Version Changes
Revised. Amendments from Version 1
We have corrected the text to specify that the moth was not sexed on collection.
Abstract
We present a genome assembly from an individual male Asteroscopus sphinx (the Sprawler moth; Arthropoda; Insecta; Lepidoptera; Noctuidae). The genome sequence has a total length of 857.30 megabases. Most of the assembly is scaffolded into 32 chromosomal pseudomolecules, including the Z sex chromosome and a putative B chromosome. The mitochondrial genome has also been assembled and is 15.35 kilobases in length.
Keywords: Asteroscopus sphinx, the Sprawler moth, genome sequence, chromosomal, Lepidoptera
Species taxonomy
Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Amphiesmenoptera; Lepidoptera; Glossata; Neolepidoptera; Heteroneura; Ditrysia; Obtectomera; Noctuoidea; Noctuidae; Amphipyrinae; Asteroscopus; Aste roscopus sphinx Hufnagel, 1766 (NCBI:txid988069).
Background
The genome of the Sprawler, Asteroscopus sphinx, was sequenced as part of the Darwin Tree of Life Project, a collaborative effort to sequence all named eukaryotic species in the Atlantic Archipelago of Britain and Ireland. Here we present a chromosomally complete genome sequence for Asteroscopus sphinx, based on one male specimen from Wytham Woods, Oxfordshire, UK.
Genome sequence report
The genome of an adult Asteroscopus sphinx ( Figure 1) was sequenced using Pacific Biosciences single-molecule HiFi long reads, generating a total of 25.79 Gb (gigabases) from 2.16 million reads, providing approximately 29-fold coverage. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data, which produced 125.57 Gbp from 831.61 million reads, yielding an approximate coverage of 146-fold. Specimen and sequencing information is summarised in Table 1.
