Abstract
Nab proteins constitute an evolutionarily conserved family of corepressors that specifically interact with and repress transcription mediated by three members of the NGFI-A (Egr-1, Krox24, zif/268) family of immediate-early gene transcription factors, which includes NGFI-C, Krox20, and Egr3. We explored the mechanism of Nab1 repression and identified structural domains required for Nab1 function. Nab1 does not act by blocking DNA binding or nuclear localization of NGFI-A. In fact, Nab1 repression is not unique to NGFI-A because multiple types of non-NGFI-A activation domains were repressed, as was a heterologous transcription factor carrying the NGFI-A R1 domain, which is required for Nab1 interaction. Additionally, Nab1 tethered directly to DNA repressed constitutively active promoters. Tethered repression was not dependent on the identity of the basal promoter elements, the presence of a distal enhancer, or the distance separating the binding sites from the promoter. These results suggest that Nab1 repression is not specific to particular activators and that Nab1 is an active repressor that works by a direct mechanism. We identified a bipartite-like nuclear localization sequence and localized the repression function to the Nab conserved domain 2 (NCD2), a region found in the carboxy-terminal half of all Nab proteins. Three small regions of homology between Nab1 and previously characterized corepressors, Dr1 and E1b 55-kDa protein, were identified within NCD2. Replacement mutagenesis of residues conserved between these proteins interfered with Nab1 repression, although Nab1 does not function by the same mechanism as Dr1. The human NAB1 genomic locus was mapped to chromosome 2q32.3-33.
Repression of gene transcription is critical to the function of cells and to the health and normal development of organisms. For example, patterns of gene expression underlying Drosophila melanogaster development are regulated by a diverse array of spatially localized transcriptional repressors (e.g., Krüppel [23]). In addition, the mitotic cell cycle is controlled by the interplay of positive and negative regulatory proteins, which regulate the transcription of genes required for S phase. E2F transcription factors promote progression of the cell cycle by stimulating transcription. Mutations that prevent repression of E2F, and the appropriate negative regulation of the cell cycle by the retinoblastoma protein (Rb), have been implicated in the process of human tumorigenesis and cause defects in mouse development (10, 26, 31, 41, 76).
Repressors function by negatively regulating transcriptional activators, the general transcription apparatus, or chromatin structure through a variety of mechanisms categorized as passive versus active (for reviews, see references 11, 25, 28, 32, and 37). Passive repressors interfere with transcription factor functions, such as DNA binding or nuclear localization, that are not directly involved in the mechanism of activation. Active repressors are characterized as working by direct or quenching mechanisms, which both require protein-protein interactions. Direct repressors interfere with the function of the general transcription apparatus (GTA) but not that of specific activating transcription factors. In contrast, quenching repressors interfere with the ability of specific activators to stimulate transcription.
Recently, a novel family of corepressors named Nab that negatively regulate the transcriptional activity of the NGFI-A family of zinc finger transcription factors was identified. NGFI-A (also called Egr-1, Krox24, and zif/268) was discovered as an immediate-early gene induced after nerve growth factor treatment of PC12 cells, which results in neuronal differentiation, and serum stimulation of fibroblasts, which results in mitosis (9, 43, 49, 66). Three structurally related proteins, Krox20, NGFI-C, and Egr3, were discovered later and also characterized as immediate-early gene transcription factors which recognize the NGFI-A DNA binding site (12, 43, 55, 70). Additional studies, including knockout analyses in mice, suggest that NGFI-A family proteins play important roles in the genetic regulation of cell growth, differentiation, and apoptosis in response to extracellular stimuli (for a review, see reference 20). For example, lack of NGFI-A resulted in female infertility due to absence of luteinizing hormone expression by pituitary cells (42), and lack of Krox20 resulted in defects in hindbrain development and differentiation of Schwann cells and osteoblasts (45, 64, 69, 71).
Mutagenesis of NGFI-A identified an inhibitory domain, named R1, which when deleted resulted in enhanced NGFI-A transcriptional activity (21, 61). The R1 domain was used as bait in a yeast two-hybrid screen to isolate a novel protein called Nab1, which could interact with NGFI-A in vitro and repressed NGFI-A-mediated transcription in cells (62). Krox20 and Egr3, but not NGFI-C, possess domains homologous to R1 and were also repressed by Nab1 (62, 64a). Subsequent work identified a mammalian protein, Nab2, that shares two large regions of homology with Nab1 and can also repress NGFI-A (38, 68). Related genes have also been identified in Caenorhabditis elegans and D. melanogaster, suggesting that Nab proteins are evolutionarily conserved corepressors, common to all multicellular organisms.
In this study, we report the results of experiments designed to explore the mechanism by which Nab1 represses NGFI-A-mediated transcription. We determined that Nab1 is an active repressor that appears to work via a direct mechanism, because repression is not specific to particular transcription factors or promoters. In addition, mutagenesis of Nab1 identified domains required for repression and nuclear localization in NCD2, the carboxy-terminal region conserved in all Nab proteins.
MATERIALS AND METHODS
Plasmids and DNA manipulation.
Standard molecular cloning reagents and techniques were used for generating all protein expression and luciferase reporter vectors. Sequencing was performed with an ABI model 373 automated sequencer. The following luciferase reporter vectors were described previously: 5 × Gal4 E1b TATA (61), 2 × A Prol Min Prom (12), and 8 × B Prol Min Prom (79). Adenovirus major late (AdML) promoter-luciferase and DNA β-polymerase (β-Pol)–Inr–simian virus 40 (SV40) enhancer (Enh)–luciferase were gifts from D. Reinberg, Howard Hughes Medical Institute University of Medicine and Dentistry of New Jersey; SV40 promoter-luciferase (pGL2 promoter) was purchased from Promega Corp. Other reporter vectors were constructed by using elements from the vectors described above. Except where noted, all protein coding sequences were placed under the control of the cytomegalovirus (CMV) immediate-early promoter in the CMVneo mammalian expression vector (6). Rapid amplification of cDNA ends (RACE) PCR demonstrated that the sequences of mouse and human Nab1 diverge from the rat sequence upstream of a conserved methionine codon (rat amino acid 86; human and mouse amino acid 1), which conforms to the Kozak consensus sequence for efficient translation initiation (40, 67a). Thus, rat Nab1 may typically begin translation at amino acid 86 (the second methionine encoded by the cDNA [62]), a form of the protein which is fully functional for NGFI-A repression. To ensure that our analysis was consistent among the different species, we studied rat Nab1 that began translation at the second methionine but retained the numbering used in the initial description of the cDNA (62). In some instances, we expressed Nab1 beginning at the first methionine (amino acid 1), so that the protein could be detected by using a polyclonal antibody recognizing residues 4 to 19 (described below). The construct for expressing Nab1 from the second methionine was created by removing all 5′ untranslated region and coding sequence up to amino acid 78 from the original cDNA. Nab1 mutations fused to the carboxy terminus of the Gal4 DNA binding domain (DBD) (amino acids 1 to 147) were generated by cloning fragments into the pM1,2,3 series of expression vectors (63). The Nab1-VP16 activation domain chimera was created by fusing VP16 amino acids 414 to 490 to the carboxy terminus of Nab1. VP16 coding sequence was a gift from S. Triezenberg, Michigan State University (73). Expression vectors for NGFI-A proteins with native and Gal4 DBD, and NGFI-B, were described previously (56, 61). Synthetic transcription factors comprised of the NGFI-A R1 domain, zinc finger DBD, and heterologous activation domains (ADs) were created by fusing AD coding sequence to the carboxy terminus of the zinc fingers in the previously described R1 domain competitor construct (61); VP16 AD was amino acids 414 to 490, E2F-1 AD was amino acids 285 to 437 (33), and NGFI-B AD was amino acids 38 to 99 (56). E2F-1 coding sequence was a gift from D. Dean, Washington University School of Medicine. Chimeric transcription factors R1–NGFI-B and R1–NGFI-A I293F point mutation (R1–NGFI-A pm) were constructed by fusing the NGFI-A R1 domain (amino acids 267 to 306) to the amino termini of NGFI-B and NGFI-A pm, respectively.
Tissue culture, transfections, and luciferase assays.
COS, CV-1, and QT-6 cells were cultured as described previously (2, 56). Transient transfections were performed by using the calcium phosphate precipitation technique (8, 67, 68). Luciferase assays were performed as described previously (12, 68, 79). The identities and amounts of DNA constructs used in each experiment are included in the figures and legends. When appropriate, reporter background activity was subtracted from the data. Transfections were internally controlled by including a constitutive lacZ gene (CMV promoter; 500 ng/well). Luciferase activity was normalized by dividing by β-galactosidase (β-Gal) activity, measured by using a chemiluminescence assay kit (Galacto-Light; Tropix). Each data point represents the average normalized activity in lysates prepared from two identically transfected wells.
