Abstract
Purified retrovirus Gag proteins or Gag protein fragments are able to assemble into virus-like particles (VLPs) in vitro in the presence of RNA. We have examined the role of nucleic acid and of the NC domain in assembly of VLPs from a Rous sarcoma virus (RSV) Gag protein and have characterized these VLPs using transmission electron microscopy (TEM), scanning TEM (STEM), and cryoelectron microscopy (cryo-EM). RNAs of diverse sizes, single-stranded DNA oligonucleotides as small as 22 nucleotides, double-stranded DNA, and heparin all promoted efficient assembly. The percentages of nucleic acid by mass, in the VLPs varied from 5 to 8%. The mean mass of VLPs, as determined by STEM, was 6.5 × 107 Da for both RNA-containing and DNA oligonucleotide-containing particles, corresponding to a stoichiometry of about 1,200 protein molecules per VLP, slightly lower than the 1,500 Gag molecules estimated previously for infectious RSV. By cryo-EM, the VLPs showed the characteristic morphology of immature retroviruses, with discernible regions of high density corresponding to the two domains of the CA protein. In spherically averaged density distributions, the mean radial distance to the density corresponding to the C-terminal domain of CA was 33 nm, considerably smaller than that of equivalent human immunodeficiency virus type 1 particles. Deletions of the distal portion of NC, including the second Zn-binding motif, had little effect on assembly, but deletions including the charged residues between the two Zn-binding motifs abrogated assembly. Mutation of the cysteine and histidine residues in the first Zn-binding motif to alanine did not affect assembly, but mutation of the basic residues between the two Zn-binding motifs, or of the basic residues in the N-terminal portion of NC, abrogated assembly. Together, these findings establish VLPs as a good model for immature virions and establish a foundation for dissection of the interactions that lead to assembly.
In retroviruses, the Gag polyprotein directs the assembly and budding of virions from the plasma membrane. Even in the absence of other viral proteins, expression of Gag leads to budding of particles resembling real virions. In wild-type viruses, Gag is cleaved late in the budding process by the viral protease (PR) to yield the mature proteins MA, CA, and NC that are common to all retroviruses plus other small proteins or peptides particular to the retrovirus species. For most retroviruses, including human immunodeficiency virus type 1 (HIV-1), Rous sarcoma virus (RSV), and murine leukemia virus (MuLV), assembly and budding occur concomitantly. These processes are not obligatorily coupled, however, since for the B- and D-type viruses assembly of immature viral cores takes place in the cytoplasm, followed by transport of the intact cores to the membrane and subsequent envelopment and proteolytic maturation. From deletion analyses, it appears that budding is dependent on the function of three short amino acid sequences in Gag, sometimes called assembly domains. The M (membrane binding) sequence, comprising approximately the N-terminal half of MA with its critical basic amino acid residues and including the N-terminal myristate group present in most retroviruses, directs binding and/or transport of Gag to the membrane (59, 61, 74, 78, 85; reviewed in reference 52). The I (interaction) sequence(s), comprising one or more clusters of basic residues in NC, somehow promotes tight interactions between Gag molecules, presumably by virtue of its binding to RNA (11, 18, 24). The L (late) motif, found either between MA and CA or near the C terminus of Gag, is believed to facilitate the pinching off and release of the budding virions (2, 21, 36, 40, 60, 63, 64, 79, 80, 81). Although budding of membrane-enveloped particles requires primarily these sequences in Gag, the particles thus produced may be grossly aberrant in morphology or size unless a functional CA sequence and the associated C-terminal spacer (SP) sequence are present (1, 51, 53).
Despite a wealth of data on the molecular genetics, biochemistry, and structural biology of Gag proteins, the principles of retrovirus assembly are incompletely understood. For example, the protein-protein interactions in assembly, the role of RNA in virion structure, the nature and timing of the morphological change underlying maturation, and the mechanism of protein-lipid interaction are aspects of assembly that remain to be clarified. Since budding cannot be synchronized, the steps in this process are difficult to follow biochemically. Retroviruses are not icosahedral and are heterogeneous in size (32, 82), and therefore a conventional three-dimensional reconstruction of immature or mature virions from cryoelectron micrographs cannot be done.
Retrovirus-like particles (VLPs) can assemble spontaneously in vitro from purified Gag protein or fragments of Gag, as studied most extensively for Mason-Pfizer monkey virus (50), RSV (14, 15, 46), and HIV-1 (13, 35, 41–44, 58, 77). Assembly of VLPs from Gag protein translated in crude extracts also has been reported (55, 67, 68, 71). To be competent to assemble into regular structures, the Gag protein must contain at least an intact CA sequence. Under most conditions, productive in vitro assembly also requires NC and RNA, although CA as an isolated protein can assemble into tubular particles (29, 35, 41, 42, 49, 77). The morphology of the VLPs is either spherical, resembling immature cores in virions lacking an active protease (PR), or tubular. In the case of HIV-1, conical particles resembling mature cores also are formed under some conditions (35), but in the presence of tubular particles. Several factors have been identified that promote Gag polymerization into one or another macromolecular shape. In HIV-1, addition of amino acid residues to the N terminus of CA-NC redirects the assembly outcome from tubes to spheres (43, 77). Deletion of the 14-amino-acid SP1 peptide between CA and NC also causes this shape change, as does alteration of the pH of the assembly reaction (44). In RSV, deletion of the p10 domain from Gag leads to tubular particle formation (15). The critical sequence element for this effect is the segment of 25 amino acid residues comprising the C-terminal segment of p10 (46).
Because it is a well-defined system, in vitro assembly promises to be a useful approach by which to unravel the protein-protein and protein-nucleic acid interactions of Gag that lead to virion formation. We have studied several parameters influencing RSV VLP assembly in vitro, using a soluble RSV Gag protein missing N- and C-terminal domains. The results showed that the appearance and density of VLPs are not affected by the length or type of nucleic acid and that the ratio of protein mass to nucleic acid mass in the purified particles is constant. The mean mass of the VLPs implies a stoichiometry of about 1,200 Gag molecules per particle. By cryoelectron microscopy (cryo-EM), the VLPs, although lacking an external lipid bilayer, otherwise closely resemble immature particles formed in vivo. The structure of the Zn-binding motifs can be disrupted without impact on assembly, and the distal parts of NC are dispensable for the assembly process.
MATERIALS AND METHODS
DNA constructs.
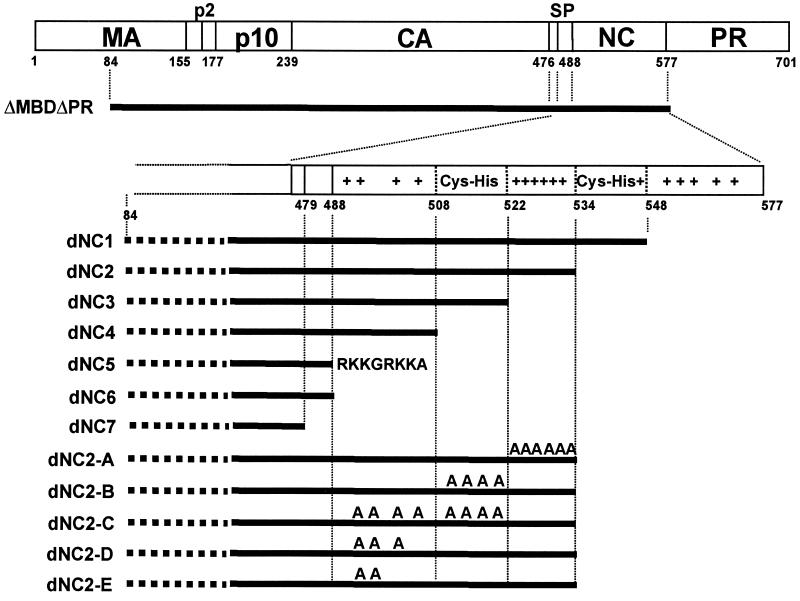
Nucleotide sequences and numbers refer to the RNA sequence of the Prague C strain of RSV. All plasmids were constructed by using common subcloning techniques and propagated in Escherichia coli DH5α. After confirmation by restriction enzyme digestion and sequence analysis, the plasmids were moved into E. coli strain BL21 DE3(pLysS) for protein expression. The expression plasmid encoding the Gag protein ΔMBDΔPR was available from previous work (15, 46) and is based on the vector pET3xc (Novagen). The N terminus of this protein begins at amino acid residue 84 of MA, and the C terminus is the last residue of NC.
