Abstract
Low abundance mRNAs are more difficult to examine using microarrays than high abundance mRNAs due to the effect of concentration on hybridization kinetics and signal-to-noise ratios. This report describes the use of low complexity representations (LCRs) of mRNA as the targets for cDNA microarrays. Individual sequences in LCRs are more highly represented than in the mRNA populations from which they are derived, leading to favorable hybridization kinetics. LCR targets permit the measurement of abundance changes that are difficult to measure using oligo(dT) priming for target synthesis. An oligo(dT)-primed target and three LCRs detect twice as many differentially regulated genes as could be detected by the oligo(dT)-primed target alone, in an experiment in which serum-starved fibroblasts responded to the reintroduction of serum. Thus, this target preparation strategy considerably increases the sensitivity of cDNA microarrays.
INTRODUCTION
cDNA and oligonucleotide microarrays are convenient for identifying changes in mRNA abundances (1–3). The proportion of transcripts that can be detected and measured and the accuracy of measurements of changes in transcript abundance determine the kinds of problems that can be addressed using microarrays. The abundance of a transcript can fall well below one copy per cell, on average, such as in cases where a transcript is rare but biologically active, in cases where a message has a brief transcription window, such as during cell cycle, or in complex clinical samples where cells with high expression are mixed with cells with low expression or no expression. However, sensitivities in the range of one or a few transcripts per mammalian cell are difficult to achieve routinely, and experimental noise at the lower limits of sensitivity complicates the quantitative assessment of changes in gene expression. While the measurement of changes in relatively abundant transcripts is appropriate for certain goals, such as in the classification of cancer types (4–9), greater sensitivity and accuracy is often desirable, if not necessary, such as in surveys for changes in individual transcript abundances that are important in diseases, or when analysis is hampered by missing data (10).
This report describes a strategy for improving microarray performance by using subsets, or low complexity representations (LCRs), of the transcriptome as microarray targets. There are several methods for producing LCRs (11–14). Here, we use arbitrarily primed PCR applied to oligo(dT)-primed first strand cDNA to generate LCRs. In contrast to a random primer, most of the positions in an arbitrary primer are specified, but its sequence need not be chosen on the basis of homology, as would be the case with a specific PCR primer. Arbitrarily primed PCR amplifies the sequences between sites in a DNA template where an arbitrary primer or a pair of arbitrary primers find approximate matches on opposite strands in close proximity. The complex class of transcripts participates in this reaction more often than the less complex class of abundant transcripts due to these requirements. As a result, arbitrary sets of rare transcripts become highly represented in the reaction product. The sequence of the arbitrary primers, the characteristics of arbitrary priming sites, their distance from one another and the characteristics of the sequences that they flank determine the sequences that are amplified and the extent of their amplification. Different primers result in the amplification of different subsets of the original mRNA sequence space, including different transcripts and different parts of mRNA isoforms. Sequences amplify reproducibly such that, when two different mRNA populations are compared, differences in expression can be detected (15). Lower complexity, over-representation of sequences from the class of rare transcripts, and differential selection of isoforms and family members suggested that LCRs may be useful for measuring changes in the abundances of rare transcripts that are difficult to measure accurately using cDNA microarrays. In previous work, LCRs made using arbitrary priming methods (11,13) allowed the measurement of abundance changes in transcripts that were difficult to detect using oligo(dT)-primed reverse-transcribed targets applied to nylon membrane cDNA arrays (16,17). Here, this approach is adapted to glass slide microarrays. Individual LCRs can detect one-third to one-half of all transcripts, and three different LCRs used in combination with an oligo(dT)-primed target can detect 80% of all genes represented on a cDNA microarray. The number of differentially regulated genes that can be detected and measured using three LCRs together with oligo(dT)-primed targets is ∼2-fold higher than can be detected and measured using oligo(dT)-primed targets alone.
MATERIALS AND METHODS
Cell lines and RNA preparation
Human fibroblast from ATCC (CRL 2091) were grown to ∼80% confluence in 150 cm dishes in DMEM with 10% fetal bovine serum (heat inactivated at 56°C for 30 min, Omega scientific), and with 200 U/ml penicillin and 200 μg/ml streptomycin. For serum starvation, cells were grown in media containing 0.01% serum for 48 h as described previously (3), and then were treated with 10% serum for 0 (i.e. no serum), 1 and 4 h. Cells were washed with ice-cold phosphate-buffered saline, and total RNA was prepared using an RNeasy Mini Kit (Qiagen, Valencia, CA). RNA concentration was determined spectroscopically, and integrity was assessed qualitatively by agarose gel electrophoresis.
