Abstract
The cellular site of herpesvirus tegument assembly has yet to be defined. We have previously used a recombinant herpes simplex virus type 1 expressing a green fluorescent protein (GFP)-tagged tegument protein, namely VP22, to show that VP22 is localized exclusively to the cytoplasm during infection. Here we have constructed a similar virus expressing another fluorescent tegument protein, YFP-VP13/14, and have visualized the intracellular localization of this second tegument protein in live infected cells. In contrast to VP22, VP13/14 is targeted predominantly to the nuclei of infected cells at both early and late times in infection. More specifically, YFP-13/14 localizes initially to the nuclear replication compartments and then progresses into intense punctate domains that appear at around 12 h postinfection. At even later times this intranuclear punctate fluorescence is gradually replaced by perinuclear micropunctate and membranous fluorescence. While the vast majority of YFP-13/14 seems to be targeted to the nucleus, a minor subpopulation also appears in a vesicular pattern in the cytoplasm that closely resembles the pattern previously observed for GFP-22. Moreover, at late times weak fluorescence appears at the cell periphery and in extracellular virus particles, confirming that YFP-13/14 is assembled into virions. This predominantly nuclear targeting of YFP-13/14 together with the cytoplasmic targeting of VP22 may imply that there are multiple sites of tegument protein incorporation along the virus maturation pathway. Thus, our YFP-13/14-expressing virus has revealed the complexity of the intracellular targeting of VP13/14 and provides a novel insight into the mechanism of tegument, and hence virus, assembly.
The herpesvirus tegument has been described as an amorphous region of the virus particle located between the capsid and the envelope. It comprises the major structural proteins VP1/2, VP13/14, VP16, and VP22 (10, 17), along with a number of other minor constituents, such as the product of the UL13 gene (4, 5, 15). While the precise functions of most of the individual tegument proteins are not yet clear, it seems likely that they perform dual roles in the virus replication cycle, providing activities both at the onset of infection, as the capsid-tegument enters the cell, and during virion assembly, as the virus matures and exits the cell. Such dual functions are best exemplified by the well-studied tegument protein VP16, which has been shown to function both as the transactivator of immediate-early (IE) gene expression (3, 14) and as an essential protein in virus assembly (20).
The incorporation of large numbers of tegument molecules into the herpesvirus particle means that this group of proteins could act as cellular markers for virus assembly, thereby helping to address a range of issues surrounding the herpesvirus maturation pathway. In particular, the cellular compartments involved in both virus envelopment and tegumentation have not yet been precisely defined. It was initially proposed that assembled capsids present within the nucleus acquired their final virion envelope at the inner nuclear membrane as they budded into the lumen of the endoplasmic reticulum (13, 16). More recently, however, there have been several reports on the localization and processing of virus glycoproteins that support an alternative model of virus maturation, whereby virions budding through the nuclear membrane proceed through a further stage of deenvelopment at the outer nuclear membrane and acquire their final envelope downstream in the secretory pathway (2, 18, 21). One significant difference between these two pathways is that while the former model absolutely requires tegument assembly to occur in the nucleus, the latter model allows for tegument assembly to occur either in the nucleus or at a later stage within the cytoplasm. Thus, studies on the subcellular localization of tegument proteins may contribute to our understanding of herpesvirus morphogenesis.
We have previously investigated tegument protein localization during infection by constructing a herpes simplex virus type 1 (HSV-1) in which a major tegument gene, namely, the UL49 gene encoding VP22, was replaced with the gene for a green fluorescent protein (GFP)-22 fusion protein (9). We have shown that this virus incorporates GFP-tagged VP22 into assembling virions, thereby allowing us to use fluorescent microscopy of live infected cells to analyze the intracellular trafficking of VP22. This virus indicated that throughout the course of a high-multiplicity infection, VP22 localized exclusively to the cytoplasm, exhibiting a diffuse localization at early stages of expression, followed by an increasingly vesicular pattern as infection progressed. At late stages fluorescent VP22 could be seen at the cell periphery and in extracellular particles, indicative of virus release from the cell. Hence, we have suggested that at least one of the major tegument proteins is incorporated into the virion at a stage downstream from the nuclear envelope.
To extend our observations on tegument protein localization, we have also initiated a study of the proteins referred to collectively as VP13/14, the differentially modified products of the UL47 gene (12, 22). The function of VP13/14 in virus infection is not known, but there is some evidence to suggest that it may be involved in the regulation of VP16 transactivation of IE genes (11, 23). Surprisingly, in spite of being a major component of the virion, VP13/14 has been shown to be dispensable for virus growth in tissue culture (1, 22, 23). However, viruses unable to express VP13/14 appear to be retarded in the early stages of virus growth, supporting a potential role for the protein in gene expression (22, 23). In a paper accompanying the present study, we show that transient expression of VP13/14 results in its efficient nuclear localization (7). Here, we have further investigated the cellular compartmentalization of VP13/14 by constructing a virus expressing fluorescently tagged VP13/14 in a manner similar to our GFP-tagged VP22 virus. We show that this virus replicates and incorporates fluorescent VP13/14 into virions as efficiently as wild-type (Wt) virus. Strikingly, VP13/14 localizes predominantly to the nucleus throughout most of the virus replication cycle, with only a small subpopulation of the protein present in the cytoplasm. However, at late time points, fluorescent particles are observed in large numbers in the extracellular medium, suggesting that, as for the GFP-22 virus, virions containing fluorescent VP13/14 are detectable by fluorescent microscopy. These results demonstrate that in infected cells the tegument protein VP13/14 exhibits a trafficking pathway very different from that for the tegument protein VP22, and the implications for virus assembly of these contrasting results will be discussed.
