Abstract
A cell wall is required to control cell shape and size to maintain growth and division. However, some bacterial species maintain their morphology and size without a cell wall, calling into question the importance of the cell wall to maintain shape and size. It has been very difficult to examine the dispensability of cell wall synthesis in rod-shaped bacteria such as Escherichia coli for maintenance of their shape and size because they lyse without cell walls under normal culture conditions. Here, we show that wall-less E. coli L-form cells, which have a heterogeneous cell morphology, can be converted to a mostly uniform oval shape solely by FtsZ-dependent division, even in the absence of cylindrical cell wall synthesis. This FtsZ-dependent control of cell shape and size in the absence of a cell wall requires at least either the Min or nucleoid occlusion systems for positioning FtsZ at mid cell division sites.
Subject terms: Cellular microbiology, Bacteriology
Wall-less E. coli L-forms with a heterogeneous cell morphology can be converted to a uniform oval shape solely by FtsZ-dependent septal cell wall synthesis, even without cylindrical cell wall synthesis.
Introduction
It is very important for cells to control their size. Several mechanisms underlying bacterial cell size homeostasis have been proposed, including the critical size model, in which cell division initiates upon reaching a size threshold, and the constant extension model, in which cell division starts when cells reach a certain length prior to cell division1,2. Almost all bacteria, including Escherichia coli, are surrounded by a cell wall or peptidoglycan, which determines the cell shape and size and protects bacterial cells from turgor pressure3. However, it is unclear how cell wall synthesis is involved in cell size homeostasis. In addition, the models described above are based on studies of walled bacterial cells. However, several species, including Mycoplasma species, maintain a constant cell shape and size without a cell wall. Therefore, there may be a mechanism(s) that controls cell shape and size, even without a cell wall.
Bacterial cells generally exhibit two modes of cell wall synthesis, one for cell elongation, mediated by the Rod complex (the elongasome), and another for cell division, mediated by the divisome3,4. The Rod complex and divisome are composed of cytoskeletal proteins such as bacterial actin MreB and bacterial tubulin FtsZ, respectively, and penicillin-binding proteins (PBP2 and PBP3). The Rod complex interacts with the divisome, and this interaction is probably important for switching cell wall synthesis from elongation to division modes5,6. FtsZ polymers assemble at mid cell to form a ring-like structure, called the Z ring, along with many proteins including ZapA, PBP3 (FtsI), FtsW, and FtsN3,7, which constitute the divisome. Divisome proteins are recruited to the Z ring in a specific order7–11, with FtsZ and ZapA being early divisome proteins, followed by later recruits such as PBP3, FtsW, and FtsN. Cell division is coupled with cell wall synthesis and is initiated by the activation of PBP3 and FtsW12–14, which are septal peptidoglycan synthases. However, studies of the bacterial divisome have focused mainly on the divisomes of E. coli and Bacillus subtilis7. Therefore, it remains unknown whether the Z ring also works in wall-less bacteria, such as Mycoplasma species, which lack most divisome proteins other than FtsZ15.
Cell elongation is associated with cell shape determination because cell shape is determined by the shape of the newly synthesized peptidoglycan cell wall16. Perturbation of several E. coli proteins, including MreB, a scaffold protein for the Rod complex, the transmembrane protein RodZ, and PBP2, results in abnormal cell shape17–22. Cell division is associated with cell size control since cells become filamentous when cell division is inhibited in E. coli. In addition, a regulatory model of cell size control has been proposed in which cells elongate to a certain length and then divide2. However, it has never been examined whether cell wall syntheses for elongation and division are necessary for cell size control as inhibition of cell wall synthesis inhibits cell proliferation.
The ftsZ gene is not only widely conserved in walled bacteria but also in most wall-less bacteria and most archaea whose cell walls are different from those in bacteria23,24. The ftsZ gene was acquired at a fairly early stage of evolution25,26, but the actual role of FtsZ in bacteria without a cell wall is uncertain. JCVI-syn3.0, a synthetic bacterium created at the Venter Institute, lacks ftsZ and shows a heterogeneous cell morphology, suggesting poor control of uniform cell size maintenance27. JCVI-syn3 + 126, which is JCVI-syn3.0 with 7 genes added back, has a relatively uniform spherical shape compared to JCVI-syn3.027,28. Interestingly, ftsZ is one of the 7 re-introduced genes. Therefore, in wall-less JCVI-syn3 + 126 cells, FtsZ may play a role in regulating cell shape and size without a cell wall. To accurately investigate the role(s) of FtsZ in regulating cell shape irrespective of the presence or absence of a cell wall, it is important to compare the function of FtsZ in cells of the same bacterial species with and without a cell wall. To this end, we compared E. coli cells with a normal cell wall with cells lacking a normal wall, called L-forms.
Bacterial L-forms, originally discovered in bacterially infected rats29, have the ability to proliferate without a wall under certain conditions. Recently, L-form cells were found in patients with recurrent bacterial infections30. In humans with bacterial infections, the addition of antibiotics with cell wall synthesis inhibitory activity inhibits cell wall synthesis and causes some bacterial cells to switch from a walled state to a wall-deficient L-forms. This switch allows the bacteria to survive in the presence of antibiotics. Upon termination of antibiotic treatment, the infecting L-form cells resume cell wall synthesis and grow normally in a walled state, causing recurrent infections30. In the laboratory, the conversion to L-forms is induced by inhibiting cell wall synthesis through the addition of antibiotics such as penicillin and fosfomycin, or by deletion of genes involved in the cell wall synthesis pathway, under conditions of high osmotic strength and high concentrations of Mg2+ 16,31,32. In addition, anaerobic conditions promote the conversion to the L-form33,34.
Whether FtsZ is required for the division of E. coli L-forms is controversial. D’Ari’s lab showed that FtsZ is required for L-form growth in the presence of cefsulodin35 whereas Errington’s lab showed that cell division depends on membrane fluidity and is independent of the Z ring machinery (divisome) in the presence of fosfomycin36,37. If L-form division is indeed independent of FtsZ, then division would likely be driven by unregulated physical forces such as membrane blebbing and tubulation. This L-form-specific cell division switches to normal cell division, which is dependent on the Z ring machinery, by restarting cell wall synthesis35,38. It is still unclear whether the Z ring can control cell division, even in L-form cells, and how FtsZ functions in wall-less cells. In B. subtilis L-forms32 or wall-less Mycoplasma genitalium39, FtsZ forms filaments or clusters instead of discrete Z rings, suggesting that a cell wall is required for Z ring formation.
In walled E. coli cells, the position of the Z ring is determined by the Min system and nucleoid occlusion40,41. The Min system inhibits Z ring formation at the cell poles via pole-to-pole oscillation of the MinCDE complex40,42. Nucleoid occlusion inhibits Z ring formation over the nucleoid through the action of the DNA-binding protein SlmA41. Thus, the placement of the Z ring is negatively regulated by both the Min system and nucleoid occlusion. The shapes of L-form cells and their nucleoids are different from those of normal-walled cells; the L-form is ameboid and has multiple nucleoids43. No clear cell poles exist in ameboid L-form cells, and the nucleoid partitioning mechanism in L-forms may differ from that in walled cells.
As mentioned above, studying the relationship between the cell size-determining mechanism and cell wall synthesis for cell division is challenging because cell wall synthesis is essential for walled bacteria; therefore, it is difficult to observe how the loss of the cell wall affects cell size regulation by using walled bacteria. In addition, it has never been examined whether the Min and nucleoid occlusion systems could function in wall-less cells that lack uniform cell shapes. Therefore, in this study, we address these questions using E. coli L-forms, which are free from the constraints of cell wall synthesis, cell shape, and cell lysis. We demonstrate that the formation of the septal cell wall only, without the side wall, is sufficient to confer uniform cell shape. We also reveal that the Min and nucleoid occlusion-mediated regulation of Z ring formation and Z ring positioning are functional in wall-less L-form cells. Furthermore, at least one of the Min or nucleoid occlusion systems is required for the conversion from the FtsZ-independent division in wall-less cells to the FtsZ-dependent division in septal walled cells and hence to form a uniform cell shape.
Results and Discussion
Confirmation of the non-essentiality of FtsZ in E. coli L-form cells
FtsZ is not essential for the viability of L-forms of B. subtilis32. In B. subtilis L-form cells, FtsZ forms filaments instead of discrete rings32. However, the essentiality of FtsZ in E. coli L-form cells is controversial35,36. We first examined whether E. coli cells could form L-form colonies without FtsZ in our experimental condition, a high-osmotic medium plate (NA/MSM) containing fosfomycin (Fos), penicillin G (PenG), or cefsulodin (Cef). Fos inhibits the activity of MurA, which catalyzes the first step to synthesize peptidoglycan44. PenG inhibits the transpeptidase activity of penicillin-binding proteins (PBPs)45,46. Cef inhibits the transpeptidase activities of PBP1A and PBP1B47. Therefore, peptidoglycan is not synthesized in L-form cells in the presence of Fos; less peptidoglycan is synthesized in L-form cells in the presence of PenG than in L-form cells in the presence of Cef48.
Since FtsZ is essential for the growth of walled E. coli cells, and therefore ftsZ gene cannot be deleted, we constructed a strain in which the expression of the ftsZ gene could be induced by sodium salicylate (NaSal) and checked whether L-form cells could grow under FtsZ depletion. RU2055 (FtsZ-depletion strain) cells, in which ftsZ expression can be controlled, formed colonies on NA/MSM plates with the inducer (Supplementary Fig. 1a, +NaSal, NA/MSM). On the other hand, without the inducer, this strain failed to form colonies as walled cells on NA/MSM plate without antibiotics (Supplementary Fig. 1a, -NaSal, NA/MSM), indicating that FtsZ was produced and depleted enough to support and inhibit growth in the presence and absence of the inducer, respectively. We confirmed protein levels of FtsZ by immunoblot using anti-FtsZ antibody and found that little was produced without the inducer (Supplementary Fig. 1b, ΔftsZ -). The amount of FtsZ produced from the plasmid in the presence of NaSal was higher than that of FtsZ produced from the chromosome. We also noticed that FtsZ produced from the plasmid showed double bands (Supplementary Fig. 1b). We previously used the same plasmid to produce FtsZ and did not detect such double bands of FtsZ in immunoblots with anti-FtsZ antibody49. However, in another paper, FtsZ was detected as double bands when glucose was added to nutrient-depleted conditions although the detailed reason for this was not discussed50. Therefore, FtsZ may be modified in some way under the specific medium or nutrient conditions. The strain could grow as L-form cells on NA/MSM plates containing antibiotics, regardless of the presence or absence of the inducer (Supplementary Fig. 1a, +NaSal or -NaSal, +Fos, +PenG, and +Cef), indicating that FtsZ is not essential for the growth of E. coli L-forms, as previously indicated36.
