Abstract
Prostate cancer (PCa) is the most common malignancy diagnosed in men. The purpose of this study was to report the molecular pathways of Homo sapiens solute carrier family 4 member 4 (SLC4A4) in the progression of PCa. Here, we report our findings from clinical specimens of prostatic acinar adenocarcinoma collected from patients. We found that low-grade prostate cancers have higher SLC4A4 expression. We investigated the role of SLC4A4 and the signaling mechanism underlying its role in modulating the PCa progression. Firstly, we reported the SLC4A4/GSK-3β/β-catenin signaling axis, which regulates the clonogenic potential, invasiveness, and metastasis. In this, we found reduced phosphorylation of GSK at serine 21 of α and serine 9 of the β subunit in shSLC4A4 cells of PCa, which ultimately relieved the activity of GSK-3β. This activated GSK-3β phosphorylates β-catenin at Ser33/37 with a subsequently reduced β-catenin level in PCa cells. Our functional analysis revealed that SLC4A4 knockdown retards tumor growth and lowers invasion and migration potential. Secondly, we investigated the SLC4A4/RB axis, which acts to drive cell proliferation. SLC4A4 knockdown decreases the interaction between these molecules with hypophosphorylation of RB protein and cell cycle arrest. Likewise, transcriptome sequencing using the SLC4A4 knockdown in DU145 cells regulates differentiated expressed genes and multiple metabolic pathways. Our results suggest that SLC4A4 may serve as a potential therapeutic target for prostate cancer patients in the future.
Subject terms: Biochemistry, Biological techniques, Cancer, Cell biology, Molecular biology, Diseases, Medical research
Introduction
Prostate cancer (PCa) is the fifth-leading cause of death among men worldwide in 2020. The International Agency for Research on Cancer (IARC) released an updated Globocan 2020 with new estimates on the global cancer burden, indicating almost 1.4 million new cases and 375,000 deaths (3.8% of all male deaths caused by cancer) related to PCa1 and accounts for nearly 26% of all cancer cases2. Death rates for PCa are down 50% from their peak due to improvements in early detection, timely diagnosis and optimal treatment3.
Homo sapiens solute carrier family 4 member 4 (SLC4A4) is one of the differentially expressed genes encoding the electrogenic Na+/HCO3− cotransporter 1 (NBCe1) that moves HCO3- into cells and compensates intracellular H+ 4. SLC4A4 transporter is of particular interest in cancer research because it regulates the invasion, migration, and proliferation of cancer cells, and its expression is higher in cholangiocarcinomas, pheochromocytomas and paragangliomas than its normal tissue counterpart5. In PCa, the acidic microenvironment is an important prognostic factor used as a robust imaging biomarker for aggressive cancer6. Accumulative evidence shows that SLC4A4 is one of the amplified genes found in patients with prostate cancer7 and is related to prostate cancer progression8.
Retinoblastoma-associated protein (RB1) is well-known across a wide spectrum of human malignancies, and mechanisms responsible for suppressing tumorigenesis have been reported9. It inhibits pro-oncogenic E2F1-mediated transcription in its hypophosphorylated state10. Defects at the RBI locus leading to the loss of functional RB protein result in a constitutive release from the G1 checkpoint11. WNT signaling is an important contributor to PCa-related processes, and protein–protein interactions in this signaling pathway led to subsequent activation of downstream cascades in PCa12–14, which enabled activation of WNT-β-catenin cascades, thereby stimulating PCa metastasis15.
The serum prostate-specific antigen (PSA) quantification has been relevant in the screening and clinical treatment of men with PCa16, however, PSA screening may have led to overdiagnosis and overtreatment of clinically insignificant cancers17. For this reason, PSA screening is still controversial, as it has limited specificity in detecting prostate cancer in asymptomatic patients18.
To this end, we obtained gene expression data from the TCGA, investigated SLC4A4 expression levels, and analyzed the correlation between the SLC4A4 levels and patient clinical staging prognosis. We reported the SLC4A4/GSK-3β/β-catenin signaling axis, which regulates the clonogenic potential, invasiveness and metastasis in PCa cells. In this, we found reduced phosphorylation of GSK at serine 21 of α and serine 9 of the β subunit in shSLC4A4 cells of PCa, which ultimately relieved the activity of GSK-3β. This activated GSK-3β phosphorylates β-catenin at Ser33/37, decreasing the β-catenin level in PCa cells. This resulted in less cell invasion and migration. We also reported the SLC4A4/RB signaling axis in PCa cells, which acts to drive cell proliferation. SLC4A4 knockdown inhibits PCa cell growth and viability. shSLC4A4- induced loss of Phospho-S608RB with cell cycle arrest was associated with loss of total RB-sequestered SLC4A4 in these cells. These findings suggested SLC4A4 as a potential therapeutic target for PCa patients and provided new insights for understanding the important role of SLC4A4 in PCa.
Materials and methods
Data process and SLC4A4 gene expression
The UALCAN data analysis portal, available at http://ualcan.path.uab.edu, was used to investigate the differential gene expression of the SLC4A4 gene in all TCGA tumors, as well as normal prostate and prostate adenocarcinoma19. Expression of SLC4A4 in prostate adenocarcinoma (PRAD) based on patient's Gleason score and its effect on patient survival were also evaluated by using the same portal.
Patient specimens, tissue microarrays and immunohistochemical staining
All the prostate specimens were collected by the Department of Surgery at Queen Mary Hospital, The University of Hong Kong, with ethics approval from the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (Ref. #: UW 20-574). All experiments were performed in accordance with Institutional guidelines and regulations. Specimens of prostatic acinar adenocarcinoma and adjacent paired normal tissue were obtained from 100 patients who underwent Robotic-assisted laparoscopic radical prostatectomy at Queen Mary Hospital, The University of Hong Kong. Archival dates of the specimen fell between 2005 and 2011. Clinicopathological features of the patients were presented in Table 1. All patients were confirmed by clinical and pathological diagnosis. Patient tissue samples with prior hormonal or radiation therapy treatment were excluded. Histological diagnoses were reviewed and graded by pathologist AHN Tang using the 2014 version of the Gleason grading system20 and the WHO/ISUP Grade group21. Four tissue microarrays (TMA) were constructed from the 100 specimens. The pathologist marked the representative areas (carcinoma and normal tissue) of each specimen. Carcinoma components were sampled in triplicates to account for tumour heterogeneity. Immunohistochemistry (IHC) was performed following the product manual and protocol. Paraffin sections (4 µm) from 4 TMA were stained with rabbit polyclonal antibody against SLC4A4 (Atlas Antibodies, Sweden, catalogue #HPA035628) at a 1:400 dilution. EnVision + system-HRP (Dako, CA, USA) was used for visualization according to the manufacturer’s protocol. Staining was revealed by counter-staining with haematoxylin. The staining intensity of SLC4A4 was scored on a 0–3 scale (0: Negative, 1: weak, 2: moderate, 3: strong) by a pathologist.The slides were digitalized by the digital slide scanner (NanoZoomer S210, Hamamatsu, Japan), and images were captured using the accompanied viewer NDP. View 2.9.29 software (Version 14.01.2022, Hamamatsu Photonics K.K., Japan).
Table 1.
Clinicopathological parameters in a cohort of prostate cancer patients.
