Abstract
We report a thorough investigation of the role of single-stranded thymidine (ssT) linkers in the stability and flexibility of minimal, multistranded DNA nanostructures. We systematically explore the impact of varying the number of ssTs in three-way junction motifs (3WJs) on their formation and properties. Through various UV melting experiments and molecular dynamics simulations, we demonstrate that while the number of ssTs minimally affects thermodynamic stability, the increasing ssT regions significantly enhance the structural flexibility of 3WJs. Utilizing this knowledge, we design triangular DNA nanoparticles with varying ssTs, all showing exceptional assembly efficiency except for the 0T triangle. All triangles demonstrate enhanced stability in blood serum and are nonimmunostimulatory and nontoxic in mammalian cell lines. The 4T 3WJ is chosen as the building block for constructing other polygons due to its enhanced flexibility and favorable physicochemical characteristics, making it a versatile choice for creating cost-effective, stable, and functional DNA nanostructures that can be stored in the dehydrated forms while retaining their structures. Our study provides valuable insights into the design and application of nucleic acid nanostructures, emphasizing the importance of understanding stability and flexibility in the realm of nucleic acid nanotechnology. Our findings suggest the intricate connection between these ssTs and the structural adaptability of DNA 3WJs, paving the way for more precise design and engineering of nucleic acid nanosystems suitable for broad biomedical applications.
Graphical Abstract
INTRODUCTION
Nucleic acid (NA) nanotechnology utilizes the complementary nature of NA to construct complex assemblies with discrete secondary structures through canonical Watson–Crick interactions.1–3 This approach has led to the creation of numerous NA-based nanoscaffolds of 1D, 2D, and 3D shapes with different connectivities and the ability to act as targeted delivery vehicles or carriers for other functionalities.4,5 Over the past two decades, significant contributions have been made toward the development of NA-based nanoscaffolds through molecular self-assembly, pioneered by the work of Nadrian Seeman in the DNA nanotechnology field6 as well as Neocles Leontis, Eric Westhof,7 and Peixuan Guo8 in the RNA nanotechnology and therapeutic RNA nanotechnology fields. This has led to the construction of diverse 1D, 2D, and 3D NA nanoscaffolds, including cubes, polygons, tetrahedra, bipyramids, dodecahedra, etc., which have been demonstrated to be effective in targeted delivery.9–11 For instance, prior research led by Seeman and collaborators utilized this concept to create innovative DNA-based nanomaterials through DNA self-assembly. This approach yielded a myriad of DNA-based nanostructures suitable for diverse applications.12–14 Similarly, this principle extends to the realm of RNA nanotechnology. Here, RNA nanotechnology involves the deliberate design and experimental assembly of complex RNA structures consisting of multiple strands.15 These structures incorporate various functionalities, some of which hold potential for therapeutic use.1,9 By employing natural or artificially selected RNA motifs and modules,7,16,17 it is possible to program RNAs to generate a wide array of compact and stable artificial 3D nanostructures termed RNA nanoparticles (RNA NPs). These RNA NPs are well suited for a wide spectrum of clinical and nanotechnological applications.18,19 Through bioconjugation techniques such as click chemistry,20,21 therapeutic nucleic acids, proteins, or small molecules can be individually attached to the programmed RNA monomers that form RNA NPs. The assembly of these monomers facilitates the merging of desired functionalities, enabling precise control over their structure, composition, and modularity.
In our previous reports,22–27 we have employed a computational approach to design an artificial nucleic acid (NA) module featuring three helical stems connected via four pyrimidines. These pyrimidines are single-stranded uridine for RNA or single-stranded thymidine for DNA and are collectively referred to as a tetra-U/T helix, which serves as the link between helices in the three-way junction motif (3WJ).27 The 3WJ-based building module has proven to be a robust structural module that facilitates the design and assembly of various other polygonal structures, including triangles, tetragons, pentagons, and hexagons.15,28,29 These nanoparticles can be effectively assembled into desired forms by varying the stoichiometry of DNA, RNA, or 2’-fluoro-modified RNA strands, resulting in hybrid nucleic acid complexes.23 Notably, the advantages of these NA polygonal nanoscaffolds are manifold: (1) their modular nature enables the incorporation of specific aptamers at precise, regiospecific positions;22 (2) their physicochemical properties can be finely tuned to enhance or reduce thermal and enzymatic stability by adjusting the ratio of DNA, RNA, or 2′F-modified RNA components;29,30 and (3) the polygonal scaffolds can be controlled to either enhance immunostimulatory activity using RNA-only strands or reduce the immune response using DNA-only strands.31 One notable limitation of these polygons is their reliance on relatively long oligonucleotides for self-assembly. In solid-phase NA synthesis, the nucleotide coupling efficiency tends to decrease with increasing oligonucleotide length. For instance, the practical yield for producing a 50 nt-long DNA strand averages only about 78%.32 Given the growing global demand for therapeutic nucleic acids, the large-scale production of functional nucleic acid complexes is directly affected by the length of oligonucleotides. Therefore, there is an imperative need to develop functional, cost-effective, and compact nanoparticles with minimal oligonucleotide lengths.
Despite the numerous advantages presented by the tetra-U/T motif listed above, there remains a notable gap in our understanding when it comes to a comprehensive exploration of this motif and whether a single-stranded linkage composed of 4 Ts surpasses the performance of those featuring, for example, 3 or 2 Ts. While the formation of artificially designed DNA polygons is fundamentally driven by helix formation, an intriguing question arises: what role do these unpaired nucleotides play and do they contribute to the stability or flexibility of nanostructures in a broader context? Our approach involves a multifaceted investigation to address these questions. First, we examine the thermodynamic stabilities and flexibilities of individual motifs using UV optical melting and molecular dynamic simulation approaches. Second, we use this knowledge to design triangular nanoparticles, systematically varying the number of single-stranded Ts (ssTs) while exploring their physicochemical characteristics. Finally, we utilize more stable and flexible motifs to construct smaller polygonal nanoparticles and assess their structural properties. Scheme 1 provides an overview of the workflow for this research project, outlining the stepwise approach to unravel the intricacies of the tetra-T motif and its implications for the design and behavior of DNA nanostructures.
Scheme 1.
Schematic Depiction of the Workflow of Evaluation of Single-Stranded Thymidine’s Impact on DNA 3WJ Motif and DNA Nanoparticle Stability and Flexibility
MATERIALS AND METHODS
DNA Oligonucleotide Design and Assembly.
DNA oligonucleotides (ODNs) including the 5′-end labeled by Alexa Fluor 488 dye used in this project were purchased from Integrated DNA Technologies (IDT DNA, Inc.) (Tables S1 and S2). The lyophilized DNA samples were diluted to 100 μM with double-deionized water (dilute H2O) and stored at −20 °C as superstocks.
DNA labeling at the 5′ end with [32P] ATP (PerkinElmer) was performed using standard T4 PNK (New England BioLabs Inc.) protocol following the manufacturer’s recommendations.
The DNA 3WJ motifs were designed to fold from two single-stranded ODNs as shown in Figure S1. Three DNA duplexes of seven base pairs each were generated, with one duplex connected to a stable GAAA tetraloop to achieve a two-stranded design. The sequence choice was aimed at ensuring that the predicted melting temperature for the resulting secondary structure fell within the 35–40 °C range, enabling appropriate transition profiles suitable for UV melting experiments. The design of the polygonal DNA nanoparticles followed procedures similar to previously published reports,26,27 with variations made in the lengths of the polygon sides, utilizing a 10-base pair duplex providing a complete helical turn (Figure S1).
The assembly experiments were performed using a “one-pot” approach, involving the heating (+90 °C) and gradual cooling (+4 °C) of samples with equimolar concentrations of oligonucleotides in the presence of TMS buffer at pH = 8.0 (comprising 50 mM TRIS, 100 mM NaCl, and 10 mM MgCl2).
Electrophoretic Mobility Shift Assay (EMSA).
The successful formation of P-32 labeled DNA 3WJs was confirmed by mixing annealed samples with loading buffer (containing 40% (w/v) sucrose, 0.1% (w/v) xylene cyanol, and 0.1% (w/v) bromophenol blue) and subjecting them to 15% polyacrylamide gel electrophoresis (Acrylamide/bis-Acrylamide 37.5:1) in TBM buffer (89 mM Tris-base, 200 mM boric acid, and 5 mM MgCl2) at a constant 80 V and ambient room temperature. Gel images were obtained by exposing the gels overnight to a storage phosphor screen (Cytiva) and scanning the screen using a Typhoon-5 (Cytiva) biomolecular imager.
The self-assembly of the polygonal DNA nanoparticles, with and without Alexa-488-labeled strands, was analyzed by 4% agarose gel electrophoresis in the presence of ethidium bromide (EB) in TAE running buffer (40 mM Tris-acetate at pH 8.3 and 10 mM EDTA). The gels were run at constant 100 V and ambient temperature. Gel images were captured using a Typhoon-5 (Cytiva) imaging system with both the EB channel (λexc = 532 nm, λem = 655–685 nm) and the Alexa 488 channel (λexc = 488 nm, λem = 515–535 nm).
UV Melting Experiments.
