Abstract
Background
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths worldwide. CRC deaths can be reduced with prevention and early diagnosis. Circulating tumor DNA-based liquid biopsies, are emerging tools for cancer detection. However, the tumor-signal-dependent nature of this approach results in low sensitivity in precancerous and early CRC stages. Here we propose the host immune response to the onset of cancer as an alternative approach for early detection of CRC.
Methods
We perform whole transcriptome analysis of peripheral blood mononuclear cells (PBMCs) isolated from individuals with CRC, precancerous lesions or negative colonoscopy in two independent cohorts using next-generation sequencing.
Results
We discover and validate novel early CRC RNA biomarkers. Taking into account, and adjusting for, the sensitivity of PBMCs transcriptomes to processing times, we report distinct transcriptomic changes in the periphery related to specific CRC stages. Activation of innate immunity is already detectable in the peripheral blood of individuals with pre-malignant advanced adenomas. This immune response is followed by signs of transient B-cell activation and sustained inhibition of T-cell responses along CRC progression, whereby at late stages, protumoral myeloid cells, wound healing and coagulation processes prevail. Moreover, some biomarkers show similar dysregulation in tumors and are implicated in known pathways of CRC pathophysiology.
Conclusions
The strong systemic immune modulation triggered during CRC progression leads to previously unnoticed alterations detectable in PBMCs, paving the way for the development of an early CRC screening blood test, incorporating 226 validated biomarkers identified through immunotranscriptomics.
Keywords: Colorectal Cancer, Next generation sequencing - NGS, Biomarker, B cell, Neutrophil
WHAT IS ALREADY KNOWN ON THIS TOPIC
Lifestyle-driven prevention and early diagnosis are the best strategies to reduce the mortality of colorectal cancer (CRC). Liquid biopsies measuring circulating tumor DNA are emerging as an alternative to the invasive colonoscopy for CRC detection. However, their sensitivity to detect precancerous stages is low.
WHAT THIS STUDY ADDS
We present an alternative approach to identify CRC onset: peripheral-blood mononuclear cells (PBMCs) whole transcriptome analysis. We identify and validate 226 RNA biomarkers that reveal strong systemic immune modulation along CRC progression. We also describe the impact of handling and storage on PBMCs transcriptome.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study paves the way for the development of a diagnostic test for early CRC detection using the transcriptome of RNA-stabilized blood samples.
Background
Colorectal cancer (CRC) is the third most common cancer type and second leading cause of cancer-related deaths worldwide. More than 1.9 million patients are diagnosed yearly and approximately half of these patients die from the disease (WHO 2020 statistics). Excessive drinking, obesity, high red or processed meat intake, high-sugar diet and low exercising are the main risk factors.1 With these habits rising globally, CRC incidence is expected to become a major health issue.
The high mortality rate is, at least in part, caused by late-stage diagnosis when patients display symptoms such as blood in stool and abdominal pains. Therefore, early diagnosis is key to improve CRC patients’ prognosis.2 CRC development is mostly slow, with a 10–15 years asymptomatic period wherein polyps or sessile serrated lesions undergo a transformation into carcinoma.3 This extended time frame provides a substantial screening window to detect the onset of the disease. However, the high invasiveness of the golden diagnostic procedure, colonoscopy,2 leads to suboptimal compliance from the population despite national screening campaigns.4 The most recommended alternative non-invasive screening procedures (fecal immunochemical test and multitarget stool DNA test) involve stool sampling, a method that fails to meet compliance with the recommended guidelines due to its lack of convenience.4
More appealing blood molecular biomarker-based tests are emerging as alternative. The majority rely on the detection of tumor-derived DNA (circulating tumor DNA).4,6 However, while they are sensitive for CRC detection, the sensitivity to detect precancerous stages (advanced adenoma (AA)) is low.7 We, instead, have developed a test based on the gene expression profile of peripheral blood mononuclear cell (PBMC) and quantification of two plasma proteins, Colox.8 Assessing the host response to the tumor might represent an advantage over capturing direct low and heterogenous tumor signals in the blood. Indeed, systemic alterations of the immune system are detected in peripheral blood9 and Colox is, so far, the most sensitive test for non-cancerous early CRC stages, with a specificity of 90% and a sensitivity of 79.5% and 55.4% for CRC and large adenomatous polyps, respectively.8 Colox signature was developed in a targeted approach by measuring the expression levels of well-known immune cell genes by high throughput but limited Real Time Quantitative PCR(RT-qPCR). Thus, whole transcriptome analysis of PBMC using next-generation sequencing is an alternative to identify unknown biomarkers of early phases of the disease that might have been missed with this approach and further understand disease progression.
In this study, we compared the full PBMC transcriptome of patients with different CRC stages, precancerous AA lesions and control subjects and validated 226 CRC biomarkers which reveal a strong systemic immune modulation driven by progression from precancerous lesions to full-fledged CRC tumors.
Material and methods
Study cohorts
In this study, we analyzed a subset of PBMC samples previously collected in a case–control study (DGNP-COL-0310) involving three South Korean and six Swiss study centers, for the development and validation of the Colox test.8 The study enrolled 1,665 subjects older than 50 years who were referred for colonoscopy by general practitioner or were scheduled for surgery. For the analysis presented here, we selected samples from the training and validation sets of the Colox study8 and with sufficient material for RNA sequencing analysis (>500 ng of RNA). The characteristics of the study cohort are summarized in table 1.
Table 1. Data sets compositions.
| Study groups | Discovery set (batches 1 and 2) | Validation set (batch 3) | Total subjects |
| Study subgroups | |||
| Colorectal cancer | 85 | 109 | 196 |
| Stage I | 24 | 37 | 61 |
| Stage II | 27 | 21 | 49 |
| Stage III | 20 | 34 | 54 |
| Stage IV | 14 | 15 | 29 |
| Unknown | 0 | 2 | 3 |
| Advanced adenoma | 64 | 50 | 114 |
| Low grade | 35 | 16 | 51 |
| High grade | 26 | 34 | 60 |
| No dysplasia | 2 | 0 | 2 |
| Unknown | 1 | 0 | 1 |
| Control subjects | 108 | 116 | 226 |
| No colorectal lesion | 92 | 98 | 192 |
| Hyperplastic polyp | 16 | 18 | 34 |
| Other cancers | 38 | 0 | 38 |
| Lung | 11 | 0 | 11 |
| Pancreas | 12 | 0 | 12 |
| Prostate | 15 | 0 | 15 |
| Total | 295 | 275 | 570 |
| % men - % women | 58–42 | 61–39 | 60–40 |
| % Swiss - % Korean | 61–39 | 48–52 | 55–45 |
| Age (min, median, max) | (39, 63, 85) | (49, 61, 91) | (39, 62, 91) |
Batches corresponds to the three sequencing batches and sets to the data set used for the biomarker analyses.
Screening design and data set generation
The analysis set consisted of 570 PBMC samples. The samples were separated into three sequencing batches. Batches 1 and 2 were combined to form a discovery data set of 295 samples, and batch 3 (n=275) was used as a validation set.
Blood collection and processing
Peripheral blood samples from all subjects were taken either up to 30 days before or up to 12 weeks after colonoscopy and prior to any surgical intervention for polyp or cancer resection. Blood samples were collected on 4×4 mL BD Vacutainer(r) CPT Tubes (Becton Dickinson, Franklin Lakes, New Jersey, USA) and kept at room temperature before PBMCs separation within 6 hours, following BD Vacutainer(r) CPT tube manufacturer’s instructions. PBMC pellets were resuspended in RNAlater® Solution (Life Technologies, Carlsbad, California, USA) and stored at −80°C. For this study, frozen PBMC samples were retrieved from the Novigenix biorepository and defrost at room temperature without direct sunlight for 2 hours.