| Clinical parameters | No. of patients | p value |
|---|---|---|
| All patients | 100 | |
| Age (years) | 0.0067 | |
| < 65 | 38 | |
| ≥ 65 | 62 | |
| Pre-operative PSA level (ng/mL) | 0.0269 | |
| Low (PSA < 10 ng/mL) | 58 | |
| High (PSA ≥ 10 ng/mL) | 42 | |
| Biochemical recurrence (PSA > 0.2 ng/mL) | 0.0001 | |
| PSA > 0.2 ng/mL | 28 | |
| PSA < 0.2 ng/mL | 72 | |
| Metastasis | 0.0041 | |
| M0 | 93 | |
| M1 | 7 | |
| Gleason grade | 0.0003 | |
| 6 | 30 | |
| 7 | 45 | |
| 8 | 10 | |
| 9 | 15 | |
| Stage | 0.0067 | |
| I | 30 | |
| II | 33 | |
| III | 12 | |
| IV | 10 | |
| V | 15 | |
Cell culture and reagents
Androgen-dependent 22RV1, LNCAP, VCAP, C4-2 and androgen-independent DU145and PC3 human tumor prostate cell lines were purchased from the American Type Culture Collection (ATCC). 22RV1, LNCAP, VCAP and C4-2 were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (Gibco, Brazil, Catalog # 10500064) and 1% antibiotic–antimycotic (Gibco USA, Catalog # 15240062) in a 5% CO2 humidified atmosphere at 37 °C. PC3 and DU145 cells were maintained in F12K (Gibco USA, Catalog # 21127022) and EMEM (Gibco USA, Catalog # 11095080) respectively supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Brazil, Catalog # 10500064) and 1% antibiotic–antimycotic (Gibco USA, Catalog # 15240062) in a 5% CO2 humidified atmosphere at 37 °C. RWPE-1 normal prostate epithelial cells were also purchased from ATCC and maintained in Keratinocyte Serum Free Medium (Gibco USA, Catalog # 17005042) supplemented with bovine pituitary extract (BPE) and human recombinant epidermal growth factor (EGF). Human embryonic kidney cells, HEK-293T/17, were grown in Dulbecco's Modified Eagle Medium (Gibco USA, Catalog # 10569010) supplemented with 10% fetal bovine serum (Gibco, Brazil, Catalog # 10500064) and 1% antibiotic–antimycotic (Gibco USA, Catalog # 15240062) in a 5% CO2 humidified atmosphere at 37 °C. Cell growth and viability were monitored with a hemocytometer and 0.4% trypan blue (Gibco USA, Catalog # 15250061) exclusion assay. PCa cells were subcultured as per the recommendation of ATCC. All cells were regularly screened and were free from mycoplasma infection by the Centre for PanorOmic Sciences (CPOS), LKS Faculty of Medicine, The University of Hong Kong.
Plasmids construction, cell transfection and Generation of stable transfectants
pLKO.1 TRC cloning vector, a replication-incompetent lentiviral vector, was used to express the SLC4A4-shRNA. Primer sequences with high mean knockdown level were chosen for use in the pLKO.1 puro (Addgene # 10878) and obtained from Integrated DNA Technologies (Coralville, IA). The sequences were;
(F1-5′-CCGGGCCTATTGAAACCTGAACTTACTCGAGTAAGTTCAGGTTTCAATAGGCTTTTTG-3′ & R1-5′-AATTCAAAAA-GCCTATTGAAACCTGAACTTACTCGAGTAAGTTCAGGTTTCAATAGGC-3′) and (F2-5′-CCGGCCCAGAGTATTTGCCAACTATCTCGAGATAGTTGGCAAATACTCTGGGTTTTTG-3′ & R2-5′- AATTCAAAAA- CCCAGAGTATTTGCCAACTATCTCGAGATAGTTGGCAAATACTCTGGG-3′)
These sequences were cloned into pLKO.1 puro following the depositor's protocol. As a control, a pLKO.1 vector encoding a non-targeting scrambled shRNA and puromycin resistance as a marker gene was used (non-targeting-puro shRNA, Catalog # SHC002, Sigma Aldrich, USA). To generate lentiviral particles, a combination of 2.5 μg pMD2.g (Addgene # 12259), 7.5 μg psPAX2 (Addgene # 12260), and 10 μg of SLC4A4-shRNA plasmids was utilized22. HEK-293T/17 cells were co-transfected with the aforementioned plasmids when they reached approximately 50–60% confluence in 10 cm cell culture dishes. The transfection was carried out using DMEM and 10% FBS, along with lipofectamine-2000 transfection reagent (Invitrogen, CA, USA, Catalog #11668019), following the instructions provided by the manufacturer. After 48 h, the medium containing viral particles was collected, and an additional 5 mL of medium was added to the dish for another 24 h before the final collection. The collected lentiviral medium was stored at − 80 °C until it was needed. Transduction of DU145, PC3 and 22RV1 cells with the lentiviral particles was performed in a 6-well plate. 600 μL viral particles were added to 5 × 104 cells. After 72 h, transduced cells were put into the 6 well plates and cultured in respective tissue culture media with 10% heat-inactivated FBS and selected for three weeks in 2 μg /mL puromycin.
Quantitative real‑time PCR (qRT‑PCR)
The extraction of total RNA was performed using the PureLink RNA Mini Kit (Life Technologies, CA, USA, Catalog # 12183025) according to the provided instructions. To synthesize complementary DNA (cDNA), 1 μg of the total RNA was subjected to reverse transcription using the PrimeScript RT Master Mix (Takara Bio Inc, CA, USA, Catalog # RR047A).The mRNA expression of the SLC4A4 target gene was examined using the PowerUp™ SYBR Green Master Mix Kit (Applied Biosystems, Vilnius, Lithuania, Catalog # A25742) by Real-Time PCR (qPCR) using the StepOnePlus™ Real-Time PCR System (Applied Biosystems, CA, USA). The following thermal cycling conditions were applied: initial pre-denaturation at 95 °C for 2 min, followed by denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s over a total of 40 cycles, and extension at 72 °C for 1 min. For normalization, GAPDH was used as an internal reference. The relative expression levels of the genes were calculated using the 2−ΔΔCt method23. All primers were synthesized by Integrated DNA Technologies (Coralville, IA) (Table 2).
Table 2.
Primer sequences for reverse transcription quantitative polymerase chain reaction.
| Gene | Primer Sequences (5′–3′) |
|---|---|
| SLC4A4 |
F: TGATGAATCCAGCAGCAGCA R: CACTCCATCTCCTGCCCATC |
| β-Catenin |
F: ATGATGGTCTGCCAAGTGGG R: TCCTGGCCATATCCACCAGA |
| SMIM22 |
F: TTCCTCACCTTCATGGGCAC R: CCACTCCCTTGGGTCTTTCC |
| ALDH3A1 |
F: AAGAGGAGATCTTCGGGCCT R: GGCAGAGAGTGCAAGGTGAT |
| SPINK4 |
F: TTGGCTGCCCTCCTTGTTG R: AGGTTGGACATCTGGGAACA |
| AQP5 |
F: CCCGCTCACTGGGTTTTCTG R: CCTCGTCAGGCTCATACGTG |
| GAPDH |
F: GGAGCGAGATCCCTCCAAAAT R: GGCTGTTGTCATACTTCTCATGG |
Western blot analysis
The PCa cells were washed, collected as pellets, and then subjected to extraction of total soluble protein using M-PER lysis buffer (Thermo Scientific, IL, USA, Catalog # 78503) supplemented with 1/100 volume of protease inhibitors (Sigma, MO, USA, Catalog # P8340) and phosphatase inhibitors (Sigma, MO, USA, Catalog # P5726). The protein concentration was determined using the Pierce BCA protein assay (Thermo Scientific, IL, Catalog # 23227) following the manufacturer's instructions. 20 μg of protein was resolved by SDS PAGE using a 10% polyacrylamide gel. The electrotransfer was done onto a PVDF membrane (Millipore, MA, Catalog # IPVH00010) at 400 mA. The membranes were blocked in dry nonfat milk before being incubated with the indicated primary antibody overnight at 4 °C. Images were captured on a GE Amersham Imager AI680. For loading control, α-tubulin was applied. Antibodies used in the study are anti‐ SLC4A4 (Proteintech, IL, USA Catalog # 11885-1-AP), anti- GSK3ß (BD Biosciences, CA, USA, Catalog # 610201), anti-Phospho-GSK-3α/β (Ser21/9) (Cell Signaling, MA, USA, Catalog # 8566), anti‐ RB (Cell Signaling, MA, USA, Catalog # 9309), anti‐ Phospho-S608-RB (Cell Signaling, MA, USA, Catalog # 2181), anti-beta-catenin (Cell Signaling, MA, USA, Catalog # 9582), anti- Phospho-S33/37-beta-catenin (Cell Signaling, MA, USA, Catalog # 2009), anti‐rabbit IgG HRP (Cell Signaling, MA, USA, Catalog # 7074), anti‐mouse IgG HRP (Cell Signaling, MA, USA, Catalog # 7076)and anti-α tubulin antibody (Santa Cruz Biotechnology, Inc. Texas, USA, Catalog # SC-8035).
Co-immunoprecipitation (Co-IP) assay
To perform the Co-IP assay, PCa cells (shCON and shSLC4A4) were collected and lysed using M-PER lysis buffer (Thermo Scientific, IL, USA, Catalog # 78503), supplemented with a 1/100 volume of protease inhibitors (Sigma, MO, USA, Catalog # P8340) and phosphatase inhibitors (Sigma, MO, USA, Catalog # P5726). For pre-clearing, 25 μg of PureProteome™ Protein G Magnetic Beads (Millipore, MA, USA, Catalog # LSKMAGG02) were added to 500 μg of total protein and rotated at 4 °C for 2 h. Subsequently, Protein G Magnetic Beads were added to the samples in each group along with 2 μg of anti-SLC4A4 antibody (Proteintech, IL, USA, Catalog # 11885-1-AP), and the mixture was rotated overnight at 4 °C. The bead/antibody/protein slurries were then washed and subjected to standard SDS-PAGE analysis.
