Abstract
The herpes simplex virus type 1 gene UL47 encodes the tegument proteins referred to collectively as VP13/14, which are believed to be differentially modified forms of the same protein. Here we show that the major product of the UL47 gene during transient expression is VP14, suggesting that some feature of virus infection is required to produce VP13. We have tagged VP13/14 with green fluorescent protein and have demonstrated that the protein is targeted efficiently to the nucleus, where it often localizes in numerous punctate domains. Furthermore, we show that removal of the N-terminal 127 residues of the protein abrogates nuclear accumulation, and we have identified a 14-amino-acid peptide from this region that is sufficient to function as a nuclear targeting signal and transport a heterologous protein to the nucleus. This short peptide contains two runs of four arginine residues, suggesting that the VP13/14 nuclear localization signal may behave in a manner similar to that of the arginine-rich nuclear localization signals of the retrovirus transactivator proteins Tat, Rev, and Rex. In addition, by using heterokaryon assays, we show that VP13/14 is capable of shuttling between the nucleus and cytoplasm of the cell, a property that may be attributed to three leucine-rich stretches in the C-terminal half of the protein that again bear similarity to the nuclear export signals of Rev and Rex. This is the first demonstration of a tegument protein that is specifically targeted to the nucleus, a feature which may be relevant both during virus entry, when VP13/14 enters the cell as a component of the tegument, and at later times, when large amounts of newly synthesized VP13/14 are present within the cell.
The herpes simplex virus type 1 (HSV-1) proteins VP13 and VP14 are major structural components of the virion tegument region, the compartment located between the capsid and the virus envelope (50). Both proteins have been shown to be encoded by the true late gene UL47 (31, 53), and they are therefore referred to collectively as protein VP13/14. They have apparent masses of 82 and 81 kDa, respectively (31) and have been shown to be posttranslationally modified by phosphorylation, nucleotidylylation, and glycosylation (2, 33). While the molecular differences between the two proteins are not yet clear, it has been suggested that differential modification may account for the differing migration of VP13 and VP14 upon analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). However, the only variance that has been reported to date in the activities of the two products is that VP13 alone binds to the nuclear matrix of infected cells (44).
Despite being a major structural component, with approximately 1,800 copies in the virion tegument (23), VP13/14 has been shown to be dispensable for virus replication in tissue culture (1, 53). Furthermore, virions made in the absence of VP13/14 appear to have increased levels of another tegument protein, VP11/12, suggesting a possible structural redundancy of these two proteins (53). Little is known about the role of HSV-1 VP13/14 during virus infection, but there is some evidence to suggest that it may be involved in the modulation of the activity of the tegument protein VP16, the transactivator of immediate-early (IE) gene expression (6, 20, 39). McKnight and coworkers (30) have shown that coexpression of the UL47 gene with the gene for VP16 modulates the ability of VP16 to activate IE promoters during transient transfection. Moreover, viruses unable to express VP13/14 appear to be retarded in the early stages of virus growth, supporting a potential role for the protein in gene expression (53, 54).
In this study we have investigated the intrinsic properties of VP13/14 in the absence of other viral proteins by fusion of the UL47 gene to the C terminus of green fluorescent protein (GFP). We show that GFP-13/14 localizes efficiently to the cell nucleus and exhibits a diverse range of intranuclear patterns. In addition, we identify a 14-residue nuclear localization signal (NLS) at the N terminus of VP13/14 which is both necessary and sufficient to direct a heterologous protein to the nucleus. Thus, VP13/14 is the first NLS-containing tegument protein to be described. Finally, by using heterokaryon assays, we show that VP13/14 not only localizes to the nucleus but also is capable of shuttling between the nucleus and cytoplasm, a feature that is consistent with a role in the regulation of gene expression.
MATERIALS AND METHODS
Cells and viruses.
Vero cells, BHK cells, and COS-1 cells were grown in Dulbecco's modified minimal essential medium supplemented with 10% newborn calf serum. The HSV-1 strain 17 was used for infections and virion purification.
Plasmids.
The UL47 open reading frame, including both the start methionine and the stop codon, was amplified by PCR from HSV-1 genomic DNA using primers which incorporated BamHI sites at both ends. (forward primer, CGCGGATCCCCCGCGTCTATCGCCACC; reverse primer; GCGGGATCCCGGCAGCACGGGCGGAGG). This product was digested with BamHI and inserted into the BamHI sites of pEGFP-C1 (Clontech), pCMV19aSV5, and pcDNA1.Amp (Invitrogen) to create plasmids pMD10, encoding GFP-13/14, pMD12, encoding SV-5-tagged VP13/14, and pMD13, encoding wild-type (Wt) VP13/14. Plasmid pMD10.ΔSac, which lacks residues 1 to 187 of VP13/14, was constructed by digesting pMD10 with SacI, removing the 3′ overhangs from the large fragment, and religating. Residues 1 to 127 of VP13/14 were deleted by digesting pMD10 with SalI and religating the larger fragment, resulting in pMD10.ΔSal, while residues 1 to 36 were removed by digesting pMD10 with KpnI and end-filling the larger fragment prior to religation to form pMD10.ΔKpn. To produce the series of plasmids expressing GFP-13/14 with mutated NLS sequences, a range of overlapping PCRs was carried out on the N terminus of the UL47 gene, and the PCR products were inserted back into plasmid pMD10. This resulted in plasmids pMD14 (R63 through R66 to G), pMD15 (R72 through R75 to G), and pMD16 (R63 through R66 to G and R72 through R75 to G). The series of GFP-NLS constructs, containing various sequences of the N terminus of VP13/14 fused to GFP, was made as follows. The N-terminal 92, 76, 68, or 22 codons of the UL47 gene were amplified by PCR and inserted into the BamHI site of pEGFPC1 to make plasmids NLS1+2+3+, NLS1+2+3, NLS1+2, and NLS1, respectively. Plasmid NLS1+3 was made by PCR amplification of the N-terminal 76 codons of UL47 from pMD14, which were inserted as an EcoRI/BamHI fragment into pEGFPC2 (Clontech). For plasmids NLS2 and NLS2+3, double-stranded oligonucleotide adapters encoding either residues 63 to 72 or residues 63 to 76 were made and inserted into the pEGFPC1 multiple cloning site. Plasmids pCDNA3 myc-hnRNPA1 and pCDNA3 myc-hnRNPC1 were kindly provided by Gideon Dreyfuss, University of Pennsylvania.
