Abstract
Soil microbes are a major source of organic residues that accumulate as soil organic matter, the largest terrestrial reservoir of carbon on Earth. As such, there is growing interest in determining the microbial traits that drive soil organic matter formation and stabilization; however, whether certain microbial traits consistently predict soil organic matter accumulation across different functional pools (e.g., total vs. stable soil organic matter) is unresolved. To address these uncertainties, we incubated individual species of fungi in soil organic matter-free model soils, allowing us to directly relate the physiological, morphological, and biochemical traits of fungi to their soil organic matter formation potentials. We find that the formation of different soil organic matter functional pools is associated with distinct fungal traits, and that ‘multifunctional’ species with intermediate investment across this key grouping of traits (namely, carbon use efficiency, growth rate, turnover rate, and biomass protein and phenol contents) promote soil organic matter formation, functional complexity, and stability. Our results highlight the limitations of categorical trait-based frameworks that describe binary trade-offs between microbial traits, instead emphasizing the importance of synergies among microbial traits for the formation of functionally complex soil organic matter.
Subject terms: Carbon cycle, Microbial ecology, Microbial ecology
Soil microbes contribute to soil organic matter. Here, the authors explore how microbial traits contribute to organic matter accumulation, finding that synergies among fungal traits promote soil organic matter formation, functional complexity, and stability.
Introduction
Microbial metabolism shapes the balance between carbon (C) loss and accrual within the soil organic matter (SOM) pool, the largest actively cycling reservoir of C on Earth1. While microbial decomposition and respiration (catabolism) are responsible for a significant proportion of annual carbon dioxide emissions from SOM to the atmosphere, these processes are accompanied by the production of microbial biomass (anabolism), and subsequent deposition of cellular residues and extracellular products (i.e., microbial necromass), thereby providing a pathway for the simultaneous accumulation of SOM2. As such, microbes are increasingly recognized for their contributions to SOM alongside direct inputs from plants3–6. Contemporary theory recognizes SOM formation and persistence as the balance between microbial catabolic and anabolic processes, further regulated by soil physicochemical properties that restrict decomposer access to substrates (e.g., pore connectivity, aggregation, mineralogy), as well as biotic factors that influence the outcome of microbe-substrate interactions (e.g., microbial traits such as enzymatic repertoire or growth efficiency)7–10. Owing to this important role of microbial anabolism in the soil C cycle, a growing number of studies attempt to link microbial traits to SOM formation and stabilization11–14; however, whether certain microbial traits consistently predict organic matter accumulation across different SOM pools is unresolved.
Soil organic matter is structurally and molecularly complex, being comprised by multiple functional pools that contribute to the myriad of ecosystem functions provided by soils, including nutrient cycling, C storage, water retention and erosion prevention15–17. Such functional pools include particulate organic matter (POM) and mineral-associated organic matter (MAOM). POM is characterized by larger organic matter fragments that are relatively accessible to microbial decomposers, promoting soil C and nutrient cycling, while MAOM is comprised of smaller biopolymers and monomers in close association with silt and clay minerals, rendering this SOM relatively protected from microbial decomposition and promoting C storage in soils18,19. POM and MAOM are integrated into the complex three-dimensional structure of soil aggregates, further protecting SOM from microbial decomposition through physical occlusion and modification of micro-scale environmental gradients (e.g., O2 availability)18,20.
While these SOM pools differ broadly in their relative stabilities, each pool can contain organic matter with a range of turnover times (e.g., “fast cycling” MAOM or occluded POM)21–23. Indeed, emerging understanding suggests that mineral sorption should be viewed as a reversible process that temporarily increases C retention in soils8 and that greater spatial heterogeneity and molecular diversity of SOM (so-called ‘functional complexity’17) promote its retention, while simultaneously maintaining other vital ecosystem functions (e.g., nutrient cycling, water retention)15–17. This view underscores the need for a holistic understanding of the role of microbial traits in the formation of SOM functional pools, and in the generation of SOM functional complexity17. Because SOM pools are formed via multiple plant and microbial pathways6,18, it is likely that their formation is controlled by distinct traits.
Trait-based frameworks adapted from plant ecology24–26 are increasingly applied in microbial ecology to distill the compositional and functional complexity of microbial communities, and to link microbial identity to emergent ecosystem functions, including SOM formation27,28. Prior frameworks have theorized the role of several microbial trait categories (e.g., physiology, growth morphology and cellular/extracellular biochemistry)27–30 in SOM formation and persistence. However, empirical evidence is limited, with a majority of studies measuring a single physiological trait—carbon use efficiency (CUE), the proportion of substrate C that microbes allocate towards growth relative to respiration31. By definition, higher microbial CUE corresponds with greater allocation of resources (per unit substrate consumed) to biomass production, and many studies thus propose a positive empirical relationship between CUE, microbial necromass production, and soil C accrual11,14,30,31. Thus, it has been proposed that microbes who maximize investment in CUE at the expense of other traits (i.e., genetic and/or physiological tradeoffs)32 will make the largest contributions to SOM formation27,28. However, evidence is mixed13,14,33,34, suggesting that current trait-based frameworks emphasizing single traits (e.g., CUE) – or binary tradeoffs between traits – are insufficient to describe the microbial controls on SOM accumulation across functional pools.
Whereas soil microbial CUE appears to positively correlate with total SOM-C14,35,36, its relationship with relatively stable pools of SOM, such as MAOM, may be positive, negative or neutral13,33,34, and understanding is limited by the paucity of studies evaluating stable subfractions of MAOM alongside CUE33. Emerging evidence suggests that CUE may be decoupled from MAOM accumulation under circumstances where MAOM-C is predominantly plant- rather than microbial-derived (e.g., in temperate forests34 or detritusphere soils37). Microbial CUE may also become decoupled from stable SOM formation if other traits strongly influence the incorporation of microbial residues into mineral-associated or otherwise protected pools12,18. It was recently hypothesized that microbial physiological traits, such as CUE and growth rate, act as initial filters on the pool of microbial residues available for incorporation into SOM, while additional microbial traits regulate the subset of those residues that become stabilized in soils (i.e., “feedstock traits” and “MAOM formation traits”, respectively)18. For example, physiological traits such as CUE may predict total microbial residue inputs to soils, while growth morphology (e.g., fungal hyphal surface area) and cellular and extracellular residue chemistries (e.g., protein contents) are likely to influence the degree and strength of resulting organo-mineral interactions18,29. A comprehensive approach that accounts for the groupings of physiological, biochemical and morphological traits of microbes involved in SOM formation would more accurately capture the multidimensional nature of microbial trait expression38, allowing for the identification of potential interactions and synergies between microbial traits that may be necessary for the formation of different SOM pools. However, empirical tests of such a framework are currently lacking.
Here, we characterized a suite of microbial physiological, biochemical, and morphological traits hypothesized to be important for the production of microbial residues and their subsequent incorporation into SOM functional pools. We took a comprehensive approach, accounting for the role of these multidimensional trait profiles in the formation of multiple SOM pools, including total SOM-C, POM, MAOM, water-stable aggregates, and chemically or biologically stable fractions of SOM. Our study focused on fungi, which dominate the microbial biomass pool39,40, necromass pool41,42, and decomposition processes43,44 in many ecosystems, and are therefore likely a major source of microbial-derived SOM29. We employed a unique study design with individual fungal species incubated axenically in SOM-free model soils. Fungal species spanned three phyla (Basidiomycota, Ascomycota, and Mucoromycotina) and were selected to represent a gradient of trait values, based on known differences in their trait profiles32. Model soils have been successfully employed to monitor microbial formation of SOM from simple C substrates45–47, providing direct evidence that microbes are a source of SOM quantity and chemical complexity14,47. Importantly, such simplified systems allow direct inferences to be made about relationships between the trait profiles of microbes and the SOM that they generate. Thus, our study system allowed us to directly relate each fungal species’ multidimensional trait profile to its SOM formation potential. We hypothesized that physiological traits such as CUE would be primary drivers of microbial residue inputs to soils, and thus would be the strongest predictors of total soil C. We expected that such physiological traits may become decoupled from MAOM and stable SOM formation if other fungal traits (e.g., hyphal morphology and biomass chemistry) influence the degree and strength of microbe-mineral interactions, and correspondingly that traits within these latter categories would be important predictors of stable SOM pools. Our results partially supported our hypothesis, illuminating a key grouping of fungal traits involved in SOM formation, with distinct traits emerging as important predictors of total soil C, MAOM-C, and persistent subfractions of the MAOM pool. We propose a holistic framework for understanding the microbial role in SOM formation, identifying synergies among microbial traits that promote SOM quantity, stability, and functional complexity.
Results
Fungal traits
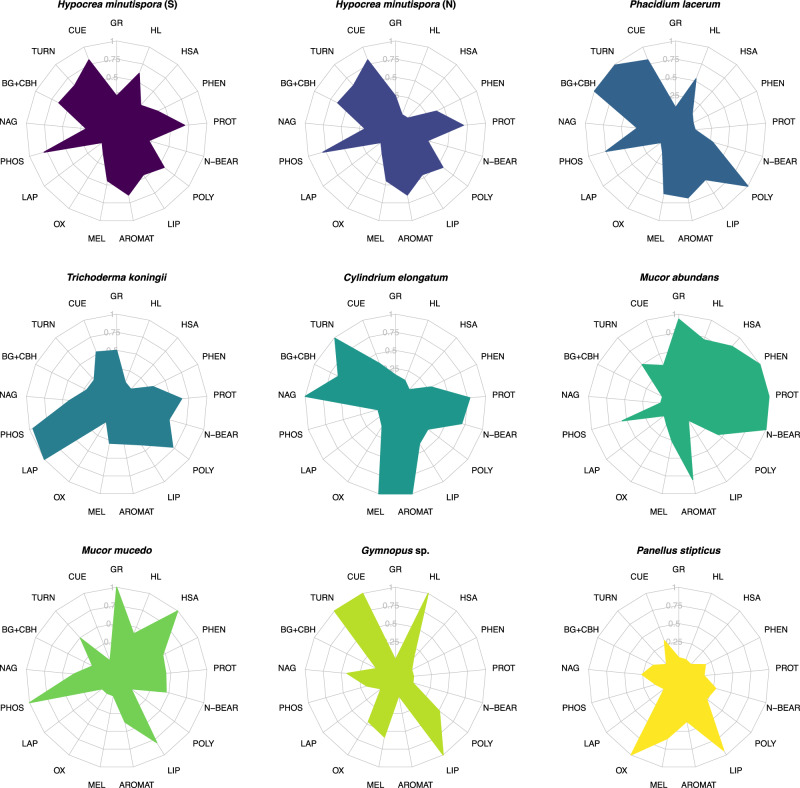
We characterized a suite of physiological (CUE, growth rate, turnover rate, extracellular enzyme production), biochemical (biomass chemistry, melanin content), and morphological (hyphal length and surface area per soil volume) traits for eight fungal species spanning three phyla (Basidiomycota, Ascomycota, Mucoromycota [subphylum Mucoromycotina]). To evaluate each species’ performance of multiple traits, we calculated a metric of ‘trait multifunctionality’ by adapting an approach for characterizing ecosystem multifunctionality48,49 (details provided in Methods). This metric accounted for both the presence of traits (value > 0) within an isolate’s trait profile and their relative values. While our focus in this experiment was on relationships between fungal trait profiles and the formation of different SOM pools, we present results at the species and phylum levels to aid interpretation and understanding of the range of trait (and SOM pool) values spanned by the fungal isolates included in this study. Relative values are not indicative of innate or immutable characteristics of the fungal isolates, but rather their relative trait values and contributions to SOM pools under the specific experimental conditions of this study. Fungal species exhibited distinct trait profiles (MANOVA: P = 0.001; Fig. 1; Supplementary Fig. 1), corresponding with differences in trait multifunctionality (one-way ANOVA; P = 0.003; Fig. 1; Supplementary Fig. 2). Certain species (e.g., H. minutispora, P. lacerum) exhibited intermediate values across a wider range of traits (Fig. 1), corresponding with higher trait multifunctionality scores (Supplementary Fig. 2), while other species (e.g., P. stipticus, Gymnopus sp.) exhibited a greater degree of specialization, with high values restricted to a more limited suite of traits (lower trait multifunctionality scores). Correspondingly, these species had relatively low values across the remaining suite of measured traits (Fig. 1; Supplementary Fig. 2).
