Abstract
We have examined the role of the human immunodeficiency virus type 1 (HIV-1) Tat protein in the regulation of reverse transcription. We show that a two-exon but not a one-exon form of Tat markedly suppressed cell-free reverse transcriptase (RT) activity. Conversely, viruses expressing two-exon Tat (pNL43 and pNL101) showed rapid replication kinetics and more efficient endogenous RT activity compared with viruses expressing one-exon Tat (pM1ex). The pM1ex virions, as well as pM1ex-infected cells, also contained higher levels of viral DNA than did either the pNL43 or pNL101 viruses, indicating that reverse transcription might have continued during later stages of viral replication in the absence of the second Tat exon. Moreover, degradation of viral genomic RNA was more apparent in the pM1ex virions. Accordingly, we propose that the two-exon Tat may help augment viral infectivity by suppressing the reverse transcription reaction during late stages of viral synthesis and by preventing the synthesis of potentially deleterious viral DNA products.
Reverse transcription of retroviral RNA into double-stranded DNA is an essential step of virus replication and is catalyzed by the viral reverse transcriptase (RT) enzyme (47). Multiple studies have shown that human immunodeficiency virus type 1 (HIV-1) reverse transcription is regulated by both viral and host factors. For example, cellular tRNA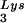 is preferentially incorporated into HIV-1 virions (21) and is used to initiate reverse transcription after binding to a complementary stretch of viral RNA termed the primer binding site (PBS) (41). The viral nucleocapsid protein (NCp7) also plays a role in this process by annealing tRNA
is preferentially incorporated into HIV-1 virions (21) and is used to initiate reverse transcription after binding to a complementary stretch of viral RNA termed the primer binding site (PBS) (41). The viral nucleocapsid protein (NCp7) also plays a role in this process by annealing tRNA to the PBS (6, 13). In addition, other viral proteins, including Nef (2, 42), Vif (51), integrase (34), and Tat (16), and other cellular proteins, such as cyclophilin A (14, 48) and DNA topoisomerase I (39, 45), which are specifically incorporated into virions, may also be involved in promoting efficient reverse transcription.
to the PBS (6, 13). In addition, other viral proteins, including Nef (2, 42), Vif (51), integrase (34), and Tat (16), and other cellular proteins, such as cyclophilin A (14, 48) and DNA topoisomerase I (39, 45), which are specifically incorporated into virions, may also be involved in promoting efficient reverse transcription.
The Tat protein, which is a transcriptional transactivator of HIV-1, is essential for viral transcription. Tat binds to the transactivation response element (TAR), a stem-loop structure located at the 5′ end of the genomic RNA, and can consequently play roles in both transcriptional initiation, including promoter clearance and elongation (20, 22). In addition, Tat is thought to have a role in maintenance of viral infectivity (17). HIV-1 virions, lacking the tat gene, displayed decreased efficiency of reverse transcription, suggesting that Tat was needed to stimulate RT activity (16). Additionally, the expression of a mutant Tat protein, which was functionally defective for activation of viral transcription, was able to complement the defective RT activity of virions lacking the tat gene (50). These results suggest that the domains of Tat responsible for the regulation of reverse transcription and viral gene expression are distinct, i.e., that Tat may regulate viral reverse transcription through either direct or indirect means, although the mechanisms involved are unknown.
To study the role of Tat in reverse transcription, we prepared recombinant Tat proteins and studied them in reconstituted cell-free RT reactions. Our results show that wild-type Tat protein, containing both Tat exons (i.e., two-exon Tat [2-exon-Tat]) was able to strongly suppress RT activity, whereas Tat molecules deleted of the second exon (i.e., 1-exon-Tat) could not. Conversely, HIV-1 molecular clones expressing 2-exon-Tat (pNL43 and pNL101) showed more rapid replication kinetics and more efficient expression of endogenous RT activity than did viruses deleted of the second Tat exon (pM1ex). Viruses deleted of the second Tat exon, as well as cytosolic fractions of infected cells, contained higher levels of viral DNA than did viruses containing wild-type Tat. Moreover, more extensive degradation of viral genomic RNA was detected in pM1ex virions. Accordingly, we propose that reverse transcription reactions may continue during the postintegration stage of viral replication, in the absence of the second Tat exon, and that this may have contributed to the degradation of genomic RNA and decreased infectivity of progeny virus. In contrast, the presence of wild-type Tat, i.e., 2-exon-Tat, may increase viral infectiousness by suppressing RT activity during the late stages of viral replication. These data further imply that excessive viral DNA synthesis and/or incorporation of such DNA into virions may be deleterious for viral infectivity.
MATERIALS AND METHODS
Reagents.
Synthetic peptides corresponding to amino acids 61 to 86 of HIV-1 Tat (Tat[61–86]) were purchased from Tecnogen S.C.p.A. (Piana di Monte Verna, Italy).
Plasmids.
Plasmids for the expression of His6-tagged HIV-1 Tat were constructed, as described previously (24). Briefly, tat cDNA molecules encoding either 72-, 86-, or 101-amino-acid (aa) polypeptides were amplified from ACH-2 (7) cellular mRNA using the following primers: 5′-CGggatccCATGGAGCCAGTAGATC-3, 5′-AActgcagCCTACTTTGATAGAGAA-3′, 5′-AActgcagCCTATTCCTTCGGGCCT-3′, and 5′-AAActgcagCTCACTAATCGAATGG-3′. For expression of the 72-aa Tat (Tat 72), the stop codon (underlining denotes the mutation in the antisense primer) was introduced at aa-73. Lowercase letters indicate the BamHI (ggatcc) and PstI (ctgcag) restriction sites. After PCR amplification, tat cDNAs were subcloned into a pQE-31 vector (Qiagen, Mississauga, Ontario, Canada). For the expression of 101-aa Tat (Tat 101), the stop codon at aa 86 in ACH-2 tat cDNA was removed (TAG→TCG) by PCR-based mutagenesis using the primer 5′-GGCCCGAAGGAATCGAAGAAGAAG-3′ (underlining denotes the mutation), after subcloning into pQE-31. cDNA sequences were amplified by PCR using Pfu DNA polymerase (Stratagene, La Jolla, Calif.), and the generated mutations were confirmed by sequencing. The pNL43 (1)-derived HIV-1 proviral molecular clones, pM1ex and pNL101, expressing 72- and 101-aa Tat, respectively (35), were kindly provided by K.-T. Jeang (Laboratory of Molecular Microbiology, National Institute of Allergy and Infectious Diseases, Bethesda, Md.).
Purification of recombinant proteins.
Recombinant Tat proteins were expressed in Escherichia coli M15(pREP4) and purified using nickel-nitrilotriacetic acid (Ni-NTA) resin as described previously (24). Heterodimeric HIV-1 RT (p66-p51) was expressed in E. coli M15(pREP4) and also purified using Ni-NTA resin and ion-exchange chromatography (29).
Templates and primers.
The RNA template, consisting of a 239-nucleotide (nt) HIV-1 RNA sequence, spanning the R region of the long terminal repeat (LTR) and the PBS, was in vitro transcribed from linearized pHIV-PBS (3). RNA template lacking the TAR region [TAR(−) template, 203 nt] and its control [TAR(+) template, 258 nt] were in vitro transcribed from PCR products corresponding to nt 512 to 711 and nt 455 to 711 of HIV-1 (pNL43), respectively, and fused with T7 promoter sequences. Recombinant human tRNA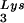 was in vitro transcribed from linearized pT7hLys3 (52). In vitro transcription reactions were carried out using the T7-MEGAshortscript In Vitro Transcription Kit (Ambion, Austin, Tex.). To prepare labeled tRNA
was in vitro transcribed from linearized pT7hLys3 (52). In vitro transcription reactions were carried out using the T7-MEGAshortscript In Vitro Transcription Kit (Ambion, Austin, Tex.). To prepare labeled tRNA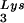 , [α-32P] UTP was added to the transcription reaction. The DNA primer, which is complementary to the PBS, was end labeled with [γ-32P]ATP using T4 polynucleotide kinase. RNA transcripts and labeled DNA primer were electrophoretically purified prior to use in RT reactions.
