Abstract
Limited information is available on the molecular mechanisms by which long-term HIV-1-infected nonprogressors suppress HIV-1 infection and maintain immune functions. The intestinal mucosal immune system is an early target for HIV-1 infection and severe CD4+ T cell depletion. We evaluated mucosal T lymphocyte subsets, virus-specific cellular responses, gene expression profiles, and viral loads in intestinal mucosal biopsies of long-term nonprogressor (LTNP) patients as compared to chronically HIV-1-infected patients with high viral loads (HVLs) and CD4+ T cell loss, as well as HIV-seronegative healthy individuals. This study aims to identify the mucosal correlates of HIV disease progression and to determine the molecular changes associated with immune and intestinal dysfunction. LTNP patients had undetectable viral loads, normal CD4+ T cell levels, and virus-specific cellular responses in peripheral blood and mucosal compartments. Microarray analysis revealed a significant increase in gene expression regulating immune activation, cell trafficking, and inflammatory response in intestinal mucosa of HVL patients as compared to LTNP patients. Genes associated with cell cycle regulation, lipid metabolism, and epithelial cell barrier and digestive functions were down-regulated in both HVL and LTNP patients. This may adversely influence nutrient adsorption and digestive functions, with the potential to impact the efficacy of antiretroviral therapy. We demonstrate that the maintenance of mucosal T cells, virus-specific responses, and distinct gene expression profiles correlate with clinical outcome in LTNP patients. However, the intestinal mucosal immune system remains an important target of HIV-1 infection in LTNP, and these effects may ultimately contribute toward disease progression.
Although most HIV-infected individuals experience progressive CD4+ T cell loss and develop AIDS, a small fraction (5–10%) of HIV-infected individuals, called long-term nonprogressors (LTNP), are able to naturally suppress viral loads, maintain normal peripheral CD4+ T cell numbers and immune functions, and remain clinically healthy for >10 years in the absence of any antiretroviral therapy (ART) (1–3). Studies have shown that LTNP patients display higher levels of HIV-1-specific responses in peripheral blood lymphocytes than chronically infected patients (1, 4, 5). However, the underlying mechanisms resulting in the development and maintenance of an LTNP phenotype remain unknown.
The intestinal mucosal tissue is an early target organ of HIV and a site of severe CD4+ T cell depletion during primary HIV-1 infection, which is not adequately reflected in peripheral blood (3). Impairment of the mucosal immune system may lead to the disruption of gastrointestinal functions frequently observed in HIV-1-infected patients. Studies in the simian immunodeficiency virus (SIV) model have demonstrated that nutrient digestive and absorptive functions can be impaired early during primary SIV infection (6, 7). Moreover, high throughput gene expression studies indicated that these disruptions are characterized by down-regulation of host genes involved in mucosal growth and enterocyte function, and that these pathological changes persist during chronic infection in the absence of antiviral therapy (8–10).
Our current study provides insights into the role of mucosal pathology in HIV-1 disease progression through a comparative investigation of pathophysiological changes in intestinal mucosa of LTNP patients and chronically HIV-1-infected patients with high viral loads and significant CD4+ T cell depletion (HVL group). Maintenance of mucosal CD4+ T cells and presence of virus-specific cellular responses and distinct expression profiles correlated with the asymptomatic clinical outcome in LTNP. A significant increase in mucosal gene expression regulating immune activation and inflammation was detected in HVL patients, but not in LTNP patients. Both LTNP and HVL patients demonstrated dysregulation of expression of genes associated with cell cycle regulation, lipid metabolism, and enterocyte functions. These findings suggest that the mucosal immune system remains an important target of pathologic effects of HIV in LTNP patients despite the suppression of viral loads and maintenance of immune functions.
Materials and Methods
Study Subjects and Sample Collection. Fifteen ART-naïve HIV-1 seropositive individuals and eight HIV-1 seronegative healthy individuals were enrolled in the study (Table 1). LTNP patients (n = 4) were identified based on the following criteria: HIV-1 seropositive for 7 or more years, undetectable plasma viral loads, >500 CD4+ T cells per μl in peripheral blood, ART-naïve, and clinically healthy. HIV-1-infected patients in the HVL group (n = 11) were enrolled by using the criteria: CD4+ T cell depletion (8–427 cells per μl of blood), >10,000 viral RNA copies per ml of plasma, and ART naïve. Endoscopic jejunal biopsy specimens were cryo-preserved for transcriptional analysis and immunohistochemistry, or collected in RPMI medium 1640 (Invitrogen) for flow cytometric analysis. Peripheral blood samples were collected at the time of biopsy. The Institutional Review Board at University of California, Davis, approved the study, and written informed consent was obtained from the participants in the study.
