Abstract
The prototype JHM strain of murine hepatitis virus (MHV) is an enveloped, RNA-containing coronavirus that has been selected in vivo for extreme neurovirulence. This virus encodes spike (S) glycoproteins that are extraordinarily effective mediators of intercellular membrane fusion, unique in their ability to initiate fusion even without prior interaction with the primary MHV receptor, a murine carcinoembryonic antigen-related cell adhesion molecule (CEACAM). In considering the possible role of this hyperactive membrane fusion activity in neurovirulence, we discovered that the growth of JHM in tissue culture selected for variants that had lost murine CEACAM-independent fusion activity. Among the collection of variants, mutations were identified in regions encoding both the receptor-binding (S1) and fusion-inducing (S2) subunits of the spike protein. Each mutation was separately introduced into cDNA encoding the prototype JHM spike, and the set of cDNAs was expressed using vaccinia virus vectors. The variant spikes were similar to that of JHM in their assembly into oligomers, their proteolysis into S1 and S2 cleavage products, their transport to cell surfaces, and their affinity for a soluble form of murine CEACAM. However, these tissue culture-adapted spikes were significantly stabilized as S1-S2 heteromers, and their entirely CEACAM-dependent fusion activity was delayed or reduced relative to prototype JHM spikes. The mutations that we have identified therefore point to regions of the S protein that specifically regulate the membrane fusion reaction. We suggest that cultured cells, unlike certain in vivo environments, select for S proteins with delayed, CEACAM-dependent fusion activities that may increase the likelihood of virus internalization prior to the irreversible uncoating process.
Enveloped virus particles introduce their genetic cargo into organisms by binding to cellular receptors and by the subsequent coalescence of the membranes surrounding virus and cell. Since glycoproteins projecting from the virus envelope perform both of these functions, it is not surprising that paradigms for protein-mediated membrane fusion reactions come from studies of viral glycoprotein structure and function. The current general view is that viral glycoproteins are metastable entities, poised to convert to alternative “fusion-competent” conformations following binding to a cellular receptor(s) and/or endocytosis into acidic intracellular vesicles (5, 6, 13, 47). For many viral glycoproteins, the precise way in which this conversion process is initiated and then completed is unclear. However, a mechanistic understanding is required to identify determinants of viral tropism and pathogenesis (31), to rationally design therapeutic antiviral agents (34), and to develop viruses as gene therapy vectors (33).
Our studies are aimed at understanding how coronaviruses perform these essential cell entry functions. A coronavirus particle projects about 200 spikes (S) from its virion membrane (15)—these projections are essential for receptor binding and membrane fusion. Each spike projection is comprised of oligomeric type I integral membrane glycoproteins. The monomeric units of each oligomer exist either as single-chain proteins or as two similarly sized endoproteolytic cleavage products (see Fig. 2). The cleavage products are designated S1 and S2; S1 is the peripheral fragment and remains noncovalently associated with the membrane-spanning S2 fragment (7). During the initial stages of infection, the peripheral S1 on some of these virion projections engages host cell receptors (37). For the murine hepatitis coronaviruses (MHVs), these receptors are members of the carcinoembryonic antigen family of cell adhesion molecules (CEACAMs) (4, 18). The CEACAMs are integral membrane proteins with immunoglobulin-like ectodomains, and their binding to spikes is thought to induce structural changes that are relevant to the fusion of virus and cell membranes (23, 32, 61). The initial virion-cell fusion reaction may take place at or near the cell surface, as acidification is not additionally required for MHV infection, although there are notable rare isolates that do require endocytosis into acid-pH vesicles for entry (25). Later during infection, another important membrane fusion reaction takes place as S proteins accumulate on infected cell surfaces and engage CEACAM receptors on opposing cells. These important intercellular fusion events generate easily identified syncytia and permit the rapid spread of virus infection.
FIG. 2.
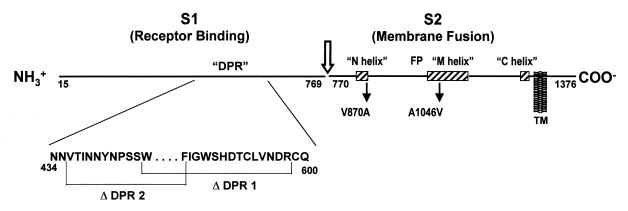
Schematic diagram of the JHM spike protein, delineation of relevant functional regions, and location of mutations eliminating MHVR (CEACAM)-independent fusion activity. Lines depict a single S protein monomer after removal of the signal sequence (residues 1 to 14) and endoproteolytic cleavage into S1 and S2 near the multibasic residues 765 to 769 (arrow). While not depicted here, the native S protein structure contains multiple noncovalent associations between S1 and S2 fragments (29). S1 is the peripheral fragment, and S1 fragments comprising the amino-terminal 330 residues will bind to receptors (37). S2 contains a single membrane-spanning region, designated TM. Membrane fusion is accomplished by S2, which has three regions predicted to form helical coiled coils (58), identified here as the N (amino acids 838 to 872), M (amino acids 1020 to 1124), and C (amino acids 1274 to 1300) helices. The M and C helices match earlier predictions (16), and they are positioned similarly to those found in fusion proteins from other virus families (59). S2 also contains a candidate fusion peptide (FP) region (41), similar to those found in fusion proteins from many other virus families (68). Mutations eliminating receptor-independent fusion include deletions ΔDPR1 and ΔDPR2, whose endpoints are indicated. These two deletions lie within S1 residues 429 to 604, a region known to accommodate omissions ranging from 29 to 159 amino acids (50, 54). S2 point mutations V870A and A1046V also eliminate receptor-independent fusion.
The coronaviruses exhibit considerable serologic and sequence variation (57), with the most extreme variability being within S genes. S protein differences are now known to impact pathogenic outcome (14, 39, 55), and in this regard it is notable that there exists a wide spectrum of coronavirus-induced diseases, including common colds, peritonitis, and gastroenteritis. This diversity in pathogenetic outcome is also evident among the MHV strains. Despite the name, some MHV strains cause gastroenteritis or encephalitis rather than hepatitis (52). This directs attention to mechanisms by which variation in MHV spikes impacts entry into distinct cells and tissues, thereby causing a particular disease pattern.
To begin to address how differences in S protein structure impact virus entry and resulting in vivo pathogenesis, we have focused attention on a particular strain of MHV termed the JHM strain (9). The JHM strain was originally discovered as the causative agent of demyelination and paralysis and was subsequently passaged several times in suckling mouse brain to generate an exceedingly neurovirulent isolate (9, 66). Neurovirulent JHM is unique with respect to its very rapid, ultimately lethal dissemination throughout the murine central nervous system. At the same time, JHM is poorly propagated in tissue culture, and variants of JHM that exhibit superior growth in culture are readily amplified (25, 26). These variants contain spike mutations, and those variants that have been tested have greatly reduced neurovirulence (20, 51), again establishing the link between spike variation and in vivo pathogenesis.
Here we have explored how different JHM-derived spike variants compare with the prototype JHM spikes in their capacities for binding to the murine CEACAM receptor and for their induction of membrane fusion. We found that many of the mutations fixed into spike genes during tissue culture adaptation had little if any effect on spike-receptor binding but that all reduced fusion. Indeed, we demonstrate here that the prototype JHM spikes induced membrane fusion even without the requirement for murine CEACAM binding. However, the tissue culture-adapted mutants could not mediate this type of “spontaneous” fusion reaction. We have further explored the impact of these mutations on the gross structural features of the spike protein. Possible relationships between spike-induced membrane fusion potential and virus entry kinetics are discussed.
MATERIALS AND METHODS
Cells.
HeLa-tTA (27), HeLa-MHVR (53), and rabbit kidney clone 13 (RK13) cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Summit Biotechnology). 293 EBNA:sMHVR-Fc cells (23) were grown in DMEM–10% FBS containing the antibiotics G418 (100 μg/ml) and hygromycin B (200 μg/ml). Murine 17 clone 1 (17 cl 1) fibroblasts were grown in DMEM containing 5% FBS and 5% tryptose phosphate broth (Difco Laboratories). All growth media were buffered with 0.01 M sodium HEPES (pH 7.4). All cell lines were propagated as adherent monolayer cultures.