, [α-32P] UTP was added to the transcription reaction. The DNA primer, which is complementary to the PBS, was end labeled with [γ-32P]ATP using T4 polynucleotide kinase. RNA transcripts and labeled DNA primer were electrophoretically purified prior to use in RT reactions.
In vitro reverse transcription reaction.
The annealed primer-template complex was prepared before incubation with RT. Mixtures containing 32P-labeled tRNA (primer) and template at a ratio of 1:2 in a buffer containing 50 mM Tris-HC1 (pH 7.8) and 50 mM NaCl were heated to 95°C for 3 min, followed by incubation at 70°C for 20 min, and then cooled to 37°C and incubated further for 20 min. The annealed tRNA-template complex was incubated with RT and Tat preparations at 37°C for 3 min in a 20-μl reaction mixture containing 50 mM Tris-Hcl (pH 7.8), 50 mM NaCl, 1 mM dithiothreitol (DTT), and 0.2 mM concentrations of deoxynucleoside triphosphates (dNTPs), and then reverse transcription was initiated by the addition of MgCl2 at a final concentration of 6 mM. The reactions were allowed to proceed at 37°C for the indicated times and were stopped by adding 2-μl aliquots of the reaction mixture to 8 μl of 95% formamide. Reaction products were analyzed on 8% polyacrylamide–7 M urea gels and visualized by autoradiography. In some experiments, end-labeled DNA primer was replaced with tRNA as a primer of these reactions.
Gel retardation experiments.
The annealed tRNA-template complex, prepared as described above, was incubated with RT and Tat preparations in a 10-μl buffer containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, 6 mM MgCl2, 1 mM DTT, 0.01% Triton X-100, and 25% glycerol at room temperature for 20 min. Thereafter, samples were separated on 5% polyacrylamide gels (0.5 × Tris-borate-EDTA [TBE]; 44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA) at 4°C, and then the gels were dried and the radioactive bands were visualized with X-ray film.
Cells, transfection, and virus infection.
Jurkat and H9/HTLV-IIIB (38) cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (complete medium). Cos-7 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Virus stocks were prepared by transfection of HIV-1 proviral molecular clones into Cos-7 cells using Lipofectamine Plus reagent (Canadian Life Technologies, Inc., Montreal, Quebec, Canada) according to the manufacturer's instructions. The production of progeny virus was assessed by measuring the levels of p24 (CA) antigen released into culture fluids at 72 h after transfection by enzyme-linked immunosorbent assay (Abbott Laboratories, Abbott Park, Ill.). For infection of Jurkat cells, 6 × 105 cells in 0.3 ml of complete medium were exposed to virus (10 ng of p24) at 37°C for 2 h. After being washed with complete medium, infected cell cultures were maintained for 18 days, and culture supernatants were collected every 3 days for determinations of the RT activity.
In addition, multinuclear-activation-of-a-galactosidase-indicator (MAGI) assays were performed using HeLa cells stably transfected with retroviral vectors expressing both CD4 and a truncated HIV-1 LTR–β-galactosidase plasmid (i.e., HeLa–CD4–LTR–β-Gal [NIH AIDS Research and Reference Reagent Program; reagent supplied by Michael Emerman]) (26a). Toward this end, cells were prepared at a concentration of 4 × 104 cells per well in a 24-well plate at 1 day before infection. The virus was diluted to determine the appropriate concentration for use in infection studies (the optimal concentration of virus produced 100 to 200 blue-stained cells per well). At 48 h after infection, cells were fixed with a solution containing 1% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline for 5 min. After an extensive washing with phosphate-buffered saline, the cells were incubated in staining solution (4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal] at 0.4 mg/ml) for 50 min. The number of blue-stained cells was scored by microscopy. For each viral preparation, three independent infections were performed, and the average number of blue-stained cells was determined.
Endogenous RT reaction.
Culture supernatants of transfected Cos-7 and H9/HTLV-IIIB cells were clarified at 3,000 rpm for 30 min at 4°C. After that, HIV-1 virions were pelleted through a 20% sucrose cushion at 40,000 rpm for 1 h at 4°C by using an SW41 rotor in a Beckman XL-80 ultracentrifuge. Endogenous RT reactions were performed as described previously (40). Briefly, pelleted virions, containing 250 ng of p24, were incubated in 100-μl reaction mixtures containing 50 mM Tris-HCl (pH 7.8), 10 mM NaCl, 60 mM KCl, 5 mM MgCl2, 10 mM DTT, 1 mM EGTA, 0.1% NP-40, 0.4 mM concentrations of dATP, dTTP, and dGTP, 10 μM dCTP, and 10 μCi of [α-32P]dCTP at 37°C for 5 h. The reactions were terminated by adding an equal amount of stop buffer (1% sodium dodecyl sulfate [SDS], 50 mM EDTA and 0.4 M NaCl). The samples were then treated with 20 μg of proteinase K at 56°C for 30 min and extracted with phenol-chloroform, followed by ethanol precipitation. Reaction products were separated on 1% denaturing agarose gels (20 mM NaOH, 1 mM EDTA) at 4°C; the gels were then dried, and radioactive bands were visualized with X-ray film.
DNA isolation from the cytosolic fraction of infected cells.
Infected Jurkat cells were washed once with phosphate-buffered saline, gently suspended onto lysis buffer containing 10 mM HEPES-KOH (pH 7.8), 10 mM KCl, 0.1 mM EDTA, and 0.1% NP-40, and then placed on ice for 3 min. The nuclei were then pelleted by centrifugation at 13,000 rpm for 10 min at 4°C. After centrifugation, the supernatants were studied as cytosolic fractions, from which DNA was isolated using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions. Extracted DNA was normalized on the basis of cell number and subjected to PCR analysis. To monitor the efficiency of DNA isolation from these cytosol preparations, the mitochondrial DNA-encoded cytochrome c-oxidase II (CytOxy II) gene was amplified by 20 cycles of PCR (93°C for 1 min and 65°C for 2 min) using the specific primer pair 5′-ATGCAGCGCAAGTAGGT-3′ and 5′-GGAAAATGATTATGAGGGCGTG-3′ (16, 50).
Extraction of virus-associated nucleic acid.
Culture supernatants of infected Jurkat or transfected Cos-7 cells were clarified at 3,000 rpm for 30 min at 4°C. In the case of the Jurkat cells, culture supernatants were harvested at 15 days postinfection and treated with 10 U of deoxyribonuclease I (Canadian Life Technologies, Inc.) per ml at 37°C for 15 min in the presence of 5 mM MgCl2 to eliminate potentially contaminating plasmid DNA and/or proviral DNA released from lysed cells. Pelleted virions were then prepared as described above. Virion-associated nucleic acids were extracted by use of a procedure modified for this purpose (4). Briefly, the pelleted virions were resuspended in 400 μl of lysis buffer (50 mM Tris-HCl, pH 7.4; 100 mM NaCl; 10 mM EDTA; 1% SDS), and 10-μl aliquots of the samples were removed for p24 determinations. The remainder was supplemented with 25 μg of tRNA and treated with 20 μg of proteinase K at 37°C for 20 min. Virion-associated nucleic acids were then extracted with phenol-chloroform, followed by ethanol precipitation. Thereafter, extracted nucleic acids were normalized on the basis of p24 content, and samples were subjected to either PCR (Jurkat samples) or Northern blot analysis (Cos-7 samples).
PCR analysis of HIV-1 DNA.