We recently developed a system to monitor the conversion of walled cells to L-form cells and the growth of L-form cells in real-time by microscopy48. Using this system, we confirmed that E. coli cells could be transformed from walled to ameboid L-forms and could grow as L-forms, even under FtsZ-depleted conditions (Supplementary Fig. 2a-c and Supplementary Movies 1-6). We first counted the number of cells 4 hours after the addition of antibiotic (Fos, PenG, or Cef). Of those cells, we counted the number of cells that were able to divide up to 12 hours later. 21.2% (22/104 cells), 55.7% (39/70 cells), or 30.4% (38/125 cells) of WT L-forms divided while 22.0% (26/118 cells), 33.3% (36/108 cells), or 44.2% (53/120 cells) of ΔftsZ L-forms divided in the presence of Fos, PenG, or Cef, respectively. Although the percentage of cells that divided when converted to L-form by Fos is lower than with other antibiotics, these results also suggest that growth and division of L-forms do not require FtsZ as previously shown36.
The phenotypes when the antibiotic was removed from the medium in which the cells were grown as L-form cells differed depending on antibiotics. Under FtsZ-depleted conditions, L-form cells grown with Fos lysed during the return to a walled state after removing Fos from the medium (Supplementary Fig. 2a ΔftsZ, and Supplementary Movie 2). L-form cells grown with PenG did not revert to rod-shaped cells but instead were abnormally elongated (Supplementary Fig. 2b ΔftsZ, and Supplementary Movie 4). This cell morphology is similar to a phenotype, called coli-flower, exhibited by a mutant strain isolated from ftsZ null L-form cells as a strain capable of growing as walled cells without ftsZ36. Unexpectedly, L-form cells grown with Cef showed the coli-flower phenotype at the beginning of re-synthesizing peptidoglycan (Supplementary Fig. 2c ΔftsZ, and Supplementary Movie 6). However, the “branches” of the cells became rod-like shaped but grew abnormally long (Supplementary Fig. 2c ΔftsZ, red arrow), probably because they could not divide due to the lack of ftsZ.
WT L-form cells generated by the addition of Fos can revert to normal walled cells when Fos is removed, but all the ΔftsZ L-form cells generated by Fos lysed during the reverting process (Supplementary Fig. 2a). These results suggest that FtsZ is required to revert to walled cells from cells completely lacking cell wall. The results resemble the observation that cell division precedes cell elongation during reversion from spheroplasts without peptidoglycan51. We also noticed that WT L-forms generated by Fos lyse in a significant percentage of cells upon resynthesizing the cell wall. In fact, when we measured how many of the L-forms present after 12 hours in antibiotics were able to revert to rod-shaped cells, only 3.0% (4/132) of the Fos L-forms were able to revert to rod-shaped cells. This is an extremely low percentage compared to the cells that reverted to rod-shaped cells from PenG L-forms and Cef L-forms (18.4%, 38/207 cells, and 21.5%, 37/172 cells, respectively). These results indicate that it is possible, but very difficult, to revert from a state of complete absence of a cell wall (Fos L-forms) to cells surrounded by a cell wall.
In our experimental system, the NB/MSM medium is constantly flowing (approximately 2.5 µL/h) and cell division could potentially occur as a result of shear forces due to flow. To demonstrate that division of L-forms occurs independent of medium flow, we filled the device with antibiotic-containing NB/MSM medium and then introduced the bacterial cells to induce conversion to L-forms while the medium flow rate was set to 0. However, it was not possible to completely eliminate the flow velocity in this system, which was about 0.1–0.3 µL/h. We observed that Z ring-independent divisions occurred under the conditions, suggesting that L-form cells can divide under almost no flow conditions (Supplementary Fig. 3a and Supplementary Movie 7). We counted L-form cells that divided without the medium flow rate and found that cell division occurred in 23.0% (50/217) of Fos L-form cells. This was higher than cells that divided with medium flow (9.9%, 17/141). Although we cannot exclude the possibility that medium flow may promote cell division of L-forms, medium flow is not necessarily required for L-form division.
Z rings are assembled but fail to constrict in L-form cells
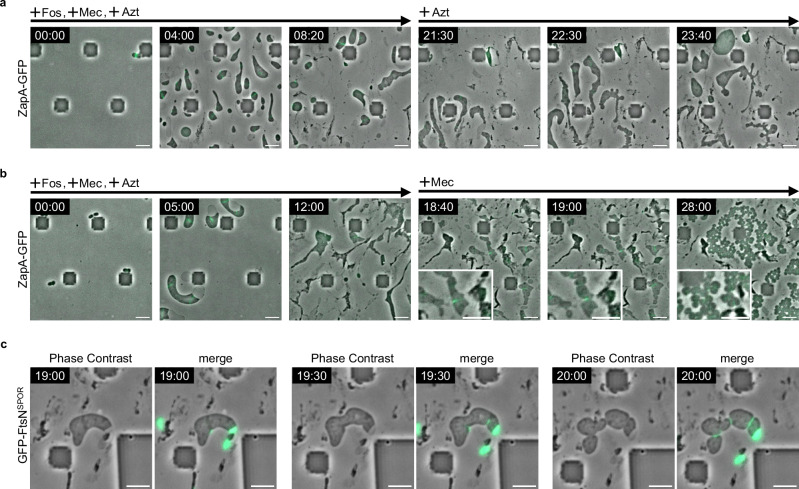
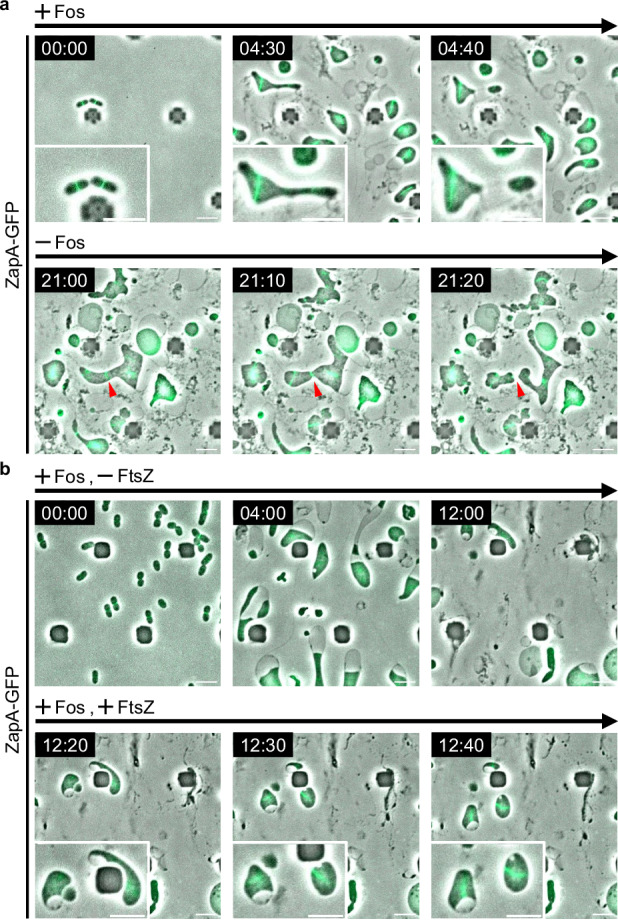
Since FtsZ was not essential in E. coli L-forms36, similar to what was observed in B. subtilis L-forms32, we predicted that, in E. coli L-forms, FtsZ might form non-ring filamentous structures similar to those observed in B. subtilis L-forms32. To visualize the Z ring, we fused sfGFP (super-folder GFP) with ZapA (denoted as ZapA-GFP), a component of the Z ring dependent on the Z ring for its localization. As expected, ZapA-GFP formed ring-like structures dependent on FtsZ in walled cells (Supplementary Fig. 3b). Surprisingly, ZapA-GFP formed ring-like structures in E. coli L-form cells, unlike in B. subtilis L-forms (Fig. 1a +Fos and Supplementary Movie 8), and ZapA-GFP rings were dependent on FtsZ in E. coli L-form cells (Fig. 1b, +Fos -FtsZ and Supplementary Movie 9). To confirm that the Z ring (ZapA) does indeed form ring structures rather than a filamentous structure, we analyzed the localization of ZapA-GFP in three dimensions. We confirmed that ZapA-GFP also localizes in a ring-like structure in L-form cells (Supplementary Fig. 3c and Supplementary Movie 10).
Fig. 1. Subcellular localization of ZapA-GFP in L-form cells.
a Time-lapse images of RU1125 (zapA-sfGFP::cat) cells grown in NB/MSM medium under anaerobic conditions containing Fos. Fos was removed from NB/MSM medium after 12 h. Images were taken every 10 min. Red arrowheads indicate cell division sites. Magnified images are also shown. Scale bars: 5 μm. b Time-lapse images of RU2057 (zapA-sfGFP ∆ftsZ::kan/pWM2765) cells grown in NB/MSM medium under anaerobic conditions containing Fos. After 12 h, 10 μM NaSal was added to the NB/MSM medium. Images were taken every 10 min. Magnified images are also shown. Scale bars: 5 μm.
We then analyzed whether L-form cells divided where the Z ring is formed. In walled cells, the position of the Z ring coincided perfectly with the division position (359/359 divisions), as expected. In contrast, most of the L-form cells divided independently of the Z ring (Fig. 1a and Supplementary Fig. 3d), while the position of the Z ring coincided with the division position in only 9.5% of these L-form cells (16/169 divisions, Supplementary Fig. 3e), suggesting that L-forms can divide independently of the Z ring. We will discuss the result that 9.5% of the cells appear to divide at the Z ring later. To exclude the possibility that Z rings found in E. coli L-form cells were formed in the original walled cells and persisted in L-form cells, we examined whether ZapA-GFP rings could be generated de novo in L-form cells when FtsZ production was restored. In the absence of FtsZ, ZapA-GFP diffused throughout the cytoplasm (Fig. 1b, +Fos, -FtsZ, and Supplementary Movie 9); however, after inducing ftsZ expression, ZapA-GFP re-assembled in L-form cells to form ring-like structures (Fig. 1b, +Fos, +FtsZ, and Supplementary Movie 9). These observations suggest that the Z ring can be generated in L-form cells and that Z ring assembly does not require a cell wall. Furthermore, when cell wall synthesis was resumed in L-form cells, the Z ring which had not constricted, began to constrict, and cell division occurred at the site where the Z ring was generated in L-form cells, suggesting that the Z ring was generated in a functional form in L-form cells (Fig. 1a, -Fos, Supplementary Fig. 4 and Supplementary Movie 8). This suggests that the Z ring is assembled normally in L-form cells but does not constrict, probably because all the components of the divisome are not assembled and/or cell wall synthesis was absent. In fact, a large number of divisome proteins must be assembled at the Z ring3,9,52.
