Abstract
The lungs are a major organ site of cytomegalovirus (CMV) pathogenesis, latency, and recurrence. Previous work on murine CMV latency has documented a high load and an even distribution of viral genomes in the lungs after the resolution of productive infection. Initiation of the productive cycle requires expression of the ie1/3 transcription unit, which is driven by the immediate-early (IE) promoter P1/3 and generates IE1 and IE3 transcripts by differential splicing. Latency is molecularly defined by the absence of IE3 transcripts specifying the essential transactivator protein IE3. In contrast, IE1 transcripts were found to be generated focally and randomly, reflecting sporadic P1/3 activity. Selective generation of IE1 transcripts implies molecular control of latency operating after ie1/3 transcription initiation. P1/3 is regulated by an upstream enhancer. It is widely assumed that the viral transcriptional program is started by activation of the enhancer through the binding of transcription factors. Accordingly, stochastic transcription during latency might reflect episodes of enhancer activation by the “noise” activity of intrinsic transcription factors. In addition to ie1/3, the enhancer controls gene ie2, which has its own promoter, P2, and is transcribed in opposite direction. We show here that ie2 is also randomly transcribed during latency. Notably, however, ie1 and ie2 were found to be expressed independently. We infer from this finding that expression of the major IE genes is regulated asymmetrically and asynchronously via the combined control unit P1/3 -E-P2. Our data are consistent with a stochastic nature of enhancer action as it is proposed by the “binary” or probability model.
The molecular regulation of viral latency and reactivation is a central unsolved issue in the understanding of cytomegalovirus (CMV) biology. Research on human CMV (hCMV) latency has so far focused on latency in hematopoietic progenitors of the myeloid differentiation lineage (18, 29, 38, 42, 46, 62, 64, 69). These studies have revealed the existence of sense and antisense latency-associated transcripts derived from the major immediate-early (MIE) region (30). The sense latency-associated transcripts, while transcribed in the same orientation, are distinct from the productive cycle ie1/2 transcripts by usage of alternative transcription start sites located in the enhancer upstream of the ie1/2 transcription start site. To date, one of four open reading frames (ORFs) specified by sense latency-associated transcripts, namely ORF94, was studied in greater detail. It encodes a protein with nuclear localization, but the ORF94 protein was found not to be critically involved in either productive or latent infection (70). There is increasing evidence to suggest that cytokine-triggered transcription factors which bind to regulatory modules of the enhancer initiate virus reactivation (for a review, see reference 15). Cytokines implicated in allogeneic immune responses and proinflammatory cytokines in general are candidates for inducing hCMV reactivation (18, 64). Specifically, tumor necrosis factor alpha (TNF-α) signaling activates NF-κB, which then binds to the enhancer resulting in MIE promoter activity (36, 54).
Like hCMV, murine CMV (mCMV) can establish a latent infection in cells of the myeloid lineage (9, 23, 47, 51). Since mCMV genome remains present in various organs after resolution of the latent infection of hematopoietic lineage cells in bone marrow and blood (4, 32, 34, 57), there must exist one or more widely distributed further cell type(s) representing the cellular site(s) of enduring latency. Sinusoidal lining cells of the spleen stroma (43, 53), renal peritubular epithelial cells (27), and capillary endothelial cells in various organs (28) are likely candidates, but the discussion is still going on.
Our own interest was focused on the lungs, because interstitial pneumonia is a critical manifestation of primary as well as recurrent hCMV disease in immunocompromised patients, in particular after bone marrow transplantation (BMT) (59, 61). Likewise, in the murine model, the lungs proved to be a major organ site of mCMV pathogenesis (58) and, after resolution of productive infection, the load of latent viral genomes was found to be particularly high in the lungs (5). In more recent reports we have established a model of mCMV infection in the specific context of hematopoietic reconstitution after experimental syngeneic BMT performed with BALB/c mice as donors and recipients. In this model, acute infection of the lungs lasts for 4 to 6 weeks. It is confined to inflammatory foci and is finally resolved by lung-infiltrating antiviral CD8 T cells (22, 49). The establishment of viral latency in these lungs was defined by the following criteria: (i) resolution of productive infection in the lungs, as well as at other organ sites of mCMV replication (34), in particular also in the salivary glands, which are known as the site at which viral replication persists for a prolonged time period (37, 40, 63) before it eventually discontinues (34, 66); (ii) clearance of viral DNA in bone marrow and blood to exclude an undetected remote site of persistent productive infection and to exclude contamination of the lungs by latently infected intravascular cells of hematopoietic origin (4, 32, 34); (iii) life long maintentance of viral DNA in lung tissue (32, 34, 66); (iv) absence of productive cycle transcripts such as spliced IE3 mRNA (32) encoding the essential transactivator protein IE3 (1, 44) and (v) reactivation of productive cycle transcription and recurrence of infectious virions after secondary immunoablative treatment (5, 33, 50, 66).
In essence, these studies have so far shown that latent viral DNA is present in lung tissue in a high load that corresponds to a high risk of recurrent infection (57, 66). Notably, episodes of focal transcriptional activity restricted to the generation of spliced IE1 transcripts were detected in latently infected lungs in a frequency of ca. 10 events at any moment (32, 33). Gene ie1 of mCMV represents exons 1 to 4 of the ie1/3 MIE transcription unit (25, 44), which corresponds to ie1/2 of hCMV (41, 67) and is controlled by a specific promoter, P1/3, and by a strong upstream enhancer (13). Exons 2 to 4 specify the IE1 protein (25, 26), which is a cotransactivator of early-phase gene expression (44), induces cellular thymidylate synthase gene expression promoting mCMV replication in quiescent cells (16), and plays a role in CD8 T-cell-mediated immunity (for a review, see reference 56). Transcripts of ie3 are generated by differential splicing and encompass exons 1 to 3 and exon 5 of which exons 2, 3, and 5 specify the E-gene transactivator protein IE3 (44), the functional analog and amino acid sequence homolog of hCMV IE2 (20, 65). Very recent work on ie3-deficient mCMV has proven that IE3 is indeed essential for viral replication (1). Accordingly, the absence of spliced IE3 mRNA in latently infected lungs explains why latency is maintained in spite of the sporadic activity of P1/3 that is indicated by the random and focal occurrence of spliced IE1 mRNA (32). In mCMV, the enhancer controls also gene ie2 that possesses its own promoter and is transcribed in a direction opposite to ie1/3 (45). Gene ie2 has no counterpart in hCMV and, according to deletion mutagenesis, the IE2 protein of mCMV is dispensable (10, 39).
Sporadic cytokine-induced activation of the enhancer by binding of transcription factors could possibly account for the observed stochastic episodes of P1/3 activity during pulmonary latency. The two-sided gene flanking of the enhancer gave us the unique chance to test whether the MIE gene control unit P1/3 -E-P2 is active during latency in a coordinated, symmetric mode.
MATERIALS AND METHODS
Experimental BMT and establishment of latent mCMV infection.
For sex-matched syngeneic BMT, female mice of the inbred strain BALB/c (major histocompatibility complex haplotype H-2d) were used at the age of 8 weeks as BM cell (BMC) donors and recipients. Hematoablative conditioning of the recipients was performed by total-body gamma irradiation with a single, sublethal dose of 6 Gy from a 137Cs-γ-ray source (OB58; Buchler, Braunschweig, Germany). Cell suspensions of femoral and tibial BMCs were prepared and depleted of CD8 T cells as described previously (22). BMT was performed by infusion of 5 × 106 donor BMCs into the tail vein of recipients at ca. 6 h after the irradiation. Recipients were infected ca. 2 h after BMT with 105 PFU of cell culture-propagated and sucrose gradient-purified murine CMV (strain Smith ATCC VR-194/1981) by subcutaneous inoculation in the left hind footpad (34). In this model, productive infection is resolved in all organs, including the salivary glands, by 6 months after inoculation (34). The previously described reverse transcription (RT)-PCR-based focus expansion assay was employed to verify the absence of infectious virus in organs with the highest possible sensitivity of detection (34). In addition, the load of viral DNA in blood leukocytes derived from tail vein blood was monitored monthly by PCR specific for a 363-bp sequence within exon 4 of the ie1 gene (32, 34). Subsequent analyses were performed after 10 months when the infection of the recipients had become latent as defined by absence of productive infection in organs and by clearance of viral DNA from blood leukocytes.
Determination of viral DNA load in lung tissue.
The viral DNA load in the lungs of each individual latently infected mouse was measured representatively for DNA isolated from the postcaval lobe. The number of viral genomes present in tissue DNA was quantitated by diluting the DNA serially and subsequent PCR specific for a 363-bp sequence within exon 4 of gene ie1 of mCMV, followed by phosphorimaging, essentially as described in previous reports (32, 66). Cellular DNA derived from postcaval lobes of uninfected mice served as the negative (mock) control. As positive control and quantification standard, this mock DNA was supplemented with defined copy numbers of plasmid pIE111 (44), which encompasses gene ie1. Amplification products (20 μl thereof) were vacuum dot blotted onto nylon membrane, hybridized with a γ-32P-end-labeled internal oligonucleotide probe, and analyzed quantitatively by digital phosphorimaging (66).
Construction of recombinant plasmids for in vitro transcription.
In vitro transcripts were generated to serve as standards for evaluating the sensitivity of RT-PCR assays. The map locations of the synthetic RNAs representing transcripts from genes ie1 and ie2 of mCMV are illustrated in Fig. 1. Recombinant plasmids containing coding sequences of genes ie1 and ie3, namely, pSP64 Poly(A)-ie1 and pBlue-ie3, respectively, have been described previously (32). Synthetic RNA transcribed from pSP64 Poly(A)-ie1 has a length of 2.17 kb, including a 30-nucleotide poly(A) tail. Synthetic RNA transcribed from pBlue-ie3 has a length of 1.045 kb and lacks a poly(A) tail.