The HIV-1 specific primers U3 (5′-CACACACAAGGCTACTTCCCT-3′ [nt 57 to 77 of pNL43]), R (5′-GGCTAACTAGGGAACCCACTG-3′ [nt 496 to 516]), U5 (5′-CTGCTAGAGATTTTCCACACTGAC-3′ [nt 635 to 612]), 5NC (5′-CCGAGTCCTGCGTCGAGAGATC-3′ [nt 701 to 680]), p7 (5′-ATTGCAGGGCCCCTAGGAAAAAGG-3′ [nt 2000 to 2023]), RT1 (5′-GTCTCAATAGGACTAATGGGAAAA ([nt 2569 to 2546]); Tat1 (5′-ATGGAGCCAGTAGATC-3′ [nt 5830 to 5845]), Tat2 (5′-TGCCATAGGAGATGCCTAA-5′ [nt 5974 to 5956]); Env1 5′-CGCAAAACCAGCAAGAAAAGAATG-3′ [nt 8160 to 8183]), and Env2 (5′-CGTTCACTAATCGAATGG-3′ [nt 8465 to 8448]) were used. Primer pair R-U5 was designed to detect the earliest RT products synthesized either before or immediately after the first template switch. With primer pairs U3-U5, Env1-Env2, Tat1-Tat2, and p7-RT1, a specific PCR product was expected only if a negative-strand DNA of increased length, relative to wild-type, had been synthesized after the first template switch. Primer pair R-5NC, which flanks the PBS, is predicted to amplify only late RT products synthesized after the second template switch (49). PCR amplification was performed for 30 cycles of denaturation at 94°C for 1 min, followed by primer annealing at 56°C for 1 min and polymerization at 72°C for 1 min. The PCR products were separated on 6% polyacrylamide gels, and then the gels were dried and the radioactive bands were visualized with X-ray film.
Nondenaturing Northern blot analysis.
Viral RNA samples corresponding to 500 ng of p24 were separated on non-denaturing 0.9% agarose gels (1× TBE) at 4°C. Thereafter, the gels were soaked in 2 volumes of 50 mM NaOH for 30 min and then soaked again in 2 volumes of 200 mM sodium acetate (pH 5.2) for 30 min. The RNA was next transferred onto a nylon membrane (Hybond-N; Amersham Pharmacia Biotech, Inc., Uppsala, Sweden). The membrane was baked at 80°C for 2 h and then prehybridized for 3 h at 42°C in hybridization buffer containing 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2 PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 0.5% SDS, 50% formamide, and 2 mg of herring sperm DNA. Hybridization with the denaturing HIV-1 probe was carried out for 16 h at 42°C in the hybridization buffer. The 32P-labeled HIV-1 probe was generated from HindIII fragments of pNL43 (nt 531 to 9609) by using a nick translation kit (Roche Molecular Biochemicals). The membrane was washed, and the radioactive bands were visualized with X-ray film.
RESULTS
The 2-exon-Tat suppresses negative-strand DNA synthesis in an in vitro RT reaction.
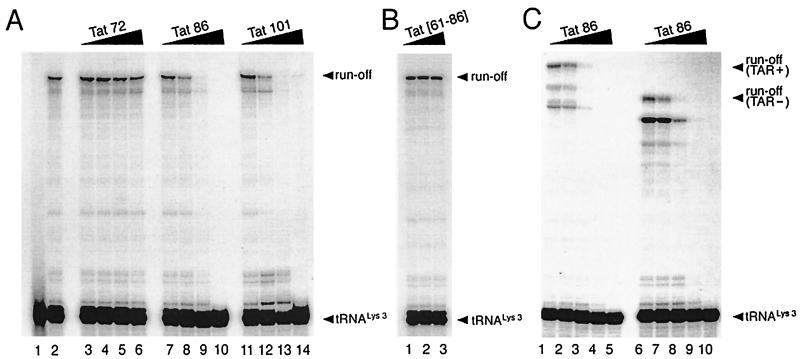
To study the role of Tat in RT reactions, we prepared the recombinant Tat proteins, i.e., Tat 72 (1-exon-Tat), Tat 86 (2-exon-Tat), and Tat 101 (a version of 2-exon-Tat in some viral isolates) (35), and employed them in a reconstituted cell-free reverse transcription reaction consisting of viral RNA template, recombinant tRNA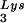 , and RT (p66-p51). As shown in Fig. 1A, the synthesis of negative-strand DNA was suppressed in the presence of either Tat 86 or Tat 101 in a dose-dependent manner (lanes 7 to 14). There was no significant difference between Tat 86 and Tat 101 in regard to this suppressive effect (compare lanes 7 to 10 to lanes 11 to 14), at either 10 pmol (lanes 8 and 12), 30 pmol (lanes 9 and 13), or 100 pmol (lanes 10 and 14) per 20-μl reaction. In contrast, Tat 72 was ineffective at 3, 10, or 30 pmol (lanes 3 to 5), and only slightly suppressed the reaction at 100 pmol (lane 6).
, and RT (p66-p51). As shown in Fig. 1A, the synthesis of negative-strand DNA was suppressed in the presence of either Tat 86 or Tat 101 in a dose-dependent manner (lanes 7 to 14). There was no significant difference between Tat 86 and Tat 101 in regard to this suppressive effect (compare lanes 7 to 10 to lanes 11 to 14), at either 10 pmol (lanes 8 and 12), 30 pmol (lanes 9 and 13), or 100 pmol (lanes 10 and 14) per 20-μl reaction. In contrast, Tat 72 was ineffective at 3, 10, or 30 pmol (lanes 3 to 5), and only slightly suppressed the reaction at 100 pmol (lane 6).
FIG. 1.
(A) Tat 86 and Tat 101, but not Tat 72, suppress DNA synthesis primed with tRNA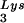 . The annealed tRNA
. The annealed tRNA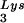 (1 pmol)-template (2 pmol) complex and RT (3 pmol) were mixed with various amounts of Tat 72 (lanes 3 to 6), Tat 86 (lanes 7 to 10), or Tat 101 (lanes 11 to 14). Reverse transcription was then initiated and allowed to proceed at 37°C for 30 min as described in Materials and Methods. Lane 1 represents the control reaction without the addition of MgCl2. The amounts of Tat used in these reactions were 0 pmol (lanes 1 and 2), 3 pmol (lanes 3, 7, and 11), 10 pmol (lanes 4, 8, and 12), 30 pmol (lanes 5, 9, and 13), and 100 pmol (lanes 6, 10, and 14) per lane. (B) The polypeptide within the second exon of Tat cannot suppress the RT reaction. RT reactions were carried out as described in panel A. The amounts of the synthetic peptide, corresponding to aa 61 to 86 of Tat (Tat[61–86]), used in these experiments were 50 pmol (lane 1), 100 pmol (lane 2), and 200 pmol (lane 3) per reaction. (C) Tat suppresses cell-free RT reactions in a TAR-independent manner. RT reactions were carried out as described in panel A, except that TAR(−) (lanes 6 to 10) and TAR(+) (lanes 1 to 5) templates were used. The amounts of Tat 86 in these reactions were 0 pmol (lanes 1 and 6), 3 pmol (lanes 2 and 7), 10 pmol (lanes 3 and 8), 30 pmol (lanes 4 and 9), and 100 pmol (lanes 5 and 10) per lane. Reaction products were analyzed on 8% polyacrylamide–7 M urea gels and visualized by autoradiography. The runoff bands in panels A and B and the runoff (TAR+) and run-off (TAR−) bands in panel C indicate full-length synthesis of DNA, i.e., 239, 258, and 208 nt, respectively. The tRNA
(1 pmol)-template (2 pmol) complex and RT (3 pmol) were mixed with various amounts of Tat 72 (lanes 3 to 6), Tat 86 (lanes 7 to 10), or Tat 101 (lanes 11 to 14). Reverse transcription was then initiated and allowed to proceed at 37°C for 30 min as described in Materials and Methods. Lane 1 represents the control reaction without the addition of MgCl2. The amounts of Tat used in these reactions were 0 pmol (lanes 1 and 2), 3 pmol (lanes 3, 7, and 11), 10 pmol (lanes 4, 8, and 12), 30 pmol (lanes 5, 9, and 13), and 100 pmol (lanes 6, 10, and 14) per lane. (B) The polypeptide within the second exon of Tat cannot suppress the RT reaction. RT reactions were carried out as described in panel A. The amounts of the synthetic peptide, corresponding to aa 61 to 86 of Tat (Tat[61–86]), used in these experiments were 50 pmol (lane 1), 100 pmol (lane 2), and 200 pmol (lane 3) per reaction. (C) Tat suppresses cell-free RT reactions in a TAR-independent manner. RT reactions were carried out as described in panel A, except that TAR(−) (lanes 6 to 10) and TAR(+) (lanes 1 to 5) templates were used. The amounts of Tat 86 in these reactions were 0 pmol (lanes 1 and 6), 3 pmol (lanes 2 and 7), 10 pmol (lanes 3 and 8), 30 pmol (lanes 4 and 9), and 100 pmol (lanes 5 and 10) per lane. Reaction products were analyzed on 8% polyacrylamide–7 M urea gels and visualized by autoradiography. The runoff bands in panels A and B and the runoff (TAR+) and run-off (TAR−) bands in panel C indicate full-length synthesis of DNA, i.e., 239, 258, and 208 nt, respectively. The tRNA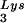 band indicates unprocessed 32P-labeled tRNA
band indicates unprocessed 32P-labeled tRNA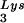 (76 nt).
