Abstract
Entry of herpes simplex virus type 1 (HSV-1) into host cells occurs through fusion of the viral envelope with the plasma membrane and involves complex and poorly understood interactions between several viral and cellular proteins. One strategy for dissecting the function of this fusion machine is through the use of specific inhibitors. We identified a peptide with antiviral activity that blocks HSV-1 infection at the entry stage and during cell-to-cell spreading. This peptide (called EB for “entry blocker”) consists of the FGF4 signal sequence with an RRKK tetramer at the amino terminus to improve solubility. The activity of EB depends exclusively but not canonically on the signal sequence. Inhibition of virus entry (hrR3) and plaque formation (KOS) strongly depend on virus concentrations and serum addition, with 50% inhibitory concentrations typically ranging from 1 to 10 μM. Blocking preadsorbed virus requires higher EB concentrations. Cytotoxic effects (trypan blue exclusion) are first noted at 50 μM EB in serum-free medium and at ≥200 μM in the presence of serum. EB does not affect gC-dependent mechanisms of virus attachment and does not block virus attachment at 4°C. Instead, EB directly interacts with virions and inactivates them irreversibly without, however, disrupting their physical integrity as judged by electron microscopy. At subvirucidal concentrations, EB changes the adhesive properties of virions, causing aggregation at high virus concentrations. This peptide may be a useful tool for studying viral entry mechanisms.
Herpes simplex virus type 1 (HSV-1) is a significant human pathogen causing mucocutaneous lesions primarily in the oral mucosa but also in other sites as well. More serious manifestations of HSV-1 infection include encephalitis and blinding keratitis. Indeed, HSV-1 is the leading cause of sporadic viral encephalitis and the leading cause of blindness due to infection in the United States (65). HSV-1 also has the capacity to latently infect the host, with important consequences for transmission, treatment, and difficulties in eradication of the latent infection. Obtaining more precise information about many aspects of the viral life cycle is critical for designing prevention and intervention strategies. One critical aspect of the life cycle is the ability of HSV to enter host cells. Previous studies have shown that entry occurs at the cell surface due to fusion of the viral envelope with the plasma membrane of the cell (54). The combined processes of attachment and entry involve a number of components and require a number of steps. This complexity makes the study of these processes exceedingly difficult.
HSV encodes at least 12 glycoproteins, 10 of which are structural components of the viral envelope (54). Of these, five are clearly involved in attachment and entry. The proteins involved in attachment and entry are gC, gB, gD, and a complex of the gH and gL proteins (4, 10, 13, 21, 24, 25, 31, 34, 38, 47, 62). Of these five, four (gB, gD, gH, and gL) are essential in that their loss results in noninfectious virus. The one nonessential protein, gC, is likely to be nonessential only in cell culture. Under circumstances of a natural infection, gC is likely to be essential because of its greater receptor binding activity (22), its ability to enhance the infectivity of the virus (21), and its role in infection via apical rather than basolateral surfaces of epithelial cells (50). Other glycoproteins (e.g., gE and gI) appear to be involved in egress or cell-to-cell spreading (54).
The initial attachment of HSV to cells occurs primarily through binding of gC to heparan sulfate proteoglycans (HSPG) (5, 21, 23, 30, 33, 52, 53, 57). In the absence of gC, gB mediates binding to HSPG, but the specific infectivity is substantially reduced (21, 22). In the absence of HSPG, virus can bind to chondroitin sulfate proteoglycan with lower efficiency, but the significance of this is not clear (2). The function of the heparin binding of gB is unknown. In addition to the glycoproteins, the product of the UL45 gene is located in the envelope (7, 60) and may also be involved in regulating the functions of gB (18).
Using a variety of methods, several studies have shown that gB, gD, and gH/gL are required for fusion (reviewed in reference 54). It appears that gB, gD, and gH/gL together are necessary and sufficient to mediate membrane fusion (59), suggesting these proteins work together as a fusion machine (64). The gL protein is required for proper folding, processing, and surface expression of gH but, to date, does not appear to be specifically involved in fusion (10, 25, 47, 62). Of the fusion proteins, the potential role of gD has been characterized best. Several studies indicate that gD binds to cell surface proteins (coreceptors). These include observations that soluble gD inhibits infection (14, 40, 57) and that cells expressing gD are resistant to infection (26). Expression cloning studies have been used to identify the host cell coreceptors that interact with gD. Four different gD binding coreceptors (HveA to HveD) have been identified and appear to function in different cell types (39, 41, 63). The Prr1 (HveC) protein may be the main cell mediator of HSV-1 entry, since it is widely distributed on several cell types (15, 29).
The molecular mechanisms by which gB and gH/gL participate in entry and fusion are unknown, except that the UL45 protein may regulate the fusogenic activity of gB during entry (18). Neutralizing monoclonal antibodies to gB or gH/gL block entry but not attachment, and virions lacking these proteins bind but do not enter cells (10, 13, 24, 31, 38, 47, 54). Cross-linking studies have also shown that during entry, interactions between gB, gH, and gD occur and are in flux during the process, but the significance of these interactions is presently not clear (20).
One strategy for dissecting the functions of gB, gH, and gD in the entry process is to use specific inhibitors. Compounds with antiviral activity have proven to be useful tools for studying the functions of their targets. For example, inhibitors of HSV polymerase and thymidine kinase have been used to identify active site residues and have contributed greatly to our understanding of catalytic mechanisms. (8, 16, 19, 28, 35, 48). There are few known inhibitors of HSV attachment and entry. The majority, such as soluble heparan sulfate, primarily block attachment and have been useful in understanding this process. If specific inhibitors of fusion and entry could be identified, discovery of their targets and mechanism of inhibition would provide important details for explaining the processes involved.
In a project originally designed to determine if peptides with the ability to enter cells could be used to deliver poorly penetrating antiviral peptides, we discovered a peptide with antiviral activity, which blocks the entry and fusion steps of HSV-1 infection. This peptide consists of the 16-amino-acid FGF4 signal peptide with an RRKK tetramer attached to its amino-terminal end to improve solubility in aqueous solutions. Earlier studies had shown that the FGF4 signal peptide (32) is one of several unrelated peptides that can penetrate membranes and deliver potentially useful covalently linked molecules into cells (1, 9, 11, 43–45, 49, 58, 61). Here we show that the modified FGF4 signal peptide, called EB (for entry blocker) inhibits HSV-1 entry whereas a scrambled sequence peptide (EBX) is significantly less active. This peptide may serve as a useful tool for studying the entry and fusion process in HSV-1 infection.
MATERIALS AND METHODS
Cell culture and virus.
The procedures for growing Vero cells and preparing high-titer stocks of HSV-1 KOS have been described previously (17). Vero cells were maintained in carbonate-buffered Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% calf serum and 5% fetal bovine serum (regular medium). For some studies, cells were switched to serum-free DMEM buffered with 25 mM HEPES (pH 7.4) and allowed to adapt to that medium for 30 min prior to experimental treatments. Vero cells were seeded into wells (0.28 cm2) of microtiter plates either at 3.5 × 104 cells/well for use 1 day later (8 × 104 cells/well) or at 1 × 104 cells/well for use 3 days later (2 × 105 cells/well). The gC-null mutant HSV-1 KOSΔgC2-1 has a deletion of 900 bp of gC coding sequence between two NaeI sites with a concurrent insertion of the Escherichia coli lacZ gene. The gC+ revertant HSV-1 KOSΔgC2-3rev was generated by replacing the lacZ insertion of HSV-1 KOSΔgC2-3 with wild-type gC sequence by marker transfer. The construction and characterization of these viruses has been described previously (2, 22).
Plaque reduction assay.
Confluent Vero cell cultures in microtiter plates were infected for 1 h at 37°C in 40 μl of medium. Except where indicated, peptide treatments in 40 μl of medium lasted from 1 h before through 1 h after infection. At the end of the adsorption period, the cultures were refed with 100 μl of regular medium. Plaque formation was scored 2 days later, and the number of plaques scored per well was normalized to the number counted in the absence of peptide. Using an ocular micrometer, plaque size (π/2 × L × S) was determined by measuring the largest plaque diameter (L) and the diameter at a 90° angle to that (S). The size of each of the first 40 scored plaques was measured, except when a plaque included less than 10 rounded cells or touched the side of the well.
Yield reduction assay.
At 3 days postinfection, Vero cell cultures in microtiter plates were frozen (−80°C) and thawed (37°C) three times. The cells were suspended by repeated pipetting, and microtiter plates were centrifuged for 10 min at 700 × g in a Beckman model TJ-6 tabletop centrifuge. The virus-containing supernatant were serially diluted in regular medium and subjected to titer determination on Vero cells. Plaques were counted after the monolayers were stained with crystal violet (17).
