Abstract
Thiophene is a privileged pharmacophore in medicinal chemistry owing to its diversified biological attributes. The thiophene moiety has been ranked 4th in the US FDA drug approval of small drug molecules, with around 7 drug approvals over the last decade. The present review covers USFDA-approved drugs possessing a thiophene ring system. Our analysis reveals that 26 drugs possessing thiophene nuclei have been approved under different pharmacological classes. The review further covers reported thiophene and its substituted analogues with diverse biological activities, including anti-diabetic, anticancer, anti-inflammatory, anticonvulsant, and antioxidant activity. Besides, a section is dedicated to appreciating the implications of structural bioinformatics in drug discovery. Additionally, the manuscript delves into structure–activity relationship studies to explore the chemical groups responsible for eliciting potential therapeutic activities. The review may provide invaluable insights for researchers working with thiophene nuclei in developing novel analogues with greater efficacy and fewer side effects.
Thiophene is a privileged pharmacophore in medicinal chemistry owing to its diversified biological attributes.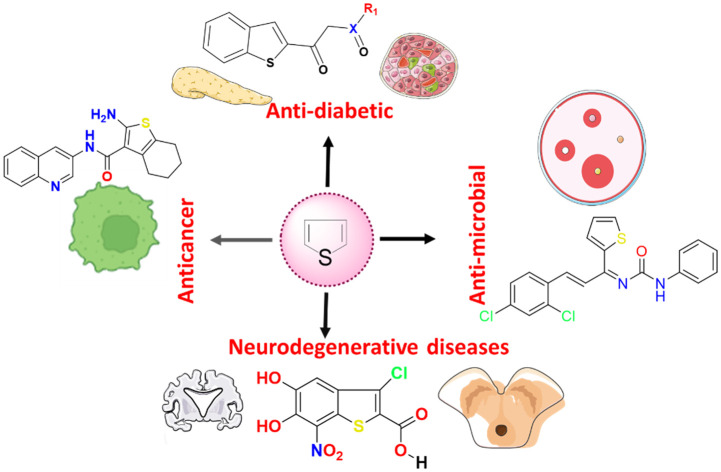
1. Introduction
Heterocyclic scaffolds have drawn the attention of medicinal and organic chemists owing to their diverse biological and medicinal properties.1 These compounds consist of heteroatoms that include sulfur, oxygen, and nitrogen.2 Incorporating heteroatoms into a compound significantly modifies its physicochemical properties.3 It improves drug–receptor interactions and alters solubility and metabolism because of the electronegativity difference and availability of unshared electron pairs between the heteroatoms and carbon.4 Among numerous heterocycles available, one such privileged heterocycle scaffold is thiophene, which contains sulfur atoms with five-member rings.5 ‘Thiophene’ is derived from the Greek words ‘theion’ and ‘phaino’. ‘Theion’ means ‘sulfur,’ while ‘phaino’ stands for ‘to show’ or ‘to appear’.6 This is owing to its discovery in 1882 by Viktor Meyer, who discovered it as a contaminant in benzene.6,7 Before this, it was believed that isatin, upon reaction with crude benzene in the presence of sulphuric acid, gives a blue-colour compound, ‘indophenin’.8 However, the reaction or the formation of indophenin did not occur when purified benzene was utilised for the reaction.9
Nonetheless, the thiophene moiety plays a significant role in drug discovery and medicinal chemistry. This is mainly owing to its versatile structural diversity and pharmacophoric properties.10 The thiophene ring not only provides synthetically accessible modification sites within itself but is also considered an important pharmacophore for replacing existing functionalities in a drug candidate,11 thus serving as an important ring and pharmacophoric system for the medicinal chemist toolbox. Moreover, the sulfur atom in the thiophene ring is an excellent atom that enhances drug–receptor interaction by participating in additional hydrogen bonding.12 Apart from this, in structure–activity relationship (SAR) studies, thiophene is explored as a bio-isosteric replacement for monosubstituted phenyl rings.13 This improves physiochemical properties, the parent compound's metabolic stability, and binding affinity.
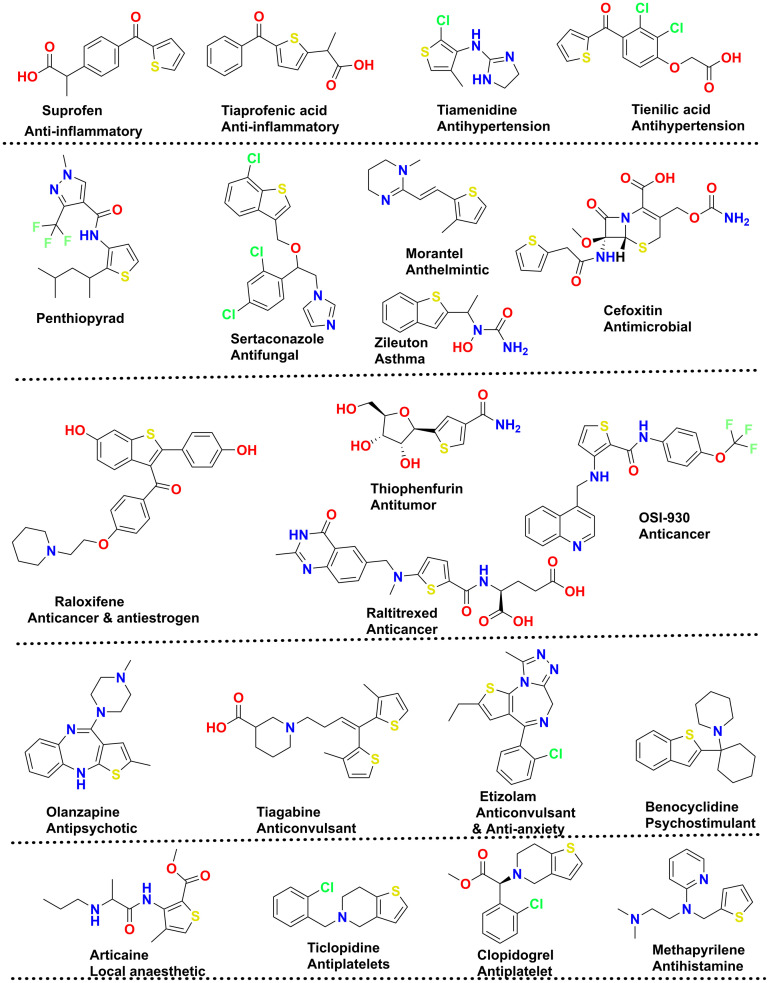
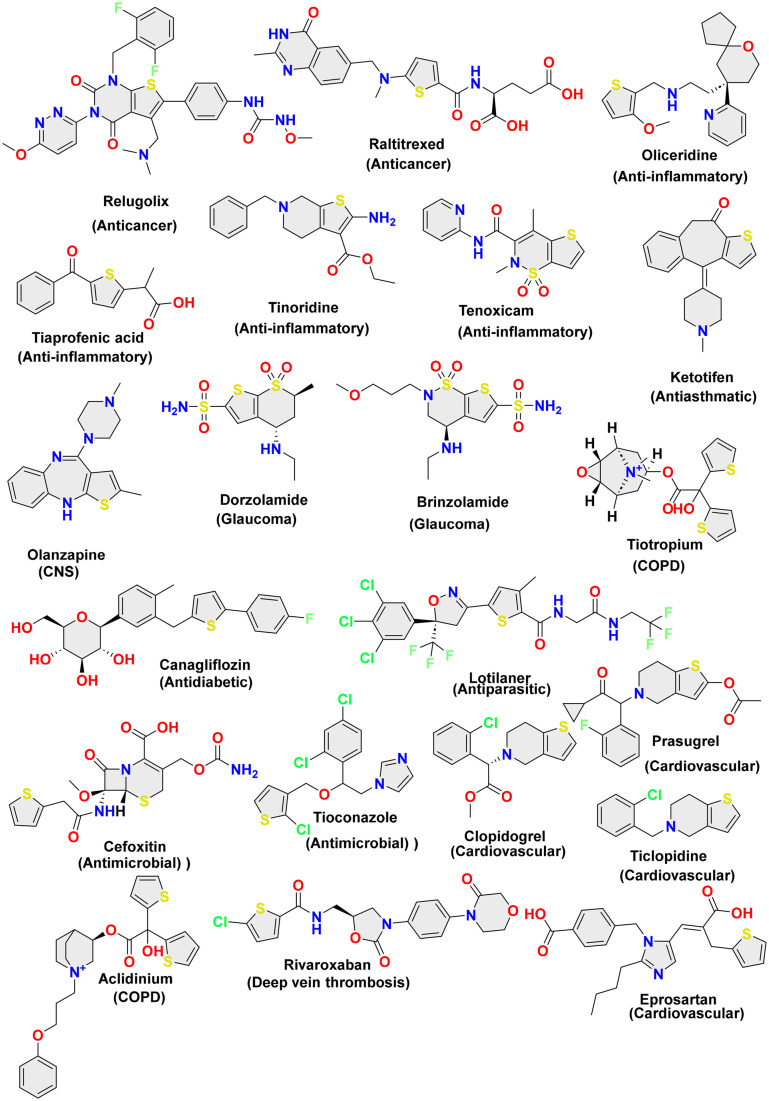
A recent analysis by Marshall and co-workers on U.S. FDA-approved pharmaceuticals for 2013–2023 ranked the thiophene moiety (with seven drugs approved) at the 4th position in the U.S. FDA sulfur-containing drug category, along with thiazoles and sulfones.14 The same research also highlighted that among sulphur-containing drugs, sulfonamide and sulfonamide-containing drugs hold rank 1 with 21 new drug approvals, followed by thioethers and disulfides with 12 and 8 drugs approved, respectively. Other than the mentioned drugs of the decade, thiophene also had its role in giving some therapeutically essential drugs that include suprofen (anti-inflammatory),15 tiaprofenic acid (anti-inflammatory),16 cefoxitin (antimicrobial),17 penthiopyrad (antifungal),18 sertaconazole (antifungal),19 morantel (anthelmintic),20 cefoxitin (antimicrobial),17 zileuton (asthma),21 tiamenidine (antihypertension),22 tienilic acid (antihypertension),23 methapyrilene (antihistamines),24 thiophenfurin (anti-tumor),25 raltitrexed (anticancer),26,27 OSI-930 (anticancer),28 raloxifene (anticancer),29 olanzapine (antipsychotic),30 tiagabine (anticonvulsant),31 etizolam (anticonvulsant & antianxiety),32 benocyclidine (psychostimulant), articaine (local anaesthetic),33 ticlopidine (antiplatelets), and clopidogrel (antiplatelets).34 The chemical structures of these analogues are illustrated in Fig. 1.
Fig. 1. Marketed drugs based on thiophene-based nucleus.
From a chemistry perspective, thiophene derivatives are used as corrosion inhibitors, organic semiconductors, organic light-emitting diodes (OLEDs) and organic field-effect transistors (OFETs).35
Though a few articles cover the medicinal attributes of thiophene in general,4,36–38 we have classified our approach in two ways to make our present work unique. The first section involves the development of thiophene derivatives via the traditional synthesis methods, also called traditional drug discovery practices. The second section involves computational techniques, including structural bioinformatics, high throughput virtual screening, molecular dynamics, and machine learning approaches. This is extended further to cover the USFDA-approved thiophene-containing drugs. Further, a medicinal chemistry-based perspective is presented on the preclinical development of thiophene-containing drugs, particularly anti-diabetic, anticancer, neurodegenerative, anti-inflammatory, anti-microbial and antioxidant agents. Additionally, we have sketched the critical synthetic methodologies for developing this versatile ring system and the commentaries on the metabolism, dosage form, route of administration, clearance, and half-life of the approved thiophene-containing drugs. A substantial weightage is also given to the elemental and structural analyses of the approved thiophene-based drugs.
2. Chemistry of thiophene and its derivatives
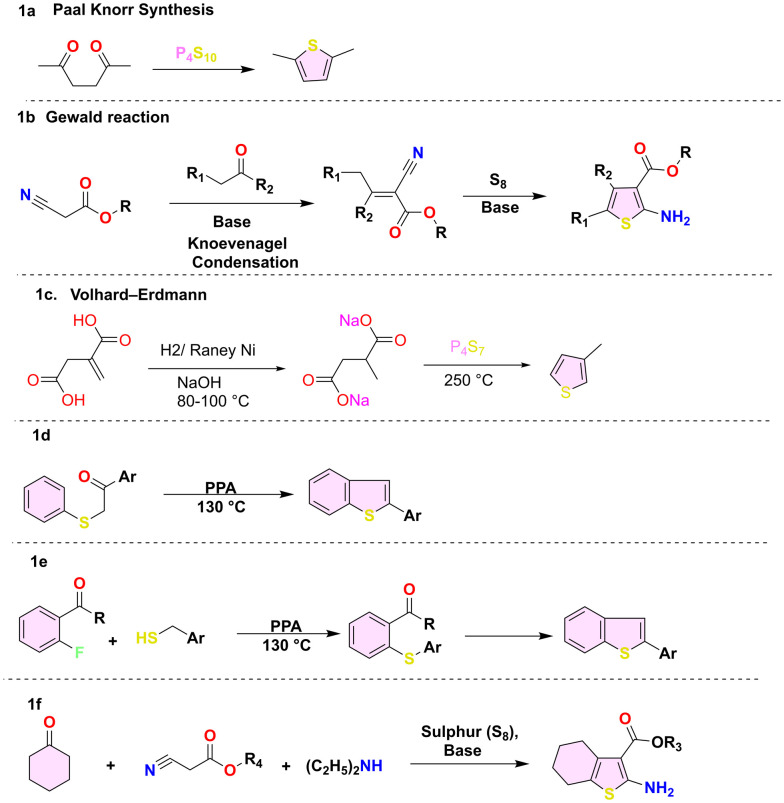
Thiophene is a five-membered ring with a molecular mass of 84.14 g mol−1 and a −38 °C melting point. Thiophene is soluble in organic solvents such as ether and alcohol. However, it is insoluble in water and forms azeotropes in organic solvents such as ethanol. The conventional approach to thiophene synthesis involved the Paal–Knorr and Gewald reactions. However, these methods were often characterised by harsh experimental conditions, limited effectiveness with numerous functional groups, and occasionally yielded low results. The well-known Paal–Knorr synthesis is a chemical reaction that converts 1,4-dicarbonyl compounds into thiophene in the presence of sulfiding reagents (e.g., Lawesson's reagent, phosphorus pentasulfide) under an acidic environment (Scheme 1a).39 In the Gewald reaction (Scheme 1b), condensation of an aliphatic aldehyde or ketone compound with an active cyano ester in the presence of a base and sulphur is performed.40 Another method is the Volhard–Erdmann cyclisation.41 This reaction involves the cyclisation of disodium succinate with 1,4-difunctional compounds like 1,4-diketones in the presence of phosphorus heptasulfide at a temperature of 250 °C (Scheme 1c). Similarly, the traditional method for synthesising benzo[b]thiophene involves cyclising a β-keto sulfide to form 2-aryl benzothiophene (Scheme 1d).42 Another method involved the Knoevenagel condensation of ortho-fluoroketone and benzyl thiol, forming S-benzyl ortho-acyl thiophenol (Scheme 1e).43 Another method for synthesis of 4,5,6,7-tetrahydrobenzothiophene derivatives is the treatment of cyclohexanone and ethyl cyanoacetate heated at 60 °C in the presence of sulphur and the addition of a base (triethylamine, dimethylamine) to form 4,5,6,7-tetrahydrobenzothiophene derivatives (Scheme 1f).44 Although historical, scientists have actively sought more efficient methods to synthesize thiophene.45,46
Scheme 1. Generalised synthetic methods (1a–1f) for thiophene synthesis.
2.1. Metal-catalysed methods
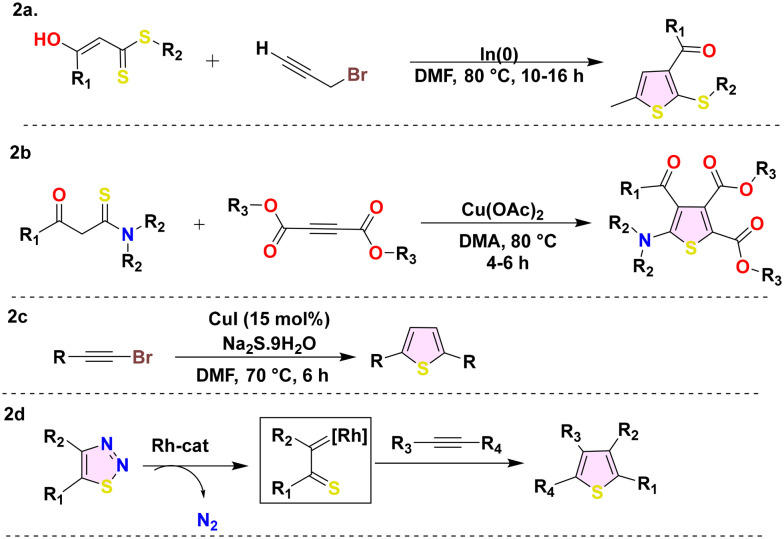
Metal-mediated synthetic approaches have extensively transformed the synthesis of thiophene. Copper is widely employed as a predominant metal, with scientists employing various copper ligands such as copper acetate and copper iodide.47 Furthermore, indium and rhodium are frequently used to synthesise thiophene derivatives. These techniques significantly promote the production of intricate thiophene derivatives by offering a high level of regioselectivity and the ability to accommodate a broad spectrum of functional groups. S. Chowdhury and colleagues developed a straightforward, effective, and practical method for producing highly substituted thiophene frameworks by interconnecting α-enolic dithioesters and propargylic bromides using the organoindium enzymatic coupling (Scheme 2a).48 Similarly, Ge et al. devised a simple synthetic technique for producing 2 amino thiophene derivatives (Scheme 2b) using the Cu(ii)-catalyzed addition/oxidative cyclization of easily accessible thioamides with alkynoates in an air environment.49 The regioselective synthetic schemes (Scheme 2c) for the substituted thiophene synthesis from haloalkynes were developed by Jiang et al., employing copper(i) as a catalyst and sodium sulfide as a sulfur source.50 Kurandina et al. introduced a novel approach to formulate rhodium-catalyzed regioselective fully substituted thiophene derivatives (Scheme 2d) by utilizing 1,2,3-thiadiazoles and alkynes as substrates.51
Scheme 2. Metal-mediated thiophene synthesis (2a–2d).
2.2. Metal-free approaches
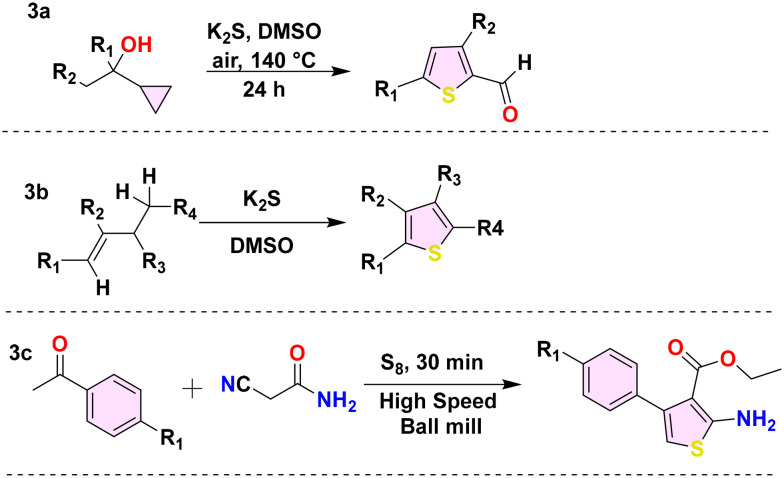
A multitude of metal-free methodologies have been devised for the synthesis of thiophene derivatives. These metal-free methods employ controlled reaction conditions and minimise metal toxicity, thus advancing the field of green chemistry. Several scientists have devised novel synthetic approaches, including using potassium sulfide or elemental sulfur or trithiocarbonate anion as sulfur sources.52,53 Common starting substrates for synthesising thiophene derivatives are cyclopropyl and buta-1-enes. Ting and colleagues synthesised substituted thiophene aldehyde (Scheme 3a) by utilizing a cyclopropyl ethanol derivative as a substrate and potassium sulfide as the sulfur source.54 Similarly, Liang and his colleague devised an innovative, cost-effective, transition-metal-free approach by substituting buta-1-enes with potassium sulfide (Scheme 3b).55 Furthermore, Shearouse and colleagues also produced substituted 2-amino thiophene derivatives (Scheme 3c) in a solvent-free environment by employing high-speed ball milling during Gewald synthesis with aryl-alkyl ketones.56
Scheme 3. Metal-free synthesis of thiophene derivatives (3a–3c).
