Abstract
The dimerization initiation site (DIS) and the dimer linkage sequences (DLS) of human immunodeficiency virus type 1 have been shown to mediate in vitro dimerization of genomic RNA. However, the precise role of the DIS-DLS region in virion assembly and RNA dimerization in virus particles has not been fully elucidated, since deletion or mutation of the DIS-DLS region also abolishes the packaging ability of genomic RNA. To characterize the DIS-DLS region without altering packaging ability, we generated mutant constructs carrying a duplication of approximately 1,000 bases including the encapsidation signal and DIS-DLS (E/DLS) region. We found that duplication of the E/DLS region resulted in the appearance of monomeric RNA in virus particles. No monomers were observed in virions of mutants carrying the E/DLS region only at ectopic positions. Monomers were not observed when pol or env regions were duplicated, indicating an absolute need for two intact E/DLS regions on the same RNA for generating particles with monomeric RNA. These monomeric RNAs were most likely generated by intramolecular interaction between two E/DLS regions on one genome. Moreover, incomplete genome dimerization did not affect RNA packaging and virion formation. Examination of intramolecular interaction between E/DLS regions could be a convenient tool for characterizing the E/DLS region in virion assembly and RNA dimerization within virus particles.
Retrovirus RNAs packaged into virions are dimeric. The association between the two RNA molecules is noncovalent because the dimeric RNA dissociable into monomers under mild denaturing conditions, such as incubation at high temperature (∼70°C) or treatment with denaturing reagents (for a review, see references 14 and 24). Electron microscopic analysis of genomic RNA dimers obtained from several species of retroviruses reveals a symmetrical form with a contact point between the RNA situated in a region near the 5′ end (4, 5, 19, 27, 31, 32, 37, 48, 57). It is likely that the presence of two genomes in single virus particles is advantageous for virus survival, facilitating recovery from physical damage to the RNA or providing genetic variety to the virus progeny (15, 28).
Synthetic RNA fragments derived from the 5′ region of retrovirus RNA can spontaneously dimerize in vitro upon incubation in appropriate buffer without protein factors (3, 6, 9, 11, 13, 16–18, 25, 26, 29, 33, 39, 51, 53, 56, 58). The contact point between the two monomers that constitute in vitro-synthesized dimers is referred to as the dimer linkage sequences (DLS). In human immunodeficiency virus type 1 (HIV-1), the 5′ untranslated region just downstream of the splicing donor (s.d.) was first reported to be a DLS, because the RNA fragments harboring deletions in this region have formed dimers in vitro at significantly reduced efficiency (3, 39, 58). Recently, several groups reported that another site within the 5′ untranslated region is also important for RNA dimerization in vitro. This site is located upstream of the 5′ s.d. and designated the dimer initiation site (DIS) (33, 47, 51, 56). The DIS consists of a stem-loop structure with a conserved palindromic sequence at the top of the loop. In their proposed model, the palindromic sequences on two RNA molecules first contact each other, forming a “kissing hairpin” interaction when dimer formation is initiated (33, 47, 51, 56). Mutation within this region also abolished in vitro RNA dimerization (13, 46).
In contrast to these in vitro data, several lines of evidence indicate that dimer formation in vivo is not as simple. The viral nucleocapsid protein (NC) appears to act as a molecular chaperon to refold viral RNA so that it has the appreciate secondary structure (16, 17, 20). We and other groups reported that mutations introduced in and around the encapsidation signal and DIS-DLS (E/DLS) region did not affect the stability of dimers in virus particles (7, 12, 54). We also found that it is likely that the regions located far from the primary E/DLS region affect the stability of the RNA dimer (54). Furthermore, electron microscopic observation indicates that the dimeric form of HIV-1 RNA contains more than one contact point in the primary E/DLS (27).
A problem that has hampered in vivo analysis of the DIS-DLS is that deletion or mutation of the dimerization site also abolishes RNA packaging, since the putative dimerization site largely overlaps the packaging signal. To try to obviate this problem, we generated mutant viral RNA carrying additional dimerization sites to see whether two dimerization sites within the same RNA molecule might interact with each other and interfere with normal intermolecular dimer formation. If such intramolecular interaction negatively affects intermolecular interaction, it might be possible to modify one dimerization site without affecting packaging efficiency and thereby functionally segregate the encapsidation and DIS-DLS regions. We report here that the duplication of a packaging-dimerization site on the same RNA molecule indeed caused the appearance of monomeric RNA in virions. Such monomers were not observed when the original packaging-dimerization site was deleted, suggesting that the duplicated region actually mediates RNA-RNA interaction in the virion. This system could be used for defining and examining the exact location of dimer linkage sites within virus particles.
MATERIALS AND METHODS
Plasmids and viral expression constructs.
