Abstract
Network pharmacology is an interdisciplinary field that utilizes computer science, technology, and biological networks to investigate the intricate interplay among compounds/ingredients, targets, and diseases. Within the realm of traditional Chinese medicine (TCM), network pharmacology serves as a scientific approach to elucidate the compatibility relationships and underlying mechanisms of action in TCM formulas. It facilitates the identification of potential active ingredients within these formulas, providing a comprehensive understanding of their holistic and systematic nature, which aligns with the holistic principles inherent in TCM theory. TCM formulas exhibit complexity due to their multi-component characteristic, involving diverse targets and pathways. Consequently, investigating their material basis and mechanisms becomes challenging. Network pharmacology has emerged as a valuable approach in TCM formula research, leveraging its holistic and systematic advantages. The manuscript aims to provide an overview of the application of network pharmacology in studying TCM formula compatibility rules and explore future research directions. Specifically, we focus on how network pharmacology aids in interpreting TCM pharmacological theories and understanding formula compositions. Additionally, we elucidate the process of utilizing network pharmacology to identify active ingredients within TCM formulas. These findings not only offer novel research models and perspectives for integrating network pharmacology with TCM theory but also present new methodologies for investigating TCM formula compatibility. All in all, network pharmacology has become an indispensable and crucial tool in advancing TCM formula research.
Keywords: active ingredients, compatibility rules, molecular mechanism, network analysis, network pharmacology, systems biology, target prediction, traditional chinese medicine
1. Introduction
Network pharmacology is an interdisciplinary field that merges systems biology, bioinformatics, and various other disciplines to comprehensively analyze the molecular interactions between drugs and therapeutic targets. This approach offers a systems-level and holistic perspective, enabling a deeper understanding of the complex mechanisms underlying drug action (Wang et al., 2021, Wang et al., 2021). By revealing the systemic pharmacological mechanisms of drugs, it guides the development of new drugs and clinical treatment. Meanwhile, it is also an emerging and original discipline for systematic drug research in the era of artificial intelligence and big data (Li, 2021, Niu et al., 2021). Utilizing computational methods, network pharmacology has the capability to predict potential mechanisms of drug action and explore both the therapeutic effects and adverse reactions associated with drugs. Presently, there is a broad application of network pharmacology by scholars in the field of traditional Chinese medicine (TCM) pharmacology and TCM formula compatibility studies (Zhang & Li, 2015). By constructing the “drug-ingredient-target” interaction network, network pharmacology can infer the interaction relationship between the chemical components in TCM formulas and disease targets, and predict the key chemical components and their target functions in the prescriptions (Zuo et al., 2018).
In clinical practice, TCM adheres to various principles regarding the compatibility of herbal medicines. These principles include theories such as the “Four Natures and Five Flavors” theory, mutual coordination, medicinal properties attributed to meridians, drug antagonism, and drug compatibility (Xiao, 2005). The concept of “Sovereign-Minister-Assistant-Courier” in TCM formulas is an important principle in the compatibility of different herbs in a TCM prescription. It involves the interactions between different herbs in the TCM prescriptions. Through network pharmacology and constructing the “drug-ingredient-target” interaction network, “Sovereign-Minister-Assistant-Courier” can be explained and elaborated to a certain extent, and key chemical components and target functions in TCM formulas can be predicted (Li and Su, 2016, Li et al., 2016).
In addition, network pharmacology can also apply molecular docking technology to predict the material basis and mechanism of action of drugs. By constructing a “drug-target-disease” network and conducting enrichment analysis on this network, it is possible to systematically explain the biological processes and signaling pathways involved in the treatment of diseases by the formula, and thus reveal its mechanism of preventing and treating complex diseases. Furthermore, network pharmacology can further verify the predicted results through in vitro and in vivo pharmacological experiments, greatly improving the efficiency of screening active ingredients in TCM and providing a prominent role in predicting new active ingredients in TCM. Therefore, the effective combination of network pharmacology research mode and TCM theory has expanded the scientific and effective interpretation framework of the holistic concept of TCM and provided a new approach for the study of the pharmacological material basis and molecular mechanism of TCM (Han et al., 2019, Zhuang et al., 2021).
2. Interpretation of compatibility rules of TCM formulas
The principle of compatibility in TCM formula is mainly based on “medicinal properties”, “seven emotional factors compatibility”, and “Sovereign-Minister-Assistant-Courier”. Given the intricate nature of the components, pathways, and targets within TCM formulas, the conventional “single target-single drug” research model faces challenges when applied to investigate the compatibility of TCM formulas. However, through the application of network pharmacology, the compatibility principles of herbs in TCM formulas can be effectively explained (Tang et al., 2023).
Fig. 1 illustrates an example of exploring the compatibility rules within a herbal formula and the association of gene targets with diseases using network pharmacology, taking Banxia Xiexin Decoction as an example. The process demonstrates a brief overview of how the principles of compatibility within the formula and the correlation with disease-related gene targets can be elucidated. By utilizing methods such as random walk with restart (RWR) based scores, it is observed that Pinelliae Rhizoma (Banxia in Chinese), Zingiberis Rhizoma (Ganjiang in Chinese), and Coptidis Rhizoma (Huanglian in Chinese) exhibit relatively high scores within Banxia Xiexin Decoction, indicating their strong association with the target genes. On the other hand, Ginseng Radix et Rhizoma (Renshen in Chinese) and Jujubae Fructus (Dazao in Chinese) demonstrate a lower correlation with the disease. In Banxia Xiexin Decoction, Pinelliae Rhizoma is considered the “Jun” (Sovereign) medicine, Zingiberis Rhizoma, Coptidis Rhizoma, and Scutellariae Radix (Huangqin in Chinese) are the “Chen” (Minister) medicine, Ginseng Radix et Rhizoma and Jujubae Fructus are the “Zuo” (Assistant) medicine, and Glycyrrhizae Radix et Rhizoma (Gancao in Chinese) is the “Shi” (Courier) medicine. The RWR scoring curves for each herb were used to calculate the AUCC (Area Under the Curve of the Receiver Operating Characteristic) values, and the AUCC values increased in the following order: Pinelliae Rhizoma > Zingiberis Rhizoma > Coptidis Rhizoma > Glycyrrhizae Radix et Rhizoma > Scutellariae Radix > Jujubae Fructus > Ginseng Radix et Rhizoma The results were consistent with the theoretical expectations, except for Glycyrrhizae Radix et Rhizoma, which showed relatively good consistency.
Fig. 1.
Case of network pharmacology analysis of Banxia Xiexin Decoction (Using NP to explore compatibility rules within Banxia Xiexin Decoction and association of gene targets with diseases).
2.1. Exploring theory of TCM pharmacology, especially concept of “medicinal properties”
The concept of “medicinal properties” in TCM refers to a systematic description of the efficacy, characteristics, and application rules of TCMs. It includes important aspects such as “four natures and five flavors”, meridian tropism, ascending and descending, floating and sinking, toxicity, non-toxicity, etc. The multi-component and multi-target characteristics of TCMs in the formulation are consistent with the research framework of network pharmacology. By using network target identification methods, different molecular groups underlying the medicinal properties of TCMs can be distinguished, and the molecular basis of the medicinal properties can be visually demonstrated.
2.1.1. Four natures and five flavors
Due to the extremely complex and large number of ingredients contained in each prescription, the application of network pharmacology methods can be used to characterize and analyze the chemical and biological properties of the medicinal properties’ theory of a certain disease-treating prescription. From the micro-level of ions, molecules, and cells, the theory of TCM properties can be explained (Lin et al., 2023) and the compatibility rules of TCM formula can be explored. Li Shao proposed the new concept of “network target” from the perspective of the interaction between drugs and the body, combined with the perspectives of network pharmacology and systems biology (Li, 2011). Through experiments, they found that the molecular networks of the hot syndrome and cold syndrome can distinguish the different biological effects of hot-type and cold-type formulas (Li, 2009). Chen Jian and his research team proposed the concept of “property-flavor network pharmacology” for the first time based on the theory of TCM properties and flavors (Chen et al., 2021, Chen et al., 2021). They constructed a network of properties and flavors of TCMs to explore the mechanism of Rujin Jiedu Powder in treating viral pneumonia. The team found that bitter and cold medicines had the most effective ingredients in common, indicating that effective ingredients screened through network pharmacology can reflect the scientific nature of the theory of TCM properties and flavors. Han et al. used network pharmacology methods to explore the molecular mechanisms by which typical cold and hot TCM combinations act on the body. The results showed that the typical cold and hot TCM groups could be found by this network, which can identify the biological signal pathways and key molecules involved (Han et al., 2021, Han et al., 2021). Previous data have shown that some researchers have used the TCM system pharmacology database and analysis platform to retrieve and screen targets of TCMs that can raise and float or sink and settle. The predicted targets from network pharmacology were confirmed by animal experiments (Zhang et al., 2023, Zhang et al., 2023).
2.1.2. Ascending and descending, floating and sinking
Li et al. applied pharmacological networks to compare the differences in the regulation of the immune-related pathways of H1N1 influenza by different aromatics, such as Pogostemonis Herba (Huoxiang in Chinese) and Atractylodis Rhizoma (Cangzhu in Chinese), which belong to different channels in TCMs (Li et al., 2016, Li and Su, 2016). Their findings indicated that Pogostemonis Herba, which is classified within the lung channel, might exhibit greater efficacy in H1N1 influenza treatment compared to Atractylodis Rhizoma, which does not belong to the lung channel. Wang et al. attempted to explore the possibility of using network pharmacology to study the relationship between liver meridian herbs and their therapeutic effects on diseases (Wang et al., 2019, Wang et al., 2019). The results revealed a correlation between disease treatment and liver meridian herbs, as determined through network pharmacology analysis. This finding aligns with the physiological role of the liver meridian according to TCM theory. Network pharmacology can effectively conduct research based on the material foundation and molecular mechanism of TCM properties and flavors, allowing research questions to be explored from different dimensions. This research method is helpful for the future development and technical summary of TCM properties and flavors. We summarized the research on some typical rising-floating, sinking-descending, cold and hot TCM and their treatment of diseases, as well as experimental verification methods using network pharmacology, as shown in Table 1, Table 2.
Table 1.
Partial typical rising and sinking TCMs explored and verified by network pharmacology and their related treatment of diseases.
| Medicinal properties | TCMs | Diseases | Verification methods | References |
|---|---|---|---|---|
| Rising | Astragali Radix | Liver cancer | In vitro cell experiment | Zhuang, Yao, Hou, Zhang, & Song, 2023 |
| Ephedrae Herba (Mahuang in Chinese) | Bronchial asthma | In vivo animal experiment | Huang et al., 2020, Huang et al., 2020, Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022 | |
| COVID-19 | In vivo animal experiment | Gao, Song, & Song, 2020 | ||
| Bupleuri Radix (Chaihu in Chinese) | Depression | In vivo animal experiment | Li et al., 2022, Li et al., 2022, Li et al., 2022, Li et al., 2022, Li et al., 2021, Li et al., 2021, Li et al., 2021, Li et al., 2021 | |
| Puerariae Lobatae Radix (Gegen in Chinese) | ||||
| Dyslipidemia | In vivo animal experiment | Lv et al., 2021 | ||
| Diabetes | In vivo animal experiment | Wei, Li, Han, Fu, & Hao, 2022 | ||
| Myasthenia gravis | In vivo animal experiment | Chen et al., 2021, Chen et al., 2021 | ||
| Liver cancer | In vitro cell experiment | Zhou et al., 2020, Zhou et al., 2020 | ||
| Colon cancer | In vitro cell experiment | Li et al., 2022, Li et al., 2022, Li et al., 2022, Li et al., 2022 | ||
| Platycodonis Radix (Jiegeng in Chinese) | Cervical cancer | In vivo animal experiment | Ma et al., 2022 | |
| Asthma | In vitro cell experiment | Zhao et al., 2021, Zhao et al., 2021 | ||
| Sinking |
Descurainiae Semen Lepidii Semen (Tinglizi in Chinese) |
Bronchial asthma | In vivo animal experiment | Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022 |
| Cardiac fibrosis | In vivo animal experiment | Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022 | ||
| Poria (Fuling in Chinese) | Gastric cancer | In vitro cell experiment | Song & Shu, 2022 | |
| Nasopharyngeal carcinoma | In vitro cell experiment | Wang and Zhu, 2023, Wang and Hai, 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023 | ||
| Breast cancer | In vitro cell experiment | Ma et al., 2020, Sun et al., 2022 | ||
| Achyranthis Bidentatae Radix (Niuxi in Chinese) | Osteoarthritis | In vitro cell experiment | Chen et al., 2020, Lin et al., 2022, Lin et al., 2022 |
Table 2.
Partial typical hot and cold TCMs explored and verified by network pharmacology and their related treatment of diseases.
| Medicinal properties | TCMs | Diseases | Verification methods | References |
|---|---|---|---|---|
| Hot | Chuanxiong Rhizoma (Chuanxiong in Chinese) | Cerebral ischemic stroke |
In vitro cell experiment In vivo animal experiment |
Bai et al., 2022, Liu et al., 2022, Liu et al., 2022, Liu et al., 2022 |
| Migraine | In vivo animal experiment | Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Zhu et al., 2022 | ||
| Citri Reticulatae Pericarpium (Chenpi in Chinese) | Hyperlipidemia | In vivo animal experiment | Zhang, 2022 | |
| Chronic obstructive pulmonary disease | In vitro cell experiment | Liu et al., 2022, Liu et al., 2022, Liu et al., 2022, Zhou et al., 2021 | ||
| Cinnamomi Cortex (Rougui in Chinese) | Depression | In vivo animal experiment | Ma et al., 2023 | |
| Euodiae Fructus (Wuzhuyu in Chinese) | Breast cancer | In vitro cell experiment | Liu et al., 2020, Liu et al., 2020, Liu et al., 2020 | |
| Gout | In vivo animal experiment | Ou, Wang, Yang, Wang, & Zhao, 2021 | ||
| Insomnia | In vivo animal experiment | Li et al., 2021, Li et al., 2021, Li et al., 2021, Li et al., 2021 | ||
| Nasopharyngeal carcinoma | In vitro cell experiment | Xu et al., 2022, Xu et al., 2022 | ||
| Cold | Lonicerae Japonicae Flos (Jinyinhua in Chinese) | Acute alcoholic liver injury | In vivo animal experiment | Liu et al., 2021, Liu et al., 2021, Liu et al., 2021, Liu et al., 2021 |
| COVID-19 | In vitro cell experiment | Li et al., 2022, Li et al., 2022, Li et al., 2022, Li et al., 2022 | ||
| Scutellariae Radix | Melanoma | In vitro cell experiment | He et al., 2022 | |
| Oral leukoplakia | In vitro cell experiment | Hou et al., 2022 | ||
| Osteoarthritis | In vitro cell experiment | Yi et al., 2021 | ||
| Anemarrhenae Rhizoma (Zhimu in Chinese) | Inflammation | In vitro cell experiment | Su, Tan, Lv, & Lei, 2022 |
2.2. Elucidating mechanism of toxicity reactions of TCMs
The toxic effects of TCMs may be exerted through multiple pathways and targets. Through the method of network pharmacology, the toxic reactions of TCM can be sorted out, and the toxic TCM-target network can be constructed to elucidate the possible toxic mechanisms in TCMs. This research method provides strong theoretical support for the safe and effective clinical application of TCM and can also guide the compatibility process of TCM formulas to some extent. Through network pharmacology analysis, a comprehensive understanding of the toxicity and efficacy mechanisms of drugs can be gained.
