Abstract
The capsid of Kaposi's sarcoma-associated herpesvirus (KSHV) was visualized at 24-Å resolution by cryoelectron microscopy. Despite limited sequence similarity between corresponding capsid proteins, KSHV has the same T=16 triangulation number and much the same capsid architecture as herpes simplex virus (HSV) and cytomegalovirus (CMV). Its capsomers are hexamers and pentamers of the major capsid protein, forming a shell with a flat, close-packed, inner surface (the “floor”) and chimney-like external protrusions. Overlying the floor at trigonal positions are (αβ2) heterotrimers called triplexes. The floor structure is well conserved over all three viruses, and the most variable capsid features reside on the outer surface, i.e., in the shapes of the protrusions and triplexes, in which KSHV resembles CMV and differs from HSV. Major capsid protein sequences from the three subfamilies have some similarity, which is closer between KSHV and CMV than between either virus and HSV. The triplex proteins are less highly conserved, but sequence analysis identifies relatively conserved tracts. In alphaherpesviruses, the α-subunit (VP19c in HSV) has a 100-residue N-terminal extension and an insertion near the C terminus. The small basic capsid protein sequences are highly divergent: whereas the HSV and CMV proteins bind only to hexons, difference mapping suggests that the KSHV protein, ORF65, binds around the tips of both hexons and pentons.
Herpesviruses constitute an extensive family of DNA viruses distinguished by the large sizes of their linear double-stranded genomes, which range from ∼125 to ∼ 250 kbp, and a common structural design (17, 20, 33, 41). All herpesviruses identified to date have thick-walled icosahedral nucleocapsids, ∼1,250 Å in diameter, surrounded by a partially ordered proteinaceous layer called the tegument, which in turn is enclosed within the envelope, a lipid bilayer studded with viral glycoproteins.
Herpesviruses cover a wide host range, infecting organisms throughout the animal kingdom, from mammals to fish and amphibia (34). Tissue tropism rather than host species was included among the biological properties used to designate the three subfamilies; e.g., the alpha subfamily includes neurotropic viruses, and the gamma subfamily includes lymphotropic viruses. This classification system predates but is largely consistent with the extensive genomic information that is now available (12). The eight herpesviruses that have been implicated in human diseases include members of all three subfamilies.
One fundamental question is the extent to which variations in herpesvirus genome size and content and in host range—which are indices of evolutionary divergence—are paralleled by variations in virion structure. The component for which most information is available is the capsid, which has been studied by cryoelectron microscopy (cryo-EM) in the cases of equine herpesvirus 1 (3) and herpes simplex virus type 1 (HSV-1) (see, e.g., references 4, 11, 30, 37, and 54), which are both alphaherpesviruses; the betaherpesviruses human cytomegalovirus (HCMV) (7, 8) and simian cytomegalovirus (SCMV) (47), and channel catfish virus (5), a distant relative not assigned to any subfamily (13). The distinctive properties of herpesvirus capsids include their T number of 16 (they are the only viruses yet found to have this property) and the “triplex” complexes that overlie their sites of threefold symmetry. Despite considerable sequence divergence, the capsid protein composition appears well conserved in terms of the number of major constituents and their molecular weights (Table 1). Its basic matrix is composed of 150 hexamers and 12 pentamers of the major capsid protein; the 320 triplexes are αβ2 heterotrimers; and the hexon tips of HSV-1, SCMV, and HCMV are occupied by an additional small basic capsid protein. The particle first assembled, the procapsid (28, 46), has a morphogenic scaffolding core that undergoes proteolysis during maturation (22, 51). The major capsid protein and the triplex proteins are essential for capsid assembly, while the small basic capsid protein of HSV-1 (and probably also of other herpesviruses) is dispensable (42, 45) and is added only after procapsid maturation (31). Although VP26 is not required for capsid assembly, it enhances HSV-1 replication in the trigeminal ganglia by an as yet unknown mechanism (14).
TABLE 1.
Masses of the herpesvirus capsid proteins and whole capsidsa
| Protein | No. of copies | Mass (kDa) of protein in:
|
||
|---|---|---|---|---|
| HSV-1 | HCMV | KSHV | ||
| Major capsid protein | 960 | 149 | 154 | 153 |
| Triplex-α | 320 | 50 | 33 | 36 |
| Triplex-β | 640 | 34 | 35 | 34 |
| Small basic capsid protein | 900a | 12 | 8 | 19 |
| Total mass | 191,600 | 188,000 | 197,260 | |
KSHV probably has 960 copies of ORF65 (see Results), with a calculated capsid mass of 198,400 kDa.
The situation outlined above is based primarily on studies of HSV and CMV. Corresponding structural information on gammaherpesviruses has been hard to come by, primarily on account of difficulty in obtaining capsids in sufficient quantity for analysis. Kaposi's sarcoma-associated herpesvirus (KSHV; also called human herpesvirus 8 [HHV-8]) is a recently discovered member of this subfamily (23, 36), which has attracted much research activity on account of its involvement in the etiology of Kaposi's sarcoma, a cancer prevalent in immunosuppressed AIDS patients (1, 16, 21, 25, 27, 38). Very recently, a cryo-EM study of KSHV appeared (53). We have also been studying this virus, and in the accompanying paper by Nealon et al. (26), the propagation of KSHV in cultured B cells and the isolation and biochemical analysis of its capsids are described. In the studies reported here, we examined the molecular architecture of these capsids by cryo-EM and image reconstruction. These observations are compared with prior data on other herpesviruses and are discussed in terms of bioinformatic analysis of the corresponding proteins. An additional motivation for this study was to investigate the status of the KSHV ORF65 protein—putatively, its small basic capsid protein—which has emerged as an important immunological marker for KSHV infection (38, 43).
MATERIALS AND METHODS
Preparation of KSHV capsids.
