ABSTRACT
Bacteria employ a number of dedicated secretion systems to export proteins to the extracellular environment. Several of these comprise large complexes that assemble in and around the bacterial membrane(s) to form specialized channels through which only selected proteins are actively delivered. Although typically associated with bacterial pathogenicity, a specialized variant of these secretion systems has been proposed to play a central part in bacterial sporulation, a primitive protective process that allows starving cells to form spores that survive in extreme environments. Following asymmetric division, the mother cell engulfs the forespore, leaving it surrounded by two bilayer membranes. During the engulfment process an essential channel apparatus is thought to cross both membranes to create a direct conduit between the mother cell and forespore. At least nine proteins are essential for channel formation, including SpoIIQ under forespore control and the eight SpoIIIA proteins (SpoIIIAA to -AH) under mother cell control. Presumed to form a core channel complex, several of these proteins share similarity with components of Gram-negative bacterial secretion systems, including the type II, III, and IV secretion systems and the flagellum. Based on these similarities it has been suggested that the sporulation channel represents a hybrid, secretion-like transport machinery. Recently, in-depth biochemical and structural characterization of the individual channel components accompanied by in vivo studies has further reinforced this model. Here we review and discuss these recent studies and suggest an updated model for the unique sporulation channel apparatus architecture.
SECRETION AND SPORULATION
Bacteria utilize sophisticated nanomachines to transport proteins, small molecules, and DNA across membranes to the extracellular environment. These transport machineries, also known as secretion systems, are involved in various cellular functions, such as adhesion to surfaces or host cells, cell-cell communication, motility (flagella), virulence effector protein secretion, and, notably, bacterial pathogenesis (1–5). Several of the identified protein secretion systems comprise large complexes that localize and assemble in and around the bacterial membrane(s), forming specialized channels through which the selected substrate(s) is actively delivered (6–9). Although exhibiting significant diversity in structure, substrate, and function, the dedicated type II, III, IV, and IV-pilus secretion systems (T2SS, T3SS, T4SS, and T4PS, respectively) in didermic Gram-negative bacteria each transport a specific subset of proteins to the extracellular milieu via passage through large stacked ring-shaped channels that span the inner membrane (IM) and outer membrane (OM).
Recently, a novel variant of these secretion systems has been proposed to play a central role during bacterial sporulation (10). Sporulation is an ancient developmental process, observed most typically in Gram-positive bacteria from the Firmicutes phylum but also observed in some Gram-negative species (Myxococcus xanthus [11], for example), that allows starving cells to differentiate into metabolically dormant spores that can survive extreme environmental conditions. The sporulation process involves multiple steps, each with corresponding morphological changes that are governed by intercellular signaling pathways through the activation of cell-specific sigma factors that control gene expression (Fig. 1). Early in sporulation, an asymmetric septum divides the rod-shaped bacterium into two cells: a smaller “forespore,” which becomes the spore, and a larger “mother cell” that contributes to the development of the forespore but ultimately dies. After asymmetric division is complete, σF is activated in the forespore and then signals for σE activation in the mother cell. σE expression triggers the start of the engulfment step, in which the mother cell membrane migrates around the forespore in a phagocytic-like process, resulting in the forespore being engulfed as a double-membraned protoplast within the mother cell, ending with σG activation in the forespore. Later on, σK is activated in the mother cell and supports spore maturation, mother cell lysis, and mature spore release to the environment.
FIGURE 1.
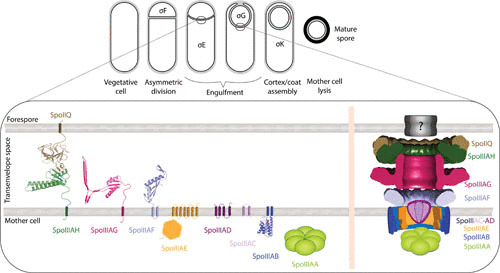
Schematic representation of the sporulation process and the active sporulation channel architecture model. (Top) Morphological changes mediated by cell-specific sigma factors that regulate gene expression in Bacillus subtilis. (Bottom) Sporulation channel assembly and function during the engulfment stage involve the expression of nine core component proteins forming a channel that crosses the mother cell membrane, the transenvelope space, and the forespore membrane. (Left) Monomeric topology and known structures of the essential core proteins. (Right) Schematic illustration of the suggested model of the assembled SpoIIIA-IIQ channel. Based on the similarities of the individual components to proteins from other bacterial secretion systems, it is predicted that the core components oligomerize into ring-like structures that are stacked to form this sporulation-specialized secretion system. In this model, the stacked rings of SpoIIIAF, SpoIIIAG, and SpoIIIAH-SpoIIQ form the main conduit in the transenvelope space connecting the mother cell and the forespore. SpoIIIAC and SpoIIIAD form a simplified version of an export apparatus through the mother cell membrane. SpoIIIAE utilizes the proton motif force for substrate transportation and also to mediate the interaction with the SpoIIIAA ATPase and its possible docking platform formed by oligmerized (?) SpoIIIAB. Any additional pore-forming protein(s) required at the forespore membrane has yet to be identified.