MATERIALS AND METHODS
Cells and virus infections.
Vero cells, COS-1 cells, and BHK-1 cells were maintained in Dulbecco's modified minimal essential medium containing 10% newborn calf serum. The parental virus used in this study was the HSV-1 strain 17. Virions and infectious virus DNA were purified from extracellular virus released into the infected cell medium as described previously (9).
Construction of YFP-13/14 virus.
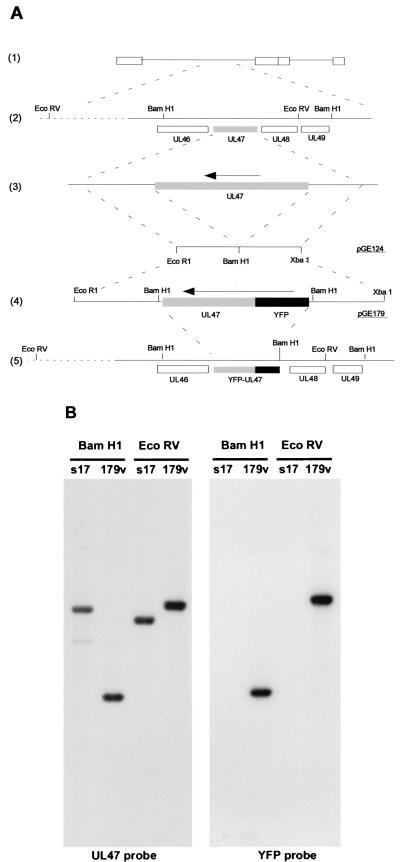
The construction of the yellow fluorescent protein (YFP)-13/14-expressing virus is detailed in Fig. 1. To construct a YFP-13/14 cassette for recombination into the genome, each of the 200-bp flanking sequences of the HSV-1 UL47 gene [Fig. 1A(3)] was amplified together by PCR from purified genomic DNA, resulting in a single 400-bp fragment incorporating an EcoRI site at one end, an XbaI site at the other, and a BamHI site engineered in place of the UL47 gene [Fig. 1A(3)]. This was inserted into plasmid pSP72 (Promega) as an EcoRI/XbaI fragment to produce plasmid pGE124 [Fig. 1A(3)]. The UL47 gene from plasmid pMD10 (7) was inserted as a BamHI fragment into the BglII site of pEYFPC1 (Clontech). The NdeI/AgeI fragment of this plasmid was then replaced with the equivalent fragment from pEGFPN1 (Clontech), resulting in a plasmid that carried YFP-UL47 on a BamHI cassette. This BamHI fragment was then inserted into the BamHI site of pGE124 to produce plasmid pGE179 [Fig. 1A(4)], which consisted of YFP-UL47 surrounded by the UL47 flanking sequences, and hence driven by the UL47 promoter.
FIG. 1.
Construction and analysis of HSV-1 expressing YFP-13/14. (A) The UL47 gene (shaded) is located on the Bam F fragment of the HSV-1 genome [(1) and (2)]. The UL47 flanking sequences were amplified by PCR and inserted into plasmid pSP72 as an EcoRI/XbaI fragment to form pGE124, containing a BamHI site in place of the UL47 gene [(3)]. YFP-UL47 on a BamHI cassette was then inserted into the BamHI site of pGE124 to form plasmid pGE179 [(4)], which was introduced back into the virus genome by homologous recombination [(5)]. Note that the downstream BamHI site in pGE179 has been lost in the recombinant virus, because recombination took place within the UL47 gene. (B) Virus DNAs from both the recombinant YFP-13/14-expressing virus (179v) and the parental virus (s17) were digested with either BamHI or EcoRV before analysis by Southern blotting. Hybridization with probes specific for UL47 or YFP indicated that the YFP-UL47 cassette had recombined in the correct location.
Equal amounts (2 μg) of DNA from plasmid pGE179 and the infectious HSV-1 strain 17 were transfected into 106 COS-1 cells grown in a 60-mm dish, using the calcium phosphate precipitation technique modified by substitution of BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline for HEPES-buffered saline. Four days later, the infected cells were harvested into the cell medium and subjected to three rounds of freeze-thawing, and the resulting virus was titrated on Vero cells. Plaques were then screened for possible recombinants by YFP fluorescence.
Virus genomic DNA screening.
Virus DNA for restriction digestion was purified from 5 × 107 infected BHK cells as described previously (9) and was digested for 8 h with either BamHI or EcoRV in the presence of RNase A. Electrophoresis was carried out overnight in 0.8% agarose, and the gel was transferred to a nylon membrane by standard procedures. The Southern blots were then hybridized with a 32P-labeled DNA probe synthesized by the random priming of fragments specific for either UL47 or YFP.
One-step growth curves.