The pET3xc.dNC series of plasmids was created using pET3xc.ΔMBDΔPR as the starting plasmid. pET3xc.dNC7 was made by replacing the SacII (nucleotide [nt] 1806 in RSV)-KpnI (nt 4995 in RSV) fragment from pET3xc.ΔMBDΔPR with two self-annealing oligonucleotides designed to delete all of NC and the nine amino acids (SP) between CA and NC from ΔMBDΔPR. pET3xc.dNC6 was created by replacing the SacII-KpnI fragment with two self-annealing oligonucleotides designed to delete all of NC from ΔMBDΔPR but to leave the SP peptide. To generate pET3xc.dNC1 through pET3xc.dNC5, PCR was performed using pATV-8K as a template, a forward primer in CA, and a reverse primer with a KpnI site that created the desired C terminus of the protein. The PCR product was digested with SacII and KpnI and subsequently subcloned into pET3xc.ΔMBDΔPR. The mutants dNC2-A to dNC2-E were created by replacing the SacII-KpnI fragment of dNC2 with a PCR-derived fragment carrying Ala substitutions in clusters of basic amino acid residues. In dNC2-A, all six K or R residues between the two CCHC motifs were mutated. In dNC2-B and dNC2-C, the three Cys residues and one His residue in the first CCHC motif were mutated. The following residues in the proximal segment of NC were mutated in the other constructs: dNC2-C, R5, R7, R16, and R18; dNC2-D, R5, R7, and R16; dNC2-E, R5 and R7.
Protein and RNA purification.
E. coli BL21 DE3(pLysS) cells were grown and induced for protein expression as previously described (14, 15). Purification of protein GagΔMBDΔPR was done using a modification of the method previously described (15). The frozen bacterial pellet was resuspended on ice in buffer A (20 mM Tris [pH 7.5], 0.5 M NaCl, 10% glycerol, 1 mM EDTA, 10 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) at 25 ml/liter of cell culture, and the cells were broken by sonication. In some experiments, 0.1% Nonidet P-40 was added to buffer A. Insoluble debris and nucleic acids were removed by addition of 0.3% (wt/vol) polyethyleneimine, followed by centrifugation (19). The protein was precipitated by adding 1 volume of room temperature saturated ammonium sulfate to 3 volumes of supernatant. After 30 min at 0°C, the precipitate was collected by centrifugation for 10 min at 10,000 rpm in an SS34 rotor. The precipitate was resuspended in buffer B (20 mM Tris [pH 7.5], 1 mM EDTA, 10 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) plus 0.5 M NaCl at 2 ml/liter of cell culture and incubated on ice for 30 min. Buffer B was slowly added until the final NaCl concentration was 0.1 M. Insoluble protein was removed by centrifugation for 2 min at 7,000 rpm in an SS34 rotor, and the supernatant was immediately applied to batch phosphocellulose (Whatman P11) at a volume ratio of 10:1. The resin with bound protein was washed with buffer B plus 0.1 M NaCl and then with buffer B plus 0.3 M NaCl. The volume of the washes was approximately 3 ml/ml of packed resin. Protein was eluted with the same buffer plus 0.5 M NaCl and then with the same buffer plus 1.0 M NaCl. The eluted protein was precipitated with 50% saturated ammonium sulfate, resuspended, and dialyzed overnight against buffer B plus 0.5 M NaCl at 4°C. Storage was at −80°C. The concentration of the purified protein was determined by spectrophotometry (A280 and A260) as described below. Purity was gauged by Coomassie blue staining after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
For purification of proteins dNC1 through dNC5, frozen cells were resuspended in buffer A and broken by sonication. Insoluble debris was removed by centrifugation at 10,000 rpm for 10 min in an SS34 rotor, and soluble protein was precipitated with 25% saturated ammonium sulfate. The precipitate was resuspended in buffer B plus 0.5 NaCl and dialyzed overnight at 4°C.
Proteins dNC6 and dNC7 were purified using a different scheme. Frozen cells were resuspended in buffer A without NP-40 and broken by sonication. Protein was precipitated with 34% saturated ammonium sulfate from the soluble crude cell extract. The precipitate was resuspended in buffer B plus 0.1 M NaCl at 5 ml/liter of cell culture and dialyzed overnight against the same buffer at 4°C. Insoluble material was removed by centrifugation, and the supernatant was applied to a DEAE-cellulose (DE-52) column. The resin was washed with buffer B plus 0.1 M NaCl. The flowthrough and wash of DEAE were pooled and precipitated with 50% saturated ammonium sulfate. The precipitate was resuspended in buffer B plus 0.1 M NaCl and dialyzed overnight at 4°C.
Total E. coli RNA was purified by phenol extraction using standard procedures (5). Tobacco mosaic virus (TMV) RNA or RNA associated with the ΔMBDΔPR particles was purified by digesting virus or particles with proteinase K in 1% SDS–10 mM Tris-HCl (pH 8.0)–1 mM EDTA at 37°C for 1 h, after which phenol-chloroform extraction and ethanol precipitation were performed. RSV RNA was synthesized in vitro by transcription with T7 RNA polymerase according to instructions from the manufacturer (Promega Corp.), using plasmids that were engineered to contain the desired RSV sequences. All purified RNA was stored in diethyl pyrocarbonate-treated water or as an ethanol precipitate at −20°C.
Analysis of particles assembled in vitro.
When the dialysis method was used for in vitro assembly, protein (typically 1 mg/ml) and RNA or DNA (typically, a 10% [wt/wt] ratio of nucleic acid to protein) were mixed in buffer B plus 0.5 M NaCl and dialyzed for 2 h at room temperature against buffer C (assembly buffer) (50 mM morpholine ethanesulfonic acid [pH 6.0], 0.1 M NaCl). For some experiments, assembly buffer D was used instead (50 mM morpholine ethanesulfonic acid [pH 6.5], 0.1 M KCl, 10 μM ZnCl2), with similar results. For the direct dilution method of in vitro assembly, protein (5 mg/ml) and RNA were mixed in buffer B plus 0.5 M NaCl and diluted with 4 volumes of buffer C without NaCl. Reaction mixtures were incubated at room temperature for 30 min. Particles formed under these conditions were negatively stained with 2% uranyl acetate (pH 5) on Formvar-carbon-coated grids. To determine the density of the particles produced and to assess the efficiency of in vitro assembly, the assembly reaction mixture was layered onto a 10 to 60% (wt/wt) linear sucrose gradient in buffer C. The gradient was centrifuged for 4 h at 50,000 rpm in a Beckman SW60 rotor. Fractions were collected from the top of each tube and analyzed by SDS-PAGE.
The apparent binding site size of ΔMBDΔPR protein on RNA or DNA was estimated from the measured A260/A280 values of purified protein, purified nucleic acids, and VLPs dissolved in 0.1% SDS in pH 6.0 buffer. The measured A260/A280 ratio was 0.7 for the purified protein and 2.0 for RNA and varied for oligonucleotides depending on base composition. The molar extinction coefficient for protein at 280 nm was calculated from the number of tryptophan, tyrosine, and cysteine residues to be 4.9 × 105. The molar extinction coefficients for oligonucleotides at 260 nm were calculated from the published values at 260 nm for dGMP, dCMP, dAMP, and TMP (5). For RNA, it was assumed that the A260 nm is 1.0 for 40 μg/ml (5).
