Abstract
Background
Helicobacter pylori (Hp) infection and metabolic syndrome (MetS) have a high prevalence of co-morbidities and both pose a significant threat to human health and survival. It has been suggested that Hp infection affects the development of MetS in the host, but the causal relationship between the two has not been confirmed.
Methods
We conducted a two-sample Mendelian randomization study to investigate the causal effect of Hp infection with MetS and its components. Summary statistics for exposure factors (Hp infection) were obtained from the GWAS Catalog (anti-Hp IgG, n = 8,735; Hp VacA antibody levels, n = 1,571; Hp GroEL antibody levels, n = 2,716; Hp OMP antibody levels, n = 2,640). Summary statistics for outcome factors (MetS) were obtained from the most comprehensive genome-wide association study (GWAS) currently available (n = 291,107) as well as from the components of MetS: fasting glucose (n = 46,186), hypertension (n = 461,880), serum triglycerides (n = 115,082), waist circumference (n = 21,949), and high-density lipoprotein (n = 400, 754). The inverse-variance weighted (IVW) method was used as the primary MR method and the robustness of the results was assessed through sensitivity analyses.
Results
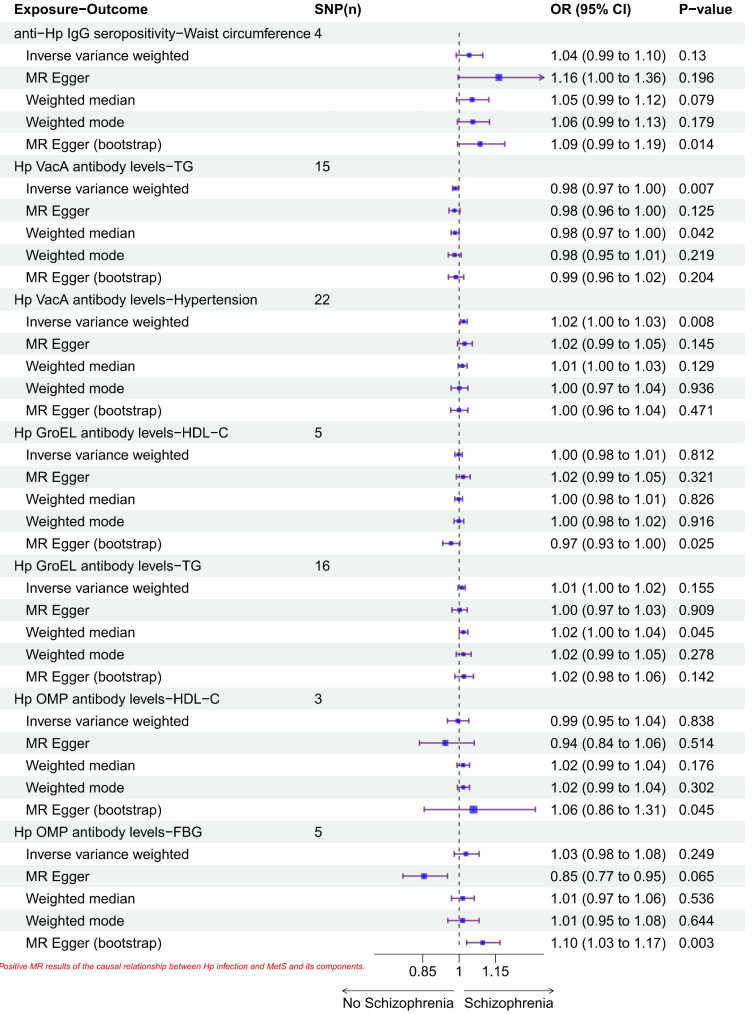
MR analysis showed that anti-Hp IgG levels were positively correlated with waist circumference (β = 0.08, P = 0.012), and GroEL antibody levels showed an opposite correlation with HDL levels (β= -0.03, P = 0.025) and TG (β = 0.02, P = 0.045). In contrast, OMP antibodies levels were positively correlated with both HDL and FBG (β = 0.064, P = 0.037 and β = 0.09, P = 0.003). In the estimation of IVW as the main causal method, VacA antibody level was positively associated with hypertension level and negatively associated with TG (β = 0.02, P = 0.008 and β= -0.02, P = 0.007). Meanwhile, the results of sensitivity analyses showed no heterogeneity or significant level pleiotropy.
Conclusions
Our study suggests that there is a causal effect between Hp infection and Mets diagnosis and its composition, and further studies are needed to understand the mechanism of its influence.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13098-024-01519-1.
