Abstract
The current classification of parvoviruses is based on virus host range and helper virus dependence, while little data on evolutionary relationships among viruses are available. We identified and analyzed 472 sequences of parvoviruses, among which there were (virtually) full-length genomes of all 41 viruses currently recognized as individual species within the family Parvoviridae. Our phylogenetic analysis of full-length genomes as well as open reading frames distinguished three evolutionary groups of parvoviruses from vertebrates: (i) the human helper-dependent adeno-associated virus (AAV) serotypes 1 to 6 and the autonomous avian parvoviruses; (ii) the bovine, chipmunk, and autonomous primate parvoviruses, including human viruses B19 and V9; and (iii) the parvoviruses from rodents (except for chipmunks), carnivores, and pigs. Each of these three evolutionary groups could be further subdivided, reflecting both virus-host coevolution and multiple cross-species transmissions in the evolutionary history of parvoviruses. No parvoviruses from invertebrates clustered with vertebrate parvoviruses. Our analysis provided evidence for negative selection among parvoviruses, the independent evolution of their genes, and recombination among parvoviruses from rodents. The topology of the phylogenetic tree of autonomous human and simian parvoviruses matched exactly the topology of the primate family tree, as based on the analysis of primate mitochondrial DNA. Viruses belonging to the AAV group were not evolutionarily linked to other primate parvoviruses but were linked to the parvoviruses of birds. The two lineages of human parvoviruses may have resulted from independent ancient zoonotic infections. Our results provide an argument for reclassification of Parvovirinae based on evolutionary relationships among viruses.
The virus family Parvoviridae comprises small animal viruses with linear single-stranded DNA genomes. The genomes of parvoviruses are about 5 kb in length and contain two large open reading frames (ORFs). The first codes for two nonstructural proteins, NS-1 and NS-2, while the second encodes coat proteins VP-1 to VP-3 (or two of them), which have substantial amino acid identity, being derived from overlapping reading frames (for a review, see reference 12).
As now classified, the family Parvoviridae contains two subfamilies: the Parvovirinae, or viruses from vertebrates, and the Densovirinae, or viruses from insects and (tentatively) other arthropods (62). The subfamily Parvovirinae contains three genera: Parvovirus, comprising most parvoviruses from vertebrates; Erythrovirus, comprising B19 and V9 parvoviruses as well as parvoviruses from rhesus and pig-tailed macaques, and Dependovirus, which comprises adeno-associated viruses (AAV). The last two genera include human viruses: the B19 and V9 parvoviruses (Erythrovirus) and AAV serotypes 1 to 6 (Dependovirus). Within Densovirinae, four genera are recognized. The current classification of parvoviruses is based primarily on their host range and their dependence on help from other viruses for replication, according to the traditional separation of parvoviruses into three types: (i) autonomous viruses of vertebrates, (ii) helper-dependent viruses of vertebrates, and (iii) autonomous viruses of insects (62).
The relationships among parvoviruses have been extensively studied by using serological methods as well as DNA hybridization and restriction mapping analyses. A relatively high sequence homology of goose and Muscovy duck autonomous parvoviruses (GPV and MDPV, respectively) with helper-dependent AAV-2, but not with other autonomous parvoviruses, has been documented (18, 68). Although direct data on sequence homology of GPV and MDPV to AAV serotypes other than AAV-2 were not available, DNA cross hybridization data have suggested that GPV is even more similar to AAV-1 and AAV-3 (18). On the other hand, little similarity between the two groups of human parvoviruses, B19 and AAV, has been observed (68). Feline panleukopenia virus, canine parvovirus, and mink enteritis virus (MEV) are highly homologous and classified as host range variants of the feline parvovirus (FelinePV) (reviewed in reference 46), but another mink parvovirus, the Aleutian mink disease virus (AMDV), has little homology with MEV (14). These observations prompted the suggestion that the original hypothesis of a host-dependent evolution of parvoviruses (8) may have limited value both within and among genera (49, 68). Furthermore, autonomous parvoviruses are dependent on helper functions that are transiently expressed in host cells, and helper viruses can substantially increase their replication, while helper-dependent viruses can replicate autonomously under certain conditions (12). These observations together with genetic homology between some autonomous and helper-dependent viruses resulted in the validity of the other main criterion for classification of parvoviruses, dependence on helper viruses, also being questioned (18, 68). The distinction that autonomous parvoviruses encapsidate primarily DNA strands that are complementary to mRNA, whereas AAV encapsidate strands of either polarity with equal frequency, is also far from absolute. For instance, bovine parvovirus encapsidates up to 30% of DNA strands with the same polarity as that of mRNA. In certain hosts, the autonomous parvovirus LuIII encapsidates strands with either polarity in equal measure (12).
Over the last decade, a massive amount of genetic information has been obtained for various virus groups. For several groups, including the human immunodeficiency viruses (HIV) and hepatitis C viruses, genetic classifications that reflect evolutionary relationships have been developed (38, 39, 57). However, no systematic and explicit study on evolutionary relationships among Parvoviridae has yet been performed, although (virtually) full-length genomes of several members of each of the recognized genera are available. Such a study is essential to elucidate the principal issues of parvovirus biology, including the evolutionary relationships both among and within subfamilies and genera, the driving forces of parvovirus evolution, and possible cross-species transmissions. In the present study, we address these basic issues and analyze currently available sequence information on parvoviruses by using phylogenetic methods.
MATERIALS AND METHODS
Sequences.
In the GenBank, we identified 472 sequences of parvoviruses for use in this study. They were retrieved by using Batch Entrez software, which allows a search for sequences belonging to a specified organism (http://www.ncbi.nlm.nih.gov/Entrez/batch.html). We specified Parvoviridae as the organism name, according to the taxonomy database at the National Center for Biotechnology Information, and performed an additional search for Parvovirus. We used the classification of parvoviruses accepted by the International Committee on Taxonomy of Viruses (http://www.ncbi.nlm.nih.gov/htbin-post/Taxonomy/). The viruses we studied are referred to by their descriptive name (e.g., FelinePV) or trivial name (e.g., B19), as it is used in the nomenclature of parvoviruses (62). The sequences are referred to by their GenBank accession numbers, and the reference information is provided in Table 1.
TABLE 1.