Vero cells grown in a six-well plate (106 cells per well) were infected at a multiplicity of 10 in 1 ml of medium per well. After 1 h (taken as 1 h postinfection), the inoculum was removed, the cells were washed with phosphate-buffered saline (PBS), and 2 ml of fresh medium was added to each well. At 1, 5, 10, 15, and 25 h postinfection, both extracellular virus from the cell medium and intracellular virus from the cells were harvested from one well of infected cells and scraped into 1 ml of PBS. Each virus sample was then titrated on Vero cells.
SDS-PAGE and Western blot analysis.
Solubilized proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were either stained with Coomassie blue or transferred to nitrocellulose filters and reacted with the appropriate primary antibody. A horseradish peroxidase-linked secondary conjugate was utilized, and reactive bands were visualized using enhanced chemiluminescence (ECL) detection reagents (Amersham).
Antibodies.
The polyclonal anti-VP13/14 antibody R220 was kindly provided by David Meredith. The polyclonal anti-VP22 antibody AGV30 has been described previously (8). Antibodies against IE110 (11060) and VP16 (LP1) were kindly provided by Roger Everett and Tony Minson, respectively. The polyclonal anti-GFP antibody and the monoclonal anti- ICP5 antibody were obtained from RDI and Autogen Bioclear, respectively.
Live-cell microscopy and time-lapse analysis.
Cells for short-term live analysis of YFP expression were plated into coverslip chambers and examined using the 488 laser on a Zeiss LSM 410 inverted confocal microscope. Images were obtained by collecting Z sections of each field and merging these to produce a single image. Resulting images were processed using Adobe Photoshop software.
Cells for long-term time lapse analysis were plated onto 42-mm coverslips contained in 60-mm dishes. Prior to analysis, the coverslip was transferred to a Bachhoffer POC chamber (obtained from Carl Zeiss) in open cultivation mode. This chamber was placed on a Saur heated frame (obtained from Carl Zeiss) seated on the microscope and was covered with a Perspex lid through which a constant supply of 5% CO2 was fed. “XYZT” software from Zeiss was used to collect a Z series of images for each time point in the time series; these were then merged to produce an individual Z image for each time point. Animation of the time series was carried out using NIH Image software and saved as a Quicktime video.
RESULTS
Construction of an HSV-1 recombinant expressing fluorescently tagged VP13/14.
The gene expressing VP13/14, gene UL47, is located on the Bam F restriction fragment in the unique long region of the HSV-1 genome [Fig. 1A(1) and (2)]. To construct a virus expressing fluorescent VP13/14, the UL47 gene was initially fused in frame with the C terminus of the gene expressing the YFP version of GFP. This entire fusion gene was then inserted as a BamHI fragment into plasmid pGE124, containing the UL47 flanking sequences joined by a BamHI site [Fig. 1A(3)]. The resulting plasmid, pGE179 [Fig. 1A(4)] was cotransfected into COS-1 cells together with purified HSV-1 strain 17 genomic DNA and incubated for 4 days until cytopathic effect was seen in all the cells. Virus was harvested and plaques were screened for YFP fluorescence to identify potential recombinants. The final virus, designated 179v [Fig. 1A(5)], was plaque purified a total of four times before production of a master stock.
The genomic structure of the recombinant virus was assessed by Southern blotting of BamHI- and EcoRV-digested samples of both Wt S17 and the 179v virus DNA. Hybridization was carried out with both a UL47- and a YFP-specific probe, and indicated that the EcoRV fragment on which UL47 is located had increased in size from 7.5 to 8.3 kb and contained the YFP sequence (Fig. 1B, EcoRV digests). Moreover, the BamHI fragment on which UL47 is located had decreased in size from 8 to 4.9 kb, confirming the introduction of one new BamHI site during the recombination process (Fig. 1B, BamHI digests).
Growth characteristics of the 179v virus.
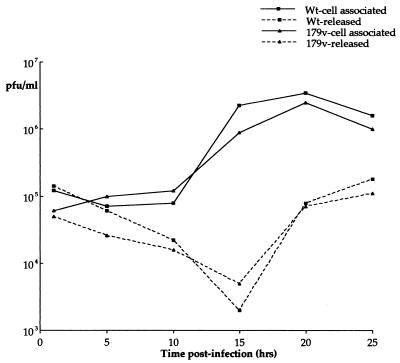
To assess the replication of 179v in comparison to that of Wt virus, one-step growth curves were carried out. Vero cells were infected at a multiplicity of 10, and both cell-associated virus and extracellular secreted virus were harvested every 5 h for 25 h. These virus stocks were then titrated on Vero cells, and the resulting growth curves were plotted (Fig. 2). The results indicate that both the cell-associated virus yield (Fig. 2, cell-associated plots) and the extracellular virus yield (Fig. 2, released plots) from the two viruses were very similar, suggesting that virus assembly and egress are not affected by the tagging of VP13/14 with YFP.
FIG. 2.
The YFP-13/14-expressing virus replicates as efficiently as Wt virus. Vero cells infected with either strain 17 (Wt) or the YFP-13/14-expressing virus (179v) at 10 PFU per cell were harvested every 5 h after infection up to 25 h. The resulting virus was then titrated on Vero cells, and one-step growth curves were plotted. Both intracellular virus (cell associated) and extracellular virus (released) yields were measured.