Table 1. Clinical characteristics of study patients.
| Study group | Patient ID | Age, years | Sex | CDC classification | Year of HIV-1 diagnosis | Peripheral CD4+ T cells counts, cells per ml |
|---|---|---|---|---|---|---|
| Group 1 (HIV-1 seronegative) | P8 | 22 | M | NA | NA | 1,346 |
| P11 | 24 | M | NA | NA | 1,465 | |
| P12 | 22 | F | NA | NA | 1,236 | |
| P15 | 52 | M | NA | NA | NA | |
| P34 | 35 | F | NA | NA | 1,046 | |
| P38 | 48 | F | NA | NA | 632 | |
| P41 | 43 | F | NA | NA | 1,203 | |
| P65 | 37 | M | NA | NA | 634 | |
| Group 2 (HVL) | P13 | 48 | F | C3 | NA | 8 |
| P14 | 36 | M | C3 | 2001 | 6 | |
| P19 | 35 | M | C3 | 2001 | 46 | |
| P20 | 45 | M | C3 | 2002 | 294 | |
| P24 | 34 | M | C3 | 2002 | 34 | |
| P43 | 30 | F | C3 | 2002 | 174 | |
| P47 | 23 | M | C2 | 2002 | 205 | |
| P48 | 42 | M | C1 | 2002 | 427 | |
| P67 | 43 | M | C1 | 1994 | 170 | |
| P106 | 37 | F | C1 | 1997 | 289 | |
| P111 | 50 | M | C1 | 1992 | 42 | |
| Group 3 (LTNP) | P16 | 49 | M | A1 | 1989 | 910 |
| P30 | 43 | M | A1 | 1997 | 587 | |
| P77 | 40 | M | A1 | 1995 | 968 | |
| P46 | 45 | M | A1 | 1985 | 787 |
Range of CD4+ T cell counts in patient study groups is shown. The normal range for peripheral CD4+ T cell is 493-1,488 cells per ml. Group 1, HIV-1-negative healthy controls; group 2, HIV-1 seropositive, plasma viral load > 10,000 copies per ml, absolute CD4T cell counts < 500 cells per ml; group 3, HIV-1 seropositive, plasma viral load < 500 copies per ml, absolute CD4T cell counts > 400 cells per ml. NA, test not performed, not relevant, or not available. A1, C1, C2, and C3 denote centers for Disease Control (CDC) classification for clinical stages of HIV-1 infection.
Cell Isolation and Flow Cytometry. Isolation of mononuclear cells from jejunal biopsies and peripheral blood and immunophenotypic analysis were performed as described (3). Cells were stained with monoclonal antibodies, CD3 FITC (Pharmingen), CD4 PE, CD4 TC, CD8 APC, and CD8 FITC (Caltag Laboratories, South San Francisco, CA), CD69 PerCP, IFN-γ phycoerythrin (PE), and IL-2 PE (Pharmingen), and analyzed by flow cytometry using a FACSCalibur (Becton Dickinson) and flowjo (Treestar, San Carlos, CA).
HIV-1-Specific T Cell Responses. Freshly isolated mononuclear cells from peripheral blood and jejunal biopsies were stimulated with HIV-1 Gag and CMV peptides (Becton Dickinson), staphylococcal enterotoxin B (Sigma), or media alone, and immunostained with monoclonal antibodies specific for CD3, CD8, CD69, IFN-γ, and IL-2 according to published methods (11). Samples were analyzed by four-color flow cytometry using a FACSCalibur, collecting at least 100,000 events. The percentage of T cells responding to viral peptide stimulation was determined by subtracting the background cytokine response detected in cells stimulated with media alone. The net antigen-specific cytokine production was required to exceed 0.05% of CD8+ T cells, and to exceed background levels (in media controls) by at least 2-fold to be considered significant.