DNA 3WJ constructs 0T–4T were prepared by mixing their corresponding equimolar single strands ssDNA 1 and ssDNA2 at equimolar concentration spanning a 15-fold dilution ranging from 0.5 to 10 μM in a total volume of 100 μL in either CB #1 (20 mM sodium cacodylate pH = 7.0, 0.5 mM sodium EDTA, 1 M NaCl) or CB #2 (20 mM sodium cacodylate pH = 7.0, 10 mM MgCl2, 0.1 M NaCl). The samples were annealed and degassed (2–3 min) in a SpeedVac prior to the optical melting, which was performed using similar methods described by Teter et al.33 Melting experiments were carried out using a Shimadzu UV–vis spectrophotometer (UV-2600i) using a 5 mm-path length multicell (Micro Cell for TMSPC-8). Absorbances at 260 nm or at 280 nm were measured as a function of temperature from 15 to 90 °C with the heating or cooling rates of 1 °C/min and equilibration time of 10 s. The melts were collected in four consecutive rounds, alternating heating–cooling-–heating–cooling. For each construct, optical melts of ssDNA1 and ssDNA2 strands were also collected (Figure S2). Graphical software OriginPro (OriginLabR) was used to fit obtained data by using a Boltzmann sigmoidal function, which describes the sigmoidal shape of the curve as a function of temperature as follows
| (1) |
where T is the temperature, is the melting temperature, w is the slope of the curve at , and are the baseline values of the curve (lower and upper asymptotes, respectfully), w is the slope of the curve, and is the amplitude of the curve.
Equation 1 assumes that the melting of DNA is a cooperative process and that the fraction of double-stranded DNA as a function of temperature can be described by a sigmoidal curve. It provides a good balance between accuracy and simplicity and can capture most of the important features of the melting process, such as the melting temperature, the melting curve shape, and the cooperativity of the melting.
Thermodynamic parameters were determined from van’t Hoff plots where the equilibrium constant for the bimolecular nonself-complementary reaction is
| (2) |
Ct is the total strand concentration.34 and van’t Hoff equation
| (3) |
where is expressed in Kelvin and R is the gas constant, 1.987 cal·mol−1 K−1. The equation follows a linear function where and are calculated from the slope and intersect, respectively.
The final free energy change is calculated at 37 °C (310.14 K) according to the following relationship
| (4) |
Molecular Dynamics Simulations.
The initial 0T d3WJ structure was built by assembling two NMR structures with pdb IDs 1DRG and 1JVE using Accelrys Discovery Studio. The junction region was extended by inserting thymine nucleotides to build nT d3WJ (n = 0–4). A short energy minimization was applied to repair atomic clashes using the Amber sander program.35 The minimized structures were solvated with the TIP3P water, Na+ neutralization ions, and additional ions (0.15 M NaCl) using Amber force fields36–38 and Amber LEaP program.35 The solvated system was minimized, while all heavy atoms of DNA were fixed. The minimized system was equilibrated at 310.15 K, while all heavy atoms in the DNA were constrained. The entire system was followed by unconstrained minimization and equilibration at a constant 1 atm pressure and temperature (301.15 K) while holding the 5′ and 3′ ends at each stem by applying distance constraints. The product MD simulations were performed with constant pressure (1 atm) using the Langevin piston method39 and constant temperature (310.15 K) using weakly coupled Langevin dynamics. The system was simulated with periodic boundary conditions with full electrostatics computed using the particle mesh Ewald method.40 Short-range nonbonded terms are evaluated at every step using a 12 Å cutoff for van der Waals interactions and a smooth switching function. The production simulations are conducted for 120 ns with a 2 fs time step. The distance constraints on the 5’ and 3’ ends were applied for the first 20 ns and then released for the remaining 100 ns. The minimization, equilibration, and production MD simulations of the solvated systems were carried out using the NAMD simulation package.41,42
Temperature Gradient Gel Electrophoresis (TGGE).
The TGGE analysis was carried out using 7% native PAGE in a buffer solution containing 50 mM TRIS at pH 8.0, 100 mM NaCl, and 0.2 mM MgCl2, as per the previously established protocol.43 To initiate the temperature gradient denaturation, temperatures ranging from 36.6 to 73.2 °C were applied perpendicular to the electrical current for a total duration of 40 min at 20 W. The TGGE assay employed a total DNA concentration of 0.2 μM for the analysis. The gel was imaged by using the ChemiDoc MP system (Bio-Rad). The apparent transition melting (Tm) values were determined by identifying the temperature at which half of the fractions of the polygons were dissociated, measured from the individual bands of the triangle, square, pentagon, and hexagon nanoparticles. The melting temperature of each polygon was determined by dividing the temperature range (30 °C) by the total number of wells. The melting point of the polygon was then determined based on the inflection point of the location of the gel bands.
Dynamic Light Scattering (DLS).
DLS measurements were performed using a Zetasizer Nano ZS instrument (Malvern Panalytical, U.K.) equipped with a helium–neon laser operating at 633 nm. The samples (1 μM) of the self-assembled DNA triangles were prepared by diluting in TMS buffer as described above. DLS measurements were performed by using a backscattering detector at an angle of 173°. The hydrodynamic size for each sample was analyzed at least three times to ensure reproducibility of the results. The hydrodynamic size distribution of the DNA nanoparticles was obtained by analyzing the correlation function using Zetasizer software. The hydrodynamic diameter indices of the DNA nanoparticles were calculated using the cumulant analysis method provided by instrument software. Statistical analyses and a presentation of the results were conducted using OriginPro software.
Dissociation Constant Measurement.
Apparent equilibrium dissociation constants (Kd) for DNA polygon formations were assessed through the titration experiment over a concentration range from 1 to 500 nM with subsequent analysis by 7% native PAGE, following established methodologies.17,29 Fixed quantities of 5’-end Alexa 488-labeled strands (1.0 nM) of the corresponding DNA polygons were combined with varying amounts of unlabeled DNA strands to achieve the specified final concentrations (ranging from 1.0 to 500 nM for each polygon). The resultant DNA strands underwent heating to 90 °C for 3 min in TMS buffer and were slowly cooled to 4 °C. Gel shift analyses were performed using ImageJ software and processed through the Origin 8.0 program.
values for each DNA complex were determined by plotting the fraction (f) of the specific polygon against the total concentration (Ct) of the corresponding DNA strands. The combined data obtained from three independent measurements underwent nonlinear curve fitting using the following equation
| (5) |
where n = number of DNA strands.34
Stability Assay in Fetal Bovine Serum.
Stability of assembled DNA nanoparticles, set at a final concentration of 1 μM in TMS buffer, was evaluated by incubating samples in a 10% aqueous solution of fetal bovine serum for 12 h at 37 °C. An equal volume of DNA polygons was mixed with a 20% FBS solution, resulting in a final FBS concentration of 10% in all tested samples.
A 2 μ M solution of ssDNA 5′ -GGCCCTCGCGTTTTCTAGGGATGGTTTTCCTCTATGCCTTTTGAATTTCGGG-3′ was used as a positive control. Following the incubation period, the reaction was halted by combining the samples with an equal volume of gel loading buffer. Samples were then subjected to analysis using 4% agarose gel electrophoresis supplemented with EB in a TAE running buffer. Gel electrophoresis was performed at a constant voltage of 100 V under ambient temperature conditions. Gel imaging was carried out using a Typhoon-5 imaging system, allowing visualization in the EB channel (λexc = 532 nm, λem = 655–685 nm) to evaluate the migration pattern of the DNA triangles. Subsequently, ImageJ software was employed for the quantification of the remaining fractions of triangle bands after the 12 h incubation period.
Immune Reporter Cells.
Reporter cell lines human embryonic kidney (HEK) Blue hTLR7 and hTLR9 (InvivoGen) were maintained at 37 °C and 5% CO2 in 1× Dulbecco’s modified Eagle medium (DMEM) with 4.5 g/L glucose, 2 mM l-glutamine (Thermo Fisher) including 10% heat-inactivated fetal bovine serum (FBS) and 100 U/mL penicillin, 100 μg/mL streptomycin, 100 μg/mL Normocin, 10 μg/mL Blasticidin, and 100 μg/mL Zeocin (invivoGen). HEK Lucia RIG-I cells were maintained at 37 °C and 5% CO2 in DMEM with 4.5 g/L glucose, 2 mM l-glutamine including 10% heat-inactivated fetal bovine serum (FBS) and 100 U/mL penicillin, 100 μg/mL streptomycin, 100 μg/mL Normocin, 30 μg/mL Blasticidin, and 100 μg/mL Zeocin. HEK Blue hTLR7 and hTLR9 cells were seeded at 40,000 cells per well in 100 μL of media in a 96-well plate. HEK Lucia RIG-I cells were seeded at 20,000 cells per well in 100 μL of media in a 96-well plate. After 24 h of incubation at 37 °C and 5% CO2, medium was aspirated from the cells and replaced with DNA triangle treatments with and without Lipofectamine 2000 (Thermo Fisher) in media. Plates were incubated once again for 24 h prior to conducting the immune reporter and cell viability assays.
Immune Reporter Assays.
For HEK Blue hTLR7 and hTLR9 cells, QUANTI-Blue (InvivoGen) solution was preheated to 37 °C. Following 24 h post-transfection, 180 μL of QUANTI-Blue and 20 μL of cell supernatant were combined in a new sterile 96-well plate. The plate was incubated (37 °C, 5% CO2) for 75 min and then read at 620 nm on a Tecan Spark microplate reader.
For HEK Lucia RIG-I cells, QUANTI-Luc (InvivoGen) solution was preheated to 37 °C. Following 24 h after transfection, 50 μL of QUANTI-Luc was added to 20 μL of cell supernatant in a new sterile black-walled 96-well plate. The plate was then immediately read for luminescence (100 ms reading time) on a Tecan Spark microplate reader. A 5 μg/mL portion of R848 (InvivoGen) was used as a positive control for hTLR7 cells, 5 μM of the recombinant peptides ODN 2006 (InvivoGen) for hTLR9 cells, and 0.5 μg/mL of 3p-hp RNA (InvivoGen) for Lucia RIG-I cells.
