ABSTRACT
Regulatory RNAs, present in many bacterial genomes and particularly in pathogenic bacteria such as Staphylococcus aureus, control the expression of genes encoding virulence factors or metabolic proteins. They are extremely diverse and include noncoding RNAs (sRNA), antisense RNAs, and some 5′ or 3′ untranslated regions of messenger RNAs that act as sensors for metabolites, tRNAs, or environmental conditions (e.g., temperature, pH). In this review we focus on specific examples of sRNAs of S. aureus that illustrate how numerous sRNAs and associated proteins are embedded in complex networks of regulation. In addition, we discuss the CRISPR-Cas systems defined as an RNA-interference-like mechanism, which also exist in staphylococcal strains.
GENERAL INTRODUCTION
Regulatory RNAs have been identified in many bacteria and in pathogenic bacteria such as Staphylococcus aureus, where they play major roles in the regulation of virulence or the synthesis of metabolic proteins, besides transcriptional factors and two-component systems (1–4). Most of them are noncoding RNAs (sRNAs), but some of them express small peptides. Certain sRNAs, acting in cis, are situated at the 5′ untranslated regions (UTRs) of mRNAs and act as sensors of metabolites, tRNA, or environmental stimuli (e.g., temperature, pH) or are situated at the 3′ UTR. In contrast, the genes encoding sRNAs, which act in trans, sit on the opposite strand of the regulated mRNA or at genomic locations distant from the mRNAs they regulate. Cis-encoded sRNAs, also called antisense RNAs (asRNAs), are fully complementary to their targets. In contrast, trans-encoded sRNAs share only partial complementarity, and as a consequence, they can regulate many mRNAs. Most of them are encoded mainly in the core genome, while a few of them are localized within mobile elements, pathogenic islands, or plasmids. In this review, we will focus on the most recent mechanisms of RNA regulation discovered in S. aureus and how regulatory RNAs are part of sophisticated networks that allow the bacteria to adapt quickly to their environment or survive in their host.
mRNA 5′ UTRs: RIBOSWITCHES, T-BOXES, AND THERMOSENSORS
5′ UTRs of mRNAs contain riboswitches, T-boxes, or thermosensors with potential impacts for novel antibiotherapy. Riboswitches and T-boxes are found in the 5′ UTR of some mRNAs and contain highly structured domains, which recognize metabolites such as cofactors, vitamins, amino-acids, nucleotides, second messenger cyclic di-GMP, Mg2+, or nonaminoacylated tRNAs (5). Binding of these metabolites induces structural changes that modify the expression of the downstream mRNA, for example, by inducing premature transcription arrest, repression/activation of translation, or cleavage (Fig. 1a). A T-box senses the aminoacylation status of tRNAs and mainly controls transcription of downstream genes that encode proteins involved in biosynthesis, transport of amino acids, or aminoacylation of tRNAs (Fig. 1b). Based on sequence and structure conservation, most of the T-boxes and riboswitches were predicted in S. aureus genomes (4). A large proportion of riboswitches control the expression of genes involved in metabolic pathways. Because these genes are often essential for growth, they represent interesting targets for the development of alternative antimicrobial drugs in the battle against multidrug-resistant S. aureus. This strategy was used with the guanine and glucosamine-6-phosphate (GlcN6P) riboswitches. Mulhbacher et al. (6) identified a pyrimidine derivative that binds to the guanine riboswitch and represses the expression of guaA. This compound significantly attenuated S. aureus infections in a mouse model. In Gram-positive bacteria, the glmS mRNA is both a ribozyme, which catalyzes its own cleavage, and a riboswitch responding to GlcN6P. Its product encodes an essential enzyme, which converts fructose-6-phosphate into GlcN6P, a building block of bacterial peptidoglycan. Tight regulation of glmS mRNA is crucial to maintain a homeostatic level of GlcN6P in the cell. At high concentrations of GlcN6P, its binding to the 5′ UTR of glmS leads to site-specific self-cleavage, which generates a 5′ hydroxylated end molecule rapidly degraded by the RNase J1 (7, 8). A recent study led to the design, synthesis, and characterization of a GlcN6P analogue, carba-GlcN6P, which constitutively activates the glmS ribozyme of vancomycin-resistant S. aureus and destabilizes its mRNA (7). This compound was experimentally shown to induce the efficient self-cleavage of the glmS mRNA in a similar fashion as the natural metabolite and thus represented an important step in the development of antibiotics with a new mode of action. Very recently, a new approach called “Term-seq” revealed that several antibiotic resistance genes are under the control of riboswitches responding to antibiotics commonly used against Gram-positive pathogenic bacteria such as Listeria spp. and Enterococcus faecalis (9). These results suggest that the same phenomenon could exist in S. aureus and that RNA-mediated regulation could play a broader role in antibiotic resistance mechanisms than has been envisioned.
FIGURE 1.
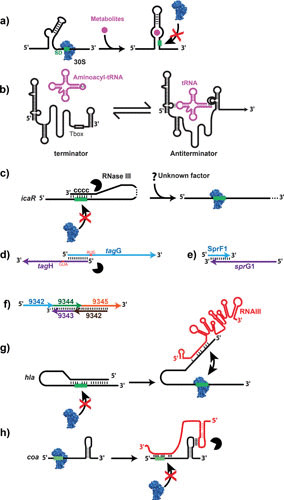
Several mechanisms of RNA regulation in S. aureus. (a) Schematic drawing of the flavin mononucleotide riboswitch. The 5′ UTR adopts a particular structure recognized by the flavin mononucleotide, which in turn leads to the stabilization of a stem-loop structure sequestrating the SD sequence to inhibit translation. 30S is for the small ribosomal subunit. (b) An example of a T-box motif as found in the 5′ UTR of many mRNAs encoding aminoacyl-tRNA synthetases. Nonaminoacylated tRNA binds to the leader region at two sites and stabilizes an antiterminator structure, allowing transcription of the downstream gene. The drawing is adapted from reference 4. (c) The 3′ UTR of the biofilm repressor IcaR possesses a cytosine-rich motif, which binds to the SD sequence and hinders ribosomes from its binding site on the mRNA (see text for details). (d) Overlapping 5′ UTRs of tagG and tagH mRNAs are processed by the endoribonuclease III (Rnase III). Shorter 5′ ends might facilitate ribosome recruitment. (e) The antitoxin RNA SprF1 interacts at the 3′ end of the toxin encoded by sprG1 and triggers its degradation. (f) A cluster of five sRNAs was sequenced in the S. aureus Newman strain that encodes a putative toxin-antitoxin system (see text for details). (g,h) sRNAs act by an antisense mechanism. Binding of the 5′ UTR of RNAIII to the 5′ UTR of hla mRNA liberates its SD and activated translation (g), whereas the 3′ domain of RNAIII acts as a repressor domain, which contains C-rich motifs for base-pairing with the SD sequence of mRNA as coa mRNA depicted in the figure (h). Green bar, SD sequence; black circle, RNase III (for references and more details, see text).