Attachment assay.
HSV-1 KOS was labeled with [32P]orthophosphate to a specific activity of 0.01 cpm/PFU. Briefly, Vero cells were infected at a multiplicity of infection (MOI) of 5.0 and [32P]orthophosphate (0.5 mCi/ml) was added 6 h postinfection. At 18 h postinfection, the cells and culture medium were harvested separately. The cells were subjected to three freeze-thaw cycles, and cell debris was pelleted by centrifugation at 2,000 × g for 10 min. The freeze-thaw supernatant was combined with the medium, and virus was pelleted by centrifugation through a 26% sucrose gradient cushion (60). The viral pellet was resuspended in phosphate-buffered saline (PBS) for use. Confluent Vero cell cultures in microtiter plates were switched to serum-free DMEM, chilled on ice, and maintained at 4°C. After 30 min, peptides were added, and 60 min later the cells were incubated for 2 h with 32P-labeled virus (2 × 104 cpm/well). After labeling, the cells were rinsed with ice-cold medium. Bound 32P was then quantitatively extracted with 1% sodium dodecyl sulfate–1% Triton X-100 in PBS and counted in a Beckman LS5801 liquid scintillation counter.
lacZ+ virus (hrR3) entry assay.
Confluent Vero cell cultures in 96-well microtiter plates were switched to HEPES-buffered serum-free DMEM, cooled on ice to 4°C for 30 min, and infected with hrR3 for 1 h at 4°C in 40 μl of medium. Unattached virus was removed by rinsing the mixture with ice-cold medium. Treatments with EB or EBX (Table 1) or mock treatments with peptide-free medium were carried out in serum-free DMEM as indicated. Virus entry was initiated by transferring cultures to 37°C. To inactivate any remaining extracellular virus, the cultures were rinsed with PBS and exposed to low-pH citrate buffer (40 mM citric acid, 10 mM KCl, 135 mM NaCl [pH 3.0]) (24) for 1 min at 23°C. The citrate was rinsed off with PBS, and cultures were maintained in serum-supplemented DMEM until they were fixed with 0.5% gluteraldehyde in 5× PBS for 30 min at 23°C; stained for β-galactosidase activity for 1 h or overnight at 23°C with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Fisher Biotech; BP1615-1) in 1× PBS containing 2 μM MgCl2, 1.3 mM K4Fe(CN)6, and 1.3 mM K3Fe(CN)6; and scored for the presence of blue lacZ+ cells.
TABLE 1.
Synthetic peptides tested for antiviral activity
| Peptide | Sequence | Purity (%) | Molecular mass |
|---|---|---|---|
| EB | RRKKAAVALLPAVLLALLAP | 95 | 2,085 |
| EBX | RRKKLAALPLVLAAPLAVLA | 93 | 2,085 |
| EBPP | RRKKAAVALLAVLLALLAPP | 91 | 2,085 |
| RRKK | RRKK | —a | 587 |
—, not retained on high-pressure liquid chromatography column.
Virucidal assay.
hrR3 (1.2 × 106 PFU/ml) was incubated with various concentrations of EB or EBX for 1 h at 37°C in 70 μl of serum-free DMEM (pH 7.4). The treated virus was diluted 200-fold with serum-supplemented DMEM and assayed for infectivity approximately 1 h later in microtiter wells seeded with Vero cells (3.5 × 104 cells/well) 1 day earlier. Then 40- or 100-μl volumes of diluted virus were adsorbed for 1 or 2 h at 37°C, and lacZ+ cells were scored 8 h later. In some experiments, aliquots of diluted virus were first dialyzed (Spectra/Por; molecular weight cutoff, 12,000 to 14,000) overnight at 4°C against a 60-fold excess volume of HEPES-buffered serum-supplemented DMEM or forced by syringe through 0.22-μm-pore-size membranes (Millex-GV; Millipore) before the remaining infectious virus was assayed.
Trypan blue exclusion assay.
Uninfected Vero cells in serum-free or serum-supplemented DMEM were treated for 1 h at 37°C with EB or EBX, rinsed with PBS, stained for 5 min at 23°C with 0.4% trypan blue in PBS, rinsed again with PBS, and air dried.
Electron microscopy.
Purified HSV-1 KOS virions (2.5 × 107 PFU/ml) (60) were treated for 5 to 60 min at 4 or 23°C with 25 μM EB or the control peptide (EBX) in 40 μl of serum-free DMEM buffered with 25 mM HEPES (pH 7.4). Aliquots (10 μl) were adsorbed to pioloform poly-l-lysine-coated grids for 5 min at 23°C. The grids were rinsed with PBS, stained with 2% phosphotungstic acid (PTA) in water adjusted to pH ≈ 6, and air dried. Alternatively, virus was preadsorbed to grids and treated with peptides thereafter. A total of 4 × 109 PFU of purified HSV-1 KOS per ml in 5 μl of PBS was applied to the coated grids for 5 min at 23°C, and the grids were rinsed once with serum-free DMEM buffered with 25 mM HEPES (pH 7.4) and treated with 15 μl of 5 mM EB or EBX in the same medium for 30 min at 37°C. The pH of highly concentrated solutions of EB and EBX was readjusted to 7.4 with NaOH prior to use. To prevent evaporation of the peptide-containing solutions, each grid was held in a Hiraoka flexible staining plate and covered with a miniature bell jar made from a 0.5-ml polypropylene microcentrifuge tube, small enough for the 15 μl to fill half of the bell jar facing the coated surface of the grid. The entire assembly was then incubated in a moist chamber for 30 min at 37°C. After treatment, grids were rinsed twice with DMEM and once with PBS before they were stained with PTA and dried. Grids were examined in a JEOL JEM-1200EX electron microscope at magnifications of ×15,000 and ×40,000.
Peptide synthesis.
Synthesis and analysis of peptides were done at the Biotechnology Center of the University of Wisconsin—Madison. Synthesis was carried out at a 25-pmol scale using an automated synthesizer (Applied Biosystems model 432A “Synergy”) following the principles initially described by Merrifield (37) with modifications by Meienhofer et al. (36) and Fields et al. (12). The cleaved peptides were precipitated with cold t-butylmethylether, dissolved in water, and examined by analytical high-pressure liquid chromatography (purity) and electrospray ionization mass spectroscopy (molecular mass) (Table 1). Peptide concentrations in solution were determined from absorbance readings at 215 and 225 nm (51).
RESULTS
Antiviral activity of peptides.
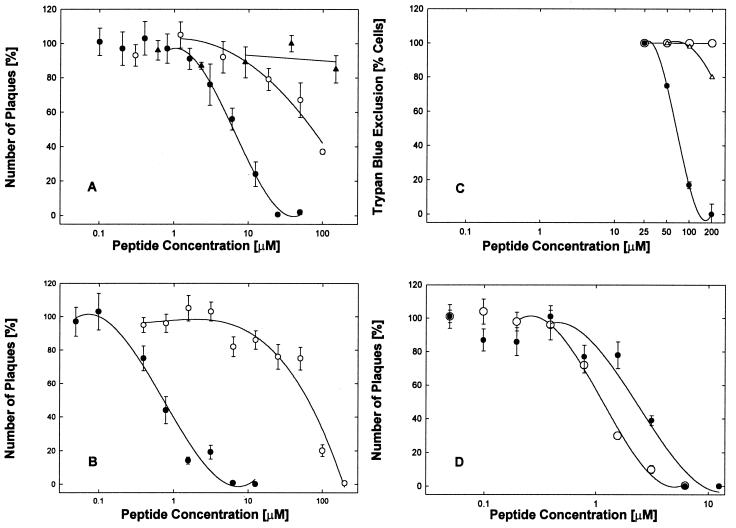
The EB peptide, consisting of the RRKK tetrapeptide attached to the FGF4 signal sequence (sequence data are given in Table 1) was an effective antiviral agent when present during infection of Vero cell cultures with HSV-1 KOS, blocking plaque formation (Fig. 1A and B, ●) and reducing virus yields by up to 8 orders of magnitude depending on the concentration (see below). Compared to the EBX peptide (Fig. 1A and B, ○), in which the RRKK tetrapeptide was attached to a scrambled version of the signal sequence, the EB peptide was a far more effective antiviral, blocking infections at 10- or 100-fold-lower concentrations depending on the presence (Fig. 1A) or absence (Fig. 1B) of serum.