Fig. 1. Radar plots illustrating trait profiles of fungal species.
Trait values are scaled between 0 to 1 across species for each individual trait measurement, where 0 represents the lowest relative value of trait expression, and 1 represents the highest relative value (n = 3). Species are ordered from high trait multifunctionality scores (top row) to low trait multifunctionality (bottom row), and the color associated with each fungal species is carried throughout subsequent plots. Text abbreviations around the radar plots represent individual traits. Trait abbreviations are as follows: GR = growth rate (average of two metrics; μg biomass-C produced biomass-C−1 hr−1); CUE = carbon use efficiency (average of three metrics; units of C); MEL = melanin (mg g−1 biomass); TURN = biomass turnover time (reflected such that a high value corresponds with high turnover rate); BG + CBH = combined activity of beta-glucosidase and cellobiohydrolase (μmol substrate h−1 g−1 dry soil; same units for all subsequent enzymes); NAG = N-acetyl-glucosaminadase activity; PHOS = acid phosphatase activity; LAP = leucine aminopeptidase activity; OX = combined activity of phenol oxidase (ABTS) and peroxidase (TMB); AROMAT = relative abundance (%) of aromatic compounds in fungal biomass; LIP = biomass lipids (%); POLY = biomass polysaccharides (%); N-BEAR = biomass N-bearing compounds (%); PROT = biomass proteins (%); PHEN = biomass phenols (%); HSA = hyphal surface area (m2 g−1 dry soil); HL = hyphal length (m g−1 dry soil). Source data are provided as a supplementary Source Data file.
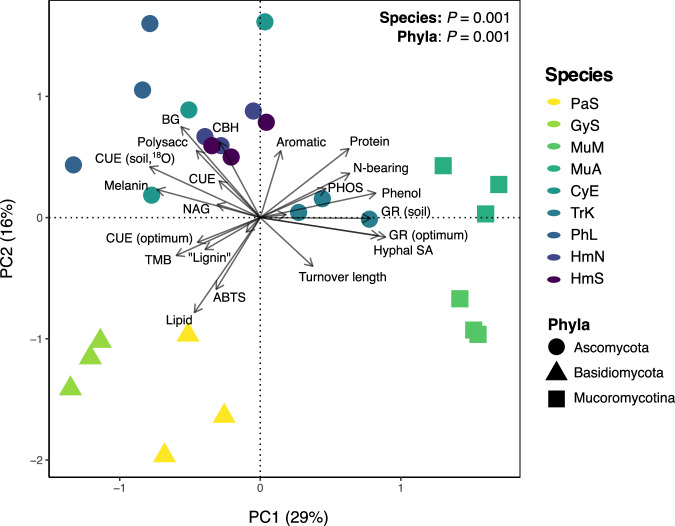
Trait profiles exhibited clear groupings at the phylum level (MANOVA: P = 0.001; Fig. 2). Higher growth rates were associated with Mucoromycotina and Ascomycota (P < 0.001, one-way ANOVA, Supplementary Fig. 1; post-hoc Tukey HSD results in Supplementary Table 1), while higher CUE was associated with Basidiomycota and Ascomycota (all P < 0.05; Supplementary Fig. 1; Supplementary Table 1). Biomass turnover time was generally shorter (i.e., faster turnover rates) among the Ascomycota and one Basidiomycota species (Gymnopus sp.; P = 0.004; Supplementary Fig. 1). Biomass protein, phenol, and N-bearing contents were highest among the Mucoromycotina and Ascomycota (all P < 0.05; Supplementary Table 1; Supplementary Fig. 1). Biomass melanin concentrations were highest among Basidiomycota and Ascomycota species (P < 0.001; SupplementaryTable 1), particularly C. elongatum and P. lacerum (Supplementary Fig. 1). Mucoromycotina exhibited the highest hyphal surface areas (P < 0.001), driven in part by their large relative hyphal diameters (P < 0.001; Supplementary Table 1; Supplementary Fig.1). Intermediate to high hyphal lengths were observed for individual species within each phylum (particularly Gymnopus sp., H. minutispora [sporulating] and M. abundans; Supplementary Fig. 1). Oxidative enzyme (OX) activities were highest among the Basidiomycota (P = 0.001), while the hydrolytic enzymes BG and CBH were highest among the Ascomycota (P < 0.001; Supplementary Table 1; Supplementary Fig. 1). PHOS activity was higher among the Ascomycota and Mucoromycotina isolates than the Basidomycota (P = 0.011; Supplementary Table 1; Supplementary Fig. 1). The potential activities of LAP and NAG did not differ significantly by phylum (P = 0.09 and P = 0.29, respectively; Supplementary Table 1), though there were significant differences at the species-level (P < 0.01; Supplementary Fig. 1).
Fig. 2. Principal components analysis (PCA) of fungal trait profiles.
The two most explanatory PC axes are presented (PC1, PC2), which collectively explained 45% of variation in fungal trait data. The color of sample points represents fungal species (n = 3), while point shape represents fungal phyla. Vectors indicate individual trait measurements. The results of separate one-way MANOVA analyses for species and phyla are presented (both P = 0.001). Fungal species abbreviations are consistent throughout this manuscript and are as follows: HmS = H. minutispora (sporulating); HmN = H. minutispora (non-sporulating); PhL = P. lacerum; TrK = T. koningii; CyE = C. elongatum; MuA = M. abundans; MuM = M. mucedo; GyS = Gymnopus sp.; PaS = P. stipticus. Source data are provided as a supplementary Source Data file.
Fungal SOM Formation
Fungi were incubated axenically in initially SOM-free, substrate-amended model soil for between 3-6 months depending on species’ individual growth dynamics. Each species received the same total quantity and rate of substrate-C amendment, while the timing of amendments and harvests were made on a species-specific basis (based on respiration rates, Suppl. Fig. 3; see Methods for details). Species’ contributions to total soil C, MAOM-C, POM-C, water-stable aggregates, and biologically stable or chemically stable subfractions of SOM were assessed. Fungal contributions to each of the measured functional pools were compared against sterile control soils (model soil mixture + substrate) to account for possible abiotic retention of substrate-C. Fungal species differed greatly in their contributions to different SOM functional pools (Fig. 3; Supplementary Fig. 4; P < 0.05 for all pools). Across all species, between 31-46% of the total substrate-C added over the course of the experiment remained as soil C (on average; Supplementary Fig. 4). The average proportion of total soil C comprised by the MAOM fraction ranged between 46-79%, while the average proportion in POM was 8-34% (Supplementary Fig. 4; all P < 0.05). We achieved ~96% soil mass recovery during the fractionation procedure, with C losses ranging from 4-36% across species (Supplementary Fig. 5; explanation provided in supplement). Total soil C varied by species (P = 0.007; Fig. 3) and was generally highest among the Ascomycota and Basidiomycota isolates, though these differences were not significant at the phylum-level (P = 0.301; Supplementary Table 2). Contributions to MAOM-C and water-stable aggregates varied by phylum (P = 0.045 and P < 0.001, respectively; Supplementary Table 2), with the highest average values observed among Ascomycota and Basidiomycota isolates (post-hoc comparisons: P < 0.05; Supplementary Table 2). Biologically and chemically stable SOM also varied by phylum (both P < 0.01; Supplementary Table 2), with the largest contributions in this case observed among the Ascomycota and Mucoromycotina (post-hoc comparisons: P < 0.05; Supplementary Table 2). We thus sought to understand how fungal contributions to different SOM functional pools were related to their respective trait profiles and functioning.
Fig. 3. Isolate contributions to SOM functional pools.
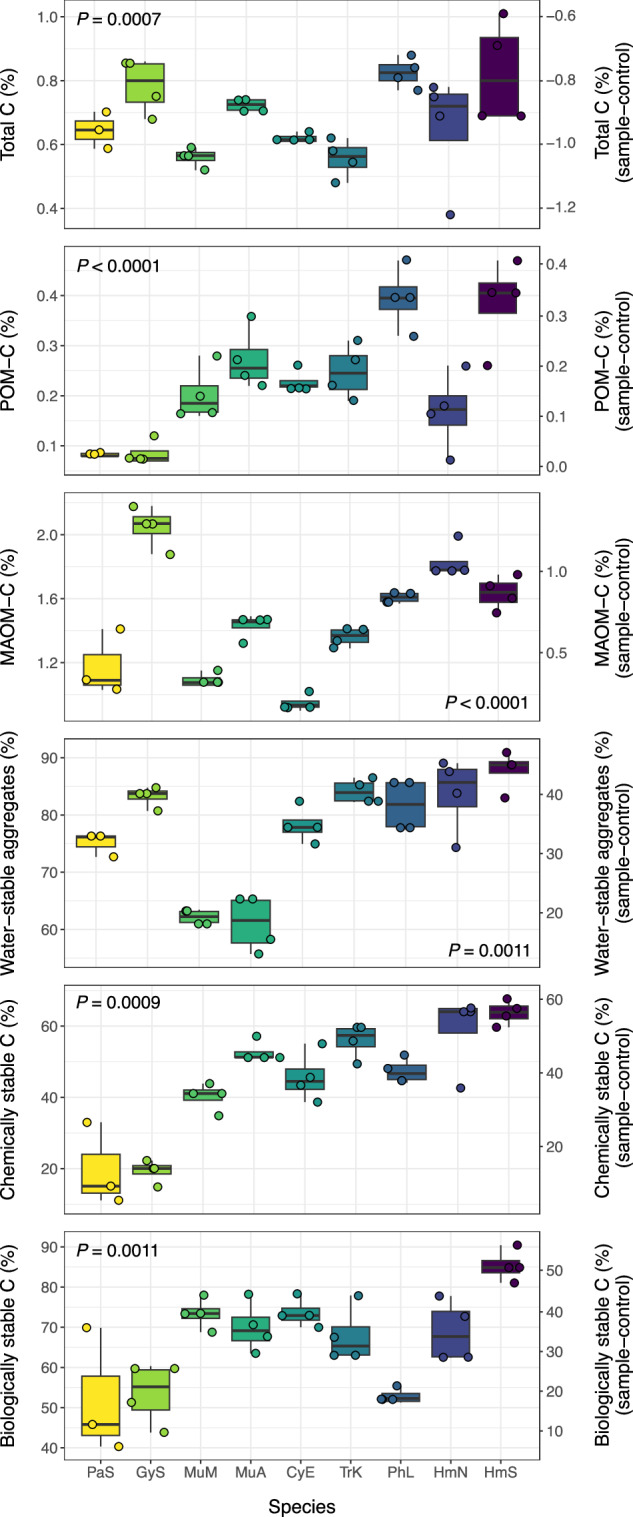
Contributions are presented as total uncorrected values (lefthand Y-axis) and corrected values (righthand Y-axis), which were calculated as the difference between inoculated (fungal) and sterile control samples (n = 4, except P. stipticus, n = 3). Corrected values for total C are negative, as the sterile control samples contained higher C concentrations than the inoculated samples, due to the absence of respiratory CO2 losses. Colors represent fungal species (carried throughout figures). Boxplots represent 25th and 75th percentile, median and outlying points. Whiskers extend from minimum to maximum values (no further than 1.5x the inter-quartile range). Results (p-values) of one-way ANOVA or Kruskal-Wallis tests are presented for each SOM functional pool (all P < 0.05). Source data are provided as a supplementary Source Data file.