Antibody production.
Nab1 polyclonal antibodies were prepared by immunizing rabbits with peptides coupled to keyhole limpet hemocyanin (Calbiochem). The amino-terminal antibody was prepared against residues 4 to 19 (CQRLPRDSALRYIIST), and the internal antibody was prepared against a peptide comprised of C followed by residues 321 to 335 (EMSDEDPHKEEEIRK). Affinity purification of the antibodies was performed as described previously (68).
Immunoblot analysis.
Protein expression levels in transfected cells was analyzed by fractionating cell lysates though sodium dodecyl sulfate-polyacrylamide gels, blotting to nitrocellulose, and detecting proteins with specific antibodies by using an enhanced chemiluminescence assay kit according to the manufacturer’s instructions (Amersham). To compare Nab1 mutations, expression vectors were transfected into QT-6 cells, and the blots were probed with the Nab1 internal antibody (1:150 dilution), or amino-terminal antibody (1:200) if a mutation altered the internal antibody epitope. To compare the Nab1-Gal4 DBD fusions, expression vectors were transfected into COS cells, and the blots were probed with monoclonal antibody RK5C1 (Santa Cruz Biotechnology) recognizing Gal4 amino acids 1 to 147 (1:1,000 dilution). As an internal control, a constitutive lacZ gene (driven by the CMV promoter) was included in the transfections, and β-Gal levels in the cell lysates were assayed by immunoblotting, using the specific monoclonal antibody D19-2F3-2 (1:3,600 dilution; Boehringer Mannheim). Prior to detecting Nab1 proteins, cell volumes were adjusted to contain approximately equal amounts of β-Gal. Secondary antibodies were rabbit anti-mouse or goat anti-rabbit antibodies conjugated to horseradish peroxidase (1:10,000 dilution).
Preparation of DNA affinity matrix.
The DNA affinity matrix was prepared by annealing 8 μg of each of two single-stranded oligonucleotides with overlapping complementary sequence and filling in with Klenow enzyme. The double-stranded product contained two tandem NGFI-A binding sites (GCGTGGGCGTGCGTGGGCG), with 38 5′ and 55 3′ flanking bp, and a 5′ EcoRI site. The DNA was digested with EcoRI, and the 5′ overhang was filled in with Klenow enzyme by using a deoxynucleoside triphosphate mix in which dATP was replaced by biotin-14-dATP (Gibco BRL) such that each oligonucleotide contained two biotin molecules. The DNA was then electrophoresed on a 3% agarose gel, visualized by ethidium bromide staining and UV light transmission, excised from the gel, weighed, and dissolved in 4 volumes (e.g., 1 g = 4 ml) of 6 M NaI. Dynabeads M-280 Streptavidin (Dynal) (80 μl) were added, and the suspension was then mixed by rocking for 20 min at 24°C. The beads were retained by a magnetic field, the supernatant was removed, and the beads were washed three times with 1 ml of 10 mM Tris (pH 8.0)–1 mM EDTA–1 M NaCl, resuspended in 80 μl of the same buffer, and stored at 4°C until use.
NGFI-A–Nab1 complex isolation by oligonucleotide affinity purification.
COS cells were transfected with Nab1 and NGFI-A expression constructs, or pGEM3 (Promega), and nuclear extracts were prepared by using a rapid technique similar to one reported previously (1). Nab1 and NGFI-A protein expression levels were assessed by immunoblot analysis. Binding reactions consisted of binding buffer (340 μl), variable volumes of nuclear extract, so as to equalize protein concentration, DNA affinity matrix bead suspension (15 μl), NaCl to a final, monovalent cation concentration of 140 mM, and distilled water to a final volume of 0.5 ml. Binding buffer consisted of 20 mM Tris (pH 7.6), 5 mM MgCl2, 20 μM ZnSO4, 5% glycerol, 1 mM dithiothreitol, 0.1% Nonidet P-40, poly(dA-dT) · (dA-dT) (20 μg/ml; Pharmacia), bovine serum albumin (100 μg/ml), 1 mM phenylmethylsulfonyl fluoride, aprotinin (10 μg/ml), pepstatin (1 μg/ml), leupeptin (1 μg/ml), and 0.1 mM NaF. One reaction also included 4 μg of double-stranded oligonucleotides containing two NGFI-A binding sites, as specific competitor. Reactions were incubated at 24°C with rocking for 50 min. Beads were magnetically separated from the supernatant and washed three times with 1 ml of wash buffer that was identical in composition to binding buffer except that poly (dA-dT) · (dA-dT) was not included. After the last wash was removed, proteins bound to the beads were eluted in 1× Laemmli protein sample buffer (100 μl). Proteins were analyzed by immunoblotting (7.5% gel) using an antibody recognizing NGFI-A (1:5,000 dilution) (14), followed by stripping of the blot as instructed by the manufacturer (Amersham) and then using the Nab1 internal-epitope antibody (1:150 dilution). For the experiment shown in Fig. 1, 90 μl of Nab1–NGFI-A and 135 μl of Nab1–NGFI-A I293F point mutation nuclear extracts were used in the binding reactions, and 30-μl aliquots of the bound and eluted proteins were analyzed and compared with 1.7% of the volume of total nuclear extracts.
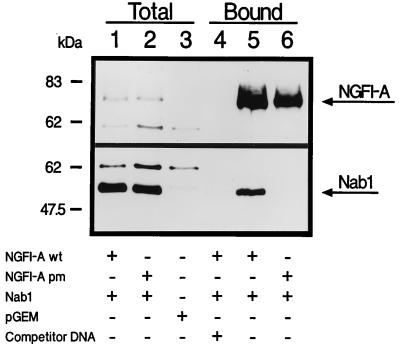
FIG. 1.
Nab1 physically interacts with NGFI-A bound to DNA. Nuclear extracts prepared from cells transfected with Nab1 and NGFI-A were incubated with a DNA affinity matrix containing NGFI-A binding sites. After washing, bound proteins were eluted and detected by using specific Nab1 and NGFI-A antibodies by immunoblotting (arrows). Total nuclear extracts are shown in lanes 1 to 3, demonstrating equivalent expression of the proteins in lanes 1 and 2. Nuclear extract from cells transfected with pGEM3 is shown in lane 3. Proteins bound to DNA are shown in lanes 4 to 6. Wild-type (wt) NGFI-A and NGFI-A pm (containing the I293F point mutation preventing Nab1 binding) both bound to DNA (lanes 5 and 6), but Nab1 was found only in the complex that included wild-type NGFI-A (lane 5). No proteins bound in the presence of excess NGFI-A site competitor oligonucleotides (lane 4). Proteins present in nuclear extracts and components in binding reactions are indicated at the bottom (+, present; −, absent).
Immunofluorescence staining.
Nab1 cellular localization was detected by indirect immunofluorescence microscopy, as described previously for detection of Nab2 (67), using the amino-terminal and internal antibodies specific for Nab1 (1:500 dilution). QT-6 cells were transfected as described previously (67), with the modification that 10 μg of Nab1 protein expression vector, which included both possible initiator methionines, was used.
Nab1 human chromosomal localization.
An approximately 100-kb human Nab1 genomic DNA fragment, derived from a P1 clone (Genome Systems), was labeled with biotin-dUTP or digoxigenin-11-dUTP (Bethesda Research Laboratories) as described previously (68). This probe was used for fluorescence in situ hybridization (FISH) of normal human chromosomes derived from synchronized peripheral lymphocyte cultures as described previously (84, 68). Eighty metaphases were analyzed.
RESULTS
Nab1 does not prevent DNA binding by NGFI-A.
To determine if Nab1 inhibition of NGFI-A activity was due to interference with binding of NGFI-A to DNA, we attempted to isolate complexes from cells in which Nab1 was bound to NGFI-A at the same time that NGFI-A was bound to DNA. Nuclear extracts prepared from COS cells transfected with Nab1 and NGFI-A were incubated with a DNA affinity matrix containing specific NGFI-A binding sites. Using this technique, NGFI-A was isolated bound to the DNA (Fig. 1), indicating that the presence of Nab1 in the reaction did not interfere with NGFI-A DNA binding. Inclusion of competitor (nonbiotinylated) NGFI-A binding site oligonucleotides blocked binding of NGFI-A to DNA, demonstrating the specificity of the NGFI-A-DNA interaction. Importantly, we could copurify Nab1 with NGFI-A, but not in the presence of competitor oligonucleotides. Furthermore, Nab1 was not copurified with a version of NGFI-A containing a point mutation (NGFI-A pm) in the R1 domain (I293F) that prevents NGFI-A interaction with Nab proteins (62, 68), although NGFI-A pm bound as efficiently as wild-type NGFI-A to the DNA (Fig. 1). These data suggest that Nab1 and NGFI-A can physically interact within cells and that Nab1 binds to NGFI-A when NGFI-A is bound to DNA, in a manner dependent on an intact R1 domain.