Antibodies.
The polyclonal anti-VP13/14 antibody R220, kindly provided by David Meredith, was used at dilutions of 1:5,000 for Western blotting and 1:400 for immunofluorescence. The polyclonal antibody against GFP (RDI) was used at a dilution of 1:1,000 for Western blotting and 1:600 for immunofluorescence. The monoclonal antibody 336 (kindly provided by Rick Randall, University of St. Andrews) against the SV5 epitope tag was used at a dilution of 1:2,000 for immunofluorescence. The monoclonal anti-myc antibody (Invitrogen) against the myc epitope tag was used at a dilution of 1:1,000 for immunofluorescence.
Transfections.
COS-1 cells were plated onto six-well dishes at a density of 3 × 105 per well for Western blotting. For live-cell analysis of GFP-expressing cells, COS-1 cells were plated into two-well coverslip chambers (Life Technologies) at a density of 105 per chamber. DNA transfection mixtures, consisting of 200 ng of expression plasmid made up to 2 μg with pUC19 DNA, were transfected by the calcium phosphate precipitation technique modified by substitution of BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline for HEPES-buffered saline. Transfected cells were analyzed 40 h posttransfection unless otherwise stated.
Western blot analysis.
Proteins were separated by electrophoresis through SDS-polyacrylamide gels cross-linked with bisacrylamide. Gels were transferred to nitrocellulose filters for Western blotting and reacted with an appropriate antibody. A horseradish peroxidase-linked secondary conjugate was used, and reactive bands were visualized by development with enhanced chemiluminescence (ECL) detection reagents (Amersham).
Virion purification.
Virions were purified from extracellular virus released into the infected cell medium as described previously (14).
Heterokaryon assays.
Interspecies heterokaryons of COS-1 and mouse NIH 3T3 cells were formed as described previously (45). Briefly, COS-1 cells seeded at 106 per 25-cm2 flask were transfected with 6 μg of plasmid DNA. Twenty-four hours posttransfection the COS-1 cells were seeded on 16-mm glass coverslips in a six-well tray, and after overnight incubation these cultures were then seeded with an equal number of untransfected NIH 3T3 cells. The coculture was incubated for 3.5 h in the presence of 50 μg of cycloheximide/ml and for 30 min in the presence of 100 μg of cycloheximide/ml. After being fused with 50% polyethylene glycol for 2 min, cells were washed and returned to medium containing 100 μg of cycloheximide/ml for a further 4 h.
Immunofluorescence and microscopy.
Cells were fixed in 4% paraformaldeyde in phosphate-buffered saline (PBS) for 20 min, washed in PBS, and then permeabilized with 0.5% Triton X-100 in PBS for 10 min before washing with PBS. Fixed cells were blocked by incubation with PBS containing 10% calf serum for 20 min at room temperature. Primary antibody was added in the same solution and was incubated for 20 min at room temperature. After a wash with PBS, secondary antibodies were added in PBS–10% calf serum and incubated for 20 min at room temperature. Coverslips were washed with PBS prior to mounting in Vectashield alone or Vectashield with added 4′,6′-diamidino-2-phenylindole (DAPI) (both from Vector Laboratories). Samples were analyzed using either a Zeiss LSM 410 inverted confocal microscope or a Photometrics Quantix digital camera on an Axiovert S100 TV inverted microscope. Images were processed using Adobe Photoshop software.
RESULTS
VP14 is the major product of the UL47 gene in transfected cells.
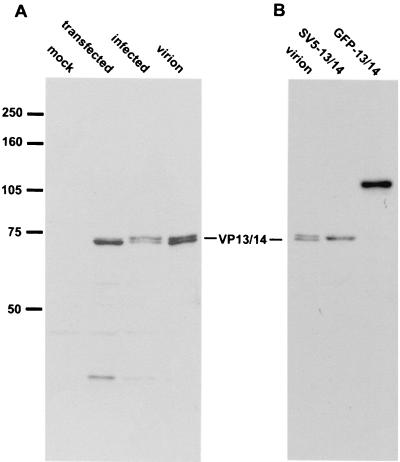
While VP13 and VP14 have previously been identified as alternative products of the UL47 gene in virus-infected cells, we wanted to determine if both proteins would be expressed from this gene in the absence of other virus gene products. Thus, COS-1 cells were first transfected with the UL47 expression vector pMD13 and then analyzed by Western blotting with the VP13/14-specific antibody R220 (Fig. 1). Comparison of the pMD13-expressing cells with both HSV-1-infected cells and purified virions revealed that the major product of the transiently transfected UL47 gene was of the same molecular weight as the faster-migrating VP14 (Fig. 1A). However, upon longer exposure, small quantities of the slower-migrating VP13 were also detectable in the UL47-transfected cells, albeit at much lower levels than in infected cells (data not shown). This suggests that some additional feature of virus infection is required in order to express high levels of VP13. It is also noteworthy that in our hands the VP13/14 doublet exhibits an apparent size of 72 to 74 kDa, somewhat smaller than the previously published size of 81 to 82 kDa (31). Interestingly, transfection and expression of the plasmid pMD12, carrying the UL47 gene with a 5′ terminal SV5 epitope tag, results in a protein of the correct size for SV5-tagged VP14 (Fig. 1B, SV5-13/14), suggesting that VP14 is not the result of an internal initiation event but is probably differentially modified in comparison to VP13.
