Abstract
Background
Patients with Triple Negative Breast Cancer (TNBC) currently lack targeted therapies, and consequently face higher mortality rates when compared to patients with other breast cancer subtypes. The tumor microenvironment (TME) cytokine Oncostatin M (OSM) reprograms TNBC cells to a more stem-like/mesenchymal state, conferring aggressive cancer cell properties such as enhanced migration and invasion, increased tumor-initiating capacity, and intrinsic resistance to the current standards of care. In contrast to OSM, Interferon-β (IFN-β) promotes a more differentiated, epithelial cell phenotype in addition to its role as an activator of anti-tumor immunity. Importantly, OSM suppresses the production of IFN-β, although the mechanism of IFN-β suppression has not yet been elucidated.
Methods
IFN-β production and downstream autocrine signaling were assessed via quantitative real-time PCR (qRT-PCR) and Western blotting in TNBC cells following exposure to OSM. RNA-sequencing (RNA-seq) was used to assess an IFN-β metagene signature, and to assess the expression of innate immune sensors, which are upstream activators of IFN-β. Cell migration was assessed using an in vitro chemotaxis assay. Additionally, TNBC cells were exposed to TGF-β1, Snail, and Zeb1, and IFN-β production and downstream autocrine signaling were assessed via RNA-seq, qRT-PCR, and Western blotting.
Results
Here, we identify the repression of Toll-like Receptor 3 (TLR3), an innate immune sensor, as the key molecular event linking OSM signaling and the repression of IFN-β transcription, production, and autocrine IFN signaling. Moreover, we demonstrate that additional epithelial-mesenchymal transition-inducing factors, such as TGF-β1, Snail, and Zeb1, similarly suppress TLR3-mediated IFN-β production and signaling.
Conclusions
Our findings provide a novel insight into the regulation of TLR3 and IFN-β production in TNBC cells, which are known indicators of treatment responses to DNA-damaging therapies. Furthermore, strategies to stimulate TLR3 in order to increase IFN-β within the TME may be ineffective in stem-like/mesenchymal cells, as TLR3 is strongly repressed. Rather, we propose that therapies targeting OSM or OSM receptor would reverse the stem-like/mesenchymal program and restore TLR3-mediated IFN-β production within the TME, facilitating improved responses to current therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13058-024-01918-2.
Keywords: Triple negative breast cancer, Tumor microenvironment, Oncostatin M, Interferon-β, Toll-like Receptor 3, Epithelial-mesenchymal transition, TGF-β1, Snail, Zeb1
Background
Breast cancer is clinically characterized by the presence or absence of estrogen receptor (ER), progesterone receptor (PR), and/or human epidermal growth factor receptor 2 (HER2). Triple Negative Breast Cancer (TNBC) lacks all three receptors and is the most aggressive subtype, with patients having a worse prognosis at every stage of disease when compared to patients with other breast cancer subtypes [1]. The lack of ER, PR, and HER2, which serve as therapeutic targets in the treatment of other breast cancer subtypes, leave patients with TNBC currently lacking targeted therapeutic strategies; thus, the standard of care for treating TNBC remains surgical resection, DNA-damaging chemotherapy, and ionizing radiation [2]. Once a patient progresses or relapses, additional treatment options remain limited. Consequently, the mortality of patients with TNBC is high due to recurrent, metastatic disease. The fact that patients with TNBC tend to be younger than patients with other breast cancer subtypes presents several unique challenges and makes TNBC an important unmet clinical need in search of new therapeutic approaches [3, 4].
The more aggressive nature of TNBC is explained biologically by an enrichment of a subpopulation of stem-like/mesenchymal cells, which are functionally defined by their enhanced tumor-initiating capacity, intrinsic resistance to DNA-damaging therapies, and enhanced migratory capacity. Importantly, acute exposure to DNA-damaging chemotherapies can drive the adaptive emergence of therapy-resistant, stem-like/mesenchymal cells, which are not evident before treatment [5, 6]. Moreover, stem-like/mesenchymal cancer cells express lower levels of MHC-I and increased levels of PD-L1, and often secrete cytokines, chemokines, and growth factors that reinforce the stem-like/mesenchymal characteristics of the cancer cells in addition to creating an immunosuppressive tumor microenvironment (TME) [7–9].
Across numerous TNBC models, the IL-6 family member Oncostatin M (OSM) induces the dedifferentiation to a stem-like/mesenchymal state, mediated primarily through STAT3 activation (in addition to PI3K/AKT and MAPK activation) [10, 11]. For example, treatment of sorted, non-stem (CD24HI/CD24LO) transformed human mammary epithelial cells (HMECs) or TNBC cells with OSM induces tumorsphere forming capacity and enhanced migration and invasion [12]. Basal breast cancer tumors have elevated expression of OSM and OSM receptor (OSMR), both of which correlate with decreased patient survival. Furthermore, syngeneic mouse models of TNBC demonstrate that the intratumoral levels of OSM influence metastatic potential and overall survival, with OSM inducing tumor cell proliferation and metastasis, while deletion of OSMR inhibits tumor development [13, 14]. Therapeutically, antibody-based neutralization of OSM/OSMR signaling in squamous cell carcinoma and ovarian cancer efficiently inhibits STAT3 activation, primary tumor growth, and metastasis [15, 16]. Success with these antibody-derived therapies demonstrates that inhibition of the OSM/OSMR/STAT3 axis is a viable therapeutic strategy across multiple models of solid cancer and may have potential in TNBC.
Another important facet of TME factors such as OSM, is their engagement with infiltrating immune cells to influence primary tumor growth and dissemination by altering tumor antigen presentation and skewing the differentiation of infiltrated immune cell population subtypes [17, 18]. OSM is produced primarily by immune cells, in particular tumor-associated macrophages and neutrophils, and it shapes the landscape of immune cells and stromal cells within the TME, in favor of an immunosuppressive environment. OSM polarizes macrophages to an M2 phenotype which promotes tumor growth and helps facilitate metastasis [19]. The potent activation by IL-6-family members in monocytic myeloid-derived suppressor cells (mMDSCs) drives an interaction between mMDSCs and tumor cells that increases tumor cell invasiveness [20]. OSM also impacts adaptive immunity in the TME by increasing the pro-tumorigenic regulatory T cell (Treg) population and simultaneously decreasing the CD4+ population [21]. Furthermore, OSM is one of four cytokines found to be most frequently expressed by circulating tumor cell-associated neutrophils [22]. As neutrophils surround circulating tumor cells, they protect them from NK or T-cell mediated lysis, and the neutrophil-secreted OSM engages OSMR on the tumor cells, providing survival signals while in circulation, ultimately enhancing metastasis [23–25].
In contrast to OSM, we previously identified Interferon-β (IFN-β) as a repressor of stem-like/mesenchymal properties. Treatment of sorted, CD24LO/CD44HI stem-like cells with IFN-β induces their differentiation into a less aggressive non-stem-like, epithelial state characterized by reduced tumorsphere-initiating capacity and migratory potential. In clinical TNBC samples, an elevated IFN-β gene signature correlated with a repressed cancer stem cell gene signature, increased numbers of tumor-infiltrating lymphocytes, and ultimately improved patient survival [26]. Consistent with our findings that IFN-β reverses EMT, other studies have demonstrated that type I IFNs also negatively correlate with metastasis. Loss of type I IFN signaling is commonly observed in metastatic breast cancer cells, and knockout of type I interferon receptor (IFNAR1) in mice drives accelerated metastasis [27, 28]. Conversely, favorable responses to frontline chemotherapy correlate with robust interferon signaling in both human and mouse studies [29–32]. Moreover, type I IFN also correlates with responses to immunotherapy as administration of IFN-β prior to surgical resection significantly improves response to anti-PD1 [33]. While interferon treatment is currently approved to treat hematological malignancies and some solid tumors (melanoma), the high doses of interferons needed to inhibit cancer cell proliferation or induce death result in side-effects that limit its effectiveness [34–36].
Like OSM, type I IFNs influence the innate and adaptive immune cell response to cancers. Type I IFNs skew monocytes toward a dendritic cell subtype and precursor macrophages into the antitumor M1 phenotype: both of these cell subtypes demonstrate the ability to induce tumor cell death [37, 38]. Similarly, type I IFNs enhance anti-tumor cytotoxicity capabilities in NK and CD8+ T cells by increasing cell-surface expression of perforin and granzyme A on these cell types [39]. Importantly, type I IFNs activate CD8+ T cells to avoid NK-cell mediated lysis, allowing for stability of an effector T cell population in the TME [40, 41]. Concomitant with the stability of effector T cells, type I IFNs also suppress the infiltration of Tregs into the TME [42, 43].
Importantly, in addition to the opposing roles of OSM and IFN-β in regulating tumor cell biology and the immune cell landscape, effectors of each pathway oppose one another molecularly. OSM signaling represses basal IFN-β production in TNBC cells and, subsequently, dismantles the autocrine activation of Interferon-Stimulated Gene Factor 3 (ISGF3) and downstream Interferon Stimulated Genes (ISGs) as it drives EMT and stem-like behaviors [12]. Given the clinical significance of IFN-β and OSM signaling in the outcomes of patients with TNBC, we were interested in understanding the mechanism responsible for OSM-mediated inhibition of IFN-β autocrine signaling. By elucidating how OSM inhibits IFN-β signaling, we could identify new prognostic biomarkers and gain new insights on therapeutic strategies aimed at reactivating IFN-β autocrine signaling.
Here, we demonstrate that OSM inhibits IFN-β production through potent downregulation of Toll-like Receptor 3 (TLR3), an extracellular double-stranded RNA (dsRNA) sensor that activates IFN-β1 transcription. Additionally, we demonstrate that additional EMT-inducing factors, including Transforming Growth Factor-β1 (TGF-β1) and the transcription factors Snail and Zeb1, also repress TLR3 and IFN-β production. The ability of OSM, TGF-β1, Snail, and Zeb1 to repress TLR3 similarly suggests that EMT itself can negatively regulate type I IFN production to help facilitate the dedifferentiation of cancer cells and dismantle anti-tumor immunity. Identifying the molecular mechanisms that prevent TLR3 and IFN-β repression would allow for the development of new therapies that can enforce cancer cell differentiation and re-engage anti-tumor immunity, helping improve outcomes for patients with TNBC.
Methods
Cell culture
BT-549 cells were purchased from ATCC and their identity was validated by STR analysis. E0771 cells were a kind gift from Dr. Ruth Keri (Cleveland Clinic, Cleveland, OH) and their identity was validated by STR analysis. 4T1.2 cells were purchased from ATCC. All cell lines underwent mycoplasma testing (#LT07–318; Lonza) approximately once every two months to confirm cell lines were free of contamination. BT-549 cells were cultured in RPMI 1640 (#12-500p; Cleveland Clinic Media Preparation Core) with 10% FBS (#S11150; Atlanta Biologicals), and 0.023 U/mL of human insulin (#I9278; Sigma Aldrich). E0771 cells were cultured in RPMI1640 (#12-500p; Cleveland Clinic Media Preparation Core) with 10% FBS (#S11150; Atlanta Biologicals). 4T1.2 cells were cultured in MEM α (#99AW500CUSTp; Cleveland Clinic Media Preparation Core) with 10% FBS (#S11150; Atlanta Biologicals) and 1X Penicillin/Streptomycin solution (#30–001-CI; Corning). 293 T cells were cultured in DMEM (#11-500p; Cleveland Clinic Media Preparation Core) with 10% FBS (#S11150; Atlanta Biologicals). All cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C. Treatment of cells was performed as follows unless otherwise specified in figure legends: 10 ng/mL human recombinant Oncostatin M (#OSM01-13; DAPCEL), 10 μM ruxolitinib (#S1378; Selleckchem), 20 ng/mL mouse recombinant Oncostatin M (#495-MO-025/CF; R&D Systems), 6 µg/mL mouse IgG2b isotype control antibody (#BE0086; Bio X Cell), 6 µg/mL mouse Oncostatin M Antibody (#AF-495-NA; R&D Systems), 10 µg/mL poly(I:C) HMW (#tlrl-pic; Invivogen), 100 IU/mL human recombinant interferon beta 1a (#11410–2; PBL Assay Science), and 100 IU/mL human recombinant interferon alpha 2a (#11101–1; PBL Assay Science) unless otherwise stated in the figure legends. All treatments were performed as denoted in the figure legend.
Viral constructs and virus production
Empty vector (Vec), pLenti CMV-Puro OSM, pLenti CMV-Puro SNAI1, and pLenti CMV-Puro ZEB1 vectors were generated as described previously [44]. Mouse Osm (NM_001013365.1) was excised from pCMV3-OSM (#MG50112-UT; Sino Biological Inc.) using KpnI and XbaI, inserted into pENTR4 no ccDB (686–1) (Plasmid #17424; Addgene), then recombined with pLenti CMV-Puro DEST (w118-1) using the LR clonase II enzyme mix, as described previously [44] to generate pLenti CMV-Puro Osm. Human TGF-β1 (NM_000660.7) RNA was isolated from Human Mammary Epithelial Cells (HMECs) treated with recombinant TGF-β1 (#100–21; PeproTech) and then converted to cDNA. TGF-β1 cDNA was amplified by PCR using the primers TGFβ1 forward 5’-GCCGCCACCATGCCGCCCTCCGGGCTGC-3’ and TGFβ1 reverse 5’-TTTAATGGGGCCCCAGGTGGGCTTGGGG-3’, which introduced EcoRV and BamHI restriction enzyme recognition sites at the ends of the sequence. Human TGF-β1 was then subcloned into the pCR8/GW/TOPO entry vector using EcoRV and BamHI. The pCR8-TGF-β1 gateway entry vector was then recombined with pLenti CMV-Puro DEST (w118-1) using the LR clonase II enzyme mix, as described previously [44] to generate pLenti CMV-Puro TGF-β1. These vectors were sequence verified before their transfection into cells. Lenti-CRISPRV2 constructs and guides used to knockout OSMR were prepared as previously described [45]. Lentiviral vectors encoding short-hairpin RNA (shRNA) targeting human TLR3 (NM_003265.2-2547s21c1; TRCN0000358586 and NM_003265.2-455s21c21; TRCN0000358648), and mouse Tlr3 (NM_126166; TRCN0000065731 and NM_126166; TRCN0000065732) were purchased from the Cleveland Clinic Lerner Research Institute Hybridoma Core repository of predesigned lentiviral shRNA vectors from Millipore Sigma. The lentiviral vector used to knockdown GFP (pLVTHM-shGFP) was generated by cleaving pLVTHM (Plasmid #12247; Addgene) and pLVTH-siGFP (Plasmid #12248; Addgene) with EcoRI and XbaI then ligating the siGFP fragment into the LVTHM vector backbone. This vector was sequence-verified before transfection into cells. Lentiviruses were produced by transfecting lentiviral vectors into HEK 293 T cells with the packaging constructs, pCMV-dR8.74 and pMD2G, kind gifts from Dr. Didier Trono (EPFL, Geneva, Switzerland), as previously described [46]. Supernatant media containing viruses were collected every 24 h for 3 days following transfection and filtered using a 0.22 µm filter (#SCGP00525; Millipore). Lentiviruses supplemented with polybrene (#sc-134220; Santa Cruz Biotechnology) were used to transduce BT-549 or E0771 cells for 24 h in a humidified atmosphere containing 5% CO2 at 37 °C. Non-transduced cells were removed with puromycin (#P8833; Sigma).
