Abstract
Transcription factor IIIA (TFIIIA) binds to the internal control region of the 5S RNA gene as the first step in the in vitro assembly of a TFIIIB-TFIIIC-TFIIIA-DNA transcription complex. An 81-amino-acid domain that is present between zinc fingers 8 and 9 of TFIIIA from Saccharomyces cerevisiae is essential for the transcription factor activity of this protein (C. A. Milne and J. Segall, J. Biol. Chem. 268:11364–11371, 1993). We have monitored the effect of mutations within this domain on the ability of TFIIIA to support transcription of the 5S RNA gene in vitro and to maintain cell viability. TFIIIA with internal deletions that removed residues 282 to 315, 316 to 334, 328 to 341, or 342 to 351 of the 81-amino-acid domain retained activity, whereas TFIIIA with a deletion of the short leucine-rich segment 352NGLNLLLN359 at the carboxyl-terminal end of this domain was devoid of activity. Analysis of the effects of double and quadruple mutations in the region extending from residue 336 to 364 confirmed that hydrophobic residues in this portion of the 81-amino-acid domain, particularly L343, L347, L354, L356, L357, and L358, and to a lesser extent F336 and L337, contributed to the ability of TFIIIA to promote transcription. We propose that these hydrophobic residues play a role in mediating an interaction between TFIIIA and another component of the transcriptional machinery. We also found that TFIIIA remained active if either zinc finger 8 or zinc finger 9 was disrupted by mutation but that TFIIIA containing a disruption of both zinc finger 8 and zinc finger 9 was inactive.
The yeast Saccharomyces cerevisiae has served as a useful organism for detailed characterization of the factors that direct accurate initiation of transcription by RNA polymerase III and for investigation of the molecular interactions involved in the assembly of stable initiation complexes (reviewed in references 27 and 29). The three accessory transcription factors of S. cerevisiae that are minimally required to promote accurate initiation of transcription of the 5S RNA gene by RNA polymerase III are TFIIIA, TFIIIB, and TFIIIC. These factors assemble sequentially onto the 5S RNA gene in vitro to form a stable preinitiation complex that recruits RNA polymerase III to the start site of transcription (reviewed in references 29 and 86). TFIIIA, a sequence-specific DNA-binding protein that contains nine zinc fingers of the Cys2-His2 type, binds to the internal control region (ICR) of the 5S RNA gene as the first step in the in vitro assembly of this multifactor complex. This is followed by incorporation of the large, multisubunit TFIIIC (or τ) into the TFIIIA-DNA complex. Formation of the TFIIIC-TFIIIA-DNA complex is necessary for recruitment of TFIIIB, a multisubunit factor that consists of TFIIIB70/Brf, TFIIIB90/Tfc5, and the TATA-binding protein, TBP (10, 46). In the TFIIIB-TFIIIC-TFIIIA-DNA complex, TFIIIB is stably bound upstream of the start site of transcription and recruits RNA polymerase III for multiple rounds of transcription (45).
TFIIIA is required only for transcription of the 5S RNA gene. On tRNA genes, TFIIIC binds directly to the intragenic A- and B-box promoter elements and acts to place TFIIIB upstream of the start site of transcription (47). Despite the requirement for TFIIIA in the assembly of a preinitiation complex on the 5S RNA gene, the relative placement of the individual subunits of TFIIIC and TFIIIB in preinitiation complexes formed on a 5S RNA gene and on a tRNA is similar (5, 6, 9).
The gene, or cDNA, coding for TFIIIA has been identified from S. cerevisiae, various amphibian species, and humans. Although the deduced sequences of these TFIIIAs indicate that they are structurally similar in that they contain nine zinc fingers of the Cys2-His2 type, the extent of sequence identity among the TFIIIAs from these organisms is low (2, 3, 23, 28, 32, 87). Moreover, the 81-amino-acid domain that interrupts the otherwise repeating nature of the zinc finger motifs between fingers 8 and 9 of yeast TFIIIA is not present in human TFIIIA or Xenopus TFIIIA. These differences among TFIIIAs are consistent with the observation that several components of the RNA polymerase III transcriptional machinery differ extensively between organisms. For example, both human and yeast TFIIIBs contain a subunit, referred to as TFIIIB90 in the human factor (82) and as TFIIIB70/Brf in the yeast factor (11, 18, 55), that is related to TFIIIB, yet it is only the TFIIB-related amino-terminal portions of the proteins that show significant identity. Another striking example is the complete absence of sequence similarity between the subunit of mammalian TFIIIC that interacts with the B-box region of tRNA genes (49, 51) and the functionally related B-box binding subunit of yeast TFIIIC (50).
Although Xenopus TFIIIB and TFIIIC are relatively uncharacterized, Xenopus TFIIIA and its interaction with the 50-bp ICR of the amphibian 5S RNA gene have been studied extensively (reviewed in reference 74). The ICR of the Xenopus 5S RNA gene contains three elements that contribute to efficient transcription of the gene: the A box, which spans nucleotides +50 to +64; the intermediate element, which spans nucleotides +67 to +72; and the C box, which spans nucleotides +80 to +97 (8, 66, 67). Xenopus TFIIIA binds to the ICR (25) such that its amino terminus is oriented towards the 3′ end of the ICR and its carboxyl terminus is positioned towards the 5′ end of the ICR (59, 80). The three amino-terminal and three carboxyl-terminal fingers of the molecule are proposed to wrap around the major groove of the DNA helix at each end of the ICR; the zinc fingers in the middle of the protein are thought to lie on one side of the helix, with finger 5 contacting the major groove and fingers 4 and 6 each crossing the minor groove (16, 26, 33, 35, 36, 58). The three amino-terminal zinc fingers interact with the C box with an affinity that is comparable to that of the intact protein (53). The interaction between the carboxyl-terminal zinc fingers and the A box (16, 35) appears to be necessary for transcription (72). Indeed, mutations that disrupt any one of the three carboxyl-terminal zinc fingers lead to reduced transcription (17, 21, 70). These carboxyl-terminal fingers may play a role in transcription by properly positioning the portion of Xenopus TFIIIA that extends beyond the ninth zinc finger. A 14-amino-acid segment that is present in this carboxyl-terminal extension is essential for the transcription factor activity of this TFIIIA (57, 76, 80). Human TFIIIA is thought to bind to the 5S RNA gene in a manner analogous to that of Xenopus TFIIIA, as both the sizes and the patterns of the DNase I footprints generated by these TFIIIAs on their respective templates are similar (62, 81).
Yeast TFIIIA, like its amphibian counterpart, binds to the 5S RNA gene with its carboxyl terminus positioned towards the 5′ end of the gene (61, 71). The ICR of the yeast 5S RNA gene, which is considerably smaller than that of the Xenopus 5S RNA gene, consists of only a C-box element between nucleotides +81 and +94 (13). The DNase I footprint obtained with yeast TFIIIA on the yeast 5S RNA gene is correspondingly smaller than that of Xenopus TFIIIA on the Xenopus 5S RNA gene; the region protected by yeast TFIIIA is 35 bp, extending from nucleotide +64 to nucleotide +99 of the 5S RNA gene (10, 71). The smaller DNase I footprint provided by yeast TFIIIA compared with the footprints generated by Xenopus TFIIIA and human TFIIIA can be accounted for by the absence of an intimate interaction of zinc fingers 6 through 9 of yeast TFIIIA with DNA (71). Site-specific DNA-protein photo-cross-linking suggests, however, that yeast TFIIIA may be positioned over a larger region of the gene than that detected by DNase I footprinting, particularly in the TFIIIB-TFIIIC-TFIIIA-DNA complex (9).
We previously analyzed a series of truncated forms of yeast TFIIIA for their ability to bind to the 5S RNA gene, incorporate TFIIIC into the TFIIIA-DNA complex, and support transcription of the 5S RNA gene (61). We found that a polypeptide containing the three amino-terminal zinc fingers binds to the ICR of the 5S RNA gene with an affinity comparable to that of intact TFIIIA and that this truncated form of TFIIIA can recruit TFIIIC (61, 71). The resultant TFIIIC-TFIIIA-DNA complex, however, is unable to support transcription of the 5S RNA gene. We found that the yeast-specific 81-amino-acid domain that is present between zinc fingers 8 and 9 is essential for the transcription factor activity of yeast TFIIIA (61). As a step towards understanding the role of this novel 81-amino-acid domain in establishing an active transcription complex, we carried out a mutational analysis to identify amino acids within this domain that are essential for its transcription factor activity. In addition, we assessed the potential role of zinc fingers 8 and 9 in the transcription factor activity of yeast TFIIIA.
MATERIALS AND METHODS
Plasmids.
pXS-TFC2, which served as the parental plasmid for introduction of mutations into the coding sequence for TFIIIA, was constructed as follows. First, an XbaI- and SspI-less derivative of pBluescript II SK(+) was generated. The unique XbaI site of pBluescript II SK(+) was destroyed by digesting the plasmid with XbaI, filling in the overhanging ends with the Klenow form of DNA polymerase in the presence of deoxynucleoside triphosphates (dNTPs), and religating the DNA. The resultant plasmid was digested with SspI to yield a 2,831-bp fragment and a 130-bp fragment that contains the promoter for the β-lactamase gene. The large fragment was gel purified and ligated with a 55-bp fragment that contains a weak bacterial promoter (kindly provided by D. E. Pulleyblank) to produce pXS, an XbaI- and SspI-less vector. pXS-TFC2 was generated by cloning a KpnI-BamHI fragment, which contains the coding region of TFIIIA obtained from pJA454 (3), between the corresponding sites in the polylinker of pXS. This places the coding sequence for TFIIIA downstream of a promoter for T7 RNA polymerase.
Plasmids coding for versions of TFIIIA with carboxyl-terminal truncations were generated by replacing the XbaI-BamHI fragment of pXS-TFC2 with DNA amplified from pJA454 by PCR. The reverse primers for the amplification reactions contained a BamHI restriction site at their 5′ ends followed by a stop codon and then sequence from TFC2. The forward primer (primer A) annealed within the coding region of the eighth zinc finger of TFIIIA. The PCR products were gel purified, digested with XbaI and BamHI, repurified, and cloned between the corresponding sites of pXS-TFC2 to generate plasmids coding for versions of TFIIIA that contained amino acids 1 to 378, 1 to 359, 1 to 353, 1 to 347, 1 to 339, and 1 to 311.