Virus sequences used in this studya
| Virus species | Abbreviated name(s) | Accession no. | Referenceb |
|---|---|---|---|
| Parvovirinae | |||
| Parvovirus | |||
| Bovine parvovirus | BovinePV, BPV | M14363 | 20 |
| Canine parvovirus | CaninePV, CPV | M19296 | 51 |
| M38245 | 47 | ||
| D26079 | 34 | ||
| Minute virus of mice | MVM | J02275 | 3 |
| X02481 | 54 | ||
| V01115 | 3 | ||
| M12032 | 2 | ||
| U34256 | 13 | ||
| Mouse parvovirus 1 | MousePV-1 | U12469 | 5 |
| Mouse parvovirus 1b | MousePV-1b | U34253 | 13 |
| Mouse parvovirus 1c | MousePV-1c | U34254 | 13 |
| LuIII | M81888 | 24 | |
| Barbarie duck parvovirus | BarbduckPV, BDPV | U22967 | 68 |
| Feline panleukopenia virus | FelinePV, FPV, FPLV | M38246 | 47 |
| X55115 | 44 | ||
| M10824 | 19 | ||
| Goose parvovirus | GoosePV, GPV | U25749 | 68 |
| Mink enteritis virus | MEV | D00765 | 36 |
| Porcine parvovirus | PorcinePV, PPV | L23427 | 11 |
| M38367 | 64 | ||
| D00623 | 50 | ||
| M32787(orf2) | 63 | ||
| U44978 | 10 | ||
| Rat parvovirus 1a | RatPV-1a | AF036710 | 6 |
| Kilham rat virus | KilhamRatPV | U79033 | Brown and Like, unpub., 1996 |
| AF036711 | 6 | ||
| Aleutian mink disease parvovirus | AMDV | M20036 | 14 |
| X97629 | 56 | ||
| Z18276 | Perryman et al., unpub., 1992 | ||
| Hamster parvovirus | HamsterPV | U34255 | 13 |
| Muscovy duck parvovirus | MuscduckPV, MDPV | X75093 | 68 |
| Raccoon parvovirus, Georgia raccoon virus | RaccoonPV | M24005 (partial) | 48 |
| Parvovirus H1 | H1 | X01457 | 52 |
| Simian parvovirus from cynomolgus monkeys (long-tailed macaque) | LTMPV | U26342 | 17 |
| Chipmunk parvovirus | ChipmunkPV | U86868 | 67 |
| Erythrovirus | |||
| B19 virus | AF162273 | Gallinella and Venturoli, unpub., 1999 | |
| AF161226 | 33 | ||
| AF161225 | 33 | ||
| AF161224 | 33 | ||
| AF161223 | 33 | ||
| AF113323 | 31 | ||
| AB030694 | Ishii et al., unpub., 1999 | ||
| AB030693 | Ishii et al., unpub., 1999 | ||
| AB030673 | 61 | ||
| Z70599 | 32 | ||
| Z70560 | 32 | ||
| M24682 | 15 | ||
| V9 virus | AX003421 | 4 | |
| Rhesus macaque parvovirus | RMPV | AF221122 | 27 |
| Pig-tailed macaque parvovirus | PTMPV | AF221123 | 27 |
| Dependovirus | |||
| Adeno-associated virus serotype 1 | AAV-1 | AF063497 | 65 |
| Adeno-associated virus serotype 2 | AAV-2 | J01901 | 55 |
| Adeno-associated virus serotype 3 | AAV-3 | U48704 | 45 |
| Adeno-associated virus serotype 3B | AAV-3b | AF028705 | 53 |
| Adeno-associated virus serotype 4 | AAV-4 | U89790 | 22 |
| Adeno-associated virus serotype 5 | AAV-5 | AF085716 | 21 |
| Adeno-associated virus serotype 6 | AAV-6 | AF028704 | 53 |
| Densovirinae | |||
| Junonia coenia densovirus | JcDNV | S47266 | 25 |
| Galleria mellonella densovirus | GmDNV | L32896 | 58 |
| Bombyx mori densovirus | BmDNV | AB042597 | Nonaka et al., unpub., 2000 |
| Aedes albopictus parvovirus | AaPV | X74945 | 16 |
| Aedes aegypti densonucleosis virus | AedesDNV | M37899 | 1 |
| Periplaneta filiginosa densovirus | PfDNV | AF192260 | Guo et al., unpub., 1999 |
| AB028936 | 66 | ||
| Diatraea saccharalis densoviruses | DsDNV | AF036333 | Boublik et al., unpub., 1997 |
| Yamanashi isolate from silkworm | S78547 | 7 | |
| Parvovirus-like virus | AB033596 | 29 | |
| Infectious hypodermal and hematopoietic necrosis virus from penaeid shrimps | AF218266 | 43 |
Sequences in boldface are used in Fig. 1.
Unpub., unpublished data.
Sequence analysis.
The BioEdit, version 4.8.6, software (28) was used to manipulate the retrieved sequences. The alignment of sequences was performed by using the ClustalW software (60). For full-length genomes as well as noncoding regions, nucleotide sequences were aligned. For coding regions, the alignment was performed for amino acid sequences.
Phylogenetic analysis was performed by using several methods. For all methods, positions containing an alignment gap were excluded from pairwise sequence comparisons. Bootstrap resampling was performed for each analysis (100 replications). Nucleotide distances were analyzed by using the neighbor-joining algorithm as implemented in the PHYLIP package (NEIGHBOR), based on the Kimura two-parameter distance estimation method or the proportion of differences (p distance). For coding regions, additional analyses of nonsynonymous and synonymous nucleotide substitutions (those which change or do not change the amino acid, respectively) was performed by using the MEGA software (37). Estimation of both synonymous distances (Ds) and nonsynonymous distances (Da) was based on the Nei-Gojobori method (37). The ratios of synonymous to nonsynonymous substitutions (Ds/Da) were calculated (41).
Recombination analysis was performed by using the bootscanning method as implemented in the SimPlot software (available at http://www.med.jhu.edu/deptmed/sray/).
Many viruses are represented in the GenBank by single full-length genome sequences, but more than one sequence are available for several viruses. For B19 virus, we used full-length sequences but also about 200 shorter sequences, typically a few hundred nucleotides in length. These partial genomes were aligned with all full-length genome sequences, and the B19 consensus sequence was calculated as the arithmetic mean of all nucleotides or amino acids at a particular position (39, 42). This consensus sequence was used in the analyses together with the individual full-length genomes.
RESULTS
Identification of phylogenetic groups within Parvovirinae.
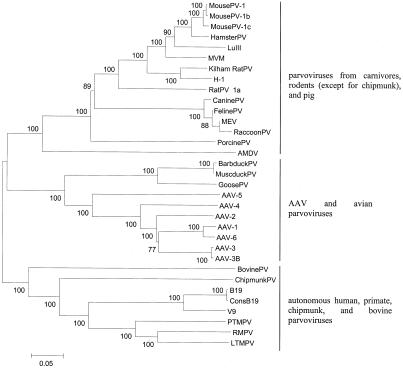
To identify groups of phylogenetically related viruses within Parvovirinae, we analyzed (virtually) full-length genomes of all members of the three genera that are distinguished by the International Committee on Taxonomy of Viruses as distinct virus species. Parvovirus species included were bovine, simian (from the cynomolgus [long-tailed] macaque), Manchurian chipmunk, canine, feline panleukopenia, Georgian raccoon (only a partial sequence of 2,410 nucleotides in length is available), porcine, mice minute, mouse 1, mouse 1b, mouse 1c, rat 1a, Kilham rat, hamster, LuIII, Barbarie duck, and H1 parvoviruses as well as MEV, AMDV, GPV, and MDPV. Erythrovirus species included an individual B19 virus and the consensus of 215 B19 sequences and V9 and rhesus and pig-tailed macaque parvoviruses. Dependovirus species included AAV serotypes 1 to 6. The list of sequences analyzed is provided in Table 1.
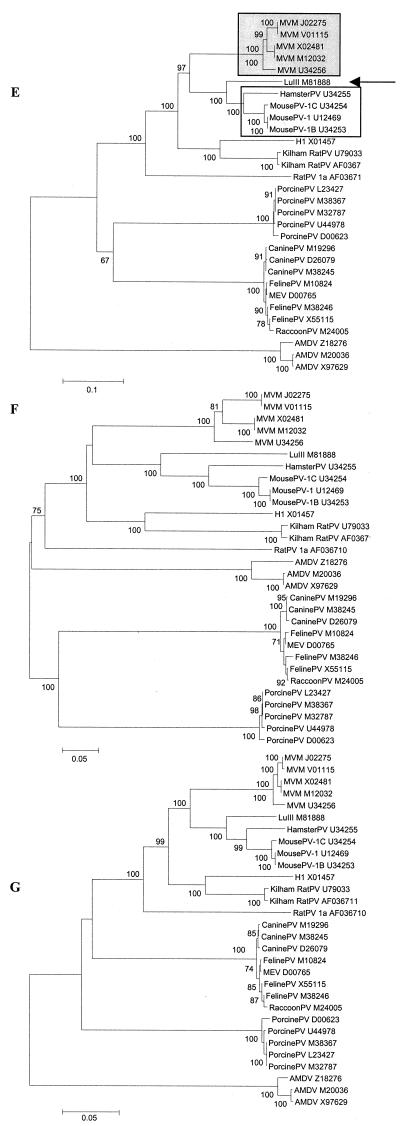
In total, genomic sequences of 32 virus species were aligned. Based on phylogenetic analysis, they fell into three groups (Fig. 1): (i) AAV serotypes 1 to 6 and GPV, Barbarie duck parvovirus, and MDPV; (ii) primate (B19, V9, and three viruses from macaques), chipmunk, and bovine parvoviruses; (iii) parvoviruses from all rodents (except for chipmunks), carnivores, and pigs.