Viruses.
All coronaviruses were plaque purified on HeLa-MHVR cells, propagated as stocks in 17 cl 1 cells, and purified by differential centrifugation as previously described (26). Vaccinia virus recombinants, each harboring spike cDNA of a particular coronavirus strain, were generated in a stepwise fashion. Total RNA from coronavirus-infected 17 cl 1 cells was isolated by phenol-chloroform extraction, and subsequent cDNA preparation, PCR amplification, and S gene sequencing were performed as previously described (23). PCR restriction fragments containing mutations (relative to JHM) were then used to replace corresponding fragments from the vaccinia virus insertion-expression vector pTM1-SJHM (23). All recombinant pTM1-S plasmids were cloned, amplified in Escherichia coli DH5α, and sequenced to confirm that only the desired S gene mutations were present in the cDNAs. Plasmids were then recombined into the thymidine kinase gene of vaccinia virus (strain WR) by standard methods (43), and the plaque-purified thymidine kinase-negative virus isolates were amplified in RK13 cells. Expression of S genes from these vaccinia virus vectors required bacteriophage T7 RNA polymerase, which was supplied by coinfection with vTF7.3 (22).
Intercellular fusion assays.
The cell fusion-dependent reporter gene (β-galactosidase) activation assay of Nussbaum et al. was adapted to our studies of spike protein-mediated membrane fusion (48). In brief, effector (spike-bearing) cells were generated by coinfection of HeLa-tTA cell monolayers (106 cells/well) with vTF7.3 and respective vTM1-S recombinants. Target cells (containing the β-galactosidase reporter gene) were generated by infection of either HeLa-tTA or HeLa-MHVR no. 5 cells (53) with vCB21R-lacZ (1). All inoculations were for 1 h at 37°C at multiplicities of 2 PFU/cell. After 6 h, target cells were trypsinized, resuspended in DMEM–10% FBS, repelleted, and resuspended in serum-free DMEM (SFM). Tests for the pH dependence of intercellular fusion involved resuspending target cells in SFM lacking HEPES and containing the following buffers at a concentration of 0.025 M: at pHs of 6.0 and 6.5, piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) or morpholineethanesulfonic acid (MES); at pHs of 7.0 and 7.5, HEPES; and at pHs of 8.0 and 8.5, Tris or Bicine. Target cells were then overlaid onto the spike-bearing (effector) cells at an effector/target ratio of 1.0 and were incubated for the indicated time periods at 37°C. Media were then removed and cells were lysed with 0.5 ml of phosphate-buffered saline (PBS) containing 0.5% NP-40/well. β-Galactosidase production was quantified in triplicate 0.05-ml aliquots using a colorimetric assay involving turnover of chlorophenyl red β-galactopyranoside (CPRG).
Preparation of 125I-labeled sMHVR-Fc.
The molecule sMHVR-Fc is a chimeric protein comprising the virus-binding sMHVR domain, also known as the N domain of murine CEACAM1a (4), covalently linked to the Fc of human immunoglobulin G1 (IgG1). The sMHVR-Fc molecule exists as an ∼100-kDa dimer and is constitutively secreted from the transfectant cell line 293 EBNA:sMHVR-Fc (23). T225 flasks containing ∼2 × 107 293 EBNA:sMHVR-Fc cells were incubated in SFM for 2 days, and then the spent media (25 ml/flask) were removed and filtered. To 100 ml of medium, 0.1 ml of Sepharose-protein G beads (Pharmacia) was added, and incubation was continued for 18 h at 4°C. Sepharose beads were pelleted by centrifugation (2,000 × g for 5 min) and were then rinsed with PBS-P (PBS [pH 7.4] containing 0.1% protease inhibitor cocktail [Sigma catalog no. P2714]) via three cycles of resuspension and repelleting. The rinsed bead pellet was resuspended and agitated for 10 min at room temperature in 1 ml of 0.1 M glycine (pH 2.7). Beads were then removed by centrifugation. Eluted sMHVR-Fc was added to 0.1 ml of 1 M Tris-HCl (pH 8.7) and was then dialyzed extensively against PBS-P. This process yielded purified sMHVR-Fc at an ∼0.3-mg/ml concentration, as measured relative to bovine IgG standards in a bicinchoninic acid-based assay (Pierce).
For radioiodination, a highly concentrated (1 mg/ml) preparation of purified sMHVR-Fc was dialyzed against Tris iodination buffer (25 mM Tris-HCl [pH 7.5], 0.4 M NaCl). One hundred micrograms (1 nmol) was then iodinated using 1 mCi of Na125I (Amersham) and IODO-GEN reagent precoated onto iodination tubes (Pierce). Radioiodination, as well as removal of unincorporated 125I by gel filtration, was carried out according to Pierce Company Protocol 28601. Following gel filtration, preparations were mixtures of sMHVR-Fc and bovine serum albumin, a carrier protein added after the iodination reaction. Therefore, specific activity measurements required capturing aliquots of 125I-labeled sMHVR-Fc onto Sepharose-protein G beads (Pharmacia) and subsequent elution of bead-associated proteins at 100°C in sample solubilizer (0.06 M Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 2.5% Ficoll, 0.01% bromphenol blue). 125I-labeled sMHVR-Fc protein concentrations were then determined by Coomassie staining after SDS-polyacrylamide gel electrophoresis in parallel with known amounts of purified sMHVR-Fc. 125I was measured using a Packard Tri-Carb gamma counter.
Binding assays.
Parallel monolayers of HeLa-tTA cells (5 × 105 cells in 5-cm2 wells) were inoculated at 2 PFU per cell with vTF7.3 and the respective vTM1-S recombinant vaccinia viruses. At various times after infection, individual cultures were chilled on ice, rinsed with ice-cold PBS, and then incubated for 2 h at 4°C in PBS containing 1% formalin. Monolayers were then rinsed extensively with PBS to remove formalin and then overlaid with 0.5 ml of HNB (50 mM Na-HEPES [pH 7.0], 100 mM NaCl, 0.01% bovine serum albumin) containing serial dilutions of 125I-labeled sMHVR-Fc (specific activity of 1.5 × 109 cpm/nmol)/well. Culture plates were incubated on rocker platforms for 2 h at room temperature, and then buffer containing unbound sMHVR-Fc was removed. Cell monolayers were rinsed extensively with ice-cold HNB before being dissolved into 0.5 ml of HNB containing 0.5% NP-40 and 0.1% SDS. Each set of binding conditions was carried out in duplicate, and the cell lysate and medium-associated radioactivities in duplicate aliquots were quantitated using a Packard Tri-Carb gamma counter.
Metabolic radiolabeling and immunoprecipitation of S proteins.
Parallel HeLa-tTA cell monolayers (∼106 cells in 10 cm2) were infected with vTF7.3 or with vTF7.3 plus vTM1-S. Cells were incubated from 6.5 to 7.5 h postinfection with methionine- and cysteine-free DMEM containing 1% dialyzed FBS and were then pulse labeled for 10 min with methionine- and cysteine-free DMEM containing [35S]translabel (50 μCi/ml). After extensive rinsing with ice-cold SFM, 1 ml of prewarmed (37°C) chase medium (DMEM–0.1% FBS containing 0.3 mg of methionine/ml and 0.6 mg of cysteine/ml) was then added to each well. After various chase periods, media were removed and saved, and cell sheets were lysed with 1 ml of PBS-P containing 0.5% NP-40/well. Nuclei were pelleted from the cell lysates (10,000 × g at 4°C for 5 min), and 0.5-ml volumes of media or clarified cell extract were incubated with Sepharose-protein G beads (Pharmacia) that had been previously conjugated with sMHVR-Fc. After 16 h at 4°C, beads were pelleted (3,000 × g at 4°C for 10 min) and washed with PBS-P containing 0.5% NP-40 via three cycles of resuspension and pelleting. The final bead pellets were mixed with sample solubilizer and heated to 100°C for 5 min, and 35S-labeled proteins were visualized by fluorography after SDS-polyacrylamide gel electrophoresis.