Keywords: Metabolic syndrome, Helicobacter pylori infection, Intestinal flora imbalance, Causal associations, Mendelian randomization
Introduction
The World Health Organization (WHO) defines metabolic syndrome (MetS) as a group of metabolic disease syndromes including elevated blood pressure, elevated blood glucose, abnormal glucose tolerance, hyperinsulinemia, hypertriglyceridemia, lowered high-density lipoprotein, and central obesity [1–3]. MetS is a useful theme that emphasizes the pathological state of metabolic abnormalities in various components of the body as a generalized and economical holistic concept for the diagnosis and treatment of a group of highly relevant diseases. The results of a recent Meta-analysis of the prevalence of MetS in 28 million adults worldwide showed that the global prevalence of MetS is largely distributed between 12.5% and 31.4% [4], even if limited by differences in diagnostic criteria between regions. Moreover, the prevalence of metabolic syndrome continues to increase year by year as the economy improves and people’s lifestyles change [5]. The danger of metabolic syndrome increases the risk of developing diabetes and coronary heart disease and other cardiovascular diseases significantly, which seriously affects the quality of people’s survival [6]. Therefore, the etiology and risk factors of MetS should be explored in order to develop pharmacological treatment or lifestyle intervention strategies.
H. pylori, as a class I carcinogen, is a common bacterium that colonizes the stomach. It has become a medical consensus that Hp infection causes gastrointestinal diseases [7]. However, in recent years, some studies have shown that Hp is also involved in the process of occurrence and development of many extra-gastrointestinal diseases [8]. H. pylori parasitization in the digestive system not only induces local inflammatory responses, but also leads to the body being in a state of chronic inflammation, and may lead to disorders of lipid metabolism in the host [9]. Meanwhile, Hp also induces macrophage and neutrophil aggregation and phagocytosis in the gastric mucosa of the host, which affects the health of the organism through immune pathways and intestinal flora dysregulation [10–12]. Increasing evidence suggests that MetS as a systemic disease and Hp infection may be intricately linked, and this close association has prompted us to explore the causal relationship between the two and their pathogenesis.
The overlap between the diagnosis of Hp infection and the diagnosis of MetS is common, with a high chance of co-morbidity, which confirms their association. Among the multiple methods of association studies, randomized controlled trial (RCT) has a higher level of evidence but is more difficult to achieve. Observational studies suffer from confounding factors and causal inversion, and inferring disease causality is often limited. Therefore, causal associations between exposure factors ( or interventions) and disease outcomes need to be inferred with the help of appropriate causal models. Mendelian randomization (MR) is an inferential method based on genetic variation that uses single nucleotide polymorphisms (SNPs) as instrumental variables (IVs) to reveal causal relationships [13]. The basic principle is to utilize the influence of genotype on phenotype in nature to infer the influence of biological factors on disease [14]. Among other things, genotypes are randomly assigned and independent of other confounding factors, overcoming the biases induced in observational studies and providing an effective way to address the above issues.
Materials and methods
Study design
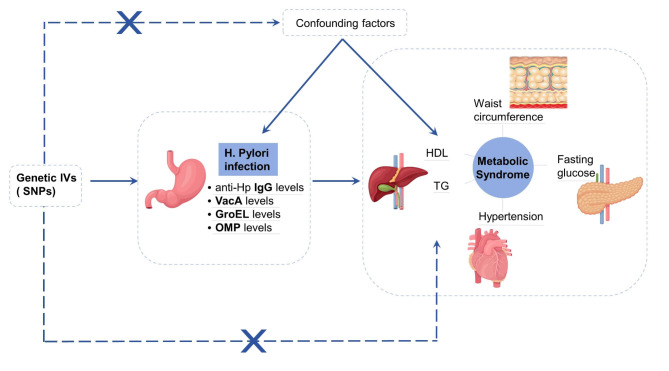
We conducted a two-sample Mendelian randomization study to investigate the causal association between Hp infection and MetS and its components. To accurately demonstrate causal effects, MR relies on three key assumptions: (1) the assumption of association: selected instrumental variables (IVs) are highly correlated with our exposure factor of interest (Hp infection); (2) the assumption of independence: included IVs are not associated with confounders of the exposure-outcome association; and (3) the assumption of exclusionary restrictions: IVs modify outcomes only through their effect on Hp infection, i.e., there is no horizontal pleiotropy. We obtained summary-level data on Hp infection and MetS from genome-wide association studies (GWAS). To improve the accuracy and standardization of the study, we strictly followed the guidelines of Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization (STRBOE-MR) for reporting study results [15]. All GWAS studies included in the analysis were approved by the relevant ethical review boards, so no additional ethical approval was required. The flowchart of the MR study design is shown in Fig. 1 below.