Figure 1. Photograph of the Asteroscopus sphinx (ilAstSphi1) specimen used for genome sequencing.
Table 1. Specimen and sequencing data for Asteroscopus sphinx.
| Project information | |||
|---|---|---|---|
| Study title | Asteroscopus sphinx (the sprawler) | ||
| Umbrella BioProject | PRJEB57684 | ||
| Species | Asteroscopus sphinx | ||
| BioSample | SAMEA110451502 | ||
| NCBI taxonomy ID | 988069 | ||
| Specimen information | |||
| Technology | ToLID | BioSample accession | Organism part |
| PacBio long read sequencing | ilAstSphi1 | SAMEA110451677 | thorax |
| Hi-C sequencing | ilAstSphi1 | SAMEA110451677 | thorax |
| RNA sequencing | ilAstSphi1 | SAMEA110451678 | abdomen |
| Sequencing information | |||
| Platform | Run accession | Read count | Base count (Gb) |
| Hi-C Illumina NovaSeq 6000 | ERR10501030 | 8.32e+08 | 125.57 |
| PacBio Sequel IIe | ERR10499364 | 2.16e+06 | 25.79 |
| RNA Illumina NovaSeq 6000 | ERR11641115 | 7.40e+07 | 11.17 |
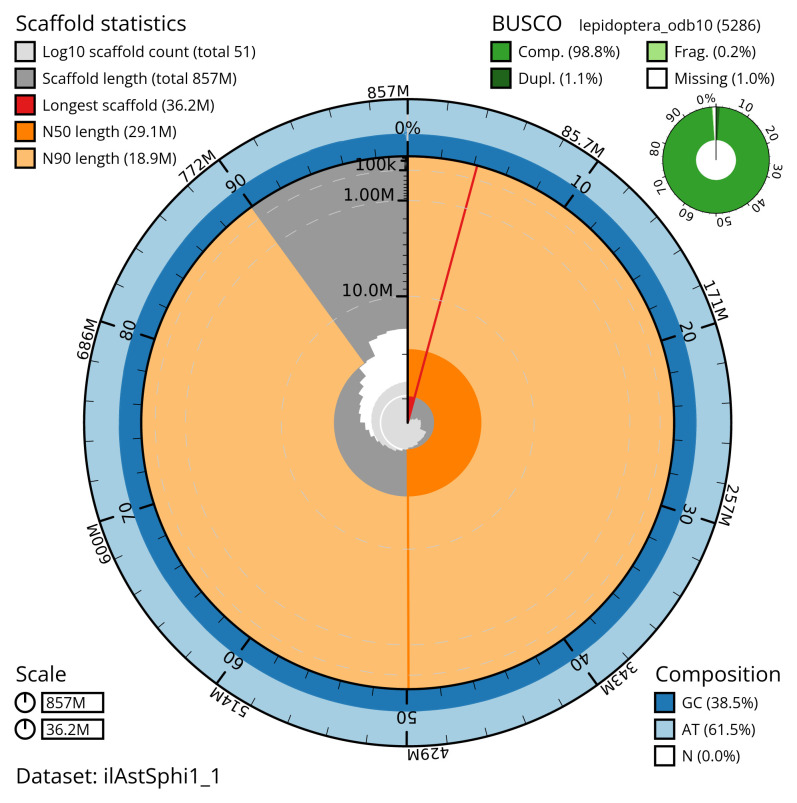
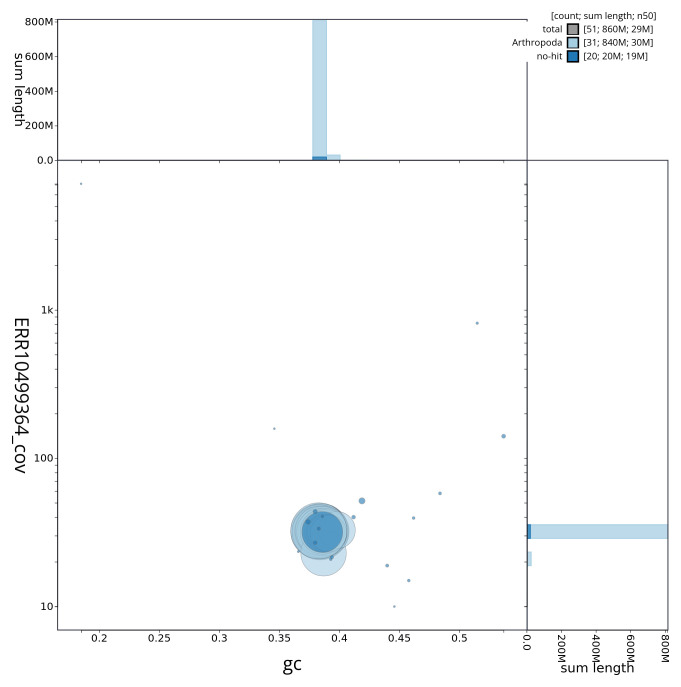
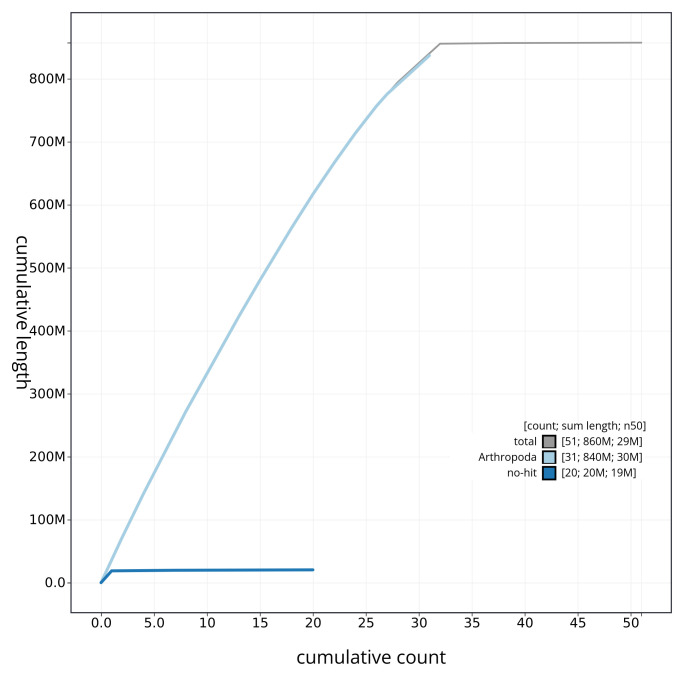
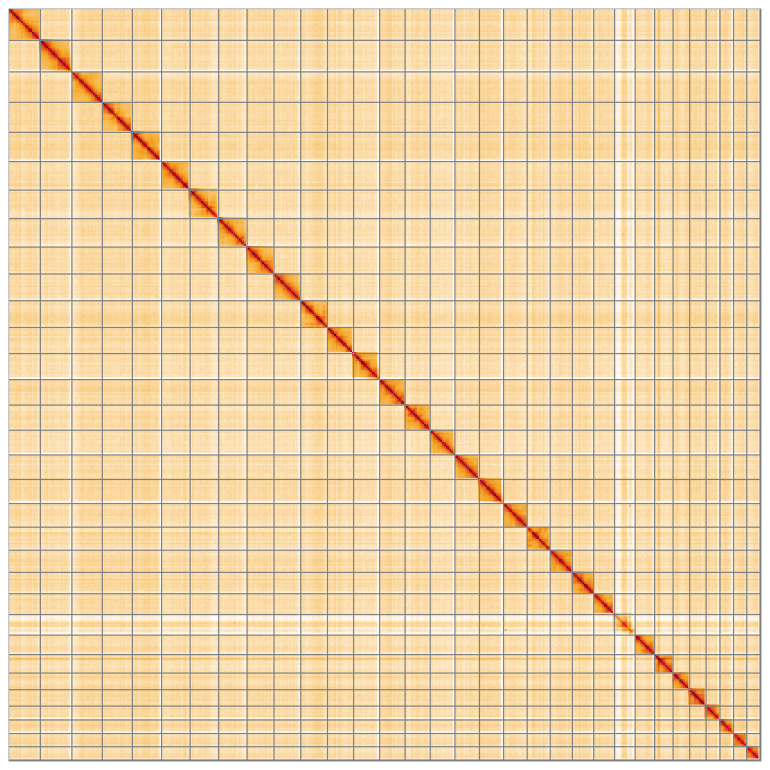
Manual assembly curation corrected 8 missing joins or mis-joins and one haplotypic duplications. The final assembly has a total length of 857.30 Mb in 50 sequence scaffolds with a scaffold N50 of 29.1 Mb ( Table 2). The snail plot in Figure 2 provides a summary of the assembly statistics, while the distribution of assembly scaffolds on GC proportion and coverage is shown in Figure 3. The cumulative assembly plot in Figure 4 shows curves for subsets of scaffolds assigned to different phyla. Most (99.8%) of the assembly sequence was assigned to 32 