ABSTRACT
Members of the phylum Bacteroidetes have many unique features, including gliding motility and the type IX protein secretion system (T9SS). Bacteroidetes gliding and T9SSs are common in, but apparently confined to, this phylum. Most, but not all, members of the phylum secrete proteins using the T9SS, and most also exhibit gliding motility. T9SSs secrete cell surface components of the gliding motility machinery and also secrete many extracellular or cell surface enzymes, adhesins, and virulence factors. The components of the T9SS are novel and are unrelated to those of other bacterial secretion systems. Proteins secreted by the T9SS rely on the Sec system to cross the cytoplasmic membrane, and they use the T9SS for delivery across the outer membrane. Secreted proteins typically have conserved C-terminal domains that target them to the T9SS. Some of the T9SS components were initially identified as proteins required for gliding motility. Gliding does not involve flagella or pili and instead relies on the rapid movement of motility adhesins, such as SprB, along the cell surface by the gliding motor. Contact of the adhesins with the substratum provides the traction that results in cell movement. SprB and other motility adhesins are delivered to the cell surface by the T9SS. Gliding and the T9SS appear to be intertwined, and components of the T9SS that span the cytoplasmic membrane may energize both gliding and protein secretion. The functions of the individual proteins in each process are the subject of ongoing investigations.
INTRODUCTION
Members of the phylum Bacteroidetes have many unique features, including novel machinery for protein secretion and gliding motility (1–3). Most members secrete proteins across the outer membrane (OM) using the type IX protein secretion system (T9SS), which is confined to this phylum. Many also crawl rapidly over surfaces by gliding motility. For these gliding bacteria, the motility machinery and T9SS appear to be intertwined. Here we explore gliding motility, the T9SS, and the connections between them.
GLIDING MOTILITY
Bacteroidetes gliding motility was observed 100 years ago (4), but the mechanism of movement was unknown until recently. These bacteria crawl rapidly over surfaces without the aid of flagella or pili. Instead, motility involves the rapid movement of cell surface adhesins (5, 6). Gliding also occurs in bacteria that belong to other phyla (myxobacteria, mycoplasmas, cyanobacteria, and others), but these have their own unique motility machineries (1, 7–10). Gliding of Flavobacterium johnsoniae is typical of members of the Bacteroidetes (Fig. 1A). The long slender cells (0.4 μm by 5 to 10 μm) move over surfaces at speeds of approximately 2 μm/s. Cells glide following their long axes and may reverse direction, with the head becoming the tail. Cells on wet surfaces often flip or pivot and may also attach by one pole and rotate at frequencies of about 2 revolutions per s (11, 12). The proton motive force (PMF) powers movement, and uncouplers that dissipate it block gliding (5, 12). Movement of cells on agar often results in thin spreading colonies (Fig. 1B).
FIGURE 1.
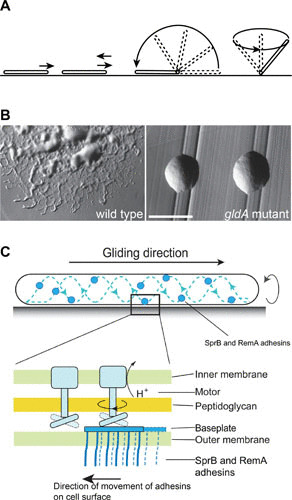
Gliding of F. johnsoniae cells. (A) Characteristic movements of cells. (B) Spreading colonies formed by wild-type cells and nonspreading colonies formed by cells of a nonmotile gldA mutant. Bar corresponds to 1 mm. (C) Model of F. johnsoniae gliding. Gld proteins in the cell envelope form the PMF-powered rotary motors that are attached to the cell wall and propel adhesins, such as SprB and RemA, along looped helical tracks on the cell surface. The action of the motors on adhesins that are attached to the substratum results in forward movement and rotation of the cell. Two rotary motors are shown. Rotation of one motor propels a baseplate carrying SprB and RemA adhesins and delivers it to the next motor. Modified from reference 13.