FIG. 1.
The three evolutionary groups of Parvovirinae. The neighbor-joining phylogenetic tree is based on the analysis of (virtually) full-length genomes of all members of the Parvovirinae subfamily that are recognized as individual virus species, one sequence per species (except for the B19 virus, for which a consensus, ConsB19, of 215 available sequences is also included). For RaccoonPV, only a shorter sequence is available. Bootstrap values are shown (100 replications). Sequences used in this analysis are in boldface in Table 1. For virus abbreviations, see Table 1.
Additionally, we analyzed viruses from the Densovirinae subfamily (Table 1). None of viruses from the Densovirinae clustered together with Parvovirinae (data not shown).
AAV and avian parvoviruses.
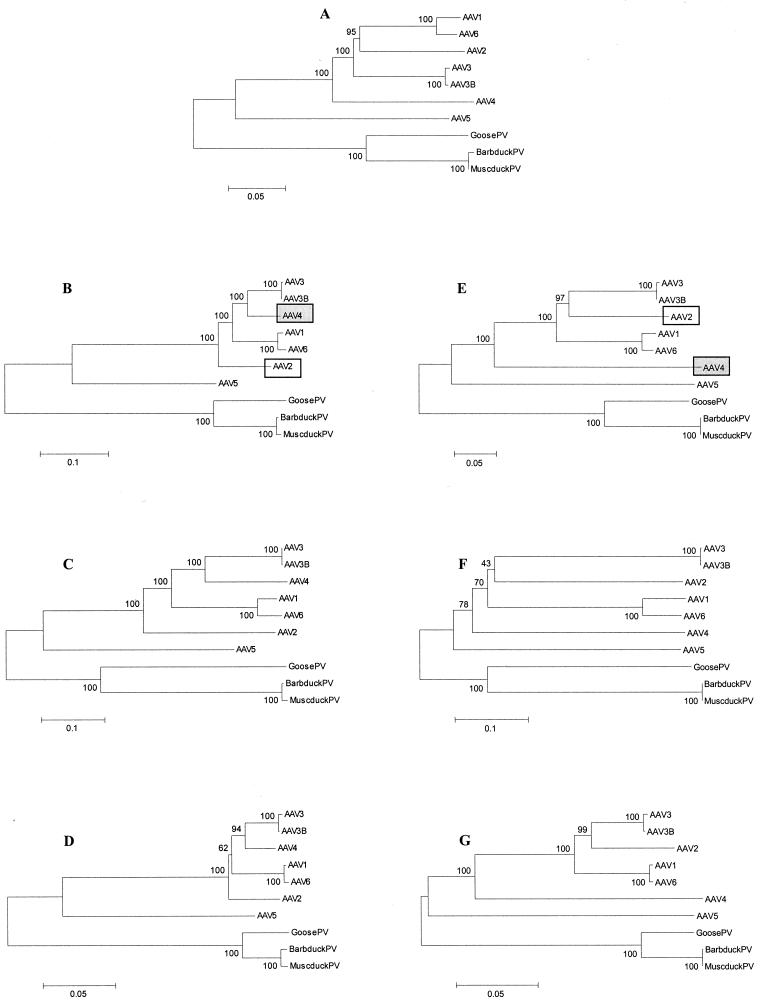
To analyze phylogenetic relationships among AAV and avian parvoviruses, we aligned sequences of viruses belonging to this phylogenetic group. The analyses were based on nucleotide distances as well as, for coding regions, Ds and Da.
Irrespective of the phylogenetic model and genomic region used, the three avian parvoviruses clustered together and separately from AAV, with a bootstrap value of 100 (Fig. 2). The two viruses from ducks were virtually identical, with their Ds and Da being 0.01 for orf1 and 0.00 for orf2. Among AAV, two pairs of closely related viruses were found. Besides the two sequences belonging to viruses from the same serotype (AAV-3 and AAV-3B), AAV-1 and AAV-6 also clustered together. Although these two viruses are defined as separate AAV serotypes, the vast majority of nucleotide substitutions between AAV-1 and AAV-6 are synonymous, with Ds being 0.07 and 0.11 for orf1 and orf2, respectively, and Da being 0.00 for both ORFs. AAV-3 and AAV-3B, AAV-1 and AAV-6, and AAV-2 were approximately equidistant from each other as well as from AAV-5, which appeared to be the most distantly related to other AAV (Fig. 2). Within this virus group, branching orders of two viruses, AAV-2 and AAV-4, varied with the genetic region analyzed. The orf1 sequence of AAV-4 clustered together with AAV-3 and AAV-3B, AAV-1 and AAV-6, and AAV-2 and was most closely related to AAV-3 (Fig. 2B to D), whereas the orf2 sequence of AAV-4 branched out between AAV-5 and the main cluster of AAV (Fig. 2E to G). The position of AAV-2 (within or outside the AAV-3 and AAV-3B and AAV-1 and AAV-6 clusters) also depended upon the genetic region (Fig. 2).
FIG. 2.
Phylogenetic relationships among the AAV serotypes 1 to 6 and parvoviruses from GPV, Barbarie duck parvovirus (BarbduckPV), and MDPV (MuscduckPV). Bootstrap values are shown (100 replications). (A) Relationships based on nucleotide p distances among full-length genome sequences; (B to D) relationships based on nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf1; (E to G) nucleotide distances, Ds, and Da, respectively, for orf2. For panels B and E, positions of AAV-2 and AAV-4 are marked. Virus abbreviations are listed in Table 1.
For all pairwise sequence comparisons, the Ds/Da ratios were markedly higher than 1. The Da between any two sequences did not exceed 0.39 (AAV-1 and AAV-6 versus avian parvoviruses; orf1), but the vast majority of pairwise Ds were higher (Fig. 2D and G versus C and F). Remarkable differences between the Ds and Da were observed for GPV versus duck parvoviruses, for which the Ds were 0.58 to 0.59, compared to Da of 0.06 to 0.07. Among AAV, the most remarkable differences between Ds and Da were observed for the comparisons of AAV-2 and AAV-4 in orf1, 0.44 versus 0.07, and AAV-1 and AAV-6 and AAV-3 and AAV-3B in orf 2, 0.52 to 0.57 versus 0.08 to 0.09 (Fig. 2).
Autonomous primate, chipmunk, and bovine parvoviruses.
