Abstract
The great pond snail Lymnaea stagnalis has served as a model organism for over a century in diverse disciplines such as neurophysiology, evolution, ecotoxicology and developmental biology. To support both established uses and newly emerging research interests we have performed whole genome sequencing (avg.176 × depth), assembly and annotation of a single individual derived from an inbred line. These efforts resulted in a final assembly of 943 Mb (L50 = 257; N50 = 957,215) with a total of 22,499 predicted gene models. The mitogenome was found to be 13,834 bp long and similarly organized as in other lymnaeid species, with minor differences in location of tRNA genes. As a first step towards understanding the hermaphroditic reproductive biology of L. stagnalis, we identified molecular receptors, specifically nuclear receptors (including newly discovered 2xDNA binding domain-NRs), G protein-coupled receptors, and receptor tyrosine kinases, that may be involved in the cellular specification and maintenance of simultaneously active male and female reproductive systems. A phylogenetic analysis of one particular family of GPCRs (Rhodopsin neuropeptide FMRFamide-receptor-like genes) shows a remarkable expansion that coincides with the occurrence of simultaneous hermaphroditism in the Euthyneura gastropods. As some GPCRs and NRs also showed qualitative differences in expression in female (albumen gland) and male (prostate gland) organs, it is possible that separate regulation of male and female reproductive processes may in part have been enabled by an increased abundance of receptors in the transition from a separate-sexed state to a hermaphroditic condition. These findings will support efforts to pair receptors with their activating ligands, and more generally stimulate deeper insight into the mechanisms that underlie the modes of action of compounds involved in neuroendocrine regulation of reproduction, induced toxicity, and development in L. stagnalis, and molluscs in general.
Keywords: Molluscan genome, Nuclear receptors, GPCRs, Gene expansion, Simultaneous hermaphroditism
Subject terms: Evolution, Molecular evolution
Introduction
The ability of animals to perceive and react to their environments is key to their survival and evolutionary fitness. The study of such sensory abilities across the metazoan tree of life has led to deep insights across many fields and has deepened our understanding of how the abilities to taste, hear, touch, see and smell evolved1–4. Similarly, highly sensitive and consistent cellular responses to a local environment are prerequisites for the operation of robust developmental and reproductive programs. With the growing library of metazoan genome sequences, it is clear that a rich diversity of molecular receptors underlies many aspects of these sensory abilities5–7. For example, the family of receptors known as G protein-coupled receptors (GPCRs) is one of the largest protein superfamilies in the human genome8. However, our general understanding of the ligands that bind these receptors in most cases is lacking, and concerted efforts to pair these receptors with their activating ligands (increasingly with new technologies such as artificial intelligence), are underway9–13. Given a history across a variety of disciplines (from behavioural, to neural and molecular–reviewed in14), the great pond snail L. stagnalis has the potential to make significant contributions to these efforts. Many of these previous investigations were focused on basic biological processes such as respiration, heart rate, egg laying, mating, feeding and more, and have also resulted in Lymnaea becoming a standard species for ecotoxicology studies (OECD TG24315).
More recently, knowledge of fundamental neuro-biological processes has provided important insight into learning and memory in L. stagnalis, and how these processes are disturbed by human and/or environmental factors (e.g.16–18). Likewise, ecotoxicological compounds and immune elicitors in the environment have been shown to affect this species in terms of behavioural, developmental, endocrine, immunological and oxidative stress responses (e.g.19–25). Importantly, some of these responses have been linked to molecular profiles in the central nervous system (e.g.26–29). For example, recent work has highlighted the role of the neurotransmitter serotonin during development in the egg (e.g.30,31), for which the extensively documented ontology development of this species has proven valuable (e.g.32–36). Lymnaea has also proven to be a valuable model for investigations focused on the cellular and molecular mechanisms of memory37–39, biomineralisation40–42, and has been instrumental in elucidating the key genes involved in body-plan chirality, thanks to a naturally occurring sinistral mutant line that could be back-crossed, and experimentally and transcriptionally investigated alongside the normal, dextral type43–46. A similar approach can be used to identify the gene that is responsible for the recently discovered shell colour variant, referred to as Ginger47.
A fascinating feature of L. stagnalis that has received increasing attention over the past decades is its reproduction. As a simultaneous hermaphrodite, these snails can mate in the male and female role, but within a single mating interaction one individual must choose which role to perform; roles can subsequently be immediately swapped with the same or a different partner48. The particulars of this behaviour allow for experimentally disentangling male and female processes and underlying motivations49–52, something that is difficult to do in many other hermaphroditic species53,54. As a result, the great pond snail has played a central role in driving evolutionary research on hermaphroditic reproductive processes including selfing, mate choice, sexual conflict, and sperm competition55–60.
To advance these fields, the Lymnaea research community requires an annotated genome sequence to facilitate the use of contemporary sequencing methods (RNA-Seq, single cell sequencing, ATACseq), proteomics, comparative genomics and functional genomics (e.g., RNAi and CRISPR-Cas9). Moreover, such a resource will be the foundation for the identification and characterisation of novel molecular receptors. The process of ligand/receptor de-orphanisation will enable the study of their role in basic biological processes and inform the ecological risk assessment of chemicals such as suspected endocrine disrupters (see the OECD framework for endocrine disruption61,62). In addition, such an inventory will provide a framework to investigate whether this hermaphrodite (and others) deviates from separate-sexed species in its use and diversity of receptors, and how such life history strategies may have evolved.
To promote further progress, we sequenced, assembled and annotated the genome of L. stagnalis, and searched for gene family expansions using orthology analyses. We focused on identifying and analysing nuclear receptors (NRs), G-protein coupled receptors (GPCRs), and receptor tyrosine kinases (RTKs). These three highly conserved superfamilies were selected based on their pivotal roles in endocrine systems (NRs63), signalling pathways and reproduction (GPCRs64 and RTKs65,66), and possible crosstalk among the signalling pathways that are activated by these different types of receptors67. For a subset of these receptors, we performed in situ hybridisation (ISH) on early developmental stages and histological sections of sex-specific organs of adult L. stagnalis to localise their expression. Our annotation effort complements a previous effort devoted to ion-channels and ionotropic receptors in the central nervous system68. Here we focus on the largest family of receptors, GPCRs, to assess the difference between simultaneously hermaphroditic gastropod molluscs (such as L. stagnalis) and gastropods that manifest one sex at a time. Given that GPCRs are involved in numerous behaviours across the Metazoa including copulation and egg-laying69, we anticipate that our approach will contribute to evolutionary hypotheses underlying the evolution and diversification of receptors and their roles in maintaining the simultaneous hermaphroditic state.
Results
Genome sequencing and assembly
The Lymnaea stagnalis genome was sequenced using a combination of both Illumina paired-end and mate-pair reads. The resulting draft assembly spans 943 Mb and comprises 6640 scaffolds with lengths ranging from 2000 to 7,106,496 bp, as outlined in Table 1. This assembly is 250 Mb lower than the previously measured genome size of 1.193 Gb (C-value = 1.22 pg70). This discrepancy is consistent with the general trend observed in eukaryotes (see71).
Table 1.
Summary of the L. stagnalis genome assembly.
| Metric | Final assembly contigs | Final assembly scaffolds |
|---|---|---|
| Assembly size | 873,879,680 | 942,996,421 |
| Number | 64,200 | 6640 |
| N50 | 35,351 | 957,215 |
| L50 | 6921 | 297 |
| N90 | 7115 | 151,064 |
| L90 | 27,651 | 1178 |
| Largest | 315,608 | 7,106,496 |
| GC% | 37.55% | |
| %N’s | 7.33% |
The Illumina and nanopore sequencing data, as well as the genome assembly and gene predictions, are available in the European Nucleotide Archive (ENA) under the umbrella project PRJEB67819 (which encompasses PRJEB67821 and PRJEB67698 subprojects). In addition, the correspondance between locus tags and protein IDs and their BLASTp best-hit, as well as gene prediction mapping onto the assembly, and sequences (full gene, transcript, protein) are provided as supplementary information (ST21–ST25).
Repeat content
Repetitive elements spanned 37.89% of the 943 Mb-genome. Using a semi-automated process (described in the materials and methods section), we annotated the L. stagnalis genome with 2643 transposable element (TE) consensus sequences (available via INRAE data repository: https://doi.org/10.57745/19M0TH), giving 883,147 TE copies (24,190 of which are full-length copies (FLCs). In the consensus library, 1517 TEs were classified as retrotransposons (Class I) and 774 as DNA transposons (Class II), 243 were not classified and 105 as Potential Host Genes (PHGs). Most of these consensus sequences (2604, i.e., 99%) have one or more full-length copy, indicating that a consensus sequence is likely a reasonable estimate of the common ancestor of any one TE. The resulting annotation was used to mask the TE landscape in the genome.
Annotation
Following repeat masking of the genome, we employed ab initio, transcriptome- and homology-based methods to predict a set of 22,499 protein-coding genes (see Table 2 for detailed gene characteristics; see also Supplementary Tables ST21–ST25 for more information). This number is somewhat lower than in other annotated gastropod genomes (e.g., 23,851 genes in L. gigantea, 28,683 in A. californica, 36,943 in B. glabrata), and lower than the average number of genes found in a panel of 20 representative Metazoa (see OrthoFinder analysis). We compared the predicted proteome with the proteomic dataset published by Wooller et al.29 using BLASTp. This comparison returned 13,104 with significant similarity between the two datasets. Furthermore, 5232 of these predicted proteins shared at least 50% similarity with 86.5% of the proteomic database (4285 of 4953 sequences).
Table 2.
Characteristics of the L. stagnalis genome annotation.
| Parameter | Value |
|---|---|
| Assembly size | 943 Mbp |
| #predicted genes | 22,499 |
| #predicted genes without intron | 3492 (15.5%) |
| #exon per gene avg:med | 7.59:5 |
| Gene size (nt) avg:med | 15,299:8,502 |
| CDS size (nt) avg:med | 1375:993 |
| Intron size (nt) avg:med | 1611:604 |
| Complete BUSCOs (vs. mollusca odb10) | 4857 (91.8%) |
| Complete and single-copy BUSCOs | 4801 (90.7%) |
| Complete and duplicated BUSCOs | 56 (1.1%) |
| Fragmented BUSCOs | 144 (2.7%) |
| Missing BUSCOs | 294 (5.5%) |
| Total BUSCO groups searched | 5295 |
| #predicted genes with domain* | 19,387 (86%) |
| #predicted genes with InterPro* domain* | 16,622 (74%) |
| #predicted genes with Pfam* domain* | 15,088 (67%) |
*At least one protein domain.
Mitochondrial genome
BLAST searches returned mitogenomic sequence similarities for the full length of Lsta_scaffold2639. As indicated by a 127 bp sequence overlap at the termini, this scaffold represents a circular sequence. Nanopore reads confirmed that the assembled sequence is contiguous, and gene annotations revealed the complete standard gene complement for a metazoan mitogenome. The 13,834 bp mitogenome of L. stagnalis (Genbank MW221941) is 28.21% GC-rich, different from the 37.55% AT content of the nuclear genome. Several protein coding genes are delimited by downstream tRNA genes, causing the ORF to terminate with a truncated stop codon (T- or TA-) that is completed by polyadenylation of the mRNA transcripts. This mitogenome sequence has 94% identity with another, independently characterised L. stagnalis mitogenome (13,807 bp, MT874495). The different lengths of these two mitogenomes are mostly due to indels in regions with tRNA- and rRNA genes, a two bp indel occurs at the 5’ start of ND4L. These mitogenome sequences align closely but contain abundant single nucleotide variants (including several non-synonymous substitutions) within protein coding genes. The organisation of the 22 tRNA genes, two rRNA genes plus 13 protein coding genes (Supplementary Fig. SF1) of the L. stagnalis mitogenome is generally similar to that of other lymnaeid (and hygrophilid) snail species, with a few minor differences in the location of tRNA genes.
Receptor identification
Receptor tyrosine kinases
A total of 30 gene predictions (including manually curated versions) were identified as Receptor Tyrosine Kinases (RTKs), distributed across 18 of the 20 families or types (see Supplementary Tables ST1 and ST18). Among these, 12 gene predictions did not contain expected (canonical) domains. We found no gene prediction representing type III or XI RTKs, while two gene predictions could not be assigned to a specific RTK type.