RNA extraction
Automated extraction of total RNA was performed on QIAcube by RNeasy Mini kit (Qiagen, Venlo, Netherlands) and included a DNase treatment. RNA concentration was measured by a fluorometer Qubit (Thermo Fisher, Waltham, Massachusetts, USA) and RNA integrity was analyzed by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA). Isolated total RNA was aliquoted and stored at −80°C.
RNA sequencing
Samples were separated into three batches for RNA sequencing, following the same protocol and performed with the same instruments and operators. Libraries were prepared by Fasteris (Genesupport SA, Geneva, Switzerland) using the Standard Protocol mRNA Stranded (TruSeq Stranded mRNA Kit) (Illumina, Baltimore, Maryland, USA) with poly(A) tails selection. Library quality controls were assessed with the 4200 TapeStation system (Agilent Technologies, Santa Clara, California, USA). Some samples were repeated in batches 1, 2 and 3 for library preparation and sequencing to ensure repeatability. Paired-end (2×75 bp) sequencing was performed with the Illumina HiSeq 3000/4000 SBS Kit and SR Flow cell version, on its HiSeq 3000/4000 platform (Illumina, Baltimore, Maryland, USA), with a depth of minimum 30 M reads/sample.
RNA sequencing data analysis
FASTQ files were aligned to the human genome reference GRCh38.p12 by HISAT2 (V.2.2.0) using the pseudo-alignment approach. MultiQC was used to perform the alignment quality control and to remove adapter sequences and low-quality bases from the 3’end of reads.
Transcripts were quantified using Salmon (V.1.4.0) and the resulting raw counts were normalized by transcripts per million (TPM). A threshold of one TPM per gene per clinical group was applied to filter out background noise and variance stabilization transformation (VST) was performed for normalization.
A descriptive analysis was performed to identify the main sources of variability. For that, VST values were used as input for principal component analysis. 21 sex chromosome-encoded genes were filtered out to avoid gender-related confounding factor (online supplemental figure S1A,B). Variance partitioning analysis was performed by variancePartition package (V.1.32.1) (online supplemental figure S1C) to identify further confounding factors.
Differential expression analysis (DEA) was performed by DESeq2 (V.1.3), using time of blood processing (TBP) and sequencing batches as covariates. Lastly, a list of 1,893 genes identified as biologically less informative, that is, genes that undergo somatic recombination (immunoglobulin (IG) and T-cell receptor (TCR) genes), highly polymorphic Human Leukocyte Antigen (HLA) genes, non-coding genes, pseudogenes and transcripts to be experimentally confirmed (TEC genes), were excluded from the DEA results (online supplemental figure S2B). Differential expressed genes (DEGs) were selected based on adjusted p value (Benjamini-Hochberg method) and log2 fold change as reported in the results section.
Multivariate analyses were performed with different machine learning methods, as described in the online supplemental methods section, on ~16,000 genes, with the goal of selecting the best genes as predictors of AA and CRC samples. The machine learning methods were applied to the gene expression values after a linear model-based correction for confounding variables (TBP and batch). The R package limma (V.3.12) was used to compute the correction.
Cell type deconvolution of PBMC transcriptome was performed using LM22 gene expression matrix10, TPM values and support vector regression method, reimplemented in granulator V.1.4.0 package.
Public RNA sequencing data analysis
The FASTQ files from Gene Expression Omnibus (GEO) accession numbers GSE156451, GSE196006, GSE89393, GSE109203, GSE136630, GSE76987 and GSE164541 were downloaded and processed in-house as described in the online supplemental methods section.
Functional analysis
Over-representation analysis (ORA) was performed on each list of DEGs using clusterProfiler (V.4.4.4)11 and ReactomePA (V.1.40.0).12 This analysis considered both Gene Ontology and Reactome databases to retrieve enriched biological processes and pathways, respectively.
Data visualization
Graphs were generated using ggplot2 (V.3.4.3), enrichplot (V.1.16.2) and VennDiagram (V.1.7.3) R packages, and figures were made on Adobe Illustrator.
Patient and public involvement
Written informed consent was obtained from all study participants but clinical research was not conducted with or by any of the participants.
Results
The time of blood processing impacts PBMCs’ transcriptome
To identify accessible biomarkers of CRC development and progression, we performed total mRNA sequencing and DEA of PBMCs from control subjects and patients with colorectal lesions. Samples were retrieved from a multicentric study conducted between 2010 and 2013, involving 570 subjects from Switzerland and Korea.8 The cohort includes control subjects (CON) without colorectal lesions or with benign hyperplastic polyps, subjects with AA with different dysplasia levels (no, low or high), patients with CRC from all stages (I-IV) and patients with other cancers (lung, prostate, pancreas) (table 1). To increase the robustness of our potential new CRC biomarkers, we split the cohort into two independent data sets: a discovery set (n=295) and a validation set (n=275) (figure 1A). Each set had similar numbers of CRC and CON samples, and both sets were matched for age, sex and patient country (table 1). Nonetheless, 21 sex chromosome-encoded transcripts were excluded for the DEA as principal component analysis of the samples from the discovery set revealed sex as a major confounding variable (online supplemental figure S1A,B). Lowly expressed genes were also filtered out before DEA (see methods).
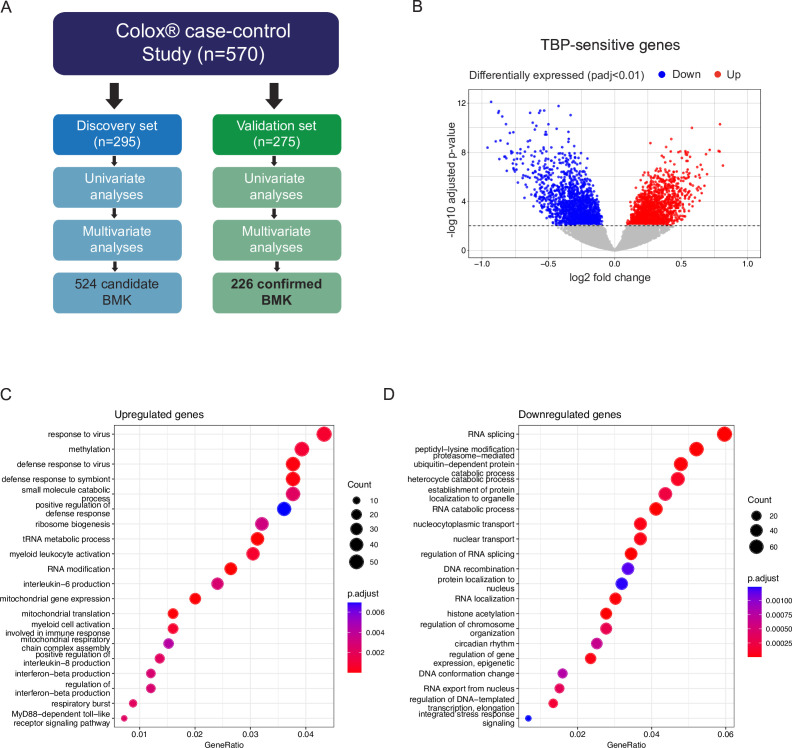
Figure 1. TBP impacts peripheral blood mononuclear cells transcriptome. (A) Study design. The cohort was split into a discovery (n=295 samples) and a validation set (n=279 samples). (B) Volcano plot showing DEGs on control subjects samples from the discovery set processed under 2 hours compared with those processed over longer durations. Dash line represents the significance threshold at adjusted p value (padj)<0.01. In total, 3229 DEGs were identified. Upregulated DEGs (padj<0.01) are colored in red (n=1547) and downregulated DEGs in blue (n=1682). Genes non-significantly differentially expressed are shown in gray. C–D. Over-representation analysis using Gene Ontology (GO) biological process (BP) for the upregulated (C) and downregulated DEGs (D). A p value cut-off of 0.01 and q value cut of 0.01 were used followed by a simplification step by padj with a 0.7 cut-off. All the significantly enriched terms were shown for the upregulated DEGs and the top 20 terms for the downregulated ones. The X-axis represents the ratio between the number of DEGs mapped to the GO BP and the total amount of DEGs. The size of the dots represents the number of genes from the DEG list that are found in each BP and the color represents the significance of the enrichment per biological pathway. DEG, differential expressed genes; TBP, time of blood processing.