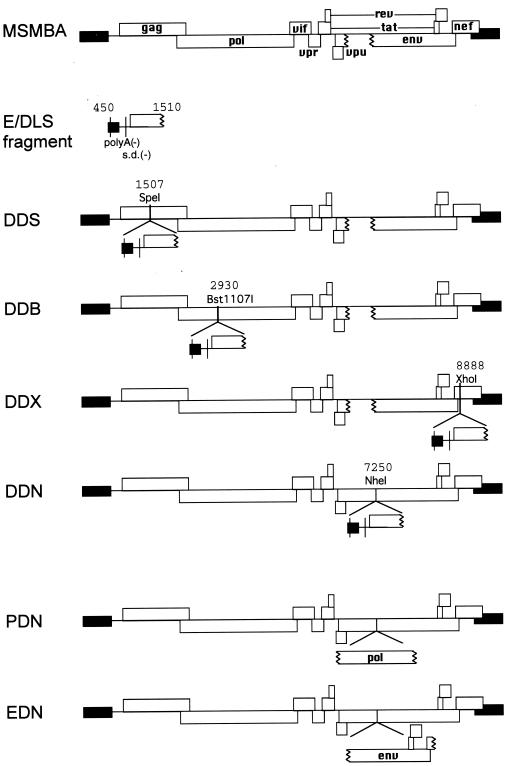
The replication-competent HIV-1 proviral clone pNL4-3 (1) and its defective derivative pMSMBA (40), which had about a 900-bp deletion in the env gene, were used as the progenitors for all the mutants listed below. The nucleotide designations refer to the DNA sequence of pNL4-3 starting from the beginning of U3. To construct a series of mutants containing two copies of the dimer linkage site, we first constructed pGEM-MM, which contains the 5′ leader region (nucleotide positions 454 to 1523) of pMSMBA with mutations in both the s.d. and the polyadenylation signal. First, two overlapping fragments were PCR amplified from pMSMBA with primers containing mutations in the s.d. Sense primer 438-464 BamHI (5′-GCTTTTTGCCGGATCCGGGTCTCTCTG-3′; underlining indicates bases that were substituted to introduce mutations) and antisense primer 756-733 BclI (5′-TTGGCGTCCTGATCAGTCGCCGCC-3′) were used to generate fragment 1, and sense primer 733-753 BclI (5′-GGCGGCGACTGATCAGGACGCCAA-3′) and antisense primer 1535-1512 BamHI (5′-CATCCTATTGGATCCTGAAGGGTA-3′) were used to generate fragment 2. Fragments 1 and 2 were digested with BclI, ligated, digested with BssHII and SpeI, and cloned into pMSMBA that had been cut with BssHII and SpeI to construct pMSMBA s.d.(−). Next, two overlapping fragments were PCR amplified from pMSMBA s.d.(−) with primers containing mutations in the polyadenylation signal. Sense primer 438-464 BamHI and antisense primer 543-520 PvuI (5′ TCAAGGCAACGATCGTTGAGGCTT-3′) were used to generate fragment 3, and sense primer 520-543 PvuI (5′-AAGCCTCAACGATCGTTGCCTTGA-3′) and antisense primer 1535-1512 BamHI were used to generate fragment 4. Fragments 3 and 4 were digested with PvuI, ligated, digested with BamHI, and cloned into the BamHI site of pGEM3Zf(+) (Promega) to construct pGEM-MM. The amplified region of pGEM-MM was analyzed by sequencing, and two base substitution mutations (G to A at nucleotide position 1175 and C to T at 1195) were detected that probably arose from errors during PCR. However, these positions are far from the known encapsidation region or putative dimerization site and were left within pGEM-MM.
The mutated fragment (fragment 5) was isolated from pGEM-MM by digesting with BamHI, and the protruding ends were converted to blunt ends using T4 DNA polymerase. Fragment 5 was ligated into the SpeI, Bst1107I, or XhoI site of pMSMBA which had been similarly blunt ended with T4 DNA polymerase to generate pDDS, pDDB, and pDDX, respectively. Fragment 5 was ligated into the NheI site of pNL4-3 which had been similarly blunt ended with T4 DNA polymerase to construct pDDN. pssS was constructed by ligating fragment 5 into the blunt-ended SpeI site of p5′ssβglob (41). The 2,000-bp NotI-ApaI fragments of pDDB and pDDX were exchanged with the corresponding fragment of p5′ssβglob to construct pssB and pssX, respectively. The SalI-XhoI env fragment of p5′ssβglob was exchanged with the corresponding fragment of pDDN to construct pssN. The MscI-MscI (2622 to 4554) fragment and the T4 DNA polymerase-treated NheI-XhoI (7251 to 8892) fragment of pMSMBA were ligated into the T4 DNA polymerase-treated NheI site of pNL4-3 to construct PDN and EDN, respectively. To construct pDDNΔPBS and pssNΔPBS, pMPΔPBS was constructed first. Plasmid pdPBS (42), carrying a 19-base deletion mutation in the primer-binding site, was digested with MfeI, blunt ended, and digested with SpeI, and an 840-bp fragment was isolated. This fragment was ligated into the EheI and SpeI sites of pGEM-MM to construct pMPΔPBS. The mutated fragment (fragment ΔP) was isolated from pMPΔPBS by digesting with BamHI and blunt ended with the T4 DNA polymerase. Fragment ΔP was ligated into the blunt-ended NheI site of pNL4-3 and p5′ssβglob to construct pDDNΔPBS and pssNΔPBS, respectively.
To construct the HIV-1 env expression vector p5′ssEnvEXSV, pNL4-3 was digested with BbvII, blunt ended with T4 DNA polymerase, and digested with XhoI, and a 2.7-kb env region fragment (6208 to 8888) was isolated. The env fragment was then inserted into the XhoI and T4 DNA polymerase-treated MluI sites of p5′ssβglob to construct p5′ssEnvEX. pGL3basic (Promega) was digested with XbaI, blunt ended with T4 DNA polymerase, and digested with SalI, and the 270-bp fragment containing the simian virus 40 polyadenylation signal was isolated. This fragment was ligated into the XhoI and T4 DNA polymerase-treated EspI sites, located in the nef gene, of p5′ssEnvEX to construct p5′ssEnvEXSV.