Cell Viability Assays.
MTS (Promega) solution was preheated to 37 °C. 24 h after transfection, 20 μL of MTS was added to 80 μL of cells. The plate was incubated (37 °C, 5% CO2) for 75 min and then read for absorbance at 515 nm on a Tecan Spark microplate reader.
Dehydration and Stability Assays.
For the dehydration and stability assays, the protocol published by Tran et al. was followed with minor modifications.44 Functionalized polygon samples (0.5 μM) were aliquoted (6 μL per tube). For the SpeedVac method, the samples were centrifuged in a CentriVap micro-IR vacuum concentrator (Labconco) (55 °C) for 1 h or until fully dry and then sealed. For lyophilization, samples were frozen in liquid nitrogen for 3 min and lyophilized in a VirTis SP Scientific Benchtop Pro with an Omnitronics freeze dryer. The samples were held at 20.5 °C shelf temperature, an ~− 91 °C condenser, and ~20 mTorr vacuum overnight. The samples were equilibrated with atmospheric air and sealed.
Control samples were kept in solution at 55 and 4 °C. All dehydrated samples were sealed with parafilm and stored in a heat block at 55 °C for 1 week. The samples stored in the sealed Eppendorf tubes were rehydrated with ddH2O (6 μL), mixed by pipetting, and centrifuged and kept in the cold room (4 °C) with the control samples until needed. Sample stability was assessed via visualization on 8% (37.5:1) polyacrylamide native-PAGE run in the presence of running buffer containing 89 mm tris-borate (pH 8.2) and 2 mM MgCl2 for 30 min at 300 V in a cold room (4 °C), using a Mini-PROTEAN Tetra system (Bio-Rad). The gel was imaged using the ChemiDoc MP system (Bio-Rad).
RESULTS
Comparative Analysis of Thermodynamics of DNA 3WJ Motifs by UV Optical Melting.
To investigate the significance of the number of ssT linkers interconnecting three helical stems, we design a DNA 3WJ (d3WJ) complex formed by two ssDNA strands (Figures 1A and S2 and Table S1).
Figure 1.
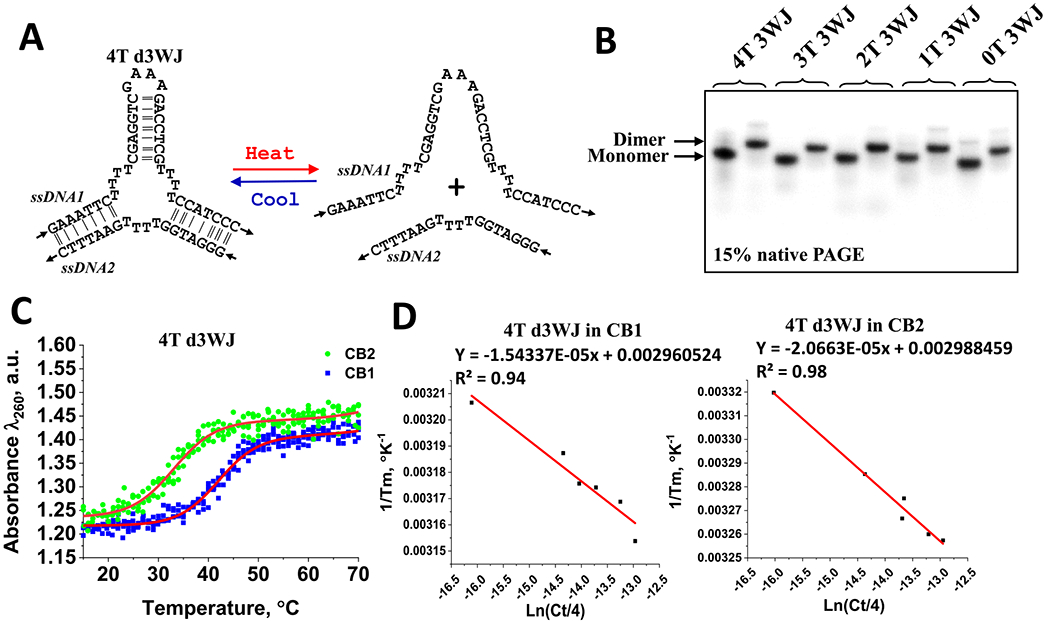
(A) DNA 4T 3WJ strand dissociation process used in UV melting analysis. (B) EMSA assay showing formation of individual d3WJs. (C) Examples of optical melts of 4T d3WJ in CB1 and CB2 buffers showing cooperative transition. Data were fit using the sigmoidal Boltzmann method in Origin Pro software to find Tm. (D) Van’t Hoff plot of 4T d3WJ melt data in CB1 and CB2 buffers.
The longer strand incorporates a stable GNRA tetraloop45,46 region (ssDNA1), which hybridizes with a complementary shorter ssDNA2 strand. The formation of the duplexes and consequently the 3WJ motif is verified through electrophoretic mobility shift assay (EMSA), as shown in Figure 1B. All constructs display a distinct dimer band with a slower migration rate than that of the monomer.
The UV melting experiments are then performed to evaluate thermodynamic stabilities of the d3WJs. The melting curves exhibit sharp transitions, supporting the assumption of a cooperative shift from the DNA duplex to single-stranded DNAs (Figure 1C). None of the single strands display a transition (Figure S2). Thermodynamic parameters for the 3WJ were determined using UV optical melting experiments at different ionic strengths: sodium cacodylate buffer containing 1 M NaCl (CB1) and sodium cacodylate buffer with 0.1 M NaCl and 10 mM MgCl2 (CB2). Table 1 summarizes the calculated parameters for each 3WJ obtained from van’t Hoff plots, where the linear fit goodness exceeded 0.90 for all melts as exemplified in Figure 1D. The free energies for the investigated 3WJs range from −9.0 to 11.0 kcal/mol in the CB1 buffer and from −7.1 to −7.7 kcal/mol in the CB2 buffer. There are no substantial variations in the melting temperatures and free energies determined in the CB2 buffer in the presence of Mg2+ ions, indicating that this ion does not significantly contribute to the stability of this motif. Divalent metal ions are known to form coordination bonds with the phosphate groups,47,48 effectively neutralizing the negative charges and reducing the electrostatic repulsion. However, as the number of thymidine connecting helices increases, the stability slightly decreases from 0T d3WJ to 4T d3WJ, as indicated by the progressively less favorable ΔG values (from −7.73 to −7.09 kcal/mol). This can be attributed to a balance between the stabilizing effect of Mg ions and the destabilizing influence of increased steric hindrance and the altered geometry introduced by the thymidine loops. The intricate interplay of electrostatic forces, coordination interactions with Mg ions, and structural features ultimately dictates the overall stability of DNA 3WJ.
Table 1.
Measured Thermodynamic Parameters for d3WJ in 1 and 0.1 M NaCla
| name | buffer | Tm (°C) for 10−5 M | ΔHapp (kcal/mol) | ΔSapp (cal/mol) | ΔG37 (kcal/mol) | ΔΔG37 (kcal/mol) |
|---|---|---|---|---|---|---|
| 4T d3WJ | CB1 | 43.9 ± 1.2 | 89.6 | −257.1 | −9.88 | −2.79 |
| CB2 | 33.9 ± 0.8 | −96.2 | −287.4 | −7.09 | ||
| 3T d3WJ | CB1 | 43.3 ± 0.7 | −104.8 | −305.1 | −10.23 | −3.08 |
| CB2 | 34.9 ± 0.6 | −111.5 | −336.5 | −7.15 | ||
| 2T d3WJ | CB1 | 44.3 ± 0.6 | −131.1 | −387.3 | −11.00 | −3.41 |
| CB2 | 36.1 ± 0.6 | −94.4 | −279.7 | −7.59 | ||
| 1T d3WJ | CB1 | 43.2 ± 0.7 | −137.8 | −410.6 | −10.50 | −2.94 |
| CB2 | 35.2 ± 0.5 | −99.2 | −295.5 | −7.56 | ||
| 0T d3WJ | CB1 | 41.1 ± 0.5 | −71.1 | −200.2 | −9.03 | −1.3 |
| CB2 | 33.5 ± 0.4 | −43.4 | −115.2 | −7.73 |
The errors in enthalpy, entropy, and free energy are estimated to be 10%, 10%, and 5%, respectively.49
On the contrary, the average stability in the presence of 1 M NaCl is enhanced by approximately −3 kcal/mol across DNA constructs. The 0T d3WJ exhibits a stabilization of −1.3 kcal/mol. The most stable d3WJ motif is found to be the construct with 2 Ts in the linking region, displaying a free energy of −11 kcal/mol, while the least stable is the 0T d3WJ with −7.7 kcal/mol.
The change in entropy (ΔS) value is related to the disorder or randomness of a system, and in the context of DNA duplex formation, a more positive (less negative value) ΔS suggests increased disorder, which can be associated with greater flexibility of the investigated molecules. The entropic value for the 4T d3WJ is calculated to be −257.1 cal/mol kcal/(mol K), significantly higher compared to the 3T d3WJ, 2T d3WJ, and 1T d3WJ in the absence of Mg2+ ions, indicating a greater dynamic flexibility for this motif.
Relative Flexibility of DNA 3WJs by MD Simulations.