During engulfment, at least nine proteins assemble into a channel apparatus that spans the two opposing membranes separating the mother cell and forespore (10). Eight of these proteins (SpoIIIAA to -AH) are encoded in a single operon (spoIIIA) expressed in the mother cell under the control of σE (12, 13) and a ninth protein, SpoIIQ, is produced in the forespore under the control of σF (14). Mutants lacking any of these channel genes display collapsed forespores that are unable to carry out gene expression either dependent or independent of the late-acting σG (14–18). Collectively, these observations suggest that the channel transports one or more substrates, yet to be identified, that support forespore physiology and the capacity to carry out macromolecular synthesis.
THE INDIVIDUAL CHANNEL COMPONENTS
Although many questions remain regarding the SpoIIIA-IIQ sporulation channel, recent studies have helped to bring into focus its evolutionary origins and some of its structural features (19). Intriguingly, the emerging picture is of a novel hybrid secretion system with proteins that share common elements with those of diverse bacterial secretion systems.
SpoIIIAA
Homology searches reveal that SpoIIIAA resembles AAA+ superfamily ATPases of the T2SS, the related T4PS, and T4SS (18 to 20% sequence similarity) (17, 20). In these systems the secretion ATPase is found closely associated with the IM complex and likely adopts a distinct hexameric structure (21, 22) (Fig. 1). The predicted SpoIIIAA hexamer is proposed to utilize energy from ATP hydrolysis to drive substrate export (17).
SpoIIIAB
SpoIIIAB is predicted to be a bitopic membrane protein with anchoring transmembrane helices (TMHs) at the N and C termini and an intervening soluble domain (Fig. 1) (23, 24). The X-ray crystallographic structure of the SpoIIIAB soluble domain (residues 27 to 153) adopts a six-helix bundle similar to that of soluble regions of the polytopic GspF/PilC membrane proteins of the T2SS and evolutionarily related T4PS (22 to 28% sequence similarity) (25–28) (Fig. 2A). The latter variants are localized to the IM platform of the T2SS/T4PS, although their specific function is unknown. Additional structural similarity was found between SpoIIIAB and the C-subunit protein from the bacterial V-ATPase complex (3.4-Å root mean square deviation [RMSD] over 108 Cα atoms) that serves as a ‘‘socket’’ attaching the cytosolic V1 central stalk subunits to the membrane-bound V0 domain (29). It was hypothesized that by analogy, SpoIIIAB could oligomerize and be positioned to serve as a structural link between the membrane-bound protein components and other soluble components of the SpoIIIA-IIQ channel, such as the SpoIIIAA ATPase (28).
FIGURE 2.