(76 nt).
The polypeptide within the second exon of Tat is not sufficient to inhibit the RT reaction.
To further assess the importance of the second exon of Tat, we next studied the ability of a synthetic peptide corresponding to aa 61 to 86 of Tat (Tat[61–86]) to interfere with RT activity in our assay. However, Tat[61–86] did not display any inhibitory effects at concentrations of between 50 to 500 pmol (Fig. 1B and data not shown). These results suggest that this synthetic peptide does not possess the natural structure of 2-exon-Tat that is needed to suppress the RT reaction.
Tat suppresses cell-free RT reactions in a TAR-independent manner.
Tat binds to the TAR RNA stem-loop structure, which comprises the first 57 nt of the R region in HIV genomic RNA (10), and regulates viral transcription (20, 22). On the other hand, Tat can also stimulate HIV-1 gene expression in a TAR-independent manner (5, 25, 26, 46, 53). Since the viral RNA template employed in our reconstituted RT reaction contains the TAR region, we next analyzed whether or not the suppressive effect of 2-exon-Tat for the RT reaction was TAR dependent through use of TAR(+) and TAR(−) RNA templates. As shown in Fig. 1C, Tat 86 suppressed the RT reaction in either the presence or the absence of the TAR region with similar efficiency (compare lanes 1 to 5 and lanes 6 to 10). Thus, 2-exon-Tat suppressed the synthesis of negative-strand DNA in a TAR-independent manner.
Tat suppresses the elongation stage of reverse transcription.
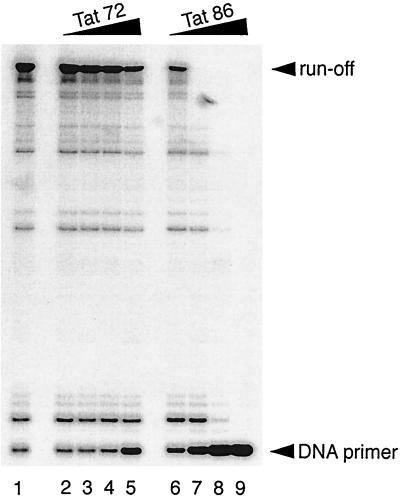
The synthesis of negative-strand DNA primed with tRNA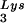 in the HIV-1 RT reaction involves both specific initiation and nonspecific elongation stages (18, 27). The results of Fig. 1A implied that 2-exon-Tat may have exerted its suppressive effect during the elongation rather than the initiation stage since the synthesis of the runoff products, but not that of short products, was suppressed by low concentrations (10 and 30 pmol) of the 2-exon-Tat preparations (lanes 8, 9, 12, and 13). Since the initiation stage of reverse transcription is not affected by negative-strand DNA synthesis primed with a DNA primer (18), we next primed our RT reactions with a DNA primer to further test whether or not the elongation of DNA synthesis was suppressed by 2-exon-Tat. As shown in Fig. 2, Tat 86 suppressed the synthesis of negative-strand DNA in this reaction in a dose-dependent manner (lanes 6 to 9). Tat 72 exerted only a slight suppressive effect in these reactions (lanes 2 to 5). The suppressive effect of Tat for reactions performed with the DNA primer was greater than that observed in reactions primed with tRNA
in the HIV-1 RT reaction involves both specific initiation and nonspecific elongation stages (18, 27). The results of Fig. 1A implied that 2-exon-Tat may have exerted its suppressive effect during the elongation rather than the initiation stage since the synthesis of the runoff products, but not that of short products, was suppressed by low concentrations (10 and 30 pmol) of the 2-exon-Tat preparations (lanes 8, 9, 12, and 13). Since the initiation stage of reverse transcription is not affected by negative-strand DNA synthesis primed with a DNA primer (18), we next primed our RT reactions with a DNA primer to further test whether or not the elongation of DNA synthesis was suppressed by 2-exon-Tat. As shown in Fig. 2, Tat 86 suppressed the synthesis of negative-strand DNA in this reaction in a dose-dependent manner (lanes 6 to 9). Tat 72 exerted only a slight suppressive effect in these reactions (lanes 2 to 5). The suppressive effect of Tat for reactions performed with the DNA primer was greater than that observed in reactions primed with tRNA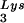 (compare Fig. 2 with Fig. 1A). These results indicate that 2-exon-Tat suppressed the elongation rather than the initiation stage of the HIV-1 RT reaction.
(compare Fig. 2 with Fig. 1A). These results indicate that 2-exon-Tat suppressed the elongation rather than the initiation stage of the HIV-1 RT reaction.
FIG. 2.
Tat 86, but not Tat 72, suppresses DNA synthesis primed with a DNA primer in an in vitro RT reaction. The annealed DNA primer (2.5 pmol)-template (5.0 pmol) complex and RT (2.5 pmol) were mixed with various amounts of Tat 72 (lanes 2 to 5) or Tat 86 (lanes 6 to 9). The amounts of Tat preparations used were 0 pmol (lane 1), 7.5 pmol (lanes 2 and 6), 25 pmol (lanes 3 and 7), 75 pmol (lanes 4 and 8), and 250 pmol (lanes 5 and 9) per reaction. Note that the ratios of Tat to primer were 3:1 (lanes 2 and 6), 10:1 (lanes 3 and 7), 30:1 (lanes 4 and 8), and 100:1 (lanes 5 and 9). RT reactions were carried out as described in the legend to Fig. 1A, except that the reactions were allowed to proceed at 37°C for 10 min. The runoff and DNA primer bands indicate full-length products and unprocessed 32P-labeled DNA primer, respectively.
2-exon-Tat forms a supershifted complex with primer-template and RT.
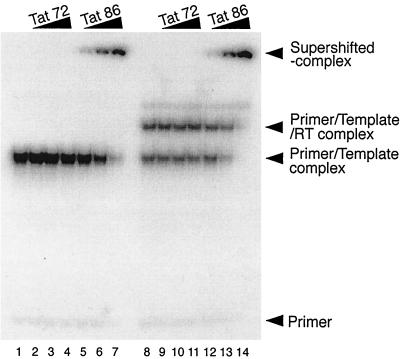
To further examine how Tat may have suppressed the RT reaction, we next studied interactions between RT and Tat by immunoprecipitation using antibodies against both of these molecules. However, we were unable to detect any direct interactions between these proteins, i.e., no coprecipitation (data not shown). Therefore, we instead performed gel retardation experiments to visualize interaction(s) between Tat and other components of the RT reaction. As shown in Fig. 3, both the annealed primer-template complex (all lanes) and a slower-migrating complex, consisting of RT and preannealed primer-template (lanes 8 to 14), were detected. Furthermore, when Tat 86, but not Tat 72, was added to the reaction, the presence of supershifted bands was detected (lanes 5 to 7 and lanes 12 to 14). The intensities of these supershifted bands increased depending on the concentration of Tat 86, and both the primer-template and the primer-template-RT complexes were supershifted in these studies (lanes 12 to 14). These results suggest that 2-exon-Tat, i.e., Tat 86, was able to form a complex with the primer-template and RT. This complex may be involved in the suppression of the cell-free RT reaction, since both the suppression of RT (Fig. 1A) and the formation of the supershifted complex (Fig. 3) occured within a similar range of Tat concentrations.