High Throughput Oligonucleotide Microarray Analysis. Total RNA was isolated from the tissue samples, and double-stranded cDNA was synthesized as described (8). Biotin-labeled cRNA was hybridized with Affymetrix U95Av2 GeneChips. GeneChips were normalized, and comparative analysis of gene expression profiles of LTNP and HVL groups was performed by using dchip analysis software (12). Statistical analysis of comparisons between each HIV-1-infected group (LTNP, n = 3; and HVL, n = 4) to the uninfected healthy control group (n = 4) was performed. Hierarchical clustering was performed to identify similar patterns of transcription. Genes were considered significantly altered if the fluorescence intensity changed by 50% (1.5-fold) or more with statistical confidence (P < 0.05). Genes in clusters were functionally classified by using ease software (version 2.0, National Institutes of Health, Bethesda) (13). Further details regarding the methodology used for gene expression analysis are available in Supporting Text, which is published as supporting information on the PNAS web site.
Real-Time PCR Analysis. Primers and probes specific to HIV-1 gag were designed for a Taqman real-time PCR assay to measure HIV-1 viral RNA levels in tissue and to validate the differential expression of specific genes observed by DNA microarray analysis. Briefly, cDNA was synthesized from extracted RNA by using random hexamers, and reactions were carried out by using PCR master mix (Applied Biosystems) on an Applied Biosystems Prism 7700 sequence detector. Data were analyzed with sequence detector software (sds) and computed by using the relative computational method (14). Real-time PCR validation of gene expression was performed by using Assays on Demand Systems (Applied Biosystems). Gene expression levels were normalized to GAPDH expression. Plasma viral loads were obtained measured by branched DNA assay (Bayer) (15).
Immunohistochemistry. Detection and localization of CD4+ and CD8+ T lymphocytes, macrophages, and HIV-1-infected cells were performed by immunohistochemistry as described (3). Briefly, jejunal tissue sections were immunostained for CD4 (Nova Castra Laboratories, Newcastle, England), CD3, CD8, and Ham56 (Macrophage marker) (Dako), and HIV-1 p24 (NIH AIDS Reagents) at a 1:100 dilutions for 1 h. Tissue sections were examined for morphological changes and inflammatory infiltrates, and cell counts were obtained by using a stage micrometer (16). Specific cells were counted within 20 square areas (within the villi), and the results were extrapolated to a 1-mm2 area.
Results
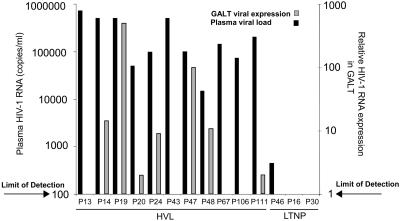
Comparison of Mucosal Viral Burden, CD4+ and CD8+ T Cells, and CTL Response in LTNP and HVL Patients. Analysis of HIV-1 expression indicated that viral loads were elevated in both plasma and jejunal biopsy samples from HVL patients, whereas viral loads were undetectable in biopsies from all LTNP patients and all but one plasma sample (439 HIV RNA copies per ml of plasma) (Fig. 1). HIV-1 p24-positive cells were also detected by immunohistochemistry in both lamina propria and intraepithelial compartments in HVL, but not in LTNP patients (data not shown), providing further evidence of viral suppression in both the mucosa and peripheral blood of LTNP patients. These results indicate that there was no correlation between the viral loads in plasma and gut biopsies in HVL patients as reported (17).
Fig. 1.
HIV-1 RNA levels in plasma and GALT. Plasma HIV-1 RNA levels were measured by branched DNA assay and are shown as copies per ml of plasma. Jejunal tissue viral RNA levels were measured by TaqMan real-time PCR assay, normalized to GAPDH expression, calibrated against patient 20 (lowest detectable HIV-1 viral RNA level), and are shown as relative expression in mucosal tissue. Arrows indicate lower limit of detection. No correlation was observed between plasma viral burden and relative expression of HIV-1 in mucosal tissue. Viral RNA was not detected in mucosa of LTNPs or healthy negative controls. Jejunal biopsy samples were unavailable for analysis from patients P13, P67, and P106.