To investigate this question, we tested whether the late divisome proteins PBP3 (FtsI) and FtsN localize to the Z ring in L-form cells. PBP3, which is a monofunctional transpeptidase important for peptidoglycan synthesis at the division site, and FtsN, which activates the divisome and is the last protein to localize to it, are both essential for septal wall synthesis in walled cells3,9,52. To observe the localization of PBP3 and FtsN in L-form cells, we constructed plasmids carrying either msGFP2-PBP3 or msGFP2-FtsN. msGFP2 is a monomeric super-folder derivative of eGFP53. For simplicity, msGFP2-PBP3 and msGFP2-FtsN are denoted as GFP-PBP3 and GFP-FtsN, respectively.
We found that in walled cells, GFP-PBP3 mainly co-localized with ZapA-mCherry, but a few dot-like foci of GFP-PBP3 were localized to the cell peripheries (Supplementary Fig. 5a). GFP-FtsN co-localized with ZapA-mCherry to the division site (Supplementary Fig. 5b). These results are consistent with those of previous studies54,55. In L-form cells, both GFP-PBP3 (Fig. 2a) and GFP-FtsN (Fig. 2b) formed ring-like structures and co-localized with ZapA-mCherry, as observed in walled cells, suggesting that a complete divisome is properly formed in L-form cells and is poised for constriction. FtsN contains a peptidoglycan-binding SPOR domain that facilitates its localization to division septa, but can also localize to Z rings independently of the SPOR domain through an interaction between its N-terminal cytoplasmic domain and the cell division protein FtsA, which itself is recruited to the Z ring56,57 (Supplementary Fig. 6a). To confirm that the co-localization of FtsN and ZapA in the L-form we observed (Fig. 2b) was not dependent on cell wall, we examined the localization of FtsN lacking a SPOR domain (FtsNΔSPOR) and a mutant composed only of the SPOR domain. Since the SPOR domain alone cannot be translocated to the periplasm, the TorA signal sequence, which is a transport signal to the periplasm, was fused to the SPOR domain (denoted as FtsNSPOR) (Supplementary Fig. 6a, GFP-FtsNSPOR). Consistent with previous studies56, FtsNΔSPOR localizes primarily to the Z-ring in walled cells, whereas FtsNSPOR localized to the cell periphery (probably because of the binding to cell wall) but was enriched at the septum (Supplementary Fig. 6b). FtsNΔSPOR colocalized with ZapA-mCherry in the L-form as it did as in walled cells, whereas FtsNSPOR localized throughout the periplasm in L-forms, as expected (Supplementary Fig. 6c and 6d). The results suggest that FtsN localizes to the division septum in a Z ring-dependent manner even in the absence of cell wall.
Fig. 2. Subcellular localization of late division proteins and ZapA-mCherry in L-form cells.
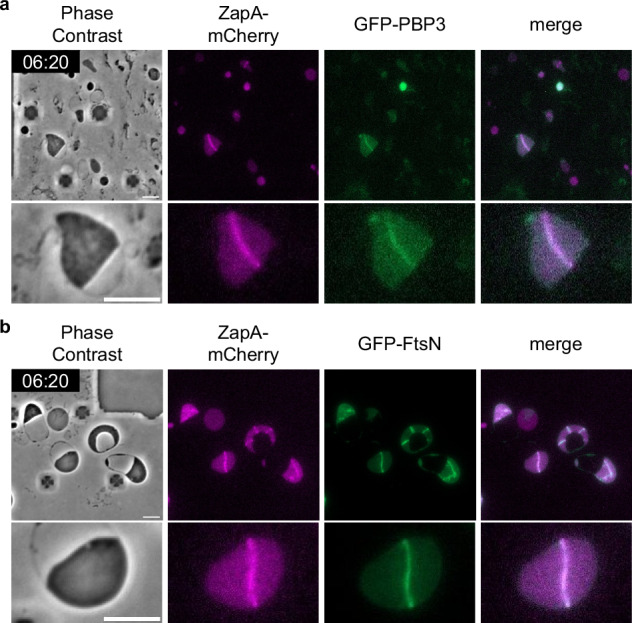
Images of RU2452 (zapA-mCherry) /pRU2282 (GFP-PBP3) (a) or pRU2281 (GFP-FtsN) (b) cells were grown in NB/MSM medium containing Fos and IPTG under anaerobic conditions. After conversion to the L-form, the growth conditions were changed from anaerobic to aerobic as mCherry could not be observed under anaerobic conditions. The magnified images are shown. Scale bars: 5 μm.
Reactivation of PBP3 changes the L-form cell division system from FtsZ-independent to FtsZ-dependent
Thus far, our data suggest that divisomes in E. coli L-form cells are fully assembled but fail to activate constriction because of the absence of a cell wall. Since it was shown that the PBP3-specific inhibitor aztreonam (Azt) inhibits Z ring constriction but does not inhibit Z ring assembly58,59, we expected that if PBP3 regains its cell wall synthesis activity in L-form cells, the Z ring might become functional, even in L-form cells. As mentioned earlier, E. coli uses either the Rod complex or the divisome for its two modes of cell wall synthesis, which are mediated by PBP2 and PBP3, the specific transpeptidases for elongation and division, respectively59,60. PBP2 and PBP3 activities are specifically inhibited by mecillinam (Mec) and aztreonam (Azt), respectively58,59. Indeed, E. coli cells exhibited oval/round or filamentous shapes, respectively, in the presence of Mec or Azt in our microfluidics chamber (Supplementary Fig. 7a, 7b, and Supplementary Movie 11).
When walled E. coli cells were simultaneously treated with three antibiotics (Fos, Mec, and Azt), they were converted to ameba-like L-form cells that divided independently of Z rings (Supplementary Fig. 8a 4:00 and Supplementary Movie 12). Under this condition, cells were converted to L-forms by inhibition of Lipid II synthesis by Fos37 and inhibition of PBP2 and PBP3 enzymatic activities by Mec and Azt, respectively. In these L-form cells, ZapA-GFP formed ring-like structures as observed in L-form cells converted by only Fos treatment (Fig. 1a and Supplementary Movie 8). Next, after conversion to L-form cells with Fos, Mec and Azt, only Mec or Azt was removed from the media, re-initiating cell wall synthesis for elongation or division in L-form cells. As expected, cells continued to grow as L-forms even though inhibition of PBP2 or PBP3 activity was released by the removal of Mec or Azt in the presence of Fos (Supplementary Fig. 8a, 8b, and Supplementary Movie 12). We also removed only Fos from medium containing Fos, Mec, and Azt. In the presence of both Mec and Azt, the cells proliferated as L-form cells (Supplementary Fig. 8c and Supplementary Movie 12). This suggests that inhibition of the activity of PBP2 and PBP3 by Mec and Azt, respectively, allows cells to grow as L-forms.
Next, after conversion to L-form cells with Fos, Mec, and Azt treatment, Fos and Mec were removed from the media, re-initiating cell wall synthesis catalyzed only by PBP2 in L-form cells. These cells, synthesizing the cell wall only for elongation (SWE cells), elongated and swelled in various directions, but the Z rings did not constrict (Fig. 3a +Azt and Supplementary Movie 13). This was probably because Azt inhibited the activity of PBP3, and Z ring constriction was specifically inhibited. In contrast, if Fos and Azt were removed from the medium, thus re-initiating cell wall synthesis catalyzed only by PBP3 in L-form cells, approximately 7 h after the removal Z rings started to constrict in cells synthesizing the cell wall only for division (SWD cells), triggering division (Fig. 3b +Mec and Supplementary Movie 14). This suggested that activation of PBP3 is sufficient to initiate Z ring constriction and cell division in SWD cells. Although it was previously thought that cell wall synthesis for division is triggered by the interaction between the Rod complex containing PBP2 and the divisome containing PBP35, PBP2-mediated synthesis of cylindrical cell wall is not required for constriction and division. This resembles the division of E. coli cells lacking rodA or mreBCD in which the activity of the Rod complex is inactivated or decreased.
Fig. 3. PBP3 is important for Z ring constriction.
a, b Time-lapse images of RU1125 (zapA-sfGFP::cat) cells grown in NB/MSM medium containing Fos, Mec and Azt under anaerobic conditions. After 12 h, Fos and Mec (a) or Fos and Azt (b) were removed from the NB/MSM medium. Magnified images are also shown. Scale bars: 5 μm. c Time-lapse images of RU2452 (zapA-mCherry) cells carrying a plasmid encoding GFP-FtsNSPOR grown in NB/MSM medium containing Fos, Mec, and Azt. GFP-FtsNSPOR encoded on pRU2624 was expressed without IPTG. After 12 h, Fos and Azt were removed from the NB/MSM medium. After 16 h, the growth conditions were changed from anaerobic to aerobic. Scale bars: 5 μm.
To confirm whether cell wall synthesis is re-initiated only at the Z ring constriction in SWD cells, cell wall synthesis was visualized by GFP-FtsNSPOR, which binds specifically to newly synthesized septal peptidoglycan61. After the removal of Fos and Azt, GFP-FtsNSPOR was localized in the periplasm in an ameba-like cell (Fig. 3c 19:00). However, at 19:30 and 20:00, cell constrictions were clearly observed in the cells and GFP-FtsNSPOR was localized at the constriction sites (Fig. 3c, Supplementary Fig. 9, Supplementary Movie 15), suggesting that the cell wall is synthesized at the constriction site. It should be noted that GFP-FtsNSPOR seemed to localize only at the constriction site in SWD cells while it localized at the constriction site and cell periphery in walled cells (Supplementary Fig. 6b), consistent with the idea that SWD cells synthesize cell wall only at division sites. These results also suggest that reactivation of PBP3 changes the division system of L-form cells from FtsZ-independent to FtsZ-dependent. Although the molecular mechanisms and signals responsible for Z ring constriction remain incomplete62–64, recent studies have shown that septal cell wall synthesis is initiated by in vivo assembly and activation of FtsW and PBP314,65–67. Using L-form cells in which the divisome is fully assembled but is blocked from constriction and septal synthesis, our results show that FtsZ-dependent cell division could be started only upon re-activation of PBP3. In walled-E. coli cells, the Rod complex containing PBP2 interacts with the divisome containing PBP3 to prepare for cell division5. However, our results show that FtsZ-dependent cell division occurs in SWD cells and does not require the synthesis of the cylindrical cell wall. This mode of division is similar to that of E. coli cells lacking rodA or mreBCD as described above.