Nuclear receptors
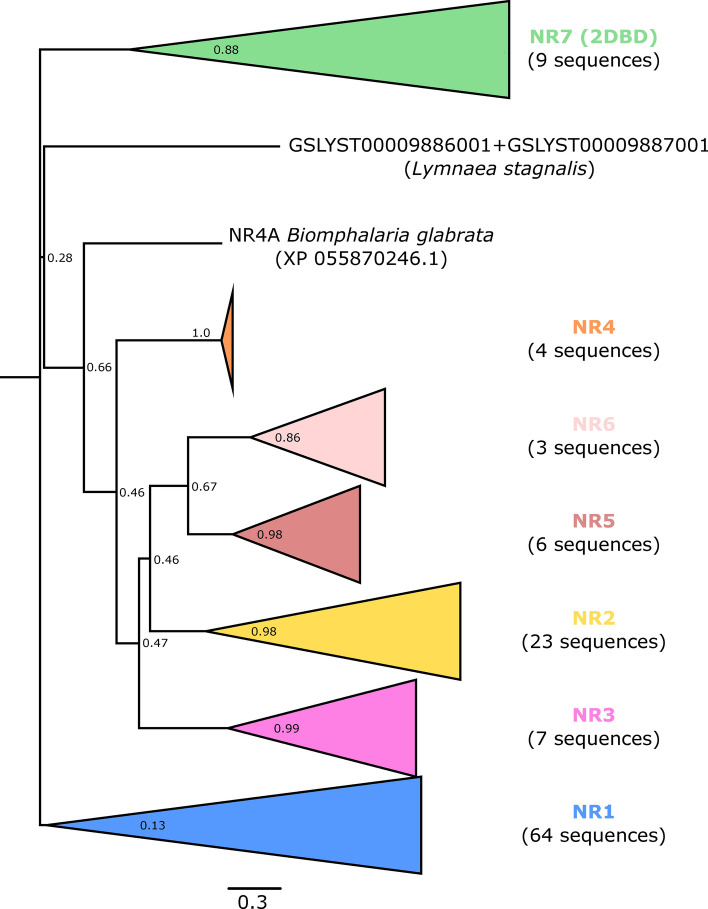
In total, 47 nuclear receptors (NRs) were identified from automatic and manual gene predictions, 39 of which containing the complete sequence of both conserved domains, i.e., the DNA-binding domain (DBD) and ligand binding domain (LBD) (Supplementary Tables ST2–ST3 andST19). All 6 traditional NR families were represented as follows: NR1 (28 predictions), NR2 (12), NR3 (3), NR4 (1), NR5 (2), NR6 (1). However, in terms of NR nomenclature, these corresponded to only 29 different NRs, as several gene predictions were found to have the same annotation (e.g., NR1F1_RORA, represented by four different gene predictions, that share low levels of domain sequence similarity (from 28.9 to 41.5%).
Significantly, we found two NRs with 2DBDs and one LBD. The occurrence of such an atypical NR structure was first reported in 2006 in the parasitic platyhelminth Schistomosa mansoni72 and further confirmed in other animal species, including protostomes and deuterostomes (both non-chordate and chordates; see73), suggesting an origin that predates the protostome-deuterostome split. Using the metazoan-scale orthology analysis performed in this study, the two sequences identified in L. stagnalis belong to the same orthogroup (OG00004255), which is composed of 33 sequences from molluscan (Gastropoda and Bivalvia), annelid, and platyhelminth taxa, plus the hemichordate Saccoglossus kowalevskii. However, in this OG, no protein other than the two L. stagnalis’ possess 2 DBDs, which may reflect conflicts in orthology search due to this very particular NR structure. Although a stable phylogeny of 2DBD-NRs is still to be established, Wu and LoVerde73 proposed to group them into a new family (NR7). Accordingly, we found higher sequence similarity among the four DBDs of these two 2DBD-NRs than with any other L. stagnalis NR or with their corresponding best hits (see green clade in Fig. 1 and Supplementary Fig. SF2).
Fig. 1.
L. stagnalis NR relationships. Phylogenetic relationships among L. stagnalis NR families: ML tree (Jones-Taylor-Thornton substitution model, Gamma distributed substitution rate with 4 categories, statistical support from 1000 bootstraps) based on ClustalW-aligned concatenated sequences of the DBD and most conserved part of the LBD of L. stagnalis NRs and their BLAST best-hits (alignment available upon request). Family NR0 was excluded from this analysis as it could not be aligned with DBD-only and LBD-only sequences. The triangles represent collapsed clades, with the number of branches/sequences indicated in brackets (see details on the full tree provided in Supplementary Fig. SF2).
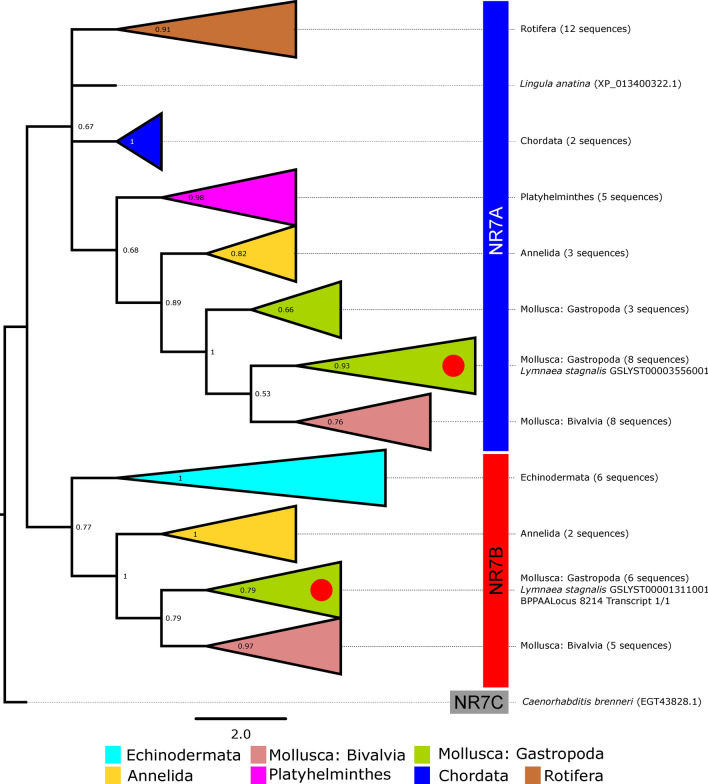
To take the analysis of the unusual 2DBD-NRs further, we concatenated the DBD and LBD domains from 60 other 2DBD-NRs representative of the metazoan groups studied by Wu and Loverde73 (see Supplementary Table ST4). A phylogenetic analysis of these domains suggests that the two L. stagnalis receptors are members of two different groups (A vs. B; Fig. 2 and Supplementary Fig. SF3), albeit with low statistical support (see branch bootstrap values). Therefore, following the recommendation of Wu et al. 2023, L. stagnalis has two NR7 genes, GSLYST00003556001 (NR7A) and GSLYST00001311001 (NR7B), corresponding respectively to C. gigas 2DBDγ and 2DBDδ74.
Fig. 2.
Phylogenetic analysis of 2DBD-NRs relationships across metazoans. Maximum Likelihood (ML) tree (same parameters as in Fig. 1; values below branches indicate statistical support based on 1000 bootstraps) based on the ClustalW-aligned concatenated conserved domains (DBD1, DBD2, and LBD) of the two sequences found in the genome of L. stagnalis (present study, indicated by red dots), and 60 sequences identified by Wu and Loverde (2023) from various lineages (Brachiopoda, Rotifera, Platyhelminthes, Annelida, Mollusca, Nematoda, Echinodermata, Cephalochordata, indicated by the different colours) and having the three domains complete (as verified using Conserved Domains DataBase). The different NR7 groups referred to in the text are indicated on the right in blue, red and grey. Alignment available upon request. See also Supplementary Fig. SF3 for detailed clade composition).
Among the incomplete NRs, four genes were found to lack either the entire LBD (1 gene) or the DBD (3 genes) and should thus be classified as members of the NR0 family (although this grouping is not monophyletic). We tentatively annotated them following their BLAST best-hit (H. sapiens and/or nr). On this basis, the single DBD-only gene was annotated as NR1A2_THRB, despite the lack of a true thyroid system in Mollusca (but see75, and the three LBD-only genes as NR1D2_REVERBB, NR1H3_LXRA, and NR2F1_COUP-TFI, respectively. Note that NR1D2 and NR1H3 annotations were shared by at least one other gene prediction which contained both the DBD and LBD. Another specific case was GSLYST00019008001 (and its related transcript). Although we could not find an open reading frame (ORF) that would generate both the DBD and LBD, two alternative ORFs would generate sequences with either of the domains. Ambiguously, each of these sequences has a different BLAST best-hit in H. sapiens: NR1C1_PPARA for the DBD-only ORF, and NR1H3_LXRA for the LBD-only ORF. In addition to the observation that NR1C1 and NR1H3 are already represented by two and three other gene predictions respectively, this suggests that GSLYST00019008001 and its corresponding transcript do not constitute a functional NR.
Despite extensive searches for alternative ORFs, 5 NRs were found to have an incomplete domain sequence (lacking part of either the DBD or the LBD). These might represent non-functional gene copies, as suggested by the observed density of nucleotide repeats in the problematic areas. While this should be confirmed through specific re-sequencing, the fact that some of these and other genes with complete domains share the same functional annotation (see NR1C3_PPARG and NR2E1_TLL, Supplementary Table ST2) tends to support the hypothesis of non-functionality. One last specific situation is that of NR2E3 (PNR), which is represented by two protein predictions, neither of which have both domains in full (an incomplete DBD, and the LBD is either complete or missing).
G protein-coupled receptors
InterProScan identified a significant GPCR hit for a total of 892 gene predictions, four of which were discarded due to incompleteness or other conflicting annotations (see Supplementary Tables ST5 and ST20). The remaining 888 gene predictions are distributed among the five main GPCR families (GRAFS system76) and the ancestral cAMP77: class A–Rhodopsin-like (784); class B–adhesion/secretin (65); class C–Glutamate (31); class E–cAMP (3); class F–Frizzled (5). Of these, 418 were members of five orthogroups (OGs) found to be expanded within the L. stagnalis genome (orthology analysed at metazoan level, Z-score > 2, Table 3; see also Supplementary Information for further details), while the remainder (470) was distributed across 135 non-expanded OGs. Most gene predictions belonging to the five expanded GPCR OGs were identified as Rhodopsin-like (class A) 7 trans-membrane (7TM; IPR17452). These five expanded OGs encompassed a high proportion (89–100%) of molluscan proteins from the six taxa included in the orthology analysis, and four of these OGs were predominantly exclusive to gastropods (85–100%; Table 3).
Table 3.
Summary statistics for GPCR Orthology Groups, with a focus on molluscan representatives.
| OrthoGroup | InterProScan | OG size | Mollusca | Gastropoda | L. stagnalis | Z-score |
|---|---|---|---|---|---|---|
| OG0000008 | Rhodopsin_R (peptide, FMRFamide) | 1215 | 1085 (89%) | 1035 (85%) | 369 (30%) | 2.537 |
| OG0001159 | Rhodopsin_R (peptide, FMRFamide) | 72* | 71 (99%) | 71 (99%) | 24 (33%) | 2.336 |
| OG0002276 | Rhodopsin _R (peptide, FMRFamide) | 49* | 48 (98%) | 48 (98%) | 17 (35%) | 2.452 |
| OG0002279 | Rhodopsin_R (2 GPCRs) | 49 | 49 (100%) | 49 (100%) | 22 (45%) | 3.106 |
| OG0002601 Absent in Deuterostoma and Arthropoda | Rhodopsin_R (peptide, FMRFamide) | 45 | 38 | 22 | 4 | < 2 |
| OG0010705 | Rhodopsin (peptide, FMRFamide) | 8 | 7 | 7 | 4 | <2 |
| OG0000001 | Rhodopsin_R (peptide) | 3444 | 911 | 561 | 139 | <2 |
| OG0004949 | Rhodopsin_R (biogenic amine) | 30 | 30 (100%) | 27 (90%) | 14 (47%) | 3.347 |
| OG0000121 | Rhodopsin_R (peptide and biogenic amine) | 286 | 268 | 115 | 29 | < 2 |
| OG0000012 | Adhesion_R | 1072 | 310 | 186 | 42 | < 2 |
Percentage values indicate for each orthogroup the proportion of proteins belonging either to Mollusca, Gastropoda, or to L. stagnalis. Abbreviations: OG, Orthogroup. A Z-score value (number of standard deviations from the mean) > 2.0 indicates significant expansion.
*In OG0001159, the single non-molluscan member was B. floridae, in OG0002276 C. elegans.