Moreover, variance partitioning analysis exposed the TBP, the time between the blood withdrawal and PBMC isolation, as another major covariate (online supplemental figure S1C). We therefore investigated the source and effect of this variable. We found significant differences in TBP among the different collection sites, but not countries, which led to significant differences among clinical groups and subgroups due to the uneven distribution of clinical groups among collection sites (online supplemental figure S1D–H).
To understand which genes were susceptible to the TBP, we performed a DEA among CON samples processed in less and more than 2 hours. We identified more than 3,000 significantly DEGs (figure 1B). Functional analysis highlighted a broad activation of cellular stress, characterized by the upregulation of mitochondrial genes, along with specific activation of myeloid cells, and a concurrent downregulation of transcriptional processes in samples processed in more than 2 hours (figure 1C,D). Previous proteomic analysis of 24 hours stored compared with freshly isolated PBMCs also revealed upregulation of myeloid cell activation processes.13 When we compared the proteins identified by Bonilauri et al to our genes sensitive to processing time, we found 9 (18% of upregulated proteins) upregulated genes in common (S100A11, HK3, MNDA, NCF2, S100A8 and CYBB; p-adjusted <0.01 for transcripts, p value<0.01 for proteins) and 6 (6.1% of downregulated proteins) shared downregulated genes (ILF2, TCP1, HNRNPU, NCL, CYTH1 and P4HB (online supplemental table 1).13 These findings indicate that PBMC, and especially myeloid cells, are highly sensitive to storage duration, as even a few hours of delay can trigger cellular stress programs. Therefore, it is critical to control technical variables during sample preparation when conducting transcriptomic analyses of PBMCs. To mitigate this issue, a linear model-based correction for TBP was performed before applying machine learning algorithms, and TBP was added as a covariate to the DEA where sequencing batch was considered as covariates too (online supplemental figure S1I and online supplemental methods).
Identification and validation of 226 CRC blood RNA biomarkers
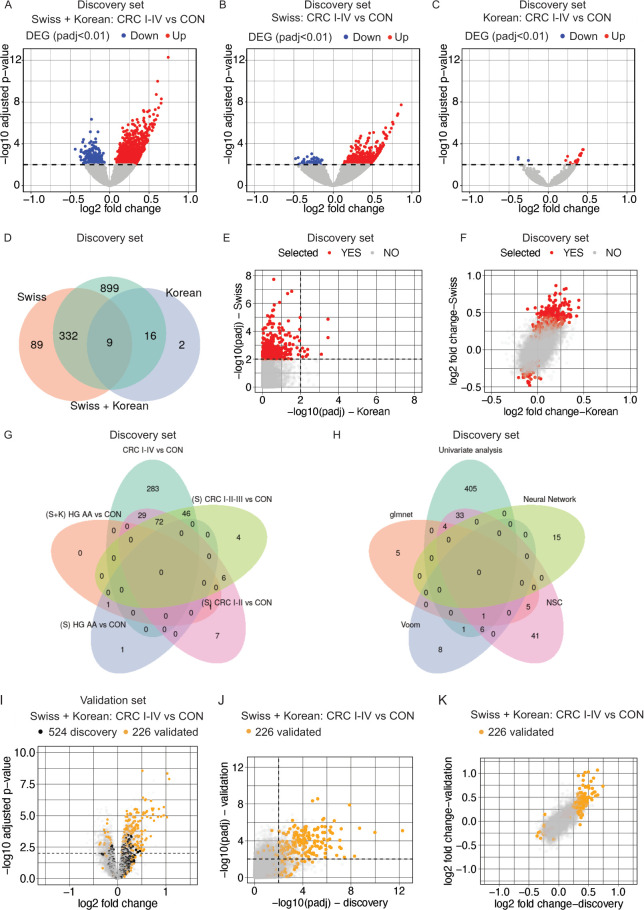
The DEA between CRC and CON samples in the discovery set revealed 1,337 DEGs (adjusted p value<0.01) (online supplemental figure S2A). Among them, we found highly polymorphic genes (IG, TCR and HLA genes), experimentally unconfirmed transcripts (TECs), non-coding genes and pseudogenes, which were excluded to prioritize the identification of prevalent biologically relevant genes involved in CRC pathogenesis (online supplemental figure S2A,B). The exclusion of these genes (online supplemental figure S2B) was performed in all consecutive DEA analyses (see methods). After filtering, 1,256 DEGs (977 up and 279 down) remained between CRC versus CON samples (figure 2A). Since the patient country was previously identified as a major source of variation (online supplemental figure S1I), we also performed DEA on the Swiss (S) and Korean (K) samples separately (figure 2B,C). As shown in figure 2B,C, stratifying the data set by the country of origin led to lower numbers of identified DEGs, with 430 (378 up and 52 down) and 27 (24 up and three down) DEGs identified in the S and K patients, respectively. Only nine genes (KCNJ15, DYSF, CCR1, AGP9, CR1, DLGAP5, S100A8, LHFPL2, SNCA) were found commonly upregulated between the different CRC versus CON DEAs (figure 2D), suggesting PBMC transcriptomic differences between the two patient populations. Comparison of the adjusted p values and fold change of the three CRC versus CON DEAs revealed good correlations (online supplemental figure S3A–F), but mainly between the S+K and S DEAs (online supplemental figure S3C,F), indicating that the S cohort is the main contributor to the significant changes observed in the global DEA. The lower proportion of DEGs observed in the K cohort may be linked to the reduced prevalence of patients with high-grade (III-IV) CRC compared with the S cohort (online supplemental figure S3G–H). Indeed, the DEA conducted within distinct CRC subgroups versus CON samples revealed a heightened prevalence of significant transcriptome alterations in PBMCs as the CRC progresses to more advanced stages (online supplemental figure S4A–F). Nonetheless, PBMC transcriptome differences between S and K samples arising from genetic background and/or lifestyle cannot be excluded.
