Significance
The mineralocorticoid receptor (MR) is critical for regulation of whole-body mineral and water homeostasis by regulating gene transcription in response to changes in circulating aldosterone levels. We show that coexpression of the closely related glucocorticoid receptor (GR) potentiates MR/aldosterone-regulated transcription, even in the absence of physiological agonists of GR. GR appears to exert its effects by stabilizing productive MR binding to chromatin, rather than altering the genome-wide distribution of the receptor. Our results implicate GR in the modulation of MR and suggest a mechanism that may underlie functional cross-modulation among other members of the subfamily 3 of nuclear receptors.
Keywords: steroid receptors, heteromerization, chromatin binding, RNA-seq, single-molecule tracking
Abstract
The glucocorticoid and mineralocorticoid receptors (GR and MR, respectively) have distinct, yet overlapping physiological and pathophysiological functions. There are indications that both receptors interact functionally and physically, but the precise role of this interdependence is poorly understood. Here, we analyzed the impact of GR coexpression on MR genome-wide transcriptional responses and chromatin binding upon activation by aldosterone and glucocorticoids, both physiological ligands of this receptor. Transcriptional responses of MR in the absence of GR result in fewer regulated genes. In contrast, coexpression of GR potentiates MR-mediated transcription, particularly in response to aldosterone, both in cell lines and in the more physiologically relevant model of mouse colon organoids. MR chromatin binding is altered by GR coexpression in a locus- and ligand-specific way. Single-molecule tracking of MR suggests that the presence of GR contributes to productive binding of MR/aldosterone complexes to chromatin. Together, our data indicate that coexpression of GR potentiates aldosterone-mediated MR transcriptional activity, even in the absence of glucocorticoids.
Adrenal glands coordinate physiological responses to cope with stress, acute injury, or prolonged deprivation of water and food. Two important categories of adrenal hormones mediate key specific homeostatic responses: glucocorticoids (cortisol and corticosterone) and mineralocorticoids (aldosterone). However, these hormones show significant promiscuity. An excess of glucocorticoid signaling produces mineralocorticoid-like effects, particularly hypertension (1). Conversely, an excess of mineralocorticoids can mimic glucocorticoid effects, such as glucose homeostasis dysregulation and development of metabolic syndrome (2). The molecular basis for this cross-talk is at least partially explained by the close evolutionary relationship between the mineralocorticoid receptor (MR) and the glucocorticoid receptor (GR), which confers poor ligand specificity and overlapping modes of action (3, 4). Both mineralocorticoid and glucocorticoid hormones potently activate MR, while glucocorticoids also activate GR (5). Since glucocorticoids circulate at concentrations several orders of magnitude higher than aldosterone, certain cells coexpress MR with 11-β-hydroxysteroid dehydrogenase type 2 (11-β-HSD2), an enzyme that metabolizes glucocorticoids into their biologically inactive 11-keto metabolites, creating a low-glucocorticoid milieu (6). In contrast to MR, GR is partially selective, with potent activation by glucocorticoids and weak activation by mineralocorticoids, even though aldosterone binds with similar high affinity [Kd ≈ 14 nM, (7)]. GR expression is essentially ubiquitous, while MR expression is also widespread but generally at a lower abundance, except in the hippocampus and aldosterone-target epithelia such as the renal collecting duct and distal colon, where MR and GR abundance is similar (8). Pharmacological approaches or the use of mouse models with selective knockout of MR or GR in tissues that coexpress both receptors conclusively demonstrate the mutual influence of MR and GR in determining glucocorticoid signaling outcomes (9–11). This, together with coexpression or not of 11-β-HSD2, generates at least three scenarios for corticosteroid hormone receptor function: GR-mediated responses to glucocorticoids; GR/MR-mediated responses to glucocorticoids; MR-mediated responses to aldosterone in the presence of presumably inactive GR.
Both MR and GR share a highly conserved DNA-binding domain (DBD), which implies that they recognize with high affinity the same DNA consensus sequence, known as “Hormone Response Element” (HRE) (12), and likely regulate a partially overlapping set of genes. The largest differences in the amino acid sequences of MR and GR proteins occur in the N-terminal domain (NTD) with Mus musculus MR containing over 100 more amino acids than M. musculus GR and only 29% similarity (SI Appendix, Fig. S1A). By contrast, the DBD and the ligand-binding domain of MR and GR have 90% and 69% similarity, respectively. The NTD of MR is important for gene regulation (13, 14). The AF1 activation regions of the MR NTD appear to be separated into two distinct amino acid sequences, AF1a and AF1b, similar to those of the androgen or progesterone receptors but unlike that of GR, which has one central AF1 domain in its NTD (14–16). In addition, MR is reported to have a region with intrinsic inhibitory function placed between AF1a and AF1b (16). The divergent structure of MR and GR NTDs may account to some extent for differential, tissue-specific transcriptional responses (17, 18).
To further complicate the picture, it has been conclusively demonstrated that MR and GR can physically interact to form heteromers (19–26). Examination of the functional properties of MR/GR interaction has produced conflicting experimental results. Gene-reporter assays or studying the expression of specific genes indicate that GR may enhance MR transcriptional activity in certain cell lines (27, 28), although it appears to be inhibitory or noninfluential in others (19, 22, 29, 30). A study performed in keratinocytes demonstrated that MR coexpression alters GR genomic binding but has a relatively low contribution to the global transcriptional response to the synthetic glucocorticoid dexamethasone (9). The fact that GR expression is ubiquitous and generally higher than MR (8) implies that MR will typically function in the presence of significant levels of GR. The functional effects of this coexpression are unclear. Data obtained in vivo suggest that both receptors may be needed for potent aldosterone biological effects (31, 32). However, there are no studies to date directly analyzing the global influence of GR on MR-mediated transcriptional responses, whether driven by aldosterone or glucocorticoids.
Given the physical interaction between MR and GR, the molecular basis for their specific physiological roles, overlapping functions and pathological consequences of dysregulation can only be understood after clearly defining the consequences of coexpression in genome-wide studies. Answering this question requires a cell model where the expression and activation of both receptors can be tightly controlled. To this end, we initially took advantage of a well-characterized cellular system to study MR chromatin binding, gene regulation, and single-molecule dynamics in the presence or absence of GR. Our results indicate that GR profoundly affects MR genome-wide chromatin binding in a locus- and ligand-specific way and generally potentiates MR/aldosterone (MR/Aldo)-mediated gene transcription. This was confirmed using colon organoids derived from wild type or inducible GR knockout (GRKO) mice, a physiologically relevant model for MR/Aldo action. Single-molecule tracking (SMT) experiments indicate that the presence of GR increases the fraction of chromatin-bound MR in the low-mobility state correlated with active transcription. Together, these data shed light on the interplay between MR and GR, with GR potentiating the activity of MR in response to aldosterone.
Results
MR Transcriptional Activity Is Potentiated by the Presence of GR.
To study the genome-wide function of mammalian MR both in the presence and absence of the closely related GR, we stably introduced GFP-tagged MR into a well-characterized GRKO C127 mouse mammary tumor cell line and its parental line that expresses GR endogenously (33). The GFP-tag of the integrated MR is inserted after residue 147 of the N terminus of the receptor, which has been shown to optimize its hormone response over an N-terminal GFP tag and it does not interfere with translocation of MR to the nucleus upon Aldo treatment [SI Appendix, Fig. S1 B and C; (34)]. We then performed genome-wide total RNA sequencing (RNA-seq) from the two MR cell lines before and after hormone treatments with 10 nM aldosterone (Aldo) or with 100 nM corticosterone (Cort), both saturating concentrations for MR (7). We compared differential expression (DE) of genes in response to 2 h of exposure to either hormone from each cell line versus vehicle. We chose a false discovery rate (FDR) cutoff of 0.01 as determined by DESeq2 (via Homer) (35) to determine which genes show a change in exon RNA levels after treatment using two to three biological replicates per condition (SI Appendix, Fig. S2 A and B).
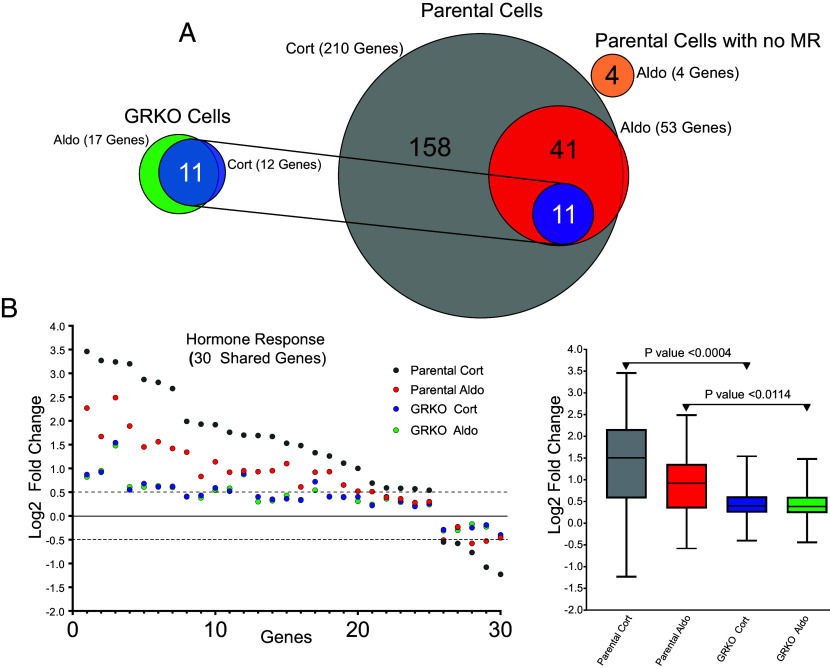
The parental cells with endogenous GR treated with Cort exhibit the strongest hormone response in both the number of up- or down-regulated genes and quantitative changes in RNA levels after treatment (Dataset S1). We categorized protein-coding genes that met our FDR cutoff and exhibited an absolute hormone-dependent log2 fold change in RNA levels of at least 0.5. The parental cells treated with Cort had 210 hormone-responsive genes compared to 53 genes when treated with Aldo, 52 of which are common to both hormone treatments (Fig. 1A). The GRKO cells exhibited a much-reduced response to both Cort and Aldo with 12 and 17 genes changed, respectively, 11 of which are common to both treatments and also overlap with the common responsive genes in the parental cells (Fig. 1A). Among all shared genes between the two cell lines that meet the FDR criterion regardless of fold change (30 genes), the hormone response in the GRKO cells is attenuated compared to the cells with GR (Fig. 1B). Among these 30 genes, the hormone-dependent fold change in the GR-expressing cell line is lower with Aldo treatment compared to Cort; however, in the GRKO cells the level of hormone response is lower compared to that in the parental cells, regardless of which hormone the cells were treated with. This indicates that MR by itself, in this cell line, is a poor transcriptional regulator with either Cort or Aldo. The higher gene response of the parental cells with Cort treatment can be primarily attributed to GR. The number of Cort-responsive genes is similar to that obtained in these cells when treated with dexamethasone (36). Most importantly, the gene response of the parental cells is also greater for Aldo treatment, suggesting that MR regulates genes better with its natural ligand in concert with Aldo-liganded GR. RNA-seq data from Aldo-treated parental cells without MR show only four genes that meet the FDR and FC cutoffs, none of which overlap with hormone-responsive genes in the MR-expressing cell lines (SI Appendix, Fig. 1A and Dataset S1). This shows that GR by itself cannot induce a significant transcriptional response when liganded to Aldo, only in conjunction with MR.