RESULTS
Differences among MHV strains in syncytium-forming ability: spikes produced by JHM strain infection are uniquely fusion active in the absence of the CEACAM receptor.
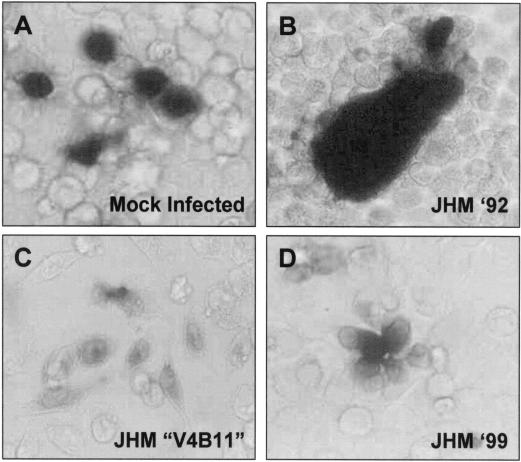
Cells infected with the JHM strain of MHV are powerful mediators of intercellular fusion, so powerful that the prototype receptor, MHVR or murine CEACAM1a (4, 18), is not required on target cells (24). This result is unusual because the murine CEACAM receptor is generally considered essential to the membrane fusion reaction; CEACAM binding to viral spikes may perform a central role in generating a fusion-active spike conformation (23, 32). We set out to further characterize JHM spikes, expecting that we might identify interesting alternative triggers for JHM-specific fusion activation. To begin the studies, a qualitative assay for intercellular fusion was performed. Multinucleated syncytia of JHM-infected murine cells were overlaid with target cells that were devoid of the murine CEACAM. Some of the target cells were made to constitutively express a β-galactosidase gene; thus their fusion with virus-infected cells could be recognized by the expansion of β-galactosidase into syncytia, as measured by in situ turnover of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) substrate (42). In performing these assays, we discovered that our more recent stocks of MHV JHM were incapable of inducing this receptor-independent fusion, while older stocks from 1992 tested positive (Fig. 1). This finding was somewhat remarkable because the 1992 and 1999 stocks were closely related; only five passages in 17 cl 1 cells separated the two. We also discovered that cells infected with many previously characterized isolates derived by serial passage of the prototype JHM were unable to fuse with receptor-negative cells (Fig. 1). This included the JHM-X strain (63) and the V4B11 and V5A13 isolates (26). These findings led us to conclude that CEACAM receptor-independent membrane fusion activity is not stably maintained as JHM viruses are propagated in tissue culture. These findings also made it clear to us that we could identify the genetic basis for murine CEACAM-independent fusion through a comparison of the RNA genomes of different JHM stocks.
FIG. 1.
Qualitative assessments of spike-induced intercellular fusion: only the prototype JHM '92 encodes S proteins that can mediate syncytia with MHVR (murine CEACAM)-negative HeLa cells. Monolayers of MHVR-negative HeLa cells were transfected with pCMV–β-galactosidase to generate ∼1% β-galactosidase-positive cells (A). At 1 day posttransfection, parallel monolayers were overlaid with HeLa-MHVR cells that were infected 4 h earlier (multiplicity of infection 0.1) with the indicated JHM strains (B to D). After 30 h, cells were fixed with formaldehyde and incubated with X-Gal to identify dissemination of β-galactosidase into virus-induced syncytia.
Tissue culture-adapted stocks of JHM have mutations in two different regions of the spike gene.
A straightforward reverse transcriptase PCR and cDNA sequencing approach was initiated to compare the S gene sequences of passaged viruses with those of the previously sequenced JHM (50). We first concentrated on the V4B11 isolate that was tested for Fig. 1. This is a previously characterized isolate that was originally generated by serial passage in the presence of neutralizing anti-S monoclonal antibody 4B11 (12). Our sequencing data confirmed the presence of a large 149-residue deletion in the receptor-binding S1 subunit (Δ437–585) (26, 50). Omissions of this sort in S1 are commonly found in JHM isolates from a number of laboratories—for example, there are JHM-X (Δ446–598) and JHM-Wurzburg (Δ454–594). We have generally found that the tissue culture yields (in PFU) of S1 deletion variants are about 100 times that of the prototype JHM, consistent with their in vitro selection. In this report we refer to this region of S1 that is missing in the tissue culture-adapted variants as the deletion-prone region (DPR). Since no deletions extend beyond residues 429 to 604, we presently designate this segment the consensus DPR of the 1,376-amino-acid S protein (Fig. 2).
We also sequenced reverse transcriptase PCR products derived from the RNA of a single plaque-purified isolate from the 1999 JHM stock that was tested for Fig. 1. Sequencing of the S gene revealed only two relatively conservative missense mutations in this isolate, V870A and A1046V. Both of these changes were within the integral membrane fusion-inducing S2 subunit. Furthermore, both of these changes were within regions identified by the Learn-Coil VMF program (58) as putative amphipathic helical coiled coils. These mutations and the three putative coiled-coil regions of S2 are schematically illustrated in Fig. 2.
Mutant spikes selected after tissue culture adaptation are poor inducers of membrane fusion.
To determine whether the spike mutations identified by sequencing (Fig. 2) would indeed eliminate CEACAM-independent membrane fusion (Fig. 1), we first constructed a series of vaccinia virus recombinants capable of expressing different S genes. These recombinants were designated vTM1-SJHM (complete JHM S gene), vTM1-SΔDPR1 (Δ446–598, JHM-X deletion), vTM1-SΔDPR2 (Δ437–585, V4B11 deletion), and vTM1-SV870A/A1046V. Expression of spike genes from vaccinia virus vectors provides many advantages for studying the structure and function of this membrane fusion protein. First, spike cDNAs are amenable to molecular genetic manipulation, and the mutations identified by sequencing of variant virus cDNAs can be easily inserted into spike cDNAs and then expressed to produce well-defined mutant proteins. Second, the vaccinia virus vectors allow us to synthesize spike proteins in a variety of cell lines that are otherwise resistant to natural infection by coronaviruses. Third, the vectors permit spike synthesis in the absence of other components of coronavirus particles, and this allows spikes to flow from the endoplasmic reticulum to the cell surface without recruitment into intracellular sites of coronavirus assembly (49). Thus, cells infected with the vaccinia virus vectors display spikes abundantly on the plasma membrane, making them potent effectors of intercellular fusion (64).
We expressed the different S genes containing defined mutations in HeLa cells and then tested their potential as “effectors” of intercellular fusion in quantitative assays of the type originally described by Nussbaum et al. (48). In brief, this involved infecting target HeLa cells with vCB21R-lacZ (1), which harbors a transcriptionally silent β-galactosidase reporter gene under T7 promoter control, and then overlaying the targets onto the various spike-bearing effector cells. Since the effector cells were infected with vTF7.3 (22), which encodes T7 RNA polymerase, intercellular fusion with target cells mixes the T7 polymerase with the lacZ reporter gene, and β-galactosidase enzyme is generated.
