To the Editor:
The ZRSR2 gene, located on the X chromosome (Xp22.1), is a member of the RNA splicing machinery family of genes which also includes SF3B1, SRSF2, and U2AF1 [1–3]. Spliceosome gene mutations are found in 35% of myelodysplastic syndrome (MDS) patients and were described as being mutually exclusive with each other [3, 4]. ZRSR2m is seen in 4% of MDS, and across different myeloid neoplasms (MN) including acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), myeloproliferative neoplasm (MPN) [1, 5–7]. Damm et al. found that mutations in ZRSR2 were evenly distributed across the entire gene, and included nonsense, frameshift, and splice site mutations [1, 2]. The only co-mutation that showed significant association with ZRSR2 was TET2 (p < 0.001) [2, 8, 9]. ZRSR2m cells showed increased precursor cells for macrophages and decreased precursor cells for erythroid cells [10]. Malcovati et al. reported that TET2, ZRSR2 co-mutation was predictive of myelomonocytic phenotype and showed higher hemoglobin (Hgb) levels and monocyte counts [11].
This is a retrospective study done with IRB approval at our institution. No interaction was done with patients. A waiver of consent was approved by our IRB due to minimal risk nature. No cases of age younger than 18 were included. This study was conducted in a single institution. All methods carried out in our study were in accordance with relevant guidelines and regulations. Next-generation sequencing (NGS) was performed in the molecular hematopathology laboratory with the NGS gene panel including 42-47 genes, between 2016-2023. BlueSky Statistics V10.3.1 was used for data analysis.
NGS was performed clinically on 9320 samples, 164 had the ZRSR2m genotype, with only 2 being female (1.2%). Median patient age was 74 (range 31–92). The most common diagnosis was MDS (n = 53, 32.3%), clonal cytopenia of undetermined significance (CCUS) (n = 39, 23.8%), MPN (n = 33, 20.1%), MDS/MPN overlap (n = 23, 14%), AML (n = 15, 9.1%) and 1 mixed phenotype acute leukemia (MPAL). Fifteen (9%) patients had concurrent non-myeloid hematological malignancies diagnosed at the time of the NGS. Only 15 patients (9.1%) received prior chemotherapy or radiotherapy, 29 patients (17.7%) received prior immunotherapy with the highest frequency among CCUS patients (n = 13, 33%) (Supplementary Table 1). Abnormal cytogenetics were found in 54 patients (33%), with +8 (16) and -Y (11) being the most common (Supplementary Table 2).
Seventy-eight patients (48%) were diagnosed before our in-house NGS (Supplementary Table 3). Out of 10 patients diagnosed with CCUS, 8 (80%) progressed to MDS by the time of the NGS, and two (7.7%) and 5 (11.5%) MDS patients progressed to AML and MDS/MPN overlap, respectively.
The most common subtype among MDS was low blast (MDS-LB) (n = 37, 69.8%). Eighteen of 23 MDS/MPN Overlap were CMML (78%) and the most common MPN subtype was myelofibrosis (MF) (n = 27, 81.8%).
The most common risk stratification among MDS patients by IPSS-M scoring was low risk (n = 16, 30.2%) (Supplementary Figs. 1, 2, 3, 4). Twenty-four (45.3%) of MDS patients were stratified as low risk according to IPSS-R (Supplementary Table 4). Twenty-seven MDS patients (51%), and 23 CCUS patients (59%) had absolute monocyte count ≥0.5 ×109/L (Supplementary Table 1).
Median Gender-corrected VAF was 35 (range, 1–66). and multiple ZRSR2 mutations were found in 7 patients (4.3%).
