Abstract
Context
Oxytocin supplementation improves obstructive sleep apnea (OSA), and animal studies suggest involvement of oxytocin in respiratory control. However, the relationship between endogenous oxytocin signaling and human sleep status remains undetermined.
Objective
In this study, we approached the contribution of the intrinsic oxytocin-oxytocin receptor (OXTR) system to OSA by genetic association analysis.
Methods
We analyzed the relationship between OXTR gene polymorphisms and sleep parameters using questionnaire data and sleep measurements in 305 Japanese participants. OSA symptoms were assessed in 225 of these individuals.
Results
The OXTR rs2254298 A allele was more frequent in those with OSA symptoms than in those without (P = .0087). Although total scores on the Pittsburgh Sleep Quality Index questionnaire did not differ between the genotypes, breathlessness and snoring symptoms associated with OSA were significantly more frequent in individuals with rs2254298 A genotype (P = .00045 and P = .0089 for recessive models, respectively) than the G genotype. A multivariable analysis confirmed these genotype-phenotype associations even after adjusting for age, sex, and body mass index in a sensitivity analysis. Furthermore, objective sleep efficiency measured by actigraph was not significantly different between genotypes; however, subjective sleep efficiency was significantly lower in the rs2254298 A genotype (P = .013) compared with the G genotype. The frequency of the A allele is higher in East Asians, which may contribute to their lean OSA phenotype.
Conclusion
The OXTR gene may contribute to OSA symptoms via the respiratory control system, although it could be in linkage disequilibrium with a true causal gene.
Keywords: oxytocin, oxytocin receptor, obstructive sleep apnea, genetic association study
Obstructive sleep apnea (OSA) is a highly prevalent breathing disorder characterized by episodes of breathing cessation during sleep, and its prevalence is expected to increase further with the global increase in obesity [1]. OSA is not only associated with poor quality of life, reduced work efficiency, and increased driving accidents [2, 3] but also with increased incidence of hypertension [4], type 2 diabetes [5], stroke [6], atrial fibrillation, heart failure, and coronary disease [7]. Despite this ever-increasing health burden caused by OSA, beneficial drug therapies against this disease are currently not yet available or approved.
The pathogenesis of OSA is formed by a complex combination of anatomical factors of maxillofacial morphology, obesity, and physiological characteristics of the upper airway collapse [8, 9]. In addition, respiratory regulatory systems are also involved in the pathogenesis of upper airway obstruction. OSA pathogenesis has been considered to be caused by the instability of the ventilatory drive from the medullary respiratory center and the loss of genioglossus muscle activity [10, 11].
Not only environmental factors but also genetic factors are involved in OSA pathogenesis [12], and genetic polymorphisms involved in the OSA development have been reported by genome-wide association study (GWAS) [13-17].
Oxytocin (OXT) is a nonapeptide produced by magnocellular neurons located in the paraventricular and supraoptic nuclei, secreted by the posterior pituitary gland, and acts as a hormone. It is also widely secreted into the brain through exocytosis from the cell bodies and dendrites of magnocellular neurons, where it plays an essential role in regulating social behaviors and cognitive processes [18, 19]. Furthermore, parvocellular OXTergic neurons in the paraventricular nuclei project to the medulla oblongata and spinal cord [20]. To date, OXT has been linked to several pathological conditions, and based on its usage in obstetrics, it is recognized for its high safety as a therapeutic agent [21]. Based on this background, its effectiveness as a treatment for various pathological conditions has been investigated. In clinical trials, the beneficial effects of nasal OXT administration on schizophrenia [22], pain [23], and acute alcohol withdrawal syndrome [24] have been reported. Of particular note is that intranasal administration of OXT was shown to have some efficacy in OSA [25, 26]. Drug targets in the pipeline with human genetic evidence of disease association are reported to be more than twice as likely to lead to an approved drug [27]. Therefore, this study aimed to clarify the association between the OXT-OXT receptor (OXTR) system and OSA from genetic association analysis, considering the therapeutic application of OXT.
The OXTR gene, which encodes the OXT receptor, a major player in OXT signaling, has been associated with various psychiatric disorders and social cognition through genetic association analysis [28]. In particular, the association between OXTR SNPs and diseases has been extensively studied, primarily in the field of autism. Among these, rs53576 and rs2254298 are the single nucleotide polymorphisms (SNPs) most frequently reported to be associated with autism and related phenotypes [29-35]. Although the neural circuits involved in OSA and psychiatric disorders, including autism, are different, rs53576 and rs2254298 have been consistently reported in large studies, suggesting their importance in influencing OXTR signaling. While most research on OXTR SNPs has focused on psychiatric disorders, these 2 SNPs have also been linked to a variety of conditions involving diverse organs and neural circuits, such as bowel disease [36], pain [37], alcohol abuse [38], and liver fibrosis [39]. These findings indicate that rs53576 and rs2254298 may have broader biological significance, potentially affecting biological systems including respiratory control.
To verify the involvement of endogenous OXT signals in sleep states, the relationship between genetic polymorphisms of OXTR and sleep-wake states measured by detailed sleep questionnaires, including OSA-related items and sleep measurements by actigraph, was investigated in a Japanese community population cohort.
Methods
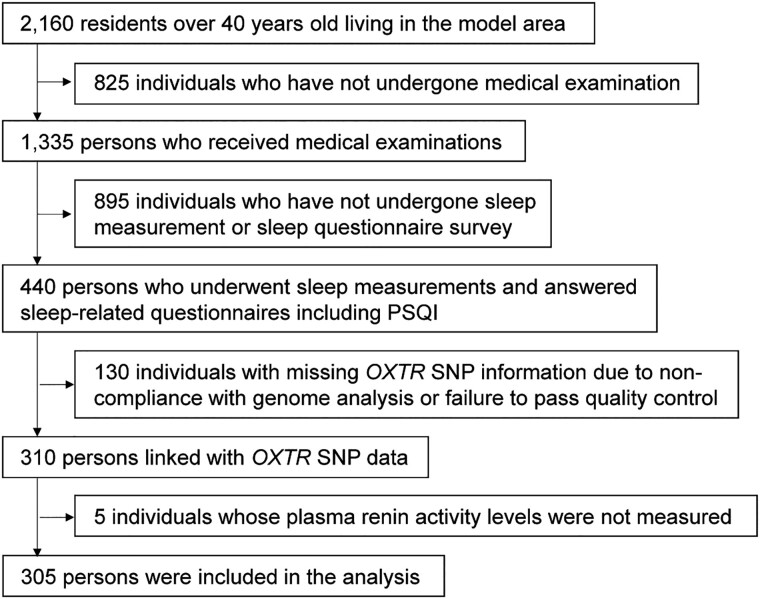
Study Participants
Data from a cross-sectional analysis known as the Shika study were obtained [40]. This project was conducted from 2013 to 2016 in the town of Shika, located in the rural sector of Ishikawa Prefecture, Japan. The primary objective was to monitor the health conditions of the local populace and explore preventive measures for lifestyle-related diseases. Cluster sampling, one of the random sampling methods, was employed as the sample extraction method. Specifically, this study was conducted on all middle-aged residents legally residing in 2 specific elementary school districts (Horimatsu Elementary School District and Higashi Masuho Elementary School District) within the town of Shiga, Ishikawa Prefecture. All 2160 residents were asked to complete a questionnaire and participate in a health check. All respondents who voluntarily agreed to participate in the health check were included in the study. From this group, 440 adults aged ≥ 40 years answered a comprehensive sleep questionnaire, including the Pittsburgh Sleep Quality Index (PSQI), and underwent sleep measurement using actigraphy, which determines sleep and wakefulness from time series data of measured physical activity. Of these, 130 individuals were excluded due to missing SNP information. Additionally, 4 individuals with missing plasma renin activity data were excluded, as plasma renin activity was used as an objective marker for the presence of OSA symptoms. Therefore, all analyses were conducted on the subset of participants with complete plasma renin activity data, to ensure consistency in the target population. Ultimately, the study analyzed 305 individuals. To enhance clarity, we have included a study flow diagram (Fig. 1) that clearly illustrates the participant selection process. OSA symptoms were defined based on observations of sleep apnea. Therefore, the association between OXTR SNPs and OSA symptoms was examined in the 225 subjects, excluding 80 subjects whose OSA status was unknown, likely due to the lack of third-party observation. To handle other missing data, pairwise deletion was utilized. Specifically, data were missing for 14 subjects for estimated glomerular filtration rate based on serum cystatin C (eGFRcys), 52 for nonspecific immunoglobulin E (IgE), 14 for N-terminal pro-brain-type natriuretic peptide (NT-proBNP), and 134 for alcohol intake as estimated by the brief-type self-administered diet history questionnaire (BDHQ).