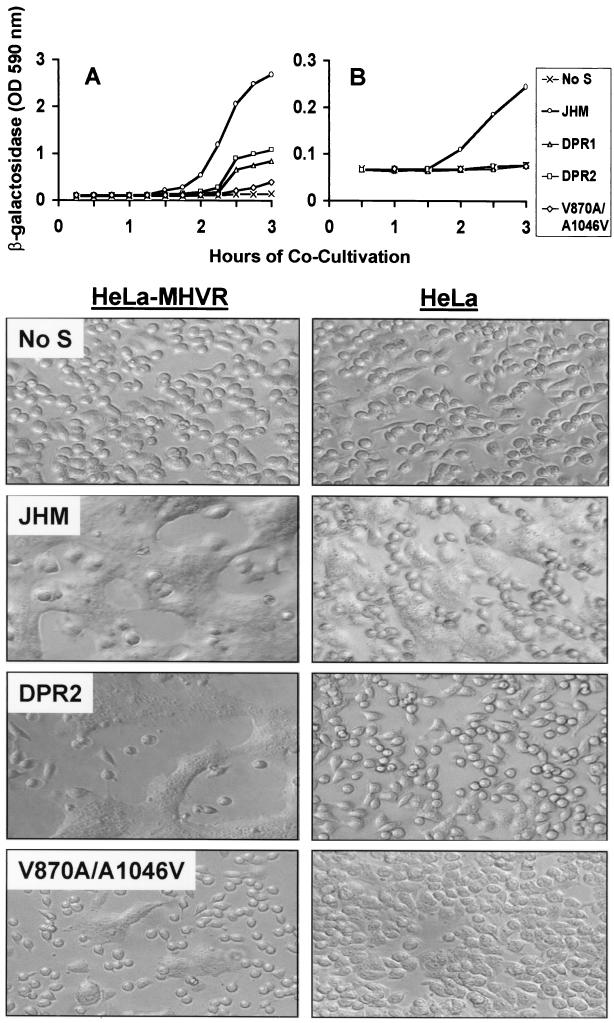
The β-galactosidase enzyme levels in lysates collected at various times after effector/target cell cocultivation facilitated estimating the relative syncytium-inducing potency of each S protein (Fig. 3). When the target cells were HeLa-MHVR, which stably express the prototype murine CEACAM1a gene at levels sufficient to display ∼100,000 receptors per cell (53), membrane fusion was observed for all S proteins (Fig. 3A). The JHM spikes were the most potent effectors, and they promoted fusion most rapidly, ∼30 min sooner than did the parallel SΔDPR proteins. The variant spikes with S2 point mutations (SV870A/A1046V) were by far the least potent effectors of syncytium formation. The data in Fig. 3 are from effector/target cell cocultivations that began at 6 h post-vaccinia virus infection. However, the observation of rapid and powerful SJHM-induced fusion was consistently observed, regardless of whether cocultivations were initiated at 6, 9, or 12 h postinfection (data not shown).
FIG. 3.
Quantitative and qualitative assessment of intercellular fusion mediated by MHV spike proteins. HeLa cell monolayers (106 cells in 10-cm2 dishes) were inoculated with vTF7.3 (No S) or with vTF7.3 plus vTM1-S recombinants encoding JHM, ΔDPR1, ΔDPR2, or V870A/A1046V spikes. At 6 h postinfection, media were removed from parallel cultures and replaced with suspensions of HeLa-MHVR (A) or HeLa (B) target cells that had been infected 6 h earlier with vCB21R-lacZ (1). Each dish received 106 target cells in a volume of 2 ml of SFM (pH 7.5). Cocultivation fuses the S-bearing effector cells with targets, a process which then initiates lacZ gene expression. At the indicated times after cocultivation, media were removed, cells were dissolved in PBS–0.5% NP-40, and β-galactosidase enzyme activities in lysates were determined using a colorimetric assay involving spectrophotometric quantitation of CPRG at 590 nm. The photographs below each graph reveal the extent of syncytium formation after 3 h of cocultivation. OD, optical density.
To assess the requirement for the MHV receptor in these intercellular fusion reactions, we replaced HeLa-MHVR target cells with the parent HeLa cell line and repeated the cocultivation (fusion) assays. Our results indicated that SJHM proteins induced fusion with HeLa cells that was ∼10% of that observed with HeLa-MHVR targets (Fig. 3B). This level of fusion with HeLa cells was variable; in 12 independent assays, the level of reporter gene activation after 3 h of cocultivation with HeLa targets ranged from 5 to 21% (average, 8%) of that observed with HeLa-MHVR targets. However, in every assay performed, effector cells displaying mutant S proteins were incapable of generating β-galactosidase on cocultivation with HeLa cells (Fig. 3B). Results of this sort were additionally documented by inspection of cultures for syncytia (Fig. 3, lower panels). These results conclusively demonstrated that hyperactive (murine CEACAM-independent) fusion activity is unique to SJHM and that the deletions in S1 or the two point mutations in S2 will eliminate this property.
Effect of spike mutations on interaction with the prototype MHV receptor.
The extraordinarily powerful fusion activity of SJHM might be explained by its relatively high presentation on the surface of effector cells or, in the case of fusion with HeLa-MHVR cells, by its relatively high affinity for the MHV receptor. To address these possibilities, a series of binding assays were performed in which 125I-labeled sMHVR-Fc was incubated with S-bearing effector cells. The sMHVR-Fc molecule is a hybrid protein that is comprised of the spike-binding N domain of murine CEACAM1a fused to the Fc portion of human IgG1 (19, 23). Interaction of spikes with sMHVR-Fc is considered to be a relevant mimic of the authentic binding between spikes and integral membrane MHV receptors.
To perform the binding assays, adherent HeLa cell cultures displaying S proteins were chilled at 12 h post-vaccinia virus infection to halt exocytic transport and to stabilize the S1-S2 complexes that had reached the plasma membrane. Parallel cultures were then rinsed, incubated at 4°C with 1% formalin in PBS, and then rinsed again before application of increasing amounts of 125I-labeled sMHVR-Fc. This formalin fixation prevented undesired detachment of cells from plastic and had no effects on the spike–sMHVR-Fc interaction, which reached equilibrium within 2 h at room temperature (data not shown).
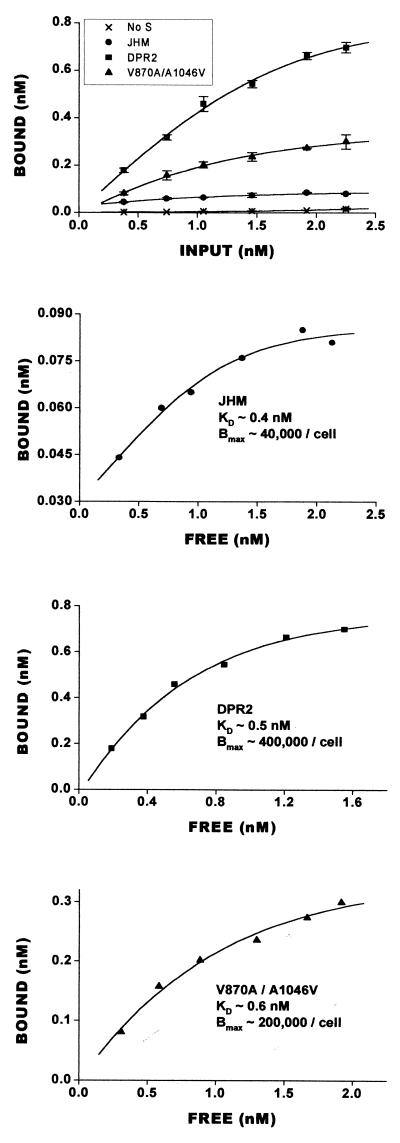
The results of the binding assays are presented in Fig. 4. In sharp contrast to earlier suspicions, the surface levels of SJHM proteins were far lower than those of any of the other mutant S proteins. The sMHVR-Fc radioligand began to exhibit saturable binding on SJHM cultures at ∼0.085 nM, which corresponds to ∼40,000 sMHVR-Fc molecules per cell. The monolayers displaying SV870A/A1046V and SΔDPR2 maximally bound about 5 and 10 times more sMHVR-Fc (∼200,000 and ∼400,000 molecules per cell), respectively. Similar differences were obtained upon sMHVR-Fc binding to cultures fixed at earlier times (8 and 10 h) postinfection (data not shown). Therefore, the powerful fusion activity of SJHM occurs even though its surface presentation is relatively low.
FIG. 4.