Western blot analysis
Whole-cell protein extracts were isolated by lysing sample pellets using cell lysis buffer (#FNN0021, Invitrogen) supplemented with 1X phosphatase inhibitor (#4906837001, Roche) and 1X protease inhibitor (#P8340, Sigma-Aldrich). Whole-cell protein extracts were mixed with Laemmli Sample Buffer (#1610747; BIORAD) supplemented with 2-mercaptoethanol (#BP176-100; Fisher Scientific), then boiled at 100 °C for 5 min, then loaded into precast protein gels (#4561094 and #4561034; BIORAD). Gels were placed in an electrophoresis cell with 1X Tris/Glycine/SDS Buffer (#1610732; BIORAD) and electricity was applied to the cell. After sufficient electrophoresis, proteins from the gel were transferred to a methanol-activated hydrophobic transfer membrane (#IPFL00010; Millipore) in a new electrophoresis chamber using 1X Tris/Glycine Buffer (#1610734; BIORAD). Transfer reactions were run at a constant of 0.1 amps overnight. Upon completion of the transfer, membranes were blocked in 1X PBS (#BP399; Fisher Bioreagents) with 0.1% Tween (#BP337-500; Fisher Bioreagents) (1X PBST) and 5% bovine serum albumin (BSA) (#BP1600; Fisher Bioreagents). Primary antibodies used include: OSM (#sc-390–253; Santa Cruz Biotechnology), phosphorylated STAT3 Y705 (#9145; Cell Signaling Technology), STAT3 (#9139; Cell Signaling Technology), Pan-actin (#MCA-5J11; Encore Biotechnology Inc.), phosphorylated STAT1 Y701 (#9167; Cell Signaling Technology), STAT1 (#14994; Cell Signaling Technology), phosphorylated STAT2 Y690 (#88410; Cell Signaling Technology), STAT2 (#72604; Cell Signaling Technology), IRF9 (#76684; Cell Signaling Technology), MX1 (#37849; Cell Signaling Technology), GP130 (#86384; Cell Signaling Technology), TLR3 (#6961; Cell Signaling Technology), OSMR (#10,982–1-AP; Proteintech), TGF-β (#3711; Cell Signaling Technology), Snail (#3879; Cell Signaling Technology), Zeb1 (#70512; Cell Signaling Technology), phosphorylated SMAD2 S465/S467 (#18338; Cell Signaling Technology), SMAD2 (#5339; Cell Signaling Technology), Claudin-1 (#4933; Cell Signaling Technology), and Vimentin (#5741; Cell Signaling Technology). Membranes were incubated overnight in primary antibody dilutions in 1X PBST with 5% BSA. Secondary antibodies used were Dylight800 anti-Rabbit (#5151S; Thermo Fisher Scientific) and Dylight680 anti-Mouse (#5470S; Thermo Fisher Scientific). Membranes were incubated for one hour in secondary antibody diluted in 1X PBST with 5% nonfat dry milk (#9999S; Cell Signaling Technology), then washed 3 times in 1X PBST. Membranes were imaged using an Odyssey CLx imaging system (LI-COR).
RNA isolation and quantitative real-time RT-PCR
Cell pellets were lysed and total RNA isolated using the RNeasy Plus kit (#74134; QIAGEN). RNA (1 µg) was then reverse-transcribed using the iSCRIPT cDNA Synthesis kit (#170–889; BIORAD). Quantitative real-time PCR (qRT-PCR) was conducted using iQ SYBR Green Supermix (#170–8880; BIORAD) on a CFX96 thermocycler (BIORAD) in quadruplicate (technical replicates) to amplify genes using the primer sequences below. Conditions for each qRT-PCR were as follows: 95 °C for 180 s followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s. Genes in human samples were normalized to β-ACTIN and genes in mouse samples were normalized to Gapdh. Error bars for qRT-PCR represent SEM and the data presented are representative of three independent, biological replicates. Primer sequences used for the amplification of human genes are as follows: β-ACTIN forward 5’-CAGCCATGTACGTTGCTATCCAGG-3’, β-ACTIN reverse 5’-AGGTCCAGACGCAGGATGGCATG-3’, IFN-β1 forward 5’-CAACTTGCTTGGATTCCTACAAAG-3’, IFN-β1 reverse 5’-TATTCAAGCCTCCCATTCAATTG-3’, MX1 forward 5’-CTTTCCAGTCCAGCTCGGCA-3’, MX1 reverse 5’-AGCTGCTGGCCGTACGTCTG-3’, OAS1 forward 5’-TGAGGTCCAGGCTCCACGCT-3’, OAS1 reverse 5’-GCAGGTCGGTGCACTCCTCG-3’, OAS2 forward 5’-AGGTGGCTCCTATGGACGGAA-3’, OAS2 reverse 5’-GGCTTCTCTTCTGATCCTGGAATTG-3’, IFNAR1 forward 5’-GAAACCACTGACTGTATATTGTGTGAAA-3’, IFNAR1 reverse 5’-CAGCGTCACTAAAAACACTGCTTT-3’, IFNAR2 forward 5’-AGTCAGAGGGAATTGTTAAGAAGCA-3’, IFNAR2 reverse 5’- TTTGGAATTAACTTGTCAATGATATAGGTG-3’, TLR3 forward 5’- CCTGGTTTGTTAATTGGATTAACGA-3’, TLR3 reverse 5’-TGAGGTGGAGTGTTGCAAAGG-3’. Primer sequences used for the amplification of mouse genes are as follows: Gapdh forward 5’-CATGGCCTTCCGTGTTCCTA-3’, Gapdh reverse 5’-CCTGCTTCACCACCTTCTTGA-3’, Ifn-β1 forward 5’-AACCTCACCTACAGGGCGGACTTCA-3’, Ifn-β1 reverse 5’-TCCCACGTCAATCTTTCCTCTTGCTTT-3’, Mx1 forward 5’-CTCTGGGTGTGGAGCAGGAC-3’, Mx1 reverse 5’-GAGGGCCACTCCAGACAGTG-3’, Oas1a forward 5’-CTTTGATGTCCTGGGTCATGT-3’, Oas1a reverse 5’-GCTCCGTGAAGCAGGTAGAG-3’, Oas2 forward 5’- TGAACAGTGCCAGGAGAAGT-3’, Oas2 reverse 5’-GAGCTGGTGCAGAAGGATGT-3’, Ifnar1 forward 5’-TCCCCGCAGTATTGATGAGT-3’, Ifnar1 reverse 5’-CTGGTCTGTGAGCTGTACTT-3’, Ifnar2 forward 5’- CTATCGTAATGCTGAAACGG-3’, Ifnar2 reverse 5’-CGTAATTCCACAGTCTCTTCT-3’, Tlr3 forward 5’- AGGATACTTGATCTCGGCCT-3’, Tlr3 reverse 5’-TGGCCGCTGAGTTTTTGTT-3’.
RNA-sequencing and gene set enrichment analysis
RNA-sequencing (RNA-seq) was performed in duplicate per biological sample by Active Motif. A total of 3000 ng of RNA was prepared and paired-end 42 base-pair sequencing reads were generated by Illumina sequencing using NextSeq 500. Sequencing reads were mapped to genome build hg38 using the STAR RNA-seq aligner 2.5.2b mapping algorithm with default parameters [47]. Each sample underwent a minimum of 42 million reads. Pairwise comparisons were performed using the DESeq2 software package [48] and a Wald test was used to determine significance: genes with an adjusted p value of less than 0.1 was considered differentially expressed. Gene set enrichment analysis (GSEA) was performed using a ranked list based on log2Ratio of the expression of each pairwise comparison. The “IFN-β Metagene Signature” was derived from microarray analysis in our previous study [26] and was used as input for the enrichment analysis. Additional pathway analysis was performed using iPathwayGuide v17.1 (Advaita Bioinformatics).
Subcutaneous injection of 4T1.2 cells and treatment with OSM-neutralizing antibody
Three 6-week-old female BALB/c mice were subcutaneously injected with 500,000 4T1.2 cells into both flanks. Mice were anesthetized with 3% isoflurane in 2 L/min oxygen throughout the procedure. When the tumors reached a palpable size, approximately 40 mm3 as measured by caliper, treatment was initiated. The treatment group received weekly intratumoral injections of 12.5 µg of a neutralizing antibody (#AF-495-NA; R&D Systems), dissolved in PBS to a final volume of 20 µL per injection. The control group received weekly intratumoral injections of 12.5 µg of an isotype control antibody (#AB-108-C; R&D Systems) dissolved in PBS to a final volume of 20 µL per injection. After 3 weeks, the mice were sacrificed by CO2 asphyxiation followed by cervical dislocation, and tumors were harvested for further analysis. Tumor volumes were measured by caliper, and the specimens were either formalin-fixed or snap-frozen for subsequent studies. Formalin-fixed, paraffin-embedded (FFPE) tumors were sectioned and stained with various markers: Ki67 to assess proliferative index and CD34 for angiogenesis. Quantification of marker-positive cells was conducted using ImageJ software. Tissue processing, paraffin embedding, and immunohistochemical staining were carried out by the Histology, Tissue Procurement, and Imaging Core Facility at Case Western Reserve University. Imaging was performed using a Hamamatsu NanoZoomer Slide Scanner at the Case Western Reserve University School of Medicine's Light Microscopy Imaging Core. All animal experiments and treatments were conducted in compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC).
Migration assays
Migration assays were performed using the IncuCyte Sx5 imaging system using 96-well chemotaxis plates (Sartorious, #4582). Briefly, cells (2000 cells/well) were suspended in their base media (BT549-RPMI with 0.023 U/mL of insulin, E0771-RPMI) containing 0.5% FBS or complete media and seeded onto 96-well ClearView-Chemotaxis plates with 8-mm pores. Complete media was added to the bottom wells (BT549-RPMI with 10% FBS and 0.023 U/mL of human insulin; E0771-RPMI with 10% FBS). BT549 and E0771 cells were stained with live cell NIR Nuclight Dye (Sartorius; #4804) at concentrations of 1:500 (E0771) and 1:1000 (BT549) before the plates were incubated and imaged over the indicated time points. Cells migrating to the bottom chamber across the pores were imaged and quantified every 2–4 h. All conditions were collected in replicates of 6 and are presented as the mean ± SD.
Kaplan–Meier survival curves
Kaplan–Meier survival curves were generated using Kaplan–Meier Plotter, a meta-analysis tool assessing the correlation between expression of 30,000 genes (using mRNA, miRNA, or protein) and survival in 25,000 samples from 21 different tumor types from Gene Expression Omnibus, European Genome-Phenome Archive, and The Cancer Genome Atlas databases [49]. All Kaplan–Meier Plots were graphed using the median as the cut off value for high and low expression.
Population doubling calculation
Population doubling assays were performed by seeding either 5,000 BT-549-derived cell lines or 100,000 E0771-derived cell lines per well in triplicate in 6-well plates. For BT-549-derived cell lines, cells underwent media changes 3 days after initial seeding and 5 days after initial seeding. Total cell number was quantified 7 days after initial seeding using a Beckman Coulter Counter. For E0771-derived cells, total cell number was quantified 3 days after initial seeding, then cells were reseeded in new 6-well plates, underwent a media change after 2 days, and total cell number was quantified again 4 days after initial seeding. These procedures were repeated for both cell line derivatives to assess population doubling across a 14 day timeframe. The total cell number at each time point was used to calculate population doubling using the formula: PD = log(cells counted/cells plated)/log(2). The population doubling at each point the cells were counted represents the mean of all triplicates counted.
Transfection of siRNAs into BT-549 cells
BT-549 cells were seeded onto tissue culture plates and incubated at 37 °C overnight. The next day, cells were washed with Opti-MEM (#31985070; Gibco). siRNAs targeting IL6ST (NM_001190981.1; siRNA ID #s7318 and #s7319; Ambion, Inc.) or a scrambled (Scram) negative control siRNA (#4390844; Ambion, Inc.) were diluted in Opti-MEM at a final concentration of 30 nM along with RNAiMax (#13778075; Invitrogen). The siRNA/lipid complexes in Opti-MEM were incubated at room temperature for 20 min before being added to BT-549 cells. Cells were incubated with a siRNA/lipid complex for 6 h at 37 °C, then media was aspirated from the plates and replaced with fresh cell culture media. Cells were incubated at 37 °C for 18 h before being treated with recombinant OSM for 5 min.
ELISA
E0771 cells were seeded on 10 cm plates and incubated in a humidified atmosphere containing 5% CO2 at 37 °C for 72 h. Media from E0771 cells was harvested and spun down at 2,000 × G for 10 min before use. OSM levels in the media were assessed following the protocol of the SimpleStep ELISA Kit (#ab263891; Abcam). Optical density was measured using a Spectra i3X Multi-Mode Microplate Reader (Molecular Devices).
Extreme limiting dilution tumorsphere assay
BT-549 and E0771 cells were live-sorted at limiting dilutions (1, 5, 10, 25, and 100 cells/well) into 96-well ultra-low attachment, polystyrene, flat bottom plates (#3474; Corning-Costar) using the FACS ARIA-SORP (BD). Media supplemented with antibiotic/antimycotic (#15–240-062; Gibco) was added to each well every 3 days for 14 days: after 14 days, tumorspheres were enumerated via bright-field microscopy observation. Stem cell frequency was calculated using Extreme Limiting Dilution Assay software [50].