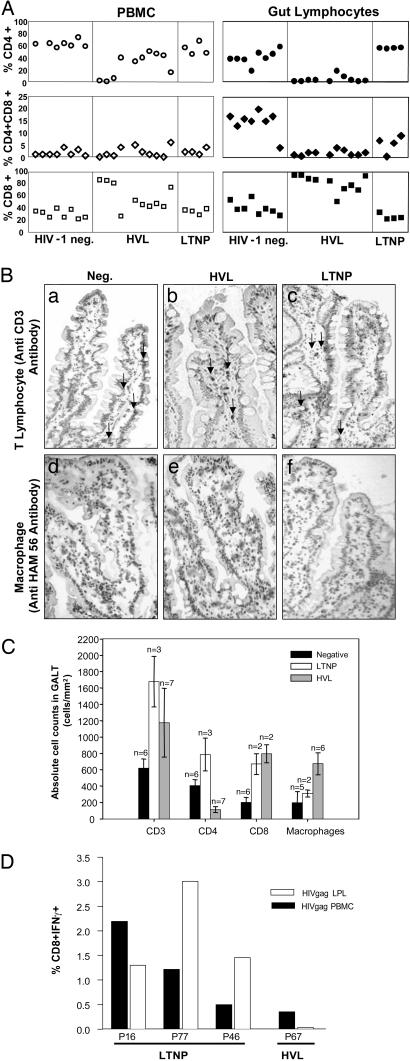
Alterations in the intestinal mucosal and peripheral blood T cell subsets in LTNP and HVL patients were evaluated by flow cytometry (Fig. 2A). In contrast to the HVL patients, LTNP patients maintained normal CD4+ T cell levels in both compartments. In support of flow cytometric data, quantitative analysis of jejunal biopsies by immunohistochemistry revealed a severe depletion of mucosal CD4+ T cells within the villi of HVL patients (<200 cells per mm3), and slightly higher CD4+ T cell levels in LTNP patients as compared to healthy HIV-1-negative controls (Fig. 2 B and C). Interestingly, an increase in the absolute number of mucosal CD8+ T cells was detected in both HVL and LTNP patients in comparison to uninfected control subjects.
Fig. 2.
Analysis of lymphocyte subsets in HIV infection. (A) Immunophenotypic analysis of isolated T lymphocytes from peripheral blood and GALT was performed by flow cytometry as described in Materials and Methods. HIV-1 infection resulted in a pronounced reduction of CD4+ T cells in GALT of HVL patients that was not observed in LTNP patients. CD8+ T cell percentages increased only in HVL patients, whereas CD4CD8 double positive T cell percentages decreased in both HIV-1-infected groups. (B) CD3+ T lymphocytes and macrophages in GALT were detected by immunohistochemistry. CD3+ T cell numbers were higher in HVL and LTNP patients as compared to HIV-1-negative subjects (compare a, b, and c). Higher numbers of macrophages were detected in GALT of HVL patients compared to both HIV-1 negative controls and LTNP patients (compare d, e, and f). (C) CD3+, CD4+, and CD8+ T cells and macrophages were identified by immunohistochemistry and counted. CD4+ T cells were severely depleted from the jejunal mucosa of HVL patients, coinciding with an increase in CD8+ T cells. Increased numbers of macrophages were detected in GALT of both LTNP and HVL patients. (D) Antigen-specific responses to HIV-1 gag were compared between LTNP and HVL patients by measuring intracellular IFN-γ production in isolated CD8+ T cells in response to stimulation with an HIV-1 Gag peptide pool. HIV-1-specific responses were consistently higher in LTNP as compared to HVL patients.
Measurement of viral antigen-specific cellular responses showed that HIV-1 Gag-specific CTL responses were higher in LTNP than in HVL patients (Fig. 2D). The percentage of CD8+ T cells from both the mucosal and peripheral compartments producing IFN-γ in response to HIV-1 Gag was consistently higher in LTNP patients than in HVL patients. In addition, mucosal CD4+ T cells also displayed elevated IFN-γ responses to Gag in comparison to HVL patients, whereas no significant differences could be detected in circulating CD4+ T cells from peripheral blood (data not shown). These data are supported by evidence of increased CD8+ and CD4+ T cells in LTNP patients and suggest that viral suppression in LTNP patients may result, in part, from efficient maintenance of CD4+ T helper and HIV-1-specific CD8+ T cell responses in both gut-associated lymphoid tissue (GALT) and circulating lymphocytes.