Not surprisingly, the presence of antibiotics can also modulate the regulatory activity of T-boxes. In S. aureus, an unusual glyS T-box regulates transcription antitermination (Fig. 1b) of the unique glycyl-tRNA synthetase (glyRS) gene responsible for catalyzing the aminoacylation of the five tRNAGly isoacceptors, independently of their anticodon (GCC or UCC) and with different binding affinities (10). Thereafter, the T-box senses the availability of glycine not only for its incorporation into nascent polypeptide chains during translation but also for the formation of pentaglycine bridges into the peptidoglycan molecule, linking two essential pathways. Antibiotics targeting the small ribosomal subunit stabilized the T-box/tRNA complex and induced a read-through of transcription, while chloramphenicol and linezolid attenuated glyS transcription (11). The outcome depended on the binding sites of the protein synthesis inhibitors (11). Although T-boxes can be direct targets for antibiotics against S. aureus, it was also reported that a high concentration of the antibiotic tigecycline might induce possible off-target inhibition of the RNA polymerase (11).
RNA thermosensors are regulatory elements often localized at the 5′ UTR of mRNAs encoding heat or cold shock proteins and virulence factors (for review see 12). Briefly, at low temperatures, the mRNA cannot be translated, since the Shine and Dalgarno (SD) sequence is trapped in a hairpin structure, which melts gradually when the temperature increases. The best-studied example in Gram-positive bacteria is the thermosensor regulating the expression of the transcriptional factor prfA, which activates the expression of most of the virulence genes in Listeria monocytogenes at high temperatures (13, 14). No such example has yet been demonstrated for other Gram-positive bacteria, including S. aureus.
3′ UTRs OF mRNAs ACT IN CIS OR ARE RESERVOIRS OF sRNAs
Transcriptome analysis of the human pathogen S. aureus revealed that at least one-third of mRNAs carry long 3′ UTRs and thus might display multiple regulatory functions (15). Some of them have direct action on the expression of their own mRNA (15, 16). Long 3′ UTRs (>100 nucleotides) can end at an intrinsic Rho-independent terminator of transcription (TT) and can be generated from a specific RNase cleavage or from a termination read through of the RNA polymerase. Remarkably, several 3′ UTRs contain riboswitches functioning in metabolite-sensing regulation (16). Indeed, the TT of the riboswitch in an OFF conformation also serves as the TT of the gene encoded upstream of the riboswitch.
A singular example of a newly identified posttranscriptional regulatory mechanism was shown for the icaR mRNA (Fig. 1c), which possesses an unusually long 3′ UTR (390 bp). In this case, the expression of the mRNA is modulated through a long-distance interaction between 3′ UTR and 5′ UTR (16). Because icaR codes for a transcriptional repressor of the icaADBC operon, encoding enzymes involved in the synthesis of PIA-PNAG, the main polysaccharides of the biofilm matrix, this regulation has a direct impact on biofilm formation. Base-pairing interactions between the long 3′ UTR of icaR mRNA and the SD sequence of the same mRNA hindered efficient translation initiation (16). This long-range RNA duplex generated a specific site for the double-stranded endoribonuclease III (RNase III) for cleavage. As a consequence of this cleavage, PIA-PNAG synthesis increased. However, the mechanism allowing IcaR translation is not known to prevent the action of the 3′ UTR as a cis-acting antisense RNA (Fig. 1c).
It is also postulated that the 3′ UTR length provides other types of transcript-specific regulation. Indeed, in Salmonella spp., the 3′ UTRs are also reservoirs for sRNAs, which originate either by transcription from an internal promoter or by processing. In both cases, the sRNA generated from the 3′ UTR regulates trans-encoded mRNA targets. For instance, the sRNA CpxQ is generated by RNase E cleavage of the 3′ UTR of cpxP and represses the translation of mRNAs encoding a family of envelope proteins, whereas the sRNA DapZ is transcribed from an internal promoter within the dapB gene and inhibits translation of the major ABC transporters, DppA and OppA (17, 18; for review see 19). Recent work has shown that such 3′ UTR-derived sRNAs also exist in S. aureus, although their functions remain to be addressed (E. Desgranges, S. Marzi, P. Romby and I. Caldelari, unpublished data).
asRNAs IN PERVASIVE TRANSCRIPTION AND ACTING AS ANTITOXINS
asRNAs are transcribed from the opposite strand of the mRNAs they regulate, so that they display perfect complementarities with their targets. Short asRNAs are often encoded on mobile elements such as plasmids, transposons, and phage-like elements. Such elements can potentially be transferred horizontally to other bacterial species or be duplicated (20). In S. aureus, they were first described to control plasmid conjugation and replication (21). The size of asRNAs can vary from 10 to thousands of nucleotides, because these RNAs can overlap with part of a gene (3′ or 5′ ends), the entire gene, or a group of genes (Fig. 1d to f). This phenomenon is called pervasive transcription. Initially, pervasive transcription was considered a nonfunctional transcriptional noise. However, considering the large number of asRNAs expressed from the entire genome and in several bacterial species, these RNAs might play an important role in the regulation of gene expression. Genome-wide analysis of S. aureus highlighted that the expression of a significant proportion (75%) of antisense transcripts to annotated open reading frames are synthesized from the complementary strand and that these sense/antisense duplexes are digested by RNase III, generating short fragments all along the genome (22). Another study using the RIP-Seq approach confirmed the involvement of RNase III in the regulation of sense/antisense transcripts and overlapping UTRs (23). The situation might be even more complex, because recent results suggested that the termination factor Rho plays a major role in preventing pervasive transcription in Bacillus subtilis, but also in S. aureus (24, 25). Although the biological outcome of pervasive transcription is not clearly understood, some of the asRNAs produced are functional and control several biological processes (15).
Type I toxin-antitoxin (TA) systems are particular cases of short asRNAs, in which the antitoxin is an asRNA regulating the translation or the fate of the toxin-encoding mRNA, whereas in the type III system, the antitoxin sequesters the toxin (reviewed in 26, 27). In S. aureus, several type I TA module systems have been described (reviewed in 28). One of them, called SprF1/SprG1, expresses SprF1 as the asRNA and sprG1 mRNA (Fig. 1e), which encodes two short secreted peptides with hemolytic and antibacterial activity (29). The antitoxin SprF1 binds to the 3′ end of sprG1 mRNA, which leads to mRNA degradation and inhibition of peptide synthesis to protect cells against lethality (29). The SprA1/asSprA1 pair is another intriguing and unconventional system. The asSprA1 is transcribed from the opposite strand of the sprA1 mRNA, producing a cytolytic peptide. Their 3′ ends overlapped by 35 nucleotides, but experimental data indicated that the functional domain of asSprA1 is outside the complementary sequence with sprA1 mRNA. Both RNAs were expressed concomitantly, and asSprA1 5′ base-paired with the ribosome-binding site (RBS) of sprA1, impairing its translation (30). Thus, asSprA1 acts as a trans regulator with its complementary target, suggesting that it can potentially interact with other RNA targets. Finally, a cluster of five genes encoding sRNAs specific to the S. aureus Newman strain contained a putative TA system (31) (Fig. 1f). Three genes were transcribed from the positive strand and two from the negative strand. Moreover, one small open reading frame was detected within one of the genes from the minus strand and coded for a secreted peptide with similarity to the RelE toxin (32). Whether these two overlapping genes corresponded to a novel TA system remains to be addressed. Interestingly, this locus was expressed in a growth-phase-dependent manner, in nutriment starvation, and in oxidative stress. Type I TA systems have been involved in many functions (e.g., membrane depolarization, plasmid maintenance), mainly in Escherichia coli, including the persistence phenomenon (for review see 33), but not in S. aureus until now (28).