Figure 2. Identification of 226 CRC blood biomarkers. (A–C) Volcano plot representing the −log10 adjusted p value (padj) as a function of the log2 fold change for the DEA between CRC and CON samples, for the entire discovery set (Swiss and Korean) (A), only the Swiss samples (B) and the Korean samples (C). Dash lines represent the significance threshold (padj<0.01). Downregulated genes are marked in blue and upregulated genes in red. (D) Venn diagram depicting the overlap between the first three DEA analyses in panels A to C. (E) Scatter plots comparing −log10 padj from the CRC versus CON DEAs of Swiss versus Korean cohorts. Red dots represent selected genes (n=430) from CRC versus CON DEA for validation. Dotted line indicates the significance threshold (padj<0.01). (F) Scatter plots comparing the log2 fold change from the CRC versus CON DEA of Swiss versus Korean cohorts. Red dots represent selected genes for validation. (G–H) Venn diagram representing the overlap between the BMKs identified by the different univariate (G) and multivariate (H) analyses performed on the discovery set. (I) Volcano plot representing the −log10 adjusted p value (padj) as a function of the log2 fold change for the DEA between CRC I-IV and CON samples for the entire validation set. (J) Scatter plots of the −log10 padj from the CRC I-IV versus CON DEA of the discovery versus validation set. (K) Scatter plots of the log2 fold change from the CRC I-IV versus CON DEA of the discovery versus validation set. Black dash lines represent the significance threshold (padj<0.01). Black dots represent the 524 BMKs identified in the discovery set, orange dots the 226 validated BMKs and gray dots the entire DEA results. AA, advanced adenoma; BMK, biomarker; CON, control subjects; CRC, colorectal cancer; DEA, differential expression analysis; DEG, differential expressed genes; HG, high grade; NSC, nearest shrunken centroids.
We therefore decided to prioritize the selection of DEGs arising from the S population (table 2), which ranked among the genes with top fold changes of the K DEA (figure 2E,F). To enrich early biomarkers of CRC, we stratified the CRC group into CRC stages and performed DEA against CON samples. No DEGs were identified when comparing all AAs with CON samples (data not shown). However, we did detect one upregulated (C3orf35) and one downregulated gene (MIR6723) in high grade (HG) AAs compared with CON (online supplemental figure S4A) in the entire cohort and another two in the S DEA (online supplemental figure S4D) (one new, OR2A9P, table 2). We performed further DEAs on the S population per CRC subgroup leading to the identification of 450 DEGs by univariate analysis (table 2, see methods). Figure 2G shows the unique and common DEGs per DEA. In addition to the univariate analyses for biomarker (BMK) identification, we performed several multivariate analyses on the entire cohort to further increase our list of candidate BMKs (figure 2H). 74 additional BMKs were identified (table 2). In total, we identified a panel of 524 genes as candidate BMKs for patient with CRC discrimination from healthy subjects (figure 1A and table 2).
Table 2. Summary of the total number of BMKs identified per differential expression analysis (padj<0.01) or multivariate analysis and those selected for validation in the discovery set or confirmed in the validation set.
| Biomarker selection in discovery set | Patients considered | Number of identified BMKs (padj<0.01) | Number of selected BMKs |
| Univariate (DEGs) | 1,016 | 450 | |
| CRC I-IV vs CON | S+K ∩ S | 341 | 341 |
| CRC I-IV vs CON | S | 430 | 89 |
| HG AA vs CON | S+K | 2* | 2 |
| HG AA vs CON | S | 2* | 1 |
| CRIC I-II vs CON | S | 115 | 13 |
| CRC I-II-III vs CON | S | 128 | 4 |
| Multivariate | 141 | 74 | |
| Voom | S+K | 16 | 8 |
| glmnet | S+K | 15 | 10 |
| NSC | S+K | 90 | 41 |
| Evolutionary random forest | S+K | 20 | 15 |
| Total unique BMKs | 524 |
| Analyses performed on validation set | Patients considered | Number of identified BMKs (padj<0.01) | Number of confirmed BMKs |
| Univariate (DEGs) | 3,733 | 212 | |
| CON vs CRC | S+K ∩ S | 614 | 124 |
| CON vs CRC | S | 715 | 3 |
| CON vs AA | S+K ∩ S | 10 | 1 |
| CON vs CRC I-II | S+K ∪ S | 601 | 24 |
| CON vs CRC I-II-III | S+K ∪ S | 852 | 19 |
| CON vs CRC | S+K ∪ S | 324** | 16 |
| CON vs CRC III-IV | S+K ∪ S | 617** | 25 |
| Multivariate | 345 | 14 | |
| Voom | S+K | 97 | 2 |
| NSC | S+K | 159 | 0 |
| glmnet | S+K | 89 | 12 |
| Total unique BMKs | 226 |
New BMKs were added to the selection pool subsequently in the discovery set. S=Swiss samples only, K=Korean samples only. *padj, **DEGs confirmed with absolute log2 fold change regardless of padj on validation set.
*padj<0.05.
**DEGs confirmed with absolute log2 fold change >0.5 or <−0.5 regardless of padj on validation set.
AAadvanced adenomaBMKbiomarkerCONcontrol subjectsCRCcolorectal cancer DEGdifferential expressed geneKKoreanNSCnearest shrunken centroidspadjadjusted p valueSSwiss
To confirm the biomarkers identified in the discovery set, we performed DEA in a validation set. DEA between CRC and healthy controls revealed 1,939 DEGs (figure 2I) and a good correlation with adjusted p values and log2 fold changes of the discovery set (figure 2J,K). By performing univariate (online supplemental figure S5) and multivariate analyses we confirmed (226/524) 43% of the BMKs identified in the discovery set (table 2). Of note, 41 genes were confirmed using a log2FC>0.5 or <−0.5 threshold on the validation set (table 2), but met the adjusted p value<0.01 selection criteria in at least one CRC versus CON DEA in the discovery or validation data set (online supplemental figure S5I,J). The confirmation of 43% of the BMKs in the validation set indicates a high robustness of the identified BMKs and a high reproducibility of the data acquisition and analysis, since both sets were independently sequenced and analyzed.
CRC blood biomarkers are involved in wound healing and myeloid cell activation processes
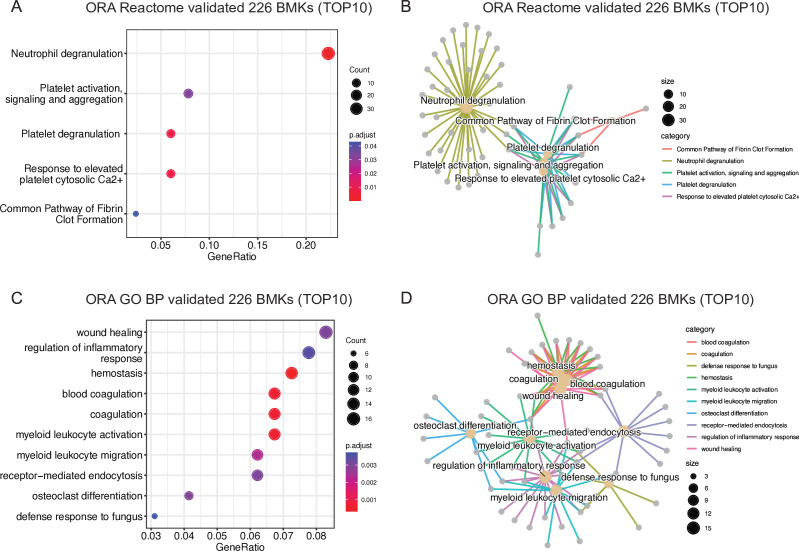
To gain insights into the biological responses involved in CRC development, we performed functional analyses of the newly discovered and validated 226 BMKs. ORA using Reactome revealed a clear enrichment of genes involved in neutrophil-mediated immune response and platelet activation (figure 3A,B, online supplemental table 2). These pathways were confirmed in a Gene Ontology ORA analysis: wound healing processes, where platelets participate, and myeloid cell activation and chemotaxis were found among the 10 top enriched pathways (figure 3C,D, online supplemental table 2). Enriched pathways were found among upregulated BMKs in CRC compared with CON samples (online supplemental figure S6A–D), whereas no significant pathway enrichment was found among downregulated BMKs (online supplemental table 2). The upregulation of myeloid cell migration pathway is also in line with the observed increase in myeloid cells (neutrophils, basophils, eosinophils and monocytes) hematological counts in patients with AA and CRC compared with healthy individuals (online supplemental figure S7A–C), which results in higher neutrophil-to-lymphocyte ratios (NLR) along CRC progression (online supplemental figure S7D–E), previously reported to be associated with worse CRC prognosis.14 Therefore, PBMC transcriptome BMKs can reveal key pathways of CRC pathophysiology.