Site-directed mutagenesis (alanine scanning) was achieved by recombinant PCR using the overlap extension procedure described in references 39 and 40. Complementary reverse and forward oligonucleotides containing the desired mutation(s) were used in separate PCRs with pJA454 as the template. The sequences upstream and downstream of the mutant codons were amplified by using the reverse mutant oligonucleotide and upstream primer A (see above) and the forward mutant oligonucleotide and downstream primer B, respectively. Primer B anneals within the coding region of TFIIIA downstream of the ninth zinc finger. The PCR products of the two reactions were gel purified, mixed, and subjected to three PCR cycles in order to allow extension of the heteroduplexes formed between the overlapping mutant sequences. The extended heteroduplexes were then amplified in the presence of primers A and B, and the resulting 502-bp fragment was gel purified, digested with XbaI and SspI, repurified, and used to replace the corresponding XbaI-SspI fragment of pXS-TFC2. Constructs made in this way directed the synthesis of versions of TFIIIA containing the following alanine-scanning mutations: K287A/K289A, L296A/V297A, D299A/H300A, K308A/H309A, D314A/E315A, R324A/K325A, F336A/L337A, D342A/E344A, L343A/L347A, K345A/R346A, E348A/E351A, N352A/N355A, L354A/L356A, L354A, L356A, L357A/L358A, N359A, and R363A/K364A. Versions of pXS-TFC2 coding for TFIIIA containing quadruple alanine-scanning mutations were made by recombinant PCR as described above by using a version of pXS-TFC2 that contained the appropriate double alanine-scanning mutation as the template in the first set of PCRs. This approach generated mutant genes encoding versions of TFIIIA with the following quadruple alanine-scanning mutations: F336A/L337A/L343A/L347A, D342A/E344A/E348A/E351A, L343A/L347A/L354A/L356A, L343A/L347A/L357A/L358A, K345A/R346A/L357A/L358A, K345A/R346A/R363A/K364A, L354A/L356A/L357A/L358A, and L357A/L358A/R363A/K364A.
Versions of pXS-TFC2 coding for TFIIIA lacking amino acids 282 to 287, 282 to 315, and 282 to 353 were constructed by taking advantage of the unique EcoRV restriction site at codon 282 and unique restriction sites introduced by alanine-scanning mutations at codons 287, 315, and 353. To construct pXS-TFC2(Δ282-287), pXS-TFC2(K287A/K289A) was digested with NdeI, and after the overhanging ends had been filled in with the Klenow form of DNA polymerase in the presence of dNTPs, the DNA was digested with EcoRV. The ∼5-kbp fragment was gel purified and religated to create pXS-TFC2(Δ282-287), which codes for a version of TFIIIA with an in-frame deletion and which retains the K289A mutation. To construct pXS-TFC2(Δ282-315), pXS-TFC2(D314A/E315A) was digested with PstI, and after the overhanging ends had been blunted by treatment with the Klenow form of DNA polymerase, first in the absence and then in the presence of dNTPs, the DNA was digested with EcoRV. The ∼4.9-kbp fragment was gel purified and religated to create pXS-TFC2(Δ282-315), which codes for a version of TFIIIA with an in-frame deletion. To construct pXS-TFC2(Δ282-353), pXS-TFC2(N352A/N355A) was digested with NheI, and after the overhanging ends had been filled in with the Klenow form of DNA polymerase in the presence of dNTPs, the DNA was digested with EcoRV. The ∼4.8-kbp fragment was gel purified and religated to create pXS-TFC2(Δ282-353), which codes for a version of TFIIIA with an in-frame deletion and which retains the N355A mutation.
Versions of pXS-TFC2 coding for TFIIIA(Δ316-334), TFIIIA(Δ328-341), TFIIIA(Δ342-351), and TFIIIA(Δ352-359) were made by recombinant PCR using a variation of the overlap extension procedure described above (39, 40). Partially overlapping oligonucleotides spanning the deletion junction were used as reverse and forward primers in separate PCRs with primers A and B (see above), respectively. The partially overlapping PCR products were gel purified, mixed, and subjected to three PCR cycles to allow extension of heteroduplexes, which were then amplified by the addition of primers A and B. The amplified DNA, which contained a deletion of the coding region of TFIIIA, was gel purified, digested with XbaI and SspI, repurified, and cloned between the corresponding sites of pXS-TFC2. This recombinant PCR approach was used previously to construct a gene encoding TFIIIA-Δ81, referred to as TFIIIA(Δ284-364) in this article, and is described in detail in reference 61.
All PCR amplifications were performed by using the high-fidelity Vent DNA polymerase as instructed by the manufacturer (New England Biolabs). The sequence of all amplified DNA was verified by DNA sequencing. The sequences of the oligonucleotides used to generate the mutations are available upon request.
Mutation of a zinc-coordinating residue in each of fingers 8 and 9 was obtained in a pilot experiment using Taq DNA polymerase under conditions of reduced fidelity (52) to introduce random mutations during PCR amplification of the sequence of TFIIIA from codons 266 to 397. A 100-μl reaction mixture contained 16.6 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 6.7 μM EDTA, 0.17 mg of bovine serum albumin/ml, 10 mM β-mercaptoethanol, 1 mM each dNTP, 6.1 mM MgCl2, 0.5 mM MnCl2, 20 pmol each of primers A and B (see above), 6 fmol of pJA454 as the template, and 2.5 U of Taq DNA polymerase. The amplified DNA was gel purified, digested with XbaI and SspI, repurified, and ligated between the corresponding sites of pXS-TFC2. The ligated products were recovered by transformation into Escherichia coli, and the DNAs of several plasmids were sequenced from the XbaI site to the SspI site. This led to the identification of a plasmid that contained mutations in both codons 272 and 367. A unique EcoRV restriction site located between these codons was used to separate the two mutations: an NcoI-EcoRV fragment containing the mutation of codon 272 and an EcoRV-BamHI fragment containing the mutation of codon 367 were separately subcloned between the corresponding sites of pXS-TFC2 to generate pXS-TFC2(H272R) and pXS-TFC2(C367Y), respectively. DNA sequencing confirmed that these plasmids contained the single mutations H272R and C367Y.
The yeast shuttle vector pG3 (73), a pUC18-derived plasmid that contains a 2μm origin of replication and the selectable marker TRP1, was used for in vivo expression of wild-type and mutant forms of TFIIIA. KpnI-BamHI fragments containing the open reading frames of the wild-type and mutant versions of TFIIIA were purified from pXS-TFC2 plasmids and inserted between the KpnI and SalI sites of pG3 after the BamHI- and SalI-generated ends had been filled in by the Klenow form of DNA polymerase I in the presence of dNTPs. This placed the coding region of TFIIIA between the constitutive promoter of the glyceraldehyde-3-phosphate dehydrogenase gene and the transcription terminator of the phosphoglycerate kinase gene.
In vitro synthesis of TFIIIA.
Wild-type and mutant versions of TFIIIA were synthesized in vitro by using the TnT coupled transcription-translation system (Promega), in which a rabbit reticulocyte lysate supports translation of transcripts synthesized by T7 RNA polymerase. The reactions were carried out according to the manufacturer’s instructions and with the addition of ZnSO4 to 0.1 mM. pXS-TFC2 or its variants were used as the template unless otherwise indicated. TFIIIA(1-397) and TFIIIA(1-365) were synthesized in vitro from pJA454 that had been linearized with SspI and from pJA454-1 that had been linearized with BamHI, respectively (61). Wild-type and mutant proteins that had been synthesized in the presence of [35S]methionine were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis to confirm that protein of the appropriate size was synthesized (data not shown). In vitro-synthesized proteins used in gel mobility shift assays and transcription assays were not radiolabeled.
EMSAs and transcription assays.
Electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (71) except that 0.25 μg of pBluescript II SK(+) was included as a competitor DNA in the reactions. A 20-μl reaction mixture contained 2 μl of an in vitro transcription-translation reaction mixture that had been programmed to produce the indicated version of TFIIIA, 2 μl of partially purified TFIIIC where indicated, and a 270-bp radioactively end-labeled DNA fragment, which was excised from p19-5S and contains the yeast 5S RNA gene (13). The partially purified TFIIIC-containing fraction derived from yeast was prepared as described for fraction j in reference 77. In vitro transcription assays were performed as described elsewhere (77) with the yeast 5S RNA gene (p19-5S) as a template. A 50-μl reaction mixture contained 4.5 μl of an in vitro transcription-translation reaction mixture that had been programmed to produce the indicated version of TFIIIA and 12 μl of a yeast-derived heparin-agarose fraction (fraction h) that contained TFIIIC, TFIIIB, and RNA polymerase III (77).
Yeast media, culture conditions, and transformations.
Rich medium (yeast extract-peptone-dextrose [YPD]) and minimal medium (synthetic dextrose [SD]) were as previously described (38). All yeast cultures were grown at 30°C. Transformation of yeast cells was performed by the lithium acetate method of Geitz et al. (30).
In vivo analysis of the mutant versions of TFIIIA.
The haploid yeast strain YRW1 (MATα can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 ade2-1 tfc2::LEU2, harboring pJA230) was constructed to test the ability of the variant forms of TFIIIA to support cell viability (60). As the first step in construction of YRW1, the plasmid pRKO (60) was digested with BssHII and BamHI to release a DNA fragment that contains the yeast LEU2 gene flanked by 213 and 300 bp of noncoding sequence from the regions upstream and downstream, respectively, of the TFC2 gene. This fragment was used to replace the entire coding region of the chromosomal TFC2 gene with the LEU2 gene by integrative transformation as follows. The diploid strain LP112 (64) was transformed with the gel-purified BssHII-BamHI fragment, and replacement of one chromosomal copy of the TFC2 gene by LEU2 was confirmed by Southern blot analysis of a Leu+ transformant. Plasmid pJA230, a CEN/ARS-based plasmid with a URA3 selectable marker and a 10-kbp insert of yeast DNA containing RPO26 and TFC2 (4), was then introduced into the TFC2/tfc2::LEU2 strain. Sporulation of a Ura+ transformant generated the haploid strain YRW1. Since TFC2 is an essential gene (3), viability of YRW1 depends on the presence of pJA230.
YRW1 was transformed with derivatives of pG3 directing expression of wild-type or mutant forms of TFIIIA. After transformants had been selected on SD medium lacking uracil and tryptophan, the pG3-containing strains were grown on SD medium lacking tryptophan and containing uracil to allow for loss of pJA230. Cells were then streaked on SD medium containing 5-fluoro-orotic acid (5-FOA) and uracil and lacking tryptophan. Because 5-FOA kills cells containing the URA3 gene, only those cells that have lost pJA230 and that contain a pG3 derivative encoding a functional version of TFIIIA will grow on the 5-FOA-containing plates.
Preparation of anti-TFIIIA and Western blot analysis.