BCBL-1 is a B-cell line derived from a primary effusion lymphoma that is latently infected with KSHV; no EBV DNA is present in this line (32). The cells were maintained as described in the accompanying paper (26). KSHV virions and released capsids were isolated essentially as described previously (26, 32). In brief, 1 to 2 liters of BCBL-1 grown to 2 × 105 to 3 × 105 cells/ml was treated with both 20 ng of 12-O-tetradecanoyl phorbol 13-acetate per ml and 0.3 mM sodium butyrate for 12 to 18 h. The cells were then changed to their standard medium and incubated for another 6 to 7 days. The medium was centrifuged (600 × g for 5 min and then 2,000 × g for 30 min) to sediment the cells, nuclei, and large debris, and then the viral and subviral particles were pelleted from this supernatant by ultracentrifugation (50,000 × g for 2 h). The pellet was resuspended in DNase buffer (10 mM MnCl2, 50 mM Tris HCl [pH 7.5]) with 0.03 U/of DNase I (Roche Molecular Biochemicals) per ml and incubated for 30 min at 37°C. The reaction was stopped on ice with 20 mM EDTA (pH 8.0) Tris HCl (pH 8.0) and NaCl were then added to give final concentrations of 20 and 250 mM, respectively, and a protease inhibitor cocktail (PILL; Roche Molecular Biochemicals) was added. Triton X-100 was then added to a final concentration of 2% and the mixture was incubated overnight at 4°C. This mixture was sonicated in a bath for 15 s and sedimented (75,000 × g for 30 min) through a 35% (wt/vol) sucrose cushion made up in 20 mM Tris HCl (pH 8)–250 mM NaCl–1 mM EDTA (MTNE). The resulting pellet was resuspended, sonicated as above, loaded (60 μl/gradient) onto a 600-μl 20 to 50% sucrose-MTNE gradient, and centrifuged at 75,000 × g for 40 min. We then collected 40-μl fractions from the top of the gradient. Fractions containing empty capsids were identified by negative-staining EM as well as by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with antibodies against the scaffolding protein (ORF17.5) and major capsid protein (ORF25) as described in the accompanying paper (26). These fractions were pooled, diluted with MTNE, pelleted (75,000 × g for 30 min), and finally resuspended in 20 μl of MTNE by brief sonication in a bath.
Cryo-EM.
Because of the low concentration of capsids in the sample, we attempted to maximize the density of particles in micrograph fields as follows. The sample was sonicated in a bath for 10 s and then centrifuged at 5,000 × g at 4°C for 6 min. Three 3-μl drops from the bottom 10 μl were applied sequentially for 10 min each to an EM grid carrying a thin continuous carbon film supported on a thick holey carbon film. Finally, the grid was blotted and the specimen was vitrified and transferred into a CM200-FEG electron microscope (FEI, Mahwah, N.J.) with a Gatan 626 cryo-holder. The grid was searched at low magnification for populated holes, using a charge-coupled device camera, and cryo-electron micrographs were recorded on film, using low-dose procedures and a nominal magnification of ×38,000 essentially as described previously (47).
Image processing.
Electron micrographs were evaluated by optical diffraction and scanned at 7 μm/pixel with a SCAI scanner (Zeiss Photogrammatics, Englewood, Colo.). The micrographs typically contained 5 to 20 usable particle images, and a total of 28 focal pairs were combined in the reconstruction. Particle images were extracted with the X3Dpreprocess program (10, 49) and binned threefold to give a nominal pixel size of 5.53 Å. The defocus was determined for each micrograph, ranging from 0.6 to 2.0 μm (first zeros at spacings of 14 to 26 Å). Images were computationally corrected for contrast transfer effects by inverting the phases in appropriate regions of their Fourier transforms, and focal pairs of images were combined with CTFMIX (9).
Particle orientations were determined using EMPFT (2), with a density map of the HSV-1 capsid at 18-Å resolution (29) as starting model. The particles selected for the final reconstruction were the 80% with the highest correlation coefficients, resulting in the inclusion of 398 particles. Resolution was estimated in terms of the Fourier shell correlation coefficient (50), with a threshold of 0.3.
To facilitate comparisons of the KSHV, HSV-1, and SCMV capsids, the reconstructions were scaled to the same size by matching the density peak at a ∼500-Å radius, corresponding to the conserved (see Results) floor region, and each map was calculated to the same resolution limit (22 Å). To assign appropriate contour levels for surface rendering and to facilitate difference imaging, each map was normalized so that its volume, thus contoured, would contain the expected mass of protein (Table 1). This procedure gave contour levels close to 1 standard deviation in each case.
Sequence analysis.
Genomic sequences from various herpesviruses were obtained from the the EMBL and GenBank databases. The capsid protein sequences were taken from the annotated genes in the database files, except the small capsid protein of HCMV, which was taken from Gibson et al. (18), and the muromegalovirus capsid proteins, where the sequences were extracted based on comparisons with HCMV. The sequences used were as follows: HSV-1, HE1CG, NC_001806; HSV-2 (strain HG52), HSV2HG52, NC_001798; varicella-zoster virus, HEVZVXX, NC_001348; equine herpesvirus 1 (strain Ab4p), NC_001491; equine herpesvirus 4 (strain NS80567), AF030027, NC_001844; bovine herpesvirus 1, BHV1CGEN, NC_001847; gallid herpesvirus 1 (serotype 2, Marek's disease), AB024414; gallid herpesvirus 2, AF147806, NC_002229; HCMV (strain AD169), HEHCMVCG, NC_001347; muromegalovirus 1, MCU68299; HHV-6 U1102 (variant A), HHV6AGNM, NC_001664; HHV-6B (strain Z29), NC_000898; HHV-7 JI, HH43400, NC_001716; Epstein-Barr virus, HEHS4B958, NC_001345; KSHV, KSU75698; rhesus rhadinovirus 17577, AF083501; ateline herpesvirus 3, NC_001987; herpesvirus saimiri 2, HSGEND, NC_001350; EHV-2, EHU20824, NC_001650; alcelaphine herpesvirus 1, AF005370; murine herpesvirus 4, (murine herpesvirus 68 strain WUMS), U97553.
Sequences were aligned using ClustalW (44) with minor hand editing. Comparisons involving CMV were performed with HCMV sequences, but the corresponding proteins of SCMV are sufficiently similar (major capsid protein, 76% identical, 88% similar; triplex-α, 72% identical. 90% similar; triplex-β, 78% identical, 92% similar; small basic capsid protein, 64% identical, 77% similar [W. Gibson, personal communication]) to justify the substitution.