Fig. 1.
MR transcriptional response in the presence or absence of GR. (A) Venn diagrams of hormone-regulated protein-coding genes (2 h treatment/vehicle) to 100 nM Cort or 10 nM Aldo. Total number of hormone-responsive genes (FDR ≤ 0.01, Log2 FC ≥ ± 0.5) denoted in parentheses for MR-expressing GRKO cells, MR-expressing parental cells or parental cells without MR (data from 2 to 3 independent replicates). Circles connected with lines denote 11 hormone-responsive genes common to the two cell lines. (B) Scatter plot of Log2 FC for all shared genes meeting the FDR 0.01 cutoff, regardless of fold change. Box and whiskers plot of the same data displays interquartile range (IQR) depicting the 25th, 50th, and 75th percentile as box with the median as black bar. The whiskers mark the most induced and repressed genes. Statistical analysis was performed using a two-tailed unpaired t test.
Coexpression of GR Alters MR Genome-Wide Binding in Locus-Specific and Ligand-Specific Ways.
To explore how receptor transcriptional activity is related to chromatin binding, we collected genome-wide datasets for MR chromatin immunoprecipitation (ChIP) before and after hormone treatments in the GRKO and parental cell lines. Additionally, the same chromatin preparations from the MR-expressing parental cells treated with Aldo or Cort were also used to ChIP endogenous GR.
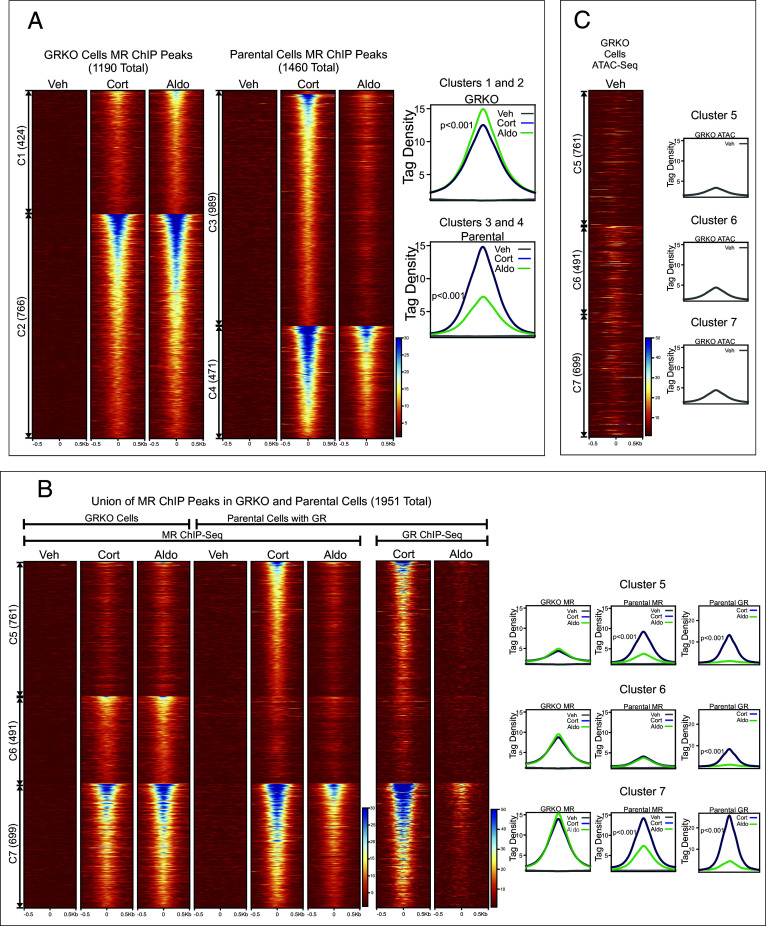
We performed two independent replicates of ChIP-seq for MR in the GRKO cells and the parental cells to determine how the presence of GR may affect MR binding across the genome. We treated cells with vehicle, Aldo, or Cort for 1 h prior to sample collection to detect differences in MR chromatin binding with Aldo (MR specific) or with Cort, which activates both MR and GR (5, 37, 38). We took the union of MR peaks from each ChIP replicate (separately for each cell line/hormone treatment) and performed a Pearson correlation (SI Appendix, Fig. S2D). We used only MR ChIP peaks separately present in both replicates of each condition for further analysis (Fig. 2A).
Fig. 2.
Chromatin binding of MR. (A) Comparison of MR binding after 1 h treatment with vehicle, 100 nM Cort, or 10 nM Aldo in two cell lines (GRKO with no GR or parental with endogenous GR; N = 2). Heatmaps of merged replicate data represent ±500 bp around the center of the MR peak. ChIP-seq intensity scale is noted Lower Right on a linear scale. Clusters of peaks (C1 to C4) are labeled on the left with the peak number in parentheses and are sorted from high to low signal for the condition with the highest overall signal. Aggregate plots represent total ChIP-seq tag density of all peaks normalized as reads per genomic content (1× normalization). Denoted P-values correspond to comparison of aggregate plots for Cort/Aldo treatments calculated using a Wilcoxon Signed-Rank test. (B) A union list of MR ChIP peaks from panel A was created (Materials and Methods) and clustered by cell line and hormone treatment. 1,951 unique MR peaks are distributed into parental-specific/Cort (C5), GRKO-specific/Aldo (C6), or shared between two cell types (C7). Aggregate plots and heatmaps are displayed as described for A for each cluster and treatment. (C) ATAC-seq data are from untreated GRKO cells with stably expressed GFP-GRwt (33). The ATAC data heatmap is sorted the same as ChIP data in B and intensity scale is noted Lower Right on a linear scale.
GRKO cells treated with Aldo have 13% higher aggregate MR chromatin binding than with Cort, as seen by normalized ChIP-seq signal intensity (Fig. 2A, aggregate plots). A union list of 1,190 called MR peaks in the GRKO cells distributes into two clusters: 424 Aldo-specific peaks (cluster 1) and 766 peaks that occur with either treatment (cluster 2). The GRKO-Cort condition (cluster 1) shows some low levels of MR binding that falls just below our peak calling criteria (Materials and Methods). MR binding in the parental cells with endogenous GR stands in contrast to its binding in the GRKO cells, as Cort induces 40% higher MR binding than Aldo (Fig. 2A, aggregate plots). A union list of 1,460 MR peaks in the parental cells distributes into two large clusters: 989 Cort-specific peaks (cluster 3), and 471 Aldo/Cort-shared peaks (cluster 4). Again, some low levels of MR binding in the parental cells can be seen in the Aldo condition that do not meet our peak-calling criteria (cluster 3).
A comparison of genome-wide MR binding between the two cell lines indicates how much GR influences MR chromatin binding and how this influence is likely related to MR–GR receptor interactions. When we combine the two sets of MR ChIP peaks across the two cell lines with either ligand, there are a total of 1,951 unique peaks that break into three clusters (Fig. 2B, clusters 5 to 7; Dataset S2). The 1,190 MR peaks in the GRKO cells distribute into clusters 6 and 7 while the 1,460 MR peaks in the parental cells distribute into clusters 5 and 7. We performed a paired Wilcoxon signed rank test and calculated the fraction of MR peaks that have a P-value ≤ 0.01 for each compared hormone condition (SI Appendix, Fig. S3A; see Materials and Methods). The Cort-treated parental cells have 761 unique MR peaks (cluster 5) that do not occur in the GRKO cells with either ligand, suggesting Cort-liganded GR is required to enable MR binding at these sites. This requirement for Cort-liganded GR does not necessarily signify direct interaction or interdependence of the two nuclear receptors. GR binding could simply cause chromatin accessibility changes at these sites which could then enable MR binding; however, binding site clusters 6 and 7 provide more compelling evidence for receptor interaction. Cluster 6 has 491 peaks specific to the GRKO cell line, indicating that in the parental cells Cort- or Aldo-liganded GR may inhibit MR binding at these sites. The cluster 6 aggregate MR binding signal in the parental cells is approximately 60% lower than the respective GRKO binding and is below our peak-calling threshold for both hormones. The cluster 6 endogenous GR ChIP-seq signal in the parental cells also has very low binding with Cort treatment and virtually no binding with Aldo, suggesting that Aldo-liganded GR cannot efficiently bind these sites (Fig. 2B). It is, thus, unlikely that GR is simply outcompeting MR for these sites (39). The 699 MR binding sites shared between the two cell lines (cluster 7) exhibit the highest signal intensity; however, the sites show 35% less aggregate intensity in the parental cells treated with Aldo versus Cort. This observation suggests that after 10 nM Aldo treatment, GR not only binds chromatin poorly but may also inhibit MR binding via receptor interaction. A 71% decrease in aggregate binding intensity (all clusters) of Aldo-liganded GR (versus Cort-liganded GR) is directly shown by ChIP-seq of the parental cells with a GR antibody (Fig. 2B).
Taken together, these data suggest that liganded GR has a dominant effect on MR interactions with chromatin. GR potentiates MR binding at some sites inaccessible to MR by itself (cluster 5) and reduces MR binding at sites it can bind when acting in the absence of GR, either with both ligands (cluster 6) or only for Aldo (cluster 7). These effects on MR binding at clusters 6 and 7 could be due to GR reducing chromatin accessibility of the binding site or, more likely, due to mixed receptor heteromers having different binding efficacies. Very few GR binding events have been shown to reduce chromatin accessibility at the site of direct receptor binding (40). It appears that the type of ligand, Aldo or Cort, also affects chromatin binding efficiency of both receptors, which is reduced when they are coexpressed and stimulated with Aldo. Despite the lower MR ChIP signal in the parental cells compared to the GRKO cells, it has the second-highest transcriptional response, behind the parental cells treated with Cort (Fig. 1). This suggests that receptor binding alone is not sufficient to induce a transcriptional response.