LCR preparation
LCRs were prepared using RNA arbitrarily primed PCR (11,16,17). Reverse transcription was performed on 5 μg total RNA using an oligo(dT)20-VN primer (Genosys Biotechnologies, The Woodlands, TX). The reactions contained 50 mM Tris–HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 20 mM DTT, 0.2 mM each dNTP, 0.5 μM primer and 20 U M-MuLV reverse transcriptase (Promega, Madison, WI), in a final volume of 200 μl. Reverse transcription was performed at room temperature for 15 min, followed by 37°C for 1 h. Reactions were stopped by heating for 5 min in boiling water and cooling on ice. cDNA was diluted to 3 ng/μl with distilled water prior to the PCRs.
Primers synthesized by Genosys Biotechnologies used to generate LCRs were (in 5′–3′ orientation) pm13 (CAGTGGGAG + AGTGAGCAC), pm14 (ACGAAGAAG + AGGGCACCAC), pm19 (RRRGACAGTG), pm20 (RRRCTGCGCT), pm21 (CAGAGGTRRR), pm22 (AACGGCGRRR), pm23 (AACGGCGACR), pm24 (GGGTGTGTAR), pm25 (GGTGAACGRR), pm28 (RTCCCCGCGA), pm29 (RRATGCCACT), pm30 (RRTTCGGAAG), pm31 (TCCGATGCTG), pm32 (TGACGTCCGATGCTG), pm33 (GTGACAGACA), pm34 (AACTGGTGACAGACA), pm35 (TGCGAAGGGGCACCA), pm36 (AACTGGAACTAGGGGCACCA), pm37 (AGGGGCACCA) and pm38 (TGCGAAGGGGCACCA). Diluted cDNA (25 μl) was mixed with an equal volume of 2× PCR mixture containing 20 mM Tris–HCl, pH 8.3, 20 mM KCl, 6 mM MgCl2, 0.35 mM each dNTP, 2 μM each primer, 2 μCi [α-32P]dCTP (ICN, Irvine, CA) and 0.5 U/μl AmpliTaq DNA polymerase Stoffel fragment (Applied Biosystems, Foster City, CA). Thermocycling used 3 min at 94°C followed by 35 cycles of 94°C for 1 min, 35°C for 1 min and 72°C for 2 min. Product was purified using a QIAquick PCR Purification Kit (Qiagen) and examined for repeatability on sequencing-style polyacrylamide gels. For fluorescent dye labeling, 1 μg of RNA arbitrarily primed PCR product was mixed with 12 μg of random hexamer and boiled for 5 min at 95°C. Reactions were performed at 37°C overnight in 50 μl of buffer containing 10 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 7.5 mM DTT, 0.025 mM dATP, dCTP, dGTP and 0.009 mM dTTP, 0.014 mM Cy3- or Cy5-linked dUTP (Amersham, Arlington Heights, IL) and 10 U of exonuclease-free Klenow (New England Biolabs Inc., Beverly, MA). The targets were purified with QIAquick PCR Purification Kit (Qiagen). Labeling was checked by spectrophotometry at 550 and 650 nm for Cy3 and Cy5, respectively.
Microarray preparation
Human cDNA clones (I.M.A.G.E) (Research Genetics/Invitrogen) were grown overnight in 96 well plates, inserts were amplified using vector-specific primers and purified using multiscreen filter plates (Millipore, Billerica, MA). Each amplified cDNA was combined 1:1 with dimethyl sulfoxide for arraying as described previously (18) and printed onto Ultra-GAPS coated glass microscope slides (Corning Inc. LifeSciences, Acton, MA) using an OmniGrid 100 printer (Genomic Solutions Ann Arbor, MI) at 40–60% relative humidity. Printed slides were UV cross-linked (250 mJ), baked for 3 h at 80°C and stored at room temperature in a desiccator.
Hybridization and washes
After incorporation of fluorescent nucleotides, the LCR and oligo(dT)-primed targets were lyophilized and brought to a final volume of 45 μl in 25% formamide, 5× SSC, 0.1% SDS and blocking agent [poly(A)15, yeast tRNA and COT-1 DNA]. The target was heated for 5 min at 95°C, centrifuged briefly, and immediately applied to the slide in a hybridization chamber. The chambers were submerged in a 42°C water bath overnight. The slides were washed for 30 min at 42°C in a 2× SSC, 0.1% SDS solution and two times for 30 min at room temperature in 0.1× SSC, 0.1% SDS and in 0.1× SSC solutions, sequentially. Fluorescence intensities were measured using a ScanArray5000 (Hewlett Packard) laser scanner.