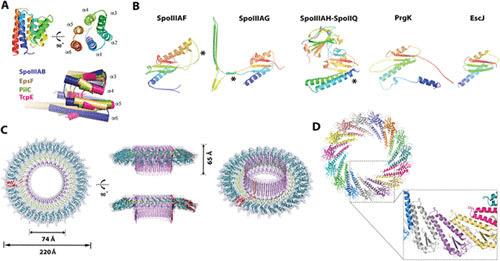
Structures of the core components of the sporulation channel share similar structural motifs with homologs from other bacterial secretion systems. (A) (Top) The SpoIIIAB soluble domain adopts a six-helix bundle fold with both N and C termini in close proximity and facing the mother cell membrane. The molecule is shown in two views, related by a 90° rotation. (Bottom) SpoIIIAB shares a fold similar to that of homologous proteins from the T2SS and T4PS. Shown is a structural overlay of SpoIIIAB with EpsF, TcpE (both from Vibrio cholerae), and PilC (Thermus thermophilus) proteins in blue, wheat, green, and pink, respectively (PDB codes 6BS9, 3C1Q, 2WHN, and 4HHX, respectively). Two regions of structural variation are seen in the helix 6 angle and the increasing dimensions of helices 4 and 5 and the loop connecting them. (B) SpoIIIAF, SpoIIIAG, and the SpoIIIAH-SpoIIQ heterodimer contain an RBM fold similar to that of the T3SS basal body proteins, EscJ (Escherichia coli) and PrgK (Salmonella Tryphimurium) (PDB codes 6DCS, 5WC3, 3UZ0, 1YJ7, and 3J6D, respectively). All five structures are displayed in cartoon representation and rainbow color scheme and for clarity are individually shown in identical orientations originating from structural superposition. SpoIIIAF is presented as an overlay of the two monomers seen in the crystal structure, with the region of alternate conformation associated with regulation marked with an asterisk. SpoIIIAG adopts the canonical RBM fold, with a large insertion of the β-triangle motif marked with an asterisk. An SpoIIIAH additional N-terminal helix is marked with an asterisk. (C) Cryo-EM structure of the SpoIIIAG soluble domain 30-meric ring. A three-dimensional reconstruction and atomic model are shown in top side, cropped, and tilted views. The SpoIIIAG ring structure is colored according to distinctive ring elements: RBM in cyan, planar β-ring in green, and vertical β-ring in pink, with the single protomer in red. (D) SpoIIIAH-SpoIIQ representative computational modeled ring, here in C15 symmetry with zoomed-in view of the predicted interaction region between the RBMs of SpoIIIAH. Ring model coordinates were obtained from Meisner et al. (46).
SpoIIIAC and SpoIIIAD
SpoIIIAC and SpoIIIAD are predicted to be small, polytopic membrane proteins with two and four TMHs, respectively (Fig. 1). Although they share no detectable similarity with any protein of known function, their size, number, and orientation of transmembrane segments do resemble components of the flagellar and T3SS proteins, namely, FliQ/SpaQ (SpoIIIAC) and FliP/SpaP (SpoIIIAD), of the IM export apparatus proposed to play a central role in substrate selection and chronological secretion. Recently the near-atomic-resolution, cryo-electron microscopic (cryo-EM) structure of the Salmonella enterica serovar Typhimurium flagellar IM export apparatus revealed that none of the three integral membrane proteins FliP, FliQ, and FliR (5:4:1 stoichiometry, respectively) adopts the canonical integral membrane protein topologies previously predicted by primary sequence analysis. Instead, the cryo-EM structure showed that the intimately associating helix-turn-helix structural elements common to all 3 flagellar proteins mediate a soluble, coiled-coil-like export apparatus complex formation that sits atop rather than directly embedded in the IM (30). Further, in the same study, the FliR monomer was shown to be a structural fusion of FliP and FliQ partners in the export apparatus assembly, suggesting that in primitive secretion systems two proteins might be functionally sufficient for export apparatus assembly. Based on the overall secondary-structure similarities, one can therefore speculate that SpoIIIAC and SpoIIIAD may also form a homologous helix-turn-helix, coiled-coil export apparatus in the sporulation channel and play a central role in substrate recognition and secretion (Fig. 1).
SpoIIIAE
Primary sequence analysis suggests that SpoIIIAE is a multipass polytopic membrane protein with 7 TMHs and an N-terminal Sec-type signal peptide that is followed by a small (∼75-residue) mother cell cytoplasmic domain (Fig. 1). Mutations of the SpoIIIAE signal peptide arrest sporulation following the engulfment stage and thereby prevent activation of σG (31). SpoIIIAE was shown to interact with SpoIIIJ, a membrane protein translocase (YidC homolog), promoting correct localization and topology in the membrane by the Sec system (15, 31). Bioinformatic analyses suggest that SpoIIIAE is similar to the permease domain of the ATP-binding cassette transporters of the type I secretion system and to electrochemical-potential-driven transporters involved in the shuttle of various drugs and other proteins across membranes (18 to 24% sequence similarity) (32). Furthermore, the SpoIIIAE N-terminal domain shares remote similarity to SD3 (see below) of InvA/FlhA proteins from the T3SS/flagella (∼8% sequence similarity). These are highly conserved, polytopic IM proteins which form an additional major cytosol-facing component of the export apparatus (described in part above—SpaPQRS and InvA or FliPQR and FlhAB in T3SS and flagellar nomenclature, respectively) and are associated with secretion regulation. InvA/FlhA have been shown to oligomerize into a nonameric ring with each protomer containing an N-terminal integral membrane domain of 7 or 8 TMHs that has been hypothesized to employ proton motive force to promote protein export, as well as a ring-forming C-terminal cytoplasmic domain (33). The latter domain is thought to comprise a central component of the cytoplasmic, substrate “docking platform” of the apparatus and was shown to interact with members of the FlhB superfamily, secretion substrates in complex with their chaperones, and the conserved ATPase and its regulators (33–36). The monomeric structure of the InvA/FlhA cytosolic domain contains four subdomains, SD1 to SD4, with SD3 shaping the inner pore surface and participating in ring-stabilizing interactions (35, 37). It is possible that the SpoIIIAE N-terminal domain and following transmembrane domain, although swapped in position along the polypeptide chain relative to InvA/FlhA, may share function and oligomerization propensity similar to that of InvA/FlhA in the assembled SpoIIIA-IIQ channel.