Genetic analyses revealed F. johnsoniae Gld proteins that are essential for gliding and Spr proteins whose absence results in severe but incomplete motility defects (13). Most of the Gld and Spr proteins are novel, complicating prediction of function. The 669-kDa, repetitive cell surface protein SprB was the first F. johnsoniae motility protein to be assigned a function. SprB is an adhesin that is required for cell movement over agar, and sprB mutants thus form nonspreading colonies (6). SprB filaments extend from the cell surface. Antibodies against SprB were used to label live cells, revealing that SprB is propelled rapidly from pole to pole following an apparent closed helical loop (5, 6). Cells lacking SprB retain the ability to move on some surfaces other than agar because of the presence of additional motility adhesins, such as RemA (14). The multiple motility adhesins allow cells to move over diverse surfaces. PMF-dependent rotary motors embedded in the cell envelope are thought to propel the motility adhesins along a helical track (13, 15–17). When the adhesins attach to the substratum, the motors pushing against them result in rotation of and forward movement of the cell in a screw-like manner (Fig. 1C). The cytoplasmic membrane (CM) proteins GldL and GldM (Fig. 2B) are candidates for the motor proteins that harvest the PMF (5, 16, 18, 19). The only other known motility proteins that span the CM are components of the GldA-GldF-GldG ATP-binding cassette (ABC) transporter. This is unlikely to be the gliding motor because the ABC transporter is presumably powered by ATP rather than PMF and because several members of the Bacteroidetes that lack this transporter exhibit gliding motility (1, 20).
FIGURE 2.
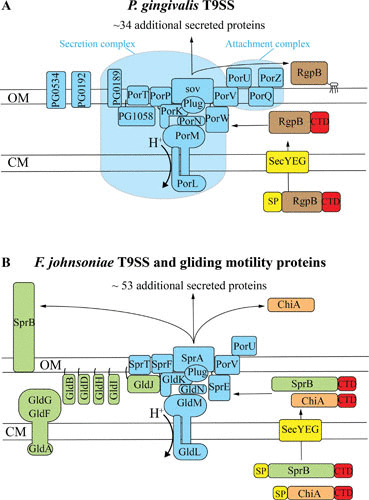
T9SS and gliding motility proteins. Proteins in blue are associated with the T9SS, and proteins in green are motility proteins that are not directly associated with the T9SS. Orthologous T9SS proteins between panels A and B are shown in the same relative positions, color, and shapes. F. johnsoniae GldK, GldL, GldM, GldN, SprA, SprE, and SprT correspond to P. gingivalis PorK, PorL, PorM, PorN, Sov, PorW, and PorT, respectively. Black lines are lipid tails on lipoproteins. Proteins secreted by the T9SS have predicted N-terminal signal peptides (yellow) that target them to the Sec system for export across the cytoplasmic membrane (CM) and C-terminal domains (red) that target them to the T9SS for secretion across the outer membrane (OM). Proteins are not drawn to scale, and stoichiometry of components is not illustrated. (A) P. gingivalis T9SS proteins. Where protein names were not available, locus tags (from P. gingivalis strain W83) were used. The gingipain protease RgpB is shown covalently attached to the outer membrane acidic lipopolysaccharide (A-LPS). Secretion complex and attachment complex are indicated by the large and small blue barrels, respectively. (B) F. johnsoniae T9SS and gliding motility proteins. SprB is a motility adhesin that is propelled by some of the other proteins shown. SprF is required for secretion of SprB but not for secretion of other proteins. SprF and nine other F. johnsoniae proteins are related to P. gingivalis PorP. F. johnsoniae PorV is required for secretion of ChiA and many other proteins, but not for secretion of SprB.
Analyses of many nonmotile gld mutants revealed surprising phenotypes. The mutants failed to digest chitin or to adhere to surfaces and they were resistant to bacteriophages (18, 19, 21). The reason for these phenotypes is now known. gldK, gldL, gldM, and gldN encode core components of the T9SS that secretes SprB and RemA to the cell surface (19, 22, 23). The unexpected phenotypes of these gld mutants were caused by failure to secrete adhesins, phage receptors, a chitinase, and dozens of other proteins.