TRANSCRIPTIONAL FACTORS AND sRNAs BUILD COMPLEX REGULATORY NETWORKS
The sRNAs belong to intricate networks of regulation, and their synthesis is often dependent on transcription factors or on two-component systems. In addition, sRNAs can also control transcription factors at the posttranscriptional level (Fig. 2). Typical examples will be described below. In addition, trans-acting sRNAs regulate mRNAs by imperfect base-pairings, which signifies that one sRNA can modulate several targets and one target can be controlled by several sRNAs. In S. aureus, the annealing region between sRNA and mRNAs are often longer than in E. coli and mostly targets the RBS of mRNAs affecting translation. In several cases, a second distinct site of interaction occurs in the coding region. Unlike Gram-negative bacteria, in which Hfq and ProQ proteins participate in sRNA regulation by stabilizing and facilitating their pairings with mRNA targets (see reviews in 34, 35), no RNA chaperones have yet been identified in S. aureus. Indeed, there is no identifiable proQ homolog, and the role of Hfq is still unclear. Recent work has shown that the rim domain of Hfq has an amino acid composition (low in arginine) incompatible with RNA annealing activity compared to E. coli Hfq (36).
FIGURE 2.
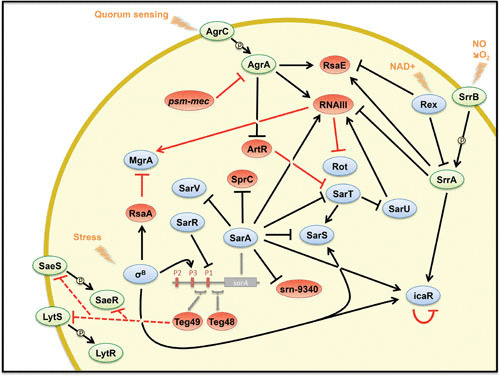
Examples of the complex network between sRNAs and transcriptional factors in S. aureus in response to stress. Arrows show activation and bars show repression. Blue, transcriptional regulators; green, two-component systems; red, regulatory sRNAs. Red lines corresponded to posttranscriptional regulation, and black lines, to transcriptional regulation. Dotted lines are for the target mRNAs that were not experimentally validated. Only sRNA-dependent mRNA targets encoding transcriptional factors are depicted in the figure.
AgrA, the response regulator of the agr quorum sensing system, activates the transcription of RNAIII. The bifunctional RNAIII codes for δ-hemolysin and regulates the expression of virulence genes at the posttranscriptional level (see below). It interacts with various mRNAs either to activate or to repress translation (Fig. 1g,h). The RBS of hla mRNA encoding α-hemolysin is embedded in a hairpin, which prevents ribosome binding and blocks the start of translation. The 5′ UTR of RNAIII possesses complementary sequences to the leader region of hla. Interaction between these RNAs enables the recruitment of ribosomes to initiate Hla translation. In its 3′ UTR, RNAIII carries conserved UCCC motifs that are used as the seed sequences to bind with the RBS of the target mRNAs coding for protein A, coagulase, Sbi protein, the transcription factor Rot (a repressor of exotoxins), and the endopeptidase LytM (for review see 37). Moreover, AgrA represses the sRNA ArtR, which inhibits translation of the SarA homolog SarT (38), and activates RsaE (see 39) (Fig. 2).
The staphylococcal accessory regulator SarA is synthesized from three distinct promoters (P1, P2, P3) and binds DNA or RNA (1). As a transcription factor, it regulates many genes involved in virulence, autolysis, biofilm formation, stress response, antibiotic resistance, and metabolism, but also two sRNAs, SprC and Srn_9340, located on the same pathogenicity island (40). SprC prevents ribosome binding to the SD of the atl mRNA coding for an autolysin. Deletion of the sprC gene causes enhanced phagocytosis of S. aureus by monocytes and macrophages, and this effect was found to be partly due to the deregulation of atl expression (41). SarA represses both sprC and srn_9340 transcription and requires an ATTTTAT sequence in its binding site (40). However, while the SarA level remains relatively constant during bacterial growth, the expression of SprC fluctuates, which suggests that additional factors might control its synthesis and that a mechanism of derepression should coexist under specific conditions. These are the first examples of two sRNAs regulated by the same transcription factor.
In the following, we will describe an sRNA whose transcription is controlled by three independent transcription factors. The sRNA RsaE possesses two consensus sequence motifs UCCC as found in RNAIII, which interact with the RBS of several mRNAs involved in central metabolism to repress their translation (39, 42). RsaE is highly conserved between the Staphylococcaceae and Bacillaceae families. Not only is the sequence of RsaE conserved between B. subtilis and S. aureus species, but so are its regulation and functions. Recent studies have shown that its transcription is activated by AgrA (see above), the two-component system SrrAB (staphylococcal respiratory response) in response to NO, and a binding site for the redox sensing repressor Rex has been predicted (39, 43, 44). In B. subtilis, the Rex-repression of RsaE (also called RoxS) has been proposed to readjust the cellular balance of NAD+/NADH on various signals (44).
The alternative sigma B factor (σB) together with RNA polymerase guides transcription of genes mainly in the stationary phase of growth and under stress conditions. Its regulon comprises more than 200 genes, including several virulence factors, transcription factors, and sRNAs (Fig. 2). Among the sRNAs induced by σB are SbrA, B, and C activated by KOH (45), and RsaA (39). The stability of RsaA depends on RNase III and the endoribonuclease RNase Y (23, 46). RsaA acts as an acute virulence attenuator in S. aureus (see below) by inhibiting translation of the MgrA transcription factor (Fig. 2) (47), which in turn causes the activation of the synthesis of several surface proteins (48). RsaA possesses two UCCC motifs, which in the case of mgrA mRNA, bind to two distant regions, involving an imperfect duplex masking the SD sequence of the mRNA and a loop-loop interaction occurring downstream in the coding region. These two distant binding sites are required for efficient repression and RNase III-dependent degradation of the repressed mRNA (47). Finally, the sRNA Teg49 is transcribed from the σB- dependent P3 promoter of sarA and is probably processed by RNase III and RNase Y with the help of the helicase CshA (49–51). Transcriptomic analyses revealed that besides genes involved in virulence and autolysis, Teg49 might posttranscriptionally affect the SaeRS and LytRS two-component systems, yet the exact mechanism is not known (51). In addition, another sRNA, Teg48, whose role is not known, is transcribed from the P1 promoter of sarA (Fig. 2). Even if the maturation process and the function of Teg48 and Teg49 are not clearly established, the long 5′ UTR of sarA mRNA encoding a master regulator of virulence in S. aureus represents a putative reservoir for novel sRNAs.