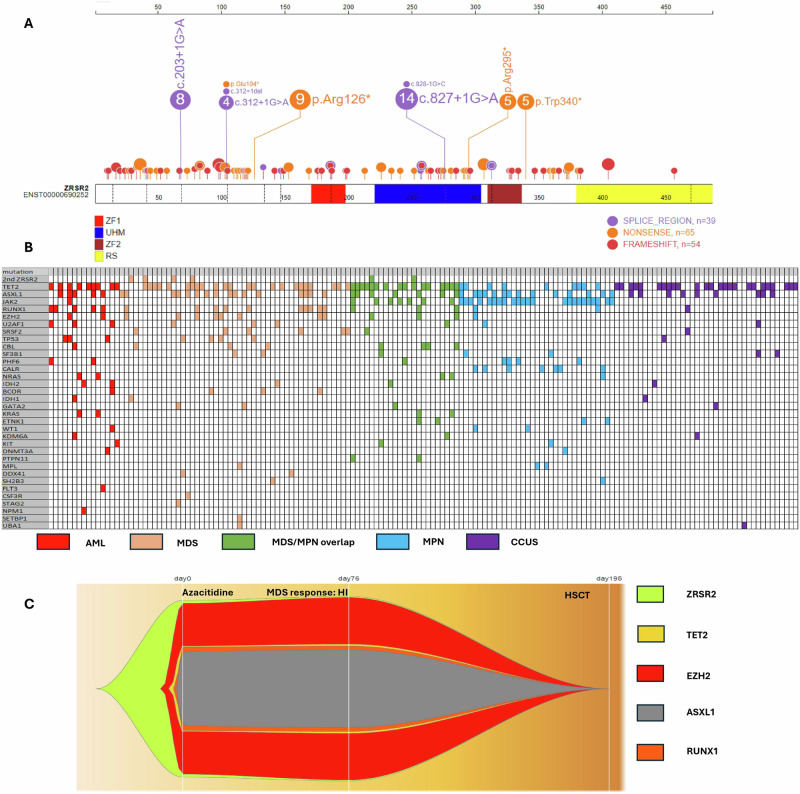
Mutations occurred in Pre-ZF1 (49%), UHM (27%), and Post-ZF2 (13%) domains and were spread across the entire length of the gene. (Fig. 1A, Supplementary Fig. 5A, Supplementary Table 5, Supplementary Table 6) The most common mutation type was nonsense (n = 69, 42%) (Supplementary Fig. 5B, Supplementary Table 5).
Fig. 1. ZRSR2 mutations, co-mutations and clonal dynamics with treatment.
A Representation of ZRSR2 mutations detected, positioned on the ZRSR2 protein and its functional domains. B The co-mutational pattern in 164 ZRSR2m MN patients. A column represents each patient. C Fishplot showing the progression of mutations VAF in ZRSR2m MDS patients throughout disease course and therapy.
The median number of co-mutations was 2 (range, 0–6). A significant correlation was found between MN classification and number of co-mutations in ZRSR2m patients (p < 0.001) (Supplementary Fig. 7).
The most common co-mutation was TET2 which was present in 84 patients (51%) (42% had multiple TET2) (Fig. 1B, Supplementary Figs. 8, 9, Supplementary Table 7) A significant correlation was found between MN classification and presence of TET2 in ZRSR2m patients (p = 0.007) (highest frequency among MDS/MPN overlap, 70%). Other common co-mutations were ASXL1 (n = 52, 32%), and JAK2 (n = 31, 19%) (Supplementary Fig. 8, Supplementary Table 8). A significant correlation was also found between MN classification and RUNX1 co-mutations (p = 0.008), and they were found with the highest prevalence among AML (n = 5, 33%) and MDS (n = 12, 23%).
Other members of the spliceosome family of genes were present in 14.7% of patients, including U2AF1 (n = 9, 5.5%), SRSF2 (n = 8, 5%), and SF3B1 (n = 7, 4%).
Only 13 patients (4.3%) had isolated ZRSR2 mutations. Median IPSS-M score was lower in patients with isolated ZRSR2m (−1.18) compared to patients with co-mutations (–0.74) (p = 0.02).
108 patients (66%) received treatment. 8 AML patients (66.7%) achieved response to therapy with 2 (16.7%) complete remissions with incomplete hematologic recovery (CRi), and 6 (50%) with CR, of whom 1 relapsed (Supplementary Fig. 10: B). Among 39 ZRSR2m MDS receiving treatment, 7 (18%) had hematological improvement (HI), 3 (7.7%) had complete remission with limited recovery (CRL), 1 CR equivalent (2.6), and 3 (7.7%) had CR, of which 2 relapsed (Supplementary Fig. 10: A). Hematopoietic stem cell transplant (HSCT) was performed in 21 patients (12.8%).
The most used medications were hypomethylating agents (HMA) (Supplementary Fig. 11, Supplementary Table 9). Fig. 1C shows the progression of mVAF in a ZRSR2m MDS patient among ZRSR2 and the co-mutations and their changes after therapy and HSCT.