Figure 1.
Flowchart of inclusion and exclusion criteria for the study population.
Data Collection
For each participant, data, including age, gender, and body metrics (height, weight), were obtained. The body mass index (BMI) was calculated by dividing an individual's weight in kilograms by the square of their height in meters. The alcohol consumption assessment leveraged the Japanese adaptation of the BDHQ, a validated instrument for determining the intake frequency of 58 prevalent Japanese food and beverage items in the last month, chosen for their representation in the National Health and Nutrition Survey of Japan. Designed for in-depth nutritional epidemiology studies in Japan, the BDHQ's reliability and repeatability have been verified in previous studies [41, 42]. Following overnight fasting, blood specimens were drawn from the forearm vein of the participants, typically between 08:00 and 12:00. These samples were thereafter forwarded to the SRL Kanazawa Laboratory and preserved at Kanazawa University at a temperature of −30 °C until the analysis.
To assess the presence of OSA symptoms, participants were asked in the questionnaire about “observed to stop breathing during sleep.” This question aligns with the “observed to stop breathing” item in the well-established STOP-Bang questionnaire, which is widely used for OSA screening [43]. The observation of cessation of breathing is one of the most effective items for OSA screening [44] and is a common component in many OSA screening questionnaires [45]. Participants who reported the “observed to stop breathing during sleep” were considered to have OSA symptoms. To assess sleep quality over the past month, a self-administered survey, the PSQI, consisting of 19 questions, was used [46]. These questions are designed to evaluate 7 distinct aspects of sleep: subjective sleep quality, sleep latency, sleep duration, subjective sleep efficiency, sleep disturbances, sleep medication use, and daytime activity dysfunction. The overall PSQI score is derived by summing the scores from these 7 areas, resulting in a total score ranging from 0 to 21. A lower score is indicative of better sleep quality. The PSQI is widely recognized as a reliable and valid tool for detecting sleep disorders and assessing sleep quality in various populations [47]. In addition, its usefulness as a screening tool for OSA has been reported [48]. The PSQI includes items related to snoring, which is also a component of the STOP-Bang score for OSA screening, as well as items related to breathlessness during sleep, which is associated with OSA. For sleep measurement, the ACOS MTN-220 actigraph was used to assess sleep/wake status. For data processing, we used an algorithm provided by the manufacturer. This algorithm determines sleep/wake states based on activity intensity data collected every 0.125 seconds and summarized over a 2-minute epoch. The algorithm employs a 5-variable linear model that includes activity intensity data for the current epoch, the 2 previous epochs, and the 2 subsequent epochs. The effectiveness of this algorithm has been demonstrated in previous studies, showing a high overall agreement with polysomnography [49].
SNP Genotyping
The Qiagen QIAamp DNA Blood Maxi Kit was used to isolate the genomic DNA from the obtained blood samples. This process adhered to the manufacturer's guidelines or was alternatively executed by a specialized clinical lab, SRL Inc. The Japonica Array v2 produced by TOSHIBA Co., Ltd was used for genotyping of genome-wide SNPs. The SNP genotype data underwent rigorous quality control measures. Quality control procedures for both SNPs and participants involved several criteria: matching gender identity as indicated in karyotypic analysis with questionnaire results, call rate thresholds, evaluation through the Hardy–Weinberg equilibrium test, analysis of inbreeding coefficients, detection of cryptic familial relationships, and population structure assessment. This process and its protocols have been comprehensively described in previous publications [50]. The 1000Genomes_30x research data are the source of allele frequency data for the rs2254298 gene for each population [51].
Statistical Analysis
All statistical analyses were performed using RStudio (version 2023.03.0 Build 386). The normality of continuous variables was assessed using both the Kolmogorov-Smirnov test and Q-Q plots. Results from the Q-Q plots were consistent with those from the Kolmogorov-Smirnov test. In our analysis of between-group differences, normally distributed continuous variables were evaluated using the 2-sample t test. The Mann-Whitney U test was employed for non-normally distributed variables. Categorical variables were evaluated using Pearson's chi-squared test. When 3 groups were compared, particularly under a codominant genetic model, the Kruskal-Wallis test was used for variables lacking normal distribution. Each SNP (rs2254298 and rs53576) was analyzed independently to assess their association with OSA symptoms. These 2 SNPs were selected based on their previous associations with psychiatric disorders and social behaviors, and importantly, they are not in significant linkage disequilibrium with each other, allowing for independent analyses. To account for multiple comparisons, a Bonferroni correction was applied based on the number of SNPs analyzed (2 SNPs: rs2254298 and rs53576) and the number of outcomes investigated after narrowing the analysis to rs2254298. For analyses involving the 2 SNPs, the corrected alpha level was set at 0.025 (0.05 ÷ 2). After focusing on rs2254298, the correction was applied based on the 3 outcomes, and the corrected alpha level was set at 0.0167 (0.05 ÷ 3). This ensured appropriate control of the type I error rate across all comparisons. Our approach to identifying the most appropriate genetic model followed the methodology outlined by Minelli et al [52] and modified by Rotman et al [53]. Univariate odds ratios (ORs) were calculated to contrast homozygous minor alleles with homozygous major alleles (represented as ORGG) and heterozygous with homozygous major alleles (represented as ORGg). The mode of inheritance was determined by the ratio of log ORGg to log ORGG, known as λ. Consistent with criteria from previous studies, λ values of < 0.25 were classified as recessive, between 0.25 and 0.75 as additive, and > 0.75 as dominant. In this study, after determining that the recessive model best fit the data, we focused solely on the recessive inheritance model for our analysis. For multivariable analysis, ordinal logistic regression and reported ORs were obtained along with their 95% CI.
Ethics
Our study strictly complied with the ethical standards laid out in the Helsinki Declaration. The study protocol was approved by the Human Studies Ethics Committee of Kanazawa University Hospital, with approval numbers 1491, 2016-376. Furthermore, our study was officially registered in the University Hospital Medical Information Network (UMIN) Clinical Trials Registry, bearing the registration number UMIN 000024915. Before participation, written consent was obtained from all of the participants after being fully informed about the study.
Results
First, to address and assess potential selection bias in the target population for which sleep and actigraph data were available, we compared sociodemographic variables and lifestyle factors between participants with and without sleep/actigraph data. Specifically, we assessed age, gender, BMI, smoking status (current, former, or nonsmoker), and alcohol consumption habits. The results of this comparison are shown in Supplementary Table S1 [54]. A statistically significant difference between the 2 groups was found in smoking status. Fewer participants with sleep data were current smokers, suggesting that sleep data might be collected from a more health-oriented group.
Table 1 details the background of patients analyzed in this study. The presence of OSA symptoms, frequency of sleeping medication use divided into 4 levels, subjective sleep quality assessed by the PSQI, and primary sleep parameters measured by actigraph were assessed. Since alcohol consumption significantly affect sleep quality [55], the amount of alcohol consumption assessed by the BDHQ was evaluated.
Table 1.
