Abstract
A nucleic acid-bound capsid protein dimer was previously identified using a Sindbis virus in vitro nucleocapsid assembly system and cross-linking reagents. Cross-link mapping, in combination with a model of the nucleocapsid core, suggested that this dimer contained one monomer from each of two adjacent capsomeres. This intercapsomere dimer is believed to be the initial intermediate in the nucleocapsid core assembly mechanism. This paper presents the purification of cross-linked dimers of a truncated capsid protein and the partial purification of cross-linked dimers of a full-length assembly-defective mutant. The assembly of core-like particles from these cross-linked capsid protein dimers is demonstrated. Core-like particles generated from cross-linked full-length mutant CP(19-264)L52D were examined by electron microscopy and appeared to have a morphology similar to that of wild-type in vitro-assembled core-like particles, although a slight size difference was often visible. Truncated cross-linked CP(81-264) dimers generated core-like particles as well. These core-like particles could subsequently be disassembled when reversible cross-linking reagents were used to form the dimers. The ability of the covalent intercapsomere cross-link to rescue capsid proteins with assembly defects or truncations in the amino-terminal region of the capsid protein supports the previous model of assembly and suggests a possible role for the amino-terminal region of the protein.
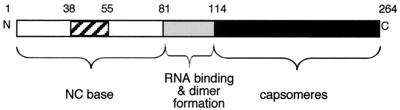
Despite numerous advances in the area of structural biology and biochemistry of the alphaviruses in recent years, the definition of the assembly pathway of the nucleocapsid core (NC) remains elusive. The NC of Sindbis virus (SINV) is a stable cytoplasmic structure composed of 240 copies of a single 264-amino-acid capsid protein (CP) arranged in a 410-Å icosahedron with T=4 symmetry surrounding the viral genomic RNA (3). Although no high-resolution structure of the NC exists, several cryoelectron microscopy image reconstructions of complete alphaviruses have been generated, demonstrating the organization of the NC in the complete virion (3, 13, 21). High-resolution structural information about the CP can be gleaned from the several atomic structures of the carboxyl-terminal chymotrypsin-like region of the protein (4–6, 19). Crystallization of the complete NC, from both in vivo and in vitro sources, has not proved fruitful, due to poor diffraction of crystals generated to date (16). Modeling of the atomic coordinates of CP(114-264) of SINV into the cryoelectron microscopy electron density of Ross River virus has provided a high-resolution model of the structure of the NC (3). Examination of the model suggested that the C-terminal domain of the CP accounts for the density observed comprising the ∼40-Å pentameric and hexameric projections on the surface of the core. Little evidence exists to position the remaining 113 residues of the CP, although this region has been proposed to form both the base of the core upon which the capsomeric projections rest and the internal protein component of the core associated with nucleic acid (Fig. 1) (3, 25). A recent 9-Å cryo-electron microscopy image reconstruction of Semliki Forest virus (SFV) has confirmed the orientation of the CP in the NC proposed in the original model (20).
FIG. 1.
Schematic of SINV nucleocapsid protein. Amino acid numbers are shown on top of the CP schematic, and the positions of the amino (N) and carboxyl (C) termini are indicated. Black shading designates the C-terminal protease domain (residues 114 to 264). The position of helix I is designated by cross-hatching. The CP can be divided into three major regions: the residues comprising the base of the NC, the residues involved in nucleic acid binding and dimer formation (gray), and the residues contained within the capsomeric projections.
An in vitro assembly system using CP purified from Escherichia coli cells has been developed with an aim towards understanding the mechanism of assembly (23). The nucleic acid and protein requirements for in vitro assembly of core-like particles (CLPs) have been characterized. Several truncations and mutants of the CP that failed to assemble CLPs in the in vitro system have been identified. Of these defective proteins, CP(81-264) and CP(19-264)L52D were found to bind nucleic acid and incorporate into CLPs when present as additives along with wild-type protein in assembly reactions. The ability to incorporate into CLPs suggested that these assembly-defective proteins, although unable to complete particle assembly, retained some of the contacts required to oligomerize during assembly. Examination of these particles by electron microscopy suggested that no defect existed in the CLPs as a consequence of truncated or mutant protein incorporation.
More recently, the ability of CP(81-264) and CP(19-264)L52D to bind nucleic acid and the oligomeric state of these proteins associated with nucleic acid have become an area of investigation. When analyzed by X-ray crystallography, the CP was found as a dimer (4, 6, 19); however, analytical ultracentrifugation and biochemical experiments indicated that the solution state of the CP was a monomer (T. L. Tellinghuisen, J. Burgner, and R. J. Kuhn, unpublished data). Combined with the observation that no assembly of NCs occurred in the absence of nucleic acid, these data implied that nucleic acid binding was an early and absolutely required step in assembly (23, 27). Early chemical cross-linking experiments with SINV cores purified from virus particles and CP extracted from purified cores demonstrated that numerous cross-linking reagents could generate oligomers of the CP (8–10). To investigate the oligomeric state of the CP associated with nucleic acid, a variety of lysine-specific imidoester cross-linking reagents were investigated for the ability to identify a nucleic acid-dependent cross-linked oligomer of the CP (24). A dimer of CP(81-264) was observed with cross-linkers having a reagent length of 11 Å, although shorter regents failed to cross-link the CP. Mapping of the location of the cross-link indicated that lysine 250 on one CP monomer was cross-linked to lysine 250 on an adjacent CP. Identical cross-linking was observed with the assembly-defective CP(19-264)L52D, as well as with in vitro-assembled CLPs of both SINV and Ross River virus. Additionally, cross-linked dimers of both CP(81-264) and CP(19-264)L52D could be incorporated into CLPs when present as minor components in assembly reactions. Examination of the existing model of the NC, in relation to the mapped location of the cross-link, both confirms the model and suggests the dimer as a building block of the NC. It has been postulated that nucleic acid-bound dimers of CP incorporate into the assembling NC by an intercapsomere oligomerization. This model of assembly is in accordance with the observed cross-linking and assembly data, as well as by the lack of detectable preformed capsomeres (23, 26, 27). The model of assembly has important implications as to the role of the amino-terminal portion of the CP in spanning the region between capsomeres when associated with nucleic acid.