Real-time RT–PCR
Real-time PCRs were performed in a solution containing SybrGreen I, 0.35 mM 6-ROX (Molecular Probes, Eugene OR), 0.2 mM dNTP, 1× PCR buffer (Qiagen), 4 mM MgCl2, 5 mM each primer and 0.025 U of HotStartTaq DNA polymerase (Qiagen), using an ABI PRISM 7900HT Sequence Detector. Thermocycling was performed with an initial 10 min incubation at 95°C followed by 50 cycles of 95°C for 15 s, 60°C for 1 min and 72°C for 30 s. This cycling reaction was followed with 2 min at 95°C, 15 s at 60°C and 15 s at 95°C. A standard curve for each gene was prepared with a four point dilution series. Each measurement was made in duplicate. Measurements were normalized to an internal glyceraldehyde-3-phosphate dehydrogenase RNA. Oligonucleotide primers used are described in the Supplementary Table 1.
RESULTS
LCRs select and amplify subsets of mRNA sequences
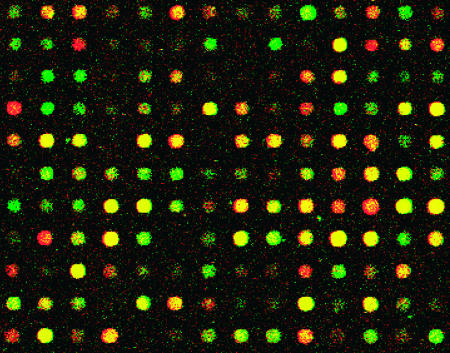
With nylon membrane arrays, it had been shown that LCRs select and amplify subsets of the mRNA represented in an oligo(dT)-primed target (16,17). To demonstrate this effect using microarrays, total RNA was isolated from growing human fibroblasts, followed by target synthesis using anchored oligo(dT) priming or RNA arbitrarily primed PCR to make an LCR. These were compared by hybridization to a glass slide cDNA microarray containing several thousand human gene sequences. The two target preparation methods resulted in different sequence-specific signal intensities (Figure 1). In many cases, the LCR-derived signal exceeded the oligo(dT)-target-derived signal and vice versa. This suggested that LCRs could be used to enhance signals for some transcripts on microarrays, consistent with previous studies using LCRs as targets for cDNA arrays printed on nylon membranes (16,17).
Figure 1.
LCRs enhance subsets of mRNA sequences. Comparison between oligo(dT) and an LCR target on a microarray. The LCM target (red) displays different sequence representation than the oligo(dT) target (green), which is seen as different ratios of red and green false coloring of fluorescence signal intensity.
LCRs can detect differential gene regulation
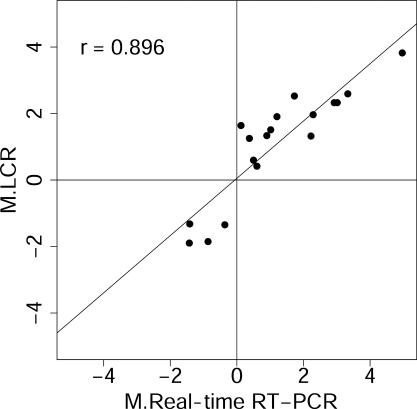
The differential amplification of sequences in an LCR, when compared with an oligo(dT)-primed target or with different LCRs, suggested that LCRs could be used to detect differential gene expression in cases where the signal from an oligo(dT)-primed target is too weak to be useful. However, first, it was necessary to demonstrate that LCRs could, indeed, be used as targets for microarrays to measure differential gene expression. Transcripts having altered abundances 4 h after the addition of serum to serum-starved fibroblasts were measured by microarray analysis of an LCR target (LCR pm22), and these measurements were compared with real-time RT–PCR measurements. Each microarray contained three identical subarrays of 4621 spotted probes, 3770 of which were human gene sequences, and the rest of which were controls of various sorts, including 96 Salmonella sequences that served as negative controls for non-specific signals. Analyses were performed using the limma package in BioConductor and the R programming environment (19–21). Fluorescence intensity measurements from human and control gene sequences were adjusted using background subtraction, print-tip loess normalization and scaling between reciprocal dye-swap pairs of chips. MA plots were constructed [M = log2R − log2G; A = (log2R + log2G)/2] on the background subtracted, normalized and scaled channel intensities for visual inspection of the data (Supplementary Table 2 and Supplementary Figure 1a). Real-time RT–PCR was performed for 17 transcripts that showed an apparent change in abundance in the microarray experiment, and had a modified t-statistic with P ≤ 0.05 (21). Primers for real-time RT–PCR spanned splice junctions to avoid amplification from unspliced mRNA or from possible genomic contamination. Agreement between the two quantitation methods was observed, with Pearson's correlation r = 0.90 (Figure 2). This indicated that LCRs can be used as targets for microarrays to discover mRNA abundance changes. Data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) (GEO GSE2655).