Mammalian two-hybrid analysis.
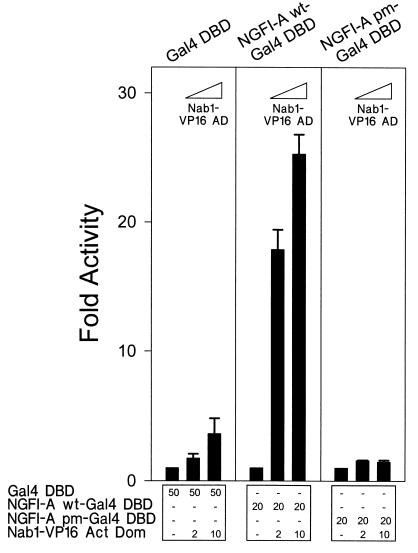
To corroborate the physical interaction experiments described above, we performed a genetic reporter experiment to detect the Nab1–NGFI-A interaction in mammalian cells. The VP16 activation domain (73) was fused to the carboxy terminus of Nab1 (Nab1-VP16). This construct was transfected into CV-1 cells with NGFI-A containing the Gal4 DBD (61) and a reporter vector containing Gal4 binding sites and a minimal promoter. The effect of the expressed proteins on transcriptional activity was measured by using the luciferase assay (Fig. 2 shows a schematic representation of luciferase reporter contructs). Nab1-VP16 had essentially no effect on the Gal4 DBD alone (Fig. 3) but dramatically stimulated transcription, in a dose-responsive manner, in cooperation with NGFI-A. Superactivation did not occur however, when NGFI-A pm, which is not repressed by Nab1, was used. These results support the idea that Nab1 binds to NGFI-A, through the R1 domain, when the latter is bound to DNA.
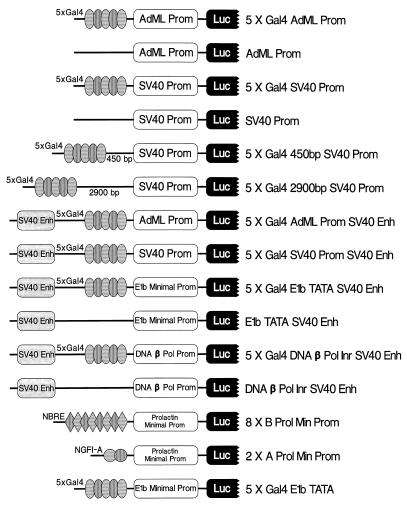
FIG. 2.
Schematic diagram and abbreviated names of luciferase reporter vectors. The arrangement of enhancer (Enh) elements, promoters (Prom), and defined transcription factor response elements is indicated, but their sizes and distances are not drawn to scale.
FIG. 3.
Mammalian two-hybrid analysis provides genetic evidence that Nab1 binds to NGFI-A within cells and does not block NGFI-A DNA binding. Nab1 fused with the VP16 AD (Nab1-VP16) was expressed in cells with NGFI-A (containing the Gal4 DBD). Nab1-VP16 superactivated a reporter gene in cooperation with wild-type (wt) NGFI-A, but not NGFI-A pm (containing the I293F point mutation preventing Nab1 binding), and did not affect Gal4 DBD alone. CV-1 cells were transfected with expression constructs and the 5 × Gal4 E1b TATA reporter vector (500 ng). The quantity (nanograms) of each expression vector is shown at the bottom. The baseline activity of each DNA binding protein was defined as 1 and the effect of Nab1-VP16 was calculated as fold activity over the activation occurring in its absence. Each bar of the graph represents the mean of three independent experiments, and the error bar represents the standard deviation.
Nab1 can repress different types of activation domains.
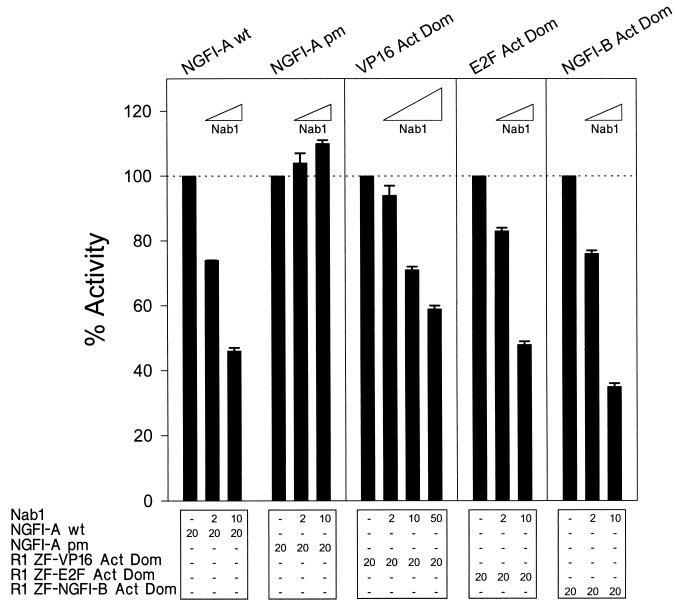
To determine if Nab1 repression was unique to members of the NGFI-A family, we tested the effect of Nab1 on activation of a minimal promoter containing NGFI-A sites by transcription factors consisting of the NGFI-A R1 domain, NGFI-A DBD, and heterologous ADs from VP16 (73), E2F (74), and NGFI-B (56). Transcription stimulated by NGFI-A and the chimeric activators, but not by NGFI-A pm, was repressed by Nab1 in a dose-dependent manner, although repression of the VP16 AD required a greater quantity of Nab1 expressor DNA (Fig. 4). These results demonstrate that Nab1 can repress different types of ADs.
FIG. 4.
Nab1 repression is not activator specific. Nab1 repressed activation of a reporter gene by wild-type NGFI-A (wt) and three synthetic activators comprised of the NGFI-A R1 and DBDs and ADs from the unrelated transcription factors VP16, E2F, and NGFI-B. NGFI-A pm was unaffected. CV-1 cells were transfected with expression constructs and the 2 × A Prol Min Prom reporter vector (500 ng). The quantity (nanogram) of each expression vector is shown at the bottom. The baseline activity of each transcription factor was defined as 100%. Each bar of the graph represents the mean of three independent experiments, and the error bar represents the standard deviation.
The R1 domain is portable.
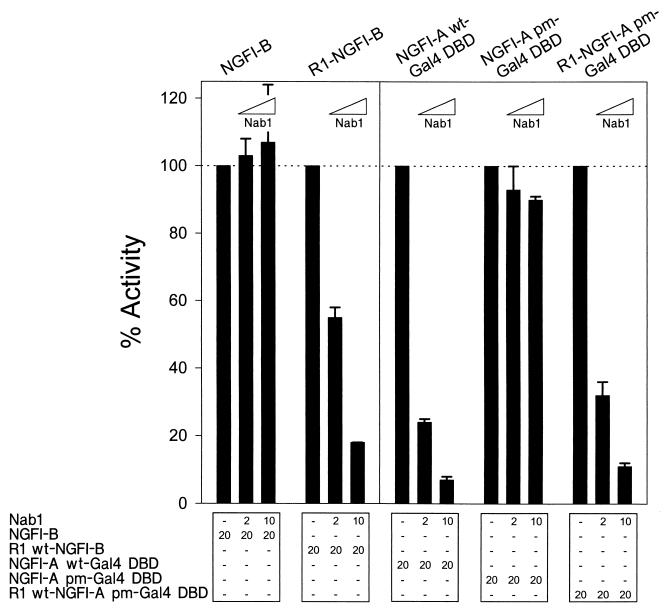
The NGFI-A R1 domain was used to isolate Nab1 by interactive cloning in yeast and can compete with full-length NGFI-A for binding to Nab1 in mammalian cells (62). These observations suggested that possession of the R1 domain by a protein may be sufficient to mediate its interaction with Nab1. We tested whether NGFI-B, an orphan nuclear receptor structurally unrelated to NGFI-A (56), could be made susceptible to Nab1 repression by fusing the R1 domain to the amino terminus of this protein (R1–NGFI-B). Activation of a minimal promoter containing NGFI-B sites by wild-type NGFI-B was unaffected by Nab1. In contrast, R1-NGFI-B was repressed in a dose-dependent manner (Fig. 5). To extend this observation, we fused an intact R1 domain to the amino terminus of NGFI-A (with the Gal4 DBD) containing the I293F point mutation in the endogenous R1 domain (R1–NGFI-A pm). Activation of a minimal promoter containing Gal4 sites by NGFI-A and R1–NGFI-A pm, but not by NGFI-A pm, was repressed by Nab1 in a dose-dependent manner (Fig. 5). Thus, the R1 domain is portable and can confer Nab1 repressibility on proteins which are otherwise not susceptible. In addition, moving the location of the R1 domain does not affect the ability of Nab1 to repress transcription.