The data presented in this work further demonstrate the importance of the observed cross-linked dimer. A method for the purification of small amounts of the cross-linked dimer of CP(81-264) and the partial purification of CP(19-264)L52D dimers, as well as methods of refolding these proteins, have been established. The rescue of in vitro CLP assembly using these normally assembly-defective CPs by the presence of the covalent intercapsomere cross-link is described. Although the rescue of CLP formation by cross-linking appears to be an inefficient process, CLPs can be generated from previously assembly-defective proteins. Analysis of CLPs rescued through cross-linking suggests these particles have a shape similar to those of normal CLPs at the level of negative-stain electron microscopy, but appear slightly smaller. The use of reversible cross-linking reagents allows the removal of the cross-link from these rescued CLPs, thereby leading to particle disassembly. The ability of the cross-link to rescue assembly of these defective proteins has important implications for the role of the amino terminus of the alphavirus CP, as well as for the mechanism of particle assembly.
MATERIALS AND METHODS
SINV CP and nucleic acids.
SINV CP(19-264), CP(19-264)L52D, and CP(81-264) were expressed and purified as described previously (23). Proteins are identified by the abbreviation CP followed by the residues expressed in parentheses. Mutant proteins are further identified by the wild-type one-letter amino acid code, the residue number, and the substituted amino acid. Typical protein purities were 90 to 95% as estimated by silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentrations reported were based on absorbance at 280 nm by using an extinction coefficient of 27,880 M−1 cm−1 for CP(19-264) and 22,190 M−1 cm−1 for CP(81-264). The standard synthetic 48-base DNA oligonucleotide used in in vitro assembly assays was 5′-CCGTTAATGCATGTCGAGATATAAAGCATAAGGGACATGCATTAACGG-3′. Transfer RNA material consisted of commercial preparations of ultrapure yeast tRNA (Boehringer Mannheim, Indianapolis, Ind.).
Cross-linking of CP.
Cross-linking of CP(81-264) was performed as described previously (24). Binding of CP(81-264) to DNA oligonucleotides or tRNA was conducted as described below. Equal molar amounts of nucleic acid and protein were mixed at room temperature in buffer A (25 mM HEPES [pH 7.4], 100 mM potassium acetate, 1.7 mM magnesium acetate) in a reaction volume of 25 μl. Binding reaction mixtures were incubated for 10 min. Mock nucleic acid-bound samples contained 12.5 μl of 1-mg/ml CP and 12.5 μl of buffer A. Following nucleic acid binding, reactions were cross-linked where indicated with 1 μl of a 0.25 mM stock of dissuccinimidyl suberate (DSS) or dithio-bis-succinimidyl propionate (DSP) (both from Pierce, Rockford, Ill.) in dry dimethyl sulfoxide. Cross-linking was allowed to proceed for 30 min at room temperature, and reactions were terminated with the addition of glycine to a final concentration of 200 mM. Cross-linking efficiency was monitored by electrophoresis of aliquots of reaction mixtures by SDS-PAGE (12% polyacrylamide gels) under nonreducing conditions and subsequent staining with Coomassie brilliant blue R-250 and densitometric analysis. Efficiency of cross-linking was typically 30%. Reversal of DSP cross-linking was performed by the addition of β-mercaptoethanol to a final concentration of 1%. Cross-linking of CP(19-264)L52D was performed as described for CP(81-264).
Purification of CP dimers.
CP(81-264) or CP(19-264)L52D cross-linking reaction mixtures were mixed with an equal volume of nonreducing SDS-PAGE sample buffer and subjected to SDS-PAGE (12% polyacrylamide gels). CP(81-264) proteins were then transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes by following the manufacturer's protocol. Efficient transfer of proteins from SDS-PAGE gels was monitored by Ponceau S staining of PVDF membranes. Bands corresponding to the cross-linked dimer of CP(81-264) were excised, and the PVDF was minced into fragments of approximately 0.5 mm2. Samples were then vortexed in a 1.5-ml Eppendorf tube in PVDF elution buffer (25 mM Tris [pH 8.5], 1% Triton X-100, 2% SDS) for 10 min. Ten microliters of PVDF elution buffer was used per square millimeter of PVDF membrane excised. After vortexing, PVDF fragments were pelleted by a 1-min centrifugation in a microcentrifuge, and liquid fractions containing the eluted protein were removed. For CP(19-264)L52D dimers, bands were cut directly from unfixed and unstained SDS-PAGE gels and, following mincing of gel slices, were placed in acrylamide elution buffer (25 mM Tris, [pH 7.3], 2% SDS, 1% Triton X-100) at room temperature for 6 h. The location of the CP dimers was determined by direct comparison to a control lane containing a CP(19-264)L52D cross-linking reaction product that was excised from the gel and stained with Coomassie brilliant blue R-250. Approximately 400 μl of elution buffer was used per 100 mg of minced acrylamide. Recovery of both CP(81-264) and CP(19-264)L52D was approximately 80% of input protein. For both CP(19-264)L52D and CP(81-264) protein samples, excess SDS was separated from the purified dimers by use of SDS-OUT reagent (Pierce) by following the manufacturer's suggested protocol. CP(81-264) could be completely purified from contaminating monomeric CP and nucleic acid by using this procedure. CP(19-264)L52D could be completely purified from nucleic acid, but monomeric CP was seen as a contaminant (approximately 25% of total eluted protein) in all preparations of CP(19-264)L52D dimers. It is not clear why complete purification was not achieved with the CP(19-264)L52D dimers, although the full-length and truncated proteins behaved differently during purification and refolding.
CP(19-264) NC assembly.