For analysis by scanning transmission EM (STEM) in vitro assembly with ΔMBDΔPR protein was carried out either with E. coli RNA or with an 80-nt oligonucleotide and with assembly buffer C. The preparations of VLPs at about 1 mg of protein per ml were adsorbed to a 3-nm carbon-supported grid, upon which TMV had been adsorbed previously to provide an internal standard. The grids were processed and analyzed as described previously for wild-type RSV virions (76). In brief, the mass of each VLP was determined by summing the number of scattered electrons over the particle, subtracting the background scattering from the support film, and calibrating with TMV particles nearby on the grid.
For analysis by cryo-EM, VLPs were assembled with ΔMBDΔPR protein and E. coli RNA at a protein concentration of 1 mg/ml. Solutions containing the particles were applied to perforated carbon-coated EM grids, which were briefly blotted to remove excess liquid and then rapidly plunged into liquid ethane. Specimens were examined using a Philips CM200 FEG transmission electron microscope equipped with a Gatan cryostage. Images were recorded on Kodak SO-163 photographic film at a nominal magnification of ×27,500 under low-dose conditions (<20 electrons/Å2). Micrographs were digitized on a Zeiss-SCAI scanner using a step size of 7 μm. The program I.C.E. (45) was used to box images of the particles. To calculate the spherically averaged density distribution for each particle, images of the particles were first circularly averaged. The origin for averaging was chosen so that the variance from exact circular symmetry was minimized. The spherically averaged density distribution was then calculated from the circularly averaged projections through the inverse Abel transform (72, 75). Inversion of Abel's integral equation was done using the method described in reference 28, which is stable in the presence of high levels of noise in the experimental data. Spherically averaged density distributions have previously been calculated from cryo-EM images of immature retroviral particles using a Fourier-Bessel method (32, 44). The mathematical equivalence of the two procedures for calculating spherically averaged density distributions follows from the fact that the Fourier transform of the Abel transform is the zero-order Hankel (Fourier-Bessel) transform (25).
RESULTS
Nucleic acid requirement for assembly.
For in vitro assembly studies, it is imperative to have a purified soluble Gag protein. The full-length RSV Gag protein with an inactivating mutation in the PR domain remains entirely in inclusion bodies in E. coli and is refractory to solubilization after lysis. A deletion mutant of Gag missing the PR domain is also found in inclusion bodies and can be solubilized and assembled in vitro, but this protein is still prone to nonspecific aggregation (15). However, further deletion of the 84-amino-acid membrane-binding domain leads to a protein that remains in the soluble fraction of E. coli lysates and can be readily purified and maintained in soluble form at several milligrams per milliliter. We have used the latter protein, ΔMBDΔPR (Fig. 1), as a model for RSV Gag. In previous experiments, partially purified ΔMBDΔPR assembled efficiently into spherical VLPs in vitro and assembly was blocked or reversed by treatment of the protein with RNase (15). UV absorption showed that the protein had an A260/A280 ratio of about 1.3, implying contamination with RNA, which was inferred to be acting as a scaffold for assembly.
FIG. 1.
Diagrammatic representation of proteins. The rectangle at the top represents the RSV Gag protein, with vertical lines marking the sites of proteolytic cleavages during maturation. Numbers indicate amino acid residues from the N terminus. The ΔMBDΔPR protein is missing the N-terminal membrane-binding domain, as well as the C-terminal PR domain. Black bars represent proteins examined in this study. The mutant NC proteins all have the same N-terminal portion as ΔMBDΔPR but are truncated in the NC domain as indicated. +, lysine or arginine residue; Cys-His, 14-amino-acid segment including the conserved zinc-binding motif; A, alanine substitution for a lysine, arginine, cysteine, or histidine residue.
In order to investigate the nucleic acid requirement for assembly, we removed residual RNA from ΔMBDΔPR by first treating the soluble cell extract in the presence of a high salt concentration with polyethyleneimine, a synthetic polycationic reagent, prior to ammonium sulfate precipitation (19). Following further chromatography on phosphocellulose, the protein had an A260/A280 ratio of 0.7, consistent with the absence of nucleic acid. After addition of E. coli RNA at a high salt concentration and dialysis for 2 h against the low-salt assembly buffer, ΔMBDΔPR formed abundant VLPs that could be visualized by negative-staining EM (Fig. 2A). In the absence of added RNA, no regular structures were observed (data not shown). The particles had a morphology similar to that of authentic immature virions and had a mean external diameter of about 70 nm (Table 1). The majority of particles appeared intact and nearly spherical, although incomplete or broken particles also were apparent. Faint striations were visible at the particle circumference. These observations are consistent with analysis of the particles using cryo-EM (see below). We interpreted the open particles to be the result of incomplete assembly. To provide a more quantitative measurement of assembly, assembly reaction mixtures were submitted to centrifugation in an isopycnic 10 to 60% (wt/wt) sucrose gradient (Fig. 3A and B). In the presence of RNA, almost all of the protein banded at a density of 1.22 to 1.25 where VLPs could also be readily found by EM (data not shown). In the absence of RNA, the protein remained near the top of the gradient. These results support the previous conclusion (15) that nucleic acid is an absolute requirement for VLP formation in this system.
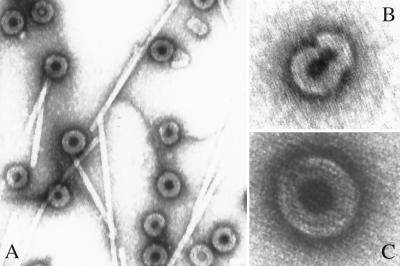
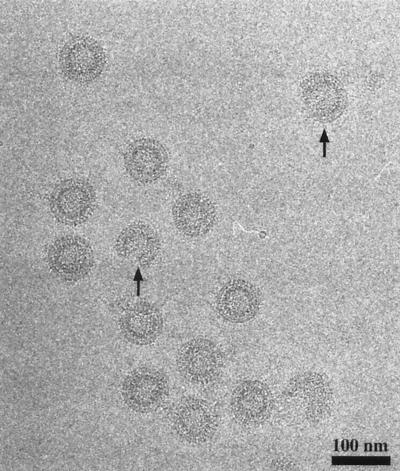
FIG. 2.
Negative-staining EM of VLPs. ΔMBDΔPR protein was assembled with E. coli RNA, and particles were visualized by negative-staining EM. TMV was added as an internal standard. Original magnifications: A, ×92,000; B, ×200,000; C, ×290,000. The structures visible in panel B are representative of incomplete particles seen at variable frequency in the assembly reaction mixtures.
TABLE 1.