Fig. 1.
Flowchart of the design of the MR study
Data sources
The diagnosis of Hp infection relies on a variety of invasive (endoscopy) and non-invasive diagnostic methods (urea breath test, serologic tests, gastric fluid PCR test) [16]. In most cases, the choice of diagnostic method depends on the clinical information sought as well as the availability and clinical cost of the test to the patient. Among them, serologic tests, which are not affected by ulcer bleeding, gastric atrophy and proton pump inhibitor (PPI) or antibiotic use, have a high diagnostic sensitivity and are commonly used in epidemiologic investigations. According to the study, Hp IgG antibody in serum is the main diagnostic indicator with a sensitivity of about 0.94 and high diagnostic accuracy [17]. In addition, Epplein et al. identified the other four serologic antibodies with the highest sensitivity associated with H. pylori infection as VacA antibodies (sensitivity: 100%; 95% CI: 84-100%), followed by GroEL (sensitivity: 95%; 95% CI: 76-100%), HcpC (sensitivity: 80%; 95% CI: 57 -82%) and HP1564 (sensitivity: 75%; 95% CI: 53-59%) [18]. Unfortunately, we did not find any GWAS data related to HcpC and HP1564 antibodies. Therefore, the final available indicators related to Hp infection include anti-H. pylori IgG, VacA, GroEL, and OMP antibody levels.
We obtained estimates of genetic associations for Hp infection from the latest GWAS study on genetic determinants of infectious agents [19]. Briefly, UKB recruited over half a million UK adults between 2006 and 2010, with a subsample of 9724 participants providing serum samples for serologic measurements of 20 different microorganisms. Finally, of the 20 original pathogens, they selected those with a seropositivity rate of > 15% as GWAS.
Summary-level data for MetS were obtained from the most comprehensive GWAS available from the UK Biobank [20]. The data relate to a comprehensive GWAS analysis of the metabolic syndrome and include information on a total of 291,107 cases (59,677 cases and 231,430 controls). Correspondingly, the definition of MetS in the Joint Interim Statement of the International Diabetes Federation’s Task Force on Epidemiology and Prevention was used in this study [21]. In order for a participant to be classified as having MetS, at least 3 of the following 5 criteria should be fulfilled: (i) serum glucose ≥ 6.1 mmol/L or antidiabetic treatment; (ii) blood pressure ≥ 130/85 mmHg or antihypertensive treatment; (iii) serum triglycerides ≥ 1.7 mmol/L; (iv) waist circumference > 102 cm in men and > 88 cm in women; and (v) HDL-cholesterol < 1.0 mmol/L in men and < 1.3 mmol/L in women.
Data sources and genetic associations for MetS interpretable components: Summary-level data on fasting glucose characteristics were obtained from the large-scale meta-analysis study of the Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC). They meta-analyzed approximately 25,000 directly genotyped or imported autosomal snp from 21 GWAS, comprising a total of 46,186 non-diabetic individuals of European ancestry [22]. For waist circumference, we extracted summary statistics from the GWAS study on overweight and obesity conducted by Wong et al. which included a total of 21.987 subjects [23]. We also obtained summary level data for HDL, TG and hypertension from other studies [24–26]. Their data sources were all from the UK Biobank, so we downloaded the required information from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). The specific details and characteristics of all the above data are shown in Table 1.
Table 1.
Details of data included in Mendelian randomization
| Phenotype | GWAS ID | Year | Sample size | Number of SNPs | Ethnicity | Author |
|---|---|---|---|---|---|---|
| anti-Hp IgG seropositivity | GCST90006910 | 2020 | 8735 | 9170,312 | European | Butler-Laporte G |
| Hp VacA antibody levels | GCST90006916 | 2020 | 1571 | 9178,635 | European | Butler-Laporte G |
| Hp GroEL antibody levels | GCST90006913 | 2020 | 2716 | 9172,299 | European | Butler-Laporte G |
| Hp OMP antibody levels | GCST90006914 | 2020 | 2640 | 10,096,615 | European | Butler-Laporte G |
| MetS | GCST009602 | 2019 | 291,107 | 9463,307 | European | Lind L |
| FBG | GCST000568 | 2010 | 46,186 | 2,445,760 | European | Dupuis J |
| Hypertension | GCST90044351 | 2021 | 461,880 | 9,851,867 | European | Jiang L |
| TG | GCST90092854 | 2022 | 115,082 | 11,590,399 | European | Richardson TG |
| Waist circumference | GCST90103753 | 2022 | 21,949 | 6,370,138 | East Asian | Wong HS |
| HDL-C | GCST90025956 | 2021 | 400,754 | 4,218,934 | European | Barton AR |
Genetic instrumental variable selection
IVs associated with exposure and outcome should meet the following requirements: (1) Single nucleotide polymorphisms (SNPs) potentially associated with exposure were extracted using P < 1 × 10− 5 as the significance threshold screening condition. (2) To fulfill the above assumptions, we set the chain imbalance coefficient R2 = 0.01 and the region width as kb = 10,000 to remove the chain imbalanced SNPs. (3) To prevent the influence of alleles on the outcome of causality between Hp infection and MetS, palindromic and incompatible SNPs were excluded. (4) The F-statistic was.