DISCOVERY OF THE T9SS
The first hint that the Bacteroidetes might have a novel protein secretion system came from studies of the nonmotile human pathogen Porphyromonas gingivalis. P. gingivalis secretes virulence factors, such as gingipain proteases, that are important in periodontal disease (24, 25). The secreted proteins typically remain attached to the P. gingivalis cell surface. A conserved carboxy-terminal domain (CTD) of secreted proteins was suggested to be linked to secretion and surface attachment (26–28). In 2005, genetic experiments revealed the first component of the secretion system, the OM protein PorT (29). Two years later, studies on F. johnsoniae motility and P. gingivalis secretion converged with the publication of papers on the F. johnsoniae OM motility protein SprA (30) and the orthologous P. gingivalis protein involved in secretion of virulence factors, Sov (31). Another OM protein involved in secretion, PorV, was recognized soon after (32). Many of the remaining T9SS proteins were discovered by comparative genome analyses that revealed genes that co-occur with the secretion gene porT (23). Targeted mutagenesis of these identified 10 P. gingivalis genes that were required for optimal secretion. Among these were orthologs of six F. johnsoniae gliding motility genes (gldK, gldL, gldM, gldN, sprA, and sprE, named porK, porL, porM, porN, sov, and porW in P. gingivalis) and porP, porQ, and porU. Mutations in any of these P. gingivalis genes resulted in defects ranging from partial to complete loss of secretion. Analysis of F. johnsoniae strains with mutations in the corresponding genes revealed similar secretion defects (19, 22, 23), except that porQ and porU mutants were largely competent for secretion (33, 34). The secretion system was initially referred to as the Por secretion system and was later named the T9SS (1, 35). T9SSs are found in most Bacteroidetes but are absent in most members of the genus Bacteroides (1, 36), which instead rely on other secretion systems (37). Some bacteria that have T9SSs also use other secretion systems (1, 38).
SECRETED PROTEINS
Proteins secreted by T9SSs have cleavable N-terminal signal peptides that are thought to target them to the Sec system for export across the CM. They also typically have conserved 70- to 100-amino-acid CTDs that are required for secretion across the OM and are often cleaved during this process (26, 27, 29, 34, 39–43). Attachment of a T9SS CTD to a foreign protein results in its efficient secretion (34, 40, 41). The structures of two CTDs have been determined (44, 45). Both have an Ig-like fold with seven β-strands forming a sandwich-like structure that is thought to interact with components of the T9SS. The conserved CTD sequences allow predictions of secreted proteins from genomic data. Most T9SSs are predicted to secrete dozens of proteins, and some secrete many more. The cellulolytic bacterium Cytophaga hutchinsonii is predicted to use its T9SS to secrete at least 147 proteins (46), many of which have been verified by proteomic analyses (43), and Fluviicola taffensis is predicted to secrete 230 proteins (36). Given the large number of proteins secreted, it is not surprising that T9SSs are important for many processes in environmental and host-associated bacteria, including virulence (23, 47, 48), polymer degradation (40, 46), adhesion (19, 22), S-layer formation (49, 50), motility (19, 22, 46, 47, 51), and biofilm formation (49–51).
Many T9SS-secreted proteins are large. F. johnsoniae SprB, for example, is 669 kDa. Some proteins are secreted in soluble form, and others become attached to the cell surface. Most proteins secreted by P. gingivalis are of the latter type and are cell surface associated as a result of covalent bonding to an acidic form of lipopolysaccharide (A-LPS) (52). This modification occurs after transit of the OM and removal of the CTD (53).
COMPONENTS AND STRUCTURE OF THE T9SS
At least 17 proteins are thought to have roles in T9SS-mediated secretion (Fig. 2; Table 1). Several regulatory proteins, not discussed here, have also been identified (23, 54, 55). P. gingivalis T9SS proteins that are essential or nearly essential for secretion include PorK, PorL, PorM, PorN, Sov, PorW, PorT, PorP, PorV, and PG1058 (23, 31, 56–58). The F. johnsoniae orthologs of the first 7 of these (GldK, GldL, GldM, GldN, SprA, SprE, and SprT) are also required for secretion (19, 22, 23), but the situation is more complicated for PorP, PorV, and PG1058. P. gingivalis has a single PorP, whereas F. johnsoniae has 10 porP-like genes (59). Requirement of these genes for F. johnsoniae secretion has likely been masked by redundancy. One F. johnsoniae PorP-like protein, SprF, is required for T9SS-mediated secretion of SprB, but it is not required for secretion of other proteins examined (59). Redundancy may also occur for the five F. johnsoniae proteins that are related to PG1058. The genes encoding these PG1058-like proteins are each adjacent to sprF-like genes. These have not yet been examined for roles in secretion. P. gingivalis PorV is required for secretion (32), whereas F. johnsoniae PorV is required only for secretion of some proteins that are targeted to its T9SS (34). PG0189 interacts with PorK and PorN and thus may be involved in secretion (60), but mutants lacking PG0189 have apparently not been examined.