Mammary fat pad injection of E0771 shGFP and shTlr3 cells
C57BL/6 J female mice (6 weeks old) were purchased from The Jackson Laboratory. E0771 cells transduced with lentiviral vectors encoding shRNAs targeting GFP or Tlr3 were injected into the mammary fat pad. Mice were anesthetized with 3% isoflurane in 2 L/min oxygen throughout the procedure. Each mouse was injected in one mammary fat pad with a total of 100,000 cells. Cells were re-suspended in a solution containing 50% culture medium and 50% matrigel (#354230; Corning) at a final concentration of 2 × 106 cells/mL so that 50 µL were injected per injection. A total of 5 mice were injected per cell line for 15 mice total. Tumors were measured twice weekly with a caliper and when tumors reached 500 mm3 in size, mice were sacrificed by CO2 asphyxiation followed by cervical dislocation. All procedures were performed in compliance with the Case Western Reserve University Institutional Animal Care and Use Committee (IACUC).
Bright-field imaging of cells
Bright-field images were obtained with oblique illumination and a 10X PlanFluor_DL 0.30/15.20 mm Ph1 objective using a Keyence BZ-X810 Microscope and BZ-X Analyzer software. Fields were captured from biological triplicates for each cell line, with one representative image per sample presented in the figures.
Statistical analyses
Statistical analyses were performed using GraphPad Prism version 10.2.3 software. All data are presented as the mean ± SEM for quantitative-PCR, flow cytometry analysis, tumor measuring, and quantification of histology staining or mean ± SD for population doubling, ELISA, limiting dilution, and migration assays. Differences between two groups were compared using an unpaired Student’s t test with a Welch’s correction. Differences among the means of more than two groups was compared using a two-way ANOVA followed by a multiple comparisons test. A p value less than 0.05 was considered to indicate statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Results
OSM inhibits IFN-β production and autocrine signaling in TNBC.
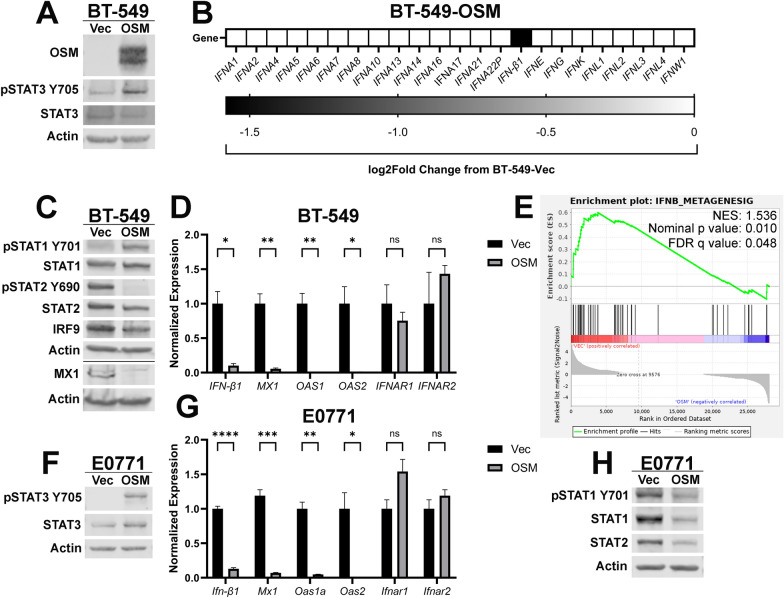
To investigate the impact of elevated OSM signaling on TNBC, BT-549 cells were infected with a lentivirus encoding either OSM (BT-549-OSM) or an empty vector (BT-549-Vec). OSM expression and downstream signaling was assessed by Western analysis, which confirmed the elevated levels of both OSM and phosphorylated STAT3 (Fig. 1A). Treatment of naïve cells with conditioned media from BT-549-OSM cells confirmed that the level of OSM being secreted by BT-549-OSM cells is physiologically-relevant. Conditioned media from BT-549-OSM cells induced STAT3 phosphorylation comparable to adding 1 ng/mL of recombinant OSM, which saturates OSMR-mediated STAT3 phosphorylation (Supplementary Fig. 1A). Importantly, comparison of BT-549-OSM and BT-549-Vec cell population doubling confirmed that elevated OSM has no impact on cell growth (Supplementary Fig. 1B).
Fig. 1.
OSM inhibits IFN-β production and autocrine signaling in TNBC. A BT-549 cells were infected with lentiviruses encoding OSM or control (Vec). Following selection, cells were assessed via Western blot analysis for OSM and STAT3 phosphorylation. B RNA-sequencing was performed on BT-549-OSM and BT-549-Vec cells and pairwise comparisons of the sequencing data were used to assess interferon-family genes. C Western blot and D qRT-PCR analyses of BT-549-OSM and BT-549-Vec cells assessing IFN-β1, ISGs, IFNARs, and ISGF3. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01. E GSEA of RNA-sequencing data of BT-549-OSM and BT-549-Vec cells was performed using an experimentally-derived IFN-β metagene signature. A false-discovery rate (FDR) correction was applied to the statistical significance. F E0771 cells were infected with lentiviruses encoding OSM or control (Vec). Following selection, cells were assessed via Western blot analysis for STAT3 phosphorylation. G qRT-PCR and H Western blot analyses of E0771-OSM and E0771-Vec cells assessing Ifn-β1, ISGs, IFNARs, and ISGF3. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via t tests where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001
RNA-sequencing analysis confirmed that BT-549-OSM cells have significantly reduced IFN-β1 RNA, as we have previously reported [12]. However, none of the other type-I, type-II, or type-III interferon transcripts were altered by OSM signaling (Fig. 1B). Consistent with the repression of IFN-β1 transcription and autocrine secretion, IFNAR1/2-mediated phosphorylation of STAT2 is markedly repressed in BT-549-OSM cells, undermining the activation of the ISGF3 complex and reducing the expression of several Interferon-Stimulated Genes (ISGs), including MX1, OAS1, and OAS2 (Fig. 1C and 1D). OSM did not significantly alter mRNA levels of IFNAR1 and IFNAR2 (Fig. 1D). Of note, STAT1 phosphorylation was increased in BT-549-OSM cells (Fig. 1C), likely due to the ability of OSMR co-receptor GP130 to phosphorylate STAT1 in addition to STAT3 [51]. In fact, we demonstrate that a 5-min pulse of BT-549 cells with recombinant OSM drove STAT1 phosphorylation, which was reduced following siRNA-mediated knockdown of IL6ST or treatment with the JAK inhibitor ruxolitinib (Supplementary Fig. 1C–E).
RNA-sequencing determined that a prominent signature altered by OSM expression involved the repression of genes related to interferon signaling. Gene Set Enrichment Analysis (GSEA) identified a significant negative correlation with our experimentally-derived IFN-β metagene signature (Fig. 1E) [26]. Based on these findings, we conclude that OSM is repressing IFN-β1 transcription, resulting in reduced IFN-β production and repression of canonical type-I IFN/ISGF3 signaling.
Our findings were confirmed in a second model of autocrine/paracrine OSM expression in mouse E0771 TNBC cancer cells. Expression of murine OSM in E0771 cells (E0771-OSM) led to ~ 350 pg/mL of secreted OSM in the medium, which led to the phosphorylation of STAT3 when compared to control cells (E0771-Vec) (Supplementary Fig. 1F and Fig. 1F). OSM concentrations in the plasma of patients with Crohn’s Disease or Inflammatory Bowel Disease (IBD) commonly reach more than 1 ng/mL [52]. OSM concentrations within diseased tissue would undoubtedly be higher; therefore, we conclude that the OSM secreted by both BT-549-OSM and E0771-OSM cells is physiologically relevant. As with the BT-549-OSM cells, OSM expression had no impact on growth kinetics in E0771 cells (Supplementary Fig. 1G).
As observed with BT-549 cells, the mRNA of Ifn-β1 and ISGs (Mx1, Oas1a, and Oas2) was significantly decreased in E0771-OSM, while Ifnar1 and Ifnar2 were unaffected (Fig. 1G). Additionally, OSM decreased STAT1 and STAT2 protein levels, as well as STAT1 phosphorylation, contrasting the increased STAT1 phosphorylation observed in the BT-549-OSM cells (Fig. 1H). However, short-term treatment of E0771 cells with recombinant OSM increased STAT1 phosphorylation, similar to the signaling observed in the BT-549 cells (Supplementary Fig. 1H). The transient OSM-mediated STAT1 phosphorylation observed in E0771 cells, in contrast with stable OSM-mediated STAT1 phosphorylation in BT-549 cells, is likely due to differences in the negative feedback regulation controlling STAT1 stability and phosphorylation between cell lines or species and remains to be determined.
Finally, we confirmed that recombinant OSM treatment also led to the repression of IFN-β1/Ifn-β1 and MX1/Mx1 mRNA, while concurrently repressing STAT2 phosphorylation and MX1 protein expression, in both BT-549 and E0771 cells (Supplementary Fig. 2A-2E). Taken together, our findings identify a conserved mechanism of OSM-mediated IFN-β signaling repression is operant in both human and mouse cancer cells.
OSM-mediated repression of TLR3 suppresses IFN-β production and its autocrine signaling.
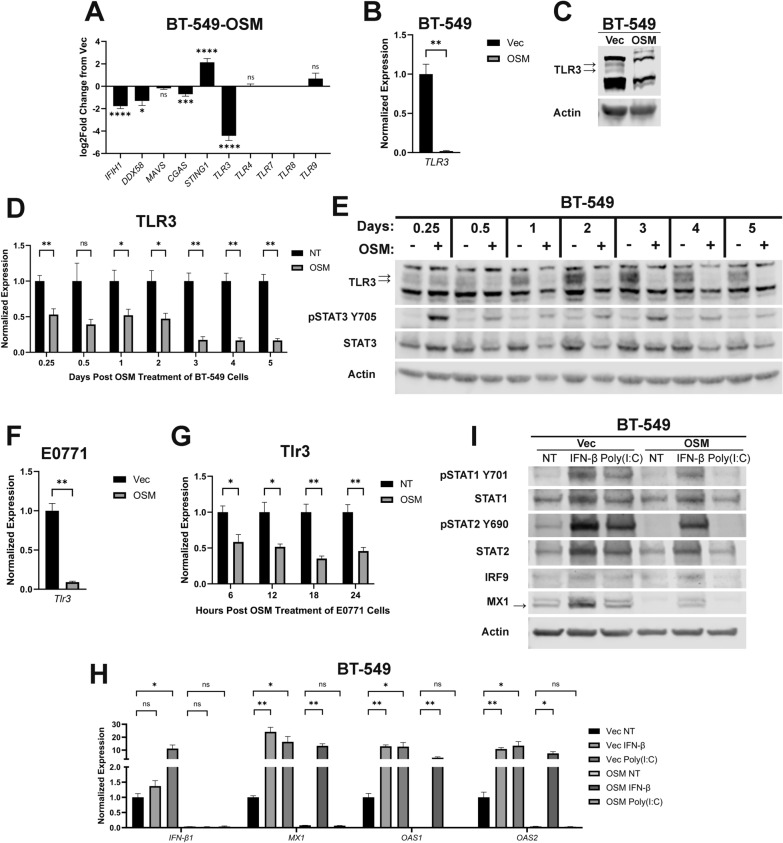
The expression of innate immune sensors, the upstream activators of IFN-β1 transcription, was next assessed following OSM expression or treatment. While transcripts for several innate immune sensors are significantly decreased in BT-549-OSM cells, Toll-like Receptor 3 (TLR3) is the most prominently decreased, comparatively (Fig. 2A). TLR3 is a member of the TLR protein family that binds to a variety of Pathogen-Associated Molecular Patterns (PAMPs) and Damage-Associated Molecular Patterns (DAMPs) of both intracellular and extracellular origin. Canonically, TLR3 binds to extracellular double-stranded RNA (dsRNA) and the activated dsRNA/TLR3 complex becomes endocytosed where it activates multiple downstream effectors; TLR3-phosphorylated IRF3 synergizes with TLR3-activated AP-1 and NF-κB to induce IFN-β1 transcription [53–55]. OSM-mediated TLR3 repression was confirmed in both BT-549 and E0771 cell models using OSM-expressing cells and naïve cells treated with recombinant OSM. Both qPCR and western analyses validate OSM-mediated TLR3 repression at the RNA and protein levels in BT-549 cells (Fig. 2B–E) while qPCR analysis demonstrates OSM-mediated Tlr3 transcript repression in E0771 cells (Fig. 2F, G).
Fig. 2.
OSM-mediated repression of TLR3 suppresses IFN-β production and its autocrine signaling. A Pairwise comparisons of genes encoding the indicated innate immune sensors were made using RNA-sequencing data of BT-549-OSM and BT-549-Vec cells. Data represents mean fold changes ± SEM, n = 2. Statistical significance was determined via Wald tests where *p < 0.05, ***p < 0.001, and ****p < 0.0001. B qRT-PCR and C Western blot analyses of BT-549-OSM and BT-549-Vec cells assessing TLR3 expression. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where **p < 0.01. D qRT-PCR and E Western blot analyses of TLR3 expression in BT-549 cells treated with recombinant OSM for the indicated time points. Cells undergoing no treatment (NT) were included as a negative control group for each time point. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01. F qRT-PCR analysis of E0771-OSM and E0771-Vec cells assessing Tlr3 expression. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t tests where **p < 0.01. G qRT-PCR analysis of Tlr3 expression in E0771 cells treated with recombinant OSM for the indicated time points. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01. H qRT-PCR and I Western blot analyses of BT-549-OSM and BT-549-Vec cells treated for 6 h with either recombinant IFN-β or poly(I:C) assessing IFN-β1, ISGs, IFNARs, and ISGF3. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01
We have demonstrated that OSM represses TLR3 and IFN-β production and autocrine signaling without impacting either type I IFN receptor (IFNAR1/2). To assess the functionality of TLR3 and type I IFN receptor in OSM-expressing TNBC cells, a synthetic dsRNA agonist of TLR3—poly (I:C)—and recombinant IFN-β were used to treat BT-549-Vec and -OSM cells. In this way, we sought to confirm the extent of OSM-mediated TLR3 repression and to understand how downstream IFN-β signaling might be reactivated in OSM-expressing cells. Indeed, poly(I:C) significantly increased IFN-β1 mRNA, STAT1 and STAT2 phosphorylation, and ISG transcription comparable to recombinant IFN-β treatment in BT-549-Vec cells (Fig. 2H, I). Importantly, poly(I:C) was incapable of inducing IFN-β1, STAT1 and STAT2 phosphorylation, and ISG transcription in OSM-expressing cells, confirming that OSM-mediated TLR3 repression undermines the detection of dsRNA that is responsible for basal IFN-β expression in TNBC cells (Fig. 2H–I). In contrast, recombinant IFN-β increased in the entire IFN-β signaling cascade in OSM-expressing cells, confirming that all signaling components downstream of IFNAR1/2 remain intact (Fig. 2H–I). Furthermore, we confirmed that treatment of OSM-expressing cells with recombinant IFN-α is also capable of activating STAT1/STAT2 phosphorylation and increasing MX1 protein expression, suggesting that all type I IFNs can reengage downstream IFN signaling in cells exposed to OSM (Supplementary Fig. 3A). These findings provide novel insight into the potential efficacy of therapies aimed at activating innate immune sensors as a means of engaging autocrine and paracrine IFN-β signaling in TNBC.
