [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Zhao X, Meng Q, Li B. Screening of potential biomarkers for polycystic ovary syndrome and identification of expression and immune characteristics. PLoS ONE. 2023;18(10): e0293447. 10.1371/journal.pone.0293447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niklaus NJ, Tokarchuk I, Zbinden M, Schläfli AM, Maycotte P, Tschan MP. The multifaceted functions of autophagy in breast cancer development and treatment. Cells. 2021. 10.3390/cells10061447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusinska R, Górniak P, Pastorczak A, Fendler W, Potemski P, Mlynarski W, Kordek R. Influence of genomic variation in FTO at 16q12.2, MC4R at 18q22 and NRXN3 at 14q31 genes on breast cancer risk. Mol Biol Rep. 2012;39(3):2915–9. 10.1007/s11033-011-1053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barber TM, Bennett AJ, Groves CJ, Sovio U, Ruokonen A, Martikainen H, Pouta A, Hartikainen AL, Elliott P, Lindgren CM, et al. Association of variants in the fat mass and obesity associated (FTO) gene with polycystic ovary syndrome. Diabetologia. 2008;51(7):1153–8. 10.1007/s00125-008-1028-6. [DOI] [PubMed] [Google Scholar]
- 40.Zago G, Veith I, Singh MK, Fuhrmann L, De Beco S, Remorino A, Takaoka S, Palmeri M, Berger F, Brandon N, et al. RalB directly triggers invasion downstream Ras by mobilizing the Wave complex. Elife. 2018. 10.7554/eLife.40474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, He Y, Qin Z, Jiang Y, Jin G, Ma H, Dai J, Chen J, Hu Z, Guan X, et al. Evaluation of functional genetic variants at 6q25.1 and risk of breast cancer in a Chinese population. Breast Cancer Res. 2014;16(4):422. 10.1186/s13058-014-0422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas C, Gustafsson J. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011;11(8):597–608. 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 43.Herzog SK, Fuqua SAW. ESR1 mutations and therapeutic resistance in metastatic breast cancer: progress and remaining challenges. Br J Cancer. 2022;126(2):174–86. 10.1038/s41416-021-01564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dustin D, Gu G, Fuqua SAW. ESR1 mutations in breast cancer. Cancer. 2019;125(21):3714–28. 10.1002/cncr.32345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castelli S, De Falco P, Ciccarone F, Desideri E, Ciriolo MR. Lipid catabolism and ROS in cancer: a bidirectional liaison. Cancers (Basel). 2021. 10.3390/cancers13215484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Q, Mao B, Cheng J, Gao Y, Jiang K, Chen J, Yuan Z, Meng S. YAP enhances autophagic flux to promote breast cancer cell survival in response to nutrient deprivation. PLoS ONE. 2015;10(3): e0120790. 10.1371/journal.pone.0120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumariya S, Ubba V, Jha RK, Gayen JR. Autophagy in ovary and polycystic ovary syndrome: role, dispute and future perspective. Autophagy. 2021;17(10):2706–33. 10.1080/15548627.2021.1938914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hassan A, Zhao Y, Chen X, He C. Blockage of autophagy for cancer therapy: a comprehensive review. Int J Mol Sci. 2024. 10.3390/ijms25137459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Pu X, Wang X, Xu M. Reprogramming of lipid metabolism in the tumor microenvironment: a strategy for tumor immunotherapy. Lipids Health Dis. 2024;23(1):35. 10.1186/s12944-024-02024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study. The datasets supporting the conclusions of this article are included within the article and its additional files.