We confirmed that the various forms of pG3-encoded TFIIIA were expressed in vivo by standard Western blot analysis. Polyclonal antibodies were generated by injection of rabbits with 500 μg of bacterially expressed and purified full-length yeast TFIIIA emulsified with an equal volume of complete Freund’s adjuvant. Purification of yeast TFIIIA was carried out as described previously (71) except that as a final step the protein band corresponding to TFIIIA was excised from an SDS-polyacrylamide gel to achieve further purification. Rabbits were boosted every 4 weeks with 100 μg of yeast TFIIIA emulsified with an equal volume of incomplete Freund’s adjuvant. Blood was collected from the rabbits 2 weeks after each boost.
YRW1 and strains of YRW1 containing pG3-derived plasmids that directed expression of mutant versions of TFIIIA were grown overnight in SD medium lacking uracil and tryptophan to an optical density at 600 nm of ∼3.0. The cells from 1 ml of culture at this density, or the appropriate volume of culture if the cell density was different, were harvested by centrifugation. The pellets of cells were resuspended in 1 ml of YPD medium, and extracts of proteins were prepared from the yeast cells as described in reference 89. The proteins were separated on an SDS–10% polyacrylamide gel and then electroblotted onto nitrocellulose filters at 4°C in transfer buffer (25 mM Tris-HCl, 194 mM glycine, 20% methanol, 0.05% SDS). The filters were blocked in phosphate-buffered saline (PBS)-milk (5% powdered skim milk in PBS containing 0.05% Tween) for 1 h to overnight and were then incubated for 1 h in PBS-milk containing a 1:2,000 dilution of crude serum containing polyclonal antibodies against yeast TFIIIA (see above). The filters were washed three times for 5 to 15 min each in PBS-milk and were then incubated in PBS-milk containing a 1:2,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit antibody. The filters were washed four times for 5 to 15 min each in TTBS (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 0.05% Tween), and the secondary antibody was detected by the ECL chemiluminescence system (Amersham).
RESULTS
The nine zinc fingers of yeast TFIIIA occur in succession except for zinc fingers 8 and 9, which are separated by an 81-amino-acid domain (Fig. 1A). We have previously shown that TFIIIA lacking the 81-amino-acid domain recruits TFIIIC to a TFIIIA-5S RNA gene complex; the resultant complex, however, is unable to promote transcription (61). We also found that the ninth zinc finger of TFIIIA, although not essential, contributes to efficient transcription (61). In this study we have defined the region of the 81-amino-acid domain that is essential for its function, which we refer to as its transcription factor activity, and we have further assessed the requirement for the adjacent zinc fingers in supporting efficient transcription.
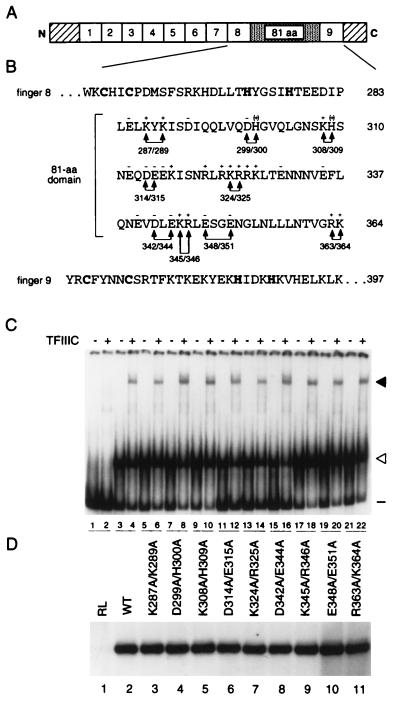
FIG. 1.
Effects of alanine replacement of charged residues within the 81-amino-acid domain on activity of TFIIIA. (A) Schematic representation of yeast TFIIIA. The numbered boxes represent the nine zinc fingers of yeast TFIIIA, the stippled region between fingers 8 and 9 represents the 81-amino-acid domain, and the diagonally striped boxes represent the 48-amino-acid amino-terminal region and the 35-amino-acid carboxyl-terminal region. (B) Amino acid sequence of yeast TFIIIA from residues 253 to 397. The sequences for zinc fingers 8 and 9, with the zinc-coordinating cysteines and histidines in boldface, and for the intervening 81-amino-acid domain are given. The residue number, relative to the initiator methionine, of the last amino acid on each line is given on the right. Minus and plus symbols above residues in the 81-amino-acid domain indicate negatively and positively charged amino acids, respectively. Double arrowheads indicate pairs of charged residues that were mutated to alanine; the positions of the residues are given below the lines connecting the arrowheads. (C) Abilities of mutant versions of TFIIIA to bind to the 5S RNA gene and to recruit TFIIIC to the TFIIIA-DNA complex, as assessed by EMSA. A radioactively labeled DNA fragment containing the yeast 5S RNA gene was incubated with in vitro-synthesized versions of TFIIIA in the absence (odd-numbered lanes) or presence (even-numbered lanes) of partially purified yeast TFIIIC prior to electrophoresis on a nondenaturing polyacrylamide gel. Lanes 1 and 2, in vitro transcription-translation reactions that were not programmed to synthesize TFIIIA; lanes 3 and 4, wild-type TFIIIA; lanes 5 to 22, TFIIIA with the following mutations: K287A/K289A (lanes 5 and 6), D299A/H300A (lanes 7 and 8), K308A/H309A (lanes 9 and 10), D314A/E315A (lanes 11 and 12), K324A/R325A (lanes 13 and 14), D342A/E344A (lanes 15 and 16), K345A/R346A (lanes 17 and 18), E348A/E351A (lanes 19 and 20), and R363A/K364A (lanes 21 and 22). The positions of free DNA (minus sign), TFIIIA-DNA complexes (open arrowhead), and TFIIIC-TFIIIA-DNA complexes (solid arrowhead) are indicated on the right. (D) Abilities of mutant versions of TFIIIA to support in vitro transcription of the 5S RNA gene. In vitro transcription reaction mixtures contained the yeast 5S RNA gene as a template; partially purified yeast TFIIIC, TFIIIB, and RNA polymerase III; and the version of in vitro-synthesized TFIIIA indicated above the lane. RL, reticulocyte lysate (in vitro transcription-translation reaction not programmed to synthesize TFIIIA); (WT, wild type. The RNAs synthesized in vitro were analyzed on a 7 M urea–10% polyacrylamide gel. The autoradiogram shows the portion of the gel containing 5S RNA.
Alanine-scanning mutagenesis through charged regions of the 81-amino-acid domain.
As a first approach towards identification of amino acids within the 81-amino-acid domain that are involved in the transcription factor activity of TFIIIA, we performed alanine-scanning mutagenesis of charged residues. A charged region of a protein is likely to be solvent exposed, and a charged amino acid(s) on this surface may be involved in interprotein contacts in a multiprotein complex. The choice of alanine for substitutions within a target region allows for a consistent series of mutations with a small amino acid that is unable to provide a side-chain interaction involving atoms beyond the β carbon. This approach has been used extensively for identification of amino acids that are critical for protein-protein interactions (15, 84).
Seventeen of the 30 charged residues (assuming that the histidines are positively charged) found within the 81-amino-acid domain of TFIIIA are clustered within three regions that extend from residues 312 to 317, residues 321 to 327, and residues 340 to 348 (Fig. 1B). The first and third of these regions are predominately acidic, whereas the second region consists entirely of basic residues. Using a PCR-based approach for site-directed mutagenesis (see Materials and Methods), we changed nine pairs of adjacent, or nearby, charged amino acids to alanine and assessed the effects of these double mutations on the ability of TFIIIA to direct in vitro transcription of the 5S RNA gene.
For these studies, the mutant forms of TFIIIA were synthesized in vitro (see Materials and Methods). We first confirmed that approximately equivalent amounts of TFIIIA of the appropriate sizes were produced, as monitored by SDS-polyacrylamide gel electrophoretic analysis of proteins synthesized in the presence of [35S]methionine (data not shown). As a preliminary test for the functional integrity of the mutant proteins, we used an EMSA to assess formation of protein-DNA complexes. Because the three amino-terminal zinc fingers of TFIIIA suffice for high-affinity binding of TFIIIA to the ICR of the 5S RNA gene and for incorporation of TFIIIC into the TFIIIA-DNA complex (61, 71), we anticipated that mutations within the 81-amino-acid domain would not impair formation of a TFIIIC-TFIIIA-DNA complex. Indeed, each of the in vitro-synthesized mutant proteins bound to the 5S RNA gene (Fig. 1C, odd-numbered lanes from 5 to 21) and recruited TFIIIC (Fig. 1C, even-numbered lanes from 6 to 22) in a manner similar to wild-type TFIIIA (Fig. 1C, lanes 3 and 4). Each form of TFIIIA had about the same activity, as measured by the ability to form a TFIIIC-TFIIIA-DNA complex in this qualitative EMSA. The in vitro-synthesized proteins were then tested for their ability to support in vitro transcription of the 5S RNA gene in the presence of a yeast fraction containing TFIIIC, TFIIIB, and RNA polymerase III (see Materials and Methods). All nine of the mutant forms of TFIIIA containing pairwise substitutions of charged residues (K287A/K289A, D299A/H300A, K308A/H309A, D314A/E315A, K324A/R325A, D342A/E344A, K345A/R346A, E348A/E351A, and R363A/K364A) directed transcription of the 5S RNA gene as efficiently as did wild-type TFIIIA (Fig. 1D, lanes 2 to 11). This analysis eliminated the possibility that any of 18 charged residues within the 81-amino-acid domain was essential for the transcription factor activity of TFIIIA. Because of the qualitative nature of this assay, we cannot exclude the possibility that minor deficiencies in the activity of TFIIIA escaped detection.
Effects of carboxyl-terminal truncations on the transcription factor activity of TFIIIA.
Because site-directed mutagenesis of charged amino acids did not identify any residues as critical for the transcription factor activity of TFIIIA, we next analyzed a series of TFIIIAs that contained carboxyl-terminal truncations that extended into the 81-amino-acid domain. We showed previously that a form of TFIIIA that is truncated at the end of the eighth zinc finger, and therefore lacks the entire 81-amino-acid domain, is unable to support transcription of the 5S RNA gene, whereas a form of TFIIIA that is truncated at the beginning of the ninth zinc finger, and therefore contains the 81-amino-acid domain, supports transcription of the 5S RNA gene, albeit less efficiently than does wild-type TFIIIA (61).