Endogenous OSM/OSMR signaling controls IFN-β production and tumor-initiating capacity.
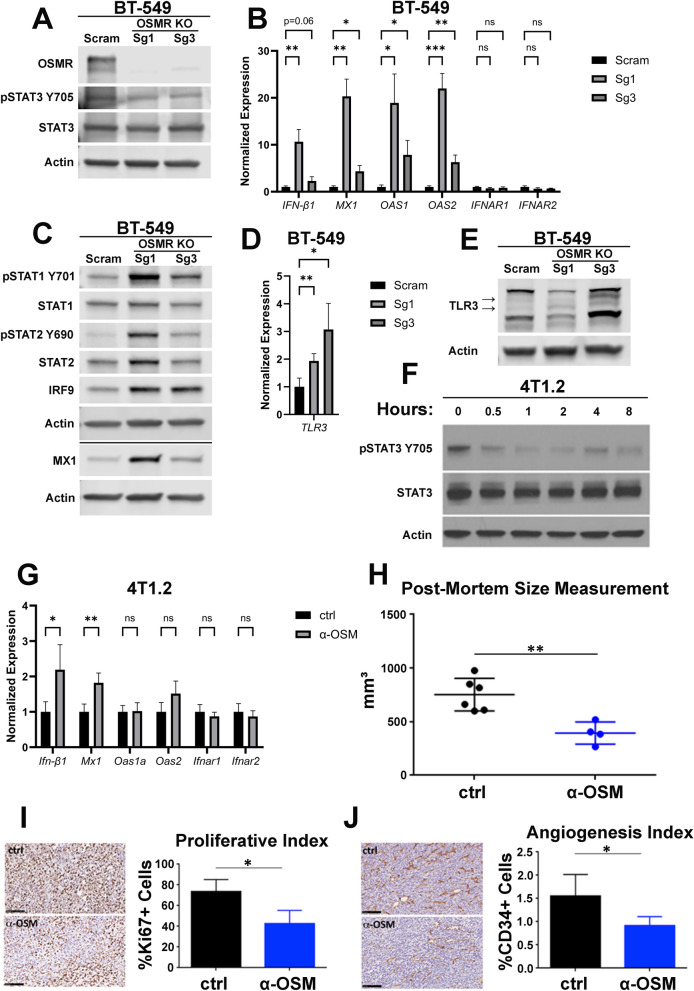
To assess how endogenous, autocrine OSM-OSMR signaling impacts basal IFN-β production, CRISPR/Cas9 was used to knockout OSMR in BT-549 cells (BT-549-OSMR-KO); a scrambled sgRNA (BT-549-Scram) was used as a control. Two different gRNAs were used to knock-out OSMR and, in both cases, basal levels of STAT3 phosphorylation were diminished, indicating a disruption of endogenous OSM/OSMR mediated JAK/STAT3 phosphorylation (Fig. 3A). Interestingly, sgRNA 1 decreased BT-549 cell proliferation while sgRNA 3 had no impact on cell growth (Supplementary Fig. 4A). Importantly, IFN-β1 expression, STAT1/STAT2 phosphorylation, and downstream ISG expression are significantly increased following OSMR knock-out; IFNAR1/2 are unchanged (Fig. 3B, C). Next, TLR3 expression was assessed in BT-549-OSMR-KO cells to further corroborate its relationship with OSM signaling. Indeed, TLR3 mRNA and protein is increased in BT-549-OSMR-KO cells compared to BT-549-Scram cells (Fig. 3D–E).
Fig. 3.
Endogenous OSM controls IFN-β production and tumor-initiating capacity. A BT-549 cells were infected with lentiviruses encoding small guide RNAs targeting OSMR or a scrambled (Scram) control. Following selection, cells were assessed by Western blot analysis for OSMR expression and STAT3 phosphorylation. B qRT-PCR and C Western blot analyses on BT-549-OSMR-KO and BT-549-Scram cells assessing IFN-β1, ISGs, IFNARs, and ISGF3. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05, **p < 0.01, and ***p < 0.01. D qRT-PCR and E Western blot analyses of BT-549-OSMR-KO and BT-549-Scram cells assessing TLR3 expression. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01. F 4T1.2 cells were treated with an OSM-neutralizing antibody for the indicated time points. STAT3 phosphorylation was assessed via Western blot analysis. G qRT-PCR of 4T1.2 cells were treated with either an OSM-neutralizing antibody (α-OSM) or an isotype control (ctrl) for 24 h. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01. Comparison of size, H Ki67 staining, I and CD34 staining J of 4T1.2-derived tumors subcutaneously injected into BALB/c mice treated with either an OSM-neutralizing antibody or an isotype control (ctrl). Data represents mean fold changes ± SEM, n ≥ 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01
Curiously, OSM overexpression and knockdown of basal OSM/OSMR signaling result in increased STAT1 phosphorylation (Figs. 1C and 3C). We hypothesize that overexpression of OSM increases STAT1 phosphorylation through OSMR/GP130-mediated JAK activation, which results in the canonical phosphorylation of STAT3 and the non-canonical phosphorylation of STAT1, while knock-out of OSMR alleviates basal OSM-mediated repression of TLR3, inducing IFN-β production and secretion, leading to canonical IFNAR-mediated STAT1 phosphorylation.
Similarly, to further assess how endogenous autocrine OSM-OSMR signaling impacts basal IFN-β production in a mouse model of TNBC, 4T1.2 cells and tumors were treated with an OSM-neutralizing antibody. Indeed, the OSM-neutralizing antibody decreased STAT3 phosphorylation, confirming interference in OSM/OSMR binding, and increased Ifn-β1 and Mx1 transcripts without changing Ifnar1/2 (Fig. 3F–G). To assess if suppressing OSM signaling and a subsequent increase in IFN-β signaling translates to less aggressive tumor development, mice were injected with 4T1.2 cells and, upon detection of palpable tumors, received intratumoral injections of an OSM-neutralizing antibody weekly for three weeks. Importantly, tumor growth was inhibited following treatment with the OSM-neutralizing antibody compared to tumors treated with an isotype control antibody (Fig. 3H). Furthermore, anti-OSM-treated tumors were less proliferative and less angiogenic than control-treated tumors, as determined by Ki67 and CD34 staining, respectively (Fig. 3I, J).
This data further supports the interplay between OSM/OSMR signaling and IFN-β production and autocrine signaling, confirming that TNBC cells establish an equilibrium of endogenous OSM and TLR3/IFN-β that can be modulated by increases or decreases in tumor OSMR expression or tumor microenvironmental OSM levels.
TLR3 is a major mediator of IFN-β production and autocrine signaling.
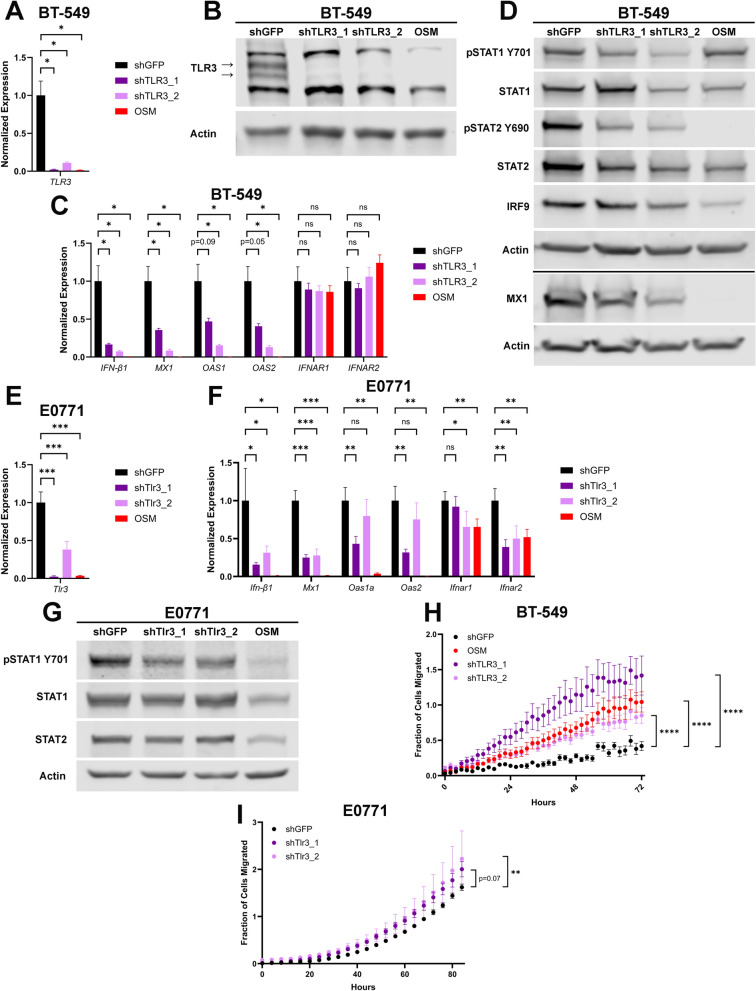
The repression of TLR3 by OSM/OSMR signaling, followed by the associated repression of IFN-β production led us to hypothesize that TLR3 is a major mediator of IFN-β production and autocrine signaling. To test this hypothesis, TLR3 was knocked down in BT-549 cells using lentiviruses encoding shRNAs targeting TLR3 (BT-549-shTLR3) or GFP (BT-549-shGFP); two unique TLR3-targeting shRNAs were used. The impact of TLR3 knockdown on cell growth kinetics was minimal (Supplementary Fig. 5A). In both BT-549-shTLR3 populations, significant repression of TLR3 mRNA and protein expression was noted, with TLR3 levels being comparable to the OSM-expressing cells (Fig. 4A, B). Importantly, in both BT-549-shTLR3 lines, IFN-β1, STAT1 and STAT2 phosphorylation, and ISGs were repressed consistent with OSM-expressing cells; again, no impact on IFNAR1/2 transcripts was noted (Fig. 4C–D). Similarly, TLR3 was knocked down in E0771 cells using lentiviruses encoding shRNAs targeting Tlr3 (E0771-shTlr3) or GFP (E0771-shGFP); two unique Tlr3-targeting shRNAs were used. TLR3 knockdown had minimal effect on the growth of E0771 cells (Supplementary Fig. 5B). Results comparable to those observed in BT-549 cells were observed: TLR3 knockdown caused repression of Ifn-β1, STAT1 phosphorylation, and ISG expression similar to OSM expression (Fig. 4E–G). Based on this data, we conclude that endogenous TLR3 is responsible for a substantial portion of IFN-β production and autocrine signaling in TNBC cells.
Fig. 4.
TLR3 is a major mediator of IFN-β production and autocrine signaling. A BT-549 cells were infected with lentiviruses encoding OSM or shRNAs targeting either TLR3 or GFP. Following selection, cells were assessed by qRT-PCR and B Western blot analyses for the efficiency of TLR3 knockdown. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05. C qRT-PCR and D Western blot analyses of BT-549-shTLR3, BT-549-shGFP, and BT-549-OSM cells assessing IFN-β1, ISGs, IFNARs, and ISGF3. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05. E E0771 cells were infected with lentiviruses encoding OSM or shRNAs targeting either Tlr3 or GFP. Following selection, cells were assessed by qRT-PCR for efficiency of Tlr3 knockdown. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where **p < 0.01. F qRT-PCR and G Western blot analyses of E0771-shTlr3, E0771-shGFP, and E0771-OSM cells assessing Ifn-β1, ISGs, IFNARs, and ISGF3. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where *p < 0.05 and **p < 0.01. H Transwell migration of BT-549-shTLR3, BT-549-shGFP, and BT-549-OSM cells was assessed for 72 h via Incucyte imager. Data is graphed as mean ± SD, n = 6. Statistical significance was determined via two-way ANOVA with multiple comparisons where ****p < 0.0001. I Transwell migration of E0771-shTlr3 and E0771-shGFP cells was assessed for 84 h via Incucyte imager. Data is graphed as mean ± SD, n = 6. Statistical significance was determined via two-way ANOVA with multiple comparisons where **p < 0.01
We have previously reported that OSM drives EMT (including increased migration and invasion) and the acquisition of stem-like properties in TNBC cells, while IFN-β suppresses these processes [12]. Thus, BT-549-shTLR3 and E0771-shTlr3 cells were used to assess whether TLR3 knockdown drives a more migratory phenotype and/or the acquisition of stem-like properties, similar to OSM. Indeed, TLR3 knockdown increases in vitro transwell migration in both BT-549 and E0771 cells, a trend consistent with the OSM-expressing cells (Fig. 4H, I). Moreover, the impact of TLR3 knockdown on tumor-initiating capacity was assessed in vitro using a tumorsphere assay and in vivo via mammary fat pad xenografts of E0771-shTlr3 cells. Whereas OSM expression increased tumorsphere-initiating capacity in both BT-549 and E0771 cells, TLR3 knockdown did not (Supplementary Fig. 5C, D). Likewise, transplantation of OSM-expressing E0771 cells into mice led to more rapid tumor growth and increased mortality, whereas TLR3 knock-down did not significantly alter tumor growth or mortality (Supplementary Fig. 5E).
Such data suggests that TLR3 knockdown and elevated OSM signaling drive one similar molecular signaling phenomenon: namely the loss of TLR3-mediated IFN-β signaling. OSM is unique, however, as it drives an additional signaling cascade in TNBC cells responsible for pro-tumorigenic increased tumor-initiating capacity.
EMT-inducing factors inhibit IFN-β production and autocrine signaling.