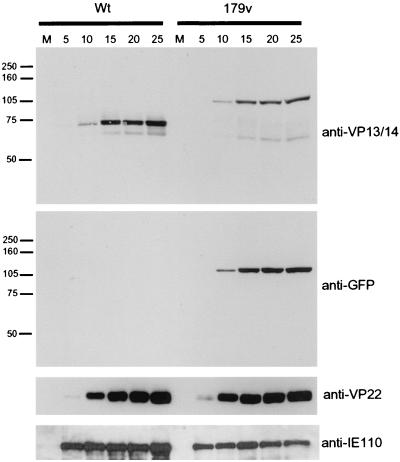
To examine 179v growth in more detail, we analyzed the kinetics of expression of several viral proteins. Vero cells were infected with either Wt or 179v virus at a multiplicity of 10, and total cell lysates were harvested every 5 h for 25 h. Following SDS-PAGE these samples were analyzed by Western blotting using a number of antibodies (Fig. 3). Analysis with the anti-VP13/14 antibody R220 indicated that 179v produced a YFP-13/14 fusion protein of the correct size, approximately 110 kDa (Fig. 3, 179v, anti-VP13/14). Moreover, comparison of the Wt time course with that of 179v indicated that the overall kinetics of VP13/14 synthesis was similar in both viruses, with the protein first detectable by Western blotting at 10 h postinfection (Fig. 3, anti-VP13/14, compare Wt and 179v). In addition, analysis of the 179v time course with both the anti-VP13/14 and anti-GFP antibodies clearly showed that the fusion protein remained intact throughout infection, with no breakdown products detectable (Fig. 3, 179v, anti-VP13/14 and anti-GFP). The kinetics of synthesis of two other viral proteins, namely, the late gene product VP22 and the IE gene product IE110, were also shown to be very similar for 179v and Wt viruses (Fig. 3, anti-VP22 and anti-IE110), confirming that the fusion of the YFP open reading frame to the N terminus of VP13/14 has no apparent deleterious effect on virus growth.
FIG. 3.
YFP-13/14 is synthesized with the same kinetics as VP13/14. Vero cells infected with either strain 17 (Wt) or the YFP-13/14-expressing virus (179v) at 10 PFU per cell were harvested every 5 h after infection, up to 25 h. Equal amounts of total cell lysates were analyzed by SDS-PAGE followed by Western blotting with antibodies R220 (anti-VP13/14), αGFP (anti-GFP), AGV30 (anti-VP22), and 11060 (anti-IE110). M, mock.
YFP-13/14 is efficiently incorporated into 179v virions.
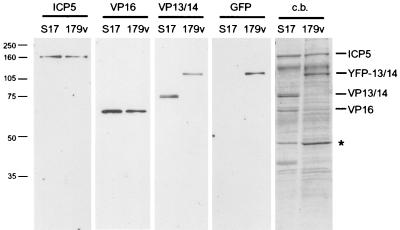
Although VP13/14 is a major structural protein of HSV-1, it has previously been shown to be dispensable for growth in tissue culture (1, 23), and hence virus particles can assemble without incorporating VP13/14. Therefore, to determine if YFP-13/14 was incorporated into the HSV-1 virions produced by 179v infection, we prepared extracellular virions from both Wt- and 179v-infected cells. These virions were purified on a 5 to 15% Ficoll gradient, and approximately equivalent numbers of particles were analyzed by SDS-PAGE followed by either Coomassie blue staining or Western blotting (Fig. 4). Western blotting of the virions with antibody against the major capsid protein ICP5 (Fig. 4, ICP5 panel) indicated that both preparations of virions were loaded in approximately equivalent amounts. The total protein profiles of the purified virions showed that the VP13/14 doublet present in the S17 virions was absent from the 179v virions (Fig. 4, c.b.). Furthermore, a novel doublet band of the correct size for YFP-13/14 was present in the 179v virions (Fig. 4, c.b.). Western blotting of the virions with antibodies against both VP13/14 and GFP confirmed that this additional species in the 179v profile was YFP-13/14 (Fig. 4, VP13/14 and GFP panels) and indicated that there was a slightly lower level of VP13/14 in 179v virions than in s17 virions (Fig. 4, VP13/14 panel). Moreover, Western blotting with an antibody against the tegument protein VP16 implied that the levels of this second tegument protein in the 179v virions may be slightly reduced in the presence of the much larger YFP-13/14 fusion protein (Fig. 4, VP16 panel), a result that is supported by the total protein profiles of the two viruses. There were also a number of other differences between the viruses, most notably a species of around 45 kDa which appears to be much more abundant in the 179v virions. This protein has yet to be identified.
FIG. 4.
YFP-13/14 is efficiently incorporated into the virus particle. Purified extracellular virions from Wt virus (s17) and the YFP-13/14-expressing virus (179v) were solubilized and analyzed by SDS-PAGE followed by either Coomassie blue staining (c.b.) or Western blotting with antibodies against ICP5, VP16, VP13/14, and GFP. ∗, an unidentified protein which is present in 179v virions in greater abundance than in Wt virions.
Cellular localization of fluorescent VP13/14 through HSV-1 infection.