TABLE 1.
T9SS componentsa
| P. gingivalis protein name or locus tagb | F. johnsoniae protein name or locus tag | Localization | Predicted role in secretion/motility | Essential for F. johnsoniae gliding? | Interaction partner | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
| Secretion complex | |||||||
| PorL | GldL | CM | Energizes secretion/motility | Essential | PorM | Forms complex with PorK, PorM, and PorN | 18, 19, 23, 61 |
| PorM | GldM | CM | Energizes secretion/motility | Essential | PorK, PorL, PorN, PorP | Forms complex with PorK, PorL, and PorN | 18, 19, 23, 61, 62 |
| PorK | GldK | OM lipoprotein | Component of periplasmic channel | Essential | PorM, PorN, PorP, PG0189 | Forms complex with PorL, PorM, and PorN | 18, 19, 23, 60, 61 |
| PorN | GldN and GldO | P | Component of periplasmic channel | Essential | PorK, PorM, PG0189 | Forms complex with PorK, PorL, and PorM | 18, 19, 22, 23, 60, 61 |
| Sov | SprA | OM | OM pore | Nearly essential | PorV, plug | 19, 30, 31, 58, 64 | |
| PorT | SprT | OM | Unknown | Nearly essential | ND | 23, 29, 71 | |
| PorP | SprF and 9 other PorP-like proteinsc | OM | Unknown | ND | PorK, PorM | 23, 46, 59, 61 | |
| PorW | SprE | OM lipoprotein | Unknown | Nearly essential | ND | 23, 72 | |
| PG1058 (PGN_1296) | Fjoh_1647,d Fjoh_2275, Fjoh_3950, Fjoh_3973, Fjoh_4540 | OM lipoprotein | Anchors T9SS to peptidoglycan | ND | ND | OmpA C-terminal peptidoglycan-binding domain | 57 |
| PG0189 (PGN_0297) | Fjoh_1692 | OM | Unknown | ND | PorK, PorN | 60 | |
| PG2092 (PGN_0144) | Fjoh_1759, plug | P/OM | Forms plug on periplasmic side of SprA pore. Prevents nonspecific leakage of periplasmic contents. | ND | SprA | 64 | |
| Attachment complex | |||||||
| PorVe | PorV | OM | Shuttles secreted proteins from secretion complex to attachment complex | Not needed for gliding | PorU, SprA | Forms complex with PorU, PorQ, and PorZ | 32, 34, 56, 64, 73, 74 |
| PorU | PorU | Cell surface | CTD cleavage; attachment of substrates to A-LPS | Not needed for gliding | PorV | Forms complex with PorV, PorQ, and PorZ | 39, 56, 73 |
| PorQ | Fjoh_2755 | OM | Unknown | Not needed for gliding | Forms complex with PorU, PorV, and PorZ | 23, 56 | |
| PorZ | Fjoh_0707 | Cell surface | Assembly of PorU on cell surface; secretion and/or modification of secreted proteins | ND | Forms complex with PorU, PorV, and PorQ | 45, 56 | |
| Other | |||||||
| PG0534 (PGN_1437) | Fjoh_0118 | OM | Secretion and/or modification of secreted proteins | ND | 65 | ||
| Omp17; PG0192 (PGN_0300) | Fjoh_0599,f Fjoh_1000, Fjoh_1688, Fjoh_1689 | OM | PorU maturation or stabilization; post-secretion processing | ND | OmpH-like | 66 | |
Abbreviations: CM, cytoplasmic membrane; OM, outer membrane; P, periplasm; ND, not determined.
Locus tags for P. gingivalis strains W83 (PG) and ATCC33277 (PGN_) are listed, with the PGN_ locus tags in parentheses.
PorP-like proteins recognized by assignment to Tigrfam “type IX secretion system membrane protein PorP/SprF family” TIGR03519.