Thirteen patients (8.7%) progressed to AML, 9 from MDS (17%, reported average is 30-40%) 2 from MDS/MPN overlap (8.7%), and 2 from MPN (6%), none of these patients had isolated ZRSR2 and only 1 MDS patient had isolated TET2 as a co-mutation. [25,26] Ten patients (8%) progressed into CMML or MDS/MPN overlap from CCUS, MDS, or MPN.
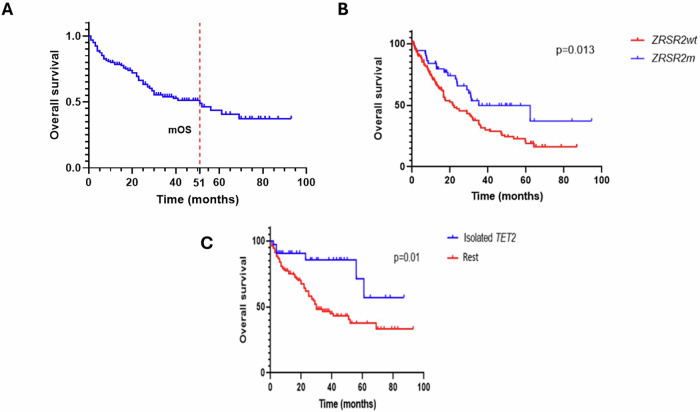
There were 68 deaths, with median overall survival (mOS) 51 months, and median follow-up of 35 months. (Fig 2A) ZRSR2m MDS patients had better mOS compared to the MDS control group with ZRSR2wt (35 months vs 22 months, p = 0.013). (Fig 2B) mOS of ZRSR2m patients varied significantly among different MNs (p = 0.004) (Supplementary Fig. 12).
Fig. 2. Overall survival of patients with myeloid neoplasm based on presence or lack of ZRSR2 mutation and other co-mutations.
A Overall survival for 164 ZRSR2m MN patients (with dotted red line showing median OS). B OS of ZRSR2m vs ZRSRwt in MDS patients. C OS according to the presence of an isolated TET2 co-mutation in ZRSR2m MN patients.
ZRSR2m with spliceosome and tumor suppressor gene (TSG) co-mutations showed worse survival (25 vs 56 months, p = 0.02 and 20 vs 51 months, respectively, p = 0.04) (Supplementary Figs. 10, 13). Patients with TET2 as an isolated co-mutation had better survival (not reached vs 30 months, p = 0.01). (Fig. 2C) Patients with RUNX1 co-mutations had worse survival (28 vs 52 months, p = 0.02) (Supplementary Fig. 13).
Improved survival was seen in patients with PB blasts <5% (52 months vs 9 months, HR = 0.027, p < 0.001) and higher Hgb concentration (HR = 0.78, p < 0.001), while patients with increased WBCs count (HR = 1.02, p < 0.001), absolute neutrophil count (ANC) (HR = 1.04, p = 0.01), absolute monocyte count (AMC) (HR = 1.2, p = 0.008), BM blasts (HR = 1.02, p < 0.001), PB blasts (HR = 1.03, p < 0.001) showed worse mOS (Supplementary Fig. 14). Patients with higher number of co-mutations (HR = 1.49, p < 0.001) and patients with abnormal cytogenetics (25 vs 61 months, p < 0.001) showed worse OS (Supplementary Fig. 15). On multivariate analysis, only higher Hgb concentration (HR = 0.8, p = 0.004), PB blasts>5% (HR = 2.2, p = 0.02), and abnormal cytogenetics (HR = 1.9, p = 0.01) retained significance (Supplementary Table 10).
Sequential NGS (S1-NGS) was performed in 55 out of the 164 patients (33.5%), 50 (91%) of them continued to have ZRSR2m. Out of the 21 patients who had HSCT, 4 (19%) performed sequential NGS post-transplant and all of them had negative NGS. mVAF for ZRSR2m was 80% and 89% for S1-NGS and S2-NGS, respectively, showing a statistically significant increase of 11% for S1-NGS from the first NGS (p = 0.01).
Our study’s cohort had only 2 female patients, data by Daichi Inoue et al. suggest that ZRSR2 escapes from X inactivation and is not pathogenic in females at heterozygous state [2, 12]. We also find that MDS patients with ZRSR2 mutations have better survival, indicating a favorable prognosis.
Our study found a notable association between ZRSR2m and a higher incidence of CCUS diagnoses, which is a novel finding. Our findings suggest that the ZRSR2m mutation carries a favorable prognosis among MNs, especially in isolated TET2m group.
