Abstract
Calcium-dependent protein kinases (CDPKs) act as key signal transduction enzymes in plants, especially in response to diverse stresses, including herbivory. In this study, a comprehensive analysis of the CDPK gene family in upland cotton revealed that GhCPKs are widely expressed in multiple cotton tissues and respond positively to various biotic and abiotic stresses. We developed a strategy for screening insect-resistance genes from a CRISPR–Cas9 mutant library of GhCPKs. The library was created using 246 single-guide RNAs targeting the GhCPK gene family to generate 518 independent T0 plants. The average target-gene coverage was 86.18%, the genome editing rate was 89.49%, and the editing heritability was 82%. An insect bioassay in the field led to identification of 14 GhCPK mutants that are resistant or susceptible to insects. The mutant that showed the clearest insect resistance, cpk33/74 (in which the homologous genes GhCPK33 and GhCPK74 were knocked out), was selected for further study. Oral secretions from Spodoptera litura induced a rapid influx of Ca2+ in cpk33/74 leaves, resulting in a significant increase in jasmonic acid content. S-adenosylmethionine synthase is an important protein involved in plant stress response, and protein interaction experiments provided evidence for interactions of GhCPK33 and GhCPK74 with GhSAMS1 and GhSAM2. In addition, virus-induced gene silencing of GhSAMS1 and GhSAM2 in cotton impaired defense against S. litura. This study demonstrates an effective strategy for constructing a mutant library of a gene family in a polyploid plant species and offers valuable insights into the role of CDPKs in the interaction between plants and herbivorous insects.
Key words: cotton, CDPKs, mutant library, CRISPR–Cas9, Ca2+ influx, insect resistance
CDPKs are crucial signal-transduction genes involved in plant defense against herbivorous insects. This study demonstrates a novel approach to screening for insect-resistance genes in the CDPK family of upland cotton using a CRISPR–Cas9 mutant library. This approach provides valuable genetic resources for functional studies of the CDPKs and creation of insect-resistant cotton germplasms.
Introduction
Cotton is a vital source of fiber, oil, and feed, making a significant contribution to global development (Chen et al., 2007). Throughout its growth period, cotton is constantly threatened by various herbivorous insects, which seriously affect yield and fiber quality. Insect pests have emerged as factors limiting the development of the cotton industry (Wu and Guo, 2005; Li et al., 2020; Quan and Wu, 2023). The traditional strategy of molecular breeding for insect resistance in cotton involves the introduction of exogenous genes encoding Bacillus thuringiensis insecticidal protein into the cotton genome to enhance its insect resistance (Wu et al., 2008; Huang et al., 2013; Qiao et al., 2017). Although this method temporarily alleviates the pressure caused by insect pests, issues such as the evolution of target pest resistance and outbreaks of non-target secondary pests have gradually emerged with the long-term cultivation of B. thuringiensis cotton (Lu et al., 2010; Heckel, 2012). There is therefore a pressing need to develop new strategies to address insect pest outbreaks.
Over millions of years, the selective pressure exerted by herbivorous insects has led to the evolution of effective defense systems in plants (Erb and Reymond, 2019). Plants respond to herbivorous insect attacks by initiating early signaling events, including depolarization of plasma transmembrane potential (Vm), increases in cytoplasmic Ca2+, generation of reactive oxygen species, and activation of mitogen-activated protein kinases. These pathways play central and conserved roles in enhancing broad-spectrum insect resistance in plants (Howe and Jander, 2008; Erb et al., 2012). Following the influx of Ca2+, Ca2+ sensors decode temporal and spatial signal transmission patterns to regulate metabolism and gene expression (Sanders et al., 2002). Typical Ca2+ sensors include calmodulin, calmodulin-like proteins, calcineurin B-like proteins, and calcium-dependent protein kinases (CDPKs). Among these, CDPK is the only sensor that combines sensing activity through a EF-hand structure with response activity through a protein kinase structure (Yip Delormel and Boudsocq, 2019). CDPK comprises four domains: the N-terminal variable domain, the Ser/Thr kinase domain, the inhibitory-junction domain, and the calmodulin-like domain (including an EF-hand calcium-binding site) (Batistic and Kudla, 2012). Its distinctive structure enables it to function concurrently as a Ca2+ receptor and an effector (Zuo et al., 2013). This specificity has generated particular research interest.
As an important class of Ser/Thr protein kinase genes, CDPKs form a large multigene family that is widely distributed in plants, green algae, protozoa, and oomycetes (Valmonte et al., 2014). CDPKs can be expressed across multiple tissues, including roots, stems, leaves, flowers, fruits, and seeds (Romeis et al., 2001; Ye et al., 2009). They can also localize to various cellular compartments, including the cytoplasm, nucleus, vacuolar membrane, endoplasmic reticulum, mitochondria, chloroplasts, and peroxisomes (Simeunovic et al., 2016). The wide distribution of CDPKs in plant tissues and multiple organelles indicates that they may have important functional roles in various biological processes of plants. For example, CDPKs play a significant role in the plant signal-transduction network in response to attack by herbivorous insects. In Arabidopsis, AtCPK3/13 regulate the transcript level of PDF1.2 by phosphorylating HsFB2a, thereby enhancing resistance against Spodoptera littoralis (Kanchiswamy et al., 2010). Simultaneous silencing of NaCPK4 and NaCPK5 results in significant accumulation of jasmonic acid (JA) in Nicotiana attenuata, enhancing its resistance against Manduca sexta (Yang et al., 2012). Knockout of GmCPK38, a soybean homolog of NaCPK4/5, alters numerous resistance-related phosphorylated proteins, genes, and metabolites, thereby enhancing soybean resistance to the common cutworm (Li et al., 2022). In addition, GmCPK17 has been shown to positively regulate the resistance of soybean hairy roots to the common cutworm (Wang et al., 2022). Various CDPKs from different plant species thus mediate responses to herbivorous insects by regulating gene expression or modulating hormone biosynthesis, exerting positive or negative effects.
The emergence of genome-editing tools has provided a tremendous opportunity for deciphering gene function, and the development of genome sequencing has accelerated the use of gene-editing tools (Pan et al., 2023). Because of its simple design, CRISPR–Cas9 can target the majority of DNA sequence fragments in the genome, enabling the rapid construction of large-scale knockout mutant libraries for forward genetic screening. Therefore, high-throughput loss-of-function screening of mutant libraries constructed using the CRISPR–Cas9 gene-editing system has been rapidly applied to plants (Shalem et al., 2014; Wang et al., 2018; Gaillochet et al., 2021). Lu et al. developed a pooled method for induction of genome-wide gene mutations in rice using a single-guide RNA (sgRNA) library (Lu et al., 2017). They constructed a pooled library of 88 541 sgRNAs targeting 34 234 genes, ultimately producing 91 004 T0 plants. Using CRISPR–Cas9, they performed comprehensive functional screening across the entire rice genome, producing a valuable resource for rice research and breeding. Small-scale functional genomic screening based on CRISPR mutant libraries has also been performed in tomato (Solanum lycopersicum), cotton (Gossypium hirsutum), soybean (Glycine max), and maize (Zea mays) through construction of libraries containing 54, 512, 70, and 1368 sgRNAs, respectively (Jacobs et al., 2017; Bai et al., 2020; Liu et al., 2020; Sun et al., 2024). These groundbreaking studies have independently demonstrated the utility of pooled CRISPR screening in plants.
CDPKs play a key role in plant defense against herbivorous insects as early signal-transduction genes (Romeis and Herde, 2014). We therefore developed a new approach to screen insect-resistance genes from the CDPK gene family in upland cotton using a CRISPR–Cas9 mutant library. First, through comprehensive analysis of the GhCPK gene family, we identified 82 GhCPKs in the upland cotton genome and predicted that this gene family was involved in multiple biological processes in cotton, laying the foundation for functional analysis of the GhCPK family. Next, we constructed a CRISPR–Cas9 mutant library for the GhCPK gene family. Through field phenotyping and insect-resistance experiments, we screened the insect-resistant cpk33/74 cotton lines from the mutant library, and we performed preliminary investigations of the insect-resistance mechanisms of the screened candidate genes, GhCPK33 and GhCPK74. This study provides valuable insights into the role of CDPKs in plant–insect interactions. In addition, the numerous mutants developed here provide a valuable genetic resource for functional study of the CDPKs and creation of pest-resistant cotton germplasms.
Results
Comprehensive analysis of the CDPK gene family in upland cotton
A total of 82 GhCPK genes were identified in the upland cotton genome, and genomic information and physicochemical properties of all GhCPK members were summarized, including gene names, chromosomal locations, amino acid residues (which varied from 331 to 907), relative molecular weights (37.40 to 101.03 kDa), and isoelectric points (4.68 to 9.48) (Supplemental Table 1). N-myristoylation and S-palmitoylation are important post-translational protein modifications; among the 82 predicted GhCPKs, 35 had N-terminal myristoylation sites, and 75 had at least one palmitoylation site. Predictions of transmembrane domains, signal peptides, and EF-hand structures revealed that all GhCPK proteins lack transmembrane domains, most lack signal peptides, and 78 have four EF-hands, whereas four have three EF-hands (Supplemental Table 1).
To examine the evolutionary relationships among the CDPK genes, we constructed an unrooted phylogenetic tree by aligning the full-length protein sequences of 34 AtCPKs, 31 OsCPKs, and 82 GhCPKs (Supplemental Figure 1). All CDPK proteins from the three species clustered into four distinct subgroups (groups 1–4), implying that these subgroups existed prior to the differentiation of monocots and dicots. We used the MEME website to predict 10 conserved motifs in the GhCPK proteins (Supplemental Figure 2). Motifs 1, 3, and 5 were present in all GhCPK proteins except GhCPK45 and GhCPK50. Motif 4 was present in all proteins except GhCPK50 and GhCPK82. There were also differences in protein motifs among different subgroups; for example, motif 7 was unique to group 3, and all members of group 4 lacked motif 6. GhCPKs from the same subgroup possessed similar numbers, types, and spatial distributions of motifs, supporting their close evolutionary relationships. Differences in motifs among different subgroups imply that CDPKs have diverse functions in upland cotton. An analysis of gene structure is shown in Supplemental Figure 2; despite variation in exon numbers (7 to 16) among family members, GhCPKs from the same subgroup exhibited very similar exon–intron structures. Most members of group 1 had seven exons, and all members of group 4 had 12 exons. Conservation of these gene structures among members of each subgroup lends support to their close evolutionary relationships.