SpoIIIAF
SpoIIIAF is a bitopic membrane protein anchored to the mother cell membrane by two N-terminal TMHs followed by a larger soluble domain (residues 60 to 206) (Fig. 1). The latter was shown to oligomerize into homomeric rings through a canonical ring-building motif (RBM) fold (residues 85 to 206) as determined by X-ray crystallography (Fig. 2B) (38). The RBM fold is defined by a growing group of characterized small mixed α/β modular domain structures that pack into oligomeric rings of large assemblies. RBMs characterized thus far share a superficial common architecture divided into two groups based on secondary-structure topology: an αββαβ fold distributed in the IM basal body proteins of the T3SS (PrgK/EscJ and PrgH from the T3SS and FliF from the flagella) (39) and a βαββα fold predominantly found in the OM secretins common to the T2SS, T3SS, and T4PS (40). While the SpoIIIAF core RBM fold shares significant similarity to the T3SS PrgK/EscJ variants (Cα RMSD < 2 Å), it also contains two unique features: an extended N-terminal helix, associated with multimerization and possible interaction with the mother cell membrane, and an 11-residue insertion within a loop region observed to adopt two distinct conformations. The ability of the same primary sequence to adopt different secondary-structure conformations is associated with protein regulation, suggesting dual structural and regulatory roles for the SpoIIIAF RBM (38).
SpoIIIAG
SpoIIIAG contains a short N-terminal mother cell-cytosolic region followed by a single TMH and a large soluble domain, shown to face the intermembrane region (Fig. 1) (41). A near-atomic-resolution single-particle cryo-EM structure of the SpoIIIAG soluble domain revealed that the monomeric form contains a long disordered region (33 residues) followed by two structural motifs: an RBM and a novel β-triangle motif insertion (Fig. 2B and C) (42). Interestingly, SpoIIIAG was found to self-assemble into a large and stable 30-mer complex comprised of three distinctive circular structures: an inner 60-strand vertical β-barrel and a 60-strand planar β-ring, both formed by the β-triangle motif repeats, and an external ring formed by the RBMs (42). Overall, the SpoIIIAG complex shares striking similarity with two major components of the T3SS/flagellar basal body: the IM rings and the secretin. The IM components of these three systems all display a high oligomerization number (30/24/26-mers of SpoIIIAG/PrgK/FliF from SpoIIIA-IIQ/T3SS/flagella, respectively), common repetitive RBM interaction interfaces (Cα RMSD < 2 Å), and large outer ring diameters (∼22/19/24 nm for SpoIIIAG/PrgK/FliF, respectively) (6, 42, 43). The SpoIIIAG complex and secretins found in the T2SS and T3SS also share distinct architectures of giant β-barrels comprised of 60 β-strands that are vertical to the membrane axis. These secretin megastructures are formed by a completely unique 15-mer repeat of a four-stranded β-sandwich that assembles the central domain of the massive double-layered β-barrel (6, 44). The outer repeating four-stranded β-sheet from each monomeric sandwich forms an overall 60-stranded anti-parallel β-barrel that constitutes the outer wall, while the inner sheet forms a vertical anti-parallel barrel that serves as the inner “periplasmic gate.” Notably, a low-resolution cryo-EM structure of the R-domain for FliF revealed a complex with a similar overall shape and with an almost identical vertical dimension of ∼6.5 to 7 nm and similar inner channel diameters of ∼7.5 nm in SpoIIIAG (and PrgK) and ∼9 nm in FliF (6, 42, 43). Although the high-resolution FliF complex structure is currently unavailable, prior model-building bioinformatic predictions have proposed that FliF is comprised of an RBM with a β-strand insertion similar to that of SpoIIIAG and therefore potentially a similar β-barrel structure (42).