chromosomal-level scaffolds, representing 31 autosomes and the Z sex chromosome. Chromosome-scale scaffolds confirmed by the Hi-C data are named in order of size ( Figure 5; Table 3). Chromosome 23 is a putative B chromosome ( Fraïsse et al., 2017). While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited. The mitochondrial genome was also assembled and can be found as a contig within the multifasta file of the genome submission.
Figure 2. Genome assembly of Asteroscopus sphinx, ilAstSphi1.1: metrics.
The BlobToolKit snail plot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 857,355,149 bp assembly. The distribution of scaffold lengths is shown in dark grey with the plot radius scaled to the longest scaffold present in the assembly (36,197,790 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 scaffold lengths (29,090,398 and 18,941,627 bp), respectively. The pale grey spiral shows the cumulative scaffold count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the lepidoptera_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilAstSphi1_1/dataset/ilAstSphi1_1/snail.
Figure 3. Genome assembly of Asteroscopus sphinx, ilAstSphi1.1: BlobToolKit GC-coverage plot.
Sequences are coloured by phylum. Circles are sized in proportion to sequence length. Histograms show the distribution of sequence length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilAstSphi1_1/dataset/ilAstSphi1_1/blob.
Figure 4. Genome assembly of Asteroscopus sphinx ilAstSphi1.1: BlobToolKit cumulative sequence plot.
The grey line shows cumulative length for all sequences. Coloured lines show cumulative lengths of sequences assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilAstSphi1_1/dataset/ilAstSphi1_1/cumulative.