Collinearity analysis of the GhCPKs was performed with MCScanX and Circos software to investigate their mechanism of amplification (Supplemental Figure 3). A total of 128 collinear gene pairs were identified, each composed of two genes located on distinct chromosomes; because these pairs were not physically adjacent, segmental duplication appears to have been the main amplification mechanism for the GhCPK gene family. Collinearity analyses of the G. hirsutum GhCPKs with CPKs of Arabidopsis thaliana, Oryza sativa, and Gossypium raimondii revealed 174 pairs of orthologous genes between G. hirsutum and G. raimondii, 84 pairs between G. hirsutum and A. thaliana, and only 13 pairs between G. hirsutum and O. sativa (Supplemental Figure 4). These results suggest a closer evolutionary relationship of G. hirsutum to other dicots. To examine the potential functions and regulatory mechanisms of the GhCPK family, we analyzed the cis-acting elements in the 2-kb promoter sequences of all GhCPKs (Supplemental Figure 5). A total of 29 elements were identified and classified into four categories based on their functions: elements related to plant hormones, plant growth and development, defense response, and light response. These results indicate that CDPKs may function in diverse biological processes, indirectly reflecting the importance of this gene family in upland cotton.
GhCPKs respond to multiple stresses at the transcriptional level
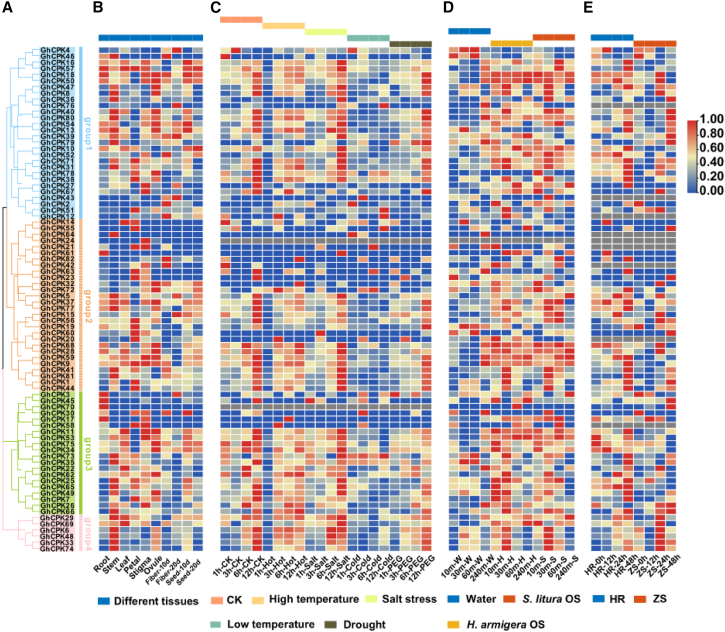
Gene expression is closely related to gene function. To explore the potential roles of the GhCPKs, the expression patterns of the 82 GhCPK genes were analyzed in the context of their phylogenetic relationships (Figure 1A). Most GhCPKs were expressed in multiple tissues, although GhCPK55 was specifically expressed in petals and GhCPK64 was specifically expressed in stems. All GhCPKs in group 4 showed relatively high expression in all tissues (Figure 1B). Expression of the cotton GhCPKs under four abiotic stress conditions (heat, salt, cold, and drought) was also analyzed (Figure 1C). A total of 73% of the GhCPKs showed significantly decreased expression after 1 h of high-temperature treatment (38°C) compared with the control group. Most GhCPKs demonstrated continuous downregulation in response to low-temperature treatment (4°C), particularly after 12 h. We next analyzed expression of the GhCPKs in cotton leaves after exposure to the oral secretions (OS) of Helicoverpa armigera and S. litura (Figure 1D). At 10 and 30 min after treatment with H. armigera or S. litura OS, at least 73% of the GhCPKs showed upregulated expression, which peaked at 30 min after treatment, and all GhCPKs in group 4 showed marked upregulation at both time points. However, after 240 min of treatment, the induced differential gene expression was no longer evident. We also studied the expression of GhCPKs at different time points in two cotton cultivars (exhibiting either strong resistance [HR] or sensitivity [ZS] to whitefly) upon whitefly infection (Figure 1E). The differential expression of GhCPKs in cotton leaves was most pronounced after 48 h of whitefly infection. In the strongly resistant cultivar, 54% of GhCPKs were significantly induced.
Figure 1.
Expression patterns of the GhCPK gene family in different tissues and under different stress conditions.
(A) Phylogenetic tree of the GhCPK gene family. Different colors represent different subgroups.
(B) Expression profiles of GhCPKs in different tissues. From left to right are root, stem, leaf, petal, anther, stigma, ovule, fiber 10 days, fiber 20 days, seed 10 days, and seed 20 days.
(C) Expression profiles of GhCPKs induced by four types of abiotic stress: heat, cold, polyethylene glycol (PEG), and salt. The induction times for each type of stress were 1, 3, 6, and 12 h.
(D) Expression patterns of GhCPK genes after exposure to OS of H. armigera and S. litura. Four time points after processing were selected: 10, 30, 60, and 240 min.
(E) Expression patterns of the GhCPK gene family at different time points (0, 12, 24, and 48 h after infection) in two cotton cultivars (with strong resistance [HR] or sensitivity [ZS] to whitefly) infected by whitefly. In all the expression profiles mentioned above, red indicates increased expression, and blue indicates decreased expression.
In summary, GhCPKs are widely expressed in different tissues and show significant changes in expression under various external stresses. These findings imply that the CDPK gene family is involved in multiple biological processes during cotton’s growth, development, and response to adverse environmental conditions, making it an important gene group for upland cotton.
Construction of a mutant library for the upland cotton CDPK gene family
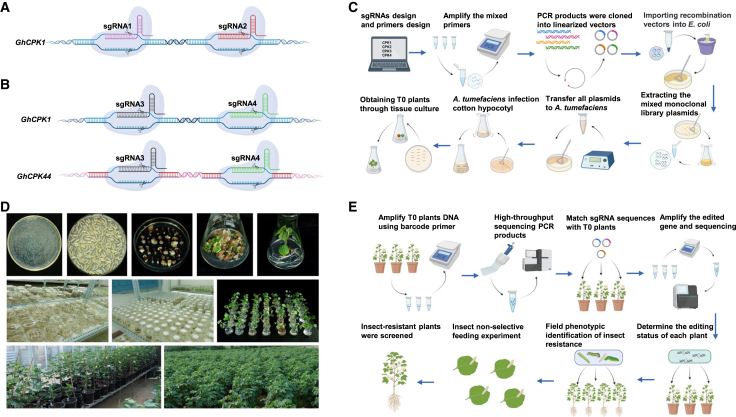
To better understand the functional roles of GhCPKs in cotton resistance to insects, we developed an insect-resistance screening method for the GhCPKs based on a CRISPR–Cas9 mutant library. First, we designed 246 sgRNAs targeting the GhCPKs (Supplemental Table 2); these sgRNAs were classified into two types (Supplemental Figure 6A). The first type was designed to target individual GhCPKs; in this case, two different sgRNAs were designed to target an exon in a specific GhCPK (Figure 2A). Because of possible functional redundancy among genes of allotetraploid cotton, we also designed a second type of sgRNA to target homologous GhCPKs in the A and D subgenomes. In this case, two sgRNAs were designed to target the exon region in a pair of homologous genes, simultaneously mutating the two homologous GhCPKs but not other genes (Figure 2B).
Figure 2.
Construction and detection procedures for the mutant library of the GhCPK gene family.
(A) Schematic diagram of sgRNA design for knockout of a single GhCPK. SgRNA1 and sgRNA2 target GhCPK1 individually and exclusively.
(B) Schematic diagram of sgRNA design for knockout of homologous GhCPKs. SgRNA3 and sgRNA4 simultaneously target both GhCPK1 and GhCPK44 and exclusively target only GhCPK1 and GhCPK44.
(C) Procedure for construction of a mutant library for the CDPK gene family in upland cotton.
(D) The process of cotton genetic transformation.
(E) Molecular detection and insect-resistance screening procedure for the GhCPK mutant library.
To rapidly and efficiently obtain a mutant library of the GhCPKs, we used a mutant library construction method based on primer mixing pools (Figure 2C). We designed 246 primers based on the sgRNA sequences and constructed a mixed vector library using six primer mixing pools (Supplemental Table 3). The mixed-vector library constructed with primer mixing pool 1 was used to validate the feasibility of the method (Supplemental Table 4). By sequencing 100 collected monoclonal clones, we found that 36 sgRNAs were detected, with a coverage rate of 90% (Supplemental Figure 6B). After matching sgRNAs with genes, we found that the 36 sgRNAs could target all 20 genes, achieving a gene coverage of 100% (Supplemental Figure 6C). The majority of sgRNAs were detected two or three times (Supplemental Figure 6D). These results indicate that the method of constructing a mixed-vector library through primer mixing pools is feasible. The plasmids extracted from the mixed-vector library were transferred into Agrobacterium tumefaciens for stable transformation of cotton to produce genetically edited plants (Figure 2D), and we obtained 518 regenerated plants in the T0 generation. The method of constructing a CRISPR–Cas9 mutant library based on a primer mixing pool saves significant time and effort, enabling the rapid acquisition of a substantial number of mutant plants.
Molecular detection for the GhCPK mutant library
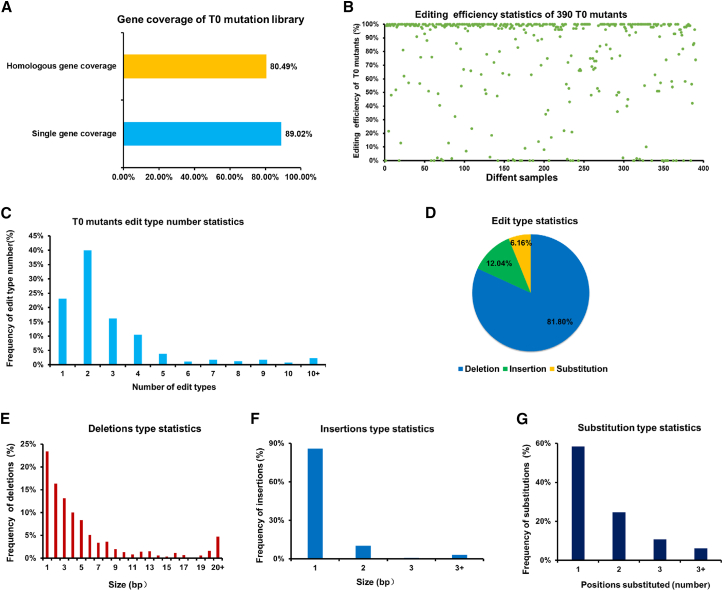
Molecular detection was performed to study the gene-editing status of each plant in the mutant library using the specific detection procedure shown in Figure 2E. Barcode high-throughput sequencing enabled us to quickly match the sgRNA sequences carried by each plant, accurately identifying the edited target gene(s). Among the 518 plants in the mutant library, 16.02% had no detected sgRNAs, 81.08% had one detected sgRNA, and 2.89% had two or three detected sgRNAs. Thus, only one (or a pair of) GhCPKs had been edited in the majority of T0 plants (Supplemental Figure 7B). Only T0 plants carrying a single sgRNA were selected for further investigation. A total of 147 sgRNAs targeting 73 individual GhCPKs and 33 pairs of homologous GhCPKs were detected. The average target gene coverage was 86.18% (Figure 3A). In general, on the basis of sgRNA detection in the T0-generation plants, we successfully obtained gene-edited materials that met our needs for establishment of a CRISPR–Cas9 mutant library of the CDPK gene family in upland cotton. This result implied that construction of a large-scale mutant library using a mixed-vector library is feasible in upland cotton.