FIG. 3.
Tat forms a supershifted complex with primer-template and RT. The annealed, 32P-labeled tRNA (1 pmol)-template (2 pmol) complex was incubated with Tat 72 (lanes 2 to 4 and lanes 9 to 11) or Tat 86 (lanes 5 to 7 and lanes 12 to 14) in the presence (lanes 8 to 14) or absence (lanes 1 to 7) of RT (3 pmol). The amounts of Tat in the reaction mixtures were 0 pmol (lanes 1 and 8), 3 pmol (lanes 2, 5, 9, and 12), 10 pmol (lanes 3, 6, 10, and 13), and 30 pmol (lanes 4, 7, 11, and 14). After incubation at room temperature for 20 min, the samples were separated on 5% polyacrylamide gels at 4°C, and then the gels were dried and the bands were visualized on X-ray film.
It should be noted that we were unable to detect the shifted bands, i.e., those denoting Tat-TAR interactions, in the presence of Tat 72 under these experimental conditions (lanes 2 to 4 and lanes 9 to 11), even though the first Tat exon contains an RNA binding region (20, 22). Therefore, the supershifted complex in the presence of Tat 86 (lanes 5 to 7 and lanes 12 to 14) may be generated not only as a result of the Tat-TAR interaction but also by nonspecific interactions between Tat and the RNA templates studied in a TAR-independent manner.
Tat suppresses the endogenous RT reaction.
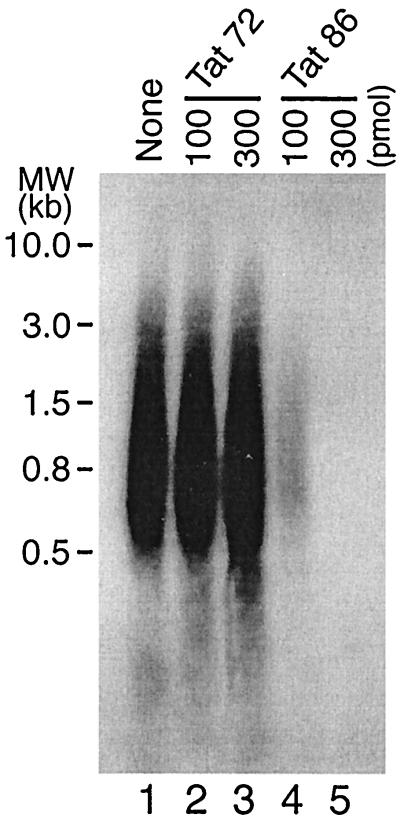
We next asked whether Tat could suppress the RT reaction under physiological conditions. To address this question, we performed endogenous RT reactions in the presence of exogenously added recombinant Tat proteins. (Permeabilized HIV-1 virions, which contain RT, viral RNA genome, natural tRNA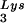 , NCp7, and other viral components can initiate reverse transcription in vitro in the presence of dNTPs). As shown in Fig. 4, exogenously added Tat 86 strongly suppressed these endogenous RT reactions at concentrations of 100 and 300 pmol per 100-μl reaction (lanes 4 and 5). In contrast, Tat 72 had no effect in this system (lanes 2 and 3). These data further testify to the ability of physiologically relevant preparations of Tat to suppress RT activity (even though it has not been demonstrated that Tat becomes incorporated into HIV-1 virions).
, NCp7, and other viral components can initiate reverse transcription in vitro in the presence of dNTPs). As shown in Fig. 4, exogenously added Tat 86 strongly suppressed these endogenous RT reactions at concentrations of 100 and 300 pmol per 100-μl reaction (lanes 4 and 5). In contrast, Tat 72 had no effect in this system (lanes 2 and 3). These data further testify to the ability of physiologically relevant preparations of Tat to suppress RT activity (even though it has not been demonstrated that Tat becomes incorporated into HIV-1 virions).
FIG. 4.
Tat 86, but not Tat 72, suppresses the endogenous RT reaction. HIV-1 virions (HTLV-IIIB strain) were pelleted through a 20% sucrose cushion at 40,000 rpm for 1 h at 4°C. The permeabilized virions containing 250 ng of p24 were then mixed with Tat 72 (100 and 300 pmol, lanes 2 and 3, respectively) or Tat 86 (100 and 300 pmol, lanes 4 and 5, respectively), and endogenous RT reactions were performed as described in Materials and Methods. Lane 1 represents a reaction performed without Tat.
Virus particles encoding only 1-exon-Tat, i.e., pM1ex, show low efficiency of endogenous RT reactions.
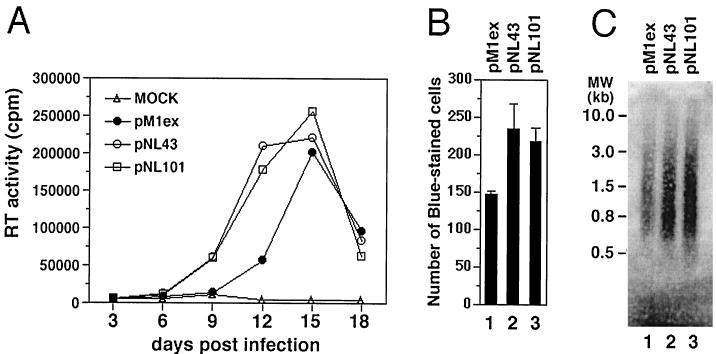
To further study the role of 2-exon-Tat in the suppression of RT activity, we compared HIV-1 proviral molecular clones that expressed either 1-exon-Tat or 2-exon-Tat. It was previously reported that an HIV-1 molecular clone expressing 1-exon-Tat (pM1ex) showed decreased replication capacity compared to viruses that expressed 2-exon-Tat (pNL101) (35). Our data confirm that pM1ex viruses possessed poorer replication kinetics than did either of two 2-exon-Tat encoding viruses, i.e., pNL101 or pNL43 (Fig. 5A). Namely, pM1ex viruses displayed an approximate 3-day delay in replication kinetics compared with either pNL43 or pNL101 in tissue culture. In concert with previous findings (9), it is conceivable that pM1ex viruses might replicate 1 to 2 logs less well than either pNL43 or pNL101.
FIG. 5.
Comparison of replication capacity and endogenous RT activity among pM1ex, pNL43 and pNL101 viruses. (A) Jurkat cells (6 × 105 cells) were infected with equivalent amounts (10 ng of p24 content) of viruses. The production of progeny virus was monitored by measuring de novo RT activity in the culture supernatant. (B) HeLa–LTR–β-Gal cells were infected with equivalent amounts (3 ng of p24 content) of viruses. After 48 h, cells were fixed and stained as described in Materials and Methods. The number of blue-stained cells was scored, and the results are expressed as the average ± the standard deviation. Three independent infections were performed for each viral preparation. (C) pM1ex, pNL43, and pNL101 virions were isolated from culture supernatants of transfected Cos-7 cells. Endogenous RT reactions were carried out using pelleted viruses containing 250 ng of p24, as described in Materials and Methods.
In addition, the replication capacity of these viruses was further analyzed in a MAGI assay. The results of Fig. 5B show that the pM1ex viruses (1-exon-Tat) generated fewer blue-stained cells than did either pNL43 or pNL101.
We also used purified virions of each type in endogenous RT reactions. Figure 5C shows that the efficiency of this reaction was lower for pM1ex than for either pNL101 or pNL43 virions. These results suggest that the decreased replication of pM1ex, compared to either pNL43 or pNL101, was caused by less-efficient RT reactions in the case of the pM1ex virions.