FIG. 1.
Expression of the UL47 gene by transient transfection. (A) COS-1 cells were transfected with either pUC19 (mock) or plasmid pMD13 carrying the UL47 gene (transfected). Total cell lysates from these transfections and from infected Vero cells (infected), together with purified extracellular virus particles (virion), were analyzed by Western blotting with the anti-VP13/14 antibody R220. (B) Total cell lysates from COS-1 cells transfected with either plasmid pMD12 (SV5-13/14) or plasmid pMD10 (GFP-13/14), together with purified extracellular virus particles (virion), were analyzed by Western blotting with the anti-VP13/14 antibody R220.
GFP-13/14 is directed to the nuclei of transfected cells.
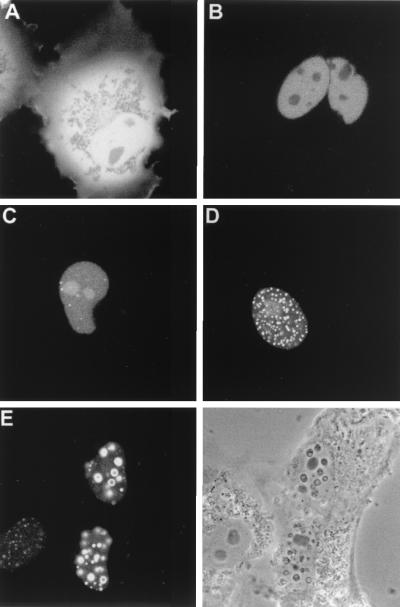
We next addressed the question of VP13/14 cellular localization by constructing plasmid pMD10, which carries the gene encoding GFP fused in frame to the 5′ terminus of the UL47 gene. Western blotting of COS-1 cells expressing this fusion protein demonstrated that the resulting product was approximately the correct molecular weight of 110 kDa (Fig. 1B, GFP-13/14). To examine the cellular localization patterns of GFP-13/14, we then transfected COS-1 cells with either the GFP-13/14 expression vector or the unfused GFP expression vector pEGFPC1 and examined the cells live (Fig. 2). As expected, COS-1 cells transfected with the plasmid encoding GFP alone displayed a pattern of diffuse fluorescence throughout both the cytoplasm and the nucleus (Fig. 2A). By contrast, in cells expressing GFP-13/14, GFP fluorescence was localized entirely within the cell nuclei (Fig. 2B to E), where it exhibited a range of patterns. In some nuclei the protein appeared diffuse and either was excluded from the nucleoli (Fig. 2B) or accumulated in the nucleoli (Fig. 2C). In other nuclei GFP-13/14 was concentrated in either multiple speckled domains (Fig. 2D) or much larger punctate domains ranging in size from 0.3 to 0.6 μm (Fig. 2E). Interestingly, these larger punctate domains could also be seen as phase-dense spherical structures when the cells were examined by light microscopy, and in some cells large “doughnut” structures were evident. We believe that the heterogeneity displayed by GFP-13/14 is a reflection of the relative expression levels in individual cells, with the diffuse patterns representing low-level expression and the punctate patterns representing high-level expression.
FIG. 2.
Subcellular localization of GFP-13/14 expressed by transient transfection. COS-1 cells were transfected with plasmids expressing either unfused GFP (A) or GFP-13/14 (B to E). The cells were examined live by confocal microscopy 40 h after transfection. (Bottom right) Phase-contrast image of the same field as in panel E.
VP13/14 contains a nuclear localization signal within its N terminus.
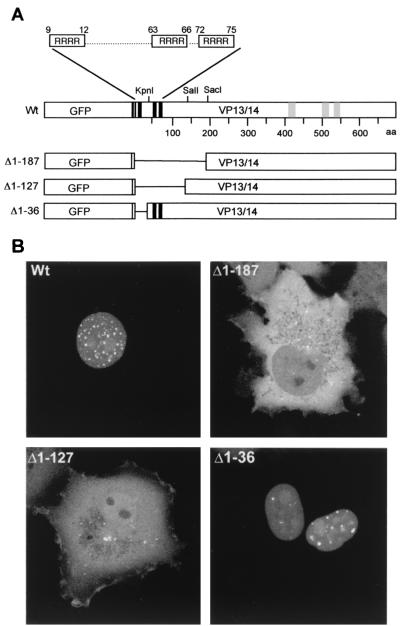
The classical monopartite and bipartite NLSs are basic, lysine-rich regions (19, 38). However, an inspection of the amino acid sequence of VP13/14 revealed that although there was no such region in VP13/14, there were three groups of four consecutive arginine residues toward the N terminus of the protein at amino acid positions 9 to 12, 63 to 66, and 72 to 75, which we have termed NLS1, NLS2, and NLS3, respectively (Fig. 3A, Wt). Moreover, in contrast to the classical NLSs described above, the nuclear import signals for human immunodeficiency virus type 1 (HIV-1) Rev, HIV-1 Tat, and human T-cell lymphotropic virus type 1 (HTLV-1) Rex have previously been shown to consist of arginine-rich domains (21, 22, 27, 48, 49). In an attempt to determine if any or all of the VP13/14 arginine clusters are involved in the nuclear localization of the protein, three gross deletions were made within the GFP-13/14 fusion protein toward the N terminus of VP13/14, resulting in plasmids which expressed proteins lacking the first 187 residues, the first 127 residues, or the first 36 residues, fused to GFP (Fig. 3A). These constructs were transfected into COS-1 cells, and the subcellular distribution of the GFP fusion proteins was examined 40 h after transfection by direct fluorescence of live cells (Fig. 3B). Strikingly, removal of the first 187 residues from VP13/14 abrogated the nuclear accumulation of the GFP fusion protein (Fig. 3B, compare Wt and Δ1–187). Moreover, the fusion protein lacking only the first 127 residues of VP13/14 was also unable to accumulate within the nucleus (Fig. 3B, Δ1–127). By contrast, however, deletion of the first 36 N-terminal amino acids from VP13/14, which removes the NLS1 sequence, did not affect the nuclear targeting of GFP-13/14, (Fig. 3B, Δ1-36), suggesting that the residues R9 through R12 are not required for nuclear localization.