Our findings clinically support Madan et al. findings as we found that over half of the ZRSR2m MDS and CCUS cohort had elevated absolute monocyte count ( ≥ 0.5 ×109/L), majority of MDS/MPN overlap patients were diagnosed as CMML, and about 8% of patients originally diagnosed as CCUS, MDS or MPN progressed or were re-diagnosed as CMML later [10].
Interestingly, MPN was found in 20% of ZRSR2m patients and consisted mainly of MF, which raises the possibility of acquiring ZRSR2m later in MPN progression into MF. Our study demonstrated a strong association between ZRSR2m and TET2m (51% of patients) and that the presence of TET2m as an isolated co-mutation was shown to be associated with longer survival and a higher prevalence among MDS/MPN overlap patients. Other studies reported enrichment of ZRSR2 in spliceosome mutated cases and our paper supports this (10.4% of our ZRSR2m MN patients had either U2AF1 (5.5%) or SRSF2 (4.9%)) [3, 13].
Survival among ZRSR2m MN patients was affected mostly by the MN diagnosis (AML showing worst survival) and expectedly by PB blasts >5%. Other factors that affected survival positively were higher Hgb concentration and the presence of isolated TET2m. On the contrary, presence of RUNX1m and cytogenetic abnormalities affected survival negatively.
Our study was limited by data collection from a single institution, the retrospective nature, shorter follow-up duration, small cohort size (due to gene rarity), and delayed NGS introduction in some cases. Furthermore, our focus was on clinical aspects of this mutation and lacked mechanistic insights into this disease.
In conclusion, the ZRSR2m was almost exclusively seen in males, with a striking increased frequency of CCUS patients. TET2m was the most common co-mutation and is linked to better survival especially as an isolated co-mutation. Over half of the patients with ZRSR2m MDS and CCUS had a higher absolute monocyte count indicating a possible association with monocytic differentiation. However, further studies are needed to confirm these findings.
Supplementary information
Acknowledgements
The protein diagram was generated using ProteinPaint (https://proteinpaint.stjude.org/). The Fishplots depicting clonal evolution were generated using https://github.com/chrisamiller/fishplot.
Author contributions
MY, BK, YJ, and AA planned the study, reviewed data, performed statistical analysis, and wrote the manuscript. RH and DV performed molecular analysis and reviewed the manuscript. PG performed cytogenetic analysis and reviewed the analysis. KB coordinated NGS data collection. DJ, JF, JP, AS, MH, KB, WH, MP, MS, and HA reviewed the paper and contributed patients.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Previous publications: Part of this manuscript was accepted for poster publication at the European Hematology Association Congress 2024.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02374-9.
References
- 1.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. [DOI] [PubMed] [Google Scholar]
- 2.Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119:3211–8. [DOI] [PubMed] [Google Scholar]
- 3.Thol F, Kade S, Schlarmann C, Löffeld P, Morgan M, Krauter J, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–84. [DOI] [PubMed] [Google Scholar]
- 4.Chiereghin C, Travaglino E, Zampini M, Saba E, Saitta C, Riva E, et al. The genetics of myelodysplastic syndromes: clinical relevance. Genes (Basel). 2021;12:1144. [DOI] [PMC free article] [PubMed]
- 5.AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov, 2017;7:818–31. [DOI] [PMC free article] [PubMed]
- 6.Hosono N. Genetic abnormalities and pathophysiology of MDS. Int J Clin Oncol. 2019;24:885–92. [DOI] [PubMed] [Google Scholar]
- 7.Togami K, Chung SS, Madan V, Booth CAG, Kenyon CM, Cabal-Hierro L, et al. Sex-biased ZRSR2 mutations in myeloid malignancies impair plasmacytoid dendritic cell activation and apoptosis. Cancer Discov. 2022;12:522–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Ruiz C, Martínez-Valiente C, Cordón L, Liquori A, Fernández-González R, Pericuesta E, et al. Concurrent Zrsr2 mutation and Tet2 loss promote myelodysplastic neoplasm in mice. Leukemia. 2022;36:2509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malcovati L, Papaemmanuil E, Ambaglio I, Elena C, Gallì A, Della Porta MG, et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124:1513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue D, Polaski JT, Taylor J, Castel P, Chen S, Kobayashi S, et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat Genet. 2021;53:707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor J, Mi X, North K, Binder M, Penson A, Lasho T, et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood. 2020;136:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.