PG1058-like proteins recognized by the presence of domains corresponding to pfam00691 (OmpA_C terminal domain), pfam07676 (WD40-like Beta propeller repeat), and pfam13620 (carboxypeptidase regulatory-like domain).
PorV may be a component of the secretion complex and the attachment complex.
Proteins related to PG0192 (PGN_0300) recognized by the presence of domains corresponding to pfam03938 (OmpH-like).
The P. gingivalis proteins described above all localize to the cell envelope and may constitute the core of the secretion complex (Fig. 2A). Only PorL and PorM span the CM, and they may be involved in energy transduction to power secretion, similar to the proposed role of GldL and GldM in gliding motility (5, 18, 19, 61). PorL/GldL proteins have a completely conserved glutamate predicted to be buried in the membrane that might facilitate transit of protons to energize both processes (19, 61).
Recent studies of protein-protein interactions, protein complexes, and protein structures have added greatly to our understanding of the T9SS secretion complex (60–63). PorL/PorM and PorK/PorN complexes were isolated, and additional protein-protein interactions were identified (Table 1), suggesting a potential envelope-spanning complex comprised of PorK, PorL, PorM, PorN, PorP, and PG0189. Stoichiometry was suggested to be PorL3/PorM2/PorN2/PorK2, with perhaps 3 or 4 copies of this basic structure forming the core of the secretion machine. Crystal structures of dimers of the periplasmic domains of PorM and GldM were recently reported (62). Despite only 22% amino acid identity, the proteins formed remarkably similar four-domain structures predicted to span most of the periplasm. In GldM the domains were linear, whereas in PorM there was a bend between domains 2 and 3. The PorM and GldM structures may represent different dynamic states associated with protein translocation. Transition between these states could involve PorL/GldL-mediated PMF-driven energy transduction. The very large (267-kDa) SprA protein appears to form the OM pore of the secretion system (64). Cryo-electron microscopic analysis revealed that a single SprA protein forms a 36-strand transmembrane β-barrel with an internal pore of approximately 70 Å in diameter, which should allow transit of folded proteins. The SprA pore appears to be alternately occluded on the periplasmic side by the Plug protein (Fjoh_1759) and on the external side by PorV. This may prevent nonspecific leakage of periplasmic contents. PorV is thought to escort proteins from the lumen of the SprA channel to the outside of the cell.
P. gingivalis PorU, PorQ, and PorZ appear to form a complex with PorV that modifies secreted proteins and attaches them to the cell surface (56). PorU is the peptidase that removes CTDs after secretion (39). It may also covalently attach the newly exposed C termini of the secreted proteins to A-LPS via a “sortase-like” mechanism (53). Cells of porU mutants secrete proteins, but these retain their CTDs and fail to attach to the cell surface. PorZ is required for proper localization and stability of PorU, and thus, porZ mutants behave similarly to porU mutants (45). PorV is thought to function as the shuttle that delivers proteins from SprA and the secretion complex to the attachment complex (56). Cells lacking PG0534 and PGN_0300 (PG0192) have phenotypes that may indicate that they also function in modification of secreted proteins (65, 66).
RELATIONSHIP BETWEEN THE T9SS AND GLIDING MOTILITY
The F. johnsoniae T9SS and gliding motility machines appear to be intertwined, since many mutations disrupt gliding and secretion (67). This is reminiscent of the bacterial flagellum, which has a type III secretion system involved in flagellar assembly at its core (68, 69). GldL and GldM have been suggested to be part of the PMF-driven rotary gliding motor, and they are also thought to energize secretion (5, 18, 19, 61). Other core components of the T9SS (GldK, GldN, SprA, SprE, and SprT) are also essential for gliding, suggesting the possibility that a transmembrane complex of these T9SS proteins may be central to gliding and secretion. Loss of some other motility proteins, GldA, GldB, GldD, GldF, GldG, GldH, GldI, and GldJ (green in Fig. 2B), also results in defects in motility and secretion (67). The secretion defects were unexpected, since these proteins are not associated with the P. gingivalis T9SS. The reason for the loss of secretion became clear when it was discovered that the F. johnsoniae motility protein GldJ is required to stabilize the T9SS protein GldK (67). It was already known that GldA, GldB, GldD, GldF, GldG, GldH, and GldI are needed to stabilize GldJ (70). Apparently, the absence of any of these proteins results in loss of GldJ, which results in loss of GldK and thus in defects in protein secretion. Truncated forms of GldJ were identified that are nonfunctional for motility but that stabilize GldK and thus allow secretion (67). This partially untangles gliding from secretion, but if current suggestions that GldL and GldM are motor proteins for secretion and motility are correct, it may be difficult to completely separate the two processes.