7TM-class A peptide GPCRs: a predominance of FMRFamide R-like
Three of the five expanded OGs with a GPCR hit (OG000008, 0G0001159, OG0002276) consisted exclusively of Rhodopsin-like receptors from the group of (7-transmembrane-domain) 7tmA_FMRFamide_R-like receptors (peptide), representing in total 424 L. stagnalis proteins. However, OG0002279, consisting of 49 exclusively Gastropoda proteins, was heterogeneous with only 2 GPCR hits for L. stagnalis proteins. The 22 other L. stagnalis proteins in this OG included BLASTp annotations against bacterial proteins, casting doubt on the statistical reliability of this OG. By contrast, OG0002601, although not expanded in L. stagnalis, was clearly an FMRFamide receptor OG (45 proteins), apparently absent in Deuterostomia and Arthropoda. Molluscs were the most represented taxa in this group, with 38 proteins, 22 of which belong to Gastropoda, and 4 to L. stagnalis. OG0010705 is also a small, non-expanded, FMRFamide receptor OG (8 proteins), with 7 molluscan representatives, all gastropods from the Heterobranchia order. However, the fact that one protein from this OG belongs to the arthropod F. candida questions the validity of this small group as a true OG. Finally, an extra-set of 44 L. stagnalis gene predictions from various and heterogeneous OGs were annotated as FMRFamide_R-like, although only 18 gene predictions had a complete 7TM motif (as determined with NCBI Conserved Domains database search tool). In summary, about 450 GPCRs could be confidently designated as FMRFamide_R-like. For completion, an OG of biogenic amine GPCRs (G0004949), which was also expanded in L. stagnalis, contained 13 receptors to muscarinic acetylcholine and one receptor to adrenaline.
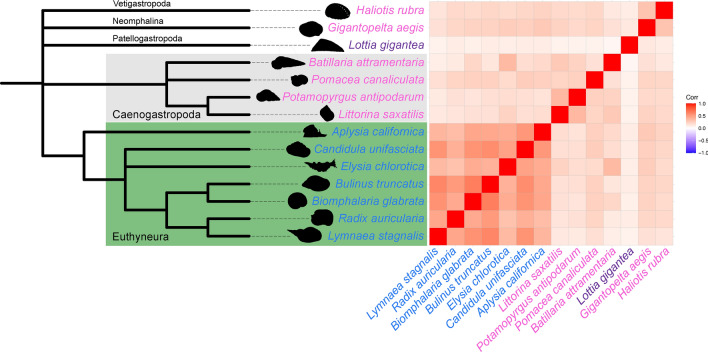
We hypothesised that FMRFamide receptor proliferation may have supported the evolution of simultaneous hermaphroditism in gastropods. To test this hypothesis, we refined the analysis of orthology by focusing on gastropods, and used available genomic resources to balance hermaphroditic species (A. californica, B. glabrata, Bulinus truncatus, Candidula unifasciata, Elysia chlorotica, L. stagnalis, Radix auricularia, Lottia gigantea) against gonochoristic species (separate genders, Batillaria attramentaria, Gigantopelta aegis, Haliotis rubra, Littorina saxatilis, Pomacea canaliculata, Potamopyrgus antipodarum) (see Supplementary Information, Orthology analyses). Note that L. gigantea is a protandrous sequential hermaphrodite while all other species are simultaneously hermaphroditic. This analysis generated nearly 29,500 OGs (Fig. 3; Supplementary Table ST6). Because all simultaneous hermaphrodites in our analysis are in the Euthyneura infraclass of molluscs, we also identified various OGs specific to this clade, including FMRFamide receptors, c-type lectins, cytochrome P450 cyclodipeptide synthase-associated, (see Supplementary Table ST7 for domain annotation of the main euthyneura-specific OGs).
Fig. 3.
Gastropod proteome orthology. Left: phylogenetic relationships among 14 gastropod species (topology taken from141) used for gene orthology assessment (colour code: blue, simultaneous hermaphrodite; purple, sequential hermaphrodite: pink, separate sexed). Right: pairwise correlation of species contribution in terms of number of proteins to 29,428 orthogroups produced by OrthoFinder. Correlations are particularly strong within the set of euthyneuran species used in the analysis. The heat map gradient on the right indicates the level of (positive) correlation.
The search for FMRFamide-R among L. stagnalis orthologs retrieved 483 genes distributed in 89 different OGs. FMRFamide-R groups previously found expanded in L. stagnalis appeared no more enriched, which is not surprising as all members of the analysis were gastropods. We excluded Radix auricularia from this analysis because that dataset was abnormally small (less than 18,000 protein predictions in total), and that species was recurrently absent from OGs otherwise well represented by other Hygrophila species (casting further doubt on the completeness of its predicted proteome). Excluding R. auricularia, the number of FMRF-R amide genes was significantly higher (ANOVA F-value [1, 11] = 10.21, P = 0.009) in hermaphrodites (simultaneous and sequential: 289.86 ± 149.10 genes) than in gonochoric gastropods (89.17 ± 36.88 genes). After excluding L. gigantea (a sequential hermaphrodite), simultaneous hermaphrodites were more significantly enriched in FMRFamide receptor genes (329.17 ± 117.04) than non-hermaphrodite species (ANOVA F-value [1, 10] = 22.95, P <0.001).
Peptide GPCRs: a high level of ligand diversity
OG0000001 is comprised of peptide GPCRs and is the second largest OG determined by OrthoFinder with 3444 genes representative of all taxa included. Although not expanded in L. stagnalis, this OG contains 139 L. stagnalis gene predictions, all assigned as peptide GPCRs, and predicted to bind a great diversity of molecules (more than 40 categories; see Supplementary TableST8). OG0000121 grouped gene predictions which shared four annotations with OG0000001 (in bold in Supplementary Table ST8) or were identified as other peptide receptors (neurotensin, somatostatin, FF neuropeptide, NPY peptide, opioid).
Among the rhodopsin-like gene predictions, a structural pattern was observed in the genome, with a spatial clustering of similar genes. Whether all members of a given cluster are functional genes or pseudogenes cannot be determined based on our data, yet it is to be noted that they do not all possess a complete 7TM motif and these clusters were also often interspersed with repetitive regions. In line with its high level of occurrence, FMRFamide_R-like was either the main or exclusive annotation of 30 of the 46 largest groups (size: 4 to 26 tandem GPCR genes), and also appeared in other groups. Among these, the proportion of transmembrane domains with at least 6 helices varied from 50 to 100%. Furthermore, 10 of the 30 groups of FMRFamide_R-like genes contained genes which also exhibited an additional viral chemokine GPCR hit (PHA02638 or PHA03087). Details on cluster annotations are given in Supplementary Table ST9.
Consistent with the levels of diversity observed in metazoan GPCR classes, classes B, C, E and F were more moderately represented in L. stagnalis. After discarding gene predictions with transmembrane domains containing less than 6 helices (considered as incomplete), we found 37 proteins representative of class B (12 B1_hormone receptors, 19 B2_adhesion, and 6 B3_Methuselah), 17 of class C (3 GABA B, 12 metabotropic glutamate, and 2 orphan GPR158), a single class E (cAMP), and five class F receptors (four Frizzled and one Smoothened protein) (see Supplementary Table ST9). Several other predicted proteins with significant matches to GPCRs from either of these classes were also identified but discarded due to incomplete or total lack of a transmembrane domain.
Spatial expression of FMRFamide receptors
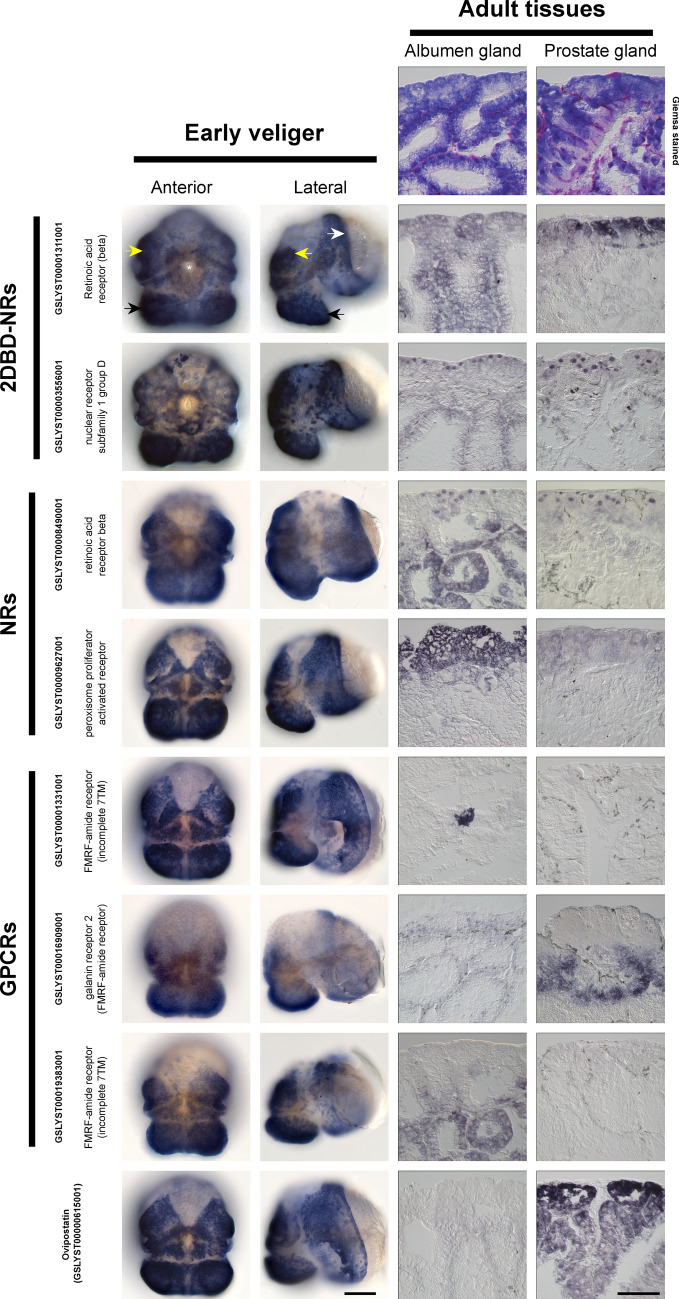
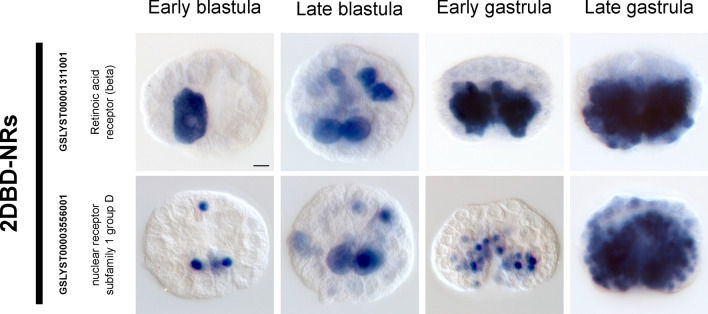
While the association between FMRFamide receptor expansion and simultaneous hermaphroditism is strong (as indicated by OG composition), it is problematic that the Euthyneura are monophyletic and are also the only simultaneous hermaphrodites used in these analyses. Therefore, to clarify whether some of these receptors are associated with the hermaphroditic condition in L. stagnalis (as opposed to other euthyneuric traits such as detorsion), we searched for differences in the expression of receptors in our transcriptomic datasets, and accordingly a selection was used for in situ expression analysis. The differential expression of some FMRFamide receptors between male (prostate) and female (albumen) reproductive organs drove our selection of 4 of these receptors (GSLYST00019397001, GSLYST00001331001, GSLYST00016909001, GSLYST00019383001). In addition, we investigated the expression patterns of three differentially expressed “traditional” nuclear receptors (GSLYST00008490001, GSLYST00009627001 and GSLYST00020139001) and the two 2DBD-NRs (GSLYST00001311001 and GSLYST00003556001). We also included ovipostatin (GSLYST00000615001) as a reference because it is known to be differentially upregulated in the prostate gland relative to the albumen gland78. The spatial expression profiles for these genes were striking in their similarity in early veligers; in general, these genes were robustly expressed in ectodermal cells of the foot, head and mantle edge (Fig. 4). The albumen and prostate glands are fully differentiated organs with separate reproductive functions, and we were able to detect qualitatively distinct spatial expression patterns for these genes in one or both reproductive organs. As expected, ovipostatin generated a strong signal in sections of the prostate gland, while effectively no signal was detected in the albumen gland (Fig. 4). In addition, the expression patterns of the two 2DBD-NRs in early embryos (early and late blastulae, early and late gastrulae) indicates that these gene products play likely critical roles in early developmental processes (Fig. 5).
Fig. 4.