Figure 3. Wound healing and myeloid cell activation pathways are triggered in blood of CRC patients. (A) Over-representation analysis (ORA) results of validated 226 CRC BMKs using Reactome, adjusted p value<0.05, q value<0.05 and Benjamini-Hochberg correction method. (B) Network showing the relationship between the enriched Reactome pathways and genes found among validated 226 CRC BMKs. (C) Top10 enriched Gene Ontology (GO) biological process (BP) among validated 226 CRC BMKs. A p value cut-off of 0.05 and q value cut off-of 0.05 were used followed by a simplification step by padj with a 0.7 cut-off. (D) Network showing the relationship between the enriched GO BP pathways and genes found among validated 226 CRC BMKs. In panels A and C, the X-axis represents the ratio between the number of genes that are part of the BP indicated in the Y-axis and the total BMK list. The size of the dots represents the number of genes from the BMKs that are found in each BP and the color represents the significance of the enrichment per biological pathway. CRC, colorectal cancer; BMK, biomarker.
PBMC RNA biomarkers from individuals with pre-malignant to late-stage CRC reveal early inflammatory signals followed by protumoral modeling of immune cells
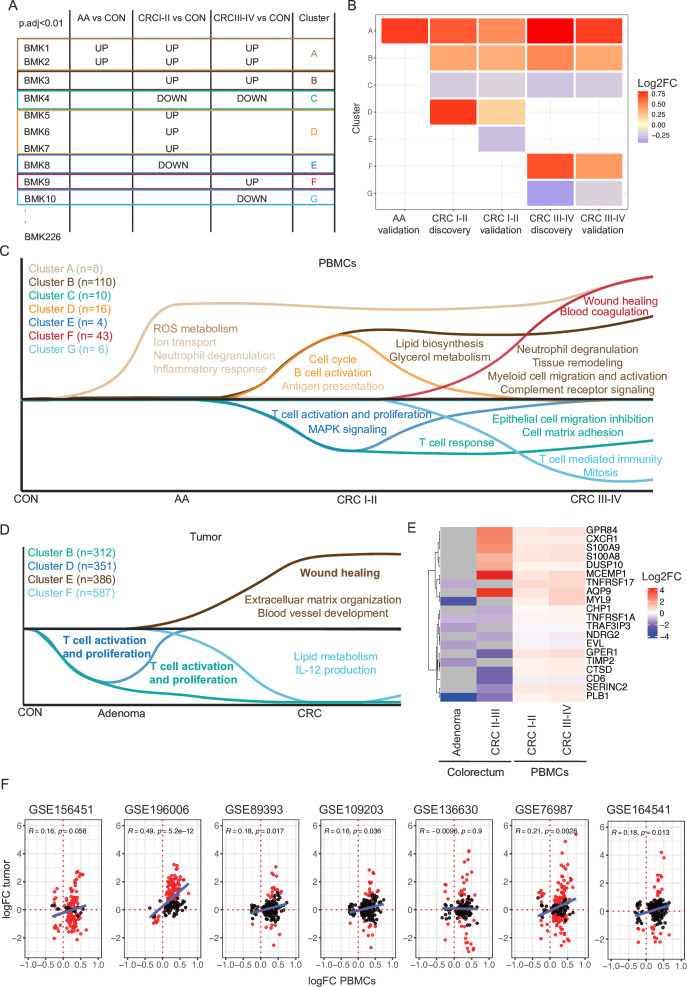
As the identified PBMC BMKs revealed established biological processes implicated in CRC pathogenesis, we sought to conduct a more in-depth analysis of BMK expression patterns throughout CRC progression, aiming to uncover potential novel pathways involved in the onset and development of CRC. To do so, we assessed the adjusted p value of each of the 226 confirmed BMKs for each of the CRC stage-stratified DEA analyses performed on the entire cohort (S+K): AA/HG versus CON, CRC I-II versus CON and CRC III-IV versus CON, in the discovery and validation sets in parallel. Genes with adjusted p value<0.01 in at least one of the two data sets were retained as BMK for the CRC stage analyzed (see online supplemental method). By grouping the BMKs with the same CRC stage expression patterns, we defined seven BMK clusters: A-G (figure 4A,B and online supplemental table 3). These clusters were composed of 8, 110, 10, 16, 4, 43 and 6 genes, respectively. Two additional genes (TRIM58 and MIR6723) showed a unique pattern, being up and downregulated in AA and CRC III-IV, respectively. 27 BMKs from the 226 BMK list were not attributed to any cluster due to the absence of significant adjusted p value in the DEAs of the entire cohort at different stages. These 27 BMKs were previously identified and validated based on other criteria: lower adjusted p value, log2 fold change-based significant thresholds, through multivariate analyses, in S population restricted DEAs or in global CRC I-IV versus CON DEAs (see table 2).
Figure 4. CRC development and progression is characterized by stage-specific biomarkers and associated biological processes. (A) Graphical representation of BMK cluster identification. (B) Heatmap representing the mean log2 fold change per cluster and DEA. (C) Graphical representation of the biological processes from GO BP and reactome where each gene cluster participates. In clusters A and D, significantly enriched pathways are in dark colors and not significantly enriched pathways in light colors. In clusters B, E and F, significantly enriched pathways are represented whereas in clusters C and G, no significantly enriched pathways are indicated, due to the low number of genes per cluster. (D) Graphical representation of the enriched biological processes from GO BP in tumor DEG clusters (padj <0.01). Pathways commonly found in patients with CRC PBMCs are marked in bold. (E) Heatmap representing the mean log2 fold change between indicated groups and control colorectum or PBMCs (mean log2FC of discovery and validation sets). (F) Scatter plots comparing the log2 fold change from the CRC versus CON DEA of the entire discovery set and CRC tumor versus normal tissue DEA of indicated public data sets. Red dots indicate DEGs (padj<0.05) of each tumor data set, while the blue line represents the linear regression fit. A paired DEA was performed for GSE196006 (n=21) and GSE89393 (n=5) data sets, whereas an unpaired DEA was performed for GSE156451 (n_tumor=72, n_normal=72), GSE109203 (n_tumor=12 and n_normal=3), GSE136630 (n_tumor=5, n_normal=5), GSE76987 (n_tumor=4 and n_normal=12) and GSE164541 (n_tumor=5, n_normal=5). GSE156451, GSE109203 and GSE164541 include samples from China, GSE196006, GSE136630 and GSE76987 from USA and GSE89393 from Poland. Pearson correlation coefficients and p values are displayed on the top. AA, advanced adenoma; BMK, biomarker; CON, control subjects; CRC, colorectal cancer; DEA, differential expression analysis; padj, adjusted p value; PBMC, peripheral blood mononuclear cell.