Motif Analyses Show That MR Binding Favors Consensus NR3C1-4/AP1 Motifs and Is Affected by GR.
We performed motif analyses on clusters 5 to 7 from the 1,951 MR binding sites that occur across the two cell lines/treatments. We queried the MR binding sites against known motifs in the Homer database (41) and selected the most commonly returned motifs (Dataset S3). This included three motifs that reflect the 13mer NR3C1-4 steroid receptor consensus sequence GnACAnnnTGTnC to use as a proxy for an MR binding motif. The three Homer MR motifs range in the stringency to the above consensus sequence primarily at the positions 3 and 5 “A”, position 9 “T,” or position 13 “C,” while the other positions are well conserved.
The 761 MR peaks of cluster 5 that only appear in the parental cells with Cort treatment had some form of MR-like binding motif between 35 to 61% of sites (Fig. 3 and Dataset S3). The prevalence of the consensus steroid hormone receptor motif likely enabled both GR and, subsequently, MR binding (39). The AP1-like nTGAnTCAn motif (Fig. 3 and Dataset S3) occurred between 3 to 5% of sites while THRb, Runx1, ZNF domain, and ETS motifs were also detected. Compared to cluster 5, the 491 GRKO-specific MR peaks of cluster 6 were less enriched for MR-like consensus motifs (10 to 23% of sites) while AP1-like motifs occurred more often (6 to 14% of sites; Fig. 3 and Dataset S3). The 699 MR peaks of cluster 7 that are shared across the two cell lines were slightly less enriched than those in cluster 5 for MR-like binding motifs (33 to 57% of sites) and cluster 6 for AP1-like motifs (5 to 12% of sites).
Fig. 3.

Motif analysis of MR binding. Figure shows position weight matrix logos for known motifs from the HOMER database. For Clusters 5 to 7, the percentage of MR binding sites that contain the designated known motif. See also Dataset S3 for numbers of sites containing a designated motif and background information.
The motif analyses align with the MR ChIP-seq data in that the GR-dependent cluster 5 MR peaks are more enriched than cluster 6 peaks for NR3C1-4 motifs. Our previous studies have shown GR to be capable of binding to inaccessible, nucleosomal sites with GRE consensus motifs prior to hormone stimulation (36, 42) whereas GRKO-specific sites (cluster 6) may more often have factors like AP1 bound prior to hormone that enable MR binding (43). This is reflected in previously published chromatin accessibility data of GRKO cells that show higher signal in the assay for transposase-accessible chromatin (ATAC) in cluster 6 prior to hormone than in cluster 5 [Fig. 2C; (33)]. Cluster 7 MR sites exhibit the highest receptor binding intensity and have relatively higher enrichment of NR3C1-4 motifs than cluster 6 and higher AP1 consensus motifs than cluster 5. Like cluster 6, cluster 7 also exhibits higher prehormone accessibility than cluster 5 (Fig. 2C).
Overall, the motif analyses show that the shared peaks (cluster 7) contain both a higher frequency of MR-like motifs and more accessible chromatin prior to hormone. These characteristics likely explain why cluster 7 has the highest ChIP signal intensities. When MR is present alone in the GRKO cells, it binds more promiscuously at sites with fewer MR-like motifs (cluster 6).
Intergenic eRNAs Correlate with MR-Mediated Gene Transcription.
The RNA-seq data are not in agreement with the MR and GR ChIP-seq if we consider overall ChIP signal (binding intensity and number of peaks) to correlate with gene response (44). Among the four experimental conditions (two cell lines and two hormones), the parental line treated with Aldo generates the fewest MR and GR peaks but exhibited the second-largest transcriptional response. The GRKO cell line shows fewer regulated genes with lower fold-changes in response to Aldo despite producing more ChIP peaks compared to that treatment in the GR-containing parental line. Within each cell line, the ChIP and the RNA results do correlate, as Aldo induces slightly more genes and more ChIP peaks in the GRKO cells while Cort does the same in the parental line, in this case likely due to the action of GR/Cort.
We used Homer to annotate the MR peaks in the three clusters of Fig. 2B to the closest gene in cis and detected if these nearby genes are among the 210 responsive genes of the parental cells treated with Cort. This is the most inclusive and repeatable set of genes defined as hormone responsive with either ligand. Among the 699 GRKO/parental shared peaks of cluster 7, 83 were closest to these 210 genes, while 56 and 27 peaks were closest to these same genes in clusters 5 and 6, respectively (Dataset S2). These results are in line with those obtained by Ueda et al., which identified 25 out of 1,414 ChIP-seq peaks placed proximal to aldosterone-regulated genes, as determined by microarray analysis, including common MR/GR targets such as Sgk1, Tsc22d3, and Tns1 (45). Hormone-responsive genes were sometimes detected near peaks from more than one cluster (ex. Tns1) or even all three clusters (ex. Ampd3, Tgm2) (Dataset S2). Of the 11 common MR responsive genes in GRKO cells, 10 are linked to annotated peaks in clusters 1, 2, or 3. The remaining gene, Ccn2 (coding for the connective tissue growth factor), a known MR/Aldo target gene in the heart (46), has a nearby distal peak in cluster 3, but with a nonhormone-responsive gene occurring closer to it detected by the Homer peak annotation. MR binding at several loci near a hormone-responsive gene likely contributes to its transcriptional response and these loci may occur in more than one of the clusters in Fig. 2A. However, the strongest MR binding sites (cluster 7), as measured by ChIP signal intensity, are associated in cis to the highest number of hormone-responsive genes.
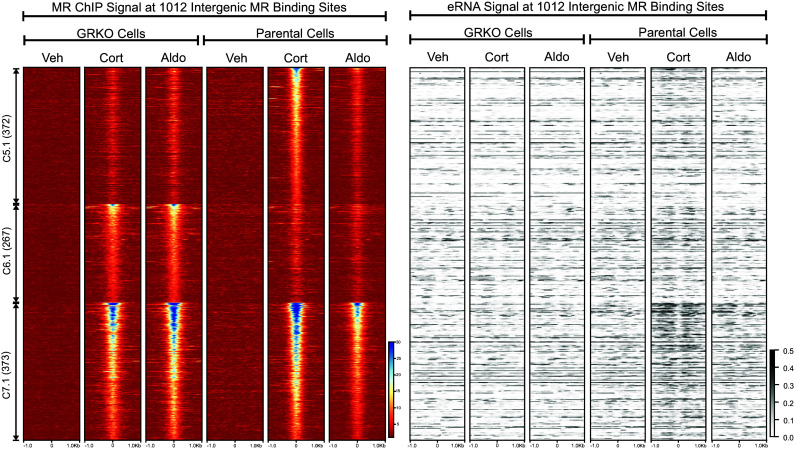
Active enhancers produce short bidirectional RNAs, known as enhancer RNAs (eRNAs), at sites of transcription factor binding near actively transcribing genes (47, 48), with previous evidence linking GR binding to eRNA transcription (49, 50). We used the total RNA-seq data to look for eRNAs at intergenic MR ChIP peaks as an indication of such activity. We left out MR peaks annotated to intron, UTR, exon, and promoter sites to avoid RNA signals made by transcription near or within gene bodies. Of the 1,951 MR peaks that make up clusters 5 to 7, 1,012 are classified as intergenic according to Homer. We plotted the intergenic eRNA signal at these peaks sorted as a subset of each cluster in Fig. 2B and by ChIP signal intensity (Fig. 4). The eRNA heatmaps show little hormone-dependent change in signal at intergenic peaks in the GRKO cells. However, a statistically significant increase in eRNA levels (SI Appendix, Fig. S3B) correlates with MR ChIP signal intensity at all three clusters, with cluster 7.1 in particular showing more change in overall eRNA signal than either cluster 5.1 or cluster 6.1. Thus, the eRNA signal correlates with MR ChIP binding signal in the parental cells with GR present, but not in the GRKO cells. The overall eRNA signal also correlates with the overall transcriptional response at the gene level.
Fig. 4.
eRNA signals at intergenic MR chromatin binding peaks. The left heatmap shows subsets of clusters 5 to 7 (Fig. 2B) representing intergenic MR ChIP peaks. Heatmaps represent ±1 kbp around the center of the MR peak. ChIP-seq intensity scale is scaled linearly (Lower Right). Clusters of peaks are labeled on the left with the peak number in parentheses and are sorted from high to low signal for the condition with the highest overall signal. The Right heatmap shows total normalized RNA-seq signal from merged replicates in the same order and breadth as the MR-ChIP heatmaps. RNA-seq intensity scale is noted Lower Right on a linear scale.
The MR NTD Contributes to Transcriptional Activity.
Comparison between the transcriptomic and chromatin binding data demonstrates that MR binding alone (ChIP signal intensity) in the absence of GR is not sufficient to induce a robust transcriptional response at the gene level or even of eRNAs at sites of MR binding. This could be due to a few possibilities. i) MR requires a cofactor that may be present in cell types that naturally express MR but is missing in our cell lines. However, the extent of the MR gene response to Aldo in the parental cells argues against this being a wide-ranging limitation to MR activity. ii) MR requires the involvement of GR and works better in concert with it to produce a transcriptional response. The greater hormone response of the parental cells that express endogenous GR agrees with this inference. iii) The unique structure of the NTD of MR may convey an inhibitory effect on steroid-responsive genes similar to that shown by Litwack and colleagues on the MR-responsive Na/K ATPase β1 gene (29) or through its recruitment of particular corepressors as shown by Lombes and colleagues (16).
To further explore how the NTD of MR affects its genome-wide hormone response, we performed RNA-seq using an NTD-truncation mutant (MR-580C) in both the GRKO and parental cell lines (SI Appendix, Fig. S4A). We categorized protein-coding genes that met our FDR cutoff (0.01) and exhibited an absolute hormone-dependent Log2 fold change of at least 0.5. Like the full-length version of MR (MRwt), the MR-580C mutant exhibited a low number of regulated genes in the GRKO cells with 11 and 20 MR-responsive genes to Cort and Aldo treatment, respectively (SI Appendix, Fig. S4B and Dataset S1). The presence of endogenous GR in the parental cells appears to potentiate the transcriptional effects of the MR-580C. The Cort-treated parental cells have 314 responsive genes that meet the FDR and FC cutoffs while the Aldo-treated have 41 genes that meet the cutoffs. Again, the larger number of responsive genes with Cort versus Aldo can be attributed mainly to the presence of endogenous GR, but MR-580C activates/represses more genes with Aldo treatment in the presence of GR than in the GRKO cells (SI Appendix, Fig. S4B and Dataset S1). A comparison of 218 Cort-responsive genes meeting only the FDR cutoff and common to the MRwt cells and MR-580C cells show overall similar hormone responses, indicating that these genes are primarily responding to GR and not MR (SI Appendix, Fig. S4C). A similar comparison of 39 genes with Aldo treatment often shows reduced gene responses with MR-580C compared to MRwt but does not meet our P-value threshold of 0.01 (SI Appendix, Fig. S4D). These data suggest that the NTD of MR is indeed functional in our model cell lines and contributing to the hormone-dependent transcriptional response, especially with Aldo treatment in the presence of GR. The MR-580C data also indicate that the NTD of MR does not have an overall inhibitory effect on hormone-dependent transcriptional activity, as MRwt response is as high or higher than MR-580C.