CONCLUSIONS
The novel machines described above, involved in protein secretion and cell movement, have remarkable properties. The T9SS efficiently secretes huge proteins across the OM, and the gliding motility machinery rapidly propels some of these along the cell surface. Rapid progress has been made in our understanding of gliding and secretion, but many mysteries remain regarding the functioning of the machines responsible for these processes.
ACKNOWLEDGMENT
This work was supported by grant MCB-1516990 from the National Science Foundation.
REFERENCES
- 1.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. 10.1128/JB.01962-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. 10.3389/fcimb.2017.00215. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veith PD, Glew MD, Gorasia DG, Reynolds EC. 2017. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106:35–53. 10.1111/mmi.13752. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Hutchinson HB, Clayton J. 1919. On the decomposition of cellulose by an aerobic organism (Spirochaeta cytophaga, n. sp.). J Agric Sci 9:143–173. 10.1017/S0021859600004755. [DOI] [Google Scholar]
- 5.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150. 10.1073/pnas.1219753110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson SS, Bollampalli S, McBride MJ. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857. 10.1128/JB.01904-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. 10.1038/nrmicro1900. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding.Annu Rev Microbiol 64:519–537. 10.1146/annurev.micro.112408.134116. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Nan B, McBride MJ, Chen J, Zusman DR, Oster G. 2014. Bacteria that glide with helical tracks. Curr Biol 24:R169–R173. 10.1016/j.cub.2013.12.034. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YW, Gonzales A, Harwood TV, Huynh J, Hwang Y, Park JS, Trieu AQ, Italia P, Pallipuram VK, Risser DD. 2017. Dynamic localization of HmpF regulates type IV pilus activity and directional motility in the filamentous cyanobacterium Nostoc punctiforme. Mol Microbiol 106:252–265. 10.1111/mmi.13761. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Lapidus IR, Berg HC. 1982. Gliding motility of Cytophaga sp. strain U67. J Bacteriol 151:384–398. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pate JL, Chang L-YE. 1979. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr Microbiol 2:59–64. 10.1007/BF02601737. [DOI] [Google Scholar]
- 13.McBride MJ, Nakane D. 2015. Flavobacterium gliding motility and the type IX secretion system. Curr Opin Microbiol 28:72–77. 10.1016/j.mib.2015.07.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. 2012. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J Bacteriol 194:3678–3688. 10.1128/JB.00588-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrivastava A, Berg HC. 2015. Towards a model for Flavobacterium gliding. Curr Opin Microbiol 28:93–97. 10.1016/j.mib.2015.07.018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrivastava A, Lele PP, Berg HC. 2015. A rotary motor drives Flavobacterium gliding. Curr Biol 25:338–341. 10.1016/j.cub.2014.11.045. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrivastava A, Roland T, Berg HC. 2016. The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys J 111:1008–1013. 10.1016/j.bpj.2016.07.043. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. 10.1128/JB.187.20.6943-6952.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. 10.1128/JB.00333-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, McBride MJ. 2016. Comparative analysis of Cellulophaga algicola and Flavobacterium johnsoniae gliding motility. J Bacteriol 198:1743–1754. 10.1128/JB.01020-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang LE, Pate JL, Betzig RJ. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol 159:26–35. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. 10.1128/JB.01495-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. 10.1073/pnas.0912010107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathirana RD, O’Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. 2007. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun 75:1436–1442. 10.1128/IAI.01627-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien-Simpson NM, Paolini RA, Hoffmann B, Slakeski N, Dashper SG, Reynolds EC. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect Immun 69:7527–7534. 10.1128/IAI.69.12.7527-7534.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol 188:6376–6386. 10.1128/JB.00731-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen KA, Travis J, Potempa J. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J Bacteriol 189:833–843. 10.1128/JB.01530-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veith PD, Talbo GH, Slakeski N, Dashper SG, Moore C, Paolini RA, Reynolds EC. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem J 363:105–115. 10.1042/bj3630105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, Nakayama K. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280:8668–8677. 10.1074/jbc.M413544200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Nelson SS, Glocka PP, Agarwal S, Grimm DP, McBride MJ. 2007. Flavobacterium johnsoniae SprA is a cell surface protein involved in gliding motility. J Bacteriol 189:7145–7150. 10.1128/JB.00892-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saiki K, Konishi K. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol Immunol 51:483–491. 10.1111/j.1348-0421.2007.tb03936.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Ishiguro I, Saiki K, Konishi K. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol Lett 292:261–267. 10.1111/j.1574-6968.2009.01489.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Shrivastava A. 2013. Cell surface adhesins, exopolysaccharides and the Por (type IX) secretion system of Flavobacterium johnsoniae. PhD dissertation. University of Wisconsin, Milwaukee, Milwaukee, WI. [Google Scholar]
- 34.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. 10.1128/JB.02085-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. 10.1111/1574-6968.12028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. 2017. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J Bacteriol 199:e00884-16. 10.1128/JB.00884-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson MM, Anderson DE, Bernstein HD. 2015. Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS One 10:e0117732. 10.1371/journal.pone.0117732. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16:227–236. 10.1016/j.chom.2014.07.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. 10.1074/jbc.M112.369223. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kharade SS, McBride MJ. 2014. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. 10.1128/JB.01170-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. 10.1371/journal.pone.0021372. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slakeski N, Seers CA, Ng K, Moore C, Cleal SM, Veith PD, Lo AW, Reynolds EC. 2011. C-terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J Bacteriol 193:132–142. 10.1128/JB.00773-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veith PD, Nor Muhammad NA, Dashper SG, Likić VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res 12:4449–4461. 10.1021/pr400487b. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, Szmigielski B, Nowak M, Nowakowska Z, Potempa B, Houston JA, Enghild JJ, Thøgersen IB, Gao J, Kwan AH, Trewhella J, Dubin G, Gomis-Rüth FX, Nguyen KA, Potempa J. 2016. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal β-sandwich domain. Sci Rep 6:23123. 10.1038/srep23123. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lasica AM, Goulas T, Mizgalska D, Zhou X, de Diego I, Ksiazek M, Madej M, Guo Y, Guevara T, Nowak M, Potempa B, Goel A, Sztukowska M, Prabhakar AT, Bzowska M, Widziolek M, Thøgersen IB, Enghild JJ, Simonian M, Kulczyk AW, Nguyen KA, Potempa J, Gomis-Rüth FX. 2016. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci Rep 6:37708. 10.1038/srep37708. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol 98:763–775. 10.1007/s00253-013-5355-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Li N, Zhu Y, LaFrentz BR, Evenhuis JP, Hunnicutt DW, Conrad RA, Barbier P, Gullstrand CW, Roets JE, Powers JL, Kulkarni SS, Erbes DH, García JC, Nie P, McBride MJ. 2017. The type IX secretion system is required for virulence of the fish pathogen Flavobacterium columnare. Appl Environ Microbiol 83:e01769-17. 10.1128/AEM.01769-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Y, Hu D, Guo J, Wang T, Xiao Y, Wang X, Li S, Liu M, Li Z, Bi D, Zhou Z. 2017. Riemerella anatipestifer type IX secretion system is required for virulence and gelatinase secretion. Front Microbiol 8:2553. 10.3389/fmicb.2017.02553. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narita Y, Sato K, Yukitake H, Shoji M, Nakane D, Nagano K, Yoshimura F, Naito M, Nakayama K. 2014. Lack of a surface layer in Tannerella forsythia mutants deficient in the type IX secretion system. Microbiology 160:2295–2303. 10.1099/mic.0.080192-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomek MB, Neumann L, Nimeth I, Koerdt A, Andesner P, Messner P, Mach L, Potempa JS, Schäffer C. 2014. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol Oral Microbiol 29:307–320. 10.1111/omi.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kita D, Shibata S, Kikuchi Y, Kokubu E, Nakayama K, Saito A, Ishihara K. 2016. Involvement of the type IX secretion system in Capnocytophaga ochracea gliding motility and biofilm formation. Appl Environ Microbiol 82:1756–1766. 