Figure 5. Genome assembly of Asteroscopus sphinx ilAstSphi1.1: Hi-C contact map of the ilAstSphi1.1 assembly, visualised using HiGlass.
Chromosomes are shown in order of size from left to right and top to bottom. An interactive version of this figure may be viewed at https://genome-note-higlass.tol.sanger.ac.uk/l/?d=FAkje8YkT_GPw9jqsE-o2Q.
Table 2. Genome assembly data for Asteroscopus sphinx, ilAstSphi1.1.
| Genome assembly | ||
|---|---|---|
| Assembly name | ilAstSphi1.1 | |
| Assembly accession | GCA_949699075.1 | |
| Accession of alternate haplotype | GCA_949699085.1 | |
| Span (Mb) | 857.30 | |
| Number of contigs | 183 | |
| Contig N50 length (Mb) | 9.6 | |
| Number of scaffolds | 50 | |
| Scaffold N50 length (Mb) | 29.1 | |
| Longest scaffold (Mb) | 36.2 | |
| Assembly metrics * | Benchmark | |
| Consensus quality (QV) | 64.9 | ≥ 50 |
| k-mer completeness | 100.0% | ≥ 95% |
| BUSCO ** | C:98.8%[S:97.6%,D:1.1%],F:0.2%,M:1.0%,n:5,286 | C ≥ 95% |
| Percentage of assembly mapped to
chromosomes |
99.8% | ≥ 95% |
| Sex chromosomes | Z | localised homologous pairs |
| Organelles | Mitochondrial genome: 15.35 kb | complete single alleles |
* Assembly metric benchmarks are adapted from column VGP-2020 of “Table 1: Proposed standards and metrics for defining genome assembly quality” from Rhie et al. (2021).
** BUSCO scores based on the lepidoptera_odb10 BUSCO set using version 5.3.2. C = complete [S = single copy, D = duplicated], F = fragmented, M = missing, n = number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/ilAstSphi1_1/dataset/ilAstSphi1_1/busco.
Table 3. Chromosomal pseudomolecules in the genome assembly of Asteroscopus sphinx, ilAstSphi1.
| INSDC accession | Name | Length (Mb) | GC% |
|---|---|---|---|
| OX453001.1 | 1 | 36.2 | 38.5 |
| OX453003.1 | 2 | 34.74 | 38.5 |
| OX453004.1 | 3 | 34.22 | 38.5 |
| OX453005.1 | 4 | 33.19 | 38.5 |
| OX453006.1 | 5 | 32.54 | 38.0 |
| OX453007.1 | 6 | 32.49 | 38.5 |
| OX453008.1 | 7 | 32.4 | 38.5 |
| OX453009.1 | 8 | 30.61 | 38.5 |
| OX453010.1 | 9 | 30.46 | 38.5 |
| OX453011.1 | 10 | 30.38 | 38.5 |
| OX453012.1 | 11 | 29.82 | 38.5 |
| OX453013.1 | 12 | 29.57 | 38.0 |
| OX453014.1 | 13 | 29.09 | 38.5 |
| OX453015.1 | 14 | 28.61 | 38.5 |
| OX453016.1 | 15 | 28.11 | 38.5 |
| OX453017.1 | 16 | 27.96 | 38.5 |
| OX453018.1 | 17 | 27.41 | 38.5 |
| OX453019.1 | 18 | 26.93 | 39.0 |
| OX453020.1 | 19 | 26.12 | 38.5 |
| OX453021.1 | 20 | 25.37 | 38.5 |
| OX453022.1 | 21 | 24.26 | 38.5 |
| OX453023.1 | 22 | 23.75 | 39.0 |
| OX453024.1 | 23 | 23.39 | 38.5 |
| OX453025.1 | 24 | 22.19 | 38.5 |
| OX453026.1 | 25 | 21.04 | 39.0 |
| OX453027.1 | 26 | 18.94 | 38.5 |
| OX453028.1 | 27 | 18.7 | 38.5 |
| OX453029.1 | 28 | 15.79 | 39.0 |
| OX453030.1 | 29 | 15.41 | 40.0 |
| OX453031.1 | 30 | 15.09 | 39.0 |
| OX453032.1 | 31 | 15.04 | 39.0 |
| OX453002.1 | Z | 35.86 | 38.5 |
| OX453033.1 | MT | 0.02 | 19.0 |
The estimated Quality Value (QV) of the final assembly is 64.9 with k-mer completeness of 100.0%, and the assembly has a BUSCO v5.3.2 completeness of 98.8% (single = 97.6%, duplicated = 1.1%), using the lepidoptera_odb10 reference set ( n = 5,286).