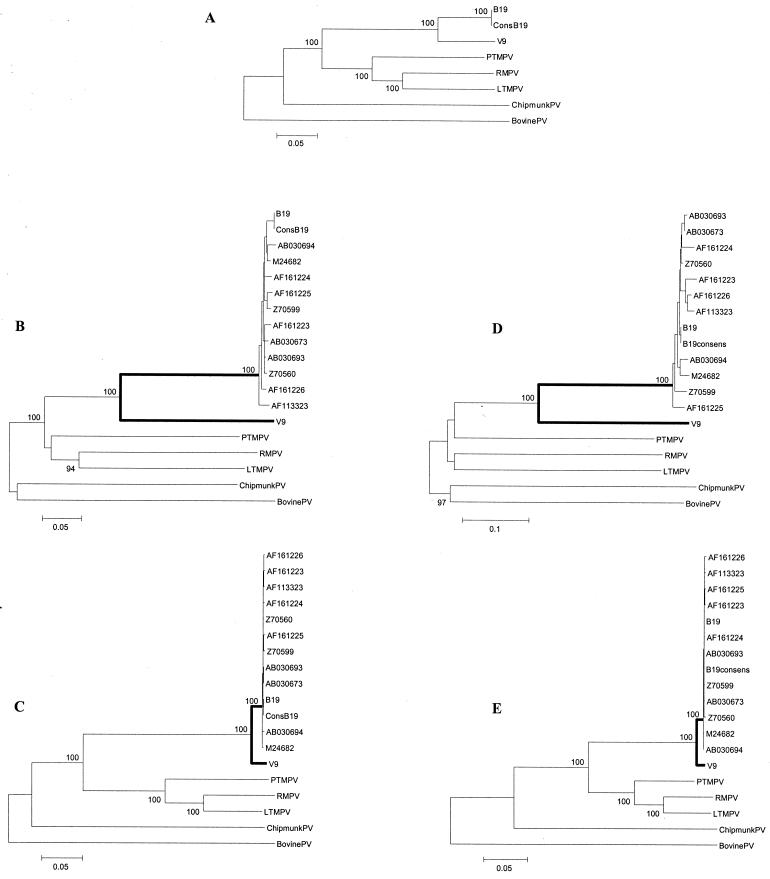
In addition to a single B19 individual sequence and the B19 consensus, we analyzed all 12 individual B19 sequences for which both orf1 and orf2 regions are available.
For this group of viruses, topologies of phylogenetic trees were virtually identical when based on full-length sequence analysis or Da in orf1 and orf2 (Fig. 3). B19 and V9 clustered together, as did the simian parvoviruses, whereas chipmunk and bovine parvoviruses (ChipmunkPV and BovinePV) were outliers. Among B19 viruses, high genetic homogeneity was observed. Within the two ORFs, the mean Ds among B19 viruses were 0.022 (range, 0.007 to 0.034) and 0.035 (range, 0.007 to 0.054) and the mean Da were 0.003 (range, 0.001 to 0.005) and 0.002 (range, 0.001 to 0.005), resulting in the mean Ds/Da ratios of 7.3 and 17.5 for orf1 and orf2, respectively. Negative selection was even more evident in our comparison of the two human parvoviruses: while the Ds between the V9 sequence and the B19 consensus were 0.40 and 0.45, the Da were 0.03 and 0.02 (Fig. 3B to E), resulting in mean Ds/Da ratios of 13.3 and 22.5 for orf1 and orf2, respectively. Pairwise Ds between human and simian parvoviruses were also markedly higher than Da (Fig. 3). Moreover, phylogenetic analysis of orf2 based on Ds resulted in virtually complete loss of tree structure as B19 and V9, the three macaque viruses, ChipmunkPV, and BovinePV were equidistant from each other (Fig. 3D).
FIG. 3.
Phylogenetic relationships among the autonomous primate, chipmunk, and bovine parvoviruses. In addition to sequences used in Fig. 1, 11 more sequences of B19 are included (labeled by their GenBank accession numbers). Bootstrap values above 70 are shown (100 replications). (A) Relationships based on nucleotide p distances among full-length genome sequences; (B and C) relationships based on Ds and Da, respectively, for orf1; (D and E) relationships based on Ds and Da, respectively, for orf2. Branches between the B19 cluster and V9 are in boldface (B to E). Virus abbreviations are in Table 1.
Parvoviruses from rodents, carnivores, and pigs.
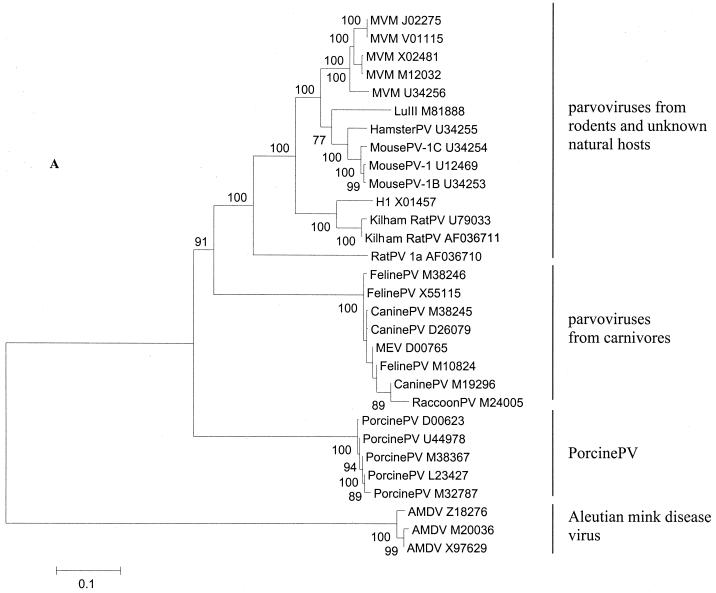
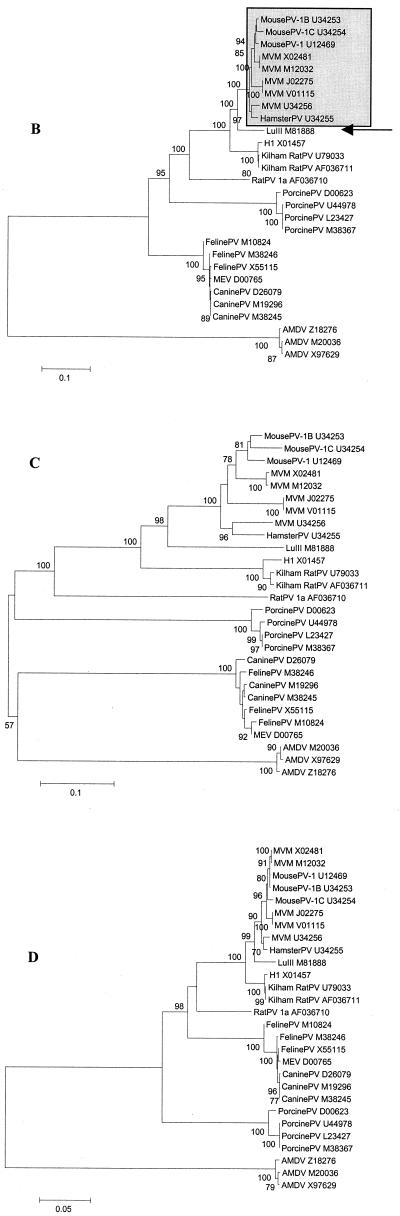
For several viruses within this evolutionary group, more than one full-length sequence were available, permitting study of genetic heterogeneity within virus species. We included five strains of minute virus of mice (MVM), two Kilham rat parvoviruses (KilhamRatPV), three FelinePV, three canine parvoviruses (CaninePV), five porcine parvoviruses (PorcinePV), and three AMDV sequences in the analysis.
Our analysis distinguished four major subgroups of evolutionarily related viruses: (i) viruses from rodents and unknown natural hosts, (ii) viruses from carnivores, except for AMDV, (iii) PorcinePV, and (iv) AMDV, which was the most distantly related to all other viruses in this group (Fig. 4A). The four subgroups were observed in all phylogenetic trees (Fig. 4), but their branching order varied. Based on Da, viruses of rodents, carnivores, and pigs clustered together and were approximately equidistant from each other (Fig. 4D and G), while AMDV appeared to be an outlier. Based on synonymous distances, all four subgroups were equidistant from each other (Fig. 4C and F).
FIG. 4.
Phylogenetic relationships among parvoviruses from rodents, carnivores, and pigs. The four phylogenetic subgroups are shown. (A) All full-length sequences available for each virus species are included (labeled by their virus names and the GenBank accession numbers). Bootstrap values above 70 are shown (100 replications). (A) relationships based on nucleotide p distances among full-length genome sequences; (B to D) relationships based on the nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf1; (E to G) relationships based on nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf2. In orf1 (B to D), MousePV, HamsterPV, and MVM form a homogeneous cluster (B, grey box), to which LuIII (arrow) is an outlier. In contrast, in orf2 (E to G), MousePV and HamsterPV (E, open box) cluster with LuIII (arrow) and not with MVM (grey box). Virus abbreviations are in Table 1.