What is the biological importance of Z ring formation in the E. coli L-form, in which the Z ring is not necessary for cell division? L-form cells have been linked to recurrent infections in patients treated with antibiotics30,68–70. The resistance to antibiotics through conversion to the L-form and re-conversion to normally growing walled cells from the L-form was observed in patients with recurrent infections30. Therefore, FtsZ-independent cell division in L-form cells may be a temporary adaptation to antibiotic treatment. If the Z ring could be formed in L-form cells surviving in a patient under antibiotic treatment, the bacterial cells would be able to resume cell wall synthesis and grow normally as walled cells through FtsZ-dependent cell division as soon as the antibiotics or cell wall synthesis inhibitors, such as lysozyme, are removed.
L-form cells can divide to yield cells of homogenous sizes by reactivating PBP3
After the removal of Fos and Azt from the culture medium in which Fos, Mec, and Azt converted walled cells to L-form cells, the E. coli SWD cells converted gradually into uniform oval shapes during repeated divisions (Fig. 3b, 28:00, and Supplementary Movie 14). In those SWD cells, ZapA-GFP was localized at the mid-cell (Fig. 3b 28:00). This suggests that Z ring-dependent cell wall synthesis and cell division occur only at the mid-cell to convert heterogeneously sized cells with ameboid morphology (L-form cells) into uniformly sized cells with an oval shape (SWD cells). This indicates that cell shape can be controlled solely by septal cell wall synthesis even if the cylindrical cell wall is not synthesized. This type of cell shape control is similar to, for example, those of E. coli ΔrodA cells and the coccoid Staphylococcus aureus. The cell division planes of E. coli ΔrodA and S. aureus are perpendicular to the previous division plane71,72 while the division plane of WT-walled E. coli remains in a consistent perpendicular orientation. To investigate whether the orientation of the division plane of SWD cells is constant or changes with each division like ΔrodA and S. aureus cells, we analyzed SWD cell divisions in detail and found that the division plane of SWD cells is perpendicular to the previous division plane (Supplementary Fig. 10 and Supplementary Movie 14). This result indicates that the division mode of SWD cells resembles those of ΔrodA and S. aureus. Because nucleoid occlusion spatially regulates division sites in SWD cells like in S. aureus73, nucleoid occlusion may regulate Z ring positioning (see below). Similarly, the wall-less synthetic bacterium JCVI-syn3.0 is irregularly shaped, but JCVI-syn3 + 126, in which several genes including ftsZ were re-introduced into this synthetic bacterium, became a uniformly sized coccus27,28. These results strongly indicate that cells can maintain a round or oval morphology even if they can divide in an FtsZ (or its functional equivalent protein) dependent manner without elongation like ΔrodA and ΔmreB cells.
The Min system and nucleoid occlusion control spatial positioning of the Z ring in both L-form and walled cells
Z ring formation is negatively regulated by two systems, Min and nucleoid occlusion, in walled E. coli cells40,41. MinC protein, a component of the Min system, inhibits the assembly of Z rings at the cell poles. In ΔminC cells, the Z ring often forms at cell poles as well as at the cell center, resulting in minicells, as previously reported74 (Supplementary Fig. 11 ΔminC). Z ring formation over nucleoids is inhibited by SlmA bound to SlmA-recognition sequences in genomic DNA75–77. This negative spatial regulation is called nucleoid occlusion75. Z ring formation in ΔslmA mutant cells was indistinguishable from that in WT cells (Supplementary Fig. 11 ΔslmA), as previously reported75. Although the minC and slmA double mutant cells have been reported to exhibit synthetic lethality in LB media75, we found that the strain was viable under high-osmotic culture conditions (NA/MSM), even as walled cells (Supplementary Fig. 11 ΔminC Δ slmA). Nonetheless, compared to the single gene deletion mutants, the ΔminC ΔslmA double mutant showed an exacerbated phenotype, with multiple Z rings and mislocalized rings (Supplementary Fig. 11 ΔminC ΔslmA).
We then examined the effect of the Min system and nucleoid occlusion on the Z ring formation in L-form cells. In wild-type L-form cells, ZapA-GFP was localized to the HupB-free (nucleoid-free) region (Fig. 4, WT), suggesting that nucleoid occlusion is functional in L-form cells. Unlike in ΔslmA walled cells, in ΔslmA L-form cells, some Z rings formed over the nucleoids (Fig. 4, ΔslmA), and some ZapA-GFP appeared to form filaments rather than ring-like structures (Fig. 4, ΔslmA; yellow arrowheads). Surprisingly, in ΔminC L-form cells, ZapA-GFP formed mesh-like structures (Fig. 4, ΔminC). However, ZapA-GFP did not localize over nucleoids, indicating that nucleoid occlusion remained functional in ΔminC L-form cells. ZapA-GFP in ΔminC L-forms seemed punctate compared to WT or ΔslmA L-forms. This suggests that MinC plays a role in forming smooth filaments or that Z-ring components may form filaments with punctate if they do not form rings correctly. In addition, a mesh-like structure was also observed for GFP-FtsN in ΔminC L-form cells, suggesting that all components of the divisome were involved, even in the mesh-like structures (Supplementary Fig. 12 ΔminC). It should be noted that ZapA-GFP appears more punctate compared to the smoother mesh observed with GFP-FtsN in ΔminC L-form cells. This may be due to a slight overproduction of GFP-FtsN expressed from the plasmid, which appears to form smooth filaments.
Fig. 4. Subcellular localization of ZapA-GFP in L-form cells lacking minC, slmA, or both.
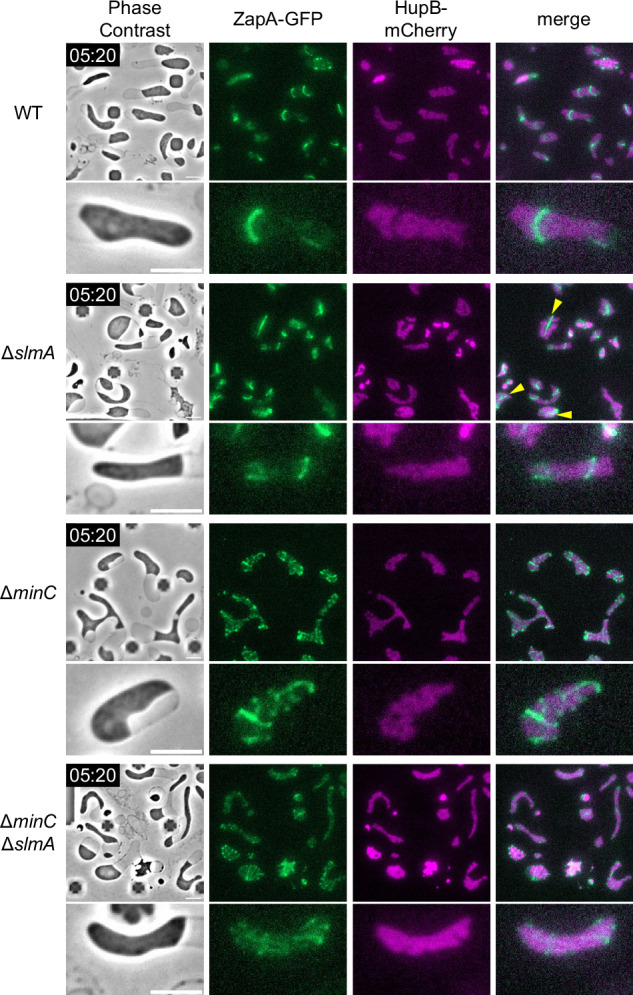
Subcellular localization of ZapA-GFP and HupB-mCherry in the L-form cells. Cells (WT: RU1974, ∆slmA: RU2401, ∆minC: RU2400, ∆minC ∆slmA: RU2409) were grown in NB/MSM medium containing Fos under anaerobic conditions. After conversion to the L-form, the growth conditions changed from anaerobic to aerobic as mCherry could not be observed under anaerobic conditions. The magnified images are shown. The yellow arrows indicate the filament structure. Scale bars: 5 μm.
It is well-known that MinC prevents the Z ring from assembling at cell poles and depolymerizes FtsZ filaments in vitro78. This inhibitory activity of MinC near the cell poles helps to restrict Z rings at midcell by preventing aberrantly localized FtsZ polymers, which are reflected by the mesh-like structures in L-forms in the absence of MinC. Consistent with this, it was previously shown that FtsZ was disorganized even in walled spherical E. coli cells lacking minCDE79. Recently, it was also shown that when FtsZ is encapsulated within lipid vesicles, it forms mesh-like structures, whereas when MinC is added, it forms a ring-like structure80. These results suggest that MinC plays not only a negative role but also a positive role in organizing the Z ring at midcell. It should be emphasized that in E. coli, MinC along with its ATPase partner MinD oscillates from one pole to the other, whereas in B. subtilis MinC and MinD are anchored to the cell poles and largely act to prevent Z rings from forming new division septa at the cell poles11,81. In addition, in E. coli SlmA interacts directly with FtsZ filaments to cause depolymerization82, whereas in B. subtilis Noc, an important protein for nucleoid occlusion in B. subtilis, binds to DNA and the cell membrane, physically inhibiting FtsZ filament formation in the vicinity of the membrane83. This difference in MinC-MinD dynamics and the different functions between SlmA and Noc may account for differences in the ability of E. coli and B. subtilis to form Z rings in L-form cells. L-form cells of double mutants (ΔslmA ΔminC) showed an additive phenotype of ΔslmA and ΔminC single deletion L-form cells (Fig. 4, ΔslmA ΔminC). These results suggest that the Min system and nucleoid occlusion, used to determine the localization of the Z ring in E. coli walled cells, are functional in E. coli L-form cells. Although L-forms divide in a Z ring-independent manner, division at a Z ring was observed in 9.5% of cells. As nucleoid occlusion is involved in determining the position of Z-rings in L-forms, we speculate that division sometimes coincides with Z rings because these cells tend to divide in spaces between nucleoids anyway.