Traits Predicting SOM Formation
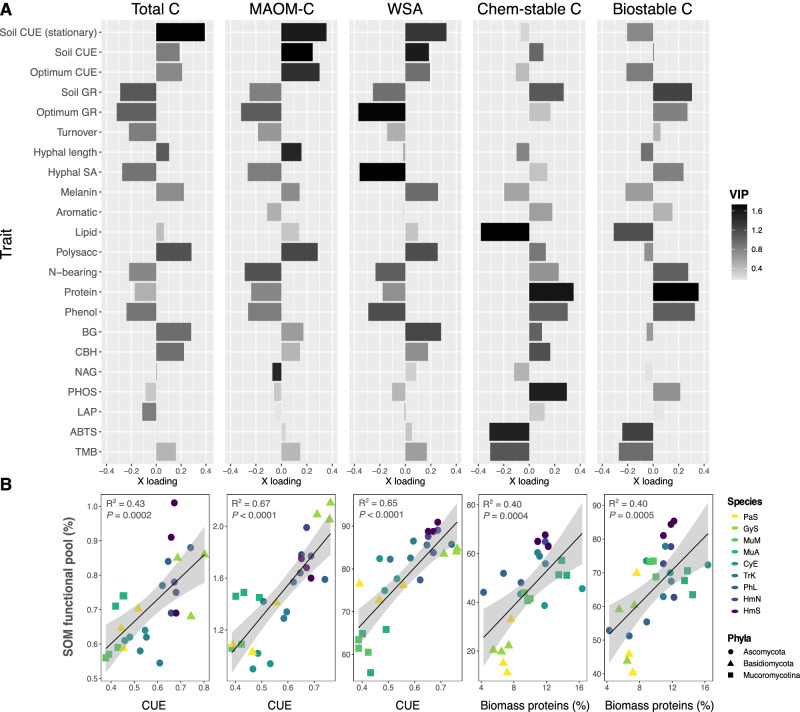
We used partial least squares regression (PLSR) to identify fungal traits associated with the formation of each SOM functional pool. Across our dataset, CUE was one of the most important traits regulating the formation of MAOM and total C (Fig. 4a; strong positive loadings, high VIP scores in PLSR models), whereas biomass protein and phenol contents were the most important traits contributing to the formation of chemically and biologically stable SOM (Fig. 4a). Growth rate was also an influential trait variable in each PLSR model, loading positively on the most explanatory latent factor for biologically stable SOM (and to a lesser extent, chemically stable SOM) versus negatively on the latent factors for total C and MAOM-C. These relationships were reflected in the significant positive correlations between CUE and total C or MAOM-C (Fig. 4b; R2 = 0.43-0.67; P ≤ 0.0002), and in the positive correlations between biomass protein content and chemically or biologically stable SOM (R2 = 0.40; P ≤ 0.0004). CUE was also positively correlated with the proportion of water-stable aggregates (R2 = 0.65; P < 0.0001).
Fig. 4. Fungal traits predicting SOM pool formation.
A Bar plots of trait loadings on the first (most explanatory) PLSR latent factor for each SOM functional pool, accounting for 50–78% of variation in each pool. From left to right: total C, MAOM-C, water-stable aggregates (WSA), chemically stable C, and biologically stable C. Bar color is shaded to represent variable importance scores (VIP) for each trait variable (n = 3), indicating the overall importance of each trait to the PLSR model, integrating latent factor 2, and in some cases, factor 3. Full PLSR model results are reported in Supplementary Table 3a, b, and a version of this plot with POM included is presented in Supplementary Fig. 6. Optimum growth rate and CUE were measured in liquid culture. All growth rate and CUE assays were conducted during log-phase, unless specified at stationary growth. B Linear regressions (n = 27) between the trait variable with the strongest loading on PLSR1 for each SOM functional pool, where regression plots correspond with the functional pool whose PLSR results are presented directly above (i.e., the first panel represents the correlation between CUE and total C, and so on). For regressions involving CUE, an average CUE value was calculated across the three CUE metrics included in this study (liquid culture log-phase, soil log-phase, soil stationary growth). Biomass proteins represent the relative abundance of proteins in the biomass of fungal isolates grown in liquid culture, as quantified by Py-GC/MS. Point color represents fungal species, as in prior plots, with trait multifunctionality and SOM formation potential generally increasing from light to dark colors (Supplementary Fig. 2). Error bands represent 95% confidence interval. Source data are provided as a supplementary Source Data file.
Principal components analysis (PCA), used to visualize differences in trait profiles across species and phyla, demonstrated similar patterns of association between fungal traits and SOM pools (Fig. 2). Higher biomass protein, phenol and N-bearing contents, as well as fungal growth rates, loaded positively on PC1 (associated with Mucoromycotina and certain Ascomycota isolates), while CUE loaded negatively on PC1 (associated with the Basidiomycota and remaining Ascomycota isolates). Correspondingly, PC1 was generally associated with greater proportions of chemically and biologically stable C (quadratic polynomial regressions in Supplementary Fig. 7; R2 = 0.32 and 0.28, and both P ≤ 0.05), whereas it was negatively correlated with total C and MAOM-C (linear regressions; R2 ≈ 0.2 and P < 0.05 for both).
The results of these analyses (Figs. 1–4) led us to hypothesize that microbial taxa with intermediate to high performance across this key grouping of traits (namely, CUE, growth rate, and biomass protein and phenol contents) would be most proficient at forming SOM across multiple functional pools. To test this hypothesis, we calculated a metric of SOM formation potential to characterize each species’ average contributions to the measured SOM functional pools. The same approach that was used to calculate trait multifunctionality48,49 was adapted for this purpose by using SOM pools as input values instead of fungal traits. We found that trait multifunctionality was strongly positively correlated with SOM formation potential (Fig. 5; R2 = 0.70; P < 0.0001), and this relationship was consistent whether all measured functional pools (Fig. 5) or only the putatively stable pools of SOM were included in the calculation (Supplementary Fig. 8; R2 = 0.65; P < 0.0001). To illustrate this point, Gymnopus sp. (Basidiomycota) exhibited a high degree of specialization in its trait profile and had a relatively low trait multifunctionality score (0.31; Supplementary Fig. 2). Correspondingly, Gymnopus sp. made large contributions to a small subset of SOM functional pools, namely MAOM-C, but had one of the lowest levels of both chemically and biologically stable C of all the isolates (Fig. 3). In contrast, H. minutispora (Ascomycota) exhibited high trait multifunctionality (0.40 for HmS; Supplementary Fig. 2), including intermediate CUE, growth rate, and biomass protein contents, and formed intermediate to high levels of SOM across all measured functional pools. Overall, the five fungal species with the highest SOM formation potentials (Fig. 5; Supplementary Fig. 2) were associated with intermediate to high trait values (>0.5 when species’ trait values were scaled 0 to 1) for CUE, turnover rates, and PHOS activity (Fig. 1). Four out of the five taxa were associated with intermediate to high values for biomass proteins, phenols, polysaccharides, and aromatics.
Fig. 5. Linear regression between trait multifunctionality and SOM formation potential.
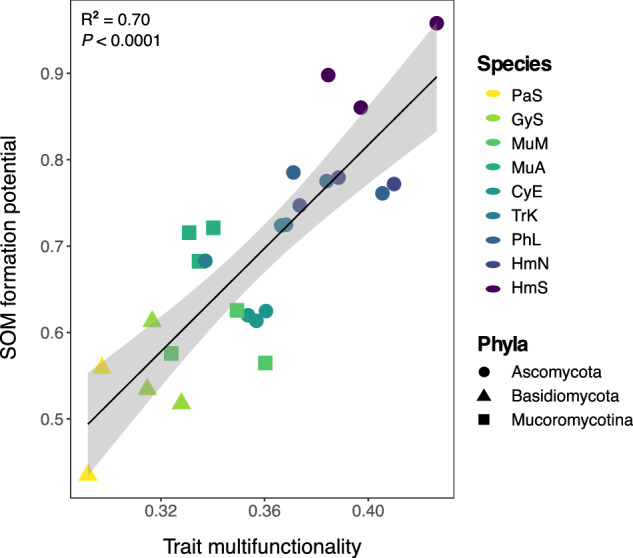
Point color represents fungal species, while point shape represents fungal phyla. The SOM pools included in the calculation of average SOM formation potential included: total C, POM, MAOM, water-stable aggregates, chemically stable C and biologically stable C. One PaS replicate with significantly lower trait multifunctionality ( ~ 0.2) is removed from the plot for ease of visualizing differences across other samples, but does not change the interpretation of the results (both P < 0.0001). One-way ANOVA results are presented for the model that included all replicates (n = 27; R2 = 0.70; P < 0.0001). Error bands represent 95% confidence interval. Source data are provided as a supplementary Source Data file.
Chemistry of fungal-derived SOM
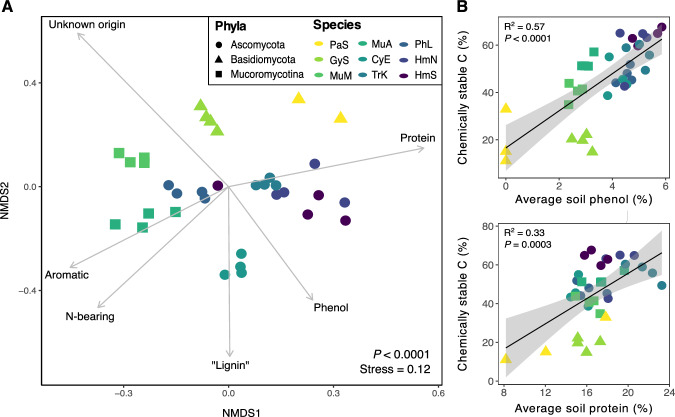
In addition to evaluating fungal contributions to SOM quantity, we sought to understand whether species identity influenced SOM chemistry. We had two main objectives: (1) evaluate whether fungal production of specific compounds is related to the stability of SOM produced, and (2) characterize the molecular diversity of SOM generated by each fungal species17. Using a ramped pyrolysis gas chromatography/mass spectrometry (Py-GC/MS) approach, we found that different fungal species generated SOM (Fig. 6a) and MAOM (Supplementary Fig. 9) with distinct chemistries (both P ≤ 0.001), and this species-level variation was apparent within each pyrolysis thermal fraction (Supplementary Fig. 10; PERMANOVA, all P ≤ 0.001). In the ramped pyrolysis approach, organic compounds that resist pyrolysis at lower temperatures, but combust at higher temperatures, are assumed to have higher thermal stabilities, and thus to comprise relatively persistent pools of SOM50. In general, the relative abundances of proteins, N-bearing compounds, and phenols increased as pyrolysis temperature increased (abundances peaked between 444–600 °C), suggesting that these fungal-derived compounds were relatively stable in soils (Supplementary Fig. 11). In contrast, the relative abundances of polysaccharides and lipids peaked between 330–396 °C and generally declined at higher pyrolysis temperatures, suggesting that these compounds were less thermally stable. Consistent with these results, we found that the average relative abundances of phenols and proteins in soils at the end of the long-term incubation were positively correlated with the proportion of chemically stable SOM produced by each species (Fig. 6b, c; R2 = 0.57 and 0.33, respectively; both P ≤ 0.0003). Additionally, fungi with relatively high SOM formation potentials tended to produce thermally stable SOM characterized by a higher relative abundance of proteins and phenols (HmS, HmN, PhL, TrK, MuA in Supplementary Fig. 10). These fungi also generally produced SOM that was more chemically diverse, and we observed a positive correlation between the chemical diversity (Shannon index) of thermally stable SOM (735 °C thermal fraction) and the proportion of chemically stable C formed by each species (Supplementary Fig. 12; R2 = 0.55; P < 0.0001).
Fig. 6. Ordination of chemical composition and correlations between abundances of soil phenol and protein.
A NMDS ordination of SOM chemical composition (individual compound-level data) after long-term incubation with fungal isolates (n = 4, except P. stipticus, n = 3). Results for a single temperature fraction (503oC) from the ramped pyrolysis GC/MS analysis are presented here for simplicity (P < 0.0001; PERMANOVA). Significant variations in species’ SOM chemistries were observed for each thermal fraction (all P < 0.001; one-way PERMANOVA), and full results are shown in Supplementary Fig. 10. Point color represents fungal species, while point shape represents fungal phyla. Environmental vectors representing broad chemical compound classes were included in the plot if significant (P < 0.05). B Correlations between the relative abundances of phenols (top) or proteins (bottom) in soils at the end of the long-term incubations and the proportion of chemically stable C produced by each fungal species (n = 27). One-way ANOVA results are presented (both P < 0.001). Error bands represent 95% confidence interval. Source data are provided as a supplementary Source Data file.
Discussion
Fungi differed considerably in their formation and stabilization of SOM based on variation in their trait profiles, with distinct fungal traits emerging as important controls on the formation of different SOM functional pools. Total SOM and MAOM formation were primarily associated with higher CUE, whereas the formation of chemically and biologically stable SOM was associated with higher fungal growth rates and biomass protein and phenol contents. Species with higher trait multifunctionality, especially those with intermediate to high performance across this key grouping of traits, exhibited the greatest capacities for SOM formation across functional pools. We present a holistic framework for understanding the role of fungal traits in SOM formation, proposing that synergies among these traits promote SOM quantity, stability, and functional complexity17.