Background, sleep parameters, and OXTR SNPs of the subjects
| Parameter | n = 305 |
|---|---|
| Age | 64.00 [55.00, 70.00] |
| BMI | 23.32 [21.31, 25.31] |
| Sex, n (%) | |
| Male | 143 (46.9) |
| Female | 162 (53.1) |
| OSA symptoms, n (%) | |
| No | 189 (62.0) |
| Yes | 36 (11.8) |
| Uncertain | 80 (26.2) |
| Sleep medication, n (%) | |
| 0 | 289 (94.8) |
| 1 | 4 (1.3) |
| 2 | 3 (1.0) |
| 3 | 9 (3.0) |
| Alcohol intake (g/day) | 2.39 [0.00, 17.85] |
| PSQIG | 4.00 [3.00, 7.00] |
| Sleep parameters by actigraph | |
| Number of days measured by sleep monitor | 7.00 [7.00, 7.00] |
| Total sleep time (min/day) | 366.29 [322.67, 412.86] |
| Wake after sleep onset (min/day) | 50.50 [26.44, 78.00] |
| Number of wakes after sleep onset (/day) | 4.00 [2.17, 5.86] |
| Number of awakenings over 9 minutes (/day) | 3.00 [0.14, 12.43] |
| Number of posture changes (/day) | 16.38 [3.57, 55.14] |
| Sleep efficiency (%) | 83.89 [76.07, 89.39] |
| Sleep latency (min) | 14.57 [9.60, 22.50] |
| OXTR SNPs frequency | |
| rs53576 genotype (%) | |
| A/A | 128 (42.0) |
| A/G | 134 (43.9) |
| G/G | 43 (14.1) |
| rs2254298 genotype (%) | |
| A/A | 33 (10.8) |
| G/A | 107 (35.1) |
| G/G | 165 (54.1) |
Categorical variables are displayed as n (%).
Continuous variables are displayed as mean (SD) or median [interquartile range].
Sleep medication is scored based on the frequency of sleep medication use in the last month as follows: 0, no use; 1, less than once per week; 2, 1-2 times per week; 3; more than 3 times per week.
Abbreviations: BMI, body mass index; OSA, obstructive sleep apnea; PSQIG, The Pittsburgh Sleep Quality Index Global score; SNP, single nucleotide polymorphism.
The number of SNPs that could be analyzed was first calculated due to the limited number of participants. Based on the allele frequency of rs2254298, a representative OXTR SNP, the proportion of patients with and without OSA symptoms, the number of alleles in the target population, and the Bonferroni-corrected α error, the number of SNPs available for analysis was calculated as 2. Two OXTR SNPs, rs2254298 and rs53576, were examined independently for their association with OSA symptoms because they have been most commonly reported to be associated with psychiatric disorders and social behaviors and are not in linkage disequilibrium with each other [29-35]. The A allele frequency of rs2254298 was significantly higher (P = .0087) in participants with OSA symptoms than in those without. With respect to rs53576, no significant difference was observed in allele frequency between patients with and without OSA symptoms (Table 2). Based on these results, subsequent analyses focused exclusively on rs2254298 to further explore its association with OSA-related symptoms.
Table 2.
Association between respiratory arrest symptoms and rs2254298 allele frequency
| SNP ID/Allele | Respiratory arrest symptoms | P value | |
|---|---|---|---|
| rs2254298 | No | Yes | |
| A | 97 (25.7%) | 30 (41.7%) | .0087a |
| G | 281 (74.3%) | 42 (58.3%) | .0087a |
| SNP ID/Allele | Respiratory arrest symptoms | P value | |
|---|---|---|---|
| rs53576 | No | Yes | |
| A | 242 (64.0%) | 52 (72.2%) | .23 |
| G | 136 (36.0%) | 20 (27.8%) | |
P values were from chi-square test.
a Significant difference (corrected α = 0.025, Bonferroni procedure).
Single nucleotide polymorphism (SNP) allele distribution significantly associates with sleep apnea severity, suggesting a genetic basis for its variability.
Factors associated with sleep quality and representative sleep indices were compared among genotypes of rs2254298 (Table 3). No significant differences were found between genotypes under recessive genetic model for items associated with sleep quality, such as age, sex, and frequency of sleep medication use. No significant differences were observed in the total PSQI scores or various sleep measurement indices assessed using actigraph, including objective sleep efficiency between genotypes (Table 3). Given that our study included 305 participants, it is possible that the lack of a significant association could be due to insufficient power.
Table 3.
Association of rs2254298 genotype with factors affecting sleep quality and sleep metrics
| Parameters | Genotypes of rs2254298 | ||||
|---|---|---|---|---|---|
| A/A | G/A | G/G | Recessive model | ||
| n | 33 | 107 | 165 | ||
| Age | 61.00 [49.00, 69.00] | 64.00 [55.00, 70.00] | 64.00 [56.00, 70.00] | 0.24 | |
| BMI | 22.51 [21.31, 24.61] | 23.87 [21.55, 25.85] | 23.12 [21.11, 24.86] | 0.559 | |
| Sex (%) | Male | 17 (51.5) | 54 (50.5) | 72 (43.6) | 0.585 |
| Female | 16 (48.5) | 53 (49.5) | 93 (56.4) | ||
| Frequency | |||||
| Sleep medication (%) | 0 | 30 (90.9) | 101 (94.4) | 158 (95.8) | 0.317 |
| 1 | 2 (6.1) | 1 (0.9) | 1 (0.6) | ||
| 2 | 0 (0.0) | 1 (0.9) | 2 (1.2) | ||
| 3 | 1 (3.0) | 4 (3.7) | 4 (2.4) | ||
| Alcohol intake (g/day) | 4.18 [0.00, 24.10] | 6.27 [0.00, 28.64] | 0.23 [0.00, 12.81] | 0.7 | |
| PSQIG | 5.00 [3.00, 7.00] | 5.00 [3.00, 7.00] | 4.00 [3.00, 7.00] | 0.23 | |
| Sleep parameters by actigraph | |||||
| Total sleep time (min/day) | 344.00 [312.00, 388.00] | 361.33 [310.41, 408.57] | 376.57 [326.57, 420.00] | 0.179 | |
| Wake after sleep onset (min/day) | 49.43 [29.43, 78.57] | 50.86 [28.57, 76.78] | 50.50 [25.50, 78.29] | 0.687 | |
| Number of wake after sleep onset (/day) | 4.17 [2.43, 6.25] | 4.00 [2.30, 5.67] | 3.86 [2.00, 5.86] | 0.621 | |
| Number of awakenings over 9 minutes (/day) | 3.29 [1.86, 4.75] | 2.86 [1.73, 4.43] | 3.00 [1.67, 4.43] | 0.665 | |
| Number of posture changes(/day) | 16.71 [12.71, 20.50] | 16.13 [11.29, 20.05] | 16.71 [12.00, 22.71] | 0.952 | |
| Sleep efficiency (%) | 83.89 [70.95, 88.46] | 82.48 [75.94, 88.82] | 84.33 [77.20, 89.58] | 0.64 | |
| Sleep latency (min) | 15.71 [7.71, 18.75] | 14.29 [9.14, 22.54] | 14.57 [10.00, 21.71] | 0.664 | |
Categorical variables are displayed as n (%). Continuous variables are displayed as median [interquartile range].
Parameters were evaluated in a univariate analysis.
Nonparametric variables were compared using the Kruskal-Wallis test in the additive model and the Mann-Whitney U test in the recessive model.
Pearson chi-square test was used for categorical parameters.
Abbreviations: BMI, body mass index; OSA, obstructive sleep apnea; PSQIG, The Pittsburgh Sleep Quality Index Global score; SNP, single nucleotide polymorphism.
Next, a recessive model was used to perform inter-genotype comparisons for individual PSQI items related to OSA (Table 4). Subjective sleep efficiency (ratio of actual sleep time/bedtime in the past month) was significantly lower in the A genotype of rs2254298 (P = .013). The questionnaire asking about the frequency of symptoms associated with OSA, the frequency of sleep difficulty due to breathlessness assessed by Q5d (P = .00045), and coughing and loud snoring assessed by Q5e (P = .0089) revealed that they were both significantly higher in the A genotype (Table 4). To account for multiple comparisons across these 3 outcomes, Bonferroni correction was applied to control for the risk of type I errors. Even after applying the correction, the differences between genotypes remained statistically significant, with the corrected alpha set at 0.0167.