For this study, we constructed a series of truncated TFIIIAs that terminated at various positions within the ninth zinc finger and the 81-amino-acid domain (Fig. 2A). We refer to these carboxyl-terminal-truncated forms of TFIIIA as TFIIIA(1-n), where n identifies the carboxyl-terminal residue of the protein. After confirming that the in vitro-synthesized forms of these TFIIIAs were of the appropriate size (data not shown) and were active in forming a TFIIIC-TFIIIA-5S RNA gene complex (Fig. 2B), we tested their ability to support in vitro transcription of the 5S RNA gene (Fig. 2C). As expected from our previous study (61), TFIIIA(1-397), which contained the ninth zinc finger but lacked the last 35 amino acids of the protein, was as active as wild-type TFIIIA in supporting accurate transcription (Fig. 2C, lanes 3 and 4). TFIIIA(1-378), which terminated within the ninth zinc finger, had modestly reduced activity, and TFIIIA(1-365), which lacked the ninth zinc finger, had further reduced activity (Fig. 2C, lanes 5 and 6, respectively). In some experiments, 5S RNA transcripts could be readily detected in reactions containing TFIIIA(1-365) (reference 61, this study, and data not shown), and in other experiments the level of 5S RNA transcripts obtained with this form of TFIIIA was just above background (Fig. 2C; compare lane 6 with lane 2). TFIIIAs with carboxyl-terminal deletions that extended into the 81-amino-acid domain [TFIIIA(1-359), TFIIIA(1-353), TFIIIA(1-347), TFIIIA(1-339), and TFIIIA(1-311)] appeared unable to support transcription of the 5S RNA gene (Fig. 2C, lanes 7 to 11). We note that because control reactions lacking TFIIIA gave rise to a trace amount of 5S RNA, due to contaminating TFIIIA present in the yeast fraction containing TFIIIC, TFIIIB, and RNA polymerase III (Fig. 2C, lanes 1 and 2, and data not shown), we could not confidently distinguish mutant forms of TFIIIA that had very low levels of activity from forms of TFIIIA that were inactive. Therefore, as an alternative approach for monitoring the activity of TFIIIA, we tested the ability of these mutant forms of TFIIIA to support cell growth. We note that the only essential function of TFIIIA in vivo is in promoting transcription of the 5S RNA gene (12).
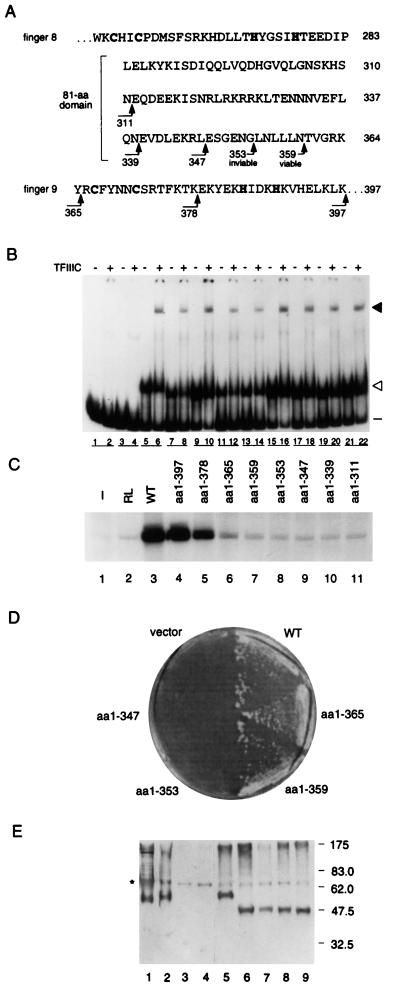
FIG. 2.
Effects of carboxyl-terminal deletions on activity of TFIIIA. (A) Amino acid sequences of zinc finger 8, the 81-amino-acid domain, and zinc finger 9 of yeast TFIIIA as shown in Fig. 1B. Arrowheads indicate the positions of the carboxyl termini of truncated forms of TFIIIA; the number of the last residue of each truncated protein is given below the bent tail. (B) Abilities of carboxyl-terminal-truncated versions of TFIIIA to bind to the 5S RNA gene and to recruit TFIIIC to the TFIIIA-DNA complex, as outlined in the legend for Fig. 1C. Lanes 1 and 2, no transcription-translation mixture; lanes 2 and 3, in vitro transcription-translation reactions that were not programmed to synthesize TFIIIA; lanes 5 and 6, wild-type TFIIIA synthesized in vitro; lanes 7 to 22, in vitro-synthesized, truncated versions of TFIIIA extending from the initiator methionine to residue 397 (lanes 7 and 8), 378 (lanes 9 and 10), 365 (lanes 11 and 12), 359 (lanes 13 and 14), 353 (lanes 15 and 16), 347 (lanes 17 and 18), 339 (lanes 19 and 20), or 311 (lanes 21 and 22). (C) Abilities of carboxyl-terminal-truncated versions of TFIIIA to support in vitro transcription of the 5S RNA gene. For details, see the legend for Fig. 1D. (D) Abilities of carboxyl-terminal-truncated versions of TFIIIA to support cell viability. A plasmid shuffle system was used to test the abilities of mutant versions of TFIIIA to replace wild-type TFIIIA in vivo. pG3-derived plasmids that expressed truncated versions of TFIIIA were transformed into the yeast strain YRW1 (see the text for details), and the abilities of these cells to grow on medium containing 5-FOA were monitored. vector, pG3 not expressing TFIIIA; WT, pG3 expressing wild-type TFIIIA; aa1-n, pG3 ex- pressing a version of TFIIIA that extends from the initiator methionine to residue n. (E) Assessment by Western blot analysis of in vivo expression of truncated versions of TFIIIA. Protein extracted from YRW1 yeast cells containing pG3-derived plasmids was separated on an SDS–10% polyacrylamide gel and electrotransferred to a nitrocellulose filter, and the filter was probed with anti-TFIIIA antibody. Lane 1, TFIIIA partially purified from yeast; lane 2, yeast TFIIIA purified from bacteria; lane 3, protein extract of YRW1 cells; lanes 4 to 9, protein extract of YRW1 cells containing pG3 expressing no TFIIIA (lane 4), wild-type TFIIIA (lane 5), TFIIIA(1-365) (lane 6), TFIIIA(1-359) (lane 7), TFIIIA(1-353) (lane 8), and TFIIIA(1-347) (lane 9). The asterisk on the left indicates a cross-reacting molecule not related to TFIIIA. The positions and sizes (in kilodaltons) of molecular mass markers are shown on the right.
To assess the ability of mutant forms of TFIIIA to function in vivo, we used a plasmid-shuffling protocol (75) to replace the wild-type gene encoding TFIIIA with a mutated version of the gene. First, we constructed strain YRW1, in which the chromosomal copy of TFC2, the gene encoding TFIIIA, has been deleted and cell viability is maintained by the presence of pJA230, a plasmid that contains a wild-type copy of TFC2 and URA3 as the selectable marker (3). Plasmids that express mutant forms of TFIIIA can then be introduced into YRW1, and the ability of these strains to survive in the absence of pJA230 can be monitored by assessing growth on medium containing 5-FOA. Because cells that grow on this medium must have lost the URA3-containing pJA230, which also contains the wild-type TFC2 gene, growth indicates that the mutant TFIIIA is active in directing transcription of the 5S RNA gene in vivo.
Using this plasmid-shuffling protocol, we found that pG3-derived, high-copy-number plasmids (see Materials and Methods) that expressed TFIIIA(1-397), TFIIIA(1-365), or TFIIIA(1-359) supported cell viability, whereas plasmids that expressed TFIIIA(1-353), TFIIIA(1-347), TFIIIA(1-339), or TFIIIA(1-311) did not (Fig. 2D and data not shown). Western blot analysis of cells containing pJA230 and pG3-derived plasmids confirmed that the truncated versions of TFIIIA, including those that did not support cell viability, were stable in vivo (Fig. 2E). We note that for the development time used for the blot shown in Fig. 2E, TFIIIA expressed from the low-copy-number plasmid pJA230 in strain YRW1 could not be readily detected (Fig. 2E, lanes 3 and 4), whereas TFIIIA expressed from the glyceraldehyde-3-phosphate dehydrogenase promoter in a high-copy-number plasmid was readily detectable (Fig. 2E, lane 5). Although transcripts of the 5S RNA gene could not be detected in an in vitro transcription reaction containing TFIIIA(1-359), this form of TFIIIA nonetheless supported cell viability. It is possible that the in vitro system lacked the sensitivity to detect a low level of TFIIIA activity and that this low level of activity was sufficient to support cell viability; it is also possible that the high level of expression of the mutant TFIIIA in vivo, as observed by Western blot analysis, compensated for its reduced activity in promoting transcription of the 5S RNA gene.
In summary, we found that TFIIIA(1-365), which lacked the ninth zinc finger and had reduced activity in vitro, and TFIIIA(1-359), which lacked an additional 6 amino acids and had no detectable activity in vitro, each supported cell viability. Deletion of the next 6 amino acids, which removed a leucine-rich segment of the 81-amino-acid domain, abolished the ability of TFIIIA to support cell viability. We therefore concluded that an amino acid(s) within the region from residue 354 to residue 359 was essential for the transcription factor activity of TFIIIA.
Effects of internal deletions within the 81-amino-acid domain of TFIIIA.
As described above, analysis of the effects of carboxyl-terminal deletions suggested that a residue(s) within the leucine-rich sequence adjacent to amino acid 359 of TFIIIA was essential for the activity of the protein. Because the hydrophobic, nonpolar nature of this segment suggested the possibility that it might play a role in folding of the 81-amino-acid domain, we assessed the effects of a series of internal deletions within the 81-amino-acid domain (Fig. 3A) on the activity of TFIIIA. We anticipated that if the leucine-rich segment did contribute to folding of the 81-amino-acid domain rather than being directly involved in its function, this approach might identify another region(s) that contributed to the function of TFIIIA.
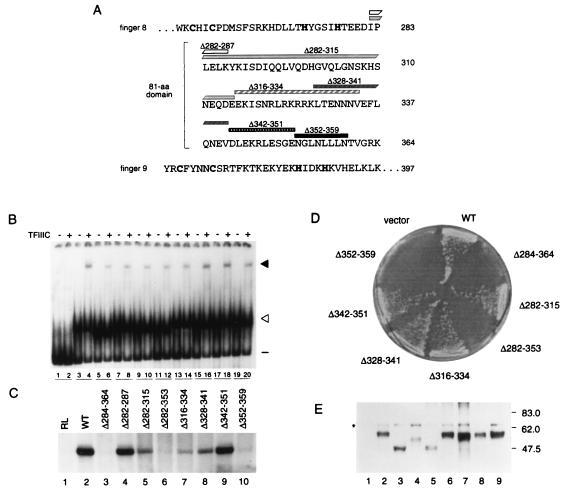
FIG. 3.