Quantitation of sMHVR-Fc binding to HeLa cells displaying S proteins. A series of HeLa cell cultures (5 × 105 cells in 5-cm2 dishes) were infected with vTF7.3 (no S) or with vTF7.3 plus one of the vTM1-S recombinants (JHM, DPR2, or V870A/A1046V). At 12 h postinfection, cells were rinsed with ice-cold PBS and fixed with 1% formalin in PBS. 125I-labeled sMHVR-Fc (1.5 × 109 cpm/nmol) was diluted in HNB buffer to the indicated concentrations, and 0.5-ml volumes were then added to the formalin-fixed cells. After a 2-h incubation at 22°C, unbound (free) 125I-labeled sMHVR-Fc was removed. Cells were rinsed extensively with ice-cold HNB buffer and were then dissolved in HNB–0.5% NP-40–0.1% SDS. Bound and free radioactivities in 0.05-ml aliqouts were counted in a Packard Tri-Carb gamma counter. Error bars represent mean standard deviations (n = 4). Bmax, maximal binding (in molecules/cell).
All of the S proteins under investigation exhibited similar affinities for sMHVR-Fc. This was evident from estimating the concentrations of free 125I-labeled sMHVR-Fc under conditions of half-maximal binding (Fig. 4B to D). Kd values were in the ∼0.5 nM range, which is similar to the affinity of human immunodeficiency virus (HIV) gp120 for sCD4-IgG (46), a soluble form of the HIV receptor that is structurally analogous to the sMHVR-Fc used in this study. Our results suggest that the potency of SJHM-induced fusion with HeLa-MHVR cells (Fig. 3) cannot be attributed to an increased affinity of this particular spike for the receptor.
Mutant spikes selected after tissue culture adaptation maintain relatively stable S1-S2 heteromeric association.
The relatively poor capture of 125I-labeled sMHVR-Fc onto cells presenting SJHM could be due to inefficient SJHM transport to the plasma membrane, low cell surface stability of the SJHM proteins, or both. These possibilities were investigated by monitoring the posttranslational fate of different S proteins via metabolic radiolabeling experiments. We pulse labeled infected HeLa cells producing the S proteins with [35S]methionine and chased (without radioactivity) and then solubilized cells at hourly intervals with nonionic detergent. The S proteins were immunoprecipitated with either polyclonal anti-S serum or with sMHVR-Fc and were then visualized by fluorography following SDS-polyacrylamide gel electrophoresis.
The electrophoretic profiles of S proteins immunoprecipitated by sMHVR-Fc are depicted in Fig. 5. The profiles obtained from immunoprecipitation with polyclonal antiserum are not shown because they paralleled those depicted in all but one parameter. Newly synthesized S proteins were not efficiently captured by sMHVR-Fc (see cell lanes for 0 h in all panels), but they were captured by polyclonal serum. This was because the receptor-binding site formed ∼30 min after synthesis, concomitant with S protein oligomerization within the endoplasmic reticulum (D. N. Lewicki and T. M. Gallagher, manuscript in preparation).
FIG. 5.
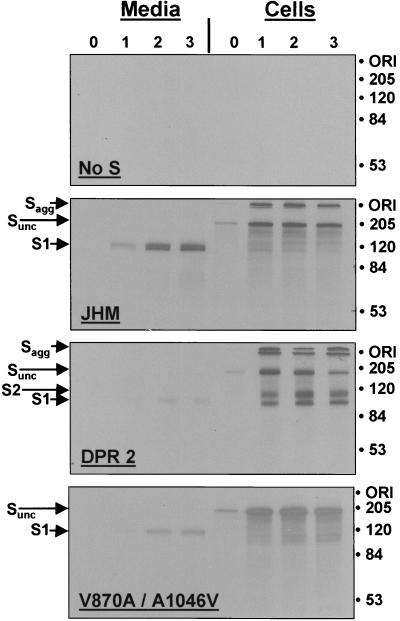
Electropherograms depicting the stability of S1-S2 interactions among S variants. HeLa-tTA cells infected with vaccinia-S recombinant viruses were pulse labeled with [35S]methionine and/or [35S]cysteine from 7.5 to 7.7 h postinfection and were then further incubated (chased) for various times in media containing unlabeled amino acids. At hourly intervals, media (1 ml per culture) were collected and remaining adherent cells were solubilized in media containing 0.5% NP-40 (1 ml per culture). Radiolabeled S proteins from 0.25-ml aliquots were bound to Sepharose-protein G beads conjugated with sMHVR-Fc, and the adsorbed 35S-labeled proteins were identified after SDS-polyacrylamide gel electrophoresis and fluorography. Lanes in each panel represent the 35S-labeled proteins precipitated after 0, 1, 2, and 3 h of chase. Molecular masses are indicated in kilodaltons. ORI, origin of resolving gel. Sagg, SDS-resistant aggregates of S proteins. Sunc, uncleaved precursors.
The electrophoretic profiles revealed additional important characteristics of S proteins. First, similar levels of the different S proteins were immunoprecipitated as the chase progressed, indicating that the various mutations did not impact S protein biosynthesis. Second, SDS-resistant aggregates of S proteins were observed (see Sagg bands in Fig. 5), and these aggregates were absent in spikes with the V870A/A1046V mutations. Third, the major cell-associated S proteins were high-molecular-weight ∼180-kDa chains. These are uncleaved precursors (Sunc), and their presence throughout the 3-h chase indicated that only a subset of spikes transported successfully to Golgi-localized sites of endoproteolysis (7). This data supported the view that all of the S proteins were similarly effective in transport through the exocytic pathway, although it is reasonable to suspect that minor differences would not be revealed by these tests.
The major effect of the S mutations was revealed by recovery of peripheral S1 posttranslational cleavage products in the various immunoprecipitates. In cultures synthesizing SJHM, the S1 cleavage products were found exclusively in the media, with no clear evidence of cell-associated S1. This pattern contrasted most sharply with the pattern from cells producing SΔDPR2, where the deletion mutant S1ΔDPR2 was primarily cell associated. In cultures producing SV870A/A1046V, the presence of cell-associated S1 was somewhat difficult to identify, although relative to SJHM there was clearly more radioactive protein in a position consistent with S1, and significantly less S1 was captured from medium samples. Therefore, the collective findings indicate that the various S proteins assemble into oligomers with similar speed and efficiency but that only a minor fraction of these oligomers advance into mature S1 and S2 cleavage products within 3 h. For SJHM, these mature S1 and S2 cleavage products separate without delay, and S1 is therefore recovered in culture media. For SΔDPR2, and to a lesser extent for SV870A/A1046V, the S1 and S2 cleavage products remained associated and both were recovered from cell extracts. Thus, under the physiological conditions employed in these experiments (37°C media at pH 7.5), the S1 deletion and the double S2 point mutation stabilized the S1-S2 hetero-oligomer and increased its life span on the infected cell surface. All of these observations were consistent with the finding that cells producing the variant S proteins bound relatively high levels of sMHVR-Fc (Fig. 4).
Consideration of JHM-specific alternative receptors.
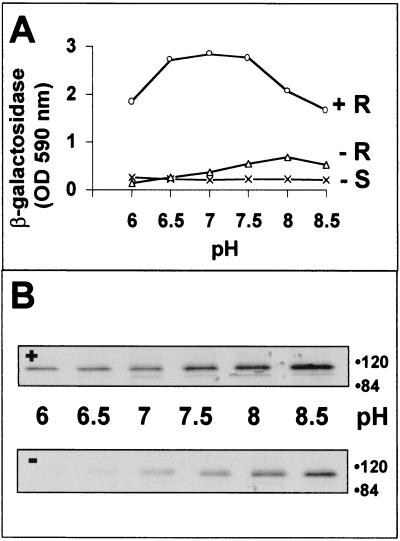
One explanation for the extended membrane fusion activity of SJHM with human (HeLa) cells appeals to the existence of a receptor(s) on nonmurine cells that is specifically recognized by JHM spikes (32). These putative receptors may trigger SJHM-induced membrane fusion. Indeed, a growing body of evidence indicates that some MHV strains can utilize nonmurine CEACAMs for cross-species transmission of infection (3, 10, 30, 35, 56). However, at present we have no evidence for the involvement of JHM-specific HeLa cell receptors in our fusion assays; we have never observed any selective binding of JHM virions to our HeLa target cells. We favor an alternative explanation for our membrane fusion data (Fig. 3B), in which we suggest that JHM spikes are highly unstable proteins that simply do not require the free energy of receptor binding to transit into the fusion-active conformation. This is not to say that there is no trigger for JHM spikes to convert into fusion-active forms. Indeed, we have found that CEACAM-independent fusion responds to changes in extracellular pH, a condition that is known to dramatically alter spike protein conformation (61). While CEACAM-induced fusion by SJHM was largely pH independent, CEACAM-independent fusion was only observed in cultures incubated at pH values above ∼7.0 (Fig. 6A). This fusion activity at elevated pH correlated with S1 separation from S2. When SJHM-bearing cells were first incubated with sMHVR-Fc at 4°C and then shifted to 37°C at various pH values, S1 release occurred, and this S1 separation increased at basic pH (Fig. 6B, + gel). Without preincubation with sMHVR-Fc, S1 was still released from the SJHM-bearing cells, but the extent of release was lower and was prominent only at basic pH (Fig. 6B, − gel). We conclude that elevated pH can to some extent replace receptor interaction as a mediator of both S1-S2 separation and fusion activation.