2.2.1. Drug-target network analysis
Constructing a network of interactions between drugs and their target proteins, it involves analyzing the network topology and associations to identify key target proteins as well as signaling pathways and biological processes associated with drug medication. This approach demonstrates the potential mechanisms of drug medication and potential toxicity.
The concept of “network toxicology” was first proposed by the team of Xiaohui Fan (Fan, Zhao, Jin, Shen, & Liu, 2011). In order to answer the urgent needs in the field of TCM toxicology and quality control, Li et al (Li & Ding, 2019) showed the compatibiliy with the strategy of “network target, multicomponent therapeutics”, while forming the hot research field together with network pharmacology. They discussed the new progress on these blooming research fields, and their integration potential with network pharmacology. Ding et al. explored the possible molecular mechanisms of the reproductive toxicity of Tripterygium wilfordii Hook.f. (Leigongteng in Chinese) from the perspective of network pharmacology and bioinformatics and screened the key targets of its reproductive toxicity (Ding, Wu, & Liu, 2021). He et al. used database retrieval to identify the targets of cardiac adverse reactions induced by Aconiti Lateralis Radix Praeparata (Fuzi in Chinese), and searched the target components of major toxic constituents in Aconitum carmichaelii Debx. (Wutou in Chinese) through the GeneCards database (He, Wu, Dong, & Gao, 2019). Then, they used Cytoscape software to construct a gene-target network associated with Aconiti Lateralis Radix Praeparata toxicity-induced cardiac adverse reactions, performed biological process enrichment analysis, and ultimately predicted the mechanism of cardiac toxicity of Aconiti Lateralis Radix Praeparata. The results showed that the quantitative structure-activity relationship model had certain accuracy in predicting the acute toxicity of TCM components. Other data suggested that the quantitative structure-activity relationship model could also predict the acute toxicity of aromatic amines with high accuracy and predictive ability (Li & Liao, 2013).
2.2.2. Network analysis based on systems biology
Integrating principles and methods of systems biology, it involves analyzing the interaction networks between drugs and genes, proteins, metabolites, and other entities. By constructing and analyzing these networks, it is possible to uncover the interactions between drugs and various levels within an organism and their impact on biological processes. This provides a deeper understanding of the mechanisms underlying the toxicity and efficacy of drugs. Li et al. investigated the hepatotoxicity of Rhei Radix et Rhizoma (Dahuang in Chinese) using a combination of UPLC-Q-TOF/MS metabolomics and network pharmacology (Li et al., 2021, Li et al., 2021, Li et al., 2021, Li et al., 2021). They identified potential targets related to the hepatotoxicity of Rhei Radix et Rhizoma and found that Rhei Radix et Rhizoma may exert its hepatotoxic effects through the cyclic adenosine monophosphate (cAMP) signaling pathway, cholinergic synapses, and modulation of TRP channels involved in inflammation. The dopamine D1 receptor (DRD1) and dopamine D2 receptor (DRD2) were among the identified targets.
2.2.3. Drug-disease network analysis
The networks consisting of drugs, diseases, and related genes, proteins, etc., are constructed to analyze the effects of drugs on diseases and their underlying mechanisms. This analysis can reveal the interaction between drugs and diseases, providing a theoretical basis for understanding the therapeutic effects and potential toxicities of drugs. Dan et al. retrieved databases and constructed a compound-target-network of TCM, analyzed their properties and distribution in meridians, and summarized the rules of anti-tumor drugs with cardiac toxicity (Dan et al., 2020).
2.2.4. Toxicity prediction and screening
Utilizing known toxicity data and drug chemical features, machine learning and prediction models are applied to predict the potential toxicity of drugs. This approach enables early-stage screening and evaluation of drugs, reducing the risk of toxicity. In the study of nephrotoxicity using network toxicology, Qiu et al. constructed a high-throughput in vitro model for evaluating renal toxicity and predicted the toxicity of a large number of compounds using a quantitative structure-activity relationship (Qiu, Zhou, & Li, 2017). Zhang et al. applied the network toxicology prediction software Mold2 to calculate molecular descriptors for 7 409 chemical components of TCM and constructed a quantitative structure-activity relationship model using the random forest algorithm to select the optimal predictive model (Zhang et al., 2014).
Network pharmacology can analyze and predict the toxic components of TCM by constructing network models, screening for toxic components in toxic TCMs, studying their toxic mechanisms and compatibility taboos, providing a new approach to the study of TCM toxicity. This research method provides new technical support for improving the safety and rational use of TCMs in the study of the compatibility rules of TCM formulas, and further promotes the modernization of TCMs (Pelkonen, Xu, & Fan, 2014).
Through the aforementioned methods, network pharmacology can provide a comprehensive and systematic analysis of drug effects and toxicities, aid in a scientific understanding of drug mechanisms, optimize drug development and clinical applications, and improve drug safety and efficacy.
2.3. Explaining basic principles of composing formula compatibility
In a TCM formula, the “Sovereign” herb represents the main therapeutic component of the formula and directly treats the primary disease. Other herbs such as the “Minister”, the “Assistant”, and the “Courier” have different roles in enhancing the formula's efficacy, treating accompanying symptoms, promoting the main component's therapeutic effect, and harmonizing the formula. The different roles and compatibility relationships among these herbs can further improve the efficacy and safety of the formula, enabling it to exert a more significant therapeutic effect in clinical applications (Wei & Wang, 2013). The interpretation of the “Sovereign-Minister-Assistant-Courier” relationship in TCMs using network pharmacology can be achieved by collecting data on the medicinal ingredients and targets related to the “Sovereign-Minister-Assistant-Courier” concept. This data can be obtained from TCM databases, chemical databases, bioinformatics databases, and other relevant sources.
Next, the medicinal ingredients of the “Sovereign”, “Minister”, and “Assistant” herbs are considered nodes in the network model, and they are connected to their corresponding targets. This network model can be constructed using network analysis tools and algorithms such as Cytoscape.
Subsequently, network pharmacology methods are applied to analyze and explore the relationships within the network model. This includes network topology analysis, module discovery, node centrality calculation, and functional enrichment analysis, etc. These analyses uncover the underlying relationships and mechanisms involved in the “Sovereign-Minister-Assistant-Courier” concept of TCMs. Finally, the results of network pharmacology analysis are interpreted and validated through experimental validation and literature research. This validation process involves verifying the biological functions and interactions of key nodes identified in the network model. Experimental validation can include in vitro experiments, animal studies, clinical trials, and etc.
Based on the results of network pharmacology analysis, conclusions regarding the “Sovereign-Minister-Assistant-Courier” relationship in TCMs can be drawn. These conclusions can be applied to the rational use of TCMs and drug development.
Tao et al. identified 58 bioactive compounds and predicted 32 potential targets associated with cardiovascular diseases in the formula containing Curcumae Radix (Yujin in Chinese) using integrated network analysis (Tao et al., 2013). The results indicated that Curcumae Radix is the main component for preventing cardiovascular diseases, and the other three drugs, Gardeniae Fructus (Zhizi in Chinese), Moschus (Shexiang in Chinese), and Borneolum Syntheticum (Bingpian in Chinese), can serve as auxiliary drugs to enhance the efficacy of the main components. Moreover, these three drugs may act synergistically to effectively prevent and treat cardiovascular diseases, which is consistent with the principle of synergistic effects in the TCM formula and is in line with the concept of “Sovereign-Minister-Assistant-Courier” in TCMs. Yang et al. clarified the molecular mechanisms of individual herbs and highlighted the synergistic effects among herbs in Banxia Xiexin Decoction, effectively interpreting the traditional theory of “Sovereign-Minister-Assistant-Courier” in TCM formula from a network perspective (Yang et al., 2018). Sheng et al. studied the thrombolytic capsule formula for treating cardiovascular diseases by using a rat model of disseminated intravascular coagulation (Sheng et al., 2014). They identified important cross-targets of the single herbal medicine in the formula, where multiple active chemical components may simultaneously interact with the same target, thus explaining the compatibility rules of “Sovereign-Minister-Assistant-Courier” in the herbal formula. The use of network pharmacology allows for a systematic exploration of the interactions between the constituents of TCMs and their targets, providing insights into their underlying mechanisms of action. It provides a scientific basis for the clinical application and rational compatibility of TCMs.
The research strategy of component-based TCMs based on network pharmacology can effectively study the rationality of the compatibility of TCM components, as well as the characteristics of drug interactions between effective components, such as synergy, addition, antagonism, etc. This is consistent with the theory of “Dan Xing (single action)”, “Xiang Shi (mutual promotion)”, “Xiang Xu (mutual restraint)”, “Xiang Wei (mutual avoidance)”, “Xiang Sha (mutual killing)”, “Xiang Wu (mutual hatred)”, and “Xiang Fan (mutual opposition)” in TCM formula compatibility. This strategy can further reveal the compatibility laws of TCM formulas and verify their scientific and rational nature. Modern research has shown that methods such as target network analysis, functional annotation analysis, and model construction based on network pharmacology can reveal the compatibility laws of TCM formulas. Li et al. demonstrated the importance of network target-based methods in evaluating synergistic drug combinations and promoting the development of combination therapy (Li, Zhang, & Zhang, 2011). They used network-based target screening to identify the synergistic drug combinations and showed that applying network pharmacology can discover and explore the rules of formula compatibility. In another study, researchers explored the network pharmacological mechanism of the effective ingredients of the TCM formula Zuojin Pills for gastric cancer patients. They used protein-protein interaction and gene ontology and genome analysis combined with previously retrieved data to identify 176 targets of Coptidis Rhizoma, respectively, with the majority of targets overlapping, indicating the possible interaction between these two herbs during the treatment process (Zhang et al., 2020, Zhang et al., 2020). Li et al. constructed a herbal network using 3 865 related formulas, which effectively explained the TCM pairing and compatibility phenomena, and also discovered new drug pairs with synergistic and antagonistic effects (Liang, Li, & Li, 2014). Han et al. established a “compound-target-pathway-disease” network diagram in a study on Yuanhu Zhitong Dropping Pill, which showed that the monarch drug, Chuanxiong Rhizoma (Chuanxiong in Chinese), played a major therapeutic role, and the minister drug, Angelicae Dahuricae Radix (Baizhi in Chinese), played an auxiliary role in prolonging the effect of Chuanxiong Rhizoma, consistent with the theory of herbal pairing and compatibility in TCMs (Han et al., 2016). The author summarized the current research on some TCM formulas and their corresponding diseases using network pharmacology, as well as the validation methods for the results obtained (Table 3).
Table 3.
Some TCM formulas, corresponding treated diseases, and validation methods for network pharmacology analysis results.
| Disease types | TCM formulas | Diseases | Verification methods | References |
|---|---|---|---|---|
| Respiratory system cisease | Mai Men Dong Decoction | Pulmonary fibrosis | In vitro cell experiment | Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022 |
| Endocrine system disease | Bu Yang Huan Wu Decoction | Diabetes | In vivo animal experiment | Liu et al., 2023, Liu et al., 2023 |
| Erchen Decoction | Olycystic ovarian syndrom | In vivo animal experiment | Cong et al., 2021 | |
| Xihuang Capsules | Granulomatous mastitis | In vivo animal experiment | Dai and Xie, 2022, Dai et al., 2022 | |
| Reproductive system disease | Chaihu Shugan Powder | Liver-qi stagnation and blood stasis type psychogenic erectile dysfunction | In vitro cell experiment | Lin et al., 2022, Lin et al., 2022 |
| Qiangjing Tablets | Male infertility | In vivo animal experiment | Shen et al., 2022 | |
| Metabolic disease | Chaihu Shugan Powder | Non-alcoholic fatty liver disease | In vivo animal experiment | Lei et al., 2022, Nie et al., 2020 |
| Central nervous system disease | MaHuang Fuzi Xixin Decoction | Migraine | In vivo animal experiment | Ge et al., 2022 |
| Yiqi Tongmai Fang | Ischemic stroke | In vivo animal experiment | Chen and Cui, 2022, Chen and Han, 2022 | |
| Circulation system disease | Xueshuan Tong Capsules | Ischemic microvascular dysfunction in the brain | In vivo animal experiment | Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022 |
| Pingyang Jianya Fang | Hypertension | In vivo animal experiment | Liu et al., 2021, Liu et al., 2021, Liu et al., 2021, Liu et al., 2021 | |
| Shexiang Xintong Ning Tablets | Coronary disease | In vitro cell experiment | Jia et al., 2021 | |
| Digestive system disease | Huangqi Jianzhong Decoction | Gastric cancer | In vivo animal experiment | Li et al., 2022, Li et al., 2022, Li et al., 2022, Li et al., 2022 |
| Huanglian Jiedu Decoction | Liver cancer |
In vitro cell experiment In vivo animal experiment |
Huang et al., 2020, Huang et al., 2020 | |
| Moluo Dan Pill | Chronic atrophic gastritis | In vitro cell experiment | Zhou et al., 2022, Zhou et al., 2022 | |
| Motor system disease | Shenxian Decoction | Myasthenia gravis | In vivo animal experiment | Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022 |
| Xianling Gubao Capsules | Osteoporosis | In vivo animal experiment | Chai et al., 2022 |
The compatibility of TCMs is not the simple sum of the effects of individual herbs. It also needs to take into account the dosage of drugs and whether they are appropriate for the symptoms. Therefore, it is necessary to consider the compatibility rules of the prescription as a whole. Network pharmacology has played an important auxiliary role in the design of TCM formulas. Through network pharmacology methods, the targets of TCMs in the formula, active ingredients, pharmacological mechanisms, and compatibility rules can be further revealed. Network pharmacology not only enables the regulation of disease systems under the guidance of TCM theory but also promotes the progress of research on TCM formulas. Therefore, in the compatibility of TCMs, the overall compatibility should be emphasized by network pharmacology, and it should be used as an auxiliary tool to design more rational TCM formulas.
3. Using network pharmacology to discover active components in TCMs
By combining multiple database software and network visualization techniques, researchers can construct network models of component-target-pathway, and further analyze the relationship between drugs, targets, and diseases. It can not only effectively predict the active components of TCM and their corresponding mechanisms of action, but can also be used in conjunction with molecular docking technology to virtually screen active components that may improve diseases.