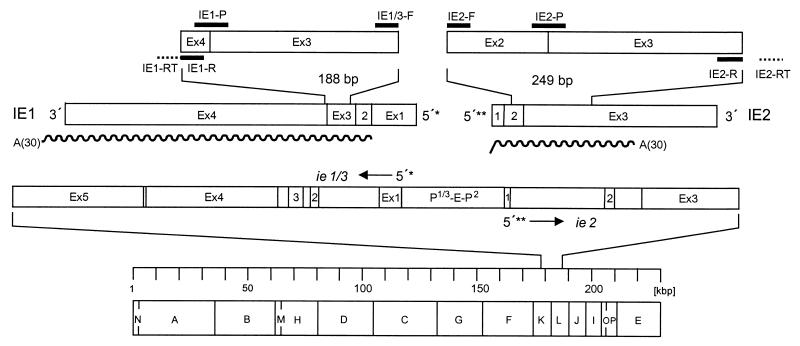
FIG. 1.
Map of the transcriptional analysis. The bottom part shows the HindIII physical map of the mCMV Smith strain genome with the IE region expanded to reveal the exon (Ex)-intron structure of transcription unit ie1/3 and gene ie2. Resolved to greater detail and drawn to scale, IE1 and IE2 transcripts, the amplified fragments therein, and the positions of all RT-PCR primers and probes (black bars) are shown at the top. The wavy lines illustrate the positions and lengths of in vitro-synthesized polyadenylated [A(30)] RNAs. The 5′-transcriptional start sites of ie1/3 and ie2 are marked by single and double asterisks, respectively. Arrows indicate the orientation of transcription. The suffix -RT denotes a specific primer for RT. The suffix -R denotes a reverse PCR primer, -F denotes a forward PCR primer, and -P denotes a hybridization probe. P-E-P, promoter-enhancer-promoter.
Recombinant plasmid pBlue-ie2, which contains coding sequences from all three exons of gene ie2, was constructed as follows. Plasmid pPoly(A)-ie2 was generated as an intermediate. For this, part of the ie2 ORF was amplified from mCMV RNA by RT-PCR. RT was performed by using an oligo(dT) primer, and the resulting cDNA was amplified by using oligonucleotides ie2ex1-Hind (5′-GCAAGCTTGCGGCTCGATACGACCCTAC-3′) and ie2ex3-Xba (5′GCTCTAGAA TGTTCTTGAGCACGGGGTC-3′) as forward and reverse primers, respectively. Throughout, PCRs involved in cloning were performed with proofreading high-fidelity VentR-DNA-polymerase (catalog no. 254S; New England Biolabs, Frankfurt, Germany). The 942-bp amplification product was digested with HindIII and XbaI and cloned into the HindIII and XbaI sites of vector pSP64Poly(A). To make use of the more efficient in vitro synthesis with RNA polymerase T7, the ie2 sequence including the poly (A) tail was excised from pPoly(A)-ie2 by HindIII and EcoRI digestion and cloned into the HindIII and EcoRI sites of vector pBluescript II SK(+). The resulting plasmid, pBlue-ie2, thus includes exon sequences from position 3288 to position 753 (positions refer to GenBank accession no. L06816). The accuracy of the RT-PCR-based cloning steps was verified by sequencing (automated DNA sequencing system, model 4000; LI-COR, Inc., Lincoln, Nebr.).
Synthetic RNA transcribed from pBlue-ie2 has a length of 1.013 kb including a 30-nucleotide poly(A) tail. In vitro synthesis and subsequent purification of transcripts were performed essentially as described for ie1 and ie3 RNA previously (32). Specifically, in the case of ie2, pBlue-ie2 was linearized with EcoRI, and transcription was performed with RNA polymerase T7 (catalog no. A008405; AGS GmbH, Heidelberg, Germany).
Analysis and quantitation of transcripts.
Poly(A)+ RNA was isolated from shock-frozen lung tissue pieces by an oligo(dT)-cellulose affinity method as described previously (3, 32, 34). Transcripts of genes ie1, ie2, and ie3 were detected by specific RT-PCRs, which have previously been described in detail for ie1 and ie3 (32). The ie2 gene transcript was reverse transcribed by using oligonucleotide 5′-n2270-2250 (with the counting starting at the 5′ transcription start site of ie2; see reference 45) located in exon 3 as a sequence-specific RT primer (IE2-RT). The resulting cDNA was amplified by using oligonucleotide 5′-n1370-1390 located in exon 2 as the forward primer (IE2-F) and oligonucleotide 5′-n1984-1964 located in exon 3 as the reverse primer (IE2-R). Oligonucleotide 5′-n1436-1448 (exon 2) n1813-1828 (exon 3) served as the probe (IE2-P) directed against the exon 2-exon 3 junction. The PCR conditions were as described previously (32, 34), except that primer annealing was performed for 1 min at 62°C. The fragment length of the correct amplificate is 249 bp, while an amplificate from contaminating viral DNA can readily be identified by its length of 613 bp. Amplification products were analyzed by gel electrophoresis, Southern blot, and hybridization with the respective γ-32P-end-labeled internal probe, followed by autoradiography. For qualitative on-or-off analysis of transcription, 200 ng of poly(A)+ RNA were subjected to RT-PCR. Since a piece of lung tissue, representing a 1/18 aliquot of a lung, gives a yield of ca. 2 μg of poly(A)+ RNA, a maximum of 10 assays could be performed for each piece.
Plasmid constructs for reporter gene assays.
Recombinant plasmids for use in Firefly luciferase assays were constructed according to established procedures. Enzyme reactions were performed as recommended by the suppliers.
Plasmid pUCAMB served to generate an ie2 promoter-enhancer containing fragment. pUCAMB was derived from plasmid pAMB25 (24) by excision of the sequence from map positions 176,441 to 187,035 of the mCMV Smith strain genome (GenBank accession no. MCU68299 [complete genome] [55]) with BamHI followed by ligation into plasmid pUC19. For generating plasmid pGL2-ie2, pUCAMB was digested with BanII and BlpI, and sticky ends were filled up with Klenow DNA polymerase. The resulting 1.328-kbp fragment containing mCMV sequences from map positions 184,272 to 182,944 was cloned into the SmaI-digested pGL2-Basic vector (Promega; GenBank accession no. 47295) that contains the Firefly luciferase gene.
Plasmid pGL2-ie1/3 was constructed as follows. Plasmid pIE111 (44) was digested with KpnI. The resulting 1.34-kbp fragment, which encompasses the ie1/3 promoter-enhancer sequences from map positions 182,896 to 184,236, was ligated into the KpnI-digested pGL2-Basic vector.
Comparison of promoter-enhancer strengths by the DLR assay system.
The Dual-Luciferase Reporter (DLR) assay (catalog no. E1910; Promega Corp. Madison, Wis.) was employed to standardize for transfection efficacy and was performed as recommended by the supplier (technical manual no. 040, revised 5/99, part TM040; Promega). Specifically, NIH 3T3 cells were plated in six-well culture plates. On the following day, the cells of the monolayers (ca. 2 × 105 cells per well) were cotransfected with 20 ng of control reporter Renilla luciferase-encoding plasmid pRL-TK (GenBank accession no. AF025846; catalog no. E2241; Promega) and 2 μg of reporter Firefly luciferase-encoding plasmid, which was either pGL2-ie1/3 or pGL2-ie2. Transfection was performed by using 3 to 4 μl of the nonliposomal transfection reagent FuGENE 6 (catalog no. 1814443; Boehringer, Mannheim, Germany). On day 2 after transfection, the cells were lysed in 500 μl of Passive Lysis Buffer (Promega), followed by two freeze-thaw cycles to get complete lysis. The assays were performed with 20 μl samples of cleared cell lysate. Luminescence, expressed as relative luminescence units (RLU), was measured with a single-sample luminometer (Lumat LB 9507; Berthold, Bad Wildbad, Germany). According to the read-inject-read format of the DLR assay, signals from Firefly and Renilla luciferase were detected sequentially for each individual sample, with Firefly luminescence being measured first. Linear ranges and assay backgrounds for the two luciferases were determined as suggested by the supplier. Background RLU were subtracted from assay RLU.
Frequency estimation of transcriptional foci in the lungs.
The previously established “mosaic approach” (32, 33) was employed to estimate the frequency of transcriptional events in latently infected lungs. In essence, lungs were cut into 18 bona fide equally sized pieces. Pieces referred to as pieces #1 to 9 are derived from the superior, middle, and inferior lobes of the right lung (three pieces per lobe), pieces #10 and #11 are derived from the postcaval lobe (and used for determining the viral DNA load, see above), and pieces #12 to 18 are derived from the left lung (for a scheme of the lung anatomy, see reference 32). The presence of transcripts from genes ie1, ie2, and ie3 was analyzed for each individual piece. The previous work has shown that the resolution defined by 18 pieces is sufficient for a Poisson distribution analysis (35). For each type of transcript evaluated separately, the respective frequency of “transcriptional foci” was estimated from the fraction of transcriptionally negative lung pieces f(0) by using the following Poisson distribution equation: λ(lambda) = −ln f(0). The fraction of pieces containing n (n = 0, 1, 2, 3, and so forth) foci, F(n), was calculated by using the formula F(n) = λn × e−λ/n! (factorial) or (for n > 0) the mathematically equivalent formula F(n) = λ/n × F(n − 1). The estimates were based on 80 pieces derived from lungs (except the postcaval lobes) of five latently infected mice and were then downextrapolated to 18 pieces of a complete prototypic “statistically generated lung.”
RESULTS
Starting hypothesis and establishment of RT-PCRs for the analysis of mCMV IE gene expression.