FIG. 6.
Effect of pH on SJHM-induced fusion activation (A) and on S1 separation from cells (B). (A) HeLa (−R) and HeLa-MHVR (+R) target cells were suspended in ice-cold SFM adjusted to the indicated pH values and were then deposited onto S-bearing effector cells by centrifugation. After a 3-h, 37°C cocultivation period, all cells were dissolved and β-galactosidase enzyme activites were determined and plotted. (B) Release of peripheral S1 fragments into media was performed. At 12 h postinfection, S-bearing HeLa cells were radiolabeled for 2 h with [35S]translabel, rinsed extensively, and then incubated for 3 h at 4°C in SFM alone (−) or in SFM containing 10 μg of sMHVR-Fc/ml (+). Cell monolayers were rinsed and were then incubated for 1 h at 37°C in buffered DMEM–0.1% FBS adjusted to the indicated pH values. Medium samples were collected and 35S-labeled S1 fragments were adsorbed onto Sepharose–sMHVR-Fc beads and visualized by fluorography after SDS-polyacrylamide gel electrophoresis. Molecular masses are given in kilodaltons. Only the relevant portions of the fluorograms are depicted.
CEACAM-independent membrane fusion by JHM spikes also required temperatures above 30°C (data not shown). Interestingly, recent fusion assays performed at highly elevated temperatures revealed that a spike point mutant (SA1046V) that we had deemed incompetent for CEACAM-independent fusion can induce syncytium formation on HeLa cell targets at temperatures above 42°C (E. Thorp and T. M. Gallagher, unpublished). These findings bring us to a theory in which many of the coronavirus spikes can mediate a membrane fusion reaction without CEACAM binding, if a sufficiently high temperature is provided to create the requisite structural changes. In this context, the JHM spikes that we have set apart as uniquely CEACAM independent would actually represent an extreme case in which the mild conditions of slightly basic pH and physiologic temperature are sufficient for the fusion reaction.
DISCUSSION
The JHM strain of MHV was selected for rapid growth within the murine central nervous system. After more than 50 serial passages in suckling mouse brain, JHM was isolated and investigated as an agent of neuropathogenesis (9, 66). The spike glycoproteins of this enveloped virus can promote an unusual intercellular membrane fusion activity that is not dependent on either acid pH exposure or binding to the murine CEACAM1a receptor. Therefore, when JHM spikes are displayed on infected-cell surfaces, adjacent cells need not express the receptor to be recruited into syncytia. This characteristic may contribute to the rapid, disseminated, and lethal panencephalitis that is unique to JHM virus infection (20, 51). In summary, the prototype JHM spikes are well suited for rapid, indiscriminate cell-cell membrane fusion.
When stocks of MHV strain JHM are generated by growth in tissue culture, variants are selected that possess a more tempered cell-cell fusion activity. A reasonable explanation for this selection may rest on the fact that some of our tissue culture cells, in particular 17 cl 1 cells, are relatively resistant to JHM-induced syncytium formation. This prompts us to speculate that prototype JHM virions may release the hyperactive fusion potential within their spikes on 17 cl 1 cell surfaces in irreversible and unproductive reactions. On serial passage in these cells, these JHM viruses are replaced by variant progeny that are less prone to releasing their potential energy in futile cell surface reactions. These variant viruses would instead release their membrane fusion potential at later times in the infection cycle, most likely after virus internalization and perhaps in response to acidification in the endosome. In summary, the variant spikes may be well suited for delayed, CEACAM- or acid-dependent virus-cell fusion reactions, and this activity may well support efficient infection of tissue culture cells by virions.
S1 deletion variants: delayed membrane fusion reactions.
In support of our views on coronavirus variation and selection, we routinely observed that spikes from the tissue culture-adapted S1 deletion variants (SΔDPR) were relatively slow in mediating intercellular fusion, ∼30 min slower than prototype SJHM spikes (Fig. 3A). This delay could not be attributed to a relatively low affinity of SΔDPR for MHV receptors, nor could it be due to poor presentation of SΔDPR on cell surfaces—variant spikes were in fact far more abundant on effector cells than was SJHM (Fig. 4). These results suggest that the efficiency of S1 deletion variant viruses in tissue culture is related to their delay of the fusion reaction. Interestingly, internalization of MHV (strain A59) into murine L2 cells was completed within 40 min (36), and we hypothesize that this endocytic process precedes the fusion reactions brought about by S1 deletion variant spikes. Such temporal regulation may improve the efficiency by which virions deliver genomes to the cytosol.
The deletion of residues within the central portion of S1 also made spike-induced fusion entirely dependent on MHV receptor (CEACAM1a) binding (Fig. 3B). One way of interpreting this finding is to suggest that this DPR can change the energy barrier between native and fusion-active S conformations such that the free energy released upon CEACAM binding becomes critical to the reaction. It is not yet clear how the DPR might affect S protein conformation, but it is notable that functional homologs of this region may exist in spike proteins of retroviruses. A proline-rich region exists within the SU portion of murine leukemia virus, and mutations in this region affect membrane fusion (38), modulating envelope protein function similarly to the way that S1 deletions impact coronavirus S protein functions. It is furthermore remarkable that mutations in the retrovirus proline-rich region also change the stability of the SU-TM heteromeric interaction (28), similar to the increased S1-S2 stability seen upon removal of the S1 DPR (Fig. 5). Although atomic structures for the retroviral proline-rich regions are not yet available, existing models suggest that the residues fold as a series of beta turns, to generate so-called polyproline beta-turn helices (21). The DPR of the coronavirus spike is also rich in proline and glycine, with 10 predicted beta turns within the ∼150-amino-acid consensus deletion region (11). This region is relatively hydrophilic and accessible to antiviral antibodies (26), leading to a view of the DPR as a surface-exposed removable module. This module regulates the membrane fusion reaction without noticeably altering the CEACAM-binding site.
S2 variants: a move toward acid pH-dependent membrane fusion reactions.
Spike protein-mediated membrane fusion is influenced by pH, and while some studies indicate increased fusion at elevated pH (61), other reports point toward endosome acidification as a component of MHV entry (44). Earlier studies of tissue culture-adapted JHM variants correlated point mutations within the putative M helix of S2 (Q1067H, Q1094H, L1114R) (Fig. 2) with a strong conversion to acid pH-dependent fusion activity (25). Such changes ensure that the membrane fusion reaction is delayed to a time after virus internalization into acidic endosomes. The V870A and A1046V mutations identified in this report greatly reduce intercellular fusion activity (Fig. 3A). Very recent findings have revealed that this low fusion potential of SV870A/A1046V is actually increased by acidification (Thorp and Gallagher, unpublished), making it likely that we have identified yet another way that S2 can change to create an acid pH sensor for the fusion reaction.