GR Significantly Potentiates MR/Aldo Transcriptional Response in Mouse Colon Organoids.
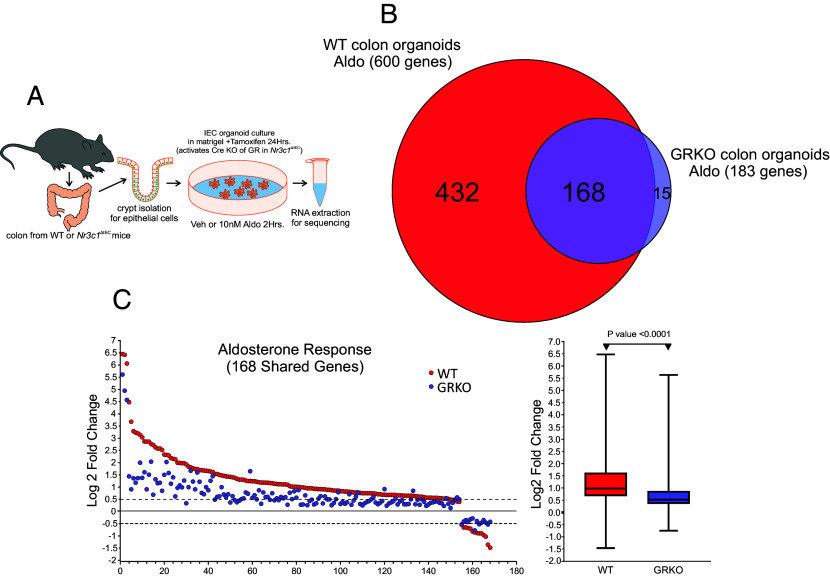
The cell lines used in our analysis do not express MR endogenously, which brings into question whether our results can be extrapolated to cells that normally coexpress MR and GR. We, therefore, tested the effect of GR coexpression on MR transcriptional activity in a physiologically relevant model with endogenous expression of both receptors. To that end, we used cultured organoids derived from the colon of wild type mice or mice with intestinal epithelium-specific tamoxifen-inducible GRKO (51). Since GRKO induces limited intestinal inflammation, we induced GRKO after establishing the organoid culture (Fig. 5A). Quantitative analysis of mRNA expression by qPCR showed that tamoxifen treatment for 24 h eliminated GR expression, without affecting MR expression (SI Appendix, Fig. S5). Cultured organoids also expressed endogenous 11-β-HSD2 and responded to Aldo with potently increased expression of the γ subunit of the epithelial Na+ channel (γENaC, Scnn1g gene) (SI Appendix, Fig. S5), a well-characterized and physiologically relevant MR/Aldo target in the colon (52, 53). GR expression was slightly repressed by aldosterone (SI Appendix, Fig. S5). Our results with C127 cell lines show that it is difficult to assign MR-specific effects to Cort treatment. In addition, the expression of 11-β-HSD2 makes it difficult to study Cort effects. Therefore, we focused on the global effects of Aldo on the transcriptome of wild-type (WT) and GRKO organoids. To that end, we collected total RNA from both cell types treated with vehicle or 10 nM Aldo for 2 h and performed total RNA-seq. (Fig. 5A). Similarly to the C127 cell lines, we performed DE analysis with an FDR cutoff of 0.01 as determined by DESeq2 (via Homer) (35), using in this case five to six biological replicates per condition (SI Appendix, Fig. S2C). Wild type organoids showed the most prominent Aldo response, both in the number of up- or down-regulated protein-coding genes and quantitative changes in RNA abundance (Log2 cut-off of ±0.5 or greater; Dataset S1). The list of regulated genes includes well-characterized Aldo-target genes in addition to Scnn1g, such as Sgk1, Tsc22d3 (GILZ), Per1, and Fkbp5 (45, 54, 55), to name a few. Wild type organoids treated with Aldo had 432 responsive genes, compared to 183 genes in GRKO organoids, 168 of which are common to both conditions (Fig. 5B). Remarkably, GRKO organoids showed a general reduction in the response within the common Aldo-responsive genes (Fig. 5C), which was even more pronounced than that detected in the C127 cell lines. These results demonstrate that the effect of GR on MR/Aldo transcriptional response is physiologically significant in a model with endogenous MR expression.
Fig. 5.
MR/Aldo transcriptional response is potently enhanced by GR in mouse colon organoids. (A) Schematic diagram of the experimental approach. Five to six replicates were used per condition. (B) Venn diagrams of hormone-regulated protein-coding genes (2 h treatment/vehicle) to 10 nM Aldo. The number of hormone-responsive genes (FDR ≤ 0.01, Log2 FC ≥ ± 0.5) denoted in parentheses for GRKO organoids or WT organoids (both express endogenous MR). (C) Scatter plot of Log2 FC for all shared Aldo-responsive genes meeting the FDR 0.01 cutoff, regardless of fold change. Box and whiskers plot of the same data displays IQR depicting the 25th, 50th, and 75th percentile as box with the median as black bar. The whiskers mark the most induced and repressed genes. Statistical analysis was performed using a two-tailed unpaired t test.
SMT of MR Suggests GR Contributes to Productive Binding of MR/Aldo Complexes.
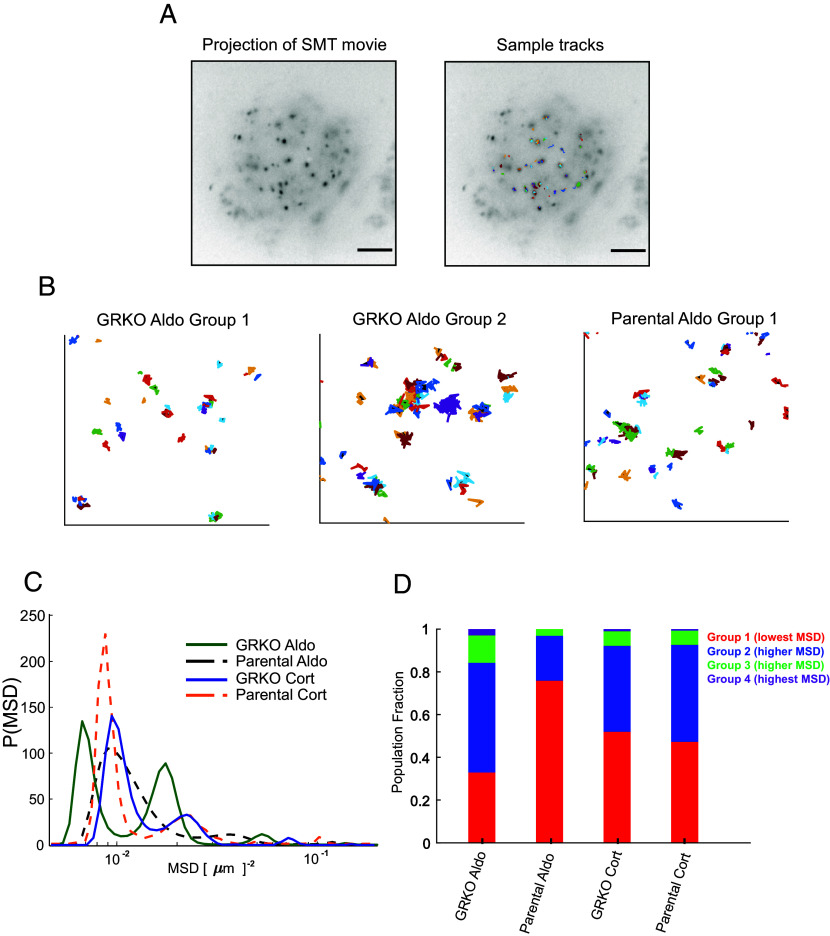
Given that our overall ChIP-seq results do not explain well the global changes in MR-mediated transcription induced by GR coexpression, we tested whether GR-induced changes in MR chromatin binding and transcriptional activity correlate with altered receptor dynamics in the nucleus. We performed SMT of transiently transfected HaloTag-MR chimeras in the GRKO and parental cells to determine the spatiotemporal dynamics of MR under these conditions. We fluorescently labeled Halo-MR with low concentrations of organic dye [Materials and Methods; (56)], and imaged cell nuclei using highly inclined laminated optical sheet (HILO) microscopy (57). We focused on the spatial mobility of molecules that remain stable on the order of tens of seconds as these have been shown to be correlated with transcriptional outcomes (58). We imaged the cells every 200 milliseconds (to minimize photobleaching), with 10 ms exposures (to minimize motion blur) (59). We note that at this frame rate, freely diffusing molecules will rapidly exit the focal plane on our analysis timescales. This method allows us to track molecules that are bound to chromatin for several seconds. The temporal projection of representative SMT movies along with overlaid tracks are shown in Fig. 6A.
Fig. 6.
SMT of MR. (A) Temporal projection of an MR SMT movie (Left) overlaid with tracks (Right). (Scale bar 5 μm.) (B) Representative tracks for MR in the dominant lowest mobility groups. (Left) MR/Aldo group 1 tracks in GRKO cells, (center) MR/Aldo group 2 tracks in GRKO cells, (Right) MR/Aldo group 1 tracks in parental cells. Note that MR/Aldo in parental cells exhibits a very small fraction of group 2 tracks (see also panel D). (C) Distribution of MR mean squared displacement (MSD) at a timelag of 0.8 s, obtained by iteratively fitting the van Hove correlation (vHc) function using the Richardson-Lucy algorithm for each of the four conditions—GRKO-Aldo (green solid line), parental Aldo (black dashed line), GRKO-Cort (blue solid line), parental-Cort (orange dashed line). (D) Population fractions for the different mobility groups for the indicated condition. Ncells/Ntracks: 51/1,878 (GRKO MR/Aldo), 51/2,000 (GRKO MR/Cort), 57/2,419 (parental MR/Aldo), 60/2,036 (parental MR/Cort). Recordings of cells were collected on two separate days for each condition.