Representative whole mount in situ hybridisation (WMISH) expression profiles for a selection of 2DBD-NRs, NRs, GPCRs and ovipostatin differentially expressed between early veliger stage and adult reproductive organs (paired albumen and prostate glands). The expression patterns of these receptors and ovipostatin in early-veliger-stage larvae are broad, and generally similar; all genes are robustly expressed in ectodermal cells covering the foot (black arrowheads), surrounding the stomodaeum (white asterisk), the mantle (white arrowhead) and the cephalic lobes (yellow arrowheads). In adult tissues ovipostatin is highly expressed in the prostate and effectively undetectable in the albumen gland as expected. Qualitative differences are also evident for many of the receptors surveyed, and hint at possible roles in maintaining these simultaneously active reproductive organs. Representative scale bars are 100 mm.
Fig. 5.
WMISH patterns of 2 2DBD-NRs during early development. Both of these receptors display dynamic, asymmetric patterns of expression prior to and during gastrulation. Expression of GSLYST00003556001 appears to largely be restricted to nuclei, other than 2 large prominent cells in late blastulae. Conversely, transcripts of GSLYST00001311001 are distributed throughout the cytoplasm. Both genes are restricted to the vegetal hemisphere in early gastrulae and are concentrated around the blastopore.
Discussion
Our characterisation of the genome of L. stagnalis expands the available range of organisms to study molluscan biology in the genomic era and provides a fundamental resource for research involving L. stagnalis in its role as a longstanding, invertebrate model organism. Specifically, our analysis revealed that the nuclear genome of L. stagnalis encodes an abundance of receptors, including DNA-binding-, 2DBD-, RTK-receptors and GPCRs. We focused our attention on the identification of receptors involved in biological processes because such information is lacking in many fields that employ Lymnaea as a model. For example, in ecotoxicology mechanistic evidence for toxicity pathways is critically lacking in L. stagnalis, even though it is used as a model for its ecological value (functional representativity within food-webs and in other ecological interactions). The resources provided in this work will be of particular interest for the study of endocrine disruptors (nuclear receptors, domain annotation and phylogeny) and more generally for xenobiotic responsive pathways involving GPCRs (see e.g.79) or RTKs66.
We have focused our analysis of the L. stagnalis genome on the identification of molecular receptors, and the expression, evolution and diversification of a selection of these. We have also characterised the mitogenome (see Supplementary Information), two unique double DNA-binding domain (2DBD) containing genes73, and investigated the in situ expression profiles of several receptors during embryogenesis and in sex-specific adult organs of this simultaneous hermaphrodite. These findings complement a previous study that concentrated on ion-channels and ionotropic receptors in the central nervous system (CNS68). Perhaps most notably, we observe an association between the evolution of simultaneous hermaphroditism and the diversification of GPCRs.
Identification of potential male/female receptors
As L. stagnalis is a simultaneous hermaphrodite, meaning each individual can execute male and female reproductive processes (see below), we searched for receptors that are potentially involved in regulating this simultaneous state. We used RNA-seq data from albumen gland (AG; female) and prostate gland (PG; male) to look for sex-specific expression patterns. Indeed, some GPCRs and NRs were exclusively expressed in one or the other of these organs (Supplementary Table ST15). These potential sex-specific receptors are particularly interesting because these are potential targets of seminal fluid proteins (LyACPs, Lymnaea Accessory Gland Proteins) which have been shown to specifically influence male or female reproductive processes in the sperm recipient (e.g.55,56). In the future, it will be fruitful to test the regulatory status of these receptor genes, how transient their expression is, and whether this is influenced by mating status (e.g., recently inseminated). We expect that such efforts will not only result in de-orphanising these receptors but will also present new opportunities for understanding the proximate mechanisms via which LyACPs induce their effects. This also argues for the generation of more well-annotated chromosome level gastropod genomes, and for the functional analysis of specific gene products.
Receptor expression
The broadly similar spatial expression patterns of the 2 NRs, 3 GPCRs and ovipostatin in early veliger larvae is intriguing (Fig. 4), and hints at potential functional commonalities for these distinct genes at this stage of development. These similarities are made more remarkable by the fact that ovipostatin is not a receptor, but rather a secreted accessory gland protein produced by the prostate gland that is understood to repress oviposition behaviours in Lymnaea78. These genes expand their expression from an apparently exclusive ectodermal pattern in early veligers, to the endodermally-derived adult reproductive organs we investigated (Fig. 4). The presence of mRNA transcripts for the selected receptors in one or both of these terminally differentiated organs implies that they are functionally required in these contexts, and that they therefore contribute to the maintenance of simultaneously functioning male and female reproductive states. As mentioned above, gene-specific (and likely reproduction-condition-specific) assays are required to elucidate the functional roles of these genes in these contexts. However, these results provide a foundation from which future investigations can be started.
The expression of the 2DBD-NR retinoic acid receptor-“beta” (as referred to in74) gene (GSLYST00001311001) in distinct cells of gastrula embryos (Fig. 5) corresponds with earlier reports on the effect of lithium (which is known to interact synergistically with retinoic acid80) on gastrulation and development in L. stagnalis81. The expression pattern of the 2DBD-NR subfamily 1 group D gene (GSLYST00003556001) in numerous nuclei of gastrula embryos also suggests an important role in early development, however, pending functional data (such as knock-downs or knock-outs) the specific ontogenetic roles of these genes remain unknown.
GPCR evolution/expansion
In general, G protein-coupled receptors (GPCRs) are an important class of membrane receptors that are involved in many biological processes, including external (e.g., pheromones, ions, odours, toxins, taste, and light) and internal (e.g., neuropeptides, proteins, lipids, neurotransmitters, and nucleotides) sensory signal detection (e.g.82). As a superfamily, GPCRs have evolved by tandem local duplication during metazoan evolution (prior to two rounds of whole-genome duplication in vertebrates83). In this context it is significant that we found around 450 FMRFamide R-like (Rhodopsin-like) GPCRs in the Lymnaea stagnalis genome. The presence of this expanded set of receptors can be explained by the fact that they are the receptors for the many FMRFamide-like (neuro)peptides that are involved in regulating many biological processes, including the hermaphroditic condition (e.g.84). These peptides belong to a class that contain the C-terminal Arg-Phe-NH2 (RFamide; RFamide-related peptides or RFRPs). The evolutionary origins of these (neuro)peptides dates back to a common bilaterian ancestor85. The first of these peptides to be discovered was the cardio-excitatory tetrapeptide FMRFamide, identified in the neural ganglia of the bivalve mollusc, Macrocallista nimbosa86. Since then, FMRFamide has been reported to be present in many species, including the ecdysozoan protostome Caenorhabditis elegans87, and various lophotrochozoan molluscs, including Cephalopods88,89 such as Sepia officinalis90 and Octopus vulgaris91,92, and Gastropods including Cornu aspersum, Biomphalaria glabrata and Biomphalaria alexandrina93,94, to name but a few. In addition, many RFRPs have since been discovered and are involved, besides cardiovascular regulation, in osmoregulation, digestion, locomotion, feeding and reproduction (e.g.95).
The mechanism that supported the apparent expansion of these receptors in the Euthyneura remains unknown. Whole genome duplication (WGD) events have been identified within several molluscan clades (at the base of the Cephalopoda, and within the Stylommatophora, the Neogastropoda, Tonnoidea, Cypraeidae, Capulidae and Heterobranchia) but apparently not at the origin of Euthyneura96–98. With respect to GPCRs, due to the high level of gene clustering observed in L. stagnalis, it seems unlikely that a WGD event at the origin of the Euthyrneura would be responsible for the FMRFamide receptor proliferation we observe in euthyneuran species. However, this hypothesis should be tested further, using appropriate tools and chromosome-level datasets in the Mollusca and elsewhere in the animal kingdom. Interestingly Rondón et al.99 recently reported an expansion of GPCR genes in two lineages of caenogastopods, one of which belongs to a lineage that underwent a whole genome duplication. As in the Lymnaea genome they also observe clustering of expanded GPCRs, which argues against a genome duplication event being a direct explanation to the observed expansion in GPCRs.
Based on our analyses of molluscan RFRPs and GPCRs, we postulate that the patterns of their abundance may in part have supported the transition from a separate-sexed state to a hermaphroditic condition. Our phylogenetic analysis supports an expansion of GPCRs in parallel with the occurrence of hermaphroditism in the molluscan species with well annotated genomes that were available at the time of writing for comparison. Supporting this observation is the fact that many molluscan reproductive processes are regulated by RFRPs and other (neuro)peptides. As in vertebrates, separate sexed molluscs can use the same substances for regulating male and female reproductive processes, because each body only contains one sex (e.g., Octopus vulgaris91; Sepia officinalis90; Sepiella japonica100; Haliotis discus hannai101). However, the evolution of simultaneous hermaphroditism means that male and female processes must be regulated concurrently within one body (e.g., Lymnaea stagnalis, Cornu aspersum, Aplysia californica:88). This requires largely nonoverlapping neurobiological wiring, regulatory peptides and accompanying receptors to avoid the simultaneous expression of conflicting reproductive processes (e.g.102). Hence, excitatory and inhibitory crosstalk between the male and female regulatory systems needs to be accompanied by an expansion of the receptors involved, so that they can be mutually exclusively used for executing one sexual role (while potentially inhibiting the other role).
It is worth illustrating this with the multitude of roles that FMRFamide and its co-expressed peptides fulfil103–106 next to being cardio-excitatory107. These tetra- and heptapeptides have been shown to be important for the fine coordination of the execution of male mating behaviour and insemination106,108. Importantly, FMRFamide has also been shown to inhibit the activity of the caudo-dorsal cells (CDC), which are normally responsible for the release of egg laying hormone during their synchronous bursting (CDCH109–111). Interestingly, the single, previously identified receptor for an RFamide is a ligand for the decapeptide TPHWRPQGRF-NH2 (LyCEP/luqin/tachykinin112). This peptide is cardioexcitatory, but also inhibits CDC activity, and by extension egg laying. Thus, when the CDCs are activated and egg laying is triggered, LyCEP release is likely suppressed, which would agree with the observed reduction in heartrate during egg laying113. This illustrates how the different RFRPs are interlinked and can collectively stimulate one physiological function while simultaneously inhibiting conflicting functions.
Conclusion
Here we report a full genome assembly and annotation for the great pond snail Lymnaea stagnalis. This snail has long served as a model organism to a wide variety of scientific disciplines, and we envisage this resource will assist traditionally distinct communities to more efficiently collaborate, provide deeper insight into long-standing questions, and to stimulate novel research directions. The annotation of this genome highlights a remarkable expansion of receptors critical for a wide variety of biological processes, and hints at a causal link with the evolution of simultaneous hermaphroditism in gastropods. The identification of such receptors is also crucial for understanding the chemosensory lifestyle that many gastropods lead and will likely provide insight into the mechanisms that underlie phenomena such as pheromone release, egg-laying, predator detection, mate location, mate-partner recognition, avoidance behaviours, memory, parasite infection and co-habitation behaviours. We envisage that our ongoing efforts to identify novel receptors, in combination with parallel advances in structural in silico modelling of protein structure, protein receptor localisation techniques and functional studies of receptors will ultimately de-orphanise the many receptors present in the genome of L. stagnalis and will provide insight not only into their mechanisms of action, but into their evolutionary histories.
Materials and methods
Snail line generation
The individual snail used for genome sequencing and assembly was derived from an inbred line previously used to map and identify the snail chirality locus46. This line was derived by alternating rounds of self-fertilisation and full-sib mating, over at least ten generations. The inbred snail line is freely available from the authors (AD) upon request and was also used for all RNA-seq.
Nucleic acid extraction
Genomic DNA was extracted with a Qiagen Puregene Core Kit A according to the manufacturers protocol, including RNAse treatment followed by resuspension in Qiagen rehydration solution. DNA quality was checked using Opgen QCards (Opgen, Rockville, MD, USA), and concentration determined via Qubit Fluorometric Quantitation (Thermo Fisher Scientific, Waltham, MA, USA).
RNAs from various tissues and organs (Supplementary Table ST11) were extracted with the NucleoSpin RNA Kit (Macherey Nalgen) and DNAse treatment according to the manufacturers protocol. Quality control was performed with the Agilent Bioanalyzer system.
Transcriptome sequencing and assembly
A set of cDNA libraries were constructed from a variety of tissues (see Supplementary ST11). For the stomach, digestive gland, reproductive system and foot stranded mRNAs were generated using Illumina® TruSeq stranded mRNA Library Prep, whereas for neural ganglia, head and mantle tissue, a kit adapted to low input RNA was used (SMARTer® Universal Low Input RNA Kit). Long-read sequencing was also performed on the ovotestis RNA using Oxford nanopore technology. To do so, two cDNA libraries were synthesized using either the SMARTer®PCR cDNA kit or the Nanopore sequencing SQK-MAP006, and purified with Ampure beads.