Cell growth Assay
PCa cells (shCON and shSLC4A4) were plated in a 100 mm petri dish and subjected to serum starvation for 24 h, following the method described by Khammanit et al.24. After the serum starvation period, the cells were passaged and induced to re-enter the cell cycle by adding serum. Cell counts were taken at 24, 48, and 72 h using a hemocytometer and 0.4% trypan blue (Gibco USA, Catalog # 15250061) exclusion staining.
Colony formation assay
A total of 750 PCa cells were seeded in petri dishes containing a complete medium and cultured for a duration of 14 days. Subsequently, the cells were washed once with PBS and then fixed using 4% paraformaldehyde (Santa Cruz Biotechnology, Inc., Texas, USA, Catalog # SC-281692). The colonies formed were stained with 1% crystal violet (Sigma, MO, USA) and scored for analysis. The ChemiDoc Imaging System (Bio-Rad) was used to image-stained plates, and the resulting images were quantified using ImageJ software (NIH, Bethesda, MD, USA).
Cell migration and invasion assays
To perform the cell migration and invasion assays, an 8 µm pore size Millicell Hanging cell culture insert (Millipore, MA, USA, Catalog # MCEP24H48) was used. PCa cells (shCON and shSLC4A4) were pre-incubated in a serum-free medium overnight. Subsequently, 5 × 104 PCa cells were placed in the insert and allowed to migrate for 20 h. The outer chamber was filled with 600 µl of complete medium. After the incubation period, non-migrating cells on the upper surface of the insert were removed using a cotton swab. The migrated cells were fixed with methanol (Merck, Darmstadt, Catalog # 106009) and stained with 1% crystal violet (Sigma, MO, USA). In the invasion assay, 5 × 104 cells were seeded in the inserts coated with Matrigel matrix (Corning, USA, Catalog # 354234) and allowed to invade for 20 h. The invasive cells were stained, and random fields of the membrane inserts were captured under a microscope. Finally, the migrated or invaded cells were quantified.
Cell cycle analysis by flow cytometry
Cell cycle analysis was performed as previously described25. Briefly, PCa cells (shCON and shSLC4A4) were harvested and washed with ice-cold PBS. The cells were then collected through centrifugation and resuspended in 200 µl of a cold hypotonic staining solution containing propidium iodide (Sigma, MO, USA, Catalog # P4170), Triton X-100 (Sigma, MO, USA, Catalog # T8787), and sodium citrate (Sigma, MO, USA, Catalog # C3674). Following a 1-h incubation at room temperature, the cells were analyzed using flow cytometry (LSR Fortessa, BD) with 488-nm excitation. FlowJo 7.6 software (FlowJo LLC, USA) was used to analyze the relative ratios of the G1, S, and G2 phases.
Hierarchical clustering analysis
The intensities of protein bands detected in western blotting were measured densitometrically using ImageJ software (NIH, Bethesda, MD, USA). To analyze the expression of proteins that were upregulated or downregulated after SLC4A4 knockdown, hierarchical clustering and heatmap analysis were conducted using OriginPro 2021 software (OriginLab Corporation, Northampton, MA, USA). All normalization, clustering, and statistical analyses were performed using OriginPro 2021 software available at https://www.originlab.com. The densitometric data regarding the expression/activation of protein molecules in PCa cells following SLC4A4 knockdown were normalized to the untreated control (shCON).
Library preparation and RNA sequencing
Total RNA extraction was performed using the PureLink RNA Mini Kit (Life Technologies, CA, USA, Catalog # 12183025) according to the manufacturer's instructions. The concentration of RNA in each sample was determined using the RNA 6000 Nano Kit (Agilent Technologies, Inc, Catalog # 5067-1511). The RNA samples were analyzed on an Agilent 6000 Bioanalyzer (Agilent Technologies, Inc.) with Nano chips, following the provided guidelines. The integrity of the RNA in the samples was assessed using the Agilent RNA 6000 Nano Kit and the 2100 expert software, which calculated the RNA integrity number (RIN).
Library preparation and Illumina sequencing (NovaSeq 6000) were done at The University of Hong Kong, LKS Faculty of Medicine, Centre for PanorOmic Sciences (CPOS), Genomics Core. cDNA libraries were prepared by KAPA mRNA HyperPrep Kit (KapaBiosystems, Cape Town, South Africa, Catalog # KK8581) according to the manufacturer's protocol. In brief, Poly-A containing mRNA was collected using poly-T oligo-attached magnetic beads. The purified mRNA was fragmented to 200–300 bp by incubating at 94 °C for 6 min in the presence of magnesium ions. The fragmented mRNA was then applied as a template to synthesize the first-strand cDNA using random hexamer-primer and reverse transcriptase. In the second strand cDNA synthesis, the mRNA template was removed, and a replacement strand was generated to form the blunt-end double-stranded (ds) cDNA. The ds cDNA underwent 3’ adenylation and indexed adaptor ligation. The adaptor-ligated libraries were enriched by 10 cycles of polymerase chain reaction (PCR). The libraries were denatured and diluted to optimal concentration. Illumina NovaSeq 6000 was used for Pair-End 151 bp sequencing.
Data quality
Using software from Illumina (bcl2fastq), sequencing reads were assigned to individual samples, with each sample having an average throughput of 13.0 Gb and a total throughput of 78.1 Gb. Regarding sequence quality, an average of 93% of the bases achieved a quality score of Q30. In NovaSeq 6000 platform, 79.18 to 89.90 M clean reads per library were generated.
Statistical analysis
Experiments were biological replicates in triplicate, and the results were shown as the mean with standard deviation (SD). Fisher's exact test and chi-square tests were performed to compare the differences in clinicopathological parameters between groups of prostate cancer patients. qRT-PCR, functional and cell cycle assays were examined using the Student’s t test or the ANOVA test, whichever is applicable. P < 0.05 was considered statistically significant. Differentially expressed genes (DEGs) were analyzed by GraphPad Prism software version 9.4.0 (GraphPad Software, San Diego, CA, USA) (https://www.graphpad.com). DEGS subjected to KEGG pathway enrichment analysis26 were analyzed by ShinyGO version 0.76 software (http://bioinformatics.sdstate.edu/go). Hierarchical clustering analysis for protein expression/activation was performed by OriginPro 2021 software (OriginLab Corporation, Northampton, MA, USA) (https://www.originlab.com). Briefly, densitometric values for each western blot band were determined using Image J version 1.53 (NIH, Bethesda, MD, USA) (https://imagej.nih.gov). The values were normalized to the loading control for that lane and put in a worksheet of the OriginPro 2021b software version 9.85 (OriginLab Corporation, Northampton, MA, USA) (https://www.originlab.com). Under cluster columns and rows setting in "HeatmapDendrogram apps" of this software, we selected "Pearson correction" in the distance type and drawn a hierarchical clustering and heatmap analysis graph.