Similar to previous experiments employing model soils to investigate SOM formation, we demonstrate that microbes can form SOM that is chemically and functionally diverse, absent complex plant inputs14,33. Additionally, we provide direct evidence that fungi contribute to SOM formation and stabilization via multiple pathways within soils, from aggregate formation and particulate hyphal residue inputs within the POM ( > 53 μm) fraction, to the formation of relatively stable fractions of mineral-associated SOM. Prior frameworks have emphasized the role of fungi in soil particle arrangement and enmeshment, suggesting that fungi are particularly central to the formation of soil macroaggregates51–53. However, these frameworks have been less clear on the potential role of fungi as agents of microaggregate and stable MAOM formation, often suggesting that bacteria are primarily responsible for the decomposition of hyphal residues and simultaneous production of biofilms (including EPS and other extracellular compounds) that become major inputs to MAOM29,54–56. Here, we demonstrate that in a bacteria-free environment, fungi generated chemically and functionally diverse SOM and MAOM, which included fractions that resisted chemical and biological destabilization. While hyphal decomposition and EPS production by bacteria surely play important roles in MAOM formation in natural soils, our results emphasize that the direct contributions of fungi should not be overlooked. Fungal inputs may include cell wall fragments, cytosolic components from lysed cells, EPS originating from hyphal coatings, as well as other extracellular metabolites (e.g., organic acids, sugars)29,57–59. In our study, fungal species differed in their contributions to SOM functional pools. Below, we detail how these differences were linked to variation in fungal traits.
We observed a pattern among our study species consistent with the theory of ecological trade-offs among microbial generalists versus specialists60,61; here, we adapt this theory in the context of microbial traits. Certain species exhibited high trait values within a limited suite of trait categories (i.e., functional specialists; e.g., P. stipticus, Gymnopus sp.) whereas other species exhibited intermediate values across a wide range of traits (functional generalists; e.g., H. minutispora), which corresponded with higher trait multifunctionality scores. We found that a species’ trait multifunctionality was an important positive predictor of its SOM formation potential. Because microbes exhibit fundamental tradeoffs in genetic and/or physiological investment in different traits32, such a relationship could exist if generalists invest at intermediate levels across a wider range of trait categories than functional specialists, increasing their likelihood of possessing the genetic and/or physiological capacity for traits important to SOM formation. In our study, certain fungal traits were positively correlated with one SOM functional pool but neutrally or negatively associated with another. Therefore, species with the ability to perform multiple traits that are positively correlated with distinct SOM pools may make the largest contributions to SOM accrual. In contrast to past studies that have focused on individual traits (e.g., CUE) or binary trade-offs between traits (e.g., high CUE, low resource acquisition), our results point towards the need to consider interactive effects among microbial traits and their net effect on SOM accumulation. Below, we discuss a key grouping of traits that emerged as synergistic for the formation and stabilization of SOM.
Microbial community CUE has been the focus of most empirical studies evaluating the relationships between microbial traits and SOM formation, with evidence generally supporting a positive correlation between CUE and total soil C storage11,13,14,35. However, the relationship between CUE and relatively persistent SOM is less clear, with some lab-based studies showing a positive empirical relationship between these two variables33 and emerging field-based evidence demonstrating that microbial CUE can become decoupled from stable SOM formation (e.g., in temperate forests34 or detritusphere soils37). Our results suggest that intermediate to high growth rates and biomass protein, phenol and N-bearing contents are stronger predictors of fungal contributions to persistent SOM than CUE, but that when fungi possess intermediate (to high) CUE alongside these other traits, SOM formation potential is amplified. Multiple species with high SOM formation potential also exhibited intermediate to high biomass turnover rates. Microbial CUE, growth and turnover rates may act as initial filters on the “feedstock”18 of microbial residues available for incorporation into SOM functional pools, while additional microbial traits (“MAOM formation traits,” e.g., cellular and extracellular residue chemistries) regulate the subset of those residues that become stabilized in soils, as proposed by Sokol et al.18. Our results suggest that this specific combination of traits (intermediate to high CUE, growth rate, biomass turnover, and biomass protein and phenol contents) is synergistic for the formation of SOM. Thus, individual species with the capacity to perform these traits (high trait multifunctionality), or communities of microorganisms where these key traits are well-represented, may possess the greatest SOM formation potentials.
An understanding of microbial growth and turnover rates alongside CUE provides a more complete picture of microbial physiology, giving insight into the amount of biomass and cellular necromass inputs to soils per unit time18,62. While high CUE has been the focus of discussions around a ‘high yield’ strategy among microbes28, CUE is typically defined per unit substrate63 and does not guarantee high biomass and necromass yield per unit time. In ecosystems or spatial microsites in soils where C inputs are relatively continuous (e.g., rhizosphere soil), it is likely that the integrative effects of microbial growth rates, turnover rates, and CUE regulate SOM-C accrual, especially microbial-derived SOM11,13,37,62. Higher growth rates and activity levels among microbes may be associated with faster biomass turnover and more extensive processing of necromass into lower molecular weight compounds, producing “feedstock” for stable MAOM formation18,64. Indeed, we observed greater SOM formation potential among species with a higher capacity for the production of hydrolytic enzymes involved in necromass decomposition (e.g., PHOS, BG, CBH), consistent with the idea that microbial processing of particulate OM can increase the reactivity and diffusion of these fragmented residues to soil minerals8.
In contrast, if fungi are efficient (i.e., high CUE) but their growth rate is slow, they may promote C retention in soils by limiting the amount of soil C lost via respiration, but this C will not necessarily be converted into stable forms. In field soils, microbial communities characterized by slower growing species may be associated with more direct stabilization of plant-derived, as opposed to microbial-derived SOM34,37. In our study, a Basidiomycota species (Gymnopus sp.) yielded the highest MAOM-C concentrations and one of the highest total soil C concentrations; however, only a small fraction of this C was biologically or chemically stable. Although incubation length was longest for this species (6 mo. compared with 3 mo. for the fastest growing taxon), this slow-growing fungus may have had sufficient access to substrate C throughout the course of the incubation, reducing its need to mine C and nutrients from existing necromass. Overall, our results suggest that more active fungal species with intermediate to high growth rates, CUE, turnover rates, and enzyme activities produce more residues with the potential for incorporation into stable SOM pools compared to taxa characterized exclusively by high CUE.
While physiological traits largely control the pool of available microbial residues, additional microbial traits have been proposed to regulate the incorporation of these residues into stable SOM pools (i.e., MAOM formation traits18), alongside characteristics of the soil matrix65,66. Our results provide empirical support for the existence of MAOM formation traits, suggesting that biomass chemistry, namely protein and phenol contents, may be one such trait influencing the formation of persistent SOM. While the chemical complexity (‘recalcitrance’) of plant and microbial residues is no longer thought to be a dominant control on their long-term retention in soils9,10, the sorptive affinity of individual compounds is an important factor influencing organo-mineral associations8,67. Proteins comprise an important fraction of MAOM because of their unique surface reactivity (e.g., abundance of polar and charged functional groups)68,69 and conformational changes in structure (unfolding) upon association with certain minerals67. Similarly, N-rich microbial metabolites and amino acids have been shown to preferentially accumulate in MAOM fractions55,70 and amino acids are strongly sorbed to Fe and Al (hydr)oxides71. Phenols also exhibit strong sorptive affinity for Fe and Al (hydr)oxides and short-range order minerals72 and while their presence in field soils is commonly attributed to plant origins (e.g., lignin)54,73, the source of many soil phenols is unknown, and a larger fraction may be microbial-derived than is currently recognized, as proposed by Whalen et al.6. (e.g., melanin-derived phenols, phenolic microbial metabolites)14,59.
We observed a positive correlation between the relative abundances of fungal-derived soil proteins and phenols and the amount of chemically stable SOM produced by fungi. Consistent with a recent field study50, these compounds also comprised an important fraction of thermally stable fungal SOM. Since fungal species generated SOM with distinct chemical fingerprints, these results suggest that microbes who produce and release higher quantities of proteins and phenols have a greater potential for the formation of persistent SOM. This finding builds on emerging evidence that microbial community composition influences the chemical composition and stability of SOM14,33. Taxa with the highest SOM formation potentials also generated SOM that was more chemically diverse, and the chemical diversity of SOM was positively correlated with its stability. While our data are currently limited to eight fungal species, these results provide preliminary support for the functional complexity model proposed by Lehmann et al. 17, wherein SOM persistence is a function of its molecular diversity and spatial heterogeneity, which likely increases with the formation of multiple SOM functional pools. Thus, microbial communities and their associated traits may play a role in the emergence of SOM functional complexity in soils, as is recognized for other characteristics of the soil habitat74.
Together, our results build on emerging evidence34,37 that the formation of different SOM functional pools is associated with distinct microbial traits. We demonstrate that SOM functional complexity and stability is promoted by more ‘multifunctional’ microbes with intermediate to high performance across a key grouping of traits (namely, CUE, growth rate, turnover rate, and biomass protein and phenol contents). We show that these traits are synergistic for the formation and stabilization of SOM across functional pools, and thus, representation of these traits among microbial communities may promote soil C storage, while also maintaining the vital ecosystem services provided by soil aggregates and actively cycling SOM pools15–17. Prior trait-based frameworks have proposed microbial life history strategies that influence SOM formation and stabilization27,28, generally emphasizing a single, defining feature associated with each strategy (e.g., high CUE or high investment in enzymatic machinery for resource acquisition28). While these frameworks are based on fundamental microbial tradeoffs in genetic and/or physiological investment in different traits32, they may (inadvertently) portray investment in strategies as if they were mutually exclusive, obscuring the continuous nature of microbial trait expression38. Such frameworks may thereby limit identification of the specific clusters of microbial traits that are involved in SOM formation, and possible synergies among traits. Where possible, future studies should consider synergistic, additive, and/or antagonistic interactions among microbial traits and their net effect on SOM accrual across multiple SOM pools.
Methods
Selection and culturing of fungal isolates
Eight saprotrophic fungal isolates spanning three phyla (Ascomycota, Basidiomycota, Mucoromycota) were selected for inclusion in our study. The Ascomycota included Phacidium lacerum (shortened in figure labels to ‘PhL’), Cylindrium elongatum (CyE), Trichoderma koningii (TrK), and Hypocrea minutispora (Hm). The Basidiomycota included Panellus stipticus (PaS) and a Gymnopus sp. (GyS), and the two Mucoromycota (subphylum Mucoromycotina) isolates were Mucor mucedo (MuM) and Mucor abundans (MuA). Fungal cultures were isolated from decomposing leaf litter, soil or sporocarps in control plots at the Harvard Forest Long-term Ecological Research (LTER) site (Petersham, MA, USA), as described by van Diepen et al.75. Species identification was confirmed using the Basic Local Alignment Search Tool (BLAST)76 and comparison to respective ITS sequences in the NCBI nucleotide database. Taxa were assigned provisional species names as in Morrison et al.32. The eight isolates were selected based on three central criteria: (1) they represent taxa that are highly abundant soil and litter saprotrophs in a model northern hardwood forest, the Harvard Forest (based on past meta-barcoding studies)32, (2) they represent a broad phylogenetic range and were confirmed in a previous liquid culture study to show substantial variation in key physiological traits (e.g., CUE, growth rate)32, and (3) their genomes have been sequenced32. The selected isolates were plated from long-term storage slants onto potato dextrose agar (PDA) plates and maintained at 4 °C for use in subsequent incubations. Prior to each incubation experiment, a fungal plug was transferred to PDA working plates and grown at 25 °C to acclimate to incubation conditions.
Model soil preparation
Model soil was created from a mixture of organic matter-free sand (Quikrete Premium Play Sand; sieved <2 mm), silt (Lab-Aids Pure Silt Soil, 500cc) and montmorillonite clay (Sigma-Aldrich K10 Montmorillonite Powder). To remove native organic matter and adjust soil pH, the sand fraction was acid washed in 1% HCl, and the silt fraction was muffled at 550oC for three hours and subsequently treated four times with concentrated 6 M HCl. The clay fraction was soaked in a bath of 0.1 M CaCl2 to make it homoionic. Soil size separates were combined 70:20:10 (sand:silt:clay) by weight to create a sandy loam texture approximating the soil texture at the Harvard Forest LTER, where fungi were isolated. The final pH of the soil mixture was confirmed with a pH probe (pH = 4.8 ± 0.01), approaching the pH of A horizon soils at Harvard Forest ( ~ 4.6 in control plots)77. Achieving a lower pH was challenging due to the high buffering capacity of the silt fraction. Once homogenized, 50 g soil was weighed into specimen cups and sterilized by autoclaving three times at 120 °C with 24 h rest periods in between each cycle.