Table 4.
Association of rs2254298 genotype with PSQI items related to OSA
| Parameters | rs2254298 recessive model | |||
|---|---|---|---|---|
| Severity | A Genotype, n = 33 | G Genotype, n = 272 | P value | |
| Subjective sleep efficiency | 0.78 [0.40, 0.97] | 0.92 [0.75, 1.00] | .013a | |
| 0 | 28 (84.8) | 265 (97.4) | .00045a | |
| C5 Q5d. (%) | 1 | 3 (9.1) | 4 (1.5) | |
| Cannot breathe comfortably | 2 | 0 (0.0) | 2 (0.7) | |
| 3 | 2 (6.1) | 1 (0.4) | ||
| 0 | 27 (81.8) | 256 (94.1) | .0089a | |
| C5 Q5e. (%) | 1 | 3 (9.1) | 11 (4.0) | |
| Cough or snore loudly | 2 | 2 (6.1) | 4 (1.5) | |
| 3 | 1 (3.0) | 1 (0.4) | ||
Categorical variables are displayed as n (%). Continuous variables are displayed as median [interquartile range].
Nonparametric variables were compared using the Mann-Whitney U test.
a Significant difference (corrected α = 0.0167, Bonferroni procedure).
C5 asks how often have you had trouble sleeping during the past month for each reason.
The scores for each item of C5 are as follows. 0, not during the past month; 1, less than once a week; 2, once or twice a week; 3, three or more times a week.
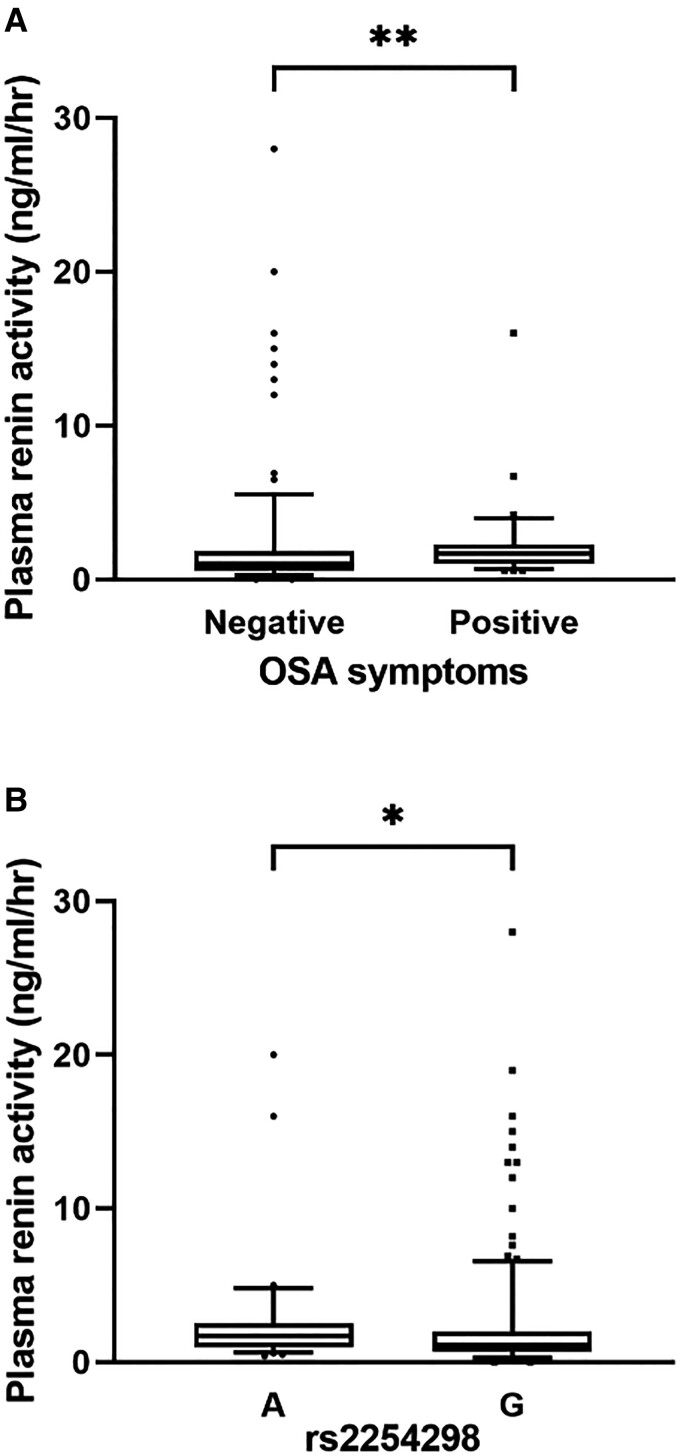
OSA activates the sympathetic activity [7], which can lead to increased plasma renin activity [56]. Indeed, plasma renin activity was significantly higher (P = .0025) in participants with OSA symptoms in our cohort (Fig. 2A). Plasma renin activity was significantly higher in the rs2254298 A genotype (P = .011, recessive model) than in the G genotype, supporting OSA symptom exacerbation in the A genotype (Fig. 2B).
Figure 2.
Association of the plasma renin activity with OSA symptoms and rs2254298 genotype. (A) Comparison of plasma renin activity with (n = 173) and without (n = 29) OSA symptoms. (B) Comparison of plasma renin activity between rs2254298 A genotype (n = 33) and rs2254298 G genotype participants (n = 272) (recessive model). **P < .01; *P < .05;
Abbreviation: ns, not significant.
In addition to OSA, chronic renal failure, chronic heart failure, and bronchial asthma are associated with paroxysmal nocturnal dyspnea [57-59]. Estimated GFR based on serum cystatin C (eGFRcys), N-terminal proBNP (NT-proBNP), and serum immunoglobulin E (IgE), which reflect the pathophysiology of these diseases, showed no significant differences between genotypes (Table 5).
Table 5.
Association of rs2254298 genotype with activity indices of diseases that can cause breathlessness during sleep
| rs2254298.recessive | A Genotype | G Genotype | P value |
|---|---|---|---|
| eGFRcys (mL/min/1.73 m2) | 79.80 [65.95, 100.25] | 82.20 [70.83, 94.30] | .826 |
| Nonspecific IgE (IU/mL) | 59.80 [17.50, 138.00] | 62.35 [18.00, 163.50] | .77 |
| NT-proBNP (pg/mL) | 38.00 [22.50, 81.00] | 42.00 [23.00, 66.00] | .963 |
Continuous variables are displayed as median [interquartile range].
Nonparametric variables were compared using the Mann-Whitney U test.
Abbreviations: eGFRcys, estimated GFR based on serum cystatin C; IgE, immunoglobulin E; NT-proBNP, N-terminal proBNP.
Although alcohol consumption is known to increase the frequency and duration of sleep apnea [60], no significant differences in alcohol consumption were observed between the rs2254298 genotypes (Table 3). Similarly, while obesity is a known risk factor for OSA [61], no significant BMI differences were observed between the rs2254298 genotypes (Table 3). We examined whether there was an interaction between rs2254298 genotype and alcohol intake or BMI with respect to OSA-related symptoms assessed by Q5d and Q5e of the PSQI. No significant interaction was observed between rs2254298 and alcohol consumption (Q5d, P = .086; Q5e,,P = .194). On the other hand, interaction analysis between rs2254298 and BMI (SNP × BMI interaction) showed a suggestive effect on OSA symptoms (Q5d; P = .356; Q5e, P = .035). However, as we simultaneously evaluated 2 outcomes (Q5d and Q5e), Bonferroni correction was applied to control for multiple comparisons, and the significance threshold was set at 0.025. None of the interactions reached this corrected significance level. Specifically, the effect of the rs2254298 genotype on OSA risk appears to decrease as BMI increases, although these results should be interpreted with caution due to the lack of statistical significance. Despite this potential interaction, multivariable analysis adjusting for BMI, sex, and age revealed that the OXTR rs2254298 A genotype remained an independent risk factor for increased sleep difficulty (items Q5d and Q5e; Table 6). Even after adjusting for smoking and alcohol intake, which can be problematic when extrapolating results from this population with lower smoking prevalence and potentially more health-conscious behaviors to the general population, the OXTR rs2254298 A genotype remained a significant risk factor for OSA-related symptoms (Table 6).