Transfection.
Approximately 7 × 106 293 cells (23) or 293tat cells (49) were seeded on 150-mm-diameter plates the day before transfection and transfected with 20 μg of plasmid DNA using the calcium phosphate precipitation method (2). The day after transfection, the supernatant was discarded and replaced with fresh medium.
Northern blotting.
At 48 to 72 h after transfection, the medium and cytoplasmic RNA were concurrently collected as described elsewhere (40). Pelleted RNA was resuspended in T-buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% sodium dodecyl sulfate [SDS], 100 mM NaCl, and 10% formamide), and the thermostability of dimeric viral RNA was determined by incubating RNA aliquots for 10 min at the temperatures indicated in the relevant figures (54). Viral RNA was electrophoresed at room temperature in nondenaturing 0.75% native agarose gels containing 0.5× Tris-borate-EDTA buffer (34). The agarose gel was then treated with 10% formaldehyde at 65°C and washed with H2O three times, and the RNA was electroblotted onto a Hybond-N+ nylon membrane (Amersham). Northern hybridization was then performed as described (34). The plasmid T7pol (54) was transcribed with T7 RNA polymerase to synthesize a 300-base RNA fragment complementary to the NL4-3 pol gene. Approximately 7 × 106 cpm of this riboprobe per blot was used in the hybridization reaction. Hybridization was carried out in the presence of Rapid-Hyb buffer (Amersham). Membranes were washed extensively with 0.1× SSC (1× SSC is 150 mM NaCl and 15 mM sodium citrate [pH 7.0]) and 0.1% SDS at 70°C. In the experiments designed to assess the conversion of dimer to monomer RNA species, the relative amount of both RNA species was quantitated by PhosphorImager analysis (Molecular Dynamics).
RNase protection assays.
An antisense RNA probe (∼108 cpm/mg) was synthesized by transcription of pGEM(600-1000) (41) with T7 RNA polymerase (New England Biolabs) or pT7HIV-1 410-910 (54) with SP6 RNA polymerase (Promega) following linearization with NotI or SalI, respectively (38). To serve as size markers for denaturing polyacrylamide gels, HpaII-digested pGEM3Zf(+) fragments were end labeled with 32P (38). One-third of the virion or cytoplasmic RNA preparation was mixed with 2 × 105 Cerenkov counts of 32P-labeled antisense RNA and precipitated with ethanol. RNase protection assays were then performed as described (40). After electrophoresis in 5% polyacrylamide–8 M urea gels, various protected RNA species were quantitated by PhosphorImager analysis (Molecular Dynamics). In this report, the packaging efficiency was determined by calculating the ratio of virus-associated RNA to p24, not by referring the level of RNA packaged divided by the level of the RNA species in the virus-producing cell.
Infection and MAGI cell assays.
293 or 293tat cells (2 × 106) were transfected with 3 μg each of p5′ssEnvEXSV and pMSMBA or the other mutant derivatives of pMSMBA. At 48 to 72 h after transfection, the medium was clarified by centrifugation, and the supernatant was used for infection. Infection was accomplished by incubating cells for 18 to 24 h (M8166) or 72 h (MAGI) with equivalent p24 units of virus in the presence of DEAE-dextran (8 μg/ml). MAGI cell assays were performed as described previously (30).
Western blotting analysis.
Lysates of pelleted virus were prepared as previously described (60). Virion proteins were resolved on 12% SDS–polyacrylamide gels and then electrophoretically transferred to polyvinylidene difluoride membranes. ECL Western blotting detection reagent (Amersham International plc, Buckinghamshire, England) was used to detect viral proteins on the membrane. Briefly, the membranes were incubated at room temperature with human anti-HIV serum for 1 h, washed, incubated with horseradish peroxidase-labeled protein A for 1 h, washed, and visualized by exposure to X-ray film.
RESULTS
Duplication of the E/DLS region of HIV-1 RNA results in the accumulation of monomeric RNA in virions.
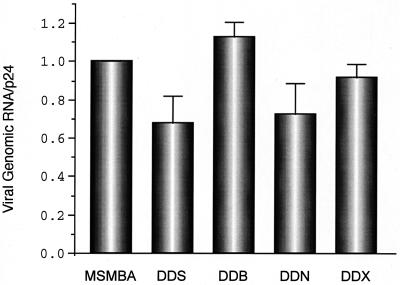
To see whether duplication of the packaging-dimerization region (E/DLS) of HIV-1 affects encapsidation of genomic viral RNA, we constructed a variety of mutants carrying two copies of the encapsidation and dimer linkage region. The duplicated region was approximately 1,000 bp in length and included TAR, R/U5, U5/L, SL1, SL2, SL3, and SL4 stem-loops and the 5′ half of the gag gene. These stem-loops are located in the 5′ region of the HIV-1 genome and play important roles in viral genome packaging and dimerization. The polyadenylation signals and the s.d. in the ectopic fragment were deleted to obviate undesired polyadenylation and ectopic splicing. We initially generated four mutants (pDDS, pDDB, pDDN, and pDDX) that contain an additional E/DLS region at various locations in the HIV-1 genome (Fig. 1). These mutants and the wild-type plasmid (pMSMBA) were transfected in parallel into 293tat cells along with pCMV259Δ21 (40). Plasmid pCMV259Δ21, which was used as a helper plasmid, efficiently produces all the viral structural proteins except Env but does not produce packageable RNA. Cytoplasmic RNA was isolated from the transfected cells, and the virion RNA was isolated from the culture supernatant (40). The amount of RNA within virus particles was then quantitated by an RNase protection assay (40) using a riboprobe which detects both wild-type and mutant RNAs. Although slight variations were observed in packaging efficiency among the various mutants, each of the mutant RNAs was packaged with an efficiency similar to that of the wild type (Fig. 2). This indicates that the presence of an additional E/DLS region at several alternative positions in the viral genome had little effect on RNA packaging.