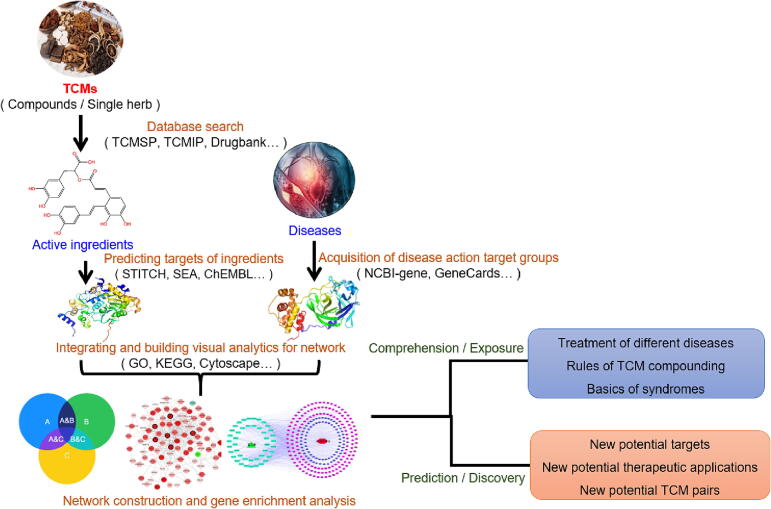
By constructing a network relationship diagram of drug-target-disease, researchers can analyze and predict the various pharmacological activities of TCMs, and explore their therapeutic mechanisms and potential clinical application values. The advantage of this method is that it can comprehensively reveal the mechanisms of action of multiple active ingredients in TCMs, and greatly shorten the time and cost of new drug development. Furthermore, network pharmacology has become an important research field in modern pharmacology. Currently, the general process of network pharmacology research on the pharmacological effects and mechanisms of TCM formulas or single herbs includes the following steps: collection and screening of active ingredients, prediction of target genes based on effective ingredients, obtaining corresponding target genes based on disease/phenotype, identifying the intersection of disease targets and compound targets, and ultimately obtaining the targets of action for TCM formulas or single herbs against a certain disease. Based on this, network construction and gene enrichment analysis are performed, the flowchart is shown in Fig. 2.
Fig. 2.
General process of network pharmacology research on TCMs.
For example, Liu et al. utilized network pharmacology in combination with molecular docking and bioinformatics analysis to investigate the active ingredients and potential pharmacological mechanisms of Huaihua San (HHS) in treating ulcerative colitis (UC) (Liu et al., 2021, Liu et al., 2021, Liu et al., 2021, Liu et al., 2021). The study identified a total of 28 bioactive components of HHS and 421 HHS-UC related target proteins. Bioinformatics analysis indicated that quercetin, hesperetin, and kaempferol are potential candidate drugs for UC. In another study, Zhang et al. employed a comprehensive network pharmacology approach combined with molecular docking to investigate the mechanism of action of Salvia miltiorrhiza Bge. in treating diabetic nephropathy (DN) (Zhang et al., 2021, Zhang et al., 2021, Zhang et al., 2021). They identified 66 active components and 189 target proteins of S. miltiorrhiza. Among them, 64 targets were found to overlap with DN-related proteins, revealing the active ingredients and potential molecular therapeutic mechanisms of S. miltiorrhiza in DN. In the analysis of Suanzaoren Decoction (SZRD) for the treatment of Parkinson's disease with a sleep disorder (PDSD), Liu et al. utilized network pharmacology tools to predict a total of 135 active ingredients and 41 corresponding target proteins for PDSD treatment (Liu et al., 2021, Liu et al., 2021, Liu et al., 2021, Liu et al., 2021). These examples strongly demonstrate that network pharmacology methods can efficiently screen and predict the active ingredients in TCMs, accelerating the drug discovery process and providing scientific evidence for the clinical application of TCMs.
During the process of discovering effective ingredients in TCMs by network pharmacology, experimental validation is a critical step to ensure data accuracy and reliability. While network pharmacology can infer the active ingredients and mechanisms of TCM through large-scale data analysis and prediction, experimental validation is still necessary to verify the predicted results.
Through in vitro cell experiments or in vitro enzyme activity assays, the interaction between active ingredients in TCMs and specific targets can be evaluated to determine the activity and efficacy of the TCM components. Zhou et al. employed network pharmacology and in vitro experiments to elucidate the action and mechanism of hesperidin against rotavirus (RV) (Zhou, Qian, Yuan, Yang, & Song, 2023). Through network pharmacology analysis, molecular docking validation, and experimental research, it was demonstrated that hesperidin not only exhibits anti-RV biosynthesis activity but also may exert its anti-RV effects through the modulation of the PI3K/Akt signaling pathway.
To determine the efficacy and safety of TCMs, animal experiments are conducted by observing the symptoms, physiological indicators, and histopathological changes in animals. Li et al. aimed to validate the key mechanisms of Honghua Buji Fang in treating vitiligo (Li, Xin, Huang, & Zhu, 2023). They established a vitiligo mice model and conducted network pharmacology studies to preliminarily screen the main active components of Honghua Buji Fang, such as mosloflavone, and key targets such as IL-6. In the animal experiment, the expression of inflammatory factors such as IL-6 in mouse serum was measured to explore the anti-inflammatory ability of Honghua Buji Fang. The results showed a significant reduction in inflammation and alleviation of superficial skin hyperplasia in the mice after treatment with Honghua Buji Fang, providing preliminary confirmation of the accuracy and reliability of the network pharmacology results.
Clinical trials, including safety assessment, efficacy evaluation, and side effect monitoring, are conducted to validate the clinical application value of TCMs. Zhang et al. utilized network pharmacology to predict the efficacy of Qianliexiao Decoction in the treatment of chronic prostatitis (Zhang et al., 2021, Zhang et al., 2021, Zhang et al., 2021). Clinical randomized non-blind trials were conducted to validate the findings. The results showed that network pharmacology can predict the corresponding targets of TCM's action on diseases to a certain extent. Qianliexiao Decoction demonstrated a significant improvement in clinical symptoms for patients with type IIIA prostatitis with damp-heat syndrome.
Experimental validation plays a vital role in obtaining pharmacological characteristics of active components in TCMs, thereby enhancing our understanding of their mechanisms and therapeutic efficacy. This crucial step in network pharmacology research ensures reliable data support for the pharmacological exploration of TCMs. Furthermore, experimental validation serves as a valuable guide and provides evidence for the clinical application and drug development of TCMs.
3.1. Collection and screening of active ingredients in TCMs
The acquisition of active compounds is the first step in TCMs network pharmacology research. Commonly used databases for obtaining active ingredients in TCM include the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://tcmsp-e.com/tcmsp.php) (Ru et al., 2014), the Traditional Chinese Medicine Integrated Pharmacology Research Platform (TCMIP, https://www.tcmip.cn/ETCM/index.php/Home/) (Xu et al., 2019b, Xu et al., 2019a) and Traditional Chinese Medicine integrative database (TCMID, http://www.megabionet.org/tcmid/) (Huang et al., 2018, Xue et al., 2013) et al.
In exploring the mechanism of the therapeutic effect of the Fuzi-Yinyanghuo (Epimedii Folium) herb pair on chronic heart failure, Huang et al. used TCMSP to collect and search for target active ingredients of Fuzi and Yinyanghuo, and obtained 28 active ingredients of the herb pair (Huang et al., 2021). Xu et al. and his research team obtained 59 candidate compounds from Zhenwu Decoction by using the TCMSP platform for network pharmacology analysis (Xu et al., 2020). Zhang et al. identified a total of 234 compounds in Zuojin Pill by searching for its main active ingredients using TCMSP (Zhang et al., 2020, Zhang et al., 2020). In a study on Yigan Kang's improvement of insulin resistance, Yu et al. obtained all the effective components of 12 TCMs contained in Yi Tang Kang through the TCMSP database, with a total of 438 effective components that met the screening criteria (Yu et al., 2023). In the study of the identification of key drug targets and molecular mechanisms of Curcumae Rhizoma (Ezhu in Chinese) in the treatment of liver cancer, Zhao et al. searched and screened the chemical components of roots and rhizomes of Curcumae Rhizoma in the TCMSP database, and finally 22 active ingredients were selected (Zhao et al., 2022). In exploring the mechanism of action of total flavonoids of Astragali Radix (Huangqi in Chinese) in treating nephrotic syndrome, Zhang et al. used the TCMSP and TCMID databases to obtain the main active ingredients of total flavonoids of Astragali Radix, and screened out 20 active ingredients (Zhang et al., 2018). The results showed that 29 active ingredients in total flavonoids of Astragali Radix acted on 50 targets, involving various biological processes, molecular functions, and cellular components, which was consistent with the literature reports.
The elucidation of the pharmacologically active substances in TCM formulas is crucial for investigating their overall efficacy and mechanisms of action. And it forms the basis and core of TCM safety and quality control. However, currently, the issue of homogenization in key component selection is particularly serious in TCM network pharmacology research. That is, regardless of different diseases, treatment methods, and TCM formulas used, the same key components such as quercetin, kaempferol, β-sitosterol, stigmasterol, and schisandrin are always screened out by TCM network pharmacology-related databases (Zeng & Zhou, 2022). The phenomenon of “homogenization” of the material basis violates and indirectly denies the holistic concept and dialectical connotation of “treating the same disease with different therapies” or “treating different diseases with the same therapy” (Wang et al., 2023, Wang and Zhu, 2023, Wang and Hai, 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023). Although current research indicates that network pharmacology research can fit in with the holistic thinking of TCM (Liu et al., 2022, Liu et al., 2022, Liu et al., 2022) due to the limitations of the TCM database and the lack of rigor in methods, different diseases and prescriptions always appear in search results with homogenized components as key active ingredients. This is likely to lead to an overestimation of the pharmacological effects of homogenized components and causes significant uncontrollable errors in identifying active ingredients, pharmacological effects of TCM, flavor and meridian tropism, toxicity-effect networks, and other factors (Xu et al., 2019b, Xu et al., 2019a). In summary, although the network pharmacology research of the “same disease with different treatments” and “different diseases with the same treatment” theories of TCM is still in its initial stage, network pharmacology methods have higher screening accuracy and predictive ability than traditional large-scale screening methods. By constructing a drug-target-pathway network model, researchers can comprehensively understand the mechanism of action and potential therapeutic effects of active ingredients in TCMs. This method can not only shorten the screening time for drugs, but also reduce experimental costs and manpower input, and improve research efficiency. It is worth mentioning that the application of network pharmacology methods is not limited to TCM research. In modern pharmacology research, network pharmacology methods have been widely used in fields such as compound screening, drug efficacy prediction, and drug reutilization. Therefore, future research will deepen people's understanding and recognition of TCMs in the treatment of complex diseases. The promotion and application of this method can not only promote the research of TCMs but also inject new vitality into the development of modern pharmacology.
3.2. Prediction of targets of active ingredients
Currently, high-throughput screening technology is rapidly developing, and the amount of pharmacological activity data for natural product drugs is increasing substantially, mainly collected by databases such as PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al., 2021, Kim et al., 2021, Kim et al., 2022, Kim et al., 2021, Kim et al., 2021). Network pharmacology can verify the targets of active ingredients in TCMs through techniques such as molecular docking, thereby further confirming the biological activity of these ingredients (Li, 2021). According to different prediction principles, the techniques and strategies for predicting drug-target interactions can be mainly divided into four categories: ligand-based methods, target-based methods, machine learning methods, and combined application methods (Table 4). The databases commonly used to predict the targets of active ingredients include STITCH (http://stitch.embl.de/) (Kuhn et al., 2010, Kuhn et al., 2008, Szklarczyk et al., 2016), TTD (https://db.idrblab.net/ttd/) (Southan et al., 2013, Wang et al., 2020, Wang et al., 2020, Yang et al., 2016, Zhou et al., 2022, Zhou et al., 2022), SEA (https://sea.bkslab.org/) (Keiser et al., 2007), SwissTargetPrediction (http://www.swisstargetprediction.ch/) (Daina, Michielin, & Zoete, 2019), BATMAN-TCM (http://bionet.ncpsb.org/batman-tcm/) (Liu et al., 2016) and others. TTD and SEA are ligand-based methods, by using the principle of chemical similarity search. Wang et al. used the SEAware 1.7 software to predict absorption, distribution, metabolism and excretion (ADME) and therapeutic targets of the chemical constituents of Aquilariae Lignum Resinatum (Chenxiang in Chinese) essential oil when studying and predicting the chemical constituents of Aquilariae Lignum Resinatum essential oil-disease treatment targets (Wang et al., 2019, Wang et al., 2019). Jia et al. used network pharmacology to study the target genes of the anti-angiogenic effect of the TCM formula Chang Fu Kang on colorectal cancer (Jia,Yang, & Sun, 2020). They searched for colorectal cancer genes in five databases including TTD, DrugBank, OMIM, GAD, and PharmGKB, and found 40, 0, 43, 336, and 4 targets, respectively. After removing duplicate targets, they obtained 339 colorectal cancer gene targets.
Table 4.
Partial drug-target interaction prediction techniques.
| Categories | Methods | Drugs | References |
|---|---|---|---|
| Ligand-based | SEA | Chenxiang essential oil | Wang et al., 2019, Wang et al., 2019 |
| Peach kernel-safflower | Han et al., 2021, Han et al., 2021 | ||
| TTD | Chang Fu Kang | Jia, Yang, & Sun, 2020 | |
| Qing Fei Yin | Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022 | ||
| Target-based | Reverse molecular docking | Quercetin | Xu et al., 2022, Xu et al., 2022 |
| Avicularin | Duan, Li, Dong, Xu, & Ma, 2019 | ||
| Machine learning | Principal component analysis | Xuefu Zhuyu Decoction | Song, Sun, Wu, & Wang, 2020 |
| SVM | Wogonin | Wang et al., 2021, Wang et al., 2021 | |
| RF | Chuanxiong Rhizoma | Yuan, Li, Chen, Song, & Wang, 2014 | |
| QSAR | LCFs | Hu et al., 2022 | |
| Evodiae Fructus | Chen and Han, 2022, Chen and Cui, 2022 | ||
| Combined | SVM, RF, AutoDOCK | Yu Jin Formula | Tao et al., 2013 |
| SVM, RF, AutoDOCK | Glycyrrhizae Radix et Rhizoma | Liu, Wang, Zhou, Wang, & Yang, 2013 | |
| TTD, SEA | Curcumae Radix, Corn Stigma | Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022 | |
| SEA, Swiss target prediction, AutoDOCK | Polygoni Cuspidati Rhizoma et Radix | Shan, Ji, Wu, & Zhao, 2022 |
The prediction principle of SwissTargetPrediction is based on the similarity of the two-dimensional and three-dimensional structures of ligands and known compounds, using pharmacophore models to obtain the targets of compounds. Zhang et al. used network pharmacology methods to study the mechanism of action of Prunellae Spica (Xiakucao in Chinese) in treating breast cancer, and through STITCH and Swiss Target Prediction, predicted 379 potential targets for 32 active compounds in Prunellae Spica (Zhang et al., 2020, Zhang et al., 2020).
The target prediction method based on receptors mainly involves reverse molecular docking, which predicts the optimal orientation of ligand-receptor complex formation to form a stable complex using molecular docking technology and scores the affinity between ligand and receptor based on a scoring function. Xu et al. conducted large-scale reverse docking experiments and molecular dynamics simulation experiments on quercetin glycoside derivatives and found that TGFBR1 may be one of the potential protein targets of quercetin glycoside derivatives, demonstrating its potential as a promising drug for the treatment of breast cancer metastasis (Xu et al., 2022, Xu et al., 2022).