Table 6.
Multivariable analysis of the association between the rs2254298 genotype and OSA-related PSQI items
| Factors | Odds ratio | P value |
|---|---|---|
| Q5d | ||
| Model 1: Adjusted for Age, Sex, and BMI | 6.62 (1.79-23.04) | .0031a |
| Model 2: Model 1+ Alcohol intake | 6.10 (1.18-30.49) | .025a |
| Model 3: Model 1+ Smoking habits | 6.76 (1.82-23.58) | .0029a |
| Model 4: Model 1+ Drinking habits | 6.85 (1.85-23.98) | .00259a |
| Q5e | ||
| Model 1: Adjusted for Age, Sex, and BMI | 3.75 (1.23-10.24) | .013a |
| Model 2: Model 1+ Alcohol intake | 3.83 (1.02-13.07) | .036a |
| Model 3: Model 1+ Smoking habits | 3.68 (1.21-10.07) | .015a |
| Model 4: Model 1+ Drinking habits | 3.65 (1.20-10.02) | .015a |
Multivariable P values were from ordinal logistic regression analyses.
a Significant difference (P < .05).
To examine the contribution of OXTR polymorphism to OSA by population, the rs2254298 A allele frequency among populations was compared, showing that the A allele frequency was higher in the Japanese and East Asian populations than in the European and other populations (Table 7).
Table 7.
Interpopulation comparison of rs2254298 SNP A allele frequency
| Shika-cohort | 1000 Genomes Project | |||||
|---|---|---|---|---|---|---|
| Japanese | East Asia | South Asia | European | American | African | |
| A allele frequency | 0.285 | 0.356 | 0.105 | 0.109 | 0.223 | 0.249 |
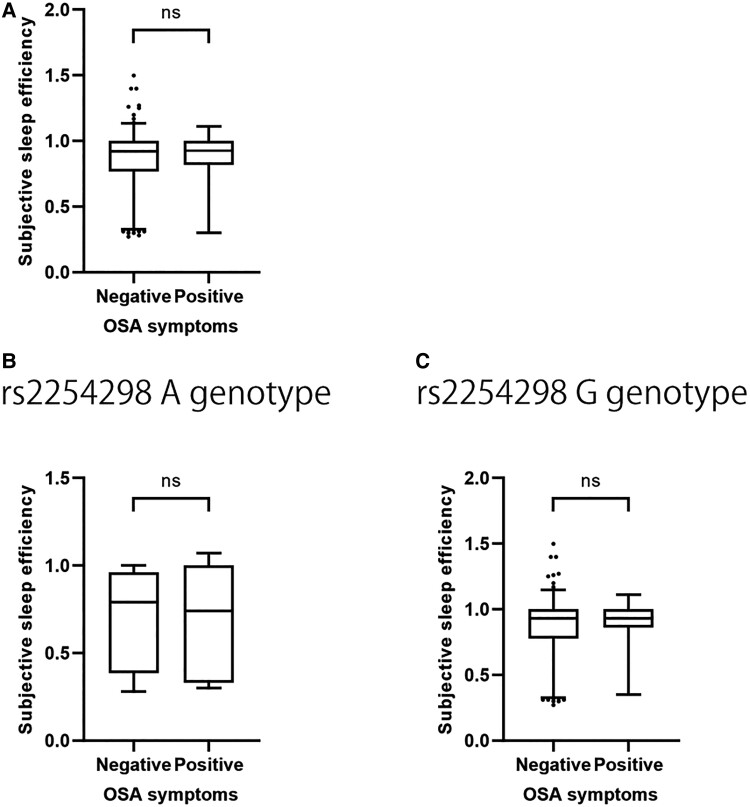
Regarding the cause of decreased subjective sleep efficiency in the A genotype, OSA symptoms might contribute to decreased subjective sleep efficiency; however, no difference in subjective sleep efficiency was observed between participants with and without OSA symptoms (P = .43) (Fig. 3A). In addition, the same analysis was performed for each rs225428 genotype, and no significant differences in subjective sleep efficiency were observed between the groups with and without OSA symptoms in either the A genotype or G genotype of the recessive model (P = .737, P = .365, respectively) (Fig. 3B and 3C).
Figure 3.
Association of subjective sleep efficiency with OSA symptoms. A, Comparison of subjective sleep efficiency between participants with (n = 189) and without (n = 36) OSA symptoms for all participants. B, Comparison of subjective sleep efficiency between participants with (n = 16) and without (n = 7) OSA symptoms in the rs2254298 A genotype (recessive model). C, Comparison of subjective sleep efficiency between participants with (n = 173) and without (n = 29) OSA symptoms in the rs2254298 G genotype (recessive model).
Abbreviation: ns, not significant.
Discussion
The present study comprehensively evaluated the OXTR polymorphism, sleep quality assessment based on questionnaires, and sleep measurements. The OXTR rs2254298 A allele was associated with the frequency of OSA symptoms and subjective sleep efficiency.
By what mechanism is OXT signaling involved in OSA? Ventilatory drives from the medullary respiratory center operate on the diaphragm and other respiratory muscles, as well as the hypoglossal nerve, which innervates the genioglossus muscle, a group of muscles involved in opening the upper airway. The instability of this ventilatory drive from the medullary respiratory center is present in OSA pathology [11]. In addition, neuromuscular electrical stimulation of the hypoglossal nerve has been used as a potential alternative to positive airway pressure therapy, suggesting the importance of upper airway collapse due to decreased genioglossus muscle activity in the OSA pathogenesis [10, 62]. Paraventricular nucleus–derived OXTnergic neurons project to the rostral ventrolateral medulla region and phrenic nucleus and exert respiratory output via diaphragmatic movements [63]. Furthermore, a recent animal study reported that OXT receptors are also abundant in hypoglossal motoneurons in the medulla and that OXT administration stimulates respiratory-related tongue muscle activity, which in turn facilitates the opening of the upper airway [64]. Indeed, clinical studies in patients with OSA have confirmed that intranasal OXT treatment significantly reduced the duration of obstructive events and the incidence of oxygen desaturation and bradycardia and increased respiratory rate during the nonobstructive period, consistent with the supposed physiological effects of OXTergic neurons [26]. Respiratory control system abnormalities have been implicated in the familial clustering of OSA; however, its specific genetic predisposition has not yet been determined [63, 65]. Combined with our findings, OXTR could be a genetic predisposition involved in the OSA pathogenesis via modulation of the respiratory control system, including the phrenic nerve and the hypoglossal nerve.
Mice deficient in the OXTR gene exhibit an obese phenotype due to impaired thermogenesis [66]. Since obesity is a typical risk factor for OSA [61] and is strongly influenced by genetic predisposition [67], the association between obesity and OXTR polymorphisms was also examined. However, no significant differences in BMI were observed among the rs2254298 genotypes, indicating that rs2254298 may not be directly associated with BMI in this population.
In the current study, the rs2254298 A genotype, the genetic predisposition for OSA, was not associated with objective sleep efficiency as measured by actigraph but with lower subjective sleep efficiency as assessed by the PSQI. Conversely, our findings indicate that reduced subjective sleep efficiency is not explicitly associated with OSA symptoms and that reduced subjective sleep efficiency in individuals with the rs2254298 A genotype may also be independent of OSA symptoms. Although the exact mechanism of subjective–objective sleep discrepancy as observed in the OXTR rs2254298 A genotype is not yet understood [68], considering that OXTR SNPs are associated with negative-emotion reactivity under a stressful context [69], A allele carriers may perceive stresses such as sleep latency and awakenings as more significant and underestimate actual sleep duration.