Long-term incubation experiment
To quantify each isolate’s potential to form SOM across different functional pools, each of the eight isolates was incubated axenically in model soils. The moisture content of sterilized soils (50 g) was adjusted to 50% water holding capacity with a sterile liquid media solution (pH = 4.5). Substrate media contained 12.6 g L−1 D-glucose (0.42 M C), 1.4 g L−1 potato infusion extract (46.6 mM C, 3.8 mM N), 8.53 g L−1 MES monohydrate buffer (40 mM), and 0.78 g L−1 ammonium nitrate (19.5 mM N). Glucose and potato infusion extract supplied 90% and 10% of substrate C, respectively, and substrate additions were made at a rate of 5.6 mg C g−1 soil (soil C:N = 20). Potato infusion extract was used to provide essential micronutrients and cofactors required for fungal growth. While other C-supplying substrates were considered, glucose was selected due to its rapid assimilation and low level of discrimination among microbial taxa, including saprotrophic fungi78–80. A detailed rationale for this decision is provided in the supplement (see Supplementary Methods and Discussion). Soils were inoculated under sterile conditions with a 5 mm diameter agar plug (50 ml PDA/plate) of the respective fungal culture and sealed in autoclaved quart-size ( ~ 946 ml) mason jars with lids containing rubber septa. Jar flushing (O2 replenishment) and gas sample collection (respiration measurements) were conducted using sterile needles and syringes fitted with 0.2 μm filters (PES membrane; Corning).
Soils were incubated at 25 °C for between 3–6 mo., depending on the growth dynamics of each fungal species. To provide C and nutrients to sustain fungal growth, two additional substrate amendments (1 ml; 5.6 mg C g−1 soil) were applied over the course of the incubation using a sterilized glass syringe and 8-inch stainless steel needle. While more frequent, less concentrated C amendments would have more realistically simulated root exudation rates into soils14, a central constraint of our study was the need to maintain axenic conditions of fungal cultures; therefore, we minimized the number of substrate additions to limit the risk of contamination. A total of 840 mg C was added to each 50 g soil unit over the course of the incubation experiment (16.8 mg C g−1 soil, prior to respiratory losses). Respiration was monitored every 12–96 h (depending on isolate growth stage) using an infrared gas analyzer (LI-COR 6252 IRGA), and the timing of substrate amendments was determined on an isolate-specific basis after each isolate had undergone an exponential growth phase (after prior substrate amendment) followed by a period of stationary growth (Supplementary Fig. 3). The second substrate addition was made once isolate respiration rate had reached ≤ 1 μg CO2-C g−1 soil h−1. Before the third substrate addition, we intentionally induced a period of C and nutrient limitation by maintaining isolates at stationary growth for ~20 days to facilitate biomass and necromass recycling. Soils were harvested once isolate respiration rates returned to <1 μg CO2-C g−1 soil h−1 following the third substrate addition. While alternative approaches to standardization were considered (e.g., standardized by time), it was ultimately determined that the selected approach would allow for the most accurate assessment of trait-SOM relationships. A central concern was ensuring that most of the added substrate media was utilized by fungi prior to incubation harvest; had all species been incubated for the same amount of time, it is more likely that unprocessed substrate would have remained in soils for slower-growing compared with faster-growing species. While it cannot be ruled out that a small fraction of media components remained in soils – especially if fungi experienced nutrient (e.g., N, P) limitation or environmental stress (e.g., space limitation) – our approach to standardization minimized the potential for such biases across fungal taxa (additional rationale provided in supplementary materials).
Experimental units were monitored regularly for contamination (visually and via respiration measurements), and contaminated samples were promptly removed from the experiment. For all but one taxon (Panellus stipticus; Basidiomycota), we achieved at least four replicates that met the criteria for inclusion in subsequent analyses; for P. stipticus, we were only able to include three replicates in subsequent analyses due to contamination challenges with this slow-growing taxa (additional details provided in Supplementary Methods and Discussion). During incubation, approximately half of the Hypocrea minutispora replicates began producing ascocarps and sporulating. We expected that this could influence SOM formation dynamics and alter the chemistry of SOM produced, and thus, samples for this taxon were separately labeled as “sporulating” (HmS) or “non-sporulating” (HmN) (four replicates each). Four sterile control samples (50 g) were established with the equivalent amount of substrate added to each sample containing fungal cultures (840 mg C total) and were harvested 24 h later to preclude the possibility of contamination. After harvest, soils were gently homogenized by passing them through a 4 mm sieve and subsequently air-dried for 72 h. Separate subsamples were used for the characterization of SOM functional pools, SOM chemistry, and a subset of the fungal trait measurements (hyphal lengths, hyphal surface area).
Trait measurements
A suite of trait measurements was conducted to characterize physiological, morphological, and biochemical traits of the fungal isolates. At the end of the long-term incubations, soil was subsampled (3 g) to directly characterize the length and diameter of hyphae produced by each isolate in the incubated soils. Fungal hyphae were extracted from soils and stained following standard methods outlined in Brundrett et al.81. Stained hyphae were evaluated at 200x magnification, and the line intersect method82 was used to estimate hyphal lengths. Hyphal diameters were assessed at 400x magnification and quantified using AmScope software (AmScope, Irvine, CA). Ten measurements of hyphal diameter were taken and then averaged for each replicate (four replicates per isolate). The hyphal surface area was calculated as follows: HSA = (2 π r h) + (2 π r), where r represents the hyphal radius (1/2 hyphal diameter), and h represents the hyphal length. It should be noted that the hyphal length and HSA measurements represent the low end of possible values for these variables, as some of the hyphae produced earlier in the experiment may have been recycled prior to incubation harvest.
For all remaining trait measurements, separate short-term incubations were conducted for each isolate in model soils, with a small subset of measurements conducted in liquid culture as required (3-4 replicates per isolate per trait measurement). All samples were analyzed immediately following incubation harvest. Carbon use efficiency (CUE) and mass-specific growth rates were assessed in both liquid culture and in model soil. We include these multiple metrics to better represent the long-term growth dynamics of fungal taxa during incubation. The liquid culture measurements were conducted in a previous study32 and data are included here as a metric of species’ optimum CUE and growth rate under experimental conditions (25 °C, C:N = 20; same media solution). Fungal biomass was collected from liquid culture conditions through vacuum filtration and weighed to assess growth, while respiration was monitored using the LI-COR infrared gas analyzer. The mass-specific growth rate was calculated as biomass-C produced biomass-C−1 hr−1 during log-phase growth by regressing the log of biomass over time (i.e., the standard approach to measure mass-specific growth). In a small lab incubation mirroring this liquid culture approach32, soil CUE and growth rate were assessed during log-phase growth. Twenty-one experimental units (10 g soil) per isolate were incubated at 25 °C for ~1 week to 1 month (depending on isolate growth rate). Experimental units were destructively harvested at seven-time points across each isolate’s respective log-phase growth curve (3 replicates x 7 sampling times), and a ~ 0.3 g soil sample was flash frozen and stored at −80 °C for downstream DNA extraction. Respiration was measured 12 h before, and then again immediately before each harvest to assess respiratory losses of CO2 (LI-COR analysis, described above).
Soil DNA was extracted as a metric of fungal biomass growth using a modified phenol-chloroform extraction procedure designed for clay- and iron oxide-rich soils83,84 and adapted for model soils (e.g., mineral packets)85. All solutions used in this procedure were nuclease-free. Briefly, soils ( ~ 0.3 g) received 1.168 ml urea extraction buffer (4.67 ml g−1) and 82.5 μl lysis buffer (4 M guanidine isothiocyanate, 1% sarkosyl, 1 M NaPO4, 10% bovine serum albumin, 10 μl L−1 2-(β)-mercaptoethanol) at pH 8, and lysing matrix E beads (MP Biomedicals, Santa Ana, CA). Tubes were shaken on a Biotech Mini Bead Beater for 2.5 min, and subsequently centrifuged. The supernatant was transferred to a new tube and combined with 325 μl detergent solution (5% sarkosyl, 5% CTAB), 168 μl 5 M potassium acetate, and an equal volume of 24:1 chloroform/isoamyl alcohol. Samples were centrifuged at 10 K x g for 10 min, and the supernatant was transferred to a new tube and combined with 0.6 volumes of isopropanol and 2 μl GlycoBlue co-precipitant (Invitrogen). Samples were incubated for 1.5 h at room temperature and subsequently centrifuged for 30 min at 16 K x g. The resulting supernatant was removed from the tube, leaving only the DNA pellet, which was washed with 1 ml 70% ice-cold ethanol (molecular grade). Samples were centrifuged for 10 min at 16 K x g, ethanol was removed, and pellets were air-dried and resuspended in 90 μl Tris EDTA buffer. DNA solutions were frozen overnight and purified by incubation with 7.5 M LiCl (30 min at 4 °C) the following day. After incubation, samples were centrifuged, supernatant removed, and combined with 80 μl isopropanol. Samples were incubated for 30 min at −20 °C, centrifuged, supernatant removed, and the DNA pellet washed with 70% ice-cold ethanol. DNA pellets were dried and then dissolved in 20 μl PCR-grade water. Lastly, DNA was quantified using a Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY) following the manufacturer’s protocol.
Microbial biomass C (MBC) was estimated from DNA concentrations using a published conversion factor of 5.0 to convert μg DNA g−1 soil to μg MBC g−1 soil86. To assess the validity of this approach for our samples, we generated a species-specific conversion factor for each isolate by extracting DNA from a known quantity (1-2 g) of fungal biomass32. Because of low replication, we calculated an average conversion factor for all eight isolates ( ~ 4.5), which was comparable to the published conversion factor86. After conversion from DNA to MBC, we regressed MBC over time (separately for each isolate) and took the slope of the linear portion of each growth curve to estimate isolate growth rates. Carbon use efficiency was subsequently calculated as (growth/growth + respiration), where respiration rates were acquired by taking the slope of the linear portion of the curve for respiration (μg CO2-C produced g−1 soil h−1) regressed over time, as in Morrison et al.32. Standard errors of CUE were calculated by pooling the standard errors of the slopes for growth and respiration32.
Soil CUE was also assessed during stationary phase growth using the 18O-water tracing method, which quantifies the incorporation of 18O-labeled water into DNA to measure gross growth63,87. Four additional replicates per isolate (10 g soil) were incubated at 25 °C and harvested when each isolate was determined to have reached stationary growth (steady state; based on respiration measurements). Steady-state was selected as the timepoint for harvest and for initiation of the 18O-water tracing assay to meet the assumptions for estimating biomass turnover. The 18O-water tracing assay was conducted following methods recommended in Geyer et al.63. Briefly, enriched ( ~ 97 at%) 18O-water was diluted with unlabeled filtered deionized water (FDIW) to achieve a target soil moisture enrichment of 20 atom%. Approximately 0.3 g soil was weighed into microcentrifuge tubes, receiving either a labeled (18O) or unlabeled (16O; control) FDIW solution. Soils were sealed in 50 ml amber vials fitted with rubber septa and flushed with CO2-free air. Soils were incubated for 48 h, whereafter respiration measurements were taken and soils were flash frozen and stored at −80 oC until DNA extraction. DNA was extracted using the modified phenol-chloroform procedure described above. DNA extracts were mixed with a diluted salmon DNA solution (20 μg ul−1) to bring total oxygen mass within detection limits and dried overnight in silver encapsulation tins at 60 °C. Tins containing dried DNA extracts were sent to the Cornell Stable Isotope Laboratory for δ18O quantification.