Figure 3.
Molecular detection results for T0-generation plants of the GhCPK mutant library.
(A) Gene coverage of T0-generation plants from the GhCPK mutant library. Yellow represents the coverage of homologous genes, and blue represents the coverage of individual genes.
(B) Editing efficiency of 390 T0-generation plants from the GhCPK mutant library. Different samples are represented by green dots.
(C) Types of edits in T0-generation plants of the GhCPK mutant library. “Edit type” refers to different types of mutated reads produced after genes have been edited by the Cas9 protein. The number of edit types varies for each plant, ranging from one to more than 10.
(D) Classification of edit types in T0-generation plants of the GhCPK mutant library. Blue represents base absence, green represents base insertion, and yellow represents base substitution.
(E) Base deletion statistics. The x axis represents the number of deleted bases, and the y axis represents the frequency.
(F) Base insertion statistics. The x axis represents the number of inserted bases, and the y axis represents the frequency.
(G) Base substitution statistics. The x axis represents the number of substituted bases, and the y axis represents the frequency.
Next, primers were designed for each mutant plant based on the position of the sgRNA in the target gene (Supplemental Table 6). By analyzing the amplified sequences, the specific editing status of each gene could be determined. Because upland cotton is an allopolyploid, various types of fragments appear in the amplified sequences after editing. Traditional Sanger sequencing cannot meet the requirements for such complex amplifications. Therefore, high-throughput sequencing was performed. To verify the accuracy of this method, two randomly selected gene-edited plants were subjected to both high-throughput sequencing and Sanger sequencing (Supplemental Figure 7C). The results of the two sequencing methods were consistent, indicating that high-throughput sequencing is suitable for the detection of gene-edited cotton. By analyzing the high-throughput sequencing results of 390 T0-generation plants, we found that 349 plants (89.49%) had undergone effective editing (editing efficiency ≥10%) (Supplemental Figure 7D). Among them, 273 plants (78.22%) showed highly efficient editing (editing efficiency ≥80%) (Figure 3B). Analysis of editing patterns in T0 plants revealed that the number of editing types varied from one to more than 10 for different plants (Figure 3C). cpk37-3, cpk36/76-1, and cpk6/48-1 demonstrated one, two, and more than 10 editing types, respectively (Supplemental Figure 7E). The editing types included deletions, insertions, and substitutions. cpk45-3 exhibited all three types of mutation simultaneously (Supplemental Figure 7F). Deletions (81.80%) accounted for the largest proportion in all editing types, followed by insertions (12.04%) (Figure 3D). The majority of deletions consisted of one to five missing bases (Figure 3E), whereas most insertion and substitution events consisted of one- or two-base insertions or substitutions (Figure 3F and 3G). These results demonstrate that the CRISPR–Cas9 system enables efficient editing of upland cotton through small-fragment insertions, deletions, and substitutions.
Editing heritability analysis of gene-edited materials in the GhCPK mutant library
To determine whether the edited genes could be stably inherited in T1 offspring, 50 gene-edited plants from the T0 generation were randomly selected for genetic analysis. Three plants from the progenies of each line were randomly chosen for high-throughput sequencing, and editing heritability analysis was performed by comparing their gene-editing types and editing efficiencies with those of the parental plants. If all the gene-editing types found in the T1 plants could be traced back to the parental plants, they were considered to be inherited from the parental generation. Among 150 T1 plants, 123 plants (82%) had gene-editing types that were completely inherited from the parental generation, 24 plants (16%) contained new gene-editing types, and three plants (2%) had not undergone gene editing (Supplemental Figure 8A and 8C). In terms of gene-editing efficiency, 101 plants (67.33%) in the T1 generation inherited 100% editing efficiency from the parental generation. Thirty-one plants (20.67%) had higher gene-editing efficiency than the parental generation, and 18 plants (12%) had lower gene-editing efficiency (Supplemental Figure 8B). These results indicate that most T1 plants inherit the parental editing patterns, but the genetic rules are relatively complicated. For instance, T1-cpk53-1-2 completely inherited the editing type and efficiency of its T0 parent; T0-cpk64-2 had an editing efficiency of only 2.91% in the T0 generation, but the offspring showed an inheritance efficiency of 100% (Supplemental Figure 8D).
Screening of insect-resistant materials from the GhCPK mutant library
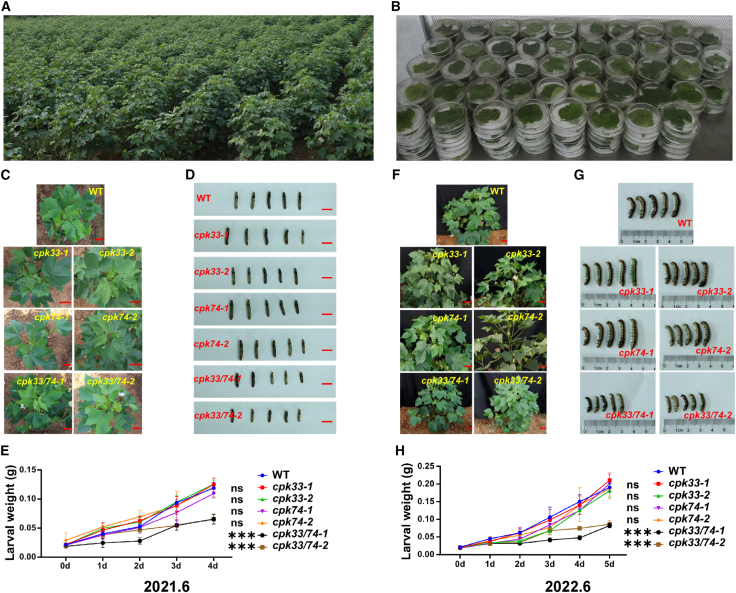
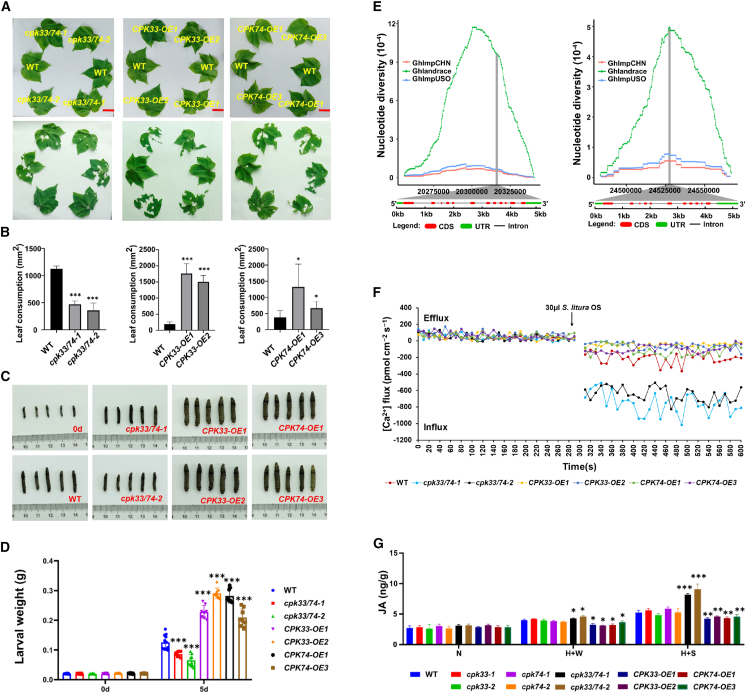
To quickly screen for insect-resistant GhCPK mutants, we measured the damage caused by chewing pests on 243 cotton lines (involving editing of 62 independent GhCPKs and 28 pairs of homologous GhCPKs) grown in the field in two consecutive years and performed no-choice feeding experiments on all cotton lines using S. littoralis larvae (Figure 4A and 4B). Based on the damage caused by chewing pests to cotton lines in the field, combined with the results of the no-choice feeding experiment with S. littoralis larvae, this serves as a reference standard. Mutant lines whose field pest-resistance phenotypes were consistent with the results of the non-selective feeding experiment were selected for pest resistance evaluation (Supplemental Figure 9A and 9B). We also required that the insect-resistance phenotypes of mutant lines could be replicated across different lines with the same gene knockout. Six stable insect-resistance genes and eight stable susceptibility genes were identified (Table 1). In particular, the cpk33/74 cotton lines with simultaneous knockout of the homologous genes GhCPK33 and GhCPK74 showed the most significant insect-resistance phenotype. Plants of cpk33/74 showed almost no trace of chewing-insect damage in the field evaluation (Figure 4C and 4F), and larvae that fed on cpk33/74 had significantly lower weights than controls in a selective feeding assay. However, cotton plants with only GhCPK33 (cpk33) or only GhCPK74 (cpk74) knockout did not exhibit insect resistance (Figure 4D, 4E, 4G, and 4H).
Figure 4.
Screening for materials that are resistant or sensitive to chewing pests in the GhCPK mutant library.
(A) Field phenotyping for insect resistance in materials from the GhCPK mutant library.
(B) Non-selective feeding experiment with S. litura larvae on leaves of materials from the GhCPK mutant library.
(C) Field phenotyping for insect resistance of T1-generation cpk33, cpk74, and cpk33/74 plants. The red line represents 5 cm.
(D) Comparison of body sizes of S. litura larvae after 4 days of continuous non-selective feeding on leaves of T1-generation cpk33, cpk74, and cpk33/74 plants. The red line represents 1 cm.
(E) Average weight changes in S. litura larvae after 4 days of continuous feeding on leaves of T1-generation cpk33, cpk74, and cpk33/74 plants.
(F) Field phenotyping for insect resistance of T2-generation cpk33, cpk74, and cpk33/74 plants. The red line represents 5 cm.
(G) Comparison of the body sizes of S. litura larvae after 5 days of continuous non-selective feeding on leaves of T2-generation cpk33, cpk74, and cpk33/74 plants.
(H) Average weight changes in S. litura larvae after 5 days of continuous feeding on leaves of T2-generation cpk33, cpk74, and cpk33/74 plants. Means ± SE (n = 12). Statistical analyses were performed using Student’s t-test. ∗∗∗p < 0.001.
Table 1.