Regarding the ethnic clustering of OSA, studies comparing the association between OSA and population within the same cohort have found higher ORs for OSA in Asians compared with European-origin individuals when adjusted for age and BMI [70, 71]. Anatomical issues such as the distribution of fat deposition and craniofacial structure have been assumed to account for these ethnic differences [72]. Still, the influence of specific genetic predisposition differences and nonanatomical factors has not been evident. The A allele of rs2254298, the risk allele identified in this study, is particularly enriched in Japanese and East Asians compared with other populations. Therefore, this risk allele could explain the genetic background of the higher risk of OSA in Asians by mechanisms other than anatomic factors.
The present study is the first to consider OXTR as a genetic predisposition of OSA. However, the following limitations of this study are inevitable: first, familial clustering has also been reported to exist in the anatomical characteristics of maxillofacial morphology in patients with OSA, and 5 loci among the genes identified in the GWAS of OSA are associated with facial morphological characteristics [73]. In the present study, data on maxillofacial morphological characteristics were not obtained, making it difficult to evaluate the association between OXTR gene polymorphisms and maxillofacial morphology. Second, although sleep apnea status was estimated in the present study based on subjective symptoms and hypoxia-responsive sympathetic nerve activity, a follow-up study is needed to further improve the design by adding polysomnographic indices, which are essential for the definitive diagnosis of OSA. The third limitation of our study is the assessment of alcohol consumption using the self-reported Japanese adaptation of the BDHQ. Self-reported measures can introduce biases, such as underreporting of alcohol consumption. Additionally, the BDHQ was not administered to all participants due to differences in implementation by year, which can be considered a form of random allocation. There were no significant differences in rs2254298 genotype, alcohol consumption habits, OSA symptom-related indicators, age, or gender between those who completed the BDHQ and those who did not (data not shown). Therefore, we believe that the results of our multivariable analyses, adjusted for alcohol consumption, can be generalized to the overall study population. A fourth limitation of this study was the low smoking rate among participants with sleep/actigraphy data. This may have resulted in selection bias. This group had fewer smokers compared to the overall population, suggesting that the overall risk of OSA may be lower due to greater health awareness. Therefore, caution should be used in generalizing the results. However, the association between rs2254298 genotype and OSA symptoms remained significant even after adjusting for potential confounders such as smoking and alcohol consumption, which are problematic in health-oriented populations. This suggests that the genotype-phenotype relationship is robust and potentially generalizable despite initial selection bias.
In conclusion, OXTR polymorphism rs2254298 was identified as a genetic polymorphism associated with OSA symptoms, subjective sleep efficiency, and sympathetic activity, independently of alcohol intake and weight. However, it is important to note that rs2254298 may not be the direct causal variant, but it could be in linkage disequilibrium with another causal variant. Further studies are needed to explore the true causal variants. The A allele of rs2254298 may contribute to the increased risk of OSA in Asians compared with other ethnic groups. The current study highlights the potential of OXT as a treatment for OSA, for which there is still no FDA-approved treatment.
Acknowledgments
We thank the participants from the Shika town and all staff for their cooperation in the Shika study. We would like to give special thanks to Sakae Miyagi, Chie Takazawa, Yukari Shimizu, Talica Marama, Tomoko Kasahara, Koichi Hayashi, and Tadashi Konoshita for their help with data collection in Shika town.
Abbreviations
- BDHQ
brief-type self-administered diet history questionnaire
- BMI
body mass index
- eGFRcys
estimated glomerular filtration rate based on serum cystatin C
- GWAS
genome-wide association study
- IgE
immunoglobulin E
- NT-proBNP
N-terminal pro-brain-type natriuretic peptide
- OSA
obstructive sleep apnea
- OXT
oxytocin
- OXTR
oxytocin receptor
- PSQI
Pittsburgh Sleep Quality Index
- SNP
single nucleotide polymorphism
Contributor Information
Hisanori Goto, Email: pitanori19890838@gmail.com, Department of Endocrinology and Metabolism, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan; Department of Biochemistry and Molecular Vascular Biology, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan.
Yasuhiko Yamamoto, Department of Biochemistry and Molecular Vascular Biology, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan.
Hiromasa Tsujiguchi, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan; Advanced Preventive Medical Sciences Research Center, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Takehiro Sato, Department of Human Biology and Anatomy, Graduate School of Medicine, University of the Ryukyus, Nishihara, Okinawa 903-0215, Japan.
Reina Yamamoto, Department of Endocrinology and Metabolism, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan.
Yumie Takeshita, Department of Endocrinology and Metabolism, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan.
Yujiro Nakano, Department of Endocrinology and Metabolism, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan.
Takayuki Kannon, Department of Biomedical Data Science, School of Medicine, Fujita Health University, Toyoake, Aichi 470-1192, Japan.
Kazuyoshi Hosomichi, Laboratory of Computational Genomics, School of Life Science, Tokyo University of Pharmacy and Life Sciences, Hachioji, Tokyo 192-0392, Japan.
Keita Suzuki, Advanced Preventive Medical Sciences Research Center, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Masaharu Nakamura, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan.
Yasuhiro Kambayashi, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan; Department of Public Health, Faculty of Veterinary Medicine, Okayama University of Science, Imabari, Ehime 794-8555, Japan.
Jiaye Zhao, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan.
Atsushi Asai, Advanced Preventive Medical Sciences Research Center, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Koji Katano, Advanced Preventive Medical Sciences Research Center, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Aya Ogawa, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan.
Shinobu Fukushima, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan.
Aki Shibata, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan.
Fumihiko Suzuki, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan; Department of Geriatric Dentistry, Ohu University School of Dentistry, Koriyama, Fukushima 963-8611, Japan.
Hirohito Tsuboi, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan; Graduate School of Human Sciences, The University of Shiga Prefecture, Hikone, Shiga 522-8533, Japan.
Akinori Hara, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan; Advanced Preventive Medical Sciences Research Center, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Mitsuhiro Kometani, Department of Health Promotion and Medicine of the Future, Kanazawa University Graduate School of Medical Sciences, Kanazawa 920-8640, Japan.
Shigehiro Karashima, Institute of Liberal Arts and Science, Kanazawa University, Kanazawa 920-1192, Japan.
Takashi Yoneda, Department of Health Promotion and Medicine of the Future, Kanazawa University Graduate School of Medical Sciences, Kanazawa 920-8640, Japan.
Atsushi Tajima, Department of Bioinformatics and Genomics, Graduate School of Advanced Preventive Medical Sciences, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Hiroyuki Nakamura, Department of Hygiene and Public Health, Faculty of Medicine, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa City 920-8640, Japan; Advanced Preventive Medical Sciences Research Center, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan.
Toshinari Takamura, Email: ttakamura@med.kanazawa-u.ac.jp, Department of Endocrinology and Metabolism, Kanazawa University Graduate School of Medical Sciences, Kanazawa, Ishikawa 920-8640, Japan.
Funding
None
Author Contributions
The authors’ responsibilities were as follows: H.G. designed the study; H.G., Y.Y., and T.T. conducted the research; T.K. and H.T. built the database for the cohort; T.S., K.H., and A.T. performed genetic analyses; H.G. analyzed the data; H.T., R.Y., Y.T., Y.N., K.S., S.M., M.N., C.T., Y.S., Y.K., J.Z., T.M., A.A., K.K., T.K., A.O., S.F., A.S., F.S., K.H., T.K., H.T., A.H., A.T., and H.N. supported the data analysis; H.G. wrote the article; H.G. and T.T. edited the article and had primary responsibility for final content; and all authors read and approved the final manuscript.
Disclosures
The authors declare no conflict of interest.
Data Availability
The datasets analyzed in the current study are not publicly available but are available from the corresponding authors upon reasonable request.
Clinical Trials Registration
This study was approved by the Ethics Committee for Human Studies at Kanazawa University Hospital (1491, 2016-376) and performed following the principles outlined in the Declaration of Helsinki.