Metadata for specimens, BOLD barcode results, spectra estimates, sequencing runs, contaminants and pre-curation assembly statistics are given at https://links.tol.sanger.ac.uk/species/988069.
Methods
Sample acquisition
An adult Asteroscopus sphinx (specimen ID Ox001963, ToLID ilAstSphi1) of unknown sex was collected from Wytham Woods, Oxfordshire, UK (latitude 51.78, longitude –1.33) on 2021-11-03 by potting. The specimen was collected and identified by James Hammond (University of Oxford) and preserved on dry ice.
The initial identification was verified by an additional DNA barcoding process according to the framework developed by Twyford et al. (2024). A small sample was dissected from the specimens and stored in ethanol, while the remaining parts of the specimen were shipped on dry ice to the Wellcome Sanger Institute (WSI). The tissue was lysed, the COI marker region was amplified by PCR, and amplicons were sequenced and compared to the BOLD database, confirming the species identification ( Crowley et al., 2023). Following whole genome sequence generation, the relevant DNA barcode region was also used alongside the initial barcoding data for sample tracking at the WSI ( Twyford et al., 2024). The standard operating procedures for Darwin Tree of Life barcoding have been deposited on protocols.io ( Beasley et al., 2023).
Nucleic acid extraction
The workflow for high molecular weight (HMW) DNA extraction at the Wellcome Sanger Institute (WSI) Tree of Life Core Laboratory includes a sequence of core procedures: sample preparation and homogenisation, DNA extraction, fragmentation, and purification. Detailed protocols are available on protocols.io ( Denton et al., 2023b). In sample preparation, the ilAstSphi1 sample was weighed and dissected on dry ice ( Jay et al., 2023). Tissue from the thorax was homogenised using a PowerMasher II tissue disruptor ( Denton et al., 2023a).
HMW DNA was extracted in the WSI Scientific Operations core using the Automated MagAttract v2 protocol ( Oatley et al., 2023). The DNA was sheared into an average fragment size of 12–20 kb in a Megaruptor 3 system ( Bates et al., 2023). Sheared DNA was purified by solid-phase reversible immobilisation, using AMPure PB beads to eliminate shorter fragments and concentrate the DNA ( Strickland et al., 2023). The concentration of the sheared and purified DNA was assessed using a Nanodrop spectrophotometer and Qubit Fluorometer using the Qubit dsDNA High Sensitivity Assay kit. Fragment size distribution was evaluated by running the sample on the FemtoPulse system.
RNA was extracted from abdomen tissue of ilAstSphi1 in the Tree of Life Laboratory at the WSI using the RNA Extraction: Automated MagMax™ mirVana protocol ( do Amaral et al., 2023). The RNA concentration was assessed using a Nanodrop spectrophotometer and a Qubit Fluorometer using the Qubit RNA Broad-Range Assay kit. Analysis of the integrity of the RNA was done using the Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Sequencing
Pacific Biosciences HiFi circular consensus DNA sequencing libraries were constructed according to the manufacturers’ instructions. Poly(A) RNA-Seq libraries were constructed using the NEB Ultra II RNA Library Prep kit. DNA and RNA sequencing was performed by the Scientific Operations core at the WSI on Pacific Biosciences Sequel IIe (HiFi) and Illumina NovaSeq 6000 (RNA-Seq) instruments. Hi-C data were also generated from thorax tissue of ilAstSphi1 using the Arima-HiC v2 kit. The Hi-C sequencing was performed using paired-end sequencing with a read length of 150 bp on the Illumina NovaSeq 6000 instrument.