10.1128/AEM.03452-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol 58:847–863. 10.1111/j.1365-2958.2005.04871.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. 10.1371/journal.ppat.1005152. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadowaki T, Yukitake H, Naito M, Sato K, Kikuchi Y, Kondo Y, Shoji M, Nakayama K. 2016. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci Rep 6:23288. 10.1038/srep23288. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincent MS, Durand E, Cascales E. 2016. The PorX response regulator of the Porphyromonas gingivalis PorXY two-component system does not directly regulate the type IX secretion genes but binds the PorL subunit. Front Cell Infect Microbiol 6:96. 10.3389/fcimb.2016.00096. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glew MD, Veith PD, Chen D, Gorasia DG, Peng B, Reynolds EC. 2017. PorV is an outer membrane shuttle protein for the type IX secretion system. Sci Rep 7:8790. 10.1038/s41598-017-09412-w. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heath JE, Seers CA, Veith PD, Butler CA, Nor Muhammad NA, Chen YY, Slakeski N, Peng B, Zhang L, Dashper SG, Cross KJ, Cleal SM, Moore C, Reynolds EC. 2016. PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One 11:e0164313. 10.1371/journal.pone.0164313. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saiki K, Konishi K. 2010. The role of Sov protein in the secretion of gingipain protease virulence factors of Porphyromonas gingivalis. FEMS Microbiol Lett 302:166–174. 10.1111/j.1574-6968.2009.01848.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol 193:599–610. 10.1128/JB.01203-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gorasia DG, Veith PD, Hanssen EG, Glew MD, Sato K, Yukitake H, Nakayama K, Reynolds EC. 2016. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog 12:e1005820. 10.1371/journal.ppat.1005820. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent MS, Canestrari MJ, Leone P, Stathopulos J, Ize B, Zoued A, Cambillau C, Kellenberger C, Roussel A, Cascales E. 2017. Characterization of the Porphyromonas gingivalis type IX secretion trans-envelope PorKLMNP core complex. J Biol Chem 292:3252–3261. 10.1074/jbc.M116.765081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leone P, Roche J, Vincent MS, Tran QH, Desmyter A, Cascales E, Kellenberger C, Cambillau C, Roussel A. 2018. Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat Commun 9:429. 10.1038/s41467-017-02784-7. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glew MD, Veith PD, Chen D, Seers CA, Chen YY, Reynolds EC. 2014. Blue native-PAGE analysis of membrane protein complexes in Porphyromonas gingivalis. J Proteomics 110:72–92. 10.1016/j.jprot.2014.07.033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. 10.1038/s41586-018-0693-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saiki K, Konishi K. 2010. Identification of a novel Porphyromonas gingivalis outer membrane protein, PG534, required for the production of active gingipains. FEMS Microbiol Lett 310:168–174. 10.1111/j.1574-6968.2010.02059.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Taguchi Y, Sato K, Yukitake H, Inoue T, Nakayama M, Naito M, Kondo Y, Kano K, Hoshino T, Nakayama K, Takashiba S, Ohara N. 2015. Involvement of an Skp-like protein, PGN_0300, in the type IX secretion system of Porphyromonas gingivalis. Infect Immun 84:230–240. 10.1128/IAI.01308-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnston JJ, Shrivastava A, McBride MJ. 2017. Untangling Flavobacterium johnsoniae gliding motility and protein secretion. J Bacteriol 200:e00362-17. 10.1128/JB.00362-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488. 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 69.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. 2008. Energy source of flagellar type III secretion. Nature 451:489–492. 10.1038/nature06497. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Braun TF, McBride MJ. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J Bacteriol 187:2628–2637. 10.1128/JB.187.8.2628-2637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen KA, Zylicz J, Szczesny P, Sroka A, Hunter N, Potempa J. 2009. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155:328–337. 10.1099/mic.0.024323-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. 2011. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol 193:5322–5327. 10.1128/JB.05480-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saiki K, Konishi K. 2014. Porphyromonas gingivalis C-terminal signal peptidase PG0026 and HagA interact with outer membrane protein PG27/LptO. Mol Oral Microbiol 29:32–44. 10.1111/omi.12043. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O’Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol Microbiol 79:1380–1401. 10.1111/j.1365-2958.2010.07530.x. [PubMed] [DOI] [PubMed] [Google Scholar]