The MIE enhancer of mCMV is flanked on one side by the ie1/3 transcription unit and on the other side by gene ie2 (for a map, see Fig. 1), which are driven by specific promoters P1/3 and P2, respectively. In the combined control unit P1/3-E-P2, activating or repressing factors could target the enhancer as well as the two specific promoters. From the detection of spliced IE1 transcripts we have previously inferred that P1/3-E is sporadically in “on-position” during mCMV latency in the lungs (32). Notably, in the absence of the enhancer, P1/3 shows a notable basal activity in reporter gene assays (31). While this basal activity in enhancerless mutants of mCMV suffices for initiating some virus replication after high-multiplicity infection of fibroblasts in vitro (2), the enhancer was found to be essential for replication in vivo (A. Angulo, M. Messerle, M. Griffiths, and P. Ghazal, Abstr. 25th Int. Herpesvirus Workshop, abstr. 6.09, 2000). Accordingly, it is widely assumed that CMV enhancers serve as the major genetic switch for the initiation of productive infection and, specifically, also for virus reactivation from latency (for a review, see reference 15). We therefore proposed as our starting hypothesis that the sporadic transcriptional activity of ie1/3 observed during latency (32) reflects episodes of enhancer activation by inducible transcription factors in response to sporadic triggering by endogenous cytokine signals. In that case, and provided that the enhancer operates symmetrically, P1/3-E and E-P2 should be coordinately either “on” or “off.” As a consequence, IE2 transcripts should be detectable whenever IE1 transcripts are generated.
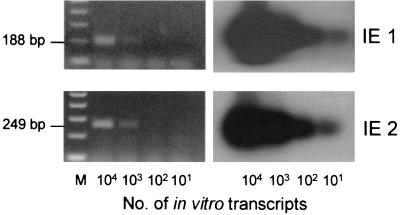
For testing this hypothesis, it is an important technical requirement that IE1 mRNA and IE2 mRNA can be detected with comparable sensitivity. In a previous report we have described the establishment of IE1-and IE3-specific RT-PCRs and the generation of the respective synthetic transcripts for evaluating the sensitivity of detection (32). Here we have completed the set of MIE genes by establishing an RT-PCR specific for IE2 and by cloning of recombinant plasmid pBlue-ie2 for the in vitro synthesis of polyadenylated IE2 RNA. Primers and probes as well as the map locations of the synthetic transcripts are compiled in Fig. 1. For each of the MIE genes, defined numbers of synthetic transcripts were subjected to the respective RT-PCRs, and the resulting cDNA amplificates were analyzed by Southern blot and hybridization with specific probes directed against the respective splice junctions (Fig. 2). The ethidium bromide-stained gel is documented to verify the predicted size of the respective amplificate (Fig. 2, left panel), and the autoradiographs reveal the sensitivity of detection (Fig. 2, right panel). In conclusion, the sensitivity of detection was identical for IE1 and IE2 transcripts. Specifically, in accordance with our previous results obtained for IE1 and IE3 (32), 10 transcripts gave a clear signal in both cases.
FIG. 2.
Sensitivity of IE1 and IE2-specific RT-PCRs. The indicated numbers of in vitro-synthesized, polyadenylated IE1 and IE2 RNA molecules were subjected to the respective RT-PCRs (for primers and probes, see the map in Fig. 1). (Left panel) Ethidium bromide-stained gels verifying the correct sizes of the amplificates. M, 100-bp size markers. (Right panel) Corresponding Southern blot autoradiographs obtained after hybridization with the γ-32P-end-labeled oligonucleotide probes IE1-P and IE2-P directed against the exon 3-exon 4 splicing junction of IE1 and the exon 2-exon 3 splicing junction of IE2, respectively.
Viral DNA load in latently infected lungs.
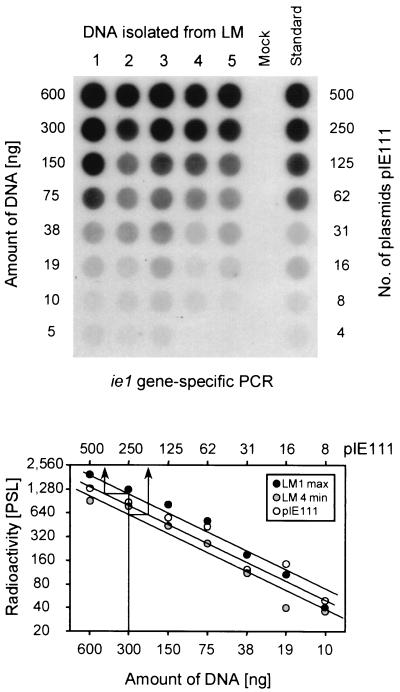
For the in vivo analysis of transcriptional activity during latency, we have used the previously described model of mCMV latency established in the lungs of infected BMT recipients (32–34, 66). In this model, productive infection is resolved in the lungs after 3 to 4 months and in the salivary glands after ca. 6 months (34). Clearance of viral genome from blood leukocytes is completed between 8 and 12 months (32, 34) and was routinely monitored by monthly PCR analysis of tail vein blood samples (not shown). According to these criteria, latency was established after 10 months in the experiments reported here. Previous work has also documented an even distribution of latent viral DNA in the lungs (32). It was therefore allowed to measure the viral DNA load in a representative fraction of the lungs, and we have here selected the postcaval lobe for that purpose. Five latently infected mice (LM) used for the assays were designated LM1 through LM5, and the viral DNA loads were determined individually by a previously established dot blot analysis (32, 66) based on an ie1-exon 4-specific PCR followed by hybridization with a radioactively labeled internal oligonucleotide probe and quantitation by digital phosphorimaging (Fig. 3). The load of viral genomes per 106 lung cells was found to range between 3,000 copies (minimum, mouse LM4) and 6,800 copies (maximum, mouse LM1). These results are in good accordance with the latent viral DNA load measured in the lungs after an analogously performed BMT for which we had originally described the sporadic and focal ie1-specific transcriptional activity (32). We had therefore good reason to expect here a similar pattern of latency-associated transcription.
FIG. 3.
Determination of the viral DNA load during mCMV latency in the lungs. For latently infected mice (LM) 1 through 5, DNA was isolated from lung tissue pieces #10 and #11 derived from the postcaval lobe. For each LM separately, the DNA was serially diluted as indicated and subjected to an ie1 exon 4-specific PCR. A negative control was provided by DNA derived from uninfected lungs (Mock). As a standard for the quantitation, the mock DNA was supplemented with defined numbers of plasmid pIE111 that encompasses gene ie1. (Top) Autoradiograph of the dot blot obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe. (Bottom) Computed phosphorimaging results of the same blot. For the sake of clarity, the computations are depicted only for the plasmid standard and for mice LM1 and LM4, representing the highest load (max) and the lowest load (min), respectively, among the five samples tested. Log-log plots of radioactivity, measured as phosphostimulated luminescence (PSL) units (ordinate) versus the amount of sample DNA (abscissa) are shown. The upper rule relates the amount of DNA to the number of plasmids in the pIE111 standard. Calculations were made from the linear portions of the graphs, as shown as examples for 300 ng of LM1 and LM4-derived lung DNA containing ca. 340 and ca. 150 copies (arrows), respectively. The loads, given as copies of the viral genome per 106 lung cells (6 μg of DNA) were thus ca. 6,800 copies for LM1 and ca. 3,000 copies for LM4.
Stochastic transcription of genes ie1 and ie2 during pulmonary latency of mCMV.
The detection and statistical quantitation of transcriptional events in the lungs was performed by a physical subdivision of the organ into pieces that were then separately analyzed for the presence of viral transcripts. In this previously established “mosaic approach,” subdivision into 18 pieces proved to give a resolution that was sufficient for a statistical analysis based on the Poisson distribution requiring a “zero fraction” of transcriptionally silent pieces for each individual type of transcript (32, 33). It should be noted that improvement of the resolution by increasing the number of pieces is technically limited by the yield of poly(A)+ RNA, which, in the case of 18 pieces, is ca. 2 μg per piece and allows for just 10 RT-PCR assays (32). The three lobes of the right lungs contributed nine pieces (pieces #1 through #9), and the left lung contributed seven pieces (pieces #12 through #18). The two remaining pieces, #10 and #11, which are derived from the postcaval lobe, were used for determining the viral DNA load (see above).
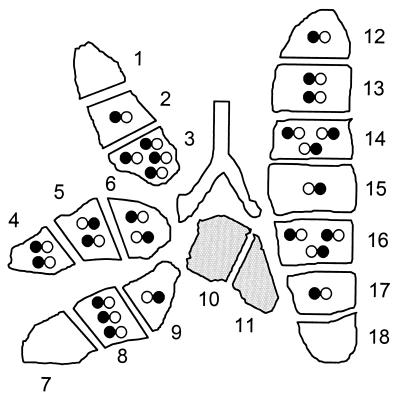
The piece-by-piece analysis of IE1 transcription (Fig. 4) confirmed the previously reported, stochastic on-or-off pattern (32, 33), which implies activity of the regulatory unit P1/3-E during mCMV latency. It should be noted that the classification of a piece as negative or positive for IE1 transcription was reproduced by testing a second sample and that transcripts of cellular hypoxanthine phosphoribosyltransferase, yielding a fragment of 163 bp (34), were detectable in comparable amounts in all pieces (data not shown here, but it is documented in reference 32). In accordance with the previous work (32), all pieces were found to be negative for IE3 transcripts as well as for glycoprotein B transcripts (not shown in Fig. 4 because the autoradiographs lack signals throughout). These results were as expected and reproduced the published characterization of the system. The exciting new information resulted from the analysis of IE2 transcription (Fig. 4). The first new message is that gene ie2 was transcribed, which implies activity of the regulatory unit E-P2 during mCMV latency. Since the RT-PCR primers were chosen to flank intron 2 of gene ie2 and since the hybridization probe IE2-P was directed against the exon 2-exon 3 junction, detection of the predicted 249-bp fragment (recall Fig. 1) shows that the latency-associated IE2 transcripts were spliced. The second new message is that latency-associated IE2 transcription, like IE1 transcription, shows a stochastic on-or-off pattern with pieces positive or negative for IE2 transcripts. According to the starting hypothesis of regulation of P1/3-E-P2 via the shared MIE enhancer, the on-or-off patterns for IE1 and IE2 were expected to be congruent. The surprising third message is that this was not the case (Fig. 4, compare the IE1 and IE2 expression patterns). Simultaneous expression of both genes is documented for a number of pieces, e.g., in mouse LM1 for the pieces #2, #5, #14, #16, and #17 (and similar examples could be listed for mice LM2, LM4, and LM5). However, there are obvious exceptions. Thus, piece #6 of mouse LM1 is strongly positive for IE1 but negative for IE2, and piece #15 of mouse LM2 is clearly positive for IE2 but negative for IE1 (and many more examples of asymmetric expression could be listed). Notably, in mouse LM3, no lung tissue piece was found to coexpress both genes. An asymmetric expression is also evident from differing strengths of signals in double-positive pieces. Specifically, in piece #14 of mouse LM1, IE1 was strongly expressed and the expression of IE2 was weak, whereas in piece #17 of the very same organ IE2 was strongly expressed and the expression of IE1 was weak. These expression data refer to steady-state levels of IE1 and IE2 poly(A)+ RNAs present at the time point of shock-freezing of the tissue, and the quantity of transcripts might thus be influenced by the in vivo mRNA half-lives. Nothing is known yet about the half-lives of IE1 and IE2 mRNAs during latency in vivo, but the examples of variegated signal intensities discussed above do not indicate a systematic bias in favor or disfavor of either type of transcript.