SpoIIIAH and SpoIIQ
Individual monomeric structures and dimerization
SpoIIIAH is anchored to the mother cell membrane by a single N-terminal TMH followed by a large soluble domain that faces the intermembrane space (45, 46) (Fig. 1). The SpoIIIAH soluble domain structure (residues 32 to 218) was determined to a 2.26-Å resolution using X-ray crystallography in complex with SpoIIQ (residues 43 to 283). The SpoIIIAH structure was shown to contain a long disordered region (∼65 residues) and a globular domain that adopts the canonical RBM fold with an additional helix packed against the first helix and β-sheet of the motif (45, 46) (Fig. 2B). The binding partner, SpoIIQ, is anchored to the forespore membrane by a single N-terminal TMH followed by a large soluble domain that also faces the intermembrane space (Fig. 1). The SpoIIQ soluble domain contains a long disordered region (∼30 residues) and a globular domain that adopts a LytM-metalloendopeptidase fold (∼20% sequence similarity). Despite its fold, SpoIIQ is not an active enzyme, as the typical catalytic and zinc-coordinating histidine residues at the active site have been evolutionarily lost. This stable heterodimer is formed through tight pairing of the third β-strands of both proteins, resulting in the formation of a continuous intermolecular five-stranded β-sheet accompanied by other auxiliary stabilizing interactions (45, 46) (Fig. 2B).
Ring formation modeling of the heterodimer
Based on the structural similarity of SpoIIIAH to the IM ring-forming proteins of the T3SS and the flagella (PrgK/EscJ and FliF, respectively), it has been speculated that SpoIIIAH in complex with SpoIIQ can also oligomerize into a circular complex, and several planar ring-shaped models have been proposed with high-order symmetries imposed (42, 45, 46) (Fig. 1 and 2D). These SpoIIIAH-SpoIIQ ring models present outer surfaces that are largely electronegative and therefore unlikely to be directly abutting the inherently anionic bacterial membranes due to charge repulsion; this suggests that additional proteins are likely required to form a continuous conduit between the mother cell and the forespore membranes.
SpoIIIAH-SpoIIQ heterodimeric complex as a protein interaction hub
In addition to their direct role as key members of the SpoIIIA-IIQ apparatus (47, 48), both proteins are at the center of a protein interaction network that may have implications for both mother cell and forespore (49). Knockouts of either SpoIIIAH or SpoIIQ result in severe cellular defects that include mislocalization of SpoIID and SpoIIP involved in engulfment (50), inactivation of σK and mislocalization of the SpoIVFA complex (48, 50), altered spore coat formation by CotE (51), altered regulation of σF and σG by SpoIIE (52), accumulation of dipicolinic acid in the forespore by SpoVV (53), and spore germination by GerM (49). It is yet to be determined if these defects are the result of direct interaction of these proteins with the core SpoIIIAH-SpoIIQ complex or rather indirect effects on downstream developmental processes.
OVERALL ARCHITECTURE AND HIERARCHY OF THE SpoIIIA-IIQ CHANNEL MODEL
To date, a full biochemical and structural elucidation of the sporulation secretion system has yet to be deduced, but significant efforts have been made to progressively assemble this puzzle piece by piece. Coimmunoprecipitation studies have demonstrated that SpoIIIAB, SpoIIIAD, SpoIIIAE, SpoIIIAF, and SpoIIIAG reside in a single complex in vivo (17). Microscopy experiments have indicated that SpoIIQ localization to the forespore membrane depends on the interaction with SpoIIIAH and GerM proteins through the thinning septal cell wall, an interaction that facilitates SpoIIIAG localization to the mother cell septal membrane (49). Taking these facts into consideration and by analogy to the architecture of other secretion systems that rely on hierarchical stacking of oligomerized rings for assembly, a composite sketch of the SpoIIIA-IIQ channel can be proposed (Fig. 1), as follows.
At the mother cell cytosol, there is an interaction between the hexameric SpoIIIAA ATPase and the possible docking platform of SpoIIIAB (oligomerization number unknown) (28).