Total microbial growth (production of new 18O-labeled DNA) was estimated using a two-pool mixing model63,88. Typically, microbial growth values are scaled to MBC using a ratio of MBC:DNA generated through chloroform fumigation extraction (CFE). Our attempts to extract and quantify biomass from model soils using the CFE approach were unsuccessful, potentially due to the high potential for cell lysates to re-sorb to clean mineral surfaces in these model soils. Therefore, we quantified DNA concentrations by Qubit and used the published conversion factor for DNA to MBC as described above86. Biomass turnover length and CUE were calculated as in Geyer et al.63. For one Basidiomycota species (Gymnopus sp.), turnover rate may be artificially inflated relative to other taxa due to especially slow linear-phase growth which may have violated assumptions of the 18O method; however, we assume that samples for most species generally met the steady-state requirement.
Soils were assayed for potential activities of hydrolytic and oxidative enzymes following standard methods89,90. Four replicates (10 g soil) were established for each isolate alongside the experimental units for measuring soil CUE, and were destructively harvested and homogenized before subsampling for enzyme assays. The hydrolytic enzymes cellobiohydrolase (CBH), ß-glucosidase (BG), acid phosphatase (PHOS), N-acetyl-ß- glucosaminidase (NAG) and leucine aminopeptidase (LAP) were assessed fluorometrically using the substrates β-D-cellobioside, β-D-glycopyranoside, phosphate, N-acetyl-β-D-glucosaminide and L-Leucine, respectively. The oxidative enzymes peroxidase (OX1) and phenol oxidase (OX2) were measured colorimetrically with the substrates 3,3’, 5,5’-Tetramethylbenzidine (TMB + H2O2)91 and 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS)92, respectively. Fluorescence (hydrolytic) and absorbance (oxidative) were measured on a BioTek Synergy HT Multi-detection Microplate Reader at 460 nm and 450 nm, respectively. Final enzyme activity values were calculated as in DeForest93 and are reported as μmol substrate h−1 g−1 dry soil.
To characterize isolate biomass chemistry and melanin content, four replicates of each isolate were grown in potato dextrose broth media to facilitate biomass collection. The liquid media contained the same concentrations of D-glucose, potato infusion extract, MES buffer, and ammonium nitrate (C:N = 20) as the substrate media used in the model soil experiments, and the pH was also adjusted to 4.5. Cultures were inoculated into sterilized glass vials containing 100 ml liquid media and shaken continuously at 75 rpm32. Cultures were grown at 25 °C for between 3–30 days depending on species’ growth rate. Fungal biomass was collected by vacuum filtration and washed three times with FDIW (100 ml/rinse). Biomass was subsequently dried at 70 °C and subsampled for biomass chemistry and melanin analyses. The chemical composition of isolate biomass (ground subsamples) was characterized by pyrolysis gas chromatography-mass spectrometry (Py-GC/MS), following methods described previously94,95, and the relative proportions of broad chemical compound classes (e.g., polysaccharides, aromatics, lipids, proteins) were determined (additional details below). Biomass melanin concentrations were measured by quantitative colorimetric analysis with Azure A dye96. Briefly, dried biomass was incubated for 90 min in an Azure A solution (initial absorbance = 0.665 at 610 nm) and subsequently filtered through a 0.45 μm nitrocellulose syringe tip filter. Absorbance of the filtrate was measured with a spectrophotometer at 610 nm and the change in absorbance was compared against a standard curve to calculate biomass melanin content. The standard curve was constructed using known amounts of melanin isolated from Cenococcum geophilum biomass, as in ref. 96.
Total C, MAOM-C and POM-C analyses
Air-dried soil (5 g) from the long-term incubation study was separated into MAOM ( < 53 μm) and POM ( > 53 μm) fractions using a standard size-based fractionation protocol97,98. Soils were dispersed via shaking with dilute (0.5%) sodium hexametaphosphate for 18 h and were subsequently rinsed through a 53 μm sieve. The MAOM fraction underwent an additional rinse step, followed by centrifugation, to remove DOC that may have been present in the water column. MAOM and POM fractions were oven dried at 105 °C and 60 °C, respectively. Subsamples of MAOM, POM and whole soil were finely ground and analyzed for total C and N by dry combustion analysis (Perkin-Elmer Series II 2400 Elemental Analyzer; Perkin Elmer Inc.).
Stable aggregate formation
The potential of each fungal species to generate water-stable aggregates was assessed using a standard wet-sieving procedure (USDA NRCS, 1999). Since the model soil mixture began as a combination of un-aggregated particle separates (sand, silt, clay), any aggregates that formed could be attributed to the growth and activity of the fungal isolates. After harvest of the long-term incubations, soils (5 g) were weighed onto a 250 μm sieve and wetted through capillary action (5 min) by positioning the surface of the sieve screen just above water-level within a plastic bin containing ~1 inch DI water. Thereafter, soils were wet-sieved for three minutes. Deionized water was added to the bin such that the soil on the sieve was completely submerged. The sieve was moved up and down over a vertical distance of 1.5 cm at a rate of 30 oscillations per minute. At the end of the three-minute period, aggregates remaining on the sieve surface were transferred to an aluminum drying pan and oven-dried at 105 °C. Water-stable aggregates were calculated as the proportion of the initial 5 g soil that remained on the 250 μm sieve after wet-sieving, minus the amount of remaining soil > 250 μm present in sterile control samples (Sample(% of soil > 250 μm) – Control(%soil > 250 um)).
Chemical and biological stability of SOM
We determined the chemical stability of fungal-derived SOM using a sequential chemical extraction procedure. Each soil sample was subjected to three sequential C-free extractants to isolate discrete pools of SOM that form distinct bonds and/or associations with soil minerals99,100. Water was used first to extract readily solubilized compounds, followed by KCl (1 M; pH = 4.8) to isolate easily exchangeable pools, and finally by sodium pyrophosphate (0.1 M; pH 10) to extract compounds bound via stronger electrostatic forces or within organo-metal complexes. For each extraction, 30 ml extraction reagent was added to 1 g soil, vortexed, and shaken for 4 h (water, KCl) or 16 h (sodium pyrophosphate) at 200 rpm99,100. Samples were centrifuged and the supernatant was filtered using 0.2 μm nylon syringe tip filters. Extracts were analyzed for total organic C (TOC) by high-temperature catalytic oxidation followed by non-dispersive infrared detection with a Shimadzu TOC analyzer (TOC-LSH; Shimadzu Corporation, Kyoto, Japan). The percent of initial soil C (pre-extraction) removed by each extractant was calculated for each isolate (full results included in supplementary materials; Supplementary Fig. 13). The proportion of initial C remaining in the soil pellet (that is, C not removed by the three extractants) was calculated as a metric of SOM chemical stability (% “chemically stable” C). These values are reported as (sample-control) to account for the small amount of C remaining in uninoculated control samples after sequential extraction, likely representing the fraction of substrate C that was directly sorbed to minerals.
We also assessed the biological stability of fungal-derived SOM in a separate 3-month incubation (sensu Kallenbach et al.14). After soils were harvested from the long-term incubation experiment (described above), 10 g soil subsamples (4 replicates/isolate) were re-inoculated with a mixed microbial community and 13C-glucose (to stimulate decomposition), and the relative stability of SOM generated by each isolate was assessed. Since unlabeled (12C) glucose was used as a substrate in the original incubation experiment, we applied 13C-labeled glucose (25 atom%; 50 μg C g−1 soil, as in Kallenbach et al.14.) in the 3-month biological stability assay, such that we could parse microbial respiration derived from the newly added glucose (13CO2) versus existing fungal-derived SOM (12CO2). The microbial inoculum was prepared from soil collected from control plots at the Harvard Forest LTER. Soil was combined with FDIW (1:25 w/v; as in ref. 101) and the viability of the inoculum was confirmed at pH 4.5 by measuring its respiration for three days following the addition of 120 mg L−1 glucose72. The microbial inoculum was added in 1 ml aliquots to each 10 g sample, and additional FDIW was added to bring soil moisture back up to 50% field capacity. Soils were incubated in mason jars at 25 °C and were hand-mixed once weekly to encourage C mineralization14. A water control was established alongside each 13C-glucose amended sample to represent background δ13C values. After 3 mo. incubation, soils were harvested and analyzed for δ13C and total C (%) at the Cornell Stable Isotope Laboratory. A standard isotope mixing model102 was used to calculate the relative proportion of isolate-derived SOM that resisted decomposition by the mixed microbial community (% “biologically stable” C). These values are reported as (sample-control) as described above for chemically stable C.
Chemical composition of fungal-derived SOM
We characterized the chemistry of fungal-derived SOM using Py-GC/MS, following methods described in refs. 94,95. Subsamples of whole soil and the MAOM fraction were analyzed separately for each isolate. MAOM fraction samples were pyrolyzed at a single temperature (600 °C) on a CDS Pyroprobe 5150 pyrolyzer (CDS Analytical Inc., Oxford, PA), following standard protocol, whereas whole soil samples were analyzed using a modified ramped pyrolysis procedure (sensu50), whereby each sample was pyrolyzed at increasing temperatures (330, 396, 444, 503, 600 and 735 °C) in a stepwise fashion. This approach characterizes the composition of organic compounds that require different activation energies for thermal decomposition as a means of assessing the relative thermal stability of SOM50. Following pyrolysis (of MAOM or whole soil samples), decomposed products were transferred to a Thermo Trace GC Ultra gas chromatograph (Thermo Fisher Scientific, Austin, TX) and subsequently to a Polaris Q mass spectrometer (Thermo Fisher Scientific). Products were ionized and detected using AMDIS (V. 2.69) and recorded peaks were classified against the National Institute of Standards and Technology (NIST) compound library. Relative percentages of organic matter compounds were summarized within broad compound classes (polysaccharides, lipids, aromatics, phenolics, proteins, N-bearing (non-proteins) or unknown source)94.
Statistical analyses
All statistics were performed in R 4.3.0 unless stated otherwise. Differences in species’ trait values and contributions to SOM functional pools were evaluated using one-way ANOVA and post-hoc Tukey HSD tests (P < 0.05). Levene’s test of homogeneity of variances and the Shapiro-Wilk normality test were used to assess homoscedasticity and normality of residuals, respectively. When normality or homoscedasticity assumptions were not met, data were evaluated using non-parametric Kruskal-Wallis and post-hoc Dunn tests, or were square-root or log-transformed when used in other analyses (e.g., regressions).
We used partial least squares regression (PLSR) to evaluate the relative importance of fungal traits associated with the formation of different SOM functional pools. A separate PLSR model was constructed for each SOM pool (response variable), and all measured fungal trait variables were included as predictors. PLSR was conducted in JMP Pro (version 16.1.0) using the NIPALS algorithm (as in ref. 103). Due to the nature of our dataset, where some of the trait measurements were collected on independent experimental units separate from the long-term incubated samples analyzed for SOM, we used a randomization approach with three iterations to construct our final PLSR model results. We ran three separate PLSR analyses, each run on a separate randomized dataset representing the three possible pairings between each trait measurement replicate (3 replicates per isolate, representing the minimum number of replicates available for certain traits) and each SOM functional pool replicate (3 replicates/isolate) (full model results in Supplementary Table 3a, b). After model results were collated, we calculated the average X loadings and variable importance in projection (VIP) scores for each trait variable across the three PLSR model iterations. Predictor variables with a VIP score greater than 0.8 were retained in the final models. Thereafter, we fit linear regressions between SOM functional pools and the trait variable with the strongest loading on each pool’s most explanatory PLSR latent factor, and we used ANOVA to assess the significance of these relationships.
Principal components analysis (PCA) was used to summarize fungal trait variables along major axes of variance. PCA was conducted using the ‘rda’ function in the ‘vegan’ package104 on z-score standardized values for each trait variable. Principle component axes were retained if eigenvalues were greater than one, and such that cumulative variation explained reached 80%. For ease of interpretation, only the two most explanatory axes (PC1 and PC2), explaining a total of 45% of variation in fungal trait data, are presented and visualized using ordination. Multivariate ANOVA (MANOVA) was used to test for statistical differences among species and phyla (‘adonis2’ function, Euclidian distance method). Additionally, we examined relationships between PC1 and SOM functional pools by fitting polynomial regression models with the lowest mean square error between each predictor and response variable. When the single-degree model (h = 1) exhibited the lowest mean square error value, linear regression was used.