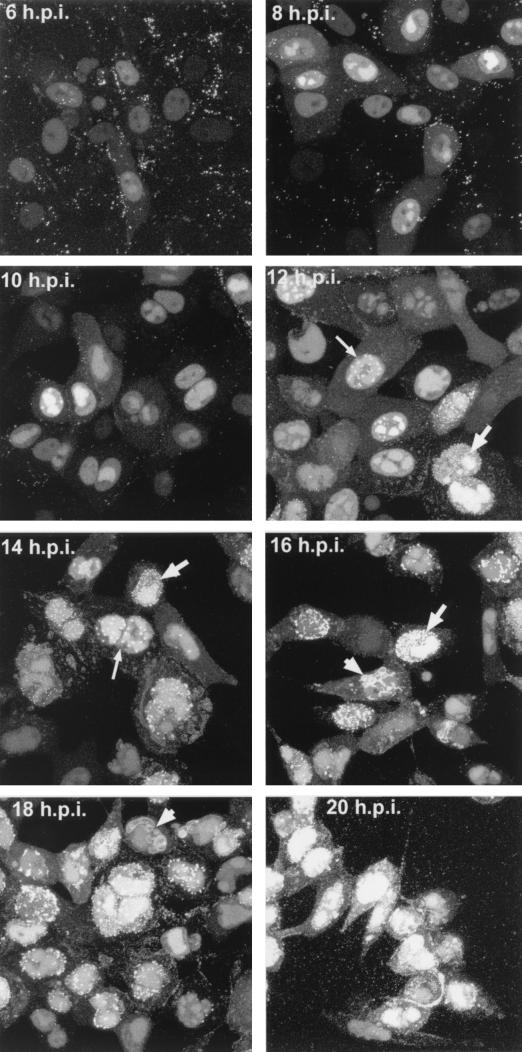
The gene encoding VP13/14, gene UL47, is known to be regulated as a true late gene and therefore is dependent on DNA replication for high levels of expression (12). Thus, we wished to investigate the time in the virus replication cycle at which YFP-13/14 was first detectable within the cell. Vero cells grown in a coverslip chamber were infected with the 179v virus at a multipicity of 10, and representative images of YFP fluorescence within the cells were then collected by confocal microscopy between the times of 4 and 20 h after infection (Fig. 5). Surprisingly, YFP-13/14 fluorescence was detectable within infected cells as early as 4 h postinfection and was clearly visible by 6 h (Fig. 5, 6 h.p.i.). Moreover, at these early stages of infection, YFP-13/14 was localized predominantly in the nucleus, with a low level of cytoplasmic fluorescence (Fig. 5, 6 and 8 h.p.i.). The nuclear pattern was initially diffuse (Fig. 5, 6 h.p.i.), but progressed quickly into a pattern reminiscent of replication compartments (Fig. 5, 8 h.p.i.). By 10 to 12 h, the intensity of YFP-13/14 fluorescence in the nucleus had greatly increased, and over the next 6 h several new patterns of YFP-13/14 localization became apparent. These included large nuclear punctate domains (Fig. 5, thin arrows in 12 and 14 h.p.i.); smaller, more numerous punctate domains at the edge of the nucleus (Fig. 5, thick arrows in 12 to 16 h.p.i.); and perinuclear membrane localization (Fig 5, arrowheads in 16 and 18 h.p.i.). Furthermore, the weak cytoplasmic fluorescence present as a diffuse pattern from 6 to 10 h began to evolve into a more locally concentrated vesicular-type pattern at later times (Fig. 5, 14 and 16 h.p.i.), and by 18 to 20 h there was weak fluorescence at the cell periphery (Fig. 5, 18 h.p.i.), and large numbers of fluorescent particles were clearly visible outside the infected cells (Fig. 5, 20 h.p.i.).
FIG. 5.
Live-cell analysis of YFP-13/14 localization during a high-multiplicity 179v infection. Vero cells grown in a coverslip chamber were infected with the YFP-13/14-expressing virus at 10 PFU per cell and were examined for YFP-13/14 fluorescence every 2 h up to 20 h postinfection (h.p.i.). Images were collected by confocal microscopy, and in each case 10 Z slices have been merged to produce the individual images. Patterns representing large punctate nuclear domains (thin arrows), micropunctate nuclear domains (thick arrows), and perinuclear membrane fluorescence (arrowheads) are indicated.