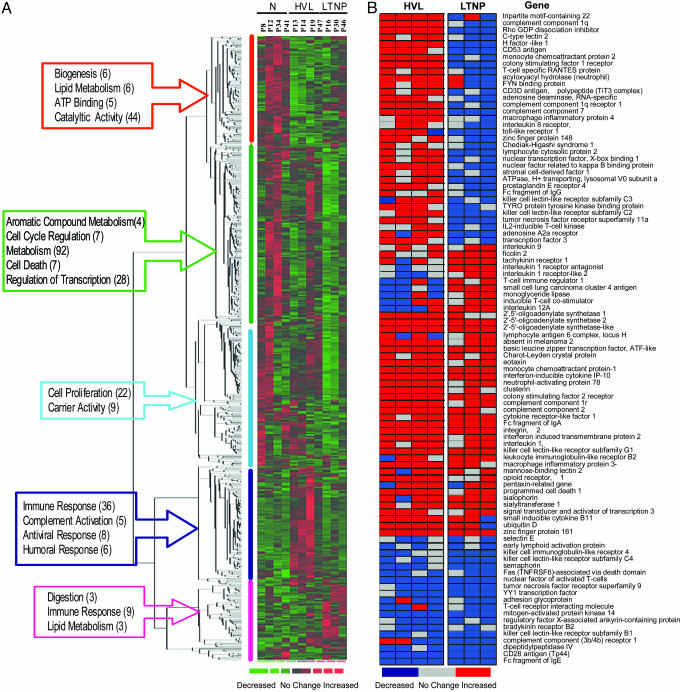
Divergence of Mucosal Gene Expression in HVL and LTNP Patients. Comparison of mucosal gene expression profiles of HVL and LTNP patients with those from uninfected healthy controls revealed an increase in expression of 369 genes in the HVL patients and 150 genes in the LTNP patients as compared to the uninfected healthy controls. Decreased expression was observed for 411 genes in LTNP patients, whereas only 196 genes were down-regulated in HVL patients as compared to uninfected controls. Surprisingly, many of these genes were commonly down-regulated.
Hierarchical cluster analysis of genes that were differentially expressed revealed an up-regulation of statistically significant number of genes in HVL patients associated with immune response, chemotaxis, signal transduction, apoptosis, and T cell activation (Fig. 3A). In addition, smaller clusters of up-regulated genes in both LTNP and HVL patients included genes associate with innate immune responses, digestion, and homeostasis. The dominance of immune response-associated transcriptional profile in the HVL group characterized the elevated localized host responses to HIV-1 infection in the microenvironment of the GALT.
Fig. 3.
Mucosal gene expression profile in HIV infection. (A) Hierarchical clustering of genes altered in GALT by HIV-1 infection in HVL and LTNP patients. Genes that were differentially expressed were clustered by using DCHIP expression analysis software. Down-regulated genes are shown in a green color scale with the largest decreases in dark green. Up-regulated genes are colored pink-red with the highest increases in red. Genes were scored as differentially expressed if their levels changed by at least 50% (P ≤ 0.05). Analysis of statistically overrepresented (P ≤ 0.05, asterisk denotes 0.05 < P < 0.10) physiological pathways within five nonoverlapping clusters indicated that defense response-associated processes such as T cell activation, innate immunity, and chemotaxis were up-regulated in HVL but not in LTNP. In addition, metabolism, signal transduction, and cell proliferation-associated gene expression was dysregulated in both LTNP and HVL patients. (B) Differential expression of genes associated with immune function was seen in HVL and LTNP patients, relative to baseline expression in HIV-1-negative controls. HVL patients displayed increased (red) expression of 34 immune response-associated genes in GALT that were down-regulated (blue) or unchanged in LTNP patients.
HVL Patients Up-Regulate Lymphocyte Activation and Inflammatory Response-Associated Genes. Expression profiling indicated that both HVL and LTNP patients increased transcription of a broad range of immune response genes in the intestinal mucosa (Fig. 3B), including those involved in chemotaxis (eotaxin, mcp-1, cytokine receptor-like factor 1, IL-9, SCYB11), IFN-induced pathways (IP-10, IFN-induced transmembrane protein-2, 2′5′-oligoadenylate synthase-1, 2′5′-oligoadenylate synthase-2), and receptors (leukocyte Ig-like receptor-B2). However, the number of genes associated with both lymphocyte activation and inflammatory/stress responses (e.g., RANTES, Toll-like receptor-1, MIP-4, CSF-1R, TNF receptor superfamily 11A, mcp-2, and prostaglandin E receptor-4) and their level of expression (data not shown) were substantially higher in HVL than in LTNP patients, suggesting that active viral replication in HVL patients resulted in chronic lymphocyte activation and inflammation in GALT. Inflammation-related gene expression has been reported in lymph nodes (18), peripheral blood (19) of chronically HIV-1-infected subjects with CD4+ T cell depletion, and in vitro HIV-I infection studies (20, 21). In summary, cell activation and inflammation are dominant features of lymphoid tissue pathology in HIV-1 infection.