Genome assembly, curation and evaluation
Assembly
The HiFi reads were first assembled using Hifiasm ( Cheng et al., 2021) with the --primary option. Haplotypic duplications were identified and removed using purge_dups ( Guan et al., 2020). The Hi-C reads were mapped to the primary contigs using bwa-mem2 ( Vasimuddin et al., 2019). The contigs were further scaffolded using the provided Hi-C data ( Rao et al., 2014) in YaHS ( Zhou et al., 2023) using the --break option. The scaffolded assemblies were evaluated using Gfastats ( Formenti et al., 2022), BUSCO ( Manni et al., 2021) and MERQURY.FK ( Rhie et al., 2020).
The mitochondrial genome was assembled using MitoHiFi ( Uliano-Silva et al., 2023), which runs MitoFinder ( Allio et al., 2020) and uses these annotations to select the final mitochondrial contig and to ensure the general quality of the sequence.
Assembly curation
The assembly was decontaminated using the Assembly Screen for Cobionts and Contaminants (ASCC) pipeline (article in preparation). Manual curation was primarily conducted using PretextView ( Harry, 2022), with additional insights provided by JBrowse2 ( Diesh et al., 2023) and HiGlass ( Kerpedjiev et al., 2018). Scaffolds were visually inspected and corrected as described by Howe et al. (2021). Any identified contamination, missed joins, and mis-joins were corrected, and duplicate sequences were tagged and removed. The process is documented at https://gitlab.com/wtsi-grit/rapid-curation (article in preparation).
Evaluation of the final assembly
A Hi-C map for the final assembly was produced using bwa-mem2 ( Vasimuddin et al., 2019) in the Cooler file format ( Abdennur & Mirny, 2020). To assess the assembly metrics, the k-mer completeness and QV consensus quality values were calculated in Merqury ( Rhie et al., 2020). This work was done using the “sanger-tol/readmapping” ( Surana et al., 2023a) and “sanger-tol/genomenote” ( Surana et al., 2023b) pipelines. The genome readmapping pipelines were developed using the nf-core tooling ( Ewels et al., 2020), use MultiQC ( Ewels et al., 2016), and make extensive use of the Conda package manager, the Bioconda initiative ( Grüning et al., 2018), the Biocontainers infrastructure ( da Veiga Leprevost et al., 2017), and the Docker ( Merkel, 2014) and Singularity ( Kurtzer et al., 2017) containerisation solutions. The genome was also analysed within the BlobToolKit environment ( Challis et al., 2020) and BUSCO scores ( Manni et al., 2021; Simão et al., 2015) were calculated.
Table 4 contains a list of relevant software tool versions and sources.
Table 4. Software tools: versions and sources.
| Software tool | Version | Source |
|---|---|---|
| BlobToolKit | 4.2.1 | https://github.com/blobtoolkit/blobtoolkit |
| BUSCO | 5.3.2 | https://gitlab.com/ezlab/busco |
| bwa-mem2 | 2.2.1 | https://github.com/bwa-mem2/bwa-mem2 |
| Gfastats | 1.3.6 | https://github.com/vgl-hub/gfastats |
| Hifiasm | 0.16.1-r375 | https://github.com/chhylp123/hifiasm |
| HiGlass | 1.11.6 | https://github.com/higlass/higlass |
| Merqury | MerquryFK | https://github.com/thegenemyers/MERQURY.FK |
| MitoHiFi | 2 | https://github.com/marcelauliano/MitoHiFi |
| PretextView | 0.2 | https://github.com/wtsi-hpag/PretextView |
| purge_dups | 1.2.3 | https://github.com/dfguan/purge_dups |
| sanger-tol/ascc | - | https://github.com/sanger-tol/ascc |
| sanger-tol/genomenote | v1.0 | https://github.com/sanger-tol/genomenote |
| sanger-tol/readmapping | 1.1.0 | https://github.com/sanger-tol/readmapping/tree/1.1.0 |
| YaHS | 1.2a | https://github.com/c-zhou/yahs |
Wellcome Sanger Institute – Legal and Governance
The materials that have contributed to this genome note have been supplied by a Darwin Tree of Life Partner. The submission of materials by a Darwin Tree of Life Partner is subject to the ‘Darwin Tree of Life Project Sampling Code of Practice’, which can be found in full on the Darwin Tree of Life website here. By agreeing with and signing up to the Sampling Code of Practice, the Darwin Tree of Life Partner agrees they will meet the legal and ethical requirements and standards set out within this document in respect of all samples acquired for, and supplied to, the Darwin Tree of Life Project.