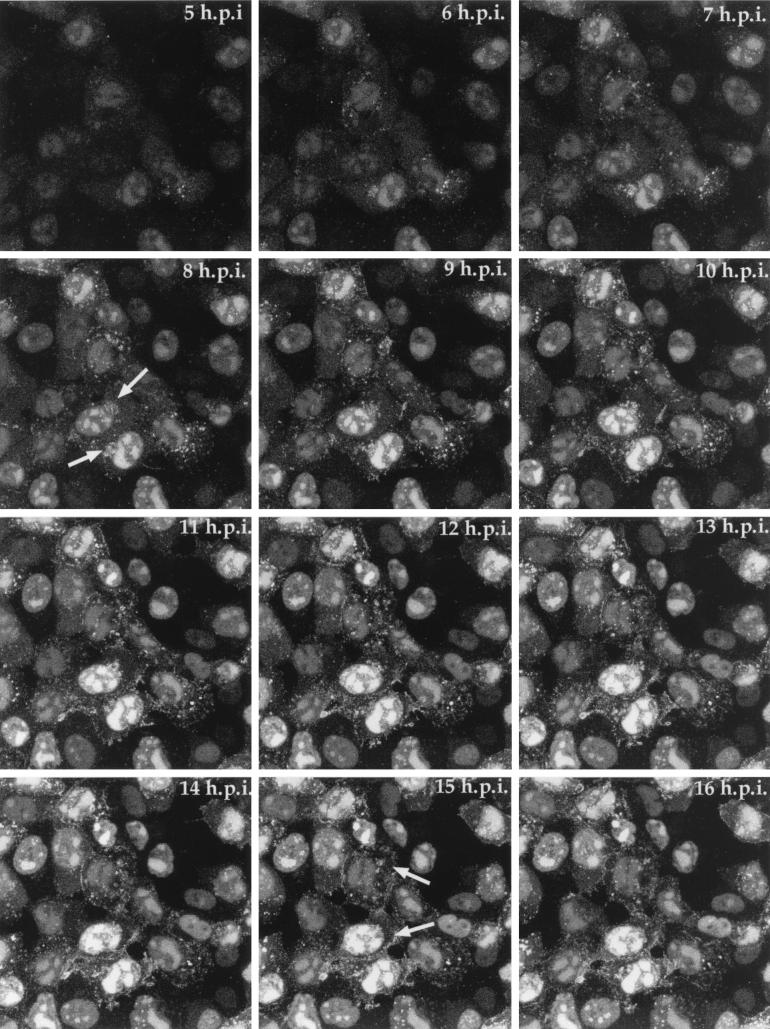
To obtain a more precise indication of YFP-13/14 progression in a single field of infected cells, we carried out time lapse confocal microscopy. Vero cells on a coverslip were infected with the YFP-13/14-expressing virus at a multiplicity of 10, and 5 h after infection the coverslip was transferred to our heated chamber. A single field of cells was chosen for time lapse analysis, and images were collected every 5 min for a further 11 h to form the time series represented both as a gallery of images (Fig. 6) and as an animation (http://www.mcri.ac.uk/VirusAssembly/figure6.mov). Over this period, the YFP-13/14 fluorescence increased greatly in intensity (Fig. 6, compare 5 h.p.i. with 16 h.p.i.), emphasizing the late expression from the UL47 promoter. Moreover, while the vast majority of fluorescence was concentrated from an early stage in the nuclei of infected cells, particularly in replication compartments, it was also possible to discern a weak subpopulation of YFP-13/14 localized in a Golgi-like compartment in several of the infected cells (Fig. 6, arrowed in 8 h.p.i.). Although the relative intensity of this cytoplasmic fluorescence was low in comparison to that of the nuclear fluorescence, the pattern closely resembles that previously observed for VP22 in the GFP-22-expressing virus (9). Moreover, the cytoplasmic fluorescence is particularly obvious in the time lapse animation, where YFP-13/14 can be seen to accumulate at the side of the nucleus and at later times at the periphery of the cell (Fig. 6, arrowed in 15 h.p.i.).
FIG. 6.
Time lapse analysis of YFP-13/14 trafficking in a high-multiplicity infection. Vero cells were infected with the YFP-13/14-expressing virus at 10 PFU per cell and were transferred to a heated chamber at 5 h postinfection (h.p.i.). A single field was chosen for analysis, and images were collected every 5 min for a further 11 h. A time point representing each hour is shown, and the corresponding animation can be found at http://www.mcri.ac.uk/VirusAssembly/figure6.mov.
Nuclear localization of YFP-13/14 during virus infection.
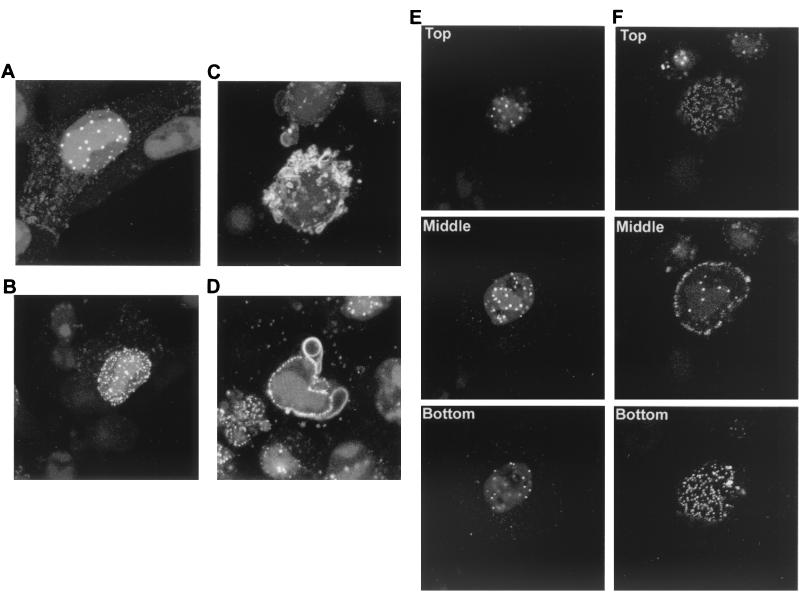
As the most striking aspect of YFP-13/14 localization at the later times shown in Fig. 5 was the range of nuclear and perinuclear locations exhibited by the protein, rather than the weak cytoplasmic fluorescence described above, we looked more closely at cells displaying these patterns. Vero cells grown in a coverslip chamber were infected with the 179v virus at a multipicity of 10, and representative high-magnification images were collected at 14 to 18 h after infection (Fig. 7). As noted above, cells with large nuclear punctate domains became apparent between 10 and 14 h (Fig. 7A). Interestingly, a large number of these intense domains appeared to be localized around the edges of the replication compartments. However, at slightly later time points, cells exhibiting smaller multiple punctate domains in the nucleus became more obvious (Fig. 7B). Furthermore, analysis of individual Z sections of cells exhibiting these two patterns revealed that while the large punctate domains were present in an intranuclear location (Fig. 7E), the small punctate domains were clearly located at the periphery of the nucleus, possibly on the nuclear membrane (Fig. 7F). In addition, the third specific pattern of localization identified in Fig. 5, where YFP-13/14 appears in membrane-like structures around the nucleus, was also evident in this experiment between 16 and 18 h after infection (Fig. 7C and D). Such structures were present either as small projections from the nuclear membrane (Fig. 7C) or as much larger whorls wrapped around part of the nucleus (Fig. 7D, shown as a single Z section for clarity). Taken together, these results reveal a complexity to the intracellular targeting of VP13/14, suggesting that analysis of the YFP-13/14-expressing virus may contribute greatly to our understanding of herpesvirus tegument assembly.
FIG. 7.
YFP-13/14 exhibits novel patterns in and around the nucleus. Vero cells grown in a coverslip chamber were infected with the YFP-13/14-expressing virus at 10 PFU per cell and were examined at around 14 h after infection for YFP-13/14 fluorescence present in either large punctate domains (A and E), micropunctate domains (B and F), or perinuclear membranes (C and D). Images were collected at high magnification and are presented either as a single merged image of 10 Z sections (A, B, and C), as a single Z section (D) or as Z sections from the top, middle, or bottom of the cell (E and F).