To further address the question whether the increasing single-stranded region will impact overall dynamics and flexibility of the DNA 3WJs, we have conducted MD simulations.41 The average structures of d3WJ complexes are illustrated in Figure 2A. Each average structure is obtained based on snapshots from 20 to 120 ns. We first perform the principal component analysis (PCA) to capture the series of vibrational motions in d3WJs using the MD trajectory. The PCA reveals that each stem within nT d3WJ (n = 1–4) exhibits distinct vibrational modes due to T–T interactions in the junction, emphasizing their heightened flexibility (Figure 2B).
Figure 2.
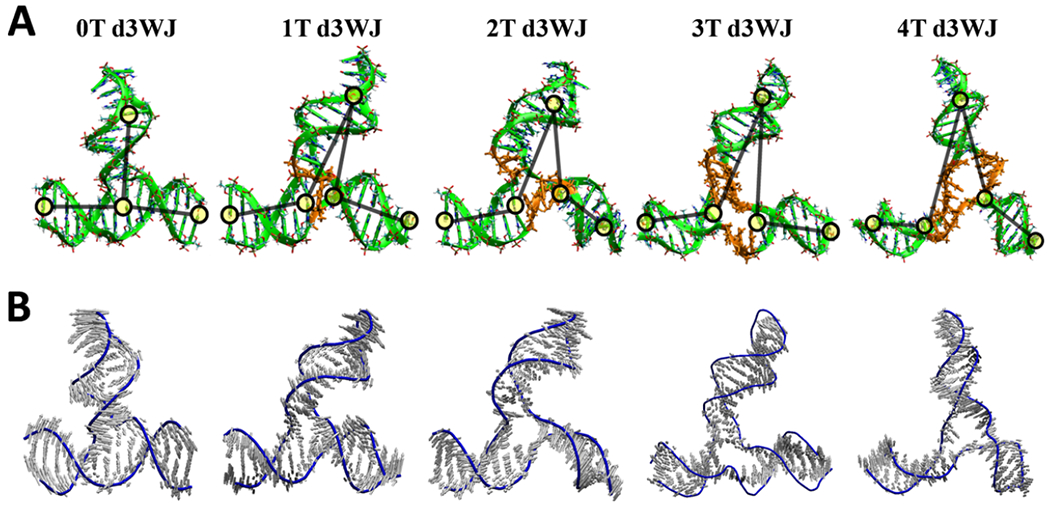
(A) Average structures of each d3WJ were derived from snapshots collected during molecular dynamics (MD) simulations on the Amber force field, spanning from 20 to 120 ns. Ts in the junction region are presented in orange. Yellow circles indicate the location of bases which are used to calculate the angle between the HP loop and each lower stems. (B) The principal component analysis mode of d3WJs normalized to 2 Å.
The root mean square deviation (RMSD) increases, indicating that complexes with more Ts exhibit greater structural variability, rendering them more flexible compared to 0T d3WJ (Figure S3). Bending angle analysis reveals that as Ts in the junction increase, each stem in relation to the hairpin (HP) undergoes more substantial angular displacements, further underscoring increased flexibility. The bending angle of each d3WJ was measured by the center of mass of two base-paired nucleotides (nt) in three different locations as shown by yellow circles in Figure 2A. The first location was the base-paired nt (C:G) next to the hairpin, the second location was the base-paired nt (C:G) in the stem next to the junction, and the third location was the base-paired nt (G:C) at the 5’ and 3’ ends of the stem. MD simulations also highlight the involvement of Ts in the junctions of 3T d3WJ and 4T d3WJ in hydrogen bond interactions and base stacking between Ts, resulting in asymmetrical conformations that enable these complexes to adopt the required vertex shape within a NANP architecture. Furthermore, while 0T d3WJ allows a larger number of sodium ions to associate with it for extended periods due to its reduced flexibility, as the number of Ts in the junction increases, d3WJ becomes more flexible, leading to increased conformational variations and decreased sodium ion association.
Despite this reduced association, sodium ions form stable solute—solvent networks that enhance the overall conformational stability of the nT d3WJ complexes (Figure S3B,C). In summary, we find that the number of Ts in nT d3WJ (n = 0–4) has a profound impact on the structural flexibility of these complexes, and increasing the number of Ts in the junction results in increasing flexibility and a broader range of conformational variations. This conclusion aligns with a previously published report on the impact of ssU on the RNA 3WJ in the context of the RNA nanocube.50 Additionally, the flexibility and T–T interactions within the junction enable 3WJ complexes to effectively adopt the necessary vertex conformation within a DNA architecture.
Collectively, our findings emphasize the intricate connection between the structural flexibility and the quantity of Ts present in the junction. The results indicate that as the number of Ts in the junction increases, the thermodynamic stabilities of the complexes remain relatively consistent. However, they do become notably more flexible, which is evident in the increased entropy observed, a conclusion supported by MD simulations.
Fabrication and Physicochemical Properties of Minimal Triangular DNA Nanoparticles Designed from d3WJs.
Data obtained by studying individual d3WJs indicate that the stability and conformational variations within these 3WJ complexes are intricately tied to the presence of Ts in the junction region. Furthermore, we explore the potential implications of these d3WJs in nanoscale architectural objects. The 2D structures of the investigated nanoparticles were conceived using the triangular model structures established in prior research by our groups.27
These structures initially feature a 22-base pair (bp) inner helical region and a 12-bp outer helical region. In the context of this research project, we opted to truncate the inner helix region of the triangular design to 10 bp, resulting in a reduced-size triangular configuration. This modification offers a cost-effective advantage by necessitating only 40 nt per strand during the synthesis process while still retaining the self-assembly capability into the desired triangular configuration. The selection of DNA sequences corresponding to these structures is conducted with precision through the utilization of secondary structure prediction tools, including UNAfold51 and NUPAC.52 Detailed information regarding the designed 3WJ sequences and their secondary structures is provided in Table S1 and Figure S1.
To validate the assembly of the designed DNA strands into the preconfigured complex, we employ the electrophoretic mobility shift assay (EMSA) under native conditions using agarose gel electrophoresis (Figure 3A). The migration of DNA complexes on the gel is influenced by the DNA’s length and, consequently, its molecular weight, with lighter fragments migrating more swiftly than heavier counterparts. Furthermore, the migration of these complexes is contingent upon the overall 3D configuration of the DNA complex. We initiated the formation of DNA complexes using the DNA 4T construct and adopted a stepwise assembly strategy. In this approach, DNA strands are sequentially introduced into four separate test tubes to generate monomer (consisting of a single strand), dimer (comprising two strands), trimer (comprising three strands), and tetramer DNA samples, which are subsequently subjected to EMSA analysis on an agarose gel (Figure 3A). Analysis of the gel images reveals distinctive migration profiles for all DNA complexes due to variations in the number of single-stranded Ts within the hinge regions, notably with the 0T triangle exhibiting more extensive migration compared to the others. Calculated yields of self-assembly, determined by quantifying the resulting triangular bands on the gel using ImageJ,53 reveal that the assembly yield for the 0T triangle is 43%, while triangles 1T, 2T, 3T, and 4T exhibit yields greater than 95% (Table 2). It is evident that in the absence of the ssT, there is structural tension in the assembling strands, leading to poor assembly of the 0T triangular complex.
Figure 3.
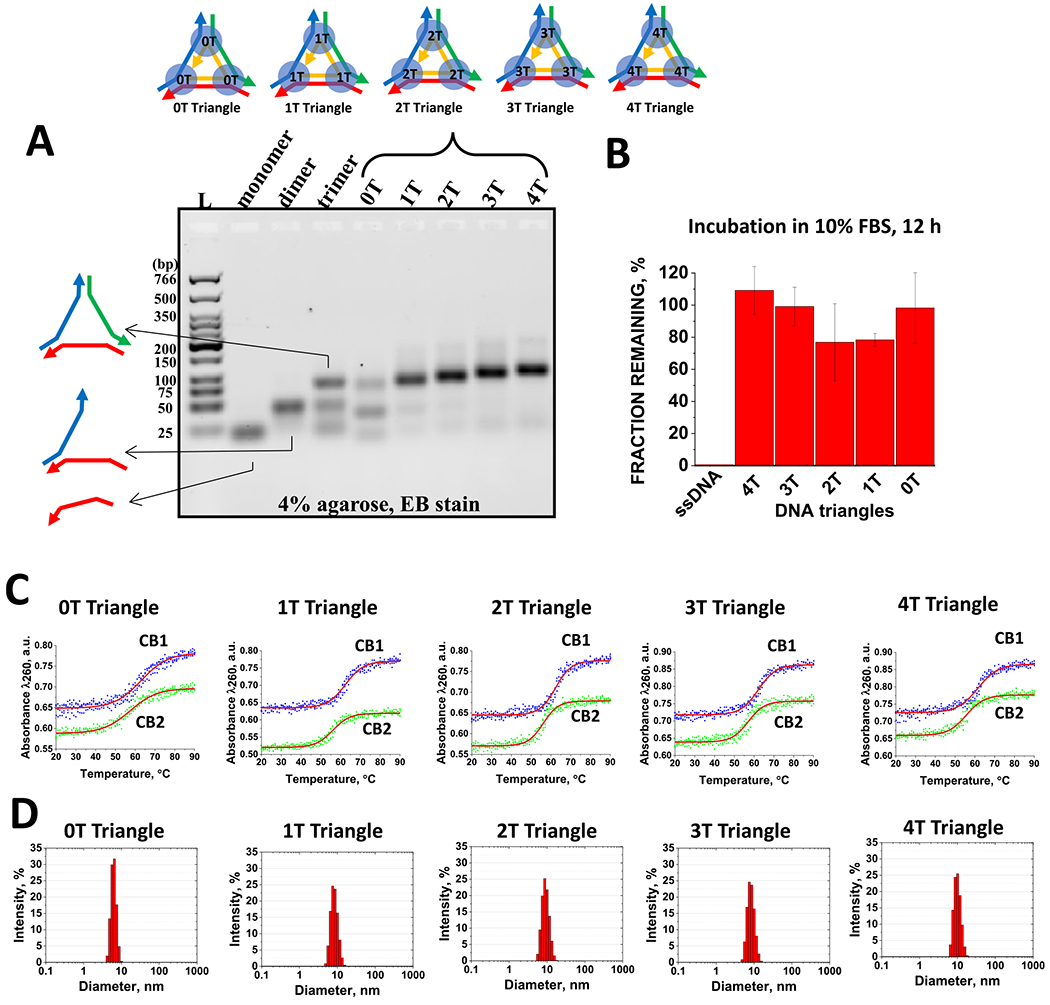
(A) Self-assembly properties of the DNA triangles evaluated by agarose gel electrophoreses. (B) FBS stability assay demonstrating intact DNA triangles after 12 h of incubation in contrast to the positive control ssDNA. (C) Examples of optical melting profiles for DNA triangles in CB1 and CB2 buffers. (D) Hydrodynamic diameter profiles for the triangular complexes measured by dynamic light scattering.