calculated to assess the strength of the IVs using the formula, 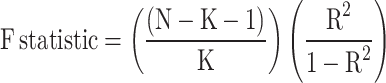 , F-statistic > 10 (excluding SNPs with weak correlation) [27]. SNPs that were independent of each other and significantly correlated with the four antibody levels were finally obtained as the final IVs.
, F-statistic > 10 (excluding SNPs with weak correlation) [27]. SNPs that were independent of each other and significantly correlated with the four antibody levels were finally obtained as the final IVs.
MR analysis and sensitivity analysis
The MR analysis in this study was run by using the TwoSampleMR algorithm package in an environment based on R Studio analysis software, version R 4.4.1. MR analysis was performed using inverse variance weighted (IVW), MR Egger, Weighted median, Weighted mode, and MR Egger (bootstrap) as analytical methods. Among these methods, IVW is generally recognized and used because it calculates a weighted average of the effect sizes of all IVs, and the results obtained through the IVW method are relatively reliable. MR Egger, Weighted median, and Weighted mode are also widely used as default methods. MR Egger (bootstrap) method is used as the MR Egger method’s bootstrap version, can be used to improve the stability of the estimation. Ultimately, we judged whether the causal association between exposure and outcome was significant by looking at the magnitude of the p-value, with a difference of p < 0.05 being considered statistically significant [28]. Heterogeneity between IVs was assessed by Cochran’s Q test, with P < 0.05 suggesting significant heterogeneity. MR Egger regression was used to test for the presence of horizontal pleiotropy. The intercept in Egger regression is a useful indicator of whether the results of the MR analysis are affected by horizontal pleiotropy, and an intercept not equal to zero indicates the presence of overall horizontal pleiotropy [29]. Sensitivity analyses were performed using the “leave-one-out” method, where all SNPs were removed one by one, and the effect values resulting from the remaining SNPs were calculated and analyzed. Sensitivity was indicated if one of the SNP removals had a significant effect on the results [30].
Results
Selection of instrumental variables
In total, we screened 81 independent SNP instrumental variables and finally selected 70 usable SNPs. For MR analysis, the SNP effect on exposure and the SNP effect on outcome had to correspond to the same allele. After harmonizing exposure and outcome data and removing duplicate and palindromic SNPs, anti-Hp IgG and waist circumference, GroEL levels and HDL and TG, OMP levels and HDL and FBG, VacA levels and hypertension levels, and TG with 4, 5, 16, 3, 5, 22, and 15 instrumental variables, respectively.
Results of causal effects between Hp infection and MetS and its components
In the estimation of IVW as the main causality method, there was a causal relationship between VacA antibody level and hypertension level (β = 0.02, P = 0.008), with an increase of 0.02 standard deviation in hypertension level for each standard deviation increase in VacA antibody level. Also, the results showed that VacA antibody levels were negatively correlated with TG (β= -0.02, P = 0.007). MR Egger (bootstrap) analysis showed that anti-Hp IgG levels were positively correlated with waist circumference (β = 0.08, P = 0.012), and GroEL antibody levels were positively correlated with HDL levels (β= -0.03, P = 0.025) and serum triglycerides showed an opposite correlation (β = 0.02, P = 0.045). Moreover, OMP antibody levels showed positive correlation with both HDL level and fasting glucose level (β = 0.064, P = 0.037 and β = 0.09, P = 0.003). The forest plot of the positive MR results for the causality between Hp infection and Mets and its composition is shown in Fig. 2 below.
Fig. 2.