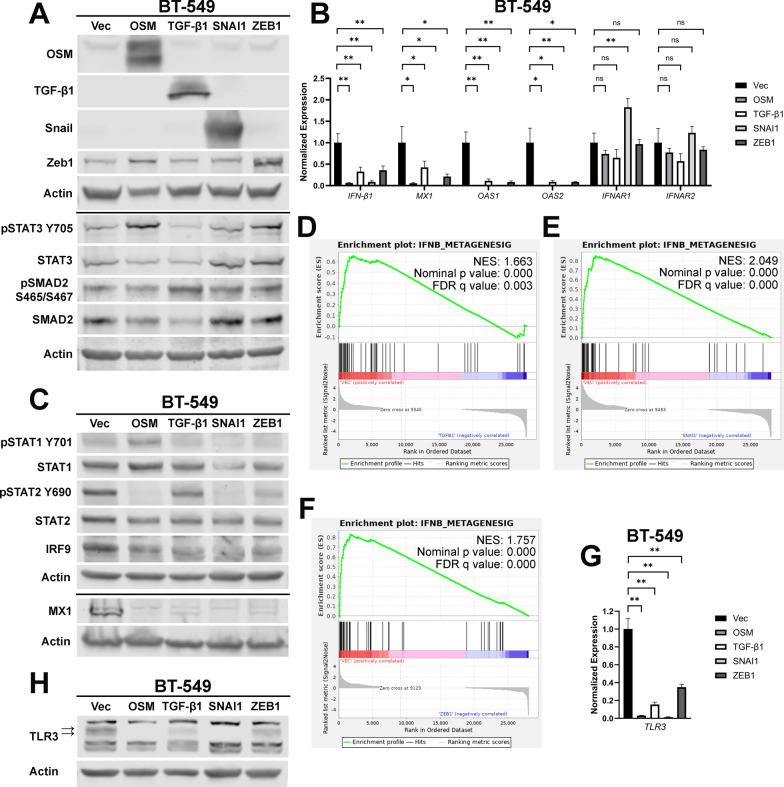
IFN-β treatment of TNBC cells increases epithelial cell markers concomitant with decreases in mesenchymal cell markers and cell migration [26]. IFN-β autocrine signaling is significantly repressed by elevated OSM, concurrent with EMT as demonstrated in this report and our previous publication [12]. This data demonstrates a causality in which the level of IFN-β signaling directly relates to the epithelial-mesenchymal character of the cells. As such, we hypothesized that, beyond OSM, additional EMT-inducing factors may also be able to suppress IFN-β autocrine signaling. To test this hypothesis, BT-549 cells were infected with a lentivirus encoding either OSM, Transforming Growth Factor-β1 (TGF-β1), Snail, Zeb1, or a control. TGF-β1 is another cytokine within the TME capable of driving EMT and both TGF-β1 and OSM mediate EMT through the activation of transcription factors Snail and Zeb1 [56–58].
The individual expression of OSM, TGF-β1, Snail, and Zeb1 induced distinct signaling cascades: OSM and, unexpectedly Zeb1, increased STAT3 phosphorylation while TGF-β1 predictably increased SMAD2 phosphorylation (Fig. 5A). RNA-sequencing was used to compare the transcriptomes of cells expressing OSM, TGF-β1, Snail, and Zeb1. Principle Component Analysis demonstrated that each genetic event drove unique changes to the transcriptomic landscape, with OSM and Snail driving the most distinctive transcriptomes (Supplementary Fig. 6A). TGF-β1 was the only EMT-inducing factor that decreased cell proliferation (Supplementary Fig. 6B). Epithelial and mesenchymal cell markers were assessed to gauge if these EMT-inducing factors were shifting cells to a more mesenchymal phenotype, as they should be. Epithelial cell marker claudin-1 is decreased by OSM, TGF-β1, and Snail expression. Additionally, expression of all these EMT-inducing factors increased vimentin, a mesenchymal cell marker (Supplementary Fig. 6C). Notably, BT-549 cells are inherently mesenchymal, and expression of each of these EMT-inducing factors drives no discernable difference to cell morphology (Supplementary Fig. 6D). Nevertheless, we demonstrate that OSM, TGF-β1, and Snail drive a significant change to the Epithelial-Mesenchymal Transition Hallmark gene signature from the Molecular Signatures database, with Zeb1 driving a near-significant alteration (Supplementary Fig. 7A–D). This data suggests that OSM, TGF-β1, Snail, and Zeb1 drive a partial EMT in an already mesenchymal cell line.
Fig. 5.
EMT-inducing factors inhibit IFN-β production and autocrine signaling. A BT-549 cells were infected with lentiviruses encoding OSM, TGF-β1, SNAI1, ZEB1, or a control vector (Vec). Following selection, cells were assessed via western blot for the indicated targets. B qRT-PCR and C Western blot analyses of cells from A assessing IFN-β1, ISGs, IFNARs, and ISGF3. D–F GSEA of RNA-sequencing data from the cells in A was performed using an experimentally-derived IFN-β metagene signature; D BT-549-TGF-β1, E BT-549-SNAI1, F and BT-549-ZEB1 cells were compared to BT-549-Vec cells. A false-discovery rate (FDR) correction was applied to the statistical significance. G qRT-PCR and H Western blot analysis of cells from A assessing TLR3 expression. Data represents mean fold changes ± SEM, n = 4. Statistical significance was determined via Welch’s t-tests where **p < 0.01
Each of the cell populations expressing EMT-inducing factors have significantly suppressed IFN-β1 and ISG expression, while IFNAR1/IFNAR2 are unchanged (Fig. 5B). Consistent with this data, all factors inactivate ISGF3 primarily by reducing STAT2 phosphorylation and IRF9 expression (Fig. 5C). GSEA confirmed that TGF-β1, Snail, and Zeb1 expression all negatively correlated with our IFN-β metagene signature, comparably to OSM expression (Fig. 5D–F, Fig. 1E). Moreover, each EMT-inducing factor significantly decreases mRNA and protein expression of TLR3 (Fig. 5G, H). This data demonstrates that the EMT-inducing factors OSM, TGF-β1, Snail, and Zeb1 all have a common transcriptomic signature: all repress TLR3 to inhibit IFN-β autocrine signaling.
A novel interferon gene signature correlates with patient prognosis
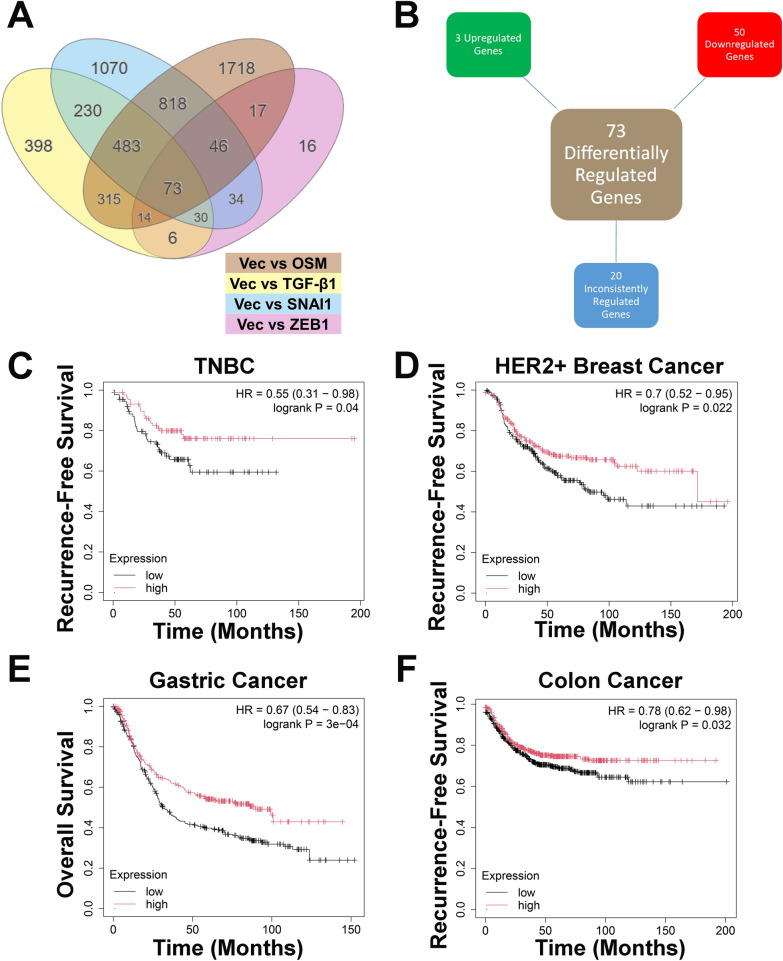
Although OSM, TGF-β1, Snail, and Zeb1 all drive unique changes to the transcriptome of TNBC cells, each also induces EMT in TNBC cells (Supplementary Fig. 6C–D, Supplementary Fig. 7A–D). The differentially regulated genes following the expression of each factor were used to create a novel gene signature. A total of 73 genes were found to be significantly differentially regulated in the OSM, TGF-β1, Snail, and Zeb1-expressing cells (relative to control cells) (Fig. 6A) of which, 50 are consistently downregulated by all factors (Fig. 6B and Table 1). Of these 50 genes, 35 are ISGs, further confirming the ability of these factors to repress interferon signaling. Two of these 50 genes, MX1 and OAS2, were demonstrated to be repressed as a result of TLR3 knockdown in BT-549 and E0771 cells (Fig. 4C, F). Considering that 35 of the 50 commonly repressed genes by OSM, TGF-β1, Snail, and Zeb1 are ISGs, and that TLR3 knockdown results in the repression of ISGs, it is likely that TLR3 is responsible for the activation of a number of these genes.
Fig. 6.
A novel interferon gene signature correlates with patient prognosis. A RNA-sequencing data was analyzed to identify common differentially expressed genes between BT-549-OSM, BT-549-TGF-β1, BT-549-SNAI1, and BT-549-ZEB1 cells relative to BT-549-Vec cells. Only genes with p values < 0.05, as determined by the DESeq2 package in R, were included. B Differentially regulated genes in BT-549-OSM, BT-549-TGF-β1, BT-549-SNAI1, and BT-549-ZEB1 cells compared to BT-549-Vec cells are organized into 3 categories: consistently upregulated in each group, consistently downregulated in each group, or inconsistently regulated between groups compared to BT-549-Vec cells. C Kaplan–Meier plot of patients with TNBC, D HER2 + breast cancer, E gastric cancer, and F colon cancer that express high or low levels of the 50 repressed genes from A using the median as the cutoff value for expression
Table 1.
Genes significantly repressed by OSM, TGF-β1, SNAI1, and ZEB1
| Gene (Average Log2Fold Change) | Vec vs OSM | Vec vs TGF-β1 | Vec vs SNAI1 | Vec vs ZEB1 | Interferon-Stimulated Gene | ||||
|---|---|---|---|---|---|---|---|---|---|
| Log2Fold Change | padj | Log2Fold Change | padj | Log2Fold Change | padj | Log2Fold Change | padj | ||
| GBP1 (− 3.7825) | − 2.85 | 5.13E–102 | − 2.40 | 1.60E–66 | − 8.42 | 1.99E–217 | − 1.46 | 5.36E–21 | Yes [26, 60–62] |
| OAS3 (− 3.5425) | − 4.68 | 1.10E–24 | − 1.60 | 1.89E–05 | − 6.96 | 4.45E–107 | − 0.93 | 1.51E–06 | Yes [26, 60, 62, 63] |
| GBP2 (− 3.4050) | − 1.27 | 3.31E–16 | − 4.87 | 3.99E–142 | − 6.48 | 2.30E–143 | − 1.00 | 8.85E–10 | Yes [60, 62] |
| IFI6 (− 3.3350) | − 4.27 | 7.18E–170 | − 1.49 | 1.34E–25 | − 5.83 | 3.73E–237 | − 1.75 | 1.95E–27 | Yes [26, 60–62] |
| IFI27 (− 3.1125) | − 4.72 | 6.93E–18 | − 2.32 | 1.15E–13 | − 4.41 | 4.07E–31 | − 1.45 | 1.48E–13 | Yes [60–62] |
| CCL5 (− 2.9575) | − 3.31 | 3.27E–12 | − 1.42 | 8.61E-11 | − 5.57 | 1.70E–73 | − 1.53 | 2.03E–17 | Yes [60, 61] |
| IFIH1 (− 2.8875) | − 1.69 | 8.58E–14 | − 1.74 | 1.88E–13 | − 6.19 | 4.19E–76 | − 1.93 | 1.09E–24 | Yes [26, 60–62, 64] |
| SLC15A3 (− 2.7850) | − 3.31 | 7.44E–27 | − 1.71 | 5.20E–09 | − 4.74 | 4.55E–44 | − 1.38 | 2.02E–12 | Yes [26, 60, 62] |
| DSP (− 2.6275) | − 3.79 | 3.30E–105 | − 1.82 | 2.18E–29 | − 2.36 | 2.37E–66 | − 2.54 | 1.02E–55 | No |
| CDH3 (− 2.5625) | − 3.35 | 1.87E–15 | − 2.24 | 4.21E–37 | − 3.11 | 2.95E–68 | − 1.55 | 1.57E–19 | No |
| GBP4 (− 2.5075) | − 3.11 | 9.10E–23 | − 2.29 | 4.27E–16 | − 3.34 | 6.15E–14 | − 1.29 | 1.38E–10 | Yes [60] |
| L1CAM (− 2.4950) | − 3.54 | 8.17E–45 | − 1.11 | 1.61E–08 | − 3.42 | 4.82E–50 | − 1.91 | 2.83E–26 | No |
| PARP14 (− 2.4425) | − 1.52 | 2.88E–18 | − 1.87 | 2.59E–26 | − 5.04 | 2.47E–52 | − 1.34 | 2.60E–13 | Yes [64] |
| S100A8 (− 2.3775) | − 2.10 | 2.69E–07 | − 2.72 | 7.29E–15 | − 3.69 | 7.08E–19 | − 1.00 | 1.26E–06 | Yes [60] |
| HERC6 (− 2.3325) | − 2.15 | 5.33E–19 | − 1.43 | 6.45E–09 | − 4.07 | 4.68E–21 | − 1.68 | 1.86E–19 | Yes [26, 60, 64] |
| MX1 (− 2.3250) | − 3.35 | 7.14E–12 | − 1.07 | 9.28E–03 | − 4.16 | 8.19E–22 | − 0.72 | 3.08E–04 | Yes [26, 60–64] |
| OAS2 (− 2.3000) | − 3.75 | 4.88E–13 | − 1.43 | 1.74E–04 | − 3.33 | 1.30E–13 | − 0.69 | 3.31E–04 | Yes [26, 60–63] |
| IFITM1 (− 2.2900) | − 1.60 | 3.12E–03 | − 2.51 | 3.92E–13 | − 4.44 | 2.00E–26 | − 0.61 | 1.10E–02 | Yes [26, 60–63] |
| SAMD9 (− 2.2775) | − 1.26 | 1.39E–08 | − 2.07 | 1.93E–20 | − 4.34 | 2.83E–63 | − 1.44 | 1.20E–14 | Yes [60] |
| GBP3 (− 2.1675) | − 2.20 | 2.20E–10 | − 1.25 | 7.88E–05 | − 4.29 | 9.72E–28 | − 0.93 | 2.30E–05 | Yes [60] |
| TRANK1 (− 2.1000) | − 2.31 | 5.76E–08 | − 1.38 | 1.19E–04 | − 3.83 | 6.08E–20 | − 0.88 | 5.10E–05 | Yes [60] |
| CFB (− 2.0825) | − 4.20 | 1.32E–38 | − 2.01 | 2.82E–13 | − 1.48 | 3.29E–08 | − 0.64 | 1.72E–02 | Yes [60] |
| EPSTI1 (− 2.0525) | − 1.91 | 3.92E–28 | − 2.14 | 1.52E–33 | − 2.72 | 9.62E–51 | − 1.44 | 1.89E–17 | Yes [26, 60] |
| PARP10 (− 2.0375) | − 2.58 | 8.92E–26 | − 1.54 | 3.02E–12 | − 2.97 | 1.29E–37 | − 1.06 | 2.57E–07 | Yes [60] |
| MUC1 (− 2.0125) | − 1.75 | 1.23E–08 | − 2.50 | 1.43E–96 | − 2.63 | 5.32E–78 | − 1.17 | 4.56E–14 | No |
| KRT81 (− 1.9750) | − 3.20 | 9.84E–32 | − 0.60 | 1.85E–02 | − 3.06 | 8.56E–32 | − 1.04 | 1.14E–07 | No |
| DDX60 (− 1.9000) | − 1.16 | 1.28E–10 | − 1.15 | 3.81E–10 | − 4.20 | 7.51E–74 | − 1.09 | 6.03E–10 | Yes [26, 60, 62] |
| APOL1 (− 1.8450) | − 2.20 | 7.13E–07 | − 1.41 | 1.90E–04 | − 3.05 | 4.85E–13 | − 0.72 | 2.01E–03 | Yes [60, 64] |
| DSC2 (− 1.8400) | − 0.91 | 2.31E–02 | − 1.82 | 2.21E–07 | − 3.84 | 1.01E–19 | − 0.79 | 6.49E–04 | No |
| PKP3 (− 1.8275) | − 2.00 | 1.67E–13 | − 1.18 | 4.44E–06 | − 3.36 | 2.16E–31 | − 0.77 | 5.28E–04 | No |
| USP18 (− 1.8275) | − 2.07 | 2.67E–11 | − 1.31 | 1.51E–05 | − 2.70 | 1.70E–09 | − 1.23 | 1.69E–09 | Yes [60, 61, 64] |
| PODXL (− 1.7875) | − 1.00 | 5.08E–05 | − 2.19 | 6.14E–06 | − 3.02 | 9.91E–33 | − 0.94 | 2.52E–19 | No |
| RSAD2 (− 1.7825) | − 1.53 | 3.66E–03 | − 1.17 | 2.84E–03 | − 2.85 | 3.92E–10 | − 1.58 | 5.36E–16 | Yes [26, 61–64] |
| PTGIS (− 1.7550) | − 1.97 | 1.19E–05 | − 1.47 | 2.87E–10 | − 2.08 | 9.05E–19 | − 1.50 | 3.62E–15 | No |
| KIF21A (− 1.7150) | − 1.98 | 1.65E–14 | − 0.97 | 8.51E–05 | − 3.08 | 1.25E–29 | − 0.83 | 3.11E–04 | No |
| DHX58 (− 1.7125) | − 1.46 | 1.32E–05 | − 0.88 | 7.91E–03 | − 3.71 | 2.38E–23 | − 0.80 | 5.97E–04 | Yes [60, 64] |
| UBE2L6 (− 1.6725) | − 1.19 | 1.04E–08 | − 1.77 | 4.29E–17 | − 3.13 | 4.30E–45 | − 0.60 | 1.62E–02 | Yes [60, 64] |