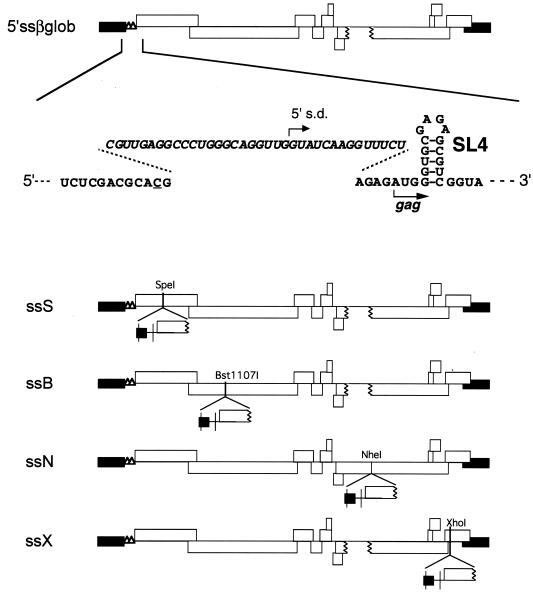
FIG. 1.
Diagram of pMSMBA and mutants containing two copies of viral sequence. Open and solid boxes indicate open reading frames and the long terminal repeat (LTR), respectively. The polyadenylation signal (polyA) and a major s.d. site on the E/DLS fragment were mutated as described in Materials and Methods. Nucleotide positions of restriction endonuclease recognition sites used for constructing these mutants are also shown.
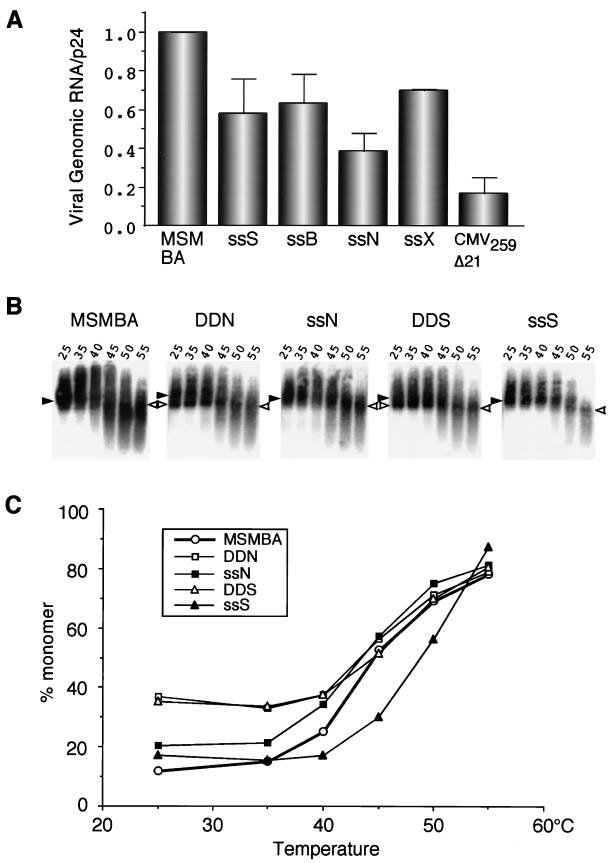
FIG. 2.
Relative encapsidation efficiency of viral RNAs containing two E regions. The relative amount of RNA from the virus particles was quantified by phosphorimage analysis, and physical virus particles were measured using a p24 antigen capture assay. The packaging efficiency was determined by calculating the ratio of virus-associated RNA to p24. The RNA/p24 ratio for the wild type was set at 1. Data points represent the means of three or more independent measurements. Error bars indicate standard errors.