Results
SLC4A4 is upregulated in PCa tissue samples and cell lines
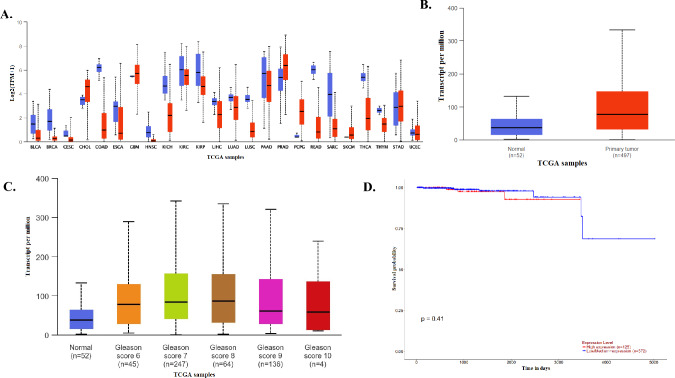
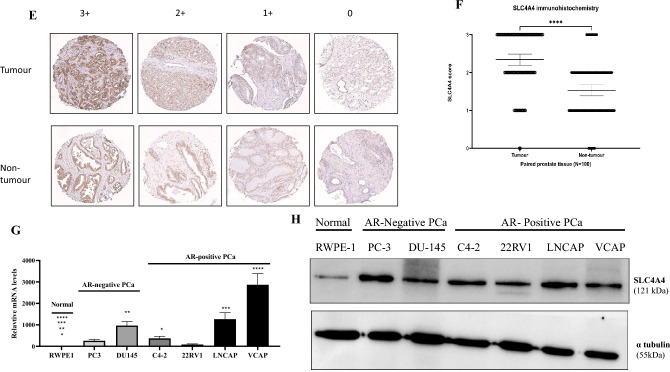
UALCAN database (http://ualcan.path.uab.edu) revealed the SLC4A4 mRNA expression in pan-cancer. Figure 1A shows SLC4A4 mRNA expressions in pan-cancer. It was highly expressed in cancers containing CHOL, PRAD, and PCPG (all P < 0.05) and poorly expressed in other cancers, including BLCA, BRCA, COAD, ESCA, HNSC, KICH, KIRP, LIHC, LUSC, READ, SARC, THCA. By applying UALCAN to analyze cancer OMICS data based on the TCGA study cohort, SLC4A4 was significantly overexpressed in human prostate cancer compared to normal tissue (p = 1.037E-09) (Fig. 1B). Analyses of a subset of prostatic adenocarcinomas based on grade showed higher SLC4A4 expression in Gleason score 7–9 tumors compared with those with Gleason score 6 and 10 (Fig. 1C). Based on TCGA data in GEPIA (http://gepia.cancer-pku.cn/), overall survival and disease-free survival analysis showed that expression level of SLC4A4 is associated with overall survival of patients with PRAD (Fig. 1D). PCa and normal prostate tissues were also collected from 100 patients, whose ages ranged between 48 and 75 years (mean age of 66.5 years). The median follow-up time was 119 months (ranging from 33 to 170 months). 58 patients (58%) had a preoperative prostate-specific antigen (PSA) less than 10 ng/mL (Table 1). Using a specific antibody, the expression of SLC4A4 in clinical PCa and normal prostate tissues was examined by IHC. As shown in Fig. 1E,F, the results of histopathological sections confirmed the localization of SLC4A4 and indicated that the expression of SLC4A4 in PCa tissues was distinctly higher than that in normal prostate tissues. SLC4A4 expression levels were determined using qPCR with SLC4A4-specific primers in human prostate cancer cell lines (PC3, DU145, C4-2, 22RV1, LNCAP, and VCAP) and normal prostate cell lines (RWPE-1). We found significantly higher mRNA levels of SLC4A4 in cancer cells than in normal prostate cells (Fig. 1G). Consistently, SLC4A4 was markedly upregulated in PCa cells compared to RWPE-1 normal prostate epithelial cells by immunoblotting (Fig. 1H and Fig. S1).
Fig. 1.
Upregulation of SLC4A4 in prostate cancer. (A, B) SLC4A4 transcript is highly expressed in several cancers, including prostate adenocarcinoma (PRAD), based on the TCGA study cohort. (C, D) A subset of PRAD showed higher SLC4A4 expression in Gleason score 7, 8, and 9 tumors and this was associated with the overall survival of patients. (E, F) Immunohistochemical (IHC) analysis of primary prostate tumor samples showed higher SLC4A4 protein expression compared to non-tumor tissues. (G, H) Compared to RWPE-1 normal prostate epithelial cells, the SLC4A4 mRNA level was higher in DU145 androgen receptor (AR)-negative prostate cancer (PCa) cells. In AR-positive PCa cells, SLC4A4 mRNA level was elevated in C4-2, followed by VCAP (*P < 0.05). SLC4A4 overexpression was also detected by Western blotting, with PC3 cells having higher SLC4A4 protein level in AR-negative PCa cells. In AR-positive PCa cells, both C4-2 and LNCAP showed higher SLC4A4 protein expression. The results are presented as means ± standard error of three independent experiments.
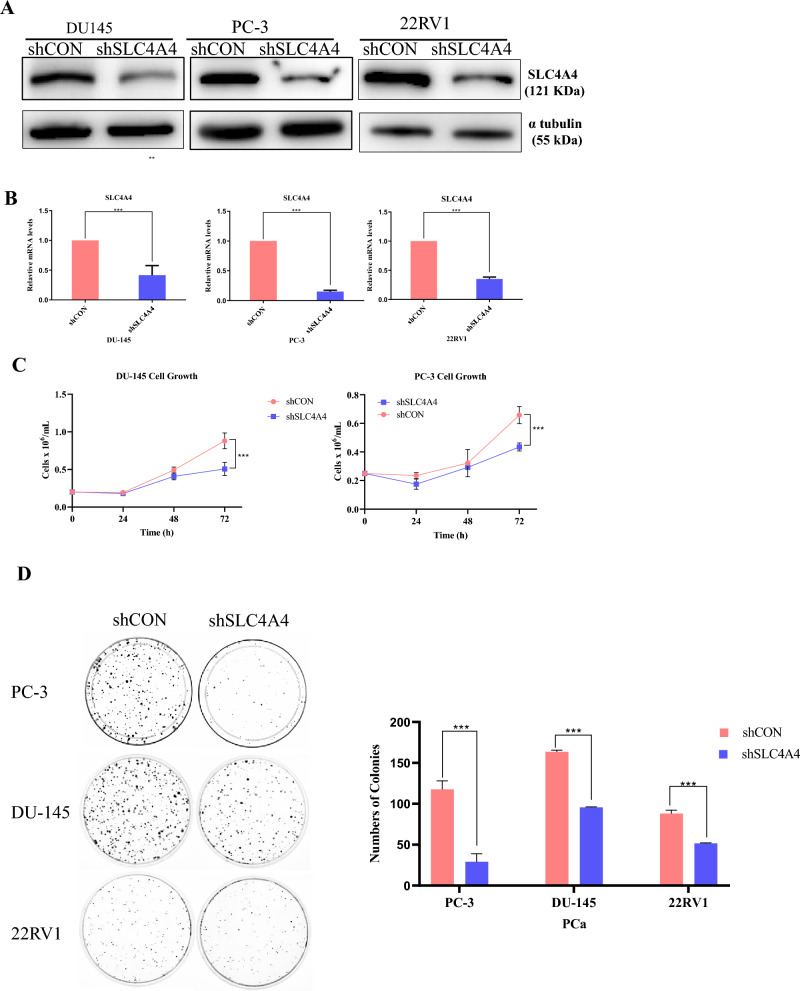
SLC4A4 knockdown reduces cell growth and clonogenic capacity in human PCa cells
Given that members of the SLC4A family mediate acid extraction in various cancer cells and are involved in cell growth and progression27,28, we studied the potential role of SLC4A4 in prostate cancer cells. We constructed a shSLC4A4-pLKO.1 plasmid and generated stably transfected prostate cancer cell lines expressing a shRNA targeted against SLC4A4. The knockdown efficiency of SLC4A4 was confirmed using western blotting (Fig. 2A and Fig. S2) and qPCR (Fig. 2B). Further, ShCON and shSLC4A4 stable transfectants were subjected to serum deprivation for 24 h and thereafter resupplied with 10% serum. The potential effects on cell growth were examined in PC-3 and DU-145 cells. The results indicated that the proliferation rates of the prostate cancer cells were significantly inhibited by SLC4A4 knockdown, compared with the control group (Fig. 2C). The aforementioned results revealed that SLC4A4 serves an important role in prostate cancer cell proliferation. To determine whether SLC4A4 is required for in vitro clonogenic growth of PCa cells, we obtained a similar result, that SLC4A4 knockdown significantly reduced the number of cell colonies (Fig. 2D). These results show that inhibiting SLC4A4 expression suppresses PCa cell growth in vitro.
Fig. 2.
SLC4A4 knockdown suppresses growth, viability and colony formation of prostate cancer (PCa) cells (A, B) Western blot analysis and quantitative reverse transcription polymerase chain reaction (qRT-PCR) confirmed the knockdown of SLC4A4 in PCa cells (C) Knockdown of SLC4A4 reduced the growth and viability of PCa cells compared to those transduced with control shRNA (shCON) (D) SLC4A4 knockdown reduced colony formation of PCa cells. Cells were previously transfected with control shRNA and SLC4A4 shRNA, seeded in 10 cm Petri dish, and allowed to grow for 2 weeks. The colonies were fixed with 4% paraformaldehyde, stained with crystal violet and imaged using a gel doc. The number of colonies in each sample was counted using Image J software. Graphic representation of the number of colonies showed a significant decrease in the SLC4A4 knockdown group compared to the control (**p < 0.05). The results are presented as means ± standard error of three independent experiments.