2-exon-Tat suppresses the RT reaction at late stages of the viral life cycle.
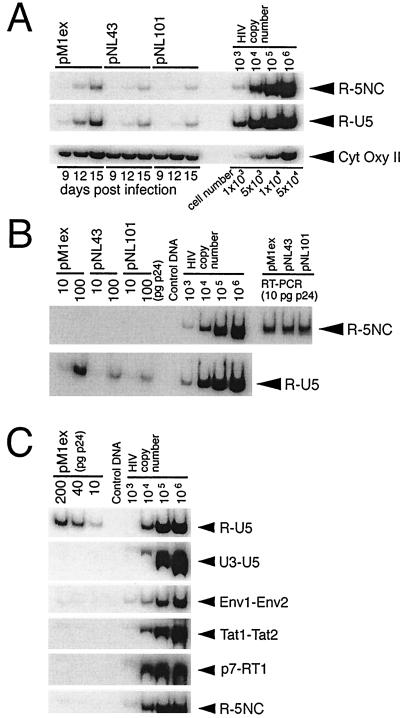
It may seem that a discrepancy exists between our in vitro results, showing the suppressive effect of 2-exon-Tat for RT activity (Fig. 1, 2, and 4), and our in vivo results, showing both superior replication capacity and more-efficient endogenous RT activity in the molecular clones that encode 2-exon-Tat (Fig. 5). One possible explanation is that 2-exon-Tat might suppress RT activity in the cytosol of infected cells at postintegration stages to promote efficient viral replication. To test this hypothesis, we performed quantitative PCR analyses of cytosolic extracts of infected cells. Figure 6A shows that the level of HIV-1 DNA in pM1ex-infected cells was markedly higher than that seen with either the pNL43 or pNL101 viruses at 12 days postinfection. Since the overall levels of HIV-1 DNA reflect the extent of ongoing de novo viral infection in an acutely infected cell culture (23), we expected that pM1ex-infected cells might have contained lower levels of HIV-1 DNA because of the poorer replication capacity of pM1ex virus (Fig. 5A) than either the pNL101 or pNL43 viruses. However, our PCR results show that levels of HIV-1 DNA, i.e., RT products, in pM1ex-infected cells were significantly higher than those seen in infections caused by either pNL43 or pNL101. The level of HIV-1 DNA in pM1ex-infected cells was even further increased, in relative terms, at 15 days postinfection (Fig. 6A). Accordingly, these results suggest the possibility that additional RT reactions may occur during the postintegration stage of viral replication in the absence of the second Tat exon.
FIG. 6.
PCR analysis of HIV-1 DNA. (A) PCR analysis of HIV-1 DNA in cytosolic fractions of infected cells. Cytosolic DNA was isolated from infected Jurkat cells at days 9, 12, and 15 postinfection, as described in Materials and Methods, and aliquots corresponding to 104 cells were subjected to quantitative PCR using the R-U5 and R-5NC primer pairs. To monitor the efficiency of cytosolic DNA isolation, the mitochondrial CytOxy II gene was amplified. (B) PCR analysis of virion-associated HIV-1 DNA. Virion-associated nucleic acids were extracted, and aliquots corresponding to 10 and 100 pg of p24 were subjected to quantitative PCR using the R-U5 and R-5NC primer pairs. To analyze the amount of viral genomic RNA in the extracted nucleic acids, samples corresponding to 100 ng of p24 were reverse transcribed with Moloney murine leukemia virus RT, and an aliquot of synthesized viral cDNA, corresponding to 10 pg of p24, was subjected to PCR, using the R-5NC primer pair (shown as RT-PCR in the figure). (C) Aliquots of pM1ex virion-associated nucleic acids, corresponding to 10, 40, and 200 pg of p24, were subjected to quantitative PCR, using the indicated primer pairs. Primer pair R-U5 was designed to detect early RT products synthesized either before or immediately after the first template switch. With primer pairs U3-U5, Env1-Env2, Tat1-Tat2, and p7-RT1, a specific PCR product was expected only if negative-strand DNA of increased length, relative to wild-type, had been synthesized after the first template switch. Primer pair R-5NC was predicted to amplify only late RT products synthesized after the second template switch. Serial 10-fold dilutions of pNL43 plasmid were used as positive controls. PCR products were separated on 6% polyacrylamide gels (0.5× TBE) at 4°C, and then the gels were dried and the bands were visualized on X-ray film.
Incorporation of negative-strand strong-stop DNA into pM1ex virions.
Since more RT reactions may occur during the synthesis of viruses lacking the second exon of Tat, the possibility exists that more HIV-1 DNA might be incorporated into pM1ex virions than into either pNL43 or pNL101. To investigate this subject, we performed quantitative PCR analyses of each of these types of virion. Figure 6B shows that the levels of HIV-1 DNA associated with pM1ex virions were markedly higher than those seen with either the pNL43 or pNL101 viruses. In contrast, similar amounts of viral genomic RNA were incorporated into each of the pM1ex, pNL43, and pNL101 virions, as assessed by RT-PCR (Fig. 6B). To further characterize the pM1ex virion-associated HIV-1 DNA, we performed PCR analysis using a variety of primer pairs. Figure 6C shows that early RT products, i.e., negative-strand strong-stop DNA, were preferentially amplified by PCR using primer pair R-U5 specifically for this purpose. In contrast, late or intermediate RT products were not nearly as detectable by PCR when primer pairs R-NC5, U3-U5, Env1-Env2, Tat1-Tat2, and p7-RT1 were employed (Fig. 6C). These results indicate that the majority of pM1ex virion-associated HIV-1 DNA was negative-strand-strong stop DNA.
Degradation of genomic RNA in pM1ex virions.
Conceivably, the poorer replication capacity of pM1ex compared with either pNL43 or pNL101 viruses (Fig. 5) might be the consequence of less-well-controlled RT activity during late stages of viral replication in the absence of wild-type Tat (Fig. 6). Although biochemical abnormalities, such as decreased packaging of RT, tRNA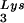 or RNA genome, have not been observed in viruses lacking the tat gene (16, 50), there is a possibility that inappropriate postintegrational initiation of reverse transcription before viral maturation might damage the viral RNA genome through RT-associated RNase H activity. To assess this possibility, we performed nondenaturing Northern blots of virion extracts. Figure 7 shows that the genomic RNA of viruses that expressed 2-exon-Tat, i.e., pNL43 and pNL101, was mainly detected as a dimer, a result consistent with expectations (lanes 2 and 3). Additionally, these dimeric RNA genomes were heat sensitive and disappeared upon incubation at 50°C for 10 min (reference 4 and Fig. 7, lane 4). In contrast, the genomic RNA of viruses encoding only 1-exon-Tat, i.e., pM1ex, was detected as a smeared band and appeared to be partially degraded (Fig. 7, lane 1). Of course, we cannot rule out the possibility that this RNA degradation of the pM1ex samples may have occurred during our extractions rather than in the intact virions, since the extracted genomes may be structurally unstable and easily degraded under inappropriate storage conditions (data not shown). However, at the very least, these data indicate that the genomic RNA of pM1ex viruses is more unstable than that of either pNL43 or pNL101.
or RNA genome, have not been observed in viruses lacking the tat gene (16, 50), there is a possibility that inappropriate postintegrational initiation of reverse transcription before viral maturation might damage the viral RNA genome through RT-associated RNase H activity. To assess this possibility, we performed nondenaturing Northern blots of virion extracts. Figure 7 shows that the genomic RNA of viruses that expressed 2-exon-Tat, i.e., pNL43 and pNL101, was mainly detected as a dimer, a result consistent with expectations (lanes 2 and 3). Additionally, these dimeric RNA genomes were heat sensitive and disappeared upon incubation at 50°C for 10 min (reference 4 and Fig. 7, lane 4). In contrast, the genomic RNA of viruses encoding only 1-exon-Tat, i.e., pM1ex, was detected as a smeared band and appeared to be partially degraded (Fig. 7, lane 1). Of course, we cannot rule out the possibility that this RNA degradation of the pM1ex samples may have occurred during our extractions rather than in the intact virions, since the extracted genomes may be structurally unstable and easily degraded under inappropriate storage conditions (data not shown). However, at the very least, these data indicate that the genomic RNA of pM1ex viruses is more unstable than that of either pNL43 or pNL101.