Effects of deletions within the 81-amino-acid domain on activity of TFIIIA. (A) Amino acid sequences of zinc finger 8, the 81-amino-acid domain, and zinc finger 9 of yeast TFIIIA, as shown in Fig. 1B. Rectangles and numbers above the amino acid sequence represent regions deleted from the 81-amino-acid domain. The deletions in TFIIIA(Δ284-364), which lacks the entire 81-amino-acid domain, and in TFIIIA(Δ282-353) are not represented. (B) Abilities of versions of TFIIIA with deletions within the 81-amino acid-domain to bind the 5S RNA gene and to recruit TFIIIC to the TFIIIA-DNA complex, as outlined in the legend for Fig. 1C. Lanes 1 to 4 were as described for Fig. 1C. Lanes 5 to 20, in vitro-synthesized versions of TFIIIA lacking residues 284 to 364 (lanes 5 and 6), 282 to 287 (lanes 7 and 8), 282 to 315 (lanes 9 and 10), 282 to 353 (lanes 11 and 12), 316 to 334 (lanes 13 and 14), 328 to 341 (lanes 15 and 16), 342 to 351 (lanes 17 and 18), or 352 to 359 (lanes 19 and 20). (C) Abilities of versions of TFIIIA with deletions in the 81-amino-acid domain to support in vitro transcription of the 5S RNA gene. For details, see the legend to Fig. 1D. (D) Abilities of versions of TFIIIA with deletions in the 81-amino-acid domain to support cell viability. A plasmid shuffle system was used to test the abilities of mutant versions of TFIIIA to support cell viability as described in the legend to Fig. 2D. Growth of cells containing pG3-derived plasmids on medium containing 5-FOA is shown. vector, pG3 not expressing TFIIIA; WT, pG3 expressing wild-type TFIIIA; Δn-n, pG3 expressing a version of TFIIIA that lacks amino acids n to n. (E) Assessment by Western blot analysis of in vivo expression of TFIIIA containing deletions in the 81-amino-acid domain. The analysis was carried out as described in the legend to Fig. 2E. Protein in lanes 1 to 9 was extracted from YRW1 cells containing pG3-derived plasmids expressing no TFIIIA, wild-type TFIIIA, TFIIIA(Δ284-364), TFIIIA(Δ282-315), TFIIIA(Δ282-353), TFIIIA(Δ316-334), TFIIIA(Δ328-341), TFIIIA(Δ342-351), and TFIIIA(Δ352-359), respectively. The asterisk on the left indicates a cross-reacting molecule not related to TFIIIA. Only the portion of the blot that contains TFIIIA is shown.
As expected, we found that TFIIIA(Δ352-359), which lacked the leucine-rich segment of the 81-amino-acid domain, was unable to support in vitro transcription of the 5S RNA gene (Fig. 3C, lane 10). Of the other internally deleted forms of TFIIIA that were tested, only TFIIIA(Δ284-364), which lacked the entire 81-amino acid domain (61), and TFIIIA(Δ282-353), which lacked all but the 11 carboxyl-terminal residues of the 81-amino-acid domain, failed to support in vitro transcription of the 5S RNA gene (Fig. 3C, lanes 3 and 6). In contrast, TFIIIA(Δ282-287) and TFIIIA(Δ282-315), which lacked 6 and 34 amino acids, respectively, at the amino-terminal end of the 81-amino-acid domain, supported in vitro transcription (Fig. 3C, lanes 4 and 5). TFIIIA(Δ282-315), however, was not as active as was wild-type TFIIIA (Fig. 3C; compare lanes 2 and 5). TFIIIA(Δ316-334) and TFIIIA(Δ328-341), which contained overlapping deletions in the central portion of the 81-amino-acid domain of TFIIIA, also supported transcription, but less efficiently than did wild-type TFIIIA (Fig. 3C, lanes 7 and 8). Finally, TFIIIA(Δ342-351), which lacked 10 amino acids just upstream of the leucine-rich region, was as active as wild-type TFIIIA in supporting in vitro transcription (Fig. 3C, lane 9).
We also tested these TFIIIAs for their ability to support cell viability, using the plasmid-shuffling protocol described above. TFIIIA(Δ284-364), TFIIIA(Δ282-353), and TFIIIA(Δ352-359), which failed to support in vitro transcription of the 5S RNA gene, also failed to support cell viability (Fig. 3D). These mutant forms of TFIIIA, although inactive, were nonetheless stable in vivo and accumulated to a level higher than that obtained with pJA230-borne TFC2 (Fig. 3E; compare lanes 3, 5, and 9 with lane 1). As expected, those forms of TFIIIA which supported in vitro transcription, TFIIIA(Δ282-315), TFIIIA(Δ316-334), TFIIIA(Δ328-341), and TFIIIA(Δ342-351), supported cell viability (Fig. 3D). The highly basic region from residue 321 to 327 is a putative nuclear localization signal (63). However, the ability of TFIIIA(Δ316-334) to support cell viability implies that this basic region is not necessary for nuclear localization, at least when TFIIIA is overexpressed.
In summary, we found that forms of TFIIIA with internal deletions that spanned the sequence from residue 282 to 351 of the 81-amino-acid domain [TFIIIA(Δ282-315), TFIIIA(Δ316-334), TFIIIA(Δ328-341), and TFIIIA(Δ342-351)] retained activity. TFIIIA(Δ352-359), which lacked 8 amino acids spanning the leucine-rich segment at the carboxyl-terminal end of the 81-amino-acid domain, however, was inactive. Because we found that TFIIIA tolerated deletions throughout most regions of the 81-amino-acid domain without loss of activity, we speculated that the leucine-rich segment was the only essential region of the 81-amino-acid domain and that this region was directly involved in the transcription factor activity of TFIIIA.
Effects of double alanine-scanning mutations of hydrophobic and nonpolar amino acids in the carboxyl-terminal portion of the 81-amino-acid domain.
We next subjected the region from residue 343 to 359, which spans the leucine-rich segment at the carboxyl-terminal end of the 81-amino-acid domain, to alanine-scanning mutagenesis (Fig. 4A). For ease of reference, forms of TFIIIA with double mutations were assigned code names beginning with the letter D. We found that TFIIIA(N352A/N355A) (D3), TFIIIA(L354A), TFIIIA(L356A), and TFIIIA(N359A) supported efficient in vitro transcription of the 5S RNA gene (Fig. 4C, lanes 6, 8, 9, and 11). TFIIIA(L343A/L347A) (D2), TFIIIA(L354A/L356A) (D4), and TFIIIA(L357A/L358A) (D5) had reduced, but readily detectable, activity (Fig. 4C, lanes 5, 7, and 10). Of these mutants, we consistently found that TFIIIA(L343A/L347A) (D2) was more active than TFIIIA(L357A/L358A) (D5) and that TFIIIA(L354A/L356A) (D4) was the most compromised for activity (Fig. 4C, lanes 5, 7, and 10). For comparison, we also monitored the effect of replacement by alanine of the hydrophobic residues at positions 296 and 297 and at positions 336 and 337. We found that both TFIIIA(L296A/V297A) and TFIIIA(F336A/L337A) (D1) were as active as wild-type TFIIIA (Fig. 4C, lanes 3 and 4).
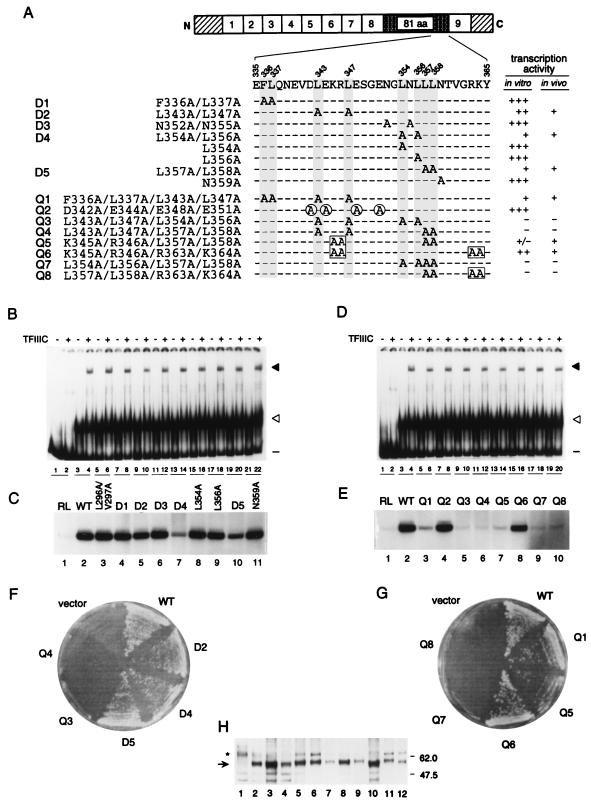
FIG. 4.
Effects of double and quadruple alanine-scanning mutations within the carboxyl-terminal portion of the 81-amino-acid domain on activity of TFIIIA. (A) Schematic representation of TFIIIA, showing the positions of mutated residues within the amino acid sequence from residue 335 to 365. The series of mutations that were introduced into the sequence from amino acid 335 to amino acid 365 are listed on the left, preceded by code names for the double (D1 to D5) and the quadruple (Q1 to Q8) alanine substitutions, and are represented by A’s aligned below the amino acid sequence. Dashes represent wild-type amino acids. For ease of alignment, Phe336 and Leu residues are shaded on all lines. A circled or boxed A indicates that alanine has replaced a negatively or positively charged residue, respectively. The two columns at the right summarize the transcription factor activity of each mutant TFIIIA with respect to its ability to support in vitro transcription of the 5S RNA gene (as shown in panels C and E) and cell viability (as shown in panels F and G). Left column: +++, the ability to support in vitro transcription was similar to that of wild-type TFIIIA; −, only a background level of in vitro transcription was observed. Right column: +, the mutant was able to support cell viability; −, the mutant was not able to support cell viability. (B and D) Abilities of versions of TFIIIA with single or double alanine-scanning mutations (B) or with quadruple alanine-scanning mutations (D) within the 81-amino-acid domain to bind to the 5S RNA gene and to recruit TFIIIC to the TFIIIA-DNA complex, as outlined in the legend to Fig. 1C. Lanes 1 to 4 were as described for Fig. 1C. (B) Lanes 5 to 22, TFIIIA with the following mutation(s): L296A/V297A (lanes 5 and 6), F336A/L337A (lanes 7 and 8), L343A/L347A (lanes 9 and 10), N352A/N355A (lanes 11 and 12), L354A/L356A (lanes 13 and 14), L354A (lanes 15 and 16), L356A (lanes 17 and 18), L357A/L358A (lanes 19 and 20), or N359A (lanes 21 and 22). (D) Lanes 5 to 20, TFIIIA with the following mutations: F336A/L337A/L343A/L347A (lanes 5 and 6), D342A/E344A/E348A/E351A (lanes 7 and 8), L343A/L347A/L354A/L356A (lanes 9 and 10), L343A/L347A/L357A/L358A (lanes 11 and 12), K345A/R346A/L357A/L358A (lanes 13 and 14), K345A/R346A/R363A/K364A (lanes 15 and 16), L354A/L356A/L357A/L358A (lanes 17 and 18), or L357A/L358A/R363A/K364A (lanes 19 and 20). (C and E) Abilities of versions of TFIIIA with single or double (C) or quadruple (E) alanine-scanning mutations in the carboxyl-terminal portion of the 81-amino-acid domain to support in vitro transcription of the 5S RNA gene. For details, see the legend to Fig. 1D. Where appropriate, the D and Q codes from panel A are given above the lanes. (F and G) Abilities of versions of TFIIIA with double and quadruple alanine-scanning mutations to support cell viability. A plasmid shuffle system was used to test the abilities of mutant versions of TFIIIA to support cell viability, as described in the legend to Fig. 2D. Cells containing pG3-derived plasmids that did not express TFIIIA (vector) or that expressed wild-type TFIIIA (WT) or versions of TFIIIA with the double or quadruple alanine-scanning mutations (D and Q codes, respectively) diagrammed in panel A were streaked onto 5-FOA-containing medium. (H) Assessment by Western blot analysis of in vivo expression of mutant forms of TFIIIA. Protein extracted from YRW1 yeast cells containing pG3-derived plasmids was analyzed as described in the legend to Fig. 2E. Protein in lanes 1 to 12 was extracted from cells containing pG3 expressing no TFIIIA (lane 1), wild-type TFIIIA (lane 2), TFIIIA(L343A/L347A) (lane 3), TFIIIA(L354A/L356A) (lane 4), TFIIIA(L357A/L358A) (lane 5), TFIIIA(L343A/L347A/L354A/L356A) (lane 6), TFIIIA(L343A/L347A/L357A/L358A) (lane 7), TFIIIA(F336A/L337A/L343A/L347A) (lane 8), TFIIIA(K345A/R346A/L357A/L358A) (lane 9), TFIIIA(K345A/R346A/R363A/K364A) (lane 10), TFIIIA(L354A/L356A/L357A/L358A) (lane 11), and TFIIIA(L357A/L358A/R363A/K364A) (lane 12). Asterisk, a cross-reacting molecule not related to TFIIIA; arrow, full-length TFIIIA.