Table 2.
Properties of Triangular DNA Nanoparticlesa
| name | buffer | Tm (°C) for 10−6 M | diameter (nm) | yield (%) |
|---|---|---|---|---|
| 4T triangle | CB1 | 61.4 ± 0.3 | 11.6 ± 0.6 | 98.1 ± 1.2 |
| CB2 | 55.6 ± 0.2 | |||
| 3T triangle | CB1 | 62.1 ± 0.2 | 10.0 ± 1.2 | 97.5 ± 2.2 |
| CB2 | 57.0 ± 0.3 | |||
| 2T triangle | CB1 | 62.3 ± 0.2 | 9.6 ± 1.1 | 97.7 ± 2.8 |
| CB2 | 56.1 ± 0.2 | |||
| 1T triangle | CB1 | 62.4 ± 0.2 | 9.8 ± 1.2 | 96.3 ± 1.5 |
| CB2 | 56.1 ± 0.2 | |||
| 0T triangle | CB1 | 63.1 ± 0.4 | 6.7 ± 1.5 | 47.2 ± 3.6 |
| CB2 | 57.4 ± 0.3 |
Errors represent standard deviations of the mean values for hydrodynamic diameters (nm) and assembly yields (%), based on data obtained from at least three independent DLS and EMSA experiments.
For prospective therapeutic applications, it is imperative that the triangular DNA constructs maintain their configurations and functionality at the physiological body temperature of 37 °C, even in the presence of a harsh nuclease-rich environment.10 To address these crucial aspects, we conduct assessments concerning the thermal stability and resistance to enzymatic degradation of DNA triangles.
One of the notable challenges associated with the biological utilization and in vitro testing of DNA nanostructures pertains to their vulnerability to nucleases present in the basal cell culture media, which is typically supplied with a growth supplement such as FBS at a concentration of 5–10%.54–57 To evaluate the resistance of DNA complexes to enzymatic degradation by exo- and endonucleases within blood serum, we conducted a fetal bovine serum (FBS) stability assay (Figures 3B and S4). In this assay, equal volumes of DNA triangles (1 μM) and 20% aqueous FBS solution was mixed and subjected to incubation in a water bath at 37 °C. Aliquots are withdrawn at intervals ranging from 1 min to 12 h. These samples are then subjected to 4% agarose gel electrophoresis, and the remaining DNA fractions are quantified using ImageJ software.53 The resulting fractions remaining are plotted as a function of the incubation time using graphical software (OriginPro2019, OriginLab Co.). The corresponding gels and the quantified percentages of the remaining fractions are summarized in Figure 3B. Data suggest only a slight variation in stability between the 4T triangle (the most stable) and the 2T triangle (slightly less stable). Remarkably, in all cases, approximately 80% of the initial fraction remains after 12 h of incubation in a 10% blood serum environment.
The UV melting technique is widely employed for the investigation of the thermal stability of nucleic acid-based complexes.33,58,59 All constructs exhibit characteristic single-transition sigmoidal curves, with the calculated melting temperatures (Tm) being remarkably consistent, ranging around 62 °C in CB1 and approximately 56 °C in CB2 as exemplified in Figure 3C and summarized in Table 2. Similar to the corresponding d3WJs, the constructs are found to be more thermally stable in the presence of high sodium ions. Importantly, these Tm values significantly exceed the body’s physiological temperature, suggesting that the complexes will indeed preserve their triangular configuration in vivo. The melting profiles of DNA triangular complexes in CB1 and CB2 are recorded at 260 nm across a temperature range of 20–90 °C, employing a ramp rate of 1 °C/min as described in the Materials and Methods.
The influence of T regions on the dimensions of DNA nanoparticles is further explored through dynamic light scattering (DLS). Native PAGE and DLS are complementary techniques, both providing valuable insights into the size of nucleic acid-based nanoparticles.25 In this study, we conduct DLS experiments utilizing a Zetasizer nano-ZS (Malvern Instruments, Ltd.) to ascertain the average dimensions of the DNA complexes. A microcuvette (Starna Cells, Inc.) is used, and all measurements are conducted at 22 °C. The results obtained reveal an increase in sizes from 0-T to 4-T, with measurements of 6.7, 9.6, 9.8, 10.0, and 11.6 nm, as shown in Figure 3D and summarized in Table 2. Notably, the addition of thymidine to the single-stranded linkage led to an increase in the size of the nanoparticles, consistent with the findings from the gel shift assay.
It is important to note that the mechanisms underlying nuclease resistance in different DNA structures remain an area of limited understanding.60 Multiple factors contribute to the stability of DNA, including the type of DNA nucleases present, DNA sequence, backbone geometry, groove width, and rigidity and flexibility of the complex. In the case of DNA triangular-shaped nanostructures, their non-native geometries and sizes may reduce enzyme binding affinity, thereby decreasing the efficiency of specific or nonspecific enzymatic cleavage. Structural conformation plays a pivotal role in the susceptibility of DNA complexes to nuclease digestion. For instance, DNA tiles and motifs comprising multiple strands hybridized together in compact forms as well as larger DNA origami structures with numerous helical domains densely packed into bundles tend to exhibit greater nuclease resistance compared to linear duplexes or circular plasmid DNA forms.61
Cell Viability and Immunorecognition of DNA Triangles.
The assessment of cell viability and immunorecognition of nucleic acid nanoparticles plays a pivotal role in ensuring the safety and efficacy of these compounds in biomedical applications.1 It is of paramount importance to verify that DNA triangles do not exhibit toxicity toward human cells and do not provoke unintended immune responses, which could potentially compromise their therapeutic effectiveness or lead to adverse reactions. A comprehensive understanding of immunostimulation during the intra- and extracellular interactions of nucleic acid nanoparticles with human cells is essential in optimizing the properties of nanomaterials for efficient therapeutic delivery while minimizing the risks associated with immune-related complications.30
In our study, we conduct a series of experiments to assess whether DNA triangles with various ssT regions would elicit an immune response or prove to be toxic to human cells. We utilize three different reporter cell lines (HEK-Blue hTLR7, HEK-Blue hTLR9, and HEK-Lucia RIG-I) engineered to turn up the activation of specific pathways related to the detection of exogenous nucleic acids.62–67 While TLR9 is a well-known key receptor for the recognition of DNA, TLR7 primarily recognizes ssRNAs. However, it has been reported that TLR7 also has the ability to respond to deoxyguanosine, independent of RNA.68,69 Furthermore, recent studies have indicated the possible recognition of DNA nanoparticles by TLR7.70 Therefore, DNA triangles could potentially activate an innate immune response through TLR7. We also included RIG-I because it can function as a cytosolic sensor for foreign DNA71,72 and DNA nanoparticles,23 particularly when RIG-I is overexpressed by the cell.
The cell viability is assessed by measuring the metabolic activity through the reduction of a tetrazolium compound into a colored formazan product in live cells. The data in Figure 4A indicate that DNA triangles do not exhibit toxicity toward the human cell lines. Also, we assess the levels of immune cell activation in response to DNA triangles. Figure 4B unequivocally shows that the DNA triangles do not elicit an immune response, whether introduced with or without the delivery agent, when compared with positive controls. The results underscore the potential of engineered nanoparticles as safe and effective carriers for medical applications.
Figure 4.