It is interesting that the mutations identified in this report, V870A and A1046V, both fit within regions predicted to form alpha-helical coiled coils (58; see also Fig. 2). In attempts to provide context, we favor a possibility in which all three of these putative helical regions collapse into a low-energy coiled-coil core during the process of membrane coalescence. This general view of stable coiled-coil formation upon membrane connection is in keeping with existing models of protein-mediated membrane fusion (65, 67). As for the effects of the alanine-valine exchanges, we note that changes in the bulk or the hydrophobicity of side chains can impact coiled-coil generation or stability (40) or alter coil conformations (45). Thus, our current aims are to determine whether alpha-helical coiled coils form in S2 fragments and whether the alanine-valine substitutions change the pH dependence of their formation.
Role of S1 separation from S2 in the fusion reaction.
Relative to the prototype JHM, spike proteins of the tissue culture-adapted variants maintained a stable S1-S2 association. This was first inferred by the relatively high levels of 125I-labeled sMHVR-Fc bound to cells displaying SΔDPR2 and SV870A/A1046V (Fig. 4) and was subsequently confirmed by monitoring the posttranslational fate of 35S-labeled spike proteins. In these studies of metabolically labelled spikes, most of the S1JHM was recovered from culture media in the absence of S2, while the majority of S1ΔDPR2 was recovered from cell lysates in conjunction with S2 (Fig. 5).
These results are remarkable in revealing the extreme instability of the SJHM proteins. That these proteins might have remained stable during transport to the plasma membrane is suggested by S1-S2 integrity at pHs of 6.0 to 6.5 (Fig. 6), values equivalent to that found in the Golgi and trans-Golgi network (17, 69). This raises the possibility that stable S1-S2 heteromers might transit through acidic intracellular organelles and then encounter the elevated pH that triggers S1 elution at the cell surface. This would then induce a CEACAM-independent membrane fusion reaction (Fig. 6). The continuous presentation of recently synthesized SJHM at infected cell surfaces would maintain this spontaneous membrane fusion activity.
It is important to remember that this correlation between S1 elution and rapid, CEACAM-independent membrane fusion does not imply that the reaction requires complete separation of S1 from S2. It is known that S proteins rendered uncleavable through site-directed mutagenesis will reach infected-cell surfaces and never elute any S1, yet they will mediate membrane fusion (60, 62). Validating these results, we found that uncleavable forms of SJHM were fully capable of both receptor-triggered and high-pH-triggered membrane fusion (data not shown). We therefore suggest that the membrane fusion reaction involves subtle displacements of S1 from S2 and that the total separation of all S1-S2 contacts is, in some cases, merely an outcome of this earlier displacement process.
This S1 displacement hypothesis is consistent with prevailing models of virus-induced membrane fusion. Models of structural transitions in HIV gp120/41, influenza virus HA1/HA2, and paramyxovirus F2/F1 all include a displacement of peripheral subunits, which allows underlying integral subunits to change conformation and link opposing membranes via extended alpha-helical coiled coils (2, 5, 8). For these proteins as well as for the MHV spikes, displacement of peripheral fragments may have to proceed along a precise pathway to activate fusion. For example, SΔDPR2 may proceed along an unproductive pathway, as S1ΔDPR2 does indeed elute to some extent from S2 (Fig. 5), yet CEACAM-independent fusion does not take place (Fig. 3B). We suggest that there are alternative pathways of conformational change that can end in S1-S2 separation but that only a subset of these pathways proceeds through the S1 displacement process that produces membrane fusion.
Role of the murine CEACAM receptor in the fusion reaction.
While changes in membrane fusion potential were observed after tissue culture adaptation, the binding sites for the prototype CEACAM1a receptor by contrast remain unaffected (Fig. 4). This maintenance of the receptor-binding site is perhaps not surprising, because the tissue cultures that we have used for JHM isolation and growth are HeLa-MHVR and 17 cl 1, both of which produce the prototype murine CEACAM1a receptors. There is apparently little pressure to amplify mutants with altered receptor specificity. This issue brings up a related question, regarding the possibility that JHM spikes have a unique receptor specificity which we have not identified in this study. Indeed, the promiscuous CEACAM1a-independent fusion activity of JHM spikes has generated suggestions that they may uniquely recognize alternative, as yet undiscovered, receptors that act as specific fusion triggers (32). However, to appeal for the existence of an alternative receptor(s) that specifically recognizes JHM spikes and triggers fusion, one must argue that two disparate types of spike variation, S1 deletions and S2 point mutations, would each render this hypothetical receptor unrecognizable. This elimination of alternative receptor-binding site(s) would have to occur without any effect on the binding site for the primary CEACAM1a receptor (Fig. 4). The hypothetical JHM-specific receptor would also have to trigger membrane fusion exclusively at elevated pH (Fig. 6) and at temperatures above 30°C. The alternative receptors would have to exist on rabbit (RK13) and hamster (BHK) cells as well as human (HeLa) cells, as these cell types also formed syncytia in response to SJHM (24). While all of these conditions might be met by a novel alternative receptor(s), we favor a view of JHM spike-induced membrane fusion that is not dependent on any receptor binding.
It appears then that the JHM strain of MHV has adapted to a lifestyle involving intercellular dissemination of infection via syncytium formation. In this context the instability of the SJHM proteins on the cell surface is not a serious disadvantage—all that is necessary is that some of the proteins continuously reach the surface near an adjacent cell and then rapidly promote membrane fusion. This type of intercellular spread to cells lacking the MHV receptor may in fact be advantageous in vivo. In other contexts, such as the tissue culture environment, serial infections may be most effective with extracellular virions containing relatively stable spikes. Virions with these stabilized spikes have the alternative advantage of delaying irreversible fusion reactions to times after internalization.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant R01-NS-31616.
REFERENCES
- 1.Alkhatib G, Broder C C, Berger E A. Cell-type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 3.Baric R S, Sullivan E, Hensley L, Yount B, Chen W. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J Virol. 1999;73:638–649. doi: 10.1128/jvi.73.1.638-649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauchemin N, Draber P, Dveksler G, Gold P, Gray-Owen S, Grunert F, Hammarstrom S, Holmes K, Karlsson A, Kuroki M, Lin S-H, Lucka L, Najjar S, Neumaier M, Obrink B, Shively J, Skubitz K, Stanners C, Thomas P. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp Cell Res. 1999;252:243–249. doi: 10.1006/excr.1999.4610. [DOI] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh D. The coronavirus surface glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 73–115. [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Cheever F S, Daniels J B, Pappenheimer A M, Bailey O T. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin: isolation and biological properties of the virus. J Exp Med. 1949;90:181–194. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D S, Asanaka M, Chen F S, Shively J E, Lai M M C. Human carcinoembryonic antigen and biliary glycoprotein can serve as mouse hepatitis virus receptors. J Virol. 1997;71:1688–1691. doi: 10.1128/jvi.71.2.1688-1691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 12.Dalziel R G, Lampert P W, Talbot P J, Buchmeier M J. Site-specific alteration of murine hepatitis virus type 4 peplomer glycoprotein E2 results in reduced neurovirulence. J Virol. 1986;59:463–471. doi: 10.1128/jvi.59.2.463-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das Sarma J, Fu L, Tsai J C, Weiss S R, Lavi E. Demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J Virol. 2000;74:9206–9213. doi: 10.1128/jvi.74.19.9206-9213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies H A, Macnaughton M R. Comparison of the morphology of three coronaviruses. Arch Virol. 1979;59:25–33. doi: 10.1007/BF01317891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Groot R J, Luytjes W, Horzinek M C, van der Zeijst B A M, Spaan W J M, Lenstra J A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demaurex N, Furuya W, D'Souza S, Bonifacino J S, Grinstein S. Mechanism of acidification of the trans-Golgi network: in situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J Biol Chem. 1998;273:2044–2051. doi: 10.1074/jbc.273.4.2044. [DOI] [PubMed] [Google Scholar]