Cell shape of SWD cells lacking minC or slmA
We showed that FtsZ formed mesh-like structures in ΔminC L-forms (Fig. 4). Next, we examined whether the Min system contributes to cell size regulation in SWD cells. Compared to wild-type (WT) SWD cells, the cell shape of ΔminC SWD cells appeared to be slightly heterogeneous, including anucleate mini cells and elongated cells. Measurement of the sizes of non-mini cells indicated that the ΔminC SWD cells were statistically significantly longer and wider than the WT SWD cells (p < 0.05, Fig. 5a, b), suggesting that cell size is regulated, at least in part, by the Min system in WT cells lacking cylindrical cell walls. However, the differences in the ratio between the length and width of WT and ΔminC SWD cells were not statistically significantly different (Fig. 5b), suggesting that the Min system controls the length and width of SWD cells synthesizing the cell wall only at mid-cell, but does not control overall cell shape and that SlmA-mediated nucleoid occlusion functions in ΔminC SWD cells.
Fig. 5. Morphologies of SWD cells lacking Min and/or nucleoid occlusion systems.
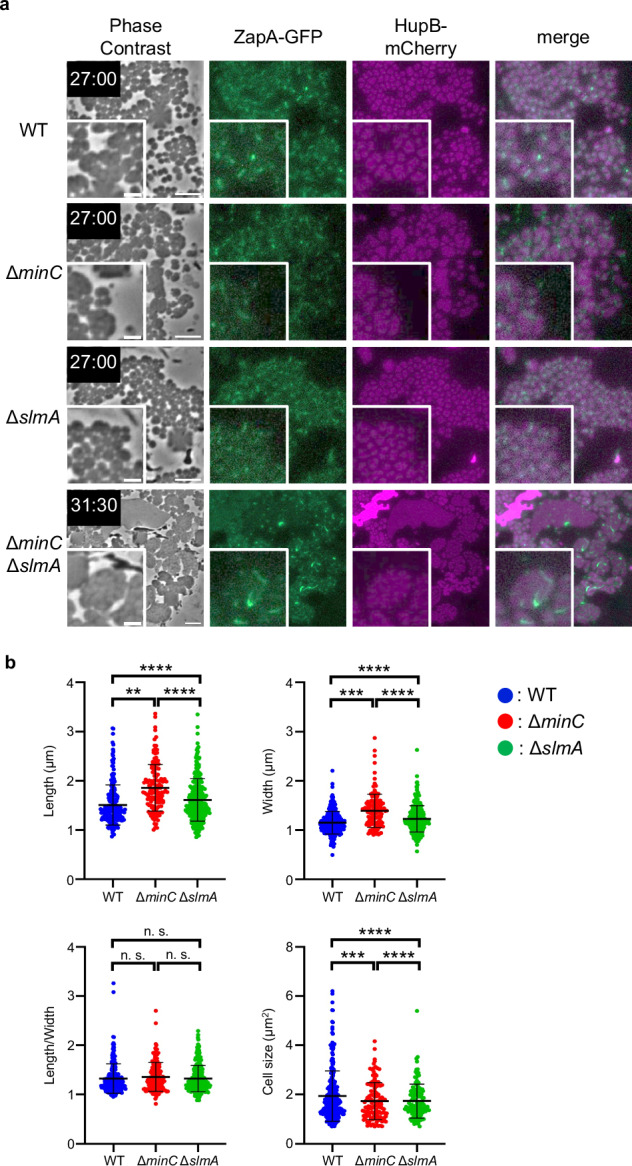
a Images of RU1974 (WT), RU2400 (∆minC), RU2401 (∆slmA), and RU2409 (∆minC ∆slmA) cells were grown in NB/MSM medium containing Fos, Mec and Azt under anaerobic conditions. After 12 h, Fos and Azt were removed from the NB/MSM medium. 12 hours after Fos and Azt removal, the growth conditions were changed from anaerobic to aerobic because mCherry could not be observed under anaerobic conditions. The magnified images are shown. Scale bars: 5 μm. b Morphological analysis of SWD cells. Cell length, width, and size were measured from pictures using MicrobeJ, a plug-in of ImageJ. Blue, red, and green dots indicate WT (n = 350 cells), ∆minC (n = 138 cells), and ∆slmA (n = 272 cells), respectively. Error bars indicate S.D. **P < 0.01, ***P < 0.005, ****P < 0.001 n.s.: not significantly different.
The shapes of ΔslmA SWD cells and ΔminC SWD cells were also homogeneous like WT SWD cells. The ratio between the length and width of ΔslmA SWD cells and ΔminC SWD cells was comparable with that of WT SWD cells, although ΔslmA SWD cells and ΔminC SWD cells were longer and wider than WT SWD cells (Fig. 5b). Z rings were formed at mid-cell in the ΔslmA SWD cells, suggesting that the Min system is functional in the cells. ΔminC cells and ΔslmA cells showed similar shapes, indicating that nucleoid occlusion and Min system contribute equally to cell division in SWD cells. To our surprise, the ΔminC ΔslmA L-form cells started to deform into an oval shape and divide in an FtsZ-dependent manner after the removal of Fos and Azt, but they could not deform completely into uniform oval shapes. Instead, they seemed to return to ameba-like morphology as if they were L-forms (Fig. 5a, Supplementary Fig. 13 and Supplementary Movie 16). These results suggest that cells lacking a cylindrical cell wall cannot maintain a uniform cell size without both the Min system and nucleoid occlusion. Previously, it was shown that the Z ring localizes to the division site independently of the Min system, nucleoid occlusion system, and the Ter-linkage in walled E. coli cells84. Considering their data together with our data, it is possible that cell wall synthesis in the cylindrical portion may facilitate the localization of or stabilize the Z ring to the division site. In other words, if there are systems that localize the Z ring to the right place in the cell, such as the Min system and/or nucleoid occlusion, FtsZ can maintain a uniform cell size without a cylindrical cell wall. These are summarized in Fig. 6. In WT walled-cells, FtsZ (green) forms the Z ring between the segregated nucleoids (magenta) (a). In the WT L-forms, the Z rings containing all components of the divisome proteins are formed in the nucleoid-free region but cannot constrict (b). WT SWD cells are divided into uniform oval cells using Z-ring by the activation of PBP3 (c). In the ΔminC ΔslmA L-forms, the short green line shows incomplete FtsZ rings/FtsZ filaments (d). ΔminC ΔslmA L-forms fail to divide in an FtsZ-dependent manner after the activation of PBP3 (e). In ΔslmA SWD cells and ΔminC SWD cells, the Z ring formation is determined by the Min system and nucleoid occlusion system, respectively. In ΔslmA cells, the Z ring is formed at mid-cell by the Min system (f). While in ΔminC cells, the Z ring is formed at cell-pole and mid-cell but not on the nucleoids. Constriction of the Z-ring at the cell poles produces mini-SWD cells without a nucleus (g). ΔminC ΔslmA walled-cells are initiated to divide and convert to rod-shaped cells by activation of both PBP2 and PBP3 (h). ΔminC ΔslmA L-form cells treated with Fos, Mec, and Azt, were able to revert to rod-shape when all antibiotics were removed (Supplementary Fig. 14 and Supplementary Movie 17). This confirms the importance of the cylindrical cell wall in determining cell size and homogeneous shape as well as the cell wall at the division site, in the absence of nucleoid occlusion.
Fig. 6. Model for cell shape control and Z ring formation in SWD cells.
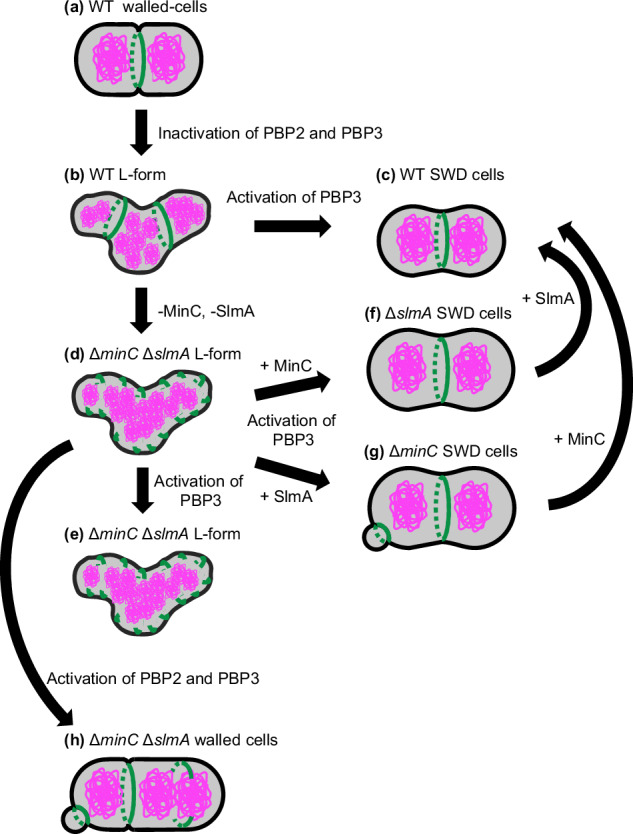
In WT walled-cells, FtsZ (green) formed the Z ring between the segregated nucleoids (magenta) (a). In the WT L-form cells, the Z rings containing all components of the divisome proteins were formed in the nucleoid-free region and could not constrict (b). WT SWD cells were divided into uniform oval cells using Z-ring by the activation of PBP3 (c). In the ∆minC ∆slmA L-form, the short green lines show incomplete FtsZ rings or filaments. ∆minC ∆slmA L-form cells failed to divide in an FtsZ-dependent manner after the activation of PBP3 (d, e). In ∆slmA SWD cells and ∆minC SWD cells, Z ring formation was determined by the Min system and nucleoid occlusion system, respectively. In ∆slmA cells, the Z ring was formed at mid-cell by using only the Min system (f). While in ∆minC cells, the Z ring was formed at cell-pole and mid-cell but not on the nucleoids. Constriction of the Z-ring at the cell poles produces minicells of SWD lacking a nucleus (g). In ∆minC ∆slmA cells, cells were initiated to divide and converted to rod-shaped cells by activation of PBP2 and PBP3 (h).