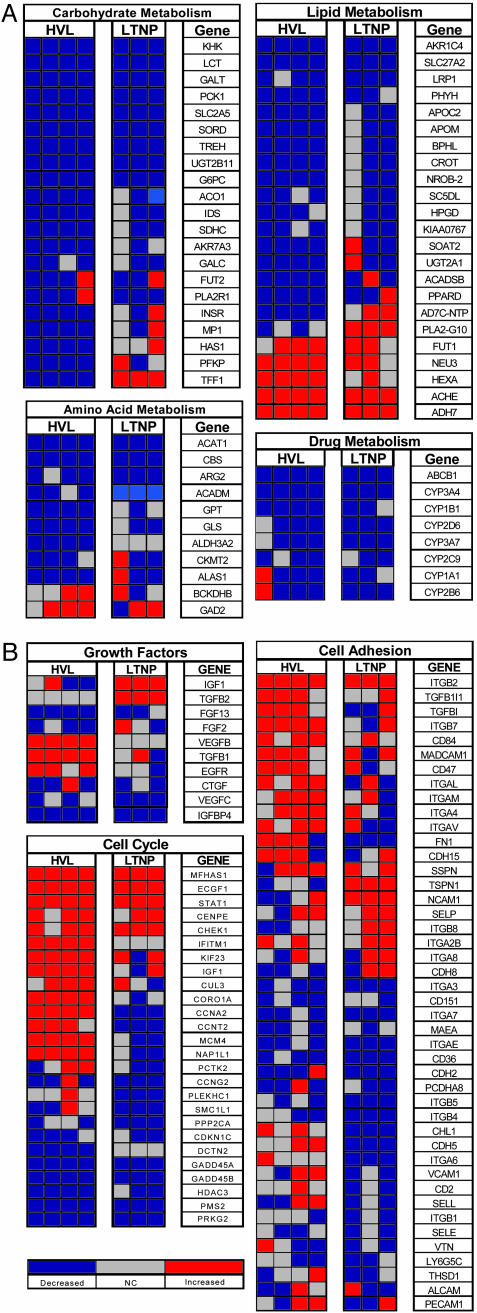
Digestion and Lipid Metabolism-Associated Gene Expression Is Down-Regulated in GALT of both LTNP and HVL Patients. Despite viral suppression and maintenance of CD4+ T cell numbers, LTNP patients displayed significant dysregulation of genes associated with lipid metabolism and nutrient absorptive functions (Fig. 3A), similar to the profile of HVL patients. The vast majority of genes involved in metabolic and digestive functions were found in clusters of genes that were down-regulated in both groups, whereas only three genes associated with digestion were up-regulated. Further analysis revealed that a broad range of genes involved in lipid and carbohydrate metabolism as well as genes mediating xenobiotic metabolism (CYP450 family) were similarly down-regulated in both LTNP and HVL patients (Fig. 4A). Genes associated with amino acid metabolism were down-regulated in only HVL patients. Collectively, these data suggest that a decrease in metabolism-associated gene expression may contribute to the development of HIV-1-associated gastrointestinal complications including nutrient malabsorption, and occur independent of the level of viral suppression.
Fig. 4.
Gene expression of selected biological processes in HIV infection. (A) Differential expression of genes involved in absorptive and metabolic functions. Multiple processes associated with enterocyte function were down-regulated (blue) in both HIV-1 infected groups. In addition to decrease in genes that function in carbohydrate, lipid, and amino acid metabolism, molecules involved in drug metabolism were also down-regulated in GALT of HVL and LTNP patients. (B) Altered transcription profiles were apparent in both patient groups in cell cycle, cell adhesion, and growth-associated factors in both HVL and LTNP patient groups. Down-regulation of fibroblast and vascular endothelial growth factors and intestinal mucosal homing marker integrin α-4 was detected in both HVL and LTNP patients.