crRNA, tracrRNA, AND THE CRISPR-Cas ADAPTIVE IMMUNITY SYSTEMS IN S. AUREUS
Phages are the most abundant forms of life on earth and the natural killers of bacteria because in most cases their lytic life cycle ends with the death of the bacterial cell. Outnumbering their microbial hosts, phages impose selective pressure for the diversification of microbial defense systems. These include various innate phage-resistance mechanisms such as restriction/modification enzymes, receptor masking, blocking DNA injection, abortive infection (52, 53), and the adaptive defense mechanism based on clustered, regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (cas) genes (54, 55). The latter RNA-interference-like mechanism relies on small noncoding RNAs, CRISPR RNA (crRNA), and in some cases trans-activating crRNA (tracrRNA), through which prokaryotic hosts (bacteria and archaea) can acquire heritable resistance to genetic parasites such as phages, but also plasmids and transposons (for reviews see 53, 56). To date, CRISPR-Cas systems have been found in about 50% of bacterial genomes and 95% of archaeal genomes (57, 58). The CRISPR and the cas loci are often located next to each other in the genomes, sometimes organized into operons, but a significant number of genomes have also isolated cas loci and/or CRISPRs (59).
Despite the large diversity of the CRISPR-cas systems, they share common features. Briefly, the CRISPR loci are characterized by an array of short and palindromic repetitive sequences interspaced by sequences called “spacers” that are derived from plasmid and viral DNAs (in some cases, also RNAs). During the initial infection with a virus or plasmid, these spacers are first integrated into the CRISPR array in the host genome to provide the host with immunity (acquisition step [60, 61]). During a second infection event they are transcribed and used as guides to inactivate the viral or plasmid genome. This two-step pathway involves a variety of Cas proteins, leading to several major types of CRISPR-cas systems (reviewed in 58, 62). However, the acquisition step involves two highly conserved Cas1 and Cas2 proteins (58, 63). The Cas1-Cas2 integrase is a heterohexameric complex of four Cas1s and two Cas2s which preferentially incorporates foreign DNA at the first CRISPR repeat and participates in the discrimination against self-DNA and in the minimization of off-targeting insertions (64–66). Both proteins have been found in several S. aureus strains (57). During the second step, activation of transcription from a promoter located in an AT-rich leader sequence preceding the first CRISPR repeat (67, 68) leads to expression of the whole array into precursor CRISPR transcripts (pre-crRNA). The pre-crRNAs are then processed into mature crRNAs consisting of partial repeat(s) and a single spacer sequence, each complementary to a unique invader sequence (69). Different endonucleases participate in the maturation step, which might vary in different bacteria. Type I and III systems perform the function by a multisubunit Cas protein complex and are characterized by Cas6 processing (69). Type II uses another sRNA, tracrRNA, to direct RNase III-dependent maturation of the pre-crRNA in the presence of Cas9, the hallmark protein of the type II system (70). Cas9 endonuclease remains associated with the dual-tracrRNA:crRNA structure, which during the interference phase, guides the cleavage of site-specific cognate target DNA (71). Type I and II CRISPR-Cas systems target DNA (72, 73), whereas type III systems provide immunity against DNA and RNA (74).
With the completion of the genome sequences of several S. aureus strains, it appears that the CRISPR-Cas systems are not highly prevalent, and only a few of them have been experimentally demonstrated. A CRISPR Finder analysis (57) of the 115 sequenced S. aureus genomes present in the CRISPRdb (http://crispr.i2bc.paris-saclay.fr) showed that the majority of CRISPR-like loci contain only a few spacers (1 or 2), a few have between 3 and 10 spacers, and only 1 genome (from methicillin-resistant S. aureus [MRSA] 08BA02176, isolated from a patient) has 15 spacers. Using CRISPRone (http://omics.informatics.indiana.edu/CRISPRone), several genes belonging to the type III-A CRISPR system (including Csm2, Cas1, Cas2, and Cas6) (Fig. 3a) have been recently predicted (72) in different isolates (MSHR1132, JS395, CIG290, and 21252). Multilocus sequence typing performed on one of these isolates (MSHR1132) has shown that it belongs to a divergent clonal complex, which appears to be closely related to Staphylococcus epidermidis and Staphylococcus lugdunensis (75), and which has been renamed Staphylococcus argenteus (76). It has thus been hypothesized that CRISPR/cas was present in a common ancestor of S. epidermidis, S. lugdunensis, and S. aureus and was later lost in most conventional S. aureus strains. It is noteworthy that numerous repeat-spacer-like structures resemble CRISPR elements but lack spacer diversity and have been classified as false-CRISPR (72). S. aureus repeat-like elements (GC-rich direct repeats) belong to this class of RNAs, for which the functions remain to be addressed (77, 78).
FIGURE 3.
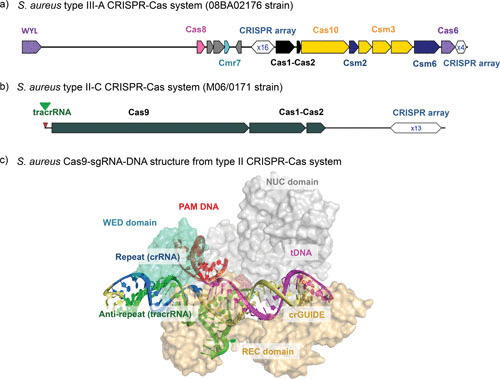
(a) Genomic organization of the loci for the type III-A CRISPR system of S. aureus strain 08BA02176. Type III is the typical S. aureus CRISPR organization. The scheme was obtained using CRISPRone (72), and the genome sequence was deposited in GenBank (accession number 08BA02176; RefSeq accession number GCF_000296595.1). (b) Genomic organization of the loci for the type II-C CRISPR system of S. aureus strain M06/0171. The CRISPR-Cas genes were found on an SCCmec inserted into the 3′ end of the chromosomally located orfX gene. The scheme was obtained using CRISPRone (72), and the SCCmec sequence was deposited in GenBank (GenBank accession number HE980450.1). (c) Cartoon (RNA and DNA) and surface (Cas9) representations of the SaCas9-sgRNA-target DNA complex (pdb file 5AXW) (80). The SaCas9 sgRNA consists of the crRNA guide region (crGUIDE represented in pale yellow) forming a heteroduplex with the target DNA strand (tDNA in magenta) and the repeat/antirepeat helix (blue, the repeat crRNA-derived strand, green, the antirepeat trascrRNA-derived strand). The protospacer adjacent region-containing DNA duplex is red. Cas9 domains are colored as follows: cyan, WED domain; pale orange, REC domain; gray, NUC domain. Molecular graphics images were prepared using PyMol.