Functional analyses on CRC stage-mapped BMKs clusters revealed stage-specific biological processes (figure 4C and online supplemental table 4). Genes involved in reactive-oxygen (ROS) metabolism already rise at the AA premalignant phase and are maintained throughout tumor progression (cluster A). Among them, we find myeloid-restricted myeloperoxidase (MPO), indicating that ROS may be produced by increasing myeloid cell numbers in blood. This is in line with previous preclinical reports15 showing that ROS production is critical for CRC initiation,16 at least in part through myeloid cell-driven oxidative DNA damage.17
An early CRC I-II stage-specific upregulation of cell cycle genes (AURKA, AURKB, EIF4G1, ECD and CDCA4) is accompanied by B-cell activation (MZB1 and IGLL5) (cluster D) but parallel downregulation of T-cell activation and proliferation genes (CD6 and RASGRF2) (cluster E). Genes involved in T-cell responses continue to be downregulated up to late CRC stages (III-IV) (BCL11B and EVL, in cluster C),18 19 while B-cell activation processes are no longer dysregulated at late stages. PBMCs from early CRC I-II stages are also characterized by a strong increase in myeloid cell migration and activation (CCR2, CD63, FCGR1A, S100A12 among others), whereby the presence of activated protumoral neutrophils (S100A8, S100A9, S100A12 among others) appears to be a hallmark of CRC progression (cluster B) as previously shown in preclinical models.20 Finally, wound healing processes, a key hallmark of cancer15 are detected in patients with late-stage CRC PBMCs along with blood coagulation activation (NRG1, MYL9, F13A1, MMRN1, SELP and THBD). The latter could result from tumor as well as neutrophil-derived cues,21 22 which in turn has been shown to promote CRC progression.23
Altogether, activation of innate immunity is detected in the periphery of individuals at pre-malignant CRC stages. This immune response is followed by a transient B-cell activation and sustained inhibition of T-cell responses along CRC progression, whereby at late stages, protumoral myeloid cells, wound healing and coagulation processes prevail.
We then compared the biological pathways found to be dysregulated along CRC progression by our analytical framework with the results obtained by digital cytometry with CIBERSORT.10 Lower CD8 T cells proportions in CRC compared with CON samples, obtained by digital cytometry (online supplemental figure S7F), are consistent with the downregulation of T-cell responses identified through our 226 BMKs (figure 4C). However, digital cytometry failed to capture the increase in monocyte frequencies in CRC compared with CON samples measured by clinical counts (online supplemental figure S7C), and indirectly by our BMKs. Moreover, cell deconvolution did not reliably capture neutrophil and B-cell activation from AA and CRCI-II stages, respectively (online supplemental figure S7F,G). Therefore, while digital cytometry results may be indicative, they exhibit limitations and alternative analytical approaches are required to capture subtle transcriptomic changes, such as specific cell activation.
Common RNA biomarkers are found in PBMCs and tumors of patients with CRC
Because identified PBMC biomarkers are involved in biological processes known to participate in tumor progression, we sought to investigate whether BMKs in the blood are also dysregulated in the colorectum during CRC progression. For that, we analyzed a publicly available RNA sequencing data set (GSE164541) including CRC, adenoma and normal adjacent tissue of five patients with CRC. Clustering of DEGs (p-adjusted <0.01 by DESeq2) between adenoma and normal tissue and CRC versus normal revealed six expression patterns (online supplemental table 5). By performing ORA of biological processes of each tumor DEG cluster, we found that, as in patients with CRC PBMCs, genes participating in T-cell activation and proliferation were downregulated in patients with CRC adenomas and tumors (figure 4D, clusters A and C showed no enrichment). Moreover, wound healing processes, which were detectable in the blood of patients with late-stage CRC, were upregulated in CRC tissues but not adenomas. Additional biological pathways dysregulated specifically in CRC included upregulation of invasion (extracellular matrix organization) and angiogenesis (blood vessel development) and further downregulation of adaptive immunity (interleukin-12 production) (figure 4D and online supplemental table 5), well-known hallmarks of cancer. Comparing the specific BMKs in PBMCs and tumors, we found 21/226 dysregulated genes in common, from which 11 showed the same pattern: AQP9, GPR84, DUSP10, S100A8, S100A9, MCEMP1 and CXCR1 were upregulated and NDRG2, EVL, TRAF3IP3 and CD8 downregulated (figure 4E and online supplemental table 5). While most commonly upregulated genes are involved in myeloid cell activation (GPR84, S100A8, S100A9 and CXCR1)24,26 downregulated genes participate in T-cell trafficking and activation (EVL, TRAF3IP3 and CD8),27 28 aligning with the well-described increase in protumoral myeloid cells and inhibition of T-cell responses along CRC carcinogenesis.29 To further evaluate the similarities between gene expression dysregulation in patients with CRC periphery and tumors, we assessed the gene expression of identified 226 CRC PBMC BMKs on six additional independent CRC tumor RNA sequencing data sets (including a total of seven data sets, 119 tumors and 132 normal tissues from three different countries). Our analysis revealed a significant correlation (p<0.05) between the logFC of identified BMKs in PBMCs of patients with CRC compared with CON subjects and tumors compared to normal tissue samples in 5/7 data sets (figure 4F). Among 226 BMKs, shared DEGs (p-adjusted <0.05 and the same dysregulation as in PBMCs) across all tumor data sets were not found. However, genes with the same dysregulation (up or downregulated regardless of adjusted p value) in PBMCs and tumors were shared across 6/7 data sets (online supplemental table 6). Given the heterogeneous molecular features of CRC tumors and their association with distinct clinical behaviors, we aimed to analyze the expression of identified BMKs across consensus molecular subtypes (CMS).30 DEA between CMS-classified tumors and normal tissue samples showed a stronger logFC correlation among 226BMKs between CMS4 tumors and PBMCs than other tumor CMSs. However, common DEGs between tumors and 226 PBMC BMKs were identified in all CMSs, with six of them being shared across all CMSs and analyzed data sets (online supplemental figure S8A,B and online supplemental table 7). Altogether, the strong immune responses triggered during CRC progression lead to systemic alterations detectable in PBMCs, wherein some genes dysregulated in tumors exhibit parallel dysregulation in the periphery.
Discussion
Tumor-driven systemic alterations in the immune system are well described in the literature in preclinical models and patients.9 However, whether those changes are strong enough to be translated into diagnostic tests is less clear. In the context of CRC, we have previously pioneered the development of a non-invasive test, Colox, based on quantification of PBMCs expression profile of 29 genes and quantification of two plasma proteins, Colox.8 This test is, so far, the most sensitive test for non-cancerous early CRC stages, with a specificity of 90% and a sensitivity of 79.5% and 55.4% for CRC and large adenomatous polyps, respectively. However, the targeted nature of its development may have hindered the identification of key immune modulations driven by CRC progression. Therefore, in this study, we performed whole transcriptome sequencing of PBMCs from patients with AA and CRC and compare it to control subjects PBMC transcriptome to reveal previously not captured CRC-driven systemic alterations.
We found that PBMCs, particularly myeloid cells, are highly sensitive to storage duration. Transcriptomic changes are observed much earlier than previously described at protein level13 when they are stored. By meticulously monitoring technical variables throughout sample processing and analysis, we were able to correct for this covariate without hindering our biomarker discovery. Nonetheless, since circadian rhythms are well-known regulators of blood immune composition,31 the effect of blood sampling time on PBMCs transcriptome should be assessed in future studies.