Cell size control in wall-less bacteria
Cell size control is an important issue in bacterial physiology, and several models have been proposed1,2,85. However, these models are mainly based on experiments using walled cells, and it would be useful for understanding how primitive cells evolved to know more about regulation of cell size in wall-less bacteria, such as Mycoplasma. The synthetic bacterium JCVI-syn3.0 has the smallest artificial genome, which is based on the Mycoplasma mycoides genome27,86. JCVI-syn3.0, which lacks both a cell wall and ftsZ, has a heterogeneous morphology and grows slowly. JCVI-syn3 + 126 is a derivative of JCVI-syn3.0 in which several genes, including ftsZ, were put back into the genome. This strain has a relatively uniform cell size and round morphology28. It should be noted that it is unknown whether the division of JCVI-syn3 + 126 cells is FtsZ-dependent or not. Nevertheless, this result strongly suggests that FtsZ or FtsZ-dependent cell division can function as a regulatory system for cell size control, even in wall-less cells. This study also reveals that wall-less L-form cells require a system of nucleoid occlusion that determines the proper localization of the FtsZ ring for cell size control to change cell division from FtsZ-independent to FtsZ-dependent. Only when cells synthesize cylindrical cell walls is the nucleoid occlusion system not necessary to regulate the cell size uniformity.
We then speculated that FtsZ could function as a factor to maintain uniform cell size and increase growth efficiency in primitive cells lacking cylindrical cell walls. We re-examined the conservations of the minC and slmA genes and again clearly indicated that minC and slmA are absent in many bacteria, as is already known87 (Supplementary Fig. 15), suggesting that minC and slmA were not acquired early during the evolutionary process, while ftsZ is highly conserved in bacteria and archaea. These findings reinforce the idea that the FtsZ-dependent cell division system is ancient and was in place before the Min and nucleoid occlusion systems evolved (Supplementary Fig. 15). If primitive cells acquired FtsZ before the acquisition of the cell wall, an alternative system to the modern nucleoid occlusion system might have served as the sole spatial regulatory system to control localization of the Z ring and to confer uniform cell morphology and size until these cells acquired the ability to elongate their cell wall. One candidate for a system to determine the localization of FtsZ is the nucleoid. FtsZ can localize between the segregated nucleoids even in walled E. coli (this study) and B. subtilis88 cells lacking the Min system and nucleoid occlusion. In these cells, nucleoids may play a role in positioning of FtsZ. It is therefore plausible to assume that the nucleoid itself served the function of nucleoid occlusion in primitive cells.
Methods
Bacterial strains and growth medium
All strains were derivatives of E. coli K-12 and are listed in Table 1. BW25113 is a WT strain. Cells were grown in NB/MSM medium32 comprising 2× nutrient broth (NB) (Oxoid, United Kingdom) mixed 1:1 with 2× magnesium-sucrose-maleic acid (MSM; 40 mM MgCl2, 1 M sucrose, and 40 mM maleic acid, pH 7.0). When the cells were converted to the L-form on the plates, 0.75% agar was added (NA/MSM). Antibiotics were added at the following concentrations: 300 μg/mL PenG, 400 μg/mL Fos, 100 μg/mL Cef, 20 μg/mL Azt, and 10 μg/mL Mec. More detailed culture conditions and other information are clearly described in the appropriate sections.
Table 1.
Strains used in this study
| Strains | Relevant genotype | Reference |
|---|---|---|
| BW25113 | rrnB ΔlacZ4787 HsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 98 |
| SN1187 | MG1655 ΔhsdR ΔendA ΔrecA(Δ2,820,759-2,821,785) | 91 |
| DH5α | F-, deoR, endA1, gyrA96, hsdR17(rk-mk + ), recA1, relA1, supE44, thi-1, Δ(lacZYA-argF)U169, (Phi80lacZΔM15) | 99 |
| WM2767 | W3110 ΔftsZ::kan/ pWM2765 (pKG110-ftsZ) | 49 |
| RU2055 | BW25113 ΔftsZ::kan/ pWM2765 (pKG110-ftsZ) | This study |
| RU1125 | BW25113 zapA-sfGFP::cat | 6 |
| RU1429 | BW25113 zapA-sfGFP | 6 |
| RU1237 | BW25113 hupB-mCherry::cat | Lab stock |
| RU2057 | BW25113 zapA-sfGFP ΔftsZ::kan/ pWM2765 (pKG110-ftsZ) | This study |
| RU1974 | BW25113 zapA-sfGFP hupB-mCherry::cat | This study |
| RU50 | ΔminC::kan | Lab stock |
| RU2332 | ΔslmA::kan | Lab stock |
| RU2400 | BW25113 zapA-sfGFP hupB-mCherry::cat ΔminC::kan | This study |
| RU2401 | BW25113 zapA-sfGFP hupB-mCherry::cat ΔslmA::kan | This study |
| RU2405 | BW25113 zapA-sfGFP hupB-mCherry ΔminC | This study |
| RU2409 | BW25113 zapA-sfGFP hupB-mCherry ΔminC ΔslmA::kan | This study |
| TH1165 | SB2/pmCherry-kan | Lab stock |
| RU2443 | BW25113 zapA-mCherry::kan | This study |
| RU2452 | BW25113 zapA-mCherry | This study |
| RU2462 | BW25113 ΔminC | This study |
Strain constructions
Detailed methods for strain constructions are described below.
RU2055 (ΔftsZ::kan)
P1 lysate, prepared from WM2767 (ΔftsZ::kan), was used to transduce ΔftsZ::kan into BW25113 to yield RU2055.
RU2057 (zapA-sfGFP ΔftsZ::kan)
P1 lysate prepared from WM2767 (ΔftsZ::kan) was used to transduce ΔftsZ::kan into RU1429 to yield RU2057.
RU1974 (zapA-sfGFP hupB-mCherry::cat)
P1 lysate prepared from RU1237 (hupB-mCherry::cat) was used to transduce hupB-mCherry::cat into RU1429 to yield RU1974.
RU2400 (zapA-sfGFP hupB-mCherry::catΔminC::kan)
A P1 lysate prepared form RU50 (ΔminC::kan) was used to transduce ΔminC::kan into RU1974 to yield RU2400.
RU2401 (zapA-sfGFP hupB-mCherry::cat ΔslmA::kan)
A P1 lysate prepared form RU2332 (ΔslmA::Kan) was used to transduce ΔslmA::kan into RU1974 to yield RU2401.
RU2409 (zapA-sfGFP hupB-mCherry ΔminC ΔslmA::kan)
A P1 lysate prepared from RU2332 (ΔslmA::kan) was used to transduce ΔslmA::kan into RU2405 to yield RU2409.
RU2452 (zapA-mCherry)
cells producing ZapA-mCherry were constructed as follows: DNA polymerase KOD One (TOYOBO) was used for polymerase chain reaction (PCR). Genomic DNA from TH1165 was amplified using primers 2470/2453, and pKD489 was amplified using primer 1679/2452. The second PCR was performed using these PCR products as templates and the primers 1679 and 2470. The PCR product was introduced into strain BW25113 carrying pKD4689 via electroporation. Cells were selected on L plates containing 25 μg/ml Kanamycin (Kan). The resulting strain (RU2443, zapA-mCherry::kan) was transformed with plasmid pCP2089 using selection for Amp resistance (AmpR) at 30 °C. The strain was then incubated at 42 °C in the absence of Amp, and colonies that grew were screened for the Amp-sensitive and Kan-sensitive phenotype at 37 °C. The resulting strain was designated as RU2452.
RU2405 (zapA-sfGFP hupB-mCherry ΔminC) and RU2462 (ΔminC)
RU2400 and RU50 were transformed with plasmid pCP20 using selection for AmpR at 30 °C. The strains were then incubated at 42 °C in the absence of Amp, and colonies that grew were screened for the Amp-sensitive, Kan-sensitive, and Cm-sensitive phenotype at 37 °C. The resulting strains were designated RU2405 (zapA-sfGFP hupB-mCherry ΔminC) and RU2462 (ΔminC).
Plasmid constructions
Detailed methods for plasmid constructions are described below. The plasmids and primers used in this study are listed in Tables 2 and 3. To construct pTrc99A-Kan, PstI fragment of pKD4 containing the Kanamycin resistance gene, which was blunted, was introduced into the SmaI fragments of pTrc99A90. Plasmids encoding msGFP2-FtsN and msGFP2-PBP3 were constructed using iVec91. To construct pTrc99A-Kan carrying msGFP2-FtsN and msGFP2-PBP3, three PCR products were amplified using pTrc99A-Kan as a template and primers 2289/2291 and 2290/2291, using synthetic DNA of msGFP2 as a template and primers 2292/2293 and 2292/2294, and using the purified BW25113 genome as a template and primers 2295/2296 and 2297/2298, respectively. The three PCR products were introduced into SN1187 cells using a modified TSS method91. Plasmids encoding msGFP2-FtsNΔSPOR and torA-msGFP2-FtsNSPOR were constructed using In-Fusion. To construct pTrc99A-Kan carrying msGFP2-FtsNΔSPOR, PCR product was amplified using pRU2281 as a template and primer 2636/2637. To construct pTrc99A-Kan carrying torA-msGFP2-FtsNSPOR, three PCR products were amplified using pRU2281 as a template and primers 2638/2639 and 2640/2641, using the purified BW25113 genome as a template and primers 2642/2643. One or three PCR products were combined with In-Fusion HD Cloning Kit (Takara Bio USA, United States) and introduced into DH5α cells using a CaCl2 method.
Table 2.
Plasmids used in this study
| Plasmids | Relevant genotype | Reference |
|---|---|---|
| pKG110 | pACYC184 derivative containing the nahG promoter, CmR | 100 |
| pWM2765 | ftsZ in pKG110, CmR | 101 |
| pTrc99A | Plac/trc, AmpR | 90 |
| pTrc99A-Kan | Plac/trc, KanR | This study |
| pRU2281 | msGFP2-ftsN in pTrc99A-Kan, KanR | This study |
| pRU2282 | msGFP2-ftsI in pTrc99A-Kan, KanR | This study |
| pRU2623 | msGFP2-ftsN∆SPOR in pTrc99A-Kan, KanR | This study |
| pRU2624 | torA-msGFP2-ftsNSPOR in pTrc99A-Kan, KanR | This study |
| pKD4 | FRT-kan-FRT, KanR, AmpR | 89 |
| pKD46 | Lambda Red recombinase, AmpR | 89 |
| pCP20 | yeast Flp recombinase gene, CmR | 89 |
Table 3.