The information content calculated here is based on the “rate of information transfer”:
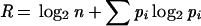 |
where n is the minimum of the number of residue types (i.e., 20) or the number of residues aligned at that position (i.e., n ≤ 21 for the 21 sequences) and pi is the fractional frequency of the occurrence of each residue at the position in the alignment (i.e., an estimate of the probability of finding each type of residue at that position).
RESULTS AND DISCUSSION
EM of isolated KSHV capsids.
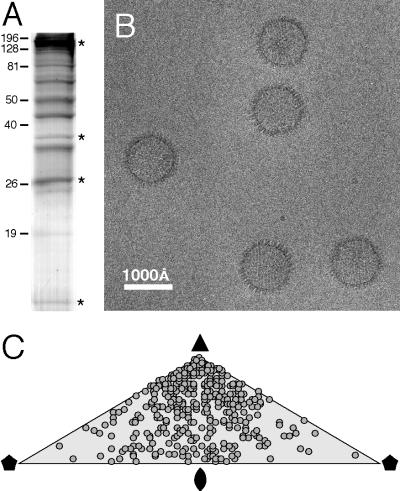
Capsids were obtained by recovering viral particles secreted from KSHV-infected B cells and removing their envelopes by detergent treatment (26). Their principal constituents are shown in Fig. 1A: a major capsid protein of ∼150 kDa, two triplex proteins of ∼35 kDa (the α subunit is ORF62, and the β subunit is ORF26), and a protein of ∼19 kDa identified by Western blotting and mass spectrometry to be ORF65 (26). To obtain cryoelectron micrographs with adequately populated fields, we found it necessary to adsorb the sample to carbon support films for extended periods prior to vitrification. Even so, to accumulate enough particles for image reconstruction, we had to combine data from many micrographs.
FIG. 1.
Cryo-EM of KSHV capsids. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie blue staining of purified capsids. The four bands marked with asterisks represent, from the top, the major capsid protein (ORF25), the triplex α-subunit (ORF62), the triplex β-subunit (ORF26), and the small basic capsid protein (ORF65). The identification of these proteins is described in the accompanying paper (26). (B) Cryo-electron micrograph showing a field containing several empty capsids (A-capsids). (C) Distribution over the icosahedral asymmetric unit of the orientations of imaged particles (2).  , twofold axis; ▴, threefold axis;
, twofold axis; ▴, threefold axis;  , fivefold axis.
, fivefold axis.
Almost all capsids seen were empty (Fig. 1B), as expected from the fractions selected for small amounts of scaffolding protein or viral DNA (see Materials and Methods). The sharp profiles of most particles indicated that most tegument proteins had been removed, although some particles were visibly “tufty,” suggesting partial retention of tegument. Because these capsids had progressed sufficiently far along the assembly pathway to be secreted, we infer that they represent mature capsids, not procapsids. This impression is supported by the angular profiles of most particles, which correspond to views along or close to the three- or twofold axes of a flat-faceted particle: in contrast, the HSV-1 procapsid is spherical and appears round from any direction (46), and it is likely that other herpesviruses share this property. When the orientations of the particles were determined, they were found to present predominantly views close to a threefold symmetry axis (Fig. 1C).
KSHV capsid structure.
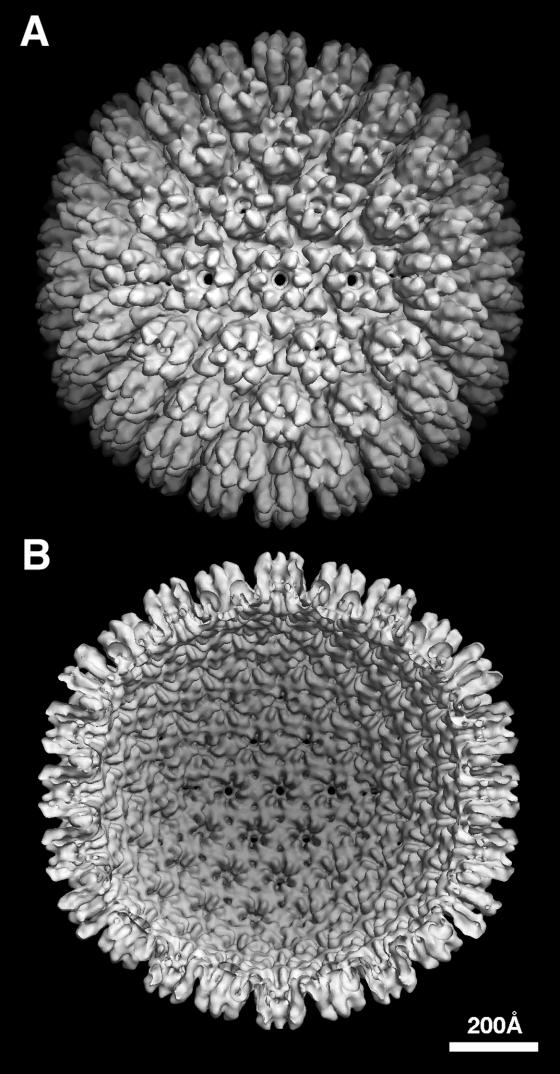
Despite the nonrandom distribution of orientations, the data sufficed to produce a density map at 24 Å resolution (Fig. 2). The KSHV capsid has a diameter of 1,260 Å along the twofold axis and 1,300 Å along the fivefold axis. As expected, it has a triangulation number of 16 and the capsomer arrangement typical of herpesviruses (Fig. 2). The radial density profile of KSHV (Fig. 3) shows the three strata previously seen for other herpesviruses: the inner “floor” region is about 30 Å thick and is composed of close-packed capsomer bases; the second zone contains the triplexes and the lower portions of the capsomer protrusions; and the outer region contains the capsomer tips. According to these profiles, the KSHV capsid is ∼15 Å thicker than that of SCMV.
FIG. 2.
The KSHV capsid at 24 Å resolution. The density map is viewed along a twofold axis of symmetry: (A) outside surface, showing the capsomer protrusions; (B) inner surface, showing the openings below each hexon and penton and the small holes under each triplex.