Machine learning methods are mainly divided into two types: supervised learning and unsupervised learning. Unsupervised learning refers to the analysis of large samples of data without any prior knowledge of the categories, to achieve sample classification, data analysis, and model construction, such as principal component analysis (PCA). Song et al. applied PCA combined with network pharmacology to explore the mechanism of Xuefu Zhuyu Decoction in the treatment of chronic heart failure and identified 588 compounds and 913 potential targets (Song, Sun, Wu, & Wang, 2020). Supervised learning methods mainly include support vector machine (SVM), random forest (RF), and MT-QSAR (multitarget quantitative structure-activity relationship) techniques. When analyzing the regulatory mechanism of baicalein on lung cancer, Wang et al. used SVM models combined with network pharmacology and immunoblotting to identify Bcl-2 and ErbB4 as the main targets of baicalein (Wang et al., 2021, Wang et al., 2021). Yuan et al. predicted the targets of 26 chemical components of Ligusticum chuanxiong Hort. (Chuanxiong in Chinese) using a RF-based drug-target interaction model and evaluated the model accuracy using 10-fold cross-validation, which was well-validated in the literature (Yuan, Li, Chen, Song, & Wang, 2014). Hu et al. used network pharmacology, 3D/2D-QSAR, molecular docking, and molecular dynamics simulations to investigate the molecular mechanism of licorice flavonoids (LCFs) against melanoma (Hu et al., 2022). They successfully identified glabridin, naringenin, and glycyrrhetinic acid as the key active components and TYR as the critical target for the anti-melanoma effect.
Although chemical similarity search and pharmacophore models have advantages such as rapidity and efficiency, their prediction accuracy is not high; whereas machine learning methods are difficult to directly discover the correlation between protein and ligand, and difficult to achieve quantitative prediction. Therefore, multiple techniques need to be combined to complement each other and design the optimal drug-target screening strategy. Tao et al. used random forest and support vector machine algorithms to establish a model for predicting potential targets of components in the compound Yujin Fang (Tao et al., 2013). Olecular docking software such as Autodock confirms the predicted results of the algorithm. Based on this, they discovered 58 active components and 32 potential targets for cardiovascular disease treatment in the compound Yujin Fang.
Currently, drug designers have developed various computational methods to assist in predicting potential drug targets. The analysis of these algorithms and their characteristics are shown in Table 5. Here, we briefly introduced their characteristics and examples of their application in predicting the active ingredients of TCMs.
Table 5.
Algorithms and features of partial network pharmacology analysis.
| Methods | Features | Applications | References |
|---|---|---|---|
| drugCIPHER | A network-based algorithm for drug target prediction, which primarily integrates three linear regression models based on different metrics in a rigorously scientific language. |
|
Liang, Li, & Li, 2014 |
| DMIM | A distance-based mutual information method for analyzing drug compatibility relationships in TCM formulas. | Study of the molecular mechanisms and interaction networks of diseases | Li, Zhang, Jiang, Wei, & Zhang, 2010 |
| LMMA | A combined method of literature mining and microarray analysis is used to construct gene networks for specific biological systems. |
|
Li, Wu, & Zhang, 2006 |
| CIPHER-SC | A regression model that integrates protein-protein interactions, disease phenotype similarity, and gene ontology similarity scould predict disease genes. |
|
Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022, Zhang et al., 2022 |
| MIClique | Identification of gene sets based on mutual information and clique filtering algorithms. |
|
Zhang, Song, Wang, & Zhang, 2009 |
| CIPHER-HIT | A method for assessing the closeness between two nodes in a phenotype-gene heterogeneity network, which can identify the genetic correspondence of disease subtypes by labeling genes and phenotypes in the phenotype-gene network. |
|
Yao, Hao, Li, & Li, 2011 |
| ComCIPHER | A novel drug-gene-disease relationship collaborative module method, characterized by closely related drugs, diseases, and genes. | Evaluation of pharmacological effects and component screening of TCM formulations; Evaluation of TCM formulations; Correlation analysis between TCM formulations and diseases |
Zhao, & Li, 2012 |
| PPA-GCN | A framework based on graph convolutional neural networks that utilize the co-directionality information of genes in specific pathways to learn graph topology patterns and gene node features, propagate node properties in the network and assist in the allocation of metabolic pathways. |
|
Lu, Li, & Li, 2022 |
| RWRNET | A network correlation algorithm based on node random walk that can combine local and global topological relationships. |
|
Liu et al., 2020, Liu et al., 2020, Liu et al., 2020 |
Drug CIPHER (Zhao, & Li, 2010) is a target prediction tool developed by the Li Shao research group, which includes three linear regression models: drug CIPHER-TS, drug CIPHER-CS, and drug CIPHER-MS. These models utilize therapeutic similarity (TS), chemical structure similarity (CS), and multiple similarity information syntheses (MS), respectively. Liang et al. used a network pharmacology approach to explore the molecular mechanisms of Liuwei Dihuang Pills in treating multiple diseases (Liang, Li, & Li, 2014). They used the drug CIPHER software for target prediction and found that Liuwei Dihuang Pills mainly act on targets such as PPARG, RARA, CCR2, ESR1, etc. The DMIM method is a distance-based mutual information method used for analyzing the compatibility relationships of drugs in TCM formulas. Li et al. conducted in vitro experiments to measure the activity of strong connections between herbs and herb pairs extracted by DMIM, to evaluate the effectiveness of the herbal network (Li, Zhang, Jiang,Wei, & Zhang, 2010). They took Liuwei Dihuang Pills as an example and proposed the concept of “synergistic modules” in a cross-herb-biomolecule-disease multi-layer network, exploring the potential combination mechanism of the herbal formula. The results showed that the herbal network constructed using DMIM technology from 3 865 related formulae not only recovered traditionally defined herb pairs and formulae well, but also generated new anti-angiogenic herbal ingredients and herbal pairs with synergistic or antagonistic effects. The TCM Network Pharmacology Analysis System(Liu et al., 2024) (TCMNPAS, http://54.223.75.62:3838/) is a network pharmacology analysis platform for TCM developed by Ming Yang from Longhua Hospital, affiliated with Shanghai University of Traditional Chinese Medicine. It integrates multiple databases such as TCMSP, TCMID, STITCH, KEGG, GeneCard, and Uniprot, and can display the mechanism of action of TCM formulas, target groups, and the network of drug-target-disease correlations based on model algorithms, multidimensional analysis, and visualization analysis. It is conducive for researchers to conduct network pharmacology analysis of TCM formulas and their active components more conveniently and efficiently.
Given the complexity of the components in TCMs and the lack of related fundamental research, predicting drug targets in TCM network pharmacology requires large-scale investigation into drug mechanisms in terms of prediction and analysis methods, molecular target mechanisms, and drug interactions, utilizing complex network models. To form high-quality integrated TCM network pharmacology data, it is necessary to establish a shared TCM molecular mechanism data resource platform, combined with modern drug molecular mechanism data resources.
3.3. Collecting target groups related to corresponding diseases and integrating them with compound targets
The target group of the disease is often queried using online databases such as Online Mendelian Inheritance in Man (OMIM, https://omim.org/) (Hamosh, Scott, Amberger, Bocchini, & McKusick, 2005), NCBI-gene (https://www.ncbi.nlm.nih.gov/gene) (Edgar et al., 2002, Piovesan et al., 2016) and GeneCards (https://www.genecards.org/) (Stelzer et al., 2016). After querying the disease targets and integrating them with the active ingredient targets, the intersection of the two can be taken to construct a network of TCM formulas/monomer-effective ingredient-target gene-disease (Table 6).
Table 6.
Treatment targets of active ingredients-diseases in some natural drugs retrieved using various databases.
| Drugs | Ingredients | Function/diseases | Main databases | Number of targets | Main targets | References |
|---|---|---|---|---|---|---|
|
Prunellae Spica |
Quercetin | Gastric cancer | OMIM, TTD, SwissTargetPrediction | 15 | AKT1, EGFR, SRC | Yang et al., 2020 |
| Ursolic acid | Osteoporosis | GeneCards | 44 | VEGFA, TP53, IL6 | Zhao et al., 2021, Zhao et al., 2021 | |
| Chlorogenic acid | COVID-19 | SwissTargetPrediction | 70 | IL6, MAPK3, CASP3 | Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022, Wang et al., 2022 | |
| Astragali Radix | Isorhamnetin | Ischemic nerve injury | DisGeNET, Swiss Target Prediction | 50 | AKT1, IL6, MMP2 | Zhang et al., 2023, Zhang et al., 2023 |
| Astragaloside IV | Diabetes retinopathy | OMIM,TTD,SwissTargetPrediction, GeneCards | 56 | AKT1, VEGFA, EGFR | Yu, Li, Li, & Xu, 2022 | |
| 7-Methoxy-4-hydroxy isoflavone | Endothelial dysfunction | OMIM, SwissTargetPrediction, GeneCards, DisGeNET | 39 | ALB, PTGS2, TNF |
Zhang et al., 2021, Zhang et al., 2021, Zhang et al., 2021 | |
| Salviae Miltiorrhizae Radix et Rhizoma | Tanshinone ⅡA | Hepatic fibrosis | GeneCards, OMIM | 29 | CCND1, MMP9, RELA | Shi et al., 2020 |
| Scutellariae Radix | Baicalin | Obesity | Swiss Target Prediction, GeneCards, SEA | 37 | TNF, NFKB1, SREBF1 | Wang et al., 2020, Wang et al., 2020 |
| Coptidis Rhizoma | Berberine | Diabetic nephropathy | GeneCards, DisGeNET | 67 | RELA, MAPK1, IL6 | Liu et al., 2023, Liu et al., 2023 |
| COVID-19 | GeneCards, DisGeNET | 23 | CCL2, IL6, STAT3 | Cao et al., 2022, Cao et al., 2022 | ||
| Hyperuricemia | DisGeNET | 15 | AKT1, IL6, VEGFA | Li et al., 2021, Li et al., 2021, Li et al., 2021, Li et al., 2021 | ||
| Rhei Radix et Rhizoma | Emodin | Rheumatoid arthritis | GeneCards, OMIM, SwissTargetPrediction, DisGeNET | 32 | CASP3, PTGS2, MAPK14 | Cao et al., 2022, Cao et al., 2022 |
| Ischemic stroke | OMIM | 10 | CASP3, KDR, PTGS1 | Jia et al., 2021 | ||
| Aloe emodin | Colon cancer | GeneCards, OMIM | 43 | HSP90AA1, JUN, SRC | Jiang, Ding, Mao, You, & Ruan, 2021 | |
| Piceatannol-3′-O-β-D-glucopyranoside | Cognitive impairment | Swiss Target Prediction, GeneCards | 71 | MAPK1, MMP9, ESR1 | Wang et al., 2023, Wang and Zhu, 2023, Wang and Hai, 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023 | |
|
Ginseng Radix et Rhizoma |
Ginsenoside Rh2 | Polycystic ovarian syndrome | GeneCards, OMIM | 13 | ESR1, CYP19A1, SRD5A1 | Wang and Hai, 2023, Wang and Zhu, 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023 |
| Ginsenoside Rg1 | Radiation enteritis | Swiss Target Prediction, GeneCards | 25 | AKT1, VEGFA, HSP90AA1 | Wang et al., 2023, Wang and Zhu, 2023, Wang and Hai, 2023, Wang et al., 2023, Wang et al., 2023, Wang et al., 2023 | |
| Ginsenoside Rg5 | Cancer | Swiss Target Prediction | 100 | STAT3, PTAFR, VEGFA | Gao et al., 2022 | |
| Glycyrrhizae Radix et Rhizoma | Glabridin | Diabetic nephropathy | GeneCards | 40 | EGFR, MAPK1, CASP8 | Tan et al., 2022 |
Liu et al. used GeneCards to search and select target genes for diabetes and obtained 1 032 targets. They also searched OMIM and obtained 60 targets (Liu et al., 2023, Liu et al., 2023). Liu et al. investigated the mechanism of Shenfu Huang Formula, which has clinical efficacy in treating COVID-19 patients with sepsis syndrome; they established a target-disease network using PharmGKB, Drugbank, and TTD databases and found that 46 targets were directly involved in immune system diseases and nine targets were associated with inflammation (Liu et al., 2020, Liu et al., 2020, Liu et al., 2020).
Despite the rapid advancement of network pharmacology, there persist inherent risks and unresolved issues that need to be addressed (Luo et al., 2020, Mao and Zhu, 2021, Yuan et al., 2022, Zeng and Zhou, 2022, Zhang et al., 2016).
The first point is that the network pharmacology of TCM mainly focuses on static theoretical analysis. However, the metabolism in organisms is a dynamic life process, in which unknown variable factors have randomness and timeliness. We cannot rely solely on a single network model to depict the entire process. The second point is that there is a homogenization phenomenon in the selection of key components in some network pharmacology studies of TCMs. In the early network pharmacology analysis, the key component results often included multiple TCM ingredients that are widely present and commonly found, such as adenosine, amino acids, etc. However, these substances were mistakenly considered as crucial components of the formula. It may be caused by the lack of precision in component screening strategies, different algorithms producing different results, the uneven quality of the information included in databases, and limitations in the information provided by public databases. The third point is that information-based prediction needs to be combined with practical application. Currently, in order to make the analysis results more reliable, it is generally necessary to conduct experiments such as in vivo or in vitro experiments, and clinical trials to verify the correctness and reliability of the analysis results. The fourth point is that information retrieval results are limited. Currently, many unknown components and disease mechanisms have not yet been recorded in the database.
From the perspective of the pharmacological characteristics of drugs, the effects of some drugs are at the macro level, while others are at the micro level. Therefore, when conducting database retrieval, conventional parameter settings may lead to the omission of some micro-level effect drug components, resulting in a reduction in the completeness and accuracy of the final retrieval results.
3.4. Network construction and gene enrichment analysis
Network construction and gene enrichment analysis of TCMs is a bioinformatics analysis method applied in the field of TCM research. This method integrates various information, such as known TCM compounds, TCM formulas, and protein targets, to construct a TCM network model, revealing the molecular mechanisms of TCM in treating diseases. In addition, through gene enrichment analysis, biological processes and pathways in the TCM network can be further explored, and genes and functional categories related to TCM treatment can be identified. Network construction is achieved through mathematical graph theory and complex network methods, describing drugs, diseases, and targets as nodes, and relationships between drugs and targets, diseases and targets as edges. Nodes and edges form complex networks with multiple entities and levels. Visualization tools such as Cytoscape (Doncheva et al., 2019, Shannon et al., 2003), Pajek (Han & Guo, 2014) and GUESS (Schwarz & Heider, 2019) can be used to integrate complex data and intuitively present the relationships between network nodes. Gene enrichment analysis refers to the functional enrichment analysis and pathway analysis of target proteins. The most commonly used methods are based on databases such as GO (Ashburner et al., 2000), KEGG (Kanehisa and Goto, 2000, Ogata et al., 1999), DAVID (Huang da, Sherman, & Lempicki, 2009) and STRING (Snel et al., 2000, Szklarczyk et al., 2019). It can associate enriched biological functions and signaling pathways with diseases and active ingredients and describe the mechanism of TCM in treating a certain disease along the main line of TCM-active compound-target/pathway-disease.