Figure 2.
LCR targets can detect differential gene regulation. LCR pm22 target was prepared from RNA from serum-starved and starved-refed fibroblasts 4 h after refeeding, and analyzed using microarrays. Differentially regulated genes detected by the LCR were quantified using real-time RT–PCR. This figure shows that the log2 ratios (M) of transcripts from the two methods correlate well and confirms that LCRs reliably report differential transcript abundance. Axis label ‘M.LCR’ is the average log2 ratio of the normalized channel intensities, I, for the 4 and 0 h treatments, i.e. M.LCR = log2(It=4/It=0), reported by the LCR target, and ‘M.Real-time RT–PCR’ is the log2 of the corresponding ratio reported by real-time RT–PCR. Pearson's correlation, r, is shown.
LCRs detect differential gene regulation not detected by oligo(dT)-primed targets
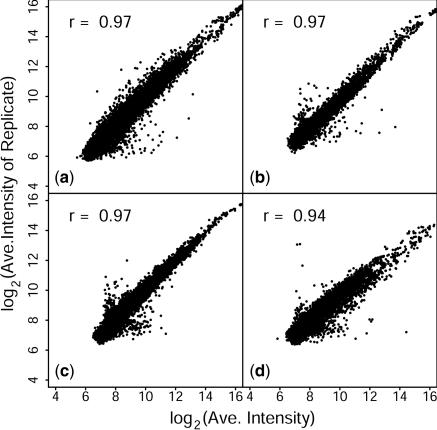
The enrichment of certain sequences in LCRs and the depletion of others suggested that LCRs might be used to detect differential gene regulation for genes that are normally difficult to study using only an oligo(dT)-primed target, due to their low representation in the mRNA population. Total RNA was harvested from serum-starved fibroblasts before and 1 h after the reintroduction of serum. Two replicate biological experiments and the corresponding reciprocal dye-swap experiments were performed for each probe type. Every scanner ‘channel’ corresponded to an independently synthesized LCR, such that four microarrays represent results from eight independent LCRs. This design was chosen so that variance in LCR preparation could be explored, but in usual practice, the dye-swap replicates would comprise technical reciprocal labeling, as is commonly done. LCRs were made using the arbitrary sequence oligonucleotide primers pm19, pm22 and pm28. Oligo(dT)-primed and LCR targets were labeled with Cye dyes and hybridized to microarrays to detect genes that were differentially regulated between the two biological conditions. LCRs made using RAP–PCR are reproducible, as shown in Figure 3. Intensities for each gene were determined from each microarray, with print-tip loess normalization and scaling between chips. Two intensity vectors were generated by averaging, gene-by-gene, one array from each biological replicate and its reciprocal from the other biological replicate, and the log2 of the resulting averages were plotted as scatter plots. Figure 3 shows that correlations of r ≥ 0.94 were achieved for all four target types. Similar analysis of any of the three subarrays gave correlations r ≥ 0.95 for all target types. Analysis of single biological replicates (i.e. from a single pair of chips) gave correlations of r ≥ 0.95 for all but pm28, which gave r = 0.93 and r = 0.91 for the two biological replicates, respectively. Background subtraction increased scatter due to variance in the background, itself, particularly for lower signals, but had only a minor effect on the overall correlation (r ≥ 0.93) (data not shown). If RAP–PCR amplified sequences only according to the occurrence of a partial match between the arbitrary primer and its target, and no other efficiency terms were involved, one may expect that subsets of mRNA sequences would be sampled, but that their representation in the final product would remain unchanged relative to their representation in an oligo(dT)-primed target. This is not the case, however. Scatter plots between different LCRs are not concentrated along the diagonal, and correlations are below r = 0.65 for all pairwise comparisons between LCRs, and between LCRs and the oligo(dT)-primed targets (Supplementary Figure 2). Combined, these observations indicate that the large differences between the oligo(dT)-primed targets and the LCR targets result from reproducible differences in the sequence representations contained within the LCR targets, and not from variance intrinsic to the LCRs. Thus, LCR synthesis can be simple and robust, and can detect differential gene regulation when hybridized to microarrays.
Figure 3.