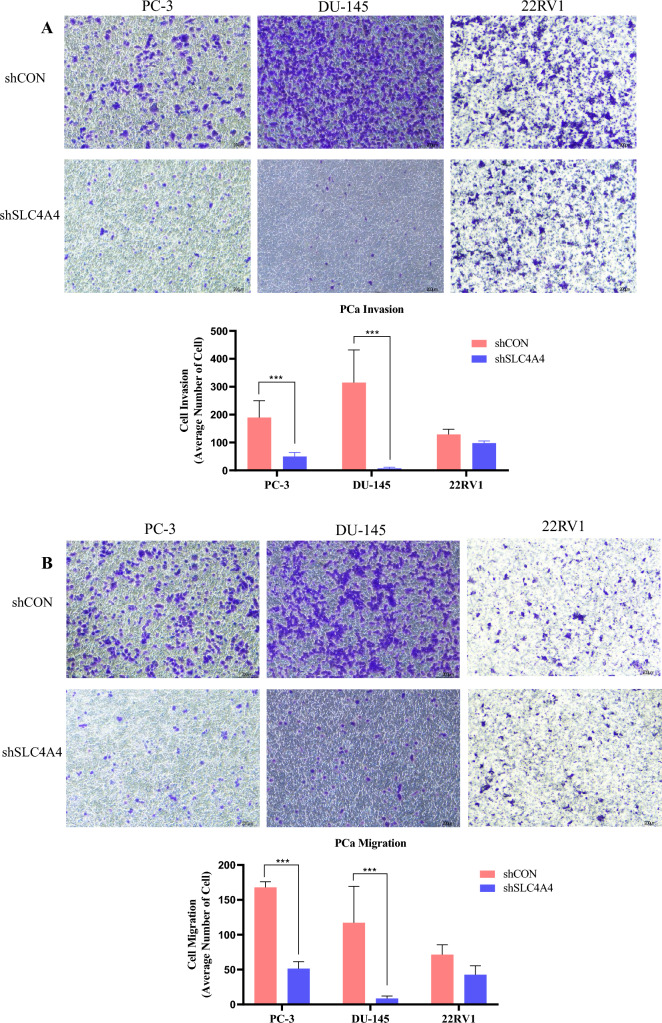
Knockdown of SLC4A4 inhibited PCa invasion and migration
To further confirm that SLC4A4 functions as a tumor promoter in PCa cells, we next conducted transwell invasion and migration assays using shSLC4A4 stable transfectants of PC-3, DU145, or 22RV1 cells. With the downregulation of SLC4A4, the invasive capability of PCa cells was notably reduced when they were assessed by transwell invasion assays (Fig. 3A). Additionally, the number of migrated cells was inhibited after SLC4A4 knockdown (Fig. 3B). Our data suggested SLC4A4 could be associated with oncogenesis in PCa cells, and that SLC4A4 knockdown restrained PCa cell invasion and migration.
Fig. 3.
Knockdown of SLC4A4 inhibited PCa invasion and migration. (A, B) PCa cells were transfected with control shRNA or SLC4A4 shRNA and subjected to matrigel invasion and migration assays. Cells were seeded into the upper chambers, and after 24 h, cells were examined under a light microscope at 20 × magnification. The results are shown as the mean number of invasive and migrated cells, which were obtained by counting cells in five randomly selected fields. The results are presented as means ± standard error of three independent experiments, with a significant decease observed in the SLC4A4 knockdown group compared to the control (**p < 0.05).
SLC4A4 regulates phosphorylation of β-catenin via SLC4A4/GSK-3β/β-Catenin axis in PCa cells
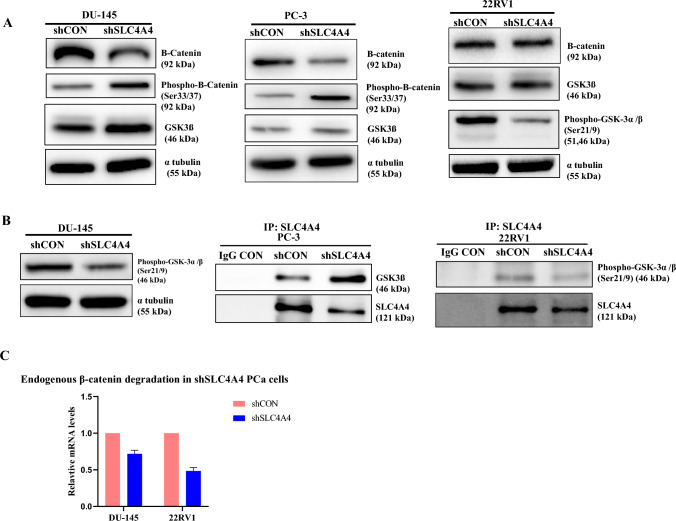
We further aimed to elucidate the potential molecular mechanism by which SLC4A4 regulates the invasion and migration of PCa cells. We conducted Western blot assays to assess the levels of β-catenin in control and SLC4A4-knockdown PCa cells. Our data demonstrated that silencing of SLC4A4 led to an obvious decline in β-catenin protein levels in PCa cells (Fig. 4A and S3A). Overall, these data proved that the activation of β-catenin signaling was suppressed by SLC4A4 knockdown in PCa cells.
Fig. 4.
SLC4A4 knockdown restores the activity of GSK-3β and enhances phosphorylation of β-catenin in Prostate Cancer cells (A) A significant decrease in the total amount of β-catenin was found in shSLC4A4 prostate cancer cells compared to shCON. Meanwhile, the level of GSK-3β was significantly increased due to its reduced phosphorylation on residues Ser21/9 which led to increase phosphorylation (S33/37) of β-catenin (B) Co-immunoprecipitation (Co-IP) experiments showed that endogenous SLC4A4 inhibited the expression of GSK-3β. However, when prostate cancer cells were transfected with the shSLC4A4 plasmid, there was hypophosphorylation of GSK-3β at Ser21/9, which activated the total GSK-3β to degrade endogenous β-catenin (C) qPCR data confirmed the degradation of endogenous β-catenin in shSLC4A4 prostate cancer cells.
Given that PCa shSLC4A4 cells have a decreased level of total B-catenin residue, we next hypothesized that SLC4A4 silencing increases the level of GSK-3β followed by hyperphosphorylation of β-catenin at Ser33/37. We performed western blot analysis using an anti- GSK-3β, phospho-GSK-3α/β (Ser 21/9) and phospho-β-Catenin (Ser33/37) antibodies. As shown in Fig. 4A and S3A, SLC4A4 knockdown reduces the phosphorylation of GSK-3β at serine 21 of the α and serine 9 of β- subunit in PCa cells, which ultimately relieved the activity of GSK-3β. This activated GSK-3β phosphorylates β-catenin at Ser33/37 with a subsequently reduced total β-catenin level in PCa cells (Fig. 4A). We performed a CO-IP experiment with an anti-SLC4A4 antibody in PCa cells to determine if SLC4A4 associates with GSK-3β and regulates its activity. The results showed that endogenous SLC4A4 inhibits the expression of GSK-3β, but as PCa cells were targeted by shSLC4A4 plasmid, the GSK-3β protein level started to increase. We also investigated that phospho-GSK-3α/β (Ser 21/9) interacts with SLC4A4 in PCa cells, but SLC4A4 KD stable cells diminished the phosphorylation of GSK-3α/β (Ser 21/9) and reduced its interaction with SLC4A4 protein (Fig. 4B, S3A and S3B). Our data strongly suggested that SLC4A4 knockdown might relieve the activity of GSK-3β by reducing its phosphorylation on residues Ser21/9, which then phosphorylated the β-catenin at Ser33/37 with the possibility of a rise in β-catenin degradation (Fig. 4C and S3B). Conclusively, the SLC4A4/GSK-3β/β-catenin signaling axis regulates the clonogenic potential, invasiveness, and metastasis of PCa cells.