To generate a metric of trait multifunctionality as well as SOM formation and stabilization potential (essentially, SOM multifunctionality) for each isolate, we adapted an approach for calculating “ecosystem multifunctionality”48,105. Broadly, multifunctionality is defined as the simultaneous performance or provisioning of multiple functions48. We used a modified version of the averaging approach48 that uses Hill numbers (q) to produce a single metric of “effective” multifunctionality, which accounts for both the total provisioning of functions as well as the evenness (e.g., relative trait values) of those provisions of function49. Trait (or SOM) multifunctionality was calculated using the ‘multifunc’ package (v. 0.9.4)48,49 as the product of the effective number of functions performed by each isolate and the arithmetic mean performance of the measured functions. Values for turnover time (originally reported in days) were reflected such that the maximum level of each trait variable in our dataset represented the “best” or highest function. We multiplied turnover time values by −1 and then added the un-reflected maximum value such that the lowest level of transformed function was zero48. All trait values were then standardized by the maximum observed value to a common scale between 0 to 1 before analysis. Effective multifunctionality was calculated at order q = 1 to accommodate information about unequal levels of functioning proportional to the relative functional performance, without making any uncomfortable decisions about upweighting high-performing functions49. Radar plots were used to visualize trait performance across isolates, with data scaled between 0 to 1 prior to plotting, and differences in effective multifunctionality were evaluated using ANOVA.
Finally, we assessed the effect of isolate identity on SOM and MAOM chemical composition using permutational multivariate analysis of variance (PERMANOVA)106 on a Bray-Curtis dissimilarity matrix calculated from the relative abundance values of individual compounds identified by Py-GC/MS. For the whole soil samples that were analyzed with the ramped pyrolysis approach, separate PERMANOVA analyses were run for each thermal fraction. To visualize the effects of species identity on SOM chemistry, we conducted non-metric multidimensional scaling (NMDS) ordination of the Bray-Curtis distance values. Environmental vectors representing broad Py-GC/MS compound classes were fitted to the NMDS using the ‘envifit’ function (‘vegan’ package) and were included in the plot if significant (P < 0.05). Lastly, we calculated SOM chemical diversity for each species within the most thermally stable fraction (735 °C) of SOM using the Shannon index (individual compound-level dataset).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Jessica Ernakovich, Marco Keiluweit, and Rich Smith for providing feedback on the earlier stages of this manuscript. Thank you also to Mel Knorr for laboratory support, as well as Andrea Jilling and Noah Sokol for feedback on experimental design and analytical approaches. This work was supported by a University of New Hampshire Dissertation Year Fellowship to E.D.W., as well as a research grant from the U.S. Department of Agriculture National Institute of Food and Agriculture through the New Hampshire Agricultural Experiment Station (NHAES; Hatch NH-00701). This is NHAES Scientific Contribution Number 3001.
Author contributions
E.D.W., A.S.G., and S.D.F. conceived of the study, and K.G. and E.W.M. contributed to early experimental design and methodological development. E.D.W. led laboratory work, sample analysis, data analysis, and writing. All authors contributed to data interpretation and editing of the manuscript.
Peer review
Peer review information
Nature Communications thanks Tessa Camenzind and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The fungal trait, SOM pool, and SOM chemistry data that support the findings of this study have been deposited in the Environmental Data Initiative repository, 10.6073/pasta/cf2a305c7d21938ddad3cb9430d22ed3. Source data associated with each main text and Supplementary Figure are also available as a supplementary Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53947-2.
References
- 1.Janzen, H. H. Carbon cycling in earth systems—a soil science perspective. Agriculture, Ecosyst. Environ.104, 399–417 (2004). [Google Scholar]
- 2.Liang, C., Schimel, J. P. & Jastrow, J. D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol.2, 17105 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Grandy, A. S. & Neff, J. C. Molecular C dynamics downstream: The biochemical decomposition sequence and its impact on soil organic matter structure and function. Sci. Total Environ.404, 297–307 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Miltner, A., Bombach, P., Schmidt-Brücken, B. & Kästner, M. SOM genesis: Microbial biomass as a significant source. Biogeochemistry111, 41–55 (2012). [Google Scholar]
- 5.Simpson, M. J., Smith, E. & Kelleher, B. P. Microbially Derived Inputs to Soil Organic Matter: Are Current Estimates Too Low? Environ. Sci. Techol.41, 8070–8076 (2007). [DOI] [PubMed]
- 6.Whalen, E. D. et al. Clarifying the evidence for microbial- and plant-derived soil organic matter, and the path toward a more quantitative understanding. Glob. Change Biol.28, 7167–7185 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Baldock, J. A. & Skjemstad, J. O. Role of the soil matrix and minerals in protecting natural organic materials against biological attack. Org. Geochem.31, 697–710 (2000). [Google Scholar]
- 8.Kleber, M. et al. Dynamic interactions at the mineral–organic matter interface. Nat. Rev. Earth Environ.2, 402–421 (2021). [Google Scholar]
- 9.Lehmann, J. & Kleber, M. The contentious nature of soil organic matter. Nature528, 60–68 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Schmidt, M. W. I. et al. Persistence of soil organic matter as an ecosystem property. Nature10.1038/nature10386 (2011). [DOI] [PubMed]
- 11.Bradford, M. A., Keiser, A. D., Davies, C. A., Mersmann, C. A. & Strickland, M. S. Empirical evidence that soil carbon formation from plant inputs is positively related to microbial growth. 271–281 10.1007/s10533-012-9822-0 (2013).
- 12.Buckeridge, K. M. et al. Environmental and microbial controls on microbial necromass recycling, an important precursor for soil carbon stabilization. Commun. Earth Environ.1, 1–9 (2020). [Google Scholar]
- 13.Kallenbach, C. M., Grandy, A. S., Frey, S. D. & Diefendorf, A. F. Microbial physiology and necromass regulate agricultural soil carbon accumulation. Soil Biol. Biochem.91, 279–290 (2015). [Google Scholar]
- 14.Kallenbach, C. M., Frey, S. D. & Grandy, A. S. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun.7, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angst, G. et al. Unlocking complex soil systems as carbon sinks: multi-pool management as the key. Nat. Commun.14, 2967 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffland, E., Kuyper, T. W., Comans, R. N. J. & Creamer, R. E. Eco-functionality of organic matter in soils. Plant Soil455, 1–22 (2020). [Google Scholar]
- 17.Lehmann, J. et al. Persistence of soil organic carbon caused by functional complexity. Nat. Geosci.13, 529–534 (2020). [Google Scholar]
- 18.Sokol, N. W. et al. Global distribution, formation and fate of mineral-associated soil organic matter under a changing climate: A trait-based perspective. Funct. Ecol.36, 1411–1429 (2022). [Google Scholar]
- 19.Lavallee, J. M., Soong, J. L. & Cotrufo, M. F. Conceptualizing soil organic matter into particulate and mineral-associated forms to address global change in the 21st century. Glob. Change Biol.26, 261–273 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Six, J. & Paustian, K. Aggregate-associated soil organic matter as an ecosystem property and a measurement tool. Soil Biol. Biochem.68, A4–A9 (2014). [Google Scholar]
- 21.Hall, S. J., McNicol, G., Natake, T. & Silver, W. L. Large fluxes and rapid turnover of mineral-associated carbon across topographic gradients in a humid tropical forest: insights from paired 14C analysis. Biogeosciences12, 2471–2487 (2015). [Google Scholar]
- 22.Heckman, K. et al. Beyond bulk: Density fractions explain heterogeneity in global soil carbon abundance and persistence. Glob. Change Biol.28, 1178–1196 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Witzgall, K. et al. Particulate organic matter as a functional soil component for persistent soil organic carbon. Nat. Commun.12, 4115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grime, J. P. Evidence for the Existence of Three Primary Strategies in Plants and Its Relevance to Ecological and Evolutionary Theory. Am. Naturalist111, 1169–1194 (1977). [Google Scholar]
- 25.McGill, B. J., Enquist, B. J., Weiher, E. & Westoby, M. Rebuilding community ecology from functional traits. Trends Ecol. Evolution21, 178–185 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Tilman, D. Community Invasibility, Recruitment Limitation, and Grassland Biodiversity. Ecology78, 81–92 (1997). [Google Scholar]
- 27.Anthony, M. A., Crowther, T. W., Maynard, D. S., van den Hoogen, J. & Averill, C. Distinct Assembly Processes and Microbial Communities Constrain Soil Organic Carbon Formation. One Earth2, 349–360 (2020). [Google Scholar]
- 28.Malik, A. A. et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. The ISME Journal10.1038/s41396-019-0510-0 (2019). [DOI] [PMC free article] [PubMed]
- 29.See, C. R. et al. Hyphae move matter and microbes to mineral microsites: Integrating the hyphosphere into conceptual models of soil organic matter stabilization. Glob. Change Biol.28, 2527–2540 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Cotrufo, M. F., Wallenstein, M. D., Boot, C. M., Denef, K. & Paul, E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Change Biol.19, 988–995 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Six, J., Frey, S. D., Thiet, R. K. & Batten, K. M. Bacterial and Fungal Contributions to Carbon Sequestration in Agroecosystems. Soil Sci. Society of Amer.2, 555–569 (2006).
- 32.Morrison, E. W. et al. Evidence for a genetic basis in functional trait tradeoffs with microbial growth rate but not growth yield. Soil Biol. Biochem.172, 108765 (2022). [Google Scholar]
- 33.Domeignoz-Horta, L. A. et al. Direct evidence for the role of microbial community composition in the formation of soil organic matter composition and persistence. ISME COMMUN1, 1–4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig, M. E. et al. Fast-decaying plant litter enhances soil carbon in temperate forests but not through microbial physiological traits. Nat. Commun.13, 1229 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao, F. et al. Microbial carbon use efficiency promotes global soil carbon storage. Nature618, 981–985 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, C. et al. Large-scale importance of microbial carbon use efficiency and necromass to soil organic carbon. Glob. Change Biol.27, 2039–2048 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Sokol, N. W. et al. The path from root input to mineral-associated soil carbon is dictated by habitat-specific microbial traits and soil moisture. Soil Biol. Biochem.193, 109367 (2024). [Google Scholar]
- 38.Aguilar-Trigueros, C. A., Powell, J. R., Anderson, I. C., Antonovics, J. & Rillig, M. C. Ecological understanding of root-infecting fungi using trait-based approaches. Trends Plant Sci.19, 432–437 (2014). [DOI] [PubMed] [Google Scholar]
- 39.He, L. et al. Global biogeography of fungal and bacterial biomass carbon in topsoil. Soil Biol. Biochem.151, 108024 (2020). [Google Scholar]
- 40.Yu, K. et al. The biogeography of relative abundance of soil fungi versus bacteria in surface topsoil. Earth Syst. Sci. Data14, 4339–4350 (2022). [Google Scholar]
- 41.Joergensen, R. G. Amino sugars as specific indices for fungal and bacterial residues in soil. Biol. Fertil. Soils54, 559–568 (2018). [Google Scholar]
- 42.Liang, C., Amelung, W., Lehmann, J. & Kästner, M. Quantitative assessment of microbial necromass contribution to soil organic matter. Glob. Change Biol.25, 3578–3590 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Schneider, T., Keiblinger, K. M., Schmid, E. & Sterflinger-gleixner, K. Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. 1749–1762 10.1038/ismej.2012.11 (2012). [DOI] [PMC free article] [PubMed]
- 44.Starke, R. et al. Niche differentiation of bacteria and fungi in carbon and nitrogen cycling of different habitats in a temperate coniferous forest: A metaproteomic approach. Soil Biol. Biochem.155, 108170 (2021). [Google Scholar]
- 45.Pronk, G. J. et al. Interaction of minerals, organic matter, and microorganisms during biogeochemical interface formation as shown by a series of artificial soil experiments. Biol. Fertil. Soils53, 9–22 (2017). [Google Scholar]
- 46.Del Valle, I., Gao, X., Ghezzehei, T. A., Silberg, J. J. & Masiello, C. A. Artificial Soils Reveal Individual Factor Controls on Microbial Processes. mSystems7, e00301–e00322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golchin, A., Clarke, P. & Oades, J. M. The Heterogeneous Nature of Microbial Products as Shown by Solid-State 13C CP/MAS NMR Spectroscopy. Biogeochemistry34, 71–97 (1996). [Google Scholar]
- 48.Byrnes, J. E. K. et al. Investigating the relationship between biodiversity and ecosystem multifunctionality: challenges and solutions. Methods Ecol. Evol.5, 111–124 (2014). [Google Scholar]
- 49.Byrnes, J. E. K., Roger, F. & Bagchi, R. Understandable multifunctionality measures using Hill numbers. Oikos2023, e09402 (2023). [Google Scholar]
- 50.Sanderman, J. & Grandy, A. S. Ramped thermal analysis for isolating biologically meaningful soil organic matter fractions with distinct residence times. SOIL6, 131–144 (2020). [Google Scholar]
- 51.Chenu, C. & G, S. Interactions Between Microorganisms and Soil Particles. in 3–40 (2002).