Similar to what was observed for AAV, avian parvoviruses, and primate parvoviruses, the mean Ds/Da ratios for pairwise comparisons within groups were above 1 (range: 1.3 to 10.3), except for PorcinePV in orf2, a reflection of its extreme genetic homogeneity (Ds = 0.002, Da = 0.003). The largest Ds/Da ratio was observed for the comparisons of mouse and hamster viruses with LuIII in orf1: 0.30/0.04 (Fig. 4C and D).
Among the rodent viruses, we identified three subclusters, which comprised (i) LuIII and viruses from mice and hamsters, (ii) KilhamRatPV and H1, and (iii) the most distantly related rat parvovirus (RatPV) (Fig. 4). The second subgroup of viruses from various natural hosts, viruses from carnivores, and the two subgroups of viruses from single hosts, PorcinePV and AMDV, were much more homogeneous. Among viruses from carnivores, the FelinePV, raccoon parvovirus (RaccoonPV), and MEV clustered together and separately from CaninePV when Da were analyzed (Fig. 4D and G). This trend was less pronounced for Ds (Fig. 4C and F). Genetic heterogeneity in FelinePV was higher than that in CaninePV. The mean Ds/Da ratios among FelinePV and CaninePV and between these two viruses were 0.016/0.004, 0.008/0.001, and 0.023/0.004, respectively, for orf1 and 0.016/0.004, 0.004/0.002, and 0.023/0.007, respectively, for orf2. Within these three subgroups, both the mean Ds and Da were generally below 0.02.
For the three subclusters of rodent parvoviruses, RatPV was an outlier in all phylogenetic trees. For the other two subclusters, remarkable patterns were identified. For the H1-KilhamRatPV subcluster, we observed a great difference in the evolutionary distances between the two viruses for the two ORFs. Within orf1, the mean Da and Ds between H1 and the two KilhamRatPV were 0.004 and 0.044, respectively, while the Da and Ds within orf2 were 34 and 9 times greater and equal to 0.135 and 0.388, respectively (Fig. 4).
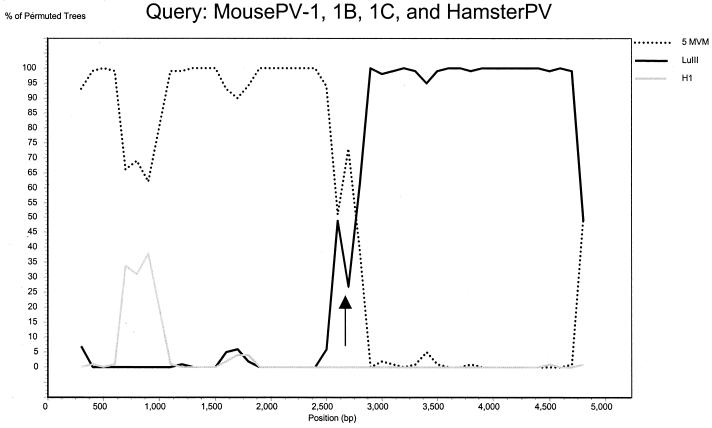
For the mouse-hamster-LuIII subcluster, even more complex relations among viruses were found. In orf1, MVM, mouse parvovirus (MousePV), and hamster parvovirus (HamsterPV) represented an extremely homogeneous (mean Da = 0.01, Ds = 0.11) monophyletic group, to which LuIII was an outlier (Fig. 4B to D; bootstrap value of 100). Within this cluster, sequences belonging to distinct virus species were intermixed (Fig. 4B). In contrast, our analysis of orf2 revealed that MousePV and HamsterPV cluster together with LuIII and are distant (mean Da = 0.18, Ds = 0.56) from MVM (Fig. 4E to G; bootstrap value of 100). In orf2, sequences belonging to all recognized virus species represented monophyletic groups (Fig. 4E to G). Yet no host-related clustering was observed, as sequences of MousePV clustered together with HamsterPV and LuIII and not with sequences of another mouse virus, MVM. The mosaicism of virus genomes within this subcluster was further supported by our analysis of full-length sequences with the bootscanning method (Fig. 5). In this analysis, MousePV-1, -1B, -1C, and HamsterPV (query group) were compared to five full-length sequences of MVM (comparison group 1) and LuIII (comparison group 2), whereas the sequence of H1 was used as an outgroup. While the left part (positions 1 to 2600) of the MousePV and HamsterPV genomes clustered together with MVM, the right part of the genomes clustered together with LuIII and not with MVM (Fig. 5).
FIG. 5.
Bootscan analysis of the phylogenetic relationships among LuIII and parvoviruses from mice and hamsters. The three MousePV and HamsterPV were used as a query sequence group in comparison to the five MVM (comparison group 1), LuIII (comparison group 2), and H1 (outgroup). Analysis settings were as follows: window size, 400 nucleotides; step 100 nucleotides; bootstrap resampling, 100; distance, Kimura two-parameter distance; transitions/transversions ratio, 2. Arrow, recombination site.
DISCUSSION
Currently, the GenBank database contains sequences of about 500 viruses from the Parvoviridae family. The vast majority of them are vertebrate viruses, while for viruses of invertebrates only a dozen sequences are available.
So far, no systematic evolutionary study has been performed on parvoviruses. Typically, earlier studies focused on describing the amino acid identities of a new virus with a few of the most closely related sequences (5, 6, 9, 27, 53, 67, 68). To the best of our knowledge, the only parvoviruses for which evolutionary issues have been specifically addressed by analyzing full-length genomes are FelinePV and CaninePV (35). Due to the lack of systematically analyzed data, genetic information was not used as the basis for parvovirus classification.
In the present study, we used all available sequence information and powerful phylogenetic methods to learn whether the Parvoviridae are evolutionarily related viruses and whether their current classification into subfamilies and genera truly reflects the evolutionary relationships among viruses. Moreover, we attempted to study how and to what degree various evolutionary factors, such as positive or negative selection, recombinations, host-dependent evolution, cross-species transmissions, and the independent evolution of genomic regions, were operational during the evolution of parvoviruses.
Toward the evolutionary classification of Parvovirinae.
Our analysis of the genomes of 32 parvoviruses, all recognized virus species for which (virtually) full-length genome sequences are available, revealed the existence of three groups of evolutionarily related viruses (Fig. 1 to 4): (i) AAV and all three known avian parvoviruses; (ii) all five known autonomous primate parvoviruses, ChipmunkPV, and the outlier BovinePV; (iii) parvoviruses from rodents (except for chipmunks), carnivores, and pigs, with AMDV being an outlier.
These findings indicate that the current classification of viruses within Parvovirinae (62) does not always reflect their evolutionary relationships. The first discrepancy was found for avian parvoviruses, now classified as members of the Parvovirus genus but revealed by our analysis to be linked evolutionarily to AAV rather than to any autonomous parvoviruses (Fig. 1 and 2). Our results concur with an observation on the relatively high homology between GPV, MDPV, and AAV-2 (18, 68) but do not show that GPV is even closer to AAV-1 and AAV-3, an earlier notion based on DNA cross-hybridization data (18). We found that all serotypes of AAV are equidistant from each of the three avian parvoviruses (Fig. 2).
Other discrepancies were found for the simian parvovirus from the long-tailed macaque, ChipmunkPV, and BovinePV. While these three viruses are classified as members of the Parvovirus genus, their evolutionary linkage to all known autonomous primate parvoviruses, and not to any known nonprimate parvoviruses, was revealed in our study (Fig. 1 and 3). Our observations concur with recent data on the genetic homology of primate (27) and chipmunk (67) parvoviruses to B19.