SLC4A4 knockdown induces cell cycle arrest in PCa cells by increasing hypophosphorylation of retinoblastoma protein (RB)
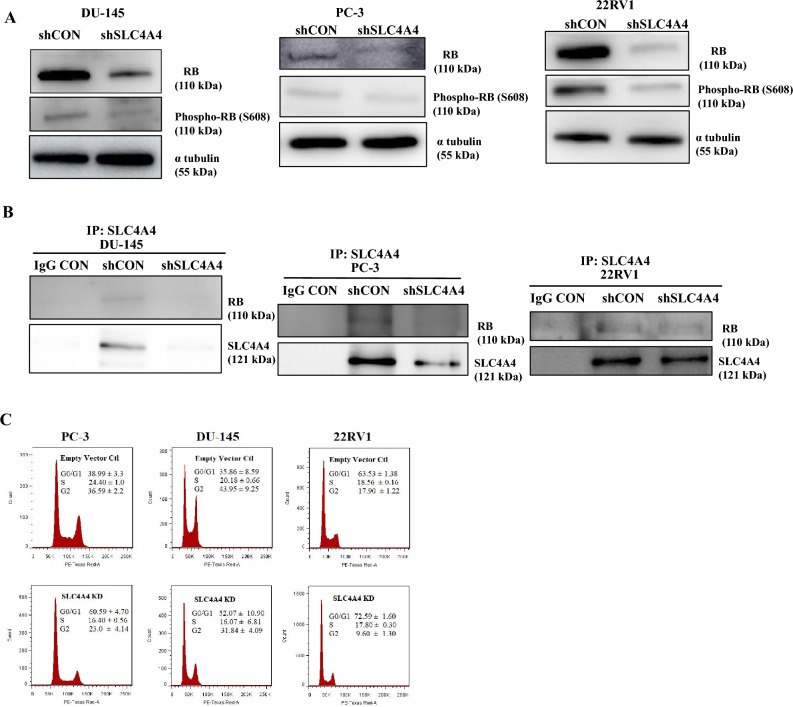
To validate whether the suppression of cell growth by SLC4A4 knockdown was related to cell cycle arrest, we performed western blot analysis using anti-RB and Phospho-Ser608(RB) antibodies. In Fig. 5A and S4A, the total RB and Phospho-Ser608 (RB) levels significantly decreased by shSLC4A4 in PCa cells compared to the shCON groups. Additionally, using co-immunoprecipitation, we found a novel interaction of SLC4A4 with RB. SLC4A4 knockdown diminished the interaction of RB and SLC4A4 in DU-145, PC-3, and 22RV1 cells (Fig. 5B and S4B) with hypophosphorylation of the RB protein.Next, to assess the effect of SLC4A4 on the cell cycle process, stable transfectants were subjected to serum deprivation for 24 h and thereafter resupplied with 10% serum. Cells were harvested and stained with PI solution for flow cytometric analysis. Figure 5C shows that G0/G1 cell cycle arrest was induced by SLC4A4 knockdown in all PCa compared to shCON groups. These findings indicate that SLC4A4 is highly associated with regulating prostate cancer proliferation. These findings demonstrate that the SLC4A4/RB signaling axis drives cell cycle progression in PCa cells.
Fig. 5.
SLC4A4 knockdown induces cell cycle arrest in PCa cells by increasing hypophosphorylation of retinoblastoma protein (RB1) (A) SLC4A4 knockdown decreased total RB expression and induced hypophosphorylation at serine 608 (S608) of RB. (B) Co-immunoprecipitation (Co-IP) results showed that SLC4A4 KD stable cells had diminished total RB interaction with the SLC4A4 protein (C) Cell cycle distribution analysis showed the percentage of PCa cells in G0/G1, S and G2/M. SLC4A4 knockdown induced G0/G1 cell cycle arrest in PCa cells. The results are presented as means ± standard error of three independent experiments.
Hierarchical clustering based on protein expression/activation in PCa cells
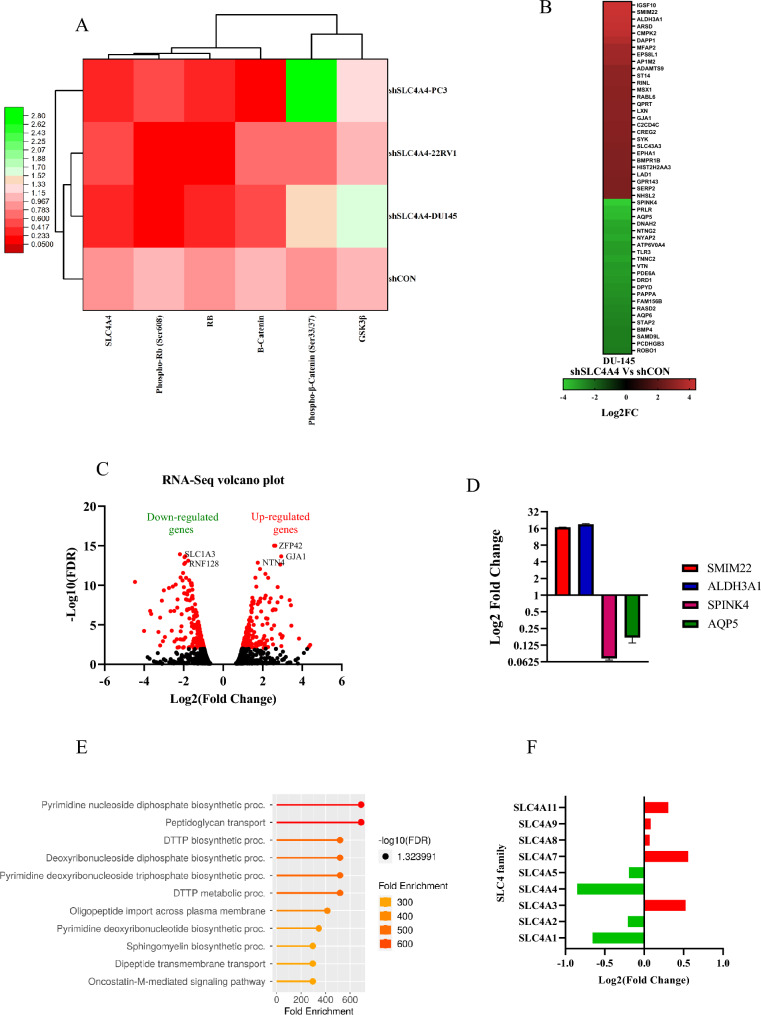
Hierarchical clustering analysis for known cell cycle and WNT/B-catenin signaling molecules found in PCa cell lysates was performed to identify coupling relationships that betray regulatory pathways driving cell proliferation. The response of signaling molecules is divided into two main clusters, one of which contains GSK-3β and Phospho-S33/37-β-catenin. The other includes four subclusters of signaling molecules (Fig. 6A). Interestingly, β-catenin is a driver for three signaling subclusters, consistent with the previous published work which suggests that the knockdown of β-catenin destabilized RB in colon cancer cells. This reduction in RB phosphorylation results from G1 arrest29. Two subclusters have the Phospho-S33/37- β -catenin and GSK-3β entity. There is a central role for GSK-3β in WNT/ β-catenin signaling. GSK-3β phosphorylated β-catenin at S33/37, causing it to destabilize and degrade, resulting in a very low level of β-catenin in the cytosol/nucleus30.
Fig. 6.
SLC4A4 knockdown aberrantly regulates different gene profiles in PCa cells (A) The intensities of protein bands detected in western blotting were measured using imageJ (NIH, Bethesda, MD, USA), OriginPro 2021 software (OriginLab Corporation, Northampton, MA, USA) was used to perform hierarchical clustering and heatmap analysis of protein expression/activation in PCa cells either control shRNA (shCON) or SLC4A4 shRNA (shSLC4A4). This analysis identified the coupling relationships that reveal the regulatory pathways driving cell proliferation. (B) A heatmap showing top 40 up‐ and downregulated genes after SLC4A4 knockdown in DU145 cells. (C) Volcano plot showing distribution of 1157 differentially expressed genes upon SLC4A4 knockdown in DU145 cells. Log2 fold change is represented in x‐axis, while y axis represents –log10 P value. Upregulated and downregulated genes are marked with dots (D) qPCR validation of the RNA Sequencing data in DU-145 cells (E) Gene Ontology (GO) biological process enrichment analysis of the genes differentially expressed upon SLC4A4 knockdown in Prostate Cancer (PCa) cells. (F) Members of the SLC4 transporter family that were altered after SLC4A4 knockdown in PCa cells.
Among the two, the subcluster consisting of GSK-3β is a driver for RB. This is consistent with the published regulatory function of GSK-3β as a driver of cell proliferation, where it has been found the GSK-3β-dependent downregulation of cyclin D1 and cyclin E1 in neuroblastoma cells31. This reduction of cyclin D1 leads to hypophosphorylation of Rb protein and regulation of E2F transcriptional activity, which is associated with cell cycle arrest. On the other side, active cyclin-CDK complexes induced the phosphorylation of RB protein, which leads to the release of transcription factors of the E2F family and S-phase entry32.