FIG. 5.
The R1 domain is portable. Nab1 did not repress NGFI-B or NGFI-A pm but was able to repress chimeric proteins comprised of the same activators fused with the R1 domain. For NGFI-A, changing the location of the R1 domain did not affect repression. The Gal4 DBD replaced the native NGFI-A DBD. CV-1 cells were transfected with expression constructs and the 5 × Gal4 E1b TATA or 8 × B Prol Min Prom reporter vector (500 ng). The quantity (nanogram) of each expression vector is shown at the bottom. The baseline activity of each transcription factor was defined as 100%. Each bar of the graph represents the mean of three independent experiments, and the error bar represents the standard deviation.
Nab1 can repress transcription when tethered directly to DNA.
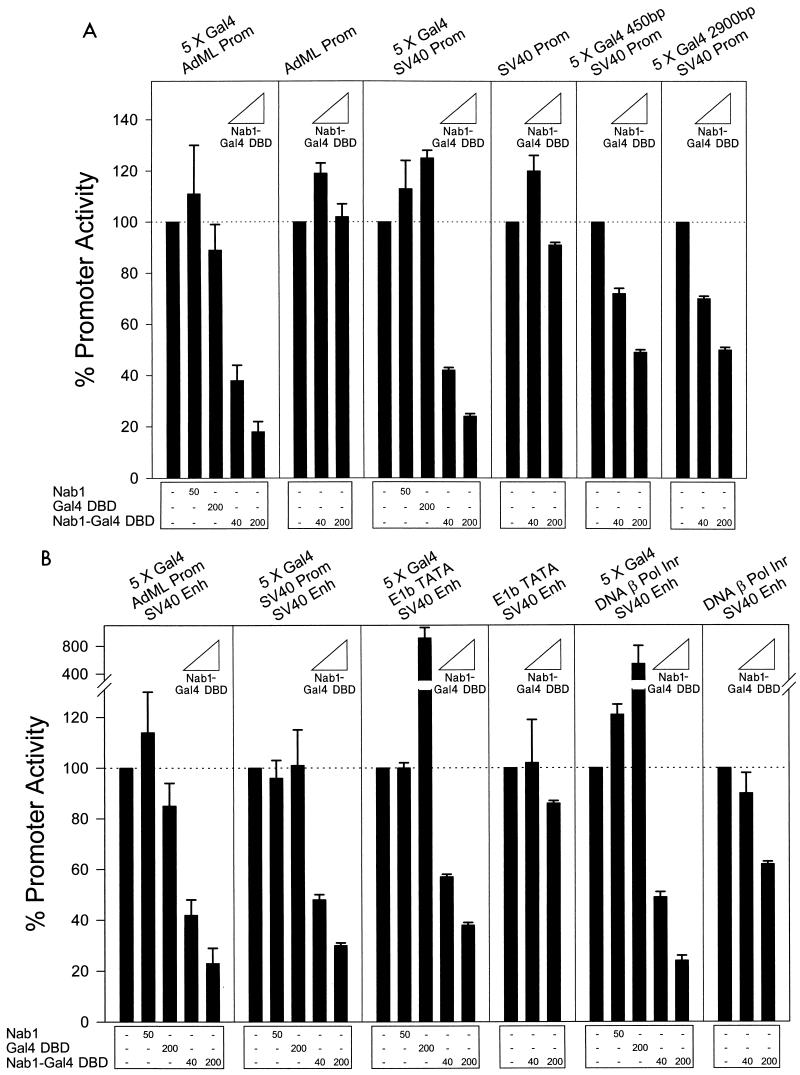
DNA binding proteins containing the R1 domain may recruit Nab1 to a promoter, where Nab1 represses by a mechanism that is not specific to particular activators. To test this idea, we determined if a different means of tethering Nab1 to a promoter would permit Nab1 to repress transcription. The Nab1 coding sequence was fused to the Gal4 DBD (tethered Nab1), and we tested the effect of this chimeric protein on transcription from the constitutively active AdML promoter containing Gal4 binding sites. The promoter was repressed in a dose-responsive manner by tethered Nab1 but not by wild-type Nab1 or by the Gal4 DBD alone. In contrast, the same promoter lacking Gal4 binding sites was not repressed (Fig. 6A). Similar results were obtained using the SV40 early promoter, indicating that repression by tethered Nab1 was not unique to the AdML promoter (Fig. 6A). We also investigated whether tethered Nab1 could repress the SV40 promoter when the distance separating the Gal4 binding sites from the promoter was increased. When the binding sites were moved approximately 450 or 2,900 bp upstream of the SV40 promoter boundary, tethered Nab1 could still repress transcription (Fig. 6A). Tethered Nab1 could also repress the AdML and SV40 promoters when their activity was stimulated by the presence of a distal (2,800-bp) SV40 Enh element (Fig. 6B). We next determined if the nature of promoter basal control elements was important for Nab1 repression. Tethered Nab1 repressed the Enh-stimulated activity of the E1b minimal promoter (46) (TATA element and transcription initiation site) and the human DNA β-Pol promoter (81) (Inr element and no TATA box) in a dose-responsive manner that depended on the presence of Gal4 binding sites (Fig. 6B). Wild-type Nab1 and Gal4 DBD alone did not repress transcription (Fig. 6B). Thus, Nab1 can repress transcription when artificially tethered to a promoter, and repression is not critically dependent on the nature of the promoter or on the distance separating it from Nab1.
FIG. 6.
Nab1 fused to the Gal4 DBD (Nab1-Gal4 DBD), and tethered directly to DNA, repressed transcription. (A) AdML and SV40 early promoters were repressed by tethered Nab1 when the promoters contained Gal4 binding sites. Nontethered Nab1 and Gal4 DBD did not repress promoter activity. Repression of the SV40 promoter also occurred when the Gal4 sites were moved 450 and 2,900 bp from the promoter boundary. (B) Tethered Nab1 repressed the AdML and SV40 promoters stimulated by a distal SV40 Enh (2.8 kbp upstream), as well as Enh-driven activity of two minimal promoters, E1b (TATA element) and DNA β-Pol (Inr element). Repression by Nab1-Gal4 DBD required Gal4 binding sites, and nontethered Nab1 and Gal4 DBD did not repress. CV-1 cells were transfected with expression constructs and the indicated luciferase reporter vector (500 ng). The quantity (nanogram) of each expression vector is shown at the bottom. The baseline activity of the promoters was defined as 100%. Each bar of the graph represents the mean of three independent experiments, and the error bar represents the standard deviation.
Identification of the Nab1 NLS.
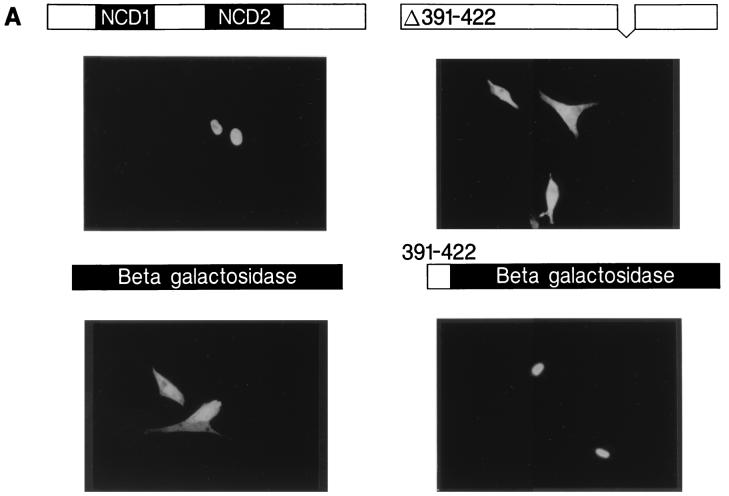
Consistent with its role as a transcriptional regulatory protein, Nab1 was found exclusively in the nucleus (62). Examination of the Nab1 protein sequence identified a consensus bipartite nuclear localization signal (NLS; Nab1 residues 334 to 350) (15, 51, 59), with critical residues conserved in Nab2 as well as in Nab homologs from C. elegans and D. melanogaster (10a). To determine if this motif controlled Nab1 nuclear localization, a deletion mutation (Δ328-388) was transfected into QT-6 cells, and cellular localization was determined by immunocytochemistry using the polyclonal antibody recognizing residues 4 to 19 (amino-terminal antibody). Unexpectedly, the mutated Nab1 protein was found exclusively within the nuclei of transfected cells (data not shown), indicating that this motif was not required for Nab1 nuclear localization. To identify the NLS, we tested successive carboxy-terminal truncations of the protein for nuclear localization. Deletion of residues 424 to 570 resulted in nuclear staining identical to that for wild-type Nab1, but further deletion (Δ391-570) caused the protein to be excluded from the nucleus (data not shown), suggesting that residues between 390 and 423 were required for nuclear targeting. We deleted residues 391 to 422 and found that this mutation abrogated Nab1 nuclear localization (Fig. 7A). To determine if these residues were also sufficient to target a protein to the nucleus, we created a chimeric molecule consisting of Nab1 residues 391 to 422 fused to Escherichia coli β-Gal. QT-6 cells were transfected with constructs for wild-type β-Gal and Nab–β-Gal, and the cellular localization of the proteins was determined by immunocytochemistry using an antibody to β-Gal. As expected, β-Gal was found in the cytoplasm, whereas Nab–β-Gal was found exclusively within the nucleus (Fig. 7A). Cellular localization of wild-type Nab1 and Nab1 Δ391-422 was also compared in CV-1 cells, with results identical to those observed in QT-6 cells (data not shown). Thus, Nab1 residues 391 to 422 are necessary for Nab1 nuclear localization and are also sufficient to direct a heterologous cytoplasmic protein to the nucleus. Sequence comparison between the rat Nab1 NLS and the homologous region from other mammalian Nab proteins demonstrated a conserved pattern of amino acids related to the bipartite NLS (Fig. 7B).