Forest plot of positive MR results for causal relationship between Hp infection and Mets and its components
Sensitivity analysis
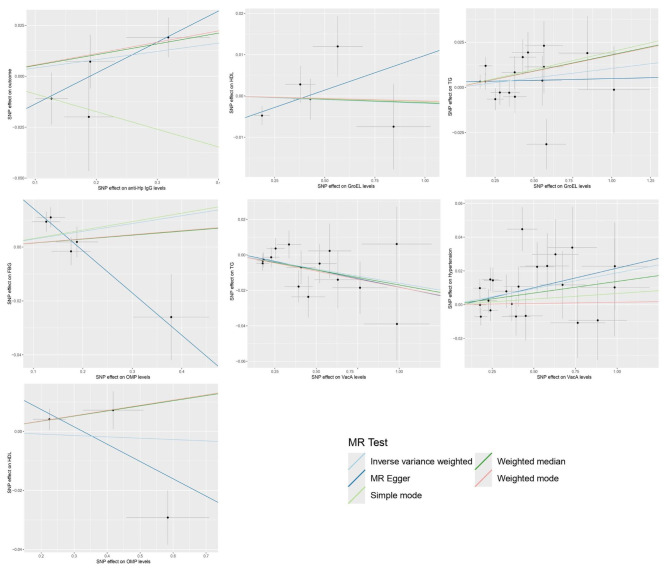
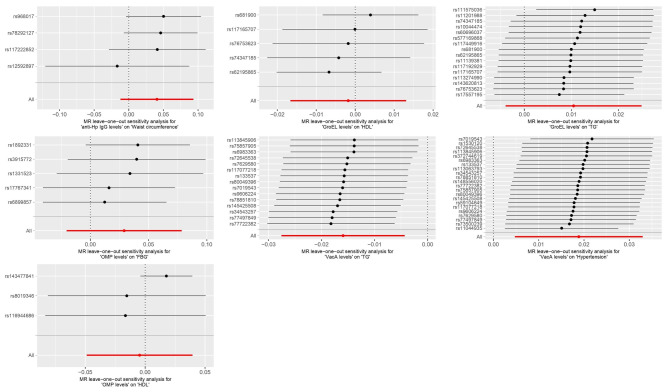
No significant heterogeneity (P > 0.05) was observed in any of the Cochran Q-test results, which are presented in Supplementary Material 1. The intercept values of the MR Egger analysis results were − 0.007, 0.019, -0.029, 0.03, 0.003, 0.001, and − 0.002. The above intercepts were close to 0, and the concomitant non-significant P-values indicate that there is no horizontal multicollinearity in the aforementioned there is no horizontal pleiotropy in the causal relationship. The scatterplot demonstrated the causal relationship between Hp infection and MetS and its components (Fig. 3). In addition, “leave-one-out” method was used to visually assess the results obtained. Specifically, we examined the results of the analysis of the remaining SNPs after removing each SNP one by one, which basically showed negligible fluctuations, confirming the stability and reliability of the results.
Fig. 3.
Scatterplot of the causal relationship between Hp infection and MetS and its components
Fig. 4.
“Leave-one-out” plots of Hp infection with MR analysis of MetS and its components
Discussion
In this two-sample MR study, we utilised large-scale and representative GWAS summary statistics to investigate the causal relationship between Hp infection and MetS and its explainable components. Notably, before translating these associations into evidence for clinical practice, it is crucial to further investigate the causal mechanisms behind both. Several previous studies have demonstrated an association between Hp infection and MetS and its interpretable components (MetS and metabolic conponents), but the results have been mixed and the causal links and directions remain to be.
confirmed [31, 32]. For example, Hp infection was significantly associated with MetS and DM in the study by Chen et al. After a period of follow-up, they emphasised even more the possibility of a high risk of DM events in Hp-infected individuals. In particular, people with different endoscopy findings also showed a correlation with cardiometabolic disease [33].
In addition, several observational studies have reported associations between serological indicators of Hp infection and MetS and its components. A study of the relationship between Hp infection and MetS conducted on 21,106 subjects in South Korea showed that anti-Hp IgG seropositivity participants had significantly higher body mass index (BMI), and waist circumference than seronegative participants (P < 0.05), which is consistent with the results of this study [34]. Not coincidentally, previous studies have shown that positive serological markers of Hp infection were significantly associated with higher systolic blood pressure (β = 1.03, P = 0.014) and lower HDL levels (β= -2.00, P < 0.001). Our results are consistent with these findings suggesting that Hp infection contributes to the development and progression of extragastrointestinal diseases [35]. Mechanistically, Hp infection causes an increase in the expression of host inflammatory factors such as interleukin (IL) and tumor necrosis factor- α (TNF-α), which puts the body in a chronic mild inflammatory state. Under the stimulation of IL-6 and TNF-α, C-reactive protein (CRP) is synthesised by liver and adipocytes, and CRP accordingly increases the production of monocyte chemoattractant protein-1 (MCP-1) [36]. And MCP-1 is a pro-inflammatory chemokine associated with diseases such as obesity, diabetes and cardiovascular diseases, which consequently causes disease development and dyslipidaemia, which is also reflected in increased body mass index, waist circumference [37].