Characteristics of VLPs formed with different nucleic acids
| Polymera | Length (nt) | Avg. diam (SD)b (nm) | Nc | A260/A280d ratio | No. of nt/proteine | Efficiencyf |
|---|---|---|---|---|---|---|
| U5-MA RNA | 580 | 71 (6) | 22 | 1.36 | ++ | |
| U5-p10 RNA | 1,110 | 72 (3) | 23 | 1.38 | ++ | |
| U5-NC RNA | 2,117 | 68 (6) | 28 | 1.33 | ++ | |
| U5-polymerase RNA | 3,118 | 72 (5) | 22 | 1.34 | ++ | |
| TMV RNA | 6,395 | 63 (5) | 16 | 1.38 | ++ | |
| E. coli RNA | ∼100–3,000 | 74 (4) | 42 | 1.35–1.40 | 12–13 | ++ |
| Poly(rA) | ∼450 | 70 (4) | 21 | ++ | ||
| Poly(U) | ++ | |||||
| Poly(rAC) | ++ | |||||
| 22-mer oligonucleotideg | 22 | 70 (4) | 14 | ++ | ||
| 12-mer oligonucleotideg | 12 | + | ||||
| 8-mer oligonucleotidegh | 8 | 74 (5) | 17 | ± | ||
| Double-stranded DNAi | 3,465 | 77 (6) | 52 | ++ | ||
| Double-stranded DNAi | 3,995 | 72 (7) | 42 | ++ | ||
| T22 oligonucleotide | 22 | 1.20 | 10 | ++ | ||
| AG22 oligonucleotide | 22 | 1.48 | 7 | ++ | ||
| GT22 oligonucleotide | 22 | 1.24 | 9 | ++ | ||
| AC22 oligonucleotide | 22 | 1.32 | 7 | ++ | ||
| TC22 oligonucleotide | 22 | 0.98 | 8 | ++ | ||
| A22 | 22 | + | ||||
| 61-mer oligonucleotide | 61 | 1.22 | 9 | ++ | ||
| Heparin | ++ |
The designations U5-MA, U5-p10, U5-NC, and U5-pol refer to the parts of the RSV genome transcribed in vitro to prepare these RNAs.
RSV ΔMBDΔPR protein at 1 mg/ml was assembled with 10% (wt/wt) different nucleic acids. Diameters of negatively stained particles were measured from photographic prints. The diameter of TMV virions measured in parallel was 18 nm (SD, 3 nm).
N, number of particles measured.
VLPs were collected by centrifugation in a microcentrifuge for 30 min, and the ratio of A260 to A280 was determined after dissolution of the particles in 0.1% SDS. The value for E. coli RNA is the range from many experiments.
Calculated from the A260/A280 ratio as described in Materials and Methods.
Efficiency was estimated by centrifugation in 10 to 60% sucrose gradients as described in the legend to Fig. 3 or by negative-staining EM. ++, 30 to 100% of the level obtained with E. coli RNA; +, 10 to 30% of the level obtained with E. coli RNA; ±, <10% of the level obtained with E. coli RNA.
The sequences of these oligonucleotides represented approximately equal numbers of A, G, T, and C residues.
Assembly efficiency was very poor for this oligonucleotide.
Two different plasmid DNAs were used.
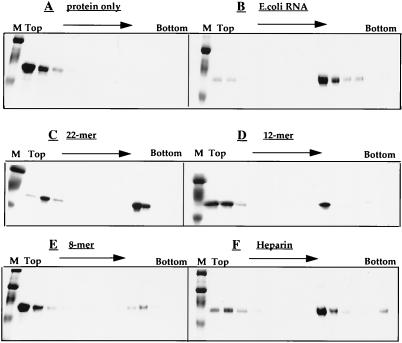
FIG. 3.
Assembly assay by sucrose gradient centrifugation. ΔMBDΔPR protein at 1 mg/ml was assembled with either E. coli RNA, DNA oligonucleotides, or heparin. The reaction mixtures were centrifuged to equilibrium through a 10 to 60% (wt/wt) sucrose gradient, and fractions were analyzed by SDS-PAGE and staining. A, protein alone; B, with E. coli RNA; C, with an oligonucleotide of 22 nt; D, with an oligonucleotide of 12 nt; E, with an oligonucleotide of 8 nt; F, with heparin. M, markers.
Most of the previous work on in vitro assembly in the RSV system has been carried out with total E. coli RNA (15, 46), which is a collection of diverse RNA species. We addressed the size and sequence dependence of assembly with ΔMBDΔPR by evaluating the products obtained with defined nucleic acids including homopolymers (Table 1). The VLPs formed with different sizes of RSV RNAs transcribed in vitro or with other RNAs were indistinguishable by morphology or size. To quantify the relative mass of RNA, the VLPs were collected by centrifugation and then dissolved in SDS and the A280 and A260 values of the resulting solutions were determined. For RNA-containing VLPs, the A260/A280 ratio ranged from about 1.35 to 1.40, implying that the RNA made up 7 to 7.5% of the mass of the protein. This estimate is based on the predicted 280-nm extinction coefficient of ΔMBDΔPR (A280 = 0.94 at 1 mg/ml) derived from number of tryptophan, tyrosine, and cyteine residues and on the approximation that RNA at 40 μg/ml has an A260 of 1.0. This percentage of RNA corresponds to 12 to 13 nt per protein molecule. The constant mass of RNA per VLP implies that the smaller the size of the RNA, the larger the number of RNA molecules in a VLP. Single-stranded DNA oligonucleotides also promoted assembly, as did linear double-stranded plasmid DNA. For DNA oligonucleotides, the A260/A280 ratio varied from 0.98 to 1.48, depending on the oligonucleotide (Table 1). Using the extinction coefficients for the relevant nucleotides, we calculated that the size of the apparent binding site for the different oligonucleotides ranges from 7 to 10 nt.
While assembly was observed for all of the nucleic acids tested, in some cases many more VLPs were evident than in others. Two assays were used to quantitate assembly efficiency, i.e., centrifugation in sucrose gradients (Fig. 3) and counting of VLPs on EM grids. By the former assay, assembly with a 22-mer oligonucleotide was nearly as efficient as with E. coli RNA but smaller oligonucleotides promoted assembly less well, with barely detectable protein banding at the position of VLPs with an 8-mer. The assembly efficiency was corroborated by counting VLPs by EM. These results are summarized in Table 1. Even heparin was an effective substitute for nucleic acid.
We drew the following conclusions about the role of nucleic acids in assembly. First, polyanions of diverse sorts promote VLP formation by ΔMBDΔPR. Thus, it is apparently electrostatic interactions with the basic NC that are required for the Gag protein to polymerize into VLPs. Second, since the ratios of RNA to protein are similar for diverse nucleic acid sizes, each Gag protein must contact nucleic acid, presumably in a similar way. We operationally defined the nucleic acid binding site size as the molar ratio of nucleotides to proteins in the VLPs, i.e., 7 to 13 nt (Table 1). By this definition, the binding site is larger than that determined for the mature retroviral NC protein in most reports, about 5 to 8 nt (30, 47, 48, 83), although in one report this size is up to 14 nt for HIV-1 NC extended at its C terminus (48). Third, there is a minimum size of oligonucleoide that can support efficient assembly. The minimum size is larger than the binding site size.
Assembly optimization and kinetics.
To establish the optimal conditions for in vitro particle formation, the effects of salt and pH on assembly were examined. Constant concentrations of protein (1 mg/ml) and E. coli RNA (10%, wt/wt) were incubated at different pH values and salt concentrations. The reaction mixtures were centrifuged for 30 min in a microcentrifuge, and fractions of the supernatant and pellet were analyzed by SDS-PAGE. Soluble proteins were expected to be in the supernatant, and VLPs were expected to be in the pellet. Samples were also visualized by EM. VLP formation was found to be pH and ionic strength dependent (data not shown), as reported less systematically previously (12). Few particles were formed above 0.3 M NaCl. The effect of a pH change (from 5.5 to 8.0) was less dramatic than that of an ionic strength change, but pH influenced both the intactness of particles and their clumping (Table 2). At pH 6.0, particles were more aggregated than at pH 7.0, as judged by EM and by increased turbidity, visible by eye or measured as A350. The number of single, unaggregated VLPs counted on EM grids increased as the pH was raised from 6.0 to 7.0. Aggregates formed at pH 6.0 could be dissociated into individual particles by increasing the pH or salt concentration. At pH 7.0, relatively more incomplete particles were observed. At pH values above 7.0, assembly was poor, with very few particles detected at pH values above 8.0.