We identified and validated 226 CRC-driven blood RNA biomarkers which reveal a strong myeloid cell, platelet and wound healing process activation in patients with CRC’s periphery. Among myeloid cells, overall neutrophils and low-density neutrophils found in PBMC isolates32 33 appear to be major players in CRC progression as neutrophil activation and degranulation genes, such as S100A8 and S100A9, are strongly upregulated in PBMCs and tumors. This is in line with previous studies where high NLR were associated with worse CRC prognosis14 and increased neutrophil numbers found in CRC tumors compared with normal tissues.34 35 This neutrophilia, probably composed of granulocytic myeloid-derived suppressor cells (PMN-MDSCs),36 might be driven by systemic secretion of CCL15 and CCL9 by CRC tumor cells that results in CCR1+neutrophil recruitment. Indeed, mouse and human CRC tumor cells have been shown to secrete CCL15 and CCL9, and deletion of either cytokine decreases the aggressiveness of CRC in preclinical models.37 38 In addition, elevated CCL15 levels are observed in patients with CRC’s plasma compared with healthy individuals.38
CCR1, which is among validated PBMC BMKs together with CCR2, is also expressed by monocytic cells (monocytes and monocytic-MDSCs) and is important for their migration into tumors. These cells and derived macrophages are mainly protumoral in several tumor types including CRC34 39 40 and several myeloid-cell targeting agents are in clinical trials for the treatment of solid tumors.36
The observed PBMC contaminating platelet41 activation in patients with CRC may be linked to direct and indirect effects of CRC as both CRC tumor cells and neutrophils have been shown to induce platelet activation in preclinical models.21 23 The resulting activation of coagulation processes further contributes to inflammation, angiogenesis, and metastatic dissemination of tumor cells in mouse models.42 43 The link between coagulation processes and CRC tumorigenesis is further evidenced by the association of polymorphisms in clotting factors and increased risk of CRC.44 Therefore, the participation of identified BMKs in key known pathways of CRC pathophysiology strengthens their reliability and robustness as BMKs.
A more comprehensive expression profile analysis of identified BMKs along CRC progression further revealed stage-specific changes in systemic immunity. Activation of innate immunity is detected in the periphery of individuals at pre-malignant CRC stages followed by modulation of adaptive immunity at later stages. Precisely, MPO, a gene involved in ROS production in myeloid cells, is upregulated at pre-malignant CRC stages. Indeed, ROS production has been shown to be critical for CRC initiation in preclinical models.16 Specifically, ROS generated by myeloid cells contribute to tumorigenesis.17 However, to our knowledge, this is the first study detecting early alterations in myeloid-specific ROS production in the peripheral blood of patients with cancer.
We also discovered B-cell activation signals in PBMCs of patients with early CRC stages (upregulation of TNFRSF17, MZB1 and IGLL5). High infiltration of B cells in tumors is associated with a better prognosis in CRC,45 46 which is in line with the loss of B-cell activation signals observed in later CRC stages in this study. Finally, while some T-cell subsets can promote tumor progression,47 the many antitumoral functions of a variety of other T-cell subsets explain the described T-cell inhibition found in PBMCs along CRC progression and association between high tumor T-cell infiltrates and better CRC prognosis in other studies.48
One of the limitations of the study is the prioritization of BMKs from Swiss individuals (table 2). In addition to cohort composition differences among K and S samples (online supplemental figure S3G,I), genetic and lifestyle differences may explain differences among S and K subjects PBMCs transcriptome.49 50 Further studies including individuals from diverse countries and ethnicities will be required to identify universal BMKs. Furthermore, although common dysregulated genes were identified between the 226 CRC PBMC BMKs and all CMS CRC tumor types30 across independent data sets, paired PBMC and CMS-classified tumor transcriptomic data is necessary to determine whether identified peripheral BMKs are universally valid across all CMS tumor types or if they are specific to certain CMS subtypes.
Altogether, we discover dynamic systemic modulation of the immune system along CRC pathogenesis that results in detectable transcriptomic changes in PBMCs. These changes reveal immune responses at early CRC stages previously only described within tumor tissues, indicating the potential of peripheral blood transcriptome analysis to detect the early onset of the disease. The involvement of identified 226 BMKs in key pathways of CRC pathophysiology strengthens their reliability to be translated into a CRC diagnostic blood test. We foresee the development of such a test using transcriptome-stabilized blood samples collected in RNA stabilizing PAXgene Blood RNA tubes, which overcome the technical challenge of working with PBMCs, in the forthcoming future.
Translational relevance
Efforts to translate untargeted immune transcriptome analysis into a diagnostic test for early CRC detection.
supplementary material
Acknowledgements
We thank Erik Vandeputte (Biolizard) and Helen Lindsay (EPFL) bioinformatic support and Davide Croci and Simona Pavan for insightful comments on the manuscript.
Footnotes
Funding: Supported by the Innosuisse CTI grant n°27752.1.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: This study involves human participants and was approved by Cantons of Bern (No. KEK 139/10), St. Gallen (No. EKSG 110/091/1B), Basel (No. EKBB 242/10) and Vaud (No. VD /10), The Severance Hospital, Yonsei University College of Medicine (No. 4-2010-0128), Asan Medical Centre, University of Ulsan College of Medicine, Seoul (No. 2010-0221) and Institutional Bioethics Review Board of Seoul National University Hospital (No. H-1004-020-315). Participants gave informed consent to participate in the study before taking part.
Data availability free text: To protect human subject privacy and rights and preserve the scope of subjects’ consent, transcriptomic data will only be shared upon request.