FIG. 3.
The N terminus of VP13/14 is required for nuclear localization. (A) Schematic diagram of the GFP-13/14 fusion protein (Wt) and the deletion mutants lacking the first 187 (Δ1–187), 127 (Δ1–127), or 36 (Δ1-36) residues of the VP13/14 open reading frame. The potential NLS and NES sequences are shown as solid and shaded boxes, respectively. (B) The four constructs diagrammed in panel A were transfected into COS-1 cells and examined live by confocal microscopy 40 h after transfection.
Point mutations in VP13/14 identify a sequence within residues 63 to 75 as necessary for nuclear localization.
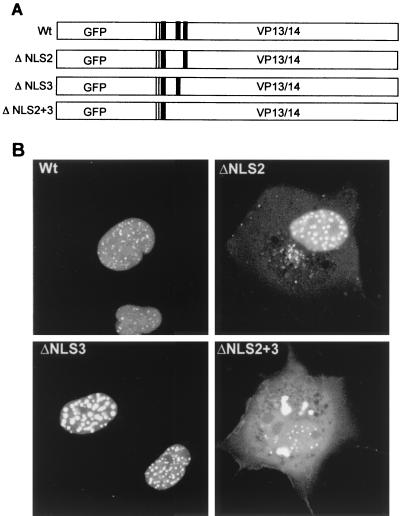
Although the above results suggest that the region between amino acids 37 and 127 of VP13/14 containing NLS1 and NLS2 is responsible for nuclear localization of the protein, we could not rule out the possibility that the entire protein conformation was disrupted by these gross deletions. In order to determine more specifically the amino acids involved, we carried out site-directed mutagenesis within this region, resulting in three constructs in which the arginine residues of NLS2, NLS3, or both NLS2 and NLS3 were mutated to glycine residues (Fig. 4A). Following transfection of COS-1 cells, the subcellular distribution of GFP-13/14 and the GFP-13/14 mutants was examined 40 h posttransfection by direct fluorescence of live cells (Fig. 4B). While mutation of NLS2 reduced the efficiency of nuclear targeting compared to that of the Wt, the fusion protein was still predominantly nuclear, with a small amount present in the cytoplasm (Fig. 4B, compare Wt and ΔNLS2). Furthermore, mutation of NLS3 had no effect on the nuclear accumulation of GFP-13/14 (Fig. 4B, compare Wt and ΔNLS3). However, mutation of both arginine motifs resulted in a fusion protein that was not targeted to the nucleus but instead accumulated within the cytoplasm (Fig. 4B, ΔNLS2+3). The majority of cells expressing this protein displayed a pattern of fluorescence in which the mutated fusion protein was excluded from the nucleus, but there was also a small population of cells that displayed a weak nuclear speckled pattern on top of the cytoplasmic material (Fig. 4B, ΔNLS2+3). This may be due to an expressing cell having progressed through cell division and hence having allowed the mutated GFP-13/14 access to nuclear material following nuclear membrane breakdown. Thus, while either NLS2 or NLS3 can be mutated without severely altering the nuclear localization of GFP-13/14, mutation of both sequences abolishes this targeting. This suggests that nuclear localization of GFP-13/14 requires at least one of these arginine motifs.
FIG. 4.
Point mutation of both NLS2 and NLS3 abrogates nuclear localization of VP13/14. (A) Schematic diagram of the GFP-13/14 fusion protein (Wt) and the mutants in which arginine residues have been mutated to glycine residues for NLS2 alone (ΔNLS2), NLS3 alone (ΔNLS3), or both NLS2 and NLS3 (ΔNLS2+3). The clusters of arginine residues are shown as solid boxes. (B) The four constructs diagrammed in panel A were transfected into COS-1 cells and examined live by confocal microscopy 40 h after transfection.