FIG. 1.
Specific antiviral and cytotoxic effects of EB and related peptides. (A and B) Antiviral activities of EB (●), EBX (○), and RRKK (▴) were compared in serum-supplemented (A) or serum-free (B) DMEM using cell cultures (2 × 105 cells/well) infected with 4000 PFU of HSV-1 KOS per well and scored for plaque formation 2 days later (control scores; 25 ± 2.5 plaques/well; n ≥ 3). Note that the effectiveness of the EB peptide (●) strongly depended on the presence of serum (IC50 = 6.4 μM [A] versus IC50 = 0.7 μM [B]) whereas that of the EBX peptide (○) did not (IC50 = 77 [A] versus IC50 = 64 [B]). (C) Cytotoxic effects were measured in uninfected cells. In serum-free medium, EB inhibited trypan blue exclusion in 50% of the cells at 68 μM (●) whereas EBX had no effect (○). In serum-supplemented medium, nearly all the cytotoxic effects of EB were alleviated (Δ). (D) Antiviral activities of EB (IC50 = 2.1 μM [●]) and EBPP (IC50 = 1.1 μM [○]) were compared in serum-free DMEM in cultures (2 × 105 cells/well) infected with 5,000 PFU of HSV-1 KOS per well. Control scores were 35 ± 3.6 plaques/well (n = 4). All points are means of three to six determinations with standard errors of the means.
The cytotoxic effects of EB, as measured by trypan blue exclusion in the absence of serum, were seen only at concentrations 100-fold higher (Fig. 1C, ●; 50% inhibitory concentration [IC50] = 68 μM) than antiviral concentrations (Fig. 1B, ●; IC50 = 0.7 μM). In the presence of serum, cytotoxic effects were seen first at 200 μM EB (Fig. 1C, Δ). No cytotoxic effects were associated with the EBX peptide (Fig. 1C, ○).
The charged amino-terminal RRKK tetramer is critical for enhancing the solubility of the otherwise hydrophobic EB peptide but does not have any important antiviral activity by itself. In the presence of serum, no antiviral activity was associated with the free RRKK tetramer at concentrations as high as 200 μM (Fig. 1A, ▴). In separate experiments, we found that free RRKK tetramer inhibited hrR3 infection of Vero cells under serum-free conditions at an IC50 of 1.3 mM (data not shown). We also found that high (up to 100-fold molar excess) but nonantiviral concentrations of the free RRKK peptide did not compete with EB activity and could not relieve the inhibition of hrR3 infections due to EB (data not shown). Together with the results obtained with the scrambled EBX control peptide, we conclude that the antiviral activity of EB is associated primarily, if not exclusively, with the FGF4 signal peptide in a sequence-specific manner.
To inquire whether the FGF4 sequence per se was important for antiviral activity, we tested a modified EB peptide in which the central proline residue was moved to the carboxy-terminal end. This EBPP peptide (Table 1) was twice as active as the original EB peptide in both plaque (Fig. 1D) and yield (data not shown) reduction assays. Since all other residues (except for proline) are compatible with α-helicity (6), this suggests that a more compact α-helical (EBPP) or partially α-helical (EB) peptide is more efficient than a peptide with two (EBX) internal proline residues.
EB acts early in the viral life cycle.
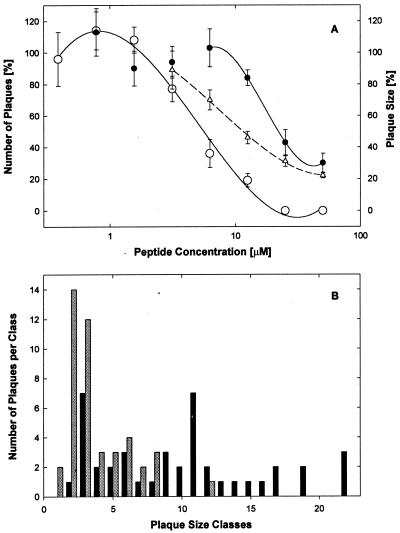
To determine whether EB acted before or after attachment and entry, we studied the effect of varying the time of peptide addition. As shown in Fig. 2A, EB was substantially more effective when present during infection and 1 h pre- and postinfection than when present continuously starting 1 hour postinfection (IC50 = 5.5 μM [○] and 24 μM [●], respectively). Furthermore, when present before and during adsorption, EB had no effect on plaque size. When the EB peptide was present continuously after infection, plaque expansion was inhibited in a dose-dependent manner (Fig. 2A, Δ; IC50 = 12 μM). To ensure that individual plaques were measured reliably, cell cultures were infected at very low multiplicity (MOI < 0.01) and plaque sizes were measured microscopically very early (1 day postinfection). As shown in Fig. 2B, in untreated control wells, the plaque size was broadly distributed (black bars; mean: 66,000 ± 6,200 μm2) whereas addition of increasing concentrations of EB 1 h postinfection progressively shifted the distribution toward smaller size classes (e.g., 25 μM EB significantly reduced the mean plaque size by 70% to 6,900 ± 2,600 μm2; t = 6.88; shaded bars). In contrast, the presence of EB up to 1 h postinfection had no effect on plaque size, even though the number of plaques was severely reduced compared to that found for postinfection treatment. Thus, the combined mean plaque size after transient treatments with 6 and 12 μM EB (68,000 ± 11,000 μm2) was indistinguishable from the controls. In conclusion, EB appeared to act at an early stage of viral infection and reduced the plaque size when added after infection.
FIG. 2.
Postinfection treatment with EB blocks viral cell-to-cell spreading. (A) Cell cultures (8 × 104 cells/well) in serum-supplemented DMEM were infected with HSV-1 KOS (700 PFU/well). EB either was added 1 h postinfection and remained present throughout the experiment (● and Δ) or was present from 1 h before until 1 h after infection (○). One day later, the cells were fixed in 10% formaldehyde in PBS and the number of plaques were scored (● and ○; control score, 13 ± 1.7; [n = 6]) and their size was determined (Δ). All data points are means of triplicate measurements with standard errors of the means. (B) The distribution of plaque size in untreated controls (black bars) and in cultures treated postinfection with 25 μM EB (shaded bars) is shown.
Pretreatment of cells with EB does not inhibit infection.
To determine whether EB also blocked infection by interacting with cell surface components of host cells, we examined the effects of pretreating cells with EB on subsequent infections with the lacZ expressing hrR3 virus. To avoid interactions of EB with serum components, pretreatments of cells were done in serum-free medium. In the first experiment, cells were pretreated with various concentrations of EB, rinsed to remove the peptide, and then infected with hrR3 in the absence of EB. lacZ+ cells were scored 8 h later. As shown in Fig. 3 (▴ and Δ), pretreatments of cells with EB inhibited subsequent virus infections in the absence of EB, with an apparent IC50 = 29 μM irrespective of the presence (▴) or absence (Δ) of serum during rinses and infections. These results are consistent with the notion that cellular interactions of EB contribute to antiviral activity. However, interpretation of the data is confounded by cytotoxic effects of EB at concentrations ≥50 μM (Fig. 1C, ●). Only the 20 to 40% inhibition seen at 25 μM EB could possibly be attributed to any specific cell-mediated antiviral activity.
FIG. 3.
Effect of pretreating cells with EB on subsequent virus infection. Cell cultures (2 × 105 cells/well) in microtiter wells were switched to serum-free HEPES-buffered DMEM and incubated for 30 min at 37°C. A first set of cells were then pretreated for 1 h with EB, rinsed three times, and infected for 1 h with 7,400 PFU of hrR3 per well before they were returned to regular medium. Rinses and infections were carried out in the absence of EB in either serum-free (Δ) or serum-supplemented (▴) DMEM. A second set of cells was pretreated for 1 h with (●) or without (○) EB and infected for 1 h with 7,400 PFU of hrR3 per well in the presence of EB before they were returned to regular medium. Triplicate counts of lacZ+ cells were performed 8 h postinfection (all points are means with standard errors of the means; control score, 265 ± 13 [n = 3]).
To avoid the problem of cytotoxicity, we reexamined the effects of cellular pretreatments with EB in cultures that were subsequently infected with hrR3 in the presence of various EB concentrations. In this second experiment, both, pretreatments and infections were done in serum-free medium. As shown in Fig. 3 (○), the presence of EB during infection alone shifted the IC50 to 7 μM. The additional pretreatment of cells with EB, however, was completely ineffective (Fig. 3, ●), indicating that interactions of EB with cells just prior to infection could at best make only a minor contribution to the antiviral activity of the peptide.
EB does not block virus adsorption.