FIG. 7.
Nondenaturing Northern blot analysis of HIV-1 genomic RNA. Viral RNA samples corresponding to 500 ng of p24 were separated on nondenaturing 0.9% agarose gels and were transferred onto a nylon membrane. Hybridization with the denaturing HIV-1 probe was carried out as described in Materials and Methods. As a control experiment, RNA samples were heated at 50°C for 10 min before electrophoresis (heated pNL101).
DISCUSSION
We have demonstrated that a 2-exon but not a 1-exon form of Tat can strongly suppress the synthesis of DNA in cell-free RT reactions primed either with tRNA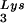 or a DNA primer (Fig. 1 and 2). A short synthetic peptide Tat[61–86], derived from the second exon of Tat, did not suppress the RT reaction (Fig. 1B), suggesting that intact Tat was necessary for the suppressive effect. In addition, this suppression was observed even when using an RNA template devoid of the TAR element, suggesting that 2-exon-Tat suppressed the RT reaction in a TAR-independent manner (Fig. 1C). These results suggest that Tat may serve an important role by limiting RT activity during the viral life cycle. Presumably, a suppressive mechanism may be required once synthesis of RT has occured, in order to limit the synthesis of inappropriately generated viral DNA, that might play an intracellular antisense function and conceivably interfere with viral assembly and/or the infectiousness of the viral progeny.
or a DNA primer (Fig. 1 and 2). A short synthetic peptide Tat[61–86], derived from the second exon of Tat, did not suppress the RT reaction (Fig. 1B), suggesting that intact Tat was necessary for the suppressive effect. In addition, this suppression was observed even when using an RNA template devoid of the TAR element, suggesting that 2-exon-Tat suppressed the RT reaction in a TAR-independent manner (Fig. 1C). These results suggest that Tat may serve an important role by limiting RT activity during the viral life cycle. Presumably, a suppressive mechanism may be required once synthesis of RT has occured, in order to limit the synthesis of inappropriately generated viral DNA, that might play an intracellular antisense function and conceivably interfere with viral assembly and/or the infectiousness of the viral progeny.
We have not yet been able to elucidate the precise mechanism by which Tat might play this inhibitory role. However, 2-exon-Tat suppressed RT reactions primed with the DNA primer (Fig. 2), indicating that Tat was able to block nonspecific elongation but not necessarily a specific initiation stage of the HIV-1 RT reaction. In addition, 2-exon-Tat was able to form a complex with primer-template RNAs and RT (Fig. 3). These results suggest that certain domains of wild-type Tat proteins can interact with the enzyme-substrate complex consisting of RT and primer-template to interfere with efficient DNA elongation. We are currently investigating this issue using a series of mutated Tat proteins in a detailed analysis of cell-free RT reactions.
In this report, the amount of viral RNA genome was analyzed by two different methods, i.e., RT-PCR (Fig. 6B) and nondenaturing Northern blot analysis (Fig. 7). It should be noted that a small region (i.e., 200 bp) can be analyzed by RT-PCR through use of the primer pair that was studied. On the other hand, the entire RNA genome (9.2 kb) can be analyzed by nondenaturing Northern blot analysis. Since degradation of pM1ex genomic RNA seems to have randomly occurred, it is likely that our RT-PCR analyses might not have been able to detect these events.
A phenotypic comparison of HIV-1 proviral molecular clones expressing either Tat 72, Tat 86, or Tat 101 revealed that viruses expressing either Tat 86 or Tat 101 (2-exon-Tat) possessed more rapid replication kinetics than viruses expressing Tat 72 (1-exon-Tat) (Fig. 5A). Conceivably, full-length 2-exon-Tat is necessary for efficient viral transactivation of the integrated genome (19), although the first exon of Tat may contain all of the domains required for this activity (20, 22). Additionally, the second Tat exon may also play a role in T-cell activation (36), which, in turn, may promote efficient viral replication (44). We have shown that viruses that encode the two-exon form of Tat, i.e., pNL43 or pNL101, had more efficient endogenous RT activity than viruses associated with the one-exon form of Tat, i.e., pM1ex (Fig. 5C). This helps to explain why the former viruses replicate better than the latter.
At first glance, a discrepancy might seem to be present between our in vitro results, showing the suppressive effect of 2-exon-Tat for RT, and our in vivo data, showing superior replication capacity and a more-efficient endogenous RT activity associated with 2-exon-Tat viruses. This is resolved by postulating that 2-exon-Tat may play a regulatory role in the suppression of RT activity in the cytoplasm of infected cells, i.e., preventing the accumulation and incorporation of prematurely synthesized viral DNA into virions. Quantitative PCR analysis revealed that the levels of HIV-1 DNA, i.e., RT products in the cytosolic fractions of cells infected with pM1ex virions that encode 1-exon-Tat were higher than was seen with either pNL43 or pNL101 that encode 2-exon-Tat (Fig. 6A). Consistently, higher levels of early RT products were also incorporated into pM1ex virions (Fig. 6B and C). However, Tat itself is presumably not incorporated into virions; this is consistent with the notion that Tat need not be present during reverse transcription in newly infected cells. We propose that a hitherto undescribed activity of Tat is to enhance viral infectivity by suppressing reverse transcription during late stages of viral production in infected cells.
Conceivably, such suppression of RT activity may also be important in preventing the premature initiation of tRNA-primed RT reactions. Of course, the existence of virion-associated viral DNA has been demonstrated (31, 49). However, the role of such DNA in the retroviral life cycle has not been elucidated, although it has been suggested that it may enhance, while not being essential for, viral infectivity (11, 54). In contrast, our PCR results indicate that excessive reverse transcription may have occured in the case of the pM1ex virus that also showed diminished replication kinetics. Moreover, nondenaturing Northern blot analysis revealed that the RNA genome of pM1ex was partially degraded (Fig. 7). Accordingly, we propose that initiation of reverse transcription before viral maturation might lead to damage of the viral RNA genome by RT-associated RNase H activity, a subject of particular importance with regard to nonspecific priming events (33). The observation that the Gag-Pol precursor polyprotein possesses some RT activity (32, 37) suggests that Tat-mediated suppression of RT is likely to be relevant during viral budding and before viral maturation.
Indeed, our data point to a possible functional analogy between the viral NCp7 and Tat proteins. NCp7 has been proposed to control the specificity of reverse transcription by inhibiting nonspecific, self-primed RT activity (28, 30). Such inhibition has also been observed for primer-specific RT reactions under certain conditions (28, 30). The fact that NCp7 can occlude viral RNA at a ratio of one NC protein per 7 nt (8) suggests that NCp7 might independently bind to residual nucleotide sequences to suppress nonspecific RT reactions. Since both Tat and NCp7 have zinc finger-like motifs (8, 15) and Tat is likely present in the cytoplasm (43), we propose that Tat might play a similar role in regard to RT activity by binding to genomic RNA in the cytosol in a TAR-independent manner.
On the other hand, the incorporation of Tat into virions has never been demonstrated, and we have also failed to detect Tat proteins in virus particles (data not shown). The mechanism whereby Tat must therefore be removed from viral genomic RNA prior to packaging remains to be elucidated. Since Tat is released from infected cells into culture fluids (12), one possible mechanism is that Tat proteins might be replaced by NCp7 during maturation of the viral core structure. We hope to demonstrate whether the affinity of mature NCp7 for viral RNA is comparable to that of Tat protein and whether NCp7 may play a role in viral assembly by potentially displacing Tat from complexes involving RT and viral genomic RNA.