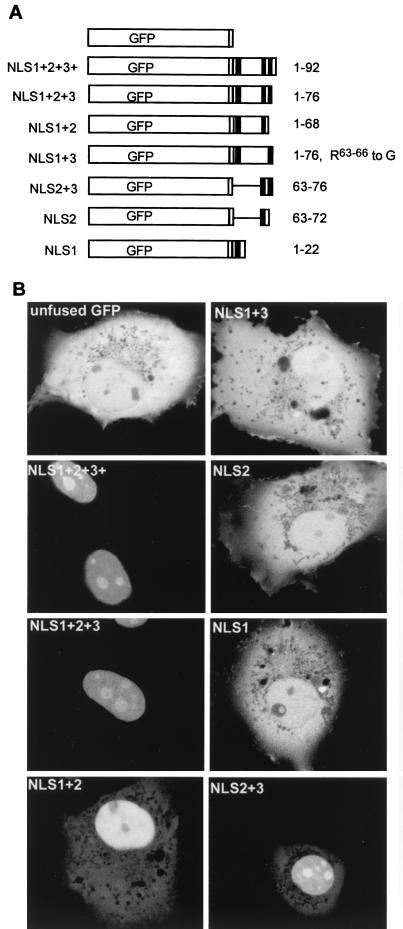
A 14-amino-acid sequence from VP13/14 is sufficient to direct GFP to the cell nucleus.
To define the minimal sequence of VP13/14 that is required for nuclear targeting, a series of plasmids encoding GFP fused to a range of short N-terminal peptides of VP13/14 was constructed (Fig. 5A). These plasmids were transfected into COS-1 cells, and the subcellular distribution of GFP fusion proteins was examined 40 h posttransfection by direct fluorescence of live cells (Fig. 5B). Fusion of GFP to residues 1 to 92 (NLS1+2+3+) or 1 to 76 (NLS1+2+3), which both include all three NLS sequences, resulted in the efficient relocalization of GFP from its characteristic pattern throughout the cell (Fig. 5B, unfused GFP) to the nucleus (Fig. 5B, NLS1+2+3+ and NLS1+2+3), confirming that residues 1 to 76 of VP13/14 contain a discrete nuclear targeting signal. Furthermore, as expected from our previous results, GFP fused to residues 1 to 22 (NLS1) was not capable of nuclear localization, confirming that the NLS1 sequence is not sufficient for nuclear targeting by itself (compare Fig. 5B, NLS1, with Fig. 4B, ΔNLS2+3). Likewise, fusion of the NLS2 sequence (residues 63 to 72) to GFP was not sufficient to direct GFP to the nucleus (Fig. 5B, NLS2). By contrast, GFP fused to either residues 1 to 68 (NLS1+2) or residues 63 to 76 (NLS2+3) was localized predominantly to the nucleus (Fig. 5B, NLS1+2 and NLS2+3), although in both cases there was still some protein in the cytoplasm. Surprisingly, however, mutation of NLS2 within the region spanning residues 1 to 76, such that only NLS1 and NLS3 are present, resulted in a protein that was unable to localize to the nucleus (Fig. 5B, NLS1+3). This is in contrast to the same mutation present in the context of the full-length protein, which was predominantly localized in the nucleus (see Fig. 4B, ΔNLS2), and may imply that this region of the protein is presented differently when expressed in the full-length protein. Alternatively, there may be additional redundant NLS sequences downstream in VP13/14 that can function in the absence of NLS2. Taken together, the above results indicate that a region as small as 14 residues originating from the N terminus of VP13/14, containing NLS2 and NLS3, is sufficient to function as a nuclear targeting signal and direct a heterologous protein to the nucleus. Furthermore, NLS2 appears to be essential for nuclear targeting in the context of the N-terminal 76 residues of VP13/14 but also requires an additional arginine motif, either NLS1 or NLS3, to cause relocalization of GFP to the nucleus. Interestingly, the GFP fusion proteins were localized to the nucleoli of expressing cells only when NLS3 was present together with NLS2 (Fig. 5B, compare NLS1+2 with NLS1+2+3+, NLS1+2+3, and NLS2+3), suggesting that NLS3 may be required for nucleolar targeting of the protein. Finally, none of these small fusion proteins localized to the characteristic punctate domains exhibited by full-length GFP-13/14, implying that the sequences involved in this intranuclear targeting are present on another region of the protein.
FIG. 5.
A 14-amino-acid peptide from VP13/14 functions as an NLS. (A) Schematic diagram of the GFP fusion constructs used to identify the VP13/14 NLS. The names of the constructs are shown on the left, and the VP13/14 residues fused to GFP are given on the right. The NLS clusters of arginines are shown as solid boxes. (B) The constructs diagrammed in panel A were transfected into COS-1 cells and examined live by confocal microscopy 40 h after transfection.
VP13/14 is capable of nuclear shuttling in an interspecies heterokaryon assay.