| HPSE (− 1.6375) | − 2.31 | 1.06E–70 | − 1.32 | 4.04E–20 | − 2.30 | 1.09E–85 | − 0.62 | 2.58E–04 | Yes [60, 62] |
| SP140L (− 1.6175) | − 2.36 | 5.04E–12 | − 1.26 | 4.08E–05 | − 1.91 | 5.13E–05 | − 0.94 | 1.86E–05 | No |
| ITPR3 (− 1.5775) | − 1.67 | 1.69E–06 | − 0.97 | 3.75E–03 | − 2.65 | 5.34E–14 | − 1.02 | 1.67E–06 | No |
| SAMHD1 (− 1.5300) | − 1.43 | 8.29E–25 | − 0.86 | 1.02E–08 | − 3.12 | 5.38E–95 | − 0.71 | 9.84E–05 | Yes [60, 63] |
| MAP3K1 (− 1.4750) | − 1.69 | 1.26E–28 | − 2.03 | 1.18E–40 | − 0.77 | 7.36E–08 | − 1.41 | 1.71E–19 | No |
| LY6E (− 1.4550) | − 1.68 | 1.34E–10 | − 1.35 | 5.82E–07 | − 1.58 | 2.34E–09 | − 1.21 | 4.56E–14 | Yes [60, 65] |
| PSMB9 (− 1.4425) | − 0.60 | 2.61E–03 | − 1.44 | 3.01E–12 | − 2.71 | 3.17E–09 | − 1.02 | 8.11E–08 | Yes [60, 64] |
| TMEM108 (− 1.4250) | − 0.70 | 1.75E–04 | − 0.79 | 2.62E–05 | − 3.47 | 1.40E–59 | − 0.74 | 3.76E–04 | No |
| ISG20 (− 1.2825) | − 1.19 | 3.93E–03 | − 1.09 | 2.88E–03 | − 2.24 | 4.72E–09 | − 0.61 | 2.58E–02 | Yes [26, 60, 61, 64, 65] |
| CXCL16 (− 1.2500) | − 1.14 | 3.13E–04 | − 0.76 | 1.84E–02 | − 2.13 | 3.72E–11 | − 0.97 | 7.50E–06 | Yes [66] |
| DDX60L (− 1.2375) | − 1.12 | 7.11E–07 | − 0.92 | 6.18E–05 | − 2.21 | 6.81E–22 | − 0.70 | 1.74E–03 | Yes [26] |
| IFIT5 (− 0.9500) | − 1.23 | 2.95E–03 | − 0.92 | 2.25E–02 | − 0.82 | 4.50E–02 | − 0.83 | 1.43E–04 | Yes [60, 62, 63] |
| MDN1 (− 0.9425) | − 0.81 | 3.11E–09 | − 0.91 | 4.72E–10 | − 1.11 | 8.79E–15 | − 0.94 | 6.63E− 07 | No |
Of 73 significantly differentially regulated genes compared to Vec cells, 50 are consistently repressed by each of these EMT-inducing factors and are listed above. Adjusted p values (padj) denote significance
Importantly, stratifying the mean expression of all 50 genes using the median values as a cutoff in the tumors of patients with TNBC demonstrates the ability to predict recurrence-free survival, with high expression of all 50 genes predicting better outcomes when compared to patients whose tumors have low expression (Fig. 6C). This correlation is conserved among other solid cancers as well, including HER2 + breast cancer, gastric cancer, and colon cancer; in each case, patients with elevated expression of these 50 genes have significantly better survival rates (Fig. 6D–F). Thus, we propose this novel gene set provides prognostic indication in TNBC and other solid cancers, similar to our previously described IFN-β metagene signature. Notably, the difference in recurrence-free survival in patients with TNBC is more distinct and significant when using a more comprehensive IFN gene signature compared to the novel IFN gene signature identified in this study [26].
Further analysis of the 73 commonly differentially regulated genes identified 3 genes (NETO1, MFAP5, and TGM2) that are upregulated by OSM-, TGF-β1-, Snail-, and Zeb1-expressing cells relative to control cells (Fig. 6B and Table 2). NETO1 and MFAP5 encode proteins expressed on the cell membrane involved in cell–cell interactions and TGM2 encodes a transglutaminase that has been shown to be involved in EMT in breast cancer [59]. Unlike the repressed genes described above, mean expression of these 3 genes does not significantly correlate with recurrence-free survival in TNBC (Supplementary Fig. 8A). However, high expression of these genes, using the median value as a cutoff, does correlate with worse prognosis for patients with gastric, colon, and ovarian cancer (Supplementary Fig. 8B–D). Based on this data, we conclude that the decrease of interferon signaling is highly predictive for TNBC prognosis, more so than an increase of EMT-related genes. Interestingly, however, both the interferon gene signature and EMT-related gene signature appear equally important for prognosis in gastric, colon, and ovarian cancer. The remaining 20 genes inconsistently regulated by OSM, TGF-β1, Snail, and Zeb1 are listed in Supplementary Table 1 (Supplementary Table 1).
Table 2.
Genes significantly induced by OSM, TGF-β1, SNAI1, and ZEB1
| Gene (Average Log2Fold Change) | Vec vs OSM | Vec vs TGF-β1 | Vec vs SNAI1 | Vec vs ZEB1 | ||||
|---|---|---|---|---|---|---|---|---|
| Log2Fold Change | padj | Log2Fold Change | padj | Log2Fold Change | padj | Log2Fold Change | padj | |
| NETO1 (2.6025) | 2.54 | 1.86E–6 | 2.94 | 2.01E–22 | 4.29 | 1.79E–27 | 0.64 | 6.87E–3 |
| MFAP5 (2.0875) | 1.61 | 5.44E–4 | 2.93 | 9.27E–24 | 2.91 | 3.07E–18 | 0.90 | 3.45E–5 |
| TGM2 (1.3675) | 2.20 | 6.63E–109 | 1.26 | 4.34E–6 | 1.35 | 2.69E–41 | 0.66 | 8.74E–5 |
Of 73 significantly differentially regulated genes compared to Vec cells, only the 3 indicated genes above are induced by each of these EMT-inducing factors. Adjusted p values (padj) denote significance
Discussion
Elevated OSM correlates with a worse prognosis for patients with diverse cancer types, including TNBC, consistent with its ability to induce the dedifferentiation of cancer cells to a stem-like/mesenchymal state [10, 11, 13, 14]. Conversely, evidence that elevated IFN-β portends improved outcomes for patients with TNBC is consistent with its ability to reduce stem cell signatures, induce an epithelial phenotype, and increase tumor-infiltrating lymphocytes [12, 26]. Here, we confirm and extend our prior research showing that OSM represses basal IFN-β production in TNBC cells, contributing to the resulting stem-like/mesenchymal properties [12]. Notably, OSM specifically represses the activation of IFN-β, with all other interferons remaining unaffected (Fig. 1B). Our studies link the OSM-mediated repression of IFN-β with the repression of TLR3 expression, since ablating TLR3 alone is sufficient to reduce IFN-β production and match the migratory phenotype of OSM (Fig. 4). Conversely, inhibition of basal OSM/OSMR signaling following OSMR knockout increased TLR3 expression, inducing IFN-β production and autocrine ISGF3 and ISG activation (Fig. 3). Our findings are defining how the balance of STAT1/2 activity from type I IFNs and STAT3 activity from IL-6 family cytokines oppose one another to impact both cancer cell and immune cell behaviors and, ultimately, influence patient outcomes. Interestingly, E0771 cells have been classified TNBC cells in some reports and as luminal B cells in other reports, suggesting that our findings could extend beyond TNBC to other breast cancer subtypes.
Consistent with our prior findings identifying IFN-β signaling as a determinant of TNBC outcomes [12, 26], TLR3 expression is also reduced in breast cancer, with TNBC tumors having the lowest expression of TLR3 relative to other subtypes [67, 68]. In fact, within TNBC, lower TLR3 expression correlates with poorer patient prognosis [68], providing an important clinical context to the repression of TLR3-mediated IFN-β production by OSM reported here. The impact of reduced TLR3-driven IFN-β production on basal tumor biology (including cancer cell differentiation state and immune cell function) undoubtedly influences the aggressiveness of TNBC (including migratory/invasive potential and outgrowth and secondary sites), as well as the response to chemotherapy. For example, Sistigu et al. identified that cancer cells stimulate the rapid production of type I IFNs following treatment with anthracyclines, a mainstay for the treatment of TNBC with ~ 75% of patients receiving an anthracycline during treatment. In tumors that respond well to anthracycline treatment, autocrine and paracrine type I IFN production was elicited by the release of RNA by stressed or dying cancer cells, activating a “viral mimicry” response. Importantly, TLR3 was a key inducer of type I IFN production, resulting in productive immune cell responses following anthracycline treatment; tumor cells lacking TLR3 or IFNARs showed no response to treatment [69]. Additional analysis by Fan et al. has also confirmed that elevated TLR3 expression positively correlates with numerous immune cell subsets—including CD4+ T cells, CD8+ T cells, neutrophils, macrophages, dendritic cells, and B cells—in TNBC [68]. By extension, our study suggests that tumors with elevated OSM levels would harbor cancer cells expressing reduced TLR3 and IFN-β, suppressing immune cell entry and activity and making TNBC less responsive to anthracycline treatment. Moreover, the presence of elevated OSM in the TME may also repress TLR3 in additional stromal cells including cancer-associated fibroblasts and endothelial cells, further reducing intra-tumoral IFN-β production.
Research presented here, together with others, suggest that dsRNA of extracellular origin is basally present and able to stimulate TLR3 to drive IFN-β production and secretion by tumor cells, yet the origin of this extracellular dsRNA remains unknown. Tumor exosomal RNAs and RNAs expelled from necrotic cells have been reported to activate TLR3 in other cancer types, providing some insight into potential mechanisms of TLR3 activity in breast cancer [70, 71]. Connecting DAMPs, TLR3 activation, and IFN-β production/secretion complements research linking intracellular DAMPs to additional innate immune sensors that lead to type I IFN production and secretion. For example, chromosomal instability and mitochondrial DNA leakage drive cGAS/STING-dependent IFN-β production, which enhances immunotherapy responses [72–75]. Changes in the expression of endogenous retroviruses (ERVs), LINEs, and SINEs, during cancer development or following treatment with therapies that induce chromatin remodeling, result in dsRNA products that induce another form of viral mimicry, engaging the intracellular dsRNA-recognition pathway consisting of MAVS, RIG-I, and MDA5, which also activate type I IFNs. Together, these DAMP-sensing pathways and the resultant type I IFNs they produce have become proposed therapeutic targets, with TLR3 agonists (Ampligen, Riboxxol, and ARNAX), cGAS/STING agonists (ADU-S100, BMS-986301, and E7766) in various stages of clinical trials. Certainly, TLR3 itself is a biomarker for the efficacy of dsRNA-focused therapies [76], but our findings also suggest that elevated OSM would also serve as a biomarker for poor TLR3 agonist activity in TNBC. Interestingly however, the OSM-mediated repression of TLR3 results in a curious increase in STING (STING1) expression (Fig. 2A), suggesting that STING agonism may be a viable therapeutic strategy in TNBC harboring repressed TLR3. Although OSM also decreases STING’s upstream regulator cGAS (CGAS) expression (Fig. 2A), which explains why an increase in STING expression does not correlate with increased IFN-β production in OSM-expressing cells, cGAS-independent STING agonists exist and have demonstrated strong anti-tumor effects in non-TNBC models [77, 78].