TABLE 2.
Effect of pH on assemblya
| pH | No. of free particles/ photographic field | % of incomplete particles |
|---|---|---|
| 6.0 | 108 | 7 |
| 6.5 | 216 | 9.5 |
| 7.0 | 325 | 20 |
ΔMBDΔPR protein at 1 mg/ml and an 80-mer oligonucleotide (10%, wt/wt) were assembled in buffer D at various pH values. Unaggregated particles were counted on two photographic prints for each reaction after negative-staining EM.
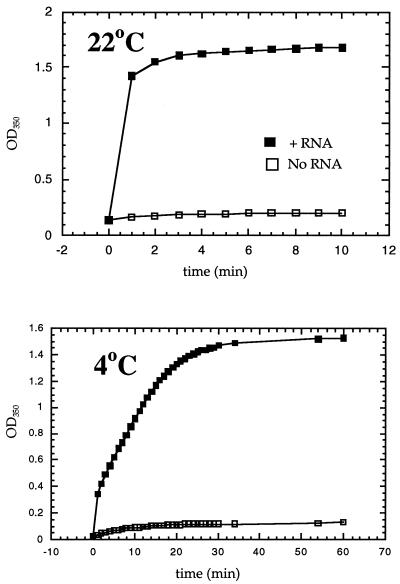
The kinetics of assembly represent an important parameter for the reaction, but one that is challenging to measure. Fully formed VLPs each contain over a thousand Gag proteins, implying a multitude of possible intermediates. Quantitating complete and partially complete particles in various stages of assembly is difficult using EM, since completed and partially completed VLPs cannot always be distinguished. Moreover, depending on the nucleic acid used and on the pH, a variable fraction of the assembly products are nonspherical aggregates. As a first step to approach the kinetics of in vitro assembly, we set up a simple light-scattering assay at a wavelength at which the protein and nucleic acid components do not absorb. ΔMBDΔPR protein and E. coli RNA were diluted rapidly in a cuvette to the standard pH and salt concentration of assembly, and then the increase in A350 was measured as a function of time (Fig. 4). By this assay, assembly was very rapid. At a concentration of 0.5 mg of protein per ml and at 22°C, the reaction was complete in a few minutes, with a half-time of less than 1 min. Assembly could be slowed by lowering the protein concentration (data not shown) or by decreasing the temperature. At 4°C, the extent of the reaction was similar, with a plateau A350 of about 1.5, but the rate was decreased at least 10-fold, with a half-time of about 7 min. Visualization of products by EM showed that by 5 min at 22°C and by 60 min at 4°C, abundant VLPs had formed. Quantitative interpretation of these results was hindered by the different scattering properties of incomplete particles, complete particles, and other aggregates that may form. Nevertheless, the simplicity and robustness of a light-scattering assay make it a potentially powerful tool with which to study assembly.
FIG. 4.
Kinetics of assembly. ΔMBDΔPR protein at 0.5 mg/ml was diluted to 0.1 M NaCl and pH 7.0 in the presence or absence of E. coli RNA, and the course of the assembly reaction was followed in a spectrophotometer at an optical density (OD) of 350 nm. The half-time represents the estimated time of half-maximal optical density. Reaction temperatures are indicated.
Mass of VLPs by STEM.
Understanding retrovirus structure requires knowledge of the stoichiometry of Gag in virions. We previously reported mass measurements of infectious RSV particles by STEM (76). STEM allows the quantitation of electrons scattered from small objects, such as virus particles, on an EM grid. Since scattering is proportional to mass, with suitable standards, it is possible to accurately determine the mass of individual particles. The mean mass of infectious RSV was found previously to be 250 MDa, with a significant variation in the mass of the particles, indicating that they are not all the same size. This conclusion is consistent with cryo-EM size measurements of HIV (32) and RSV (R. Kingston, unpublished data). Based on a number of assumptions about virion composition, the mean mass of RSV was used to calculate that virions contain ∼1,500 Gag molecules (76).
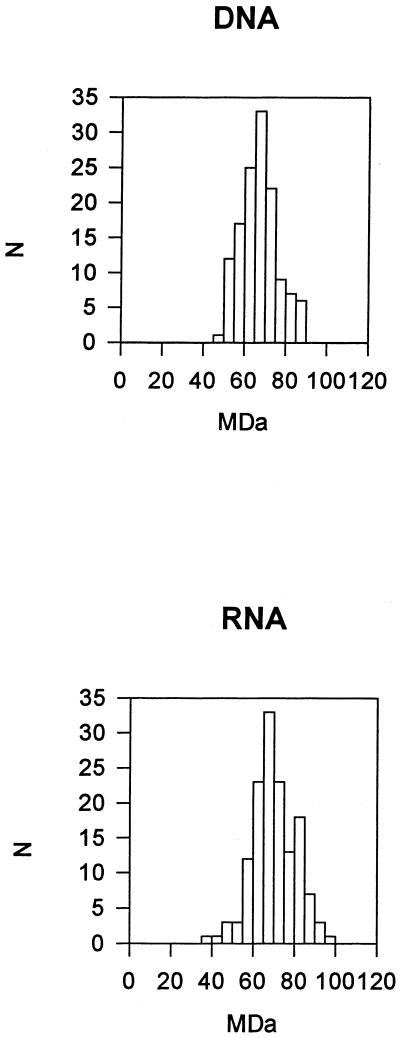
We used STEM to determine the mass of VLPs formed in vitro in the presence of RNA or of a DNA oligonucleotide. To more closely mimic intracellular ionic conditions, in this case, the assembly buffer contained KCl instead of NaCl and included a low concentration of Zn2+ ions to bind to the Cys-His motifs in NC. In addition, a slightly higher pH of 6.5, instead of 6.0, was used to reduce the tendency of VLPs to aggregate without greatly compromising particle integrity (Table 2). The DNA-containing and RNA-containing particles had similar mean masses of 6.7 × 107 and 7.0 × 107 Da (standard deviations, 0.9 × 107 and 1.1 × 107 Da), respectively (Fig. 5). These values correspond to ∼1,200 ΔMBDΔPR molecules per particle. In contrast to the calculations of Gag stoichiometry for infectious virions, the calculation for VLPs requires few assumptions, since only two macromolecular components are used in the assembly reaction. However, meaningful comparison of the calculated in vitro and in vivo stoichiometry would rely on the assumption that the VLPs are all complete. By negative-staining EM, about 90% of the VLPs appeared intact, but this estimate must be considered approximate, since incomplete particles might appear intact in some orientations on the grid and since small, incomplete sectors in the particles might not be visible by negative-staining EM.
FIG. 5.
Mass distributions of RNA-containing and DNA oligonucleotide-containing VLPs. ΔMBDΔPR protein at 1 mg/ml was mixed with E. coli RNA or with an oligonucleotide of 80 nt and dialyzed against assembly buffer D. The particles formed were adsorbed to a carbon-coated grid, and the masses of individual particles were analyzed by STEM. N, number of particles.
Cryo-EM analysis of VLPs.
Virion ultrastructure is best visualized in the absence of staining and fixation, since both of these procedures can give rise to artifacts and distortions. To gain a preliminary understanding of their structure, we analyzed unstained VLPs by cryo-EM. In vitreous ice at liquid nitrogen temperature, the particles appeared spherical and relatively homogeneous in size but exhibited various degrees of closure (completeness), as also seen by negative staining (Fig. 6). Incomplete particles resembled sectors of a sphere, having approximately the same curvature as the fully closed particles. The most striking feature of the images of the in vitro-assembled particles was the track-like striations near the particle circumference. These are characteristic of immature retrovirus particles (32, 44, 82) and are believed to arise from the side-by-side packing of the radially arranged Gag proteins. Also evident in images of the VLPs was a series of concentric electron-dense and electron-sparse annuli which result from the several globular domains of Gag that are connected by extended polypeptides.