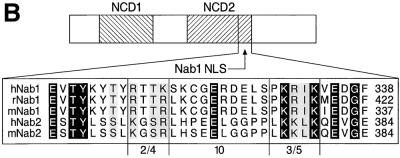
FIG. 7.
Identification of the Nab1 NLS. (A). Wild-type Nab1 was found exclusively within the nucleus of transfected cells (upper left), but deletion of amino acids 391 to 422 caused the mutated protein to remain in the cytoplasm (upper right). In transfected cells, β-Gal was located in the cytoplasm (lower left), but fusion of Nab1 amino acids 391 to 422 to the amino terminus of β-Gal resulted in nuclear localization of the chimeric protein (lower right). QT-6 cells were transfected with expression constructs, and proteins were detected by indirect immunofluorescence using antibodies recognizing the Nab1 amino-terminal epitope or β-Gal. Nontransfected cells served as internal negative controls for antibody staining (present but not visible in these photographs). Magnification, ×400. (B). Comparison of the rat Nab1 NLS (amino acids 391 to 422) with homologous sequence from other mammalian Nab proteins. At the top is a schematic of Nab1 showing the locations of the two conserved domains, NCD1 and NCD2, that define the Nab gene family. The NLS is located at the carboxy terminus of NCD2 (arrow). Conserved residues have a black background, and conservatively substituted residues have a gray background. A conserved pattern related to the bipartite NLS, in which two groups of basic residues (2 of 4 and 3 of 5) are separated from each other by any 10 amino acids, is delineated.
Mutagenesis and identification of Nab1 repression domains.
Discovery of Nab2, and related genes from D. melanogaster and C. elegans, enabled the identification of domains conserved within the Nab gene family (10a, 74). NCD1, located near the amino terminus of all Nab proteins, is required for binding NGFI-A (62, 68), but the locus of the Nab repression function was unknown. We performed extensive mutagenesis of Nab1 to identify regions of the protein necessary for repression of NGFI-A.
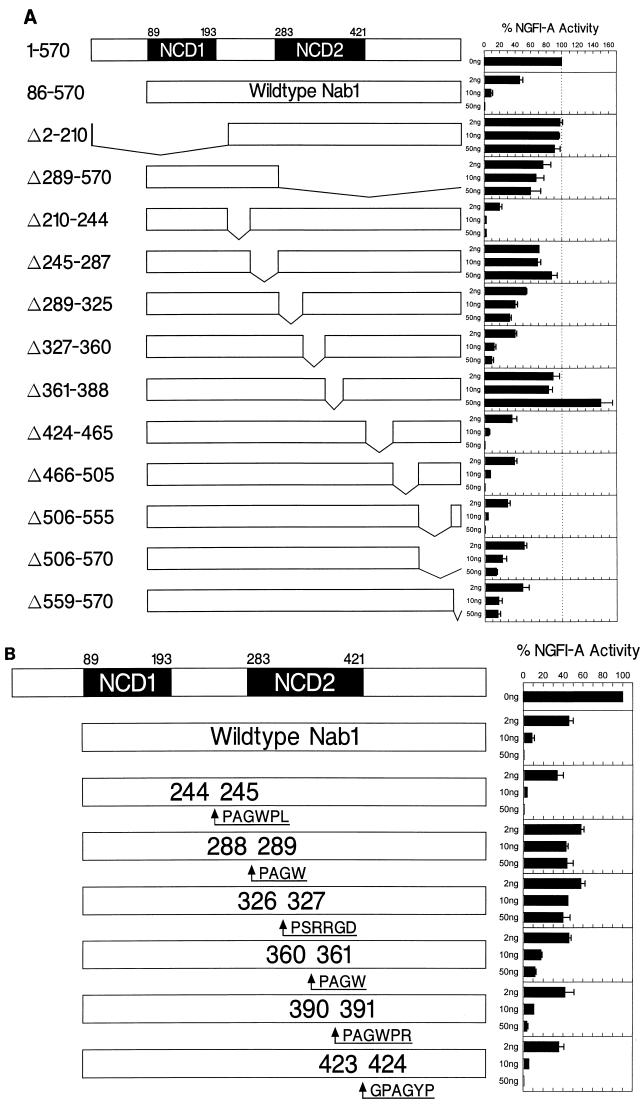
A Nab1 mutation lacking NCD1, Δ2-210, did not repress NGFI-A activity (Fig. 8A), although it was confirmed that this protein was nuclear (data not shown). However, a more extensive amino-terminal truncation, Nab1 Δ1-244, was able to repress when it was fused to the Gal4 DBD and tethered to the AdML promoter (data not shown). These mutations removed the R1 interaction domain (NCD1), confirming that Nab1 must bind to NGFI-A to repress its activity but that NCD1 is otherwise dispensable for repression. An extensive carboxy-terminal truncation, Nab1 Δ289-570, preserved NCD1 but removed the Nab1 NLS. To compensate, we fused the SV40 T-antigen NLS (34) to the carboxy terminus and confirmed that the protein was nuclear (data not shown). The Nab1 Δ289-570 mutation had minimal repressive effect on NGFI-A activity, suggesting that regions required for repression were located in the carboxy-terminal half of the protein (Fig. 8A). The amino-terminal half of the region separating NCD1 and NCD2 was dispensable for Nab1 repression (Nab1 Δ210-244), whereas the other half was required (Nab1 Δ245-287). Deletions in NCD2 (Nab1 Δ289-325, Δ327-360, and Δ361-388) were tested and found to disrupt repression, although the extent varied. The NCD2 deletion lacking the NLS, Nab1 Δ391-422, was fused to the Gal4 DBD and found to be partially defective for repression when tethered to the AdML promoter (data not shown), suggesting that this region of Nab1 contributes to repression as well as to nuclear localization. Within the nonconserved region following NCD2, three deletions (Nab1 Δ424-465, Δ466-505, and Δ506-555) did not affect repression. However, a carboxy-terminal truncation, Nab1 Δ506-570, which removed an additional 15 residues compared to Nab1 Δ506-555, was partially defective. Deletion of the final 12 residues, Nab1 Δ559-570, confirmed that the extreme carboxy terminus contributes to repression, and comparison of Nab1 and Nab2 demonstrated protein sequence similarity over the last 20 amino acids (data not shown).
FIG. 8.
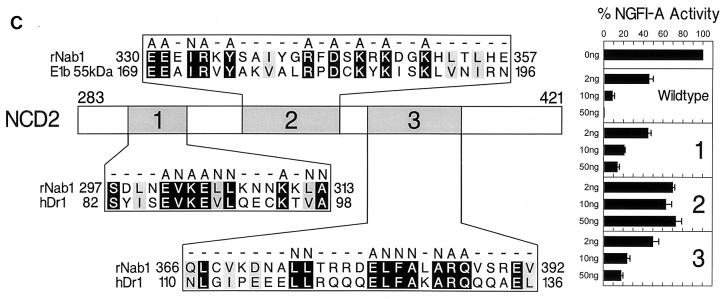
Mutagenesis of Nab1 identified regions required for repression. (A) Deletion mutagenesis. The schematic diagram shows the locations and boundaries of NCD1 and NCD2 in wild-type Nab1 and the location and relative size of each deletion. The effects of Nab1 deletions, compared to wild-type Nab1, on NGFI-A repression are shown to the right. Regions contributing to repression include all of NCD2, sequence amino terminal to NCD2, and the carboxy-terminal end of the protein. (B) Insertion mutagenesis. The schematic diagram shows the sequence of the exogenous amino acids inserted into Nab1 and the numbers of the endogenous residues between which each insertion was made. The effect of the insertions, compared to wild-type Nab1, on NGFI-A repression is shown to the right. Insertions affecting repression were located in the amino-terminal half of NCD2 (amino acids 283 to 360). (C) Replacement mutagenesis. The schematic diagram shows the locations and sequences of three regions of homology between NCD2 and the repressors Dr1 (motifs 1 and 3) and E1b 55k (motif 2). Conserved residues have a black background, and conservatively substituted residues have a gray background. Amino acids replacing conserved residues in NCD2 are indicated above the mutated residue. The effect of the replacements, compared to wild-type Nab1, on NGFI-A repression is shown to the right. For all experiments, CV-1 cells were transfected with expression constructs for Nab1 (2, 10, and 50 ng), NGFI-A with the Gal4 DBD (40 ng), and the 5 × Gal4 E1b TATA reporter vector (500 ng). Baseline NGFI-A activity was defined as 100%. Each bar of the graph represents the mean of three or more independent experiments, and the error bar represents the standard deviation.