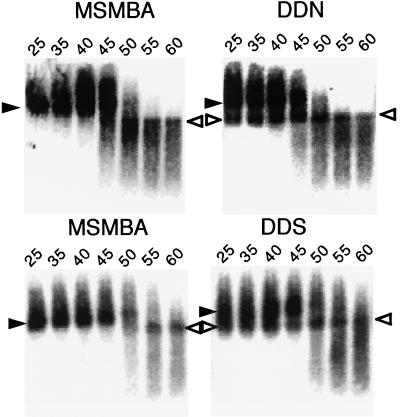
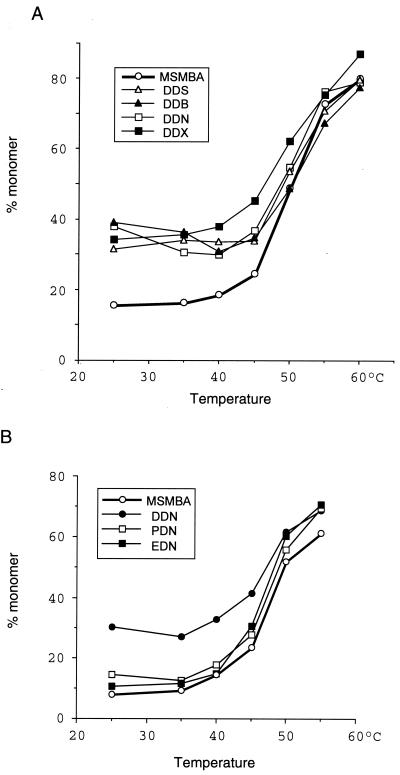
We next tried to determine whether the presence of a second E/DLS affects RNA dimer formation by analyzing virion-associated RNA on native agarose gel electrophoresis. As shown previously, both the dimeric and monomeric RNAs migrated heterogeneously on such gels (10, 26, 34, 54, 55). Nonetheless, it was possible to distinguish between these two RNA populations and to determine whether the mutations affected RNA dimerization. As expected, pMSMBA RNA isolated from virus particles was dimeric and exhibited a denaturation profile with conversion to single-stranded RNA with increasing temperature (Fig. 3). However, the RNA from the particles of the mutants with two E/DLS regions contained a readily detectible amount of monomeric RNA (25 to 40% of total signal) even under nondenaturing conditions (Fig. 3A). This indicates that duplication of the 5′ region of HIV-1 RNA causes the appearance of monomeric RNA in virions. These data also indicate that the duplication that resulted in this effect was position independent, since each of these mutants harbored monomeric RNA within virus particles.
FIG. 3.
Representative phosphorimage analysis of RNA detected by Northern blotting. Aliquots of RNA extracted from virions were resuspended in T-buffer (see Materials and Methods), incubated for 10 min in parallel reactions at various temperatures, and then analyzed on a native agarose gel. The membrane was hybridized with a probe corresponding to the pol region. The positions of dimer (solid arrowheads) and monomer (open arrowheads) viral RNAs are indicated. The temperatures (degrees Celsius) at which aliquots were incubated prior to electrophoresis are also indicated.
To determine whether duplication of a region outside of the primary dimer linkage region could result in an increase in monomeric RNA in virus particles, we constructed two mutants that had a duplication of a segment of the pol or env gene (PDN and EDN, respectively) (Fig. 1) and compared dimer formation of these mutants with that of the wild type and pDDN. DDN particles, which contained viral RNAs with two dimer linkage regions, contained detectable monomeric RNA, as expected. However, the other two mutants, which contained an additional copy of either pol or env, exhibited an RNA profile similar to that of the wild type (Fig. 4).
FIG. 4.
Thermal dissociation kinetics of dimeric viral RNA. The relative amounts of monomeric and total RNA in each lane were quantitated with a PhosphorImager, and the percentage of monomer RNA was calculated for each RNA sample. Similar results were obtained in three separate experiments. (A) Comparison of wild-type (pMSMBA) and mutant viruses containing two E regions. (B) Comparison of pMSMBA and mutants containing two copies of various viral sequences.
A single ectopic E/DLS can mediate efficient encapsidation and RNA dimerization.
Since the mutants containing RNAs with two E/DLS regions were encapsidated efficiently, we wanted to determine whether the ectopic E/DLS sequences located in different positions throughout the genome were sufficient for packaging. The HIV-1 construct p5′ssβglob, which has a large deletion in the primary encapsidation (E/psi) region and nearby cis sequences, exhibits severely reduced RNA packaging efficiency compared to the wild type (41). However, p5′ssβglob is able to express viral genes via spliced mRNAs, since this construct contains the splice donor from the first intron of the human β-globin gene. The leader region of p5′ssβglob was introduced in place of the primary E/DLS region of DD series plasmids to create pssS, pssB, pssN, and pssX (Fig. 5). 293tat cells were transfected with these plasmids, and the amounts of cellular and virus particle-associated p24 and virus-specific RNA contents were measured. As shown in Fig. 6A, the efficiency with which the various mutants were packaged was within 40 to 70% of the wild-type value, whereas that of pCMV259Δ21 and p5′ssβglob was only 10 to 20% of wild-type levels. These results indicate that the ectopic E/psi region can at least partially function as a packaging signal in the absence of the authentic E/psi site. They also indicate that E/psi can function at several different locations in the genome.
FIG. 5.
Diagram of p5′ssβglob and mutants lacking the normal E region but containing an ectopic E region. The origin and construction of these plasmids are described in Materials and Methods. β-Globin sequences are italicized. Underlined is a single nucleotide substitution generated for inserting the β-globin sequence.
FIG. 6.
Analysis of mutants containing a single ectopic E region. (A) Encapsidation efficiency. The relative amount of RNA from virus particles was quantified by phosphorimage analysis, and encapsidation was quantitated as for Fig. 2. The value for the wild-type control was set at 1. pCMV259Δ21 was included as a negative control. The data are derived from at least three independent experiments. Error bars indicate standard errors. (B) Representative phosphorimage analysis of RNA detected by Northern blotting. Experiments were performed as in Fig. 3. Positions of dimeric (solid arrowheads) and monomeric (open arrowheads) viral RNAs are indicated. The temperatures (degrees Celsius) at which aliquots were incubated are indicated for each lane. (C) Thermal dissociation kinetics of RNA dimers. The thermal stability of RNA dimers and monomers was measured as in Fig. 4. Similar results were obtained in three separate experiments.