The significance level obtained through Fisher's exact test or hypergeometric test can be utilized in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis to screen and identify genes or pathways with significant biological functions. Based on the degree of expression differences in genes or pathways, we typically use the regulatory factor (Fold Change) to select genes or pathways that exhibit significant changes. By analyzing the topological structure and node connectivity of gene regulatory networks, we utilize gene regulatory networks to identify genes with critical regulatory roles. Additionally, we use disease association to select genes or pathways associated with specific diseases or disease-related genes. By calculating centrality measures such as betweenness centrality and degree centrality, which reflect the importance of genes or pathways in a network, topological centrality is considered as a criterion to select genes or pathways with significant regulatory roles. The selection of these filtering criteria depends on the specific research objectives and analysis methods. By considering disease associations and topological centrality, researchers can ensure the selection of genes or pathways that are biologically meaningful and functionally relevant to the study. These criteria help in focusing on genes or pathways that are directly linked to diseases or have significant regulatory roles in the network, providing insights into disease mechanisms and potential therapeutic targets.
Zhou et al. retrieved the chemical components of Qingfei Dayuan Granules from the TCMSP, ETCM, and YATCM databases, and then used the STRING database to obtain protein-protein interaction (PPI) networks (Zhou et al., 2020, Zhou et al., 2020). Niu et al. constructed a molecular biology network for the hot syndrome of rheumatoid arthritis and a drug target network for the syndrome-related TCMs (Niu et al., 2012). After retrieving relevant information from the database, they used the visualization software Cytoscape 2.8.2 for visualization processing and presented it in the form of a network diagram. Li et al. used the GO and KEGG databases to explore the TCM network analysis of a famous doctor's formula for treating rheumatoid arthritis and conducted gene ontology and pathway enrichment analysis on the top 15 commonly used herbal targets (Li, Li, Ouyang, & Li, 2015). Zeng et al. used Cytoscape 3.7.2 software and Network Analyzer tool to perform topological analysis in their study on the mechanism of active ingredients in Qingfei Paidu Decoction against COVID-19 and constructed a network diagram of TCM-compound-disease-target (Zeng, Tong, & Li, 2020). Gong et al. constructed a PPI network based on the STRING database to investigate the pharmacological mechanism of “Huanglian-Ganjiang” on colorectal cancer and conducted network topology analysis and GO and KEGG pathway enrichment analysis using Cytoscape 3.2.1 software (Gong et al., 2019). Zhu et al. identified key targets for the effective active ingredients in Hedyotidis Herba (Baihua she She cao in Chinese) that act on liver and colon cancer by constructing a PPI network of collected targets using the STRING database and performing GO and KEGG pathway enrichment analysis using DAVID 6.8 database (Zhu,Sang,Ji,Li, & Wu, 2020). Huang et al. investigated the mechanism of Huanglian Jiedu Decoction in treating liver cancer by performing further analysis on the obtained genes using the DAVID 6.8 database and constructing networks of compound-target, target-pathway, and target-disease using Cytoscape 3.3.0 software (Huang et al., 2020, Huang et al., 2020).
The ultimate goal of network construction technology is to build a multi-level, highly accurate, dynamic, comprehensive, and realistic molecular network based on a large amount of experimental data, which reflects the complex interactions between molecules and drugs in the organism. This lays the foundation for using various complex molecular network algorithms to mine important information such as targets (combinations), drugs (combinations), key pathways, and so on. In network pharmacology research, disease networks, drug networks, and molecular interaction networks can reflect the occurrence and development of diseases, the interactions between drugs, and the interactions between molecules. Therefore, these networks are of great significance for clarifying disease mechanisms, explaining drug action mechanisms, and achieving drug repositioning.
4. Conclusion and prospects
Network pharmacology is in line with the characteristics of the research ideas of TCM in terms of integrity, systematicity, and emphasis on effective components. This technology provides data from a holistic perspective for the scientific explanation of TCM formula, which helps to systematically study TCM and scientifically interpret the pharmacological mechanisms and compatibility relationships of TCM formula, providing new ideas and methods for the development of TCM.
Network pharmacology can explain the compatibility relationships in TCMs involving restraint and incompatibility relationships by analyzing the pharmacological characteristics and interactions of different herbal medicines. The relationships of “restraint” and “incompatibility” among various herbs in TCM formulations can be elucidated by constructing drug-target networks, and drug-drug interaction networks. Integrating predictive research methods contributes to a deeper understanding of the pharmacological mechanisms of TCM, guiding the rational dosage and optimizing the design of TCM formulas.
In summary, network pharmacology can explain the relationships between single herbs, including restraint and incompatibility relationships, by constructing drug networks, target networks, and drug-drug interaction networks. This comprehensive research approach helps to deepen understanding of the pharmacological mechanisms of herbal medicine and guides its rational application and the optimization of herbal formulations.
Currently, the use of artificial intelligence (AI) and machine learning (ML) techniques to predict drug-target interactions is a fast and efficient method, which can be applied to discover drug mechanisms, optimize drug design, and accelerate the drug discovery process. It is based on extensive biological informatics data and chemical information and involves building prediction models to forecast the interactions between drugs and targets (Dai et al., 2022, Dai and Xie, 2022, Li et al., 2022, Li et al., 2022, Li et al., 2022, Li et al., 2022, Wang, 2019). With the advent of the big data era, AI technology is being utilized to uncover general patterns of drug-target interactions from large-scale biomedical datasets, and some researchers have already achieved notable results using this technique (Ding et al., 2022, You et al., 2022).
Despite the limitations and shortcomings of current network pharmacology, it is believed that this discipline, with the integration of other disciplines such as metabolomics, genomics, and transcriptomics, along with more advanced algorithms and constant iteration and updates, will lay a solid foundation for the study of the compatibility law of TCM formulas and the discovery of new active components in TCMs.
In addition, it will provide new ideas and methods for the prevention and treatment of complicated diseases with TCM formulas, provide guarantees for the innovative development of TCM, and promote the modernization and internationalization of TCM.
CRediT authorship contribution statement
Yishu Liu: Data curation, Writing – original draft, Writing – review & editing. Xue Li: Writing – review & editing. Chao Chen: Writing – review & editing. Nan Ding: Writing – review & editing. Shiyu Ma: Supervision, Conceptualization, Data curation, Writing – review & editing. Ming Yang: Conceptualization, Data curation, Project administration, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Natural Science Foundation of Shanghai (No. 22ZR1462100), Shanghai Municipal Education Commission Collaborative Innovation Center, Clinical Evaluation Platform for Traditional Chinese Medicine and Chinese Patent Medicine (No. A1-U21-205-902), LongHua Hospital Research Projects (No. YM2021016 and RC-2020-02-04), Shanghai Sailing Program (No. 22YF1440300), Shanghai “Rising Stars of Medical Talents” Youth Development Program (Clinical Pharmacist Program), Shanghai Jiaotong University “Jiaotong University Star” Program (medical-engineering cross-research, No. YG2022QN015), Shanghai Flagship Hospital of Traditional Chinese and Western Medicine [No. ZY(2021-2023)-0205-01], Shanghai Traditional Chinese Medicine Colorectal Cancer Speciality Alliance [No. ZY(2021-2023)-0302], Shanghai Municipal Health Commission Project (No. 2020LZ007), Traditional Chinese Medicine Research Project of Shanghai Health Commission (No. 2022QN097), and Demonstration-oriented Research Ward Construction Project of Shanghai Hospital Development Center (No. SHDC2022CRW006).
Contributor Information
Shiyu Ma, Email: may7679@163.com.
Ming Yang, Email: yangpluszhu@sina.com.
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M.…Sherlock G. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M., Liu S., Zhang J.L., Ma Y., Huang S.J., Zou J.B.…Ding Y. Study on the mechanism of drug pair Ligusticum chuanxiong-Salvia miltiorrhiza in treating cardiac cerebrovascular diseases based on network pharmacology and molecular docking. China Pharmacist. 2022;25(1) [Google Scholar]
- Cao C.H., Zeng L., Rong X.F. Therapeutic mechanism of emodin for treatment of rheumatoid arthritis: A network pharmacology-based analysis. Journal of Southern Medical University. 2022;42(6):913–921. doi: 10.12122/j.issn.1673-4254.2022.06.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Li L., Xiong L., Wang C., Chen Y., Zhang X. Research on the mechanism of berberine in the treatment of covid-19 pneumonia pulmonary fibrosis using network pharmacology and molecular docking. Phytomedicine Plus: International Journal of Phytotherapy and Phytopharmacology. 2022;2(2) doi: 10.1016/j.phyplu.2022.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai S., Yang Y.B., Ma J.T., Guo Y.P., Teng J.Y., Qin N.…Zhang H. Bioinformatics and network pharmacology analysis of Xianling Gubao Capsules in the treatment of postmenopausal osteoporosis and experimental verification. World Chinese Medicine. 2022;17(24):3456–3461. [Google Scholar]
- Chen G.T., Cui Y.L. Mechanism of Yiqi Tongmai Formula against ischemic stroke: Based on network pharmacology and experimental validation. Pharmacology and Clinics of Chinese Materia Medica. 2022;38(5):83–92. [Google Scholar]
- Chen J., Tang C.C., Zhao J.P., Zhang C.Y., Lu M.Z., Xu C.Q.…Chen Q.L. Exploring the rationality and feasibility of “property and flavor network pharmacology” based on the theory of property and flavor of chinese medicine: Taking the property and flavor network pharmacology of Rujin Jiedu Powder in treating viral pneumonia. Academic Journal of Shanghai University of Traditional Chinese Medicine. 2021;35(6):1–11. [Google Scholar]
- Chen M., Wen Y.J., Yang J.C., Wu Q., Peng G.Q., Tong W.N. Potential mechanism and preliminary verification of Gegen (Puerariae Lobatae Radix) and its compound compatibility in the treatment of myasthenia gravisabout based on network pharmacology. Journal of Hunan University of Chinese Medicine. 2021;41(11):1717–1725. [Google Scholar]
- Chen P.Y., Han L.T. Study on the molecular mechanism of anti-liver cancer effect of Evodiae Fructus by network pharmacology and qsar model. Frontiers in Chemistry. 2022;10 doi: 10.3389/fchem.2022.1060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wu G., Zheng R. A systematic pharmacology and in vitro study to identify the role of the active compounds of Achyranthes bidentata in the treatment of osteoarthritis. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2020;26 doi: 10.12659/MSM.925545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong P.W., Zhang L.N., Wang D., Zhang Y.S., Chen W.N., Miao L.Y.…Zhao D.Y. Effects of Erchen Decoction on endocrine and glucose and lipid metabolism of pcos rats with phlegm-dampness syndrome. Chinese Archives of Traditional Chinese Medicine. 2021;39(8) [Google Scholar]
- Dai Q., Yu J., Li G. Recent advances in deep learning aided drug discovery. Progress in Pharmaceutical Sciences. 2022;46(1):60–70. [Google Scholar]
- Dai X., Xie M.Y. Exploration of the mechanism of Xihuang Capsules in the treatment of granulomatous mastitis based on network pharmacology and experimental verification. Chinese Journal of Hospital Pharmacy. 2022;42(9):889–895. [Google Scholar]
- Daina A., Michielin O., Zoete V. Swisstargetprediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Research. 2019;47(W1):W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan W., Liu J., Guo X., Zhang B., Qu Y., He Q. Study on medication rules of traditional Chinese medicine against antineoplastic drug-induced cardiotoxicity based on network pharmacology and data mining. Evidence-Based Complementary and Alternative Medicine: eCAM. 2020;2020:7498525. doi: 10.1155/2020/7498525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Wu Y.H., Liu W. Molecular mechanism of reproductive toxicity induced by Tripterygium wilfordii based on network pharmacology. Medicine. 2021;100(27):e26197. doi: 10.1097/MD.0000000000026197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Tang J., Guo F., Zou Q. Identification of drug-target interactions via multiple kernel-based triple collaborative matrix factorization. Briefings in Bioinformatics. 2022;23(2) doi: 10.1093/bib/bbab582. [DOI] [PubMed] [Google Scholar]
- Doncheva N.T., Morris J.H., Gorodkin J., Jensen L.J. Cytoscape stringapp: Network analysis and visualization of proteomics data. Journal of Proteome Research. 2019;18(2):623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C., Li Y., Dong X., Xu W., Ma Y. Network pharmacology and reverse molecular docking-based prediction of the molecular targets and pathways for avicularin against cancer. Combinatorial Chemistry & High Throughput Screening. 2019;22(1):4–12. doi: 10.2174/1386207322666190206163409. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: Ncbi gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.H., Zhao Y.P., Jin Y.C., Shen X.P., Liu C.X. Network toxicology and its application to traditional Chinese medicine. China Journal of Chinese Materia Medica. 2011;36(21):2920–2922. [PubMed] [Google Scholar]
- Gao K., Song Y.P., Song A. Exploring active ingredients and function mechanisms of Ephedra-Bitter almond for prevention and treatment of corona virus disease 2019 (covid-19) based on network pharmacology. BioData mining. 2020;13(1):19. doi: 10.1186/s13040-020-00229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.F., Zhang J.J., Gong X.J., Li K.K., Zhang L.X., Li W. Ginsenoside rg5: A review of anticancer and neuroprotection with network pharmacology approach. The American journal of Chinese Medicine. 2022;50(8):2033–2056. doi: 10.1142/S0192415X22500872. [DOI] [PubMed] [Google Scholar]