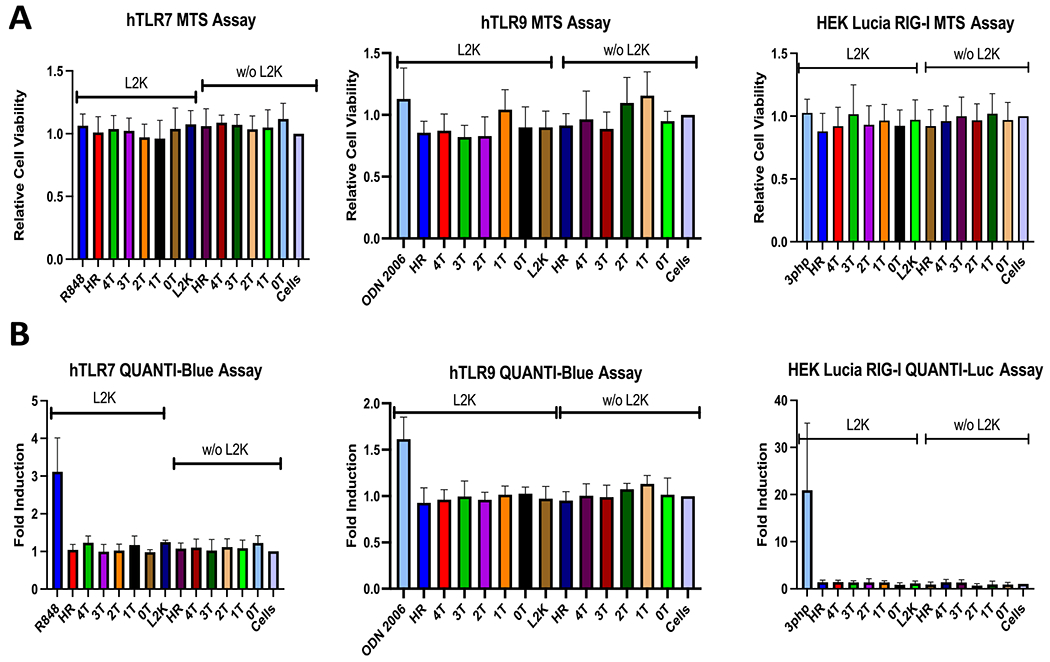
Toxicity (A) and immunostimulatory properties (B) of DNA triangles tested with and without lipofectamine 2000 (L2K). 0T–4T indicate the specific count of single-stranded thymines inserted within the junction of DNA triangles. HR refers to 4T DNA triangles with a hairpin region included. Each bar represents the mean of N = 3 biological repeats, and error bars denote mean ± SD.
Self-Assembly Properties and Characteristics of Minimal DNA Polygons Designed from 4T d3WJ.
Collectively, we observe that all triangular DNA nanoparticles, except for the 0T variant, exhibit remarkably similar physicochemical properties. However, driven by the enhanced flexibility of the 4T 3WJ region, we selected 4T d3WJ as a foundational building block for polygonal nanoparticles. Subsequently, we performed several experiments to assess their physicochemical characteristics, with the aim of extending their application to other geometric shapes, including squares, pentagons, and hexagons.
The design principles for the polygonal DNA nanoparticles are implemented by adding extra strands to enhance the complexity of the polygons and extending the length of the central DNA strand, as detailed in previous publications.15,22 All DNA polygons effectively assemble in “one pot”, displaying high efficiency, as confirmed by agarose gel electrophoresis in Figure 5A.
Figure 5.
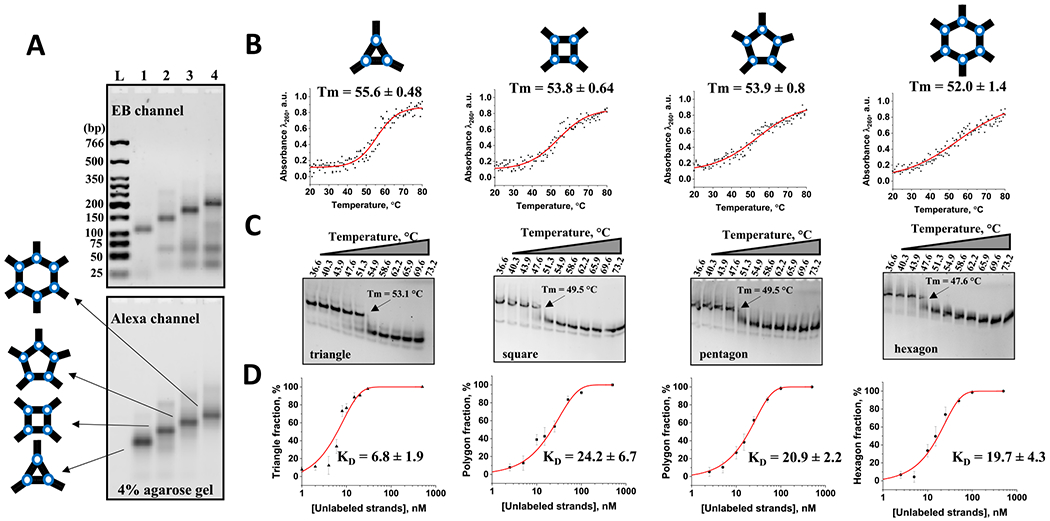
(A) Alexa-488-labeled polygonal nanoparticle assembly properties assayed by 4% agarose gel electrophoreses. Gel scanned with the ethidium bromide (EB) channel to detect all DNA strands within the gel as well as Alexa channel to visualize labeled DNAs. (B) Normalized optical melt curves of 0.2 μM polygons in TMS buffer (50 mM TRIS pH = 8.0, 100 mM NaCl, and 10 mM MgCl2). (C) TGGE assay of the Alexa-488-labeled polygons showing their melting temperature points. (D) Titration curve fitting data of DNA polygon assembly showing apparent Kd values in nM (error represents standard deviation of the mean obtained from at least two independent experiments).
In addition, the assembled DNA polygons retained their structural integrity following prolonged (12 h) incubation in a solution containing 10% FBS (Figure S5).
To assess the thermal stabilities of the DNA nanoparticles, we conduct evaluations in solution, using UV optical melting, and in gel, using temperature gradient gel electrophoresis (TGGE), as shown in Figure 5B,C, respectively. Both methods consistently indicate that Tm for the triangular nanoparticles is higher than hexagon Tm values by ~4 °C (measured by UV melting) and ~6 °C (measured by TGGE). Notably, the data from TGGE and UV optical melting profiles for the polygons are highly similar, with minor discrepancies that may arise from differences in the underlying principles of the two approaches.
The nucleic acid folding process is significantly influenced by the concentration of the participating strands. Therefore, we also measure the apparent dissociation constants (Kd) for each DNA polygon as described in the Materials and Methods (Figure S6). The Kd for the DNA triangle is determined to be approximately 7 nM, while squares, pentagons, and hexagons exhibited Kd values of around 20 mM, which is approximately 3 times higher (Figure 5D). Differences in the apparent dissociation constants (Kd) suggest distinct stability levels in their structures, indicating varying affinities for the folded forms. The notably higher Kd values for squares, pentagons, and hexagons compared to triangles imply reduced stability, likely due to the entropically unfavorable structural complexity between DNA intermolecular interactions. The formation of the square, pentagon, and hexagon complexes requires participation of 5, 6, and 7 ssDNA strands to hybridize accordingly. This contributes to a decrease in the entropic change during their formation, causing the reduction of the overall stability.
Stability Assessment for Long-Term Handling of DNA Polygons.
Long-term handling, storage, and transportation of nucleic acid nanoparticles as a potential therapeutic or diagnostic agent are essential considerations directly impacting the stability and functionality of these nanostructures.3,10,66,73 Maintaining their structural integrity during storage and transportation ensures that nanoparticles can reliably serve their intended purposes, especially in applications where DNA polygons are used as biomedical agents. Therefore, we explore the stability of the DNA polygons to assess their resilience during extended storage. In our protocol, we subject the DNA polygons to complete dehydration and store them for 1 week at a temperature of +55 °C, which exceeds their respective melting temperatures. The dehydration process is achieved using two different methods, a vacuum concentrator (SpeedVac) and lyophilization, as described in the Materials and Methods section. For comparison, the control group includes two sets of DNA polygons stored at +55 and + 4 °C in solution.
Subsequently, the dehydrated samples are rehydrated in deionized water, and the retention of polygons’ structural integrity is assessed using native PAGE (Figure S7). The results demonstrate that lyophilization is a more effective method for preserving the structural integrity after rehydration. The band patterns of the lyophilized samples closely resemble those of the control samples stored at 4 °C. Conversely, the control samples stored at 55 °C in solution exhibit reduced band intensity and a loss of pattern, consistent with denaturation of the samples when stored above their respective melting temperatures in the solution form.
Samples dried via a SpeedVac also displayed significant alterations in band patterns and intensity compared to the control. Additionally, the SpeedVac-dried samples exhibit more noticeable aggregation in the wells of the native PAGE gel, in comparison to the lyophilized and control samples stored at 4 °C.
Collectively, the results suggest that lyophilization provides robust protection for the DNA polygons against temperature fluctuations and maintains their structural integrity more effectively than the SpeedVac does when stored under similar conditions. These findings align with results previously reported by Tran et al.66
CONCLUSIONS
In this account, we tackle multiple fundamental questions regarding the design and application of DNA nanostructures. Our experimental approach used here is to first study the influence of ssT linkers in DNA three-way junctions. The 3WJ motif is often regarded as a cornerstone in RNA and DNA nanotechnology, offering a versatile platform for constructing intricate nanostructures.74–78 Its strength lies in its structural robustness, providing a stable framework for assembling complex architectures. This modularity empowers researchers to tailor the 3WJ to their needs, altering nucleic acid strand sequences and lengths with precision. It also allows the introduction of specific functional elements, such as aptamers or ligands, for custom-designed functionalities. With remarkable thermal stability and predictable self-assembly behavior, the 3WJ simplifies the design and fabrication of nanostructures, making it an essential tool in various fields, including drug delivery, diagnostics, and therapeutics.