Reproducibility. Scatter plots of average intensities from technical and biological replicates. Each average was calculated from loess adjusted, normalized intensities from dye-swap replicate microarrays, one from each biological replicate. (a) oligo(dT) target, (b) LCR pm19, (c) LCR pm22, (d) LCR pm28.
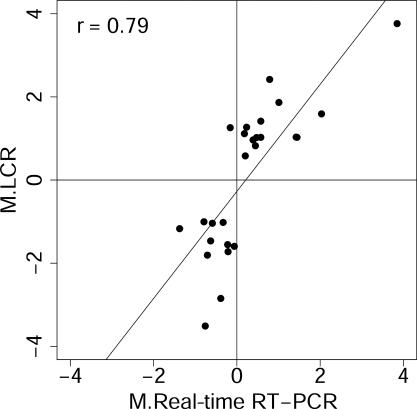
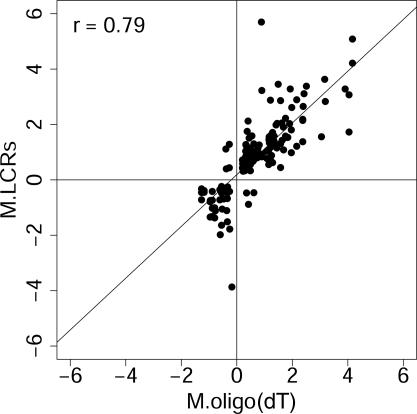
LCR targets can detect changes in transcript levels that are missed by oligo(dT)-primed targets. The modified t-statistics and associated P-values calculated for the four target types, using limma, BioConductor and R (19–21) as described above (Supplementary Table 2), were used to assess differential gene regulation in response to introduction of serum to serum-starved fibroblasts (see Table 1). Using P ≤ 0.05 as a threshold, the oligo(dT) target detected changes in 325 transcripts out of the 3770 represented on the chip, and 213 of these were unique to the oligo(dT) target, while the remaining 112 were also detected as changes by one or more of the LCR targets. The three LCRs, combined, detected changes in 416 transcripts, 304 of which were missed by the oligo(dT) target. LCRs from pm19, pm22 and pm28 contributed 123, 149 and 41 of these 304 changes, respectively, with some overlap. Figure 4 shows the correlation between those mRNA abundance changes detected only by the LCRs and the same changes measured by quantitative RT–PCR. High Pearson's correlation (r = 0.79) indicates that LCRs are able to detect differential gene expression that is largely invisible to oligo(dT)-derived targets. When the changes in these transcripts measured using oligo(dT)-derived targets were compared with the real-time RT–PCR measurements, correlation was lower (r = 0.55), as would be expected from their higher P-values. Individual gene results, accession numbers and descriptions are available in Supplementary Tables 3 and 4.
Table 1.
Detection of differential regulation by oligo(dT) and LCR targetsa
| P-value threshold | 0.05 |
| All changes detected | 629 |
| Detected with more than one target | 121 |
| Detected by oligo(dT) target and possibly by LCR targets | 325 |
| Detected only by oligo(dT) target | 213 |
| Detected by LCR targets and possibly by oligo(dT) target | 416 |
| Detected by LCR targets only | 304 |
| Detected by oligo(dT) target and one or more LCR targets | 112 |
aDifferential gene expression detected by an oligo(dT) target and three LCR targets, comparing serum-starved fibroblasts before and 1 h after reintroduction of serum.
Figure 4.
LCR targets detect differential expression that is missed by oligo(dT)-primed targets. Differential gene expression detected by LCRs but not by oligo(dT) targets are confirmed by quantitative RT–PCR. This result indicates that LCRs can be used to detect changes in gene expression that cannot be detected using an oligo(dT) target on these microarrays. Limma output for these genes is in Supplementary Table 3, while accession numbers, current Unigene designations, and descriptions are in Supplementary Table 4. Axis label ‘M.LCRs’ is the average log2 ratio of the normalized channel intensities, I, for the 1 and 0 h treatments reported by the LCR targets, i.e. M.LCRs = log2(It=1/It=0), and ‘M.Real-time RT–PCR’ is the log2 of the corresponding ratio reported by real-time RT–PCR. Pearson's correlation, r, is shown.