DISCUSSION
We have previously described the use of a fluorescently tagged tegument protein in a study of a recombinant herpes simplex virus expressing GFP-tagged VP22 (9). In the present report we have extended these studies to include the tegument proteins VP13/14, the gene products of the true late gene UL47 (12). Here, the UL47 gene was fused in frame to the YFP version of GFP in order to produce a virus that synthesizes YFP-13/14 in place of Wt VP13/14. As with the GFP-22 virus, we have shown that the YFP-13/14 virus replicates as efficiently as Wt virus, confirming that the addition of YFP onto VP13/14 has no effect on virus kinetics. Moreover, YFP-13/14 was efficiently incorporated into virions that were observed as fluorescent particles outside the cell. Thus, the herpesvirus tegument has the capacity to package large fusion proteins into its structure without any detectable effect on the infectivity of the virus.
Our major purpose for developing such fluorescent viruses is to examine the intracellular trafficking of virus structural proteins in live cells and hence to investigate the subcellular compartments involved in virus assembly. We have previously used time lapse confocal microscopy of infected live cells to show that the tegument protein VP22 localizes exclusively to the cytoplasm through the course of a high-multiplicity infection, with little if any protein detectable in the nucleus (9). Such studies revealed a reproducible trafficking pathway of VP22 through the replication cycle, beginning with diffuse cytoplasmic GFP-22 and progressing into particulate VP22 detectable in a perinuclear location that we believe to be the Golgi apparatus. GFP-22 then moves to the cell surface in vesicle-like structures, culminating in the release of fluorescent GFP-22-containing virions into the extracellular space. By contrast, we show here that YFP-13/14 is clearly targeted to the nucleus during the early stages of its expression. Moreover, as infection progresses, the concentration of YFP-13/14 in the nucleus increases greatly, suggesting that even at later times in infection VP13/14 subcellular targeting remains largely unaltered. While a subpopulation of VP13/14 does appear in the cytoplasm in the latter half of infection, the behavior of this low-level cytoplasmic fluorescence is difficult to discern, and it is most obvious in the time lapse experiment from which images are shown in Fig. 6. However, fluorescent YFP-13/14-containing particles, similar to those observed in the GFP-22 virus infection, do appear in the extracellular space, confirming that at some stage along the YFP-13/14 trafficking pathway, the protein is packaged into assembling virions.
The exclusive cytoplasmic location of GFP-22 had previously led us to propose that tegument proteins are incorporated into the virion at a nonnuclear location, probably within the secretory pathway of the cell. The fact that only a small proportion of infected-cell VP13/14 is located within the cytoplasm may imply that there are two populations of VP13/14 in the infected cell, one which is targeted to the nucleus for a specific, nonstructural purpose and one which is targeted to the cytoplasm at a later stage of infection for virus assembly. It is noteworthy that in a paper accompanying this report we have demonstrated that the nuclear localization of VP13/14 is an intrinsic property of the protein determined by a 14-residue nuclear localization signal present at its N terminus (7). Thus, if VP13/14 is differentially targeted to the cytoplasm for the purpose of virus assembly, it would have to be retained there by a dominant interaction with another virus protein or by a differential modification causing an alteration in its subcellular targeting. Alternatively, the nuclear targeting of VP13/14 may imply that a subset of tegument proteins (represented by VP13/14) is acquired by the capsid in the nucleus, while the remaining tegument proteins (represented by VP22) are acquired within the cytoplasm. In this case the cytoplasmic population of YFP-13/14 fluorescence would truly represent virus particles in the process of assembly. Hence, our YFP-13/14-expressing virus has further revealed the complexity of herpesvirus assembly and maturation, and provides us with a new tool for investigating the mechanisms involved in these processes.