Illumina reads were quality filtered and trimmed using the procedure described in Alberti et al.114. In addition, for RNA-Seq data, ribosomal RNA-like reads were detected using SortMeRNA115 and the non-ribosomal short reads were assembled with Oases 0.2.08116, using a k-mer size of 63 bp. Then, short reads were mapped back to the contigs with BWA-mem and coverage was computed based on solely consistent paired-end reads. Uncovered regions were extracted and used to identify hypothetic chimeric contigs. In addition, open reading frames (ORF) and domains were searched using respectively TransDecoder (Haas, BJ. https://github.com/TransDecoder/TransDecoder) and CDsearch117. Uncovered regions that do not contain an ORF or a domain were tagged as mis-joins and contigs were split up. Read strand information was used to correctly orient RNA-Seq contigs.
Error-correction of cDNA nanopore reads
Raw nanopore reads were corrected using Illumina RNA-Seq reads and the NaS software118 with the following parameters: --ilmn_size 150 --mode fast --nb_proc 4 --t 2 --k 32 --untgl_seq_size 300. As the computation time is correlated with the number of Illumina reads, we decided to reduce the complexity by adding a normalisation step of the Illumina short-reads using the normalise-by-median.py script of the khmer package119 with the following parameters: -C 20 -k 20 -N 4 -x 10e9. Summary information on Nanopore reads is given in Supplementary Table ST12.
DNA sequencing and genome assembly
Library preparation and whole-genome sequencing
Four Illumina PE libraries were prepared starting from 100 ng to 250 ng DNA. Four independent DNA fragmentations were performed using the E210 Covaris instrument (Covaris, Inc., USA) to generate fragments mostly around 400 and 500 bp. Libraries were constructed using the NEBNext DNA Sample Prep Master Mix Set (New England Biolabs, MA, USA). DNA fragments were PCR-amplified using Platinum Pfx DNA polymerase (Invitrogen) and P5 and P7 primers. Amplified library fragments were size selected on 3% agarose gel around 400 bp or on 2% agarose gel around 600z bp and 700 bp. Library traces were validated on Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and quantified by qPCR using the KAPA Library Quantification Kit (KapaBiosystems) on a MxPro instrument (Agilent Technologies, USA). The paired end (PE) libraries were sequenced with 250bp paired-end reads on an Illumina HiSeq 2500 instrument (Illumina, San Diego, CA, USA).
The Mate Pair (MP) libraries were prepared using the Nextera Mate Pair Sample Preparation Kit (Illumina, San Diego, CA). Briefly, genomic DNA (4 µg) was simultaneously enzymatically fragmented and tagged with a biotinylated adaptor. Tagmented fragments were size selected on a Sage Science Electrophoretic Lateral Fractionator or SageELF (Sage Science, MA, USA). This system allows production of narrowly sized MP libraries, isolating 12 different discrete size fractions from a single sample loading. We selected 8 fractions (from 3 to 15 Kb) to continue the MP library preparation. Size-selected fractions were circularised overnight with a ligase. Linear, non-circularized fragments were digested, and circularised DNA was fragmented to 300–1000 bp size range using Covaris E210. Biotinylated DNA was immobilised on streptavidin beads, end-repaired, 3`-adenylated, and Illumina adapters were added. DNA fragments were PCR-amplified using Illumina adapter-specific primers and purified. Finally, libraries were quantified by qPCR and libraries profiles were evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies, USA). Each library was sequenced using 100 base-length read chemistry on a paired-end flow cell on the Illumina HiSeq2000 (Illumina, USA). Genomic DNA library coverage is summarized in Supplementary Table ST13.
Genome profile and assembly
K-mers were counted with jellyfish1120 to draw the genomic profile. Illumina paired-end and mate-pair reads were assembled in contigs using spades v. 3.5.02121 with kmer = 127. Contigs larger than 500 bp were assembled in scaffolds with sspace2.03122 and gaps were filled with gapfiller4123. Scaffolds longer than 2 kb were kept for genome annotation.
Genome masking, repeats and transposable elements
A strategy was developed to predict and then mask repetitive DNA before gene annotation. Due to the characteristics of the obtained assembly (~943 Mbps; 6640 scaffolds), Transposable Elements (TEs) were de novo annotated with a method specifically designed to overcome computational challenges of TE discovery in large genomes and using an automated process, as implemented in the package REPET V2.5124,125. The process started with focusing on a subset of the genome sufficient to identify “easy-to-find” TEs. In brief, we eliminated stretches of ns > 11 from the scaffolds and kept contigs larger than 16 kbps. The resulting subset contained ~655 Mbps (representing 69.44% of the initial genome) and 16475 contigs, further used as effective DNA sequences to build TE consensus without N stretches.
The de novo detection was first self-aligned onto the genome subset and blasted against itself using the TEdenovo pipeline similarity approach implemented in REPET and based on Blaster default parameters. Matching sequences were clustered using programs specific to interspersed repeats (Grouper, Recon, Piler; see REPET package). A consensus sequence was then built for each cluster, based on the multiple alignment of all members of that cluster. All consensus sequences were then classified using the classification system of Wicker et al.126, as implemented in PASTEClassifier tool in REPET package. Redundancy was removed and TEs were grouped by family, resulting in a filtered library of 5085 non-redundant consensus sequences representative of “easy-to-find” TEs.
TE annotation was performed using an iterative process with REPET (Blaster, Censor, RepeatMasker, see REPET package). First, the complete “easy-to-find” TE library was aligned by similarity searching on the initial genome (943 Mbps), obtaining a TE coverage of 42.6%. This led to the retrieval of 23211 TE full length copies (FLC, i.e., TE annotations aligned on more than 95% of their cognate consensus). From these, we used the corresponding consensus sequences and built a FLC TE sub-library made of 2643 sequences. Next, the FLC TE library was aligned against the initial genome (943 Mbps). Despite lower coverage (37.89% of the assembly), this alignment identified a larger number of FLCs (24190) than did the previous one. Repeats detected ab initio and annotated TEs, representing 326 Mbps in total, were masked on the genome for further annotation.
The genome was first self-aligned and blasted against itself (similarity approach, using Blaster default parameters). Matches were clustered using programs specific to interspersed repeats (Grouper, Recon, Piler). A consensus sequence was built for each cluster, based on the multiple alignment of all sequences thereof. All consensus sequences were classified using the classification system of Wicker et al.126, as implemented in PASTEClassifier. Redundancy was removed and TEs were grouped by family, resulting in a filtered library of 5085 non-redundant consensus sequences (easy-to-find TEs).
TE annotation was performed using an iterative process with TEannot pipeline (Blaster, Censor, RepeatMasker, see REPET package). First, the complete library of easy-to-find TEs was aligned by similarity searching on the initial genome (943 Mb), with a coverage of 42.6%. This led to the retrieval of 23,211 full length copies (FLC, i.e., TE annotations aligned on more than 95% of their cognate consensus). Next, the sub-library of FLC consensus sequences (n = 2643) was aligned against the initial genome (943 Mb). Despite lower coverage (37.89%), this alignment identified a larger number of FLCs (24,190) than the previous one. Summary statistics on TE annotation are given in supplementary Tables ST14 and ST15.
Repeats detected ab initio and annotated TEs, representing 326 Mb in total (34.6% of the assembly), were masked (hard masking) on the genome for further annotation. Nevertheless, repeat sequences are fully accessible on the final assembly provided. Gene annotation was based on the search for exon/intron structure on the genome assembly and using both ab initio (using SNAP) and evidence driven (RNA-seq) gene prediction.
Genome annotation
The general annotation workflow is summarised on Supplementary Fig. SF4. Following repeat masking (as described above), gene prediction was performed using both ab initio and evidence-driven (RNA-Seq and conserved proteins) methods. Transcript contigs (obtained as described above) were first aligned against the repeat-masked genome with BLAT. For each contig, we selected a single alignment with the highest BLAT score having at least an identity percent of 90%. Additionally, any alignment containing an intron larger than 100Kb was divided into two sub-alignments. The BLAT alignments defined genomic regions where we performed spliced alignments with the corresponding RNA-Seq contig to accurately identify splice sites. These alignments were performed using est2genome127 and those with at least 90% identity and a length ratio of aligned contig ≥ 85% were selected.
We used error-corrected cDNA nanopore reads generated as described above to calibrate the SNAP ab initio gene prediction tool128. First, ORFs were detected in corrected nanopore reads using TransDecoder (default parameters, build r20131117), and these were mapped onto the genome assembly with the same workflow used to align RNA-Seq contigs. For each genomic locus, we selected the longest nanopore read as the representative isoform, and all the representative isoforms were used to train SNAP and to generate a L. stagnalis configuration file. Finally, gene structures were predicted on the masked genome assembly using SNAP and the newly created parameter file.
Gastropod protein sequences extracted from Uniprot were mapped on the masked genome using a strategy similar to the one applied to RNA-Seq contigs. Protein sequences were first masked for low complexity using segmasker129 and aligned using BLAT. The best match and matches with a score ≥ 90% of the best match score, were retained and alignments containing introns ≥ 100 kb were split to create distinct alignments. Alignments were then refined using Genewise (default parameters), which is more precise for intron/exon boundary detection130. Alignments were selected if more than 80% of the length of the protein was aligned to the genome. In order to catch poorly conserved genes, we also relaunched the same workflow in which the first alignment step was performed using BLASTx instead of BLAT and only the 5 best matches per genomic locus were retained.
Gene model prediction was performed using Gmove131. This annotation tool combines various evidence from RNA-seq and conserved protein alignments, as well as ab initio predictions. It produces a consensus on gene structure without any prior calibration. Gmove creates an oriented graph (with exons as nodes and introns as vertex) from which all possible paths are extracted, representing potential gene models. Coding frames consistent with the available protein evidence were searched and in cases where no protein alignment exists, the longest coding frame is selected. Single-exon genes and genes located in the intron of another gene were rejected if their coding sequence was shorter than 300 nt. Then, genes that overlap TEs on 50% of their lengths were also removed. Among the remaining genes, we retained all genes with a coding sequence greater than 300 nt as well as shorter genes with at least one match to UniProt proteins (e-value < 10–10).
We analysed the completeness of the predicted gene catalogue with BUSCO software (v5.2.2)132, using mollusca_odb10 as reference dataset. Annotation of the predicted genes were done via BLAST (E-value threshold of 1E- 5) search against Nr, SwissProt and KOG databases and search against InterPro using RunIprScan (http://michaelrthon.com/runiprscan/), a command line utility for interfacing with the InterProScan server at the European Bioinformatics Institute. Protein domain annotation was completed with the Conserved Domain Database (NCBI CD search tool). We also compared the predicted set of proteins with the LC-MS-based proteomic database currently available29 using BLASTp between the two datasets. The analysis showed a significant hit of the latter with 13,104 of the present predicted proteins. Furthermore, 5232 of these predicted proteins shared at least 50% similarity with 86.5% of the proteomic dataset (4285 of 4953 sequences).
Orthology and gene family analysis
An orthology analysis of the predicted proteome of L. stagnalis was performed using OrthoFinder (V1 and V2.5.5) at two phylogenetic levels. First, as a broad level of investigation, a set of 19 specific proteomes was chosen from Genbank to represent a large diversity of metazoan lineages: Placozoa (Trichoplax adhaerens), Porifera (Amphimedon queenslandica), Platyhelminthes (Schistosoma mansoni), Mollusca (Biomphalaria glabrata, Aplysia californica, Lottia gigantea, Crassostrea gigas, Octopus bimaculoides), Annelida (Helobdella robusta), Nematoda (Caenorhabditis elegans), Arthropoda (Daphnia pulex, Hyalella azteca, Folsomia candida, Drosophila melanogaster), Echinodermata (Apostichopus japonicus), Hemichordata (Saccoglossus kowalevskii), and Chordata (Branchiostoma floridae, Ciona intestinalis, Homo sapiens). Orthogroups were identified at this scale of diversity. Gene family expansion was statistically assessed in L. stagnalis using a Z-score based test133,134.