FIG. 3.
Radial density profiles of the capsids of KSHV and SCMV (47). The profiles were calculated by averaging the respective density maps in concentric shells.
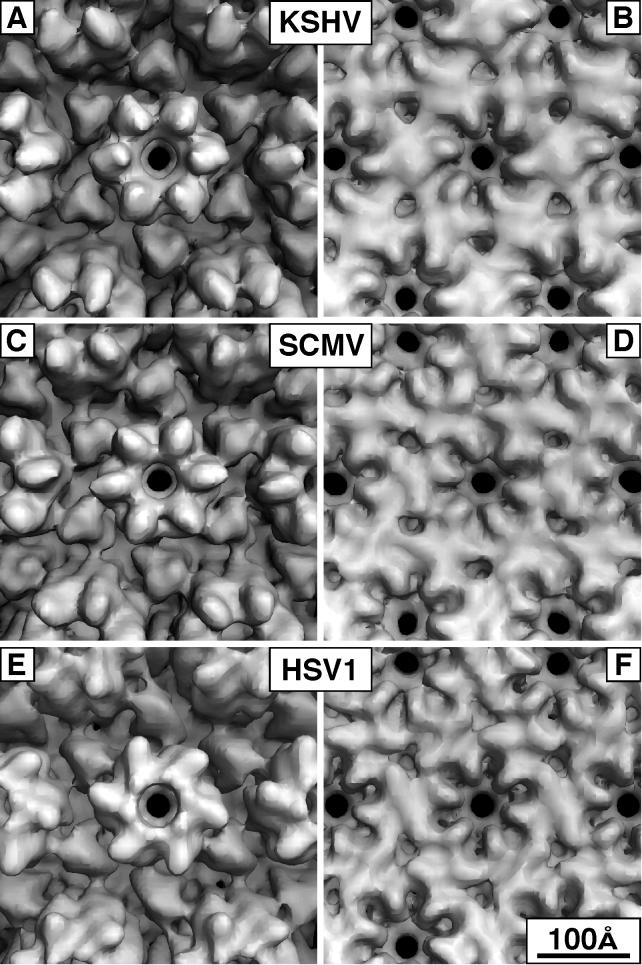
In Fig. 4 and 5, the KSHV capsid is compared to those of HSV-1 and SCMV at similar resolution. The inner surface of the floor is closely conserved among all three capsids (Fig. 4B, D, and F). The opening of the transhexon channel is scalloped in outline and surrounded by six diamond-shaped features, each of which has a local twofold axis. This dyad axis separates two copies of the major capsid protein—one subunit from the hexon in question and one from a neighboring hexon or penton. These dimer contacts are particularly evident for the KSHV capsid, but very similar arrangements are also found in HSV-1 and SCMV. They represent the major interaction between neighboring capsomers and are formed only after maturation of the procapsid (there are no direct contacts between adjacent hexons in the HSV-1 procapsid: instead, the hexons interact only indirectly via the triplexes [46]).
FIG. 4.
Comparison of hexon morphology for KSHV, SCMV, and HSV-1. In each case, the E-hexon, i.e., the hexon on the icosahedral twofold axis, is viewed from the outside (A, C, and E) and inside (B, D, and F). The maps are contoured to enclose 100% of the expected mass. In the outside views, six triplexes surround each hexon. The mutual resemblance of the KSHV and SCMV protrusions is evident.
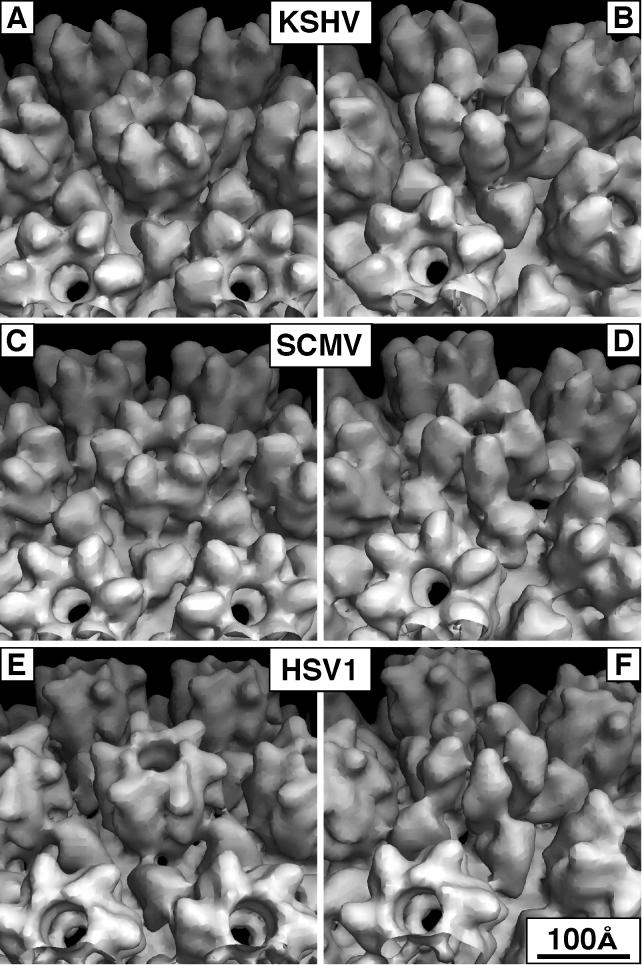
FIG. 5.
Comparison of E-hexon and penton morphology for KSHV, SCMV, and HSV-1. In this montage, both capsomers are viewed obliquely from the outside. (A, C, and E) The respective hexons; (B, D, and F) the respective pentons. The structural differences between the hexon and penton subunits of SCMV and HSV-1 are apparent.
KSHV hexons and pentons have the same height (∼160 Å), as measured from the part of the floor next to the channel, to their outer tip. The channels that run through each capsomer have two constrictions: the one close to the base is ∼15 Å wide in hexons and almost closed in pentons, and the one near the middle of the capsomer is ∼30 Å wide.