Evaluation of insect resistance in plants of the GhCPK mutant library for defense against chewing pests.
| Mutant line | Mutated gene ID | Chewing pest resistance | Average editing efficiency |
|---|---|---|---|
| cpk1 | Gh_A01G0621 | ns | 23.76% |
| cpk6 | Gh_A02G1635 | + | 100.00% |
| cpk15 | Gh_A05G1571 | + | 100.00% |
| cpk16 | Gh_A05G2355 | ns | 99.08% |
| cpk18 | Gh_A05G3246 | ns | 78.81% |
| cpk24 | Gh_A09G1033 | − − − | 93.88% |
| cpk26 | Gh_A09G1157 | ns | 100.00% |
| cpk27 | Gh_A09G1248 | ns | 100.00% |
| cpk29 | Gh_A10G0886 | ns | 100.00% |
| cpk33 | Gh_A11G1615 | ns | 100.00% |
| cpk37 | Gh_A12G2686 | − | 100.00% |
| cpk39 | Gh_A13G0563 | − | 76.02% |
| cpk40 | Gh_A13G0566 | ns | 100.00% |
| cpk45 | Gh_D02G0183 | − − | 70.21% |
| cpk46 | Gh_D02G0663 | ns | 99.28% |
| cpk47 | Gh_D02G1973 | ns | 100.00% |
| cpk48 | Gh_D03G0087 | ns | 100.00% |
| cpk51 | Gh_D04G0895 | ns | 100.00% |
| cpk52 | Gh_D04G0900 | ns | 99.69% |
| cpk53 | Gh_D04G1271 | ns | 100.00% |
| cpk61 | Gh_D07G1198 | − − − | 100.00% |
| cpk63 | Gh_D08G0142 | ns | 100.00% |
| cpk64 | Gh_D09G1054 | − | 100.00% |
| cpk67 | Gh_D09G1249 | ns | 27.64% |
| cpk71 | Gh_D10G2029 | − − | 100.00% |
| cpk74 | Gh_D11G1774 | ns | 100.00% |
| cpk75 | Gh_D11G3329 | ns | 100.00% |
| cpk76 | Gh_D12G0121 | ns | 100.00% |
| cpk77 | Gh_D12G2743 | ns | 100.00% |
| cpk79 | Gh_D13G0560 | + | 99.78% |
| cpk80 | Gh_D13G0561 | ns | 96.52% |
| cpk6/48 | Gh_A02G1635 and Gh_D03G0087 | + | 100.00% |
| cpk8/47 | Gh_A03G1505 and Gh_D02G1973 | ns | 100.00% |
| cpk10/52 | Gh_A04G0467 and Gh_D04G0900 | ns | 100.00% |
| cpk13/54 | Gh_A04G1429 and Gh_D04G1486 | +++ | 100.00% |
| cpk20/60 | Gh_A06G1772 and Gh_D06G2206 | ns | 98.45% |
| cpk21/61 | Gh_A07G1099 and Gh_D07G1198 | ns | 100.00% |
| cpk29/69 | Gh_A10G0886 and Gh_D10G0863 | ns | 99.54% |
| cpk30/70 | Gh_A10G1195 and Gh_D10G1303 | ns | 100.00% |
| cpk33/74 | Gh_A11G1615 and Gh_D11G1774 | +++ | 100.00% |
| cpk36/76 | Gh_A12G0109 and Gh_D12G0121 | ns | 100.00% |
| cpk37/77 | Gh_A12G2686 and Gh_D12G2743 | − | 58.89% |
Note: “+” indicates insect resistance, “−” indicates insect susceptibility, and “ns” indicates no difference.
Overexpression of GhCPK33 and GhCPK74 reduces cotton defense against S. litura larvae
To investigate the role of GhCPK33 and GhCPK74 in the interaction between cotton and herbivorous insects, we separately constructed overexpression vectors for GhCPK33 and GhCPK74 (Supplemental Figure 10A and 10B). Through Agrobacterium-mediated transformation, we created independent transgenic plants overexpressing GhCPK33 (CPK33-OE) and GhCPK74 (CPK74-OE). On the basis of GhCPK33 and GhCPK74 expression levels, four independent lines (CPK33-OE1, CPK33-OE2, CPK74-OE1, and CPK74-OE3) were selected for further study from 22 CPK33-OE and 14 CPK74-OE lines (Supplemental Figure 10C and 10D). The T2 generation of mutants (cpk33/74-1 and cpk33/74-2) and the T1 generation of overexpression plants (CPK33-OE1, CPK33-OE2, CPK74-OE1, and CPK74-OE3), as well as the wild type (WT), were sown simultaneously. After 40 days, selective feeding and non-selective feeding experiments were performed on all materials using S. litura larvae. After 24 h of selective feeding by S. litura larvae, leaf consumption was significantly higher for the CPK33-OE and CPK74-OE lines than the WT but significantly lower for the cpk33/74 lines (Figure 5A and 5B). In the non-selective experiment, after feeding for 5 days, the weight of the S. litura larvae fed with CPK33-OE and CPK74-OE leaves was significantly higher than that of the larvae fed with WT leaves, whereas the weight of larvae fed with cpk33/74 leaves was significantly lower (Figure 5C and 5D). CPK33-OE and CPK74-OE thus exhibited significantly reduced defense against S. litura, whereas cpk33/74 plants showed significantly enhanced defense. We hypothesize that GhCPK33 and GhCPK74 play a negative regulatory role in cotton defense against S. litura larvae.
Figure 5.
Evidence for the involvement of GhCPK33 and GhCPK74 in pest defense of upland cotton.
(A) Choice feeding assay with S. litura larvae and cpk33/74, CPK33-OE, CPK74-OE, and WT plants. The third-instar S. litura larvae were pre-starved for 6 h and photographed at 0 and 24 h. The red line represents 3 cm.
(B) Leaf consumption in the choice feeding assay with S. litura. Means ± SE (n = 6).
(C) Comparison of the body sizes of S. litura larvae after 5 days of continuous non-selective feeding on leaves of cpk33/74, CPK33-OE, CPK74-OE, and WT plants.
(D) Average body weight changes of S. litura larvae after 5 days of continuous feeding on leaves of cpk33/74, CPK33-OE, CPK74-OE, and WT plants. Means ± SE (n = 12).
(E) Nucleotide diversity of the Ghlandraces, GhImpUSO, and GhImpCHN populations. The x axis represents the 20 kb upstream and downstream of GhCPK33 or GhCPK74, and the y axis represents nucleotide diversity.
(F) NMT was used to detect dynamic changes in cytoplasmic Ca2+ flux in cpk33/74, CPK33-OE, CPK74-OE, and WT leaf mesophyll cells before and after OS induction.
(G) JA content of cpk33/74, cpk33, cpk74, CPK33-OE, CPK74-OE, and WT leaves. N, not treated; H + W, mechanical damage; H + S, S. litura feeding. Means ± SE (n = 3). Statistical analyses were performed using Student’s t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Evolution and domestication analysis of the GhCPK33 and GhCPK74 loci
Resequencing data from 1623 cotton germplasm resources from previous studies was used to investigate the evolution and domestication of the GhCPK33 and GhCPK74 loci (Li et al., 2021). The 1623 germplasms included 256 G. hirsutum landraces (Ghlandraces), which are naturally insect-resistant varieties; 438 elite G. hirsutum cultivars from the USA and other countries (GhImpUSO); and 929 elite G. hirsutum cultivars from China (GhImpCHN). The results revealed significant single-nucleotide polymorphism (SNP) variation around the GhCPK33 and GhCPK74 loci (20 kb upstream and downstream) in Ghlandraces (Figure 5E). To further investigate the evolution of GhCPK33 and GhCPK74 in the Ghlandraces population, we constructed genotype maps for the GhCPK33 and GhCPK74 loci (Supplemental Figure 11A and 11B). Haplotype analysis of the GhCPK33 locus in the Ghlandraces population revealed the presence of four SNPs, two of which were located in introns and two in the 3′ UTR. The major SNP variants were observed at positions 24 527 886 (A to G) and 24 531 788 (G to A), accounting for 24.22% (Supplemental Figure 11A). Haplotype analysis of the GhCPK74 locus in the Ghlandraces population revealed SNPs at nine positions: three in exons, five in introns, and one in the 5′ UTR. The main SNP variants were observed at positions 20 320 372 (A to G) and 20 319 885 (A to T), accounting for 6.64% (Supplemental Figure 11B). In summary, we speculate that GhCPK33 and GhCPK74 may have influenced the insect resistance of upland cotton during evolution and domestication processes.
GhCPK33 and GhCPK74 negatively regulate Ca2+ influx induced by S. litura OS
Elevation of cytoplasmic Ca2+ is an important early signal-transduction step in plants for perceiving attacks from herbivorous insects and initiating defense responses (Howe and Jander, 2008). Here, we used non-invasive micro-test technology (NMT) to investigate dynamic changes in cytoplasmic Ca2+ flux in cotton leaf mesophyll cells induced by the OS of S. litura and to explore the involvement of GhCPK33 and GhCPK74 in mediating post-ingestion Ca2+ influx by the insects (Figure 5F). In the untreated state, Ca2+ in the leaf mesophyll cells of WT, cpk33/74, CPK33-OE, and CPK74-OE showed an efflux state, with no significant differences in flow rate. The average flow rate within 5 min ranged from 40 to 100 pmol cm−2 s−1. After treatment with the OS of S. litura, the leaf mesophyll cells of cpk33/74 exhibited a strong and rapid Ca2+ influx, with an average flow rate of 701 pmol cm−2 s−1 within 5 min. However, this ability was impaired in CPK33-OE and CPK74-OE compared with the WT. The influx rate of Ca2+ was slowed in these lines, with average flow rates of only 52 and 104 pmol cm−2 s−1 within 5 min, respectively (Supplemental Figure 11C and 11D). These results indicate that GhCPK33 and GhCPK74 negatively regulate the Ca2+ influx induced by S. litura.
GhCPK33 and GhCPK74 negatively regulate JA synthesis induced by OS of S. litura
JA is an important hormone related to plant insect resistance. To investigate the underlying mechanism of ChCPK33/GhCPK74-regulated insect resistance, we measured JA content in insect-induced cotton leaves. We cultured T2 generation plants of cpk33, cpk74, cpk33/74, CPK33-OE, CPK74-OE, and WT in the same environment. After 6 weeks, the leaf surfaces of all materials were scratched using a pattern wheel, and the injured leaf area was then coated with OS of S. litura (H + S) to simulate feeding by this insect. Water was applied to the injured area of the leaf (H + W) to simulate mechanical damage, and no treatment was applied to control leaves (N). After a 30-min treatment, JA content was measured in the leaves of all materials. Both H + W and H + S treatments induced JA production in cotton leaves. However, compared with WT leaves, cpk33/74 leaves showed a more significant increase in JA content after H + S treatment. By contrast, the JA content of CPK33-OE and CPK74-OE leaves was lower than that of WT leaves after H + S treatment (Figure 5G). These results suggest that GhCPK33 and GhCPK74 can negatively regulate the JA synthesis induced by S. litura OS. Therefore, the cpk33/74 material, with simultaneous knockout of GhCPK33 and GhCPK74, shows enhanced resistance against insects. However, JA content in cpk33 and cpk74 leaves did not differ significantly from that of WT leaves after H + S treatment. Thus, we hypothesize that GhCPK33 and GhCPK74 are functionally redundant in the negative regulation of JA synthesis.