Differential expression analysis by RNA sequencing data
To investigate the signaling molecules that are regulated by SLC4A4, transcriptome sequencing was performed using RNA from SLC4A4 knockdown (shSLC4A4) and shCON cells. Figure 6B shows the log2-fold change of the top 40 up-and down-regulated genes upon SLC4A4 knockdown in DU145 cells (FDR < 0.05). Our volcano plot analysis revealed that when SLC4A4 was knockdown, 637 protein‐coding genes were downregulated, and 517 protein‐coding genes were upregulated (Fig. 6C). We validated the RNA sequencing data in SLC4A4 knockdown (shSLC4A4) and shCON cells by qPCR. Consistently, SMIM22 and ALDH3A1 genes were upregulated, and SPINK4 and AQP5 were downregulated in shSLC4A4 cells (Fig. 6D). Furthermore, differentially expressed genes (DEGs) were subjected to KEGG pathway enrichment analysis26. 11 metabolic pathways were identified and included the pyrimidine nucleoside diphosphate biosynthetic pathway, the peptidoglycan transport pathway, and the DTTP biosynthetic process (Fig. 6E). We also observed the differential expression of SLC4 transporter family members after SLC4A4 knockdown in PCa cells (Fig. 6F).
Discussion
SLC4A4 is expressed in many tissues throughout the body but is particularly abundant in the kidney, pancreas, eye, and brain33. However, pan-cancer analysis revealed that SLC4A4 expression is higher in cholangiocarcinoma and prostate adenocarcinoma34. The potential molecular mechanisms underlying SLC4A4 regulation in the proliferation, invasion and metastasis of PCa remain unknown. Utilizing an immunohistochemistry (IHC) assay from clinical specimens of prostatic acinar adenocarcinoma, we identified the SLC4A4 as being significantly overexpressed. Further analysis by western blotting and qPCR revealed that SLC4A4 was significantly overexpressed in AR-negative and positive PCa cell lines compared to RWPE-1 normal prostate epithelial cells. Notably, we found the significance of SLC4A4 in the tumor progression and reported a new signaling axis, SLC4A4/GSK-3β/β-catenin, that regulates the clonogenic potential, invasiveness and metastasis of PCa cells by regulation of GSK-3β and subsequently phosphorylation of β-catenin at S33/37. Using co-immunoprecipitation, we also reported a novel interaction of SLC4A4 with RB that drives cell cycle progression in PCa cells via SLC4A4/RB signaling axis. Both signaling axes illuminate novel targets for therapeutic intervention in prostate cancer therapy. Lastly, SLC4A4 knockdown in prostate cancer cells regulates differentiated expressed genes and multiple metabolic pathways.
SLC4A4 transcripts were higher in prostate tumour tissues than normal by TCGA analysis using the UALCAN web portal (Fig. 1A). Especially the lower Gleason grade tumors (6 and 7) showed a high SLC4A4 expression (Table 1). This was further confirmed by western blot analysis and qPCR, showing SLC4A4 expression to be high in prostate cancer cells. Our results for the SLC4A4 expression in patient-derived specimens and PCa cell lines are also consistent with the pan-cancer profiling of SLC4A4 for PRAD. However, it is poorly expressed in various cancers, including breast invasive cancer, colon adenocarcinoma, liver hepatocellular carcinoma and squamous cell carcinoma of the lung. So, there is a correlation between gene expression with copy number variations (CNVs). Shao et al.35 have reported strong evidence to support the high correlation between CNVs and differential gene expression across multiple cancer types and cancer cell lines.
Cancer cell metastasis involves a multi-step process that requires cell invasion and migration, which are to be the most critical factors in primary tumor metastasis36. Aberrant Wnt/ß-catenin signaling is a late event in tumorigenesis, and approximately a third of prostate tumors have this signaling37. In addition, low expression of GSK-3ß is correlated significantly with advanced clinicopathological characteristics and poor prognosis of HCC patients38. Here, we showed the novel signaling axis, SLC4A4/GSK-3β/β-catenin, which regulates the clonogenic potential, invasiveness and metastasis of PCa cells. Wnt signaling prevents GSK-3β from phosphorylating β-catenin, causing its stable expression in various human tumor cells39. To evaluate the relationship between SLC4A4 and β-catenin, we first investigated the levels of total β-catenin. We then constructed a pLKO.1-SLC4A4shRNA plasmid and generated stably transfected PCa cell lines expressing a shRNA targeted against SLC4A4. The results showed that knockdown of SLC4A4 attenuated β-catenin protein expression (Fig. 4A). This was associated with reduced phosphorylation of GSK-3α/β on residues Ser21/9 with restoration of GSK3ß level. This activated GSK3ß phosphorylated the β-catenin at S33/37 in PCa cells. These results showed that GSK-3β is mainly regulated by the SLC4A4. The metastatic effect of cancer of SLC4A4 can be attributed to an increase in β-catenin expression.
The present study reported that SLC4A4 was overexpressed in prostate cancer, and knockdown of SLC4A4 inhibited cell growth and colony formation. RB tumor suppressor protein is known to be central in cell cycle regulation. RB protein is hypophosphorylated in early G1 and associated with the E2F proteins to prevent the expression of cell cycle progression genes40. In SLC4A4 knockdown PCa cell lysates, hypophosphorylated RB protein expression was detected that could potentially prevent entry into the S phase and establish an irreversible cell cycle exit. This was further confirmed by flow cytometry analysis in which SLC4A4 stable transfectants were marked by G0/G1 cell cycle arrest compared to shCON PCa cells. So, SLC4A4/RB signaling axis regulates the cell proliferation in PCa cells.
Since SLC4A4 regulates bicarbonate influx/efflux at the basolateral membrane of cells, RNA sequencing results using the SLC4A4 knockdown prostate cancer cell lines DU-145 showed regulation of multiple metabolic pathways. We validated this data by qPCR (Fig. 6D). KEGG pathway analysis revealed the potential involvement of various metabolic pathways in different molecules, including the biosynthetic and transmembrane transport processes. Furthermore, our RNA sequencing data shows the differential expression of other members of the SLC4 family after the SLC4A4 knockdown. Of those, SLC4A3, SLC4A7, SLC4A8, SLC4A9 and SLC4A11 were upregulated and SLC4A1, SLC4A2 and SLC4A5 were downregulated. Most members mediate HCO3 transport across the plasma membrane in different cells41.
Conclusion
To sum up, the present study findings indicated that SLC4A4 was highly expressed in PCa tissues and played a pivotal role in PCa cell proliferation by targeting the GSK-3β and subsequently hyperphosphorylation of β-catenin. We reported a novel signaling axis, SLC4A4/GSK-3β/β-catenin, that regulates the proliferation, invasiveness and metastasis of PCa cells. Additionally, via the SLC4A4/RB signaling axis, PCa cells regulate the cell cycle progression.We found that SLC4A4 knockdown induced cell cycle arrest by decreasing the total amount of RB protein with subsequent hypophosphorylation at its S608 site. To the best of our knowledge, this study is the first to report that SLC4A4/GSK-3β/β-catenin and SLC4A4/RB signaling axes in PCa cells that regulates the cell proliferation and ß-catenin stabilization. Both signaling axes illuminate novel targets for therapeutic intervention in prostate cancer therapy.
Supplementary Information
Acknowledgements
This study was supported by the Seed Fund for Basic Research of the University of Hong Kong to AHN Tang (Grant number: 201904159002). The authors would like to acknowledge the Centre of PanorOmic Sciences, LKS Faculty of Medicine, The University of Hong Kong for the technical support. The authors thank Prof. Yam Wai Ping, Judy, Dr. Kristy Chan, Dr. NG Tung Him, Tony, Mr. Kai Wong, Ms. Vicky Tin and Ms. Annie Chan at the Department of Pathology, School of Clinical Medicine, The University of Hong Kong, for discussion and technical advice.