KSHV triplexes have a three-bladed cloverleaf shape; i.e., each triplex has three lobes of approximately equal size (Fig. 4A and 5A and B). As viewed from the outside, HSV-1 triplexes have more the shape of a distorted triangle (31, 35) (Fig. 4E and 5E and 5F). However, when sectioned lower down, they also reveal three equally spaced subunits diverging from a central cavity as they extend toward the capsid floor (Fig. 6A), giving the triplex a tripod-like form. The triplexes of KSHV and SCMV are similar in this respect, and they are also set in the same orientation relative to the surface lattice (Fig. 6).
FIG. 6.
: Sections, 5 Å thick, through the capsids of KSHV, SCMV, and HSV-1. The protein section is shown dark. (A to C) The sections are perpendicular to a twofold symmetry axis, about 70 Å below the outer surface. They pass through the lower portion of the hexon protrusions and approximately midway through the surrounding triplexes. In these sections, each triplex appears as three approximately threefold symmetric patches of density. (D to F) Similar features in a plane perpendicular to an axis of fivefold symmetry. Again, the three nodules of a triplex are circled.
The most disparate feature among the three viruses is their hexon protrusions (cf. Fig. 4A, C, and E and 5A, C, and E). In this respect, SCMV differs markedly from HSV-1 (47), whose hexons are dominated by their capping rings of VP26 subunits. On SCMV, the corresponding protein (smallest capsid protein [SCP]) is smaller (8 rather than 12 kDa) and binds in a somewhat different site on the outer wall of the hexon protrusions (Fig. 6a of reference 47). Thus, for both HSV-1 and SCMV, the protrusions of the penton subunit are visibly distinct from those of the hexon subunits (cf. Fig. 5C to E). With KSHV, the penton tips appear rounder than the hexon tips (cf. Fig. 5A and B), but this distinction is less marked than those seen with HSV-1 and SCMV and may be considered marginal at the current resolution. However, the hexons of KSHV resemble those of SCMV quite closely, both in their orientation and in the overall shape of the subunits (cf. Fig. 4A to C and 5A to D), although those of KSHV protrude further radially (Fig. 3).
Small Protein(s) capping both hexons and pentons.
The ORF65 protein of KSHV is thought to be its counterpart of VP26 and SCP of SCMV, although it is 50% larger than VP26 and twice the size of SCP and has little sequence similarity to either of them (see below). VP26 and SCP bind to their respective major capsid proteins only in their hexon conformations (6, 48, 55). In particular, VP26 binds to hexons in vitro but fails to bind to pentons, even at a 100-fold molar excess (52). ORF65 has been shown by immuno-EM to be a component of the KSHV capsid and estimated to have a stoichiometry of ∼80% compared to the major capsid protein in these preparations (26) (Fig. 1A). Although we do not have KSHV capsids lacking ORF65 that would allow this protein to be localized directly by difference imaging, we exploited the resemblance between KSHV and SCMV for this purpose.
The resulting difference map (Fig. 7), obtained by subtracting the SCMV map from the KSHV map, shows compact connected densities of about the right size for a protein of 10 to 20 kDa, overlying the positions of both the hexon and the penton subunits. Since similar densities are seen in all quasiequivalent sites, including those which are not related by icosahedral symmetry, it follows that they do not arise from imposing icosahedral symmetry on residual noise. The penton-associated densities appear larger, presumably reflecting the fact that the SCP was not subtracted. In the reverse experiment, i.e., when the KSHV was subtracted from SCMV, no such positive densities were seen. A plausible explanation is that they represent ORF65 subunits, although it has not been ruled out that they may reflect structural differences between the respective major capsid proteins. We tentatively conclude that ORF65 binds to the tips of KSHV capsomers and, unlike its counterparts in CMV and HSV-1, appears to bind to pentons as well as hexons. Alternatively, the penton-associated densities might represent some other minor capsid or tegument protein of about the same size.
FIG. 7.
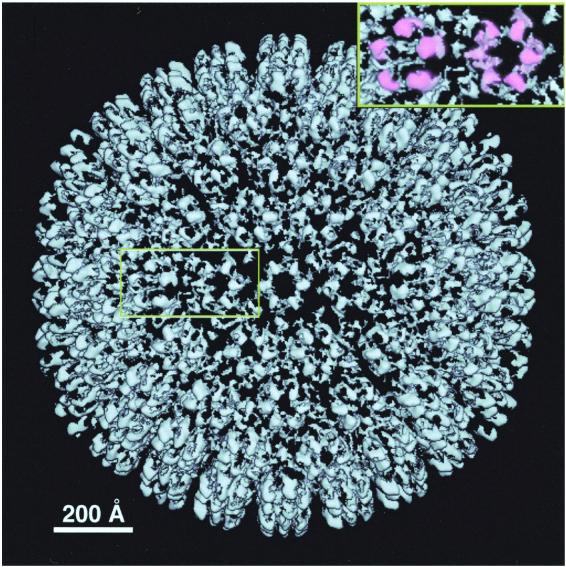
Difference map calculated between the KSHV and SCMV capsids. The map is at the same magnification and is contoured at the same level as the KSHV capsid shown in Fig. 2. Connected densities overlie the position of each hexon and penton subunit and may represent subunits of ORF65 (see Results). As seen in side view around the capsid periphery, this protein appears to consist of two domains. The other features of the difference map are much smaller and less regular; they represent either residual noise or small surface features on the KSHV major capsid protein or triplex subunits that have no counterparts on SCMV. The region around one penton and one hexon is blown up (top right), with the features tentatively assigned as ORF65 subunits (or, for the penton, possibly some other protein) shaded in pink.
Just after completion of this work, a paper appeared by Wu et al., reporting a cryo-EM structure of KSHV capsid at the same resolution (53). The two structures look very similar (compare Fig. 2 of this paper and Fig. 3 of reference 53). Although they presented no biochemical data on composition of their capsids, Wu et al. interpreted the structural similarity of the hexon and penton subunits to imply that ORF65 was not present (53). In the accompanying paper (26), Nealon et al. demonstrate that capsids identical to those studied here contain ORF65 in the amount (∼800 copies per capsid) expected for a small basic capsid protein. Since the capsids studied by Wu et al. were obtained from the same cell line and isolated by a quite similar procedure, their protein composition is likely to be the same. Moreover, there is a close resemblance between the respective radial profiles calculated for KSHV capsids (compare Fig. 4 of reference 53 and Fig. 3 of this paper). Since the shape of this profile at outer radii is sensitive to the presence of proteins at the tips of the capsomer protrusions, this observation further supports the hypothesis that ORF65 is also present in the capsid preparations of Wu et al. (53).