Next, a refined analysis was performed at the level of Gastropoda, with a specific focus on GPCRs, to test for the possible implication of FMRFamide receptor expansion (as inferred from the first analysis) in the evolution of simultaneous hermaphroditism in this class of molluscs. The analysis was based on 14 gastropod species, including 6 species with separate sexes (Haliotis rubra, Gigantopelta aegis, Batillaria attramentaria, Pomacea canaliculata, Potamopyrgus antipodarum, and Littorina saxatilis), one sequential hermaphrodite (Lottia gigantea), and 6 simultaneous hermaphrodites (Aplysia californica, Candidula unifasciata, Elysia chlorotica, Bulinus truncatus, Biomphalaria glabrata, Radix auricularia, and Lymnaea stagnalis). From this analysis, the gene contribution of each species to all 89 orthogroups with L. stagnalis members annotated as “FMRFamide_receptor-like" was summed up and used to compare the mean number of genes per species across reproductive modes (simultaneous hermaphroditism vs. other; hermaphroditism vs. separate sexes) using an analysis of variance (see Supplementary Information, orthology analyses).
Mitochondrial genome
Sequence similarity searches of the genome assembly identified a scaffold containing the complete mitochondrial genome of L. stagnalis. Constituent NaS Nanopore reads were checked to evaluate contiguity of the scaffold. The initial automated annotation of the mitogenome (MITOS; http://mitos.bioinf.uni-leipzig.de/index.py;135 was manually checked using Snapgene software (www.snapgene.com) and applying criteria for annotation of molluscan mitogenomes as recommended by Ghiselli et al.136. The gene order of the mitogenome was compared among L. stagnalis and representative hygrophylid gastropods available in GenBank. Unless specified otherwise all bioinformatic tools were applied using default parameters.
Whole mount in-situ hybridisation
A total of 10 genes were selected for whole mount in situ hybridisation (WMISH) against male (prostate gland) and female (albumen) reproductive organs. We also surveyed late developmental stages for all genes, and gastrulation stages for the two 2DBD-NRs. These included two DNA-binding-domain containing nuclear receptors (GSLYST00001311001 and GSLYST00003556001), 4 GPCR genes (GSLYST00019397001, GSLYST00001331001, GSLYST00016909001, GSLYST00019383001) and 3 nuclear receptors (GSLYST00008490001, GSLYST00009627001 and GSLYST00020139001), and as a positive control the seminal fluid protein ovipostatin (GSLYST0000061500178. Fragments of these genes were amplified from cDNA using previously described standard procedures137 and the primers described in Supplementary Table ST16 (see also ST17 for complete sequence information on amplified regions). PCR fragments were cloned into a TA vector (pGEM-T Easy) and verified via Sanger sequencing. DIG labelled anti-sense riboprobes were synthesised as previously described137. A paraffin Tissue Microarray was 3D printed and constructed using STL files as described in Pazaitis and Kaiser138. Snails for these experiments were maintained as previously described41. The albumen and prostate glands of mature L. stagnalis were dissected out as previously described139 and fixed for 2 hours in 0.1% glutaraldehyde in PBS with 0.1% Triton at room temperature. These tissues were then washed twice with PBS + 0.1% Tween-20, and then dehydrated through a series of ethanol with an overnight incubation in 100% ethanol. The following day the tissue was exchanged into 100% xylene and incubated for a further 24 hours before being perfused with molten paraffin overnight. Tissue samples were placed into the recipient paraffin block and allowed to set for several hours to overnight (the pairing of albumen and prostate glands from individual snails was always maintained). Microtome sections (10 μm) were collected onto coated microscope slides and allowed to adhere at 37°C overnight. Following de-paraffinisation in xylene and a rehydration series, in situ hybridisation was performed as previously described137.
Snails and experimental conditions for mating observations and reproductive organ RNA-seq
Ten adult snails of three months old and with a shell length of 30 mm were collected from an age-synchronised mass rearing culture at Vrije Universiteit (VU) in Amsterdam. These snails were isolated for 8 days by placing each individual in a transparent, plastic container with a volume of 625 mL filled up to 460 mL. These containers were perforated with slits on opposing sides to let water flow through and were placed in a laminar flow tank with running, low-copper water. The water temperature was 20 ± 1 °C and the light conditions were 12h:12h, as per standard culturing conditions at VU Amsterdam. These snails were each fed one disk of broad-leaf lettuce, measuring approximately 19 cm2, daily. On day seven the containers were replaced with clean ones to induce egg laying, so that this behaviour would not interfere during the mating observations on the next day. On day eight, snails were randomly placed in pairs in the same type of containers, but without slits, that were filled up to 460 mL with water of the same temperature. These containers were places on a lab bench for mating observations (6h, e.g.140. Once unilateral copulation was completed, the sperm donor (male-acting individual) and the sperm recipient (female-acting) were euthanised by injecting 2–3 mL of 50 mM MgCl2 through the foot (needle gauge: 0.3 mm × 13 mm). The (male) prostate gland and the (female) albumen gland of donors and recipients were dissected out and snap frozen for RNA-seq and differential gene expression analyses. Reads (see earlier RNA-seq description) were quality controlled using FastQC and MultiQC. Genome-guided alignments of this data was performed using HISAT2, followed by a quality control of the alignment using QualiMap. Reads that mapped to annotated genes were counted using HTseq, after which specific receptors (NR and GPCR) were selected for further investigation.
Access to sequence information
The Illumina and nanopore sequencing data, as well as the genome assembly and gene predictions, are available in the European Nucleotide Archive (ENA) under the umbrella project PRJEB67819 (which encompasses PRJEB67821 and PRJEB67698 subprojects). In addition, the correspondance between locus tags and protein IDs and their BLASTp best-hit, as well as gene prediction mapping onto the assembly and their sequences (full gene, cDNA, AA) are provided as supplementary information (ST21–ST25). Also, all RTK, NR, and GPCR predicted protein sequences (including manually curated sequences) are gathered in SSF2–SF4, respectively.
Supplementary Information
Acknowledgements
JMK and YN thank J. Mariën, C. Popelier and O. Bellaoui for technical assistance. MAC thanks M. Collinet and the INRAE-U3E staff for technical support.
Author contributions
Conceptualisation/study design: JMK, DJJ, MAC, AD, CMA, ZPF Resources: JMK, DJJ, MAC, AD, AM, BN, VJ, JP, KL, CDS, CK, JMA, PW Data collection/curation: JMK, DJJ, MAC, AM, BN, VJ, CDS, CK, JMA Bioinformatic investigation: JMK, DJJ, MAC, YN, AM, BN, VJ, CMA, CDS, CK, JMA Data analysis: JMK, DJJ, MAC, YN, AM, BN, VJ, CDS, CK, JMA Experimental investigation: DJJ, NC, JMK Visualisation: JMK, DJJ, MAC, CMA Writing (original draft): JMK, DJJ, MAC Writing (review and editing): JMK, DJJ, MAC, AD, CMA, YN, AM, BN, VJ, JP, KL, CDS, CK, JMA, PW Funding acquisition: JMK, DJJ, MAC, AD, ZPF, PW.
Funding
This work was supported by the Genoscope, the Commissariat à l'Énergie Atomique et aux Énergies Alternatives (CEA) and France Génomique (ANR-10-INBS-09–08). JMK and YN were funded by Open Competition Grant OCENW.KLEIN.062. DJJ was funded by Deutsche Forschungsgemeinschaft (DFG project #528314512).
Data availability
The Illumina and nanopore sequencing data, as well as the genome assembly and gene predictions, are available in the European Nucleotide Archive (ENA) under the project PRJEB67819.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: J. M. Koene and D. J. Jackson.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78520-1.
References
- 1.Adema, C. M. et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun.8, 15451. 10.1038/ncomms15451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlosser, G. A short history of nearly every sense-the evolutionary history of vertebrate sensory cell types. Integr. Comp. Biol.58, 301–316. 10.1093/icb/icy024 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Robertson, H. M. Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol.10.1146/annurev-ento-020117-043322 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Rigon, F. et al. Evolutionary diversification of secondary mechanoreceptor cells in Tunicata. BMC Evol. Biol.4, 12. 10.1186/1471-2148-13-112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santini, S. et al. The compact genome of the sponge Oopsacas minuta (Hexactinellida) is lacking key metazoan core genes. BMC Biol.21, 139. 10.1186/s12915-023-01619-w (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moggioli, G. et al. Distinct genomic routes underlie transitions to specialised symbiotic lifestyles in deep-sea annelid worms. Nature10.1038/s41467-023-38521-6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, J. A. et al. The dynamic genome of Hydra. Nature464, 592. 10.1038/nature08830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawasawa, Y., McKenzie, L. M., Hill, D. P., Bono, H. & Yanagisawa, M. G protein-coupled receptor genes in the FANTOM2 database. Genome Res.13, 1466. 10.1101/gr.1087603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Özçelik, R., van Tilborg, D., Jiménez-Luna, J. & Grisoni, F. Structure-based drug discovery with deep learning. Chembiochem24, e202200776. 10.1002/cbic.202200776 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Hauser, A. S., Gloriam, D. E., Bräuner-Osborne, H. & Foster, S. R. Novel approaches leading towards peptide GPCR de-orphanisation. Br. J. Pharmacol.177, 961. 10.1111/bph.14950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Civelli, O. et al. G protein-coupled receptor deorphanizations. Annu. Rev. Pharmacol. Toxicol.53, 127. 10.1146/annurev-pharmtox-010611-134548 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beets, I. et al. System-wide mapping of neuropeptide-GPCR interactions in C. elegans. Cell Rep.42, 113058. 10.1016/j.celrep.2023.113058 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel, D. et al. Large-scale deorphanization of Nematostella vectensis neuropeptide G protein-coupled receptors supports the independent expansion of bilaterian and cnidarian peptidergic systems. Elife12, 90674. 10.7554/eLife.90674 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor, I., AA, H. A., Benjamin, P. R., Koene, J. M. & Zsolt, P. The unlimited potential of the great pond snail Lymnaea stagnalis. Life9, e56962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ducrot, V. et al. Development and validation of an OECD reproductive toxicity test guideline with the pond snail Lymnaea stagnalis (Mollusca, Gastropoda). Regul. Toxicol. Pharmacol.70, 605. 10.1016/j.yrtph.2014.09.004 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hussein, A. A. A. et al. Effect of photoperiod and light intensity on learning ability and memory formation of the pond snail Lymnaea stagnalis. Invertebr. Neurosc.10.1007/s10158-020-00251-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korneev, S. A. et al. A CREB2-targeting microRNA is required for long-term memory after single-trial learning. Sci. Rep.10.1038/s41598-018-22278-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivi, V. et al. What can we teach Lymnaea and what can Lymnaea teach us. Biol. Rev. Camb. Philos. Soc.96, 1590. 10.1111/brv.12716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amorim, J. et al. Lymnaea stagnalis as a freshwater model invertebrate for ecotoxicological studies. Sci. Total Environ.669, 11. 10.1016/j.scitotenv.2019.03.035 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Brix, K. V., Esbaugh, A. J., Munley, K. M. & Grosell, M. Investigations into the mechanism of lead toxicity to the freshwater pulmonate snail Lymnaea stagnalis. Aquat. Toxicol.106–107, 147. 10.1016/j.aquatox.2011.11.007 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Bouétard, A., Besnard, A.-L., Vassaux, D., Lagadic, L. & Coutellec, M.-A. Impact of the redox-cycling herbicide diquat on transcript expression and antioxidant enzymatic activities of the freshwater snail Lymnaea stagnalis. Aquat. Toxicol.126, 256. 10.1016/j.aquatox.2012.11.013 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Côte, J. et al. Genetic variation of Lymnaea stagnalis tolerance to copper: A test of selection hypotheses and its relevance for ecological risk assessment. Environ. Pollut.205, 209. 10.1016/j.envpol.2015.05.040 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Karakaş, S. B. & Otludil, B. Accumulation and histopathological effects of cadmium on the great pond snail Lymnaea stagnalis Linnaeus, 1758 (Gastropoda: Pulmonata). Environ. Toxicol. Pharmacol.78, 103403. 