The calculated thermodynamic parameters have revealed that d3WJ is more stable in 1 M NaCl, and the presence of the Mg2+ ions does not significantly affect the stabilities of the tested d3WJ. The larger entropical contribution is observed for 4T d3WJ, suggesting a more dynamic structure. This is also confirmed by MD simulation where the increasing number of thymidine bases in the single-stranded linker regions of DNA 3WJ complexes significantly enhanced their structural flexibility and conformational adaptability. These findings are instrumental in the design of triangular DNA nanoparticles, which showed remarkable stability and the ability to resist enzymatic degradation. The high self-assembly efficiency of >90% was determined for all DNA triangles with the exception of 0T (no linkage). The absence of ssTs creates structural tension within the assembling strands as supported by MD simulation and thus results in poor assembly. Moreover, our assessment of cell viability and immunorecognition demonstrates the nontoxic and nonimmunogenic nature of DNA polygons, underscoring their potential for safe and effective applications in biomedicine.
The 4T 3WJ was chosen as the foundational building block for other polygonal shapes because it exhibits greater flexibility in the junction region, which allowed for the construction of polygons with enhanced stability and versatility. This flexibility, as revealed by molecular dynamics simulations, enables the polygons to adopt various conformational variations, making them suitable for different geometric shapes such as triangles, squares, pentagons, and hexagons. The study further highlights the importance of long-term handling and storage considerations in maintaining the structural integrity of DNA polygons and established lyophilization as an effective preservation method.
Overall, the study not only provides valuable insights into the role of ssTs in DNA nanostructures but also expands the scope of DNA polygons by introducing a versatile building block, the 4T 3WJ, with wide-ranging applications in nanotechnology, nanomedicine, and beyond. These findings contribute to our understanding of nucleic acid-based agents and their suitability for diverse biomedical and nanotechnological purposes, further advancing the field of DNA nanotechnology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (NIH) under Award Number 1R15EB031388-01 and National Science Foundation MRI Award Number 2214573 (to E.F.K.). Also, research reported in this publication was supported by the National Institute of General Medical Sciences of the NIH under Award Number R35GM139587 (to K.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This project has been funded in whole or in part with internal BSU ASPiRE Advance grant proposal.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.3c00615.
DNA nanoparticle secondary structures and sequences, annotated electrophoretic gel images, and MD simulation data (PDF)
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.3c00615
Contributor Information
Ross Brumett, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Leyla Danai, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States.
Abigail Coffman, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Yasmine Radwan, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States.
Megan Teter, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Hannah Hayth, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Erwin Doe, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Katelynn Pranger, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Sable Thornburgh, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Allison Dittmer, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Zhihai Li, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
Tae Jin Kim, Department of Physical Sciences, West Virginia University Institute of Technology, Beckley, West Virginia 25801, United States.
Kirill A. Afonin, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States
Emil F. Khisamutdinov, Department of Chemistry, Ball State University, Muncie, Indiana 47306, United States.
REFERENCES
- (1).Chandler M; Johnson B; Khisamutdinov E; Dobrovolskaia MA; Sztuba-Solinska J; Salem AK; Breyne K; Chammas R; Walter NG; Contreras LM; Guo P; Afonin KA The International Society of RNA Nanotechnology and Nanomedicine (ISRNN): The Present and Future of the Burgeoning Field. ACS Nano 2021, 15, 16957–16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rolband L; Beasock D; Wang Y; Shu YG; Dinman JD; Schlick T; Zhou Y; Kieft JS; Chen SJ; Bussi G; Oukhaled A; Gao X; Sulc P; Binzel D; Bhullar AS; Liang C; Guo P; Afonin KA Biomotors, viral assembly, and RNA nanobiotechnology: Current achievements and future directions. Comput. Struct. Biotechnol. J 2022, 20, 6120–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Afonin KA; Dobrovolskaia MA; Church G; Bathe M Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2020, 14, 9221–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ralph SF Frontiers in Nucleic Acid Nanotechnology. Nanomaterials 2015, 5, 750–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Seeman NC; Mao C; Yan H Guest editorial: nucleic acid nanotechnology. Acc. Chem. Res 2014, 47, 1643–1644. [DOI] [PubMed] [Google Scholar]
- (6).Seeman NC DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct 1998, 27, 225–248. [DOI] [PubMed] [Google Scholar]
- (7).Jaeger L; Westhof E; Leontis NB TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Guo P; Zhang C; Chen C; Garver K; Trottier M Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell 1998, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- (9).Guo P The emerging field of RNA nanotechnology. Nat. Nanotechnol 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Afonin KA; Dobrovolskaia MA; Ke W; Grodzinski P; Bathe M Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv. Drug Delivery Rev 2022, 181, No. 114081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bandy TJ; Brewer A; Burns JR; Marth G; Nguyen T; Stulz E DNA as supramolecular scaffold for functional molecules: progress in DNA nanotechnology. Chem. Soc. Rev 2011, 40, 138–148. [DOI] [PubMed] [Google Scholar]
- (12).Knappe GA; Wamhoff EC; Bathe M Functionalizing DNA origami to investigate and interact with biological systems. Nat. Rev. Mater 2023, 8, 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lo PK; Metera KL; Sleiman HF Self-assembly of three-dimensional DNA nanostructures and potential biological applications. Curr. Opin. Chem. Biol 2010, 14, 597–607. [DOI] [PubMed] [Google Scholar]
- (14).Kaviani S; Fakih HH; Asohan J; Katolik A; Damha MJ; Sleiman HF Sequence-Controlled Spherical Nucleic Acids: Gene Silencing, Encapsulation, and Cellular Uptake. Nucleic Acid Ther. 2023, 33, 265–276. [DOI] [PubMed] [Google Scholar]
- (15).Khisamutdinov EF; Li H; Jasinski DL; Chen J; Fu J; Guo P Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res. 2014, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Chworos A; Severcan I; Koyfman AY; Weinkam P; Oroudjev E; Hansma HG; Jaeger L Building programmable jigsaw puzzles with RNA. Science 2004, 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- (17).Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yang L; Li Z; Binzel DW; Guo P; Williams TM Targeting oncogenic KRAS in non-small cell lung cancer with EGFR aptamer-conjugated multifunctional RNA nanoparticles. Mol. Ther. Nucleic Acids 2023, 33, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ellipilli S; Wang H; Binzel DW; Shu D; Guo P Ligand-displaying-exosomes using RNA nanotechnology for targeted delivery of multi-specific drugs for liver cancer regression. Nanomedicine 2023, 50, No. 102667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Doe E; Hayth HL; Brumett R; Khisamutdinov EF Effective, Rapid, and Small-Scale Bioconjugation and Purification of ”Clicked” Small-Molecule DNA Oligonucleotide for Nucleic Acid Nanoparticle Functionalization. Int. J. Mol. Sci 2023, 24, 4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Doe E; Hayth HL; Khisamutdinov EF Bioconjugation of Functionalized Oligodeoxynucleotides with Fluorescence Reporters for Nanoparticle Assembly. Methods Mol. Biol 2023, 2709, 105–115. [DOI] [PubMed] [Google Scholar]
- (22).Hartung J; McCann N; Doe E; Hayth H; Benkato K; Johnson MB; Viard M; Afonin KA; Khisamutdinov EF Toehold-Mediated Shape Transition of Nucleic Acid Nanoparticles. ACS Appl. Mater. Interfaces 2023, 15, 25300–25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Johnson MB; Halman JR; Miller DK; Cooper JS; Khisamutdinov EF; Marriott I; Afonin KA The immunorecognition, subcellular compartmentalization, and physicochemical properties of nucleic acid nanoparticles can be controlled by composition modification. Nucleic Acids Res. 2020, 48, 11785–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Johnson MB; Halman JR; Burmeister AR; Currin S; Khisamutdinov EF; Afonin KA; Marriott I Retinoic acid inducible gene-I mediated detection of bacterial nucleic acids in human microglial cells. J. Neuroinflammation 2020, 17, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Lett. 2018, 18, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA; Khisamutdinov EF; Marriott I; Afonin KA Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small 2017, 13, No. 1701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Bui MN; Johnson MB; Viard M; Satterwhite E; Martins AN; Li Z; Marriott I; Afonin KA; Khisamutdinov EF Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine 2017, 13, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Goldsworthy V; LaForce G; Abels S; Khisamutdinov EF Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials 2018, 8, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Khisamutdinov EF; Jasinski DL; Guo P RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 2014, 8, 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Durbin JK; Miller DK; Niekamp J; Khisamutdinov EF Modulating Immune Response with Nucleic Acid Nanoparticles. Molecules 2019, 24, 3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia M; Afonin KA RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic Acids Res. 2019, 47, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hughes RA; Ellington AD Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harbor Perspect. Biol 2017, 9, No. a023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Teter M; Brumett R; Coffman A; Khisamutdinov EF Thermodynamic Characterization of Nucleic Acid Nanoparticles Hybridization by UV Melting. Methods Mol. Biol 2023, 2709, 151–161. [DOI] [PubMed] [Google Scholar]