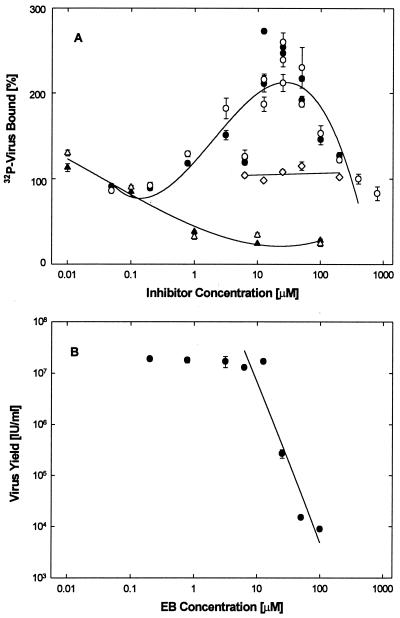
To measure the effect of EB on adsorption, the binding of 32P-labeled virus to live or formaldehyde-fixed cells was measured in binding assays carried out at 4°C essentially as we described previously (2, 22). As expected (22, 67), soluble heparin inhibited virus adsorption by as much as 75% in this assay (Fig. 4A, Δ and ▴). EB, however, had no inhibitory effect. Instead, it actually enhanced binding, maximally about 2.5-fold, at concentrations between 10 and 50 μM (Fig. 4A, ○ and ●), although in comparable assays viral yields were severely reduced at these concentrations of EB (Fig. 5B). The EBX peptide neither inhibited nor enhanced 32P-labeled virus binding to cells (Fig. 4A, ◊).
FIG. 4.
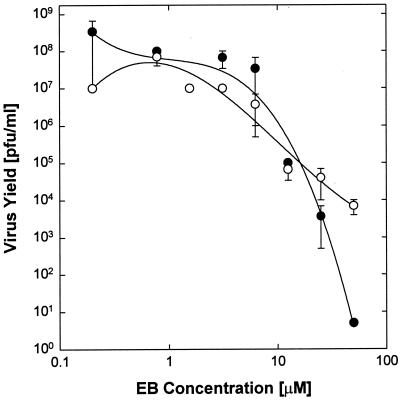
Effect of EB on virus adsorption. (A) Live (solid symbols) and formaldehyde-fixed (open symbols) cells (2 × 105 cells/well) were exposed to 32P-labeled HSV-1 KOS (2 × 104 cpm/well; 0.01 cpm/PFU) in the presence of EB (● and ○), EBX (◊), or heparin (▴ and Δ), and the bound labeled was extracted and counted (100% corresponds to ∼1.8 × 103 cpm/well). (B) Under identical conditions, cells were also infected with unlabeled HSV-1 KOS (2 × 106 PFU/well) and then returned to regular medium for 3 days before the effect of EB on virus yields was measured. Inhibition of virus production (IC50 ≈ 10 μM) coincides with maximal EB-induced virus binding (10 to 50 μM). All data points in are means of triplicate measurements with standard errors of the means (error bars are mostly smaller than the symbols).
FIG. 5.
EB inhibits gC-deficient virus. Cell cultures (8 × 104 cells/well) in serum-supplemented DMEM were infected for 1 h with 4.7 × 103 PFU of the gC-deficient HSV-1 KOSΔgC2-3 per well (○) or 3.5 × 103 PFU of a wild-type revertant, HSV-1 KOSΔgC2-3 rev per well (●), and viral yields were measured 3 days later. EB was added to the cultures 1 h prior to infection and remained present throughout the experiment. All data points represent means of triplicate measurements with standard errors of the means.
Since the initial attachment of HSV-1 to cells occurs by gC binding to heparan sulfate on the cell surface, we measured the antiviral activity of EB on a gC-null virus (HSV-1 KOSΔgC2-3) previously constructed in our laboratory (2, 22). A revertant, HSV-1 KOSΔgC2-3rev, served as a control. As shown in Fig. 5, with increasing concentration of EB, the yield reductions of the gC mutant and the revertant were essentially the same (IC50 ≈ 5 μM) and comparable to the results obtained in plaque reduction assays with wild-type virus (Fig. 1A, ●). It seems clear, therefore, that EB does not require gC for blocking infection, which supports the general conclusion that EB does not target virus adsorption to exert its inhibitory effect.
EB can aggregate virus.
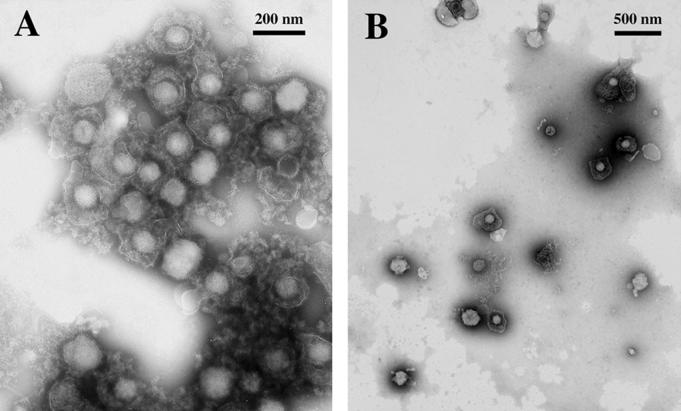
To test whether the enhancing effects of EB on virus binding were due to aggregation of virus, we examined EB-treated virions by electron microscopy. When purified virus particles at high concentrations, as required for efficient visualization, were incubated with 25 μM EB, adsorbed to coated grids, and stained with PTA, nearly all of the particles were seen in relatively few large aggregates (Fig. 6A). In contrast, untreated virus (data not shown) or virus particles treated with 25 μM EBX were nearly all found individually and uniformly scattered over the grid surface (Fig. 6B). The individual PTA-stained virus particles within aggregates were virtually indistinguishable from control particles, indicating that EB did not induce gross structural abnormalities in the virus particles. The EB-induced aggregates were formed rapidly (<5 min) at room temperature as well as at 4°C (data not shown).
FIG. 6.
EB promotes virus aggregation. Purified HSV-1 KOS (2.5 × 107 PFU) was exposed to 25 μM EB (A) or EBX (B) for 1 h at 23°C in serum-free DMEM. Aliquots (10 μl) were applied to coated grids, and the grids were stained with PTA. The electron micrographs compare EB-treated aggregated virus (A) with nonaggregated EBX-treated virus (B) particles.
The antiviral activity of EB varies with virus input and is distinct from aggregation effects.
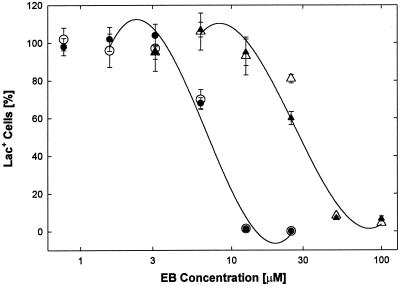
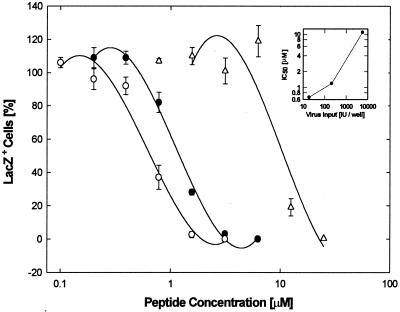
Initial results with wild-type virus had shown that the antiviral activity of EB was strongly dependent on the virus titer used for infection (data not shown). To further examine the effect of viral input, cultures were infected with hrR3 at inputs of 19,210, and 5,700 PFU/well in the presence of various concentrations of EB and scored 8 h later for lacZ+ cells; the IC50s obtained were 0.66, 1.2, and 11 μM, respectively (Fig. 7).
FIG. 7.
The antiviral activity of EB depends on virus input. Cell cultures (2 × 105 cells/well) were switched to serum-free DMEM and infected with 19 (○), 210 (●), or 5,700 PFU (Δ) of hrR3 per well EB was present from 1 h before through 1 h after infection. lacZ+ cells were scored 8 h postinfection. All data points are means of triplicate measurements with standard errors of the means. The insert shows the dependence of IC50s (0.66, 1.2, and 11 μM, respectively) on the virus inputs.
Significantly, above the intermediate input of 210 PFU/well, there was a greater increase in the IC50 with increasing virus titer than below that input (inset in Fig. 7). The inverse relationship between IC50 and virus titer would be expected if EB merely acted as an aggregation agent, which should operate more efficiently, i.e., with lower IC50, at the higher virus input. We conclude, therefore, that viral aggregation does not make any major contribution to the antiviral activity of EB in these experiments. Furthermore, the fact that the antiviral activity of EB strongly depended on the virus concentrations suggests that the EB peptide interacts with viral components.