Recent SMT studies have identified two distinct mobility groups for chromatin and chromatin-bound transcriptional regulators (60–63). Transcriptionally active steroid receptors and other transcription factors show a substantially higher proportion of binding in the lowest mobility state and binding in this state requires an intact DBD as well as domains necessary to recruit cofactors (63). We hypothesized that the substantial gene response to Aldo in parental cells could result from an increased association of MR in the lowest mobility group. To test this, we collected SMT data of MR under the two different hormone stimulation conditions in both GRKO and parental cell lines. We then iteratively fit the jump distance histogram at a specified time lag using an algorithm developed by Richardson (64) and Lucy (65) to uncover the distribution of mean-squared displacements (MSD) or equivalently, the distribution of diffusivities, as has been done for chromatin (60, 63) and steroid receptor (63) trajectories recently (Materials and Methods). We can use the MSD distribution to classify each MR trajectory into distinct mobility groups (SI Appendix, Fig. S6; see Materials and Methods). Representative tracks for the two lowest mobility groups are shown for MR/Aldo in the GRKO cells (Fig. 6 B, Left and Center) and MR/Aldo in the parental cells (Fig. 6 B, Right). Applying this analysis to MR trajectories for the four experimental conditions, we find that MR molecules exhibit multiple mobility groups exemplified by the distinct peaks in the MSD distribution (Fig. 6C). As can be seen from the areas under the peaks, the two lowest mobility groups account for most of the trajectories (Fig. 6C). Strikingly, MR/Aldo in the parental cells exhibits a substantially higher proportion of molecules in the lowest mobility group as compared to any other condition (Fig. 6D). This lowest mobility state is precisely the state in which nuclear receptors, upon activation, show a higher propensity to bind (63). Taken together, the presence of GR leads to an ~2.3-fold increase in the population fraction of MR/Aldo in the lowest mobility group as compared to that in the absence of GR (Fig. 6D; CI for each mobility group are described in SI Appendix, Table S2). Binding in this lowest mobility group is markedly higher for other steroid receptors (GR, androgen, progesterone, and estrogen receptors) in their transcriptionally active (liganded) state as compared to that in their inactive state (63). Even though parental cell MR/Aldo presents modestly lower binding as measured by ChIP-seq (Fig. 2 A and B), our live cell studies suggest that a higher proportion of MR binding in the transcriptionally active group (group 1) could contribute to the higher transcriptional output as compared to that of Aldo- and Cort-treated GRKO cells (Fig. 1). However, we cannot discount the presence of additional mechanisms such as differential coregulator recruitment.
Discussion
In this study, we have demonstrated the global importance of GR to enhance aldosterone-driven transcriptional function of MR. Because both receptors respond to Cort, we cannot distinguish the contribution of MR/GR interactions with this treatment using RNA-seq alone. We have shown that when liganded to Aldo or Cort, MR by itself binds to HREs but cannot efficiently elicit a transcriptional response. In contrast, when acting in the presence of GR, Aldo-liganded MR can increase transcription of genes and at intergenic enhancers that it cannot efficiently induce on its own. Further, GR by itself cannot elicit a significant gene response when liganded to Aldo, where the hormone acts much like an antagonist by binding the receptor without imparting a functional response (Fig. 1A) (66). Thus, in the presence of Aldo, both receptors appear to act together, enhancing MR-mediated gene response. This effect is even more apparent in a physiologically relevant mouse-derived colonic organoid model with endogenous receptor expression. The transcriptional potentiation detected in the cell line model (from 17 to 53 MR/Aldo regulated genes) correlates well with GR-dependent altered single-molecule dynamics of MR, with Aldo-liganded MR showing an ~2.3-fold increase in the population fraction of group 1 in the presence of GR (Fig. 6D).
The MR activation of genes in the GRKO cell line is low (just over 10 genes) despite comparable levels of receptor binding in cells with both MR and GR (Fig. 2 A and B). This suggests that genomic binding of MR (as measured by ChIP) is not sufficient to promote a full gene response, an effect that may underlie GR effects on recruitment of specific transcriptional coregulators and/or the kinetics of coregulator recruitment. GR also has modestly reduced binding when liganded to Aldo suggesting that the two receptors are not competing for binding at the same response elements, in agreement with previous results obtained using forebrain-specific MR knockout mice (67). When Cort is the ligand, the level of MR binding increases simultaneously with GR binding and is higher than binding with Aldo. This may indicate that the interaction of the two closely related receptors likely imposes its effects by the recruitment of coregulators necessary for modulating transcription and not via higher levels of binding. Having a heteromultimer of Aldo-liganded GR and MR may more effectively recruit cofactors than MR can accomplish by itself. Further studies on cofactor recruitment and transcriptional response are needed to answer this question.
Starting with the original studies by Trapp et al. (26) and Liu et al. (22), it has long been known that MR and GR are able to form heterocomplexes, although the functional impact of this interaction has been elusive (19–26, 30). The functional effect of GR on MR action has been mainly studied in the context of glucocorticoid signaling, based on the common assumption that in the presence of 11-β-HSD2, local glucocorticoid levels are very low and thus GR would be inactive and not affect Aldo-mediated MR activity. Reporter gene transactivation assays using low levels of cortisol stimulation (up to 10 nM, thus favoring MR over GR binding) showed increased transcriptional responses when both receptors were present (26). However, this result seems to be dependent on the promoter context, since opposite results were obtained with a reporter assay using a different promoter (22). Evidence for a direct MR/GR interaction was later expanded to a negative GRE, with data suggesting that heteromerization of MR and GR directly mediates corticosteroid-induced trans-repression of the 5-HT1A receptor promoter (27). Further work performed with rainbow trout MR and GR receptors using gene reporter assays suggested that MR–GR interaction may be involved in cortisol responses, with a dominant-negative role of MR in the process (68). Interestingly, this study found that the inhibitory role of MR persists even in the presence of its antagonist eplerenone, suggesting that MR transcriptional activity is not important in the process. Other reports agree with this notion, where MR plays a dominant-negative role on GR-mediated glucocorticoid-regulated gene expression, further suggesting that the NTD of MR is the domain involved in this effect through heterodimerization (69). The importance of this domain is confirmed by our data, as shown in SI Appendix, Fig. S4. Mifsud et al. tested the relevance of MR and GR interaction in a more physiological context, testing MR and GR binding to GREs of common glucocorticoid-target genes (Fkbp5, Per1, and Sgk1) in hippocampal neurons after exposure of rats to environmental stressors (70). Their results are consistent with gene-dependent binding of MR and GR to GREs as homo- and/or heterodimers. GR binding seemed to facilitate MR binding to GREs in Fkbp5 and Per1 genes under high-glucocorticoid conditions. Taken together, these studies generally indicate that MR and GR coexpression may impact glucocorticoid-mediated gene expression but are limited by the lack of genome-wide binding or transcriptional analyses. More recently, Rivers et al investigated the global effect of MR on GR genomic binding in transfected neuroblastoma N2a cells using ChIP-nexus (25). Their results show that MR and GR bind to overlapping, highly similar sites (58% of them with GRE motifs). RT-qPCR experiments measuring expression of selected genes (Syt2, Sgk1, Dusp4, and Ddc) showed that MR expression alone produced modest changes in expression upon 100 nM Cort stimulation, while GR coexpression induced more potent changes. This last experiment does not allow differentiating between MR-mediated and GR-mediated transcriptional changes. The authors proposed a tethering mechanism where GR mediates MR interaction with chromatin (25).
Few studies have directly investigated the impact of GR on Aldo-mediated MR transcriptional activity. Tsugita et al. examined this question with neuroblastoma and colon carcinoma cell lines expressing MR in the absence or presence of cotransfected GR and using reporter gene assays (28). This study demonstrated a lack of Aldo-induced luciferase activity unless GR is cotransfected. This MR-rescuing effect is specific for GR, since other steroid receptors such as PR, or AR did not have any effect. Interestingly, mutations in the DBD of GR prevented the potentiation of MR activity, suggesting that GR DNA binding is critical for the effect (28). Our data at the whole-genome level are consistent with these previous studies, corroborating the importance of GR in potentiating MR/Aldo transcriptional activity, not only in cell lines but also in colon organoids, which according to our data retain gene expression characteristics of the Aldo-sensitive distal colon epithelium.
What is the molecular basis for the modulation of MR transcriptional activity by GR? Our data indicate that global, steady-state binding of MR to chromatin is not predictive of transcriptional activity. Interestingly, MR seems to be intrinsically more stable in its interaction with DNA than GR, as shown by hormone washout experiments (24, 25), but this does not explain the changes in transcription seen upon GR coexpression. It may be argued that the proposed tethering mechanism, where GR mediates MR indirect binding to DNA may play a role in explaining our results (25). In this scenario, the MR ChIP peaks detected in our experiments in GRKO and parental cell lines would not be directly comparable, since the latter would correspond to a different mode of interaction that is more productive transcriptionally. However, the fact that GR binds DNA poorly when Aldo is the ligand but still has a prominent effect on potentiating MR activity rules out this possibility. Another intriguing possibility is that GR, by interacting with MR and modulating its final oligomeric state as we recently reported (21), changes MR conformation to a more active state. Interestingly, our SMT data support the idea of GR-induced differences in the kinetics of MR interaction with DNA. This is consistent with a previous report showing higher in vitro stability of MR/GR-DNA complexes when compared to MR alone (26). The situation may be more complicated, since MR and GR appear to interact with a specific GRE (a known binding site in the Per1 gene) in a cyclical way, possibly alternating homo- and heterocomplexes (71). Unfortunately, that study did not address MR dynamic interaction with chromatin in cells where GR is absent, precluding a more detailed analysis of the impact of GR on MR kinetics. On the other hand, MR and GR’s cyclical interaction with chromatin also apply when Aldo is used as the agonist (71), consistent with our findings at a genome-wide level. This further reinforces the idea that GR participates in modulating MR-mediated transcriptional responses even when Aldo is the agonist. A limitation of our study is that we did not investigate chromatin binding or single-molecule dynamics in the organoid culture model. These are challenging experiments to perform due to the inability to transfect tagged proteins into the organoids (for SMT), and the low quantity of cells recovered from culture and the lack of ChIP-grade antibodies for wildtype MR expressed in the organoids (for ChIP-seq). However, the transcriptional response is fully consistent with the data obtained in cell lines, suggesting that the same basic mechanisms may be operating in Aldo-target epithelia.