GhCPK33 and GhCPK74 interact with GhSAMS1 and GhSAMS2
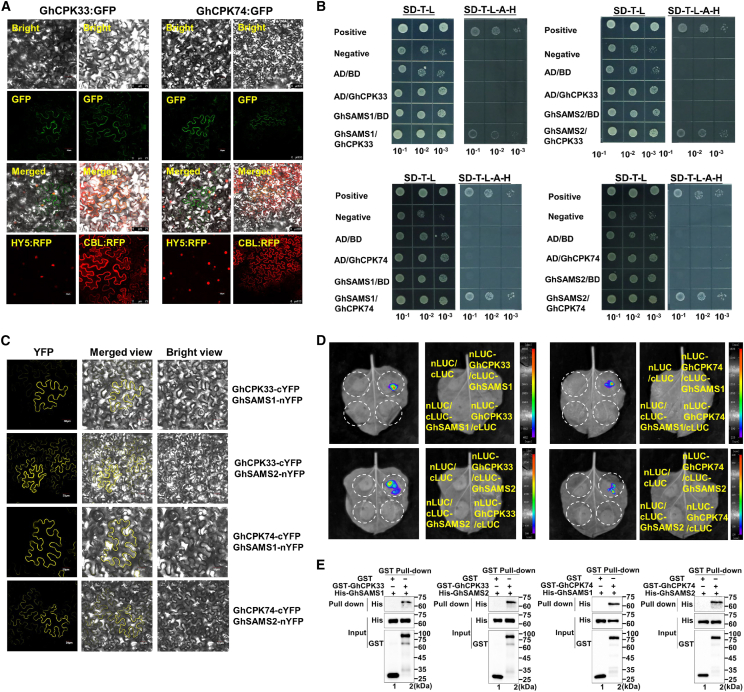
The subcellular localization of GhCPK33 and GhCPK74 was examined by fusing their full-length coding sequences (CDSs) with N-terminal green fluorescent protein (GFP) and transiently expressing the fusion proteins in tobacco epidermal cells. Fluorescence was observed in both the cell membrane and the nucleus using confocal microscopy (Figure 6A). Yeast two-hybrid (Y2H) assays were then performed to screen interacting proteins of GhCPK33 or GhCPK74. The two S-adenosylmethionine synthases (SAMSs) GhSAMS1 and GhSAMS2 were identified as interacting proteins of GhCPK33 and GhCPK74 (Figure 6B). SAMS is an important protein involved in multiple plant stress responses and is the only enzyme that catalyzes the synthesis of S-adenosyl methionine (Markham et al., 1983; He et al., 2019). Bimolecular fluorescence complementation (BiFC) and luciferase complementation imaging (LCI) assays were performed to verify the protein interactions of the GhSAMS with the GhCPKs. Strong yellow fluorescent protein signals were observed on the membranes of tobacco cells co-expressing GhSAMS1 and GhCPK33, GhSAMS2 and GhCPK33, GhSAMS1 and GhCPK74, and GhSAMS2 and GhCPK74 (Figure 6C). In the LCI tests, interaction signals between the four pairs of proteins were also detected (Figure 6D). GhSAMS1, GhSAMS2, GhCPK33, and GhCPK74 proteins were expressed and purified in prokaryotic cells, and pull-down experiments were performed to validate their interactions (Figure 6E). The results showed that GhSAMS1 and GhSAMS2 could interact with GhCPK33 and GhCPK74 in vivo and in vitro.
Figure 6.
GhCPK33 and GhCPK74 interact directly with GhSAMS1 and GhSAMS2.
(A) Subcellular localization of GhCPK33:GFP protein and GhCPK74:GFP protein. GFP fluorescence was observed after transient expression of GhCPK33:GFP protein and GhCPK74:GFP protein in tobacco epidermal cells. Calcineurin B-Like (CBL) and HY5 were used as plasma membrane and nuclear markers, respectively.
(B) Y2H assay for interactions of GhCPK33 and GhCPK74 with GhSAMS1 and GhSAMS2. SD-T-L medium lacks Trp and Leu, and SD-T-L-H-A medium lacks Trp, Leu, His, and Ade.
(C) BiFC assay of the interaction of GhCPK33-cYFP and GhCPK74-cYFP with GhSAMS1-nYFP and GhSAMS2-nYFP in tobacco epidermal cells. Bars, 30 μm.
(D) LCI analysis of the interaction of GhCPK33-nLUC and GhCPK74-nLUC with GhSAMS1-cLUC and GhSAMS2-cLUC in tobacco leaves.
(E) Pull-down assay verifying the interaction of GhCPK33 and GhCPK74 with GhSAMS1 and GhSAMS2.
Silencing GhSAMS1 and GhSAMS2 impairs cotton resistance to S. litura larvae
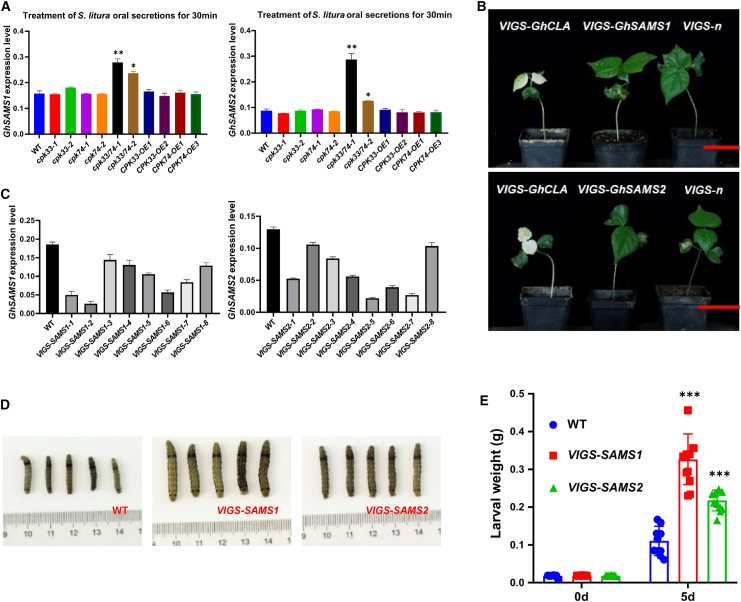
To investigate the relationship of GhCPK33 and GhCPK74 with GhSAMS1 and GhSAMS2, expression levels of GhSAMS1 and GhSAMS2 were examined after 30 min of H + S treatment in cpk33, cpk74, cpk33/74, CPK33-OE, CPK74-OE, and WT materials. The expression levels of GhSAMS1 and GhSAMS2 were significantly increased in the cpk33/74 leaves treated with H + S; however, there was no significant difference in GhSAMS1 and GhSAMS2 expression in cpk33 and cpk74 leaves treated with H + S compared with the WT (Figure 7A). To investigate the role of GhSAMS1 and GhSAMS2 in defense against insects, we performed virus-induced gene-silencing (VIGS) of GhSAMS1 and GhSAMS2, resulting in VIGS-GhSAMS1 and VIGS-GhSAMS2 seedlings (Figure 7B and 7C). Their insect resistance was assessed through a no-choice feeding experiment using S. litura larvae. The results showed a significant increase in the weight of S. litura larvae fed with VIGS-GhSAMS1 and VIGS-GhSAMS2 leaves compared with the control group (Figure 7D and 7E), suggesting that silencing of GhSAMS1 and GhSAMS2 can result in decreased resistance of upland cotton to S. litura larvae.
Figure 7.
Expression levels of GhSAMS1 and GhSAMS2 after H + S treatment and assessment of insect resistance.
(A) qRT-PCR detection of GhSAMS1 and GhSAMS2 in cpk33, cpk74, cpk33/74, CPK33-OE, CPK74-OE, and WT materials after H + S treatment. GhUB7 was used as the internal control. Means ± SE (n = 3).
(B) VIGS of GhSAMS1 and GhSAMS2. VIGS-GhCLA was the positive control, and VIGS-n was the negative control. The red line represents 5 cm.
(C) Expression levels of GhSAMS1 and GhSAMS2 in leaves of VIGS-GhSAMS1 and VIGS-GhSAMS2 plants.
(D) Comparison of the body size of S. litura larvae after 5 days of non-selective feeding on WT, VIGS-GhSAMS1, and VIGS-GhSAMS2 plants.
(E) Average weight changes of S. litura larvae after 5 days of continuous feeding on WT, VIGS-GhSAMS1, and VIGS-GhSAMS2 leaves. Means ± SE (n = 12). Statistical analyses were performed using Student’s t-test. ∗∗∗p < 0.001.
Discussion
The CDPK gene family has a long evolutionary history, dating back to the earliest land plants, such as ferns and mosses (Hamel et al., 2014). With the development of biotechnology, an increasing number of CDPKs have been identified in different species. The A. thaliana, O. sativa, Cucumis sativus, Populus alba, and Cucumis melo genomes harbor 34, 31, 19, 30, and 18 CDPK genes, respectively (Sanders et al., 2002; Ray et al., 2007; Zuo et al., 2013; Xu et al., 2015; Zhang et al., 2017). The release of upland cotton genome data has provided valuable information for a comprehensive analysis of upland cotton CDPKs. As an allopolyploid, upland cotton originated from a polyploidization event approximately 1–2 million years ago, which combined the Gossypium arboreum A subgenome and the G. raimondii D subgenome (Wendel, 1989). An assembly of the cotton A and D genomes has been published, and the genome draft of upland cotton was released in 2015, followed by several updated versions, all focusing on TM-1 as the research subject (Wang et al., 2012; Li et al., 2014; Yang et al., 2019). To obtain more comprehensive results, we retrieved CDPK family members from two protein databases (NBI and HAU) (Zhang et al., 2015; Wang et al., 2019b). Although there were some differences in alignment results between the two databases, such as the presence of GhCPK63 in only the NBI database, the majority of the results were consistent. A total of 82 GhCPKs were identified. GhCPKs were widely expressed in multiple tissues of cotton and were induced by various biotic and abiotic stresses (Figure 1), indicating that they participate in various plant biological processes in cotton. In particular, multiple GhCPKs exhibited significant differential expression induced by H. armigera, S. litura, and whitefly, indirectly confirming the feasibility of searching for insect-resistance genes among the GhCPKs. Identification and analysis of the CDPK gene family in upland cotton thus provide valuable information for subsequent research.