Herpesvirus phylogeny based on capsid proteins.
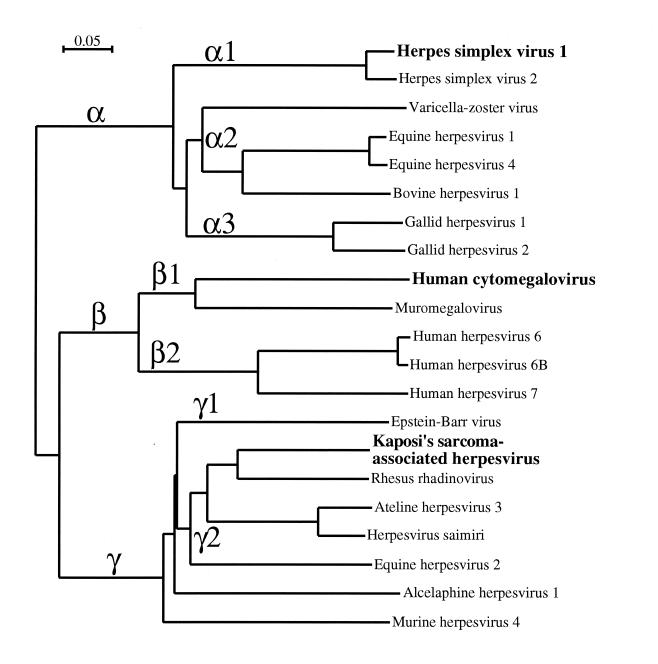
Herpesvirus phylogeny has been studied in some depth (24). However, evolutionary constraints on capsid proteins might differ from those affecting other viral gene products, and an evolutionary dendrogram based on 12 major capsid protein has been presented (36). We made a similar calculation based on a larger data set of 21 such sequences (Fig. 8), which turned out to be in good agreement with the earlier analyses (24, 36). In particular, it is apparent that the alphaherpesviruses diverged first. This observation is consistent with the closer morphological resemblance between KSHV and SCMV capsids than between either virus and HSV-1 (Fig. 4). Their relative proximity is further reflected in higher percentages of identical and similar amino acids in pairwise comparisons of the aligned sequences (Table 2). Wu et al. (53) also reported sequence similarity values calculated by ClustalW (44), comparing the capsid proteins of KSHV and HSV-1. Our numbers (Table 2), based on multisequence alignments involving 21 herpesviruses, agree quite well with theirs.
FIG. 8.
Phylogenetic dendrogram of herpesvirus major capsid proteins. The three subfamilies are indicated by α, β, and γ, and the notation for subgroupings (α1, α2, etc.) is that of McGeoch and Davison (24). Among the gammaherpesviruses, which include KSHV, the major capsid proteins of alcelaphine herpesvirus 1 and murine herpesvirus 4 appear distinct from the rest of the γ2 group.
TABLE 2.
Identities and similarities between the herpesvirus capsid proteins
| Pairwise comparison | % Identity (% similarity) between viruses fora:
|
|||
|---|---|---|---|---|
| Major capsid protein | Triplex α-subunit | Triplex β-subunit | Small basic capsid protein | |
| HSV-1 and HCMV | 24 (40) | 14 (25) | 18 (31) | 7 (20) |
| HSV-1 and KSHV | 28 (43) | 17 (28) | 16 (34) | 6 (19) |
| HCMV and KSHV | 30 (50) | 16 (30) | 19 (39) | 5 (15) |
Identity is expressed as the percentage of identical residues in the aligned positions where residues occur in both sequences. Similarity is expressed as the percentage of residues in the aligned positions with a positive score in the BLOSUM62 amino acid substitution matrix (19).
Similar analyses were also done on both triplex components (data not shown), leading to generally similar conclusions to the dendrogram based the major capsid proteins (Fig. 8). Because the triplex proteins are shorter and less closely conserved than the major capsid proteins (Table 2), the phylogeny based on the latter proteins should be more reliable.
Conserved features of herpesvirus major capsid proteins.
The major capsid proteins show a significant degree of sequence similarity, with 23 to 30% identical and 40 to 50% similar residues for proteins from different subfamilies (Table 2). Furthermore, the protein size is well conserved, and when the sequences are aligned, there are few gaps (Fig. 9A), suggesting that structural modules are also conserved. Taken together with the similar three-dimensional shapes depicted in the respective cryo-EM density maps, it is likely that they also have largely similar folds.
FIG. 9.
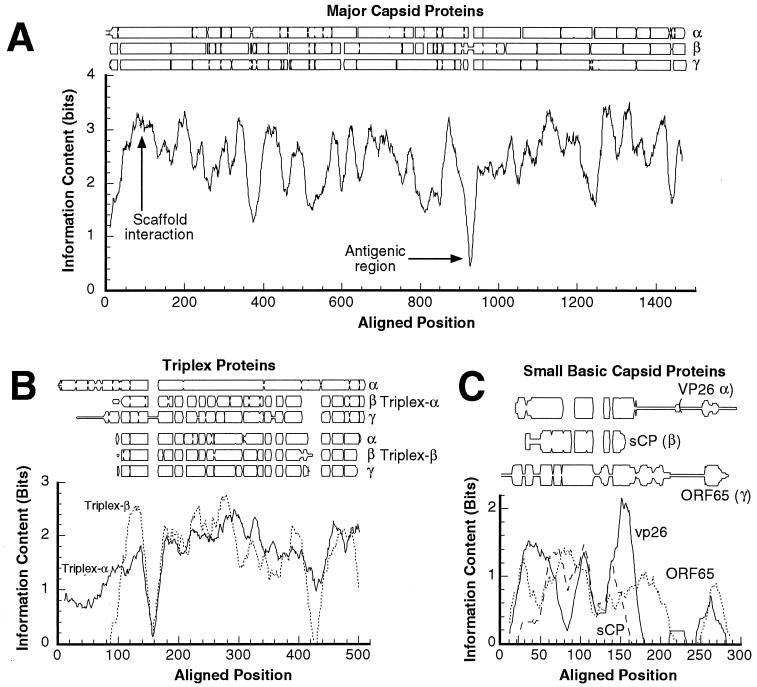
Sequence conservation among the four most abundant herpesvirus capsid proteins. The degree of conservation is plotted both in terms of the occurrence of different residues at aligned positions (top part of each panel) and in terms of “information content” (see Materials and Methods) (bottom graph in each panel). (A) Major capsid proteins. (B) Triplex α-subunits (VP19c homologs) and β-subunits (VP23 homologs). (C) Small basic capsid proteins. Information content is a measure of residue type distribution at each point in the alignment. As used here, higher values of this variable correspond to less variability. The profiles were smoothed with a 20-residue window.