Our data suggest that MR has likely evolved to work in concert with its more transcriptionally active sibling receptor, GR, which is present in most tissues, including those where MR plays important cellular functions. However, it is important to point out that neurons in the CA2 region of the hippocampus or presympathetic neurons in the paraventricular nucleus of the hypothalamus express MR in the absence of GR (18, 72), Moreover, MR is essential for establishing the phenotype of CA2 neurons (73, 74), indicating that GR coexpression is not an absolute requirement for MR function. Our study is particularly relevant for MR function in mammalian tissues where GR is present but, via Cort inactivation by 11-β-HSD2, can bind only, or mainly to Aldo, which is likely a physiologically relevant scenario. Ackermann et al. showed that while MR is constitutively nuclear in the Aldo-sensitive distal nephron, GR responds to fluctuations in Aldo circulating levels, at least in rats (75). Specifically, when Aldo levels are lowered by dietary NaCl loading, GR is localized to the cytosol, while MR remains nuclear. It is necessary to abrogate Aldo synthesis by adrenalectomy to achieve cytosolic localization for both MR and GR (75). Given the high circulating glucocorticoid levels during the peak of the circadian rhythm, it is possible that small amounts of glucocorticoids reach MR, which has high affinity for them. However, low doses of glucocorticoids would not activate GR and therefore the situation would result in relatively low MR activity. Only an increase in Aldo, which would be sensed by MR and also partially by GR would result in a more prominent MR-mediated response. This is consistent with a mechanism where GR plays an important role in the Aldo response, as originally proposed by Geering et al. (32) and indirectly corroborated by experiments in the renal collecting duct using either targeted knockout of the MR (53) or overexpression of the GR (76). In general, the present study leads us to hypothesize that progressive recruitment of GR may contribute to the modulation of MR in the Aldo-sensitive epithelia and other cells where MR and GR are coexpressed with 11-β-HSD2. This mechanism may have an impact in situations of altered glucocorticoid and mineralocorticoid signaling, including those induced under pathological situations or by pharmacological treatment of patients. Since there is evidence pointing toward aldosterone-specific gene regulation in cardiomyocytes under MR-overexpressing conditions (46), it is tempting to speculate that GR may also play a role in Aldo/MR signaling outside epithelia.
It appears that, in addition to MR/GR, other combinations of receptors within the estrogen and steroid receptor subfamily (NR3) give rise to functionally altered heteromers. These “atypical” interactions, including association of GR with the progesterone, estrogen, or androgen receptors (77, 78) are increasingly recognized as important factors in determining transcriptional outcomes of hormone signaling, although the underlying mechanisms remain poorly understood (77). The apparent stabilization of productive chromatin binding of MR by GR suggests a more general mechanism that may underlie cross talk among members of the NR3 subfamily of nuclear receptors.
Materials and Methods
Plasmids Constructs and Mutagenesis.
A fully functional mouse MR fluorescent derivative with insertion of eGFP after amino acid 147 has been previously described (34). eGFP-MR was subcloned in plasmid Donor-Rosa26_Puro_CMV (33), with CMV promoter-driven expression, a puromycin resistance cassette and homology recombination arms specific for the mouse Gt(ROSA)26Sor locus. pX330 CRISPR/Cas9 plasmid, containing a guide RNA sequence to target the Gt(ROSA)26Sor locus, was a gift from Feng Zhang [Addgene plasmid #42230; (79)]. Halo-tagged MR was constructed using In-Fusion cloning. The entire NTD of MR was deleted using the Quickchange XL mutagenesis kit, generating construct MR-580C. All constructs and mutations were confirmed by DNA sequencing.
Cell Culture and Generation of Cell Lines by CRISPR/cas9.
Cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 5 μg/mL tetracycline (Sigma-Aldrich #T7660), 10% fetal bovine serum (FBS, Gemini), sodium pyruvate, nonessential amino acids, and 2 mM glutamine. Cells were maintained in a humidifier at 37 °C and 5% CO2. Cells were plated for experiments in DMEM supplemented with 10% charcoal/dextran-treated serum for 24 h prior to hormone treatment. Cell lines used in this study derive from mouse mammary carcinoma cell line C127 (RRID: CVCL_6550). Knockout of endogenously expressed GR to generate GRKO cells has been previously described (33). Transient transfections were performed using Jetprime (Polyplus) according to the manufacturer’s instructions. eGFP-tagged MR was stably integrated in the genome using CRISPR/Cas9. To that end, cells were cotransfected with pX330 CRISPR/Cas9 plasmid with a donor plasmid containing eGFP-MR driven by the CMV promoter. Donor plasmid insertion was selected by puromycin treatment followed by fluorescence-activated cell sorting (FACS). Expression of MR in sorted polyclonal cell declined with time and therefore we selected stable lines by one additional round of FACS, followed by single-cell cloning. GFP-MR expression in individual clones was confirmed by confocal microscopy and western blot using monoclonal antibody rMR1-18 1D5 [developed by Gomez-Sanchez et al. (80), and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD, NIH and maintained at The University of Iowa, Department of Biology] as previously described (30). MR agonists aldosterone and corticosterone were obtained from Sigma and dissolved in ethanol. Cells were plated for experiments in DMEM growth medium supplemented with 10% charcoal/dextran-treated serum for 48 h prior to hormone treatment. Subsequently, cells were left untreated or treated with 10 nM aldosterone or 100 nM corticosterone for the indicated periods of time. Control cells were treated with ethanol at the same dilution used for treatments (1:1,000).
Mouse Model, Colon Crypt Isolation, and Organoid Culture.
B6-Cg-Nr3c1-Vil-Cre/ERT2 mice, referred to in the present paper as Nr3c1ΔIEC mice, were obtained as previously described (51). Briefly, C57BL/6J mice carrying lox sequences flanking the GR were crossed with transgenic mice expressing the tamoxifen-inducible CRE recombinase under the control of the villin promoter. C57BL/6J. B6.Cg- Nr3c1<tm1.1Jda.>/J were used as controls and referred to as WT. Until being killed, mice were maintained at the University of Granada Animal Facility (Biomedical Research Center, University of Granada, Spain) under specific pathogen-free conditions in air-conditioned animal modules with a 12-h light–dark cycle. Mice were given free access to autoclaved tap water and standard chow (Harlan-Teklad 2014, Harlan Ibérica, Barcelona, Spain).
Colonic intestinal organoids were obtained by crypt isolation from WT and Nr3c1ΔIEC mice. Briefly, the mouse colon was dissected and gently flushed with cold PBS then incubated twice with PBS with 2 mM ethylenediaminetetraacetic acid and 1 mM dithiothreitol for 30 min. After shaking, colon fragments were passed through a 70 µm filter and crypts were counted for seeding. Crypts were seeded in Corning-Matrigel® (Fisher Scientific, Madrid, Spain) and IntestiCult® (StemCell, Grenoble, France) supplemented with penicillin–streptomycin, gentamicin, and amphotericin (Sigma-Aldrich), with a 1:1 ratio in 24-well plates. After organoids were obtained, GR deletion was induced by adding 1 µM tamoxifen (Sigma-Aldrich) in culture media one day after passage. Tamoxifen was also added to WT organoids. 24 h later, tamoxifen was removed with fresh IntestiCult®. To perform the experiment, DMEM-F12 medium supplemented with 10% charcoal-preadsorbed FBS was substituted for Intesticult®. This medium is devoid of corticoids. Forty-eight h later, organoids were stimulated with aldosterone 10 nM. 2 h after stimulation, organoids were collected for RNA extraction.
RNA Isolation, qPCR, and RNA-seq Analysis.
Cells and organoids were treated with vehicle, 10 nM aldosterone, or 100 nM corticosterone as indicated for 2 h prior to RNA isolation. Total RNA was isolated using commercially available kits (Macherey-Nagel NucleoSpin RNA isolation in the case of cell lines, Qiagen RNeasy minikit in the case of organoids), which included an in-column DNase digestion step. Purified RNA was quantified using spectrophotometry and frozen in aliquots at −80 °C. One aliquot of organoid RNA samples was used to synthesize single-stranded cDNA starting from 1 µg of total RNA using a commercially available kit (iScript cDNA Synthesis Kit, Biorad). Specific DNA sequences were amplified with a Bio-Rad CFX connect real-time PCR device (Alcobendas, Madrid, Spain) using GoTaq® qPCR master mix (Promega) using 18 s, Hprt (Hypoxanthine-Guanine Phosphoribosyltransferase), and Ppib (Peptidylprolyl Isomerase B) as reference genes. Primers used are shown in SI Appendix, Table S1.
RNA-seq included two to three biological replicates of each condition in the case of cell lines and five to six biological replicates for organoids and used Illumina Novaseq with 150 bp stranded reads. RTA 2.4.11 was used for Base calling and Bcl2fastq 2.20 was used for demultiplexing allowing 1 mismatch. Cutadapt 1.18 was used for adapter removal and quality control. RNA-seq alignment to mouse mm10 genome was performed by STAR 2.70 using the default parameters with the following modifications: “--genomeDir mm10-125 --outSAMunmapped Within --outFilterType BySJout --outFilterMultimapNmax 20 --outFilterMismatchNmax 999 --outFilterMismatchNoverLmax 0.04 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --alignSJoverhangMin 8 --limitSjdbInsertNsj 2500000 --alignSJDBoverhangMin 1 --sjdbScore 1 --sjdbFileChrStartEnd mm10-125/sjdbList.out.tab --sjdbGTFfile UCSC_mm10_genes.gtf --peOverlapNbasesMin 10 --alignEndsProtrude 10 ConcordantPair.” All RNA-seq biological replicates correlated well with each other. Subsequent downstream analysis was performed using HOMER pipeline. Briefly, we obtained raw count data using analyzeRepeats.pl, and then the raw counts were normalized by default size factors from DESeq2 routine 23 provided via getDiffExpression.pl. We obtained differential genes using DESeq2, which fits negative binomial generalized linear models for each gene and uses the Wald test for significance testing, based on the criteria of a FDR cutoff <0.01 and absolute log2 fold change (FC) > 0.5 between no treatment and 2 h hormone treatment. We included only protein-coding genes that are annotated in the RefSeq database and included no noncoding RNA species.
ChIP-seq and Analysis.
Cells were treated with vehicle, 10 nM aldosterone, or 100 nM corticosterone for 1 h. Two independent replicates were analyzed. Chromatin crosslinking, preparation, and immunoprecipitation was performed essentially as described (36). Briefly, chromatin crosslinking was performed using 1% formaldehyde added to culture medium for 5 min. After glycine quenching and washing with PBS, cells were recovered and chromatin extracted and sonicated (Bioruptor, Diagenode) to an average DNA length of 500 bp. For immunoprecipitation of GFP-MR, 600 μgs of chromatin were incubated with 25 µg anti-GFP antibody (Abcam #ab290).
The ChIP-seq data were aligned to the mouse reference mm10 genome using Bowtie 2 with command Bowtie2 –p 8 –x bowtie2_ref/genome_prefix –U read1.fastq –S result.sam. Subsequent downstream analysis was performed using HOMER (81). Peaks in each dataset were called using the findPeaks function with style factor for TFs and the no treatment condition used as a control. Peak filtering was done with the following parameters; FDR < 0.001, >5 FC over control, >5 FC over local background, and ntagThreshold >5 Peak clusters were identified by the mergePeaks command and sorted by cell type and treatment. Predefined motif searches were performed with findMotifsGenome.pl using -m known5.motif -mscore. Gene annotation of peaks used annotatePeaks.pl mm10 -gene.