FIG. 6.
Cryo-EM of VLPs. ΔMBDΔPR protein at 1 mg/ml was mixed with E. coli RNA and dialyzed against assembly buffer C, and the resulting particles were prepared for cryo-EM as described in Materials and Methods. Arrows highlight incomplete particles.
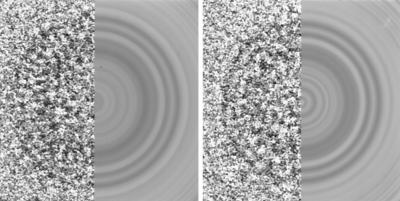
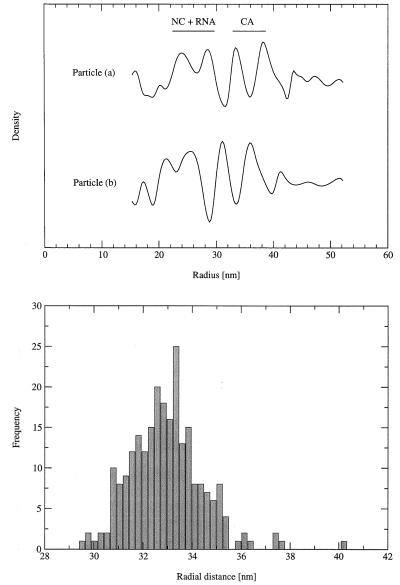
Spherically averaged density distributions were computed from images of the particles using the inverse Abel transform. At radial distances of greater than ∼20 nm, the density distributions calculated from different particles were largely consistent. A typical example is shown in Fig. 7. Based on earlier results for HIV-1 and MuLV (32, 44, 82), we can assign the features in the density distribution to the domains within the Gag polyprotein with some certainty. NC, in complex with RNA, gives rise to a broad interior peak. CA, which encompasses two spatially separated domains in RSV (16, 49), as was shown earlier for HIV (10, 33, 34, 38, 57), is associated with a double peak in the density distribution. Commensurate with the size of the two domains, the outer peak (due to the N-terminal domain) is broader than the inner peak (due to the C-terminal domain). Finally, the p10 domain and the truncated matrix protein are likely to be responsible for the weak features seen at higher radii.
FIG. 7.
Analysis of cryoelectron micrographs. (Top) Composite view of two typical particles (left) and their circular averages (right). (Middle) Spherically averaged density distribution calculated from particles in the top panels. The horizontal axis shows the distance (in nanometers) from the particle center. The vertical axis shows the spherically averaged density (on an arbitrary scale). The assignment of the consistent features of the distribution to the domains within the Gag polyprotein is indicated. (Bottom) Histogram of particle sizes. Radial distances to the innermost of the peaks due to CA are shown.
In the absence of a lipid bilayer, there are no strong features delineating the particle exterior. Hence, as a measure of the particle size, we have taken the radial distance to an interior feature, the inner of the two peaks due to CA. This is the sharpest and most conserved peak in the spherically averaged density distribution. The mean distance to this feature is 33.0 nm (measured from 244 particles without obvious defects; standard deviation, 1.5 nm) (Fig. 7). It is unlikely that the particle sizes are normally distributed when the radius is used as a size metric. The microscope magnification was not calibrated, so the true size distribution may be slightly shifted in either direction. However, the VLPs are significantly smaller than equivalent particles assembled from a truncated HIV-1 Gag polyprotein, where the mean distance to the same feature was ∼55 nm (44). The difference in the sizes of HIV-1 Gag particles and RSV GagΔPR particles is also obvious in thin sections of virus pellets prepared from a baculovirus-insect cell expression system (M. C. Johnson, H. M. Scobie, and V. M. Vogt, unpublished data).
Role of NC sequences in assembly.
Since RNA (or a substitute polyanion) is necessary for in vitro assembly, we assume that interaction between NC and RNA is the first step in the formation of VLPs in vitro. The two conserved features of retroviral NC sequences are the Cys-His zinc-binding motifs and numerous basic amino acid residues. To begin to define the sequences within NC which are important for in vitro assembly, we constructed a series of C-terminal truncations of ΔMBDΔPR, named dNC1 to dNC7 (Fig. 1). One of the proteins, dNC5, has an artificial stretch of basic residues (RKKGRKKA) replacing all of NC. The rationale for this construct is the observation that an almost identical sequence (RKKGRKK) was found to be able to replace NC in RSV assembly in vivo (11).
To determine whether these NC mutant proteins were competent to assemble, the RNA-free proteins at 1.5 mg/ml were dialyzed against the assembly buffer in the presence of RNA and then examined by both negative-staining EM and sucrose density gradient centrifugation (data not shown). The two proteins with the smallest deletions, dNC1 and dNC2, assembled into regular spherical structures at a slightly lower efficiency than ΔMBDΔPR (Table 3). Despite the lower efficiency, however, the VLPs were indistinguishable by morphology. In contrast to these two deletion mutant proteins, all of the deletions mutant proteins dNC3 to dNC7 failed to produce identifiable particles, with or without added RNA and at a variety of protein concentrations.
TABLE 3.
Assembly for site-directed NC mutant proteins
| Protein | No. of particles/photographic field |
|---|---|
| Expt 1a | |
| ΔMBDΔPR | 600–800 |
| dNC1 | 400–500 |
| dNC2 | 100–150 |
| dNC3 | 0 |
| dNC4 | 0 |
| dNC5 | 0 |
| dNC6 | 0 |
| dNC7 | 0 |
| Expt 2 | |
| dNC2 | >100 |
| dNC2-A | 0 |
| dNC2-B | >100 |
| dNC2-C | 0 |
| dNC2-D | 0 |
| dNC2-E | 0 |
In experiment 1, proteins at 1.5 mg/ml were assembled and analyzed by negative-staining EM by counting particles from five photographic prints for each reaction.
To better delineate sequence elements in NC that are important for assembly, we constructed site-directed mutant proteins in the context of dNC2, containing the minimal 46-amino-acid N-terminal NC segment. Clustered alanine substitutions were introduced to change the four Zn-coordinating residues, the six basic residues between the two Cys-His motifs, or the four basic residues in the N-terminal part of NC (Fig. 1). Each protein was partially purified and tested for the ability to form VLPs in vitro with E. coli RNA. The results showed that while the Cys and His residues are dispensable for assembly, the two clusters of six or four basic residues, respectively, are essential (Table 3). In a second set of alanine mutant proteins, only two or three of the basic residues in the proximal segment of NC were mutated. Both mutant proteins failed to assemble. Taken together, these results imply that the positively charged residues in RSV NC are the critical determinants for VLP formation, as they also are for assembly in vivo and for nonspecific RNA binding (11, 18, 24, 31, 62, 69, 84).
DISCUSSION
In this study, we used a partially purified soluble RSV Gag protein, ΔMBDΔPR, to more fully characterize the assembly of VLPs in vitro. By cryo-EM, VLPs closely resemble immature HIV-1 virions and immature MuLV virions in the absence of membranes, as well as HIV-1 VLPs formed in vitro (32, 44, 82). RNA, DNA, single-stranded oligonucleotides, and even a sulfated polysaccharide can promote the assembly of VLPs. Such promiscuity suggests that the determining interactions are predominantly electrostatic, between the basic NC domain and the acidic phosphodiester backbone. This conclusion is consistent with the results of our mutagenesis of NC, which demonstrated that the Cys-His motifs are dispensable for assembly while the clusters of positively charged amino acid residues, at least those in the N-terminal and middle portions of NC, are essential. A more detailed mutagenesis in the context of the full NC domain is needed to uncover if particular Arg or Lys residues are critical in this process.