An alternative therapeutic strategy would involve the direct delivery of type I IFN rather than stimulation of upstream innate immune sensors, as the expression of type I interferon receptors (IFNAR1/2) remained intact following OSM exposure (Fig. 1D, G). Indeed, treatment of OSM-expressing cells with type I IFNs successfully re-engaged ISGF3-mediated activation of ISGs and reduced cancer cell migration (Fig. 2H, I; Supplementary Fig. 3A) [26]. Yet, systemic delivery of type I IFNs has not proven effective due to challenging side effects of excessive immune cell activation. Rather, fusing recombinant type I IFNs to a TNBC antigen-specific antibody may be a viable therapeutic strategy, with some successes already reported in various cancer types, including a fusion of type I IFNs to anti-PDL1, anti-CD20, anti-EGFR, or anti-leptin receptor [79–82]. Such fusion proteins allow for the targeted delivery of type I IFN to tumors, the reengagement of IFN signaling in cancer cells, which would reverse the stem-like/mesenchymal cell state responsible for the aggressive nature of TNBC cells, and the enhancement of anti-tumor immune cell activities [83, 84]. Likewise, our findings further validate the OSM-OSMR interaction as a valuable therapeutic target in TNBC, since knockout of OSMR increased TLR3 and IFN-β production and treatment of tumors with an OSM-neutralizing antibody reduced proliferation and angiogenesis, and inhibited tumor growth (Fig. 3B–E, F–J). While this data suggests that cancer cells produce a basal level of OSM that is capable of autocrine signaling and repressing IFN-β, a number of recent studies have demonstrated that myeloid cells recruited to the TME produce substantial levels of OSM, which, in turn, activates cancer cells, reprogramming them into stem-like/mesenchymal variants, and driving the production of immunosuppressive factors [14, 85].
Importantly, although exposure to OSM and shRNA-mediated repression of TLR3 repressed IFN-β production and autocrine signaling, resulting in a common increase in cell migration, only OSM was able to increase tumor-initiating capacity (Fig. 4H, I, Supplementary Fig. 5C–E). This observation suggests that OSM signaling drives additional changes that the repression of IFN-β is unable to recapitulate; we conclude that the OSM-mediated acquisition of stem-like properties occurs independently of IFN-β repression. Indeed, OSM is known to induce numerous cytokines and chemokines (referred to as the OSM inflammatory module), with many of these factors implicated in promoting stem-like properties [86]. This finding provides further evidence that inhibiting OSM-OSMR signaling may have therapeutic potential for patients with TNBC. Indeed, complementing our findings using an OSM-neutralizing antibody, recent studies have identified OSMR targeting antibodies capable of preventing OSM-OSMR interaction and oncogenic signaling that can also inhibit ovarian cancer cell growth [16].
Beyond OSM, we expanded our findings to an additional TME cytokine, TGF-β1, and two EMT-inducing effectors, Snail and Zeb1, as repressors of TLR3 and IFN-β production and autocrine signaling (Fig. 5B–H). Thus, TGF-β1, Snail, and Zeb1 may also serve as biomarkers indicating that TLR3 stimulation will not be an effective therapeutic strategy. Taken together, our findings suggest that a broader EMT program may undermine a tumor cell’s ability to produce and secrete IFN-β. Of the 50 commonly downregulated genes by OSM, TGF-β1, Snail, and Zeb1, 35 are ISGs and high expression of all 50 genes correlates with better prognosis of TNBC and other solid cancers (Table 1 and Fig. 6C–F). Therefore, we propose that elevated OSM or TGF-β1 in the TME of TNBC induces the reprogramming of cancer cells into more aggressive stem-like/mesenchymal variants, engaging Snail and/or Zeb1 to repress TLR3-mediated IFN-β production, and driving the expression of immunosuppressive factors that undermine anti-tumor immunity [14, 85]. If a broader EMT program is responsible for repressing TLR3 and IFN-β, then numerous cytokines, chemokines, and growth factors within the TME that can promote EMT may also function like OSM and TGF-β1. Additional research into which EMT inducing factors can repress TLR3 are currently underway.
Only three genes were commonly induced by OSM, TGF-β1, Snail, and Zeb1 expression, and while these genes are not significantly correlated with TNBC prognosis, they are correlated with prognosis of other solid cancers (Table 2 and Supplementary Fig. 8A–D). Two of these three genes (NETO1 and MFAP5) encode proteins responsible for cell–cell interactions while the third (TGM2) is linked with EMT [59]. Interestingly, BT-549 cells are inherently mesenchymal, and the addition of each of these EMT-inducing factors, individually, increases the mesenchymal cell marker vimentin and 3/4 of them decrease the epithelial cell marker claudin-1: additionally, using RNA-seq data, we demonstrate that OSM, TGF-β1, and Snail significantly altered the EMT Hallmark gene set (Supplementary Fig. 6C and Supplementary Fig. 7A–C). Nevertheless, we propose that even in the absence of a significant change in cell state, the fact that four known EMT-inducing factors with unique signaling cascades and transcriptomes (Fig. 5A, Supplementary Fig. 6A) all repress TLR3 and IFN-β suggests that perhaps an effector common to EMT, rather than the individually tested EMT-inducing factors, is sufficient in suppressing TLR3-mediated IFN-β production.
Conclusions
Herein, we have elucidated a mechanism of pathogenesis for aggressive recurrent and metastatic TNBC consistent across diverse signaling cascades: loss of TLR3-mediated IFN-β production. Loss of TLR3 allows cancer cells to migrate more readily and if OSM is responsible for this loss of IFN-β production, then cancer cells concomitantly increase their tumor-initiating capacity. These pathogenic insights suggest that a two-pronged therapeutic approach consisting of reactivating downstream IFN-β signaling and inhibiting OSM-OSMR binding may prove efficacious for patients with TNBC. Such a strategy may provide a targeted therapy for a disease without any current targeted therapies.
Supplementary Information
Acknowledgements
This research was supported by the Hybridoma Shared Laboratory Resource of the Lerner Research Institute, the Case Western Reserve University School of Medicine Histology and Tissue Procurement Facility and Light Microscopy Imaging Core, the Translational Research Shared Resource and the Cytometry & Imaging Microscopy Shared Resource of the Case Comprehensive Cancer Center (P30CA043703), the Imaging Research Core of Case Western Reserve University. The Hybridoma Shared Laboratory Resource in the Lerner Research Institute is supported by the Cleveland Clinic and the Shared Resource of the Case Comprehensive Cancer Center (P30CA043703). The Case Western Reserve University School of Medicine Histology and Tissue Procurement Facility is supported by the Shared Resource of the Case Comprehensive Cancer Center (P30CA043703). The Case Western Reserve University School of Medicine Light Microscopy Imaging Core was is supported by an NIH shared instrumentation grant (ORIP S10OD024981). The Imaging Research Core is funded in part by the Case Comprehensive Cancer Center (P30CA043703), the Cleveland Digestive Diseases Research Core Center (P30DK097948), and the Cystic Fibrosis Research and Development Program (DRUMM19I0).
List of Abbreviations
- dsRNA
Double-stranded RNA
- EMT
Epithelial-Mesenchymal Transition
- ER
Estrogen Receptor
- GSEA
Gene Set Enrichment Analysis
- HER2
Human Epidermal Growth Factor Receptor 2
- IFNAR1
Interferon Receptor 1
- IFNAR2
Interferon Receptor 2
- IFN-β
Interferon-β
- OSM
Oncostatin M
- OSMR
Oncostatin M Receptor
- PR
Progesterone Receptor
- qRT-PCR
Quantitative Real-Time PCR
- RNA-seq
RNA-sequencing
- shRNA
Short-hairpin RNA
- siRNA
Small interfering RNA
- TGF-β1
Transforming Growth Factor-β1
- TLR3
Toll-like Receptor 3
- TME
Tumor Microenvironment
- TNBC
Triple Negative Breast Cancer
- Treg
Regulatory T Cells
Author contributions
N.M.C. and M.W.J. conceived and designed the study. N.M.C. performed experiments including qRT-PCR, Western blotting, population doubling assays, ELISA, cell migration, tumorsphere formation, and animal studies. I.T. designed and performed experiments including tumorsphere formation and animal studies. K.L.P. and X.Y. analyzed cell migration data. E.R.C. analyzed RNA-seq data and performed GSEAs and other pathway analyses. X.Y. validated results. N.M.C. and M.W.J. contributed to the critical revision and editing of the manuscript for publication. All authors read and approved the final manuscript.
Funding
MWJ is supported by the US National Institutes of Health (R01CA252224 and P01CA272161) and the US Department of Defense Breast Cancer Research Program funding (W81XWH-20–1-0464; BC191534).
Availability of data and materials
Raw RNA-sequencing data files and an Excel file with computed fold changes of genes were deposited at the Gene Expression Omnibus, with accession number GSE282447.
Declarations
Ethics approval and consent to participate
Experiments involving animals were performed in compliance with the guidelines approved by the Case Western Reserve University Institutional Animal Care and Use Committee (IACUC).
Consent for publication
All authors agree to publish the data presented in this manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC, O’Regan R, Torres MA, Meisel JL. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161(2):279–87. [DOI] [PubMed] [Google Scholar]
- 2.Kay C, Martinez-Perez C, Meehan J, Gray M, Webber V, Dixon JM, Turnbull AK. Current trends in the treatment of HR+/HER2+ breast cancer. Future Oncol. 2021;17(13):1665–81. [DOI] [PubMed] [Google Scholar]
- 3.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina breast cancer study. JAMA. 2006;295(21):2492–502. [DOI] [PubMed] [Google Scholar]
- 4.Parise CA, Bauer KR, Caggiano V. Variation in breast cancer subtypes with age and race/ethnicity. Crit Rev Oncol Hematol. 2010;76(1):44–52. [DOI] [PubMed] [Google Scholar]
- 5.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci U S A. 2014;111(50):E5429-5438. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–9. [DOI] [PubMed] [Google Scholar]
- 7.Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, Azidis-Yates E, Vassiliadis D, Bell CC, Gilan O, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019;36(4):385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Chen M, Wu P, Chen C, Xu ZP, Gu W. Increased PD-L1 expression in breast and colon cancer stem cells. Clin Exp Pharmacol Physiol. 2017;44(5):602–4. [DOI] [PubMed] [Google Scholar]
- 9.Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40(18):e108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junk DJ, Bryson BL, Smigiel JM, Parameswaran N, Bartel CA, Jackson MW. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene. 2017;36(28):4001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty MR, Parvani JG, Tamagno I, Junk DJ, Bryson BL, Cheon HJ, Stark GR, Jackson MW. The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer. Breast Cancer Res. 2019;21(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muñoz Caffarel M, Araujo A, Lawrie C, Álvarez López I, Rezola R, Abaurrea A. New targets in triple negative breast cancer: role of oncostatin M receptor pathway. Ann Oncol. 2017;28:v2. 10.1093/annonc/mdx361.002. [Google Scholar]
- 14.Araujo AM, Abaurrea A, Azcoaga P, López-Velazco JI, Manzano S, Rodriguez J, Rezola R, Egia-Mendikute L, Valdés-Mora F, Flores JM, et al. Stromal oncostatin M cytokine promotes breast cancer progression by reprogramming the tumor microenvironment. J Clin Investig. 2022. 10.1172/JCI148667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucia-Tran JA, Tulkki V, Scarpini CG, Smith S, Wallberg M, Paez-Ribes M, Araujo AM, Botthoff J, Feeney M, Hughes K, et al. Anti-oncostatin M antibody inhibits the pro-malignant effects of oncostatin M receptor overexpression in squamous cell carcinoma. J Pathol. 2018;244(3):283–95. [DOI] [PubMed] [Google Scholar]
- 16.Geethadevi A, Nair A, Parashar D, Ku Z, Xiong W, Deng H, Li Y, George J, McAllister DM, Sun Y, et al. Oncostatin M receptor-targeted antibodies suppress STAT3 signaling and inhibit ovarian cancer growth. Cancer Res. 2021;81(20):5336–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, Li J, Li F, Tan HB. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–33. [DOI] [PubMed] [Google Scholar]
- 18.Chernosky NM, Tamagno I. The role of the innate immune system in cancer dormancy and relapse. Cancers. 2021;13(22):5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrivastava R, Asif M, Singh V, Dubey P, Ahmad Malik S, Lone MU, Tewari BN, Baghel KS, Pal S, Nagar GK, et al. M2 polarization of macrophages by Oncostatin M in hypoxic tumor microenvironment is mediated by mTORC2 and promotes tumor growth and metastasis. Cytokine. 2019;118:130–43. [DOI] [PubMed] [Google Scholar]
- 20.Panni RZ, Sanford DE, Belt BA, Mitchem JB, Worley LA, Goetz BD, Mukherjee P, Wang-Gillam A, Link DC, Denardo DG, et al. Tumor-induced STAT3 activation in monocytic myeloid-derived suppressor cells enhances stemness and mesenchymal properties in human pancreatic cancer. Cancer Immunol Immunother. 2014;63(5):513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son HJ, Lee SH, Lee SY, Kim EK, Yang EJ, Kim JK, Seo HB, Park SH, Cho ML. Oncostatin M suppresses activation of IL-17/Th17 via SOCS3 regulation in CD4+ T Cells. J Immunol. 2017;198(4):1484–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C, Guo L, Cai Q, Xi W, Yuan F, Zhang H, Yan C, Huang L, Zhu Z, Zhang J. Circulating neutrophils activated by cancer cells and M2 macrophages promote gastric cancer progression during PD-1 antibody-based immunotherapy. Front Mol Biosci. 2023;10:1081762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, Scheidmann MC, Donato C, Scherrer R, Singer J, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566(7745):553–7. [DOI] [PubMed] [Google Scholar]