2.3. Multicomponent reactions (MCRs)
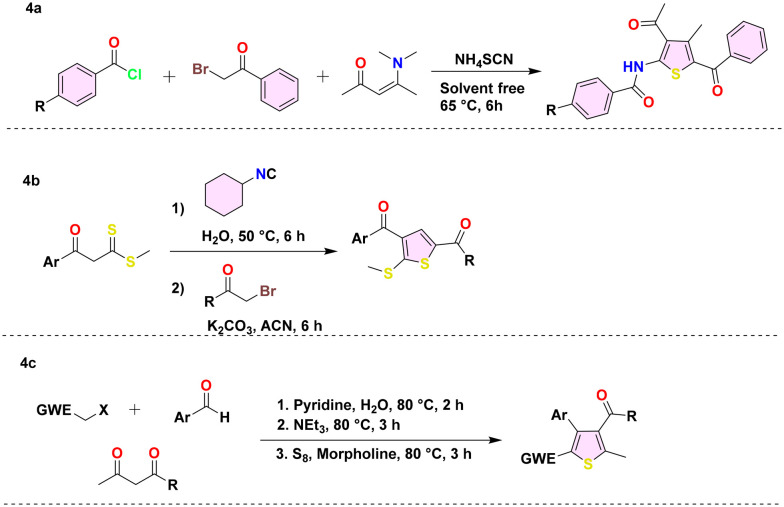
Modifications have been made to multicomponent reactions (MCRs), such as the Gewald reaction, to develop the one-pot synthesis of thiophene derivatives with varied substitution patterns. These approaches decrease the quantity of synthetic processes, thereby reducing waste and enhancing overall efficiency. Hossaini et al. devised a highly effective method for producing thiophene derivatives (Scheme 4a) by a single-step reaction involving ammonium thiocyanate, acyl chlorides, α-halo carbonyls, and enaminones. This reaction is conducted without solvents and occurs at a temperature of 65 °C.57 Likewise, Moghaddam and his colleagues devised an MCR for thiophene synthesis (Scheme 4b) by combining β-ketodithioesters, α-haloketones, and cyclohexylisocyanide in an aqueous solution.58 Using the Domino method, Adib et al. devised ways to synthesize thiophene derivatives. They synthesised one-, two-, and three-cycle thiophenes (Scheme 4c) in water by mixing aldehydes, activated methylene halides, and elemental sulfur with 1,3-dicarbonyl compounds.59
Scheme 4. Thiophene derivative synthesis (4a–4c) using multicomponent reactions.
Considering the chemical reactivities of these thiophene derivatives, the ring readily undergoes electrophilic aromatic substitution, like sulfonation and halogenation. However, alkylation and oxidation are difficult. The reactivity of thiophene towards electrophilic substitution reactions is higher than that of benzene.60 Thiophene ring sulphur atom has two lone pairs, where one electron pair of atoms is involved in the aromatic sextet and makes the thiophene aromatic in nature.
3. Exploring thiophene as a versatile pharmacophore in the USFDA drugs
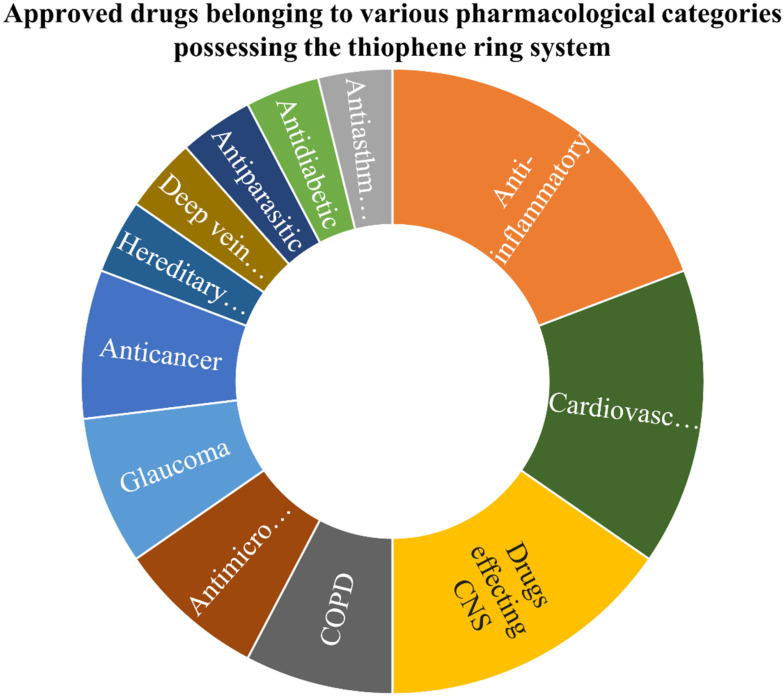
Thiophenes are recognised for their electron-rich characteristics and bioisosteric properties, which augment their capacity to interact with diverse biological targets. Thiophene, as discussed previously, remains one of the privileged pharmacophores in drug discovery. The analysis of USFDA-approved drugs revealed (Fig. 2) that 26 drugs bearing the thiophene ring system have been approved so far under numerous pharmacological categories. The deeper analysis revealed that out of 26 approved drugs, 5 were approved for treating inflammation conditions (oliceridine, tiaprofenic acid, tinoridine, tenoxicam, and suprofen). The prime targets for these drugs are cyclooxygenases (COX), lipoxygenases (LOX) and other enzymes involved in the biosynthesis of inflammatory proteins. The aromaticity and hydrophobicity of thiophenes likely enhance membrane permeability, thereby augmenting their efficacy as anti-inflammatory agents. This was followed by 4 approved drugs in the cardiovascular category (eprosartan, prasugrel, clopidogrel (platelet aggregation inhibitor), and ticlopidine). Four thiophene-based drugs have also been approved for the treatment of neurological disorders that include Parkinson's (1), antipsychotic (1), antiepileptic (1) and antianxiety (1). All the drugs targeting neurological disorders have to cross the blood–brain barrier (BBB), and the lipophilicity of thiophene contributes to its penetration. In the anticancer domain, thiophene-based drugs target kinases or apoptosis modulators. The planarity of the thiophene ring may contribute to the binding of ligands with receptors. Besides these 2 drugs, each was approved under anticancer (relugolix, raltitrexed), glaucoma (dorzolamide, brinzolamide), antimicrobial (cefoxitin, tioconazole) and COPD (tiotropium, aclidinium) categories. Moreover, one drug each was found to be approved as antidiabetic, antiparasitic, treatment of deep vein thrombosis and hereditary angioedema infection. In summary, thiophene-based FDA-approved drugs have exhibited extensive applicability in various therapeutic domains, prominently in cardiovascular, anti-inflammatory, and neurological indications.
Fig. 2. Sunburst chart revealing the approved drugs possessing the thiophene ring system under different pharmacological categories.
The detailed analysis of these drugs is compiled in Table 1. The compiled data (from the Drug Bank61) includes information on the drug name and approved year, followed by their biological target, mechanism of action, approved dosage, metabolism, and excretion information. The analysis of the compiled data indicated that most thiophene-based pharmaceuticals are excreted via urine following hepatic metabolism. Analysis of the reactive metabolites of thiophene-containing drugs revealed that only tiaprofenic acid and suprofen generate reactive metabolites devoid of pharmacological activity. In contrast, clopidogrel, prasugrel, and ticlopidine produce reactive metabolites with pharmacological effects. Table 1 indicates that icatibant is the sole thiophene-derived drug utilised to treat hereditary angioedema, a rare disease.
Pharmacological and some physiochemical parameters of thiophene-based USFDA-approved drugs.
| S no | Drug name | Category | Biological target | Mechanism | Dosage type and route | Half-life (in h) and excretion | Phase 1 metabolism | DB ID |
|---|---|---|---|---|---|---|---|---|
| (Brand name) | ||||||||
| (Approval year) | ||||||||
| 1. | Brinzolamide | Small molecule | Carbonic anhydrase-2 (CA-II) | Inhibition of CA-II reduces the formation of bicarbonates in the ciliary process and reduces the intraocular pressure | Solution and ophthalmic | 111 days, primarily eliminated through urine | Drug metabolised to N-desethylbrinzolamide | DB01194 |
| 1989 | ||||||||
| 2. | Dorzolamide | Small molecule | Carbonic anhydrase-2 (CA-II) | Inhibition of CA-II reduces the formation of bicarbonates in the ciliary process and reduces the intraocular pressure | Solution (2.0% w/v) and ophthalmic | Half-life >4 months, excreted out primarily through urine | Drug metabolised to N-desethyldorzolamide | DB00869 |
| 1995 | ||||||||
| 3. | Olanzapine | Small molecule | Dopamine receptor and serotonin receptors | Blocks dopamine in the mesolimbic pathway for its post-synaptic action | Tablet (15 mg) orally | Half-life is about 21 to 54 h (average 30 h), mainly excreted in the urine | Drug metabolized in the liver and most of the activity through cytochrome P450 (CYP-450) | DB00334 |
| 1996 | ||||||||
| 4. | Ketotifen | Small molecule | Histamine-receptor blocker (H1R) | Non-competitive antagonist of H1 histamine receptors | Solution, syrup, tablet | Half-life is about 3 to 5 h, excreted in the urine, primarily as metabolite | Drug metabolised in the liver and form three metabolites; major one is the N-glucuronide derivative | DB00920 |
| 1999 | ||||||||
| 5. | Suprofen | Small molecule | Cyclooxygenase (COX) inhibitor | The drug inhibits the prostaglandin synthesis with COX inhibition, resulting in the analgesic effect | Tablet orally | Half-life 72 h | Metabolised in the liver via CYP-450 isozyme 2C9 | DB00870 |
| 1998 | ||||||||
| 6. | Tioconazole | Small molecule | Ergosterol inhibition | Prevents the synthesis of ergosterol, which increases the cellular permeability | Ointment via topical route | NA | Metabolization via glucuronide conjugation | DB01007 |
| 1983 | ||||||||
| 7. | Duloxetine | Small molecule | Inhibition of sodium-dependent serotonin, dopamine, and noradrenaline transporters | Drug inhibits the serotonin (5-HT2, 5-HT3), dopamine, and norepinephrine uptake | Tablet orally | Half-life 12 h, excreted mainly in urine | Metabolised in the liver via CYP2D6 and CYP1A2 | DB00476 |
| 1998 | ||||||||
| 8. | Canagliflozin | Small molecule | Sodium–glucose co-transporter 2 (SGLT-2) inhibitors | Prevents SGLT-2 cotransporters, resulting in reduction | Tablet orally | Half-life 10.6 h, mainly excreted through faeces | Metabolised via O-glucuronidation (UGT1A9 and UGT2B4) | DB0890 |
| 2013 | ||||||||
| 9. | Rotigotine | Small molecule | Dopamine receptor agonist | Drugs mimic the dopamine receptor and activate neurotransmitter | Patch via transdermal route | Half-life 5 to 7 h, route of elimination is urine (71%), fecal (23%) | Metabolised via CYP isoenzyme, sulfotransferase | DB05271 |
| 2007 | ||||||||
| 10. | Ticlopidine | Small molecule | Adenosine diphosphate inhibitor | Inhibition activation of platelet and aggregation (P2Y12 ADP receptors) | Tablet orally | Half-life 7.9 h, mostly in the urine | Metabolised by the liver | DB00208 |
| 1991 | ||||||||
| 11. | Clopidogrel | Small molecule | Adenosine diphosphate inhibitor | Inhibition activation of platelet and aggregation (P2Y12 ADP receptors) | Tablet orally | Half-life 6 h, excreted 50% in the urine and 46% in the feces | Metabolised by cytochromes enzymes (CYP1A2, CYP2C9, CYP2B6) | DB00758 |
| 1997 | ||||||||
| 12. | Prasugrel | Small molecule | Adenosine diphosphate inhibitor | Inhibition activation of platelet and aggregation (P2Y12 ADP receptors) | Tablet orally | Half-life 7.4 h, mainly excreted in urine | Metabolised by CYP-450 enzymes | DB06209 |
| 2009 | ||||||||
| 13. | Eprosartan | Small molecule | Angiotensin II receptor antagonist (AT1 inhibitor) | The drug inhibits the AT1 receptor, which results in vasodilation | Tablet orally | Half-life 5–9 h, NA | Metabolised by CYP-450 enzymes | DB00876 |
| 1997 | ||||||||
| 14. | Icatibant | Small molecule | Bradykinin B2 receptor (BB2 receptor) inhibitor | Selective BB2 receptor inhibitor reduces the incidence of hereditary angioedema | Solution via parenteral route | Half-life 1.4 h, primarily excreted in the urine | Metabolised by CYP-450 enzymes | DB06196 |
| 2011 | ||||||||
| 15. | Rivaroxaban | Small molecule | Inhibits the coagulation factor | The drug inhibits the free and clot-bound factor Xa | Tablet orally | Half-life 5–9 h, excreted into urine | Metabolised by cytochrome enzymes | DB06228 |
| 2011 | ||||||||
| 16. | Raltitrexed | Small molecule | Thymidylate synthase (TS) inhibition | The drug is a folic acid antagonist via inhibiting TS, which results in cell death | Solution via parenteral route | Half-life 192 h, NA | Metabolised into folyl polyglutamate synthetase | DB00293 |
| 1998 | ||||||||
| 17. | Tenoxicam | Small molecule | Cyclooxygenase (COX) inhibitor | The drug inhibits prostaglandin synthesis with COX inhibition, resulting in the analgesic effect | Tablet orally | Half-life 72 h, NA | Metabolised in the liver to 5-hydroxy tenoxicam | DB00469 |
| 2000 | ||||||||
| 18. | Tiagabine | Small molecule | Gamma-aminobutyric acid (GABA) inhibition | Drugs block the sodium and chloride-dependent GABA transporter | Tablet orally | Half-life 7–9 h, mainly through faeces | Drug excreted in the urine (25%) and faeces (63%) and other remaining unchanged | DB00906 |
| 1997 | ||||||||
| 19. | Aclidinium | Small molecule | Muscarinic acetylcholine receptor | The drug inhibits the M3 receptor located in the lungs, resulting in the relaxation of smooth muscles | Capsule for respiratory inhalation; oral | Half-life 24–44 h, excreted in urine | 74% of drugs excreted in urine, and others cleaved into dithienylglycolic acid and inactive metabolite N-methylscopine | DB08897 |
| 2012 | ||||||||
| 20. | Tiotropium | Small molecule | Muscarinic acetylcholine receptor | The drug inhibits the M3 receptor located in the lungs, resulting in the relaxation of smooth muscles | Capsule for respiratory inhalation; oral | Half-life 24 h (chronic obstructive pulmonary disease) COPD and 44 h in asthma, excreted unchanged in urine | 74% of drugs excreted in the urine, and others cleaved into dithienylglycolic acid and inactive metabolite N-methylscopine | DB01409 |
| 2004 | ||||||||
| 21. | Cefoxitin | Small molecule | Cell wall inhibition | d-Alanyl-d-alanine carboxypeptidase DacC inhibition | Solution with intravenous and intramuscular injection | Half-life is 41 to 59 minutes, excreted in urine | Most of the drugs excreted from the kidney unchanged | DB01331 |
| 1973 | ||||||||
| 22. | Lotilaner | Small molecule | Gamma-aminobutyric acid (GABA)-gated chloride channel | Non-competitive inhibition of GABA-gated chloride channel | Ophthalmic solution | Half-life 11 days, NA | Metabolised by cytochrome enzyme | DB17992 |
| 23. | Oliceridine | Small molecule | μ-Opioid receptor agonist | Activate the μ-opioid receptor | Intravenous injection | Half-life is 1.3 to 3 h, mainly through urine | Metabolised by CYP3A4 and CYP2D6 to oxy-oliceridine | DB14881 |
| 24. | Relugolix | Small molecule | Gonadotropin-releasing hormone (GnRH) receptor agonist | GnRH receptor reduces the release of luteinising hormone | Tablet orally | Half-life 25 h, mainly through faeces | Metabolised by CYP3A | DB11853 |
| 25. | Avatrombopag | Small molecule | Agonist thrombopoietin (TPO) receptor | TPO stimulates the production of platelets | Tablet orally | Half-life 19 h, mainly faecal excretion | Metabolised by CYP3A and CYP2C9 | DB11995 |
| 26. | Tiaprofenic acid | Small molecule | COX and LOX inhibitor | Inhibit prostaglandin synthesis | Capsule, tablet orally | 1.5–2.5 hours, NA | Hepatic | DB01600 |
The predominant route of administration for most drugs is oral (15 drugs), while three are ophthalmic, two are topical, two are inhalation, and three are injectable preparations. We also explored the route of elimination of all drugs and found that the primary route of elimination of thiophene-based drugs is through urine and then faeces.
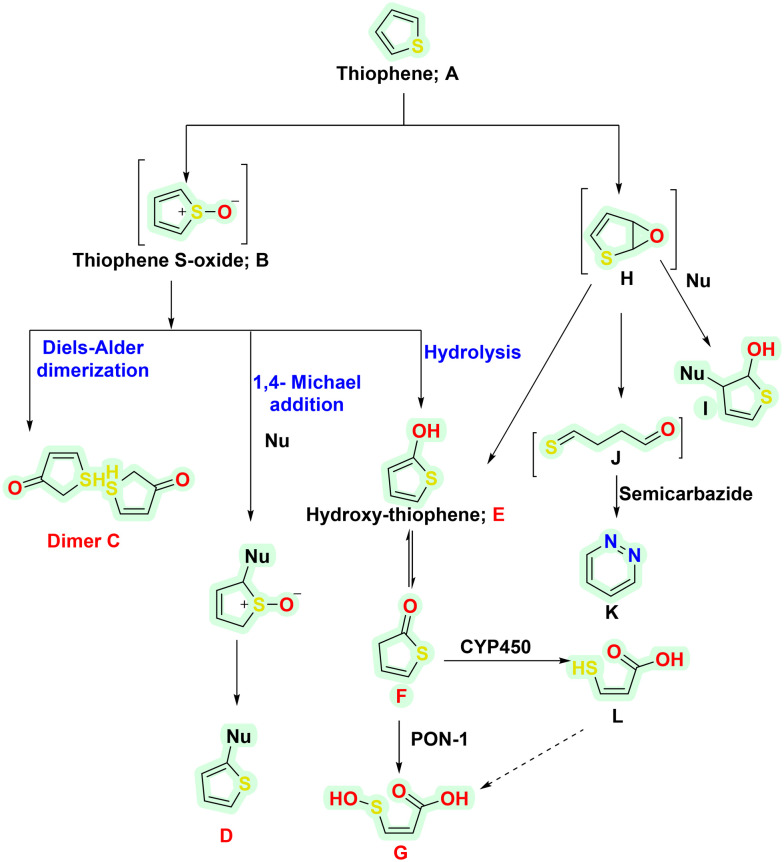
The implication of thiophene metabolism is a well-known phenomenon that generates reactive metabolites.62 Some reactive metabolites are also associated with toxicities and intended off-target effects. The ill effects are known to occur chiefly via the CYP450-mediated oxidation reaction.63,64 Though most of the metabolites are detoxified and excreted eventually, some possess toxicity threats. Thiophene-containing drugs are majorly metabolised via S-oxidation and epoxidation pathways, as briefly shown in Scheme 2. In S-oxidation, thiophene (A) is oxidised to thiophene S-oxide (B) upon its interaction with CYP450, NADPH and molecular oxygen (O2). B further undergoes dimerisation via the Diels–Alder reaction to form a dimeric adduct (C) or reacts with nucleophiles (Nu-) via 1,4 Michael addition to form other adducts D. The generated adduct poses a toxic threat as it is chemically reactive and reacts with protein nucleophiles such as cysteine, serine, lysine, histidine, and tyrosine. Besides this, D also interacts with CYP450s, making them inactive. However, upon its reaction with glutathione (GSH), the adduct D forms a GSH-adduct (phase 2 product), thus undergoing renal excretion. Besides this, B undergoes hydrolysis, leading to hydroxy-thiophene (E) formation. E may further form sulphenic acid (F) or Gvia the catalytic activity of paraoxonase 1 (PON-1). The epoxidation pathway mediates the second reaction. The initial intermediate formed is thiophene epoxide (H). It reacts with nucleophiles or undergoes hydrolysis to form I and E, respectively. Also, epoxide may be oxidised to form 4-thioxobutanal (J), followed by carbazide stable intermediate K (Scheme 5).