In this study, we screened for insect-resistance genes using a strategy based on a CRISPR–Cas9 mutant library. The entire study was based on a breakthrough in upland cotton regeneration technology and the development of the CRISPR–Cas9 system for upland cotton (Jin et al., 2006; Wang et al., 2018). In plant species that can be transformed and regenerated, the use of CRISPR for gene mutation to validate gene function is practicable (Bortesi and Fischer, 2015). However, if gene knockout for the 246 sgRNAs were performed by constructing 246 individual vectors for genetic transformation, this would be a labor-intensive and time-consuming process. By creating a mixed pool of sgRNAs, constructing a vector library, and performing Agrobacterium-mediated transformation, we demonstrated a new approach for the creation of a large mutant population (Liu et al., 2023). This method is efficient, as it avoids the construction of individual vectors and the performance of individual transformations. It requires only batch transformation of mixed Agrobacteria containing the plasmid library, thereby quickly generating the desired gene-edited plants. However, drawbacks of this method are also apparent. Because the sgRNA sequences carried by each transgenic plant are unknown, subsequent molecular detection of the T0-generation materials is relatively complex. First, it is necessary to detect the sgRNA sequences carried by each material, a process that can be simplified by barcode-based high-throughput sequencing. Through our statistical analysis, 147 target sites were detected, targeting 73 individual GhCPKs and 33 pairs of homologous GhCPKs. The average gene coverage was 86.18%. Although our sgRNA coverage was much lower (59.76%) than the nearly 100% sgRNA coverage reported in soybean and tomato (Jacobs et al., 2017; Bai et al., 2020), we designed two sgRNAs for each gene and each homologous gene pair, thus compensating for the low sgRNA coverage and ultimately achieving a gene coverage of at least 85%. Expanding the population size of the transformation groups is another possible approach for enhancing sgRNA coverage. We also failed to rule out the potential influence of knocking out GhCPKs related to cotton regeneration, which would affect the total population of the GhCPK mutant library.
Through detection of gene editing in T0 plants of the GhCPK mutant library, we found that the CRISPR–Cas9 system in upland cotton was remarkably effective, with over 70% of plants exhibiting editing efficiencies >80%. Deletion was the predominant editing type in most plants, accounting for 81.80% of all edits, followed by insertion, which accounted for 12.04%. Through genetic analysis of the T0 and T1 generations, we observed that the heredity of the CRISPR–Cas9 gene edits was complex. Although most T1 plants inherited the editing pattern of their parents, a few produced new editing types owing to the presence of CRISPR–Cas9 expression cassettes. The presence of non-homozygous editing in the T0 generations further contributed to the complexity. When selecting T1 generations, we believe that individuals with a single editing type and without the CRISPR–Cas9 expression cassettes are more suitable for seed preservation.
A small number of T0 plants in the GhCPK mutant library exhibited unique phenotypes, such as yellow leaves and dwarf stature (Supplemental Figure 13), and most of these T0 plants were sterile. These plants were valid mutants in which a specific GhCPK had been edited. However, not all mutants of the corresponding GhCPK showed these phenotypes. We re-transformed CRISPR–Cas9 vectors into cotton to knock out the corresponding GhCPKs individually, and the unique phenotypes were not reproducible. We believe that these phenotypes were related to mutations caused by transfer-DNA insertion or the long-term tissue culture process and were not related to GhCPKs. To ensure the stability and accuracy of phenotypic identification, it is necessary to select at least two independent knockout lines in the T0 generation of each GhCPK for phenotypic screening.
We identified GhSAMS1 and GhSAMS2 as interacting proteins of GhCPK74 and GhCPK33 through BiFC, LCI, and pull-down assays. SAMS is widely involved in the regulation of stress responses and growth/development across plant species. In Arabidopsis, AtSAMS3 is involved in pollen formation (Chen et al., 2016). In cucumber, CsSAMS is induced by salt stress and participates in related regulation (Zhu et al., 2021). In tobacco, overexpression of soybean GmSAMS1 enhances resistance to S. litura (Fan et al., 2018). We examined the expression levels of GhSAMS1 and GhSAMS2 in cotton leaves induced by OS of S. litura. Both GhSAMS1 and GhSAMS2 were significantly upregulated in cpk33/74 materials but showed no obvious changes in cpk33 and cpk74 compared with WT plants. We therefore hypothesize that GhCPK74 and GhCPK33 act redundantly in the negative regulation of GhSAMS1 and GhSAMS2 expression and in cotton resistance to insects. We next performed insect-resistance assays with VIGS-GhSAMS1 and VIGS-GhSAMS2 plants and found that silencing of GhSAMS1 or GhSAMS2 enhanced the feeding preference of S. litura larvae. On the basis of these results, a schematic diagram of the regulation of cotton pest defense by GhCPK33 and GhCPK74 is shown in Supplemental Figure 14. We speculate that GhCPK33 and GhCPK74, as upstream signaling factors, may regulate multiple pathways in response to herbivory stress. In this study, we provide evidence to support the negative regulation of cotton insect resistance by GhCPK33 and GhCPK74. However, the roles of GhCPK33 and GhCPK74 in the regulation of JA synthesis are still unclear, and further study is needed to reveal the underlying mechanism of GhCPK-regulated insect resistance.
Methods
Genome-wide comprehensive analysis of the GhCPK gene family
Genomic data for G. hirsutum and G. raimondii were obtained from Cottongen (https://www.cottongen.org/). Genomic data for A. thaliana and O. sativa were acquired from Ensembl Plants (http://plants.ensembl.org/index.html) and the Rice Genome Annotation Project (http://rice.uga.edu/downloads_gad.shtml), respectively. CDPK protein sequences from A. thaliana, O. sativa, and G. raimondii were used as query sequences to search the genomes of G. hirsutum (NBI, Zhang et al., 2015; and HAU, Wang et al., 2019b) with the BLASTP program. In addition, a local hidden Markov model–based search (HMMER) was developed based on the CDPK protein sequences to identify GhCPKs. The results from BLASTP and HMMER were compared to identify and exclude redundant sequences. The remaining candidate genes were then uploaded to SMART (http://smart.embl.de/) and CDD (https://www.ncbi.nlm.nih.gov/cdd/) to confirm the presence of conserved domains. Amino acid residues, molecular weights, and isoelectric points of the GhCPKs were predicted using the ProtParam tool (https://web.expasy.org/protparam/). DeepLoc-2.0 (https://services.healthtech.dtu.dk/services/DeepLoc-2.0/) was used to predict subcellular localization of the GhCPKs, DeepTMHMM (https://dtu.biolib.com/DeepTMHMM/) to predict transmembrane domains, and SignalP-6 (https://biolib.com/DTU/SignalP-6/) to predict signal peptides. A phylogenetic tree based on the full-length protein sequences of A. thaliana, O. sativa, and G. hirsutum was constructed using the neighbor-joining method in MEGA 7.0 software and visualized with EvolView (http://www.evolgenius.info/evolview/#/). Conserved motifs of the GhCPKs were analyzed using the MEME program and visualized with TBtools (Chen et al., 2020). Gene structures of the GhCPKs were analyzed by comparing CDSs with their corresponding genome sequences using GSDS software (http://gsds.gao-lab.org/). Cis-acting elements in the GhCPK promoters were analyzed using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). A collinearity analysis between G. hirsutum and three other species was performed using MCScanX software. Results of all the aforementioned analyses were visualized with TBtools. Images of GhCPK gene locations on chromosomes and genome-wide gene duplication events were produced with Circos software.
Expression profiling of GhCPKs in different tissues and stress conditions
Gene expression data for different tissues and abiotic stresses were obtained from TM-1 transcriptome data (Zhang et al., 2015). Transcriptome data for S. litura and H. armigera OS-induced gene expression at different time points were obtained from the NCBI Sequence Read Archive (NCBI: PRJNA522889) (Si et al., 2020). Transcriptomic data for ZS and HR cultivars at four time points after whitefly infestation were obtained from previous research in our lab (Li et al., 2016). For different RNA sequencing reads, low-quality reads were filtered using Trimmomatic (v.0.39), and clean reads were mapped to the TM-1 reference genome using HISAT2 (v.2.2.1) (Kim et al., 2019). The expression levels (transcripts per million) of genes were calculated using StringTie (v.2.1.4). If a gene had a value of transcripts per million >0, it was considered to be expressed. Differentially expressed genes were identified using the DESeq2 package with false discovery rate <0.05 and |log2(fold change)| ≥ 1 (Varet et al., 2016). TBtools was used for heatmap visualization.
sgRNA design
To construct a mutant library of the GhCPK gene family, we used CRISPR-P 2.0 software (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR) to design sgRNAs for 82 GhCPK genes and 41 pairs of homologous GhCPK genes, with the G. hirsutum (v1.1) genome as the reference sequence. A total of 246 sgRNAs were designed (Supplemental Table 2). Genome-wide comparison screening was performed, and the principles for sgRNA design were as follows: priority was given to sgRNAs that had more than two mismatches with other non-target genes, sgRNAs with a CG content between 40% and 60% were preferred, and sgRNAs in the key CDS region were prioritized.
Construction of a knockout vector library for the GhCPK gene family
We used the efficient CRISPR–Cas9 knockout vector pRGEB32-GhU6.7, which was developed specifically for upland cotton (Wang et al., 2018). The two BstBI restriction sites were used for complete enzymatic digestion of the vector, and the linearized plasmid was used as the ligation vector. The 246 selected sgRNA sequences were reverse complemented, and the reverse-complemented sequences were used to synthesize primers with the TTCTAGCTCTAAAAC adapter added to the 5′ end and the TGCACCAGCCGGGAAT adapter added to the 3′ end. To ensure coverage and ease of operation, the 246 synthetic primers were divided into six groups, each containing 40 or 41 primers. The primers in each group were mixed in equal amounts to form a primer pool, and amplification was performed using the PGTR plasmid as the template. The PCR products were then cloned into the linearized vector pRGEB32-GhU6.7 using In-Fusion cloning technology. The cloned plasmid was heat-shock-transformed into the Escherichia coli strain Trans5α at 42°C and incubated at 37°C for 15 h. All E. coli colonies were collected, and plasmids were extracted using the TIANprep Mini Plasmid Kit (TIANGEN, Beijing, China). The concentration of each group of plasmids was determined using a spectrophotometer, and equal amounts of plasmids from each group were mixed into one plasmid pool for Agrobacterium-mediated transformation. Multiple electroporations were performed for Agrobacterium GV3101 transformation to ensure a sufficient number of colonies and plasmid coverage. After 2 days of growth, all Agrobacterium colonies were scraped and stored at −80°C for cotton genetic transformation. All steps are illustrated in Figure 2C.