FIG. 4.
Random patterns of IE1 and IE2 transcription in latently infected lungs. For latently infected mice LM1 through LM5, poly(A)+ RNA was isolated piece by piece from pieces #1 through #9 derived from the three lobes (superior, middle, and inferior lobe) of the right lung and from pieces #12 through #18 derived from the left lung (ventral view). The postcaval lobes (shaded) were spent for determining the latent viral DNA load (recall Fig. 3). Aliquots (200 ng) of the poly(A)+ RNA, that is, ca. a tenth of the yield from each piece, were subjected to IE1- and IE2-specific RT-PCRs. (Left panels) Autoradiographs obtained after gel electrophoresis of the amplificates, Southern blotting, and hybridization with the γ-32P-end-labeled oligonucleotide probes IE1-P and IE2-P, respectively. Faint signals (asterisks) are clearly visible on the original autoradiograph and were confirmed by testing of a further aliquot of the poly(A)+ RNA. (Right panels) Compilation of the data in topographical maps of the lungs. In accordance with the location of the ie1 and ie2 genes in the genetic map (recall Fig. 1), the left and right circles of the paired-circle (infinity) symbol correspond to IE1 and IE2 transcripts, respectively. The presence of transcripts is indicated by a closed circle, and the absence of transcripts is indicated by an open circle.
As a synopsis of these, at least at first glance, rather complicated expression patterns, all data are compiled and illustrated as topographical maps of the lungs (Fig. 4, right panel). Paired circles, filled and empty, symbolize the presence and absence of transcripts, respectively, with IE1 depicted on the left and IE2 depicted on the right according to the gene locations relative to the enhancer (recall Fig. 1). Out of 80 pieces tested altogether for mice LM1 through LM5, 28 pieces displayed the double-negative “off-off” and 13 pieces displayed the double-positive “on-on” pattern. The remaining 39 pieces, that is, the majority of the 52 transcriptionally active pieces, showed asymmetric patterns. Specifically, transcription was “on-off” in 30 pieces and “off-on” in 9 pieces. Taking into account the “on-on” cases, IE1 transcripts were thus present in altogether 43 pieces, while only 22 pieces contained IE2 transcripts. In conclusion, the genes ie1 and ie2 are not coordinately expressed during mCMV latency in the lungs, and there is an imbalance in the expression incidence of ca. 2:1 (on the basis of positive pieces) in favor of ie1 gene expression.
Number and expression pattern of transcriptional foci in an individual latently infected lung.
The experimental molecular data compiled in Fig. 4 are sufficient to conclude that genes ie1 and ie2 were not coordinately transcribed in the majority of the pieces, since otherwise it would not have been possible to observe the two asymmetric patterns “on-off” and “off-on” at all. It is much more sophisticated to interpret the minority case of the 13 pieces with a virtual “on-on” pattern. This symmetric pattern could result from a truly coordinated expression of both genes from the latent viral genome. Alternatively, it could be mimicked by a superposition of two or more asymmetric transcriptional events. A statistical analysis is needed to decide between these two possibilities.
For the interpretation of the “on-on” observations, we have to consider the nature of the Poisson law (reference 35 and reference 12 cited therein). It is self-evident that a tissue piece must harbor at least one “transcriptional event,” previously defined by us as a “focus” of transcription (32, 33), whenever we can detect transcripts. However, such a positive piece can also harbor two or more foci. This basic understanding bears an important consequence: if, for instance, a tissue piece contained one “on-off” focus and one “off-on” focus, it was classfied in Fig. 4 as a tissue piece with an “on-on” pattern, even though it actually did not contain a true “on-on” focus.
The distribution of foci was determined by employing the Poisson distribution function. As an example, such a statistical calculation is shown for an individual mouse LM2 (Table 1), and the resulting distribution and expression pattern of foci is illustrated in Fig. 5. The total number of transcriptional foci in the lungs of mouse LM2 was estimated from the “zero fraction” f(0) = 3/16 = 0.1875, that is, from the number of transcriptionally negative (ie1 and ie2 negative) pieces divided by the total number of pieces tested. According to the formula λ = −ln f(0), the Poisson distribution parameter λ(ie1&2) = 1.674. In other words, each of the 16 pieces contained 1.674 foci on average and, accordingly, the 16 pieces contained 26 foci (= λ × 16; rounded off) altogether. The fraction of tissue pieces containing n (n = 1, 2, 3, and so forth) foci, F(n), can then be calculated by using the formula F(n) = λ(ie1&2)/n × F(n − 1). In like manner, the experimentally observed zero fractions for ie1 and ie2 separately, namely, 5/16 and 10/16, give us λ(ie1) and λ(ie2), respectively, with their associated distributions (Table 1). Note that there is a fairly good match between the statistically calculated distribution of foci (Fig. 5) and the molecular data (Fig. 4, mouse LM2). The calculations led to an estimate of 19 foci of transcription from gene ie1 and 7 from gene ie2, within a total of 26 foci. Notably, while the experimental observation based on pieces had suggested the existence of 4 “on-on” events (Fig. 4, pieces #5,#6, #14, and #16 of LM2), the distribution of foci allowed us to interpret each of these four cases as a superposition of “on-off” and “off-on” foci in the respective pieces. In fact, the data gave so far no evidence for the existence of true “on-on” events and, at this stage, one may therefore even ask whether the expression of ie1 and ie2 is mutually exclusive in that ie1 expression might suppress ie2 expression and vice versa.
TABLE 1.
Poisson distribution of transcriptional foci in the lung of individual mouse LM2a
| Foci per piece | IE1&IE2 (λ = 1.67)
|
IE1 (λ = 1.16)
|
IE2 (λ = 0.47)
|
|||
|---|---|---|---|---|---|---|
| Pieces | Foci | Pieces | Foci | Pieces | Foci | |
| 0 | 3 | 0 | 5 | 0 | 10 | 0 |
| 1 | 5 | 5 | 6 | 6 | 5 | 5 |
| 2 | 4 | 8 | 3 | 6 | 1 | 2 |
| 3 | 3 | 9 | 1 | 3 | 0 | 0 |
| 4 | 1 | 4 | 1 | 4 | 0 | 0 |
| Sum | 16 | 26 | 16 | 19 | 16 | 7 |
Experimental data for the presence or absence of IE1 and IE2 transcripts were obtained for 16 lung tissue pieces of individual mouse LM2 (Fig. 4), and the Poisson distribution parameter λ was calculated for each of the indicated constellations of transcription from the respective zero fraction f(0) by using the formula λ = −ln f(0). The number of pieces containing n (n = 0, 1, 2, 3, and so forth) foci was calculated by multiplying the fraction of pieces containing n foci, F(n), with the number of tested pieces. F(n) is derived from the Poisson distribution function.
FIG. 5.
Distribution and expression pattern of transcriptional foci in individual mouse LM2. Experimental data on the expression of genes ie1 and ie2 in lung tissue pieces of LM2 (see Fig. 4) were used to calculate F(n), that is, the fraction of pieces containing n (n = 0 , 1, 2, 3, and so forth) transcriptional foci, by employing the Poisson distribution function. By multiplication with the number of pieces tested (in this case 16), the fractions are converted into the number of pieces containing n transcriptional foci. The results of the statistical calculations summarized in Table 1 are illustrated by a topographical map of the lung. The map was constructed by the “puzzle method” so as to be consistent with all information given in Table 1, that is, with the total number of transcriptional foci, the total numbers of ie1 and ie2 foci separately, and the numbers of pieces with n (n = 0, 1, 2, 3, and so forth) foci of either type. Note that, unlike in Fig. 4, the symbols here represent individual transcriptional foci (most likely individual cells) instead of the overall expression in a whole tissue piece. Since, by definition, a transcriptional focus is characterized by transcription from either ie1 or ie2 or both, the absence of transcription is not indicated by a symbol. In fact, all pieces contain ca. 16,000 latent mCMV genomes on average (Fig. 3), and thus many “silent” double-negative “foci” are present in every tissue piece. Transcription from genes ie1 and ie2 is indicated by a closed circle on the left and on the right part of the infinity symbol, respectively. Open circles symbolize the absence of the respective transcripts. Shaded pieces #10 and #11 are exempt from the transcriptional analysis because they were used for determining the viral DNA load.
Number and expression pattern of transcriptional foci in a statistically generated, prototypic lung.