A connecting link between the ATPase docking platform and the mother cell membrane export apparatus can be mediated by SpoIIIAE soluble and transmembrane domains.
A simplified export apparatus of only two components, SpoIIIAC and SpoIIIAD, is embedded at the mother cell membrane.
At the intermembrane space, a model of three stacked RBM-containing protein rings (SpoIIIAF, SpoIIIAG, and SpoIIIAH in complex with SpoIIQ) that are closely associated, with the length of the flexible polypeptide linkers connecting the TMH anchors to large C-terminal domains, is suggested. In this stacked ring model, SpoIIIAF is proposed to have a regulatory role via its observed alternate structural conformations, SpoIIIAG acts as the primary stable scaffold for the complex, and the SpoIIIAH-SpoIIQ ring acts as the mediator between the mother cell and forespore membranes (38, 42, 45, 46).
Additional accessory proteins could provide mechanical support and/or be involved in signal transduction, such as the soluble GerM proteins that have been shown to interact with SpoIIQ and SpoIIIAG in vivo (49).
Based on the highly electronegative charge of the modeled SpoIIIAH-SpoIIQ rings and the length of the disordered linker connecting the soluble domain to the forespore membrane, it is postulated that SpoIIQ is unlikely to serve as the forespore membrane translocon but rather an additional, yet to be identified, protein(s) is required to allow cargo transport.
For SpoIIIA-IIQ complex disassembly, cleavage of SpoIIQ by the SpoIVB forespore-produced protease serves as a signal for the end of the engulfment stage (54, 55).
HINTS FOR SpoIIIA-IIQ CHANNEL SUBSTRATE(S)
The striking similarity between the SpoIIIAG assembled 30-mer to the IM and OM components of the T2SS, T3SS, and T4PS (6, 40, 44) suggests that the SpoIIIA-IIQ complex may also support the transport of large macromolecules rather than simply small-molecule nutrients as previously supposed (18). One possibility is that the SpoIIIAG pore, like the T3SS basal body equivalent, provides a hyperstable shell or “locknut” for an additional inner channel that, in turn, modulates the final pore size for smaller substrates (in the case of the T3SS, the IM export apparatus channel described above and an extended hollow polymerized needle that serves as a direct conduit for partially unfolded virulence effector protein substrates). Alternatively, as in the case of the T4PS and T4SS, the substrates themselves may be large polymers (pilus or DNA). Given the likelihood of large-macromolecule passage suggested by the dimensions of the SpoIIIAG conduit, additional components would be required to maintain and regulate unidirectional transport of specific substrates from the mother cell to the forespore while at the same time preventing potentially deleterious uptake as seen in other well-characterized translocation machineries.
CONCLUDING REMARKS
The SpoIIIA-IIQ sporulation channel represents a hybrid, secretion-like machinery, involving homologous proteins from different bacterial secretion systems that support forespore physiology, including the capacity to carry out macromolecular synthesis. The remarkable homology to other secretion systems accompanied by the available component structures represents major pieces of the forespore development puzzle and sets a solid foundation for better understanding of the sporulation channel machinery. Notably, RBM-containing components of the channel have broadened our understanding the motif plasticity, which supports diverse functions in large assemblies with only subtle variations in sequence, extensions, or insertions. Still, many questions remain unanswered and can fuel future research, including the following questions. What is the transported substrate(s)? What are the additional components required, especially on the forespore side? Does the sporulation channel represent an ancient ancestor to all secretion systems, or has it evolved from multiple parts of different secretion systems?
ACKNOWLEDGMENTS
We thank Liam Worrall, Bronwyn Lyons, and Dorothy Majewski for fruitful discussions and technical support.
This work was supported by a Banting fellowship to N.Z. from the Canadian Institutes of Health Research (CIHR), operating grants from CIHR and the Howard Hughes International Senior Scholar program to N.C.J.S. N.C.J.S. is a Tier I Canada Research Chair in Antibiotic Discovery.