EB inhibits viral entry.
Additional studies with preadsorbed hrR3 virus demonstrated that the antiviral effect or effects of EB are related neither to virus adsorption nor to virus aggregation but, rather, to inhibition of virus entry. In these studies, the hrR3 virus was preadsorbed to cells for 1 h at 4°C before ice-cold 25 μM EB or EBX was added in serum-free DMEM. After an additional 1 h at 4°C, the cultures were shifted to 37°C to initiate virus entry. At 15-min intervals following the temperature shift, any virus remaining outside the cells was inactivated by washing the cultures with low-pH citrate buffer. The cultures were then rinsed and returned to peptide-free serum-supplemented DMEM until they were fixed and stained for β-galactosidase 8 h after the temperature shift.
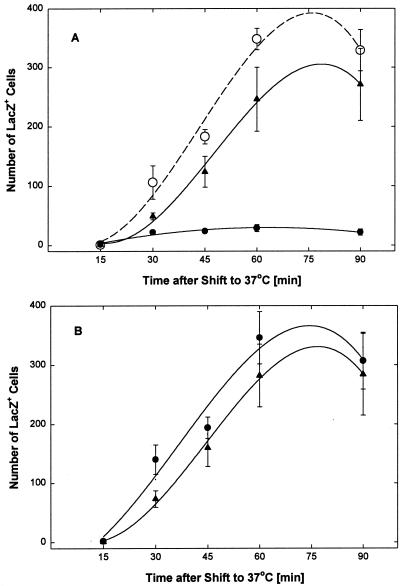
As shown in Fig. 8A, virus entry in mock-treated control cultures (○) was initiated 15 to 30 min after transfer to 37°C and was completed by about 60 min at a level of about 340 lacZ+ cells per 6.5 mm2 (or 1,450 lacZ+ cells/well). In cultures treated with the EB peptide, the number of lacZ+ cells was reduced by >90% (●). The EBX peptide did not significantly reduce the number of lacZ+ cells (▴). Essentially the same results were obtained when EB and EBX were added prior to virus adsorption (data not shown). When peptide was added immediately after each citrate treatment, EB no longer had any effect on the development of lacZ+ cells (Fig. 8B, ●; cf. Fig. 8A, ○). EBX also did not significantly inhibit the development of lacZ+ cells when added immediately after the citrate treatments (Fig. 8B, ▴). Thus, EB had no effect on the expression of the lacZ gene from the early ICP6 promoter but selectively blocked viral entry.
FIG. 8.
EB blocks virus entry. Virus (2.6 × 103 PFU of hrR3 per well) was preadsorbed for 1 h at 4°C to Vero cells (2 × 105 cells/well in 96-well microtiter strip plates), and the cultures were switched to ice-cold serum-free DMEM and kept for an additional 1 h at 4°C before they were transferred to 37°C to initiate virus entry. Every 15 min following the temperature switch, strips of wells were treated for 1 min with low-pH citrate buffer to inactivate any remaining extracellular virus. After each citrate treatment, cultures were returned to serum-supplemented medium. (A) EB (●) or EBX (▴) at 25 μM was added immediately after preadsorption of the virus and remained present until the citrate treatment. Mock-treated controls were kept in peptide-free medium (○). (B) EB (●) or EBX (▴) at 50 μM was added immediately after each citrate treatment and remained present until all cultures were fixed and stained for β-galactosidase 8 h after the temperature shift to 37°C. Blue cells in areas of 6.5 mm2 were counted in triplicate wells. All data points are means with standard errors of the means.
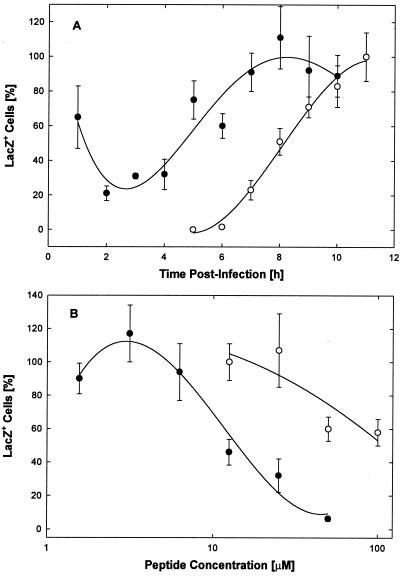
This conclusion is strengthened by the finding that the EB-sensitive phase of infection with preadsorbed virus clearly precedes the expression of lacZ genes in hrR3-infected cells (Fig. 9A). Again, hrR3 was preadsorbed to cells for 1 h at 4°C, unattached virus was rinsed off, and the cells were kept for an additional 1 h at 4°C. Cultures were then transferred to 23°C for 30 min before they were switched to 37°C. The more gradual change to 37°C allowed cell layers to remain intact through subsequent frequent medium changes. Immediately following viral adsorption, cells were treated with 50 μM EB for 1-h periods at consecutive 1-h intervals. Between 1 and 4 h postinfection, virus entry was inhibited by 70 to 80%. Thereafter, infection was no longer significantly inhibited (Fig. 9B, ●). Parallel cultures were immediately fixed after mock treatments and stained with X-Gal. In these cultures, blue (lacZ+) cells first appeared 7 h postinfection and their number increased nearly linearly for the next 3 h (Fig. 9A, ○). By 7 h postinfection, EB ceased to be inhibitory. Thus, EB blocked virus entry only during an early brief sensitive period and had no effect on the expression of the lacZ gene and the development of β-galactosidase activity once the virus had entered the cell. As shown in Fig. 9B, EB inhibited the entry of preadsorbed virus in a dose-dependent manner with an IC50 = 15 μM (●), whereas EBX was less effective (IC50 ≈ 100 μM; ○).
FIG. 9.
Entry phase and dose response. (A) Cell cultures (2 × 105 cells/well) in 96-well strip plates were switched to HEPES-buffered serum-supplemented DMEM, cooled on ice for 30 min, and infected at 4°C with 6 × 103 PFU of hrR3 per well. Cells were kept at 4°C for an additional 1 h before they were warmed to 23°C for 30 min and 37°C for the rest of the experiment. At 1-h intervals immediately following infection, strips of wells were treated for 1-h periods with 50 μM EB in serum-free DMEM and returned to regular medium. Before and after each treatment, the cells were rinsed three times with serum-free DMEM and serum-supplemented DMEM, respectively. The number of lacZ+ cells was scored 11 h postinfection (●) and normalized to the number counted in mock-treated controls (maximally 187 ± 20 [n = 3]). Separate strips of infected control cells were fixed and stained immediately following each mock treatment to monitor β-galactosidase expression over time (○). Data points represent means of triplicate measurements with standard errors of the means. (B) Cell cultures (2 × 105 cells/well) were switched to serum-free DMEM, cooled on ice, and infected at 4°C with 2.4 × 103 PFU of hrR3 per well. After infection, the cells were rinsed and treated with EB (●) or EBX (○) for 1 h at 4°C and for additional 30-min periods at 23 and 37°C. Triplicate counts of lacZ+ cells were performed 6 h later (all points are means and standard errors of means; control score, 141 ± 5.9).
Virucidal effects of EB.
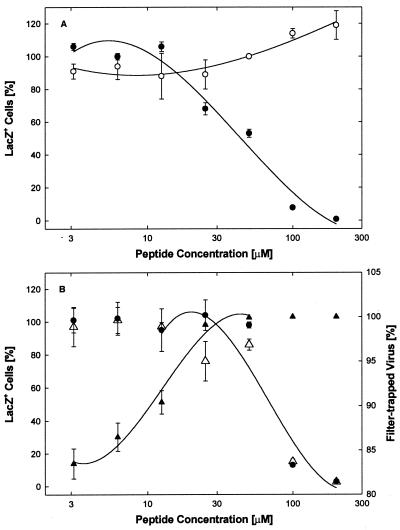
To determine if binding of EB to virus particles leads to irreversible virus inactivation, routine virucidal assays were performed with hrR3. In the first experiment (Fig. 10A), EB inhibited the infectivity of virions in a concentration-dependent manner with an IC50 of 44 μM (●) whereas EBX had no inhibitory effect ( ). In the second experiment, in which slightly higher concentrations of EB were required to achieve inhibition (Fig. 10B, ●; IC50 = 69 μM), we also found that the treated virions were irreversibly inactivated. That is, aliquots of EB-treated and then diluted virions could not be reactivated during overnight dialysis against serum-containing medium that could have trapped any reversibly bound EB (cf. Fig. 1A and B, ●). Instead, virions recovered after dialysis (31% at any EB concentration) remained inactivated, exactly like the nondialyzed controls (Fig. 10B, Δ and ●).