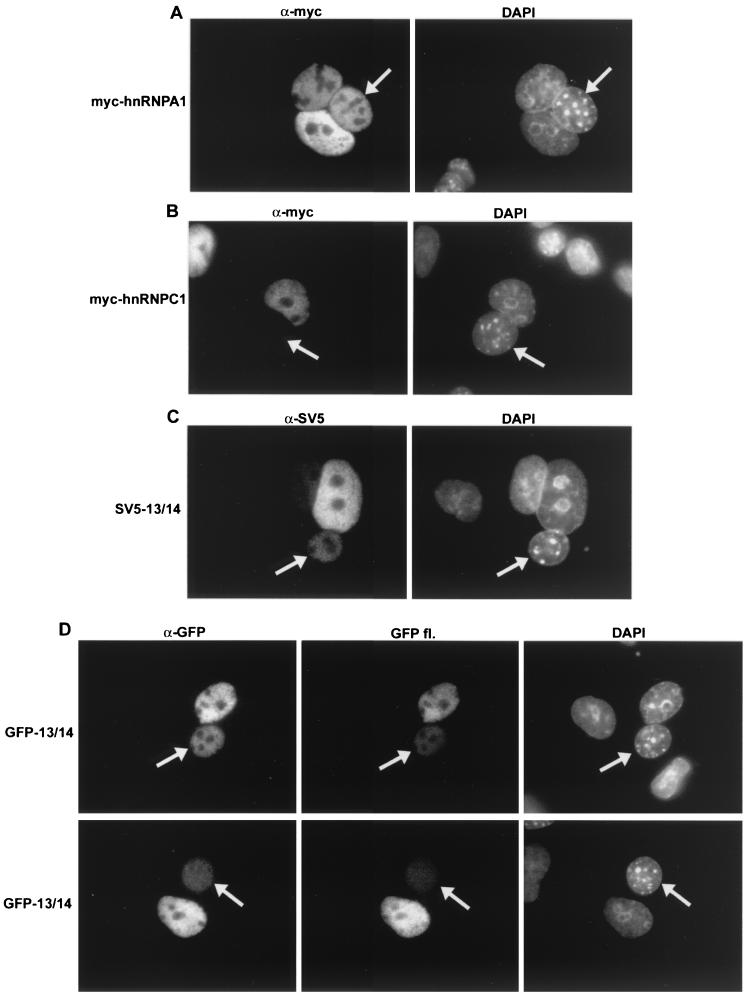
The similarity of the VP13/14 nuclear targeting signal to that of the HIV-1 Rev protein, a known nuclear shuttling protein (46), prompted us to examine the VP13/14 sequence for potential nuclear export signals (NESs). We found several leucine-rich regions toward the C terminus of VP13/14 (Fig. 3A, Wt) which were similar to the NES of Rev (34), and we therefore examined the activity of GFP-13/14 for potential nuclear export. This was carried out using a heterokaryon assay, whereby COS-1 cells expressing the protein of interest were fused to an equivalent number of NIH 3T3 cells to form interspecies heterokaryons, in the presence of the protein synthesis inhibitor cycloheximide. At the end of the assay the cells were stained with DAPI to differentiate mouse and monkey nuclei; then they were examined for the presence of our test protein in the mouse nuclei (Fig. 6), and cell counts were carried out (Table 1). To determine that the assay was working correctly, COS-1 cells were first transfected with either an expression vector for the cellular protein hnRNPA1, which is known to shuttle efficiently (45), or an expression vector for the cellular protein hnRNPC1, which does not shuttle (37); both of these were Myc tagged. Fusion of the Myc-hnRNPA1 expressing cells to the mouse cells resulted in a large number of mouse cell nuclei containing the Myc-hnRNPA1 protein (Fig. 6A and Table 1). By contrast, the vast majority of mouse cell nuclei fused to Myc-hnRNPC1 expressing cells contained no Myc-tagged hnRNPC1 protein, confirming that our assay was functioning correctly (Fig. 6B and Table 1). When the same experiment was carried out with SV5-tagged VP13/14, we were able to detect the virus protein in a high percentage of mouse cell nuclei, albeit at a low level in comparison to that with the positive control, hnRNPA1 (Fig. 6C and Table 1). Moreover, fusion of mouse cells to GFP-13/14-expressing cells also resulted in the presence of GFP-13/14 in the nonexpressing mouse nuclei, as detected by either immunofluorescence with an anti-GFP antibody (Fig. 6D, α-GFP) or direct GFP fluorescence (Fig. 6D, GFP fl., and Table 1). These results suggest that the HSV-1 tegument protein VP13/14 is capable of nuclear shuttling, and while the level of protein detectable in the mouse nuclei of the heterokaryons was somewhat low in comparison to that with the positive control, the number of nuclei in which the protein was detected was consistently high (Table 1).
FIG. 6.
VP13/14 is capable of nuclear shuttling. Heterokaryon assays were carried out between mouse NIH 3T3 cells and monkey COS-1 cells expressing either Myc-hnRNPA1 (A), Myc-hnRNPC1 (B), SV5-13/14 (C), or GFP-13/14 (D). In each case the cells were fixed and stained with either anti-Myc (A and B), anti-SV5 tag (C), or anti-GFP (D) antibodies. In the case of GFP-13/14, the cells were also examined for intrinsic GFP fluorescence (D, GFP fl.). Mouse cells (arrowed in all examples) were identified by their speckled nuclei when stained with DAPI.
TABLE 1.
Heterokaryon assay cell counts
| Transiently expressed proteina | No. of mouse nuclei
|
|
|---|---|---|
| Positiveb | Negativec | |
| Myc-hnRNPA1 | 63 | 0 |
| Myc-hnRNPC1 | 11 | 46 |
| GFP-13/14 | 51 | 2 |
| SV5-13/14 | 24 | 2 |
COS-1 cells were transiently transfected with expression vectors for each of these proteins.
Positive mouse nuclei were counted as those that were present in a fused heterokaryon with monkey nuclei from expressing cells and contained detectable levels of the expressed protein.
Negative mouse nuclei were counted as those that were present in a fused heterokaryon with monkey nuclei from expressing cells but did not contain detectable levels of the expressed protein.
DISCUSSION
The group of proteins which make up the tegument of the herpesvirus particle have been poorly studied with regard to both their individual functions during virus replication and the roles they may play in virus entry and/or egress. Here we have investigated the cellular localization of the tegument protein VP13/14 by means of transient expression of a GFP-13/14 fusion protein and have shown that this protein localizes efficiently to the nucleus. Moreover, in a paper accompanying this report, we have shown that VP13/14 is also targeted to the nucleus throughout the major part of the virus infectious cycle, confirming that this subcellular localization is reproduced during infection (11). The intrinsic targeting of VP13/14 may be indicative of a function for this protein within the nucleus, as has been suggested by the range of indirect evidence supporting a role for the protein in IE gene expression (30, 53, 54). Furthermore, our results are in agreement with the findings of previous immunofluorescence studies of infected cells which have also shown VP13/14 localized in the nucleus at early stages of infection (35).