The data discussed thus far have already made clear that there is no positive linkage between ie1 and ie2 gene expression. It was still open to question therefore, whether transcription from these two genes is mutually exclusive or is independent of each other. In the case of independent transcription, true “on-on” events should exist and should occur by random coincidence with a probability defined by the frequencies of ie1 and ie2 transcription. Because ie1 was found to be more frequently transcribed than ie2, random coincidences are predictably very rare. We have therefore compiled the data from all five mice to repeat the analysis with a higher statistical confidence based on 80 pieces. Again, the total number of transcriptional foci can be calculated from the “zero fraction” which in this case was f(0) = 28/80 = 0.35. According to the formula λ = −ln f(0), the Poisson distribution parameter was λ(ie1&2) = 1.05. Thus, admittedly more by chance than by clever design, the average number of transcriptional foci was ca. one focus per tissue piece. In absolute terms, 84 foci (= λ × 80) were present in the 80 pieces tested. The fraction of tissue pieces containing n(n = 1, 2, 3, and so forth) foci, F(n), can then be calculated by using the formula F(n) = λ (ie1&2)/n × F(n − 1). In like manner, the experimentally observed zero fractions for ie1 and ie2 separately, namely, 37/80 and 58/80, give us λ(ie1) and λ(ie2), respectively, with their associated distributions (Table 2). The calculations thus led to an estimate of 61 events of transcription from gene ie1 and 25 from gene ie2, within a total of 84 foci. Thus, the imbalance in the expression incidences is 61:25, that is, ca. 2.5:1 in favor of ie1 gene expression. Because the sum of transcriptional events, namely, 86, exceeds the number of foci, namely, 84, two foci must represent true “on-on” events. In conclusion, genes ie1 and ie2 are transcribed independently of each other during latency, and the few true “on-on” events reflect random coincidence. For a summary, the data obtained from 80 pieces derived from the lungs of LM1 through LM5 were downextrapolated to 18 pieces, and the distribution of transcriptional foci is illustrated for a prototypic, statistically generated complete lung (Fig. 6).
TABLE 2.
Poisson distribution of transcriptional foci in lungs of mice LM1 through LM5a
| Foci per piece | IE1&IE2 (λ = 1.05)
|
IE1 (λ = 0.77)
|
IE2 (λ = 0.32)
|
|||
|---|---|---|---|---|---|---|
| Pieces | Foci | Pieces | Foci | Pieces | Foci | |
| 0 | 28 | 0 | 37 | 0 | 58 | 0 |
| 1 | 29 | 29 | 29 | 29 | 19 | 19 |
| 2 | 16 | 32 | 11 | 22 | 3 | 6 |
| 3 | 5 | 15 | 2 | 6 | 0 | 0 |
| 4 | 2 | 8 | 1 | 4 | 0 | 0 |
| Sum | 80 | 84 | 80 | 61 | 80 | 25 |
FIG. 6.
Distribution and expression pattern of transcriptional foci in a statistically generated, prototypic lung. Data from all five mice LM1 through LM5 (see Fig. 4) were compiled, fractions F(n) were calculated on the basis of all 80 pieces tested (Table 2) and down extrapolated to 18 pieces of one complete prototypic lung. Specifically, by multiplication with 18, the fractions F(n) are converted into the number of pieces containing n (n = 0, 1, 2, 3, and so forth) transcriptional foci. Alternatively, one can arrive at the same result by multiplying the data presented in Table 2 by 18/80 and with appropriate rounding to whole numbers. As in Fig. 5, the results are illustrated by a topographical map, and all symbols are likewise defined.
The imbalance in the incidences of IE1 and IE2 transcription during latency is not explained by different enhancer-promoter strengths.
That expression of gene ie2 is less frequent but not generally weaker than expression of gene ie1 and that transcript stability does not disfavor detection of IE2 transcripts was already suggested by the signal intensities seen in Fig. 4. Specifically, in some of the double-positive pieces, for instance in pieces #5 and #17 of mouse LM1 and in piece #14 of mouse LM2, the amount of IE2 transcripts apparently exceeded the amount of IE1 transcripts.
To address this aspect more directly, the strengths of the separated regulatory units P1/3-E (cloned into plasmid pGL2-ie1/3) and E-P2 (cloned into plasmid pGL2-ie2) were compared by their capacities to drive the expression of the Firefly luciferase reporter gene upon transfection of NIH 3T3 cells. To normalize for transfection efficacy, plasmid pRL-TK, which contains the Renilla luciferase gene as a control reporter driven by the herpes simplex virus thymidine kinase promoter region, was cotransfected with either of the two test plasmids. The results of two independently performed DLR assays are shown in Fig. 7A as original luminescence data and in Fig. 7B after normalization for transfection efficacy. Considering the variance between replicates and the variance between the two independent experiments, there was no significant difference between the two promoter-enhancer constructs. We infer from this result that the imbalance between IE1 and IE2 transcription during latency is caused by a higher incidence of P1/3-E activity and not by quantitative differences in enhancer-promoter strengths.
FIG. 7.
Comparison of promoter-enhancer strengths by DLR assay. (A) NIH 3T3 cells were cotransfected in duplicates with Renilla luciferase-encoding expression plasmid pRL-TK (open bars) and with a Firefly luciferase-encoding expression plasmid (filled bars) alternatively with pGL2-ie1/3 and pGL2-ie2 containing the regulatory elements P1/3-E and E-P2, respectively. Two independent experiments were performed under slightly different conditions. In experiment 1, transfection was performed with 3 μl of FuGENE 6 reagent, and the DNA-FuGENE 6 mixture was preincubated for 5 min. Experiment 2 was performed with 4 μl of FuGENE 6 and a preincubation time of 15 min. Activities are expressed as RLU with the background subtracted. (B) Data were normalized for transfection efficacy by forming the ratio between the Firefly luciferase activity (RLUFL) and the Renilla luciferase activity (RLURL). Shaded bars represent the mean value of the duplicates, and the error bars indicate the range.
DISCUSSION
Latency of mCMV in the lungs is defined by maintenance of the viral genomes after resolution of acute, productive infection and by their property to reenter the productive program of viral gene expression upon induction, which results in the recurrence of infectious virions (34). The criterion of absence of infectious virions during latency, originally proposed for latency of herpes simplex viruses (60), had previously raised a debate on whether CMVs establish true molecular latency or rather a low-level productive infection below the detection limit. Since limited sensitivity of detection is a cheap argument that applies always, the competing hypotheses of “molecular latency” and “low-level persistence” could not be proven or disproven, respectively, and thus had resulted in a “dead heat” for over two decades. Recent progress in enhancing the sensitivity of detection strongly supported the “molecular latency” hypothesis for mCMV (34, 52). Specifically, an assay based on centrifugal infection of permissive cells in cell culture and subsequent detection of viral transcripts by RT-PCR lowered the detection level to a genome-to-infectivity ratio of 5, which exceeded the sensitivity of the conventional virus plaque assay by a factor of 100 (34). Using this improved assay, infectious virus remained undetectable in homogenates of latently infected lungs (32, 34).
The absence of infectious virus does not imply that the viral genome is transcriptionally silent during latency, because there exist numerous possibilities of how the highly coordinated viral replication program could be interrupted before the assembly of progeny virions. In fact, earlier reports had noted the presence of IE1 RNA, but since IE1 transcripts were then falsely considered as transcripts defining the initiation of the productive cycle, the finding was misinterpreted as being indicative of low-level persistent productive infection (19, 71). In two recent reports we have documented the existence of sequential checkpoints in the transition from mCMV latency to recurrence (32, 33). Specifically, latency in the lungs was found to be maintained in the presence of IE1 transcripts because of the absence of IE3 transcripts (32). That protein IE3 instead of IE1 is the key transactivator of mCMV E-gene expression was concluded from earlier work by Messerle et al. (44), who had demonstrated in reporter gene assays an efficient activation of the e1 promoter by IE3 alone but not by IE1 alone, whereas IE1 did enhance the activity when coexpressed with IE3. More recently, the replication deficiency of an ie3 gene (exon 5 of ie1/3) deletion mutant of mCMV and its transcomplementation in IE3-expressing cells confirmed the essential role of IE3 in viral replication (1). Thus, in conjunction with the presence of functionally competent genomes, the absence of IE3 transcripts is presently the best criterion for mCMV latency.
As outlined in greater detail above (see introduction and Fig. 1), ie1 and ie3 form a transcription unit controlled by an upstream MIE promoter-enhancer, here referred to as the P1/3-E. Accordingly, differential expression of IE1 and IE3 mRNA during latency implies a regulation operating after transcription initiation, that is, either by selective usage of a polyadenylation signal in exon 4 of ie1 or by selective splicing of a proposed 5.1-kb IE1/3 (exons 1 to 5) precursor RNA (24, 44) or by rapid degradation of spliced IE3 transcripts. Since IE1 transcripts are generated during latency, albeit with low frequency, P1/3-E must be sporadically active. It appeared to us reasonable to propose that sporadic activity results from transcription factor binding to the enhancer reflecting the “noise” of background signaling in a latently infected cell, for instance, by cytokines such as TNF-α (54). Since, in mCMV, the same enhancer is part of the regulatory element E-P2 that controls the expression of gene ie2 (45), sporadic activity of the enhancer should lead to coordinated transcription from its two flanking genes ie1/3 and ie2.
This was a testable prediction, but the result was not what we had expected. While the proposed transcription from ie2 was indeed found, and while this transcription indeed also displayed a focal and random on-or-off pattern, it was less frequent and it did not coincide with transcription from ie1. Instead, most transcriptional foci showed the alternative patterns “on-off” or “off-on”. This observation even might have raised the speculation that expression from both flanks of the enhancer is mutually exclusive. However, the in-depth statistical analysis gave evidence for the existence of true “on-on” events in accordance with a random coincidence predicted for an independent expression of the two genes. We therefore conclude that genes ie1 and ie2 are expressed neither in conjunction nor in a mutually exclusive manner but are expressed independently during mCMV latency in the lungs.