Construction of overexpression and VIGS vectors
The full-length CDS sequences of GhCPK33 and GhCPK74 were amplified using the cDNA sequence of Jin668 as a template. The amplified sequences were inserted into the pK2GW7 overexpression vector using Gateway cloning technology (Karimi et al., 2002). The recombinant overexpression plasmid was transformed into Agrobacterium by electroporation and reserved for genetic transformation in cotton. VIGS fragments of GhSAMS1 and GhSAMS2 were amplified using the cDNA sequence of Jin668 as a template. The PCR product was cloned into the TRV:00 enzyme linear products using In-Fusion cloning technology (Gao et al., 2013). The recombinant VIGS vector plasmid was transformed into Agrobacterium by electroporation and used for VIGS experiments.
Barcode high-throughput sequencing detects sgRNAs in T0-generation plants of the GhCPK mutant library
To identify the sgRNA sequences carried by each mutant plant, we used a barcode high-throughput sequencing strategy. Because only sgRNA sequences in pRGEB32-GhU6.7 vectors carried by different plants were different, we designed primer pairs for the common vector sequences upstream and downstream of the sgRNAs according to the location of sgRNAs on the pRGEB32-GhU6.7 vectors (Supplemental Figure 7A). A 9-nucleotide barcode was then added to the 5′ end of each primer pair. A total of 28 barcode primers were designed, which, depending on the combination of primer pairs, could simultaneously detect 192 samples (Supplemental Table 5). After mixing all amplified sequences, high-throughput sequencing was performed, and the sgRNA sequences carried by each plant were determined on the basis of their different barcodes.
High-throughput sequencing for detection of gene editing
According to the sgRNA sequence carried by each plant in the GhCPK mutant library, the gene ID of the edited gene was determined. Primers were then designed upstream and downstream of the target sequence on the basis of their positions within the full-length sequence of the target gene (Supplemental Table 6). The CRISPR–Cas9 system can introduce a large number of indels at the target editing site, and the upstream and downstream primers should therefore not be designed too close to the target site and should have a minimum spacing of at least 30 bp. The amplified fragments are generally not longer than 280 bp. To maintain consistency in PCR amplification conditions, all primer-annealing temperatures in this experiment were designed to be between 58°C and 60°C. After amplification, all PCR products were mixed for high-throughput sequencing, and the gene-editing status of each plant in the GhCPK mutant library was determined.
Identification of insect-resistance phenotypes in plants of the GhCPK mutant library
We performed a statistical analysis of the damage caused by chewing pests on T1-generation positive gene-edited materials from the mutant library in our experimental field in Wuhan. All plants were planted on April 18, 2021, and during the following weeks they were infested by chewing pests such as H. armigera, Spodoptera exigua, and S. litura that occurred naturally in the environment. On June 10, 2021, we performed a statistical analysis of the extent of damage caused by chewing insect pests. We repeated the same field trial and statistical analyses in 2022. We also performed no-choice feeding experiments on all materials using S. litura larvae. On June 15, 2021, we performed a no-choice feeding experiment with S. litura larvae on 243 field plant lines. Twelve S. litura larvae with a body weight of about 0.018–0.022 g were selected from each plant line for feeding. Each larva was placed in a glass dish (9 cm in diameter) and fed fresh cotton leaves every day for four consecutive days. The weights of the S. litura larvae were recorded daily. The experiment was repeated in 2022.
Evolution and domestication of the GhCPK33 and GhCPK74 loci
Resequencing data and RNA sequencing data were obtained from previous studies (Li et al., 2021). VCFtools (v0.1.16) was used to extract SNP variants from the samples, perform quality control, and calculate nucleotide diversity (π) with the specified parameters (--maf 0.01, --hwe 0.01) (Danecek et al., 2011). Plink (version 1.9) was used to convert file formats (Purcell et al., 2007). Gene structures were visualized using the gene structure files through the GSDS website (http://gsds.gao-lab.org/). Haplotype analysis was performed by analyzing SNP variants with R scripts.
Measurement of Ca2+ flux in cotton leaf epidermal cells and quantification of JA content in cotton leaves
Seeds of T1-generation cpk33, cpk74, cpk33/74, CPK33-OE, CPK74-OE, and WT materials were sown at the same time and cultivated under the same conditions (Supplemental Figure 12A). High-throughput sequencing was performed on all T2-generation plants, and all exhibited 100% editing (Supplemental Figure 12B). Expression levels of GhCPK33 and GhCPK74 were measured in the T2-generation overexpression materials, CPK33-OE1, CPK33-OE2, CPK74-OE1, and CPK74-OE3 (Supplemental Figure 12C), confirming the sustained high expression of GhCPK33 and GhCPK74 in these plants.
Fresh leaves were collected from cotton plants and immediately soaked in test buffer (0.1 mM CaCl2) for 2 h. Ca2+ flux was measured using the non-invasive micro-test technique (NMT-YG-100, YoungerUSA, Amherst, MA, USA) as described previously (Wang et al., 2019a). Before treatment of leaves with S. litura OS, the steady-state flux of leaf mesophyll cells was recorded continuously for 5 min. Subsequently, 30 μl of S. litura OS was slowly added to the measuring buffer, and the transient flux of Ca2+ was recorded and sustained for 5 min. At least three leaf mesophyll cells were tested for each line.
To measure the concentration of JA in cotton leaf samples, approximately 100 mg of leaf material was extracted twice with 80% cold methanol (v/v) overnight at 4°C. For each sample, 10 ng (±)-9,10-dihydro-JA (Olchemim) was added as an internal standard. The two extracts were combined, and the mixture was evaporated to the aqueous phase using N2, dissolved in 0.4 mL of methanol, and filtered using a syringe-facilitated filter (Nylon 66; Jin Teng Experiment Equipment, Tianjin, China). The samples were stored at −80°C before measurement. JA levels were quantified using a high-pressure liquid chromatography–tandem mass spectrometry system (AB SCIEX Triple Quad 5500 LC/MS/MS) with JA (Sigma) as the external standard (Sun et al., 2014).
Subcellular localization
The CDSs of GhCPK33 and GhCPK74 were cloned into the C-terminal fusion GFP vector pGWB405. The recombinant vectors pGWB405-GhCPK33 and pGWB405-GhCPK74 were transiently expressed in tobacco epidermal cells after Agrobacterium-mediated transfection. Protein localization was observed using an Olympus FV1200 confocal microscope 2 days after Agrobacterium transfection. CBL1:RFP was used as a plasma membrane marker, and HY5:RFP was used as a nuclear marker.
Y2H, LCI, and BiFC assays
The CDSs of GhCPK33 and GhCPK74 were cloned into pGBKT7 to generate a bait vector and transformed into yeast strain Y2H. The CDSs of GhSAMS1 and GhSAMS2 were cloned into pGADT7 to generate prey vectors and transformed into yeast strain Y187. The interactions between bait and prey were assessed by growth on SD-Leu-Trp (SD-2) medium and SD-Leu-Trp-His-Ade (SD-4) medium. For the LCI assays, the CDSs of GhCPK33 and GhCPK74 were cloned into the JW771 vector, and the CDSs of GhSAMS1 and GhSAMS2 were cloned into the JW772 vector. The recombinant vectors were transformed into A. tumefaciens GV3101 and transiently expressed in Nicotiana benthamiana leaves by injection with needleless syringes. Fluorescence signals of luciferase (LUC) luminescence in LCI were observed with a cryogenically cooled charge-coupled device camera (NightSHADE LB 985). For BiFC assays, the CDSs of GhCPK33 and GhCPK74 were cloned into the pxy104-cYFP vector, and the CDSs of GhSAMS1 and GhSAMS2 were cloned into the pxy106-nYFP vector. The vectors were transformed into A. tumefaciens strain GV3101 and transiently expressed in N. benthamiana leaves by injection with needleless syringes. Fluorescence in the N. benthamiana epidermal cells was observed 60 h later using a confocal microscope (Olympus FV1200).
qRT–PCR
For each sample, 3 μg of RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega). The real-time qRT–PCR reactions were performed using the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems), and GhUB7 was used as the internal control gene (Tu et al., 2007).
In vitro pull-down assays
The full-length CDSs of GhCPK33 and GhCPK74 were separately cloned into the pGEX-6P-1 vector (fused with the GST tag) using the Gateway technique. The full-length CDSs of GhSAMS1 and GhSAMS2 were separately cloned into the pet-SUMO vector (fused with the His tag) using the Gateway technique. In total, four vectors were constructed, GST-GhCPK33, GST-GhCPK74, His-GhSAMS1, and His-GhSAMS2. These constructs were transformed into E. coli BL21 (DE3) for prokaryotic expression. The GST fusion protein and the target His fusion protein were co-incubated in pull-down buffer at 4°C for 2 h. GST protein was used as a blank control. After 2 h, 20 μl of the incubation system was taken as the input sample to detect the interaction between GST and His proteins. GST beads were added and incubated at 4°C for 2 h. After four washes with pull-down buffer, samples were prepared for western blot analysis (Chen et al., 2023).
Availability of materials
The GhCPK mutants/transgenic lines generated in this study can be obtained by contacting the corresponding author.
Funding
This work was supported by Biological Breeding of Stress Tolerant and High Yield Cotton Varieties (2023ZD04040) to L.M., the National Natural Science Fund of China for Distinguished Young Scholars (32325039) and the National Natural Science Foundation of China (32272128) to S.J., the National Natural Science Foundation of China (32401780) and the Key Scientific and Technological Project of Henan Province (222102110151) to S.L., and the Major Science and Technology Project of Xinjiang Uygur Autonomous Region (2023A02003-2) to B.L.
Acknowledgments
No conflict of interest is declared.
Author contributions
S.J., X.Z., W.G., L.M., and B.L. provided the experimental design and supervision. F.W., S.L., and L.L. performed the experiments and manuscript preparation. Q.W. and Z.X. provided assistance with data analysis. G.W., T.H., C.F., Y.F., and L.C. provided suggestions and modified the manuscript.
Published: August 12, 2024
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Bo Li, Email: libo@xaas.ac.cn.
Lu Long, Email: lulong1826@163.com.
Wei Gao, Email: gaowei021@163.com.
Shuangxia Jin, Email: jsx@mail.hzau.edu.cn.