To support the deletion analysis, which suggested that NCD2 is important for the repression function, we created insertion mutations at the boundaries of the deletions that we made within and upstream of NCD2. Each insertion consisted of four or six amino acids, including at least one proline and one glycine, both of which disrupt α-helices. An insertion about halfway between NCD1 and NCD2, between residues 244 and 245, did not interfere with repression, but insertions between 288 and 289, near the amino-terminal boundary of NCD2, and between 326 and 327 markedly disrupted repression (Fig. 8B). The next insertion, between 360 and 361, modestly affected repression, while the last two insertion mutations, between 390 and 391 and 423 and 424, repressed as effectively as wild-type Nab1.
The mutational analysis strongly suggested that NCD2 is necessary for Nab1 repression. We compared the sequence in this region to that of previously characterized transcriptional repressor proteins and identified two that share small regions of homology with Nab1 NCD2 (Fig. 8C). Dr1 shares two homology motifs (motifs 1 and 3) with Nab1, and adenovirus E1b 55-kDa protein (E1b 55k) shares one homology motif (motif 2), located between those of Dr1. Dr1 is a global repressor that blocks interaction between TATA binding protein (TBP) and TFIIA and TFIIB (30, 81). The first Nab1-Dr1 homology motif (motif 1) corresponds to the TBP binding domain of Dr1 (residues 85 to 99), which is required for Dr1 repression (81). The other homology motif (motif 3) corresponds to a region (residues 113 to 140) necessary for TBP binding, but not repression, located between the TBP binding and Dr1 repression domains (81). We mutated conserved amino acids in each of the two homology motifs in Nab1 and found that the mutated Nab1 proteins were partially defective for repression of NGFI-A (Fig. 8C). We then tested if Nab1 could interact with TBP in a fashion analogous to that of Dr1. Using the yeast two-hybrid method, we demonstrated a genetic interaction between Dr1 and TBP but not between Nab1 and TBP (data not shown). Furthermore, exogenous TBP was ineffective in reversing Nab1 repression in mammalian cell transfection experiments (data not shown), as opposed to the reported effect of TBP on Dr1 repression (81). These results indicate that despite the sequence similarities, the mechanism of Nab1 repression is different from that of Dr1. The second homology motif in Nab1 NCD2 corresponds to an inhibition domain in E1b 55k (83). E1b 55k binds to an activation domain in the p53 tumor suppressor protein to repress p53 target genes by passive and direct mechanisms (35, 50, 75, 83). Replacement mutagenesis of Nab1 residues conserved between Nab1 and E1b 55k abrogated Nab1 repression of NGFI-A (Fig. 8C).
It was possible that mutations in Nab1 which disrupted repression operated through an indirect mechanism (e.g., preventing interaction with NGFI-A). To control for this, we fused Nab1 mutations to the Gal4 DBD and tested them for repression when tethered to the AdML promoter. In all cases, mutations defective for repression of NGFI-A were also defective when tethered to the promoter (data not shown). All of the Nab1 mutations described above (nontethered and fused to the Gal4 DBD) were examined by immunoblot analysis, which confirmed that the proteins were stable, expressed at comparable levels, and of the expected sizes (data not shown).
Chromosomal localization of the human NAB1 gene.
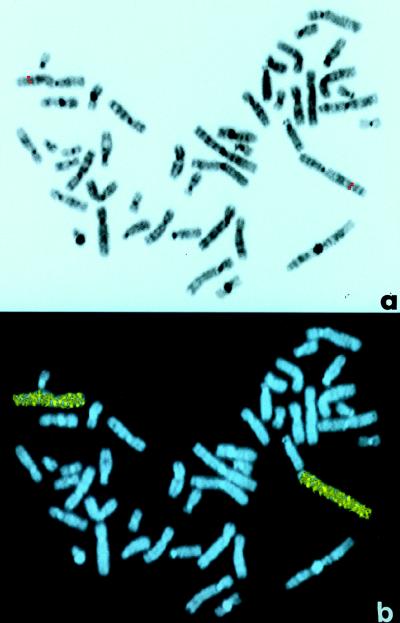
To determine if Nab1 could be implicated in the etiology of known genetic diseases, we determined the chromosomal location of the human gene by using the FISH technique. Biotinylated or digoxigenin-labeled NAB1 genomic probes were hybridized to normal human metaphase chromosomes. In the majority (75 of 80) of such chromosome spreads, we observed dual, symmetrical, fluorescent signals associated with the sister chromatids of two homologous chromosomes, corresponding to chromosomal region 2q32.3-33 (Fig. 9a). The fluorescent hybridization signal was precisely located in 20 chromosome pairs with distinctive diaminophenylindole (DAPI)-generated, and enhanced, G-like banding. The identity of the chromosomes exhibiting a specific Nab1 signal was confirmed by hybridization with a chromosome painting probe specific for chromosome 2 (Fig. 9b).
FIG. 9.
Mapping of human NAB1 genomic locus by FISH analysis. (a) Digital image of a metaphase chromosome spread derived from synchronized normal human lymphocytes after FISH with a NAB1 genomic probe. DAPI counterstaining generated an enhanced G-like banding pattern that was used to precisely locate the probe fluorescent signal to chromosome 2q32.3-33. The symmetrical, probe-derived fluorescent signals on both sister chromatids of the two chromosomes 2 are visible as red dots toward the telemore of the long arms. (b) Rehybridization of the same metaphase spread with chromosome 2-painting probe confirms the identity of the labeled chromosomes (green signal over blue background).
DISCUSSION
Previous work identified Nab1 as a nuclear protein that physically interacts with and represses transcription mediated by NGFI-A and the related transcription factors Krox20 and Egr3 (62, 64b). It is now known that Nab1 is the prototypical member of a gene family that includes mammalian Nab2 as well as Nab genes found in D. melanogaster and C. elegans (10a, 68). In this study, we explored the mechanism of Nab1 repression and identified structural regions required for this function.
Repressors are categorized as passive or active, and active repressors are further divided into those that quench and those that work by a direct mechanism (for reviews, see references 11, 25, 28, 32, and 37). Passive repressors interfere with activator functions (e.g., DNA binding) that are not directly related to the mechanism by which the activating signal is transmitted to the GTA. Active repressors interfere with the basic process of transcriptional activation or with subsequent stages required for the transcription reaction. Repressors that work by quenching prevent specific activators from transmitting their activating signal to the GTA or, alternatively, by preventing the GTA from receiving that signal. Repressors that work through a direct mechanism repress transcription in general and are not activator specific. Direct repressors interfere with the assembly or function of the GTA by contacting general transcription factors comprising the preinitiation complex (e.g., TFIID) or the RNA polymerase II holoenzyme.
Nab1 does not repress by a passive mechanism. We confirmed that Nab1 resides exclusively within the nucleus, indicating that Nab1 does not interfere with NGFI-A nuclear localization and that Nab1 does not prevent NGFI-A from binding to DNA. In fact, Nab1 requires NGFI-A to gain access to DNA. These results indicate that Nab1 does not function like IκB, which prevents both nuclear localization and DNA binding by Rel protein dimers (3, 4, 48) or like knirps, which competes with the activator bicoid for binding to overlapping DNA sites (29).
Nab1 repression is not activator specific and can repress transcription in general. This implies that Nab1 is an active repressor that works by a direct mechanism, as opposed to quenching. Evidence suggesting that the mechanism of Nab1 repression is not activator specific includes the finding that Nab1 repressed NGFI-B, a transcription factor unrelated to NGFI-A, when it contained the R1 domain required for Nab binding. In addition, repression occurred when the NGFI-A ADs were replaced by activation domains from three heterologous transcription factors (VP16, E2F, and NGFI-B) which stimulate transcription by different mechanisms. For example, VP16 targets TBP (65), TFIIB (47), and TAF40 (22), whereas E2F targets the coactivator CBP (74). Other experiments demonstrate that interaction of Nab1 with activation domains is not required for repression, which is how the quenching repressors Gal80 and mdm2 inactivate Gal4 and p53, respectively (44, 52). These include the previous findings that no individual NGFI-A AD was uniquely required for Nab1 repression (61) and that the R1 domain competitively blocked Nab1 repression of NGFI-A (62) and our observation that the R1 domain is portable. The latter two observations indicate that R1 is the sole point of contact between Nab1 and an activator and that it is sufficient by itself to confer Nab1 repressibility. R1 portability also demonstrates that changing the location at which Nab1 binds to an activator does not prevent repression, as would be expected if Nab1 was binding to, or sterically interfering with, ADs.