- 52.Lehmann, A., Leifheit, E. F. & Rillig, M. C. Mycorrhizas and Soil Aggregation. Mycorrhizal Mediation of Soil: Fertility, Structure, and Carbon Storage 241–262 10.1016/B978-0-12-804312-7.00014-0 (2017).
- 53.Oades, J. M. Soil organic matter and structural stability: mechanisms and implications for management. Plant Soil76, 319–337 (1984). [Google Scholar]
- 54.Angst, G., Mueller, K. E., Nierop, K. G. J. & Simpson, M. J. Plant- or microbial-derived? A review on the molecular composition of stabilized soil organic matter. Soil Biol. Biochem.156, 108189 (2021). [Google Scholar]
- 55.Keiluweit, M. et al. Nano-scale investigation of the association of microbial nitrogen residues with iron (hydr)oxides in a forest soil O-horizon. Geochimica et. Cosmochimica Acta95, 213–226 (2012). [Google Scholar]
- 56.Kleber, M. et al. Mineral-Organic Associations: Formation, Properties, and Relevance in Soil Environments. Adv. Agronomy130, 1–140 (2015).
- 57.Klink, S. et al. Stable isotopes reveal that fungal residues contribute more to mineral-associated organic matter pools than plant residues. Soil Biol. Biochem.168, 108634 (2022). [Google Scholar]
- 58.Op De Beeck, M., Persson, P. & Tunlid, A. Fungal extracellular polymeric substance matrices – Highly specialized microenvironments that allow fungi to control soil organic matter decomposition reactions. Soil Biol. Biochem.159, 108304 (2021). [Google Scholar]
- 59.Wang, T., Tian, Z., Bengtson, P., Tunlid, A. & Persson, P. Mineral surface-reactive metabolites secreted during fungal decomposition contribute to the formation of soil organic matter. Environ. Microbiol.19, 5117–5129 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Kneitel, J. M. & Chase, J. M. Trade-offs in community ecology: linking spatial scales and species coexistence. Ecol. Lett.7, 69–80 (2004). [Google Scholar]
- 61.Nunan, N., Schmidt, H. & Raynaud, X. The ecology of heterogeneity: soil bacterial communities and C dynamics. Philos. Trans. R. Soc. B: Biol. Sci.375, 20190249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prommer, J. et al. Increased microbial growth, biomass, and turnover drive soil organic carbon accumulation at higher plant diversity. Glob. Change Biol.26, 669–681 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geyer, K. M., Dijkstra, P., Sinsabaugh, R. & Frey, S. D. Clarifying the interpretation of carbon use efficiency in soil through methods comparison. Soil Biol. Biochem.128, 79–88 (2019). [Google Scholar]
- 64.Horsch, C. C. A., Antunes, P. M., Fahey, C., Grandy, A. S. & Kallenbach, C. M. Trait-based assembly of arbuscular mycorrhizal fungal communities determines soil carbon formation and retention. N. Phytologist239, 311–324 (2023). [DOI] [PubMed] [Google Scholar]
- 65.Creamer, C. A. et al. Mineralogy dictates the initial mechanism of microbial necromass association. Geochimica et. Cosmochimica Acta260, 161–176 (2019). [Google Scholar]
- 66.Mikutta, R. et al. Microbial and abiotic controls on mineral-associated organic matter in soil profiles along an ecosystem gradient. Sci. Rep.9, 10294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleber, M., Sollins, P. & Sutton, R. A conceptual model of organo-mineral interactions in soils: Self-assembly of organic molecular fragments into zonal structures on mineral surfaces. Biogeochemistry85, 9–24 (2007). [Google Scholar]
- 68.Rillig, M. C., Caldwell, Æ. B. A., Wo, H. A. B. & Sollins, Æ. P. Role of proteins in soil carbon and nitrogen storage: controls on persistence. 25–44 10.1007/s10533-007-9102-6 (2007).
- 69.Dümig, A., Häusler, W., Steffens, M. & Kögel-Knabner, I. Clay fractions from a soil chronosequence after glacier retreat reveal the initial evolution of organo–mineral associations. Geochimica et. Cosmochimica Acta85, 1–18 (2012). [Google Scholar]
- 70.Kopittke, P. M. et al. Nitrogen-rich microbial products provide new organo-mineral associations for the stabilization of soil organic matter. Glob. Change Biol.24, 1762–1770 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Kaiser, K., Guggenberger, G. & Haumaier, L. Changes in dissolved lignin-derived phenols, neutral sugars, uronic acids, and amino sugars with depth in forested Haplic Arenosols and Rendzic Leptosols. Biogeochemistry70, 135–151 (2004). [Google Scholar]
- 72.Mikutta, R. et al. Biodegradation of forest floor organic matter bound to minerals via different binding mechanisms. Geochimica et. Cosmochimica Acta71, 2569–2590 (2007). [Google Scholar]
- 73.Sanderman, J., Maddern, T. & Baldock, J. Similar composition but differential stability of mineral retained organic matter across four classes of clay minerals. Biogeochemistry121, 409–424 (2014). [Google Scholar]
- 74.Philippot, L., Chenu, C., Kappler, A., Rillig, M. C. & Fierer, N. The interplay between microbial communities and soil properties. Nat. Rev. Microbiol22, 226–239 (2024). [DOI] [PubMed] [Google Scholar]
- 75.Van Diepen, L. T. A., Frey, S. D., Landis, E. A., Morrison, E. W. & Pringle, A. Fungi exposed to chronic nitrogen enrichment are less able to decay leaf litter. Ecology98, 5–11 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 77.Whalen, E. D., Smith, R. G., Grandy, A. S. & Frey, S. D. Manganese limitation as a mechanism for reduced decomposition in soils under atmospheric nitrogen deposition. Soil Biol. Biochem.127, 252–263 (2018).
- 78.Jones, D. L. et al. Role of substrate supply on microbial carbon use efficiency and its role in interpreting soil microbial community-level physiological profiles (CLPP). Soil Biol. Biochem.123, 1–6 (2018). [Google Scholar]
- 79.Hill, P. W., Farrar, J. F. & Jones, D. L. Decoupling of microbial glucose uptake and mineralization in soil. Soil Biol. Biochem.40, 616–624 (2008). [Google Scholar]
- 80.Rousk, J. & Bååth, E. Growth of saprotrophic fungi and bacteria in soil. FEMS Microbiol. Ecol.78, 17–30 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Brundrett, M. et al. Extraction and staining of hyphae from soil. Practical Methods in Mycorrhizal Research. 71–80 (Mycologue Publications, Waterloo, 1994).
- 82.Tennant, D. A Test of a Modified Line Intersect Method of Estimating Root Length. J. Ecol.63, 995–1001 (1975). [Google Scholar]
- 83.Griffiths, R. I., Whiteley, A. S., O’Donnell, A. G. & Bailey, M. J. Rapid Method for Coextraction of DNA and RNA from Natural Environments for Analysis of Ribosomal DNA- and rRNA-Based Microbial Community Composition. Appl Environ. Microbiol66, 5488–5491 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi, S. et al. Successional Trajectories of Rhizosphere Bacterial Communities over Consecutive Seasons. mBio6, 10.1128/mbio.00746-15 (2015). [DOI] [PMC free article] [PubMed]
- 85.Whitman, T. et al. Microbial community assembly differs across minerals in a rhizosphere microcosm. Environ. Microbiol.20, 4444–4460 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Anderson, T.-H. & Martens, R. DNA determinations during growth of soil microbial biomasses. Soil Biol. Biochem.57, 487–495 (2013). [Google Scholar]
- 87.Blazewicz, S. J. & Schwartz, E. Dynamics of 18O Incorporation from H218O into Soil Microbial DNA. Micro. Ecol.61, 911–916 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Spohn, M., Klaus, K., Wanek, W. & Richter, A. Microbial carbon use efficiency and biomass turnover times depending on soil depth – Implications for carbon cycling. Soil Biol. Biochem.96, 74–81 (2016). [Google Scholar]
- 89.Saiya-Cork, K. R., Sinsabaugh, R. L. & Zak, D. R. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem.34, 1309–1315 (2002). [Google Scholar]
- 90.Van Diepen, L. T. A. et al. Changes in litter quality caused by simulated nitrogen deposition reinforce the N-induced suppression of litter decay. Ecosphere6, 1–16 (2015). [Google Scholar]
- 91.Johnsen, A. R. & Jacobsen, O. S. A quick and sensitive method for the quantification of peroxidase activity of organic surface soil from forests. Soil Biol. Biochem.40, 814–821 (2008). [Google Scholar]
- 92.Floch, C., Alarcon-Gutiérrez, E. & Criquet, S. ABTS assay of phenol oxidase activity in soil. J. Microbiol. Methods71, 319–324 (2007). [DOI] [PubMed] [Google Scholar]
- 93.DeForest, J. L. The influence of time, storage temperature, and substrate age on potential soil enzyme activity in acidic forest soils using MUB-linked substrates and l-DOPA. Soil Biol. Biochem.41, 1180–1186 (2009). [Google Scholar]
- 94.Grandy, A. S., Strickland, M. S., Lauber, C. L., Bradford, M. A. & Fierer, N. The influence of microbial communities, management, and soil texture on soil organic matter chemistry. Geoderma150, 278–286 (2009). [Google Scholar]
- 95.Wickings, K., Grandy, A. S., Reed, S. & Cleveland, C. Management intensity alters decomposition via biological pathways. Biogeochemistry104, 365–379 (2011). [Google Scholar]
- 96.Fernandez, C. W. & Koide, R. T. Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biol. Biochem.77, 150–157 (2014). [Google Scholar]
- 97.Cotrufo, M. F., Ranalli, M. G., Haddix, M. L., Six, J. & Lugato, E. Soil carbon storage informed by particulate and mineral-associated organic matter. Nat. Geosci.12, 989–994 (2019). [Google Scholar]
- 98.Poeplau, C. et al. Isolating organic carbon fractions with varying turnover rates in temperate agricultural soils – A comprehensive method comparison. Soil Biol. Biochem.125, 10–26 (2018). [Google Scholar]
- 99.Jilling, A., Keiluweit, M., Gutknecht, J. L. M. & Grandy, A. S. Priming mechanisms providing plants and microbes access to mineral-associated organic matter. Soil Biol. Biochem.158, 108265 (2021). [Google Scholar]
- 100.Wagai, R., Mayer, L. M., Kitayama, K. & Shirato, Y. Association of organic matter with iron and aluminum across a range of soils determined via selective dissolution techniques coupled with dissolved nitrogen analysis. Biogeochemistry112, 95–109 (2013). [Google Scholar]
- 101.Domeignoz-Horta, L. A. et al. Microbial diversity drives carbon use efficiency in a model soil. Nat. Commun.11, 3684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gearing, P. J., Gearing, J. N., Maughan, J. T. & Oviatt, C. A. Isotopic distribution of carbon from sewage sludge and eutrophication in the sediments and food web of estuarine ecosystems. Environ. Sci. Technol.25, 295–301 (1991). [Google Scholar]
- 103.Smith, R. G. et al. Environmental Correlates with Germinable Weed Seedbanks on Organic Farms across Northern New England. Weed Sci.66, 78–93 (2018). [Google Scholar]
- 104.Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.4-4https://CRAN.R-project.org/package=vegan (2017).
- 105.Hooper, D. U. & Vitousek, P. M. Effects of Plant Composition and Diversity on Nutrient Cycling. Ecol. Monogr.68, 121–149 (1998). [Google Scholar]
- 106.Anderson, M. J. A new method for non parametric multivariate analysis of variance. Austral Ecol.26, 32–46 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The fungal trait, SOM pool, and SOM chemistry data that support the findings of this study have been deposited in the Environmental Data Initiative repository, 10.6073/pasta/cf2a305c7d21938ddad3cb9430d22ed3. Source data associated with each main text and Supplementary Figure are also available as a supplementary Source Data file.