Supplemental information
References
- Bai M., Yuan J., Kuang H., Gong P., Li S., Zhang Z., Liu B., Sun J., Yang M., Yang L., et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 2020;18:721–731. doi: 10.1111/pbi.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Kudla J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta. 2012;1820:1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Bortesi L., Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen L., Zhang B., Xia L., Yue D., Han B., Sun W., Wang F., Lindsey K., Zhang X., Yang X. The GhMAP3K62-GhMKK16-GhMPK32 kinase cascade regulates drought tolerance by activating GhEDT1-mediated ABA accumulation in cotton. J. Adv. Res. 2023;51:13–25. doi: 10.1016/j.jare.2022.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zou T., McCormick S. S-Adenosylmethionine Synthetase 3 Is Important for Pollen Tube Growth. Plant Physiol. 2016;172:244–253. doi: 10.1104/pp.16.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Scheffler B.E., Dennis E., Triplett B.A., Zhang T., Guo W., Chen X., Stelly D.M., Rabinowicz P.D., Town C.D., et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007;145:1303–1310. doi: 10.1104/pp.107.107672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M., Meldau S., Howe G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012;17:250–259. doi: 10.1016/j.tplants.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M., Reymond P. Molecular Interactions Between Plants and Insect Herbivores. Annu. Rev. Plant Biol. 2019;70:527–557. doi: 10.1146/annurev-arplant-050718-095910. [DOI] [PubMed] [Google Scholar]
- Fan R., Li X., Wang S., Yu D., Wang H. The soybean S-adenosylmethionine synthetase gene GmSAMS1 confers resistance to common cutworm in transgenic tobacco. Soybean Sci. 2018;37:268–274. [Google Scholar]
- Gaillochet C., Develtere W., Jacobs T.B. CRISPR screens in plants: approaches, guidelines, and future prospects. Plant Cell. 2021;33:794–813. doi: 10.1093/plcell/koab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Long L., Zhu L.F., Xu L., Gao W.H., Sun L.Q., Liu L.L., Zhang X.L. Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics. 2013;12:3690–3703. doi: 10.1074/mcp.M113.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L.P., Sheen J., Séguin A. Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 2014;19:79–89. doi: 10.1016/j.tplants.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M.W., Wang Y., Wu J.Q., Shu S., Sun J., Guo S.R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019;141:431–445. doi: 10.1016/j.plaphy.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Heckel D.G. Ecology. Insecticide resistance after Silent spring. Science. 2012;337:1612–1614. doi: 10.1126/science.1226994. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Huang Y., Wan P., Zhang H., Huang M., Li Z., Gould F. Diminishing returns from increased percent Bt cotton: the case of pink bollworm. PLoS One. 2013;8:e68573. doi: 10.1371/journal.pone.0068573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T.B., Zhang N., Patel D., Martin G.B. Generation of a Collection of Mutant Tomato Lines Using Pooled CRISPR Libraries. Plant Physiol. 2017;174:2023–2037. doi: 10.1104/pp.17.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Zhang X., Nie Y., Guo X., Liang S., Zhu H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. (Prague) 2006;50:519–524. [Google Scholar]
- Kanchiswamy C.N., Takahashi H., Quadro S., Maffei M.E., Bossi S., Bertea C., Zebelo S.A., Muroi A., Ishihama N., Yoshioka H., et al. Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 2010;10:97. doi: 10.1186/1471-2229-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., Li Q., Ma Z., Lu C., Zou C., et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014;46:567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- Li J., Yuan D., Wang P., Wang Q., Sun M., Liu Z., Si H., Xu Z., Ma Y., Zhang B., et al. Cotton pan-genome retrieves the lost sequences and genes during domestication and selection. Genome Biol. 2021;22:119. doi: 10.1186/s13059-021-02351-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhu L., Hull J.J., Liang S., Daniell H., Jin S., Zhang X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly) Plant Biotechnol. J. 2016;14:1956–1975. doi: 10.1111/pbi.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hu D., Cai L., Wang H., Liu X., Du H., Yang Z., Zhang H., Hu Z., Huang F., et al. CALCIUM-DEPENDENT PROTEIN KINASE38 regulates flowering time and common cutworm resistance in soybean. Plant Physiol. 2022;190:480–499. doi: 10.1093/plphys/kiac260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hallerman E.M., Wu K., Peng Y. Insect-Resistant Genetically Engineered Crops in China: Development, Application, and Prospects for Use. Annu. Rev. Entomol. 2020;65:273–292. doi: 10.1146/annurev-ento-011019-025039. [DOI] [PubMed] [Google Scholar]
- Liu H.J., Jian L., Xu J., Zhang Q., Zhang M.L., Jin M., Peng Y., Zhang M., Han B., Liu J., et al. High-Throughput CRISPR/Cas9 Mutagenesis Streamlines Trait Gene Identification in Maize. Plant Cell. 2020;32:1397–1413. doi: 10.1105/tpc.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Z., Zhang X.N., Li K., Yao Q., Zhong D.T., Deng Q., Lu Y.M. Curr Opin Biotech; 2023. Large-scale Genome Editing in Plants: Approaches, Applications, and Future Perspectives; p. 79. [DOI] [PubMed] [Google Scholar]
- Lu Y., Wu K., Jiang Y., Xia B., Li P., Feng H., Wyckhuys K.A.G., Guo Y. Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science. 2010;328:1151–1154. doi: 10.1126/science.1187881. [DOI] [PubMed] [Google Scholar]
- Lu Y., Ye X., Guo R., Huang J., Wang W., Tang J., Tan L., Zhu J.K., Chu C., Qian Y. Genome-wide Targeted Mutagenesis in Rice Using the CRISPR/Cas9 System. Mol. Plant. 2017;10:1242–1245. doi: 10.1016/j.molp.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Markham G.D., Hafner E.W., Tabor C.W., Tabor H. S-Adenosylmethionine Synthetase (Methionine Adenosyltransferase) (Escherichia-Coli) Method Enzymol. 1983;94:219–222. doi: 10.1016/s0076-6879(83)94037-5. [DOI] [PubMed] [Google Scholar]
- Pan C., Li G., Bandyopadhyay A., Qi Y. Guide RNA library-based CRISPR screens in plants: opportunities and challenges. Curr. Opin. Biotechnol. 2023;79:102883. doi: 10.1016/j.copbio.2022.102883. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F., Huang J., Wang X. Fifteen Years of Bt Cotton in China: Results from Household Surveys. World Dev. 2017;98:351–359. [Google Scholar]
- Quan Y., Wu K. Managing Practical Resistance of Lepidopteran Pests to Bt Cotton in China. Insects. 2023;14:179. doi: 10.3390/insects14020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Agarwal P., Arora R., Kapoor S., Tyagi A.K. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica) Mol. Genet. Genomics. 2007;278:493–505. doi: 10.1007/s00438-007-0267-4. [DOI] [PubMed] [Google Scholar]
- Romeis T., Herde M. From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 2014;20:1–10. doi: 10.1016/j.pbi.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Romeis T., Ludwig A.A., Martin R., Jones J.D. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001;20:5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J.F. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si H., Liu H., Sun Y., Xu Z., Liang S., Li B., Ding X., Li J., Wang Q., Sun L., et al. Transcriptome and metabolome analysis reveal that oral secretions from Helicoverpa armigera and Spodoptera litura influence wound-induced host response in cotton. Crop J. 2020;8:929–942. [Google Scholar]
- Simeunovic A., Mair A., Wurzinger B., Teige M. Know where your clients are: subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016;67:3855–3872. doi: 10.1093/jxb/erw157. [DOI] [PubMed] [Google Scholar]
- Sun L., Alariqi M., Wang Y., Wang Q., Xu Z., Zafar M.N., Yang G., Jia R., Hussain A., Chen Y., et al. Construction of Host Plant Insect-Resistance Mutant Library by High-Throughput CRISPR/Cas9 System and Identification of A Broad-Spectrum Insect Resistance Gene. Adv. Sci. 2024;11:e2306157. doi: 10.1002/advs.202306157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhu L., Xu L., Yuan D., Min L., Zhang X. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 2014;5:5372. doi: 10.1038/ncomms6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L., Zhang X., Liu D., Jin S., Cao J., Zhu L., Deng F., Tan J., Zhang C. Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chinese Sci Bull. 2007;22:3110–3117. [Google Scholar]
- Valmonte G.R., Arthur K., Higgins C.M., MacDiarmid R.M. Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 2014;55:551–569. doi: 10.1093/pcp/pct200. [DOI] [PubMed] [Google Scholar]
- Varet H., Brillet-Guéguen L., Coppée J.Y., Dillies M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li X., Su F., Liu H., Hu D., Huang F., Yu D., Wang H. Soybean CALCIUM-DEPENDENT PROTEIN KINASE17 Positively Regulates Plant Resistance to Common Cutworm (Spodoptera litura Fabricius) Int. J. Mol. Sci. 2022;23:15696. doi: 10.3390/ijms232415696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu X., Zhang A., Ren Y., Wu F., Wang G., Xu Y., Lei C., Zhu S., Pan T., et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019;29:820–831. doi: 10.1038/s41422-019-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S., et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., Liu F., Pei L., Wang P., Zhao G., et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang J., Sun L., Ma Y., Xu J., Liang S., Deng J., Tan J., Zhang Q., Tu L., et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018;16:137–150. doi: 10.1111/pbi.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. New World Tetraploid Cottons Contain Old-World Cytoplasm. P Natl Acad Sci USA. 1989;86:4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.M., Guo Y.Y. The evolution of cotton pest management practices in China. Annu. Rev. Entomol. 2005;50:31–52. doi: 10.1146/annurev.ento.50.071803.130349. [DOI] [PubMed] [Google Scholar]
- Wu K.M., Lu Y.H., Feng H.Q., Jiang Y.Y., Zhao J.Z. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321:1676–1678. doi: 10.1126/science.1160550. [DOI] [PubMed] [Google Scholar]
- Xu X., Liu M., Lu L., He M., Qu W., Xu Q., Qi X., Chen X. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genomics. 2015;290:1403–1414. doi: 10.1007/s00438-015-1002-1. [DOI] [PubMed] [Google Scholar]
- Yang D.H., Hettenhausen C., Baldwin I.T., Wu J. Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol. 2012;159:1591–1607. doi: 10.1104/pp.112.199018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Ge X., Yang Z., Qin W., Sun G., Wang Z., Li Z., Liu J., Wu J., Wang Y., et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat. Commun. 2019;10:2989. doi: 10.1038/s41467-019-10820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Wang L., Xie W., Wan B., Li X., Lin Y. Expression profile of calcium-dependent protein kinase (CDPKs) genes during the whole lifespan and under phytohormone treatment conditions in rice (Oryza sativa L. ssp. indica) Plant Mol. Biol. 2009;70:311–325. doi: 10.1007/s11103-009-9475-0. [DOI] [PubMed] [Google Scholar]
- Yip Delormel T., Boudsocq M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019;224:585–604. doi: 10.1111/nph.16088. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wei C., Yang X., Chen H., Yang Y., Mo Y., Li H., Zhang Y., Ma J., Yang J., et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.) PLoS One. 2017;12:e0176352. doi: 10.1371/journal.pone.0176352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., Zhang J., Saski C.A., Scheffler B.E., Stelly D.M., et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- Zhu H., He M., Jahan M.S., Wu J., Gu Q., Shu S., Sun J., Guo S. CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco. Int. J. Mol. Sci. 2021;22:11133. doi: 10.3390/ijms222011133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo R., Hu R., Chai G., Xu M., Qi G., Kong Y., Zhou G. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa) Mol. Biol. Rep. 2013;40:2645–2662. doi: 10.1007/s11033-012-2351-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.