The strongest evidence that Nab1 repression is general, not activator specific, derives from the observation that Nab1 can repress when directly tethered to DNA through the Gal4 DBD, which obviates the need for a DNA binding activator to function as an intermediary. Tethered Nab1 repressed two different complex promoters, even when transcription was further stimulated by the addition of a distal Enh element. In addition, Nab1 repressed transcription both from a TATA box and from an Inr element, the two main types of promoter basal control elements, as well as when the binding sites for tethered Nab1 were moved far upstream from the SV40 promoter. The latter finding indicates that Nab1 is a long-range repressor, a type which has been proposed to function by directly interfering with the GTA (7). The tethering experiments also suggest that NGFI-A recruitment of Nab1 does not merely negate NGFI-A-mediated activation but converts NGFI-A binding sites to silencer elements that can repress multiple activators bound to promoters and enhancers. This model is similar to the one describing how Rb functions to repress E2F target genes involved in cell growth, although evidence suggests that Rb represses by quenching, as opposed to a direct mechanism (24, 77, 78).
Earlier work demonstrated that NCD1 is responsible for binding to R1 domains of NGFI-A family proteins. Our results indicate that the Nab1 NLS and the majority of repression function are both located in NCD2. The Nab1 NLS is contained between amino acids 391 and 422, which are located at the carboxy terminus of NCD2. This sequence is required for Nab1 to enter the nucleus and is sufficient to direct nuclear entry of the cytoplasmic β-Gal protein. Comparison of the rat Nab1 NLS to homologous sequence in other mammalian Nab1 and Nab2 proteins demonstrated a pattern of conserved amino acids similar to the bipartite NLS, which consists of two basic residues separated by a spacer of any 10 amino acids from a cluster of residues in which 3 of the next 5 must be basic (15, 51, 59). In Nab1 and Nab2, the first two basic residues of the pattern are separated by two nonbasic amino acids. Curiously, the Nab1 NLS is not conserved in invertebrate Nab proteins, and further work will be necessary to identify the NLS for D. melanogaster and C. elegans Nab.
Extensive mutagenesis of Nab1 localized the majority of repression function to NCD2, although a small, weakly conserved region at the carboxy terminus of the protein contributes as well. Examination of the NCD2 sequence demonstrated that it is highly charged, with a preponderance of Arg, Lys, Asp, and Glu residues compared to the entire Nab1 protein (38% versus 26%), but there is no obvious homology between NCD2 and that of other charged repression domains (25). However, we did identify three regions of similarity between NCD2 and two noncharged repressors, Dr1 and E1b 55k. One region of similarity with Dr1 corresponded to the Dr1 TBP binding domain, which is required for repression by Dr1. Replacement mutagenesis of amino acids conserved between Nab1 and Dr1 caused Nab1 to be partially defective for repression. However, Nab1 did not bind TBP, suggesting that Dr1 and Nab1 do not repress by the same mechanism. It is possible, however, that the conserved regions contribute to similar secondary or tertiary structures which serve different functions in the two proteins. Replacement mutagenesis of amino acids conserved between Nab1 and E1b 55k dramatically impaired Nab1 repression. An insertion mutation in the corresponding motif in 55k interfered with repression by that protein when it was tethered to a promoter (82, 83), suggesting that Nab1 and E1b 55k may repress by similar mechanisms.
Karyotypic analysis localized the human NAB1 gene to chromosomal region 2q32.3-33. Several diseases or syndromes that pertain to the NAB1 locus have been identified. A characteristic syndrome has been described for patients with a deletion in chromosome 2 [del(2)(q31q33)] that is typified by growth deficiency, developmental delay, mental retardation, minor facial anomalies, and limb defects (reviewed in references 54 and 58). Curiously, patients with duplication of the distal 2q region that includes NAB1 demonstrate some of the same features (13; reviewed in reference 58). Karyotypic analysis of tumor samples, including lung carcinomas, cervical carcinomas, ependymomas, and pediatric hepatoblastomas, suggests that loss or gain of genetic loci including NAB1 contributes to the genesis of the primary tumors or their metastases (17, 39, 53, 57, 60). Related to this, a human colon cancer cell line with a high propensity for liver metastasis in nude mice demonstrated gain of a portion of chromosome 2 that includes NAB1 (80). Finally, linkage analysis of two hereditary neurological diseases, autosomal recessive familial amyotrophic lateral sclerosis type 2 (27) and paroxysmal dystonic choreoathetosis (16, 18), has identified markers associated with these disease loci that map to chromosomal regions containing NAB1.
The Nab1–NGFI-A pairing is the latest of a number of corepressor-zinc finger transcription factor partnerships, suggesting that this association represents a flexible gene regulatory system that has been adapted to serve multiple purposes in different cell types and in organisms of varying evolutionary complexity. Examples include the yeast Ssn6/Tup1 repressor that binds to the Mig1 protein (72) and KAP-1/KRIP-1 (19, 36), a novel corepressor that interacts with the KRAB (Krüppel-associated box) domain, an evolutionarily conserved motif found at the amino termini of about one-third of all proteins that contain TFIIIA-Krüppel class Cys2-His2 zinc finger modules at their carboxy termini (5). NGFI-A family proteins do not contain KRAB domains, and database searches for additional proteins containing R1-like domains have thus far been unsuccessful. This suggests that repression mediated by Nab proteins is uniquely required to modulate transcription by NGFI-A and related activators and has an essential role in defining the biological function of this transcription factor family.
ACKNOWLEDGMENTS
We are grateful to D. Dean, D. Reinberg, and S. Triezenberg, who generously contributed constructs and reagents for use in our experiments. We also thank P. Crawford for critically reviewing the manuscript and S. Audrain for capable technical assistance.
This work was supported by NIH grant 5 P01 CA49712-08 and a CaP CURE Research Award. J.S. was supported by NIH NRSA 1F32GM18058-01. J.M. is an established investigator of the American Heart Association.
REFERENCES
- 1.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apel E D, Roberds S L, Campbell K P, Merlie J P. Rapsyn may function as a link between the acetylcholine receptor and the agrin-binding dystrophin-associated glycoprotein complex. Neuron. 1995;15:115–126. doi: 10.1016/0896-6273(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 3.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S J. I kappa B interacts with the nuclear localization sequences of the subunits of NF kappa B: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 4.Bell S, Matthews J R, Jaffray E, Hay R T. Iκ Bγ inhibits DNA binding of NF-κ B p50 homodimers by interacting with residues that contact DNA. Mol Cell Biol. 1996;16:6477–6485. doi: 10.1128/mcb.16.11.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellefroid E J, Poncelet D A, Lecocq P J, Revelant O, Martial J A. The evolutionarily conserved Kruppel associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci USA. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer C B. Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol. 1994;43:233–245. doi: 10.1016/s0091-679x(08)60606-8. [DOI] [PubMed] [Google Scholar]
- 7.Cai H N, Arnosti D N, Levine M. Long-range repression in the Drosophila embryo. Proc Natl Acad Sci USA. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christy B A, Lau L F, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with zinc finger sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke A R, Maandag E R, van Roon M, van der Lugt N M T, van der Valk M, Hooper M L, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 10a.Clements, M., and J. Milbrandt. Unpublished data.