We next analyzed the conformation of RNA within virus particles by native agarose gel electrophoresis followed by Northern blot hybridization. No apparent monomer RNAs were observed in virus particles from the mutants containing a single ectopic E/DLS (Fig. 6B and C). These results are consistent with a requirement for two intact E/DLS regions for the generation of monomeric RNA. In addition, most of the mutant RNA dimers exhibited thermostability similar to that of the wild-type virus. It is noteworthy that the mutant ssS dimer RNA showed slightly higher stability than the wild-type and other mutant RNAs. The ssS mutant has a tandemly repeated E/DLS. It is possible that four E/DLS sites positioned closely within two RNA molecules result in a more stable dimer.
Presence of an ectopic E/DLS region affects the infectivity of virus.
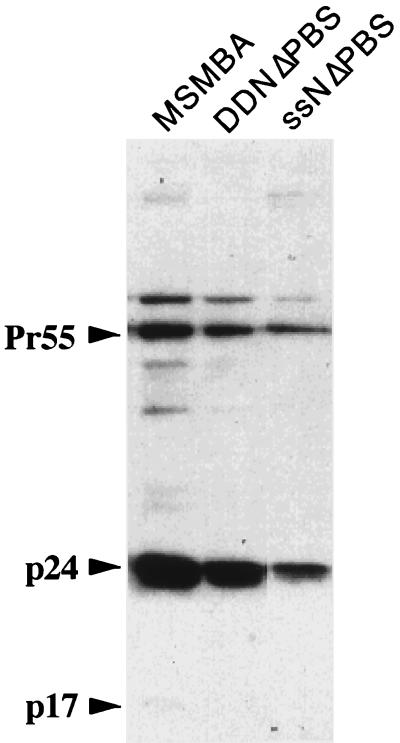
To determine whether the duplication of the E/DLS region affects the viral life cycle, we analyzed the infectivity of mutant viruses using the MAGI cell assay (30). Since the presence of an ectopic E/DLS region would also result in a virus with an extra primer-binding site (PBS), it was highly unlikely that those mutants retained infectivity. To overcome this complication, we constructed two more mutants, pDDNΔPBS and pssNΔPBS, that contained only naturally occurring PBS. The profiles and packaging efficiency of these mutant RNAs were first examined by native agarose gel electrophoresis followed by Northern blot hybridization and an RNase protection assay. This showed that DDNΔPBS and ssNΔPBS were similar to their progenitors, DDN and ssN, respectively, in packaging efficiency and dimerization or lack of dimerization (data not shown). Western blot analysis showed that the proteins of both viruses were properly expressed and processed (Fig. 7). We then compared the infectivity of those mutants with that of the wild-type, PDN, EDN, and 5′ssβGlob viruses. Since all these constructs carried a mutation in the env gene, HIV-1 Env proteins were supplied by cotransfection of env expression vector p5′ssEnvEXSV to produce infectious virions by complementation. Seventy-two hours after transfection, culture supernatants were assayed for the levels of viral p24 protein, and equivalent amounts of virus in p24 were used to infect MAGI cells (30). Forty-eight hours after infection, cells were fixed and stained, and the cells that were successfully infected, as evidenced by bacterial β-galactosidase expression, were enumerated. As shown in Table 1, there was a more than 100-fold reduction in infectivity of DDNΔPBS and ssNΔPBS compared to the wild-type virus, while the duplication of the pol and env regions had a very little or no effect on viral infectivity. Since ssNΔPBS carried only one intact E/DLS region and formed dimeric RNA as efficiently as wild-type virus, it seemed unlikely that the loss of infectivity of DDNΔPBS and ssNΔPBS was not due simply to the lack of dimeric RNA in virions. Instead, it seemed more likely that the presence of an ectopic E/DLS site affected one or more steps between virus penetration and gene expression. In particular, it seemed likely that some step in viral nucleic acid replication was arrested.
FIG. 7.
Western blot analysis of virion proteins. Positions of the Gag precursor (Pr55) and gag products p24 and p17 are indicated.
TABLE 1.
Infectivity of DD and ss mutantsa
| Virus | Relative titer |
|---|---|
| MSMBA | +++ |
| DDNΔPBS | ± |
| ssNΔPBS | − |
| PDN | ++ |
| EDN | +++ |
| 5′ssβglob | − |
The infections were performed on CD4+ long terminal repeat/β-galactosidase indicator cells as described. Titers: +++, >103; ++, 103 to 102; ±, 101 to 100; −, <100 β-galactosidase-inducing units per ml.