More recently, a type II-C CRISPR-Cas system was found in an MRSA strain isolated from an Irish patient (Fig. 3b). This CRISPR element is located on a pseudo staphylococcal cassette chromosome mec (SCCmec) composite island, which was probably horizontally acquired from an S. epidermidis strain (79). The peculiarity of this system (SaCas9) is that it contains a shorter version of Cas9 protein. The crystal structure of a complex containing SaCas9, the sgRNA (single-guide RNA; an artificial fusion product of a crRNA and a tracrRNA [71]) and its target DNA provided a model to understand how crRNA and tracrRNA guide Cas9 on the target DNA and prepare it for double-stranded DNA cleavages (80). In this structure, the DNA duplex of the protospacer-adjacent region contains the signals recognized by the Cas9 protospacer-adjacent region-interacting (PI) domain, which discriminates the invader DNA against self-DNA and facilitates unwinding of the target DNA, leading to the formation of a heteroduplex. The beginning of the RNA-DNA heteroduplex (“seed” region) adopts a distorted structure critical for Cas9-catalyzed DNA cleavage. This conformation results from the RNA-DNA helix structure, the interactions with protein residues, and the RNA helix formed by the repeat/antirepeat regions mimicking the tracrRNA-crRNA interactions (Fig. 3c). The discovery of the smaller Cas9 protein led to recent improvements in genome editing (81). The seminal works of Jennifer Doudna and Emmanuelle Charpentier on the use of CRISPR-Cas systems for genome editing have inspired many studies showing the incredible potency of the system (82–84). Thousands of publications have reported the use of Streptococcus pyogenes Cas9 (85) directed by the sequence of a sgRNA for site-specific genome modifications, gene knockouts or replacements, gene expression control, and functional genome screenings in over 40 species. Interestingly, SaCas9 has been successfully used for eukaryotic genome editing since its smaller size makes it easier to be delivered via adeno-associated virus vectors to somatic tissues (81).
The fact that CRISPR-Cas has been mapped on mobile elements in MRSA confirms the importance of horizontal gene transfer from other cocolonizing bacteria in the acquisition of novel functions and in the evolution of S. aureus strains. Interestingly, transcription of the CRISPR-Cas genes can be highly regulated and induced upon infection (86–88) by membrane stress (89) and, in Gram-negative bacteria, by quorum sensing signaling (90, 91). Recent work has developed genetic engineering tools to apply the CRISPR/Cas9 system as an antimicrobial strategy against S. aureus (92). Clearly, the CRISPR/Cas9 system offers an alternative therapy to conventional antibiotics.
ROLE OF sRNAs IN PHYSIOPATHOLOGY
S. aureus pathogenesis can take different forms depending on the infected tissue and the invading bacterial strain. This is often accompanied by the expression of various virulence factors involved in the colonization and the alteration of the tissue but also by the capacity to escape from the host immune response. Among key regulators of virulence, several sRNAs have been shown to modulate the synthesis of virulence factors in a dynamic manner, and some of them contribute to specific aspects of bacterial virulence in animal models of infection (1).
The most studied RNA in S. aureus, agr-RNAIII, is the main intracellular effector of the quorum sensing system agr. As described above, RNAIII regulates expression of virulence factors known to be associated with infectious diseases. For instance, it represses the synthesis of protein A, which triggers inflammatory signaling pathways and contributes to evasion of the immune response. Conversely, RNAIII induces the synthesis of a battery of toxins, which contribute to the degradation of tissues and subversion of host defenses, such as the pore-forming toxins, and peptides with proinflammatory and lytic activities. Recent modeling of the quorum sensing system and of its regulators has illustrated the importance of the agr system in promoting dissemination of the bacteria from biofilms or dense populations (93, 94). Nevertheless, despite the fact that many clinical isolates from acute infections express RNAIII, its steady state level varies considerably among them (95, 96). In particular, a higher level of RNAIII has been observed in the community-acquired MRSA strains with increased virulence compared to other S. aureus lineages (97, 98). Interestingly, a recent study showed that the level of RNAIII is lower in strains isolated from patients with sepsis than in those from commensal carrier patients (99). Perhaps more surprisingly, heterogeneity has been reported in patients in which agr-positive and agr-negative strains coexisted. This has been proposed as a factor that might modulate the outcome of the infections (100, 101).
S. aureus is also frequently exposed to other microbes during colonization and infection, providing opportunities to acquire mobile genetic elements that contribute to the evolution of the genome. Some of these genomic islands play key roles in pathogenesis through their possession of new virulence factors (pathogenicity islands) or through the synthesis of novel regulators modulating the expression of genes of the core genome. As an example, SprD is an important small regulatory sRNA (142 nucleotides) expressed from a pathogenicity island, which significantly promoted S. aureus diseases in a mouse sepsis model of infection (102, 103). SprD interacts through base-pairings with the sbi mRNA, which encodes an immune evasion molecule protecting the bacteria from the host immune responses (104, 105). However, the phenotype of the ΔsprD mutant strain was not linked to the SprD-dependent regulation of sbi since the Δsbi mutant strain behaved like the wild-type strain in the mouse sepsis model (102). Therefore, these data strongly suggest that SprD might regulate the expression of other proteins important for infection.
SSR42 (for small stable RNAs) is an 891-nucleotide-long sRNA whose stability is greatly enhanced in the stationary phase of growth (106, 107). It regulated the expression of approximately 80 mRNAs in 2 genetically different S. aureus strain backgrounds. While it increased the expression of capsule Cap5a, SSR42 downregulated the expression of protein A, α and γ hemolysin, and Panton-Valentin leukocidin (107). Because no direct binding was evidenced between SSR42 and mRNAs encoding virulence determinants, the effect was predicted to be indirect through the modulation of the expression of a transcriptional regulator. Phenotypically, the deletion of the SSR42 gene affected erythrocyte lysis, resistance to opsonization killing, and pathogenesis in a murine model of skin and soft tissue infections (107). More recently, SSR42 was identified as an important effector of intracellular virulence by screening of a transposon mutant library pool. After internalization in epithelial cells, the ΔSSR42 mutant strain was significantly enriched in the intracellular fraction, most likely due to an attenuated cytotoxicity (108).
In contrast to RNAIII, SprD, and SSR42, which contribute to enhanced virulence of S. aureus, other regulatory RNAs behave as attenuators of virulence. This is, for instance, the case of another encoded pathogenicity island sRNA, the so-called SprC (103). Indeed, the virulence of the isogenic strain lacking SprC was significantly and reproducibly enhanced in a mouse systemic model. Furthermore, SprC reduces S. aureus susceptibility to phagocytosis by human monocytes and macrophages (41). Another example is the psm-mec RNA, which is a bifunctional RNA located in the SCCmec mobile genetic element. It encodes a phenol-soluble modulin (PSMmec) cytolytic toxin and acts as a translational repressor through direct binding with agrA mRNA (109). The transcription of all psm genes is positively regulated by AgrA. This activation is linked to a specific binding of the phosphorylated form of AgrA to the promoter sequences upstream of the psm genes, except that this binding was not yet demonstrated for the psm-mec gene (reviewed in 110). The deletion of the psm-mec RNA increased the expression of AgrA, which resulted in an increase of toxin and PSMα production and enhanced virulence in mice (109). Interestingly, community-acquired MRSA that does not carry the psm-mec gene has been shown to be more virulent than the hospital-associated MRSA that harbors the gene (109, 111). Finally, a mutant strain that did not express the sRNA, RsaA, was attenuated in the severity of acute systemic infection in a mouse model (47). This deletion is linked to the deregulation of MgrA, a master regulator of capsule synthesis and clumping (112). This phenotype in pathogenesis is probably linked to the high sensitivity of the mutant strain to opsonophagocytosis by host polymorphonuclear leukocytes. Because the expression of these three sRNAs is detrimental for bacterial spreading within colonized host organisms, one may suggest that during evolution they have favored commensalism with the host.