We have previously defined a transcriptional focus operationally as a transcriptional event that is detectable with an RT-PCR that has the sensitivity to detect ca. 10 transcripts (32, 33). It was open to discussion whether such a focus physically represents a single cell or a group of cells. Since a single cell can produce many more transcripts than just 10, it is in principle possible that a transcriptional focus indeed represents a single latently infected cell in a state of temporary “latency-associated” transcriptional activity. Although we admit that there is no final proof for the single-cell conjecture, we believe that this interpretation is most likely on the basis of the new data presented here. Let us consider the following argument that is fairly related to the problem of tissue pieces versus individual foci discussed above. If a focus would represent a group of cells, these cells must express either ie1 or ie2 in a synchronized fashion, because otherwise we would always have observed “on-on” patterns, even though the individual cells in such a proposed cohort displayed the patterns “on-off” and “off-on.” Along the same line of argument, the observed transcriptions may even result from single viral genomes, because unsynchronized transcription from several genomes within a single latently infected cell would likewise have resulted in an “on-on” pattern. In fact, the experimentally observed asymmetric patterns can be explained only by singular molecular events or by synchronism of groups of molecular events. We intuitively would favor the first interpretation, but we do not exclude the possibility that a local signaling milieu, for instance, one defined by cytokines, might synchronize transcription from several genomes present in one cell or even in groups of cells.
The on-or-off transcription patterns observed here as well as in our previous work (32, 33) reflect an underlying stochastic process. What is the molecular basis for such a stochastic phenomenon? The answer might be given by a recent review on the function of enhancers by Fiering et al. (14). Two contrasting, but not mutually exclusive, views of how enhancer elements act are currently discussed (for reviews, see references 6 and 14). The textbook view is that an enhancer functions by increasing the rate of transcription of a cognate gene, a mechanism referred to as “rheostatic” model, “rate” model, or a “progressive-response” model. More recently, there is accumulating evidence in support of an alternative interpretation, namely, that an enhancer does not affect the level of transcription of a gene that is already switched-on but rather increases the probability that a gene is transcribed. This mechanism is referred to as the “binary” model, the “on-or-off” model, or the “probability” model. Apparently, in experiments where transcriptional activity is measured with populations of cells, such as in the transient-transfection reporter gene assays originally used to demonstrate the “classical” enhancer effect by an increase in the total assay product, it is not possible to discern between the two proposed modes of enhancer action. Likewise, bulk biochemical assays of gene expression in infected cells, in stably transfected cell lines, or in transgenic mice cannot differentiate the two models. Evidence in support of the binary model was provided by cell-by-cell analysis in tissue culture assay systems, demonstrating that the presence of an enhancer increased the number of expressing cells rather than the amount of transcripts per cell. Similarly, single cell analysis of transgene expression in mice frequently revealed mosaic expression, also known as “variegation”, with transcriptionally active and silent cells all carrying the transgene (for examples, see reference 14).
The binary model of enhancer action is relevant to the interpretation of our data in that it can explain the observed stochastic distributions of silent and active foci, that is, the apparent variegation of latency-associated transcription. To our knowledge, this is the first example of mosaic expression in an in vivo model of viral latency. Unfortunately, the predicted enhancement of transcription probability by the mCMV enhancer cannot be demonstrated in our in vivo latency model, because the enhancer appears to be essential. While enhancer swap mutants have shown that the mCMV enhancer can be replaced by the paralogous hCMV enhancer for virus replication in vitro (2) and in vivo (17), enhancerless mutants of mCMV proved to be not viable in vivo (Angulo et al., 25th Int. Herpesvirus Workshop). The binary model of enhancer action proposes an effect of the enhancer on chromatin opening or nuclear compartmentalization (for reviews, see references 6 and 14). This mechanism primarily refers to enhancers of cellular genes and to viral enhancers integrated into the cellular genome. Regarding the effects of a viral enhancer on chromatin, it would be important to know the physical state of the viral genome during latency. Our knowledge is still incomplete, but studies on hCMV latency in CD14-positive peripheral blood mononuclear cells have shown that, at least in this particular cell type, the latent viral genome is not integrated into the cellular genome but is maintained as a circular plasmid or episome (7). It is likely that latent episomal CMV genomes are not naked but persist in a chromatin-like structure, and therefore the ideas discussed for integrated enhancers may also apply to enhancers in nonintegrated viral genomes. In fact, an association with the induction of DNase I-hypersensitive sites has been observed for IE1 transcription of hCMV (48).
Usually, enhancer function is studied in systems with a single cognate gene (14). An intriguing novel aspect contributed by our work results from the fact that the mCMV MIE enhancer (13), unlike the hCMV MIE enhancer (8), governs two cognate genes by which it is flanked, namely, ie1/3 and ie2. This gave us the unique opportunity to test the symmetry of enhancer action, that is, to test whether both genes are expressed synchronously resulting in a congruent pattern of variegation. Notably, transcription proved to occur asynchronously from both ends of the enhancer and proved to be asymmetrical in that the in vivo expression probabilities were different for the two flanking genes. This result was not expected. The distribution of transcription factor binding sites on the enhancer is not symmetrical (8, 13), and such an intrinsic structural asymmetry might predilect for the observed asymmetry in gene expression. However, the luciferase reporter gene assays demonstrated that the asymmetry was not based on a difference in the basal strengths of the separated regulatory units P1/3-E and E-P2. Possibly, asymmetry only becomes apparent when the enhancer has to aid the formation of two competing transcription initiation complexes.
If the function of the activated enhancer is to open a chromatin-like structure for the transcription initiation complex, one may wonder why ie1 and ie2 are transcribed independently and with different probabilities. However, one has to consider that the distance between the two 5′ start sites is 1,375 bp, which is equivalent to the distance of seven nucleosomes in cellular DNA. Although the precise organization of the episomal latent viral DNA is not yet known, this comparison can help us to understand that the two ends of the enhancer are not necessarily simultaneously open or closed, but we admit that at the moment we can only speculate and that there may be other explanations for our data.
There exists a provocative alternative interpretation that we would like to discuss frankly. Previous work by Angulo et al. (2) has shown that an enhancerless mutant of mCMV can replicate in cell culture after high-multiplicity infection, which implies expression of the essential transactivator IE3 (1) due to a basal activity of P1/3. While this basal activity does not suffice for viral replication in vivo (Angulo et al., 25th Int. Herpesvirus Workshop), our observation of sporadic transcription from the ie1/3 transcription unit under the control of P1/3-E might reflect stochastic basal activity of the promoter involving basal transcription factors rather than stochastic activation of the enhancer by induced transcription factors, and the same may apply to E-P2. The dilemma is that the in vivo replication deficiency of the enhancerless mCMV mutant precludes the establishment of high-load latency in tissues, and therefore the contributions of the shared enhancer and the two specific promoters to latency-associated MIE gene expression cannot be distinguished. Our results certainly allow the conclusion that the enhancer does not synchronize the transcription from its two flanking genes during latency. This could be either because the enhancer is not involved at all in the process or because it operates by itself in a stochastic fashion in accordance with the binary model of enhancer action.
Why do we prefer the second interpretation? First, we consider it unlikely that the enhancer is completely silent throughout latency. It is well established that cytokine-induced transcription factors activate MIE gene expression by binding to sequence motifs on the enhancer (for a review, see reference 15), and NF-κB induced by TNF-α has been implicated in the reactivation of CMV MIE gene expression (see the introduction). A basal level of cytokine signaling must be expected in the latently infected host in an environment that is neither germfree nor devoid of antigens. Moreover, in the absence of negative autoregulation by the mCMV IE3 protein (1, 44), which is not expressed during latency (32), ie1 gene expression, once initiated, may be self-maintaining by positive autoregulation, a mechanism that is not yet formally proven for IE1 of mCMV but is established for IE1 of hCMV (11, 68). Finally, there is evidence for iterative restimulations of tissue-resident IE1-specific memory CD8 T cells during mCMV latency in the lungs (21). Thus, CD8 T-cell-derived cytokines, with TNF-α being a premier candidate, may induce transcription factors that target the enhancer and promote ie1 transcription in a regulatory circuit. Second, if one proposes sporadic transcription due to basal activity of promoters with no participation of an enhancer, this should not be a speciality of MIE promoters but should apply to any promoter. To date, the database is too small to exclude that possibility, but at least the E-phase gene M55 (gB) is not sporadically expressed during mCMV latency in the lungs (32), and there is preliminary indirect evidence that the early-late gene M83 is not expressed either (21).
At present we have no clue regarding a possible functional role for IE2 expression during latency or reactivation. The idea that IE2 protein may substitute for IE3 protein in initiating the viral replicative cycle by transactivation of E-phase genes can be refuted, because previous work has documented a lack of transactivation by IE2 of mCMV in reporter gene assays (31). Accordingly, we did not detect expression of the E-phase gene M55 (gB) during latency (32), and the transactivator IE3 was found to be indispensable for gene expression during the productive cycle (1). Recent work on the immune response to mCMV infection of the lungs has revealed an enrichment of activated (CD62Llo), tissue-resident IE1 (YPHFMPTNL)-peptide-specific memory-effector CD8 T cells during latency (21). This finding implies iterative restimulation of memory cells by presentation of the IE1 peptide during the episodes of latency-associated IE1 transcription. Our data presented here therefore raise the obvious questions as to whether the episodes of IE2 transcription have a similar immunological consequence. A search for an H-2d-restricted antigenic peptide in IE2 is under way.
Conclusion.
The data have identified ie2 as the second gene of mCMV that is expressed in a stochastic manner during viral latency in the lungs. Notably, MIE genes ie1 and ie2, which flank the same enhancer, were found to be expressed asynchronously and with different incidences, which revealed an asymmetry in the action of the MIE regulatory unit P1/3-E-P2. Apparently, the enhancer element did not synchronize transcription from its two cognate genes. The different probabilities of in vivo gene expression from both flanks of the enhancer are not explained by different basal strengths of the separated promoter-enhancer combinations P1/3-E and E-P2 and thus neither by the basal promoter activities nor by the intrinsic asymmetry in the linear distribution of transcription factor binding sites in the enhancer. We must rather speculate that asynchronism and asymmetry are imposed by the three-dimensional context of the latent viral genome. The experimentally observed random transcription is consistent with a stochastic nature of enhancer action.
ACKNOWLEDGMENTS
Sabine K. Kurz (now at The Scripps Research Institute, La Jolla, Calif.) made contributions in earlier stages of this work. We are particularly grateful to Steven N. Fiering (Department of Microbiology, Dartmouth Medical Center, Lebanon, N.H.) for critical reading of the manuscript. We thank Rafaela Holtappels (Institute for Virology, Mainz, Germany) for help with the graphics and Michael Reddehase (PMS-Elektronik GmbH, Waibstadt, Germany) for computing the LDA shown in the Appendix.