The observation that the relative mass of RNA in VLPs is constant, independent of RNA length, implies that in formation of the particle, each NC domain of Gag binds to a constant length of nucleic acid. This length can be calculated using the A260/A280 ratio of VLPs after dissolution in SDS. From the values in Table 1, for diverse RNAs, each Gag molecule appears to bind to about 12 to 13 nt. For DNA oligonucleotides, this value is somewhat smaller, about 7 to 10 nt. The binding site size for retrovirus NC proteins maximally packed on RNA has been estimated from titration experiments with fluorescence or fluorescence quenching to be 6 to 8 nt (47, 48, 83). We showed recently by STEM that RSV virions contain about 1,500 of each of the Gag molecules (76). Assuming two copies of genomic RNA of 10 kb in infectious RSV, each NC domain of Gag would thus cover about 13 nt during assembly in vivo, a value that is similar to the present estimate for VLPs formed in vitro with RNA. The significance of the difference in A260/A280 ratios for VLPs made with RNA or with DNA oligonucleotides is unclear. Perhaps the VLPs assembled with a heterogeneous mixture of RNAs contain stretches of RNA not bound by protein. Thus, the Gag proteins may be bound in clusters, reflecting independent nucleation events or secondary structures, leaving stretches of the RNA free of protein because the growing particle has reached a maximum density of protein molecules. On the other hand, the difference in A260/A280 ratios may reflect a difference in packing on ribonucleotide as opposed to deoxyribonucleotide polymers. A limitation of quantitative interpretations of the A260/A280 ratios we have measured is that it is impossible to exclude the possibility that nonspecific protein-nucleic acid aggregates contaminated the VLPs. However, EM showed no evidence of such aggregates.
The lack of specificity of the RSV NC protein in promoting assembly in vitro is in accord with the observations that retrovirus NC proteins can bind to diverse nucleic acids (47, 48, 70, 83; reviewed in references 23 and 66) and can act as general RNA chaperones (27, 65, 66, 73). However, sequence-specific in vitro binding of HIV-1 NC or NC-containing proteins to viral sequences (8, 9, 22, 37), as well as to RNAs selected from a pool of random sequences (3, 6, 20, 56), has also been reported. NC also has a preference for some DNA sequences over others (30). In vivo, the NC domain of Gag is inferred to play the central role in the specific recognition of psi, the RNA packaging sequence, and the Cys-His motifs are essential for this packaging function (reviewed in reference 7). However, certain NC mutants with subtle changes in the Cys-His motif still package RNA although they lack infectivity (39). Packaging is also affected by other mutations in NC. Notably, in the RSV system, mutation of two basic residues downstream of the second Cys-His motif dramatically reduces specific RNA packaging (54). This sequence is in the part of NC deleted in our dNC1 and dNC2 constructs, which are competent for assembly in vitro, suggesting that the sequence-specific binding and nonspecific binding are, in part, due to structurally distinct portions of the protein. This notion is also suggested by the solution structures of HIV-1 NC bound to portions of the packaging sequence (4, 26), which show phosphodiester contacts with basic residues in the N-terminal portion of NC upstream of the first Cys-His motif. It is perhaps not coincidental that we observed basic residues in the same portion of RSV NC to be critical for in vitro assembly in the context of deletion mutant proteins dNC1 and dNC2. A full understanding of the sequence-specific and nonspecific properties of NC and the NC domain remains elusive.
The manner in which NC promotes budding in vivo in the absence of genomic RNA clearly is largely nonspecific, just as we have found in vitro. That is, diverse RNAs can be packaged in the absence of any RNAs carrying the packaging sequence psi, and recent evidence suggests that the mass of RNA in particles lacking a genome is similar to the mass in those containing a genome (D. Muriaux and A. Rein, personal communication). The NC domain is not an absolute requirement for budding in vivo, since Gag proteins without NC can bud into the medium of transfected cells, although at lower efficiency than Gag proteins with an NC domain. However, NC-lacking particles have a lower density than proper retrovirus particles, suggesting that NC somehow mediates close packing of Gag molecules (11, 24, 69, 84). The sequence elements in NC that promote proper density are called interaction domains, or I domains, which have been inferred to consist of clusters of basic residues (11, 18, 24, 69, 84). However, the connection between density and basic residues in NC has been challenged recently (17). It remains to be established exactly how I-domain function correlates with assembly in vitro. However, at least in one respect, in vitro assembly is more stringent than budding. A short polybasic peptide, engineered to replace the entire NC domain, can provide I-domain function in RSV (11). A similar finding has been reported for HIV-1 (69). But the same peptide used by Bowzard et al. (11) cannot substitute for NC in our in vitro assembly assay. This discrepancy might be accounted for by Gag-membrane interactions and possibly Gag-host protein interactions in vivo, which themselves aid in the assembly process. Detailed dissection of the amino acid sequences in NC that are involved in VLP formation in vitro, in dense particle formation in vivo, and in specific and nonspecific nucleic acid binding should lead to a better definition of NC function.
Perhaps the most puzzling question about the role of nucleic acid in assembly in vitro is by what mechanism nucleic acid binding leads to polymerization of the Gag protein. We originally hypothesized that by binding to RNA, NC domains gather together the attached CA domains and thereby concentrate them in space, promoting what are otherwise weak CA-CA interactions leading to polymerization of the protein (14). How, then, is an oligonucleotide as small as a 22-mer, which binds only about two Gag molecules, efficiently incorporated into the the VLP? According to one model, NC-nucleic acid interaction leads to a conformational change in the upstream CA and SP domains, facilitating CA-CA interactions. The observation that a dimer-forming coiled-coil domain can substitute for NC in budding assays (2, 84) can be interpreted to support this model and suggests that Gag dimers are the building blocks in retrovirus assembly. In an alternative model, one NC domain forms a bridge between two oligonucleotides, functionally tying them together as if they comprised a contiguous chain. According to this notion, a portion of one NC domain would bind one oligonucleotide and a different portion would bind a second. To help distinguish between these models, it is important to fully characterize the nature and size of the oligonucleotide binding site for the NC domain in the context of proteins like ΔMBDΔPR and of the effect of DNA sequence on binding affinity, as has been done, at least in part, for HIV-1 NC (30).
Mass measurement by STEM and visualization by cryo-EM provide solid evidence that VLPs are an excellent model for immature virions formed in vivo. Since the VLPs are composed of only two components that contribute in a measurable way to VLP mass, the mean mass obtained, 65 MDa, should be reliable to within 5%. The only major uncertainty in calculating Gag stoichiometry comes from the assumption that the particles are intact. In the preparations used for STEM analysis, about 10% of the particles were judged by negative staining to be incomplete, i.e., not completely spherical or with missing sectors. Given the lack of knowledge of orientation on the EM grid and the possibility that small missing sectors would not be discernible, this value is an underestimate. However, since the distribution of mass values does not fall off more sharply at the higher values than at the lower values, it is impossible to say to what degree the calculated 65 MDa, corresponding to 1,200 Gag proteins, underestimates the mass of a complete VLP. Thus, it remains uncertain if Gag stoichiometry in vitro is different from that estimated in vivo, i.e., 1,500 (76). Considering the several assumptions required for the latter calculation, in particular, contents of lipid and of proteins other than Gag, we favor the hypothesis that the stoichiometry in vitro and that in vivo are the same. In this context, it is important to note that the radius measurements of VLPs by cryo-EM are much more tightly clustered than the mass values (and, indeed, are more tightly clustered than the measurements of diameters of infectious RSV [Johnson et al., unpublished data; R. Kingston, unpublished data]). Since radii can be determined even if a particle is not fully intact, the tighter clustering of radius measurements is not inconsistent with the mass determinations.