A reliable analysis of phylogenetic relationships among the three identified groups of Parvovirinae (Fig. 1) was obstructed by the high evolutionary distances. The topology of the phylogenetic tree, in which all three groups of Parvovirinae branch out from basically a single phylogenetic node, is likely to reflect the saturation of nucleotide substitutions among the groups. In contrast to intergroup relationships, intragroup relationships could be analyzed in detail.
Another important issue of parvovirus classification is related to the recognition of individual virus species. For several other viruses, such as HIV type 1 (HIV-1), genetic distances among isolates can be higher than 0.3 (38, 39, 41) and biological and immunological characteristics of virus isolates are highly variable (for review, see reference 40). Nevertheless, all HIV-1 strains are considered to belong to the same virus species based on their common evolutionary origin. This principle is used for some parvoviruses but not for others. For example, all five available full-length genome sequences of MVM are considered to be derived from a single species, while the three available genome sequences of MousePV are classified as belonging to three different species, MousePV-1, -1b, and -1c, even though genetic heterogeneity in MVM is actually much higher than in MousePV (Fig. 4). Similarly, the genetic distances among AMDV isolates, which are considered to belong to a single species, are not different from or even higher than those between the two duck parvoviruses or between AAV-3 and -3B or among parvoviruses from carnivores: FelinePV, CaninePV, RaccoonPV, and MEV. The parvoviruses of carnivores have been considered as (host range) variants of a single virus species (46), and our data suggest similar consideration for MousePV-1, -1b, and -1c, the two duck parvoviruses, AAV-3 and -3b, and possibly AAV-1 and -6.
Driving forces of parvovirus evolution.
To study evolutionary forces that are operational among parvoviruses, we analyzed synonymous versus nonsynonymous nucleotide substitutions in the two ORFs. Nonsynonymous substitutions, as they change the amino acids, are generally subjected to strong positive or negative selection pressure. In contrast, synonymous substitutions, which preserve amino acids, are supposed to be subjected to a weaker selection pressure or to none. Since the mutation rates at synonymous and nonsynonymous sites should be the same, Ds and Da, as well as their ratios, indicate the direction and intensity of selection in the evolutionary history of a group of species. For instance, the Ds/Da ratios among HIV-1 polymerase sequences are well above 1, since most nonsynonymous substitutions within this gene are deleterious (23). In contrast, short-term intrahost evolution of the HIV-1 envelope gene is characterized by mean Ds/Da ratios of 0.4, reflecting the advantageous character of amino acid changes in this immunogenic region (41). For long-term evolution, as the separation among HIV-1 subtypes, the Ds/Da ratios within the env gene are generally above 1 (39), reflecting accumulation of synonymous substitutions with time (26).
For all pairwise sequence comparisons, except for the extremely homogeneous PorcinePV, we found Ds/Da ratios above 1. The most extreme case of negative selection was observed for the separation between the B19 and V9 lineages, apparently an ancient event. While these two viruses were extremely homologous at the amino acid level, with the mean Da between them not exceeding 0.03, the mean Ds were 0.40 to 0.45 resulting in Ds/Da ratios of up to 22.5 (Fig. 3). The Ds/Da ratios were above 1 even for recent evolutionary events, such as the cross-species transmission of FelinePV to dogs (35, 46, 49), when an increase of nonsynonymous substitutions during virus adaptation to a new host could be expected. We did observe a two- to four-times-lower genetic heterogeneity among CaninePV than among FelinePV, a likely indication of a recent transmission bottleneck. At the same time, the mean Ds/Da ratios were 5.8 for orf1 and 3.3 for orf2 for the comparisons of CaninePV to FelinePV. Among CaninePV, the mean Ds/Da ratios were 8.0 for orf1 and 2.0 for orf2, compared to a mean ratio of 4.0 for both ORFs among FelinePV. While these data do not exclude the possibility that certain nonsynonymous substitutions were selected for during the adaptation of FelinePV to dogs (35), they indicate that the influence of positive selection during this recent cross-species transmission was extremely limited.
In contrast to that for the cross-species transmissions of FelinePV to dogs (46), the time scales for separation between and diversification within other virus species are not known. Since little is known about the evolution rate of parvoviruses, precise dating of those events is currently not possible. CaninePV has been shown to evolve in a linear fashion over time with the mean evolution rate of 10−4 nucleotides per year (35), which would require 100 years of independent evolution for the evolutionary distance of 0.01 between two lineages. Apparently, this evolution rate has to be considered maximal, since it is measured during a short period of virus adaptation to a new host. Moreover, we demonstrated that the evolution rate of parvoviruses is far from being uniform for synonymous and nonsynonymous positions.
Our analysis provided evidence for both host-dependent and independent evolution in the history of parvoviruses. Within two phylogenetic groups, the autonomous primate parvoviruses and AAV and avian parvoviruses, the phylogenetic relationships were host dependent. For instance, the relationships among B19, V9, and parvoviruses from three macaque species matched exactly the relationships among their hosts, according to an earlier analysis of primate mitochondrial DNA (30). On the other hand, human B19 and V9 viruses and AAV were evolutionarily related to simian and avian parvoviruses, respectively, rather than to each other. For the third phylogenetic group, in the homogeneous subgroup of viruses from carnivores and, most remarkably, the heterogeneous subgroup of rodent viruses, no host-specific clusters were observed (Fig. 4). While, unlike what was found for viruses from carnivores (46), there is no epidemiological evidence for cross-species transmissions of rodent parvoviruses, many of them are able to experimentally infect different hosts. For instance, LuIII can establish infection in hamsters (59). The absence of host-specific clusters and genetic mosaicism of rodent parvoviruses suggest that cross-species transmissions may have occurred among rodents.
For most virus comparisons, the topologies of phylogenetic trees were similar in both ORFs, with two exceptions. First, the positions of AAV-2 and AAV-4 in the phylogenetic trees varied in relation to the genomic region analyzed (Fig. 2B to D versus E to G). Taken with our observations of pairwise Da among various viruses being drastically higher or lower within orf1 than within orf2, this finding suggests that the selection pressure on the two genomic regions differs among distinct lineages, which could be related to functional difference between the two ORFs.
The second case of tree incongruity was observed among parvoviruses from mice and hamsters. MousePV and HamsterPV were evolutionary related to MVM within orf1 and to LuIII within orf2 (Fig. 4). Since this incongruity was observed for both nonsynonymous and synonymous substitutions, it is unlikely to be the result of convergent evolution. We demonstrated that this genomic mosaicism is likely to be the result of a recombination that occurred among lineages within this group (Fig. 5). Traditionally, MousePV and HamsterPV should be considered the recombinants between MVM and LuIII-related viruses. However, one cannot exclude the possibility that the recombination event involved a yet-undiscovered parvovirus.
ACKNOWLEDGMENT
We thank Lucy Phillips for editorial review.
REFERENCES
- 1.Afanasiev B N, Galyov E E, Buchatsky L P, Kozlov Y V. Nucleotide sequence and genomic organization of Aedes densonucleosis virus. Virology. 1991;185:323–336. doi: 10.1016/0042-6822(91)90780-f. [DOI] [PubMed] [Google Scholar]
- 2.Astell C R, Gardiner E M, Tattersall P. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J Virol. 1986;57:656–669. doi: 10.1128/jvi.57.2.656-669.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astell C R, Thomson M, Merchlinsky M, Ward D C. The complete DNA sequence of minute virus of mice, an autonomous parvovirus. Nucleic Acids Res. 1983;11:999–1018. doi: 10.1093/nar/11.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auguste, V., A., Garbarg-Chenon, and Q. T. Nguyen. 1999. Erythrovirus and its applications. French patent WO9928439.