It is noteworthy that VP13/14 is the first tegument protein shown to possess an NLS within its structure. By contrast, we have previously shown that the tegument protein VP22 localizes predominantly to the cytoplasm during both virus infection and transient expression (14, 15), although it does enter the nucleus under certain conditions (15). In addition, the tegument protein VP16, which is known to transactivate the virus IE genes within the nucleus, exhibits no particular targeting to the nucleus when expressed transiently (13, 25). Nonetheless, VP16 does localize efficiently to the nucleus during the early stages of its expression in virus infection (26, 35), a feature which has been attributed to its interaction with the host cell protein HCF (25, 26). Our demonstration that VP13/14 is targeted to the nucleus, together with the evidence that VP13/14 may modulate the transactivation function of VP16 (30, 54), raises the possibility that VP13/14 may in some way be involved in VP16 import into the nucleus. For example, Morrison and coworkers have suggested that as the virus enters the cell both VP16 and VP13/14 dissociate from the tegument by means of phosphorylation by a virus-encoded kinase (36). Thus, input VP13/14 could help to direct input VP16 to the nucleus, where it would be free to transactivate the IE genes. However, it remains to be determined whether VP13/14 can directly interact with VP16.
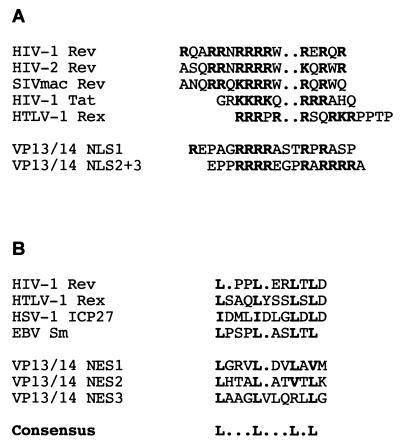
The NLS of VP13/14 is located within the N-terminal 127 residues of the protein. In this region there are at least three clusters of arginine residues, none of which can function as a nuclear targeting signal by itself. However, we have shown that two of these clusters located within the 14 residues between amino acids 63 and 76 are sufficient to direct either VP13/14 or a heterologous protein to the nucleus. We also believe that there may be an element of redundancy between these arginine clusters, as various combinations of the sequences are able to function as nuclear targeting signals, at least in the context of the full-length protein. The arginine-rich nature of the VP13/14 NLS suggests that it is not related to the classical mono- and bipartite NLS sequences first defined for proteins such as the simian virus 40 (SV40) large T antigen, most of which are made up of lysine-rich regions (19, 38), but that it is a member of the class of NLSs described for proteins such as HIV-1 Rev and Tat and HTLV-1 Rex (21, 22, 27, 48, 49). Moreover, the VP13/14 NLSs share homology with those from the retrovirus transactivators (Fig. 7A). While classical NLSs import into the nucleus via interaction with the cellular importin α/β complex (19, 38), the arginine-rich NLSs have recently been shown to import by direct interaction with importin β, with no apparent requirement for importin α (42, 52). Furthermore, Ojala and coworkers have recently suggested that HSV-1 capsid-tegument structures are directed to the nucleus by an importin β binding protein during virus entry into the cell (40). Thus, as VP13/14 is a major component of the virus tegument and has the characteristics of an importin β binding protein, it would be a good candidate for such a role during infection.
FIG. 7.
(A) Alignment of the arginine-rich clusters of VP13/14 with the known NLS sequences from HIV-1 and HIV-2 Rev (21), HIV-1 Tat (22), and HTLV-1 Rex (49). Arginine and lysine residues are boldfaced. (B) Alignment of the three leucine-rich regions of VP13/14 with the known NES sequences from HIV-1 Rev (34), HTLV-1 Rex (3), HSV-1 ICP27 (32), and the Epstein-Barr virus (EBV) Sm protein (5). Leucine residues are boldfaced.
An additional feature of the arginine-rich regions of retroviral NLSs is their ability to bind directly to RNA (9, 10, 28, 47). Furthermore, Rev and Rex shuttle between the nucleus and the cytoplasm (24, 46), thereby transporting their bound RNA out of the nucleus (18). Export of the retroviral proteins from the nucleus is controlled by their leucine-rich NESs (8, 17, 34), which have been shown to bind the cellular protein exportin 1 (CRM-1) to facilitate transport through the nuclear pore complex (51). In our studies on VP13/14 we have not only identified several leucine-rich regions in the C-terminal half of the protein but have shown from our preliminary heterokaryon assays that VP13/14 is also capable of nuclear shuttling. While it will be necessary to define the export signal more closely by means of mutational analysis, the VP13/14 leucine-rich regions share some homology with other known NESs (Fig. 7B) and may therefore be functional. Thus, VP13/14 joins a growing number of proteins, both viral and cellular, whose steady-state localization is nuclear but which have been shown by various assays to shuttle rapidly between the nucleus and cytoplasm (3–5, 32, 43, 45, 46). As the vast majority of these proteins appear to be involved in the transport of RNA from the nucleus, it would be of interest to determine if VP13/14 also has a role as an RNA binding protein and is involved in virus-specific RNA transport.