10.1016/j.etap.2020.103403 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Seppälä, O., Çetin, C., Cereghetti, T., Feulner, P. G. D. & Adema, C. M. Examining adaptive evolution of immune activity: Opportunities provided by gastropods in the age of ‘omics’. Philos. Trans. R. Soc. B Biol. Sci.376, 20200158. 10.1098/rstb.2020.0158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seppälä, O. et al. Transcriptome profiling of Lymnaea stagnalis (Gastropoda) for ecoimmunological research. BMC Genom.22, 144. 10.1186/s12864-021-07428-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng, Z.-P. et al. Transcriptome analysis of the central nervous system of the mollusc Lymnaea stagnalis. BMC Genom.10, 451. 10.1186/1471-2164-10-451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosato, M., Hoelscher, B., Lin, Z., Agwu, C. & Xu, F. Transcriptome analysis provides genome annotation and expression profiles in the central nervous system of Lymnaea stagnalis at different ages. BMC Genom.22, 637. 10.1186/s12864-021-07946-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Nierop, P. et al. Identification and functional expression of a family of nicotinic acetylcholine receptor subunits in the central nervous system of the mollusc Lymnaea stagnalis. J. Biol. Chemi.281, 1680. 10.1074/jbc.m508571200 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Wooller, S. et al. A combined bioinformatics and LC-MS-based approach for the development and benchmarking of a comprehensive database of Lymnaea CNS proteins. J. Exp. Biol.225, 243753. 10.1242/jeb.243753 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Ivashkin, E. et al. Serotonin mediates maternal effects and directs developmental and behavioral changes in the progeny of snails. Cell Rep.12, 1144. 10.1016/j.celrep.2015.07.022 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Ivashkin, E. et al. Transglutaminase activity determines nuclear localization of serotonin immunoreactivity in the early embryos of invertebrates and vertebrates. ACS Chem. Neurosci.10, 3888. 10.1021/acschemneuro.9b00346 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Morrill, J. B., Blair, C. A. & Larsen, W. J. Regulative development in the pulmonate gastropod, Lymnaea palustris, as determined by blastomere deletion experiments. J. Exp. Zool.183, 47. 10.1002/jez.1401830106 (1973). [Google Scholar]
- 33.Raven, C. P. The development of the egg of Limnaea Stagnalis L. from oviposition till first cleavage. Arch. Néerl. Zool.7, 91. 10.1163/187530146x00050 (1946). [Google Scholar]
- 34.Raven, C. P. Morphogenesis in Limnaea stagnalis and its disturbance by lithium. J. Exp. Zool.121, 1. 10.1002/jez.1401210102 (1952). [Google Scholar]
- 35.Raven, C. P. & Van der Wal, U. P. Analysis of the formation of the animal pole plasm in the eggs of Limnaea stagnalis. J. Embryol. Exp. Morphol.12, 123 (1964). [PubMed] [Google Scholar]
- 36.van den Biggelaar, J. A. Timing of the phases of the cell cycle during the period of asynchronous division up to the 49-cell stage in Lymnaea. J. Embryol. Exp. Morphol.26, 367 (1971). [PubMed] [Google Scholar]
- 37.Lukowiak, K., Sunada, H., Teskey, M., Lukowiak, K. & Dalesman, S. Environmentally relevant stressors alter memory formation in the pond snail Lymnaea. J. Exp. Biol.217, 76. 10.1242/jeb.089441 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Martens, K. R. et al. Stressful stimuli modulate memory formation in Lymnaea stagnalis. Neurobiol. Learn. Mem.87, 391. 10.1016/j.nlm.2006.10.005 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Swinton, E., Swinton, C. & Lukowiak, K. Shell damage leads to enhanced memory formation in Lymnaea. J. Exp. Biol.10.1242/jeb.207571 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Cerveau, N. & Jackson, D. J. A survey of miRNAs involved in biomineralization and shell repair in the freshwater gastropod Lymnaea stagnalis. Discov. Mater.10.1007/s43939-021-00007-x (2021). [Google Scholar]
- 41.Herlitze, I., Marie, B., Marin, F. & Jackson, D. J. Molecular modularity and asymmetry of the molluscan mantle revealed by a gene expression atlas. GigaScience10.1093/gigascience/giy056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hohagen, J. & Jackson, D. J. An ancient process in a modern mollusc: Early development of the shell in Lymnaea stagnalis. BMC Dev. Biol.13, 27. 10.1186/1471-213X-13-27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abe, M. & Kuroda, R. The development of CRISPR for a mollusc establishes the formin Lsdia1 as the long-sought gene for snail dextral/sinistral coiling. Development10.1242/dev.175976 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Kuroda, R. et al. Diaphanous gene mutation affects spiral cleavage and chirality in snails. Sci. Rep.10.1038/srep34809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibazaki, Y., Shimizu, M. & Kuroda, R. Body handedness is directed by genetically determined cytoskeletal dynamics in the early embryo. Curr. Biol.14, 1462. 10.1016/j.cub.2004.08.018 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Davison, A. et al. Formin is associated with left-right asymmetry in the pond snail and the frog. Curr. Biol.26, 654. 10.1016/j.cub.2015.12.071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledder, M., Nakadera, Y., Staikou, A. & Koene, J. M. Dominant gingers—Discovery and inheritance of a new shell polymorphism in the great pond snail Lymnaea stagnalis. Ecol. Evol.13, e10678. 10.1002/ece3.10678 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koene, J. M. & Ter Maat, A. Coolidge effect in pond snails: Male motivation in a simultaneous hermaphrodite. BMC Evol. Biol.7, 212. 10.1186/1471-2148-7-212 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boer, P., Jansen, R., Koene, J. & Maat, A. Nervous control of male sexual drive in the hermaphroditic snail Lymnaea stagnalis. J. Exp. Biol.200, 941. 10.1242/jeb.200.5.941 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Moussaoui, R., Verdel, K., Benbellil-Tafoughalt, S. & Koene, J. M. Female behaviour prior to additional sperm receipt in the hermaphroditic pond snail Lymnaea stagnalis. Invertebr. Reprod. Dev.62, 82. 10.1080/07924259.2017.1415988 (2018). [Google Scholar]
- 51.Daupagne, L. & Koene, J. M. Disentangling female postmating responses induced by semen transfer components in a simultaneous hermaphrodite. Anim. Behav.166, 147. 10.1016/j.anbehav.2020.06.009 (2020). [Google Scholar]
- 52.Álvarez, B. & Koene, J. M. Cognition and its shaping effect on sexual conflict: Integrating biology and psychology. Front. Integr. Neurosci.16, 826304. 10.3389/fnint.2022.826304 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charnov, E. L. Simultaneous hermaphroditism and sexual selection. Proc. Natl. Acad. Sci. USA76, 2480. 10.1073/pnas.76.5.2480 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anthes, N. et al. Bateman gradients in hermaphrodites: An extended approach to quantify sexual selection. Am. Nat.176, 249. 10.1086/655218 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Nakadera, Y. et al. Receipt of seminal fluid proteins causes reduction of male investment in a simultaneous hermaphrodite. Curr. Biol.24, 859. 10.1016/j.cub.2014.02.052 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Koene, J. M. et al. Male accessory gland protein reduces egg laying in a simultaneous hermaphrodite. PLoS ONE5, e10117. 10.1371/journal.pone.0010117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Álvarez, B., Koene, J. M., Hollis, K. L. & Loy, I. Learning to anticipate mate presence shapes individual sex roles in the hermaphroditic pond snail Lymnaea stagnalis. Anim. Cogn.25, 1417. 10.1007/s10071-022-01623-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puurtinen, M., Emily Knott, K., Suonpää, S., Nissinen, K. & Kaitala, V. Predominance of outcrossing in Lymnaea stagnalis despite low apparent fitness costs of self-fertilization. J Evol Biol20, 901. 10.1111/j.1420-9101.2007.01312.x (2007). [DOI] [PubMed] [Google Scholar]
- 59.Escobar, J. S. et al. Patterns of mating-system evolution in hermaphroditic animals: Correlations among selfing rate, inbreeding depression, and the timing of reproduction. Evolution65, 1233. 10.1111/j.1558-5646.2011.01218.x (2011). [DOI] [PubMed] [Google Scholar]
- 60.Coutellec, M.-A. & Caquet, T. Heterosis and inbreeding depression in bottlenecked populations: A test in the hermaphroditic freshwater snail Lymnaea stagnalis. Genetic load in a freshwater snail. J. Evol. Biol.24, 2248. 10.1111/j.1420-9101.2011.02355.x (2011). [DOI] [PubMed] [Google Scholar]
- 61.10.1787/20777876
- 62.Fodor, I. et al. Studies on a widely-recognized snail model species (Lymnaea stagnalis) provide further evidence that vertebrate steroids do not have a hormonal role in the reproduction of mollusks. Front. Endocrinol.13, 1. 10.3389/fendo.2022.981564 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertrand, S. et al. Evolutionary genomics of nuclear receptors: From twenty-five ancestral genes to derived endocrine systems. Mol. Biol. Evol.21, 1923–1937. 10.1093/molbev/msh200 (2004). [DOI] [PubMed] [Google Scholar]
- 64.de Mendoza, A., Sebé-Pedrós, A. & Ruiz-Trillo, I. The evolution of the GPCR signaling system in eukaryotes: Modularity, conservation, and the transition to metazoan multicellularity. Genome Biol Evol6, 606. 10.1093/gbe/evu038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morel, M., Vanderstraete, M., Hahnel, S., Grevelding, C. G. & Dissous, C. Receptor tyrosine kinases and schistosome reproduction: New targets for chemotherapy. Front. Genet.10.3389/fgene.2014.00238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell141, 1117. 10.1016/j.cell.2010.06.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Zein, N., D’Hondt, S. & Sariban, E. Crosstalks between the receptors tyrosine kinase EGFR and TrkA and the GPCR, FPR, in human monocytes are essential for receptors-mediated cell activation. Cell. Signal.22, 1437. 10.1016/j.cellsig.2010.05.012 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Dong, N. et al. Ion channel profiling of the Lymnaea stagnalis ganglia via transcriptome analysis. BMC Genom.22, 18. 10.1186/s12864-020-07287-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frooninckx, L. et al. Neuropeptide GPCRs in C. elegans. Front. Endocrinol.10.3389/fendo.2012.00167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinogradov, A. E. Variation in ligand-accessible genome size and its ecomorphological correlates in a pond snail. Hereditas128, 59. 10.1111/j.1601-5223.1998.00059.x (1998). [Google Scholar]
- 71.Elliott, T. A. & Gregory, T. R. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philoso. Trans. R. Soc. Lond. Ser. B Biol. Sci.370, 20140331. 10.1098/rstb.2014.0331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, W., Niles, E. G., El-Sayed, N., Berriman, M. & LoVerde, P. T. Schistosoma mansoni (Platyhelminthes, Trematoda) nuclear receptors: Sixteen new members and a novel subfamily. Gene366, 303. 10.1016/j.gene.2005.09.013 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Wu, W. & LoVerde, P. T. Updated knowledge and a proposed nomenclature for nuclear receptors with two DNA binding domains (2DBD-NRs). PLoS ONE18, e0286107. 10.1371/journal.pone.0286107 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogeler, S., Galloway, T. S., Lyons, B. P. & Bean, T. P. The nuclear receptor gene family in the Pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genom.15, 369. 10.1186/1471-2164-15-369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morthorst, J. E., Holbech, H., De Crozé, N., Matthiessen, P. & LeBlanc, G. A. Thyroid-like hormone signaling in invertebrates and its potential role in initial screening of thyroid hormone system disrupting chemicals. Integr. Environ. Assess Manag.19, 63. 10.1002/ieam.4632 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schiöth, H. B. & Fredriksson, R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gener. Comp. Endocrinol.142, 94. 10.1016/j.ygcen.2004.12.018 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Krishnan, A., Almén, M. S., Fredriksson, R. & Schiöth, H. B. The origin of GPCRs: Identification of mammalian like rhodopsin, adhesion, glutamate and frizzled GPCRs in fungi. PLoS ONE7, e29817. 10.1371/journal.pone.0029817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swart, E. M. et al. Temporal expression profile of an accessory-gland protein that is transferred via the seminal fluid of the simultaneous hermaphrodite Lymnaea stagnalis. J. Molluscan Stud.85, 177. 10.1093/mollus/eyz005 (2019). [Google Scholar]
- 79.Kim, D.-H., Park, J. C. & Lee, J.-S. G protein-coupled receptors (GPCRs) in rotifers and cladocerans: Potential applications in ecotoxicology, ecophysiology, comparative endocrinology, and pharmacology. Comp. Biochem. Physiol. C Toxicol. Pharmacol.256, 109297. 10.1016/j.cbpc.2022.109297 (2022). [DOI] [PubMed] [Google Scholar]