Scheme 5. Proposed mechanism for the metabolism of thiophene-containing drugs.
The chemical structures of the USFDA-approved drugs containing the thiophene ring are sketched in Fig. 3.
Fig. 3. Chemical structures of USFDA-approved drugs containing the thiophene ring.
4. Role of bioinformatics tool in thiophene drug design
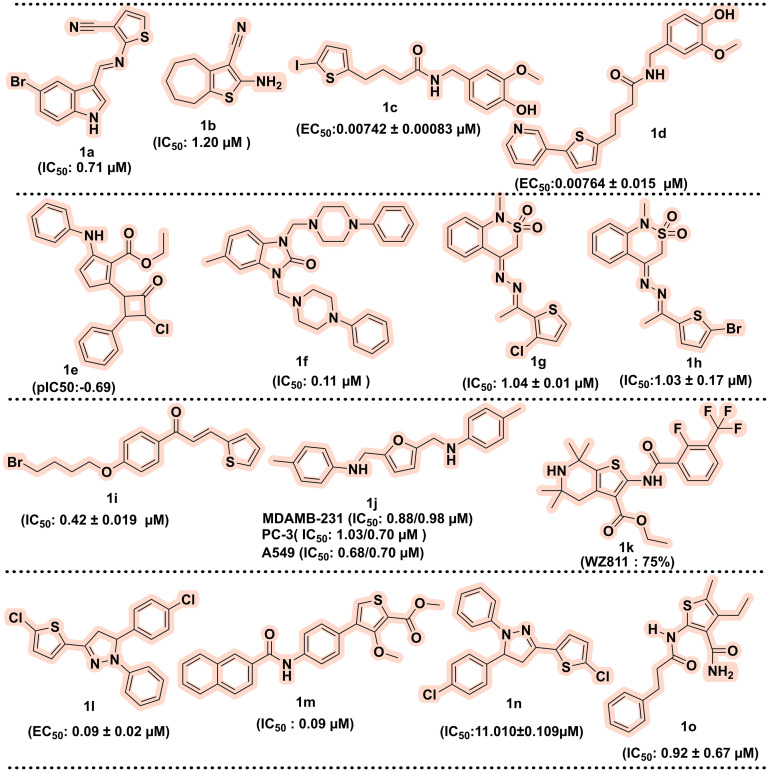
Nowadays, structural bioinformatics is one of the crucial components of modern drug design, and it uses in silico tools that assist in rational design, optimisation, and target identification. Structural bioinformatics involves the three-dimensional (3D) structures of biological targets (such as enzymes, proteins, or nucleic acids) in identifying the interaction of small molecules with these targets. Various computational approaches such as molecular docking using structure-based drug design (SBDD) and ligand-based drug design (LBDD), homology modelling, machine learning (ML) predicted models, and quantitative structure–activity relationship (QSAR) model and molecular dynamics facilitate the identification and optimisation of lead compounds and aid in streamlining the drug development process.65,66 This section discusses the contribution of structural bioinformatics in optimising and identifying new thiophene nuclei as potential drug candidates. In 2023, Luna et al. screened in silico anti-leishmanial agents using 2-aminothiophene derivatives. In this study, the machine learning (ML) model, along with the quantitative structure–activity relationship (QSAR) model, was used for screening the ChEMBL database (1862 compounds). The top eleven 2-amino thiophene molecules that exhibited good drug-likeness profiles, such as following Lipinski's rules, were selected and subsequently evaluated against Leishmania amazonensis. Among all selected compounds, 1a and 1b showed the most potent activity against amastigote and promastigote forms of the parasite with IC50 values of 0.71 and 1.20 μM.67 Next, Romeo et al. (2023) designed novel 4-(thiophen-2-yl) with the help of molecular docking, molecular dynamics simulation, and thermodynamic studies based on the in silico drug designing transient receptor potential vanilloid 1 (TRPV1) agonist. Based on the in silico drug design, at position 5 of the thiophene moiety, lipophilic iodine group, benzene, and pyridine were added. Docking studies indicated that 1c and 1d are involved in the essential hydrophobic interaction with Leu547 and Phe543. Also, the same compounds 1c and 1d showed good TRPV1 with EC50 values of 0.00742 ± 0.00083 and 0.00764 ± 0.015 μM and protection against ROS-induced oxidative stress among all synthesised compounds. Further, compound 1c was found to have a good neuroprotective effect and exhibited optimal physiochemical properties.68 In the same year, Natarajan et al. designed novel thiophene derivatives connected with lactam via the 2-dimensional quantitative structure–activity relationship (2D-QSAR) method and checked in silico for their binding affinity and interactions for antimicrobial agents against E. coli. The fifteen newly designed thiophene derivatives obtained from 2D-QSAR were further evaluated using Autodock via molecular docking, Swiss ADME, and Protox-II software for the physiochemical properties and toxicity of small molecules. Among fifteen, 1e exhibited a pIC50 value of −0.69, and other compounds had pIC50 values of 0.87 to 1.46. The docking study concluded that the chloro group at the 4th and 2nd positions of benzene interacted well with the E. coli gyrase complex (PDB ID: 6YD9). Further, ADMET and toxicity profile of molecules showed good bioavailability and could be a good lead as an antibacterial agent.69 Next, Güven et al. designed and synthesised thiophene derivatives and evaluated them against urease inhibition in vitro. All synthesised compounds showed IC50 values in the range of 0.64 ± 0.099 and 0.11 ± 0.017 μM compared to the standard drug IC50 of 0.51 ± 0.028 μM. Molecular docking revealed the crucial amino acids His221, Asp223, His322, Glu222, Arg338, and metal ion Ni2+. The compounds showed good activity, such as compound 1f (IC50: 0.11 μM), which showed interactions with these amino acids.70 Next, Javid et al. performed molecular docking of 2,1-benzothiazine derivatives and evaluated them against monoamine oxidase (MAO A and B) using PDB ID: 2Z5Y and 2V5Z. Among all designed compounds, 1g was found to be the most potent, with an IC50 value of 1.04 ± 0.01 μM for MAO-A and 1h for the MAO B inhibitor with an IC50 value of 1.03 ± 0.17 μM.71 In 2022, Hasan et al. designed novel thiophene chalcones/coumarins using molecular docking followed by molecular dynamics and ADMET studies and evaluated them against acetylcholinesterase inhibition. Compound 1i is the most active compound with an IC50 value of 0.42 ± 0.019 μM for inhibiting the enzyme acetylcholinesterase.72 In 2019, Murugavel et al. designed a thiophene derivative linked to triazole and pyridine moieties using an in silico quantum study. The compound 1j was investigated via a molecular docking study and ADMET parameters against topoisomerase IIα. The further molecule was synthesised and tested against MTT assay using three cell lines PC-3 (1.03/0.70 μM), MDAMB-231 (0.88/0.98 μM) and A549 (0.68/0.70 μM), and the crystal structure was determined by single crystal XRD.73 Gaines et al. synthesised a new CXCR4 antagonist with the help of molecular modelling. Among fifteen hits, compound 1k (75%) was more potent against metastatic cells than reference WZ811 (62%).74 In 2018, Rodriguez et al. synthesised novel thiophene derivatives with a structure–activity relationship (SAR) and tested in vitro against L. major (promastigotes). Among the synthesised compounds, 1l showed potent activity with an EC50 value of 0.09 ± 0.02 μM.75 In 2017, Gulipalli et al. designed a novel series of 3-methoxythiophene-2-carboxylate compounds and evaluated it for anticancer activity. The series of 3-methoxythiophene-2-carboxylate derivatives were designed and docked to the active site of protein tyrosine phosphatase 1B. These designed derivatives were also subjected to ADMET properties via in silico techniques. The best-scored compounds synthesised and checked in vitro for anticancer activity, among which 1m was the most potent with IC50 value in the range of 5.25 μM, and for the MCF-7 cell line, IC50 is 0.09 μM.76 In 2019, Murugavel et al. developed thiophene pyridine based compounds using computational quantum chemical study and evaluated the anticancer property against three human cancer cell lines A549, PC-3, MDAMB-231 and compound 1n was found most potent derivative.77 Among the designed and synthesised compounds 1n, the highest inflammatory effect with the aid of molecular docking study, SAR was established.78 In 2007, Louise-May et al. used virtual screening of 5,4-dialkyl thiophene substituted derivatives against the HCV assay. A SAR study revealed that di-alkyl thiophene derivatives inhibit single and double-strand RNA in the 0.92 ± 0.67 μM range for compound 1o.79 The chemical structures of the thiophene-containing compounds identified via structural bioinformatics are illustrated in Fig. 4.
Fig. 4. Chemical structures of thiophene-containing compounds identified via structural bioinformatics.
5. Exploring thiophene derivatives in preclinical trials
In the present section, we have compiled information on the preclinical development of thiophene-containing drugs, particularly as an anti-diabetic, anticancer, neurodegenerative, anti-inflammatory, anti-microbial agent and antioxidant. The idea is to provide readers with a summary of ongoing research in these particular areas about the thiophene nucleus as a core ring system.
5.1. Thiophene nucleus in the development of anti-diabetic agents
Diabetes mellitus (DM) is a chronic and progressive metabolic disorder characterised by insufficient insulin production or resistance.80 According to the International Diabetes Federation, approximately 403 million people with diabetes were diagnosed in 2019 worldwide, and its frequency was assessed to be 700 million up to 2045. Diabetes enhances the risk of various diseases such as cardiovascular diseases, kidney diseases, retinopathy, cancer, and peripheral neuropathy. The primary strategy used for targeting diabetes includes the modulation of insulin receptors and signalling. The diverse strategies for controlling the blood glucose level include the use of 1) glucose transporters (GLUTs) to increase the glucose uptake through cells by promoting the GLUT expression, particularly GLUT4 or facilitating its translocation through the cell membrane and improving the cellular insulin response;81 2) incretin, gut-derived peptide hormones, such as glucagon-like peptide (GLP-1), play a significant role in regulating insulin secretion and glucagon release;82 3) pancreatic β-cell maintains the optimum insulin levels;83 4) alpha-glucosidase and amylase inhibition is one of the potential strategies to slow down glucose absorption or reduce the postprandial blood glucose level; 5) sodium–glucose-co-transporter 2 (SGLT-2) inhibition is another attractive target that enhances glucose excretion via urine; 6) dipeptidyl peptidase-4 (DPP-4) is another druggable target that deactivates incretin hormones, which possess a vital role in regulating blood glucose levels. Inhibition of DPP-4 enhances insulin secretion and lowers blood sugar, making it a target for type 2 diabetes; 7) peroxisome proliferator-activated receptors (PPARs) are the nuclear receptors that regulate the gene expression associated with glucose homeostasis. Thus, PPAR agonists also serve as an important target and maintain insulin sensitivity, and many more targets are available for diabetes.84
Considering thiophene in antidiabetic drug discovery, canagliflozin was approved by the US FDA for treating diabetes in 2013. The drug was again approved in 2018 for treating cardiovascular events associated with type 2 diabetes. Besides this, ipragliflozin is another thiophene-containing drug in clinical trial phase III explored for its antidiabetic potential via SGLT-2 inhibition.85 Ipragliflozin is presently approved for its clinical use in Japan (2014) and Russia (2019) and is awaiting global approval by the US FDA.
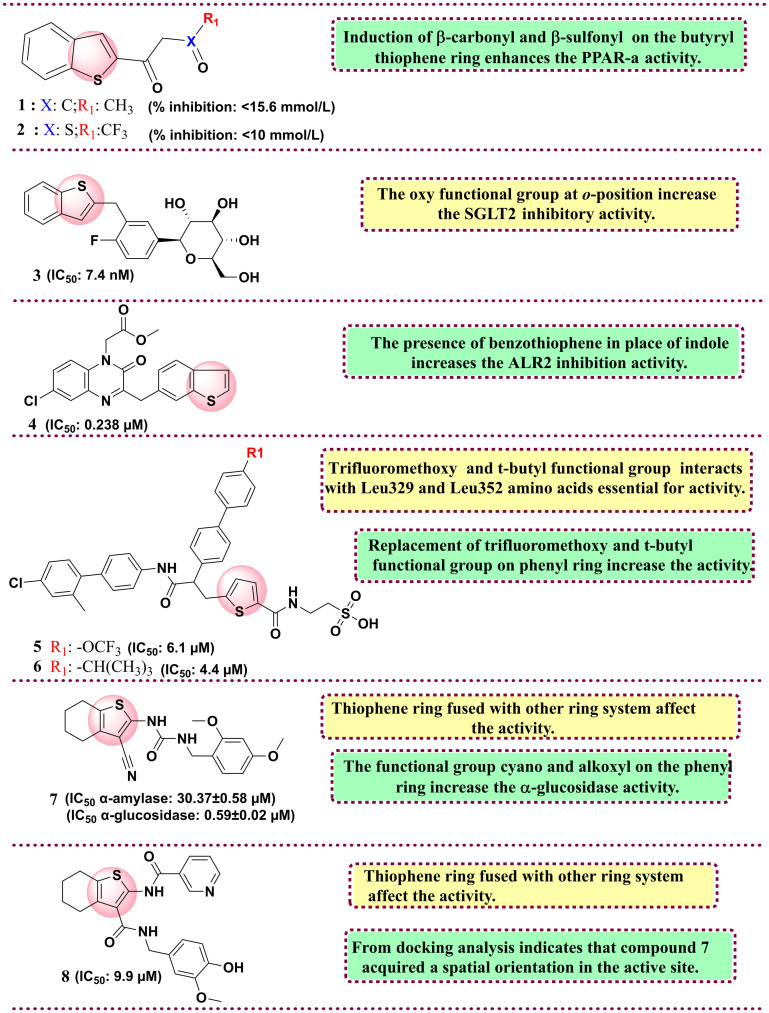
Regarding the preclinical arena, the important studies include the research by Shao and group. The group synthesised thiophene and benzothiophene analogues and screened them in vitro and in vivo against peroxisome proliferator-activated receptor γ (PPAR γ) in human hepatoma (HepG2) cells. The molecules were developed considering the associated detrimental effects of thiazolidine drugs and in the quest to develop safer PPAR γ agonists. Among all synthesised derivatives, compounds 1 and 2 showed the highest transactivation activity, with 120% and 102.14%, compared to reference rosiglitazone, with 311.53%. In their in vivo study, both compounds displayed reduced blood glucose levels for compounds 1 < 15.6 mmol L−1 and 2 < 10 mmol L−1. SAR studies showed that β-carbonyl and β-sulfonyl substituents on the thiophene ring increase the PPAR γ activity.86 In 2012, Imamura et al. synthesised a class of heteroaromatics linked with C-glucosides. The molecules were chemosynthetically derived from phlorizin, a naturally occurring O-glucoside, which is reported as a potential SGLT inhibitor but possesses selectivity issues towards SGLT isoforms and has poor metabolic stability. Among the synthesised heteroaromatics, benzothiophene with C-glucosides derivative (3) showed potent activity against SGLT-2 with an IC50 value of 7.4 nM with the highest selectivity of 254-fold towards SGLT-1. SAR studies showed that the induction of an ether linkage at the ortho position enhances the SGLT2 inhibitory activity.87 In 2015, Qin et al. reported the screening of quinoxaline derivatives linked with benzothiophene or indole against an aldose reductase (ALR2) inhibitor. The rationale behind the development of ALR2 inhibitors was owing to the peculiar feature of the ALR2 enzyme that is associated with chronic diabetic complications and, at the same time, increases cardiovascular risks. ALR2 plays a vital rate-determinant role in the polyol pathway, particularly in hyperglycemia. The activation of this pathway allows the conversion of glucose to sorbitol and its accumulation owing to its higher polarity than glucose, reversing the redox potential and giving rise to oxidative stress, a main culprit in diabetes progression. The group rationally designed and synthesised the quinoxalinone-linked phenol compounds with the potential to act as aldose reductase inhibitors and antioxidants. The synthesised compounds show good activity, among which compound 4 showed the highest selectivity and potential with an IC50 value of 0.238 μM linked with benzothiophene. SAR studies investigated the induction of benzothiophene in place of indole, which increases the activity. Induction of o-hydroxyl or p-hydroxyphenyl increases the ALR2 inhibition activity.88
In 2018, Li et al. reported a new thiophene-linked biaryl amide derivatives and evaluated them against glucagon bindings to the glucagon receptor (GCGR). The molecules were rationally designed considering the pharmacophoric features of Bay 27-9955, a glucagon receptor antagonist and MK-0893, which was found to possess an allosteric inhibition on GCGR. Among all synthesised compounds, two compounds, 5 and 6, were found to have promising activity against GCGR with IC50 values of 6.1 and 4.4 μM and cAMP functional activities of 4.4 and 14.4 μM, respectively. SAR studies indicate that the 4th position over the phenyl substitution on compounds 5 and 6 is essential. Compound 5 has trifluoromethoxy moiety, and compound 6 contains a t-butyl functional group. Both are in hydrophobic interactions with Leu329 and Leu352 amino acids and thiophene rings with Leu399. The trifluoromethoxy has more significant activity than the t-butyl functional group on the phenyl ring.89 In 2021, Xie et al. reported novel thiophen- and thiazole-urea derivatives and evaluated them against α-glucosidase and amylase inhibition. The molecules were rationally developed after performing lead optimisation of their previously in-house developed molecule (5a) with a potent α-glucosidase inhibitory potential (87.76%). Among the synthetics, compound 7 was found to have promising activity against α-amylase (IC50 = 30.37 ± 0.58 μM) and α-glucosidase (IC50 = 0.59 ± 0.02 μM). SAR analysis indicates that the thiophene ring fused with other ring systems affects the activity. The functional group cyano and alkoxyl on the phenyl ring increase the α-glucosidase activity. From docking analysis, it was deduced that compound 7 bound to the allosteric site showed non-competitive α-glucosidase inhibition.90 In 2023, Wang et al. reported the cycloalkylthiophenyl nicotinamide analogues evaluated against α-glucosidase in vitro and in vivo. The molecules were rationally developed through the lead optimisation strategies applied on their in-house developed and potent α-glucosidase inhibitors derived from 1-(3-cyano-4,5,6,7-tetrahydrobenzo[b]thiophen-2-yl)-3-(2,4-dimethoxybenzyl)urea and N-(3-cyano-4,5,6,7-tetrahydrobenzo[b]thiophen-2-yl)hydrazine-1-carboxamide. The important pharmacophoric groups from both derived series were retained to rationally design and develop a series of cycloalkyl[b]thiophen-2-ylnicotinamide derivatives. The synthesised compounds exhibit good activity as compared to reference acarbose. Among all the synthesised derivatives, compound 8 showed the highest α-glucosidase activity with an IC50 value of 9.9 μM over the reference acarbose with an IC50 value of 258.5 μM with higher selectivity against α-glucosidase than amylase by 7.4-fold. The fluorescence quenching method determined that compound 5 directly binds to α-glucosidase and reduces the blood glucose in rats. Docking analysis indicates that compound 8 acquired a spatial orientation in the active site and interacts with primary amino acid residues essential for activity, such as Phe601, Arg552, Trp432 and Asp568.91 All reported thiophene derivatives as anti-diabetic inhibitors are shown in Fig. 5. The summary of antidiabetic drugs bearing the thiophene ring is compiled in Table 2.