This analysis indicated that none of the hydrophobic and nonpolar residues analyzed was essential for transcription factor activity. However, the double mutation L357A/L358A (Fig. 4C, lane 10) led to a modest reduction in the activity of TFIIIA, and the double mutation L354A/L356A led to an even more severe reduction in the activity of TFIIIA (Fig. 4C, lane 7); neither of the single mutations L354A and L356A affected activity (Fig. 4C, lanes 8 and 9).
Effects of quadruple alanine-scanning mutations within the carboxyl-terminal portion of the 81-amino-acid domain.
Although we found that TFIIIA(Δ352-359) was inactive both in vitro and in vivo, double substitutions in this region (N352A/N355A, L354A/L356A, and L357A/L358A) did not abolish the transcription factor activity of TFIIIA (see above). We therefore next assessed the effects of quadruple mutations in this region, anticipating that replacement of four leucines by alanine would be sufficiently deleterious to abolish function. For ease of reference, forms of TFIIIA with quadruple mutations were assigned code names beginning with the letter Q. Indeed, combining the double mutations L354A/L356A and L357A/L358A, both of which compromised the activity of TFIIIA (Fig. 4C, lanes 7 and 10, respectively), with each other or with the double mutation L343A/L347A, which on its own led to a modest decrease in activity of TFIIIA (Fig. 4C, lane 5), completely inactivated TFIIIA. We found that TFIIIA(L354A/L356A/L357A/L358A) (Q7), TFIIIA(L343A/L347A/L354A/L356A) (Q3), and TFIIIA(L343A/L347A/L357A/L358A) (Q4) failed to support in vitro transcription of the 5S RNA gene (Fig. 4E, lanes 5, 6, and 9) and failed to support cell viability (Fig. 4F and G). For comparison, we combined the double mutation L343A/L347A, which led to a modest decrease in the activity of TFIIIA (Fig. 4C, lane 5), with mutation of the hydrophobic residues F336 and L337, which as a double mutation had no effect on the activity of TFIIIA (Fig. 4C, lane 4). The resultant TFIIIA(F336A/L337A/L343A/L347A) (Q1) supported a very low level of 5S RNA gene transcription in vitro (Fig. 4E, lane 3) and supported cell viability (Fig. 4G). We also monitored the effect of mutation of four negatively charged residues (D342, E344, E348, and E351) in this region. TFIIIA(D342A/E344A/E348A/E351A) (Q2) was as active as wild-type TFIIIA in supporting in vitro transcription of the 5S RNA gene (Fig. 4E, lane 4).
Analysis of the effects of carboxyl-terminal deletions (Fig. 2) implicated the region just carboxyl-terminal to L358 as contributing to, although not being essential for, the transcription factor activity of TFIIIA. We therefore examined the effect of combining mutation of the basic residues R363 and K364, which as a double mutation did not affect the in vitro activity of TFIIIA (Fig. 1D, lane 11), with the double mutation L357A/L358A, which reduced the activity of TFIIIA (Fig. 4C, lane 10). We found that TFIIIA(L357A/L358A/R363A/K364A) (Q8) was unable to support transcription of the 5S RNA gene in vitro (Fig. 4E, lane 10) and failed to support cell viability (Fig. 4G). For comparison, we also combined mutation of the pair of basic residues K345 and R346, which as a double mutation did not affect the in vitro activity of TFIIIA (Fig. 1D, lane 9), with the double mutation L357A/L358A. Although the in vitro transcription factor activity of TFIIIA(K345A/R346A/L357A/L358A) (Q5) was just above background (Fig. 4E, lane 7), this form of TFIIIA was nonetheless able to support cell viability (Fig. 4G). Growth of this strain, however, appeared to be slightly impaired. Finally, we tested the effect of combining the double mutation K345A/R346A and the double mutation R363A/K364A. Substitution of alanine for these four basic residues had only a minimal effect on the in vitro activity of TFIIIA (Fig. 4E, lane 8), and TFIIIA(K345A/R346A/R363A/K364A) (Q6) supported cell viability (Fig. 4G).
In summary, analysis of the effects of double and quadruple mutations in the region extending from residue 336 to 364 of TFIIIA confirmed that hydrophobic residues in this segment are critical for the function of TFIIIA. In particular, L343, L347, L354, L356, L357, and L358, and to a lesser extent F336 and L337, contribute to the function of this region. Although charged residues are less important, R363 and K364, and to a lesser extent K345 and R346, also make a contribution to the activity of TFIIIA.
Zinc fingers 8 and 9 contribute to the transcription factor activity of TFIIIA.
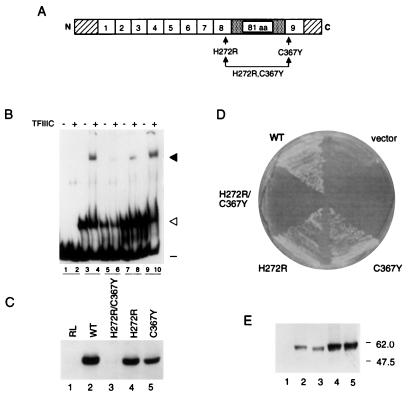
We fortuitously obtained by error-prone PCR mutagenesis a gene encoding TFIIIA with a mutation in a zinc-coordinating residue of finger 8 (H272R) and in a zinc-coordinating residue of finger 9 (C367Y) (see Materials and Methods) (Fig. 5A). This combination of mutations (H272R/C367Y), which would be expected to disrupt the structure of zinc fingers 8 and 9, abolished the ability of TFIIIA to support in vitro transcription of the 5S RNA gene (Fig. 5C, lane 3) and to support cell viability (Fig. 5D). We found, however, that TFIIIA that contained H272R or C367Y as a single mutation (see Materials and Methods) was only modestly compromised in its ability to support in vitro transcription of the 5S RNA gene (Fig. 5C, lanes 4 and 5) and was able to support cell viability (Fig. 5D). These data suggest that zinc fingers 8 and 9 make redundant contributions to the transcription factor activity of TFIIIA; the activity of TFIIIA is maintained on disruption of finger 8 or finger 9, but its activity is abolished on disruption of both fingers. We note that the assessment of protein-DNA complex formation by the EMSA shown in Fig. 5B suggests that TFIIIA(H272R/C367Y) was less active than wild-type TFIIIA in the formation of TFIIIA-DNA and TFIIIC-TFIIIA-DNA complexes. Because of the qualitative nature of this assay, however, we do not know if this apparent difference is significant.
FIG. 5.
Effects of mutations that disrupt the structure of zinc fingers 8 and 9 on activity of TFIIIA. (A) Schematic representation of yeast TFIIIA as described for Fig. 1A. The approximate positions of mutations H272R and C367Y within zinc fingers 8 and 9, respectively, are indicated by arrows. (B) Abilities of versions of TFIIIA with disruptions of zinc fingers 8 and 9 to bind to the 5S RNA gene and to recruit TFIIIC to the TFIIIA-DNA complex, as outlined in the legend to Fig. 1C. Lanes 1 to 4 were as described for Fig. 1C. Lanes 5 to 10, in vitro-synthesized versions of TFIIIA containing the following mutation(s): H272R/C367Y (lanes 5 and 6), H272R (lanes 7 and 8), or C367Y (lanes 9 and 10). (C) Abilities of versions of TFIIIA with disruptions of zinc fingers 8 and 9 to support in vitro transcription of the 5S RNA gene. For details, see the legend to Fig. 1D. (D) Abilities of versions of TFIIIA with disruptions of zinc fingers 8 and 9 to support cell viability. A plasmid shuffle system was used to test the abilities of mutant versions of TFIIIA to support cell viability, as described in the legend to Fig. 2D. (E) Assessment by Western blot analysis of in vivo expression of versions of TFIIIA with disruptions in zinc fingers 8 and 9. The analysis was carried out as described in the legend to Fig. 2E. Protein was extracted from YRW1 cells containing pG3-derived plasmids expressing no TFIIIA (lane 1), wild-type TFIIIA (lane 2), TFIIIA(H272R/C367Y) (lane 3), TFIIIA(H272R) (lane 4), or TFIIIA(C367Y) (lane 5).
DISCUSSION
We previously found that an 81-amino-acid domain that is present between zinc fingers 8 and 9 of yeast TFIIIA is essential for the transcription factor activity of the protein. TFIIIA lacking this domain binds efficiently to the 5S RNA gene and recruits TFIIIC, but the resultant TFIIIC-TFIIIA-DNA complex is unable to promote transcription (61). As a step towards understanding the role of this region in assembly of a functional preinitiation complex, we have defined specific residues within the 81-amino-acid domain that are critical for TFIIIA-mediated transcription. We tested versions of TFIIIA containing carboxyl-terminal deletions, internal deletions, and substitutions within the 81-amino-acid domain for their ability to support transcription of the 5S RNA gene in vitro and in vivo. We note that the only essential function of TFIIIA in vivo is as a transcription factor for the 5S RNA gene (12). In this study, we found that a short hydrophobic segment within the 81-amino-acid domain, from residue 352 to 359, was crucial for TFIIIA-mediated transcription.