Further, the Wellcome Sanger Institute employs a process whereby due diligence is carried out proportionate to the nature of the materials themselves, and the circumstances under which they have been/are to be collected and provided for use. The purpose of this is to address and mitigate any potential legal and/or ethical implications of receipt and use of the materials as part of the research project, and to ensure that in doing so we align with best practice wherever possible. The overarching areas of consideration are:
• Ethical review of provenance and sourcing of the material
• Legality of collection, transfer and use (national and international)
Each transfer of samples is further undertaken according to a Research Collaboration Agreement or Material Transfer Agreement entered into by the Darwin Tree of Life Partner, Genome Research Limited (operating as the Wellcome Sanger Institute), and in some circumstances other Darwin Tree of Life collaborators.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute [206194, <a href=https://doi.org/10.35802/206194>https://doi.org/10.35802/206194</a>] and the Darwin Tree of Life Discretionary Award [218328, <a href=https://doi.org/10.35802/218328>https://doi.org/10.35802/218328 </a>].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 4 approved]
Data availability
European Nucleotide Archive: Asteroscopus sphinx (the sprawler). Accession number PRJEB57684; https://identifiers.org/ena.embl/PRJEB57684 ( Wellcome Sanger Institute, 2023). The genome sequence is released openly for reuse. The Asteroscopus sphinx genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. The genome will be annotated using available RNA-Seq data and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1 and Table 2.
Author information
Members of the University of Oxford and Wytham Woods Genome Acquisition Lab are listed here: https://doi.org/10.5281/zenodo.12157525.
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.12158331
Members of the Wellcome Sanger Institute Tree of Life Management, Samples and Laboratory team are listed here: https://doi.org/10.5281/zenodo.12162482.
Members of Wellcome Sanger Institute Scientific Operations: Sequencing Operations are listed here: https://doi.org/10.5281/zenodo.12165051.
Members of the Wellcome Sanger Institute Tree of Life Core Informatics team are listed here: https://doi.org/10.5281/zenodo.12160324.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.12205391.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783558.
References
- Abdennur N, Mirny LA: Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36(1):311–316. 10.1093/bioinformatics/btz540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, et al. : MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 2020;20(4):892–905. 10.1111/1755-0998.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates A, Clayton-Lucey I, Howard C: Sanger Tree of Life HMW DNA fragmentation: diagenode Megaruptor ®3 for LI PacBio. protocols.io. 2023. 10.17504/protocols.io.81wgbxzq3lpk/v1 [DOI] [Google Scholar]
- Beasley J, Uhl R, Forrest LL, et al. : DNA barcoding SOPs for the Darwin Tree of Life project. protocols.io. 2023; [Accessed 25 June 2024]. 10.17504/protocols.io.261ged91jv47/v1 [DOI] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit – interactive quality assessment of genome assemblies. G3 (Bethesda). 2020;10(4):1361–1374. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18(2):170–175. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley L, Allen H, Barnes I, et al. : A sampling strategy for genome sequencing the British terrestrial arthropod fauna [version 1; peer review: 2 approved]. Wellcome Open Res. 2023;8:123. 10.12688/wellcomeopenres.18925.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Veiga Leprevost F, Grüning BA, Aflitos SA, et al. : BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics. 2017;33(16):2580–2582. 10.1093/bioinformatics/btx192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton A, Oatley G, Cornwell C, et al. : Sanger Tree of Life sample homogenisation: PowerMash. protocols.io. 2023a. 10.17504/protocols.io.5qpvo3r19v4o/v1 [DOI] [Google Scholar]
- Denton A, Yatsenko H, Jay J, et al. : Sanger Tree of Life wet laboratory protocol collection V.1. protocols.io. 2023b. 10.17504/protocols.io.8epv5xxy6g1b/v1 [DOI] [Google Scholar]