In vitro assembly of CP(19-264) capsids was performed as described previously (23). Briefly, equal molar amounts of CP and nucleic acid were mixed in a final volume of 100 μl in buffer A at room temperature. Typical reaction mixtures contained 50 μl of 1-mg/ml CP (1.8 nmol) and 50 μl of 1-mg/ml 48-base assembly oligonucleotide (2 nmol) or 50 μl of 500-μg/ml tRNA (1.8 nmol). Reaction mixtures were incubated for 30 min at room temperature, and reaction mixtures were assayed for the presence of CLPs as described elsewhere.
CP(81-264) dimer and CP(19-264)L52D dimer NC assembly.
One hundred microliters of approximately 200 μg of purified DSS- or DSP-cross-linked dimers of CP(81-264) per ml, following SDS removal, was mixed with 100 μl of 200 μg of tRNA/ml in buffer A. Reaction mixtures were incubated for 30 min at room temperature. CP(19-264)L52D reactions were performed identically to those described for CP(81-264) with the exception that dimers were present at approximately 75 μg/ml and tRNA was at 75 μg/ml. Reversal of DSP cross-linking in assembled CLPs was performed by the addition of β-mercaptoethanol to a final concentration of 1%.
Sucrose gradient sedimentation and Western blotting.
Following dimer-only assembly, the presence of CLPs was assayed by sucrose gradient sedimentation. Assembly reaction mixtures (200 μl) were loaded onto 12-ml 25% freeze-thaw sucrose gradients prepared with buffer A. Samples were then centrifuged in an SW-41 rotor (Beckman, Palo Alto, Calif.) at 4°C for 105 min at 38,000 rpm. Following sedimentation, gradients were fractionated into 1-ml aliquots for analysis. For Western blot assays, fractions were separated by SDS-PAGE (12% polyacrylamide gels) and transferred to nitrocellulose membranes. The membranes were exposed to a polyclonal rabbit anti-capsid antibody, followed by exposure to a secondary goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase. Blots were subjected to chemoluminescent detection using WestFempto chemoluminescent reagents (Pierce) and procedures and were exposed to Kodak X-ray film.
Electron microscopy.
Following assembly of CLPs, a 3.5-μl sample was placed on a freshly glow-discharged, 400-mesh copper grid coated with Formvar and carbon. Following 2 min of sample absorption and extensive washing with water, 7 μl of a 2% (wt/vol) uranyl acetate stain was applied. After 4 min of staining, grids were wick dried with Whatman no. 1 filter paper and allowed to air dry for a minimum of 20 min. Samples were then viewed on a Philips EM420 electron microscope using an acceleration voltage of 100 kV at a magnification of ×105,000. Images were captured on Kodak SO-163 EM film.
RESULTS
CP cross-linking.
Previous experiments demonstrating the biological activity of the cross-linked dimer were limited by the inability to purify the cross-linked dimer free of contaminating monomer (24). This inability to generate pure dimer free of the assembly-inhibiting monomeric forms of the defective proteins CP(81-264) and CP(19-264)L52D eliminated the possibility of examining if the cross-link could rescue the defects seen with CP(81-264) and CP(19-264)L52D. Therefore, a method for the purification of cross-linked dimers was required.
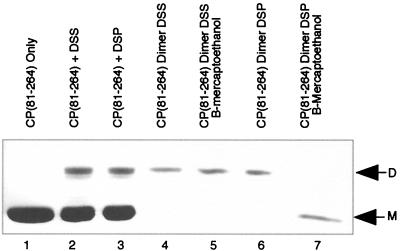
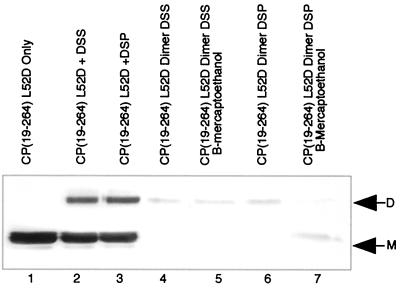
Initial cross-linking experiments with DSS were performed essentially as described previously (24). Figures 2 and 3 present the observed cross-linking with DSS using CP(81-264) and CP(19-264)L52D, respectively. In lanes 1 of both Fig. 2 and 3, the migrations of the monomeric forms of CP(81-264) and CP(19-264)L52D are indicated. Lanes 2 of both figures present the production of a dimeric species of either CP(81-264) (Fig. 2) or CP(19-264)L52D (Fig. 3) with DSS cross-linking. Lanes 3 of both figures present the observed cross-linking of CP with the reversible reagent DSP. No detectable differences in either overall cross-linking efficiency or species cross-linked were observed when comparing DSS and DSP cross-linking Fig. 2 and 3, compare lanes 2 and 3), thereby allowing the use of the reversible cross-linking reagent to investigate the role of the cross-link in particle assembly and stability.
FIG. 2.
Cross-linking, dimer purification, and cross-link reversal of CP(81-264). Lane 1 demonstrates the electrophoretic mobility of CP(81-264) on an SDS–12% polyacrylamide gel stained with Coomassie brilliant blue R-250, with only monomeric CP(M) evident. Lane 2 contains CP(81-264) treated with DSS cross-linker in the presence of tRNA. Monomeric and dimeric (D) forms of C(81-264) are visible. Lane 3 contains CP(81-264) treated with DSP cross-linker in the presence of tRNA. Similar to lane 2, monomeric and dimeric forms of CP(81-264) are present. Lane 4 contains purified DSS-cross-linked dimers of CP(81-264). Only dimeric CP is present. Lane 5 contains samples identical to those of lane 4, but samples were first treated with β-mercaptoethanol; only dimeric CP was detectable in this sample. Lane 6, purified DSP-cross-linked dimers of CP(81-264). Lane 7, β-mercaptoethanol-treated samples identical to those in lane 6. No detectable dimeric form of the CP remained following treatment.
FIG. 3.