Essential role of a hydrophobic patch within the 81-amino-acid domain.
We found that the 81-amino-acid domain of yeast TFIIIA was surprisingly tolerant to mutation. We were unable to identify any charged residue within this domain that was essential for its function (Fig. 1). Moreover, forms of TFIIIA with extensive deletions within the 81-amino-acid domain retained the ability to support transcription of the 5S RNA gene (Fig. 3). We found that TFIIIA that lacked the sequence from residue 282 to 315, from residue 316 to 334, from residue 328 to 341, or from residue 342 to 351 supported in vitro transcription of the 5S RNA gene, although in some instances to a reduced level, and supported cell viability (Fig. 3). However, deletion of an asparagine- and leucine-rich segment (NGLNLLLN; residue 352 to 359) within the carboxyl-terminal portion of the 81-amino-acid domain destroyed the activity of TFIIIA. Our data suggest that no other segment within the 81-amino-acid domain can substitute for this asparagine- and leucine-rich segment: TFIIIA lacking the carboxyl-terminal portion of the protein beyond residue 359 supported cell viability, whereas TFIIIA lacking the carboxyl-terminal portion beyond residue 353 did not support viability (Fig. 2).
We note that TFIIIA that had been truncated at the beginning of the ninth zinc finger was much less active in its ability to support in vitro transcription of the 5S RNA gene than was TFIIIA that had been truncated in the middle of the ninth zinc finger (Fig. 2). This suggests that a carboxyl-terminal extension to TFIIIA, even if unstructured, as would be expected for a partial zinc finger, enhances the activity of the asparagine- and leucine-rich segment.
Requirement for hydrophobic residues within the carboxyl-terminal portion of the 81-amino-acid domain.
Alanine-scanning mutagenesis indicated that no single amino acid in the region from residue 354 to 359 (LNLLLN) was essential for the activity of TFIIIA (Fig. 4). Analysis of the effect of double mutations in the segment 352NGLNLLLN359 indicated that the leucine residues were more important for function than the asparagine residues. TFIIIA containing the double mutation N352A/N355A appeared to be as active as wild-type TFIIIA, whereas TFIIIA containing the double mutation L354A/L356A or L357A/L358A had reduced activity. TFIIIA in which L354, L356, L357, and L358 were all mutated was unable to support cell viability. The double mutation L343A/L347A, which consists of residues amino-terminal to the hydrophobic 352NGLNLLLN359 segment, had only a very minor effect on the in vitro activity of TFIIIA. Interestingly, combining this double mutation with either the double mutation L354A/L356A or the double mutation L357A/L358A led to forms of TFIIIA that were unable to support cell viability. This observation suggests that the overall hydrophobicity of the region that encompasses 352NGLNLLLN359 is more important for its function than is any single residue. In support of this notion, we found that combining the double mutations F336A/L337A and L343A/L347A, which are in residues upstream of the 352NGLNLLLN359 hydrophobic segment and which by themselves had little effect on the activity of TFIIIA, led to a dramatic decrease in the activity of TFIIIA. It is interesting that TFIIIA with an internal deletion that eliminated residues 342 to 351 was just as active in vitro as wild-type TFIIIA. In this case, however, L337 occupied the same position, at least at the primary sequence level, as L347 normally does relative to residue 352, and consequently L337 may have fulfilled the role of L347.
In contrast to the importance of hydrophobic residues in the carboxyl-terminal portion of the 81-amino-acid domain, the charged nature of this region appeared to be less important for its function. For example, TFIIIA in which the acidic residues D342, E344, E348, and E351 had all been replaced by alanine was as active as wild-type TFIIIA (Fig. 4). Similarly, TFIIIA in which the basic residues K345, R346, R363, and K364 had all been replaced by alanine was almost as active as wild-type TFIIIA (Fig. 4). Nonetheless, these charged residues appeared to play a role in the activity of TFIIIA; combining mutation of the basic residues K345 and R346 with mutation of L357 and L358 dramatically reduced the ability of TFIIIA to support in vitro transcription of the 5S RNA gene, and combining mutation of the basic residues R363 and K364 with mutation of L357 and L358 abolished the ability of TFIIIA to support cell viability.
We note that our alanine-scanning mutagenesis was not exhaustive and that residues that contributed to function may have escaped detection. Additionally, we would not have detected residues outside of the 352NGLNLLLN359 segment that contributed to function in a redundant manner.
Role of the leucine-rich segment of the 81-amino-acid domain in TFIIIA-mediated transcription.
We previously presented a model for the role of yeast TFIIIA in the assembly of a transcription complex on the yeast 5S RNA gene (61) in which we took into consideration the fact that TFIIIC (or τ) consists of two domains, τA and τB, which interact with the A box and B box, respectively, of tRNA genes (27). It is the interaction of the τA domain with the A box of a tRNA gene that is responsible for appropriate positioning of TFIIIB upstream of the start site of transcription (43; also reviewed in reference 86). In our model, we proposed that the amino-terminal zinc fingers of yeast TFIIIA interact with the τB domain of TFIIIC and we speculated that once TFIIIC had been recruited to a TFIIIA-DNA complex by this interaction, the 81-amino-acid domain of TFIIIA interacted with the τA domain of TFIIIC (61). The latter interaction was predicted to be responsible for docking the multisubunit TFIIIC on the TFIIIA-DNA complex with the appropriate topography so that TFIIIB could be properly positioned to fulfill its role as an initiation factor. The present study implicates a hydrophobic segment in the carboxyl-terminal portion of the 81-amino-acid domain as essential for its function and, according to our model, leads to the suggestion that this segment interacts with a region of TFIIIC.
In this context, it is interesting that hydrophobic side chains are a key characteristic of many protein-protein interfaces (44, 90). One of the best-studied examples of the role of hydrophobic surfaces in mediating a protein-protein interaction is the binding of human growth hormone to its receptor. As assessed by mutational and structural analyses, the critical interactions are between well-packed hydrophobic and sterically complementary surfaces. Although peripheral contacts mediated by electrostatic interactions do contribute to binding, they are less important than the core hydrophobic interactions (15, 20, 22; reviewed in reference 85).
Although it was initially postulated that acidic residues of some transcriptional regulators were key to their activity, more detailed studies have indicated that the function of these activation surfaces in fact depends on hydrophobic residues. For example, mutational analyses have revealed that it is a specific pattern of aromatic and large hydrophobic amino acids that plays a critical role in activation by such transactivators as VP16 (19, 69), Rta (34), RelA (7), Sp1 (31), Gcn4 (24, 42), p53 (54), and the glucocorticoid receptor (1). Similarly, a leucine-rich motif, LXXLL, present in several coactivators is responsible for mediating their interaction with nuclear receptors (37, 79).
Various studies have led to the conclusion that many transcriptional activators function by contacting a component(s) of the basal transcriptional machinery that is associated with RNA polymerase II (reviewed in reference 68). In several cases, the extent to which a mutation in an activator reduces transactivation has been shown to correlate with the extent to which it weakens an interaction with a component(s) of the basal transcriptional machinery (for examples, see references 19, 41, and 88). An interesting example of the role of hydrophobic surfaces in mediating protein-protein interactions by transcriptional regulators is provided from studies of the tumor suppressor transactivator p53. The same hydrophobic residues that are exposed on one face of an amphipathic α helix of p53 (48) and that are required for activation of transcription (14, 54) are also required for interaction of p53 with TBP and TBP-associated factors (14, 56, 78). Moreover, a recent structural analysis of a peptide of p53 bound to a domain of Mdm-2, a cellular oncogene, revealed that a deep hydrophobic cleft in Mdm-2 provides steric complementarity for the hydrophobic face of the transactivation helix of p53 (48). The fact that hydrophobic residues of p53 that are known to be crucial for its ability to transactivate are buried in the interface with Mdm-2 provides an explanation for inactivation of p53 by Mdm-2 (14, 48, 54, 65).
In view of the increasing number of examples that demonstrate a role for a hydrophobic surface in mediating a protein-protein interaction, our finding that hydrophobic residues within the carboxyl terminus of the 81-amino-acid domain are of primary importance for its transcription factor activity is consistent with the notion that this region of TFIIIA interacts with TFIIIC.
Comparison of yeast and Xenopus TFIIIAs.
A 14-amino-acid segment (KRSLASRLTGYIPP) that is present in the portion of Xenopus TFIIIA that extends beyond the ninth zinc finger is essential for the transcription factor activity of this TFIIIA (57). Zinc fingers 8 and 9 also contribute to the transcription factor activity of Xenopus TFIIIA (21, 70); these fingers may have a direct role in assembly of a functional transcription complex or they may act indirectly, through their interaction with the A box of the ICR, to appropriately position the 14-amino-acid segment to fulfill its role in promoting transcription. We find no sequence similarity between this 14-amino-acid segment of Xenopus TFIIIA and the leucine-rich region of the 81-amino-acid domain of yeast TFIIIA that we have defined as being important for its transcription factor activity. This lack of sequence similarity is not surprising, as components of the RNA polymerase III transcriptional machinery appear to be quite divergent among species (see the introduction and reference 83). It is also possible that the molecular details of assembly of a functional transcription complex differ between yeast and Xenopus. For example, the activity of the transcription-activating region of Xenopus TFIIIA appears to be more position dependent (57) than the activity of the transcription-activating region of yeast TFIIIA (this study), as assessed by monitoring the effects of deletion of adjacent sequences. Zinc fingers 8 and 9 of both Xenopus TFIIIA (21, 70) and yeast TFIIIA (this study) appear to contribute to the transcription factor activity of TFIIIA. However, we found that yeast TFIIIA remained active if either finger 8 or finger 9 was disrupted by mutation; disruption of both these fingers was required in order to abolish the ability of TFIIIA to promote transcription of the 5S RNA gene. This suggests that these zinc fingers play a redundant role in promoting transcription. These fingers may help establish the overall topography of the preinitiation complex either by extending the interaction of TFIIIA with DNA (9), by interacting with another component of the complex, or by contributing to the function of the 81-amino-acid domain.
Further studies will elucidate the role of the hydrophobic segment of the 81-amino-acid domain of yeast TFIIIA in generating an active transcription complex on the yeast 5S RNA gene. Potential roles include the previously proposed function of locking TFIIIC into position in the TFIIIA-DNA complex subsequent to its recruitment by the amino-terminal zinc fingers of TFIIIA (29, 61), a contribution to the changes in architecture and properties of the transcription complex that occur on incorporation of TFIIIB (9, 45), and, perhaps, an interaction with DNA upstream of the ICR.