Abbreviations
- PCa
Prostate cancer
- SLC4A4
Solute carrier family 4 member 4
- RB
Retinoblastoma-associated protein
- PSA
Prostate-specific antigen
- GEPIA
Gene expression profiling interactive analysis
- BPE
Bovine pituitary extract
- EGF
Human recombinant epidermal growth factor
- SD
Standard deviation
- CHOL
Cholangiocarcinoma
- PRAD
Prostate adenocarcinoma
- PCPG
Pheochromocytoma and paraganglioma
- BLCA
Bladder urothelial Carcinoma
- BRCA
Breast cancer
- COAD
Colon adenocarcinoma
- ESCA
Esophageal Carcinoma
- HNSC
Head-neck squamous cell Carcinoma
- KICH
Kidney chromophobe
- KIRP
Kidney renal papillary cell carcinoma
- LIHC
Liver hepatocellular Carcinoma
- LUSC
Lung squamous cell carcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- THCA
Thyroid Cancer
- CNV
Copy number variation
- EMEM
Eagle's minimum essential medium
- PBS
Phosphate-buffered saline
- FDR
False discovery rate
Author contributions
1. A.R.: Conceptualization; data curation; formal analysis validation, investigation; visualization; methodology; writing-original draft; project administration; writing—review and editing. 2. H.L.F.: Data curation; investigation; methodology; writing—review and editing. 3. A.H.N.T.: Conceptualization; resources; data curation; supervision; funding acquisition; investigation; methodology; writing-original draft; project administration; writing—review and editing.
Data availability
The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through accession number GSE212677 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212677).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Asif Rashid, Email: arashid2@hku.hk.
Alexander Hin Ning Tang, Email: alextang@pathology.hku.hk.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-72408-w.
References
- 1.Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71(3), 209–249. 10.3322/caac.21660 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Siegel, R. L., Miller, K. D., Fuchs, H. E., & Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin.71(1), 7–33. 10.3322/caac.21654 (2021). Epub 2021 Jan 12. Erratum in: CA Cancer J Clin. 2021 Jul;71(4):359. PMID: 33433946. [DOI] [PubMed]
- 3.Etzioni, R. et al. Quantifying the role of PSA screening in the US prostate cancer mortality decline. Cancer Causes Control.19, 175–181 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, I. SLC4A transporters. Curr. Top. Membr.70, 77–103. 10.1016/B978-0-12-394316-3.00003-X (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber, J. M. et al. Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations. Oncotarget.4(5), 715–728. 10.18632/oncotarget.990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korenchan, D. E. et al. Hyperpolarized in vivo pH imaging reveals grade-dependent acidification in prostate cancer. Oncotarget10, 6096–6110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang, C. et al. Whole-genome sequencing of prostate cancer reveals novel mutation-driven processes and molecular subgroups. Life Sci.1(254), 117218. 10.1016/j.lfs.2019.117218 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Liu, Z. et al. SLC4A4 promotes prostate cancer progression in vivo and in vitro via AKT-mediated signalling pathway. Cancer Cell Int.22(1), 127. 10.1186/s12935-022-02546-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyquist, M. D. et al. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep.31, 107669. 10.1016/j.celrep.2020.107669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyson, N. J. RB1: A prototype tumor suppressor and an enigma. Genes Dev30, 1492–1502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tricoli, J. V. et al. Alterations of the retinoblastoma gene in human prostate adenocarcinoma. Genes Chromosom. Cancer15, 108–114 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Murillo-Garzon, V. & Kypta, R. WNT signalling in prostate cancer. Nat. Rev. Urol.14, 683–696 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Bovolenta, P., Esteve, P., Ruiz, J. M., Cisneros, E. & Lopez-Rios, J. Beyond Wnt inhibition: New functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci.121, 737–746 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol.13, 767–779 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Wang, L. et al. GIPC2 interacts with Fzd7 to promote prostate cancer metastasis by activating WNT signaling. Oncogene41(18), 2609–2623. 10.1038/s41388-022-02255-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gretzer, M. B. & Partin, A. W. PSA markers in prostate cancer detection. Urol. Clin. N. Am.30(4), 677–686. 10.1016/s0094-0143(03)00057-0 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Tabayoyong, W. & Abouassaly, R. Prostate cancer screening and the associated controversy. Surg. Clin. N. Am.95(5), 1023–1039. 10.1016/j.suc.2015.05.001 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Duffy, M. J. Biomarkers for prostate cancer: Prostate-specific antigen and beyond. Clin. Chem. Lab. Med.58(3), 326–339. 10.1515/cclm-2019-0693 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Chandrashekar, D. S. et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia25, 18–27. 10.1016/j.neo.2022.01.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epstein, J. I. et al. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: Definition of grading patterns and proposal for a new grading system. Am. J. Surg. Pathol.40(2), 244–252. 10.1097/PAS.0000000000000530 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Epstein, J. I. et al. A Contemporary prostate cancer grading system: A validated alternative to the Gleason score. Eur Urol.69(3), 428–435. 10.1016/j.eururo.2015.06.046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid, A. et al. Dissecting the novel partners of nuclear c-Raf and its role in all-trans retinoic acid (ATRA)-induced myeloblastic leukemia cells differentiation. Exp Cell Res.394(1), 111989. 10.1016/j.yexcr.2020.111989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res.29(9), e45. 10.1093/nar/29.9.e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khammanit, R., Chantakru, S., Kitiyanant, Y. & Saikhun, J. Effect of serum starvation and chemical inhibitors on cell cycle synchronization of canine dermal fibroblasts. Theriogenology70(1), 27–34 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Shen, M. & Yen, A. c-Cbl interacts with CD38 and promotes retinoic acid-induced differentiation and G0 arrest of human myeloblastic leukemia cells. Cancer Res.68(21), 8761–8769. 10.1158/0008-5472.CAN-08-1058 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28(1), 27–30. 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks, S. K., Cormerais, Y. & Pouyssegur, J. Hypoxia and cellular metabolism in tumour pathophysiology. J. Physiol.595, 2439–2450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. & Hong, J. H. The fundamental role of bicarbonate transporters and associated carbonic anhydrase enzymes in maintaining Ion and pH homeostasis in non-secretory organs. Int. J. Mol. Sci.21, 3392020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han, J. et al. Nuclear expression of β-catenin promotes RB stability and resistance to TNF-induced apoptosis in colon cancer cells. Mol. Cancer Res.11(3), 207–218. 10.1158/1541-7786.MCR-12-0670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, J. et al. Activation of Wnt/β-catenin signalling via GSK3 inhibitors direct differentiation of human adipose stem cells into functional hepatocytes. Sci. Rep.7, 40716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S. L. et al. GSK3β-dependent cyclin D1 and cyclin E1 degradation is indispensable for NVP-BEZ235 induced G0/G1 arrest in neuroblastoma cells. Cell Cycle16(24), 2386–2395. 10.1080/15384101.2017.1383577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarducci, C. et al. Cyclin E1 and Rb modulation as common events at time of resistance to palbociclib in hormone receptor-positive breast cancer. NPJ. Breast Cancer28(4), 38. 10.1038/s41523-018-0092-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seki, G., Yamada, H. & Horita, S. Activation and Inactivation Mechanisms of Na-HCO3 Cotransporter NBC1. J. Epithel. Biol. Pharmacol.1, 35–39 (2008). [Google Scholar]
- 34.Chen, X., Chen, J., Feng, Y. & Guan, W. Prognostic value of SLC4A4 and its correlation with immune infiltration in colon adenocarcinoma. Med. Sci. Monit.26, e925016. 10.12659/MSM.925016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao, X. et al. Copy number variation is highly correlated with differential gene expression: a pan-cancer study. BMC Med. Genet.20(1), 175. 10.1186/s12881-019-0909-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leber, M. F. & Eferth, T. Molecular principles of cancer invasion and metastasis (review). Int. J. Oncol.34, 881–895 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Chesire, D. R., Ewing, C. M., Sauvageot, J., Bova, G. S. & Isaacs, W. B. Detection and analysis of beta-catenin mutations in prostate cancer. Prostate45(4), 323–334. 10.1002/1097-0045(20001201)45:4%3c323::aid-pros7%3e3.0.co;2-w (2000). [DOI] [PubMed] [Google Scholar]
- 38.Hu, Y., Lin, X., Zuo, S., Luo, R. & Fang, W. Elevated GSK3β expression predicts good prognosis in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol.11(5), 2776–2783 (2018). [PMC free article] [PubMed] [Google Scholar]
- 39.Amit, S. et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev.16(9), 1066–1076. 10.1101/gad.230302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du, W. & Searle, J. S. The rb pathway and cancer therapeutics. Curr. Drug Targets10(7), 581–589. 10.2174/138945009788680392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorbatenko, A., Olesen, C. W., Boedtkjer, E. & Pedersen, S. F. Regulation and roles of bicarbonate transporters in cancer. Front Physiol.16(5), 130. 10.3389/fphys.2014.00130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through accession number GSE212677 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212677).