FIG. 10.
Virucidal assay. (A) hrR3 virus (1.2 × 106 PFU/ml) was treated with EB (●) or EBX (○) at the indicated concentrations for 1 h at 37°C in serum-free DMEM, diluted 200-fold with serum-supplemented DMEM, and assayed for remaining infectious virus. (B) hrR3 virus was again treated with EB and assayed exactly as in panel A. In addition, aliquots of diluted virus were dialyzed overnight at 4°C against serum-supplemented DMEM (Δ) or filtered through 0.22-μm-pore-size membranes (▴) before they were assayed for remaining infectious virus. Virions recovered after dialysis (31 ± 1.8% [n = 8]), irrespective of the EB concentration, remained inactivated exactly as in nondialyzed controls (Δ, normalized to untreated control [left scale]). Retention of virions on membranes depended on EB concentrations (▴, percentage of controls [right scale]). Triplicate counts of lacZ+ cells were performed 8 h after infection. All points are means with standard errors of the means. Scores of untreated controls were 1,245 ±11 (n = 3) (A) and 1,876 ±67 (n = 3) (B).
To assess possible contributions of viral aggregation to viral inactivation, additional aliquots of EB-treated and subsequently diluted virions were filtered through 0.22-μm-pore-size membranes before they were assayed for remaining infectivity. In the absence of or at low concentrations of EB (≤3 μM), 80 to 85% of the virions were trapped on the membranes. The remaining virions, however, were retained only once they were exposed to higher EB concentrations, which enhanced membrane adhesion and/or caused viral aggregation (Fig. 10B, ▴). Such changes in the adhesive properties of virions were induced well below the EB concentrations required for virus inactivation (Fig. 10B, ▴ versus ● and Δ; also cf. Fig. 4A and B, ●).
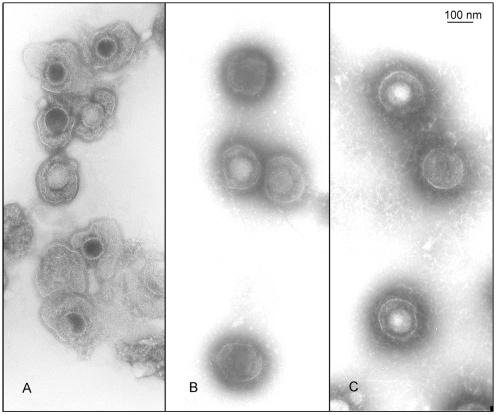
The effects of the most severe EB treatments were examined by electron microscopy of PTA-stained virions that had been preadsorbed to grids (to avoid aggregation) and exposed to 5 mM peptide. The EB-treated virions (Fig. 11A) looked essentially the same as mock-treated virions (Fig. 11B), except that the contours of the viral envelopes in the EB-treated particles were less pleomorphic, suggesting that EB stabilized the virions. At 5 mM, EBX had the same effect as EB (Fig. 11C). No attempt was made to find peptide concentrations at which the effects of EB and EBX on virion structure might be distinguishable.
FIG. 11.
Structural integrity of peptide-treated virions. Purified HSV-1 KOS (5 μl of 4 × 109 PFU/ml) was preadsorbed to grids under conditions allowing 28% of the virus to bind. The adsorbed virus was then treated for 30 min at 37°C with 15 μM serum-free DMEM alone (A) or with 5 mM EB (B) or 5 mM EBX (C) dissolved in the same medium. Electron micrographs of PTA-stained material are shown at the same magnification.
DISCUSSION
We report that the FGF4 signal peptide, with an RRKK tetramer attached to its amino terminus to improve solubility, inhibits both the entry and cell-to-cell spreading of HSV-1 and that the inhibition depends largely or entirely on direct irreversible interaction of EB with virions. The conclusion that the EB peptide acts to inhibit viral entry is supported by several findings. (i) Most important, EB specifically blocked infection by preadsorbed virus prior to inactivation of extracellular virus by treatment with low-pH citrate buffer but had no effect after such inactivation. (ii) EB blocked infection by preadsorbed virus at a stage that clearly preceded lacZ gene expression from the early ICP6 promoter in hrR3-infected cells. (iii) When added after infection, EB did not inhibit immediate-early promoters or early-gene expression from the ICP6 promoter, nor did it inhibit β-galactosidase activity in hrR3-infected cells. (iv) EB did not block attachment of virus to cells, as indicated by the fact that gC− and gC+ viruses were inhibited equally well and by the finding that binding of radiolabeled virus at 4°C was not inhibited. (v) In studies varying the time of peptide exposure, EB inhibited infection most efficiently when present during the adsorption period, indicating that it acted early in the life cycle.
The finding that EB was less effective in blocking infection by preadsorbed rather than free virus is also consistent with the conclusion that EB blocks the entry step. When corrected for different virus inputs, the IC50 of EB obtained for preadsorbed hrR3 (15 μM; Fig. 8B, ●) was approximately 10 times higher than expected on the basis of the overall antiviral activity of EB measured under comparable conditions, notably, in the absence of serum (IC50 = 1.2 μM; Fig. 7, ●). The hrR3 virus had been preadsorbed at an input of 2,400 PFU/well. However, since maximally only about 10% of the virus was bound to cells at 4°C (see below), the concentration of infectious virus was closer to 240 PFU/well, comparable to the 210 PFU/well used in the routine antiviral assay. Thus, higher concentrations of EB appear to be required to block the entry of preadsorbed virus than to inhibit infection by free virus. This seems reasonable, if only because EB must act more rapidly and binding sites may be less accessible in preadsorbed than in free virions.
The finding that EB reduced plaque size when added after infection (Fig. 2) could be due either to EB blocking secondary infections of neighboring cells or to EB blocking cell-to-cell spreading. Since the infection was allowed to proceed for only 24 h, there was little time for plaques to develop from secondary infections, suggesting that EB primarily blocked cell-to-cell spreading. The mechanism by which EB might block infection at this late stage in the viral life cycle is not known, but it seems reasonable to suggest that it involves shared functions of the virus entry and spreading (see below).
Our studies testing the attachment of radiolabeled virus to cells at 4°C showed that at EB concentrations from 10 to 50 μM, attachment was actually enhanced. Analysis by electron microscopy confirmed that EB was capable of inducing aggregation (Fig. 6), suggesting that enhancement of binding of the labeled viruses was due to aggregation. Presumably, attachment was artificially enhanced under the conditions of the binding assay, because, for technical reasons, we had to infect cell cultures with 2 × 106 PFU/well (MOI ≈ 10) rather than the ≤6,000 PFU/well (MOI ≤ 0.03) used in our antiviral assays. Infection with 2 × 106 PFU of 32P-labeled virus per well was required because the detection of binding was limited by the specific activity of the virus preparation (0.01 cpm/PFU), as well as by the efficiency of virus adsorption (live and fixed cells bound 9.7 and 8.9% of the input counts, respectively).
Some of the antiviral activity of EB may well be an artifact of viral aggregation. However, even under conditions of high virus input, aggregation would seem to play only a minor role in virus inactivation, because neither enhanced virus binding (Fig. 4) nor enhanced virus retention on filters (Fig. 10B) was closely coupled to any loss of infectivity. These findings are most readily explained by the assumption that EB-induced changes in the adhesive properties of virions lead not only to self-adhesion (aggregation) but also to enhanced cell adhesion. Such artificial cell attachment could compensate for any loss of infectivity due to virus aggregation. In any case, it seems clear that EB-induced changes in the adhesive properties of virons are merely a prelude to final virus inactivation. For unknown reasons, EB seems to enhance virus adhesion only within an intermediate concentration range (1 to 50 μM) but not at higher concentrations (Fig. 4A).
The fact that EB can inactivate preadsorbed virus clearly establishes that the antiviral activity of EB depends neither on virus adsorption nor on virus aggregation. Instead, the virucidal assays show that EB can directly interact with virions and inactivate them irreversibly (Fig. 10B, ● and Δ). Interactions of EB with host cells seem to make little or no contribution to viral inactivation (Fig. 3). Whether inactivation is due to irreversible binding of EB or to irreversible modifications of virions is not known. The electron micrographs of preadsorbed PTA-stained virus leave no doubt, however, that even at a concentration as high as 5 mM, EB does not cause any gross structural abnormalities such as the collapse of the viral envelope, stripping of the envelope, or disruption of the nucleocapsid (Fig. 11). Instead, the images suggest that EB may actually stabilize viral envelopes and render them more resistant to surface denaturation and dehydration during specimen preparation. Conceivably, such changes in the viral envelope may interfere with proper membrane fusion during infection.