Our study showed a causal link between Hp infection and the components of MetS, but we did not reach a statistically significant result for the idea that Hp infection has a direct causal relationship with the diagnosis of MetS (anti-Hp IgG seropositivity levels-MetS, P = 0.065). This does not affect our understanding of the association, and the reason for the negativity suggests that these SNP tools may not be an optimal alternative to MR analysis. For factors influencing the development of MetS, previous studies have found associations with gender, lifestyle, exercise, and eradication of Hp infection [38–40]. In a multicentre randomised controlled trial by Liou et al. there was a significant improvement in metabolic indices after Hp eradication, with a decrease in insulin resistance, triglycerides and LDL, and an increase in HDL, although there was a slight increase in BMI. In addition to the positive effects on metabolic parameters, eradication of H. pylori infection had minimal disruption of the microbiota and no effect on antibiotic resistance. Their conclusions provide evidence-based support for the long-term safety of Hp eradication therapy and are favourable for further research on MetS and risk factors for Hp infection.
Limitations
Our study is valid in that it highlights the fact that there is a correlation between Hp infection and MetS and its components and focuses our attention on this issue. Despite our fruitful discussion, our study has some limitations. Firstly, our use of serological indicators such as IgG, VacA, GroEL, and OMP to represent Hp infection is limited and further studies using histological or other indicators are needed in the future. Second, although the above metrics were derived from the most comprehensive GWAS analysis currently available for Hp infection, we chose a more lenient criterion (P < 1 × 10− 5) in the significance threshold section to obtain a positive result because of the relatively small number of patients, and this adjustment introduced some bias into our study. In addition, we acknowledge that there may have been overlap between the sample participants for exposure and outcome, but we attempted to reduce the bias present by having a larger IV, with all F-statistics in the study > 10. Finally, it should be recognised that the conclusions reached in the present study have not been externally validated in any clinical setting, and that further substantial studies are still required to provide the link between Hp infection and the MetS and its components Evidence of association.
Conclusion
To our knowledge, this is the first MR study aimed at investigating the causal relationship between Hp infection and MetS diagnosis. And, we had a fruitful discussion. Our study demonstrated a causal relationship between Hp infection and MetS and its components, which may occur through a variety of mechanisms such as inflammatory immune response and microbiota dysbiosis. Further studies are needed in the future to understand the influencing mechanisms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank the Department of Science and Technology of Shanxi Province and the Second Hospital of Shanxi Medical University for their support of this work.
Author contributions
Wang and Yang contributed equally to this work. Wang and Yang designed the study, run the program and wrote the manuscript. Cui, Zhao and Wang contributed significantly to revise and manuscript preparation. Tian, Ding and Ge guided the whole process of this work with constructive discussions. All authors approved the final version.
Funding
We were supported by the Natural Science Foundation of Shanxi Province (202303021211121), the Postgraduate Education Innovation Plan of Shanxi Province in 2024 - Graduate Practice Innovation Project (2024SJ196), the Science and Technology Innovation Project of Colleges and Universities of Shanxi Province (2024L090) and the General Project of Soft Science of Shanxi Province (2017041037-2).
Data availability
Data were available on request.
Declarations
Ethics approval and consent to participate
Ethical approval was not required, as all data used in this study were taken from publicly available summary statistics.
Consent for publication
All authors have reviewed the final version of the manuscript and approved its submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongwei Wang and Caizheng Yang contributed equally to this work and share first authorship.
Contributor Information
Fangying Tian, Email: tfy8048@163.com.
Shanshan Ge, Email: shiwang_0405@qq.com.