- Ge F., Zhang Y., Hou J.C., Luo Y.M., Dong R.J., Ge D.Y.…Tao X.H. Mechanism of Mahuang Xixin Fuzitang against migraine based on network pharmacology and experimental validation. Chinese Journal of Experimental Traditional Medical Formulae. 2022;28(22):106–115. [Google Scholar]
- Gong B., Kao Y., Zhang C., Zhao H., Sun F., Gong Z. Exploring the pharmacological mechanism of the herb pair “Huanglian-Ganjiang” against colorectal cancer based on network pharmacology. Evidence-Based Complementary and Alternative Medicine. 2019;2019 doi: 10.1155/2019/2735050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Research. 2005;33(Database issue):514–517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Wang X., Hou J., Liu Y., Liu P., Zhao T. Using network pharmacology to explore the mechanism of Peach Kernel-Safflower in the treatment of diabetic nephropathy. BioMed Research International. 2021;2021:6642584. doi: 10.1155/2021/6642584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Guo J. The application analysis of visual software. Journal of Medical Informatics. 2014;35(2):55–58. [Google Scholar]
- Han S., Lv A.P., Li J., Jiang M. Application review of network pharmacology in the study of properties theory of traditional Chinese medicines. Chinese Journal of Basic Medicine in Traditional Chinese Medicine. 2019;25(1):127–130. [Google Scholar]
- Han S., Wang D.X., Wei P.H., Li L., Zhao Y.K., Li J.…Jiang M. Specific molecular mechanism of cold/heat herbal medicines based on network pharmacology methodology. Liaoning Journal of Traditional Chinese Medicine. 2021;48(8):186–189. [Google Scholar]
- Han Y.Q., Xu J., Zhang X.M., Zhang T.J., Ren Y.J., Liu C.X. Network pharmacology-based study on mechanism of Yuanhu Zhitong Dropping Pills in the treatment of primary dysmenorrhea. Acta Pharmaceutica Sinica. 2016;51(3):380–387. [PubMed] [Google Scholar]
- He J., Wu P., Dong Y., Gao R. Adverse reactions analysis of Aconiti Lateralis Radix Praeparata and mechanism prediction of cardiac toxicity by network pharmacology. China Journal of Chinese Materia Medica. 2019;44(5):1010–1018. doi: 10.19540/j.cnki.cjcmm.20181205.003. [DOI] [PubMed] [Google Scholar]
- He R.Z., Zhao X.L., Zhou Y.J., Liu J.M., Wu L.L., Chen F. Molecular mechanism of Scutellaria baicalensis Georgi in the treatment of melanoma based on network pharmacology and molecular docking technology. Journal of Tianjin Normal University (Natural Science Edition) 2022;42(6):29–36. [Google Scholar]
- Hou F., Yu Z., Cheng Y., Liu Y., Liang S., Zhang F. Deciphering the pharmacological mechanisms of Scutellaria baicalensis Georgi on oral leukoplakia by combining network pharmacology, molecular docking and experimental evaluations. Phytomedicine: International Journal of Phytotherapy and Phytopharmacology. 2022;103 doi: 10.1016/j.phymed.2022.154195. [DOI] [PubMed] [Google Scholar]
- Hu Y., Wu Y., Jiang C., Wang Z., Shen C., Zhu Z.…Liu Q. Investigative on the molecular mechanism of Licorice flavonoids anti-melanoma by network pharmacology, 3D/2D-QSAR, molecular docking, and molecular dynamics simulation. Frontiers in Chemistry. 2022;10 doi: 10.3389/fchem.2022.843970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Guo W., Cheung F., Tan H.Y., Wang N., Feng Y. Integrating network pharmacology and experimental models to investigate the efficacy of coptidis and scutellaria containing Huanglian Jiedu Decoction on hepatocellular carcinoma. The American Journal of Chinese Medicine. 2020;48(1):161–182. doi: 10.1142/S0192415X20500093. [DOI] [PubMed] [Google Scholar]
- Huang J.Y., Ma Z.L., Chen T.Y., Zhou Z.Y., Zhu C., Tang J.Y. Network pharmacology-based analysis on mechanism of Aconiti Lateralis Radix Praeparata and Epimedii Folium in treatment of chronic heart failure. Chinese Journal of Experimental Traditional Medical Formulae. 2021;27(10):142–151. [Google Scholar]
- Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., Wen C. TCMID 2.0: A comprehensive resource for TCM. Nucleic Acids Research. 2018;46(D1):D1117–D1120. doi: 10.1093/nar/gkx1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.F., Cheng W.B., Jiang Y., Liu Q., Liu X.H., Xu W.F., Huang H.T. A network pharmacology−based strategy for predicting anti-inflammatory targets of Ephedra in treating asthma. International Immunopharmacology. 2020;83(C) doi: 10.1016/j.intimp.2020.106423. [DOI] [PubMed] [Google Scholar]
- Jia C.X., Chen J.X., Pang X.H., Gao K., Li J.Z., Zhang F.L.…Zhao H.H. Mechanism research of emodin in the treatment of ischemic stroke based on network pharmacology and molecular docking. World Chinese Medicine. 2021;16(6):878–886. [Google Scholar]
- Jia X.L., Yang Y., Sun Y.X. Pharmacological study on target gene network of traditional Chinese medicine compound Changfukang against angiogenesis in colorectal cancer. Chinese Archives of Traditional Chinese Medicine. 2020;38(3) 246–251+279. [Google Scholar]
- Jiang D., Ding S., Mao Z., You L., Ruan Y. Integrated analysis of potential pathways by which aloe-emodin induces the apoptosis of colon cancer cells. Cancer Cell International. 2021;21(1):238. doi: 10.1186/s12935-021-01942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser M.J., Roth B.L., Armbruster B.N., Ernsberger P., Irwin J.J., Shoichet B.K. Relating protein pharmacology by ligand chemistry. Nature Biotechnology. 2007;25(2):197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S.…Bolton E.E. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research. 2021;49(D1):D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Gindulyte A., Zhang J., Thiessen P.A., Bolton E.E. PubChem Periodic Table and Element Pages: Improving access to information on chemical elements from authoritative sources. Chemistry Teacher International. 2021;3(1):57–65. doi: 10.1515/cti-2020-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Cheng T., He S., Thiessen P.A., Li Q., Gindulyte A.…Bolton E.E. Pubchem protein, gene, pathway, and taxonomy data collections: Bridging biology and chemistry through target-centric views of pubchem data. Journal of Molecular Biology. 2022;434(11) doi: 10.1016/j.jmb.2022.167514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., Szklarczyk D., Franceschini A., Campillos M., von Mering C., Jensen L.J.…Bork P. STITCH 2: An interaction network database for small molecules and proteins. Nucleic Acids Research. 2010;38(Database issue):D552–D556. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Research. 2008;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S., Zhao S., Huang X., Feng Y., Li Z., Chen L.…Chen B. Chaihu Shugan Powder alleviates liver inflammation and hepatic steatosis in nafld mice: A network pharmacology study and in vivo experimental validation. Frontiers in Pharmacology. 2022;13 doi: 10.3389/fphar.2022.967623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li Y., Luo C., Liang X., Liu Z., Liu Y.…Ling Y. New approach for targeted treatment of mild COVID-19 by honeysuckle through network pharmacology analysis. Computational and Mathematical Methods in Medicine. 2022;2022 doi: 10.1155/2022/9604456. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li J.F., Liao L.M. Structural characterization and acute toxicity prediction of substituted aromatic compounds by using molecular vertexes correlative index. Chinese Journal of Structural Chemistry. 2013;32(4):557–563. [Google Scholar]
- Li, l., Kou, S., Zhao, J., Lv, C., He, X. J., Jiang, M., ... Lv, A. P. (2016). Bioinformatics analysis of immunoregulation differences between Huoxiang (Agastache Rugosus) and Cangzhu (Rhizoma Atractylodis) in influenza a (H1N1). Journal of Traditional Chinese Medicine Management, 57(12), 1011−1014.
- Li L., Lu J.J., Wang W.T., Wang N., Wang L.X., Ma Y.M.…Liu S. Potential targets of Euodiae Fructus in treatment of insomnia based on network pharmacology. China Journal of Chinese Materia Medica. 2021;46(12):3016–3023. doi: 10.19540/j.cnki.cjcmm.20210419.401. [DOI] [PubMed] [Google Scholar]
- Li P.L., Su W.W. Recent progress in applying network pharmacology to research of Chinese materia medica. Chinese Traditional and Herbal Drugs. 2016;47(16):2938–2942. [Google Scholar]
- Li Q., Huang Z., Liu D., Zheng J., Xie J., Chen J.…Li Y. Effect of berberine on hyperuricemia and kidney injury: A network pharmacology analysis and experimental validation in a mouse model. Drug Design, Development and Therapy. 2021;15:3241–3254. doi: 10.2147/DDDT.S317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhang X., Wang S. Research progress in artificial intelligence for predicting drug-target interactions. Chinese Journal of Pharmacology and Toxicology. 2022;36(1):1–10. [Google Scholar]
- Li Q.F., Lu W.T., Zhang Q., Zhao Y.D., Wu C.Y., Zhou H.F. Proprietary medicines containing Bupleurum chinense DC. (Chaihu) for depression: Network meta-analysis and network pharmacology prediction. Frontiers in Pharmacology. 2022;13 doi: 10.3389/fphar.2022.773537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Network systems underlying traditional Chinese medicine syndrome and herb formula. Current Bioinformatics. 2009;4(3):188–196. [Google Scholar]
- Li S. Network target: A starting point for traditional Chinese medicine network pharmacology. China Journal of Chinese Materia Medica. 2011;36(15):2017–2020. [PubMed] [Google Scholar]
- Li S. Network pharmacology evaluation method guidance-draft. World. Journal of Traditional Chinese Medicine. 2021;7(1):146–154. doi: 10.19540/j.cnki.cjcmm.20210914.702. [DOI] [PubMed] [Google Scholar]
- Li S., Wang Y., Li C., Yang N., Yu H., Zhou W.…Li Y. Study on hepatotoxicity of Rhubarb based on metabolomics and network pharmacology. Drug Design, Development and Therapy. 2021;15:1883–1902. doi: 10.2147/DDDT.S301417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wu L., Zhang Z. Constructing biological networks through combined literature mining and microarray analysis: A LMMA approach. Bioinformatics (Oxford, England) 2006;22(17):2143–2150. doi: 10.1093/bioinformatics/btl363. [DOI] [PubMed] [Google Scholar]
- Li, S., Zhang, B., Jiang, D., Wei, Y., & Zhang, N. (2010). Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinformatics, 11(Suppl 11), S6. [DOI] [PMC free article] [PubMed]
- Li S., Zhang B., Zhang N. Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Systems Biology. 2011;5(Suppl 1):S10. doi: 10.1186/1752-0509-5-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ding Q.Y. New progress of interdisciplinary research between network toxicology, quality markers and TCM network pharmacology. Chinese Herbal Medicines. 2019;11(4):347–348. [Google Scholar]
- Li X., Qin X.M., Tian J.S., Gao X.X., Du G.H., Zhou Y.Z. Integrated network pharmacology and metabolomics to dissect the combination mechanisms of Bupleurum chinense Dc-Paeonia lactiflora Pall herb pair for treating depression. Journal of Ethnopharmacology. 2021;264 doi: 10.1016/j.jep.2020.113281. [DOI] [PubMed] [Google Scholar]
- Li X.L., Xin R.J., Huang H., Zhu Q.G. Role and mechanism of Fufang Honghua Buji Granules in the treatment of vitiligo based on network pharmacology and animal experiments. Journal of Shanxi Medical University. 2023;54(6):814–822. [Google Scholar]
- Li Y., Li R., Ouyang Z., Li S. Herb network analysis for a famous TCM doctor's prescriptions on treatment of rheumatoid arthritis. Evidence-Based Complementary and Alternative Medicine. 2015;2015 doi: 10.1155/2015/451319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang C., Ma X., Yang L., Ren H. Identification of the potential mechanism of Radix Pueraria in colon cancer based on network pharmacology. Scientific Reports. 2022;12(1):3765. doi: 10.1038/s41598-022-07815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Li H., Li S. A novel network pharmacology approach to analyse traditional herbal formulae: The Liu-Wei-Di-Huang Pill as a case study. Molecular BioSystems. 2014;10(5):1014–1022. doi: 10.1039/c3mb70507b. [DOI] [PubMed] [Google Scholar]
- Lin J.Q., Cao J., Dong Y., Zhao Y.F., Li F.H., Ye Y.…Zhang P.H. Molecular mechanism explore of Chaihu Shugan San in the treatment of psychological erectile dysfunction caused by liver stagnation and qi stagnation based on network pharmacology and experimental verification. Chinese Journal of Andrology. 2022;36(6):84–93. [Google Scholar]
- Lin Q., Wang N.B., Li X.Y., Ye Q.Y., Wang H.Y., Huang J.J.…Zhang R.H. Discuss on mechanism of Angelicae Sinensis Radix-Achyranthis Bidentatae Radix in treating osteoarthritis based on network pharmacology. Modernization of Traditional Chinese Medicine and Materia Medica-World ScienceA. Nd Technology. 2022;24(9):3532–3547. [Google Scholar]
- Liu B., Zhan S.K., Li K.N., Kuang J.H., Xian M.H., Wang S.M. The mechanism of action of the active components of Chuanxiong Rhizoma in the treatment of ischemic stroke based on network pharmacology and in vitro experiments. Traditional Chinese Drug Research and Clinical Pharmacology. 2022;33(11):1536–1544. [Google Scholar]
- Liu C., Ding J.X., Zhou Y., Yin Z.G., Luo H.T., Kong W.T. Protective effects of Lonicerae Japonicae Flos against acute alcoholic liver injury in rats based on network pharmacology. China Journal of Chinese Materia Medica. 2021;46(17):4531–4540. doi: 10.19540/j.cnki.cjcmm.20210624.401. [DOI] [PubMed] [Google Scholar]
- Liu D.G., Li Z.R., Chen Q.H., Wang Y.H., Xiao C.J. Mechanism of Pingyang Jiangya Formula in treating hypertension based on network pharmacology and in vivo study. Digital Chinese Medicine. 2021;4(3):214–228. [Google Scholar]
- Liu H., Wang J., Zhou W., Wang Y., Yang L. Systems approaches and polypharmacology for drug discovery from herbal medicines: An example using licorice. Journal of Ethnopharmacology. 2013;146(3):773–793. doi: 10.1016/j.jep.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Liu H.R., Li J.G., Yuan L., Wang J.Z., Lei P., Zhang Y.Z. To investigate the molecular mechanism of Buyang Huanwu Decoction in the treatment of diabetes based on network pharmacology and experimental verification. Chinese Journal of Hospital Pharmacy. 2023;43(11):1201–1208. [Google Scholar]
- Liu J., Liu J., Tong X., Peng W., Wei S., Sun T.…Li W. Network pharmacology prediction and molecular docking-based strategy to discover the potential pharmacological mechanism of Huai Hua San against ulcerative colitis. Drug Design, Development and Therapy. 2021;15:3255–3276. doi: 10.2147/DDDT.S319786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Guo Y., Zhao J., He S., Bai Y., Wang N.…Xu X. Systems pharmacology and verification of Shenfuhuang Formula in zebrafish model reveal multi-scale treatment strategy for septic syndrome in COVID-19. Frontiers in Pharmacology. 2020;11 doi: 10.3389/fphar.2020.584057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Lin W., Feng M., Yang Y., Liu T.T., Zhang M. Analysis of the effect of berberine on diabetic retinopathy in the immune microenvironment based on network pharmacology and experimental verification. Journal of Otolaryngology and Ophthalmology of Shandong University. 2023;37(1):94–104. [Google Scholar]