The cytotoxicity of EB for host cells seems to be unrelated to inactivation of virions, since EB could cause aggregation (Fig. 10) or inactivation (Fig. 6) of virions not only at room temperature and at 37°C but also at 4°C. In contrast, the loss of cellular trypan blue exclusion due to EB (cf. Fig. 1C, ●) was highly temperature dependent between 18 and 28°C. Below 18°C, at concentrations up to 100 μM, EB had no effect on trypan blue exclusion, and above 28°C, the level of cytotoxicity was no longer enhanced as the temperature was raised to 37°C (unpublished results). It would appear, therefore, that the viral and cellular effects of EB involve quite different mechanisms.
Compared to cells (IC50 = 68 μM for the loss of trypan blue exclusion in serum-free medium [Fig. 1C, ●]), virions (hrR3 in serum-free medium) could be inactivated at the same or at 100-fold-lower EB concentrations (IC50 = 69 μM [Fig. 10B, ● and Δ] versus IC50 = 0.66 μM [Fig. 7, ○]) depending on the virus concentration (1.2 × 106 and 475 PFU/ml, respectively). Given such a wide range of EB effects, the therapeutic index is clearly of limited value. Still, it seems useful to point out that the therapeutic index for EB may be as large as 100.
Our results are consistent with three possible mechanisms of action: (i) EB could interact with the lipid bilayer of the envelope; (ii) EB could interact with one or more envelope proteins; or (iii) EB could interact with some combination of the above. Studies with the free RRKK tetramer and the scrambled EBX peptide (Fig. 1A and B) indicate that the antiviral activity of EB resides primarily, if not exclusively, with the FGF4 signal sequence. The finding that the EBPP peptide is twice as active as the EB peptide (Fig. 1D) implies that the entire FGF4 signal sequence is not a canonical requirement and that some secondary-structure features may play a role in antiviral activity. In any case, the fact that some aspect of the FGF4 signal sequence is important for antiviral activity and that signal sequences are membrane interactive invites the speculation that EB may act on the lipid bilayer of the viral envelope. It should be noted, however, that addition of four positively charged residues (RRKK) at the amino terminus may have substantially altered any lipid-interactive properties of the signal sequence. The membrane-transiting properties of the FGF4 leader may also have been altered by the RRKK addition.
Alternatively, EB may block entry by interacting with one or more glycoproteins on the virus envelope. Since in addition to inhibiting entry, EB may inhibit cell-to-cell spreading, it could block a common step in both processes. Initial entry through the plasma membrane and cell-to-cell spreading involve shared functions of the viral glycoproteins gB, gD, and gH/gL during membrane fusion (54). These three glycoproteins could therefore be implicated as targets of EB action. So far, this is supported only by indirect evidence. We are currently attempting to isolate EB-resistant mutant viruses in order to map any viral locus involved and are arranging to obtain purified viral glycoproteins for formal binding studies.
A BLAST search for homologies to the EB sequence identified a number of matches (data not shown), as expected for such a short peptide. Comparisons with the HSV-1 sequence failed to reveal any matches, suggesting that EB does not mimic any viral proteins in their interactions with a cellular component (i.e., EB is not a sequence-specific mimic of a viral protein). A number of partial matches with cellular proteins were identified. These proteins constituted a diverse set, and we were unable to discern a common functional theme (e.g., not all were membrane proteins). Notably, no homologies were identified in the HveA to HveD HSV coreceptor proteins, suggesting the EB may not affect the binding of gD to HveA to HveD.
Several peptides that inhibit viral entry have been described. Srinivas et al. (56) found that amphipathic helical segments derived from the HIV-1 gp 120 inhibited fusion. This peptide clearly had membrane-disrupting activity and probably functioned by altering the lipid environment (56). Another amphipathic peptide derived from apolipoprotein A-1 has been shown to inhibit HSV entry (55) and may also be acting at the level of the envelope or cell membrane to destroy lipid organization. Peptides derived from the heptad repeat sequences in the human parainfluenza virus F protein inhibit viral entry and cell-to-cell spreading. These peptides do not appear to be amphipathic, however, and may act by interfering directly with the function of the F protein (68). A pseudopeptide (HB-19) that blocks human immunodeficiency virus type 1 (HIV-1) attachment has also been described (42). Finally, a peptide (T-20) derived from the transmembrane region of the HIV gp41 protein, which also has amphipathic properties, is a potent inhibitor of HIV entry (46, 66) and is currently in clinical trials (27). A comparison of these peptides with EB revealed no sequence homologies. In addition, EB is not predicted to have amphipathic properties, suggesting that it is acting through a different mechanism(s). The EB peptide also has no homology to membrane-disrupting peptides such as the magainins, cecropins, and defensins (3).
In summary, we have identified a novel peptide with potent inhibitory activity against HSV-1 and demonstrated that it acts to inhibit entry of the virus and possibly cell-to-cell spreading. Although the peptide, or some derivatives thereof, may be useful therapeutically as one of the only two known peptidic inhibitors of HSV-1 entry, it may be more valuable as a tool to study the poorly understood process of viral entry. Future studies on the mechanism of action of EB may potentially allow us to dissect the events underlying this important step in viral infection.
ACKNOWLEDGMENTS
We thank Gary L. Case and Amy C. Harms, Biotechnology Center at the University of Wisconsin in Madison, for the synthesis and analysis of peptides and for helpful discussions; Andrew Hitt for technical assistance; Inna Larsen for administrative assistance; and Teresa Compton and Donna Peters for critical comments on the manuscript.
This work was supported by grants from the Defense Advanced Research Projects Agency (DARPA MDA 972-97-1-0005) and the NEI (EY07736) to Curtis Brandt.