- 24.Lim M, Park S, Jeong HO, Park SH, Kumar S, Jang A, Lee S, Kim DU, Cho YK. Circulating tumor cell clusters are cloaked with platelets and correlate with poor prognosis in unresectable pancreatic cancer. Cancers. 2021;13(21):5272. 10.3390/cancers13215272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cools-Lartigue J, Spicer J, McDonald B, Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P, Ferri L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest. 2013;123(8):3446–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty MR, Cheon H, Junk DJ, Vinayak S, Varadan V, Telli ML, Ford JM, Stark GR, Jackson MW. Interferon-beta represses cancer stem cell properties in triple-negative breast cancer. Proc Natl Acad Sci U S A. 2017;114(52):13792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamsal A, Andersen SB, Johansson I, Vietri M, Bokil AA, Kurganovs NJ, Rylander F, Bjorkoy G, Pettersen K, Giambelluca MS. Opposite and dynamic regulation of the interferon response in metastatic and non-metastatic breast cancer. Cell Commun Signal. 2023;21(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rautela J, Baschuk N, Slaney CY, Jayatilleke KM, Xiao K, Bidwell BN, Lucas EC, Hawkins ED, Lock P, Wong CS, et al. Loss of host type-I IFN signaling accelerates metastasis and impairs NK-cell antitumor function in multiple models of breast cancer. Cancer Immunol Res. 2015;3(11):1207–17. [DOI] [PubMed] [Google Scholar]
- 29.Legrier ME, Bieche I, Gaston J, Beurdeley A, Yvonnet V, Deas O, Thuleau A, Chateau-Joubert S, Servely JL, Vacher S, et al. Activation of IFN/STAT1 signalling predicts response to chemotherapy in oestrogen receptor-negative breast cancer. Br J Cancer. 2016;114(2):177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockwell NK, Rautela J, Owen KL, Gearing LJ, Deb S, Harvey K, Spurling A, Zanker D, Chan CL, Cumming HE, et al. Tumor inherent interferon regulators as biomarkers of long-term chemotherapeutic response in TNBC. NPJ Precis Oncol. 2019;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan Q, Peyvandi S, Duffey N, Huang YT, Barras D, Held W, Richard F, Delorenzi M, Sotiriou C, Desmedt C, et al. Type I interferon/IRF7 axis instigates chemotherapy-induced immunological dormancy in breast cancer. Oncogene. 2019;38(15):2814–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergato C, Doshi KA, Roblyer D, Waxman DJ. Type-I interferon signaling is essential for robust metronomic chemo-immunogenic tumor regression in murine breast cancer. Cancer Res Commun. 2022;2(4):246–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brockwell NK, Owen KL, Zanker D, Spurling A, Rautela J, Duivenvoorden HM, Baschuk N, Caramia F, Loi S, Darcy PK, et al. Neoadjuvant interferons: critical for effective PD-1-based immunotherapy in TNBC. Cancer Immunol Res. 2017;5(10):871–84. [DOI] [PubMed] [Google Scholar]
- 34.Bracarda S, Eggermont AM, Samuelsson J. Redefining the role of interferon in the treatment of malignant diseases. Eur J Cancer. 2010;46(2):284–97. [DOI] [PubMed] [Google Scholar]
- 35.Baldo BA. Side effects of cytokines approved for therapy. Drug Saf. 2014;37(11):921–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.How J, Hobbs G. Use of interferon alfa in the treatment of myeloproliferative neoplasms: perspectives and review of the literature. Cancers. 2020;12(7):1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papewalis C, Jacobs B, Wuttke M, Ullrich E, Baehring T, Fenk R, Willenberg HS, Schinner S, Cohnen M, Seissler J, et al. IFN-alpha skews monocytes into CD56+-expressing dendritic cells with potent functional activities in vitro and in vivo. J Immunol. 2008;180(3):1462–70. [DOI] [PubMed] [Google Scholar]
- 38.Muller E, Speth M, Christopoulos PF, Lunde A, Avdagic A, Oynebraten I, Corthay A. Both type I and type II interferons can activate antitumor m1 macrophages when combined with TLR stimulation. Front Immunol. 2018;9:2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guillot B, Portales P, Thanh AD, Merlet S, Dereure O, Clot J, Corbeau P. The expression of cytotoxic mediators is altered in mononuclear cells of patients with melanoma and increased by interferon-alpha treatment. Br J Dermatol. 2005;152(4):690–6. [DOI] [PubMed] [Google Scholar]
- 40.Crouse J, Bedenikovic G, Wiesel M, Ibberson M, Xenarios I, Von Laer D, Kalinke U, Vivier E, Jonjic S, Oxenius A. Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity. 2014;40(6):961–73. [DOI] [PubMed] [Google Scholar]
- 41.Xu HC, Grusdat M, Pandyra AA, Polz R, Huang J, Sharma P, Deenen R, Kohrer K, Rahbar R, Diefenbach A, et al. Type I interferon protects antiviral CD8+ T cells from NK cell cytotoxicity. Immunity. 2014;40(6):949–60. [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto H, Ueda R, Narumi K, Heike Y, Yoshida T, Aoki K. Type I IFN gene delivery suppresses regulatory T cells within tumors. Cancer Gene Ther. 2014;21(12):532–41. [DOI] [PubMed] [Google Scholar]
- 43.Anz D, Rapp M, Eiber S, Koelzer VH, Thaler R, Haubner S, Knott M, Nagel S, Golic M, Wiedemann GM, et al. Suppression of intratumoral CCL22 by type i interferon inhibits migration of regulatory T cells and blocks cancer progression. Cancer Res. 2015;75(21):4483–93. [DOI] [PubMed] [Google Scholar]
- 44.Bryson BL, Tamagno I, Taylor SE, Parameswaran N, Chernosky NM, Balasubramaniam N, Jackson MW. Aberrant induction of a mesenchymal/stem cell program engages senescence in normal mammary epithelial cells. Mol Cancer Res. 2021;19(4):651–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polak KL, Tamagno I, Parameswaran N, Smigiel J, Chan ER, Yuan X, Rios B, Jackson MW. Oncostatin-M and OSM-receptor feed-forward activation of MAPK induces separable stem-like and mesenchymal programs. Mol Cancer Res. 2023;21(9):975–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junk DJ, Cipriano R, Stampfer M, Jackson MW. Constitutive CCND1/CDK2 activity substitutes for p53 loss, or MYC or oncogenic RAS expression in the transformation of human mammary epithelial cells. PLoS ONE. 2013;8(2):e53776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–8. [DOI] [PubMed] [Google Scholar]
- 51.Regis G, Pensa S, Boselli D, Novelli F, Poli V. Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Semin Cell Dev Biol. 2008;19(4):351–9. [DOI] [PubMed] [Google Scholar]
- 52.Guo A, Ross C, Chande N, Gregor J, Ponich T, Khanna R, Sey M, Beaton M, Yan B, Kim RB, et al. High oncostatin M predicts lack of clinical remission for patients with inflammatory bowel disease on tumor necrosis factor alpha antagonists. Sci Rep. 2022;12(1):1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sen GC, Sarkar SN. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 2005;16(1):1–14. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly (I:C). Adv Drug Deliv Rev. 2008;60(7):805–12. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Lin J, Zhao Y, Ma X, Yi H. Toll-like receptor 3 (TLR3) regulation mechanisms and roles in antiviral innate immune responses. J Zhejiang Univ Sci B. 2021;22(8):609–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S, Lee J, Jeon M, Nam SJ, Lee JE. Elevated TGF-beta1 and -beta2 expression accelerates the epithelial to mesenchymal transition in triple-negative breast cancer cells. Cytokine. 2015;75(1):151–8. [DOI] [PubMed] [Google Scholar]
- 57.Agajanian M, Runa F, Kelber JA. Identification of a PEAK1/ZEB1 signaling axis during TGFbeta/fibronectin-induced EMT in breast cancer. Biochem Biophys Res Commun. 2015;465(3):606–12. [DOI] [PubMed] [Google Scholar]
- 58.Smigiel JM, Parameswaran N, Jackson MW. Potent EMT and CSC Phenotypes Are Induced By Oncostatin-M in pancreatic cancer. Mol Cancer Res. 2017;15(4):478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He W, Sun Z, Liu Z. Silencing of TGM2 reverses epithelial to mesenchymal transition and modulates the chemosensitivity of breast cancer to docetaxel. Exp Ther Med. 2015;10(4):1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kane M, Zang TM, Rihn SJ, Zhang F, Kueck T, Alim M, Schoggins J, Rice CM, Wilson SJ, Bieniasz PD. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe. 2016;20(3):392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pervolaraki K, Rastgou Talemi S, Albrecht D, Bormann F, Bamford C, Mendoza JL, Garcia KC, McLauchlan J, Hofer T, Stanifer ML, et al. Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 2018;14(11):e1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1(6):519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoggins JW. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol. 2014;6:40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017;15(12):e2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mar KB, Rinkenberger NR, Boys IN, Eitson JL, McDougal MB, Richardson RB, Schoggins JW. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat Commun. 2018;9(1):3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wuttge DM, Zhou X, Sheikine Y, Wagsater D, Stemme V, Hedin U, Stemme S, Hansson GK, Sirsjo A. CXCL16/SR-PSOX is an interferon-gamma-regulated chemokine and scavenger receptor expressed in atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2004;24(4):750–5. [DOI] [PubMed] [Google Scholar]
- 67.Shi S, Xu C, Fang X, Zhang Y, Li H, Wen W, Yang G. Expression profile of toll-like receptors in human breast cancer. Mol Med Rep. 2020;21(2):786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan L, Sui XY, Jin X, Zhang WJ, Zhou P, Shao ZM. High expression of TLR3 in triple-negative breast cancer predicts better prognosis-data from the Fudan university Shanghai cancer center cohort and tissue microarrays. BMC Cancer. 2023;23(1):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remedios C, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20(11):1301–9. [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, Huang B, Xu X, Zheng J, Cao X. Tumor Exosomal RNAs Promote Lung Pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016;30(2):243–56. [DOI] [PubMed] [Google Scholar]
- 71.Vasiljevic T, Tarle M, Hat K, Luksic I, Mikulandra M, Busson P, Matijevic Glavan T. Necrotic cells from head and neck carcinomas release biomolecules that are activating toll-like receptor 3. Int J Mol Sci. 2023;24(20):15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beernaert B, Parkes EE. cGAS-STING signalling in cancer: striking a balance with chromosomal instability. Biochem Soc Trans. 2023;51(2):539–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng X, Li X, Zhang Y, Cao C, Zhou Q. IL6 Induces mtDNA leakage to affect the immune escape of endometrial carcinoma via cGAS-STING. J Immunol Res. 2022;2022:3815853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokac M, Ahrenfeldt J, Litchfield K, Watkins TBK, Knudsen M, Dyrskjot L, Jakobsen MR, Birkbak NJ. Classifying cGAS-STING activity links chromosomal instability with immunotherapy response in metastatic bladder cancer. Cancer Res Commun. 2022;2(8):762–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu M, Zhou M, Bao X, Pan D, Jiao M, Liu X, Li F, Li CY. ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. J Clin Investig. 2021. 10.1172/JCI139333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salaun B, Zitvogel L, Asselin-Paturel C, Morel Y, Chemin K, Dubois C, Massacrier C, Conforti R, Chenard MP, Sabourin JC, et al. TLR3 as a biomarker for the therapeutic efficacy of double-stranded RNA in breast cancer. Cancer Res. 2011;71(5):1607–14. [DOI] [PubMed] [Google Scholar]
- 77.Chipurupalli S, Ganesan R, Dhanabal SP, Kumar MS, Robinson N. Pharmacological STING activation is a potential alternative to overcome drug-resistance in Melanoma. Front Oncol. 2020;10:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi J, Liu C, Luo S, Cao T, Lin B, Zhou M, Zhang X, Wang S, Zheng T, Li X. STING agonist and IDO inhibitor combination therapy inhibits tumor progression in murine models of colorectal cancer. Cell Immunol. 2021;366:104384. [DOI] [PubMed] [Google Scholar]
- 79.Guo J, Xiao Y, Iyer R, Lu X, Lake M, Ladror U, Harlan J, Samanta T, Tomlinson M, Bukofzer G, et al. Empowering therapeutic antibodies with IFN-alpha for cancer immunotherapy. PLoS ONE. 2019;14(8):e0219829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang X, Zhang X, Fu ML, Weichselbaum RR, Gajewski TF, Guo Y, Fu YX. Targeting the tumor microenvironment with interferon-beta bridges innate and adaptive immune responses. Cancer Cell. 2014;25(1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.anti-CD20 monoclonal antibody-interferon alpha fusion protein IGN002 [https://www.cancer.gov/publications/dictionaries/cancer-drug/def/eramkafusp-alfa?redirect=true]
- 82.Garcin G, Paul F, Staufenbiel M, Bordat Y, Van der Heyden J, Wilmes S, Cartron G, Apparailly F, De Koker S, Piehler J, et al. High efficiency cell-specific targeting of cytokine activity. Nat Commun. 2014;5:3016. [DOI] [PubMed] [Google Scholar]
- 83.Ozzello L, Blank EW, De Rosa CM, Ceriani RL, Tolo H, Kauppinen HL, Cantell K. Conjugation of interferon alpha to a humanized monoclonal antibody (HuBrE-3vl) enhances the selective localization and antitumor effects of interferon in breast cancer xenografts. Breast Cancer Res Treat. 1998;48(2):135–47. [DOI] [PubMed] [Google Scholar]
- 84.Huang TH, Chintalacharuvu KR, Morrison SL. Targeting IFN-alpha to B cell lymphoma by a tumor-specific antibody elicits potent antitumor activities. J Immunol. 2007;179(10):6881–8. [DOI] [PubMed] [Google Scholar]
- 85.Lee BY, Hogg EKJ, Below CR, Kononov A, Blanco-Gomez A, Heider F, Xu J, Hutton C, Zhang X, Scheidt T, et al. Heterocellular OSM-OSMR signalling reprograms fibroblasts to promote pancreatic cancer growth and metastasis. Nat Commun. 2021;12(1):7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.West NR, Hegazy AN, Owens BMJ, Bullers SJ, Linggi B, Buonocore S, Coccia M, Gortz D, This S, Stockenhuber K, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-sequencing data files and an Excel file with computed fold changes of genes were deposited at the Gene Expression Omnibus, with accession number GSE282447.