It is clear that newly synthesized VP13/14 in the infected cell is immediately targeted to the nucleus, where it appears initially diffuse, but rapidly localizes to replication compartments. Furthermore, at later times in infection, between 12 and 16 h, fluorescent VP13/14 appears in large punctate domains within the nucleus, a pattern that is also observed during transient expression (7). While these domains may represent one of several previously identified intranuclear sites in the infected cell, their location at the periphery of the replication compartments, together with the timing of their appearance, suggests that they resemble the structures previously described by Ward and coworkers, which they termed assemblons (19). Immunofluorescence has been used to show that these structures contain several capsid proteins, and thus they have been designated sites of capsid assembly (19). Moreover, Desai and Person have also reported the localization of GFP-tagged capsid protein, VP26c, to similar compartments in the infected cell nucleus (6). The presence of VP13/14 in these structures may imply that it plays a role in capsid assembly within the nucleus and/or that VP13/14 is assembled into the virus particle at these sites. At even later times in infection, VP13/14 becomes predominantly present in small punctate domains localized around the outside of the nucleus, and these are gradually replaced by a pattern of fluorescent cytoplasmic membranous structures encircling the nucleus. While we do not yet understand the relationship between these various nuclear and membrane-like patterns of VP13/14 localization, it is tempting to speculate that one or more of them may represent sites of incorporation of this particular tegument protein into the virus particle.
It is now apparent that the two tegument proteins VP22 and VP13/14 traffic quite differently within the infected cell. Moreover, analysis of a recently constructed virus expressing GFP-tagged VP16 may add to this complexity of tegument assembly. Preliminary results indicate that GFP-VP16 exhibits localization properties combining those of GFP-22 and YFP-13/14, whereby GFP-VP16 is nuclear at early stages of infection but becomes increasingly cytoplasmic, in what appears to be a Golgi-like location, at later time points (P. O'Hare, personal communication). At some stage in the infected cell, these individual tegument proteins must converge to form the full tegument of the newly assembling virion, and it is not until this point is reached that viral envelopment can occur. With the knowledge that many of these tegument proteins can be tagged with a fluorescent protein without any detrimental effect on virus growth, it may now be possible to investigate herpesvirus tegumentation in more detail by constructing viruses labeled with two or more fluorescent proteins.
ACKNOWLEDGMENTS
We thank Tony Minson, Roger Everett, and David Meredith for the antibodies LP1, 11060, and R220, respectively.
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Barker D E, Roizman B. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology. 1990;177:684–691. doi: 10.1016/0042-6822(90)90534-x. [DOI] [PubMed] [Google Scholar]
- 2.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M E, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 4.Coulter L J, Moss H W, Lang J, McGeoch D J. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham C, Davison A J, Dolan A, Frame M C, McGeoch D J, Meredith D M, Moss H W, Orr A C. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J Gen Virol. 1992;73:303–311. doi: 10.1099/0022-1317-73-2-303. [DOI] [PubMed] [Google Scholar]
- 6.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly M, Elliott G. Nuclear localization and shuttling of the herpes simplex virus tegument protein VP13/14. J Virol. 2001;75:2566–2574. doi: 10.1128/JVI.75.6.2566-2574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 9.Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson W, Roizman B. Proteins specified by herpes simplex virus. X. Staining and radiolabeling properties of B capsid and virion proteins in polyacrylamide gels. J Virol. 1974;13:155–165. doi: 10.1128/jvi.13.1.155-165.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKnight J L C, Pellett P E, Jenkins F J, Roizman B. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing-factor-dependent activation of α genes. J Gen Virol. 1987;61:1531–1574. doi: 10.1128/jvi.61.4.992-1001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 13.Nii S, Morgan C, Rose H M. Electron microscopy of herpes simplex virus. II. Sequence of development. J Virol. 1968;2:517–536. doi: 10.1128/jvi.2.5.517-536.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Hare P, Goding C R, Haigh A. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overton H A, McMillan D J, Klavinskis L S, Hope L, Ritchie A J, Wong-kai-in P. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology. 1992;190:184–192. doi: 10.1016/0042-6822(92)91204-8. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz J, Roizman B. Concerning the egress of herpes simplex virus from infected cells. Electron microscope observations. Virology. 1969;38:42–49. doi: 10.1016/0042-6822(69)90126-3. [DOI] [PubMed] [Google Scholar]
- 17.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Genderen I L, Brandimarti R, Torrisi M R, Campadelli G, van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 19.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R., II Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Mcknight J L. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Sirko D A, McKnight J L. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J Virol. 1991;65:829–841. doi: 10.1128/jvi.65.2.829-841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]