- 5.Ball-Goodrich L J, Johnson E. Molecular characterization of a newly recognized mouse parvovirus. J Virol. 1994;68:6476–6486. doi: 10.1128/jvi.68.10.6476-6486.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball-Goodrich L J, Leland S E, Johnson E A, Paturzo F X, Jacoby R O. Rat parvovirus type 1: the prototype for a new rodent parvovirus serogroup. J Virol. 1998;72:3289–3299. doi: 10.1128/jvi.72.4.3289-3299.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bando H, Hayakawa T, Asano S, Sahara K, Nakagaki M, Iizuka T. Analysis of the genetic information of a DNA segment of a new virus from silkworm. Arch Virol. 1995;140:1147–1155. doi: 10.1007/BF01315423. [DOI] [PubMed] [Google Scholar]
- 8.Bando H, Kusuda J, Gojobori T, Maruyama T, Kawase S. Organization and nucleotide sequence of a densovirus genome imply a host-dependent evolution of the parvoviruses. J Virol. 1987;61:553–560. doi: 10.1128/jvi.61.2.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bantel-Schaal U, Delius H, Schmidt R, zur Hausen H. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J Virol. 1999;73:939–947. doi: 10.1128/jvi.73.2.939-947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergeron J, Hebert B, Tijssen P. Genome organization of the Kresse strain of porcine parvovirus: identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J Virol. 1996;70:2508–2515. doi: 10.1128/jvi.70.4.2508-2515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergeron J, Menezes J, Tijssen P. Genomic organization and mapping of transcription and translation products of the NADL-2 strain of porcine parvovirus. Virology. 1993;197:86–98. doi: 10.1006/viro.1993.1569. [DOI] [PubMed] [Google Scholar]
- 12.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2173–2197. [Google Scholar]
- 13.Besselsen D G, Pintel D J, Purdy G A, Besch-Williford C L, Franklin C L, Hook R R, Jr, Riley L K. Molecular characterization of newly recognized rodent parvoviruses. J Gen Virol. 1996;77:899–911. doi: 10.1099/0022-1317-77-5-899. [DOI] [PubMed] [Google Scholar]
- 14.Bloom M E, Alexandersen S, Perryman S, Lechner D, Wolfinbarger J B. Nucleotide sequence and genomic organization of Aleutian mink disease parvovirus (ADV): sequence comparisons between a nonpathogenic and a pathogenic strain of ADV. J Virol. 1988;62:2903–2915. doi: 10.1128/jvi.62.8.2903-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blundell M C, Beard C, Astell C R. In vitro identification of a B19 parvovirus promoter. Virology. 1987;157:534–538. doi: 10.1016/0042-6822(87)90296-0. [DOI] [PubMed] [Google Scholar]
- 16.Boublik Y, Jousset F X, Bergoin M. Complete nucleotide sequence and genomic organization of the Aedes albopictus parvovirus (AaPV) pathogenic for Aedes aegypti larvae. Virology. 1994;200:752–763. doi: 10.1006/viro.1994.1239. [DOI] [PubMed] [Google Scholar]
- 17.Brown K E, Green S W, O'Sullivan M G, Young N S. Cloning and sequencing of the simian parvovirus genome. Virology. 1995;210:314–322. doi: 10.1006/viro.1995.1348. [DOI] [PubMed] [Google Scholar]
- 18.Brown K E, Green S W, Young N S. Goose parvovirus: an autonomous member of the dependovirus genus? Virology. 1995;210:283–291. doi: 10.1006/viro.1995.1345. [DOI] [PubMed] [Google Scholar]
- 19.Carlson J, Rushlow K, Maxwell I, Maxwell F, Winston S, Hahn W. Cloning and sequence of DNA encoding structural proteins of the autonomous parvovirus feline panleukopenia virus. J Virol. 1985;55:574–582. doi: 10.1128/jvi.55.3.574-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen K C, Shull B C, Moses E A, Lederman M, Stout E R, Bates R C. Complete nucleotide sequence and genome organization of bovine parvovirus. J Virol. 1986;60:1085–1097. doi: 10.1128/jvi.60.3.1085-1097.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiorini J A, Kim F, Yang L, Kotin R M. Cloning and characterization of adeno-associated virus type 5. J Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiorini J A, Yang L, Liu Y, Safer B, Kotin R M. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J Virol. 1997;71:6823–6833. doi: 10.1128/jvi.71.9.6823-6833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornelissen M, van den Burg R, Zorgdrager F, Lukashov V, Goudsmit J. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol. 1997;71:6348–6358. doi: 10.1128/jvi.71.9.6348-6358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diffoot N, Chen K C, Bates R C, Lederman M. The complete nucleotide sequence of parvovirus LuIII and localization of a unique sequence possibly responsible for its encapsidation pattern. Virology. 1993;192:339–345. doi: 10.1006/viro.1993.1040. [DOI] [PubMed] [Google Scholar]
- 25.Dumas B, Jourdan M, Pascaud A M, Bergoin M. Complete nucleotide sequence of the cloned infectious genome of Junonia coenia densovirus reveals an organization unique among parvoviruses. Virology. 1992;191:202–222. doi: 10.1016/0042-6822(92)90182-o. [DOI] [PubMed] [Google Scholar]
- 26.Goudsmit J, Lukashov V V. Dating the origin of HIV-1 subtypes. Nature. 1999;400:325–326. doi: 10.1038/22454. [DOI] [PubMed] [Google Scholar]
- 27.Green S W, Malkovska I, O'Sullivan M G, Brown K E. Rhesus and pig-tailed macaque parvoviruses: identification of two new members of the erythrovirus genus in monkeys. Virology. 2000;269:105–112. doi: 10.1006/viro.2000.0215. [DOI] [PubMed] [Google Scholar]
- 28.Hall T A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Res Symp Ser. 1999;41:95–98. [Google Scholar]
- 29.Hayakawa T, Kojima K, Nonaka K, Nakagaki M, Sahara K, Asano S, Iizuka T, Bando H. Analysis of proteins encoded in the bipartite genome of a new type of parvo-like virus isolated from silkworm. Virus Res. 2000;66:101–108. doi: 10.1016/s0168-1702(99)00129-x. [DOI] [PubMed] [Google Scholar]
- 30.Hayasaka K, Fujii K, Horai S. Molecular phylogeny of macaques: implications of nucleotide sequences from an 896-base pair region of mitochondrial DNA. Mol Biol Evol. 1996;13:1044–1053. doi: 10.1093/oxfordjournals.molbev.a025655. [DOI] [PubMed] [Google Scholar]
- 31.Hemauer A, Beckenlehner K, Wolf H, Lang B, Modrow S. Acute parvovirus B19 infection in connection with a flare of systemic lupus erythematodes in a female patient. J Clin Virol. 1999;14:73–77. doi: 10.1016/s1386-6532(99)00038-4. [DOI] [PubMed] [Google Scholar]
- 32.Hemauer A, von Poblotzki A, Gigler A, Cassinotti P, Siegl G, Wolf H, Modrow S. Sequence variability among different parvovirus B19 isolates. J Gen Virol. 1996;77:1781–1785. doi: 10.1099/0022-1317-77-8-1781. [DOI] [PubMed] [Google Scholar]
- 33.Hokynar K, Brunstein J, Soderlund-Venermo M, Kiviluoto O, Partio E K, Konttinen Y, Hedman K. Integrity and full coding sequence of B19 virus DNA persisting in human synovial tissue. J Gen Virol. 2000;81:1017–1025. doi: 10.1099/0022-1317-81-4-1017. [DOI] [PubMed] [Google Scholar]
- 34.Horiuchi M, Goto H, Ishiguro N, Shinagawa M. Mapping of determinants of the host range for canine cells in the genome of canine parvovirus using canine parvovirus/mink enteritis virus chimeric viruses. J Gen Virol. 1994;75:1319–1328. doi: 10.1099/0022-1317-75-6-1319. [DOI] [PubMed] [Google Scholar]