- 80.Holtz, K. M., Rice, A. M. & Sartorelli, A. C. Lithium chloride inactivates the 20S proteasome from WEHI-3B D+ leukemia cells. Biochem. Biophys. Res. Commun.303, 1058. 10.1016/s0006-291x(03)00473-x (2003). [DOI] [PubMed] [Google Scholar]
- 81.Raven, C. P. Lithium as a tool in the analysis of morphogenesis in Limnaea stagnalis. Experientia8, 252. 10.1007/BF02301379 (1952). [DOI] [PubMed] [Google Scholar]
- 82.Heo, J. et al. Prediction of the 3D structure of FMRF-amide neuropeptides bound to the mouse MrgC11 GPCR and experimental validation. Chembiochem8, 1527. 10.1002/cbic.200700188 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Yun, S. et al. Prevertebrate local gene duplication facilitated expansion of the neuropeptide GPCR superfamily. Mol. Biol. Evol.32, 2803. 10.1093/molbev/msv179 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Kriegsfeld, L. J. Driving reproduction: RFamide peptides behind the wheel. Horm. Behav.50, 655. 10.1016/j.yhbeh.2006.06.004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elphick, M. R. & Mirabeau, O. The evolution and variety of RFamide-type neuropeptides: Insights from deuterostomian invertebrates. Front. Endocrinol. (Lausanne)5, 93. 10.3389/fendo.2014.00093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Price, D. A. & Greenberg, M. J. Structure of a molluscan cardioexcitatory neuropeptide. Science197, 670. 10.1126/science.877582 (1977). [DOI] [PubMed] [Google Scholar]
- 87.Ubuka, T. & Tsutsui, K. Comparative and evolutionary aspects of gonadotropin-inhibitory hormone and FMRFamide-like peptide systems. Front. Neurosci.10.3389/fnins.2018.00747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koene, J. M., Jansen, R. F., Ter Maat, A. & Chase, R. A conserved location for the central nervous system control of mating behaviour in gastropod molluscs: Evidence from a terrestrial snail. J. Exp. Biol.203, 1071. 10.1242/jeb.203.6.1071 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Rőszer, T. & Kiss-Tóth, É. D. FMRF-amide is a glucose-lowering hormone in the snail Helix aspersa. Cell Tissue Res.358, 371. 10.1007/s00441-014-1966-x (2014). [DOI] [PubMed] [Google Scholar]
- 90.Henry, J., Zatylny, C. & Boucaud-Camou, E. Peptidergic control of egg-laying in the cephalopod Sepia officinalis: Involvement of FMRFamide and FMRFamide-related peptides. Peptides20, 1061. 10.1016/s0196-9781(99)00102-3 (1999). [DOI] [PubMed] [Google Scholar]
- 91.Di Cristo, C., Paolucci, M., Iglesias, J., Sanchez, J. & Di Cosmo, A. Presence of two neuropeptides in the fusiform ganglion and reproductive ducts of Octopus vulgaris: FMRFamide and gonadotropin-releasing hormone (GnRH). J. Exp. Zool.292, 267. 10.1002/jez.90000 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Di Cristo, C. Nervous control of reproduction in Octopus vulgaris: A new model. Invertebr. Neurosci.13, 27. 10.1007/s10158-013-0149-x (2013). [DOI] [PubMed] [Google Scholar]
- 93.Acker, M. J. et al. An immunohistochemical analysis of peptidergic neurons apparently associated with reproduction and growth in Biomphalaria alexandrina. Gener. Comp. Endocrinol.280, 1. 10.1016/j.ygcen.2019.03.017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rolón-Martínez, S. et al. FMRF-NH2 -related neuropeptides in Biomphalaria spp., intermediate hosts for schistosomiasis: Precursor organization and immunohistochemical localization. J. Comp. Neurol.529, 3336. 10.1002/cne.25195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zatylny-Gaudin, C. et al. Characterization of a novel LFRFamide neuropeptide in the cephalopod Sepia officinalis. Peptides31, 207. 10.1016/j.peptides.2009.11.021 (2010). [DOI] [PubMed] [Google Scholar]
- 96.Hallinan, N. M. & Lindberg, D. R. Comparative analysis of chromosome counts infers three paleopolyploidies in the mollusca. Genome Biol. Evol.3, 1150. 10.1093/gbe/evr087 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farhat, S., Modica, M. V. & Puillandre, N. Whole genome duplication and gene evolution in the hyperdiverse venomous gastropods. Mol. Biol. Evol.40, 171. 10.1093/molbev/msad171 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu, C. et al. Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic-terrestrial transition. Mol. Ecol. Resour.21, 478. 10.1111/1755-0998.13261 (2021). [DOI] [PubMed] [Google Scholar]
- 99.Rondón, J. J. et al. Comparative genomic analysis of chemosensory-related gene families in gastropods. Mol. Phylogenet. Evol.192, 107986. 10.1016/j.ympev.2023.107986 (2023). [DOI] [PubMed] [Google Scholar]
- 100.Li, Y. et al. Identification, characterization, and expression analysis of a FMRFamide-like peptide gene in the common Chinese Cuttlefish (Sepiella japonica). Molecules23, 742. 10.3390/molecules23040742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sukhan, Z. P., Cho, Y., Hossen, S., Lee, W. K. & Kho, K. H. Identification and characterization of Hdh-FMRF2 gene in Pacific abalone and its possible role in reproduction and larva development. Biomolecules13, 109. 10.3390/biom13010109 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Di Cristo, C. & Koene, J. M. The Oxford handbook of invertebrate neurobiology. In Mechanisms and Evolution 615–662 (Oxford University Press, 2017). [Google Scholar]
- 103.Schot, L. P. C. & Boer, H. H. Immunocytochemical demonstration of peptidergic cells in the pond snail Lymnaea stagnalis with an antiserum to the molluscan cardioactive tetrapeptide FMRF-amide. Cell Tissue Res.10.1007/bf00214687 (1982). [DOI] [PubMed] [Google Scholar]
- 104.Bright, K. et al. Mutually exclusive expression of alternatively spliced FMRFamide transcripts in identified neuronal systems of the snail Lymnaea. J. Neurosci.13, 2719. 10.1523/JNEUROSCI.13-06-02719.1993 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Santama, N. et al. Processing of the FMRFamide precursor protein in the snail Lymnaea stagnalis: Characterization and neuronal localization of a novel peptide, ‘SEEPLY’. Eur. J. Neurosci.5, 1003. 10.1111/j.1460-9568.1993.tb00952.x (1993). [DOI] [PubMed] [Google Scholar]
- 106.van Golen, F. A., Li, K. W., de Lange, R. P., Jespersen, S. & Geraerts, W. P. Mutually exclusive neuronal expression of peptides encoded by the FMRFa gene underlies a differential control of copulation in Lymnaea. J. Biol. Chem.270, 28487. 10.1074/jbc.270.47.28487 (1995). [DOI] [PubMed] [Google Scholar]
- 107.Buckett, K. J., Peters, M., Dockray, G. J., Van Minnen, J. & Benjamin, P. R. Regulation of heartbeat in Lymnaea by motoneurons containing FMRFamide-like peptides. J. Neurophysiol.63, 1426. 10.1152/jn.1990.63.6.1426 (1990). [DOI] [PubMed] [Google Scholar]
- 108.El Filali, Z. et al. Single-cell analysis of peptide expression and electrophysiology of right parietal neurons involved in male copulation behavior of a simultaneous hermaphrodite. Invertebr. Neurosci.10.1007/s10158-015-0184-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brussaard, A. B., Kits, K. S., Ter Maat, A., Van Minnen, J. & Moed, P. J. Dual inhibitory action of FMRFamide on neurosecretory cells controlling egg laying behavior in the pond snail. Brain Res.447, 35. 10.1016/0006-8993(88)90963-8 (1988). [DOI] [PubMed] [Google Scholar]
- 110.Brussaard, A. B., Schluter, N. C. M., Ebberink, R. H. M., Kits, K. S. & Maat, A. T. Discharge induction in molluscan peptidergic cells requires a specific set of autoexcitatory neuropeptides. Neuroscience39, 479. 10.1016/0306-4522(90)90284-b (1990). [DOI] [PubMed] [Google Scholar]
- 111.Croll, R. P., Van Minnen, J., Kits, K. S. & Smit, A. B. APGWamide: Molecular, histological and physiological examination of the novel neuropeptide involved with reproduction in the snail, Lymnaea stagnalis. In Molluscan neurobiology (eds Kits, K. S. et al.) 248–254 (North-Holland, Amsterdam, 1991). [Google Scholar]
- 112.Tensen, C. P. et al. The Lymnaea cardioexcitatory peptide (LyCEP) receptor: A G-protein-coupled receptor for a novel member of the RFamide neuropeptide family. J. Neurosci.18, 9812. 10.1523/JNEUROSCI.18-23-09812.1998 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smelik, W. F. E. The Natural Firing Pattern of Peptidergic Neurones in the Pondsnail Lymnaea stagnalis 93 (Vrije Universiteit Amsterdam, Amsterdam, 1995). [Google Scholar]
- 114.Alberti, A. et al. Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci. Data4, 170093. 10.1038/sdata.2017.93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kopylova, E., Noé, L. & Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics28, 3211. 10.1093/bioinformatics/bts611 (2012). [DOI] [PubMed] [Google Scholar]
- 116.Schulz, M. H., Zerbino, D. R., Vingron, M. & Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics28, 1086. 10.1093/bioinformatics/bts094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marchler-Bauer, A. & Bryant, S. H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res.32, W327. 10.1093/nar/gkh454 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Madoui, M. A. et al. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genom.16, 327. 10.1186/s12864-015-1519-z (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Crusoe, M. R. et al. The khmer software package: Enabling efficient nucleotide sequence analysis [version 1; peer review: 2 approved, 1 approved with reservations]. F1000esearch10.12688/f1000research.6924.1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics27, 764. 10.1093/bioinformatics/btr011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol.19, 455. 10.1089/cmb.2012.0021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics27, 578. 10.1093/bioinformatics/btq683 (2011). [DOI] [PubMed] [Google Scholar]
- 123.Boetzer, M. & Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol.13, R56. 10.1186/gb-2012-13-6-r56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quesneville, H., Nouaud, D. & Anxolabéhère, D. Detection of new transposable element families in Drosophila melanogaster and Anopheles gambiae genomes. J. Mol. Evol.57(1), 50. 10.1007/s00239-003-0007-2 (2003). [DOI] [PubMed] [Google Scholar]
- 125.Jamilloux, V., Daron, J., Choulet, F. & Quesneville, H. D. Novo annotation of transposable elements: Tackling the fat genome issue. Proc. IEEE10.1109/jproc.2016.2590833 (2016). [Google Scholar]
- 126.Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet8, 973. 10.1038/nrg2165 (2007). [DOI] [PubMed] [Google Scholar]
- 127.Mott, R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Bioinformatics13, 477. 10.1093/bioinformatics/13.4.477 (1997). [DOI] [PubMed] [Google Scholar]
- 128.Korf, I. Gene finding in novel genomes. BMC Bioinform.5, 59. 10.1186/1471-2105-5-59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform.10, 421. 10.1186/1471-2105-10-421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res.14, 988. 10.1101/gr.1865504 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dubarry, M. et al. Gmove a tool for eukaryotic gene predictions using various evidences. In Next generation sequencing conference (NGS) 2016. 10.7490/f1000research.1111735.1 (2016).
- 132.Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210. 10.1093/bioinformatics/btv351 (2015). [DOI] [PubMed] [Google Scholar]
- 133.Cao, Z. et al. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun4, 2602. 10.1038/ncomms3602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Faddeeva-Vakhrusheva, A. et al. Coping with living in the soil: The genome of the parthenogenetic springtail Folsomia candida. BMC Genom18, 493. 10.1186/s12864-017-3852-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Donath, A. et al. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res47, 10543. 10.1093/nar/gkz833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ghiselli, F. et al. Molluscan mitochondrial genomes break the rules. Philos Trans R Soc Lond B Biol Sci376, 20200159. 10.1098/rstb.2020.0159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jackson, D. J. Mantle modularity underlies the plasticity of the molluscan shell: Supporting data from Cepaea nemoralis. Front. Genet.10.3389/fgene.2021.622400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pazaitis, N. & Kaiser, A. TMA-Mate: An open-source modular toolkit for constructing tissue microarrays of arbitrary layouts. HardwareX14, e00419. 10.1016/j.ohx.2023.e00419 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.van Iersel, S., Swart, E. M., Nakadera, Y., van Straalen, N. M. & Koene, J. M. Effect of male accessory gland products on egg laying in gastropod molluscs. J. Vis. Exp.10.3791/51698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Álvarez, B. & Koene, J. M. Cognition and its shaping effect on sexual conflict: Integrating biology and psychology. Front. Integr. Neurosci.10.3389/fnint.2022.826304 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Vienne, D. M. Lifemap: Exploring the entire tree of life. PLoS Biol.14, e2001624. 10.1371/journal.pbio.2001624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina and nanopore sequencing data, as well as the genome assembly and gene predictions, are available in the European Nucleotide Archive (ENA) under the project PRJEB67819.