- (34).Marky LA; Breslauer KJ Calculating Thermodynamic Data for Transitions of Any Molecularity from Equilibrium Melting Curves. Biopolymers 1987, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- (35).Case DA; Aktulga HM; Belfon K; Cerutti DS; Cisneros GA; Cruzeiro VWD; Forouzesh N; Giese TJ; Gotz AW; Gohlke H; Izadi S; Kasavajhala K; Kaymak MC; King E; Kurtzman T; Lee TS; Li P; Liu J; Luchko T; Luo R; Manathunga M; Machado MR; Nguyen HM; O’Hearn KA; Onufriev AV; Pan F; Pantano S; Qi R; Rahnamoun A; Risheh A; Schott-Verdugo S; Shajan A; Swails J; Wang J; Wei H; Wu X; Wu Y; Zhang S; Zhao S; Zhu Q; Cheatham TE 3rd; Roe DR; Roitberg A; Simmerling C; York DM; Nagan MC; Merz KM Jr. AmberTools. J. Chem. Inf. Model 2023, 63, 6183–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Galindo-Murillo R; Robertson JC; Zgarbova M; Sponer J; Otyepka M; Jurecka P; Cheatham TE 3rd. Assessing the Current State of Amber Force Field Modifications for DNA. J. Chem. Theory Comput 2016, 12, 4114–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- (38).Sengupta A; Li Z; Song LF; Li P; Merz KM Jr. Correction to “Parameterization of Monovalent Ions for the OPC3, OPC, TIP3P-FB, and TIP4P-FB Water Models”. J. Chem. Inf. Model 2021, 61, 3734–3735. [DOI] [PubMed] [Google Scholar]
- (39).Feller SE; Zhang YH; Pastor RW; Brooks BR Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J. Chem. Phys 1995, 103, 4613–4621. [Google Scholar]
- (40).Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG A Smooth Particle Mesh Ewald Method. J. Chem. Phys 1995, 103, 8577–8593. [Google Scholar]
- (41).Phillips JC; Hardy DJ; Maia JDC; Stone JE; Ribeiro JV; Bernardi RC; Buch R; Fiorin G; Henin J; Jiang W; McGreevy R; Melo MCR; Radak BK; Skeel RD; Singharoy A; Wang Y; Roux B; Aksimentiev A; Luthey-Schulten Z; Kalé LV; Schulten K; Chipot C; Tajkhorshid E Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys 2020, 153, No. 044130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kalé L; Schulten K Scalable molecular dynamics with NAMD. J. Comput. Chem 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Benkato K; O’Brien B; Bui MN; Jasinski DL; Guo P; Khisamutdinov EF Evaluation of Thermal Stability of RNA Nanoparticles by Temperature Gradient Gel Electrophoresis (TGGE) in Native Condition. Methods Mol. Biol 2017, 1632, 123–133. [DOI] [PubMed] [Google Scholar]
- (44).Tran AN; Chandler M; Halman J; Beasock D; Fessler A; McKeough RQ; Lam PA; Furr DP; Wang J; Cedrone E; Dobrovolskaia MA; Dokholyan NV; Trammell SR; Afonin KA Anhydrous Nucleic Acid Nanoparticles for Storage and Handling at Broad Range of Temperatures. Small 2022, 18, No. 2104814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Blose JM; Lloyd KP; Bevilacqua PC Portability of the GN(R)A hairpin loop motif between RNA and DNA. Biochemistry 2009, 48, 8787–8794. [DOI] [PubMed] [Google Scholar]
- (46).Fiore JL; Nesbitt DJ An RNA folding motif: GNRA tetraloop-receptor interactions. Q. Rev. Biophys 2013, 46, 223–264. [DOI] [PubMed] [Google Scholar]
- (47).Shu D; Shu Y; Haque F; Abdelmawla S; Guo P Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Hill AC; Schroeder SJ Thermodynamic stabilities of threeway junction nanomotifs in prohead RNA. RNA 2017, 23, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Xia T; SantaLucia J Jr.; Burkard ME; Kierzek R; Schroeder SJ; Jiao X; Cox C; Turner DH Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 1998, 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- (50).Afonin KA; Kasprzak W; Bindewald E; Puppala PS; Diehl AR; Hall KT; Kim TJ; Zimmermann MT; Jernigan R; Jaeger L; Shapiro BA Computational and experimental characterization of RNA cubic nanoscaffolds. Methods 2014, 67, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Markham NR; Zuker M UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol 2008, 453, 3–31. [DOI] [PubMed] [Google Scholar]
- (52).Zadeh JN; Steenberg CD; Bois JS; Wolfe BR; Pierce MB; Khan AR; Dirks RM; Pierce NA NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem 2011, 32, 170–173. [DOI] [PubMed] [Google Scholar]
- (53).Schneider CA; Rasband WS; Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Li C; Luo S; Wang J; Shen Z; Wu ZS Nuclease-resistant signaling nanostructures made entirely of DNA oligonucleotides. Nanoscale 2021, 13, 7034–7051. [DOI] [PubMed] [Google Scholar]
- (55).Meng S; Zhan S; Li J Nuclease-resistant double-stranded DNA controls or standards for hepatitis B virus nucleic acid amplification assays. Virol. J 2009, 6, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Cánovas D; Bird N Human AB serum as an alternative to fetal bovine serum for endothelial and cancer cell culture. ALTEX 2012, 29, 426–428. [DOI] [PubMed] [Google Scholar]
- (57).Chelladurai KS; Christyraj JDS; Rajagopalan K; Yesudhason BV; Venkatachalam S; Mohan M; Vasantha NC; Christyraj JRSS Alternative to FBS in animal cell culture - An overview and future perspective. Heliyon 2021, 7, No. e07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Schroeder SJ; Turner DH Optical melting measurements of nucleic acid thermodynamics. Methods Enzymol. 2009, 468, 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Khisamutdinov EF; Sweeney BA; Leontis NB Context-sensitivity of isosteric substitutions of non-Watson-Crick basepairs in recurrent RNA 3D motifs. Nucleic Acids Res. 2021, 49, 9574–9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hahn J; Wickham SF; Shih WM; Perrault SD Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Chandrasekaran AR Nuclease resistance of DNA nanostructures. Nat. Rev. Chem 2021, 5, 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Panigaj M; Skelly E; Beasock D; Marriott I; Johnson MB; Salotti J; Afonin KA Therapeutic immunomodulation by rationally designed nucleic acids and nucleic acid nanoparticles. Front. Immunol 2023, 14, No. 1053550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Beasock D; Ha A; Halman J; Panigaj M; Wang J; Dokholyan NV; Afonin KA Break to Build: Isothermal Assembly of Nucleic Acid Nanoparticles (NANPs) via Enzymatic Degradation. Bioconjugate Chem. 2023, 34, 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Chandler M; Jain S; Halman J; Hong E; Dobrovolskaia MA; Zakharov AV; Afonin KA Artificial Immune Cell, AI-cell, a New Tool to Predict Interferon Production by Peripheral Blood Monocytes in Response to Nucleic Acid Nanoparticles. Small 2022, 18, No. 2204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Chandler M; Rolband L; Johnson MB; Shi D; Avila YI; Cedrone E; Beasock D; Danai L; Stassenko E; Krueger JK; Jiang J; Lee JS; Dobrovolskaia MA; Afonin KA Expanding Structural Space for Immunomodulatory Nucleic Acid Nanoparticles (Nanps) via Spatial Arrangement of Their Therapeutic Moieties. Adv. Funct. Mater 2022, 32, No. 2205581, DOI: 10.1002/adfm.202205581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Tran AN; Chandler M; Halman J; Beasock D; Fessler A; McKeough RQ; Lam PA; Furr DP; Wang J; Cedrone E; Dobrovolskaia M; Dokholyan NV; Trammell SR; Afonin KA Anhydrous Nucleic Acid Nanoparticles for Storage and Handling at Broad Range of Temperatures. Small 2022, 18, No. 2104814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Halman JR; Kim KT; Gwak SJ; Pace R; Johnson MB; Chandler MR; Rackley L; Viard M; Marriott I; Lee JS; Afonin KA A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution. Nanomedicine 2020, 23, No. 102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Davenne T; Bridgeman A; Rigby RE; Rehwinkel J Deoxyguanosine is a TLR7 agonist. Eur. J. Immunol 2020, 50, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Lind NA; Rael VE; Pestal K; Liu B; Barton GM Regulation of the nucleic acid-sensing Toll-like receptors. Nat. Rev. Immunol 2022, 22, 224–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Hong E; Halman JR; Shah A; Cedrone E; Truong N; Afonin KA; Dobrovolskaia MA Toll-Like Receptor-Mediated Recognition of Nucleic Acid Nanoparticles (NANPs) in Human Primary Blood Cells. Molecules 2019, 24, 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Choi MK; Wang Z; Ban T; Yanai H; Lu Y; Koshiba R; Nakaima Y; Hangai S; Savitsky D; Nakasato M; Negishi H; Takeuchi O; Honda K; Akira S; Tamura T; Taniguchi T A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 17870–17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Cheng G; Zhong J; Chung J; Chisari FV Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 9035–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Lam PA; Furr DP; Tran A; McKeough RQ; Beasock D; Chandler M; Afonin KA; Trammell SR The Application of Light-Assisted Drying to the Thermal Stabilization of Nucleic Acid Nanoparticles. Biopreserv. Biobanking 2022, 20, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Guo S; Piao X; Li H; Guo P Methods for construction and characterization of simple or special multifunctional RNA nanoparticles based on the 3WJ of phi29 DNA packaging motor. Methods 2018, 143, 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Khisamutdinov EF; Bui MN; Jasinski D; Zhao Z; Cui Z; Guo P Simple Method for Constructing RNA Triangle, Square, Pentagon by Tuning Interior RNA 3WJ Angle from 60 degrees to 90 degrees or 108 degrees. Methods Mol. Biol 2015, 1316, 181–193. [DOI] [PubMed] [Google Scholar]
- (76).Zhang H; Endrizzi JA; Shu Y; Haque F; Sauter C; Shlyakhtenko LS; Lyubchenko Y; Guo P; Chi YI Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 2013, 19, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Hao C; Li X; Tian C; Jiang W; Wang G; Mao C Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nat. Commun 2014, 5, No. 3890. [DOI] [PubMed] [Google Scholar]
- (78).Takezawa Y; Shionoya M Supramolecular DNA Three-Way Junction Motifs With a Bridging Metal Center. Front. Chem 2020, 7, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