- Liu W., Sun X., Peng L., Zhou L., Lin H., Jiang Y. RWRNET: A gene regulatory network inference algorithm using random walk with restart. Frontiers in Genetics. 2020;11 doi: 10.3389/fgene.2020.591461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., An T., Wan D., Yu B., Fan Y., Pei X. Targets and mechanism used by cinnamaldehyde, the main active ingredient in Cinnamon, in the treatment of breast cancer. Frontiers in Pharmacology. 2020;11 doi: 10.3389/fphar.2020.582719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Dong H.R., Tian Y.G., Liu X.F., Zhao D., Feng S.X. Based on network pharmacology and molecular docking technology to study the mechanism of Panax ginseng and Citrus Reticulata Pericarpium in the treatment of chronic obstructive pulmonary disease. Modernization of Traditional Chinese Medicine and Materia Medica-World Science A. Nd Technology. 2022:1–13. [Google Scholar]
- Liu Y., Li X., Chen C., Ding N., Zheng P., Chen X., Ma S., Yang M. TCMNPAS: A comprehensive analysis platform integrating network formulaology and network pharmacology for exploring traditional Chinese medicine. Chinese Medicine. 2024;19(1):50. doi: 10.1186/s13020-024-00924-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.Y., Yu L.H., Zhang J., Xie D.J., Zhang X.X., Yu J.M. Network pharmacology-based and molecular docking-based analysis of Suanzaoren Decoction for the treatment of parkinson's disease with sleep disorder. BioMed Research International. 2021;2021 doi: 10.1155/2021/1752570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Guo F., Wang Y., Li C., Zhang X., Li H.…He F. BATMAN-TCM: A bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Scientific Reports. 2016;6:21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.R., Lin S.H., Miu S.Y., Zheng L., Lin W.J., Ma S.Y.…Bian X.L. TCM theory of “homotherapy for heteropathy” based on network pharmacology. China Pharmaceuticals. 2022;31(13):1–7. [Google Scholar]
- Lu Y., Li Q., Li T. PPA-GCN: A efficient gcn framework for prokaryotic pathways assignment. Frontiers in Genetics. 2022;13 doi: 10.3389/fgene.2022.839453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T.T., Lu Y., Yan S.K., Xiao X., Rong X.L., Guo J. Network pharmacology in research of Chinese medicine formula: Methodology, application and prospective. Chinese Journal of Integrative Medicine. 2020;26(1):72–80. doi: 10.1007/s11655-019-3064-0. [DOI] [PubMed] [Google Scholar]
- Lv X.M., Song N., Wang Q., Chen S., Zhai Y.R., Wang J.…Yang G.L. Research on the mechanism of Guizhi Jiagegen Decoction in preventing and treating dyslipidemia based on network pharmacology and experimental verification. Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology. 2021;23(11):3932–3940. [Google Scholar]
- Ma L.L., Yang X.M., Shi S.M., Guo Y., Han F.J., Yang Y., Li Q.W. Mechanism of platycodin D on cervical carcinoma by regulating Hippo signal pathway based on network pharmacology and targeted protein verification. Central South Pharmacy. 2022;20(12):2708–2714. [Google Scholar]
- Ma T.Y., Ping Y., Shen M.T., Li K., Wang L.H., Su J. Investigation on the mechanism of antidepressant of Cinnamomi Cortex based on network pharmacology and experimental verification. Chinese Journal of Modern Applied Pharmacy. 2023:1–10. [Google Scholar]
- Ma X., Wu J., Liu C., Li J., Dong S., Zhang X.…Sun C. Deciphering of key pharmacological pathways of Poria Cocos intervention in breast cancer based on integrated pharmacological method. Evidence-Based Complementary and Alternative Medicine: eCAM. 2020;2020 doi: 10.1155/2020/4931531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Zhu X. Application progress of network pharmacology in traditional Chinese medicine. Journal of Traditional Chinese Medicine Management. 2021;29(13):98–102. [Google Scholar]
- Nie H., Deng Y., Zheng C., Pan M., Xie J., Zhang Y., Yang Q. A network pharmacology-based approach to explore the effects of Chaihu Shugan Powder on a non-alcoholic fatty liver rat model through nuclear receptors. Journal of Cellular and Molecular Medicine. 2020;24(9):5168–5184. doi: 10.1111/jcmm.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M., Zhang S.Q., Zhang B., Yang K., Li S. Interpretation of network pharmacology evaluation method guidance. Chinese Traditional and Herbal Drugs. 2021;52(14):4119–4129. [Google Scholar]
- Niu X., Li J., Lv C., Li L., Guo H., Liu C.…Lv A. The molecular mechanism of 'herbs-pattern correspondence' in RA with heat pattern based on network pharmacology. Chinese Journal of Experimental Traditional Medical Formulae. 2012;18(8):299–303. [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S.P., Wang S., Yang J.W., Wang Y.H., Zhao C.R. Prediction and validation of anti gout effect of Polygonum cuspidatum and Evodia rutaecarpa by network pharmacology. Journal of Basic Chinese Medicine. 2021;27(12):1943–1948. [Google Scholar]
- Pelkonen O., Xu Q., Fan T.P. Why is research on herbal medicinal products important and how can we improve its quality? Journal of Traditional and Complementary Medicine. 2014;4(1):1–7. doi: 10.4103/2225-4110.124323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan, A., Caracausi, M., Antonaros, F., Pelleri, M. C., & Vitale, L. (2016). Genebase 1.1: A tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database: The Journal of Biological Databases and Curation, 2016, baw153. [DOI] [PMC free article] [PubMed]
- Qiu X., Zhou X.B., Li B. Progress in research on in vitro models for assessment of drug-induced nephrotoxicity. Chinese Journal of New Drugs. 2017;26(15):1792–1796. [Google Scholar]
- Ru J., Li P., Wang J., Zhou W., Li B., Huang C.…Yang L. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J., Heider D. GUESS: Projecting machine learning scores to well-calibrated probability estimates for clinical decision-making. Bioinformatics (Oxford, England) 2019;35(14):2458–2465. doi: 10.1093/bioinformatics/bty984. [DOI] [PubMed] [Google Scholar]
- Shan C., Ji X., Wu Z., Zhao J. Network pharmacology combined with GEO database identifying the mechanisms and molecular targets of Polygoni Cuspidati Rhizoma on peri-implants. Scientific Reports. 2022;12(1):8227. doi: 10.1038/s41598-022-12366-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D.…Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.F., Zhu K., Dong L., Li G.S., Huang X.P., Chang D.G.…You Y.D. Mechanism of Qiangjing Tablets in the treatment of male infertility with ureaplasma urealyticum infection based on network pharmacology and experimental verification. Pharmacology and Clinics of Chinese Materia Medica. 2022;38(4):34–40. [Google Scholar]
- Sheng S., Wang J., Wang L., Liu H., Li P., Liu M.…Su W. Network pharmacology analyses of the antithrombotic pharmacological mechanism of Fufang Xueshuantong Capsule with experimental support using disseminated intravascular coagulation rats. Journal of Ethnopharmacology. 2014;154(3):735–744. doi: 10.1016/j.jep.2014.04.048. [DOI] [PubMed] [Google Scholar]
- Shi M.J., Yan X.L., Dong B.S., Yang W.N., Su S.B., Zhang H. A network pharmacology approach to investigating the mechanism of tanshinone IIA for the treatment of liver fibrosis. Journal of Ethnopharmacology. 2020;253 doi: 10.1016/j.jep.2020.112689. [DOI] [PubMed] [Google Scholar]
- Snel B., Lehmann G., Bork P., Huynen M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Research. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.Y., Sun X.Y., Wu D.N., Wang Q.F. Mechanism of Xuefu Zhuyu Decoction in treating chronic heart failure based on bioinformatics. Chinese Traditional Patent Medicine. 2020;42(8):2179–2186. [Google Scholar]
- Song S.Y., Shu P. Mechanism of Poria-Atractylodis Macrocephalae Rhizoma drug pair in the treatment of gastric cancer based on network pharmacology and in vitro experimental verification. World Chinese Medicine. 2022;17(20):2836–2841. [Google Scholar]
- Southan C., Sitzmann M., Muresan S. Comparing the chemical structure and protein content of chembl, drugbank, human metabolome database and the therapeutic target database. Molecular Informatics. 2013;32(11–12):881–897. doi: 10.1002/minf.201300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S.…Lancet D. The genecards suite: From gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2013;54:1–30. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Su M., Tan F.X., Lv J., Lei L.Y. Study on the material basis of Anemarrhenae Rhizoma-Phellodendri Chinensis Cortex against inflammation based on network pharmacology, molecular docking and experimental verification. Natural Product Research and Development. 2022;34(4):687–698. [Google Scholar]
- Sun M.J., Wang Y.Y., Yao L., Wang L., Lv T.T., Hao L.J.…Chen W.D. Mechanism of anti-breast cancer action of Poria cocos (Schw.) Wolf based on UPLC-Q-TOF-MS/MS and network pharmacology. Natural Product Research and Development. 2022;34(10):1764–1773. [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J.…Mering C.V. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research. 2016;44(D1):D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Chen J., Li Y., Li Y., Zhong Y., Li G.…Li Y. Glabridin, a bioactive component of Licorice, ameliorates diabetic nephropathy by regulating ferroptosis and the VEGF/AKT/ERK pathways. Molecular medicine (Cambridge, Mass.) 2022;28(1):58. doi: 10.1186/s10020-022-00481-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.L., Gong Y., Feng Y.L., Yang T.L., Zhang M., Shen J.L., Cheng Y. Analysis of medication regularities of epidemic disease prescription in Treatise on Febrile Diseases and mechanism of core drugs in treatment of COVID-19. Chinese Traditional and Herbal Drugs. 2023;54(1):192–209. [Google Scholar]
- Tao W., Xu X., Wang X., Li B., Wang Y., Li Y.…Yang L. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae Formula for application to cardiovascular disease. Journal of Ethnopharmacology. 2013;145(1):1–10. doi: 10.1016/j.jep.2012.09.051. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Wang S., Guo D.Q., Yu Z.X., Liu Y.Y., Yang Y.…Wei J.H. Prediction and analysis of the components and therapeutic targets of Agarwood essential oil. Chinese Pharmaceutical Journal. 2019;54(23):1958–1964. [Google Scholar]
- Wang F., Zhu Z.H. Discussion on mechanism and verification of Ginseng Radix et Rhizoma-Poria-Atractylodis Macrocephalae Rhizoma in the treatment of nasopharyngeal carcinoma by geo gene microarray combined with network pharmacology and molecular docking technology. Chinese Journal of Information on Traditional Chinese Medicine. 2023;30(1):24–31. [Google Scholar]
- Wang G.R., Chen Z.Y., Wu H., Liu Y.P., Chen M., Lai S.S.…Wang Z.T. Xueshuan Tong improves cerebral microcirculation disorder: Action mechanism based on network pharmacology and experimental validation. Acta Pharmaceutica Sinica. 2022;57(7):2077–2086. [Google Scholar]
- Wang J.Y., Chen J.X., Zhao H.H. Relationship between Chinese medicine of liver meridian and treatment based on network pharmacology. World Chinese Medicine. 2019;14(3):581–584. [Google Scholar]
- Wang L., Zhang J., Shan G., Liang J., Jin W., Li Y.…Chen C.L. Establishment of a lung cancer discriminative model based on an optimized support vector machine algorithm and study of key targets of wogonin in lung cancer. Frontiers in Pharmacology. 2021;12 doi: 10.3389/fphar.2021.728937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. New strategies for investigating the molecular mechanisms of traditional Chinese medicine. Chinese Journal of Pharmacology and Toxicology. 2019;33(9):647. [Google Scholar]
- Wang W., Hai X. Role and mechanism of ginsenoside Rh2 in the inflammatory response of ovarian granulosa cells based on network pharmacology and molecular biology. China Pharmaceuticals. 2023;32(1):46–51. [Google Scholar]
- Wang W.X., Zhang Y.R., Luo S.Y., Zhang Y.S., Zhang Y., Tang C. Chlorogenic acid, a natural product as potential inhibitor of COVID-19: Virtual screening experiment based on network pharmacology and molecular docking. Natural Product Research. 2022;36(10):2580–2584. doi: 10.1080/14786419.2021.1904923. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang Z.Y., Zheng J.H., Li S. TCM network pharmacology: A new trend towards combining computational, experimental and clinical approaches. Chinese Journal of Natural Medicines. 2021;19(1):1–11. doi: 10.1016/S1875-5364(21)60001-8. [DOI] [PubMed] [Google Scholar]
- Wang Y.Y., Kuang H.X., Su F.Z., Qiao W.J., Wang Y., Su Y.P., Yang B.Y. Clinical value of four natures of traditional Chinese medicine and its relationship with five flavors. Chinese Traditional and Herbal Drugs. 2023;54(4):1329–1347. [Google Scholar]
- Wang Y., Liu Q., Lv Q., Yang K., Wu X., Zheng Y.…He C. Investigating the chemical profile of Rheum lhasaense and its main ingredient of piceatannol-3'-O-β-D-glucopyranoside on ameliorating cognitive impairment. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2023;160 doi: 10.1016/j.biopha.2023.114394. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Z., Wang C., Ma D.F. Mechanism of Carthami Flos and Lepidii Semen drug pair in inhibition of myocardial fibrosis by improving cardiac microenvironment based on network pharmacology and animal experiment. China Journal of Chinese Materia Medica. 2022;47(3):753–763. doi: 10.19540/j.cnki.cjcmm.20210929.401. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yuan Y., Wang W., He Y., Zhong H., Zhou X.…Liu L.Q. Mechanisms underlying the therapeutic effects of Qingfeiyin in treating acute lung injury based on GEO datasets, network pharmacology and molecular docking. Computers in Biology and Medicine. 2022;145 doi: 10.1016/j.compbiomed.2022.105454. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z.…Zhu F. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Research. 2020;48(D1):D1031–D1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.G., Dou Y.Q., Yan Z.Q., Gao Y. Molecular mechanism of ginsenoside Rg1 against radiation enteritis: Based on network pharmacology and in vitro experiment. China Journal of Chinese Materia Medica. 2023;48(10):2810–2819. doi: 10.19540/j.cnki.cjcmm.20221223.401. [DOI] [PubMed] [Google Scholar]
- Wang Y.M., Xu M.X., Luo M., Zhao F.K., Su M., Li M.Y. Preliminary study on mechanism of Maimendong Decoction in treating pulmonary fibrosis based on network pharmacology and experimental verification. Modernization of Traditional Chinese Medicine and Materia Medica-World ScienceA; Nd Technology. 2022;24(10):3932–3940. [Google Scholar]
- Wang Y.Y., Wang Q., Liu M.S., Zuo Y., Wang L.W., Zhao S.B.…Zhao Y.L. Mechanism of Chuanxiong Rhizoma in the treatment of migraine based on network pharmacology and molecular docking. World Journal of Integrated Traditional and Western Medicine. 2022;17(6):1133–1142. [Google Scholar]
- Wang Y.Y., Zhang F.F., Li X., Wang Y.M., Li Y.B. Mechanism of Yiyi Fuzi Powder in homotherapy for rheumatoid arthritis and chronic heart failure based on network pharmacology and molecular docking. Drug Evaluation Research. 2023;46(2):321–329. [Google Scholar]