References
- 1. Benjafield AV, Ayas NT, Eastwood PR, et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med. 2019;7(8):687‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Terán-Santos J, Jiménez-Gómez A, Cordero-Guevara J. The association between sleep apnea and the risk of traffic accidents. Cooperative group Burgos-Santander. N Engl J Med. 1999;340(11):847‐851. [DOI] [PubMed] [Google Scholar]
- 3. Engleman HM, Douglas NJ. Sleep. 4: sleepiness, cognitive function, and quality of life in obstructive sleep apnoea/hypopnoea syndrome. Thorax. 2004;59(7):618‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lavie P, Herer P, Hoffstein V. Obstructive sleep apnoea syndrome as a risk factor for hypertension: population study. BMJ. 2000;320(7233):479‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagayoshi M, Punjabi NM, Selvin E, et al. Obstructive sleep apnea and incident type 2 diabetes. Sleep Med. 2016;25:156‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Munoz R, Duran-Cantolla J, Martínez-Vila E, et al. Severe sleep apnea and risk of ischemic stroke in the elderly. Stroke. 2006;37(9):2317‐2321. [DOI] [PubMed] [Google Scholar]
- 7. Javaheri S, Barbe F, Campos-Rodriguez F, et al. Sleep apnea: types, mechanisms, and clinical cardiovascular consequences. J Am Coll Cardiol. 2017;69(7):841‐858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watanabe T, Isono S, Tanaka A, Tanzawa H, Nishino T. Contribution of body habitus and craniofacial characteristics to segmental closing pressures of the passive pharynx in patients with sleep-disordered breathing. Am J Respir Crit Care Med. 2002;165(2):260‐265. [DOI] [PubMed] [Google Scholar]
- 9. Eckert DJ, White DP, Jordan AS, Malhotra A, Wellman A. Defining phenotypic causes of obstructive sleep apnea. Identification of novel therapeutic targets. Am J Respir Crit Care Med. 2013;188(8):996‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Remmers JE, deGroot WJ, Sauerland EK, Anch AM. Pathogenesis of upper airway occlusion during sleep. J Appl Physiol Respir Environ Exerc Physiol. 1978;44(6):931‐938. [DOI] [PubMed] [Google Scholar]
- 11. Hudgel DW, Gordon EA, Thanakitcharu S, Bruce EN. Instability of ventilatory control in patients with obstructive sleep apnea. Am J Respir Crit Care Med. 1998;158(4):1142‐1149. [DOI] [PubMed] [Google Scholar]
- 12. Redline S, Tosteson T, Tishler PV, Carskadon MA, Millman RP. Studies in the genetics of obstructive sleep apnea. Familial aggregation of symptoms associated with sleep-related breathing disturbances. Am Rev Respir Dis. 1992;145(2 Pt 1):440‐444. [DOI] [PubMed] [Google Scholar]
- 13. Campos AI, Ingold N, Huang Y, et al. Discovery of genomic loci associated with sleep apnea risk through multi-trait GWAS analysis with snoring. Sleep. 2023;46(3):zsac308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel SR, Goodloe R, De G, et al. Association of genetic loci with sleep apnea in European Americans and African-Americans: the candidate gene association resource (CARe). PLoS One. 2012;7(11):e48836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen H, Cade BE, Gleason KJ, et al. Multiethnic meta-analysis identifies RAI1 as a possible obstructive sleep apnea-related quantitative trait locus in men. Am J Respir Cell Mol Biol. 2018;58(3):391‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cade BE, Chen H, Stilp AM, et al. Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. Am J Respir Crit Care Med. 2016;194(7):886‐897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farias Tempaku P, Leite Santoro M, Bittencourt L, D’Almeida V, Iole Belangero S, Tufik S. Genome-wide association study reveals two novel risk alleles for incident obstructive sleep apnea in the EPISONO cohort. Sleep Med. 2020;66:24‐32. [DOI] [PubMed] [Google Scholar]
- 18. Grinevich V, Knobloch-Bollmann HS, Eliava M, Busnelli M, Chini B. Assembling the puzzle: pathways of oxytocin signaling in the brain. Biol Psychiatry. 2016;79(3):155‐164. [DOI] [PubMed] [Google Scholar]
- 19. Higashida H, Hashii M, Tanaka Y, et al. CD38, CD157, and RAGE as molecular determinants for social behavior. Cells. 2019;9(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuura T, Kawasaki M, Hashimoto H, et al. Fluorescent visualisation of oxytocin in the hypothalamo-neurohypophysial/-spinal pathways after chronic inflammation in oxytocin-monomeric red fluorescent protein 1 transgenic rats. J Neuroendocrinol. 2015;27(7):636‐646. [DOI] [PubMed] [Google Scholar]
- 21. Zeng Y, Zhang Y, Zhen M, et al. Side-effects of oxytocin in postpartum hemorrhage: a systematic review and meta-analysis. Am J Transl Res. 2022;14(3):1934‐1951. [PMC free article] [PubMed] [Google Scholar]
- 22. Feifel D, Macdonald K, Nguyen A, et al. Adjunctive intranasal oxytocin reduces symptoms in schizophrenia patients. Biol Psychiatry. 2010;68(7):678‐680. [DOI] [PubMed] [Google Scholar]
- 23. Mameli S, Pisanu GM, Sardo S, et al. Oxytocin nasal spray in fibromyalgic patients. Rheumatol Int. 2014;34(8):1047‐1052. [DOI] [PubMed] [Google Scholar]
- 24. Pedersen CA, Smedley KL, Leserman J, et al. Intranasal oxytocin blocks alcohol withdrawal in human subjects. Alcohol Clin Exp Res. 2013;37(3):484‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain V, Marbach J, Kimbro S, et al. Benefits of oxytocin administration in obstructive sleep apnea. Am J Physiol Lung Cell Mol Physiol. 2017;313(5):L825‐L833. [DOI] [PubMed] [Google Scholar]
- 26. Jain V, Kimbro S, Kowalik G, et al. Intranasal oxytocin increases respiratory rate and reduces obstructive event duration and oxygen desaturation in obstructive sleep apnea patients: a randomized double blinded placebo controlled study. Sleep Med. 2020;74:242‐247. [DOI] [PubMed] [Google Scholar]
- 27. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47(8):856‐860. [DOI] [PubMed] [Google Scholar]
- 28. Kohlhoff J, Cibralic S, Hawes DJ, Eapen V. Oxytocin receptor gene (OXTR) polymorphisms and social, emotional and behavioral functioning in children and adolescents: a systematic narrative review. Neurosci Biobehav Rev. 2022;135:104573. [DOI] [PubMed] [Google Scholar]
- 29. Wu S, Jia M, Ruan Y, et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol Psychiatry. 2005;58(1):74‐77. [DOI] [PubMed] [Google Scholar]
- 30. Jacob S, Brune CW, Carter CS, Leventhal BL, Lord C, Cook EH. Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neurosci Lett. 2007;417(1):6‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X, Kawamura Y, Shimada T, et al. Association of the oxytocin receptor (OXTR) gene polymorphisms with autism spectrum disorder (ASD) in the Japanese population. J Hum Genet. 2010;55(3):137‐141. [DOI] [PubMed] [Google Scholar]
- 32. Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, Ebstein RP. Association between the oxytocin receptor (OXTR) gene and autism: relationship to vineland adaptive behavior scales and cognition. Mol Psychiatry. 2008;13(10):980‐988. [DOI] [PubMed] [Google Scholar]
- 33. Wermter A-K, Kamp-Becker I, Hesse P, Schulte-Körne G, Strauch K, Remschmidt H. Evidence for the involvement of genetic variation in the oxytocin receptor gene (OXTR) in the etiology of autistic disorders on high-functioning level. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):629‐639. [DOI] [PubMed] [Google Scholar]