REFERENCES
- 1.Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. 2005. Mechanisms of protein export across the bacterial outer membrane. J Bacteriol 187:4306–4314. 10.1128/JB.187.13.4306-4314.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. 10.1038/nrmicro3456. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Korotkov KV, Sandkvist M, Hol WGJ. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10:336–351. 10.1038/nrmicro2762. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie PJ, Whitaker N, González-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. 10.1016/j.bbamcr.2013.12.019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkinshaw BJ, Strynadka NCJ. 2014. Assembly and structure of the T3SS. Biochim Biophys Acta 1843:1648–1693. [DOI] [PubMed] [Google Scholar]
- 6.Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JR, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NC. 2016. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540:597–601. 10.1038/nature20576. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. 2015. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523:555–560. 10.1038/nature14667. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Reichow SL, Korotkov KV, Hol WGJ, Gonen T. 2010. Structure of the cholera toxin secretion channel in its closed state. Nat Struct Mol Biol 17:1226–1232. 10.1038/nsmb.1910. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. 10.1038/nature13081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawshaw AD, Serrano M, Stanley WA, Henriques AO, Salgado PS. 2014. A mother cell-to-forespore channel: current understanding and future challenges. FEMS Microbiol Lett 358:129–136. 10.1111/1574-6968.12554. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Rosenbluh A, Rosenberg E. 1989. Sporulation of Myxococcus xanthus in liquid shake flask cultures. J Bacteriol 171:4521–4524. 10.1128/jb.171.8.4521-4524.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillot C, Moran CP, Jr. 2007. Essential internal promoter in the spoIIIA locus of Bacillus subtilis. J Bacteriol 189:7181–7189. 10.1128/JB.00915-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Illing N, Errington J. 1991. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol Microbiol 5:1927–1940. 10.1111/j.1365-2958.1991.tb00816.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Londoño-Vallejo J-A, Fréhel C, Stragier P. 1997. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol 24:29–39. 10.1046/j.1365-2958.1997.3181680.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Camp AH, Losick R. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69:402–417. 10.1111/j.1365-2958.2008.06289.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellner EM, Decatur A, Moran CP, Jr. 1996. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol 21:913–924. 10.1046/j.1365-2958.1996.461408.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP,Jr, Rudner DZ. 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5:e1000566. 10.1371/journal.pgen.1000566. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camp AH, Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23:1014–1024. 10.1101/gad.1781709. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morlot C, Rodrigues CDA. 2018. The new kid on the block: a specialized secretion system during bacterial sporulation. Trends Microbiol 26:663–676. 10.1016/j.tim.2018.01.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A 98:2503–2508. 10.1073/pnas.051436598. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu C, Turley S, Marionni ST, Park YJ, Lee KK, Patrick M, Shah R, Sandkvist M, Bush MF, Hol WG. 2013. Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure 21:1707–1717. 10.1016/j.str.2013.06.027. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancl JM, Black WP, Robinson H, Yang Z, Schubot FD. 2016. Crystal structure of a type IV pilus assembly ATPase: insights into the molecular mechanism of PilB from Thermus thermophilus. Structure 24:1886–1897. 10.1016/j.str.2016.08.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33(Suppl 2):W244–W248. 10.1093/nar/gki408. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A. 2007. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23:3403–3405. 10.1093/bioinformatics/btm477. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Py B, Loiseau L, Barras F. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep 2:244–248. 10.1093/embo-reports/kve042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abendroth J, Mitchell DD, Korotkov KV, Johnson TL, Kreger A, Sandkvist M, Hol WG. 2009. The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae. J Struct Biol 166:303–315. 10.1016/j.jsb.2009.03.009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karuppiah V, Hassan D, Saleem M, Derrick JP. 2010. Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins 78:2049–2057. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Zeytuni N, Flanagan KA, Worrall LJ, Massoni SC, Camp AH, Strynadka NCJ. 2018. Structural characterization of SpoIIIAB sporulation-essential protein in Bacillus subtilis. J Struct Biol 202:105–112. 10.1016/j.jsb.2017.12.009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, Yokoyama K, Iwata S. 2004. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci U S A 101:59–64. 10.1073/pnas.0305165101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhlen L, Abrusci P, Johnson S, Gault J, Deme J, Caesar J, Dietsche T, Mebrhatu MT, Ganief T, Macek B, Wagner S, Robinson CV, Lea SM. 2018. Structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 25:583–590. 