References
- 1.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet (London England). 2005;365(9468):1415–28. 10.1016/S0140-6736(05)66378-7 [DOI] [PubMed] [Google Scholar]
- 2.Huang PL. A comprehensive definition for metabolic syndrome. Dis Models Mech. 2009;2(5–6):231–7. 10.1242/dmm.001180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Chen D, Peng Y, Lu Z, Kwan MP, Tse LA. Association between metabolic syndrome and mortality: prospective cohort study. JMIR Public Health Surveillance. 2023;9:e44073. 10.2196/44073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noubiap JJ, Nansseu JR, Lontchi-Yimagou E, Nkeck JR, Nyaga UF, Ngouo AT, et al. Geographic distribution of metabolic syndrome and its components in the general adult population: a meta-analysis of global data from 28 million individuals. Diabetes Res Clin Pract. 2022;188:109924. 10.1016/j.diabres.2022.109924 [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Wang L, Li M, Xu Y, Jiang Y, Wang W, et al. Metabolic syndrome among adults in China: the 2010 China Noncommunicable Disease Surveillance. J Clin Endocrinol Metab. 2017;102(2):507–15. 10.1210/jc.2016-2477 [DOI] [PubMed] [Google Scholar]
- 6.Saklayen MG. The global epidemic of the metabolic syndrome. Curr Hypertens Rep. 2018;20(2):12. 10.1007/s11906-018-0812-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong TC, El-Omar EM, Kuo YT, Wu JY, Chen MJ, Chen CC, et al. Primary antibiotic resistance of Helicobacter pylori in the Asia-Pacific region between 1990 and 2022: an updated systematic review and meta-analysis. The lancet. Gastroenterol Hepatol. 2024;9(1):56–67. 10.1016/S2468-1253(23)00281-9 [DOI] [PubMed] [Google Scholar]
- 8.Tsay FW, Hsu PI. H. pylori infection and extra-gastroduodenal diseases. J Biomed Sci. 2018;25(1):65. 10.1186/s12929-018-0469-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M, Huang X, Gao M, Yang Z, Fang Z, Wei J, et al. Helicobacter pylori promotes inflammatory factor secretion and lung injury through VacA exotoxin-mediated activation of NF-κB signaling. Bioengineered. 2022;13(5):12760–71. 10.1080/21655979.2022.2071011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blüher M, Mantzoros CS. From leptin to other adipokines in health and disease: facts and expectations at the beginning of the 21st century. Metab Clin Exp. 2015;64(1):131–45. 10.1016/j.metabol.2014.10.016 [DOI] [PubMed] [Google Scholar]
- 11.Xu W, Xu L, Xu C. Relationship between Helicobacter pylori infection and gastrointestinal microecology. Front Cell Infect Microbiol. 2022;12:938608. 10.3389/fcimb.2022.938608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CC, Liou JM, Lee YC, Hong TC, El-Omar EM, Wu MS. The interplay between Helicobacter pylori and gastrointestinal microbiota. Gut Microbes. 2021;13(1):1–22. 10.1080/19490976.2021.1909459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson SC, Butterworth AS, Burgess S. Mendelian randomization for cardiovascular diseases: principles and applications. Eur Heart J. 2023;44(47):4913–24. 10.1093/eurheartj/ehad736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekula P, Del Greco M, Pattaro F, C., Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrology: JASN. 2016;27(11):3253–65. 10.1681/ASN.2016010098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. 2021;326(16):1614–21. 10.1001/jama.2021.18236 [DOI] [PubMed] [Google Scholar]
- 16.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720–41. 10.1128/CMR.10.4.720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosch DE, Krumm N, Wener MH, Yeh MM, Truong CD, Reddi DM, et al. Serology is more sensitive than urea breath test or stool antigen for the initial diagnosis of Helicobacter pylori gastritis when compared with histopathology. Am J Clin Pathol. 2020;154(2):255–65. 10.1093/ajcp/aqaa043 [DOI] [PubMed] [Google Scholar]
- 18.Butt J, Blot WJ, Shrubsole MJ, Varga MG, Hendrix LH, Crankshaw S, et al. Performance of multiplex serology in discriminating active vs past Helicobacter pylori infection in a primarily African American population in the southeastern United States. Helicobacter. 2020;25(1):e12671. 10.1111/hel.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler-Laporte G, Kreuzer D, Nakanishi T, Harroud A, Forgetta V, Richards JB. Genetic determinants of antibody-mediated immune responses to infectious diseases agents: a genome-wide and HLA association study. Open Forum Infect Dis. 2020;7(11):ofaa450. 10.1093/ofid/ofaa450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lind L. Genome-wide association study of the metabolic syndrome in UK Biobank. Metab Syndr Relat Disord. 2019;17(10):505–11. 10.1089/met.2019.0070 [DOI] [PubMed] [Google Scholar]