ACKNOWLEDGMENTS
We thank K.-T. Jeang, Laboratory of Molecular Microbiology, National Institute of Allergy and Infectious Diseases, for a generous gift of the HIV-1 proviral molecular clones, pM1ex and pNL101, and for critical review of the manuscript. We also thank Cesar Collazos, Mervi A. Detorio, and Maureen Oliveira for technical assistance.
This work was supported by a grant from the Canadian Institutes for Health Research and by the National Institute of Allergy and Infectious Diseases (grant R01 AI43878-01).
REFERENCES
- 1.Adachi A, Gendelman H G, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C, Trono D. nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts E J, Li X, Gu Z, Kleiman L, Parniak M A, Wainberg M A. Comparison of deoxyoligonucleotide and tRNALys-3 as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J Biol Chem. 1994;269:14672–14680. [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas D K, Tius M A, Zhuo J, Pardee A B. Canventol inhibits HIV-1 replication by Tat-induced Tar-independent mechanism. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12:120–127. doi: 10.1097/00042560-199606010-00004. [DOI] [PubMed] [Google Scholar]
-
6.Cen S, Huang Y, Khorchid A, Darlix J-L, Wainberg M A, Kleiman L. The role of Pr55gag in the annealing of tRNA
 to human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
to human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar] - 7.Clouse K A, Powell D, Washington I, Poli G, Strebel K, Farrar W, Barstad P, Kovacs J, Fauci A S, Folks T M. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol. 1989;142:431–438. [PubMed] [Google Scholar]
- 8.Darlix J-L, Lapadat-Taposky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 9.Dimitrov D S, Willey R L, Sato H, Chang L-J, Blumenthal R, Martin M A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dornadula G, Yang S, Pomerantz R J, Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensoli B, Barillari G, Salahuddin S Z, Gallo R C, Wong-Staal F. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature. 1990;345:84–86. doi: 10.1038/345084a0. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y-X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 15.Frankel A D, Bredt D S, Pabo C O. Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science. 1988;240:70–73. doi: 10.1126/science.2832944. [DOI] [PubMed] [Google Scholar]
- 16.Harrich D, Ulich C, García-Martínez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L-M, Joshi A, Willey R, Orenstein J, Jeang K-T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modification of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 19.Jeang K-T, Berkhout B, Dropulic B. Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J Biol Chem. 1993;268:24940–24949. [PubMed] [Google Scholar]
- 20.Jeang K-T, Xiao H, Rich E A. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 21.Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;51:470–478. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 23.Kameoka M, Kimura T, Okada Y, Fujinaga K, Nakaya T, Takahashi H, Kishi M, Ikuta K. High susceptibility of U937-derived subclones to human immunodeficiency virus type 1 infection correlates with accumulation of unintegrated circular viral DNA. Virus Genes. 1996;12:117–129. doi: 10.1007/BF00572950. [DOI] [PubMed] [Google Scholar]
- 24.Kameoka M, Tanaka Y, Ota K, Itaya A, Yamamoto K, Yoshihara K. HIV-1 Tat protein is poly(ADP-ribosyl)ated in vitro. Biochem Biophys Res Commun. 1999;261:90–94. doi: 10.1006/bbrc.1999.0964. [DOI] [PubMed] [Google Scholar]
- 25.Kashanchi F, Agbottah E T, Pise-Masison C A, Mahieux R, Duvall J, Kumar A, Brady J N. Cell cycle-regulated transcription by the human immunodeficiency virus type 1 Tat transactovator. J Virol. 2000;74:652–660. doi: 10.1128/jvi.74.2.652-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y-S, Panganiban A T. The full-length Tat protein is required for TAR-independent, posttranscriptional trans activation of human immunodeficiency virus type 1 env gene expression. J Virol. 1993;67:3739–3747. doi: 10.1128/jvi.67.7.3739-3747.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanchy J M, Ehresmann C, Le Grice S F, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 1996;15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 28.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J-L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 29.Le Grice S F J, Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990;187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
-
30.Li X, Quan Y, Arts E J, Li Z, Preston B D, De Rocquigny H, Roques B P, Darlix J-L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA
 in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar] - 31.Lori F, di Marzo Veronese F, De Vico A L, Lusso P, Reitz M S, Jr, Gallo R C. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lori F, Scovassi A I, Zella D, Achilli G, Cattaneo E, Casoli C, Bertazzoni U. Enzymatically active forms of reverse transcriptase of the human immunodeficiency virus. AIDS Res Hum Retrovir. 1988;5:393–398. doi: 10.1089/aid.1988.4.393. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y, Planelles V, Li X, Palaniappan C, Day B, Challita-Eid P, Amado R, Stephans D, Kohn D B, Bakker A, Fay P, Bambara R A, Rosenblatt J D. Inhibition of HIV-1 replication using a mutated tRNALys-3 primer. J Biol Chem. 1997;272:14523–14531. doi: 10.1074/jbc.272.23.14523. [DOI] [PubMed] [Google Scholar]
- 34.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuveut C, Jeang K-T. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J Virol. 1996;70:5572–5581. doi: 10.1128/jvi.70.8.5572-5581.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott M, Emiliani S, Van Lint C, Herbein G, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science. 1997;275:1481–1485. doi: 10.1126/science.275.5305.1481. [DOI] [PubMed] [Google Scholar]
- 37.Peng C, Chang N T, Chang T W. Identification and characterization of human immunodeficiency virus type 1 Gag-Pol fusin protein in transfected mammalian cells. J Virol. 1991;65:2751–2756. doi: 10.1128/jvi.65.5.2751-2756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 39.Priel E, Showalter S D, Roberts M, Oroszlan S, Segal S, Aboud M, Blair D G. Topoisomerase I activity associated with human immunodeficiency virus (HIV) particles and equine infectious anemia virus core. EMBO J. 1990;9:4167–4172. doi: 10.1002/j.1460-2075.1990.tb07640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quan Y, Inouye P, Liang C, Rong L, Götte M, Wainberg M A. Dominance of the E89G substitution in HIV-1 reverse transcriptase in regard to increased polymerase processivity and pattern of pausing. J Biol Chem. 1998;273:21918–21925. doi: 10.1074/jbc.273.34.21918. [DOI] [PubMed] [Google Scholar]
- 41.Ratner L, Haseltine W, Ratarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz O, Marechal V, Danos O, Heard J M. Human immunodeficiency virus type 1 nef increases the efficiency of reverse transcription in the infected cells. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stauber R H, Pavlakis G N. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology. 1998;252:126–136. doi: 10.1006/viro.1998.9400. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi H, Matsuda M, Kojima A, Sata T, Andoh T, Kurata T, Nagashhima K, Hall W W. Human immunodeficiency virus type 1 reverse transcriptase: enhancement of activity by interaction with cellular topoisomerase I. Proc Natl Acad Sci USA. 1995;92:5694–5698. doi: 10.1073/pnas.92.12.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor J P, Pomerantz R, Bagasra O, Chowdhury M, Rappaport J, Khalili K, Amini S. TAR-independent transactivation by Tat in cells derived from the CNS: a novel mechanism of HIV-1 gene regulation. EMBO J. 1992;11:3395–3403. doi: 10.1002/j.1460-2075.1992.tb05418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Telesnitsky A, Goff S P. Reverse transcriptase and generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 48.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Gottlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 49.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulich C, Dunne A, Parry E, Hooker C W, Gaynor R B, Harrich D. Functional domains of Tat required for efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1999;73:2499–2508. doi: 10.1128/jvi.73.3.2499-2508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Schwedler U, Song J, Aiken C, Trono D. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei X, Götte M, Wainberg M A. Human immunodeficiency virus type-1 reverse transcription can be inhibited in vitro by oligonucleotides that target both natural and synthetic tRNA primers. Nucleic Acids Res. 2000;28:3065–3074. doi: 10.1093/nar/28.16.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Morris G F, Lockyer J M, Lu M, Wang Z, Morris C B. Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat. Virology. 1997;235:48–64. doi: 10.1006/viro.1997.8672. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Zhang Y, Spicer T P, Abbott L Z, Abbott M, Poiesz B J. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retrovir. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]