In vitro assembly with purified components continues to be a promising avenue by which to study the principles of retrovirus assembly. This system is independent of the known and putative cellular factors important for assembly in vivo—for example, chaperonins, membrane lipids, viral and nonviral membrane proteins, and cellular proteins interacting with late domains. The ability to control the concentration of components and the other reaction conditions should facilitate a biochemical dissection of the steps in assembly.
ACKNOWLEDGMENTS
We acknowledge Stephen Campbell for carrying out much of the preliminary analysis of the assembly properties of the ΔMBDΔPR protein; Marc Johnson and Deborah Lynn for critical reading of the manuscript; and Alan Rein, John Wills, Rebecca Craven, and Stephen Campbell for discussions. We thank Michael Rossmann and Timothy Baker (Purdue University) for allowing the cryo-EM to be carried out in their laboratories and Paul Chipman for technical assistance with this work.
The cryo-EM was performed with the support of NIH grants GM33050 (to Timothy Baker) and AI34216 (to Michael Rossmann). The STEM measurements were performed at Brookhaven National Laboratory and were supported by NIH resource center grant P41-RR01777. The work at Cornell University was supported by NIH grant CA20081 (to V.M.V.).
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola M A, Strack B, Göttlinger H G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen P, Collins B, Brown D, Hostomsky Z, Gold L. A specific RNA structural motif mediates high affinity binding by the HIV-1 nucleocapsid protein (NCp7) Virology. 1996;225:306–315. doi: 10.1006/viro.1996.0605. [DOI] [PubMed] [Google Scholar]
- 4.Amarasinghe G K, De Guzman R N, Turner R B, Chancellor K J, Wu Z R, Summers M F. NMR structure of the HIV-1 nucleocapsid protien bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 6.Berglund J A, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berthet-Colominas C, Monaco S, Novelli A, Sibal G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell S. PhD thesis. Ithaca, N.Y: Cornell University; 1997. [Google Scholar]
- 13.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campos-Olivas R, Newman J L, Summers M F. Solution structure of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 17.Cimarelli A, Luban J. Human immunodeficiency virus type virion density is not determined by nucleocapsid basic residues. J Virol. 2000;74:6734–6740. doi: 10.1128/jvi.74.15.6734-6740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cimarelli A, Sandin S, Höglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark P K, Ferris A L, Miller D A, Hizi A, Kim K-W, Deringer-Boyer S M, Mellini M L, Clark A D, Jr, Arnold G F, Lebherz III W B, Arnold E, Muschik G M, Hughes S H. HIV-1 reverse transcriptase purified from a recombinant strain of Escherichia coli. AIDS Res Hum Retroviruses. 1990;6:753–764. doi: 10.1089/aid.1990.6.753. [DOI] [PubMed] [Google Scholar]
- 20.Clever J L, Taplitz R A, Lochrie M A, Polisky B, Parslow T G. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J Virol. 2000;74:541–546. doi: 10.1128/jvi.74.1.541-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to psi RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 24.Dawson L, Yu X-F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 25.Deans S R. Radon and Abel transforms. In: Poularikas A D, editor. The transforms and applications handbook. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 631–717. [Google Scholar]
- 26.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-like RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 27.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M C, Roques B, Darlix J L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutsch M, Beniaminy I. Inversion of Abel's integral equation for experimental data. J Appl Phys. 1983;54:137–143. [Google Scholar]
- 29.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher R J, Rein A, Fivash M, Urbenja M A, Casas-Finet J R, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu X, Katz R A, Skalka A M, Leis J. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein pp12. A mutation which affects RNA binding in vitro blocks viral replication. J Biol Chem. 1988;263:2140–2145. [PubMed] [Google Scholar]
- 32.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 33.Gamble T L, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 34.Gamble T L, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 35.Ganser B K, Li S, Klishdo V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 36.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 37.Geigenmueller U, Linial M L. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J Virol. 1996;70:667–671. doi: 10.1128/jvi.70.1.667-671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 39.Gorelick R J, Fu W, Gagliardi T D, Bosche W J, Rein A, Henderson L E, Arthur L O. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grättinger M, Hohenberg H, Thomas D, Wilk T, Müller B, Kräusslich H-G. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology. 1999;257:247–260. doi: 10.1006/viro.1999.9668. [DOI] [PubMed] [Google Scholar]
- 42.Gross I, Hohenberg H, Kräusslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 43.Gross I, Hohenberg H, Huckhagel C, Kräusslich H-G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gross I, Hohenberg H, Wilk T, Wiegers K, Grättinger M, Müller B, Fuller S, Kräusslich H-G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardt S, Wang B, Schmid M F. A brief description of I. C. E.: the integrated crystallographic environment. J Struct Biol. 1996;116:68–70. doi: 10.1006/jsbi.1996.0012. [DOI] [PubMed] [Google Scholar]
- 46.Joshi S M, Vogt V M. Role of the Rous sarcoma virus p10 domain in shape-determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J Virol. 2000;74:10260–10268. doi: 10.1128/jvi.74.21.10260-10268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karpel R L, Henderson L E, Oroszlan S. Interaction of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 48.Khan R, Giedroc D P. Nucleic acid binding properties of recombinant Zn2 HIV-1 nucleocapsid protein are modulated by COOH-terminal processing. J Biol Chem. 1994;269:22538–22546. [PubMed] [Google Scholar]
- 49.Kingston R, Eisenmesser E Z, Fitzon-Ostendorp T, Schatz G W, Vogt V M, Post C B, Rossmann M G. Structure and self-association of the Rous sarcoma virus capsid protein. Structure. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 50.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immnodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kräusslich H-G, Welker R. Intracellular transport of retroviral capsid components. Curr. Top. microbiol. Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 53.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee E G, Yeo A, Kraemer B, Wickens M, Linial M L. The Gag domains required for avian retroviral RNA encapsidation determined by using two independent assays. J Virol. 1999;73:6282–6292. doi: 10.1128/jvi.73.8.6282-6292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lochrie M A, Waugh S, Pratt-Dickson Jr G, Clever J, Parslow T G, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 Gag polyprotein. Nucleic Acids Res. 1997;25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 58.Morikawa Y, Toshiyuki T, Sano K. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J Biol Chem. 1999;274:27997–28002. doi: 10.1074/jbc.274.39.27997. [DOI] [PubMed] [Google Scholar]
- 59.Ono A, Freed E O. Binding of human immunodeficiency virus type 1 Gag to membrane. Role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parent L J, Bennet R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the RSV and HIV Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parent L J, Wilson C B, Resh M D, Wills J W. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J Virol. 1996;70:1016–1026. doi: 10.1128/jvi.70.2.1016-1026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puffer B A, Watkins S C, Montelaro R C. Equine infectious anemia virus Gag polyprotein late domain specifically recruits the cellular AP-2 adapter protein complexes during virion assembly. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prats A C, Sarih L, Gabus C, Litvak S, Keith G, Darlix J L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rein A, Henderson L E, Levin J G. Nucleic acid chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 67.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakalian M, Hunter E. Separate assembly and transport domains within the Gag precursor of Mason-Pfizer monkey virus. J Virol. 1999;73:8073–8082. doi: 10.1128/jvi.73.10.8073-8082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stallmeyer M J, Aizawa S-I, Macnab R M, DeRosier D J. Image reconstruction of the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1989;205:519–528. doi: 10.1016/0022-2836(89)90223-4. [DOI] [PubMed] [Google Scholar]
- 73.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vest C M. Formation of images from projections: radon and Abel transforms. J Opt Soc Am. 1974;64:1215–1218. [Google Scholar]
- 76.Vogt V M, Simon M N. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy (STEM) J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.You J C, McHenry C S. HIV nucleocapsid protein: expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 84.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]