One of the most striking features of VP13/14 localization is its targeting to multiple punctate domains within the nuclei of expressing cells. This pattern is displayed only by the full-length protein and not by any of the GFP-NLS constructs, suggesting that the signal for targeting to these domains is present outside the arginine-rich region. Moreover, the relationship between the punctate domains and nuclear shuttling is not yet known. It is also noteworthy that in our accompanying paper we show that VP13/14 localizes to similar domains during virus infection, albeit fewer than are usually seen in a transiently expressing cell (11). This punctate pattern is similar to that observed during transient expression of the HSV IE gene IE110 (7, 41), where it has been shown that a subset of the IE110 nuclear domains represent the cellular ND10s (16, 29). However, immunostaining of our VP13/14-expressing cells with an antibody against PML, a component of ND10s (12), has revealed that none of the VP13/14 punctate domains correspond to ND10s (data not shown). It therefore remains to be determined if the VP13/14 domains are in any way related to the rest of the non-ND10 IE110 domains, either in transient expression or during virus infection.
From our results presented here it is clear that many aspects of VP13/14 behavior remain to be addressed. However, the fact that this protein is not essential to virus growth in tissue culture (1, 54) suggests that its role may be that of an accessory protein that enables the virus to replicate efficiently by the enhancement of certain steps in the virus life cycle without being absolutely required.
ACKNOWLEDGMENTS
We thank David Meredith for the anti-VP13/14 antibody, Rick Randall for the anti-SV5 epitope tag antibody, and Gideon Dreyfuss for the hnRNPA1- and hnRNPC1-encoding plasmids.
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Barker D E, Roizman B. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology. 1990;177:684–691. doi: 10.1016/0042-6822(90)90534-x. [DOI] [PubMed] [Google Scholar]
- 2.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22 and 27 predicts the nucleotidylation of the UL21, UL31, UL47 and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 3.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 5.Boyle S M, Ruvolo V, Gupta A K, Swaminathan S. Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J Virol. 1999;73:6872–6881. doi: 10.1128/jvi.73.8.6872-6881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell M E, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 7.Ciufo D M, Mullen M A, Hayward G S. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J Virol. 1994;68:3267–3282. doi: 10.1128/jvi.68.5.3267-3282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 9.Daly T J, Cook K S, Gray G S, Maione T E, Rusche J R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall, Ernberg C I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 Tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly M, Elliott G. Fluorescent tagging of the herpes simplex virus tegument protein VP13/14 in virus infection. J Virol. 2001;75:2575–2583. doi: 10.1128/JVI.75.6.2575-2583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyck J A, Maul G G, Miller W J, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 13.Elliott G, Mouzakitis G, O'Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott G, O'Hare P. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J Virol. 2000;74:2131–2141. doi: 10.1128/jvi.74.5.2131-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 20.Greaves R, O'Hare P. Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J Virol. 1989;63:1641–1650. doi: 10.1128/jvi.63.4.1641-1650.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmid M, Palmeri D, Ruhl M, Jaksche H, Weichselbraun I, Bohnlein E, Malim M H, Hauber J. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev trans-activator. J Virol. 1994;68:7329–7335. doi: 10.1128/jvi.68.11.7329-7335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauber J, Malim M H, Cullen B R. Mutational analysis of the conserved basic domain of human immunodeficiency virus Tat protein. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota S, Hatanaka M, Pomerantz R J. Nucleo-cytoplasmic redistribution of the HTLV-I Rex protein: alterations by coexpression of the HTLV-I p21x protein. Virology. 1996;220:502–507. doi: 10.1006/viro.1996.0339. [DOI] [PubMed] [Google Scholar]
- 25.LaBoissiere S, Hughes T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBoissiere S, O'Hare P. Analysis of HCF, the cellular cofactor of VP16, in herpes simplex virus-infected cells. J Virol. 2000;74:99–109. doi: 10.1128/jvi.74.1.99-109.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malim M H, Bohnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 28.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 29.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 30.McKnight J L C, Pellet P E, Jenkins F J, Roizman B. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing-factor-dependent activation of α genes. J Gen Virol. 1987;61:1531–1574. doi: 10.1128/jvi.61.4.992-1001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 32.Mears W E, Rice S A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 33.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 34.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison E E, Stevenson A J, Wang Y-F, Meredith D M. Differences in the intracellular localisation and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2515–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 36.Morrison E E, Wang Y, Meredith D. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nigg E A. Nucleocytoplasmic transport: mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 39.O'Hare P, Goding C R, Haigh A. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojala P M, Sodeik B, Ebersold M W, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20:4922–4931. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Rourke D, Elliott G, Papworth M, Everett R, O'Hare P. Examination of determinants for intranuclear localization and transactivation within the RING finger of herpes simplex virus type 1 IE110k protein. J Gen Virol. 1998;79:537–548. doi: 10.1099/0022-1317-79-3-537. [DOI] [PubMed] [Google Scholar]
- 42.Palmeri D, Malim M H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phelan A, Clements J B. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 44.Pinard M F, Simard R, Bibor-Hardy V. DNA-binding proteins of herpes simplex virus type 1-infected BHK cell nuclear matrices. J Gen Virol. 1987;68:727–735. doi: 10.1099/0022-1317-68-3-727. [DOI] [PubMed] [Google Scholar]
- 45.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 46.Richard N, Iacampo S, Cochrane A. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology. 1994;204:123–131. doi: 10.1006/viro.1994.1516. [DOI] [PubMed] [Google Scholar]
- 47.Roy S, Delling U, Chen C H, Rosen C A, Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- 48.Siomi H, Shida H, Maki M, Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siomi H, Shida H, Nam S H, Nosaka T, Maki M, Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988;55:197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- 50.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 52.Truant R, Cullen B R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, McKnight J L. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Sirko D A, McKnight J L. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J Virol. 1991;65:829–841. doi: 10.1128/jvi.65.2.829-841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]