Primers used in this study
| Name | Sequence |
|---|---|
| 2289 | GGCTCGCCGCCGGGGGTTGATCTAGAGTCGACCTGCAGGCATG |
| 2290 | GGACAGGTGGCAGATCGTAATCTAGAGTCGACCTGCAGGCATG |
| 2291 | CTCTCAGTGCTATCCATGGTCTGTTTCCTGTGTGAAATTG |
| 2292 | CAATTTCACACAGGAAACAGACCATGGATAGCACTGAGAGCCTG |
| 2293 | ACATAATCTCGTTGTGCCACGGATCCGCCACTGCCTCCAGTG |
| 2294 | GTTTTCGCCGCTGCTTTCATGGATCCGCCACTGCCTCCAGTG |
| 2295 | CTGGAGGCAGTGGCGGATCCGTGGCACAACGAGATTATGTACG |
| 2296 | GCCTGCAGGTCGACTCTAGATCAACCCCCGGCGGCGAGCCG |
| 2297 | CTGGAGGCAGTGGCGGATCCATGAAAGCAGCGGCGAAAACG |
| 2298 | GCCTGCAGGTCGACTCTAGATTACGATCTGCCACCTGTCCCCTCG |
| 1679 | CAGAGAAATTTTGTCTTCACGGTTACTCTACCACAGTAAACCGAAAAGTGCATATGAATATCCTCCTTA |
| 2452 | TGGACGAGCTGTACAAGTAGGTGTAGGCTGGAGCTGCTTC |
| 2453 | GAAGCAGCTCCAGCCTACACCTACTTGTACAGCTCGTCCA |
| 2470 | CGTTACTTGAACAAGGTCGCATCACCGAAAAAACTAACCAAAACTTTGAAGGTGGCTCTGGTGGCGGTTCTGGTGGCATGGTGAGCAAGGGCGAGG |
| 2636 | CTCTAGATCATTTTTTCTCCGCCGTCGGTTTTGG |
| 2637 | GGAGAAAAAATGATCTAGAGTCGACCTGCAGG |
| 2638 | TGGCGGATCCGAGAAAAAAGACGAACGCCGC |
| 2639 | CGTTATTGTTCATGGTCTGTTTCCTGTGTG |
| 2640 | AGCGGCGGAATTCATGGATAGCACTGAGAGCCTG |
| 2641 | CTTTTTTCTCGGATCCGCCACTGCCTCCAGTG |
| 2642 | ACAGACCATGAACAATAACGATCTCTTTC |
| 2643 | TATCCATGAATTCCGCCGCTTGCGCCGCAGTCG |
L-form conversion on NA/MSM plates
L-form conversion on NA/MSM plates was performed following a previously published method48. Cells were grown overnight in NB/MSM at 30 °C under aerobic conditions. The cell density was then adjusted to OD600 = 1.0 NB/MSM using Nanodrop One (Thermo Fisher Scientific, Waltham, MA, United States). Serial dilutions ranging from 10–1 to 10–5 by NB/MSM were performed, and 5 μL of the 10–2 and 10–5 dilutions were inoculated on NA/MSM plates with or without antibiotics, respectively. The plates were incubated at 30 °C for 3 days under anaerobic conditions using an Anaeropack (MGC, Tokyo, Japan). Then, the colonies, along with some agar, were picked, placed on a slide, and covered with a coverslip. Samples were observed under an inverted microscope (Axio Observer; Zeiss, Jena, Germany) equipped with an Objective Plan-Apochromat 100×/1.40 Ph3 (Zeiss) and filter sets 38HE and 63HE (Zeiss). Images were taken and processed using ZEN (Zeiss), Adobe Photoshop 2022 (Adobe, United States), and ImageJ (NIH, United States) software.
L-form/walled cells conversions in a microfluidic device
The L-form/walled cell conversion in the microfluidic device was performed following a previously published method48. Cells were grown to the early log phase (OD600 = 0.35–0.45) in NB/MSM at 30 °C under aerobic conditions. A 50 μL aliquot of the cell culture was centrifuged, and the pellet was resuspended in 1 mL NB/MSM. Next, 50 μL of cell suspension was loaded into B04A microfluidic plates (CellASIC ONIX, Merck, United States). Microfluidic plate ceilings had a stairs-like structure with the following heights: 0.7, 0.9, 1.1, 1.3, 2.3 and 4.5 μm. In this study, only 0.7 μm was used for all of observations. The air in the microfluidic plates was replaced with N2 using an N2 gas-generating device (SIC, Tokyo, Japan). In L-form cells, mCherry fluorescence was observed under aerobic conditions because mCherry was not fluorescent under anaerobic conditions. Once the cells were converted to the L-form under anaerobic conditions, the L-form cells were viable under aerobic conditions for several hours. The microfluidic device was controlled using the software supplied by the manufacturer (Merck). The microfluidic plates were placed under a microscope and observed under an inverted microscope (Zeiss) equipped with Objective Plan-Apochromat 100×/1.40 Ph3 (Zeiss) and filter sets 38HE and 63HE (Zeiss). Images were taken and processed using ZEN (Zeiss), Adobe Photoshop 2022 (Adobe, United States), and ImageJ (NIH, United States) software. Microscopic images were analyzed using ImageJ (National Institutes of Health) and MicrobeJ software92. Graphs were created using GraphPad Prism10.
Microscopic observation
Walled cells were grown to the log phase under aerobic conditions and mounted on 2% agarose in M9 medium. The cells were observed using an Axio Observer (Zeiss, Oberkochen, Germany), and the images were processed using ZEN (Zeiss) and ImageJ.
SDS-PAGE and immunoblotting
Cells were plated on NA/MSM plates in the presence or absence of antibiotics. Colonies were suspended by NB/MSM media and suspension cultures were adjusted by the value of OD600. SDS-PAGE and Immunoblotting were performed following a previously published method93. Anti-FtsZ antibody93 as the primary antibody and horseradish peroxidase-conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch) as the secondary antibody were used. The signal was detected on ImageQuant 800 (Cytiva) using ECL Select Western Blotting Detection Reagent (Cytiva).
Phylogenetic distribution of proteins in bacterial genomes
Of the 254,090 bacterial genomes included in the Genome Taxonomy Database Release 202 (GTDB R202)94, 4,326 genomes meeting two criteria were selected: 1) designated as GTDB representative strains (type strains or high-quality genomes), and 2) having complete genome sequences. Proteomes of these genomes were used as reference databases. As search queries, for the MinC protein, we used the HMM profiles registered in Pfam (PF03775 and PF05209)95, and for the SlmA protein, we used hmmbuild to construct the HMM profile from the corresponding protein sequence in E. coli (NP_418098.4). To identify these three proteins in the 4,326 genomes, we used hmmsearch in the HMMER 3.2.196. From the search results, a genome was considered to contain a protein when the hit e-value was 1e-8 or lower. To visualize the distribution of proteins in the bacterial phylogenetic tree, we used the phylogenetic tree file provided by GTDB R202 and performed tree annotation and rendering using Interactive Tree Of Life (iTOL)97.
Statistics and reproducibility
Statistical analysis was performed with GraphPad Prizm 10.2.2. The mean and SD of the data were used to create the graphs, and p-values were obtained by unpaired t-test. p-value < 0.05 was defined as a significant difference. All the experiments were repeatedly carried out and typical results are shown. Sample sizes are shown in Figure legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
We thank Dr. Yoshikazu Kawai (The University of Sydney) for his helpful suggestions and comments. We also thank all members of the Oshima and Shiomi laboratories for their helpful discussions and suggestions. This work was supported by the JST CREST (grant number JPMJCR19S5), Japan, to D.S., grant GM131705 from the National Institutes of Health, USA, to W.M., NIG-JOINT 57A2024 from National Institutes of Genetics, Japan, to T.O. and LA-2024-005 from Institute for Fermentation, Osaka (IFO), Japan, to T.O.
Author contributions
M.H. conceived the project, performed experiments, analyzed, interpreted the data, and wrote the manuscript. C.T. performed experiments and analyzed and interpreted the data. K.H. and K.K. performed bioinformatic analyses, interpreted the data, and wrote the manuscript. W.M. interpreted the data and wrote the manuscript. T.O. and D.S. conceived the project, analyzed, interpreted the data, and wrote the manuscript.
Peer review
Peer review information
Communications Biology thanks Zhixin Lyu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris. A peer review file is available.
Data availability
All relevant data supporting the findings are available within the paper and the Supplementary Materials. Source data used for generating the plots in Fig. 5b are available in the Supplementary Data file associated with the manuscript. Uncropped and unedited blot images are included in the Supplementary Material file. All other data and materials supporting this publication are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Taku Oshima, Email: taku@pu-toyama.ac.jp.
Daisuke Shiomi, Email: dshiomi@rikkyo.ac.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07279-y.