DISCUSSION
We have found that duplication of the E/DLS region of HIV-1 RNA results in the appearance of monomers in virions without markedly affecting encapsidation efficiency. In contrast, no monomers were observed in virions of mutants that have only one E/DLS region at an ectopic position. Duplication of viral RNA per se does not interfere with dimarization, since monomers were not observed when a segment of the pol or env region was duplicated. We speculate that the presence of an additional E/DLS region at the ectopic position results in interaction between those two regions and competitively interferes with intermolecular dimer formation. Consistent with this notion, monomeric RNAs from mutant virions that had not been subjected to heat treatment exhibited less variation in mobility following electrophoresis through native gels than heated samples. This result is consistent with the idea that the mutant monomeric RNA forms a particular secondary or tertiary structure that results in relatively uniform migration, similar to that observed for dimeric RNA from the wild-type particles. In contrast, monomeric RNA profiles from protease-deficient particles were heterogenous (21, 22, 50), suggesting a more disordered secondary or tertiary structure of the viral RNA in immature particles. Alternatively, it is possible that a higher proportion of monomers in the mutant virus than in the wild-type virus reflects the relative fragility of the dimers, since it is hard to exclude the possibility that the monomers observed in RNA extracted from virions came from dissociated dimers. In fact, some RNA is observed in the monomer position in all of the unheated RNA profiles (Fig. 4 and 6C). Although this possibility could not be excluded, it is still reasonable to conclude that duplication of the E/DLS site affected dimerization of viral RNA and that the E/DLS site plays an important role in dimer formation.
We and other groups have found that mutation of the DIS loop does not affect dimer formation in vivo (7, 12, 54) and a region other than E/DLS affected dimer formation of retrovirus RNA (54, 59), also indicating a limited role for the DIS-DLS region in dimer formation. There are probably multiple sites on the virus genome that contribute to RNA dimerization. It is possible that the dimer linkage site observed by electron microscopy is the final dissociating point of RNA dimer under denaturing conditions and that such a point might not coincide with the primary contact point of RNA following virion assembly.
Although the presence of two E/DLS sites results in the presence of monomeric RNA in particles, it is still unclear whether one or two monomeric RNAs are encapsidated in each virion. Determination of the relative amounts of Gag and RNA molecules in particles might help to resolve this question. Recent studies with a Rous Sarcoma Virus MA mutant that packages monomeric RNA suggests the presence of only one RNA molecule for that mutant (52). However, it appears that only 10% of HIV-1 particles contain virus RNA (M. S. McBride, personal communication). Moreover, several reports describe data indicating that significant quantities of cellular RNA are incorporated into retrovirus particles (for a review, see reference 8). Therefore, it is difficult to deduce whether one or two monomeric RNA molecules are encapsidated in individual mutant virions.
We observed some dimeric RNA along with monomeric RNA for mutants containing two E/DLS regions. The authentic and ectopic E/DLS regions on individual RNAs may be effectively located closer to each other than those on separate RNA molecules. This might facilitate intramolecular contact but not completely eliminate intermolecular interaction. Thus, intermolecular interaction between two native DIS-DLS regions might compete with the intramolecular E/DLS interaction.
Mutants containing an E/DLS site at an ectopic position but lacking the natural E/psi site were packaged efficiently but not as efficiently as wild-type RNA. It is possible that the ectopic E/psi site was fully functional but that the mutated E/psi site at the original position had a negative effect on packaging. It is also likely that the context of the E/psi region affects packaging efficiency. However, the ectopic E/DLS region appeared to be fully functional for dimer formation, since mutants containing a single E/DLS region at a single novel location formed RNA dimers with stability similar to that of the wild type.
Mutants that had an ectopic E/DLS region were profoundly reduced in infectivity even though they fully retained a E/psi site, splice site, and PBS (Table 1). Surprisingly, duplication of the pol and env regions had only a small or slight effect on infectivity (Table 1). One explanation for this specific defect is that the 5′ region containing E/DLS may be more recombinagenic than the pol or env region. This may reflect a general higher efficiency of recombination associated with regions of the RNA normally located near the RNA termini. There may be an intrinsic feature of the termini that promotes transfer of minus-strand strong-stop DNA from the 5′ terminus of the viral RNA. In fact, a series of studies from Pedersen's group (35, 36, 43–45) showed site-specific recombination within the highly structured 5′ leader region of murine leukemia virus.
The generation of monomeric RNA due to the presence of a second E/DLS might provide a useful system for characterizing the requirements for in vivo dimer formation. Genetic analysis of one or both sites and the effect of mutations on these sites could be examined by assaying for the presence of monomers in virus particles.
ACKNOWLEDGMENTS
We thank Diccon Fiore for technical assistance and members of the Panganiban laboratory and Dan Loeb for helpful discussions. J.S. also thanks Shigeharu Ueda and Aikichi Iwamoto for helpful discussion and advice and Sayuri Sakuragi for encouragement.