Fig. 5. Reported thiophene derivatives as anti-diabetic agents.
Compilation of ongoing research on thiophene derivatives for the development of antidiabetic agents.
| Compound code | Indication | Biological target | Mechanism | Status | Mode of drug discovery | Ref. |
|---|---|---|---|---|---|---|
| 1 | Antidiabetic agent | Peroxisome proliferator-activated receptor γ (PPAR γ) | Inhibits PPAR γ, which plays a role in lipid metabolism | Preclinical | Traditional | 86 |
| 2 | Antidiabetic agent | Peroxisome proliferator-activated receptor γ (PPAR γ) | Inhibits PPAR γ, which plays a role in lipid metabolism | Preclinical | Traditional | 86 |
| 3 | Antidiabetic agent | SGLT-2 | Inhibits SGLT-2 and reduces blood glucose | Preclinical | Traditional | 87 |
| 4 | Antidiabetic agent | Aldose reductase (ALR2) inhibitor | Reduced glucose to sorbitol | Preclinical | Traditional | 88 |
| 5 | Antidiabetic agent | Glucagon binding to the glucagon receptor (GCGR) | GCGR is used to maintain blood glucose | Preclinical | Traditional | 89 |
| 6 | Antidiabetic agent | Glucagon binding to the glucagon receptor (GCGR) | GCGR is used to maintain blood glucose | Preclinical | Traditional | 89 |
| 7 | Antidiabetic agent | α-Glucosidase and amylase inhibition | Enzyme responsible for breaking down carbohydrates | Preclinical | Traditional | 90 |
| 8 | Antidiabetic agent | α-Glucosidase inhibition | Enzyme responsible for breaking down carbohydrates | Preclinical | Traditional | 91 |
5.2. Thiophene derivatives as anticancer agents
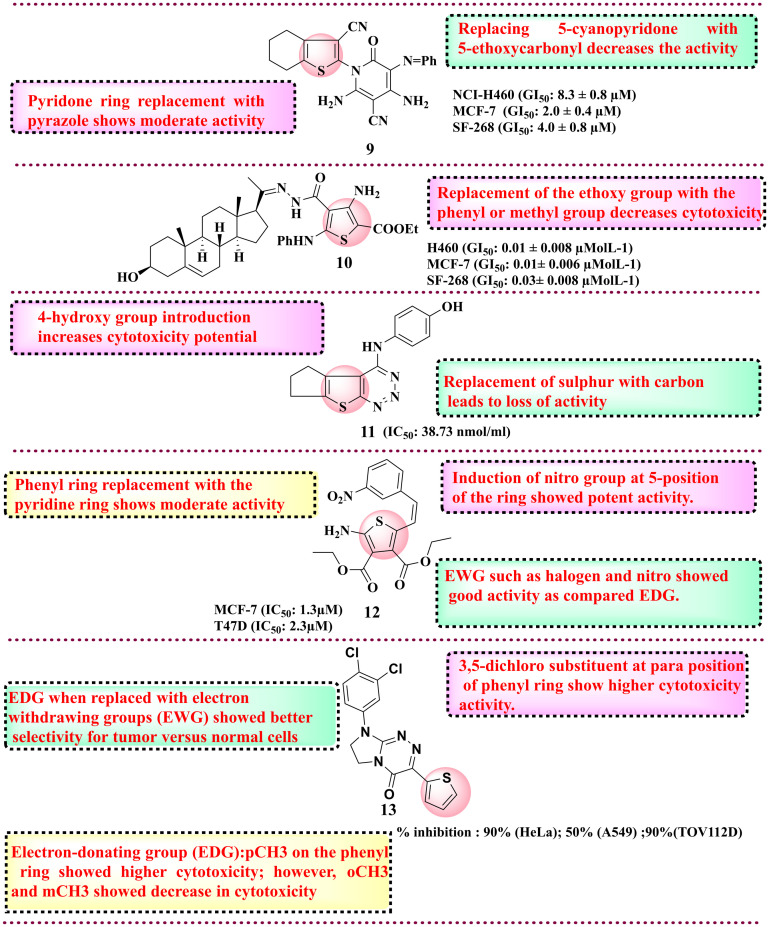
Cancer is one of the most challenging diseases affecting worldwide with an increase in the death rate. Cancer is the uncontrolled growth of cells resulting in a solid mass known as tumour. In the past three decades, the number of patients diagnosed with tumours has more than doubled, and it is anticipated that by 2030, approximately 27 million individuals worldwide will be living with cancer, with 17 million deaths occurring annually. The disease has become a significant global health issue, attracting widespread attention and concern. Currently, available therapeutics, such as chemotherapy and radiotherapy, have a very narrow or limited therapeutic index due to acquired toxicity and resistance for different types of cancers. This occurred due to complex physiological changes in the cell function, metastasis and apoptosis. Medicinal chemists have paid significant attention to thiophene-substituted derivatives because of their anticancer properties. Apart from providing molecular flexibility and substituent versatility, thiophene-based compounds target various over-expressed cancer proteins, such as tyrosine kinase receptors, cyclin-dependent kinases (CDKs), topoisomerases, HDAC and others. Thiophene derivatives have increasing utility as anticancer agents. Here, a few reports are covered to portray the diversification of thiophene derivatives in anticancer drug discovery. Various reports are available where thiophene derivatives have shown potential chemotherapeutic activity. In 2011, Shams et al., in the quest to develop new potential anticancer compounds, reported novel 2-cyano-N-(3-cyano-4,5,6,7-tetrahydrobenzothipohen-2-yl)-acetamide derivatives.92 The group explored and evaluated their synthetics against anti-tumour activities on three cell lines such as CNS cancer (SF-268), breast adenocarcinoma (MCF-7) and non-small lung cancer (NCI-H460). Out of 22 synthesised derivatives, compound 9 displayed potent activity against three human cell lines in vitro NCI-H460 cell line (GI50: 8.3 ± 0.8 μM), MCF-7 cell line (GI50: 2.0 ± 0.4 μM) and SF-268 cell line (GI50: 4.0 ± 0.8 μM) as compared to the control drug doxorubicin, as shown in Fig. 6. SAR studies showed that the cyano group at the 5th position imparts more activity than 5-ethoxycarbonyl. Subsequently, in 2012, the same group explored new pregnenolone-based thiophene derivatives for their anticancer potential. From all the synthesised derivatives, compound 10 showed potent activity against the H460 lung cancer cell line (GI50: 0.01 ± 0.008 μmol L−1), MCF-7 cell line (GI50: 0.01 ± 0.006 μmol L−1) and SF-268 cell line (GI50: 0.03 ± 0.008 μmol L−1) respectively, as compared to doxorubicin. SAR analysis indicates that the presence of phenyl and methyl groups does not affect activity, whereas the presence of the ethoxy group increases cytotoxicity.93 In 2014, Said and Elshihawy synthesised thieno(3,2-d)(1,2,3)-triazine derivatives to target tyrosine kinase. The molecules were rationally designed considering the catalytic binding site of the kinase receptor where the ATP binds. The idea was to potentially inhibit the binding of ATP within the kinase domain and thereby obstruct downstream signaling pathways, such as PI3K/Akt and MAPK, which are essential for cancer cell survival and angiogenesis. The molecules were further evaluated for their anti-proliferative activity against the MCF-7 cell line. From all synthesised derivatives, compound 11 (IC50: 38.73 nmol ml−1) showed potent activity against the breast cancer cell line. SAR studies revealed that the presence of a sulphur group is essential for activity. In addition, the induction of the –OH group on the phenyl ring showed an increase in the cytotoxicity potential.94 Bozorov et al. reported the evaluation of diethyl 2,5-diaminothiophene-3,4-dicarboxylate derivatives for their anticancer activity. They evaluated their compounds for anticancer, antidiabetic, and antimicrobial efficacy. Upon assessing their anticancer potential, they discovered that their compounds exhibited considerable toxicity against the breast cancer cell line (MCF-7), particularly compound 12. Compound 12 exhibited superior anticancer potential among all synthesised compounds compared to the positive control. Compound 12 exhibited significant anticancer efficacy against MCF-7 (1.3 μM) and T47D (2.3 μM), in contrast to the control doxorubicin, which demonstrated IC50 values of 6.75 μM for MCF-7 and 15.5 μM for T47D. SAR studies revealed that the presence of electron-withdrawing groups at the phenyl ring has higher cytotoxicity activity than electron-donating groups. In addition, the furfural ring in place of the phenyl ring with the nitro group at the 5th position increases the cytotoxicity activity.95 In 2015, Sztanke and colleagues synthesized thiophene-based derivatives by substituting the heteroaromatic furan moiety in previously established anticancer compounds that exhibit activity against T47D and HeLa cells with an equivalent thiophene ring. This modification aimed to enhance the biological properties of the newly synthesized congeners while maintaining the integrity of their structures. They evaluated them for anticancer potential and among all synthesised compounds, compound 13 showed potential cytotoxicity towards various cancer cell lines such as human negroid cervix epitheloid carcinoma cells (HeLa) with 90% inhibition, human lung carcinoma cells (A549) with 50% inhibition, and human ovarian adenocarcinoma cells (TOV112D) with more than 90% inhibition, as compared to the control drug pemetrexed for a 24 h incubation time. SAR studies revealed that electron-donating methyl substituent at the para position of the phenyl ring showed higher cytotoxicity activity.96 In 2016, Gomha et al. (https://doi.org/10.3390/ijms17091499) developed bis-pyrazolyl-thiazole derivatives and evaluated them against the HepG2 cell line for anticancer activity. Among all synthesised compounds, compound 14 was the most potent derivative with an IC50 value of 1.37 ± 0.15 μM against HepG2 compared to the positive control doxorubicin. SAR studies indicate that alkyl substituent at the 4 position of the thiazole ring showed potent cytotoxicity. Adding an electron-withdrawing group, i.e., nitro group at the 4th position of the phenyl ring, which is placed on the 5th position of the thiazole ring, increases the anti-tumour activity.97 In 2016, Ghorab et al. employed a quinoline scaffold in their quest for anti-breast cancer derivatives, recognizing it as a well-established pharmacophore with notable anticancer properties. The isoquinoline and quinoline-fused thiophene derivatives have been reported, suggesting their potential efficacy against multiple target proteins. They evaluated them for anticancer potential. Compound 15 displayed significant anticancer potential with an IC50 of 33.1 μmol L−1 against MCF-7 compared to the standard doxorubicin IC50 value of 47.9 μmol L−1. SAR studies indicate tetrahydrobenzo[b]thiophene is essential for anticancer activity. A cyclohepta[b]thiophene ring in place of cyclopenta[b]thiophene increases the anticancer activity.98 In 2017, Mohareb et al. developed pyrano[2,3-d]thiazole-2-yl-benzo[b]thiophene derivatives and evaluated their antiproliferative against six cancer cell lines, such as colon cancer (DLD-1), HA22T and HEPG-2 (liver cancer cell line), gastric cancer cell line (NUGC), breast cancer cell line (MCF-7), nasopharyngeal carcinoma (HONE-1) and normal fibroblast cells (WI-38).99
Fig. 6. Reported thiophene derivatives with anticancer activity (compounds 9–13).
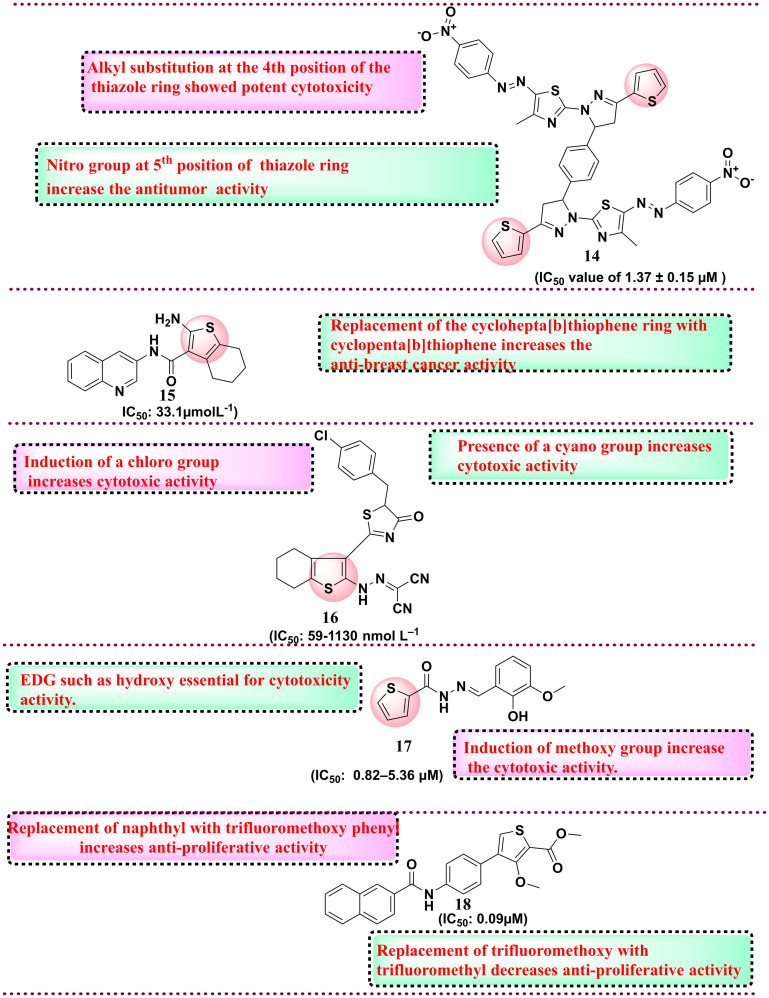
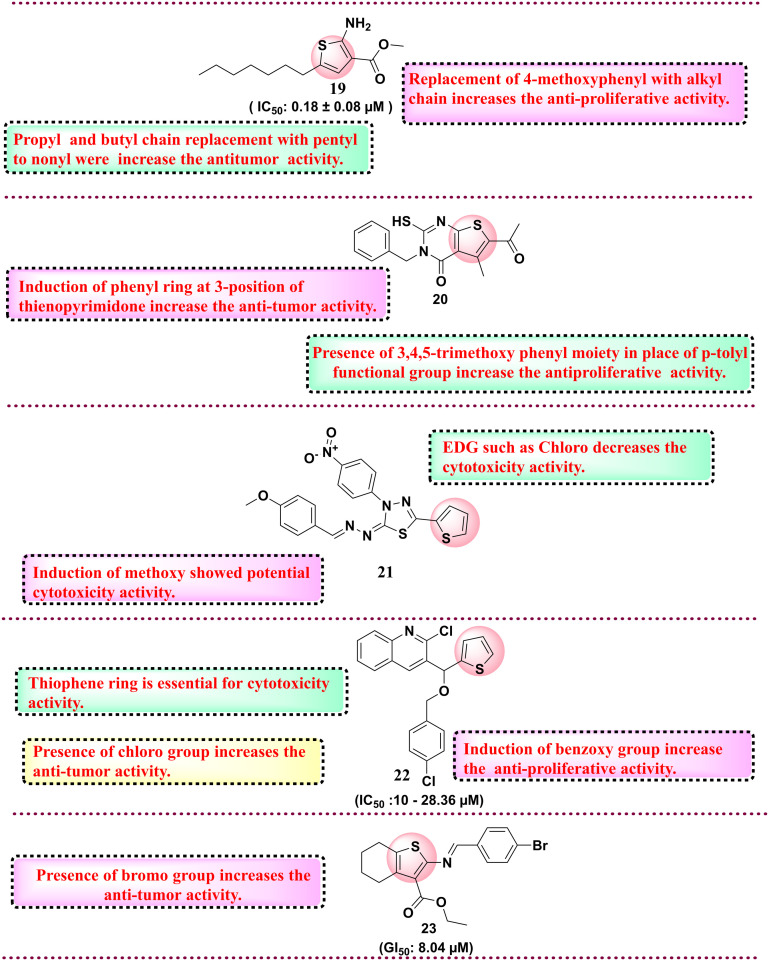
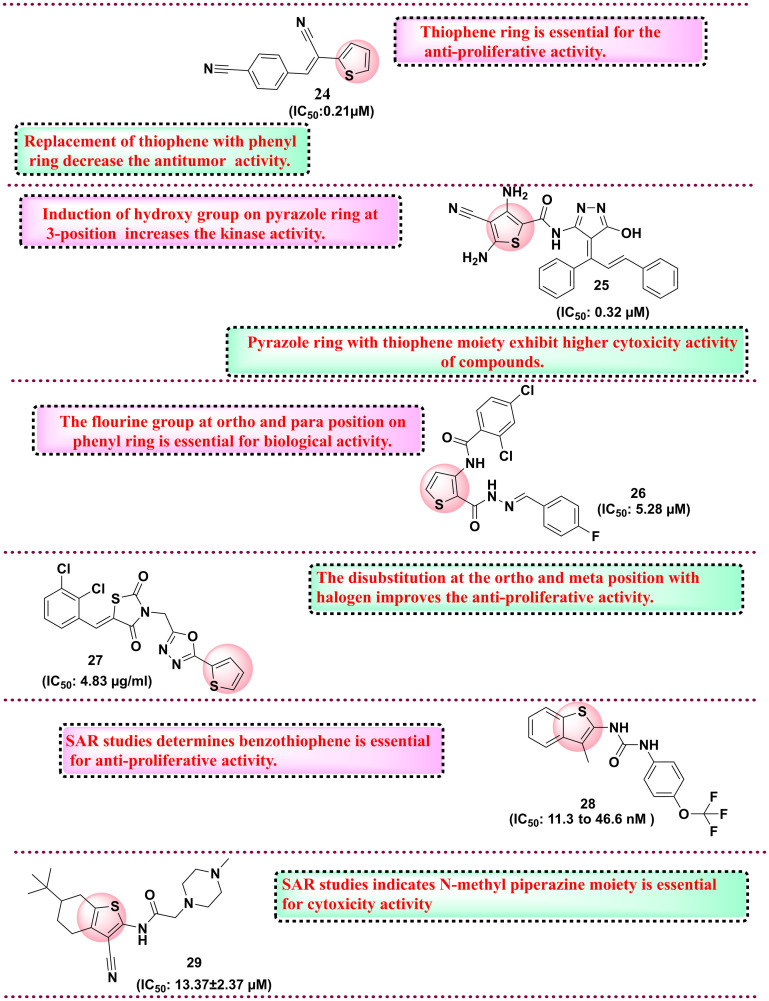
Among all synthesised derivatives, compound 16 was found to have potent antiproliferative activity against six cancer cell lines with IC50 in the range of 59–1130 nmol L−1 as compared to the reference compound. SAR studies revealed that the presence of electron-withdrawing groups, such as the chloro group and the presence of the cyano group, increase the cytotoxicity against the HEPG-2 cell line.99 In 2017, Cardoso et al. reported 57 N-acyl hydrazone-substituted thiophene derivatives and evaluated them against the panel of four different cancer cell lines such as colon cell line (HCT-116), glioblastoma cancer cell line (SF-295), leukaemia cell line (HL-60) and human ovary cell line (OVCAR-8). Among all synthesised compounds, compound 17 was the most potent derivative with an IC50 value of 0.82–5.36 μM against all four cancer cell lines. SAR analysis revealed that a hydroxy group at the phenyl ring is essential for activity.100 Protein tyrosine phosphatase 1B (PTP1B) exhibits elevated expression levels in various malignancies, notably breast, colorectal, and lung cancers. Inhibiting PTP1B has surfaced as a compelling therapeutic approach for cancer treatment. In a separate cohort, Gulipalli et al. designed compounds through in silico methodologies aimed at the PTP1B enzyme, utilizing molecular docking and ADMET predictions to refine their pharmacological characteristics. They synthesized methyl 4-(4-amidoaryl)-3-methoxythiophene-2-carboxylate derivatives and evaluated their efficacy against protein tyrosine phosphate B (PTP1B) as well as conducting an MTT assay to assess the antiproliferative activity. Among all synthesised compounds, compound 18 with IC50: 0.09 μM showed activity against MCF-7 and PTP1B. The SAR analysis disclosed that replacing the naphthyl ring with the thiophene ring diminished the activity. However, the replacement of naphthyl with trifluoromethoxy phenyl increases the antiproliferative activity, while in the place of trifluoromethoxy phenyl, the replacement of trifluoromethyl group reduces the cytotoxicity activity.76 Thomas et al. (2017) developed novel 5-alkyl-2-amino-3-carboxylate thiophene derivatives, which possess the structural capacity to engage with crucial molecular targets in cancer cells, including topoisomerases, kinases, and receptors, which govern cell growth and survival. To improve the selectivity for tumor cells, modifications were made at the alkyl and carboxylate positions. The researchers thoroughly evaluated all the derivatives for their anticancer activity against a range of cancer cell lines, including T-lymphoma CEM and Molt/4, kidney Caki-1, prostate PC-3, and hepatoma Huh-7 tumor cells. Compound 19 showed the highest cytotoxicity activity with an IC50 value of 0.18 ± 0.08 μM for lymphoma CEM and 0.057 ± 0.026 μM for Molt/4. SAR studies indicate that higher alkyl chains, such as pentyl to nonyl, in place of propyl and butyl derivatives, display better antiproliferative activity against the tumour cell lines (Fig. 7).101 In 2018, Fouad et al. developed a series of thienopyrimidine-based compounds targeting DNA polymerase, thymidylate synthase and tyrosine kinase. All the synthesized derivatives were evaluated against five different cancer cell lines (HepG-2, Hep-2, MCF-7, PC-3, and HeLa). Compound 20 showed potent activity against HEPG-2 as compared to the control doxorubicin. SAR studies revealed that the presence of the 3,4,5-trimethoxy phenyl moiety in place of the p-tolyl functional group increases the antiproliferative activity.102 In 2018, Gomha et al. synthesised 5-thiophen-2-yl 1,3,4-thiadiazoles derivatives and evaluated their anti-proliferative activity. Among the synthesised compounds, compound 21 showed the highest activity against HepG-2 and A-549 cell lines compared to the reference drug cisplatin.