In protein sequence alignments, contiguously aligned blocks are candidates to be conserved contiguous secondary-structure elements. The pattern of gaps may therefore constitute a “fingerprint” of secondary structure. In Fig. 9A, conservation as a function of position along the sequence is expressed as an information content measure, with high values indicating a greater degree of conservation (maximum is log2 20 = 4.3 bits). Local minima in this curve correspond to gaps, as well as to differences between the three subfamilies. Some of them are likely to denote surface loops, which are freer to mutate. One such minimum, around position ∼930 in the major capsid protein of HSV-1, coincides with a region, A862 to H880, that is an antigenic determinant on the tips of hexons and pentons (40) and may represent such a loop.
The only other peptide of a capsid shell protein for which positional information is available is around residue 78 (no. 93 in the alignment), a conserved region near the N terminus of VP5. A second-site revertant to a lethal point mutation in the scaffolding protein was mapped to this position (15), suggesting that it resides on the inner surface of the procapsid. A binding site for scaffolding protein localized on the inner surface of the mature capsid (56) may mark the position of this peptide. However, the significance of this interaction with the mature capsid is unclear in the sense that the scaffold and shell should disengage on maturation of the shell, producing either small-cored B capsids or scaffold-less A capsids or C capsids.
Similarity of triplex proteins across the three subfamilies: the α-subunit of alphaherpesviruses has extra segments.
The observation that the two triplex subunits form trimers at sites of local threefold symmetry suggests that they may have similar structures. This proposition is supported by the approximate threefold symmetry of the triplexes (Fig. 4A and 5A) and the nearly equal sizes of the two triplex proteins (∼ 35 kDa) in beta- and gammaherpesviruses. Such similarity might also be reflected in their sequences. To explore this proposition further, we aligned all the triplex protein sequences and compared this global alignment with separate alignments for α-subunits and β-subunits. The triplex proteins are less conserved than the major capsid proteins, with identities and similarities between subfamilies of 10 to 15% and 20 to 35%, respectively. However, in two tracts—positions 215 to 304 and 455 to 510 in the alignment—relatively close correspondence (>20% identity) is observed for the two triplex subunits (Fig. 9B), indicative of significant similarity, which is somewhat stronger among the β-subunits (Table 2, Fig. 9B).
The gap patterns in Fig. 9B suggest that the additional size of triplex α-subunits in alphaherpesviruses (50 versus 35 kDa) is due mainly to an N-terminal extension of ∼100 residues and a 20 to 30-residue insertion near the C terminus. The inference of an N-terminal extension is consistent with the observation that substantial deletions in this portion of VP19c do not abrogate HSV-1 capsid assembly (39).
As to the question whether triplex α-subunits and β-subunits are related, the average similarity scores between these sequences for the 21 herpesviruses analyzed are relatively low (9% identity, 22% similarity). However, these numbers do not preclude a relationship since there are many cases on record of capsid proteins that have similar folds despite having unrecognizably different sequences. Moreover, the fact that the sequences lined up consistently, whether they were aligned with other subunits of the same kind, with subunits of the other subfamily, or globally (data not shown), supports the idea that they are related.
The small basic capsid proteins are highly divergent.
In terms of sequence, the small basic capsid proteins show the lowest level of mutual similarity of any herpesvirus capsid proteins. Although the alignment of these proteins from different subfamilies is tenuous, it suggests that the central parts (in the region from 50 to 150 in the alignment) are weakly similar (Fig. 9C). One shared feature of this family of proteins is that its members tend to have a cluster of positive residues close to the C terminus. The C termini of VP26 and ORF65 differ widely in sequence and length, with a preponderance of prolines, serines, and glycines, and these peptides may be unstructured. The last 20 residues of ORF65 are a major antigenic determinant of KSHV (43).
In conclusion, it emerges that of the four abundant herpesvirus capsid proteins, the small basic capsid protein is the least constrained in sequence variability and the least conservative in capsid-binding properties. For HSV and CMV, this protein binds only to the major capsid protein subunits of hexons, whereas our data suggest that the KSHV protein, ORF65, may also bind to pentons. Its functional attributes are also enigmatic. On one hand, the HSV-1 protein, VP26, is dispensable both for capsid assembly (42, 45) and for microtubule-based transport of capsids along the neuronal cytoplasm (14), and channel catfish virus appears to lack such a protein altogether (5). On the other hand, VP26 markedly enhances the production of virus in trigeminal ganglia of mice (14) and most herpesviruses have contrived to retain such a protein (Fig. 9C), presumably reflecting exploitation—perhaps in different ways—of an opportunity for evolutionary advantage.
ACKNOWLEDGMENTS
We thank A. Philippsen for discussions and assistance in using his visualization program, Dino (http://www.biozentrum.unibas.ch/∼xray/dino).
This work was supported in part by the NIH Targeted Antiviral Program (IATAP) to A.C.S.; by NIH grants P30-CA44579 and R-01 5-23924, the Pew Memorial Trust, and the Doris Duke Charitable Foundation 20000355 to D.H.K; and by NIH grant R-01 AI41644-04 and NSF grant MCB-9904879 to J.C.B.