Further confirmation that LCRs reliably report differential gene regulation can be seen in the comparison of changes in expression reported by LCR targets with those reported by oligo(dT) targets for those cases where both target types reported changes. Figure 5 shows the correlation between M-values calculated for these transcripts. Recall that M is the log2 of the ratio of normalized channel intensities, such that M = 1 corresponds to a 2-fold change, and so forth. A correlation of r = 0.79 was obtained when both measurements had P ≤ 0.05, and using lower P-values tended to make the correlation better (e.g. r = 0.84 for both measurements having P ≤ 0.02; data not shown). In this experiment, up-regulated genes outnumber down-regulated genes, which might be expected, given that down-regulation must be accompanied by mRNA decay before it can be detected by microarray hybridization. This result indicated that LCRs are able to detect many of the same differentially regulated genes that can be detected by an oligo(dT)-primed target and agrees with the findings reported in Figures 2 and 4, where real-time RT–PCR was used to confirm that changes in gene expression can be detected using LCRs.
Figure 5.
LCR targets and oligo(dT)-primed targets report similar changes where overlap occurs. LCR targets and oligo(dT) targets report similar log ratios of differential expression, in those cases where both methods detect a change. This graph shows differential gene expression discovered using microarrays and oligo(dT) priming for target synthesis, compared with the corresponding measurement from the LCRs. Only those genes detected as changed with P ≤ 0.05 in an LCR are included. Axis label ‘M.LCRs’ is the average log2 ratio of the normalized channel intensities, I, for the 1 and 0 h treatments, i.e. M.LCRs = log2(It=1/It=0), and ‘M.oligo dT’ is the log2 of the ratio for the same gene reported by the oligo(dT) targets. Pearson's correlation, r, is shown.
Fraction of genes for which LCRs and oligo(dT) targets were able to detect differential expression
A point of interest is the number of array probes that had intensities large enough relative to background that a change, had it occurred, would have been observed. The data used were that described above for fibroblast serum starvation and refeeding, involving targets from oligo(dT), pm19, pm22 and pm28. For a randomly chosen set of 300 genes, the background was subtracted from the intensity values, followed by division of the signals from one channel (i.e. the 1 h time point) by small factors to artificially reduce the mean intensity value from that channel, mimicking down-regulation. Since variance may not scale with the mean, other probe intensities were searched to find the one with a mean channel intensity closest to the artificial values, and the channel intensities from these were substituted in for each of the 300 randomly chosen genes. The other channel (i.e. the 0 h time point) was left unchanged. Those differentially regulated transcripts that were normally detected without the artificial change were excluded. Modified t-tests were performed using limma after these artificial changes, as described earlier, and P ≤ 0.05 was used as the criterion for the detection of a change; the results are shown in Table 2. The columns labeled ‘4-fold’ and ‘2-fold’ show the number and percent of the 300 genes that were detected as changed at P ≤ 0.05. This experiment was performed several times with different random sets of 300 and gave similar results (data not shown). One limitation of this procedure is that background determined from the area of the chip surrounding a spot does not necessarily reflect the variance within the spot due to other nuisance factors, such as cross-hybridization. However, 81% of the genes had A-values (i.e. average of the log2 of intensities) exceeding the largest A-value from among all Salmonella controls for at least one of the four target types, indicating that signal intensity due to foreground nuisance factors other than cross-hybridization of related sequences, such as family members, was generally low. With these caveats, 43% of the probes on the array were sufficiently represented in the dT target that a 2-fold decrease would have been detected had it occurred, and 55% would have been detected after a 4-fold decrease, using the modified t-statistic and P ≤ 0.05. The best LCR may be able to detect as many as 31% of 2-fold decreases and 64% of 4-fold decreases. Overall, the combined use of oligo(dT) targets and the three LCRs may be able to detect as many as 55% of 2-fold decreases and 85% of 4-fold decreases. Transcripts that might be detected after a hypothetical increase cannot be addressed in this manner, and it remains unknown how many sequences are actually represented, but are too low to be detected without an increase due to induction or transcript stabilization.
Table 2.
Simulated down-regulation for 300 random genesa
| 4-fold | 2-fold | |
|---|---|---|
| Oligo(dT) | 166 (55%) | 130 (43%) |
| pm19 | 191 (64%) | 49 (16%) |
| pm22 | 119 (40%) | 92 (31%) |
| pm28 | 71 (24%) | 27 (9%) |
| Combined | 256 (85%) | 165 (55%) |
aSignal intensities for 300 randomly chosen genes from refed fibroblasts were divided by 2 and 4. These channel intensity values were then replaced with the channel intensities from the probe with a mean nearest these divided means, thereby mimicking 2- and 4-fold down-regulation, and including the appropriate variance. This table contains the number of genes that were scored as differentially regulated by limma with P ≤ 0.05 (% of 300 is indicated in parentheses).