Heatmap and Aggregate Plot Generation.
We used Deeptools to generate ChIP-seq and eRNA heatmaps and aggregate plots. We first generated read-normalized bigwig files from bam files using the bamCoverage -b [inputfile] -o [output.bigWig] -of bigwig --binSize 20 --effectiveGenomeSize 2652783500 --normalizeUsing RPGC. We generated matrix files using computeMatrix reference-point --referencePoint center -S [input.bigWig files] -R [peakfile.bed] -a 500 -o [matrix.gz] --sortRegions keep. We then generated heatmaps using plotHeatmap -m [matrix.gz] -o [HM.pdf] --sortRegions no --zMin --zMax --refPointLabel “0” --yAxisLabel “Tag Density.” The eRNA heatmaps used merged replicate RNA bam files from the RNA seq data to make bigwig files. Bam files were merged using samtools. Percent changes between aggregate plots were calculated using the area-under-the-curve function in Prism 10 (GraphPad). Statistical analyses of the data presented in Fig. 2C and SI Appendix, Fig. S3 were done as follows: A paired Wilcoxon signed rank test followed by the Benjamini–Hochberg correction to calculate the adjusted P-values from multiple pairwise comparisons was used to compare the ChIP tag densities for each site (row in the heatmap) for the respective conditions that are compared. For the comparison of Aldo-liganded MR in GRKO and parental cells, the same procedure was used to test whether the fold enrichment of Aldo versus the respective EtOH control were significantly different. The swarmcharts in SI Appendix, Fig. S3A represent the distributions of adjusted P-values obtained from this analysis and an adjusted P-value threshold of 0.01 was used to identify sites with significantly different MR binding. Aggregate plot statistical comparison was performed as previously described (82) using the unpaired Wilcoxon rank sum/Mann–Whitney test. Box-and-whiskers plots resulting from the added the values for each row in each condition and cell line in the eRNA heatmap shown in Fig. 4 were compared using the Kruskal–Wallis test followed by Dunn’s multiple comparisons test for the indicated pairs (SI Appendix Fig. S3B).
SMT.
Transient transfections.
GRKO or parental cell lines were plated in complete medium in two-well LabTek II chamber slides. The next day, Halo-MR was transfected into the cells using jetOPTIMUS (Polyplus) following the manufacturer’s protocol. After incubation for 4 h with the jetOPTIMUS reaction mix, the medium was replaced with DMEM supplemented with charcoal/dextran-stripped FBS. 24 h later, cells were incubated for 20 min with 5 nM of the cell-permeant HaloTag ligand Janelia Fluor 646 (JF646). After labeling, cells were washed three times for 15 min with phenol red-free DMEM (Gibco) supplemented with charcoal/dextran-stripped FBS, followed by one last wash after 10 min, to remove unbound JF646. Cells were then treated with 10 nM Aldo or 100 nM Cort for 30 min before imaging.
Microscopy.
All SMT was performed on a custom-built HILO microscopy described previously (59). The microscope is equipped with a 150×, 1.45 NA objective, (Olympus Scientific Solutions, Waltham, MA, USA), an Evolve 512 EM-CCD camera (Photometrics, Tucson, AZ, USA), a 647 nm laser (Coherent OBIS 647LX) and an Okolab stage-top incubator which was set to 37 °C and 5% CO2. Images were collected every 200 ms with an exposure time of 10 ms and laser power of 0.85 mW at the objective. The pixel size for this microscope is 104 nm.
Tracking.
Tracking was performed using TrackRecord v6, a custom MATLAB software freely available at Zenodo (https://doi.org/10.5281/zenodo.7558712) and has been described previously (83, 84). We allowed a maximum jump of 4 pixels, shortest track of 6 frames, and a gap of 1 frame. For details, see Wagh et al. (63).
Estimating the MSD distribution from single-molecule trajectories.
We calculate the self-part of the van Hove correlation function (vHc) , from the single-molecule trajectories. Here, ri is the position of the ith molecule and is a normalization constant. The vHc can be approximated as a superposition of Gaussian basis functions such that . P(M) is the distribution of mean-squared displacements of the population of MR molecules. The Richardson-Lucy algorithm is then used the extract P(M) by iteratively fitting the vHc and updating the estimate of P(M). We refer the reader to refs. 60 and 63 for further details.
Classifying tracks into different mobility groups.
Once the MSD distribution is calculated, the peaks in the distribution can be used to classify trajectories into different mobility groups. As shown in SI Appendix, Fig. S6, the local minima in the MSD distribution can be used to define four different mobility groups. The MSD of each track is then calculated at a timelag of 0.8 s, and by comparing this MSD to the four bins, the track is assigned to one of the mobility groups. The population fractions are calculated as the ratio of the number of tracks in a particular group to the total number of tracks. To estimate the 95% CI of the population fractions (SI Appendix, Table S2), the tracks were resampled with replacement to generate 10,000 bootstrapped ensembles. The distributions of population fractions obtained from these ensembles were used to estimate the 95% CI using the MATLAB function bootci.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
We thank the National Cancer Institute Advanced Technology Program Sequencing Facility for sequencing services. This research used the NIH high-performance computing systems (Biowulf) for genomics analyses. The researchers also thank Tatiana Karpova and David Ball of the Optical Microscopy Core at the NCI, NIH for assistance with the SMT experiments, and Diego M. Presman for his comments on the manuscript. Research was supported by grants from the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by grants PID2019-105339RB-I00, PID2020-112768RB-I00, and PID2022-138788NB-I00_22091 (funded by MCIN/AEI/10.13039/501100011033 and “ERDF A way of making Europe,” MICINN, Spain) and by grant PI21/00952 from Instituto de Salud Carlos III (ISCIII, Spain), cofunded by the European Union. Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd) is funded by ISCIII, Spain. D.A.d.l.R. was partially supported by PRX18/00498 (funded by Programa Estatal de Promoción del Talento y su Empleabilidad en I + D + i, Subprograma Estatal de Movilidad, del Plan Estatal de I + D + I, MICINN, Spain). D.C.-H. was supported by a fellowship from the Spanish DCH Ministry of Science, Education and Universities. A.U. was supported by NIH R35-145313 and NSF 2132922.
Author contributions
T.A.J., F.S.d.M., O.M.-A., G.L.H., and D.A.d.l.R. designed research; T.A.J., G.F., D.C.-H., and D.A.d.l.R. performed research; M.K., F.S.d.M., O.M.-A., and A.U. contributed new reagents/analytic tools; T.A.J., G.F., K.W., and A.U. analyzed data; and T.A.J., K.W., G.L.H., and D.A.d.l.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Gordon L. Hager, Email: hagerg@dce41.nci.nih.gov.
Diego Alvarez de la Rosa, Email: dalrosa@ull.edu.es.
Data, Materials, and Software Availability
The datasets produced in this study are available in the following databases: ChIP-seq and RNA-seq data have been deposited in Gene Expression Omnibus (GEO, accession number GSE232089) (85); SMT data has been deposited in Zenodo (DOI: 10.5281/zenodo.12570960) (86).
Supporting Information
References
- 1.Hunter R. W., Ivy J. R., Bailey M. A., Glucocorticoids and renal Na+ transport: Implications for hypertension and salt sensitivity. J. Physiol. 592, 1731–1744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fallo F., et al. , Prevalence and characteristics of the metabolic syndrome in primary aldosteronism. J. Clin. Endocrinol. Metab. 91, 454–459 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Baker M. E., Steroid receptors and vertebrate evolution. Mol. Cell Endocrinol. 496, 110526 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Bridgham J. T., Carroll S. M., Thornton J. W., Evolution of hormone-receptor complexity by molecular exploitation. Science 312, 97–101 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Arriza J. L., et al. , Cloning of human mineralocorticoid receptor complementary DNA: Structural and functional kinship with the glucocorticoid receptor. Science 237, 268–275 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Chapman K., Holmes M., Seckl J., 11beta-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 93, 1139–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellal-Levy C., et al. , Specific hydroxylations determine selective corticosteroid recognition by human glucocorticoid and mineralocorticoid receptors. FEBS Lett. 464, 9–13 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Farman N., Rafestin-Oblin M. E., Multiple aspects of mineralocorticoid selectivity. Am. J. Physiol. Renal. Physiol. 280, F181–F192 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Carceller-Zazo E., et al. , The mineralocorticoid receptor modulates timing and location of genomic binding by glucocorticoid receptor in response to synthetic glucocorticoids in keratinocytes. FASEB J. 37, e22709 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Joels M., de Kloet E. R., Mineralocorticoid receptor-mediated changes in membrane properties of rat CA1 pyramidal neurons in vitro. Proc. Natl. Acad. Sci. U.S.A. 87, 4495–4498 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oakley R. H., et al. , Cardiomyocyte glucocorticoid and mineralocorticoid receptors directly and antagonistically regulate heart disease in mice. Sci. Signal 12, eaau9685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson W. H., Youn C., Ortlund E. A., Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA. PLoS One 9, e107000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer K., Kelly S. M., Watt K., Price N. C., McEwan I. J., Conformation of the mineralocorticoid receptor N-terminal domain: Evidence for induced and stable structure. Mol. Endocrinol. 24, 1935–1948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuse H., Kitagawa H., Kato S., Characterization of transactivational property and coactivator mediation of rat mineralocorticoid receptor activation function-1 (AF-1). Mol. Endocrinol. 14, 889–899 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Lavery D. N., McEwan I. J., Structure and function of steroid receptor AF1 transactivation domains: Induction of active conformations. Biochem. J. 391, 449–464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallec L. P., et al. , Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: Implication of small ubiquitin-related modifier 1 modification. Mol. Endocrinol. 17, 2529–2542 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Sanchez E., Gomez-Sanchez C. E., The multifaceted mineralocorticoid receptor. Compr. Physiol. 4, 965–994 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijer O. C., et al. , Transcriptional glucocorticoid effects in the brain: Finding the relevant target genes. J. Neuroendocrinol. 35, e13213 (2022), 10.1111/jne.13213. [DOI] [PubMed] [Google Scholar]