- 35.Horiuchi M, Yamaguchi Y, Gojobori T, Mochizuki M, Nagasawa H, Toyoda Y, Ishiguro N, Shinagawa M. Differences in the evolutionary pattern of feline panleukopenia virus and canine parvovirus. Virology. 1998;249:440–452. doi: 10.1006/viro.1998.9335. [DOI] [PubMed] [Google Scholar]
- 36.Kariatsumari T, Horiuchi M, Hama E, Yaguchi K, Ishiguro N, Goto H, Shinagawa M. Construction and nucleotide sequence analysis of an infectious DNA clone of the autonomous parvovirus, mink enteritis virus. J Gen Virol. 1991;72:867–875. doi: 10.1099/0022-1317-72-4-867. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Tamura K, Nei M. Molecular evolutionary genetics analysis (MEGA). Institute of Molecular Evolutionary Genetics. University Park, Pa: Pennsylvania State University; 1993. [Google Scholar]
- 38.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy G A, van der Groen G, Fransen K, Gershy-Damet G M, Deleys R. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Lukashov V V, Goudsmit J. Evolution of the human immunodeficiency virus type 1 subtype-specific V3 domain is confined to a sequence space with a fixed distance to the subtype consensus. J Virol. 1997;71:6332–6338. doi: 10.1128/jvi.71.9.6332-6338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukashov V V, Goudsmit J. HIV heterogeneity and disease progression in AIDS: a model of continuous virus adaptation. AIDS. 1998;12:S43–S52. [PubMed] [Google Scholar]
- 41.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukashov V V, Kuiken C L, Vlahov D, Coutinho R A, Goudsmit J. Evidence for HIV type 1 strains of U.S. intravenous drug users as founders of AIDS epidemic among intravenous drug users in northern Europe. AIDS Res Hum Retroviruses. 1996;12:1179–1183. doi: 10.1089/aid.1996.12.1179. [DOI] [PubMed] [Google Scholar]
- 43.Mari J, Bonami J R, Lightner D. Partial cloning of the genome of infectious hypodermal and haematopoietic necrosis virus, an unusual parvovirus pathogenic for penaeid shrimps; diagnosis of the disease using a specific probe. J Gen Virol. 1993;74:2637–2643. doi: 10.1099/0022-1317-74-12-2637. [DOI] [PubMed] [Google Scholar]
- 44.Martyn J C, Davidson B E, Studdert M J. Nucleotide sequence of feline panleukopenia virus: comparison with canine parvovirus identifies host-specific differences. J Gen Virol. 1990;71:2747–2753. doi: 10.1099/0022-1317-71-11-2747. [DOI] [PubMed] [Google Scholar]
- 45.Muramatsu S, Mizukami H, Young N S, Brown K E. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology. 1996;221:208–217. doi: 10.1006/viro.1996.0367. [DOI] [PubMed] [Google Scholar]
- 46.Parrish C R. Emergence, natural history, and variation of canine, mink, and feline parvoviruses. Adv Virus Res. 1990;38:403–450. doi: 10.1016/S0065-3527(08)60867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parrish C R. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology. 1991;183:195–205. doi: 10.1016/0042-6822(91)90132-u. [DOI] [PubMed] [Google Scholar]
- 48.Parrish C R, Aquadro C F, Carmichael L E. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink, and raccoon parvoviruses. Virology. 1988;166:293–307. doi: 10.1016/0042-6822(88)90500-4. [DOI] [PubMed] [Google Scholar]
- 49.Parrish C R, Truyen U. Parvovirus variation and evolution. In: Domingo E, Webster R G, Holland J, editors. Origin and evolution of viruses. San Diego, Calif: Academic Press; 2000. pp. 421–439. [Google Scholar]
- 50.Ranz A I, Manclus J J, Diaz-Aroca E, Casal J I. Porcine parvovirus: DNA sequence and genome organization. J Gen Virol. 1989;70:2541–2553. doi: 10.1099/0022-1317-70-10-2541. [DOI] [PubMed] [Google Scholar]
- 51.Reed A P, Jones E V, Miller T J. Nucleotide sequence and genome organization of canine parvovirus. J Virol. 1988;62:266–276. doi: 10.1128/jvi.62.1.266-276.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhode S L, III, Paradiso P R. Parvovirus genome: nucleotide sequence of H-1 and mapping of its genes by hybrid-arrested translation. J Virol. 1983;45:173–184. doi: 10.1128/jvi.45.1.173-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rutledge E A, Halbert C L, Russell D W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sahli R, McMaster G K, Hirt B. DNA sequence comparison between two tissue-specific variants of the autonomous parvovirus, minute virus of mice. Nucleic Acids Res. 1985;13:3617–3633. doi: 10.1093/nar/13.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samulski R J, Srivastava A, Berns K I, Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 56.Schuierer S, Bloom M E, Kaaden O R, Truyen U. Sequence analysis of the lymphotropic Aleutian disease parvovirus ADV-SL3. Arch Virol. 1997;142:157–166. doi: 10.1007/s007050050066. [DOI] [PubMed] [Google Scholar]
- 57.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 58.Simpson A A, Chipman P R, Baker T S, Tijssen P, Rossmann M G. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure. 1998;6:1355–1367. doi: 10.1016/s0969-2126(98)00136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soike K F, Iatropoulis M, Siegl G. Infection of newborn and fetal hamsters induced by inoculation of LuIII parvovirus. Arch Virol. 1976;51:235–241. doi: 10.1007/BF01318027. [DOI] [PubMed] [Google Scholar]
- 60.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umene K, Nunoue T. The genome type of human parvovirus B19 strains isolated in Japan during 1981 differs from types detected in 1986 to 1987: a correlation between genome type and prevalence. J Gen Virol. 1990;71:983–986. doi: 10.1099/0022-1317-71-4-983. [DOI] [PubMed] [Google Scholar]
- 62.van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 2000. [Google Scholar]
- 63.Vasudevacharya J, Basak S, Srinivas R V, Compans R W. Nucleotide sequence analysis of the capsid genes and the right-hand terminal palindrome of porcine parvovirus, strain NADL-2. Virology. 1989;173:368–377. doi: 10.1016/0042-6822(89)90549-7. [DOI] [PubMed] [Google Scholar]
- 64.Vasudevacharya J, Basak S, Srinivas R V, Compans R W. The complete nucleotide sequence of an infectious clone of porcine parvovirus, strain NADL-2. Virology. 1990;178:611–616. doi: 10.1016/0042-6822(90)90364-w. [DOI] [PubMed] [Google Scholar]
- 65.Xiao W, Chirmule N, Berta S C, McCullough B, Gao G, Wilson J M. Gene therapy vectors based on adeno-associated virus type 1. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamagishi J, Hu Y, Zheng J, Bando H. Genome organization and mRNA structure of Periplaneta fuliginosa densovirus imply alternative splicing involvement in viral gene expression. Arch Virol. 1999;144:2111–2124. doi: 10.1007/s007050050626. [DOI] [PubMed] [Google Scholar]
- 67.Yoo B C, Lee D H, Park S M, Park J W, Kim C Y, Lee H S, Seo J S, Park K J, Ryu W S. A novel parvovirus isolated from Manchurian chipmunks. Virology. 1999;253:250–258. doi: 10.1006/viro.1998.9518. [DOI] [PubMed] [Google Scholar]
- 68.Zadori Z, Stefancsik R, Rauch T, Kisary J. Analysis of the complete nucleotide sequences of goose and muscovy duck parvoviruses indicates common ancestral origin with adeno-associated virus 2. Virology. 1995;212:562–573. doi: 10.1006/viro.1995.1514. [DOI] [PubMed] [Google Scholar]