Cross-linking, dimer purification, and cross-link reversal of CP(19-264)L52D. Lane 1 demonstrates the electrophoretic mobility of CP(19-264)L52D on an SDS–12% polyacrylamide gel stained with Coomassie brilliant blue R-250, with only monomeric CP (M) evident. Lane 2 contains CP(19-264)L52D treated with DSS cross-linker in the presence of tRNA. Monomeric and dimeric (D) forms of CP(19-264)L52D are visible. Lane 3 contains CP(19-264)L52D treated with DSP cross-linker in the presence of tRNA. Lane 4 contains purified DSS-cross-linked dimers of CP(19-264)L52D. Only dimeric CP is present. Lane 5 contains samples identical to those of lane 4, but samples were first treated with β-mercaptoethanol; only dimeric CP is seen. Lane 6, DSP-cross-linked and purified dimers of CP(19-264)L52D. Lane 7, β-mercaptoethanol-treated samples identical to those in lane 6. A small amount of the dimeric form of the CP remained following reduction.
Dimer purification.
The purification of the dimeric form of CP(81-264) and CP(19-264)L52D was a prerequisite step to investigating the ability of the cross-link to rescue particle assembly. Purification of cross-linked dimers of CPs was attempted by a variety of chromatographic and filtration techniques, with little success. The insolubility of the dimer when removed from nucleic acid and associated monomeric proteins prevented purification. A method of purification of the dimer of the CP by separation on SDS-PAGE gels and transfer to and elution from PVDF membranes was developed. The inclusion of SDS to 2% and Triton X-100 to 1% final concentrations in the PVDF elution buffer eliminated the observed solubility problems seen with more conventional separation techniques, but most likely denatured the CP. Partial refolding of the CPs by removal of SDS was possible, although the bulk of the CP appeared to be in an inactive form as monitored by nucleic acid binding competence. Figure 2, lane 4, shows purified dimers of CP(18-264) cross-linked with DSS. Lane 6 shows the purified dimers of this protein cross-linked with the reversible reagent DSP. The dimeric form of CP(81-264) could be completely separated from contaminating monomeric protein, yielding only dimeric CP, as assayed by Western blot analysis. Figure 3, lane 4, presents the partially purified dimer of CP(19-264)L52D generated with DSS. Lane 6 represents partially purified DSP-cross-linked dimers. Although CP(19-264)L52D dimers appear pure at the level of Coomassie brilliant blue R-250 staining (Fig. 3), the dimer of this protein could not be completely isolated from monomeric CP, but preparations containing 75 to 90% dimeric CP could be generated. The dimeric form of both proteins could be completely purified from nucleic acid, as monitored by agarose gel electrophoresis and ethidium bromide staining (data not shown).
Reversal of cross-linking.
The reversal of DSP cross-linking, by reduction of the internal disulfide of this reagent, was performed by treatment of purified dimers with β-mercaptoethanol. The ability to specifically eliminate the DSP cross-link allowed for evaluation of the effects of the cross-link on the oligomerization and assembly of defective CPs. Figures 2 and 3, lanes 5, show the treatment of purified DSS-cross-linked dimers of CP(18-264) and CP(19-264)L52D with β-mercaptoethanol, respectively. No loss of the dimeric form of the CP was observed, as DSS lacks an internal disulfide. Lanes 7 in Fig. 2 and 3 show the reversal of DSP-cross-linked dimers with β-mercaptoethanol. No dimeric form of CP(81-264) remained after reversal of the cross-link, with only a monomeric form of CP visible by Coomassie brilliant blue R-250 staining. It should be noted that trace amounts of CP(81-264) dimer could be detected after reduction by Western blotting using polyclonal anti-capsid antisera (corresponding to less than 5% of input dimer; data not shown). Reversal of CP(19-264)L52D DSP dimers by reduction led to a partial loss of the dimeric form of the CP, although the dimer could not be completely converted to monomeric CP. In all instances, approximately 25% of CP(19-264)L52D protein remained dimeric. The ability to cross-link CP and at least partially reverse the cross-link provided a convenient method to allow analysis of the effect of cross-linking on the rescue of assembly-defective proteins.
Analysis of cross-linked particle assembly.
To determine if the observed cross-linking of CP(81-264) and CP(19-264) could rescue the assembly competence of these defective CPs, in vitro assembly using purified dimers was performed essentially as described previously for wild-type CP(19-264) in vitro CLP assembly (23). These dimeric protein-only assembly reaction mixtures were then sedimented on sucrose gradients in a manner identical to that described previously for the detection of CP(19-264) CLPs. Gradients were fractionated, samples were applied to SDS-PAGE gels, and following electrophoretic separation, samples were transferred to nitrocellulose membranes, and Western blotting was performed with anti-capsid antisera as described in Materials and Methods.
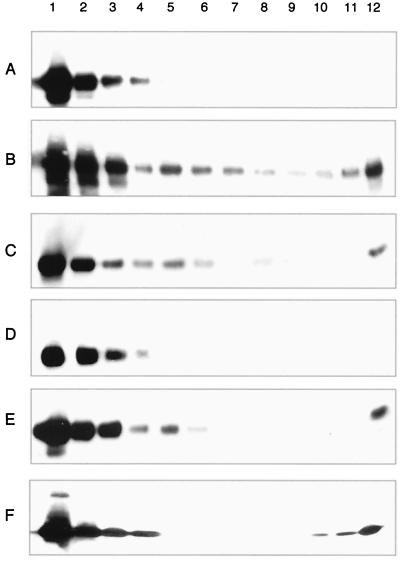
Figure 4 presents Western blot analysis results for CP(81-264) dimer-only assembly reactions. Gradient fractions in each of the panels are presented from the top (lane 1) to the bottom (lane 12) of the gradient. As described previously, authentic CP(19-264) CLPs sedimented with fractions 6 and 7 under the conditions utilized for analysis of dimer assembly (23). Figure 4A represents the gradient sedimentation profile of purified CP(81-264) dimers generated by cross-linking CP with DSS and incubating without nucleic acid. CP(81-264) dimers remained on the top of the gradient and were distributed in the first four fractions, indicating that no CLP formation had occurred. Figure 4B represents CP(81-264) dimers incubated with an equal molar amount of tRNA prior to gradient sedimentation. A peak corresponding to the migration of CP(81-264) CLPs is present in fractions 5 and 6. Figure 4C shows treatment of DSS-cross-linked CP(81-264) dimer CLPs identical to those shown in Fig. 4B with 1% β-mercaptoethanol. As the DSS reagent did not contain a disulfide, no loss of CLP sedimentation was observed with these samples. Figure 4D shows the Western blot analysis of gradient sedimentation of CP(81-264) dimers cross-linked with DSP. As seen for DSS dimers of CP(81-264), no CLP migration is evident in this panel. Figure 4E demonstrates the formation of CLPs by CP(81-264) cross-linked with DSP when protein samples were incubated with tRNA. As with CLPs generated by the CP(81-264) DSS dimers, dimers generated with CP(81-264) plus DSP sedimented in a position slightly higher than CLPs generated with CP(19-264). The reversal of cross-linking by treatment of DSP CP(81-264) dimers is shown in Fig. 4F. Samples identical to those shown in Fig. 4E were incubated for 30 min in the presence of 1% β-mercaptoethanol and were analyzed as described for samples in the other panels. No CLP sedimentation in fraction 5 or 6 is evident, suggesting that reversal of the cross-link leads to disassembly of CLPs of CP(81-264).