Support was provided by a grant to M.J.R. from the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 490, individual project B1 “Immune control of latent cytomegalovirus infection.” Publication costs were defrayed in part by the “Fonds der chemischen Industrie (FCI),” Frankfurt, Germany.
Appendix
The distributions of transcriptional foci in the latently infected lungs (Fig. 5 and 6, as well as Tables 1 and 2, in the main body of the text) were based on the mathematical conjecture of a Poisson distribution of the experimental RT-PCR data. The interpretation of previous work on mCMV latency and reactivation in the lungs also relied on this conjecture (32, 33). Since the experimental data were discrete in that transcription was found to be on-or-off in pieces of lung tissue, and since the on-or-off patterns observed in the lungs of several individual mice were apparently random with no recognizable clustering or preferred anatomical localization (recall Fig. 4), it was reasonable to apply a discrete probability distribution, such as the Poisson distribution. In a formal sense, however, the conformity to the Poisson law was not documented. The problem lies in the fact that the experiment can only give the information of whether or not transcripts are present in a tested piece of lung tissue, but in the positive case it does not reveal whether the transcripts were derived from a single transcriptional focus or from two or more foci. In other words, what we get from the molecular data is the so-called “zero fraction” of transcriptionally negative tissue pieces, f(n = 0), and the fraction of transcriptionally positive tissue pieces, f(n > 0), but not the complete distribution with the fractions for n = 1, 2, 3, and so forth. As a consequence, we cannot compare an experimentally observed, empirical distribution with the theoretical Poisson distribution for applying a χ2 (chi-squared) test for goodness of fit.
A solution to the problem is given by the limiting dilution analysis (LDA), long known in cellular immunology as a statistical approach for the quantitation of rare cells with a specific and detectable functional competence within a large pool of unrelated cells. In essence, serial dilutions are made from the pool of cells, and many samples for each dilution are tested for the absence or presence of the functional cells in question (for an overview and lecture book, see reference 35). Thus, as in our problem with the rare transcriptional events in the lungs, LDA gives on-or-off data, with the notable difference that experimental zero fractions, f(n = 0), are obtained from several appropriate dilutions. In case that the zero fractions belong to Poisson distributions, a semilogarithmic plot with the dilutions in a linear scale on the abscissa and the zero fractions plotted as −ln f(0) on the ordinate will result in a straight line (single hit curve) passing through the origin. One can then use statistical tools, such as the maximum likelihood method (see reference 34 and references therein), for linear regression, for the determination of the confidence intervals and for testing the null hypothesis, id est the conformity to the Poisson law.
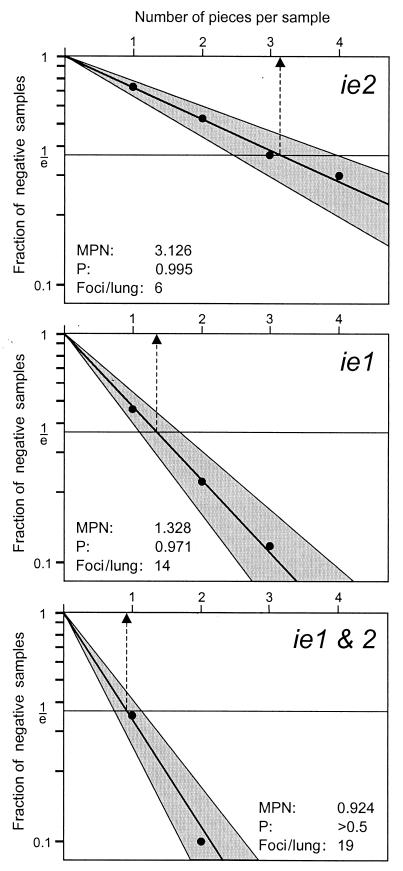
However, how can the LDA strategy be applied to transcriptional events in latently infected lungs with no serial dilutions having been performed? Obviously, the experimental data need to be transformed in such a way that the data format resembles an LDA setting. We would like to demonstrate this in greater detail for the ie2 transcription on the basis of the molecular data shown in Fig. 4. In that experiment, we had tested 16 lung tissue pieces for each of the mice LM1 through LM5, that is, n = 80 pieces altogether, for the presence of viral transcripts. With respect to ie2, 58 pieces out of 80 proved to be negative, which gives the zero fraction f(0) = nneg/n = 58/80 = 0.725. Since our assay was done with the high resolution of 80 tissue pieces, we know the result that we would have obtained if only n = 40 pieces had been tested. For instance, in the lung of mouse LM1, piece #1 is negative for ie2, while piece #2 is positive. If we had not cut this portion of the lung into two pieces but had instead tested the double-sized piece #1 + 2 as a whole, it obviously would have been positive. Likewise piece #3 + 4 and piece #5 + 6 would have been positive too, while pieces #7 + 8 and #9 + 12 would have been negative. Continuing with this strategy to the positive sample #17 + 18 of mouse LM5, we find altogether 21 negative samples out of 40, which gives the zero fraction f(0) = 21/40 = 0.525. In like manner, when three and four pieces were grouped, the zero fractions were found to be f(0) = 10/27 (rounded up) = 0.370 and f(0) = 6/20 = 0.300, respectively. The semilogarithmic LDA plot (Fig. A1) of these zero fractions shows a perfect conformity to the Poisson law with a probability value for the null hypothesis of P = 0.995 (χ2 = 0.07, degrees of freedom = 3). For the Poisson parameter λ = −ln 1/e = 1, the most probable number is 3.126. This tells us that, on average, one focus of ie2 transcription is contained in 3.126 pieces, which equals a frequency of one focus per ca. 11 million lung cells, because one piece of tissue in our molecular assays represented a 1/18 fraction of the lungs containing ca. 3.5 million lung cells. Accordingly, the number of ie2 foci in a prototypic, latently infected lung is 18 divided by 3.126 = 5.758. Since the number of foci necessarily must be a whole number, this value is rounded-up to six foci per lung. Note that this result is in accordance with the distribution of foci shown in Fig. 6.
FIG. A1.
Frequency of transcriptional foci in latently infected lungs determined by LDA. Molecular data derived from the RT-PCR analysis of 80 lung pieces from mice LM1 through LM5 (Fig. 4) were transformed into LDA format as described in greater detail in the Appendix. In essence, n samples were defined with the size of 1/18 of a lung (equivalent to 1 piece, n = 80), 1/9 of a lung (two pieces, n = 40), 1/6 of a lung (three pieces, n = 26.67 rounded up to 27 in the case of ie2 and ie1&2 or rounded off to 26 in the case of ie1), and 1/4.5 of a lung (four pieces, n = 20). The fractions of transcriptionally negative samples (for ie2 or ie1 or both), f(0) = nneg/n, were plotted as −ln f(0) on the ordinate versus the number of pieces per sample in a linear scale on the abscissa. The respective regression lines were calculated by using the maximum likelihood method. Shaded areas represent the 95% confidence intervals. The most probable number (MPN; dashed arrow) is derived from the point of intersection between the regression line and f(0) = 1/e. By convention, the null hypothesis (i.e., here the conformity to the Poisson law) is accepted for P > 0.05, which was fulfilled in all three cases. The number of foci per lung is calculated as the quotient between the number of pieces per lung and the MPN, 18/MPN, and is rounded to the closest whole number.
In like manner, the analysis was made for ie1 transcription as well as for transcription from either of the two genes, ie1&2, and the resulting zero fractions were plotted accordingly (Fig. A1). Again, the LDA graphs show perfect conformity to the Poisson law, with P values of 0.971 (χ2 = 0.06, degrees of freedom = 2) and P > 0.5 (χ2 = 0.13, degrees of freedom = 1) for ie1 and ie1&2, respectively. Based on the most-probable-number values of 1.328 and 0.924, the numbers of foci per lung are 14 and 19, respectively. This is minor difference compared to the numbers of foci shown in Fig. 6, which were 13 and 18, respectively. The explanation is that the distribution shown in Fig. 6 was based on the Poisson distribution parameter λ derived from the experimental data of 80 pieces (sample size of 1 piece in the LDA plots of Fig. A1), whereas the LDA regression line leads to a slightly different λ∗. Notably, because the number of ie1 foci plus the number of ie2 foci, namely, 20, exceeds the number of ie1&2 foci, namely, 19, the existence of true, albeit rare, “on-on” events (see Discussion) is predicted also on the basis of the LDA.
In conclusion, the analysis has provided firm evidence for a Poisson distribution of transcriptionally active foci in latently infected lungs. Thus, the previous conjecture of a Poisson distribution (32, 33) is now verified.