To evaluate the impact of sRNA expression in the context of host infection, two studies explored the possible relationships between infection severity and RNA expression levels. In the first study, the expression levels of five sRNAs (RNAIII, RsaA, RsaE, RsaG, and RsaH) were analyzed in samples from acute cutaneous infection, cystic fibrosis sputum, or nasal colonization. The expression profiles did not correlate with the type of infection, but the authors noticed that the expression of these five RNAs was more homogeneous in the nasal colonization isolates than in those responsible for infection (96). More recently, the expression levels of RNAIII and SprD were measured in 40 strains cultivated from patients with sepsis or septic shock and compared to 21 strains isolated from asymptomatic colonized carriers. It appeared that strains from septic shock had significantly lower levels of RNAIII and to a lesser extent for SprD (99). It is important to note that this analysis was performed on clinical isolates cultured in vitro and does not necessarily reflect the expression of these RNAs during infection within the host. In fact, it is very difficult to obtain reliable data from in vivo studies to assess the role and importance of RNAs in the establishment or evolution of infection. The great variability of S. aureus strains, the difficulty in obtaining highly controlled cohorts of patients, and the reliability of sampling protocols, sample processing, and RNA expression analysis are all obstacles to be overcome. Moreover, biological variables may influence the analysis since the relationships between host immune systems and microbes seem to be particularly individualized and can influence the disease outcome (113, 114). Furthermore, interspecies interactions between bacterial pathogens and the commensal microbiota, as well as limited nutrients, play major roles in promoting or preventing S. aureus colonization (113). Interestingly, it was shown that the agr system is repressed by high concentrations of hemoglobin in the nasal fluids, leading to the expression of several cell surface proteins and favoring nasal colonization (115). Similar data were obtained when S. aureus was cocultivated with the nasal strain of Corynebacterium striatum (116). A recent study also demonstrated that the commensal S. epidermidis can influence the expression of one ncRNA of S. aureus (117).
Clearly, we are just beginning to better appreciate the roles of regulatory sRNAs during colonization and in the pathophysiology of S. aureus infections.
ACKNOWLEDGMENTS
This work was supported by the Centre National de la Recherche Scientifique (CNRS) (to P.R.), by the Agence Nationale de la Recherche (ANR, ANR-16-CE11-0007-0-RIBOSTAPH) (to P.R.), and by LABEX: ANR-10-LABX-0036 NETRNA (to P.R.), funding from the state managed by the French National Research Agency as part of the investments for the future program.
REFERENCES
- 1.Bischoff M, Romby P. 2016. Genetic regulation, p 301–334. In Staphylococcus: Genetics and Physiology. Caister Academic Press, Poole, United Kingdom. [PubMed] [Google Scholar]
- 2.Tomasini A, François P, Howden BP, Fechter P, Romby P, Caldelari I. 2014. The importance of regulatory RNAs in Staphylococcus aureus. Infect Genet Evol 21:616–626 10.1016/j.meegid.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Guillet J, Hallier M, Felden B. 2013. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog 9:e1003767 10.1371/journal.ppat.1003767. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldelari I, Fechter P, Lioliou E, Romilly C, Chevalier C, Gaspin C, Romby P. 2011. A current overview of regulatory RNAs in Staphylococcus aureus, p 51–75. In Marchfelder A, Hess W (ed), Regulatory RNAs in Prokaryotes. Springer-Verlag, Vienna, Austria. [Google Scholar]
- 5.Quereda JJ, Cossart P. 2017. Regulating bacterial virulence with RNA. Annu Rev Microbiol 71:263–280 10.1146/annurev-micro-030117-020335. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Mulhbacher J, Brouillette E, Allard M, Fortier L-C, Malouin F, Lafontaine DA. 2010. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog 6:e1000865 10.1371/journal.ppat.1000865. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lünse CE, Schmidt MS, Wittmann V, Mayer G. 2011. Carba-sugars activate the glmS-riboswitch of Staphylococcus aureus. ACS Chem Biol 6:675–678 10.1021/cb200016d. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Collins JA, Irnov I, Baker S, Winkler WC. 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev 21:3356–3368 10.1101/gad.1605307. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. 2016. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:aad9822 10.1126/science.aad9822. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apostolidi M, Saad NY, Drainas D, Pournaras S, Becker HD, Stathopoulos C. 2015. A glyS T-box riboswitch with species-specific structural features responding to both proteinogenic and nonproteinogenic tRNAGly isoacceptors. RNA 21:1790–1806 10.1261/rna.052712.115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatopoulou V, Apostolidi M, Li S, Lamprinou K, Papakyriakou A, Zhang J, Stathopoulos C. 2017. Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors. Nucleic Acids Res 45:10242–10258 10.1093/nar/gkx663. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 10:255–265 10.1038/nrmicro2730. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561 10.1016/S0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 14.Loh E, Memarpour F, Vaitkevicius K, Kallipolitis BH, Johansson J, Sondén B. 2012. An unstructured 5′-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res 40:1818–1827 10.1093/nar/gkr850. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasa I, Toledo-Arana A, Gingeras TR. 2012. An effort to make sense of antisense transcription in bacteria. RNA Biol 9:1039–1044 10.4161/rna.21167. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, Romby P, Valle J, Solano C, Lasa I, Toledo-Arana A. 2013. Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet 9:e1004001 10.1371/journal.pgen.1004001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019 10.1038/emboj.2012.229. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao Y, Vogel J. 2016. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 61:352–363 10.1016/j.molcel.2015.12.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Miyakoshi M, Chao Y, Vogel J. 2015. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J 34:1478–1492 10.15252/embj.201490546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fozo EM, Hemm MR, Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72:579–589 10.1128/MMBR.00025-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novick RP, Iordanescu S, Projan SJ, Kornblum J, Edelman I. 1989. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell 59:395–404 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- 22.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, Solano C, Gingeras TR. 2011. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A 108:20172–20177 10.1073/pnas.1113521108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lioliou E, Sharma CM, Caldelari I, Helfer A-C, Fechter P, Vandenesch F, Vogel J, Romby P. 2012. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8:e1002782 10.1371/journal.pgen.1002782. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidnenko V, Nicolas P, Grylak-Mielnicka A, Delumeau O, Auger S, Aucouturier A, Guerin C, Repoila F, Bardowski J, Aymerich S, Bidnenko E. 2017. Termination factor Rho: from the control of pervasive transcription to cell fate determination in Bacillus subtilis. PLoS Genet 13:e1006909 10.1371/journal.pgen.1006909. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mäder U, Nicolas P, Depke M, Pané-Farré J, Debarbouille M, van der Kooi-Pol MM, Guérin C, Dérozier S, Hiron A, Jarmer H, Leduc A, Michalik S, Reilman E, Schaffer M, Schmidt F, Bessières P, Noirot P, Hecker M, Msadek T, Völker U, van Dijl JM. 2016. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet 12:e1005962 10.1371/journal.pgen.1005962. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goeders N, Chai R, Chen B, Day A, Salmond GP. 2016. Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins (Basel) 8:282 10.3390/toxins8100282. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coray DS, Wheeler NE, Heinemann JA, Gardner PP. 2017. Why so narrow: distribution of anti-sense regulated, type I toxin-antitoxin systems compared with type II and type III systems. RNA Biol 14:275–280 10.1080/15476286.2016.1272747. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brielle R, Pinel-Marie M-L, Felden B. 2016. Linking bacterial type I toxins with their actions. Curr Opin Microbiol 30:114–121 10.1016/j.mib.2016.01.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Pinel-Marie M-L, Brielle R, Felden B. 2014. Dual toxic-peptide-coding Staphylococcus aureus RNA under antisense regulation targets host cells and bacterial rivals unequally. Cell Reports 7:424–435 10.1016/j.celrep.2014.03.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Sayed N, Jousselin A, Felden B. 2011. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat Struct Mol Biol 19:105–112 10.1038/nsmb.2193. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Bronsard J, Pascreau G, Sassi M, Mauro T, Augagneur Y, Felden B. 2017. sRNA and cis-antisense sRNA identification in Staphylococcus aureus highlights an unusual sRNA gene cluster with one encoding a secreted peptide. Sci Rep 7:4565 10.1038/s41598-017-04786-3. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neubauer C, Gao Y-G, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. 2009. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139:1084–1095 10.1016/j.cell.2009.11.015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berghoff BA, Wagner EGH. 2017. RNA-based regulation in type I toxin-antitoxin systems and its implication for bacterial persistence. Curr Genet 63:1011–1016 10.1007/s00294-017-0710-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Updegrove TB, Zhang A, Storz G. 2016. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30:133–138 10.1016/j.mib.2016.02.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attaiech L, Glover JNM, Charpentier X. 2017. RNA chaperones step out of Hfq’s shadow. Trends Microbiol 25:247–249 10.1016/j.tim.2017.01.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Zheng A, Panja S, Woodson SA. 2016. Arginine patch predicts the RNA annealing activity of Hfq from Gram-negative and Gram-positive bacteria. J Mol Biol 428:2259–2264 10.1016/j.jmb.2016.03.027. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, Moreau K, Vandenesch F, Caldelari I, Romby P. 2016. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol 70:299–316 10.1146/annurev-micro-102215-095708. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Xue T, Zhang X, Sun H, Sun B. 2014. ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5′ UTR of sarT mRNA. Med Microbiol Immunol (Berl) 203:1–12 10.1007/s00430-013-0307-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Geissmann T, Chevalier C, Cros M-J, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, Romby P. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37:7239–7257 10.1093/nar/gkp668. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mauro T, Rouillon A, Felden B. 2016. Insights into the regulation of small RNA expression: SarA represses the expression of two sRNAs in Staphylococcus aureus. Nucleic Acids Res 44:10186–10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Pabic H, Germain-Amiot N, Bordeau V, Felden B. 2015. A bacterial regulatory RNA attenuates virulence, spread and human host cell phagocytosis. Nucleic Acids Res 43:9232–9248 10.1093/nar/gkv783. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée-Durant E, Barbet R, Jacquet E, Jacq A, Gautheret D, Felden B, Vogel J, Bouloc P. 2010. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res 38:6620–6636 10.1093/nar/gkq462. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durand S, Tomasini A, Braun F, Condon C, Romby P. 2015. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol Rev 39:316–330 10.1093/femsre/fuv007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Durand S, Braun F, Helfer A-C, Romby P, Condon C. 2017. sRNA-mediated activation of gene expression by inhibition of 5′-3′ exonucleolytic mRNA degradation. eLife 6:e23602 10.7554/eLife.23602. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen JS, Christiansen MHG, Bonde M, Gottschalk S, Frees D, Thomsen LE, Kallipolitis BH. 2011. Searching for small σB-regulated genes in Staphylococcus aureus. Arch Microbiol 193:23–34 10.1007/s00203-010-0641-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Marincola G, Schäfer T, Behler J, Bernhardt J, Ohlsen K, Goerke C, Wolz C. 2012. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol 85:817–832 10.1111/j.1365-2958.2012.08144.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Romilly C, Lays C, Tomasini A, Caldelari I, Benito Y, Hammann P, Geissmann T, Boisset S, Romby P, Vandenesch F. 2014. A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog 10:e1003979 10.1371/journal.ppat.1003979. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomasini A, Moreau K, Chicher J, Geissmann T, Vandenesch F, Romby P, Marzi S, Caldelari I. 2017. The RNA targetome of Staphylococcus aureus non-coding RNA RsaA: impact on cell surface properties and defense mechanisms. Nucleic Acids Res 45:6746–6760 10.1093/nar/gkx219. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5:e10725 10.1371/journal.pone.0010725. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, Reyes D, Beaume M, Francois P, Cheung A. 2014. Contribution of teg49 small RNA in the 5′ upstream transcriptional region of sarA to virulence in Staphylococcus aureus. Infect Immun 82:4369–4379 10.1128/IAI.02002-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manna AC, Kim S, Cengher L, Corvaglia A, Leo S, Francois P, Cheung AL. 2018. Small RNA teg49 Is derived from a sarA transcript and regulates virulence genes independent of SarA in Staphylococcus aureus. Infect Immun 86:e00635-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327 10.1038/nrmicro2315. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, van der Oost J. 2012. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet 46:311–339 10.1146/annurev-genet-110711-155447. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 10.1126/science.1138140. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7 10.1186/1745-6150-1-7. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338 10.1038/nature10886. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172 10.1186/1471-2105-8-172. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736 10.1038/nrmicro3569. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol 1:e60 10.1371/journal.pcbi.0010060. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 10.1126/science.1179555. [PubMed] [DOI] [PubMed] [Google Scholar]
- 61.Nuñez JK, Lee ASY, Engelman A, Doudna JA. 2015. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519:193–198 10.1038/nature14237. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61 10.1038/nature15386. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40:5569–5576 10.1093/nar/gks216. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. 2015. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520:505–510 10.1038/nature14302. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Modell JW, Jiang W, Marraffini LA. 2017. CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544:101–104 10.1038/nature21719. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright AV, Liu J-J, Knott GJ, Doxzen KW, Nogales E, Doudna JA. 2017. Structures of the CRISPR genome integration complex. Science 357:1113–1118 10.1126/science.aao0679. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575 10.1046/j.1365-2958.2002.02839.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol 77:1367–1379 10.1111/j.1365-2958.2010.07265.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358 10.1126/science.1192272. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607 10.1038/nature09886. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821 10.1126/science.1225829. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Q, Ye Y. 2017. Not all predicted CRISPR-Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18:92 10.1186/s12859-017-1512-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 10.1126/science.1159689. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strutt SC, Torrez RM, Kaya E, Negrete OA, Doudna JA. 2018. RNA-dependent RNA targeting by CRISPR-Cas9. eLife 7:e32724 10.7554/eLife.32724. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holt DC, Holden MTG, Tong SYC, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. 2011. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3:881–895 10.1093/gbe/evr078. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tong SYC, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 2015. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22 10.1099/ijs.0.062752-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cramton SE, Schnell NF, Götz F, Brückner R. 2000. Identification of a new repetitive element in Staphylococcus aureus. Infect Immun 68:2344–2348 10.1128/IAI.68.4.2344-2348.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Purves J, Blades M, Arafat Y, Malik SA, Bayliss CD, Morrissey JA. 2012. Variation in the genomic locations and sequence conservation of STAR elements among staphylococcal species provides insight into DNA repeat evolution. BMC Genomics 13:515 10.1186/1471-2164-13-515. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinnevey PM, Shore AC, Brennan GI, Sullivan DJ, Ehricht R, Monecke S, Slickers P, Coleman DC. 2013. Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob Agents Chemother 57:524–531 10.1128/AAC.01689-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. 2015. Crystal structure of Staphylococcus aureus Cas9. Cell 162:1113–1126 10.1016/j.cell.2015.08.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191 10.1038/nature14299. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823 10.1126/science.1231143. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. 2013. RNA-programmed genome editing in human cells. eLife 2:e00471 10.7554/eLife.00471. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826 10.1126/science.1232033. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. 2015. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep 4:5400. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol 395:270–281 10.1016/j.jmb.2009.10.057. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Quax TEF, Voet M, Sismeiro O, Dillies M-A, Jagla B, Coppée J-Y, Sezonov G, Forterre P, van der Oost J, Lavigne R, Prangishvili D. 2013. Massive activation of archaeal defense genes during viral infection. J Virol 87:8419–8428 10.1128/JVI.01020-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, Verberkmoes NC. 2012. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS One 7:e38077 10.1371/journal.pone.0038077. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, DeLisa MP. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol 79:584–599 10.1111/j.1365-2958.2010.07482.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, Staals RHJ, Fineran PC. 2016. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell 64:1102–1108 10.1016/j.molcel.2016.11.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, Bassler BL. 2017. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci U S A 114:131–135 10.1073/pnas.1617415113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang YK, Kwon K, Ryu JS, Lee HN, Park C, Chung HJ. 2017. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjug Chem 28:957–967 10.1021/acs.bioconjchem.6b00676. [PubMed] [DOI] [PubMed] [Google Scholar]
- 93.Nitzan M, Fechter P, Peer A, Altuvia Y, Bronesky D, Vandenesch F, Romby P, Biham O, Margalit H. 2015. A defense-offense multi-layered regulatory switch in a pathogenic bacterium. Nucleic Acids Res 43:1357–1369 10.1093/nar/gkv001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Audretsch C, Lopez D, Srivastava M, Wolz C, Dandekar T. 2013. A semi-quantitative model of quorum-sensing in Staphylococcus aureus, approved by microarray meta-analyses and tested by mutation studies. Mol Biosyst 9:2665–2680 10.1039/c3mb70117d. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Jelsbak L, Hemmingsen L, Donat S, Ohlsen K, Boye K, Westh H, Ingmer H, Frees D. 2010. Growth phase-dependent regulation of the global virulence regulator Rot in clinical isolates of Staphylococcus aureus. Int J Med Microbiol 300:229–236 10.1016/j.ijmm.2009.07.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Song J, Lays C, Vandenesch F, Benito Y, Bes M, Chu Y, Lina G, Romby P, Geissmann T, Boisset S. 2012. The expression of small regulatory RNAs in clinical samples reflects the different life styles of Staphylococcus aureus in colonization vs. infection. PLoS One 7:e37294 10.1371/journal.pone.0037294. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis 198:561–570 10.1086/590157. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177 10.1371/journal.pone.0015177. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bordeau V, Cady A, Revest M, Rostan O, Sassi M, Tattevin P, Donnio P-Y, Felden B. 2016. Staphylococcus aureus regulatory RNAs as potential biomarkers for bloodstream infections. Emerg Infect Dis 22:1570–1578 10.3201/eid2209.151801. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. 2014. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol 22:676–685 10.1016/j.tim.2014.09.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Pollitt EJG, West SA, Crusz SA, Burton-Chellew MN, Diggle SP. 2014. Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus. Infect Immun 82:1045–1051 10.1128/IAI.01216-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chabelskaya S, Gaillot O, Felden B. 2010. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog 6:e1000927 10.1371/journal.ppat.1000927. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pichon C, Felden B. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci U S A 102:14249–14254 10.1073/pnas.0503838102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haupt K, Reuter M, van den Elsen J, Burman J, Hälbich S, Richter J, Skerka C, Zipfel PF. 2008. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement factor H and C3b. PLoS Pathog 4:e1000250 10.1371/journal.ppat.1000250. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L. 1998. A second IgG-binding protein in Staphylococcus aureus. Microbiology 144:985–991 10.1099/00221287-144-4-985. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol 188:6739–6756 10.1128/JB.00609-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morrison JM, Miller EW, Benson MA, Alonzo F III, Yoong P, Torres VJ, Hinrichs SH, Dunman PM. 2012. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J Bacteriol 194:2924–2938 10.1128/JB.06708-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Das S, Lindemann C, Young BC, Muller J, Österreich B, Ternette N, Winkler A-C, Paprotka K, Reinhardt R, Förstner KU, Allen E, Flaxman A, Yamaguchi Y, Rollier CS, van Diemen P, Blättner S, Remmele CW, Selle M, Dittrich M, Müller T, Vogel J, Ohlsen K, Crook DW, Massey R, Wilson DJ, Rudel T, Wyllie DH, Fraunholz MJ. 2016. Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Proc Natl Acad Sci U S A 113:E3101–E3110 10.1073/pnas.1520255113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. 2013. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog 9:e1003269 10.1371/journal.ppat.1003269. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qin L, McCausland JW, Cheung GYC, Otto M. 2016. PSM-Mec-A virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front Microbiol 7:1293 10.3389/fmicb.2016.01293. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara-Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. 2011. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog 7:e1001267 10.1371/journal.ppat.1001267. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Crosby HA, Schlievert PM, Merriman JA, King JM, Salgado-Pabón W, Horswill AR. 2016. The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog 12:e1005604 10.1371/journal.ppat.1005604. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krismer B, Weidenmaier C, Zipperer A, Peschel A. 2017. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol 15:675–687 10.1038/nrmicro.2017.104. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Brown AF, Leech JM, Rogers TR, McLoughlin RM. 2014. Staphylococcus aureus colonization: modulation of host immune response and impact on human vaccine design. Front Immunol 4:507 10.3389/fimmu.2013.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pynnonen M, Stephenson RE, Schwartz K, Hernandez M, Boles BR. 2011. Hemoglobin promotes Staphylococcus aureus nasal colonization. PLoS Pathog 7:e1002104 10.1371/journal.ppat.1002104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. 2016. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 7:1230 10.3389/fmicb.2016.01230. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hermansen GMM, Sazinas P, Kofod D, Millard A, Andersen PS, Jelsbak L. 2018. Transcriptomic profiling of interacting nasal staphylococci species reveals global changes in gene and non-coding RNA expression. FEMS Microbiol Lett 365: 10.1093/femsle/fny004. [DOI] [PubMed] [Google Scholar]