Correction notice: This article has been corrected since it was first published online. Affiliation 4 has been updated to: Institute for Information and Communication Technologies (IICT), School of Engineering and Management Vaud, HES-SO, University of Applied Sciences and Arts Western, Yverdon-les-Bains, Switzerland.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1.Bishehsari F, Mahdavinia M, Vacca M, et al. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. WJG . 2014;20:6055. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bretthauer M, Løberg M, Wieszczy P, et al. Effect of Colonoscopy Screening on Risks of Colorectal Cancer and Related Death. N Engl J Med. 2022;387:1547–56. doi: 10.1056/NEJMoa2208375. [DOI] [PubMed] [Google Scholar]
- 3.Keum NN, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol . 2019;16:713–32. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 4.Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19:521–31. doi: 10.1038/s41575-022-00612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedland S, Watson D, Pan J, et al. Development and Clinical Validation of a Blood Test for Early Detection of Colorectal Adenomas and Cancer for Screening and Postpolypectomy Surveillance. Gastro Hep Adv . 2022;1:223–30. doi: 10.1016/j.gastha.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–91. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 7.Chung DC, Gray DM, 2nd, Singh H, et al. A Cell-free DNA Blood-Based Test for Colorectal Cancer Screening. N Engl J Med. 2024;390:973–83. doi: 10.1056/NEJMoa2304714. [DOI] [PubMed] [Google Scholar]
- 8.Ciarloni L, Ehrensberger SH, Imaizumi N, et al. Development and Clinical Validation of a Blood Test Based on 29-Gene Expression for Early Detection of Colorectal Cancer. Clin Cancer Res. 2016;22:4604–11. doi: 10.1158/1078-0432.CCR-15-2057. [DOI] [PubMed] [Google Scholar]
- 9.Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat Rev Cancer. 2021;21:345–59. doi: 10.1038/s41568-021-00347-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 13.Bonilauri B, Santos MDM, Camillo-Andrade AC, et al. The impact of blood-processing time on the proteome of human peripheral blood mononuclear cells. Biochim et Biophys Acta (BBA) - Proteins and Proteomics. 2021;1869:140581. doi: 10.1016/j.bbapap.2020.140581. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Zhao R, Cui Y, et al. The dynamic change of neutrophil to lymphocyte ratio can predict clinical outcome in stage I-III colon cancer. Sci Rep. 2018;8:1–8. doi: 10.1038/s41598-018-27896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 16.Myant KB, Cammareri P, McGhee EJ, et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12:761–73. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canli Ö, Nicolas AM, Gupta J, et al. Myeloid Cell-Derived Reactive Oxygen Species Induce Epithelial Mutagenesis. Cancer Cell. 2017;32:869–83. doi: 10.1016/j.ccell.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Waldman MM, Rahkola JT, Sigler AL, et al. Ena/VASP Protein-Mediated Actin Polymerization Contributes to Naïve CD8+ T Cell Activation and Expansion by Promoting T Cell-APC Interactions In Vivo. Front Immunol. 2022;13:856977. doi: 10.3389/fimmu.2022.856977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha VL, Luong A, Li F, et al. The T-ALL related gene BCL11B regulates the initial stages of human T-cell differentiation. Leukemia. 2017;31:2503–14. doi: 10.1038/leu.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackstadt R, van Hooff SR, Leach JD, et al. Epithelial NOTCH Signaling Rewires the Tumor Microenvironment of Colorectal Cancer to Drive Poor-Prognosis Subtypes and Metastasis. Cancer Cell. 2019;36:319–36. doi: 10.1016/j.ccell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guglietta S, Chiavelli A, Zagato E, et al. Coagulation induced by C3aR-dependent NETosis drives protumorigenic neutrophils during small intestinal tumorigenesis. Nat Commun. 2016;7:11037. doi: 10.1038/ncomms11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J-G, Geddings JE, Aleman MM, et al. Tumor-derived tissue factor activates coagulation and enhances thrombosis in a mouse xenograft model of human pancreatic cancer. Blood. 2012;119:5543–52. doi: 10.1182/blood-2012-01-402156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rees PA, Castle J, Clouston HW, et al. The effects of coagulation factors and their inhibitors on proliferation and migration in colorectal cancer. Cancer Med. 2023;12:17184–92. doi: 10.1002/cam4.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Li A, Tian Y, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytok Grow Factor Rev. 2016;31:61–71. doi: 10.1016/j.cytogfr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebhardt C, Németh J, Angel P, et al. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72:1622–31. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Forsman H, Dahlgren C, Mårtensson J, et al. Function and regulation of GPR84 in human neutrophils. Br J Pharmacol. 2024;181:1536–49. doi: 10.1111/bph.16066. [DOI] [PubMed] [Google Scholar]
- 27.Zou Q, Jin J, Xiao Y, et al. T cell development involves TRAF3IP3-mediated ERK signaling in the Golgi. J Exp Med. 2015;212:1323–36. doi: 10.1084/jem.20150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estin ML, Thompson SB, Traxinger B, et al. Ena/VASP proteins regulate activated T-cell trafficking by promoting diapedesis during transendothelial migration. Proc Natl Acad Sci U S A. 2017;114:E2901–10. doi: 10.1073/pnas.1701886114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021;21:653–67. doi: 10.1038/s41577-021-00534-x. [DOI] [PubMed] [Google Scholar]
- 30.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–8. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silvestre-Roig C, Fridlender ZG, Glogauer M, et al. Neutrophil Diversity in Health and Disease. Trends Immunol. 2019;40:565–83. doi: 10.1016/j.it.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardisty GR, Llanwarne F, Minns D, et al. High Purity Isolation of Low Density Neutrophils Casts Doubt on Their Exceptionality in Health and Disease. Front Immunol. 2021;12:625922. doi: 10.3389/fimmu.2021.625922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelka K, Hofree M, Chen JH, et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021;184:4734–52. doi: 10.1016/j.cell.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toor SM, Syed Khaja AS, El Salhat H, et al. Increased Levels of Circulating and Tumor-Infiltrating Granulocytic Myeloid Cells in Colorectal Cancer Patients. Front Immunol. 2016;7:560. doi: 10.3389/fimmu.2016.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grover A, Sanseviero E, Timosenko E, et al. Myeloid-Derived Suppressor Cells: A Propitious Road to Clinic. Cancer Discov. 2021;11:2693–706. doi: 10.1158/2159-8290.CD-21-0764. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura T, Fujishita T, Loetscher P, et al. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc Natl Acad Sci U S A. 2010;107:13063–8. doi: 10.1073/pnas.1002372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inamoto S, Itatani Y, Yamamoto T, et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Accumulation of Myeloid-Derived Suppressor Cells through the CCL15-CCR1 Chemokine Axis. Clin Cancer Res. 2016;22:492–501. doi: 10.1158/1078-0432.CCR-15-0726. [DOI] [PubMed] [Google Scholar]
- 39.Lee H-O, Hong Y, Etlioglu HE, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet. 2020;52:594–603. doi: 10.1038/s41588-020-0636-z. [DOI] [PubMed] [Google Scholar]
- 40.Pittet MJ, Michielin O, Migliorini D. Clinical relevance of tumour-associated macrophages. Nat Rev Clin Oncol. 2022;19:402–21. doi: 10.1038/s41571-022-00620-6. [DOI] [PubMed] [Google Scholar]
- 41.Rausser S, Trumpff C, McGill MA, et al. Mitochondrial phenotypes in purified human immune cell subtypes and cell mixtures. Elife. 2021;10:e70899. doi: 10.7554/eLife.70899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang HS, Swaim MW, Huang TF. Characterization of platelet aggregation induced by human colon adenocarcinoma cells and its inhibition by snake venom peptides, trigramin and rhodostomin. Br J Haematol. 1994;87:325–31. doi: 10.1111/j.1365-2141.1994.tb04917.x. [DOI] [PubMed] [Google Scholar]
- 43.Leslie M. Cell biology. Beyond clotting: the powers of platelets. Science. 2010;328:562–4. doi: 10.1126/science.328.5978.562. [DOI] [PubMed] [Google Scholar]
- 44.Vossen CY, Hoffmeister M, Chang-Claude JC, et al. Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol. 2011;29:1722–7. doi: 10.1200/JCO.2010.31.8873. [DOI] [PubMed] [Google Scholar]
- 45.Berntsson J, Nodin B, Eberhard J, et al. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int J Cancer. 2016;139:1129–39. doi: 10.1002/ijc.30138. [DOI] [PubMed] [Google Scholar]
- 46.Meshcheryakova A, Tamandl D, Bajna E, et al. B cells and ectopic follicular structures: novel players in anti-tumor programming with prognostic power for patients with metastatic colorectal cancer. PLoS One. 2014;9:e99008. doi: 10.1371/journal.pone.0099008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito T, Nishikawa H, Wada H, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–84. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 48.Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 49.Tabassum R, Nath A, Preininger M, et al. Geographical, environmental and pathophysiological influences on the human blood transcriptome. Curr Genet Med Rep. 2013;1:203–11. doi: 10.1007/s40142-013-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soldati L, Di Renzo L, Jirillo E, et al. The influence of diet on anti-cancer immune responsiveness. J Transl Med. 2018;16:75. doi: 10.1186/s12967-018-1448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.