FIG. 4.
Gradient sedimentation of cross-linked CP(81-264) dimer assembly reaction mixtures. In vitro CLP assembly with CP(81-264) cross-linked dimers and tRNA were assayed by sucrose density gradient sedimentation followed by fractionation and Western blot analysis. Numbers across the top of this panel correspond to gradient fractions presented from top (lane 1) to bottom (lane 12). CLPs generated in vitro with CP(19-264) and tRNA sedimented with fractions 6 and 7 under these conditions. (A) CP(81-264) DSS-cross-linked dimers. (B) CP(81-264) cross-linked dimers incubated with equal molar amounts of tRNA. Sedimentation of CP into the gradient (fractions 5 to 7) is evident. (C) Sample identical to that in panel B, but treated with β-mercaptoethanol. (D) CP(81-264) DSP-cross-linked dimers. (E) CP(81-264) DSP-cross-linked dimers incubated with equal molar amounts of tRNA. As in panels B and C, sedimentation of CP into the gradient (fractions 5 to 7) is evident. (F) Samples identical to those in panel E, but treated with β-mercaptoethanol; no CLP sedimentation was observed.
Experiments identical to those described above for CP(81-264) dimers were performed with CP(19-264)L52D dimers, yielding similar results. Figure 5 presents data for the rescue of CP(19-264)L52D assembly by cross-linking and the reversal of this phenotype by the reduction of the labile DSP reagent. Figure 5A is Western blot data for the sedimentation of CP(19-264)L52D dimers generated by DSP cross-linking. No sedimentation in the position consistent with CLPs is seen in the absence of nucleic acid, although the protein does seem to enter the gradient more than expected for free dimer (Fig. 5A, lane 5). In Fig. 5B, the sedimentation of CP(19-264)L52D dimers incubated with tRNA is shown, with CLPs visible in fractions 6 and 7, as is normally seen for the wild-type CP(19-264). Figure 5C shows samples identical to those in Fig. 5B, but samples were treated with 1% β-mercaptoethanol. DSS-cross-linked dimers of CP(19-264)L52D sedimented in gradients in a position consistent with CLPs when mixed with tRNA in the absence and presence of reducing agent (data not shown). These data suggested that cross-linking of assembly-defective CPs generated sucrose sedimentation profiles similar to those produced using wild-type full-length CP following in vitro assembly. Cross-linked samples generated significantly less CLP formation (approximately 1 to 5% of input protein) than typical wild-type CP(19-264) assembly reaction mixtures, which yielded approximately 80 to 90% of input protein assembled into CLPs (data not shown). The low efficiency of assembly observed for the cross-linked dimeric proteins compared to that of normal wild-type assembly reaction mixtures was likely due to the inefficiency of dimeric protein refolding following purification.
FIG. 5.
Gradient sedimentation of cross-linked CP(19-264)L52D dimer assembly reaction mixtures. In vitro CLP assembly with CP(19-264)L52D cross-linked dimers and tRNA assayed by sucrose density gradient sedimentation followed by Western blot analysis. Numbers across the top of this panel correspond to gradient fractions presented from top (lane 1) to bottom (lane 12). CLPs generated in vitro with wild-type CP(19-264) and tRNA sedimented with fractions 6 and 7 under these conditions. (A) CP(19-264)L52D DSP-cross-linked dimers. (B) CP(19-264)L52D DSP-cross-linked dimers incubated with equal molar amounts of tRNA. Sedimentation of CP into the gradient (fractions 5 to 7) is evident. (C) Sample identical to that in panel B, but treated with β-mercaptoethanol; no CLP sedimentation was observed in this sample.
Analysis of assembly rescued particles by electron microscopy.