- 34. Parker KJ, Garner JP, Libove RA, et al. Plasma oxytocin concentrations and OXTR polymorphisms predict social impairments in children with and without autism spectrum disorder. Proc Natl Acad Sci U S A. 2014;111(33):12258‐12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uzefovsky F, Bethlehem RAI, Shamay-Tsoory S, et al. The oxytocin receptor gene predicts brain activity during an emotion recognition task in autism. Mol Autism. 2019;10(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao T, Zhang Y, Lee J, Starkweather AR, Young EE, Cong X. The associations of single nucleotide polymorphisms with risk and symptoms of irritable bowel syndrome. J Pers Med. 2022;12(2):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lucas R, Zhang Y, Walsh SJ, Starkweather A, Young E. OXTR rs53576 variation with breast and nipple pain in breastfeeding women. Pain Manag Nurs. 2021;22(3):369‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vaht M, Kurrikoff T, Laas K, Veidebaum T, Harro J. Oxytocin receptor gene variation rs53576 and alcohol abuse in a longitudinal population representative study. Psychoneuroendocrinology. 2016;74:333‐341. [DOI] [PubMed] [Google Scholar]
- 39. Goyal NP, Rosenthal SB, Nasamran C, et al. Nonalcoholic fatty liver disease risk and histologic severity are associated with genetic polymorphisms in children. Hepatology. 2023;77(1):197‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura H, Hara A, Tsujiguchi H, et al. Relationship between dietary n-6 fatty acid intake and hypertension: effect of glycated hemoglobin levels. Nutrients. 2018;10(12):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kobayashi S, Murakami K, Sasaki S, et al. Comparison of relative validity of food group intakes estimated by comprehensive and brief-type self-administered diet history questionnaires against 16 d dietary records in Japanese adults. Public Health Nutr. 2011;14(7):1200‐1211. [DOI] [PubMed] [Google Scholar]
- 42. Kobayashi S, Honda S, Murakami K, et al. Both comprehensive and brief self-administered diet history questionnaires satisfactorily rank nutrient intakes in Japanese adults. J Epidemiol. 2012;22(2):151‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chung F, Yegneswaran B, Liao P, et al. STOP questionnaire: a tool to screen patients for obstructive sleep apnea. Anesthesiology. 2008;108(5):812‐821. [DOI] [PubMed] [Google Scholar]
- 44. Pearson F, Batterham AM, Cope S. The STOP-bang questionnaire as a screening tool for obstructive sleep apnea in pregnancy. J Clin Sleep Med. 2019;15(5):705‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gauld C, Baillieul S, Martin VP, et al. Symptom content analysis of OSA questionnaires: time to identify and improve relevance of diversity of OSA symptoms? J Clin Sleep Med. 2024;20(7):1105‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buysse DJ, Reynolds CF, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh sleep quality index: a new instrument for psychiatric practice and research. Psychiatry Res. 1989;28(2):193‐213. [DOI] [PubMed] [Google Scholar]
- 47. Mollayeva T, Thurairajah P, Burton K, Mollayeva S, Shapiro CM, Colantonio A. The Pittsburgh sleep quality index as a screening tool for sleep dysfunction in clinical and non-clinical samples: a systematic review and meta-analysis. Sleep Med Rev. 2016;25:52‐73. [DOI] [PubMed] [Google Scholar]
- 48. Mustač F, Matovinović M, Mutak T, Barun B, Šagud M, Marčinko D. Utility of screening questionnaires to detect obstructive sleep apnea in patients with obesity. Psychiatr Danub. 2022;34(Suppl 10):72‐78. [PubMed] [Google Scholar]
- 49. Nakazaki K, Kitamura S, Motomura Y, et al. Validity of an algorithm for determining sleep/wake states using a new actigraph. J Physiol Anthropol. 2014;33(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nomura A, Sato T, Tada H, et al. Polygenic risk scores for low-density lipoprotein cholesterol and familial hypercholesterolemia. J Hum Genet. 2021;66(11):1079‐1087. [DOI] [PubMed] [Google Scholar]
- 51. Byrska-Bishop M, Evani US, Zhao X, et al. High-coverage whole-genome sequencing of the expanded 1000 genomes project cohort including 602 trios. Cell. 2022;185(18):3426‐3440.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34(6):1319‐1328. [DOI] [PubMed] [Google Scholar]
- 53. Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52(3):894‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goto H. Oxytocin Receptor Polymorphism Is Associated with Sleep Apnea Symptoms (Supplemental information). Accessed November 19, 2024. http://hdl.handle.net/2297/0002001445 [DOI] [PMC free article] [PubMed]
- 55. Thakkar MM, Sharma R, Sahota P. Alcohol disrupts sleep homeostasis. Alcohol. 2015;49(4):299‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johns EJ, Kopp UC, DiBona GF. Neural control of renal function. Compr Physiol. 2011;1(2):731‐767. [DOI] [PubMed] [Google Scholar]
- 57. Yumino D, Redolfi S, Ruttanaumpawan P, et al. Nocturnal rostral fluid shift: a unifying concept for the pathogenesis of obstructive and central sleep apnea in men with heart failure. Circulation. 2010;121(14):1598‐1605. [DOI] [PubMed] [Google Scholar]
- 58. Calhoun WJ. Nocturnal asthma. Chest. 2003;123(3 Suppl):399S‐405S. [DOI] [PubMed] [Google Scholar]
- 59. Nicholl DDM, Ahmed SB, Loewen AHS, et al. Declining kidney function increases the prevalence of sleep apnea and nocturnal hypoxia. Chest. 2012;141(6):1422‐1430. [DOI] [PubMed] [Google Scholar]
- 60. Kolla BP, Foroughi M, Saeidifard F, Chakravorty S, Wang Z, Mansukhani MP. The impact of alcohol on breathing parameters during sleep: a systematic review and meta-analysis. Sleep Med Rev. 2018;42:59‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Park D-Y, Kim J-S, Park B, Kim HJ. Risk factors and clinical prediction formula for the evaluation of obstructive sleep apnea in Asian adults. PLoS One. 2021;16(2):e0246399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eastwood PR, Barnes M, Walsh JH, et al. Treating obstructive sleep apnea with hypoglossal nerve stimulation. Sleep. 2011;34(11):1479‐1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mack SO, Kc P, Wu M, Coleman BR, Tolentino-Silva FP, Haxhiu MA. Paraventricular oxytocin neurons are involved in neural modulation of breathing. J Appl Physiol (1985). 2002;92(2):826‐834. [DOI] [PubMed] [Google Scholar]
- 64. Dergacheva O, Polotsky VY, Mendelowitz D. Oxytocin mediated excitation of hypoglossal motoneurons: implications for treating obstructive sleep apnea. Sleep. 2023;46(4):zsad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Redline S, Leitner J, Arnold J, Tishler PV, Altose MD. Ventilatory-control abnormalities in familial sleep apnea. Am J Respir Crit Care Med. 1997;156(1):155‐160. [DOI] [PubMed] [Google Scholar]
- 66. Takayanagi Y, Kasahara Y, Onaka T, Takahashi N, Kawada T, Nishimori K. Oxytocin receptor-deficient mice developed late-onset obesity. Neuroreport. 2008;19(9):951‐955. [DOI] [PubMed] [Google Scholar]
- 67. Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr Rev. 2006;27(7):710‐718. [DOI] [PubMed] [Google Scholar]
- 68. Stephan AM, Siclari F. Reconsidering sleep perception in insomnia: from misperception to mismeasurement. J Sleep Res. 2023;32(6):e14028. [DOI] [PubMed] [Google Scholar]
- 69. Rodrigues SM, Saslow LR, Garcia N, John OP, Keltner D. Oxytocin receptor genetic variation relates to empathy and stress reactivity in humans. Proc Natl Acad Sci U S A. 2009;106(50):21437‐21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen X, Wang R, Zee P, et al. Racial/ethnic differences in sleep disturbances: the multi-ethnic study of atherosclerosis (MESA). Sleep. 2015;38(6):877‐888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mehra R, Stone KL, Blackwell T, et al. Prevalence and correlates of sleep-disordered breathing in older men: osteoporotic fractures in men sleep study. J Am Geriatr Soc. 2007;55(9):1356‐1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hnin K, Mukherjee S, Antic NA, et al. The impact of ethnicity on the prevalence and severity of obstructive sleep apnea. Sleep Med Rev. 2018;41:78‐86. [DOI] [PubMed] [Google Scholar]
- 73. Liu F, van der Lijn F, Schurmann C, et al. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012;8(9):e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the current study are not publicly available but are available from the corresponding authors upon reasonable request.