- 11.Cowell I. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 12.Crosby S D, Puetz J J, Simburger K S, Fahrner T J, Milbrandt J. The early response gene NGFI-C encodes a zinc finger transcriptional activator and is a member of the GCGGGGGCG (GSG) element binding protein family. Mol Cell Biol. 1991;11:3835–3841. doi: 10.1128/mcb.11.8.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallapiccola B, Forabosco A, Calabro A. Trisomy 2q. Acta Genet Med Gemellol. 1975;24:307–310. doi: 10.1017/s0001566000010424. [DOI] [PubMed] [Google Scholar]
- 14.Day M L, Fahrner T J, Aykent S, Milbrandt J. The zinc finger protein NGFI-A exists in both nuclear and cytoplasmic forms in nerve growth factor stimulated PC12 cells. J Biol Chem. 1990;265:15253–15260. [PubMed] [Google Scholar]
- 15.Dingwall C, Laskey R A. Nuclear targeting sequences-a consensus. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 16.Fink J K, Rainier S, Wilkowski J, Jones S M, Kume A, Hedera P, Albin R, Mathay J, Girbach L, Varvil T, Otterud B, Leppert M. Paroxysmal dystonic choreoathetosis: tight linkage to chromosome 2q. Am J Hum Genet. 1996;59:140–145. [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher J A, Kozakewich H P, Pavelda K, Grier H E, Shamberger R C, Korf B, Morton C C. Consistent cytogenetic aberrations in hepatoblastoma: a common pathway of genetic alterations in embryonal liver and skeletal muscle malignancies. Genes Chromosomes Cancer. 1991;3:37–43. doi: 10.1002/gcc.2870030107. [DOI] [PubMed] [Google Scholar]
- 18.Fouad G T, Servidei S, Durcan S, Bertini E, Ptacek L J. A gene for familial paroxysmal dyskinesia (FPD1) maps to chromosome 2q. Am J Hum Genet. 1996;59:135–139. [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X-P, Neilson E G, Rauscher F J., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 20.Gashler A, Sukhatme V P. Early growth response protein 1 (Egr-1): prototype of a zinc finger family of transcription factors. Prog Nucleic Acids Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 21.Gashler A L, Swaminathan S, Sukhatme V P. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. 1993;13:4556–4571. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 23.Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 24.Hagemeier C, Bannister A J, Cook A, Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 27.Hentati A, Bejaoui K, Pericak-Vance M A, Hentati F, Speer M C, Hung W-Y, Figlewicz D A, Haines J, Rimmler J, Hamida C B, Hamida M B, Brown R H, Siddique T. Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33-q35. Nat Genet. 1994;7:425–428. doi: 10.1038/ng0794-425. [DOI] [PubMed] [Google Scholar]
- 28.Herschbach B M, Johnson A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 29.Hoch M, Gerwin N, Taubert H, Jackle H. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel. Science. 1992;256:94–97. doi: 10.1126/science.1348871. [DOI] [PubMed] [Google Scholar]
- 30.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D A. Dr1, a TATA binding protein associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 31.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 32.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 33.Kaelin W G, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma binding protein with E2F like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 34.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 35.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1b 55k proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 36.Kim S-S, Chen Y-M, O’Leary E, Witzgall R, Vidal M, Bonventre J V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 38.Kirsch K H, Korradi Y, Johnson J P. Mader: a novel nuclear protein over expressed in human melanomas. Oncogene. 1996;12:963–971. [PubMed] [Google Scholar]
- 39.Kohno T, Morishita K, Takano H, Shapiro D N, Yokota J. Homozygous deletion at chromosome 2q33 in human small-cell lung carcinoma identified by arbitrarily primed PCR genomic fingerprinting. Oncogene. 1994;9:103–108. [PubMed] [Google Scholar]
- 40.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and hematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 42.Lee S L, Sadovsky Y, Swirnoff A H, Polish J A, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 43.Lemaire P, Relevant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leuther K K, Johnston S A. Nondissociation of Gal4 and Gal80 in vivo after galactose induction. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 45.Levi G, Topilko P, Schneider-Maunoury S, Lasagna M, Mantero S, Cancedda R, Charnay P. Defective bone formation in Krox20 mutant mice. Development. 1996;122:113–120. doi: 10.1242/dev.122.1.113. [DOI] [PubMed] [Google Scholar]
- 46.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 47.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 48.Matthews J R, Watson E A, Buckley S L, Hay R T. Interaction of the C-terminal region of p105 with the nuclear localization signal of p50 is required for inhibition of NF-kappa B DNA binding activity. Nucleic Acids Res. 1993;21:4516–4523. doi: 10.1093/nar/21.19.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milbrandt J. A nerve growth factor induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 50.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human Bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 51.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms, and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 52.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 53.Otsuka T, Kohno T, Mori M, Noguchi M, Hirohashi S, Yokota J. Deletion mapping of chromosome 2 in human lung carcinoma. Genes Chromosomes Cancer. 1996;16:113–119. doi: 10.1002/(SICI)1098-2264(199606)16:2<113::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 54.Palmer C G, Heerema N, Bull M. Deletions in chromosome 2 and fragile sites. Am J Med Genet. 1990;36:214–218. doi: 10.1002/ajmg.1320360215. [DOI] [PubMed] [Google Scholar]
- 55.Patwardhan S, Gashler A, Siegel M G, Chang L C, Joseph L J, Shows T B, Le Beau M M, Sukhatme V P. Egr3, a novel member of the Egr family of genes encoding immediate early transcription factors. Oncogene. 1991;6:917–928. [PubMed] [Google Scholar]
- 56.Paulsen R E, Weaver C A, Fahrner T J, Milbrandt J. Domains regulating transcriptional activity of the inducible orphan receptor NGFI-B. J Biol Chem. 1992;267:16491–16496. [PubMed] [Google Scholar]
- 57.Rader J S, Kamarasova T, Huettner P C, Li L, Li Y, Gerhard D S. Allelotyping of all chromosomal arms in invasive cervical cancer. Oncogene. 1996;13:2737–2741. [PubMed] [Google Scholar]
- 58.Ramer J C, Mowrey P N, Robins D B, Ligato S, Towfighi J, Ladda R L. Five children with del (2)(q31q33) and one individual with dup (2)(q31q33) from a single family: review of brain, cardiac, and limb malformations. Am J Med Genet. 1990;37:392–400. doi: 10.1002/ajmg.1320370320. [DOI] [PubMed] [Google Scholar]
- 59.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 60.Rogatto S R, Casartelli C, Rainho C A, Barbieri-Neto J. Chromosomes in the genesis and progression of ependymomas. Cancer Genet Cytogenet. 1993;69:146–152. doi: 10.1016/0165-4608(93)90093-2. [DOI] [PubMed] [Google Scholar]
- 61.Russo M W, Matheny C, Milbrandt J. Transcriptional activity of the zinc finger protein NGFI-A is influenced by its interaction with a cellular factor. Mol Cell Biol. 1993;13:6858–6865. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russo M W, Sevetson B R, Milbrandt J. Identification of Nab1, a repressor of NGFI-A and Krox20 mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 64.Schneider-Maunoury S, Topilko P, Seitanidou T, Levi G, Cohen-Tannoudji M, Pournin S, Babinet C, Charnay P. Disruption of Krox20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- 64a.Sevetson, B., J. Svaren, and J. Milbrandt. Unpublished data.
- 64b.Sevetson, B., and J. Milbrandt. Unpublished data.
- 65.Stringer K F, Ingles C J, Greenblatt Direct and selective binding of an acidic transcriptional activation domain to the TATA box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 66.Sukhatme V P, Cao X, Chang L C, Tsai-Morris C-H, Stamenkovich D, Ferreira P C P, Cohen D R, Edwards S A, Shows T B, Curran T, Le Beau M M, Adamson E D. A zinc finger encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 67.Svaren J, Apel E D, Simburger K S, Jenkins N A, Gilbert D J, Copeland N A, Milbrandt J. The Nab2 and Stat6 genes share a common transcription termination region. Genomics. 1997;41:33–39. doi: 10.1006/geno.1997.4609. [DOI] [PubMed] [Google Scholar]
- 67a.Svaren, J., and J. Milbrandt. Unpublished data.
- 68.Svaren J, Sevetson B R, Apel E D, Zimonjic D B, Popescu N C, Milbrandt J. Nab2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swiatek P J, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 70.Swirnoff A H, Milbrandt J. DNA binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Topilko P, Schneider-Maunoury S, Levi G, Baron-van Evercooren A, Chennoufi A B Y, Seitanidou T, Babinet C, Charnay P. Krox20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 72.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 74.Trouche D. E2F1 and E1a12s have homologous activaition domain regulated by Rb and CBP. Proc Natl Acad Sci USA. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogelstein B, Kinzler K W. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 76.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 77.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 78.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 79.Wilson T E, Fahrner T J, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1297–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 80.Yeatman T J, Cher M L, Mao W, Wloch M, Tedesco T. Identification of genetic alterations associated with the process of human experimental colon cancer liver metastasis in the nude mouse. Clin Exp Metastasis. 1996;14:246–252. doi: 10.1007/BF00053898. [DOI] [PubMed] [Google Scholar]
- 81.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 82.Yew P N, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1b 55k polypeptide by suppressor-linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 83.Yew P R, Lie X, Berk A J. Adenovirus E1b oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 84.Zimonjic D B, Rezanka L, Di Paolo J A, Popescu N C. Refined localization of the erbB-3 proto-oncogene by direct visualization of FISH signals on LUT-inverted and contrast-enhanced digital images of DAPI-banded chromosomes. Cancer Genet Cytogenet. 1995;80:100–110. doi: 10.1016/0165-4608(94)00161-4. [DOI] [PubMed] [Google Scholar]