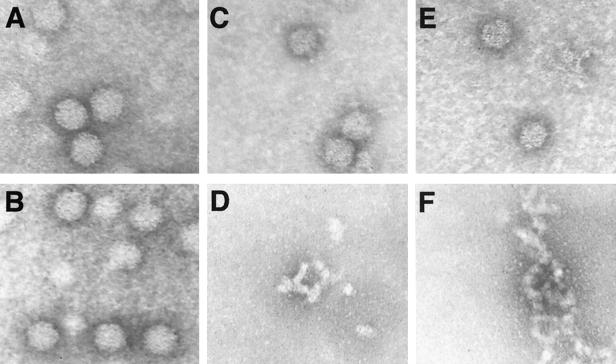
Although the data strongly suggested that CLPs were formed using dimeric forms of CP(81-264) and CP(19-264)L52D, negative-stain electron microscopy was utilized to confirm the presence of small amounts of CLPs in dimer-only assembly reactions. Figure 6 demonstrates analysis of DSP-cross-linked rescued particles of CP(81-264) and CP(19-264)L52D, the reversal of cross-linking in the particles, and control samples of wild-type SINV CP(19-264) in vitro-assembled particles. Figure 6A shows CLPs assembled in vitro using wild-type CP(19-264) for comparison with dimer-rescued CLPs. Treatment of these wild-type CLPs with β-mercaptoethanol had no effect on the particle morphology (Fig. 6B). Figure 6C shows particles with the size and morphology of wild-type CLPs (compare to Fig. 6A) assembled from cross-linked CP(81-264) dimers. A sample identical to the one shown in Fig. 6C, except for being treated with 1% (final concentration) β-mercaptoethanol, is shown in Fig. 6D. No particles are visible in this field, with only large aggregates detectable. Although no particles are seen in this field, CLPs were occasionally visible in CP(81-264) cross-link-reversed samples, possibly due to the incomplete reversal of cross-linking. Figure 6E demonstrates CLPs formed from cross-linked dimers of CP(19-264)L52D. As seen with CP(81-264) dimer assembly reaction mixtures (Fig. 6C), particles similar to wild-type in vitro-assembled CLPs were clearly visible. The reversal of the cross-link by reduction of the disulfide with β-mercaptoethanol eliminated particles from the sample (Fig. 6F). As mentioned for CP(81-264) cross-linking reaction mixtures, particles were rarely seen in these reduced samples. No dimer-only particles were observed without nucleic acid in a variety of low and high ionic strength buffers (data not shown).
FIG. 6.
Negative-stain electron microscopy of CLPs generated with cross-linked CPs. (A) In vitro CP(19-264) CLPs demonstrating normal capsid morphology. (B) CP(19-264) CLPs following treatment with β-mercaptoethanol. (C) CP(81-264) DSP-cross-linked dimer CLPs. (D) CP(81-264) DSP-cross-linked dimer CLPs identical to those in panel C, but treated with β-mercaptoethanol. (E) CP(19-264)L52D DSP-cross-linked dimer CLPs. (F) CP(19-264)L52D DSP-cross-linked dimer CLPs identical to those in panel E, but treated with β-mercaptoethanol. All micrographs are shown at a magnification of ×105,000.
DISCUSSION
Assembly studies using the SINV in vitro assembly system have suggested the importance of nucleic acid binding in initiation of the early stages of the core assembly process (23, 27). Indeed, no particle formation or oligomerization of the CP was detectable in the absence of nucleic acid. By the examination of the CP associated with nucleic acid using cross-linking reagents, a putative intermediate in the assembly process was observed (24). A nucleic acid-dependent dimerization of the CP was identified, and the protein and nucleic acid requirements for dimer formation were described. The biological relevance of the nucleic acid-bound dimer of the CP was confirmed by the ability of these dimers to incorporate into CLPs during in vitro assembly. Additionally, identical dimers were detectable in assembled CLPs, indicating that the dimer was also present in the final product of the NC assembly mechanism. More recently, identical cross-linking of in vivo-isolated core particles has indicated that the cross-link observed in the in vitro cores is not an artifact of the system (S. Mukhopadhyay, M. G. Rossmann, and R. J. Kuhn, unpublished results). Mapping of the location of the observed cross-link together with the previous results suggested the location of the dimer in the assembled NC (24). This allowed for an assembly model based on the binding of the monomeric CP to nucleic acid, followed by an oligomerization into intercapsomere dimers and subsequent polymerization of multiple dimers into an icosahedral core particle.
An interesting observation in the preliminary cross-linking work was the fact that truncated and mutant CPs capable of binding nucleic acid, but lacking assembly competence by themselves, could be cross-linked efficiently into dimers (24). These CPs, containing deletions or mutations in the N-terminal region (amino acids 1 to 80) of the CP, were capable of incorporating into CLPs when added to assembly reactions but were incapable of proceeding along the assembly pathway into complete CLPs. It was hypothesized that the N-terminal region of the CP was involved in bridging the intercapsomere space between the two monomers that compose an intercapsomere dimer (Fig. 1). The importance of nucleic acid in establishing this contact was also suggested. Examination of the putative location of the N-terminal region of CP in the fitted model of the NC, combined with this putative role of the amino terminus, suggested that the N-terminal region stabilizes the interactions of the intercapsomere dimer, allowing the oligomerization of 120 dimers into an icosahedral core particle (3, 24). It was proposed that truncations of and mutations in this N-terminal region of the CP were still competent for nucleic acid binding and subsequent dimer formation, but lacked sufficient stability to allow oligomerization into a complete NC (24). As the cross-link previously described lies in the intercapsomere space and covalently links CPs into an intercapsomere dimer, it was hypothesized that the presence of the cross-link may suppress the stability defect in these mutant CPs and lead to the rescue of assembly of CLPs. To this end, a procedure for the purification of cross-linked CP(81-264) dimers and the partial purification of CP(19-264)L52D dimers was developed. Although the purification procedure developed allowed the production of dimeric CPs, the protein produced was denatured and required refolding by SDS removal prior to conducting assembly experiments. It has previously been demonstrated that CP can be refolded to generate active protein, but the dimeric protein, containing an 11-Å hydrophobic spacer tethering two unfolded proteins together, cannot be refolded efficiently. Based on monitoring of the nucleic acid binding activity of purified dimeric protein, the refolding process generated mostly inactive CP dimers (data not shown). Despite the production of mostly inactive dimeric protein, the purification procedure allowed generation of enough active CP dimers to ascertain the ability of the cross-link to rescue defective CLP assembly in vitro. Both purified cross-linked dimers of CP(81-264) and CP(19-264)L52D were capable of assembling small amounts of CLPs in vitro. In addition, nonpurified mixtures of monomeric and dimeric CP(81-264) or CP(19-264)L52D were capable of assembling CLPs in vitro in the presence of nucleic acid, but were assembly defective in the absence of cross-linking. The CLPs rescued from dimeric CPs appeared similar to wild-type in vitro-assembled CLPs at the level of negative-stain electron microscopy. However, rescued particles often appeared to have a slightly smaller diameter than wild-type CLPs, suggesting a defect in the particles possibly due to the constrained nature of the covalently cross-linked dimer. It is important to remain cautious in the interpretation of the size and morphology of particles based solely on negative-stain electron microscopy. Although rescued CLPs appeared similar in size and shape to wild-type in vitro CLPs by negative-stain electron microscopy, no evidence exists that either of these particles possesses icosahedral symmetry.