- Diesh C, Stevens GJ, Xie P, et al. : JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biol. 2023;24(1): 74. 10.1186/s13059-023-02914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral RJV, Bates A, Denton A, et al. : Sanger Tree of Life RNA extraction: automated MagMax TM mirVana. protocols.io. 2023. 10.17504/protocols.io.6qpvr36n3vmk/v1 [DOI] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, et al. : MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels PA, Peltzer A, Fillinger S, et al. : The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38(3):276–278. 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- Formenti G, Abueg L, Brajuka A, et al. : Gfastats: conversion, evaluation and manipulation of genome sequences using assembly graphs. Bioinformatics. 2022;38(17):4214–4216. 10.1093/bioinformatics/btac460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraïsse C, Picard MAL, Vicoso B: The deep conservation of the Lepidoptera Z chromosome suggests a non-canonical origin of the W. Nat Commun. 2017;8(1): 1486. 10.1038/s41467-017-01663-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B, Dale R, Sjödin A, et al. : Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018;15(7):475–476. 10.1038/s41592-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36(9):2896–2898. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E: PretextView (Paired REad TEXTure Viewer): a desktop application for viewing pretext contact maps.2022; [Accessed 19 October 2022]. Reference Source
- Howe K, Chow W, Collins J, et al. : Significantly improving the quality of genome assemblies through curation. GigaScience. 2021;10(1): giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay J, Yatsenko H, Narváez-Gómez JP, et al. : Sanger Tree of Life sample preparation: triage and dissection. protocols.io. 2023. 10.17504/protocols.io.x54v9prmqg3e/v1 [DOI] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19(1): 125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzer GM, Sochat V, Bauer MW: Singularity: scientific containers for mobility of compute. PLoS One. 2017;12(5): e0177459. 10.1371/journal.pone.0177459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. : BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021;38(10):4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel D: Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014;2014(239): 2, [Accessed 2 April 2024]. Reference Source [Google Scholar]
- Oatley G, Denton A, Howard C: Sanger Tree of Life HMW DNA extraction: automated MagAttract v.2. protocols.io. 2023. 10.17504/protocols.io.kxygx3y4dg8j/v1 [DOI] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592(7856):737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, et al. : Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21(1): 245. 10.1186/s13059-020-02134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Strickland M, Cornwell C, Howard C: Sanger Tree of Life fragmented DNA clean up: manual SPRI. protocols.io. 2023. 10.17504/protocols.io.kxygx3y1dg8j/v1 [DOI] [Google Scholar]
- Surana P, Muffato M, Qi G: sanger-tol/readmapping: sanger-tol/readmapping v1.1.0 - Hebridean Black (1.1.0). Zenodo. 2023a. 10.5281/zenodo.7755669 [DOI] [Google Scholar]
- Surana P, Muffato M, Sadasivan Baby C: sanger-tol/genomenote (v1.0.dev). Zenodo. 2023b. 10.5281/zenodo.6785935 [DOI] [Google Scholar]
- Twyford AD, Beasley J, Barnes I, et al. : A DNA barcoding framework for taxonomic verification in the Darwin Tree of Life project [version 1; peer review: awaiting peer review]. Wellcome Open Res. 2024;9:339. 10.12688/wellcomeopenres.21143.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uliano-Silva M, Ferreira JGRN, Krasheninnikova K, et al. : MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinformatics. 2023;24(1): 288. 10.1186/s12859-023-05385-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasimuddin M, Misra S, Li H, et al. : Efficient architecture-aware acceleration of BWA-MEM for multicore systems.In: 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS).IEEE,2019;314–324. 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- Wellcome Sanger Institute: The genome sequence of the Sprawler moth, Asteroscopus sphinx Hufnagel, 1766. European Nucleotide Archive.[dataset], accession number PRJEB57684,2023.
- Zhou C, McCarthy SA, Durbin R: YaHS: yet another Hi-C scaffolding tool. Bioinformatics. 2023;39(1): btac808. 10.1093/bioinformatics/btac808 [DOI] [PMC free article] [PubMed] [Google Scholar]