J.S. was supported by a Japan Society for Promotion of Science fellowship for research abroad. This work was supported by R01 AI34733 from the NIH.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Walker B D. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. [Google Scholar]
- 3.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 4.Bender W, Chien Y H, Chattopadhyay S, Vogt P K, Gardner M B, Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, Essink B B, Schoneveld I. In vitro dimerization of HIV-2 leader RNA in the absence of PuGGAPuA motifs. FASEB J. 1993;7:181–187. doi: 10.1096/fasebj.7.1.8422965. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout B, van Wamel J L. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 9.Bieth E, Gabus C, Darlix J L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M, Garon C F, Papas T S. Native ribonucleoprotein is an efficient transcriptional complex of avian myeloblastosis virus. Proc Natl Acad Sci USA. 1980;77:1296–1300. doi: 10.1073/pnas.77.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J. Genome structure. In: Weiss R, Teich N, Vermus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. pp. 261–368. [Google Scholar]
- 15.Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 16.Darlix J L, Gabus C, Nugeyre M T, Clavel F, Barré-Sinoussi F. cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 17.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M-C, Roques B, Darlix J-L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:9373–9377. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Tapia M, Metzler V, Mougel M, Ehresmann B, Ehresmann C. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 19.Dube S, Kung H J, Bender W, Davidson N, Ostertag W. Size, subunit composition, and secondary structure of the Friend virus genome. J Virol. 1976;20:264–272. doi: 10.1128/jvi.20.1.264-272.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 24.Greatorex J, Lever A. Retroviral RNA dimer linkage. J Gen Virol. 1998;79:2877–2882. doi: 10.1099/0022-1317-79-12-2877. [DOI] [PubMed] [Google Scholar]
- 25.Greatorex J S, Laisse V, Dockhelar M C, Lever A M. Sequences involved in the dimerisation of human T cell leukaemia virus type-1 RNA. Nucleic Acids Res. 1996;24:2919–2923. doi: 10.1093/nar/24.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haddrick M, Lear A L, Cann A J, Heaphy S. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 27.Höglund S, Öhagen Å, Goncalves J, Panganiban A T, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 28.Jones J S, Allan R W, Temin H M. One retroviral RNA is sufficient for synthesis of viral DNA. J Virol. 1994;68:207–216. doi: 10.1128/jvi.68.1.207-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh I, Yasunaga T, Yoshinaka Y. Bovine leukemia virus RNA sequences involved in dimerization and specific Gag protein binding: close relation to the packaging sites of avian, murine, and human retroviruses. J Virol. 1993;67:1830–1839. doi: 10.1128/jvi.67.4.1830-1839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kung H, Hu S, Bender W, Bailey J M, Davidson N. RD-114, baboon, and woolly monkey viral RNAs compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 32.Kung H J, Bailey J M, Davidson N, Vogt P K, Nicolson M O, McAllister R M. Electron microscope studies of tumor virus RNA. Cold Spring Harb Symp Quant Biol. 1975;39:827–834. doi: 10.1101/sqb.1974.039.01.096. [DOI] [PubMed] [Google Scholar]
- 33.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 34.Lear A L, Haddrick M, Heaphy S. A study of the dimerization of Rous sarcoma virus RNA in vitro and in vivo. Virology. 1995;212:47–57. doi: 10.1006/viro.1995.1452. [DOI] [PubMed] [Google Scholar]
- 35.Lund A H, Mikkelsen J G, Schmidt J, Duch M, Pedersen F S. The kissing-loop motif is a preferred site of 5′ leader recombination during replication of SL3–3 murine leukemia viruses in mice. J Virol. 1999;73:9614–9618. doi: 10.1128/jvi.73.11.9614-9618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund A H, Schmidt J, Luz A, Sorensen A B, Duch M, Pedersen F S. Replication and pathogenicity of primer binding site mutants of SL3–3 murine leukemia viruses. J Virol. 1999;73:6117–6122. doi: 10.1128/jvi.73.7.6117-6122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maisel J, Bender W, Hu S, Duesberg P H, Davidson N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J Virol. 1978;25:384–394. doi: 10.1128/jvi.25.1.384-394.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniatis T, Fritsch E F, Sambrook J, editors. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 39.Marquet R, Baudin F, Gabus C, Darlix J L, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikkelsen J G, Lund A H, Duch M, Pedersen F S. Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo. J Virol. 2000;74:600–610. doi: 10.1128/jvi.74.2.600-610.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikkelsen J G, Lund A H, Duch M, Pedersen F S. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region. J Virol. 1998;72:6967–6978. doi: 10.1128/jvi.72.9.6967-6978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikkelsen J G, Lund A H, Dybkaer K, Duch M, Pedersen F S. Extended minus-strand DNA as template for R-U5-mediated second-strand transfer in recombinational rescue of primer binding site-modified retroviral vectors. J Virol. 1998;72:2519–2525. doi: 10.1128/jvi.72.3.2519-2525.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 47.Muriaux D, Girard P M, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength: an autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 48.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Negrini M, Rimessi P, Sabbioni S, Caputo A, Balboni P G, Gualandri R, Manservigi R, Grossi M P, Barbanti-Brodano G. High expression of exogenous cDNAs directed by HIV-1 long terminal repeat in human cells constitutively producing HIV-1 tat and adenovirus E1A/E1B. Biotechniques. 1991;10:344–353. [PubMed] [Google Scholar]
- 50.Ortiz-Conde B A, Hughes S H. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J Virol. 1999;73:7165–7174. doi: 10.1128/jvi.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 52.Parent L J, Cairns T M, Albert J A, Wilson C B, Wills J W, Craven R C. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J Virol. 2000;74:164–172. doi: 10.1128/jvi.74.1.164-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakuragi J I, Panganiban A T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen N, Jette L, Liang C, Wainberg M A, Laughrea M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol. 2000;74:5729–5735. doi: 10.1128/jvi.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stokrova J, Korb J, Riman J. Electron microscopic studies on the structure of 60-70S RNA of avian myeloblastosis virus. Acta Virol. 1982;26:417–426. [PubMed] [Google Scholar]
- 58.Sundquist W I, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tchénio T, Heidmann T. The dimerization/packaging sequence is dispensable for both the formation of high-molecular-weight RNA complexes within retroviral particles and the synthesis of proviruses of normal structure. J Virol. 1995;69:1079–1084. doi: 10.1128/jvi.69.2.1079-1084.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]