Examination of the atomic model of the SINV NC suggests that the observed cross-linking is likely between pentamers and hexamers, rather than a hexamer-hexamer contact, as lysine 250 distances are more favorable between pentamers and hexamers (10.5 Å for pentamer-hexamer contacts versus 14.7 Å for hexamer-hexamer contacts) in the model. The assembly of CLPs entirely from proteins constrained by an 11-Å cross-link separation may account for the slightly smaller size often observed for the rescued particles as well as the inefficient process of CLP formation, as the CPs in these particles would have difficulty making the 14.7-Å contact between hexameric capsomeres. It should be noted that the distances between capsomeres in the model represent measurements of a model of the NC and not an actual atomic structure of the NC. These measurements, therefore, may not represent actual distances in the NC and may not provide an explanation for the differences in particle sizes.
Although inefficient, the rescue of CLP formation for these CPs has several important implications for the role of the N-terminal region and the assembly mechanism itself. The N-terminal 100 amino acids of the CP are poorly conserved among the alphaviruses. This region is highly charged and has been hypothesized to be involved in nonspecific nucleic acid binding in the interior of the NC (3, 25). A short stretch of uncharged amino acids (residues 38 to 55) interrupts this charged region and has been proposed to form an α-helix (helix I) based on modeling of the primary sequence into helical wheel plots. The presence of two conserved leucine residues at positions 45 and 52 (SINV amino acid numbering) suggests that helix I may form a leucine zipper interaction motif between two CPs (1, 7, 14, 15, 18). Mutation of leucine 52 to aspartic acid [CP(19-264)L52D] eliminated cytoplasmic NC formation in vivo and abolished in vitro assembly (22). Based on the present results, the role of helix I may be to stabilize intercapsomere dimeric interactions following the formation of these dimer intermediates. Mutagenesis of helix I of the SINV CP has suggested that this region is not absolutely required for virus viability, but is directly involved in virion stability, presumably through stabilization of the NC (22). The ability of cross-linked dimers of CP(19-264)L52D to assemble CLPs suggests that the physical cross-link overcomes the lack of stabilization due to disruption of helix-helix interactions by mutation of the conserved leucine at position 52. This appears to be similar to the proposed ability of the glycoproteins to stabilize both SINV and SFV mutant CPs, which do not accumulate stable cytoplasmic NCs, and to allow NC formation at the plasma membrane (11). The trimeric glycoprotein spikes make direct contact with three individual CPs in three separate capsomeres, possibly providing intercapsomere stabilization similar to the role proposed for helix I and the artificial cross-link. These data provide the first direct evidence for the role of the proposed helix I in the assembly and structure of the NC. It remains to be demonstrated if this region of the CP is indeed helical and also what specific contacts are made in the assembly of the NC.
The ability of proteins having deletions in their N-terminal region to assemble into CLPs when present as cross-linked dimers, as exemplified by CP(81-264), demonstrates that the cross-link can substitute for the first 80 amino acids. This suggests that, apart from the helix described above, no absolutely required contacts exist in this region of the CP for ordered spherical particle assembly. Previous deletion analysis of the N-terminal region of SFV had demonstrated that large portions of the N-terminal region of the CP (individual deletions of amino acids 11 to 29, 11 to 63, and 66 to 78, SFV numbering) could be deleted without completely eliminating virus formation in infected cells (12). This is in agreement with the hypothesis that this region is primarily involved in nonspecific contacts with the viral RNA (3, 25). Despite the deletion of this RNA interaction region of the CP, no empty CLPs were observed and no CLP assembly was seen in the absence of nucleic acid in a variety of high-salt buffers. The exact nucleic acid composition of CP(81-264) dimer particles remains to be demonstrated, but the assembly of CLPs from this protein appears to require the same nucleic acid-to-protein ratio required for CP(19-264) assembly (23). The N-terminal region of the CP clearly does have a role in CLP assembly, as truncations of the protein past amino acid 32 abolish in vitro assembly, although these truncations may merely eliminate the stabilization provided by helix I (23).
The ability to generate CLPs using only cross-linked dimers has important implications for understanding the mechanism of assembly. Even with CP(19-264)L52D partially purified dimer preparations, only dimeric CP is observed in the peak corresponding to CLPs. The definition of a single dimeric intermediate severely limits the possible mechanisms of assembly (2, 17). Additional assembly limitations are imposed if the in vitro-assembled CLP is modeled as an icosahedral lattice based on observations of the icosahedral nature of the NC in the mature virion. Although an important component of the assembly model, the icosahedral nature of the in vitro-assembled CLP remains to be demonstrated experimentally. Despite these limitations, the presently available data suggest a model of NC assembly. The current intercapsomere dimer assembly model places 120 identical dimers into a hypothetical T=4 icosahedral lattice, imposing quasi-symmetry on the dimers as particle assembly progresses. In this model, a dimer is composed of one monomer from each of two adjacent capsomeres of the particle in a manner described previously (23). This mechanism of assembly, at least in principle, is similar to that observed with several small RNA plant viruses, such as cowpea chlorotic mottle virus (CCMV) (28), in which polymerization of multiple dimers in an intercapsomeric manner leads to the assembly of an icosahedral particle. Although the in vitro CLP is predicted to be a T=4 icosahedron, and CCMV is a T=3 virion, some similarity in the mechanism of assembly appears to exist. Both particles are made from dimeric forms of their respective CPs and appear to assemble using an intercapsomeric placement of dimers. In the case of the CCMV, intercapsomere dimers can form in the absence of nucleic acid, whereas alphavirus dimers are only observed when bound to nucleic acid. Preliminary analytical ultracentrifugation experiments suggest that the SINV CP is at least 95% monomer in solution at concentrations of up to 2 mg/ml (T. L. Tellinghuisen, J. Burgner, and R. J. Kuhn, unpublished data). The placement of identical dimers into quasi-symmetrical positions in the proposed NC icosahedron raises important questions as to what controls pentamer and hexamer formation (2, 17). For CCMV, a model based largely on the structure of the virion has postulated the importance of a hexamer of dimers as an assembly nucleus, onto which intercapsomere dimers are polymerized to generate a complete particle (28). More recent work measuring polymerization kinetics by light scattering has confirmed the polymerization of dimers to complete assembly, but has suggested a pentamer of dimers as an assembly nucleus (29). Similarly, hepatitis B virus assembly is believed to proceed by addition of preformed dimeric assembly intermediates to a trimeric nucleus (30).