Fig. 7. Reported thiophene derivatives with anticancer activity (compounds 14–18).
Overall, SAR analysis indicates that the electron donating group, i.e., methoxy, shows potential anti-tumour activity compared to the electron-withdrawing group, such as the chloro group.103 In 2019, Othman et al. synthesised a series of thiophene-based compounds where isoxazole, triazole, or phenyl moieties are anchored through a hydroxyl (OH) functionality. All the substituted derivatives were initially evaluated for anticancer activity and the lead compound was further evaluated for inhibitory activity against EGFR-TK, Topo II enzymes and cancer cell lines. Compound 22, with benzyloxy substituents and IC50 value in the range of 10.00–28.36 μM, showed potential activity against MCF-7. SAR analysis revealed that the presence of the benzyloxy group is essential for anticancer activity. In addition, the electron-withdrawing group, such as chloro, tri fluorocarbon, and fluoro, is at the 4th position of the phenyl ring, which also imparts anticancer activity. Further, the active compound 22 tested against EGFR-TK and Topo II showed potential activity against the EGFR-TK enzyme with an IC50 value of 3.66 μM.104 Next, Shah et al. and his coworkers synthesised thiophene derivatives and evaluated their anticancer activity. Among all synthesised compounds, compound 23, with a GI50 value of 8.04 μM, showed potential activity against lung cancer cell lines compared to the reference drug adriamycin.105 In 2020, Perin et al. synthesised naptho[2,1-b]thiophene derivatives and evaluated them for anticancer activity against various cancer cell lines. All synthesised compounds were tested against five cancer cell lines such as HeLa, HepG2, CFPAC-1, SW620, and A549, among which compound 24 (IC50: 0.21 μM) was found the most potent against HeLa cell lines. SAR studies showed that if the thiophene ring is replaced with a phenyl ring, there is a decrease in the activity, and also, the compound screened in silico against various targets in which androgen receptor and tyrosine kinases showed the highest affinity towards synthesised compounds indicated the anticancer profile.106 The same year, another group of Megally Abdo and coworkers developed new thiophene derivatives and evaluated them against the panel of cell lines such as A549, HT-29, H460, MKN-45, SMMC-7721, and U87MG. Among all synthesised compounds, compound 25 with the IC50 range of 0.32 μM against SMMC-7721 cell line with the control drug foretinib with the IC50 value 0.44 μM. Compound 25 was further evaluated against various kinases such as VEGFR-2, PDGFR, Pim-1 kinase, and EGFR and the compound exhibits potential activity with an IC50 value of 0.32 μM. SAR studies indicate the pyrazole ring with thiophene moiety exhibits higher cytotoxicity activity. In addition, introducing hydroxy and methoxy groups increases the cytotoxicity activity against various kinases.107 In 2023, Şenol et al. synthesised 3-aminothiophene-2-carboxylic acid methyl ester derivatives and evaluated them against HUVEC and HCT116 cell lines. Compound 26 was the most potent compound with an IC50 value of 5.28 μM on the HCT116 cell line with 33% selectivity. SAR studies suggest that the fluorine group is essential for cytotoxicity activity. The fluorine group at the ortho and para position of the phenyl ring is essential for biological activity.108 In 2024, Doddagaddavalli et al. synthesised thiophene-1,3,4,-oxadiazole-thiazolidinedione derivatives and evaluated them against a breast cancer cell line (MCF-7). Among the synthesised derivatives, compound 27 showed promising antiproliferative activity with an IC50 value of 4.83 μg ml−1 as compared to reference drugs. SAR studies indicate that the functional group at the para position of phenyl did not show any change in the activity. However, ortho and meta position halogen disubstitution increases the antiproliferative activity.109 The same year, Eldehna et al. worked on developing dual inhibitors targeting EGFR and VEGFR-2 by replacing the quinoline ring system of sorafenib with benzothiophene in the hinge region and aryl ring system to interact with the hydrophobic back pocket. They reported new benzo[b] thiophene-linked aryl urea derivatives, which were evaluated against cell line cancers (MCF-7, PanC-1 and HepG2). Among the synthesised compounds, 28 showed potent VEGFR-2 and EGFR inhibitory activities with IC50 values of 11.3 and 46.6 nM. SAR studies showed that benzothiophene is essential for anti-proliferative activity. The functional group trifluoromethoxy showed better anti-proliferative activity.110 Next, Rahman et al. synthesised thiophene derivatives linked with a carbonitrile moiety. The objective of their research was to tackle the issue of drug resistance associated with quinazoline-based EGFR inhibitors such as gefitinib. Their molecule was carefully designed utilizing the basic pharmacophoric characteristic of EGFR inhibitors, which encompasses a heterocyclic ring system that engages with critical amino acids within the binding cavity, including methionine-793 (MET), glutamine-791 (GLN), leucine-718 (LEU), and LEU844, thereby occupying the adenine binding site. The quinazoline heterocyclic ring system was substituted with tetrahydrobenzothiophene. In addition, a hydrophobic head and tail terminal are situated within the hydrophobic zones I and II. The researchers synthesized conjugates featuring these three structural modifications to gefitinib and assessed their anticancer efficacy by screening against the Mia PaCa-2 cell line and EGFR inhibitory activity. Compound 29 showed the highest cytotoxicity with IC50 of 13.37 ± 2.37 μM and suppressed EGFR2 enzyme with 0.68 μM IC50 value. SAR studies indicate that N-methyl piperazine moiety is essential for cytotoxicity activity.111 The chemical structures of the reported molecules as anticancer agents are presented in Fig. 8 and 9. A brief compilation of all the discussed anticancer agents bearing thiophene rings is done in Table 3.
Fig. 8. Reported thiophene derivatives with anticancer activity (compounds 19–23).
Fig. 9. Reported thiophene derivatives with anticancer activity (compounds 24–29).
Compilation of ongoing research on thiophene derivatives for the development of anticancer agents.
| Compound code | Indication | Cell lines/enzymes | Outcome | Status | Ref. |
|---|---|---|---|---|---|
| 9 | Antitumor activity | Inhibition of SF-268, MCF-7 and H460 cell lines | Prevent the proliferation and growth of tumor | Preclinical | 94 |
| 10 | Anticancer agent | Inhibition of H460 and MCF-7 cell lines | Prevent the proliferation and growth of tumor | Preclinical | |
| 11 | Ant-proliferative activity | Inhibition of MCF-7 cell line | Inhibits the proliferation and growth of tumor | Preclinical | |
| 12 | Antitumor activity | Inhibition of MCF-7 cell line | Prevent the proliferation and growth of tumor | Preclinical | 95 |
| 13 | Anticancer agent | Inhibition of human negroid cervix epitheloid carcinoma cells (HeLa) | Inhibits the proliferation and growth of tumor | Preclinical | 96 |
| 14 | Antitumor activity | HepG2 cell line | Inhibits the proliferation and growth of tumor | Preclinical | 97 |
| 15 | Anticancer agent | MCF-7 | Inhibits the proliferation and growth of tumor | Preclinical | 98 |
| 16 | Antitumor activity | Colon cell line (HCT-116), glioblastoma cancer cell line (SF-295), leukaemia cell line (HL-60) and human ovary cell line (OVCAR-8) | Prevent the proliferation and growth of tumor | Preclinical | 99 |
| 17 | Anticancer agent | MCF-7 | Inhibits the proliferation and growth of tumor | Preclinical | 100 |
| 18 | Anticancer agent | Protein tyrosine phosphate B (PTP1B) | Inhibits the proliferation and growth of tumor | Preclinical | 76 |
| 19 | Antitumor activity | Cell lines like T-lymphoma CEM and Molt/4, kidney Caki-1, prostate PC-3, and hepatoma Huh-7 tumour cells | Prevent the proliferation and growth of tumor | Preclinical | 101 |
| 20 | Antitumor activity | Hepatocellular panel of cell lines. Compound 20 showed potent activity against HEPG-2 | Prevent the proliferation and growth of tumor | Preclinical | 102 |
| 21 | Antitumor activity | HepG-2 and A-549 cell lines | Prevent the proliferation and growth of tumor | Preclinical | 103 |
| 22 | Antitumor activity | EGFR-TK and Topo II | Prevent the proliferation and growth of tumor | Preclinical | 104 |
| 23 | Antitumor activity | Lung cancer cell line | Inhibits the proliferation and growth of tumor | Preclinical | 112 |
| 24 | Anticancer agent | HeLa, HepG2, CFPAC-1, SW620, and A549 | Prevent the proliferation and growth of tumor | Preclinical | 106 |
| 25 | Anticancer agent | VEGFR-2, PDGFR, Pim-1 kinase | Prevent the proliferation and growth of tumor | Preclinical | 107 |
| 26 | Anticancer agent | Inhibition of HUVEC and HCT116 cell lines | Prevent the proliferation and growth of tumor | Preclinical | 108 |
| 27 | Anticancer agent | Inhibition of MCF-7 cell line | Prevent the proliferation and growth of tumor | Preclinical | 109 |
| 28 | Anticancer agent (MCF-7, PanC-1 and HepG2) | Inhibition of VEGFR-2 and EGFR | Inhibits the proliferation and growth of tumor | Preclinical | 110 |
| 29 | Anticancer agent (human pancreatic cell line) | Inhibition of EGFR2 enzyme | Prevent the proliferation and growth of tumor | Preclinical | 111 |
5.3. Thiophene ring in the development of drugs for neurodegenerative diseases with a focus on the development of anti-Alzheimer's and anti-Parkinson agents
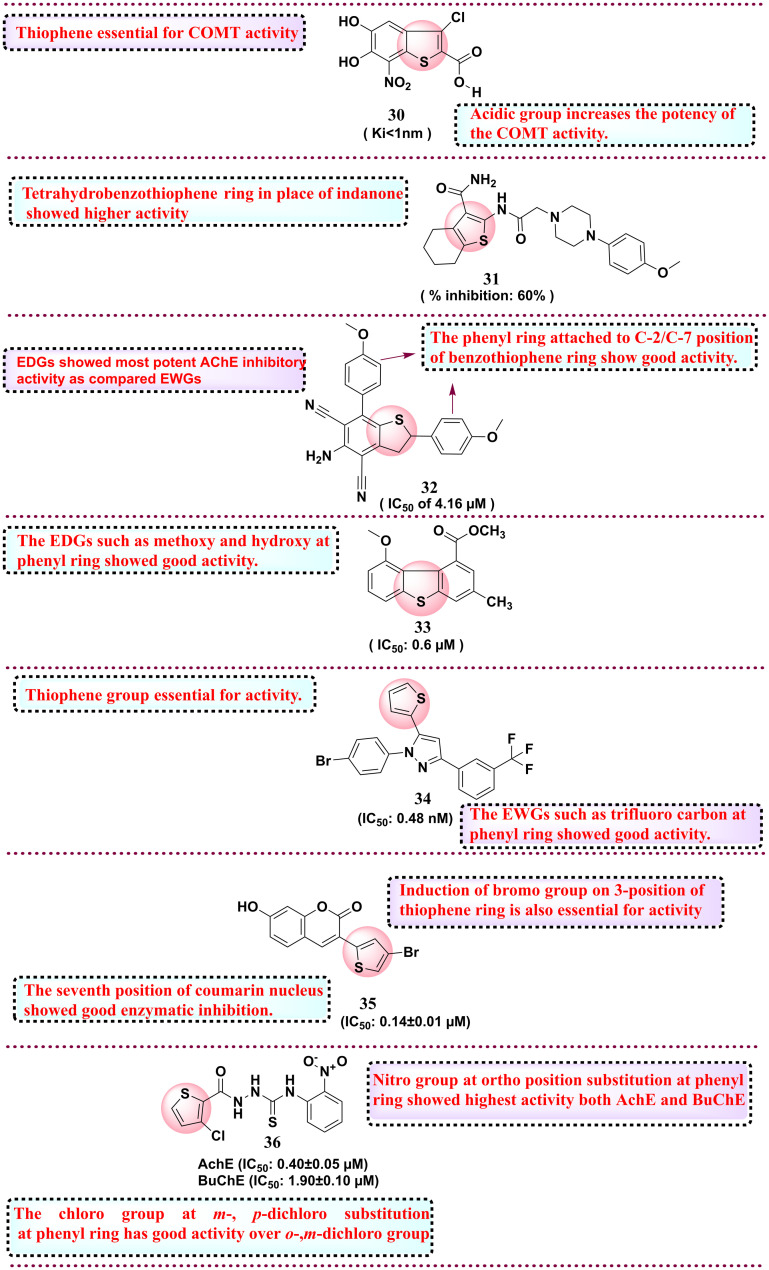
In 2010, Rautio et al. worked on improving the oral bioavailability of an investigational, very potent nanomolar inhibitor, COMT. They utilised the prodrug concept to improve the bioavailability and synthesised alkyl esters, acyloxy alkyl esters, amino acyloxy alkyl esters and cyclic carbonate esters and evaluated their COMT inhibition potential. Among all synthesised derivatives, compound 30 showed the most potential activity towards COMT with Ki < 1 nm. Furthermore, the compound is optimised to improve poor oral bioavailability by attempting various lipophilic groups as prodrugs. The two synthesised prodrugs 30a and 30b showed a two-fold increase in rat plasma levels for a 6 h exposure. The SAR study reveals that thiophene is essential for COMT activity.113 In 2012, Ismail et al. synthesised novel analogues of donepezil by substituting the indanone ring with a thiophene ring featuring a carbonyl group at position 3 utilising an acetamido group as a spacer to link the thiophene ring with arylpiperidines and piperazines. The derivatives were assessed for AChE activity. Compound 31 was the most potent among the synthesised compounds, with 60% inhibition, compared to the standard donepezil, with 40% inhibition. SAR studies revealed that tetrahydrobenzothiophene ring replacing indanone showed good activity.114 In 2013, Jeyachandran et al. synthesised dihydrobenzo[b]thiophene-4,6-dicarbonitriles and tested their (AChE) activity. Compound 32 showed the most potent activity as an AChE inhibitor with an IC50 of 4.16 μM. The SAR study showed that the substitution at C-2 and C-7 positions with electron-donating groups, such as 4-methoxyphenyl and 4-methylphenyl groups, showed the most potent AChE inhibitory activity as compared with electron-withdrawing groups such as 4-chloro, 4-bromo, 4-fluoro or 4-isopropylphenyl.115 In 2016, Funicello and co-workers, in a quest to their lead optimisation studies on 4- and 5-derivatives, developed new analogues and tested them for the beta-secretase activity (BACE1) for their plausible use in treating Alzheimer's disease (AD). Among all synthesised compounds, compound 33 showed potent activity against BACE1 with an IC50 value of 0.6 μM. SAR studies determine that methoxy (–OCH3) and hydroxy (–OH) substituents at the phenyl ring increase biological activity.116 In 2019, Cetin et al. reported new thiophene derivatives owing to the inefficacy of the reported drug candidates in treating AD in the long term. The group evaluated their synthetics against carbonic anhydrase I and II (hCA I and II) and acetylcholinesterase (AChE) enzymes, which are commonly overexpressed in AD. Compound 34 showed potential activity towards AChE with an IC50 value of 0.48 nM. SAR studies determined that the thiophene group is essential for activity. The EWGs, such as trifluoro carbon at the phenyl ring, showed good activity.117 In 2020, in a quest to develop more potent antiparkinsonian drugs, Rodriguez-Enriquez and group synthesised twelve 3-thiophenol coumarin derivatives. These molecules were rationally developed from their identified lead 3-aryl coumarins. To the lead compound, the group introduced a catechol ring system to portray the antioxidant mechanism and position 3 of the coumarin ring was substituted with a thiophene ring so that pi-electrons of sulfur may show additional interaction at the MAO-B catalytic site. The designed and synthesised derivatives were screened against monoamine oxidase B (MAO-B) inhibitors. Among all the derivatives, compound 35 was found to be the most potent inhibitor for MAO-B with an IC50 value of 0.14 ± 0.01 μM over the reference selegiline, with the IC50 of 0.017 ± 0.002 μM. SAR studies investigated the 7 position of the coumarin nucleus, which showed good enzymatic inhibition compared to the 8 position of the coumarin nucleus. The presence of the bromo group on the 3-position of the thiophene ring is also essential for activity.118 Very recently, in 2024, Ullah et al. reported new thiophene-linked thiourea analogues and tested them against butyrylcholinesterase (BuChE) and acetylcholinesterase (AChE) inhibitory activity. The molecules were rationally constructed owing to recent evidence that BuChE is chiefly found in the plaques created by AD. Among eleven synthesised derivatives, compound 36 is the most potent against both AChE with the IC50 value of 0.40 ± 0.05 μM and BuChE with the IC50 value of 1.90 ± 0.10 μM over the reference donepezil (IC50 value of 2.16 ± 0.12 μM) and BuChE with the IC50 value of 4.5 ± 0.11 μM. SAR studies have revealed that the nitro group at the ortho position of the phenyl ring show the highest activity against both AChE and BuChE over the m- (meta) and p- (para) nitro. In the case of halogen, the chloro group at m- and p-dichloro substitution at the phenyl ring has good activity compared to the ortho and m-dichloro groups. Next, electron-donating groups, such as methyl at the m-position, have good activity compared to p-methyl at the phenyl ring.119 All the reported thiophene derivatives as neurodegenerative disease inhibitors are shown in Fig. 10. A summary of the ongoing research on thiophene derivatives for developing CNS disorder inhibitors is presented in Table 4.