The number of genes whose transcripts can be detected using multiple LCRs
We determined the fraction of genes for which transcripts could be detected by one or more LCRs from a set of 20 different LCRs, relative to a collection of Salmonella negative hybridization control sequences. We assumed that these controls would provide a good measurement of the distribution of nuisance signal intensities, and that signal intensities exceeding 95% of the negative control signals represent bona fide hybridization. The microarrays used were essentially as described above, except that those used later in the screen had a greater number of genes, which were excluded from further analysis. Each microarray was hybridized with two distinct LCR targets, and each hybridization was performed in a single pair of dye-swap replicates. Normalization by total channel intensity was performed, and data from the same LCR from the two technical replicates were then compared, without background subtraction. Supplementary Figure 3 displays scatter plots of this data and shows reproducibility in the replicate hybridizations. The density distributions of signal intensities from the 96 negative controls in each of three subarrays per chip were estimated using logsplines (22), and 2327 out of 3010 (77%) human sequence probes had signals exceeding 95% of these controls in both replicates, for at least one of the 20 LCRs tested. If data from an oligo(dT) target were included, coverage increased to 2579 out of 3010 (86%) with oligo(dT) contributing an additional 9%. Detection by individual primers ranged between 55 and 5%, with most transcripts being detected by multiple LCRs. The apparent limit of ∼86% was reached approximately asymptotically. Very similar results were obtained using a Wilcoxon rank sum test. We performed RT–PCR experiments on mRNA sequences spanning splice junctions for 20 of the 14% of genes that remained undetected by any target. Fourteen out of these twenty gave PCR products of the predicted size. This suggests that neither LCRs nor oligo(dT)-primed probes are able to detect the rarest mRNAs using only a single pair of microarrays.
In this large survey, three LCRs made using the primers pm19, pm22 and pm28 detected 55% of the probes with signal intensities >95% of the negative controls, and the oligo(dT) target detected 65%, but the combined coverage using the three LCRs plus the oligo(dT) target was 80%. Individually, LCRs from primers pm19, pm22 and pm28 detected transcripts for 50, 40 and 39% of the genes represented on the array, respectively.
Enhanced signal explains some but not all of the enhanced detection of change by LCRs
LCRs have lower complexity than the mRNA population from which they are derived because some sequences amplify more efficiently than others during the PCR step, depending on how well the arbitrary primers match, the length of the sequence between the arbitrary priming sites, and other sequence-specific factors. Consequently, some sequences are more highly represented in the LCR than in the original mRNA population. In addition, the complexity of LCRs is much lower than the complexity of the original mRNA, because, on average, only a subsequence about one-sixth of the length of each mRNA is amplified. These two factors probably lead to better signal-to-noise behavior for these sequences in microarray experiments when compared with the more complex oligo(dT)-primed targets. However, LCRs detected changes in some transcripts that were not detected as changes by the oligo(dT)-primed targets even though the signal intensities from the oligo(dT)-primed targets were higher. When signal intensities for proven differentially regulated genes detected by LCRs with confidence P ≤ 0.05 (Figure 4) were compared with the corresponding intensities from oligo(dT) targets, with which differential regulation was not detected, 17 probes had higher intensities from LCR targets, while 12 had higher intensities from oligo(dT) targets (Supplementary Figure 4). For these 12 genes, relative abundance of the sequence in the target cannot alone account for the fact that LCRs out-performed the oligo(dT)-primed targets. The different ways in which oligo(dT) priming and arbitrary priming sample mRNA isoforms may explain some of this aspect of the enhanced performance of LCR targets, but we have not confirmed this possibility. Discussion sequences from rare mRNAs can be highly represented in LCRs, leading to higher signal intensities in microarray experiments. The experiments presented above show that LCRs detected differential gene expression that was not detected in parallel experiments with oligo(dT)-primed targets on PCR product cDNA microarrays. Three LCRs plus oligo(dT)-primed targets increased the detection of differentially regulated genes by 2-fold relative to oligo(dT)-primed targets alone. This is surprising because the number of genes for which transcripts are detected increases, but only by a factor of ∼1.5. A possible explanation is that the relatively lower complexity of LCRs simply reduces foreground nuisance fluorescence sufficiently that differential regulation is uncovered. Alternatively, recent estimates suggest that about half of human genes produce alternatively spliced products, with an average of 2.5–3.5 different mRNA splicing isoforms per gene (23,24), and oligo(dT) targets lead to a microarray signal that is a weighted average of all of the poly(A)-tailed isoforms that share the exon sequences represented in a probe. Arbitrary priming samples different mRNA isoforms with different efficiencies, depending on where the arbitrary primers find sufficient homology, such that different LCRs can contain sequences from one isoform and not another. If one isoform of an mRNA is regulated differently than its other isoforms, the difference is less likely to be obscured by the weighted average signal from all of the other isoforms from the gene. However, we do not have a quantitative estimate of the extent of this effect on LCR performance.