- 21.Alberti KGMM, Blood Institute; American Heart Association. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16);1640–5. World Heart Federation. 10.1161/CIRCULATIONAHA.109.192644 [DOI] [PubMed]
- 22.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–16. 10.1038/ng.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong HS, Tsai SY, Chu HW, Lin MR, Lin GH, Tai YT, et al. Genome-wide association study identifies genetic risk loci for adiposity in a Taiwanese population. PLoS Genet. 2022;18(1):e1009952. 10.1371/journal.pgen.1009952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton AR, Sherman MA, Mukamel RE, Loh PR. Whole-exome imputation within UK Biobank powers rare coding variant association and fine-mapping analyses. Nat Genet. 2021;53(8):1260–9. 10.1038/s41588-021-00892-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson TG, Leyden GM, Wang Q, Bell JA, Elsworth B, Smith D, G., et al. Characterising metabolomic signatures of lipid-modifying therapies through drug target Mendelian randomisation. PLoS Biol. 2022;20(2):e3001547. 10.1371/journal.pbio.3001547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53(11):1616–21. 10.1038/s41588-021-00954-4 [DOI] [PubMed] [Google Scholar]
- 27.Xue H, Shen X, Pan W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am J Hum Genet. 2021;108(7):1251–69. 10.1016/j.ajhg.2021.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S, Thompson SG, CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64. 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 29.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Niu Q, Wu A, Zhang Y, Hong L, Wang H. Causal relationship between circulating immune cells and the risk of type 2 diabetes: a Mendelian randomization study. Front Endocrinol. 2023;14:1210415. 10.3389/fendo.2023.1210415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Shuai P, Chen W, Liu Y, Li D. Association between Helicobacter pylori infection and metabolic syndrome and its components. Front Endocrinol. 2023;14:1188487. 10.3389/fendo.2023.1188487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kountouras J, Boziki M, Kazakos E, Theotokis P, Kesidou E, Nella M, et al. Impact of Helicobacter pylori and metabolic syndrome on mast cell activation-related pathophysiology and neurodegeneration. Neurochem Int. 2024;175:105724. 10.1016/j.neuint.2024.105724 [DOI] [PubMed] [Google Scholar]
- 33.Chen YY, Fang WH, Wang CC, Kao TW, Chang YW, Wu CJ, et al. Helicobacter pylori infection increases risk of incident metabolic syndrome and diabetes: a cohort study. PLoS ONE. 2019;14(2):e0208913. 10.1371/journal.pone.0208913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim SH, Kim N, Kwon JW, Kim SE, Baik GH, Lee JY, et al. Positive association between Helicobacter pylori infection and metabolic syndrome in a Korean population: a multicenter nationwide study. Dig Dis Sci. 2019;64(8):2219–30. 10.1007/s10620-019-05544-3 [DOI] [PubMed] [Google Scholar]
- 35.Gunji T, Matsuhashi N, Sato H, Fujibayashi K, Okumura M, Sasabe N, et al. Helicobacter pylori infection is significantly associated with metabolic syndrome in the Japanese population. Am J Gastroenterol. 2008;103(12):3005–10. 10.1111/j.1572-0241.2008.02151.x [DOI] [PubMed] [Google Scholar]
- 36.Popko K, Gorska E, Stelmaszczyk-Emmel A, Plywaczewski R, Stoklosa A, Gorecka D, et al. Proinflammatory cytokines Il-6 and TNF-α and the development of inflammation in obese subjects. Eur J Med Res 15 Suppl. 2010;2(Suppl 2120–2. 10.1186/2047-783x-15-s2-120 [DOI] [PMC free article] [PubMed]
- 37.Gögebakan Ö, Osterhoff MA, Schüler R, Pivovarova O, Kruse M, Seltmann AC, et al. GIP increases adipose tissue expression and blood levels of MCP-1 in humans and links high energy diets to inflammation: a randomised trial. Diabetologia. 2015;58(8):1759–68. 10.1007/s00125-015-3618-4 [DOI] [PubMed] [Google Scholar]
- 38.Park J, Kim N, Kim WS, Lim SH, Choi Y, Jo HH, et al. Long-term effects of the eradication of Helicobacter pylori on metabolic parameters, depending on sex, in South Korea. Gut Liver. 2023;17(1):58–68. 10.5009/gnl210588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liou JM, Chen CC, Chang CM, Fang YJ, Bair MJ, Chen PY, et al. Long-term changes of gut microbiota, antibiotic resistance, and metabolic parameters after Helicobacter pylori eradication: a multicentre, open-label, randomised trial. Lancet Infect Dis. 2019;19(10):1109–20. 10.1016/S1473-3099(19)30272-5 [DOI] [PubMed] [Google Scholar]
- 40.Chen TP, Hung HF, Chen MK, Lai HH, Hsu WF, Huang KC, et al. Helicobacter pylori infection is positively associated with metabolic syndrome in Taiwanese adults: a cross-sectional study. Helicobacter. 2015;20(3):184–91. 10.1111/hel.12190 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were available on request.