Fig. 10. Reported thiophene derivatives as neurodegenerative disease inhibitors.
Compilation of ongoing research on thiophene derivatives for the development of CNS disorder inhibitors.
| Compound code | Indication | Biological target | Mechanism | Status | Ref. |
|---|---|---|---|---|---|
| 30 | Neurodegenerative agent | Catechol-O-methyltransferase (COMT) | Reduce the action of dopamine | Preclinical | 113 |
| 31 | Alzheimer disease | Acetylcholinesterase (AChE) enzyme | Block the dopamine | Preclinical | 114 |
| 32 | Neurodegenerative agent | Acetylcholinesterase (AChE) enzyme | Reduce the action of dopamine | Preclinical | 115 |
| 33 | Neurodegenerative agent | Beta-secretase activity (BACE1). | Block the dopamine | Preclinical | 120 |
| 34 | Neurodegenerative agent | Carbonic anhydrase I and II (hCA I and II) and acetylcholinesterase (AChE) enzyme | Reduce the action of dopamine | Preclinical | 117 |
| 35 | Parkinson's disease | Monoamine oxidase B (MAO-B) inhibitors | Block the dopamine | Preclinical | 118 |
| 36 | Neurodegenerative agent | Butyrylcholinesterase (BuChE) and acetylcholinesterase (AChE) | Reduce the action of dopamine | Preclinical | 119 |
5.4. Thiophene derivatives as anti-inflammatory agents
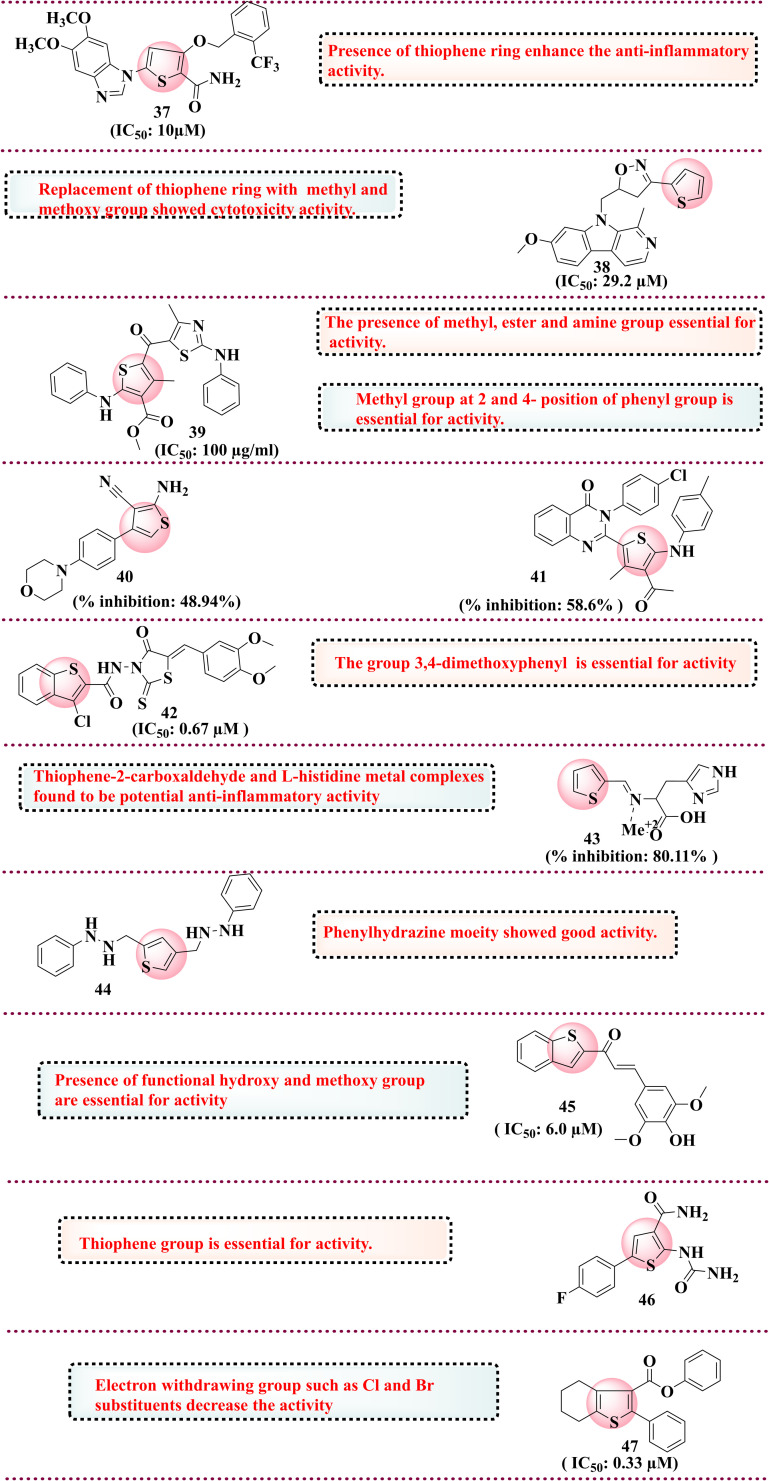
In 2015, Filali et al. disclosed the development of the isoxazoline series and its evaluation as an anti-inflammatory agent. The isoxazoline molecules were developed considering the biological interest, including anti-inflammatory and anti-viral, in the quest to develop safer anti-inflammatory antagonists. Among the synthesised compounds, the thiophene derivative (38) showed good activity against the 5-lipoxygenase enzyme with IC50 = 29.2 μM. SAR studies determined that methyl and methoxy groups in place of the thiophene ring showed cytotoxicity activity in MCF7 and HCT116 cell lines.121 In 2015, Patil et al. reported a new thiazole-thiophene series evaluated against 5-LOX for the treatment of asthma. The molecules were rationally designed and developed as per their lead optimisation studies involved in improving the thiazole-based pharmacophoric features for their potential as AR1-AR receptor antagonists. Among all synthesised compounds, 39 was the most potent against 5-LOX in vitro at 100 μg ml−1 concentration and in vivo prevent mass cell degranulation (>63%). SAR studies indicate that methyl, ester and amine groups are essential for activity.122 In 2015, Helal et al. and groups reported a series of thiophene analogues and evaluated them for anti-inflammatory activity. The rationale for selecting thiophene derivatives is due to the growing interest in this scaffold and existing approved drugs based on the same scaffold. Among all the synthesised compounds screened for anti-inflammatory activity via paw edema induced through carrageenan by albino rats compared with the standard indomethacin. Compound 40 showed potent activity with 58.6% oedema inhibition over indomethacin (47.73%). The SAR study determined that replacing the morpholine ring with the thiophene ring enhances the anti-inflammatory activity.123 In 2018, Chaudhari et al. reported thiophene derivatives and evaluated their anti-inflammatory activity. The thiophene derivatives were selected based on the existing reports available on the scaffold and possibly considering their embarking activities performed by the group in the past. Among the synthesised compounds, 41 showed 48.94% inhibition against carrageenan-induced paw edema compared to the reference diclofenac.124 In 2017, El-Miligy et al. reported a benzothiophene hybrid with rhodamine that was tested against COX-2/5-LOX dual inhibitors. Based on findings from the literature that rhodamine and benzothiophene derivatives possess good anti-inflammatory properties, the group linked both the scaffold with two atom linkers, which could result in good activity based on silico findings. Compound 42 was found to have promising activity towards COX-2 with an IC50 value of 0.67 μM compared to the reference celecoxib (1.14 μM) and LOX with an IC50 value of 2.33 μM over the reference 5.64 μM. Further, in vivo, compound 42 was more effective against formalin-induced paw edema than the reference drug celecoxib. SAR studies indicate that the 3,4-dimethoxyphenyl group is essential for activity.125 In 2019, John et al. reported thiophene-2-carboxylate linked l-histidine and its metal complexes and screened their anti-inflammatory activity. The rationale behind linking l-histidine residues is their strong affinity towards metal complexes and good docking scores against COX-2 inhibitors. The synthesised complex was tested against the albumin denaturation model in vitro, and thiophene metal complexes inhibited it dose-dependently. Complex 43 showed 80.11% inhibition as compared to the standard aspirin 76.89%. The molecular docking studies via Autodock software showed that copper II (Cu) and zinc II (Zn) showed the highest binding energy, −7.42 and −7.22 kcal mol−1, respectively. The thiophene-2-carboxaldehyde and l-histidine metal complexes were found to have potential anti-inflammatory activity.126 Next, Gaines et al., in the same year, synthesised thiophene and furan bioisostere derivatives and evaluated their anti-inflammatory properties. Compound 44 was found to be most potent against the CXCR4 receptor. SAR studies of phenylhydrazine showed good anti-inflammatory activity.74 In 2020, Chiasson et al. developed a new series of hybrid benzothiophene derivatives and evaluated them against 5-LOX inhibitors in the HEK293 cell line in vitro. The rationale behind selecting the benzothiophene nucleus is the key pharmacophore in the zileuton drug. Among the synthesised compounds, 45 showed potential activity with an IC50 of 6.0 μM. SAR studies have indicated that functional hydroxy and methoxy groups are essential for activity.127 In the same year, da Cruz et al. reported that 2-aminothiophene was evaluated for anti-inflammatory activity. Among all synthesised compounds, 46 was a potential inhibitor of various inflammatory factors. SAR studies reveal that the thiophene group is essential for activity.37 In the same year, Khatri et al. reported 2-phenyltetrahydro[b]benzothiophene derivatives screened against COX-2 inhibitors. Based on the existing literature, the tetrahydro benzothiophene derivatives were found to have excellent anti-inflammatory activity to maintain the structural integrity at the phenyl ring attached at the 2-position. Among all synthesised compounds, 47 was found to be potent towards COX-2 inhibitors with IC50 values of 0.33 μM, respectively. SAR studies reveal that electron-withdrawing groups such as Cl and Br substituents decrease the activity.128 All the reported thiophene derivatives as neurodegenerative disease inhibitors are shown in Fig. 11. The compilation of thiophene derivatives as anti-inflammatory agents is done in Table 5.
Fig. 11. Reported thiophene derivatives as anti-inflammatory agents.
Compilation of ongoing research on thiophene derivatives for the development of anti-inflammatory agents.
| Compound code | Indication | Biological target | Mechanism | Status | Ref. |
|---|---|---|---|---|---|
| 37 | Anti-inflammatory agent | Inhibiting the pro-inflammatory agents such as ERK, THP-1 and NF-κB | Reduce the inflammatory mediators | Preclinical | 129 |
| 38 | Anti-inflammatory (anticancer) | Inhibition of 5-lipoxygenase enzyme | Decrease the production of leukotrienes | Preclinical | 121 |
| 39 | Anti-inflammatory agent (asthma) | Inhibition of 5-lipoxygenase enzyme | Decrease the production of leukotrienes | Preclinical | 122 |
| 40 | Anti-inflammatory agent | LOX | Decrease the production of leukotrienes | Preclinical | |
| 41 | Anti-inflammatory agent | LOX | Decrease the production of leukotrienes | Preclinical | 123 |
| 42 | Anti-inflammatory agent | COX-2/5-LOX dual inhibitors | Decrease the production of leukotrienes | Preclinical | 124 |
| 43 | Anti-inflammatory agent | LOX | Decrease the production of leukotrienes | Preclinical | 125 |
| 44 | Anti-inflammatory agent | CXCR4 receptor | Decrease the production of leukotrienes | Preclinical | 74 |
| 45 | Anti-inflammatory agent | 5-LOX inhibitors | Reduce the inflammatory mediators | Preclinical | 127 |
| 46 | Anti-inflammatory agent | COX-2 inhibitors | Decrease the production of leukotrienes | Preclinical | 37 |
| 47 | Anti-inflammatory agent | COX-2 inhibitors | Decrease the production of leukotrienes | Preclinical | 128 |
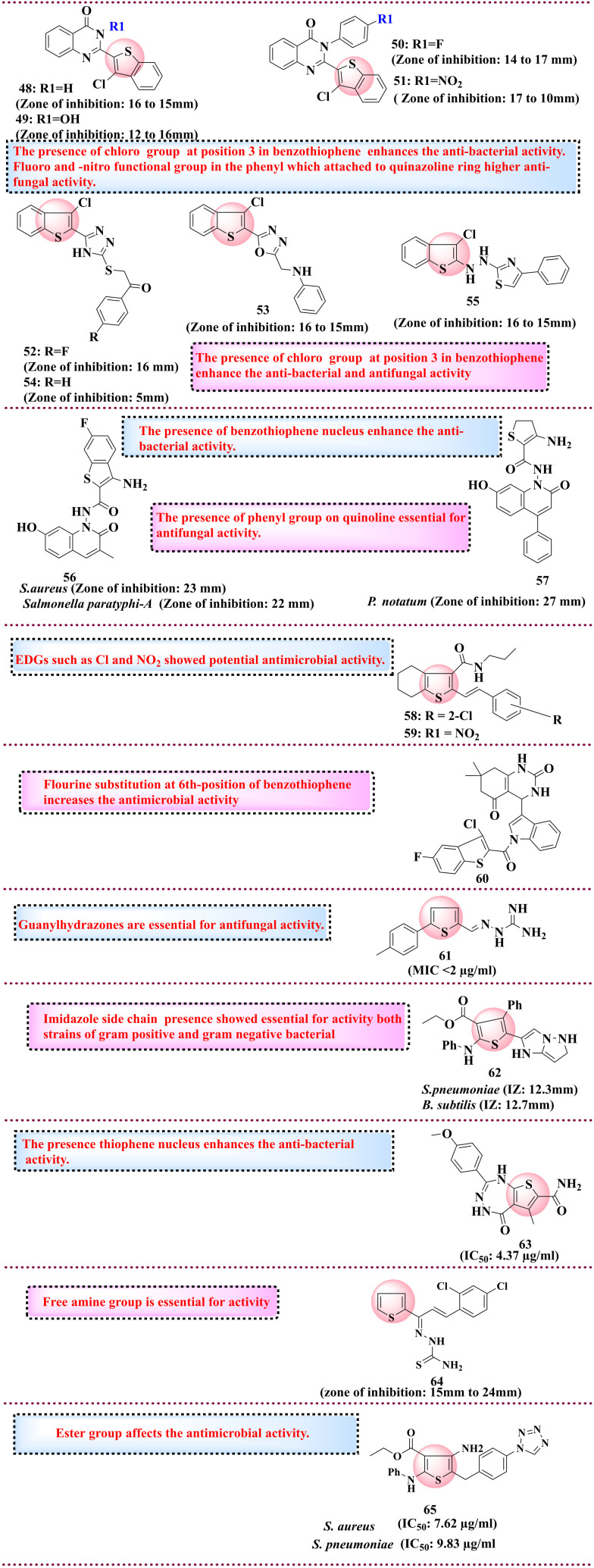
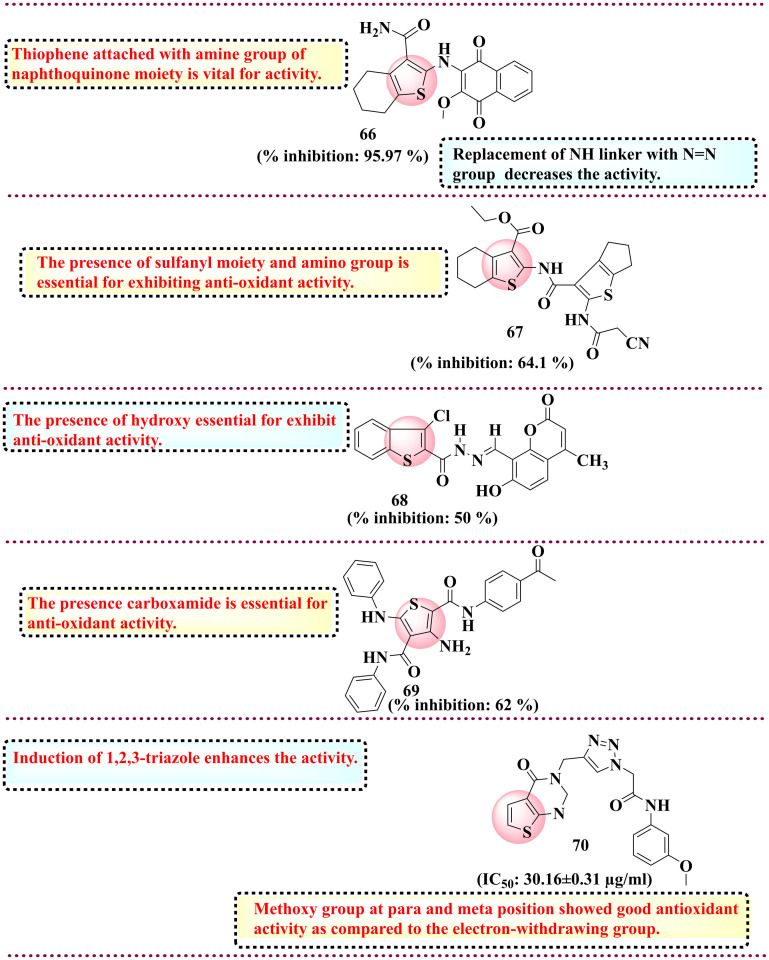
5.5. Thiophene derivatives as anti-microbial agents
In 2011, Petsom et al. and coworkers synthesised the benzothiophene-linked quinazoline-4-one series and evaluated it for antibacterial and antifungal activity. The rationale behind linking quinazoline moiety to thiophene is that it possesses varied biological activities such as anti-inflammatory and antimicrobial. Among synthesised compounds, 48 and 49 had promising antibacterial properties against B. subtilis (16 and 15 mm) and E. coli with 12 and 16 mm inhibition zones. Compounds 50 and 51 showed good antifungal activity, such as C. pannical (with an inhibition zone (IZ) of 14 and 17 mm) and A. niger (with IZ of 17 and 10 mm). The SAR study determined that the presence of a chloro functional group substituent at position 3 in the benzothiophene nucleus enhanced the anti-bacterial activity and -fluoro and -nitro functional groups in the phenyl attached to the quinazoline ring led to higher anti-fungal activity.130 The same group also developed new benzothiophene-derived compounds and evaluated them for their antimicrobial activity. Among the synthesised compounds, 52 and 53 showed good antibacterial activity against B. subtilis with an IZ of 16 mm and S. aureus 15 mm, and 54 and 55 showed antifungal activity against C. albicans with an IZ of 16 mm and 15 mm, respectively. The presence of a chloro group at position 3 in benzothiophene enhances the anti-bacterial and antifungal activity.131 In 2014, Padmashali et al. reported coumarins, pyrimidines and pyrazole-linked benzothiophene derivatives, which were screened for antimicrobial activity. The rationale behind linking thiophene with nitrogen is that it contains a heterocyclic ring due to its diverse biological activity. The thiopyrimidine nucleus is a structural analogue of biogenic purines. Among all synthesised compounds, 56 showed potential activity towards antibacterial strains of S. aureus (IZ = 23 mm), Salmonella paratyphi-A (IZ = 22 mm), and B. subtilis (IZ = 25 mm) as compared to the reference chloramphenicol and 57 showed activity towards P. notatum (IZ = 27 mm) and A. niger (IZ = 26 mm) strains of antifungal compared to the reference fluconazole.132 In 2011, Srivastava et al. reported 4,5,6,7-tetrahydro-1-benzothiophene-attached Schiff base derivatives and evaluated them for antimicrobial activity. The rationale behind developing Schiff bases linked with thiophene is due to its remarkable antibacterial activity and identification of new antimicrobial molecules. Among all synthesised compounds, compounds 58 and 59 showed excellent anti-microbial activity against both Gram-positive and Gram-negative bacteria and compound 58 was found to have potent activity against A. niger. SAR studies reveal that electron-donating and electron-withdrawing groups on the phenyl ring affect the activity. The EDGs, such as Cl and NO2, showed potential antimicrobial activity.133 In 2015, Sharma et al. synthesised tetrahydroquinazoline indole benzothiophene derivatives and evaluated their antibacterial and antifungal activity. Existing reports show that tetrahydroquinazoline was found to have antimicrobial activity. Compound 60 showed potent antibacterial and antifungal activity against various strains such as P. aeruginosa, A. clavatus, and A. niger via the broth method. SAR studies have indicated that fluorine substitution at the 6th position of benzothiophene increases the antimicrobial activity compared to other substitutions such as –OH, CH3 and Cl.134 In 2016, Ajdačić and co-workers developed a series of new guanylhydrazone-based thiophene derivatives screened for antifungal activity. The rationale for selecting guanylhydrazones is to explore their antibacterial potential. Compound 61 showed excellent antifungal activities with MIC < 2 μg ml−1. SAR studies have revealed that the guanylhydrazone moiety is essential for antifungal activity.135 In 2017, Mabkhot et al. reported thiophene derivatives and screened their anti-microbial activity. Based on the in silico findings, the compounds were designed based on their affinity towards antibacterial strains. All synthesised compounds were screened against Gram-positive and Gram-negative bacterial strains, compared to the reference gentamicin, amphotericin, and ampicillin drugs. Compound 62 showed excellent antimicrobial activity with an IZ value of 12.3 mm for S. pneumoniae and 12.7 mm for B. subtilis. SAR studies reveal that the presence of imidazole side chains was essential for the activity of both Gram-positive and Gram-negative bacteria.136 The same year, Rizk et al. reported thiophene-2-carboxamide derivatives and evaluated their in vitro antimicrobial activity. Thiophene nuclei possess diverse biological activities and are linked with pyrazole, thiazole and oxadiazole to enhance the anti-bacterial potency. Among all synthesised compounds, 63 showed potential antimicrobial activity with an IC50 value of 4.37 μg ml−1.137 Mohamed et al. reported the thiophene derivatives and evaluated their antimicrobial activity. Based on the existing reports on thiophene, 2-carboxamide exhibited antimicrobial activity in the thiophene nucleus selected for study. Among the synthesised compounds, 64 showed potential anti-microbial activity against E. coli with 15 mm to 24 mm zone inhibition. The docking analysis showed that compound 63 had binding affinity against C. albicans and A. niger with values of −6.9 kcal mol−1 and −7.4 kcal mol−1, respectively.138 In the same year, Abualnaja et al. reported the design and synthesis of the tetrazole-linked thiophene derivatives and evaluated them for the anti-microbial activity. The linked tetrazole moiety was found to be responsible for increasing the bioavailability and lowering the metabolic sensitivity. Additionally, the combination of dual heterocycles, i.e., tetrazole-linked thiophene derivative, enhanced the drug interaction within the receptor site. Compound 65 showed potential activity against S. aureus and S. pneumoniae with an IC50 value of 7.62 μg ml−1 and 9.83 μg ml−1. SAR studies revealed that the ester group affects the antimicrobial activity due to hydrolysis in the microbial system.93 The chemical structures of the reported antimicrobial agents are shown in Fig. 12.