10.1038/s41594-018-0086-9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano M, Vieira F, Moran CP, Jr, Henriques AO. 2008. Processing of a membrane protein required for cell-to-cell signaling during endospore formation in Bacillus subtilis. J Bacteriol 190:7786–7796. 10.1128/JB.00715-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr 4:215–239. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minamino T, Morimoto YV, Hara N, Namba K. 2011. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat Commun 2:475. 10.1038/ncomms1488. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minamino T. 2014. Protein export through the bacterial flagellar type III export pathway. Biochim Biophys Acta 1843:1642–1648. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. 10.1038/nsmb.2452. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B, Lara-Tejero M, Kong Q, Galán JE, Liu J. 2017. In situ molecular architecture of the Salmonella type III secretion machine. Cell 168:1065–1074.e10. 10.1016/j.cell.2017.02.022. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worrall LJ, Vuckovic M, Strynadka NCJ. 2010. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci 19:1091–1096. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeytuni N, Flanagan KA, Worrall LJ, Massoni SC, Camp AH, Strynadka NCJ. 2018. Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis. J Struct Biol 204:1–8. 10.1016/j.jsb.2018.06.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Bergeron JRC, Worrall LJ, Sgourakis NG, DiMaio F, Pfuetzner RA, Felise HB, Vuckovic M, Yu AC, Miller SI, Baker D, Strynadka NC. 2013. A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly. PLoS Pathog 9:e1003307. 10.1371/journal.ppat.1003307. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korotkov KV, Gonen T, Hol WGJ. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci 36:433–443. 10.1016/j.tibs.2011.04.002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigues CDA, Henry X, Neumann E, Kurauskas V, Bellard L, Fichou Y, Schanda P, Schoehn G, Rudner DZ, Morlot C. 2016. A ring-shaped conduit connects the mother cell and forespore during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 113:11585–11590. 10.1073/pnas.1609604113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeytuni N, Hong C, Flanagan KA, Worrall LJ, Theiltges KA, Vuckovic M, Huang RK, Massoni SC, Camp AH, Yu Z, Strynadka NC. 2017. Near-atomic resolution cryoelectron microscopy structure of the 30-fold homooligomeric SpoIIIAG channel essential to spore formation in Bacillus subtilis. Proc Natl Acad Sci U S A 114:E7073–E7081. 10.1073/pnas.1704310114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki H, Yonekura K, Namba K. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol 337:105–113. 10.1016/j.jmb.2004.01.034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Yan Z, Yin M, Xu D, Zhu Y, Li X. 2017. Structural insights into the secretin translocation channel in the type II secretion system. Nat Struct Mol Biol 24:177–183. 10.1038/nsmb.3350. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. 2012. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci U S A 109:5441–5445. 10.1073/pnas.1120087109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meisner J, Maehigashi T, André I, Dunham CM, Moran CP, Jr. 2012. Structure of the basal components of a bacterial transporter. Proc Natl Acad Sci U S A 109:5446–5451. 10.1073/pnas.1120113109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev 18:2916–2928. 10.1101/gad.1252704. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doan T, Marquis KA, Rudner DZ. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol 55:1767–1781. 10.1111/j.1365-2958.2005.04501.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Rodrigues CDA, Ramírez-Guadiana FH, Meeske AJ, Wang X, Rudner DZ. 2016. GerM is required to assemble the basal platform of the SpoIIIA-SpoIIQ transenvelope complex during sporulation in Bacillus subtilis. Mol Microbiol 102:260–273. 10.1111/mmi.13457. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. 2007. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol 65:1534–1546. 10.1111/j.1365-2958.2007.05887.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenney PT, Eichenberger P. 2012. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol 83:245–260. 10.1111/j.1365-2958.2011.07936.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flanagan KA, Comber JD, Mearls E, Fenton C, Wang Erickson AF, Camp AH. 2016. A membrane-embedded amino acid couples the SpoIIQ channel protein to anti-sigma factor transcriptional repression during Bacillus subtilis sporulation. J Bacteriol 198:1451–1463. 10.1128/JB.00958-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramírez-Guadiana FH, Meeske AJ, Rodrigues CDA, Barajas-Ornelas RDC, Kruse AC, Rudner DZ. 2017. A two-step transport pathway allows the mother cell to nurture the developing spore in Bacillus subtilis. PLoS Genet 13:e1007015. 10.1371/journal.pgen.1007015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiba S, Coleman K, Pogliano K. 2007. Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J Biol Chem 282:2576–2586. 10.1074/jbc.M606056200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang X, Rubio A, Chiba S, Pogliano K. 2005. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control σ activity. Mol Microbiol 58:102–115. 10.1111/j.1365-2958.2005.04811.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