- 18.Dveksler G S, Pensiero M N, Cardellichio C B, Williams R K, Jiang G-S, Holmes K V, Dieffenbach C W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dveksler G S, Pensiero M N, Dieffenbach C W, Cardellichio C B, Basile A A, Elia P E, Holmes K V. Mouse coronavirus MHV-A59 and blocking anti-receptor monoclonal antibody bind to the N-terminal domain of cellular receptor MHVR. Proc Natl Acad Sci USA. 1993;90:1716–1720. doi: 10.1073/pnas.90.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazakerley J K, Parker S E, Bloom F, Buchmeier M J. The V5A13.1 envelope glycoprotein deletion mutant of mouse hepatitis virus type-4 is neuroattenuated by its reduced rate of spread in the central nervous system. Virology. 1992;187:178–188. doi: 10.1016/0042-6822(92)90306-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontenot J D, Tjandra N, Ho C, Andrews P C, Montelaro R C. Structure and self assembly of a retrovirus (FeL V) proline rich neutralization domain. J Biomol Struct Dyn. 1994;11:821–837. doi: 10.1080/07391102.1994.10508035. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher T M. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J Virol. 1997;71:3129–3137. doi: 10.1128/jvi.71.4.3129-3137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallagher T M, Buchmeier M J, Perlman S. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology. 1992;191:517–522. doi: 10.1016/0042-6822(92)90223-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher T M, Escarmis C, Buchmeier M J. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J Virol. 1991;65:1916–1928. doi: 10.1128/jvi.65.4.1916-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallagher T M, Parker S E, Buchmeier M J. Neutralization-resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990;64:731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosse B, Siddell S G. Single amino acid changes in the S2 subunit of the MHV surface glycoprotein confer resistance to neutralization by S1 subunit-specific monoclonal antibody. Virology. 1994;202:814–824. doi: 10.1006/viro.1994.1403. [DOI] [PubMed] [Google Scholar]
- 30.Hensley L E, Baric R S. Human biliary glycoproteins function as receptors for interspecies transfer of mouse hepatitis virus. Adv Exp Med Biol. 1998;440:43–52. doi: 10.1007/978-1-4615-5331-1_6. [DOI] [PubMed] [Google Scholar]
- 31.Hensley L E, Holmes K V, Beauchemin N, Baric R S. Virus-receptor interactions and interspecies transfer of a mouse hepatitis virus. Adv Exp Med Biol. 1998;440:33–41. doi: 10.1007/978-1-4615-5331-1_5. [DOI] [PubMed] [Google Scholar]
- 32.Holmes K V, Dveksler G S. Specificity of coronavirus/receptor interactions. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 403–443. [Google Scholar]
- 33.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 34.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 35.Koetters P J, Hassanieh L, Stohlman S A, Gallagher T M, Lai M M C. Mouse hepatitis virus strain JHM infects a human hepatocellular carcinoma cell line. Virology. 1999;264:398–409. doi: 10.1006/viro.1999.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kooi C, Mizzen L, Alderson C, Daya M, Anderson R. Early events of importance in determining host cell permissiveness to mouse hepatitis virus infection. J Gen Virol. 1988;69:1125–1135. doi: 10.1099/0022-1317-69-6-1125. [DOI] [PubMed] [Google Scholar]
- 37.Kubo H, Yamada Y K, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavillette D, Maurice M, Roche C, Russell S J, Sitbon M, Cosset F-L. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol. 1998;72:9955–9965. doi: 10.1128/jvi.72.12.9955-9965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leparc-Goffart I, Hingley S T, Chua M M, Phillips J, Lavi E, Weiss S R. Targeted recombination within the spike gene of murine coronavirus mouse hepatitis virus-A59: Q159 is a determinant of hepatotropism. J Virol. 1998;72:9628–9636. doi: 10.1128/jvi.72.12.9628-9636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu M, Ji H, Shen S. Subdomain folding and biological activity of the core structure from human immunodeficiency virus type 1 gp41: implications for viral membrane fusion. J Virol. 1999;73:4433–4438. doi: 10.1128/jvi.73.5.4433-4438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Z, Weiss S R. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology. 1998;244:483–494. doi: 10.1006/viro.1998.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacGregor G R, Nolan G P, Fiering S, Roederer M, Herzenberg L. Use of E. coli lacZ as a reporter gene. In: Murray E J, editor. Gene transfer and expression protocols. Vol. 7. Clifton, N.J: Humana Press; 1991. [Google Scholar]
- 43.Mackett M, Smith G L, Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizzen L, Hilton A, Cheley S, Anderson R. Attenuation of murine coronavirus infection by ammonium chloride. Virology. 1985;142:378–388. doi: 10.1016/0042-6822(85)90345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monera O D, Zhou N E, Lavigne P, Kay C M, Hodges R S. Formation of parallel and antiparallel coiled-coils controlled by the relative positions of alanine residues in the hydrophobic core. J Biol Chem. 1996;271:3995–4001. doi: 10.1074/jbc.271.8.3995. [DOI] [PubMed] [Google Scholar]
- 46.Moore J P, McKeating J A, Huang Y, Ashkenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mothes W, Boerger A L, Narayan S, Cunningham J M, Young J A T. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 48.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Opstelten D-J E, Raamsman M J B, Wolfs K, Horzinek M C, Rottier P J M. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker S E, Gallagher T M, Buchmeier M J. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein of several strains of murine hepatitis virus. Virology. 1989;173:664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearce B D, Hobbs M V, McGraw T S, Buchmeier M J. Cytokine induction during T-cell-mediated clearance of mouse hepatitis virus from neurons in vivo. J Virol. 1994;68:5483–5495. doi: 10.1128/jvi.68.9.5483-5495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perlman S. Pathogenesis of coronavirus-induced infections. Adv Exp Med Biol. 1998;440:503–513. [PubMed] [Google Scholar]
- 53.Rao P V, Gallagher T M. Intracellular complexes of viral spike and cellular receptor accumulate during cytopathic murine coronavirus infections. J Virol. 1998;72:3278–3288. doi: 10.1128/jvi.72.4.3278-3288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowe C L, Baker S C, Nathan M J, Fleming J O. Evolution of mouse hepatitis virus: detection and characterization of spike deletion variants during persistent infection. J Virol. 1997;71:2959–2969. doi: 10.1128/jvi.71.4.2959-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sánchez C M, Izeta A, Sánchez-Morgado J M, Alonso S, Sola I, Balasch M, Plana-Durán J, Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schickli J H, Zelus B D, Wentworth D E, Sawicki S G, Holmes K V. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J Virol. 1997;71:9499–9507. doi: 10.1128/jvi.71.12.9499-9507.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siddell S G. The Coronaviridae: an introduction. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–10. [Google Scholar]
- 58.Singh M, Berger B, Kim P S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane fusion proteins. J Mol Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 60.Stauber R, Pfleiderara M, Siddell S. Proteolytic cleavage of the murine coronavirus surface glycoprotein is not required for fusion activity. J Gen Virol. 1993;74:183–191. doi: 10.1099/0022-1317-74-2-183. [DOI] [PubMed] [Google Scholar]
- 61.Sturman L S, Ricard C S, Holmes K V. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37°C correlates with virus aggregation and virus-induced cell fusion. J Virol. 1990;64:3042–3050. doi: 10.1128/jvi.64.6.3042-3050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taguchi F. Fusion formation by the uncleaved spike protein of murine coronavirus JHMV variant cl-2. J Virol. 1993;67:1195–1202. doi: 10.1128/jvi.67.3.1195-1202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taguchi F, Fleming J O. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology. 1989;169:233–235. doi: 10.1016/0042-6822(89)90061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vennema H, Heignen L, Zijderveld A, Horzinek M C, Spaan W J M. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Sollner T H, Rothman J E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 66.Weiner L P. Pathogenesis of demyelination induced by a mouse hepatitis virus (JHM virus) Arch Neurol. 1973;28:298–303. doi: 10.1001/archneur.1973.00490230034003. [DOI] [PubMed] [Google Scholar]
- 67.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 68.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 69.Wu M M, Llopis J, Adams S, McCaffery J M, Kulomaa M S, Machen T E, Moore H P, Tsien R Y. Organelle pH studies using targeted avidin and fluorescein-biotin. Chem Biol. 2000;7:197–209. doi: 10.1016/s1074-5521(00)00088-0. [DOI] [PubMed] [Google Scholar]