Fig. 12. Reported thiophene derivatives as anti-microbial agents.
5.6. Thiophene derivatives and antioxidant activity
Antioxidant compounds play a crucial role in protecting cells from free radicals. It has been reported that antioxidants help reduce the risk of various chronic diseases such as cardiovascular diseases and cancer. Researchers have recently concentrated on developing new synthetic, effective, safe antioxidants that can withstand oxidative stress.139 Thiophene derivatives also exhibit antioxidant properties, which are discussed in this section.
In 2013, Gouda et al. synthesised tetrahydrobenzothiophene derivatives linked to naphthoquinone and screened them for antioxidant activity. The rationale behind linking naphthoquinone with thiophene is its free radical scavenging properties, which enhance the antioxidant activity. Further, the synthesised compounds were tested against DNA damage induced by bleomycin. Compound 66 was found to have potential antioxidant activity with a percentage inhibition of 95.97% among all synthesised compounds, additionally protecting against DNA damage caused by bleomycin. SAR studies indicated that naphthoquinone and thiophene moiety were necessary for activity. Replacement of the NH linker with the N N group decreases the activity, and hydroxy group replacement with methoxy increases the activity.140 The same group reported new carboxamide benzothiophene-2-amine derivatives in which compound 67 showed potential antioxidant activity (64.1%). SAR studies have revealed that the presence of sulfanyl moiety and amino group is essential for exhibiting antioxidant activity.141 In 2014, Mruthyunjayaswamy synthesised metal complexes linked with coumarin and benzothiophene derivatives and screened them for antioxidant activity. The rationale behind selecting the coumarin scaffold is its effect as a scavenger of reactive oxygen species, which helps protect tissues. Among all metal complexes, the Cu(ii) complex 68 showed the highest radical scavenging activity towards the DPPH radical. SAR investigation deduced that presence of phenol group with metal complex enhanced the antioxidant activity. The Cu(ii) was found to form a coordinate bond with the nitrogen of azomethine and carbonyl group of amide attached to benzothiophene at 2 position.142 In 2023, Metwally et al. reported thiophene 2-carboxamide derivatives, which were screened for antioxidant activity based on the previously reported literature on thiophene-2-carboxamide lead compound against IKK-2 selected from the same studies. Compound 69 showed potential activity with a percentage inhibition of 62%. SAR studies have indicated that carboxamide is essential for activity.143 In 2024, Kumari et al. reported 1,2,3-triazoleacetamide-attached thienopyrimidine derivatives, which were screened for antioxidant activity. 1,2,3-Triazoles are appealing bioisosteres, which are present in various pharmaceutical drugs in clinical trials linked to thienopyridine. Among all the synthesised compounds, 70 with an IC50 value of 30.16 ± 0.31 μg ml−1 was found to have potential antioxidant activity. SAR studies reveal that the induction of 1,2,3-triazole enhances the activity and incorporation of electron-donating groups, such as the methoxy group at the para and meta position, which showed good antioxidant activity compared to the electron-withdrawing group.144 The chemical structures of thiophene-based drugs with antioxidant potential are compiled in Fig. 13, and the brief generated data is given in Table 6.
Fig. 13. Reported thiophene derivatives with antioxidant activity.
Compilation of ongoing research on thiophene derivatives for the development of anti-oxidant agents.
| Compound code | Indication | Biological target assay | Mechanism | Status | Ref. |
|---|---|---|---|---|---|
| 66 | Antioxidant agent | DNA damage caused by bleomycin | By scavenging free ROS (reactive oxygen species) and reactive nitrogen species | Preclinical | 140 |
| 67 | Antioxidant agent | DNA damage caused by bleomycin | By scavenging free ROS (reactive oxygen species) and reactive nitrogen species | Preclinical | 141 |
| 68 | Antioxidant agent | DPPH radical | By preventing the formation of ROS (reactive oxygen species) | Preclinical | 142 |
| 69 | Antioxidant agent | ABTS method | By neutralising free radicals | Preclinical | 143 |
| 70 | Antioxidant agent | DPPH, H2O2, and NO assay | By scavenging free ROS (reactive oxygen species) | Preclinical | 144 |
6. Conclusion
Thiophene derivatives represent a fascinating area of study within medicinal chemistry, offering many opportunities for developing new therapeutic agents. The five-membered sulfur-containing ring is a versatile scaffold that is easily synthesised and modified to produce moieties with diverse pharmacological activities. The literature review underscores thiophene's pivotal role and analogues across diverse disease realms, including antimicrobial, anticancer, anti-inflammatory, and antioxidant applications. The strategic fusion of thiophene rings with other moieties has successfully enhanced the biological activity of these compounds. This review highlights the potential of thiophene derivatives as a rich source of pharmacologically active compounds.
Additionally, it delves into the impact of substituents on the thiophene ring, elucidating the structure–activity relationship. Their antimicrobial properties make them valuable scaffolds in the fight against infectious diseases. As anticancer agents, thiophene-based compounds displayed significant antiproliferative activity. Similarly, in diabetes, neurological, and anti-inflammatory disorders, they are proven leading scaffolds for compound design. Thus, in the quest for new thiophene-based compounds, this review will help medicinal chemists understand the effect of different substitutions and explore new substituents. As the pharmaceutical industry continues seeking novel compounds, the study of thiophene derivatives will likely remain at the forefront of medicinal chemistry, contributing to the discovery and development of drugs that can address unmet medical needs.
Data availability
No primary research results, software or code has been included and no new data were generated or analysed as part of this review.
Author contributions
Ashish Ranjan Dwivedi, Hemant R. Jadhav: conceptualization, supervision methodology, data curation, Shikha Thakur, Kapil Kumar Goel, Shivani Jaiswal, Sonia Dhiman: writing – original draft preparation. Vivek Srivastava: visualization, Pramod Rawat. Devendra Kumar: writing – reviewing and editing.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
The authors thank their respective institutes for their support in carrying out the present work.
References
- Taylor A. P. Robinson R. P. Fobian Y. M. Blakemore D. C. Jones L. H. Fadeyi O. Org. Biomol. Chem. 2016;14:6611–6637. doi: 10.1039/c6ob00936k. [DOI] [PubMed] [Google Scholar]
- Sperry J. B. Wright D. L. Curr. Opin. Drug Discovery Dev. 2005;8:723–740. [PubMed] [Google Scholar]
- Gomtsyan A. Chem. Heterocycl. Compd. 2012;48:7–10. doi: 10.1007/s10593-012-0960-z. [DOI] [Google Scholar]
- Pathania S. Narang R. K. Rawal R. K. Eur. J. Med. Chem. 2019;180:486–508. doi: 10.1016/j.ejmech.2019.07.043. [DOI] [PubMed] [Google Scholar]
- Shah R. Verma P. K. Chem. Cent. J. 2018;12:1–22. doi: 10.1186/s13065-018-0511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer V. Ber. Dtsch. Chem. Ges. 1883;16:1465–1478. [Google Scholar]
- Cameron M. D. J. Chem. Educ. 1949;26:521. [Google Scholar]
- Baeyer A. Ber. Dtsch. Chem. Ges. 1879;12:1309–1319. [Google Scholar]
- Sumpter W. C. Chem. Rev. 1944;34:393–434. doi: 10.1021/cr60109a003. [DOI] [Google Scholar]
- Jha K. K. Kumar S. Tomer I. Mishra R. J. Pharma Res. 2012;5:560–566. [Google Scholar]
- Peng X. Yin G. Wu K. Wu G. Chen J. Wang Z. Asian J. Org. Chem. 2024;13:e202300631. doi: 10.1002/ajoc.202300631. [DOI] [Google Scholar]
- Hudson B. M. Nguyen E. Tantillo D. J. Org. Biomol. Chem. 2016;14:3975–3980. doi: 10.1039/C6OB00254D. [DOI] [PubMed] [Google Scholar]
- Subbaiah M. A. M. Meanwell N. A. J. Med. Chem. 2021;64:14046–14128. doi: 10.1021/acs.jmedchem.1c01215. [DOI] [PubMed] [Google Scholar]
- Marshall C. M. Federice J. G. Bell C. N. Cox P. B. Njardarson J. T. J. Med. Chem. 2024;67:11622–11655. doi: 10.1021/acs.jmedchem.4c01122. [DOI] [PubMed] [Google Scholar]
- Todd P. A. Heel R. C. Drugs. 1985;30:514–538. doi: 10.2165/00003495-198530060-00004. [DOI] [PubMed] [Google Scholar]
- Sorkin E. M. Brogden R. N. Drugs. 1985;29:208–235. doi: 10.2165/00003495-198529030-00002. [DOI] [PubMed] [Google Scholar]
- Brogden R. N. Heel R. C. Speight T. M. Avery G. S. Drugs. 1979;17:1–37. doi: 10.2165/00003495-197917010-00001. [DOI] [PubMed] [Google Scholar]
- Yanase Y. Katsuta H. Tomiya K. Enomoto M. Sakamoto O. J. Pestic. Sci. 2013;38:167–168. doi: 10.1584/jpestics.J13-01. [DOI] [Google Scholar]
- Croxtall J. D. Plosker G. L. Drugs. 2009;69:339–359. doi: 10.2165/00003495-200969030-00009. [DOI] [PubMed] [Google Scholar]
- Harrow I. D. Gration K. A. F. Pestic. Sci. 1985;16:662–672. doi: 10.1002/ps.2780160612. [DOI] [Google Scholar]
- Berger W. De Chandt M. T. M. Cairns C. B. Int. J. Clin. Pract. 2007;61:663–676. doi: 10.1111/j.1742-1241.2007.01320.x. [DOI] [PubMed] [Google Scholar]
- Kao C.-D. Chang J.-B. Chen J.-T. Wu Z.-A. Shan D.-E. Liao K.-K. Ann. Pharmacother. 2004;38:1840–1843. doi: 10.1345/aph.1E161. [DOI] [PubMed] [Google Scholar]
- Lemieux G. Beauchemin M. Gougoux A. Vinay P. Can. Med. Assoc. J. 1978;118:1074. [PMC free article] [PubMed] [Google Scholar]
- Mirsalis J. C. Mutat. Res., Rev. Genet. Toxicol. 1987;185:309–317. doi: 10.1016/0165-1110(87)90022-4. [DOI] [PubMed] [Google Scholar]
- Franchetti P. Cappellacci L. Perlini P. Jayaram H. N. Butler A. Schneider B. P. Collart F. R. Huberman E. Grifantini M. J. Med. Chem. 1998;41:1702–1707. doi: 10.1021/jm970772e. [DOI] [PubMed] [Google Scholar]
- Kandeel M. Suganuma K. Processes. 2022;10:2158. doi: 10.3390/pr10112158. [DOI] [Google Scholar]
- Singh R. K., Key Heterocyclic Cores for Smart Anticancer Drug–Design Part I, Bentham Science Publishers, 2022 [Google Scholar]
- Garton A. J. Crew A. P. A. Franklin M. Cooke A. R. Wynne G. M. Castaldo L. Kahler J. Winski S. L. Franks A. Brown E. N. Cancer Res. 2006;66:1015–1024. doi: 10.1158/0008-5472.CAN-05-2873. [DOI] [PubMed] [Google Scholar]
- Madvar R. R. Taher M. A. Environ. Res. 2024;240:117449. doi: 10.1016/j.envres.2023.117449. [DOI] [PubMed] [Google Scholar]
- Bymaster F. P. Rasmussen K. Calligaro D. O. Nelson D. L. DeLapp N. W. Wong D. T. Moore N. A. J. Clin. Psychiatry. 1997;58:28–36. [PubMed] [Google Scholar]
- Suzdak P. D. Jansen J. A. Epilepsia. 1995;36:612–626. doi: 10.1111/j.1528-1157.1995.tb02576.x. [DOI] [PubMed] [Google Scholar]
- Al Bahri A. A. Hamnett H. J. J. Anal. Toxicol. 2023;47:216–226. doi: 10.1093/jat/bkac096. [DOI] [PubMed] [Google Scholar]
- Malamed S. F. Gagnon S. Leblanc D. J. Am. Dent. Assoc., JADA. 2000;131:635–642. doi: 10.14219/jada.archive.2000.0237. [DOI] [PubMed] [Google Scholar]
- Sharis P. J. Cannon C. P. Loscalzo J. Ann. Intern. Med. 1998;129:394–405. doi: 10.7326/0003-4819-129-5-199809010-00009. [DOI] [PubMed] [Google Scholar]
- Turkoglu G., Cinar M. E. and Ozturk T., Sulfur Chemistry, 2019, pp. 79–123 [Google Scholar]
- Shah R. Verma P. K. Chem. Cent. J. 2018;12:137. doi: 10.1186/s13065-018-0511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz R. M. D. Mendonça-Junior F. J. B. de Mélo N. B. Scotti L. de Araújo R. S. A. de Almeida R. N. de Moura R. O. Pharmaceuticals. 2021;14:692. doi: 10.3390/ph14070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla S. Sharma S. Kashid S. Verma P. K. Sapra A. Mini-Rev. Med. Chem. 2023;23:1514–1534. doi: 10.2174/1389557523666230206104257. [DOI] [PubMed] [Google Scholar]
- Khaghaninejad S. and Heravi M. M., in Advances in heterocyclic chemistry, Elsevier, 2014, vol. 111, pp. 95–146 [Google Scholar]
- Gewald K. Schinke E. Böttcher H. Chem. Ber. 1966;99:94–100. [Google Scholar]
- Volhard J. Erdmann H. Ber. Dtsch. Chem. Ges. 1885;18:454–455. doi: 10.1002/cber.18850180199. [DOI] [Google Scholar]
- Yugandar S. Konda S. Ila H. Org. Lett. 2017;19:1512–1515. doi: 10.1021/acs.orglett.7b00273. [DOI] [PubMed] [Google Scholar]
- Kuhn M. Falk F. C. Paradies J. Org. Lett. 2011;13:4100–4103. doi: 10.1021/ol2016093. [DOI] [PubMed] [Google Scholar]
- Livingstone R., in Rodd's Chemistry of Carbon Compounds, Elsevier, 1964, pp. 219–328 [Google Scholar]
- Paveliev S. A. Segida O. O. Bityukov O. V. Tang H. T. Pan Y. M. Nikishin G. I. Terent'ev A. O. Adv. Synth. Catal. 2022;364:3910–3916. [Google Scholar]
- Zheng B. Li X. Song Y. Meng S. Li Y. Liu Q. Pan L. J. O. L. Org. Lett. 2021;23:3453–3459. doi: 10.1021/acs.orglett.1c00915. [DOI] [PubMed] [Google Scholar]
- Ostapiuk Y. V. Shehedyn M. Barabash O. V. Demydchuk B. Batsyts S. Herzberger C. Schmidt A. J. S. Synthesis. 2021;54:732–740. doi: 10.1055/s-0040-1719849. [DOI] [Google Scholar]
- Chowdhury S. Chanda T. Koley S. Ramulu B. J. Jones R. C. F. Singh M. S. Tetrahedron. 2015;71:1844. [Google Scholar]
- Ge L.-S. Wang Z.-L. An X.-L. Luo X. Deng W.-P. J. O. Chemistry B. Org. Biomol. Chem. 2014;12:8473–8479. doi: 10.1039/c4ob01534g. [DOI] [PubMed] [Google Scholar]
- Jiang H. Zeng W. Li Y. Wu W. Huang L. Fu W. J. Org. Chem. 2012;77:5179–5183. doi: 10.1021/jo300692d. [DOI] [PubMed] [Google Scholar]
- Kurandina D. Gevorgyan V. Org. Lett. 2016;18:1804–1807. doi: 10.1021/acs.orglett.6b00541. [DOI] [PubMed] [Google Scholar]
- Huang G. Li J. Li J. Li J. Sun M. Zhou P. Chen L. Huang Y. Jiang S. Li Y. J. Org. Chem. 2020;85:13037–13049. doi: 10.1021/acs.joc.0c01733. [DOI] [PubMed] [Google Scholar]
- Paixão D. B. Rampon D. S. Salles H. D. Soares E. G. Bilheri F. N. Schneider P. H. J. Org. Chem. 2020;85:12922–12934. doi: 10.1021/acs.joc.0c01516. [DOI] [PubMed] [Google Scholar]
- Wang T. An Z. Qi Z. Zhuang D. Chang A. Yang Y. Yan R. Org. Chem. Front. 2019;6:3705–3709. [Google Scholar]
- Chen L. Min H. Zeng W. Zhu X. Liang Y. Deng G. Yang Y. Org. Lett. 2018;20:7392–7395. doi: 10.1021/acs.orglett.8b03078. [DOI] [PubMed] [Google Scholar]
- Shearouse W. C. Shumba M. Z. Mack J. Appl. Sci. 2014;4:171–179. doi: 10.3390/app4020171. [DOI] [Google Scholar]
- Hossaini Z. Rostami-Charati F. Soltani S. Mirzaei A. Berijani K. J. M. d. Mol. Diversity. 2011;15:911–916. doi: 10.1007/s11030-011-9322-5. [DOI] [PubMed] [Google Scholar]