REFERENCES
- 1.Andre S, Schatz O, Bogner J R, Zeichhardt H, Stoffler-Meilicke M, Jahn H U, Ullrich R, Sonntag A K, Kehm R, Haas J. Detection of antibodies against viral capsid proteins of human herpesvirus 8 in AIDS-associated Kaposi's sarcoma. J Mol Med. 1997;75:145–152. doi: 10.1007/s001090050099. [DOI] [PubMed] [Google Scholar]
- 2.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 3.Baker T S, Newcomb W W, Booy F P, Brown J C, Steven A C. Three-dimensional structures of maturable and abortive capsids of equine herpesvirus 1 from cryoelectron microscopy. J Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like, packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booy F P, Trus B L, Davison A J, Steven A C. The capsid architecture of channel catfish virus, an evolutionarily distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology. 1996;215:134–141. doi: 10.1006/viro.1996.0016. [DOI] [PubMed] [Google Scholar]
- 6.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12 kDa VP26) in a large complex (the 200 MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butcher S J, Aitken J, Mitchell J, Gowen B, Dargan D J. Structure of the human cytomegalovirus B capsid by electron cryomicroscopy and image reconstruction. J Struct Biol. 1998;124:70–76. doi: 10.1006/jsbi.1998.4055. [DOI] [PubMed] [Google Scholar]
- 8.Chen D H, Jiang H, Lee M, Liu F, Zhou Z H. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 9.Conway J F, Steven A C. Methods for reconstructing density maps of “single particles” from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 10.Conway J F, Trus B L, Booy F P, Newcomb W W, Brown J C, Steven A C. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 11.Conway J F, Trus B L, Booy F P, Newcomb W W, Brown J C, Steven A C. Visualization of three-dimensional density maps reconstructed from cryoelectron micrographs of viral capsids. J Struct Biol. 1996;116:200–208. doi: 10.1006/jsbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- 12.Davison A. Herpesvirus genes. Rev Med Virol. 1998;3:237–244. [Google Scholar]
- 13.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 14.Desai P, DeLuca N A, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 15.Desai P, Person S. Second site mutations in the N-terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology. 1999;261:357–366. doi: 10.1006/viro.1999.9877. [DOI] [PubMed] [Google Scholar]
- 16.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 17.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 18.Gibson W, Clopper K S, Britt W J, Baxter M K. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J Virol. 1996;70:5680–5683. doi: 10.1128/jvi.70.8.5680-5683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 21.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 22.Liu F Y, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGeoch D J, Davison A J. The descent of human herpesvirus 8. Semin Cancer Biol. 1999;9:201–209. doi: 10.1006/scbi.1999.0093. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch D J, Davison A J. The molecular evolutionary history of the herpesviruses: origins and evolution of viruses. London, United Kingdom: Academic Press, Ltd.; 1999. [Google Scholar]
- 25.Moore P S, Kingsley L A, Holmberg S D, Spira T, Gupta P, Hoover D R, Parry J P, Conley L, Jaffe H W, Chang Y. KSHV infection prior to onset of Kaposi's sarcoma. AIDS. 1996;10:175–180. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Nealon K, Newcomb W W, Pray T R, Craik C S, Brown J C, Kedes D H. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J Virol. 2001;75:2866–2878. doi: 10.1128/JVI.75.6.2866-2878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neipel F, Fleckenstein B. The role of HHV-8 in Kaposi's sarcoma. Semin Cancer Biol. 1999;9:151–164. doi: 10.1006/scbi.1999.0129. [DOI] [PubMed] [Google Scholar]
- 28.Newcomb W W, Homa F L, Booy F P, Thomsen D R, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: Characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 29.Newcomb W W, Homa F L, Thomsen D R, Trus B L, Cheng N, Steven A, Booy F, Brown J C. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 31.Newcomb W W, Trus B L, Cheng N, Steven A C, Sheaffer A K, Tenney D J, Weller S K, Brown J C. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 34.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2221–2230. [Google Scholar]
- 35.Saad A, Zhou Z H, Jakana J, Chiu W, Rixon F J. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J Virol. 1999;73:6821–6830. doi: 10.1128/jvi.73.8.6821-6830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarid R, Olsen S J, Moore P S. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv Virus Res. 1999;52:139–232. doi: 10.1016/s0065-3527(08)60299-7. [DOI] [PubMed] [Google Scholar]
- 37.Schrag J D, Prasad B V, Rixon F J, Chiu W. Three-dimensional structure of the HSV1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 38.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V, Weiss R A, Moore P S. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 39.Spencer J V, Newcomb W W, Thomsen D R, Homa F L, Brown J C. Assembly of the herpes simplex virus capsid: performed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spencer J V, Trus B L, Booy F P, Steven A C, Newcomb W W, Brown J C. Structure of the herpes simplex capsid: peptide A862–H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–235. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 41.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 42.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses, J. Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 43.Tedeschi R, De Paoli P, Schulz T F, Dillner J. Human serum antibodies to a major defined epitope of human herpesvirus 8 small viral capsid antigen, J. Infect Dis. 1999;179:1016–1020. doi: 10.1086/314657. [DOI] [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 47.Trus B L, Gibson W, Cheng N, Steven A C. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trus B L, Kocsis E, Conway J F, Steven A C. Digital image processing of electron micrographs: the PIC system-III. J Struct Biol. 1996;116:61–67. doi: 10.1006/jsbi.1996.0011. [DOI] [PubMed] [Google Scholar]
- 50.van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–124. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 51.Welch A R, Woods A S, McNally L M, Cotter R J, Gibson W. A herpesvirus maturational protease, assemblin: Identification of its gene, putative active site domain, and cleavage site. Proc Natl Acad Sci USA. 1991;88:10792–10796. doi: 10.1073/pnas.88.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wingfield P T, Stahl S J, Thomsen D R, Homa F L, Booy F P, Trus B L, Steven A C. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L, Lo P, Yu X, Stoops J K, Forghani B, Zhou Z H. Three-dimensional structure of the human herpesvirus 8 capsid. J Virol. 2000;74:9646–9654. doi: 10.1128/jvi.74.20.9646-9654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z H, Dougherty M, Jakana J, He J, Rixon F J, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Z H, Macnab S J, Jakana J, Scott L R, Chiu W, Rixon F J. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging, Proc. Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]