REFERENCES
- 1.Aldrian-Herrada G, Desarménien M G, Orcel H, Boissin-Agasse L, Méry J, Brugidou J, Rabié A. A peptide nucleic acid (PNA) is more rapidly internalized in cultured neurons when coupled to a retro-inverso delivery peptide. The antisense activity depresses the target mRNA and protein in magnocellular oxytocin neurons. Nucleic Acids Res. 1998;26:4910–4916. doi: 10.1093/nar/26.21.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banfield B W, Ledue Y, Esford L, Visalli R J, Brandt C R, Tufaro F. Evidence for an interaction of herpes simplex virus with chondroitin sulfate proteoglycans during infection. Virology. 1995;208:531–539. doi: 10.1006/viro.1995.1184. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz B A, Bevins C L, Zasloff M A. Magainins: a new family of membrane-active host defense peptides. Biochem Pharmacol. 1990;39:625–629. doi: 10.1016/0006-2952(90)90138-b. [DOI] [PubMed] [Google Scholar]
- 4.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type I in viral entry and fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Stirpe D, Boscano A, Avitabile E, Foa-Tomasi L, Barker D, Roizman B. Glycoprotein C-dependent attachment of herpes simplex virus to susceptible cells leading to productive infection. Virology. 1990;178:213–222. doi: 10.1016/0042-6822(90)90396-9. [DOI] [PubMed] [Google Scholar]
- 6.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–147. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 7.Cockrell A S, Muggeridge M I. Herpes simplex virus type 2 UL45 is a type II membrane protein. J Virol. 1998;72:4430–4433. doi: 10.1128/jvi.72.5.4430-4433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen D M, Aschman D P, Gelep P T, Retondo M J, Weller S K, Schaffer P A. Fine mapping and molecular cloning of mutations in the herpes simplex virus DNA polymerase locus. J Virol. 1984;49:236–247. doi: 10.1128/jvi.49.1.236-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derossi D, Joliot A H, Chassaing G, Prochiantz A. The third helix of the antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:1044–1050. [PubMed] [Google Scholar]
- 10.Desai P J, Schaffer P A, Minson A C. Excretion of non-infectious virus particles lacking gH by a temperature-sensitive mutant of herpes simplex virus type I: evidence that gH is essential for virion infectivity. J Gen Virol. 1988;69:1147–1156. doi: 10.1099/0022-1317-69-6-1147. [DOI] [PubMed] [Google Scholar]
- 11.Fawell S, Seery J, Daikh Y, Moore C, Chen L L, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields C G, Lloyd D H, Macdonald R L, Otteson K M, Noble R L. HBTU activation for automated Fmoc solid-phase peptide synthesis. Peptide Res. 1991;4:95–101. [PubMed] [Google Scholar]
- 13.Fuller A D, Spear P G. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc Natl Acad Sci USA. 1987;84:5454–5458. doi: 10.1073/pnas.84.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller A O, Lee W-C. Herpes simplex virus type I entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J Virol. 1992;66:5002–5012. doi: 10.1128/jvi.66.8.5002-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geraghty R J, Krummenacher C, Cohen G, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs J S, Chiou H C, Hall J D, Mount D W, Retondo M J, Weller S K, Coen D M. Sequence and mapping analysis of the herpes simplex virus DNA polymerase gene predicts a c-terminal substrate binding domain. Proc Natl Acad Sci USA. 1985;82:7969–7973. doi: 10.1073/pnas.82.23.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grau D R, Visalli R J, Brandt C R. Herpes simplex virus stromal keratitis is not titer-dependent and does not correlate with neurovirulence. Investig Ophthalmol Visual Sci. 1989;30:2474–2480. [PubMed] [Google Scholar]
- 18.Haanes E J, Nelson C M, Soule C L, Goodman J L. The UL45 gene product is requires for herpes simplex virus type 1 glycoprotein B-induced fusion. J Virol. 1994;68:5825–5834. doi: 10.1128/jvi.68.9.5825-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall J D, Woodward S. Aphidicolin resistance in herpes simplex virus type 1 appears to alter substrate specificity in the DNA polymerase. J Virol. 1989;63:2874–2876. doi: 10.1128/jvi.63.6.2874-2876.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handler C G, Cohen G, Eisenberg R J. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold B C, WuDunn D, Soltus N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold B C, Visalli R J, Susmarski N, Brandt C R, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 23.Herold B C, Gerber S I, Belval B J, Siston A M, Shulman N. Differences in the susceptibility of Herpes simplex virus types 1 and 2 to modified heparan compounds suggest serotype differences in viral entry. J Virol. 1996;70:3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Highlander S, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchinson L, Goldsmith L K, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex virus type 1 glycoprotein forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson D C, Spear P G. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J Virol. 1989;63:819–827. doi: 10.1128/jvi.63.2.819-827.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilby M J, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;11:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 28.Knopf C W. The herpes simplex virus type 1 DNA polymerase gene: site of phosphonoacetic acid resistance mutation in strain Angelotti is highly conserved. J Gen Virol. 1987;68:1429–1433. doi: 10.1099/0022-1317-68-5-1429. [DOI] [PubMed] [Google Scholar]
- 29.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laquerre S, Argnani R, Anderson D B, Zucchini S, Manservigi R, Glorioso J C. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell to cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to, but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y-Z, Yao S-Y, Veach R A, Torgerson T R, Hawiger J. Inhibition of nuclear translocation of transcription factor NF-κβ by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J Biol Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 33.Lycke E, Johansson M, Svennerholm B, Lindahl U. Binding of herpes simplex virus to cellular heparan sulfate, an initial step in the adsorption process. J Gen Virol. 1991;72:1131–1137. doi: 10.1099/0022-1317-72-5-1131. [DOI] [PubMed] [Google Scholar]
- 34.Manservigi R, Spear P G, Buchan A. Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc Natl Acad Sci USA. 1977;74:3913–3917. doi: 10.1073/pnas.74.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews J T, Terry B J, Field A K. The structure and function of the HSV DNA replication proteins: defining novel antiviral targets. Antiviral Res. 1993;20:89–114. doi: 10.1016/0166-3542(93)90001-y. [DOI] [PubMed] [Google Scholar]
- 36.Meienhofer J, Waki M, Heimer E P, Lambros T J, Makofske R C, Chang C D. Solid phase synthesis without repetitive acidolysis: preparation of leucyl-alanyl-glycyl-valine using 9-fluorenylmethyloxycarbonylamino acids. Int J Pept Protein Res. 1979;13:35–42. [PubMed] [Google Scholar]
- 37.Merrifield R B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85:7129–7133. [Google Scholar]
- 38.Minson A C, Hodgman T C, Digard P, Hancock D C, Bell S E, Buckmaster E A. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J Gen Virol. 1986;67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- 39.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 40.Nicola A V, Willis S H, Naidoo N, Eisenberg R J, Cohen G. Structure function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nisole S, Krust B, Callebaut C, Guichard G, Muller S, Briand J-P, Hovanessian A G. The anti-HIV pseudopeptide HB-19 forms a complex with the cell-surface-expressed nucleolin independent of heparan sulfate proteoglycans. J Biol Chem. 1999;274:27875–27884. doi: 10.1074/jbc.274.39.27875. [DOI] [PubMed] [Google Scholar]
- 43.Oehlke J, Krause E, Wiesner B, Beyermann M, Bienert M. Nonendocytic, amphipathicity dependent cellular uptake of helical model peptides. Protein Pept Lett. 1996;3:393–398. [Google Scholar]
- 44.Oehlke J, Krause E, Wiesner B, Beyermann M, Bienert M. Extensive cellular uptake into endothelial cells of an amphipathic β-sheet forming peptide. FEBS Lett. 1997;415:196–199. doi: 10.1016/s0014-5793(97)01123-x. [DOI] [PubMed] [Google Scholar]
- 45.Pooga M, Hällbrink M, Zorko M, Langel Ü. Cell penetration by transportan. FASEB J. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 46.Rimsky L T, Shugars D C, Matthews T J. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasadeusz J J, Tufaro F, Safrin S, Schubert K, Hubinette M M, Cheung P K, Sacks S L. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J Virol. 1997;71:3872–3878. doi: 10.1128/jvi.71.5.3872-3878.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarze S R, Ho A, Vocero-Akbani A, Dowdy S F. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 50.Sears A E, MeGwire B S, Roizman B. Infection of polarized MDCK cells with herpes simplex virus 1: two asymetrically distributed cell receptors interact with different viral proteins. Proc Natl Acad Sci USA. 1991;88:5087–5091. doi: 10.1073/pnas.88.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segel I H. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1976. [Google Scholar]
- 52.Shieh M T, WuDunn D, Montgomery R I, Esko J D, Spear P G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shieh M T, Spear P G. Herpes virus-induced cell fusion that is dependent on cell surface heparan sulfate or soluble heparan. J Virol. 1994;68:1224–1228. doi: 10.1128/jvi.68.2.1224-1228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 55.Srinivas R V, Birkedal B, Owens R J, Anantharamaiah G M, Segrest J P, Compans R W. Antiviral effects of apolipoprotein A-I and its synthetic amphipathic peptide analogs. Virology. 1990;176:48–57. doi: 10.1016/0042-6822(90)90229-k. [DOI] [PubMed] [Google Scholar]
- 56.Srinivas S K, Srinivas R V, Anantharamaiah G M, Segrest J P, Compans R W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 57.Tal-Singer R, Peng C, Ponce de Leon M, Abrams W R, Banfield B W, Tufaro F, Cohen G H, Eisenberg R J. Interaction of herpes simplex virus glycoprotein C with mammalian cell surface molecules. J Virol. 1995;69:4471–4483. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Théodore L, Derossi D, Chassaing G, Llirbat B, Kubes M, Jordan P, Chneiweiss H, Godement P, Prochiantz A. Intraneuronal delivery of protein kinase C pseudosubstrate leads to growth cone collapse. J Neurosci. 1995;15:7158–7167. doi: 10.1523/JNEUROSCI.15-11-07158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Visalli R J, Brandt C R. The HSV-1 UL45 18 kDa gene product is a true late protein and a component of the virion. Virus Res. 1993;29:167–178. doi: 10.1016/0168-1702(93)90057-t. [DOI] [PubMed] [Google Scholar]
- 61.Vivès E, Brodin P, Lebleu B. A truncated HIV-1 tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 62.Westra D F, Glazenburg K L, Harmsen M C, Tiran A, Jan Scheffer A, Welling G W, Hauw The T, Welling-Wester S. Glycoprotein H of herpes simplex virus type 1 requires glycoprotein L for transport to the surfaces of insect cells. J Virol. 1997;71:2285–2291. doi: 10.1128/jvi.71.3.2285-2291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White J. Membrane fusion. Science. 1992;258:917–923. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 65.Whitley R J. Epidemiology of herpes simplex viruses. In: Roizman B, editor. The herpesviruses. Vol. 3. New York, N.Y: Plenum Press; 1982. pp. 1–44. [Google Scholar]
- 66.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao Q, Compans R W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]