- Moghaddam F. M. Khodabakhshi M. R. Latifkar A. Tetrahedron Lett. 2014;55:1251–1254. [Google Scholar]
- Adib M. Rajai-Daryasarei S. Pashazadeh R. Jahani M. Yazzaf R. Amanlou M. Eur. J. Org. Chem. 2018;2018:3001–3016. [Google Scholar]
- Gronowitz S. Adv. Heterocycl. Chem. 1963;1:1–124. doi: 10.1016/s0065-2725(08)60523-1. [DOI] [PubMed] [Google Scholar]
- Knox C. Wilson M. Klinger C. M. Franklin M. Oler E. Wilson A. Pon A. Cox J. Chin N. E. Strawbridge S. A. Nucleic Acids Res. 2024;52:D1265–D1275. doi: 10.1093/nar/gkad976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramec D. Peterlin Mašič L. Sollner Dolenc M. Chem. Res. Toxicol. 2014;27:1344–1358. doi: 10.1021/tx500134g. [DOI] [PubMed] [Google Scholar]
- Hu Y. Yang S. Shilliday F. B. Heyde B. R. Mandrell K. M. Robins R. H. Xie J. Reding M. T. Lai Y. Thompson D. C. Drug Metab. Dispos. 2010;38:1522–1531. doi: 10.1124/dmd.110.032581. [DOI] [PubMed] [Google Scholar]
- Zuniga F. I. Loi D. Ling K. H. J. Tang-Liu D. D. S. Expert Opin. Drug Metab. Toxicol. 2012;8:467–485. doi: 10.1517/17425255.2012.668528. [DOI] [PubMed] [Google Scholar]
- Prieto-Martínez F. D., López-López E., Juárez-Mercado K. E. and Medina-Franco J. L., In silico drug design, 2019, pp. 19–44 [Google Scholar]
- Dao N. A., Vu T.-D. and Chu D.-T., in Advances in Bioinformatics, Springer, 2024, pp. 239–248 [Google Scholar]
- Luna I. S. de Souza T. A. da Silva M. S. da Franca Rodrigues K. A. Scotti L. Scotti M. T. Mendonça-Junior F. J. B. Eur. J. Med. Chem. 2023;250:115223. doi: 10.1016/j.ejmech.2023.115223. [DOI] [PubMed] [Google Scholar]
- Romeo I. Brizzi A. Pessina F. Ambrosio F. A. Aiello F. Belardo C. Carullo G. Costa G. De Petrocellis L. Frosini M. J. Med. Chem. 2023;66:6994–7015. doi: 10.1021/acs.jmedchem.3c00447. [DOI] [PubMed] [Google Scholar]
- Natarajan R. Sivaperuman A. Samuel A. Patel D. H. Jain N. Veerappan M. Kumar N. K. Chem. Biodiversity. 2023;20:e202300331. doi: 10.1002/cbdv.202300331. [DOI] [PubMed] [Google Scholar]
- Güven O. Menteşe E. Emirik M. Sökmen B. B. Akyüz G. Arch. Pharm. 2023;356:2300336. doi: 10.1002/ardp.202300336. [DOI] [PubMed] [Google Scholar]
- Javid N. Jalil S. Munir R. Zia-ur-Rehman M. Sahar A. Arshad S. Iqbal J. RSC Adv. 2023;13:1701–1710. doi: 10.1039/d2ra07045f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A. H. Murugesan S. Amran S. I. Chander S. Alanazi M. M. Hadda T. B. Shakya S. Pratama M. R. F. Das B. Biswas S. Bioorg. Chem. 2022;119:105572. doi: 10.1016/j.bioorg.2021.105572. [DOI] [PubMed] [Google Scholar]
- Murugavel S. Ravikumar C. Jaabil G. Alagusundaram P. Comput. Biol. Chem. 2019;79:73–82. doi: 10.1016/j.compbiolchem.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Gaines T. Garcia F. Virani S. Liang Z. Yoon Y. Oum Y. H. Shim H. Mooring S. R. Eur. J. Med. Chem. 2019;181:111562. doi: 10.1016/j.ejmech.2019.111562. [DOI] [PubMed] [Google Scholar]
- Rodriguez F. Iniguez E. Pena Contreras G. Ahmed H. Costa T. E. M. M. Skouta R. Maldonado R. A. Molecules. 2018;23:1626. doi: 10.3390/molecules23071626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulipalli K. C. Bodige S. Ravula P. Endoori S. Vanaja G. R. Babu G. S. Chandra J. N. N. S. Seelam N. Bioorg. Med. Chem. Lett. 2017;27:3558–3564. doi: 10.1016/j.bmcl.2017.05.047. [DOI] [PubMed] [Google Scholar]
- Murugavel S. Ravikumar C. Jaabil G. Alagusundaram P. Comput. Biol. Chem. 2019;79:73–82. doi: 10.1016/j.compbiolchem.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Prabhudeva M. G. Bharath S. Kumar A. D. Naveen S. Lokanath N. K. Mylarappa B. N. Kumar K. A. Bioorg. Chem. 2017;73:109–120. doi: 10.1016/j.bioorg.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Louise-May S. Yang W. Nie X. Liu D. Deshpande M. S. Phadke A. S. Huang M. Agarwal A. Bioorg. Med. Chem. Lett. 2007;17:3905–3909. doi: 10.1016/j.bmcl.2007.04.103. [DOI] [PubMed] [Google Scholar]
- Blair M. Urol. Nurs. 2016;36:363–374. doi: 10.1007/s40265-016-0541-z. [DOI] [PubMed] [Google Scholar]
- Navale A. M. Paranjape A. N. Biophys. Rev. 2016;8:5–9. doi: 10.1007/s12551-015-0186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T. D. Finan B. Bloom S. R. D'Alessio D. Drucker D. J. Flatt P. R. Fritsche A. Gribble F. Grill H. J. Habener J. F. Mol. Metab. 2019;30:72–130. doi: 10.1016/j.molmet.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cersosimo E. Solis-Herrera C. Trautmann M. E. Malloy J. Triplitt C. L. Curr. Diabetes Rev. 2014;10:2–42. doi: 10.2174/1573399810666140214093600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artasensi A. Pedretti A. Vistoli G. Fumagalli L. Molecules. 2020;25:1987. doi: 10.3390/molecules25081987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku K. Isaka H. Sakatani T. Toyoshima J. J. Diabetes Invest. 2020;11:662–671. doi: 10.1111/jdi.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H. Li D. Yang Y. Guo H.-F. Liu Z.-Y. Si S.-Y. Yang Z. Li Z.-R. J. Enzyme Inhib. Med. Chem. 2010;25:282–289. doi: 10.1080/14756360903179369. [DOI] [PubMed] [Google Scholar]
- Imamura M. Nakanishi K. Suzuki T. Ikegai K. Shiraki R. Ogiyama T. Murakami T. Kurosaki E. Noda A. Kobayashi Y. Bioorg. Med. Chem. 2012;20:3263–3279. doi: 10.1016/j.bmc.2012.03.051. [DOI] [PubMed] [Google Scholar]
- Qin X. Hao X. Han H. Zhu S. Yang Y. Wu B. Hussain S. Parveen S. Jing C. Ma B. J. Med. Chem. 2015;58:1254–1267. doi: 10.1021/jm501484b. [DOI] [PubMed] [Google Scholar]
- Li J. Feng Y. Li H. Shu S. Dai A. Cai X. Wang J. Yang D. Ma D. Wang M. W. Chem. Biol. Drug Des. 2018;92:1241–1254. doi: 10.1111/cbdd.13184. [DOI] [PubMed] [Google Scholar]
- Xie H.-X. Zhang J. Li Y. Zhang J.-H. Liu S.-K. Zhang J. Zheng H. Hao G.-Z. Zhu K.-K. Jiang C.-S. Bioorg. Chem. 2021;115:105236. doi: 10.1016/j.bioorg.2021.105236. [DOI] [PubMed] [Google Scholar]
- Wang K.-M. Ge Y.-X. Zhang J. Chen Y.-T. Zhang N.-Y. Gu J.-S. Fang L. Zhang X.-L. Zhang J. Jiang C.-S. Bioorg. Med. Chem. Lett. 2023;79:129069. doi: 10.1016/j.bmcl.2022.129069. [DOI] [PubMed] [Google Scholar]
- Shams H. Z. Mohareb R. M. Helal M. H. Mahmoud A. E. Molecules. 2011;16:52–73. doi: 10.3390/molecules16010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abualnaja M. M. Alalawy A. I. Alatawi O. M. Alessa A. H. Qarah A. F. Alqahtani A. M. Bamaga M. A. El-Metwaly N. M. Saudi Pharm. J. 2024;32:101962. doi: 10.1016/j.jsps.2024.101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said M. Elshihawy H. Pak. J. Pharm. Sci. 2014;27:885–892. [PubMed] [Google Scholar]
- Bozorov K. Ma H.-R. Zhao J.-Y. Zhao H.-Q. Chen H. Bobakulov K. Xin X.-L. Elmuradov B. Shakhidoyatov K. Aisa H. A. Eur. J. Med. Chem. 2014;84:739–745. doi: 10.1016/j.ejmech.2014.07.065. [DOI] [PubMed] [Google Scholar]
- Sztanke M. Rzymowska J. Sztanke K. Bioorg. Med. Chem. 2015;23:3448–3456. doi: 10.1016/j.bmc.2015.04.037. [DOI] [PubMed] [Google Scholar]
- Gomha S. M. Edrees M. M. Altalbawy F. M. A. Int. J. Mol. Sci. 2016;17:1499. doi: 10.3390/ijms17091499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorab M. M. Alsaid M. S. Al-Dosari M. S. Ragab F. A. Al-Mishari A. A. Almoqbil A. N. Acta Pharm. 2016;66:155–171. doi: 10.1515/acph-2016-0016. [DOI] [PubMed] [Google Scholar]
- Mohareb R. M. Abdallah A. E. M. Ahmed E. A. Acta Pharm. 2017;67:495–510. doi: 10.1515/acph-2017-0040. [DOI] [PubMed] [Google Scholar]
- Cardoso L. N. F. Nogueira T. C. M. Rodrigues F. A. R. Oliveira A. C. A. Luciano M. C. d. S. Pessoa C. de Souza M. V. N. Med. Chem. Res. 2017;26:1605–1608. doi: 10.1007/s00044-017-1832-y. [DOI] [Google Scholar]
- Thomas J. Jecic A. Vanstreels E. van Berckelaer L. Romagnoli R. Dehaen W. Liekens S. Balzarini J. Eur. J. Med. Chem. 2017;132:219–235. doi: 10.1016/j.ejmech.2017.03.044. [DOI] [PubMed] [Google Scholar]
- Fouad M. M. El-Bendary E. R. Shehata I. A. El-Kerdawy M. M. Bioorg. Chem. 2018;81:587–598. doi: 10.1016/j.bioorg.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Gomha S. M. Edrees M. M. Muhammad Z. A. El-Reedy A. A. M. Drug Des., Dev. Ther. 2018;12:1511–1523. doi: 10.2147/Dddt.S165276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman D. I. A. Selim K. B. Magda A. A. Tantawy A. S. Amen Y. Shimizu K. Okauchi T. Kitamura M. Bioorg. Med. Chem. 2019;27:115026. doi: 10.1016/j.bmc.2019.07.042. [DOI] [PubMed] [Google Scholar]
- Shah R. Verma P. K. BMC Chem. 2019;13(1–13):54. doi: 10.1186/s13065-019-0569-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin N. Rep V. Sović I. Juričić Š. Selgrad D. Klobučar M. Pržulj N. Gupta C. L. Malod-Dognin N. Pavelić S. K. Eur. J. Med. Chem. 2020;185:111833. doi: 10.1016/j.ejmech.2019.111833. [DOI] [PubMed] [Google Scholar]
- Megally Abdo N. Y. Samir E. M. Mohareb R. M. J. Heterocyclic Chem. 2020;57:1993–2009. [Google Scholar]
- Şenol H. Çakır F. ChemistrySelect. 2023;8:e202302448. [Google Scholar]
- Doddagaddavalli M. A. Kalalbandi V. K. A. Seetharamappa J. Joshi S. D. Bioorg. Chem. 2024;143:107003. doi: 10.1016/j.bioorg.2023.107003. [DOI] [PubMed] [Google Scholar]
- Eldehna W. M. Al-Ansary G. H. Al-Warhi T. Jaballah M. Y. Elaasser M. Rashed M. Bioorg. Chem. 2024;143:107037. doi: 10.1016/j.bioorg.2023.107037. [DOI] [PubMed] [Google Scholar]
- Rahman A. Jain R. S. K. Meghana P. Nippu B. N. Manjunatha K. S. Rajaput P. S. Kumaraswamy H. M. Satyanarayan N. D. Bioorg. Chem. 2024;143:106968. doi: 10.1016/j.bioorg.2023.106968. [DOI] [PubMed] [Google Scholar]
- Shah R. Verma P. K. BMC Chem. 2019;13:1–13. doi: 10.1186/s13065-019-0569-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautio J. Leppänen J. Lehtonen M. Laine K. Koskinen M. Pystynen J. Savolainen J. Sairanen M. Bioorg. Med. Chem. Lett. 2010;20:2614–2616. doi: 10.1016/j.bmcl.2010.02.057. [DOI] [PubMed] [Google Scholar]
- Ismail M. M. Kamel M. M. Mohamed L. W. Faggal S. I. Galal M. A. Molecules. 2012;17:7217–7231. doi: 10.3390/molecules17067217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyachandran V. Kumar R. R. Ali M. A. Choon T. S. Bioorg. Med. Chem. Lett. 2013;23:2101–2105. doi: 10.1016/j.bmcl.2013.01.122. [DOI] [PubMed] [Google Scholar]
- Funicello M. Cerminara I. Chiummiento L. Med. Chem. 2016;6:377–384. [Google Scholar]
- Cetin A. Türkan F. Taslimi P. Gulçin İ. J. Biochem. Mol. Toxicol. 2019;33:e22261. doi: 10.1002/jbt.22261. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez F. Vina D. Uriarte E. Fontenla J. A. Matos M. J. Bioorg. Chem. 2020;101:103986. doi: 10.1016/j.bioorg.2020.103986. [DOI] [PubMed] [Google Scholar]
- Ullah H. Bashir M. Khan F. Iqbal I. Iqbal A. Rahim F. Chem. Data Collect. 2024;50:101113. doi: 10.1016/j.cdc.2024.101113. [DOI] [Google Scholar]
- Funicello M. Cerminara I. Chiummiento L. Med. Chem. 2016;6:e384. [Google Scholar]
- Filali I. Bouajila J. Znati M. Bousejra-El Garah F. Ben Jannet H. J. Enzyme Inhib. Med. Chem. 2015;30:371–376. doi: 10.3109/14756366.2014.940932. [DOI] [PubMed] [Google Scholar]
- Patil D. Dash R. P. Thakur S. K. Pandya A. N. Venkatesh P. Vasu K. K. Nivsarkar M. J. Enzyme Inhib. Med. Chem. 2015;30:229–239. doi: 10.3109/14756366.2014.913035. [DOI] [PubMed] [Google Scholar]
- Helal M. H. Salem M. A. Gouda M. A. Ahmed N. S. El-Sherif A. A. Spectrochim. Acta, Part A. 2015;147:73–83. doi: 10.1016/j.saa.2015.03.070. [DOI] [PubMed] [Google Scholar]
- Chaudhari P. S. Chitlange S. S. Nanda R. K. Anti-Inflammatory Anti-Allergy Agents Med. Chem. 2018;17:102–114. doi: 10.2174/1871523017666180910105609. [DOI] [PubMed] [Google Scholar]
- El-Miligy M. M. M. Hazzaa A. A. El-Messmary H. Nassra R. A. El-Hawash S. A. M. Bioorg. Chem. 2017;72:102–115. doi: 10.1016/j.bioorg.2017.03.012. [DOI] [PubMed] [Google Scholar]
- John L. Dasan A. Joseyphus R. S. Joe I. H. J. Mol. Struct. 2019;1198:126934. doi: 10.1016/j.molstruc.2019.126934. [DOI] [Google Scholar]
- Chiasson A. I. Robichaud S. Ndongou Moutombi F. J. Hébert M. P. A. Mbarik M. Surette M. E. Touaibia M. Molecules. 2020;25:4686. doi: 10.3390/molecules25204686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri C. K. Indalkar K. S. Patil C. R. Goyal S. N. Chaturbhuj G. U. Bioorg. Med. Chem. Lett. 2017;27:1721–1726. doi: 10.1016/j.bmcl.2017.02.076. [DOI] [PubMed] [Google Scholar]
- Hu J. Wang G. Liu X. Zhou L. Jiang M. Yang L. PLoS One. 2013;8:e78832. doi: 10.1371/journal.pone.0078832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganagowda G. Petsom A. J. Sulfur Chem. 2011;32:223–233. [Google Scholar]
- Naganagowda G. Meijboom R. Phosphorus, Sulfur Silicon Relat. Elem. 2014;186(10):2112–2121. doi: 10.1080/10426507.2011.586905. [DOI] [Google Scholar]
- Nagesh H. K. Padmashali B. Sandeep C. Yuvaraj T. C. M. Siddesh M. B. Mallikarjuna S. M. Int. J. Pharm. Sci. Rev. Res. 2014;28:6–10. [Google Scholar]
- Srivastava S. Das B. Der Pharma Chemica. 2011;3:103–111. [Google Scholar]
- Bhatt R. Sharma S. Patidar A. K. Rathore K. K. Senwar R. C. Mehta A. Int. J. Pharm. Sci. Drug Res. 2015;7(5):417–420. [Google Scholar]
- Ajdačić V. Senerovic L. Vranić M. Pekmezovic M. Arsic-Arsnijevic V. Veseli A. Bioorg. Med. Chem. 2016;24:1277. doi: 10.1016/j.bmc.2016.01.058. [DOI] [PubMed] [Google Scholar]
- Mabkhot Y. N. Al-Showiman S. S. Soliman S. M. Ghabbour H. A. AlDamen M. A. Mubarak M. S. Chem. Cent. J. 2017;11:1–9. doi: 10.1186/s13065-017-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk O. H. Shaaban O. G. Wahab A. E. A. Open Med. Chem. J. 2017;11:38. doi: 10.2174/1874104501711010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M. A. F. Roy N. Benjamin I. Joo S. W. Musthafa Y. M. M. Ghfar A. A. Obunukwu G. M. Akor F. O. Louis H. J. Mol. Struct. 2024;1306:137810. doi: 10.1016/j.molstruc.2024.137810. [DOI] [Google Scholar]
- Small D. M. Coombes J. S. Bennett N. Johnson D. W. Gobe G. C. Nephrology. 2012;17:311–321. doi: 10.1111/j.1440-1797.2012.01572.x. [DOI] [PubMed] [Google Scholar]
- Gouda M. A. Eldien H. F. Girges M. M. Berghot M. A. Med. Chem. 2013;3:2228–2232. [Google Scholar]
- Gouda M. A. El Bialy S. A. A. Eur. J. Chem. 2014;5:644–651. [Google Scholar]
- Raj K. M. Mruthyunjayaswamy B. H. M. J. Mol. Struct. 2014;1074:572–582. doi: 10.1016/j.molstruc.2014.06.039. [DOI] [Google Scholar]
- Metwally H. M. Khalaf N. A. Abdel-Latif E. Ismail M. A. BMC Chem. 2023;17:6. doi: 10.1186/s13065-023-00917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari T. L. Robert A. R. Karunakar P. Maddila S. Jonnalagadda S. B. J. Mol. Struct. 2024;1307:137883. doi: 10.1016/j.molstruc.2024.137883. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No primary research results, software or code has been included and no new data were generated or analysed as part of this review.