REFERENCES
- 1.Angulo A, Ghazal P, Messerie M. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J Virol. 2000;74:11129–11136. doi: 10.1128/jvi.74.23.11129-11136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo A, Messerie M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badley J E, Bishop G A, St. John T, Frelinger J A. A simple, rapid method for the purification of poly A + RNA. BioTechniques. 1988;6:114–116. [PubMed] [Google Scholar]
- 4.Balthesen M, Dreher L, Lucin P, Reddehase M J. The establishment of cytomegalovirus latency in organs is not linked to local virus production during primary infection J. Gen Virol. 1994;75:2329–2336. doi: 10.1099/0022-1317-75-9-2329. [DOI] [PubMed] [Google Scholar]
- 5.Balthesen M, Messerle M, Reddehase M J. Lungs are a major organ site of cytomegalovirus latency and recurrence. J Virol. 1993;67:5360–5366. doi: 10.1128/jvi.67.9.5360-5366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 7.Bolovan-Fritts C A, Mocarski E S, Wiedeman J A. Peripheral blood CD14+ cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood. 1999;93:394–398. [PubMed] [Google Scholar]
- 8.Boshart M, Weber F, Jahn G, Dorsch-Häsler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 9.Brautigam A R, Dutko F J, Olding L B, Oldstone M B. Pathogenesis of murine cytomegalovirus infection: the macrophage as a permissive cell for cytomegalovirus infection, replication, and latency. J Gen Virol. 1979;44:349–359. doi: 10.1099/0022-1317-44-2-349. [DOI] [PubMed] [Google Scholar]
- 10.Cardin R D, Abenes G B, Stoddard C A, Mocarski E S. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology. 1995;209:236–241. doi: 10.1006/viro.1995.1249. [DOI] [PubMed] [Google Scholar]
- 11.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke R D. An application of the Poisson distribution. J Inst Actuaries. 1946;72:481. [Google Scholar]
- 13.Dorsch-Häsler K, Keil G M, Weber F, Jasin M, Schaffner W, Koszinowski U H. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc Natl Acad Sci USA. 1985;82:8325–8329. doi: 10.1073/pnas.82.24.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiering S, Whitelaw E, Martin D I K. To be or not to be active: the stochastic nature of enhancer action. Bioessays. 2000;22:381–387. doi: 10.1002/(SICI)1521-1878(200004)22:4<381::AID-BIES8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Ghazal P, Nelson J A. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis. In: Becker Y, Darai G, Huang E S, editors. Molecular aspects of human cytomegalovirus diseases. Heidelberg, Germany: Springer-Verlag Publishers; 1993. pp. 360–383. [Google Scholar]
- 16.Gribaudo G, Riera L, Lembo D, De Andrea M, Gariglio M, Rudge T L, Johnson L F, Landolfo S. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J Virol. 2000;74:4979–4987. doi: 10.1128/jvi.74.11.4979-4987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grzimek N K A, Podlech J, Steffens H-P, Holtappels R, Schmalz S, Reddehase M J. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J Virol. 1999;73:5043–5055. doi: 10.1128/jvi.73.6.5043-5055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry S C, Hamilton J D. Detection of murine cytomegalovirus immediate early 1 transcripts in the spleens of latently infected mice. J Infect Dis. 1993;167:950–954. doi: 10.1093/infdis/167.4.950. [DOI] [PubMed] [Google Scholar]
- 20.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtappels R, Pahl-Seibert M-F, Thomas D, Reddehase M J. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62Llo memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J Virol. 2000;74:11495–11503. doi: 10.1128/jvi.74.24.11495-11503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtappels R, Podlech J, Geginat G, Steffens H-P, Thomas D, Reddehase M J. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan M C, Mar V L. Spontaneous activation of latent cytomegalovirus from murine spleen explants: role of lymphocytes and macrophages in release and replication of virus. J Clin Investig. 1982;70:762–768. doi: 10.1172/JCI110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keil G M, Ebeling-Keil A, Koszinowski U H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J Virol. 1984;50:784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keil G M, Ebeling-Keil A, Koszinowski U H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keil G M, Fibi M R, Koszinowski U H. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J Virol. 1985;54:422–428. doi: 10.1128/jvi.54.2.422-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klotman M E, Henry S C, Greene R C, Brazy P C, Klotman P E, Hamilton J D. Detection of mouse cytomegalovirus nucleic acid in latently infected mice by in vitro enzymatic amplification. J Infect Dis. 1990;161:220–225. doi: 10.1093/infdis/161.2.220. [DOI] [PubMed] [Google Scholar]
- 28.Koffron A J, Hummel M, Patterson B K, Yan S, Kaufman D B, Fryer J P, Stuart F P, Abecassis M I. Cellular localization of latent murine cytomegalovirus. J Virol. 1998;72:95–103. doi: 10.1128/jvi.72.1.95-103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo K, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koszinowski U H, Keil G M, Volkmer H, Fibi M R, Ebeling-Keil A, Münch K. The 89,000-Mr murine cytomegalovirus immediate-early protein activates gene transcription. J Virol. 1986;58:59–66. doi: 10.1128/jvi.58.1.59-66.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurz S K, Rapp M, Steffens H-P, Grzimek N K A, Schmalz S, Reddehase M J. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J Virol. 1999;73:482–494. doi: 10.1128/jvi.73.1.482-494.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurz S K, Reddehase M J. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J Virol. 1999;73:8612–8622. doi: 10.1128/jvi.73.10.8612-8622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurz S K, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefkovits I, Waldman H. Limiting dilution analysis of cells in the immune system. Cambridge, England: Cambridge University Press; 1979. pp. 38–59. [Google Scholar]
- 36.Löser P, Jennings G S, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NF-κB. J Virol. 1998;72:180–190. doi: 10.1128/jvi.72.1.180-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucin P, Pavic I, Polic B, Jonjic S, Koszinowski U H. Gamma interferon-dependent clearance of cytomegalovirus infection in salivary glands. J Virol. 1992;66:1977–1984. doi: 10.1128/jvi.66.4.1977-1984.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maciejewski J P, Bruening E E, Donahue R E, Mocarski E S, Young N S, St Jeor S C. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood. 1992;80:170–178. [PubMed] [Google Scholar]
- 39.Manning W C, Mocarski E S. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent α gene (ie2) is dispensable for growth. Virology. 1988;167:477–484. [PubMed] [Google Scholar]
- 40.Manning W C, Stoddart C A, Lagenaur L A, Abenes G B, Mocarski E S. Cytomegalovirus determinant of replication in salivary glands. J Virol. 1992;66:3794–3802. doi: 10.1128/jvi.66.6.3794-3802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meier J L, Stinski M F. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology. 1996;39:331–342. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 42.Mendelson M, Monard S, Sissons J G P, Sinclair J H. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 43.Mercer J A, Wiley C A, Spector D H. Pathogenesis of murine cytomegalovirus infection: identification of infected cells in the spleen during acute and latent infections. J Virol. 1988;62:987–997. doi: 10.1128/jvi.62.3.987-997.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messerle M, Bühler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Messerle M, Keil G M, Koszinowski U H. Structure and expression of murine cytomegalovirus immediate-early gene 2. J Virol. 1991;65:1638–1643. doi: 10.1128/jvi.65.3.1638-1643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minton E J, Tysoe C, Sinclair J H, Sissons J G P. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell B M, Leung A, Stevens J G. Murine cytomegalovirus DNA in peripheral blood of latently infected mice is detectable only in monocytes and polymorphonuclear leukocytes. Virology. 1996;223:198–207. doi: 10.1006/viro.1996.0468. [DOI] [PubMed] [Google Scholar]
- 48.Nelson J A, Groudine M. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol Cell Biol. 1986;6:452–461. doi: 10.1128/mcb.6.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Podlech J, Holtappels R, Pahl-Seibert M-F, Steffens H-P, Reddehase M J. Murine model of interstitial cytomegalovirus pneumonia in syngeneic bone marrow transplantation: persistence of protective pulmonary CD8–T-cell infiltrates after clearance of acute infection. J Virol. 2000;74:7496–7507. doi: 10.1128/jvi.74.16.7496-7507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Lucin P, Jonjic S, Koszinowski U H. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J Exp Med. 1998;21:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollock J L, Presti R M, Paetzold S, Virgin H W., IV Latent murine cytomegalovirus infection in macrophages. Virology. 1997;227:168–179. doi: 10.1006/viro.1996.8303. [DOI] [PubMed] [Google Scholar]
- 52.Pollock J L, Virgin H W., IV Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J Virol. 1995;69:1762–1768. doi: 10.1128/jvi.69.3.1762-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pomeroy C, Hilleren P J, Jordan M C. Latent murine cytomegalovirus DNA in splenic stromal cells of mice. J Virol. 1991;65:3330–3334. doi: 10.1128/jvi.65.6.3330-3334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prösch S, Staak K, Liebenthal J, Stamminger T, Volk H-D, Krüger D H. Stimulation of the human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNFα is mediated via induction of NF-κB. Virology. 1995;208:197–206. doi: 10.1006/viro.1995.1143. [DOI] [PubMed] [Google Scholar]
- 55.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddehase M J. The immunogenicity of human and murine cytomegaloviruses. Curr Opin Immunol. 2000;12:390–396. doi: 10.1016/s0952-7915(00)00106-0. [DOI] [PubMed] [Google Scholar]
- 57.Reddehase M J, Balthesen M, Rapp M, Jonjic S, Pavic I, Koszinowski U H. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J Exp Med. 1994;179:185–193. doi: 10.1084/jem.179.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddehase M J, Weiland F, Münch K, Jonjic S, Lüske A, Koszinowski U H. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985;55:264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riddell S R. Pathogenesis of cytomegalovirus pneumonia in immunocompromised hosts. Semin Respir Infect. 1995;10:199–208. [PubMed] [Google Scholar]
- 60.Roizman B, Sears A E. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–571. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- 61.Salomon N, Perlman D C. Cytomegalovirus pneumonia. Sem Respir Infect. 1999;14:353–358. [PubMed] [Google Scholar]
- 62.Slobedman B, Mocarski E S. Quantitative analysis of latent human cytomegalovirus. J Virol. 1999;73:4806–4812. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith M. Propagation of salivary gland virus of the mouse in tissue cultures. Proc Soc Exp Biol Med. 1954;86:435–440. doi: 10.3181/00379727-86-21123. [DOI] [PubMed] [Google Scholar]
- 64.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 65.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 66.Steffens H-P, Kurz S, Holtappels R, Reddehase M J. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J Virol. 1998;72:1797–1804. doi: 10.1128/jvi.72.3.1797-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 68.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans-activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor-Wiedeman J, Sissons J G P, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 70.White K L, Slobedman B, Mocarski E S. Human cytomegalovirus latency-associated protein pORF94 is dispensable for productive and latent infection. J Virol. 2000;74:9333–9337. doi: 10.1128/jvi.74.19.9333-9337.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuhasz S A, Dissette V B, Cook M L, Stevens J G. Murine cytomegalovirus is present in both chronic active and latent states in persistently infected mice. Virology. 1994;202:272–280. doi: 10.1006/viro.1994.1343. [DOI] [PubMed] [Google Scholar]