References
- 1.Si, F. et al. Mechanistic origin of cell-size control and homeostasis in bacteria. Curr. Biol.29, 1760–1770.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos, M. et al. A constant size extension drives bacterial cell size homeostasis. Cell159, 1433–1446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan, A. J. F., Errington, J. & Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol.18, 446–460 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Rohs, P. D. A. & Bernhardt, T. G. Growth and division of the Peptidoglycan Matrix. Annu. Rev. Microbiol.75, 1–22 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Ploeg, R.V.D. et al. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol. Microbiol.87, 1074–1087 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Yoshii, Y., Niki, H. & Shiomi, D. Division-site localization of RodZ is required for efficient Z ring formation in Escherichia coli. Mol. Microbiol.111, 1229–1244 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Levin, P. A. & Janakiraman, A. Localization, assembly, and activation of the Escherichia coli cell division machinery. Ecosal9, eESP-0022–eESP-2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du, S. & Lutkenhaus, J. At the heart of bacterial cytokinesis: the Z ring. Trends Microbiol27, 781–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du, S. & Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol.105, 177–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goehring, N. W. & Beckwith, J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Cur. Biol.15, R514–R526 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Cameron, T. A. & Margolin, W. Insights into the assembly and regulation of the bacterial divisome. Nat. Rev. Microbiol.22, 33–45 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, K.-T., Pichoff, S., Du, S. & Lutkenhaus, J. FtsA acts through FtsW to promote cell wall synthesis during cell division in Escherichia coli. Proc. Natl Acad. Sci.118, e2107210118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Y. et al. Genetic analysis of the septal peptidoglycan synthase FtsWI complex supports a conserved activation mechanism for SEDS-bPBP complexes. Plos Genet17, e1009366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park, K.-T., Du, S. & Lutkenhaus, J. Essential role for FtsL in activation of septal peptidoglycan synthesis. mBio11, e03012–e03020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Megrian, D., Taib, N., Jaffe, A. L., Banfield, J. F. & Gribaldo, S. Ancient origin and constrained evolution of the division and cell wall gene cluster in Bacteria. Nat. Microbiol.7, 2114–2127 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Kawai, Y., Mercier, R. & Errington, J. Bacterial cell morphogenesis does not require a preexisting template structure. Cur. Biol.24, 863–867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wachi, M. et al. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol.169, 4935–4940 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiomi, D. et al. Mutations in cell elongation genes mreB, mrdA and mrdB suppress the shape defect of RodZ-deficient cells. Mol. Microbiol.87, 1029–1044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi, H. et al. Deep phenotypic mapping of bacterial cytoskeletal mutants reveals physiological robustness to cell size. Cur. Biol.27, 3419–3429.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Vinella, D., Joseleau-Petit, D., Thévenet, D., Bouloc, P. & D’Ari, R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J. Bacteriol.175, 6704–6710 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiomi, D., Sakai, M. & Niki, H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J.27, 3081–3091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ago, R. et al. Relationship between the Rod complex and peptidoglycan structure in Escherichia coli. MicrobiologyOpen12, e1385 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subedi, B. P. et al. Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry. FEMS Microbes2, xtab012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ithurbide, S., Gribaldo, S., Albers, S.-V. & Pende, N. Spotlight on FtsZ-based cell division in Archaea. Trends. Microbiol. 30, 665–678 (2022). [DOI] [PubMed]
- 25.Santana-Molina, C. et al. Early origin and evolution of the FtsZ/tubulin protein family. Front. Microbiol.13, 1100249 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis, B. K. Molecular evolution before the origin of species. Prog. Biophys. Mol. Biol.79, 77–133 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Hutchison, C. A. et al. Design and synthesis of a minimal bacterial genome. Science351, aad6253–aad6253 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Pelletier, J. F. et al. Genetic requirements for cell division in a genomically minimal cell. Cell184, 2430–2440 (2021). [DOI] [PubMed]
- 29.Klieneberger, E. The natural occurrence of pleuropneumonia‐like organism in apparent symbiosis with Streptobacillus moniliformis and other bacteria. J. Path. Bacteriol.40, 93–105 (1935). [Google Scholar]
- 30.Mickiewicz, K. M. et al. Possible role of L-form switching in recurrent urinary tract infection. Nat. Commun.10, 4379–4379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lederberg, J. & StClair, J. Protoplasts and L-type growth of Escherichia coli. J. Bacteriol.75, 143–160 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leaver, M., Domínguez-Cuevas, P., Coxhead, J. M., Daniel, R. A. & Errington, J. Life without a wall or division machine in Bacillus subtilis. Nature457, 849–853 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Kawai, Y. et al. Cell growth of wall-free L-form bacteria is limited by oxidative damage. Curr. Biol.25, 1613–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai, Y. et al. On the mechanisms of lysis triggered by perturbations of bacterial cell wall biosynthesis. Nat. Commun.14, 4123 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseleau-Petit, D., Liébart, J.-C., Ayala, J. A. & D’Ari, R. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J. Bacteriol.189, 6512–6520 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercier, R., Kawai, Y. & Errington, J. Wall proficient E. coli capable of sustained growth in the absence of the Z-ring division machine. Nat. Microbiol.1, 16091–16095 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Mercier, R., Kawai, Y. & Errington, J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife3, 642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billings, G. et al. De novo morphogenesis in L-forms via geometric control of cell growth. Mol. Microbiol.93, 883–896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Torró, C. et al. Functional characterization of the cell division gene cluster of the wall-less Bacterium Mycoplasma genitalium. Front. Microbiol.12, 695572 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih, Y.-L. & Zheng, M. Spatial control of the cell division site by the Min system in Escherichia coli: Spatial control of cell division by the Min system. Environ. Microbiol15, 3229–3239 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Schumacher, M. A. Bacterial nucleoid occlusion: multiple mechanisms for preventing chromosome bisection during cell division. Sub-Cell. Biochem.84, 267–298 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Rothfield, L., Taghbalout, A. & Shih, Y.-L. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol.3, 959–968 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Wu, L. J. et al. Geometric principles underlying the proliferation of a model cell system. Nat. Commun.11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silver, L. L. Fosfomycin: mechanism and resistance. Csh Perspect. Med7, a025262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erlanger, B. F. & Goode, L. Mode of action of penicillin. Nature213, 183–184 (1967). [DOI] [PubMed] [Google Scholar]
- 46.Waxman, D. J. & Strominger, J. L. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem.52, 825–869 (1983). [DOI] [PubMed] [Google Scholar]
- 47.Ghosh, A. S., Chowdhury, C. & Nelson, D. E. Physiological functions of D-alanine carboxypeptidases in Escherichia coli. Trends Microbiol16, 309–317 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Chikada, T. et al. Direct observation of conversion from walled cells to wall-deficient L-form and vice versa in Escherichia coli indicates the essentiality of the outer membrane for proliferation of L-form cells. Front. Microbiol.12, 645965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiomi, D. & Margolin, W. Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol. Microbiol.67, 558–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekar, K. et al. Synthesis and degradation of FtsZ quantitatively predict the first cell division in starved bacteria. Mol. Syst. Biol.14, e8623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranjit, D. K., Jorgenson, M. A. & Young, K. D. PBP1B glycosyltransferase and transpeptidase activities play different essential roles during the de novo regeneration of rod morphology in Escherichia coli. J. Bacteriol.199, 181–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol.10, 123–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valbuena, F. M. et al. A photostable monomeric superfolder green fluorescent protein. Traffic21, 534–544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Addinall, S. G., Cao, C. & Lutkenhaus, J. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol.25, 303–309 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Weiss, D. S., Chen, J. C., Ghigo, J. M., Boyd, D. & Beckwith, J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol.181, 508–520 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerding, M. A. et al. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol.191, 7383–7401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Busiek, K. K. & Margolin, W. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol. Microbiol.92, 1212–1226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Georgopapadakou, N. H., Smith, S. A. & Sykes, R. B. Mode of action of azthreonam. Antimicrob. agents Chemother.21, 950–956 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spratt, B. G. & Pardee, A. B. Penicillin-binding proteins and cell shape in E. coli. Nature254, 516–517 (1975). [DOI] [PubMed] [Google Scholar]
- 60.Spratt, B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl Acad. Sci.72, 2999–3003 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yahashiri, A., Jorgenson, M. A. & Weiss, D. S. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl Acad. Sci.112, 11347–11352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Söderström, B., Chan, H., Shilling, P. J., Skoglund, U. & Daley, D. O. Spatial separation of FtsZ and FtsN during cell division. Mol. Microbiol.107, 387–401 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Boes, A., Olatunji, S., Breukink, E. & Terrak, M. Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. mBio10, e01912–e01918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, X. et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol.6, 584–593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu, B., Persons, L., Lee, L. & Boer, P. A. J. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol95, 945–970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsang, M. & Bernhardt, T. G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol.95, 925–944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Britton, B. M. et al. Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism. Nat. Commun.14, 4585 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutman, L. T., Turck, M., Petersdorf, R. G. & Wedgwood, R. J. Significance of bacterial variants in urine of patients with chronic bacteriuria. J. Clin. Invest.44, 1945–1952 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beaman, B. L., Burnside, J., Edwards, B. & Causey, W. Nocardial infections in the United States, 1972-1974. J. Infect. Dis.134, 286–289 (1976). [DOI] [PubMed] [Google Scholar]
- 70.Zou, J., Peng, B., Qu, J. & Zheng, J. Are bacterial persisters dormant cells only? Front Microbiol12, 708580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monteiro, J. M. et al. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun.6, 8055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Begg, K. J. & Donachie, W. D. Division planes alternate in spherical cells of Escherichia coli. J. Bacteriol.180, 2564–2567 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Veiga, H., Jorge, A. M. & Pinho, M. G. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol. Microbiol.80, 1366–1380 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Yu, X. C. & Margolin, W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol.32, 315–326 (1999). [DOI] [PubMed] [Google Scholar]
- 75.Bernhardt, T. G. et al. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol. Cell18, 555–564 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tonthat, N. K. et al. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J.30, 154–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tonthat, N. K. et al. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc. Natl Acad. Sci.110, 10586–10591 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu, Z., Mukherjee, A., Pichoff, S. & Lutkenhaus, J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci.96, 14819–14824 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corbin, B. D., Yu, X. & Margolin, W. Exploring intracellular space: function of the Min system in round‐shaped Escherichia coli. EMBO J.21, 1998–2008 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kohyama, S., Merino-Salomón, A. & Schwille, P. In vitro assembly, positioning and contraction of a division ring in minimal cells. Nat. Commun.13, 6098 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marston, A. L. & Errington, J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol.33, 84–96 (1999). [DOI] [PubMed] [Google Scholar]
- 82.Cho, H., McManus, H. R., Dove, S. L. & Bernhardt, T. G. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Natl Acad. Sci.108, 3773–3778 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adams, D. W., Wu, L. J. & Errington, J. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J.34, 491–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bailey, M. W., Bisicchia, P., Warren, B. T., Sherratt, D. J. & Männik, J. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet.10, e1004504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chien, A.-C., Hill, N. S. & Levin, P. A. Cell size control in bacteria. Cur. Biol.22, R340–R349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science329, 52–56 (2010). [DOI] [PubMed] [Google Scholar]
- 87.Pinho, M. G., Kjos, M. & Veening, J.-W. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol.11, 601–614 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Rodrigues, C. D. A. & Harry, E. J. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet.8, e1002561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci.97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amann, E., Ochs, B. & Abel, K.-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene69, 301–315 (1988). [DOI] [PubMed] [Google Scholar]
- 91.Nozaki, S. & Niki, H. Exonuclease III (XthA) enforces in vivo DNA cloning of Escherichia coli to create cohesive ends. J. Bacteriol.201, e00660–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol.1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shiomi, D. & Niki, H. A mutation in the promoter region of zipA, a component of the divisome, suppresses the shape defect of RodZ-deficient cells. MicrobiologyOpen2, 798–810 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res.50, D785–D794 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res.49, gkaa913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eddy, S. R. Accelerated Profile HMM Searches. Plos Comput. Biol.7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, gkab301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol.2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol.166, 557–580 (1983). [DOI] [PubMed] [Google Scholar]
- 100.Burón-Barral, M., del, C., Gosink, K. K. & Parkinson, J. S. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli Aerotaxis Transducer Aer. J. Bacteriol.188, 3477–3486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shiomi, D. & Margolin, W. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol.66, 1396–1415 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
All relevant data supporting the findings are available within the paper and the Supplementary Materials. Source data used for generating the plots in Fig. 5b are available in the Supplementary Data file associated with the manuscript. Uncropped and unedited blot images are included in the Supplementary Material file. All other data and materials supporting this publication are available from the corresponding author upon reasonable request.