These experiments used microarrays constructed with cDNA sequences having average lengths of ∼1000, 2.4-fold shorter than the average human transcript (25,26), and the products in these LCRs have a median length of ∼400 nt. Thus, about half of the amplified products for a typical mRNA in an LCR contain sequences that are not represented in the clone from which the corresponding array probe was prepared. This suggests that microarrays could be tailored to match the sequences represented in the LCRs, thereby further improving the performance by a factor of ∼2. Microarrays with genes represented in specific LCRs could be printed on separate arrays or isolated in separate hybridization chambers on the same slide to improve throughput and efficiency. The use of oligonucleotide arrays may reduce, and in some cases eliminate, ambiguities that are certain to arise from LCRs due to interference between mRNA isoforms and close gene family members. Thus, the use of LCRs opens up several avenues for increasing the sensitivity of cDNA microarrays for identifying differential gene regulation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Mr Sidney Kimmel, Ms Eileen Haag and Mr Ira Lechner for their generous support. This work was funded by grants to J.W. (R33 CA091358) from the National Institutes of Health and to M.M. (R01 CA68822-13 and DAMD17-03-1-0022) from the National Institutes of Health and the US Department of Defense. G.R. was partially supported by a fellowship from Association pour la Recherche contre le Cancer (ARC). M.J. was partially supported by a fellowship from Deutsche Gesellschaft fuer Naturforscher Leopoldina (BMBF-LPD 9901/8-62). Funding to pay the Open Access publication charges for this article was provided by a gift from Mr Sidney Kimmel.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Brown E.L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 3.Iyer V.R., Eisen M.B., Ross D.T., Schuler G., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr, Boguski M.S., et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 4.Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee A., Richards W.G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramaswamy S., Tamayo P., Rifkin R., Mukherjee S., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl Acad. Sci. USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., Korsmeyer S.J. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nature Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 8.Shipp M.A., Ross K.N., Tamayo P., Weng A.P., Kutok J.L., Aguiar R.C., Gaasenbeek M., Angelo M., Reich M., Pinkus G.S., et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nature Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 9.Ramaswamy S., Ross K.N., Lander E.S., Golub T.R. A molecular signature of metastasis in primary solid tumors. Nature Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 10.Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 11.Welsh J., Chada K., Dalal S.S., Cheng R., Ralph D., McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClelland M., Ralph D., Cheng R., Welsh J. Interactions among regulators of RNA abundance characterized using RNA fingerprinting by arbitrarily primed PCR. Nucleic Acids Res. 1994;22:4419–4431. doi: 10.1093/nar/22.21.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang P., Pardee A.B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 14.Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ralph D., McClelland M., Welsh J. RNA fingerprinting using arbitrarily primed PCR identifies differentially regulated RNAs in mink lung (Mv1Lu) cells growth arrested by transforming growth factor beta 1. Proc. Natl Acad. Sci. USA. 1993;90:10710–10714. doi: 10.1073/pnas.90.22.10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trenkle T., Welsh J., Jung B., Mathieu-Daude F., McClelland M. Non-stoichiometric reduced complexity probes for cDNA arrays. Nucleic Acids Res. 1998;26:3883–3891. doi: 10.1093/nar/26.17.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trenkle T., Welsh J., McClelland M. Differential display probes for cDNA arrays. Biotechniques. 1999;27:554–560. doi: 10.2144/99273rr03. 562, 564. [DOI] [PubMed] [Google Scholar]
- 18.Hegde P., Qi R., Abernathy K., Gay C., Dharap S., Gaspard R., Hughes J.E., Snesrud E., Lee N., Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–550. doi: 10.2144/00293bi01. 552–544, 556 passim. [DOI] [PubMed] [Google Scholar]
- 19.Smyth G.K., Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 20.Smyth G.K., Yang Y.H., Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol. Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 21.Smyth G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 22.Kooperberg C., Stone C.J. A Study of Logspline Density Estimation. Comput. Stat. Data Anal. 1991;12:327–347. [Google Scholar]
- 23.Kim H., Klein R., Majewski J., Ott J. Estimating rates of alternative splicing in mammals and invertebrates. Nature Genet. 2004;36:915–916. doi: 10.1038/ng0904-915. author reply 916–917. [DOI] [PubMed] [Google Scholar]
- 24.Brett D., Pospisil H., Valcarcel J., Reich J., Bork P. Alternative splicing and genome complexity. Nature Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 25.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 26.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.