ACKNOWLEDGMENTS
We thank Catherine Milne for providing a yeast-derived heparin-agarose fraction h, Randall Willis for construction of the yeast strain YRW1, John Hwang for assistance in the construction of a number of the plasmids used in this study, and Shelley Hepworth for valuable comments regarding the manuscript.
O.R. was supported by an Ontario Graduate Scholarship. This work was supported by a Medical Research Council (Canada) grant (MA-6826) to J.S.
REFERENCES
- 1.Almlöf T, Gustafsson J-A, Wright A P H. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Mol Cell Biol. 1997;17:934–945. doi: 10.1128/mcb.17.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa H, Nagase H, Hayashi N, Ogawa M, Nagata M, Fujiwara T, Takahashi E, Shin S, Nakamura Y. Molecular cloning, characterization, and chromosomal mapping of a novel human gene (GTF3A) that is highly homologous to Xenopus transcription factor IIIA. Cytogenet Cell Genet. 1995;70:235–238. doi: 10.1159/000134041. [DOI] [PubMed] [Google Scholar]
- 3.Archambault J, Milne C A, Schappert K T, Baum B, Friesen J D, Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J Biol Chem. 1992;267:3282–3288. [PubMed] [Google Scholar]
- 4.Archambault J, Schappert K T, Friesen J D. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol Cell Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair W S, Bogerd H P, Madore S J, Cullen B R. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogenhagen D F. The intragenic control region of the Xenopus 5S RNA gene contains two factor A binding domains that must be aligned properly for efficient transcription initiation. J Biol Chem. 1985;260:6466–6471. [PubMed] [Google Scholar]
- 9.Braun B R, Bartholomew B, Kassavetis G A, Geiduschek E P. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5S RNA gene. J Mol Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 10.Braun B R, Riggs D L, Kassavetis G A, Geiduschek E P. Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc Natl Acad Sci USA. 1989;86:2530–2534. doi: 10.1073/pnas.86.8.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 12.Camier S, Dechampesme A-M, Sentenac A. The only essential function of TFIIIA in yeast is the transcription of the 5S rRNA genes. Proc Natl Acad Sci USA. 1995;92:9338–9342. doi: 10.1073/pnas.92.20.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Challice J M, Segall J. Transcription of the 5S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J Biol Chem. 1989;264:20060–20067. [PubMed] [Google Scholar]
- 14.Chang J, Kim D-H, Lee S W, Choi K Y, Sung Y C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 15.Clackson T, Wells J A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 16.Clemens K R, Liao X, Wolf V, Wright P E, Gottesfeld J M. Definition of the binding sites of individual zinc fingers in the transcription factor IIIA-5S RNA gene complex. Proc Natl Acad Sci USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemens K R, Wolf V, McBryant S J, Zhang P, Liao X, Wright P E, Gottesfeld J M. Molecular basis for specific recognition of both RNA and DNA by a specific zinc finger protein. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 18.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 19.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham B C, Wells J A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 21.Del Rio S, Setzer D R. The role of zinc fingers in transcriptional activation by transcription factor IIIA. Proc Natl Acad Sci USA. 1993;90:168–172. doi: 10.1073/pnas.90.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vos A M, Ultsch M, Kossiakoff A A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 23.Drew P D, Nagle J W, Canning R D, Ozato K, Biddison W E, Becker K G. Cloning and expression analysis of a human cDNA homologous to Xenopus TFIIIA. Gene. 1995;159:215–218. doi: 10.1016/0378-1119(95)00145-v. [DOI] [PubMed] [Google Scholar]
- 24.Drysdale C M, Duenas E, Jackson B M, Reusser U, Braus G H, Hinnebusch A G. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelke D R, Ng S-Y, Shastry B S, Roeder R G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- 26.Fairall L, Rhodes D. A new approach to the analysis of DNase I footprinting data and its application to the TFIIIA/5S DNA complex. Nucleic Acids Res. 1992;20:4727–4731. doi: 10.1093/nar/20.18.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabrielsen O S, Sentenac A. RNA polymerase III (C) and its transcription factors. Trends Biochem Sci. 1991;16:412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- 28.Gaskins C J, Hanas J S. Sequence variation in transcription factor IIIA. Nucleic Acids Res. 1990;18:2117–2123. doi: 10.1093/nar/18.8.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 30.Geitz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginsberg A M, King B O, Roeder R G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984;39:479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- 33.Hansen P K, Christensen J H, Nyborg J, Lillelund O, Thøgerson H C. Dissection of the DNA-binding domain of Xenopus laevis TFIIIA. Quantitative DNase I footprinting analysis of specific complexes between a 5S RNA gene fragment and N-terminal fragments of TFIIIA containing three, four, or five zinc-finger domains. J Mol Biol. 1993;233:191–202. doi: 10.1006/jmbi.1993.1499. [DOI] [PubMed] [Google Scholar]
- 34.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes J J, Clemens K R. Location and contacts between individual zinc fingers of Xenopus laevis transcription factor IIIA and the internal control region of a 5S RNA gene. Biochemistry. 1992;31:11600–11605. doi: 10.1021/bi00161a045. [DOI] [PubMed] [Google Scholar]
- 36.Hayes J J, Tullius T D. Structure of the TFIIIA-DNA complex. J Mol Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 37.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 38.Hepworth S R, Ebisuzaki L K, Segall J. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3934–3944. doi: 10.1128/mcb.15.7.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 40.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 41.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature (London) 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 42.Jackson B M, Drysdale C M, Natarajan K, Hinnebusch A G. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 44.Jones S, Thornton J M. Principles of protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 46.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 49.Lagna G, Kovelman R, Sukegawa J, Roeder R G. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol Cell Biol. 1994;14:3053–3064. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefebvre O, Carles C, Conesa C, Swanson R N, Bouet F, Riva M, Sentenac A. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor IIIC. Proc Natl Acad Sci USA. 1992;89:10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.L’Etoile N D, Fahnestock M L, Shen Y, Aebersold R, Berk A J. Human transcription factor IIIC box B binding subunit. Proc Natl Acad Sci USA. 1994;91:1652–1656. doi: 10.1073/pnas.91.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 53.Liao X, Clemens K R, Tennant L, Wright P E, Gottesfeld J M. Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5S RNA gene. J Mol Biol. 1992;223:857–871. doi: 10.1016/0022-2836(92)90248-i. [DOI] [PubMed] [Google Scholar]
- 54.Lin J, Chen J, Elennbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 55.López-de-León A, Librizzi M, Puglia K, Willis I M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 56.Lu H, Levine A J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao X, Darby M K. A position-dependent transcription-activating domain in TFIIIA. Mol Cell Biol. 1993;13:7496–7506. doi: 10.1128/mcb.13.12.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBryant S J, Gedulin B, Clemens K R, Wright P E, Gottesfeld J M. Assessment of major and minor groove DNA interactions by the zinc fingers of Xenopus transcription factor IIIA. Nucleic Acids Res. 1996;24:2567–2574. doi: 10.1093/nar/24.13.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller J, McLachlan A D, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milne C A. Ph.D. thesis. Toronto, Ontario, Canada: University of Toronto; 1994. [Google Scholar]
- 61.Milne C A, Segall J. Mapping regions of yeast transcription factor IIIA required for DNA binding, interaction with transcription factor IIIC, and transcription activity. J Biol Chem. 1993;268:11364–11371. [PubMed] [Google Scholar]
- 62.Moorefield B, Roeder R G. Purification and characterization of human transcription factor IIIA. J Biol Chem. 1994;269:20857–20865. [PubMed] [Google Scholar]
- 63.Osborne M A, Silver P A. Nucleocytoplasmic transport in the yeast Saccharomyces cerevisiae. Annu Rev Biochem. 1993;62:219–254. doi: 10.1146/annurev.bi.62.070193.001251. [DOI] [PubMed] [Google Scholar]
- 64.Petko L, Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- 65.Picksley S M, Vojtesek B, Sparks A, Lane D P. Immunochemical analysis of the interaction of p53 with MDM2; −fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 66.Pieler T, Hamm J, Roeder R G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987;48:91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- 67.Pieler T, Oei S-L, Hamm J, Engelke U, Erdmann V A. Functional domains of the Xenopus laevis 5S gene promoter. EMBO J. 1985;4:3751–3756. doi: 10.1002/j.1460-2075.1985.tb04144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 69.Regier J L, Shen F, Triezenberg S J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rollins M B, Del Rio S, Galey A L, Setzer D R, Andrews M T. Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol Cell Biol. 1993;13:4776–4783. doi: 10.1128/mcb.13.8.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowland O, Segall J. Interaction of wild-type and truncated forms of transcription factor IIIA from Saccharomyces cerevisiae with the 5S RNA gene. J Biol Chem. 1996;271:12103–12110. doi: 10.1074/jbc.271.20.12103. [DOI] [PubMed] [Google Scholar]
- 72.Sakonju S, Brown D D, Engelke D, Ng S-Y, Shastry B S, Roeder R G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981;23:665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- 73.Schena M, Picard D, Yamamoto K R. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- 74.Shastry B S. Transcription factor IIIA (TFIIIA) in the second decade. J Cell Sci. 1996;109:535–539. doi: 10.1242/jcs.109.3.535. [DOI] [PubMed] [Google Scholar]
- 75.Sikorski R S, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 76.Smith D R, Jackson I J, Brown D D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984;37:645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- 77.Taylor M J, Segall J. Characterization of factors and DNA sequences required for the accurate transcription of the Saccharomyces cerevisiae 5S RNA gene. J Biol Chem. 1985;260:4531–4540. [PubMed] [Google Scholar]
- 78.Thut C J, Chen J-L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 79.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 80.Vrana K E, Churchill M E A, Tullius T D, Brown D D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988;8:1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waldschmidt R, Jahn D, Teichmann M, Jahn M, Meissner W, Seifart K H. Physical and immunological characterization of human transcription factor IIIA. Eur J Biochem. 1990;194:167–176. doi: 10.1111/j.1432-1033.1990.tb19441.x. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z, Roeder R G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 84.Wells J A. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 85.Wells J A. Binding in the growth hormone receptor complex. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.White R J. RNA polymerase III transcription. Austin, Tex: Molecular Biology Intelligence Unit, R. G. Landes Company; 1994. [Google Scholar]
- 87.Woychik N A, Young R A. Genes encoding transcription factor IIIA and the RNA polymerase common subunit RPB6 are divergently transcribed in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3999–4003. doi: 10.1073/pnas.89.9.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yaffe M P, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young L, Jernigan R L, Covell D G. A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 1994;3:717–729. doi: 10.1002/pro.5560030501. [DOI] [PMC free article] [PubMed] [Google Scholar]