The identification of an assembly nucleus and determination of how the pentamer and hexamer formation is determined during dimer addition for the alphavirus NC are important in further defining the mechanism of particle formation, but these questions remain unanswered. However, the identification of the interesting properties of cross-linked dimers using assembly-defective CPs has led to new insights and understanding of the mechanism for the assembly of the alphavirus NC.
ACKNOWLEDGMENTS
We acknowledge Thomas Smith for valuable assistance in the development of cross-linking conditions. Additionally, critical discussions with Michael Rossmann, Sergei Pletnev, Suchetana Mukhopadhyay, John Burgner, and Chris Jones are gratefully acknowledged.
This research was supported by Public Health Service grant no. GM56279 from the National Institutes of Health. Additional funding from the Lucille Markey Foundation for structural studies at Purdue University is acknowledged. T.L.T. was supported, in part, by an NIH biophysics training grant (GM98296).
REFERENCES
- 1.Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 2.Caspar D L D, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 3.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi H-K, Lee S, Zhang Y-P, McKinney B R, Wengler G, Rossmann M G, Kuhn R J. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J Mol Biol. 1996;262:151–167. doi: 10.1006/jmbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 5.Choi H-K, Lu G, Lee S, Wengler G, Rossmann M G. The structure of Semliki Forest virus core protein. Proteins Struct Funct Genet. 1997;27:345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 6.Choi H-K, Tong L, Minor W, Dumas P, Boege U, Rossmann M G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 7.Cohen C, Parry D A. Alpha-helical coiled coils: more facts and better predictions. Science. 1994;263:488–489. doi: 10.1126/science.8290957. . (Erratum, 264:1068, 1994.) [DOI] [PubMed] [Google Scholar]
- 8.Coombs K, Brown B, Brown D T. Evidence for a change in capsid morphology during Sindbis virus envelopment. Virus Res. 1984;1:297–302. doi: 10.1016/0168-1702(84)90018-2. [DOI] [PubMed] [Google Scholar]
- 9.Coombs K, Brown D T. Organization of the Sindbis virus nucleocapsid as revealed by bifunctional cross-linking agents. J Mol Biol. 1987;195:359–371. doi: 10.1016/0022-2836(87)90657-7. [DOI] [PubMed] [Google Scholar]
- 10.Coombs K, Brown D T. Topological organization of Sindbis virus capsid protein in isolated nucleocapsids. Virus Res. 1987;7:131–149. doi: 10.1016/0168-1702(87)90075-x. [DOI] [PubMed] [Google Scholar]
- 11.Forsell K, Griffiths G, Garoff H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996;15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsell K, Suomalainen M, Garoff H. Structure-function relation of the NH2-terminal domain of the Semliki Forest virus capsid protein. J Virol. 1995;69:1556–1563. doi: 10.1128/jvi.69.3.1556-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 14.Harbury P B, Tidor B, Kim P S. Repacking protein cores with backbone freedom: structure prediction for coiled coils. Proc Natl Acad Sci USA. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbury P B, Zhang T, Kim P S, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 16.Harrison S C, Strong R K, Schlesinger S, Schlesinger M J. Crystallization of Sindbis virus and its nucleocapsid. J Mol Biol. 1992;226:277–280. doi: 10.1016/0022-2836(92)90141-6. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J E, Speir J A. Quasi-equivalent viruses: a paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 18.Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Owen K E, Choi H K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid protein and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 20.Mancini E J, Clarke M, Gowen B E, Rutten T, Fuller S D. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 21.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perera R, Owen K E, Tellinghuisen T L, Gorbalenya A E, Kuhn R J. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J Virol. 2001;75:1–10. doi: 10.1128/JVI.75.1.1-10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tellinghuisen T L, Hamburger A E, Fisher B R, Ostendorp R, Kuhn R J. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tellinghuisen T L, Kuhn R J. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J Virol. 2000;74:4302–4309. doi: 10.1128/jvi.74.9.4302-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wengler G. The mode of assembly of alphavirus cores implies a mechanism for the disassembly of the cores in the early stages of infection. Arch Virol. 1987;94:1–14. doi: 10.1007/BF01313721. [DOI] [PubMed] [Google Scholar]
- 26.Wengler G, Boege U, Wengler G, Bischoff H, Wahn K. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology. 1982;118:401–410. doi: 10.1016/0042-6822(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 27.Wengler G, Wengler G, Boege U, Wahn K. Establishment and analysis of a system which allows assembly and disassembly of alphavirus core-like particles under physiological condition in vitro. Virology. 1984;132:401–412. doi: 10.1016/0042-6822(84)90045-x. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Fox J M, Olson N H, Baker T S, Young M J. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in E. coli and in vitro-transcribed viral cDNA. Virology. 1995;207:486–494. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnick A, Aldrich R, Johnson J M, Ceres P, Young M J. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 30.Zlotnick A, Johnson J M, Wingfield P W, Stahl S J, Endres D. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry. 1999;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]