- Wang Z.Y., Jiang Z.M., Xiao P.T., Jiang Y.Q., Liu W.J., Liu E.H. The mechanisms of baicalin ameliorate obesity and hyperlipidemia through a network pharmacology approach. European Journal of Pharmacology. 2020;878 doi: 10.1016/j.ejphar.2020.173103. [DOI] [PubMed] [Google Scholar]
- Wei S., Li J., Han D.W., Fu Q., Hao F. Mechanism of Astragali Radix-Puerariae Lobatae Radix combination in regulating type 2 diabetes mellitus through AMPK signaling pathway: Based on network pharmacology and experimental verification. China Journal of Chinese Materia Medica. 2022;47(10):2738–2749. doi: 10.19540/j.cnki.cjcmm.20211216.703. [DOI] [PubMed] [Google Scholar]
- Wei Y.L., Wang Z.G. Theoretical research on compatibility of traditional Chinese medicines. World Chinese Medicine. 2013;8(5):509–511. [Google Scholar]
- Xiao Y. Research considerations on compatibility rules of traditional Chinese medicine formulas. Sichuan Traditional Chinese Medicine. 2005;11:111–112. [Google Scholar]
- Xu H.B., Qi X.J., Fang C.S., Deng J.Y., Shi P.Y., Mo J.H.…Wu H. Study on the mechanism of Zhenwu Decoction in treating chronic heart failure based on network pharmacology. Chinese Journal of Modern Applied Pharmacy. 2020;37(15):1801–1811. [Google Scholar]
- Xu H.Y., Hou W.B., Li K., Shen Y., Tang S.H., Guo F.F.…Liu C.X. Discovery and application of quality marker of traditional Chinese medicine based on integrative pharmacology. Chinese Master's Theses Full-Text Database. 2019;25(6):1–8. [Google Scholar]
- Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H.…Huang L.Q. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Research. 2019;47(D1):D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang S., Wu T., Fang X., Zhao L. Discovery of TGFBR1 (ALK5) as a potential drug target of quercetin glycoside derivatives (QGDs) by reverse molecular docking and molecular dynamics simulation. Biophysical Chemistry. 2022;281 doi: 10.1016/j.bpc.2021.106731. [DOI] [PubMed] [Google Scholar]
- Xu R., Yang X., Tao Y., Luo W., Xiong Y., He L.…He Y. Analysis of the molecular mechanism of Evodia rutaecarpa fruit in the treatment of nasopharyngeal carcinoma using network pharmacology and molecular docking. Journal of Healthcare Engineering. 2022;2022:6277139. doi: 10.1155/2022/6277139. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xue R., Fang Z., Zhang M., Yi Z., Wen C., Shi T. TCMID: Traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Research. 2013;41(Database issue):D1089–D1095. doi: 10.1093/nar/gks1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y.…Chen Y.Z. Therapeutic target database update 2016: Enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Research. 2016;44(D1):D1069–D1074. doi: 10.1093/nar/gkv1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Hu Z., Zhu J., Liang Q., Zhou H., Li J.…Fei B. Systematic elucidation of the mechanism of quercetin against gastric cancer via network pharmacology approach. BioMed Research International. 2020;2020 doi: 10.1155/2020/3860213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Chen J., Xu L., Shi X., Zhou X., An R., Wang X. A network pharmacology approach to uncover the molecular mechanisms of herbal formula Ban-Xia-Xie-Xin-Tang. Evidence-based Complementary and Alternative Medicine: eCAM. 2018;2018 doi: 10.1155/2018/4050714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Hao H., Li Y., Li S. Modularity-based credible prediction of disease genes and detection of disease subtypes on the phenotype-gene heterogeneous network. BMC Systems Biology. 2011;5:79. doi: 10.1186/1752-0509-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Mi Y., Xu X., Li N., Zeng F., Yan K.…Lu M. Baicalein alleviates osteoarthritis progression in mice by protecting subchondral bone and suppressing chondrocyte apoptosis based on network pharmacology. Frontiers in Pharmacology. 2021;12 doi: 10.3389/fphar.2021.788392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Lai X., Pan Y., Zheng H., Vera J., Liu S.…Zhang L. Artificial intelligence in cancer target identification and drug discovery. Signal Transduction and Targeted Therapy. 2022;7(1):156. doi: 10.1038/s41392-022-00994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.X., Zhang H.W., Zhang W.S., Han X.Y., Qu C., Ji T.W.…Shi Y. Virtual screening of active components of Yitangkang ameliorating insulin resistance based on network pharmacology and molecular docking technology. Journal of Liaoning University of Traditional Chinese Medicine. 2023;25(2):170–183. [Google Scholar]
- Yu N.X., Li G.Q., Li B., Xu Y. Mechanism of astragaloside IV in treating diabetic retinopathy based on network pharmacology and molecular docking. Chinese Journal of Experimental Traditional Medical Formulae. 2022;28(13):209–216. [Google Scholar]
- Yuan J., Li X.J., Chen C., Song X.G., Wang S.M. Study on prediction of compound-target-disease network of Chuanxiong Rhizoma based on random forest algorithm. China Journal of Chinese Materia Medica. 2014;39(12):2336–2340. [PubMed] [Google Scholar]
- Yuan Z., Pan Y., Leng T., Chu Y., Zhang H., Ma J.…Ma X. Progress and prospects of research ideas and methods in the network pharmacology of traditional Chinese medicine. Journal of Pharmacy & Pharmaceutical Sciences a Publication of the Canadian Society for Pharmaceutical Sciences, Societe Canadienne des Sciences Pharmaceutiques. 2022;25:218–226. doi: 10.18433/jpps32911. [DOI] [PubMed] [Google Scholar]
- Zeng P., Zhou H. Homogenization of key components screening of “different diseases and different prescriptions” in network pharmacology. Chinese Journal of Experimental Traditional Medical Formulae. 2022;28(18):177–191. [Google Scholar]
- Zeng X., Tong L., Li Y. Analysis on the mechanism of Shegan-Ephedra in Qingfeipaidu Decoction in the treatment of COVID-19 based on network pharmacology. Practical Clinical Journal of Integrated Traditional Chinese and Western Medicine. 2020;20(17):115–118. [Google Scholar]
- Zhang B., Hao Z., Zhou W., Zhang S., Sun M., Li H.…Zhao M. Formononetin protects against ox-LDL-induced endothelial dysfunction by activating PPAR-γ signaling based on network pharmacology and experimental validation. Bioengineered. 2021;12(1):4887–4898. doi: 10.1080/21655979.2021.1959493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.B., Zeng M.N., Wang Y.X., Zhang Q.Q., Wang R., Fu Y.…Zheng X.K. Network pharmacology and in vivo validation of potential targets of typical ascending and floating, descending and sinking Chinese medicinals. Journal of Traditional Chinese Medicine. 2023;64(2):174–185. [Google Scholar]
- Zhang B.B., Zeng M.N., Zhang Q.Q., Wang R., Jia J.F., Guo P.L.…Zhen X.K. Mechanism of “Ephedrae Herba-Descurainiae Semen Lepidii Semen” combination in treatment of bronchial asthma based on network pharmacology and experimental verification. China Journal of Chinese Materia Medica. 2022;47(18):4996–5007. doi: 10.19540/j.cnki.cjcmm.20220211.403. [DOI] [PubMed] [Google Scholar]
- Zhang H., Jiang H., Zhao M., Xu Y., Liang J., Ye Y.…Chen H. Treatment of gout with TCM using turmeric and corn silk: A concise review article and pharmacology network analysis. Evidence-Based Complementary and Alternative Medicine: eCAM. 2022;2022:3143733. doi: 10.1155/2022/3143733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Song X., Wang H., Zhang X. MIClique: An algorithm to identify differentially coexpressed disease gene subset from microarray data. Journal of Biomedicine & Biotechnology. 2009;2009 doi: 10.1155/2009/642524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. The hypolipidemic mechanism of Citrus Reticulata-Arum Ternatum Thunb based on network pharmacology and experimental verification. Chinese Journal of Integrative Medicine on Cardio-Cerebrovascular Disease. 2022;20(17):3118–3128. [Google Scholar]
- Zhang L., Han L., Wang X., Wei Y., Zheng J., Zhao L.…Tong X. Exploring the mechanisms underlying the therapeutic effect of Salvia miltiorrhiza in diabetic nephropathy using network pharmacology and molecular docking. Bioscience Reports. 2021;41(6) doi: 10.1042/BSR20203520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang S., Liu S., Song J.K., Du G.H. Evaluation and verification of effect of isorhamnetin against ischemic nerve injury based on network of disease and active compound targets. Chinese Journal of Bioinformatics. 2023;21(1):20–28. [Google Scholar]
- Zhang W.N., Gao Y., Li K., Chao J.B., Qin X.M., Li A.P. Exploration into the mechanism of total flavonoids of Astragali Radix in the treatment of nephrotic syndrome based on network pharmacology. Acta Pharmaceutica Sinica. 2018;53(9):1429–1441. [Google Scholar]
- Zhang X., Shen T., Zhou X., Tang X., Gao R., Xu L.…Hu Y. Network pharmacology based virtual screening of active constituents of Prunella vulgaris L. and the molecular mechanism against breast cancer. Scientific Reports. 2020;10(1):15730. doi: 10.1038/s41598-020-72797-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.L., Zhou Z.X., Liu Y.H., Fan X.L., Li H.D., Wang J.T. Predicting the acute toxicity of aromatic amines by linear and nonlinear regression methods. Chinese Journal of Structural Chemistry. 2014;33(2):244–252. [Google Scholar]
- Zhang Y., Chen L., Li S. CIPHER-SC: Disease-gene association inference using graph convolution on a context-aware network with single-cell data. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2022;19(2):819–829. doi: 10.1109/TCBB.2020.3017547. [DOI] [PubMed] [Google Scholar]
- Zhang Y.B., Wang B.T., Lv Z.G., Xu P., Zhang D.M., Li Q.Y.…Wang J. Molecular mechanism of Shengxiantang in treating myasthenia gravis: Based on network pharmacology and molecular docking. Chinese Journal of Experimental Traditional Medical Formulae. 2022;28(6):142–150. [Google Scholar]
- Zhang Y.Q., Li S. Progress in network pharmacology for modern research of traditional Chinese medicine. Chinese Journal of Pharmacology and Toxicology. 2015;29(6):883–892. [Google Scholar]
- Zhang Y.Q., Mao X., Guo Q.Y., Lin N., Li S. Network pharmacology-based approaches capture essence of Chinese herbal medicines. Chinese Herbal Medicines. 2016;8(2):107–116. [Google Scholar]
- Zhang Z., Li B., Huang J., Huang S., He D., Peng W.…Zhang S. A network pharmacology analysis of the active components of the traditional Chinese medicine Zuojinwan in patients with gastric cancer. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2020;26:e923327. doi: 10.12659/MSM.923327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.C., Zhu M., Lin Z.S., Mai P.Y., Liang J.H., Peng J.…Shi X. Mechanism of Chuanxiong Rhizoma in the treatment of migraine based on network pharmacology and molecular docking decoction on the expression of IL-17 and FOXP3 in type IA prostatitis based on network pharmacology and clinical trials. World Science and Technology—Modernization of Traditional Chinese Medicine and Materia Medica. 2021;23(12):4519–4529. [Google Scholar]
- Zhao J., Sun J.L., Liu H.L., Liu G.W., Li X.P., Cang H.Q.…Sui Z.G. Molecular mechanism of ursolic acid in the treatment of osteoporosis based on network pharmacology and molecular docking. China Pharmacy. 2021;32(17):2066–2073. [Google Scholar]
- Zhao M., Fu Y., Liu L., Hou Y., Shi M., Zhou H.…Zhang G. Identification of key drug targets and molecular mechanisms of Curcumae Rhizoma acting on HBV-related HCC: Weighted correlation network and network pharmacological analyses. Evidence-Based Complementary and Alternative Medicine: eCAM. 2022;2022:5399766. doi: 10.1155/2022/5399766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li S. Network-based relating pharmacological and genomic spaces for drug target identification. PLoS One. 2010;5(7) doi: 10.1371/journal.pone.0011764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li S. A co-module approach for elucidating drug-disease associations and revealing their molecular basis. Bioinformatics (Oxford, England) 2012;28(7):955–961. doi: 10.1093/bioinformatics/bts057. [DOI] [PubMed] [Google Scholar]
- Zhao Y.Y., Zhao Y.Y., He H.J., Li X.M., Cao L.H. Mechanism of Jiegeng in improving asthma: An exploration based on network pharmacology and experimental verification. Pharmacology and Clinics of Chinese Materia Medica. 2021;37(6):82–89. [Google Scholar]
- Zhou L., Gu W., Kui F., Gao F., Niu Y., Li W., Du G. The mechanism and candidate compounds of Aged Citrus Peel (Chenpi) preventing chronic obstructive pulmonary disease and its progression to lung cancer. Food & Nutrition Research. 2021;65 doi: 10.29219/fnr.v65.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Zhou Y., Chen Y., Xu N., Li W.B., Wu C.Y.…Chen Y. Molecular mechanism of Puerariae Lobatae Radix in treatment of hepatocellular carcinoma based on network pharmacology. China Journal of Chinese Materia Medica. 2020;45(17):4089–4098. doi: 10.19540/j.cnki.cjcmm.20200427.402. [DOI] [PubMed] [Google Scholar]
- Zhou S., Li W., Ai Z., Wang L., Ba Y. Investigating mechanism of Qingfei Dayuan Granules for treatment of COVID-19 based on network pharmacology and molecular docking. Chinese Traditional and Herbal Drugs. 2020;51(7):1804–1813. [Google Scholar]
- Zhou W., Zhang H., Wang X., Kang J., Guo W., Zhou L.…Li S. Network pharmacology to unveil the mechanism of Moluodan in the treatment of chronic atrophic gastritis. Phytomedicine: International Journal of Phytotherapy and Phytopharmacology. 2022;95 doi: 10.1016/j.phymed.2021.153837. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F.…Chen Y. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Research. 2022;50(D1):D1398–D1407. doi: 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.J., Qian Y.P., Yuan Y., Yang S.Y., Song L.J. Anti-rotavirus mechanism of hesperidin based on network pharmacology and in vitro experiments. Central South Pharmacy. 2023;21(6):1509–1515. [Google Scholar]
- Zhu H., Sang T., Ji Y., Li W., Wu M. Study on target of treating tumors based on active constituents of Baihuasheshecao (Hedyotis Diffusa) in Xiaoai Jiedu Decoction. Chinese Archives of Traditional Chinese Medicine. 2020;38(5) 132–135+276−277. [Google Scholar]
- Zhu Z., Wu S., Wang Y., Wang J., Zhang Y. Reveal the antimigraine mechanism of Chuanxiong Rhizoma and Cyperi Rhizoma based on the integrated analysis of metabolomics and network pharmacology. Frontiers in Pharmacology. 2022;13 doi: 10.3389/fphar.2022.805984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y.L., Yao F., Hou W.Y., Zhang L.R., Song Q. Investigating on the mechanism of anti-hepatocellular carcinoma of astragalus based on network pharmacology and cell experiments. World Chinese Medicine. 2023;18(23):3311–3316. [Google Scholar]
- Zhuang Y.S., Cai B.C., Zhang Z.L. Application progress of network pharmacology in traditional Chinese medicine research. Journal of Nanjing University of Traditional Chinese Medicine. 2021;37(1):156–160. [Google Scholar]
- Zuo J., Wang X., Liu Y., Ye J., Liu Q., Li Y., Li S. Integrating network pharmacology and metabolomics study on anti-rheumatic mechanisms and antagonistic effects against methotrexate-induced toxicity of Qing-Luo-Yin. Frontiers in Pharmacology. 2018;9:1472. doi: 10.3389/fphar.2018.01472. [DOI] [PMC free article] [PubMed] [Google Scholar]