- 19.Bigas J., Sevilla L. M., Carceller E., Boix J., Perez P., Epidermal glucocorticoid and mineralocorticoid receptors act cooperatively to regulate epidermal development and counteract skin inflammation. Cell Death Dis. 9, 588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarisse D., et al. , Crosstalk between glucocorticoid and mineralocorticoid receptors boosts glucocorticoid-induced killing of multiple myeloma cells. Cell Mol. Life Sci. 80, 249 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fettweis G., et al. , The mineralocorticoid receptor forms higher order oligomers upon DNA binding. Protein Sci. 33, e4890 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W., Wang J., Sauter N. K., Pearce D., Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 12480–12484 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishi M., Tanaka M., Matsuda K., Sunaguchi M., Kawata M., Visualization of glucocorticoid receptor and mineralocorticoid receptor interactions in living cells with GFP-based fluorescence resonance energy transfer. J. Neurosci. 24, 4918–4927 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pooley J. R., et al. , Beyond the heterodimer model for mineralocorticoid and glucocorticoid receptor interactions in nuclei and at DNA. PLoS One 15, e0227520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivers C. A., et al. , Glucocorticoid receptor-tethered mineralocorticoid receptors increase glucocorticoid-induced transcriptional responses. Endocrinology 160, 1044–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapp T., Rupprecht R., Castren M., Reul J. M., Holsboer F., Heterodimerization between mineralocorticoid and glucocorticoid receptor: A new principle of glucocorticoid action in the CNS. Neuron 13, 1457–1462 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Ou X. M., Storring J. M., Kushwaha N., Albert P. R., Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J. Biol. Chem. 276, 14299–14307 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Tsugita M., et al. , Glucocorticoid receptor plays an indispensable role in mineralocorticoid receptor-dependent transcription in GR-deficient BE(2)C and T84 cells in vitro. Mol. Cell Endocrinol. 302, 18–25 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Derfoul A., Robertson N. M., Hall D. J., Litwack G., The N-terminal domain of the mineralocorticoid receptor modulates both mineralocorticoid receptor- and glucocorticoid receptor-mediated transactivation from Na/K ATPase beta1 target gene promoter. Endocrine 13, 287–295 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Jimenez-Canino R., Fernandes M. X., Alvarez de la Rosa D., Phosphorylation of mineralocorticoid receptor ligand binding domain impairs receptor activation and has a dominant negative effect over non-phosphorylated receptors. J. Biol. Chem. 291, 19068–19078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaeggeler H. P., et al. , Mineralocorticoid versus glucocorticoid receptor occupancy mediating aldosterone-stimulated sodium transport in a novel renal cell line. J. Am. Soc. Nephrol. 16, 878–891 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Geering K., Claire M., Gaeggeler H. P., Rossier B. C., Receptor occupancy vs. induction of Na+-K+-ATPase and Na+ transport by aldosterone. Am. J. Physiol. 248, C102–C108 (1985). [DOI] [PubMed] [Google Scholar]
- 33.Paakinaho V., Johnson T. A., Presman D. M., Hager G. L., Glucocorticoid receptor quaternary structure drives chromatin occupancy and transcriptional outcome. Genome. Res. 29, 1223–1234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilar-Sanchez C., et al. , Identification of permissive insertion sites for generating functional fluorescent mineralocorticoid receptors. Endocrinology 153, 3517–3525 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson T. A., Paakinaho V., Kim S., Hager G. L., Presman D. M., Genome-wide binding potential and regulatory activity of the glucocorticoid receptor’s monomeric and dimeric forms. Nat. Commun. 12, 1987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arriza J. L., Simerly R. B., Swanson L. W., Evans R. M., The neuronal mineralocorticoid receptor as a mediator of glucocorticoid response. Neuron 1, 887–900 (1988). [DOI] [PubMed] [Google Scholar]
- 38.Lombes M., Kenouch S., Souque A., Farman N., Rafestin-Oblin M. E., The mineralocorticoid receptor discriminates aldosterone from glucocorticoids independently of the 11 beta-hydroxysteroid dehydrogenase. Endocrinology 135, 834–840 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Voss T. C., et al. , Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146, 544–554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John S., et al. , Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson T. A., et al. , Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res. 46, 203–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biddie S. C., et al. , Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll J. S., et al. , Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Ueda K., et al. , Genome-wide analysis of murine renal distal convoluted tubular cells for the target genes of mineralocorticoid receptor. Biochem. Biophys. Res. Commun. 445, 132–137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Messaoudi S., et al. , Aldosterone-specific activation of cardiomyocyte mineralocorticoid receptor in vivo. Hypertension 61, 361–367 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Kim T. K., et al. , Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu C., et al. , A non-canonical role of YAP/TEAD is required for activation of estrogen-regulated enhancers in breast cancer. Mol. Cell 75, 791–806.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greulich F., et al. , Enhancer RNA expression in response to glucocorticoid treatment in murine macrophages. Cells 11, 28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffman J. A., et al. , Multimodal regulatory elements within a hormone-specific super enhancer control a heterogeneous transcriptional response. Mol. Cell 82, 803–815.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aranda C. J., et al. , Intestinal epithelial deletion of the glucocorticoid receptor NR3C1 alters expression of inflammatory mediators and barrier function. FASEB J. 33, 14067–14082 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Asher C., Wald H., Rossier B. C., Garty H., Aldosterone-induced increase in the abundance of Na+ channel subunits Am. J. Physiol. 271, C605–C611 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Ronzaud C., et al. , Impairment of sodium balance in mice deficient in renal principal cell mineralocorticoid receptor. J. Am. Soc. Nephrol. 18, 1679–1687 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Petrovich E., Asher C., Garty H., Induction of FKBP51 by aldosterone in intestinal epithelium. J. Steroid Biochem. Mol. Biol. 139, 78–87 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Soundararajan R., Zhang T. T., Wang J., Vandewalle A., Pearce D., A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. J. Biol. Chem. 280, 39970–39981 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Grimm J. B., et al. , A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250, 243 p following 250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tokunaga M., Imamoto N., Sakata-Sogawa K., Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Stavreva D. A., et al. , Transcriptional bursting and co-bursting regulation by steroid hormone release pattern and transcription factor mobility. Mol. Cell 75, 1161–1177.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paakinaho V., et al. , Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat. Commun. 8, 15896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashwin S. S., Nozaki T., Maeshima K., Sasai M., Organization of fast and slow chromatin revealed by single-nucleosome dynamics. Proc. Natl. Acad. Sci. U.S.A. 116, 19939–19944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia D. A., et al. , An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol. Cell 81, 1484–1498.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagh K., Stavreva D. A., Hager G. L., Transcription dynamics and genome organization in the mammalian nucleus: Recent advances. Mol. Cell, 10.1016/j.molcel.2024.09.022 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagh K., et al. , Dynamic switching of transcriptional regulators between two distinct low-mobility chromatin states. Sci. Adv. 9, eade1122 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson W. H., Bayesian-based iterative method of image restoration*. J. Opt. Soc. Am. 62, 55–59 (1972). [Google Scholar]
- 65.Lucy L. B., An iterative technique for the rectification of observed distributions. Astronom. J. 79, 745 (1974). [Google Scholar]
- 66.Savory J. G., et al. , Glucocorticoid receptor homodimers and glucocorticoid-mineralocorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces. Mol. Cell Biol. 21, 781–793 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Weert L., et al. , Mechanistic insights in NeuroD potentiation of mineralocorticoid receptor signaling. Int. J. Mol. Sci. 20, 1575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kiilerich P., et al. , Interaction between the trout mineralocorticoid and glucocorticoid receptors in vitro. J. Mol. Endocrinol. 55, 55–68 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Planey S. L., Derfoul A., Steplewski A., Robertson N. M., Litwack G., Inhibition of glucocorticoid-induced apoptosis in 697 pre-B lymphocytes by the mineralocorticoid receptor N-terminal domain. J. Biol. Chem. 277, 42188–42196 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Mifsud K. R., Reul J. M., Acute stress enhances heterodimerization and binding of corticosteroid receptors at glucocorticoid target genes in the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 113, 11336–11341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le Billan F., et al. , Corticosteroid receptors adopt distinct cyclical transcriptional signatures. FASEB J. 32, 5626–5639 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Chen J., Gomez-Sanchez C. E., Penman A., May P. J., Gomez-Sanchez E., Expression of mineralocorticoid and glucocorticoid receptors in preautonomic neurons of the rat paraventricular nucleus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 306, R328–R340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCann K. E., et al. , Novel role for mineralocorticoid receptors in control of a neuronal phenotype. Mol. Psychiatry 26, 350–364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oakley R. H., et al. , Combinatorial actions of glucocorticoid and mineralocorticoid stress hormone receptors are required for preventing neurodegeneration of the mouse hippocampus. Neurobiol. Stress 15, 100369 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ackermann D., et al. , In vivo nuclear translocation of mineralocorticoid and glucocorticoid receptors in rat kidney: Differential effect of corticosteroids along the distal tubule. Am. J. Physiol. Renal. Physiol. 299, F1473–F1485 (2010). [DOI] [PubMed] [Google Scholar]
- 76.A. Nguyen Dinh Cat et al. , Conditional transgenic mice for studying the role of the glucocorticoid receptor in the renal collecting duct. Endocrinology 150, 2202–2210 (2009). [DOI] [PubMed] [Google Scholar]
- 77.De Bosscher K., Desmet S. J., Clarisse D., Estebanez-Perpina E., Brunsveld L., Nuclear receptor crosstalk–Defining the mechanisms for therapeutic innovation. Nat. Rev. Endocrinol. 16, 363–377 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Ogara M. F., et al. , The glucocorticoid receptor interferes with progesterone receptor-dependent genomic regulation in breast cancer cells. Nucleic Acids Res. 47, 10645–10661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gomez-Sanchez C. E., et al. , Development of a panel of monoclonal antibodies against the mineralocorticoid receptor. Endocrinology 147, 1343–1348 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffman J. A., Trotter K. W., Ward J. M., Archer T. K., BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome. Elife 7, e35073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garcia D. A., et al. , Power-law behavior of transcription factor dynamics at the single-molecule level implies a continuum affinity model. Nucleic Acids Res. 49, 6605–6620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mazza D., Abernathy A., Golob N., Morisaki T., McNally J. G., A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40, e119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson T. A., et al. , The glucocorticoid receptor potentiates aldosterone-induced transcription by the mineralocorticoid receptor. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232089. Deposited 23 July 2024. [DOI] [PMC free article] [PubMed]
- 86.Johnson T. A., et al. , Data for “The glucocorticoid receptor potentiates aldosterone-induced transcription by the mineralocorticoid receptor.” Zenodo. 10.5281/zenodo.12570960. Deposited 27 June 2024. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
The datasets produced in this study are available in the following databases: ChIP-seq and RNA-seq data have been deposited in Gene Expression Omnibus (GEO, accession number GSE232089) (85); SMT data has been deposited in Zenodo (DOI: 10.5281/zenodo.12570960) (86).