Dysregulation of Cell Adhesion Molecule, Cell Cycle, and Growth Factor-Associated Gene Expression. HVL patients also displayed increased expression of numerous cell adhesion molecules in the intestinal mucosa, including integrins β7, β2, and αL (Fig. 4B). This was not observed in LTNP patients. Because integrin α4 combines with β7 to form the lymphocyte homing receptor known as LPAM-1 (lymphocyte Peyers Patch adhesion molecule 1), up-regulation in the HVL group may indicate an increase in cell trafficking into the intestinal mucosa. Taken together, these findings suggest that enhanced lymphocyte trafficking to GALT may lead to an increased number of potential HIV-1 targets and impact on the rate of disease progression.
Cell cycle-associated gene expression was substantially altered in association with HIV-1 infection in both HVL and LTNP patients (Fig. 4B). Whereas STAT-1 was up-regulated in both patient groups, indicating an increased signaling, transcription of cyclin A2 was increased in HVL patients but down-regulated in LTNP patients. However, cyclin G2 and CDKNC1, GADD45B, and PRKG2 were down-regulated in both patient groups. Expression of growth factors in GALT was similarly discordant between HVL and LTNP patients, although both groups displayed down-regulation of vascular endothelial and fibroblast growth factors, in agreement with previously published reports in the SIV model (4). Given the complexity of resident cells in the small intestine, these data suggest that dysregulation of cell cycle progression may impact proliferation and maturation of multiple cell types within the intestinal mucosa, which could contribute to disruption of both the structure and function of GALT.
Supporting Information. For further details, see Supporting Text, Data Set, and Fig. 5, which are published as supporting information on the PNAS web site.
Discussion
We have investigated clinical and molecular characteristics of the mucosal immune system of LTNP patients that distinguished them from HVL patients and uninfected healthy controls. We detected slightly elevated levels of CD4+ T cells and HIV-1 Gag-specific CD8+ T cell responses in intestinal mucosal tissue of LTNP patients, indicating a functional mucosal immune system (Fig. 2) that may contribute to the viral suppression observed at the mucosal site. In comparison, the mucosal immune system appeared to be impaired in HVL patients, as evidenced by severe CD4+ T cell depletion and low HIV-1-specific mucosal CD8+ T cell responses. Loss of mucosal CD4+ T cell helper activity may contribute substantially to the impairment of virus-specific CD8+ T cell activity. Expression of genes regulating lymphocyte activation and inflammation was dominant in the mucosal tissue of HVL, but not LTNP patients, suggesting that chronic immune activation was ongoing in GALT, potentially leading to mucosal tissue injury and dysfunction (22). Thus, elevated immune and inflammatory responses characterize chronic HIV-1 infection in vivo.
Although severe CD4+ T cell depletion is well documented at mucosal effector sites and not in peripheral blood during primary HIV-1 infection, it is not known whether LTNP patients experience this early loss of mucosal CD4+ T cells. Given the maintenance of mucosal CD4+ T cells and viral suppression in LTNP patients, it was surprising to find that numerous genes associated with active transport, lipid, amino acid, and carbohydrate metabolism, epithelial barrier maintenance, and digestive functions were down-regulated in both LTNP and HVL patients (Fig. 4A). The observation of down-regulation in transcription of metabolic factors in both patient groups may underscore a general decrease in digestive and adsorptive functions that results from chronic HIV-1 infection-associated damage to the gastrointestinal tissue. Multiple members of the cytochrome P450 family were down-regulated in both groups, indicating that metabolism of toxic compounds may have also been impaired. This finding suggests that optimal metabolism and efficacy of many antiretroviral drugs may be substantially compromised in patients receiving ART as many of these gene products are involved in drug metabolism (23, 24).
The loss of expression of metabolic genes may be a downstream effect of an altered expression of growth factors (Fig. 4B) in intestinal mucosa that are required for normal maturation of enterocytes. In the SIV model, multiple growth factors as well as genes mediating intestinal epithelial (enterocyte) repair and regeneration have been shown to be dysregulated in GALT during primary acute and chronic stages of infection (6–10). Moreover, SIV-infected macaque studies have demonstrated impaired nutrient digestive and adsorptive functions during primary SIV infection (6). Down-regulation of growth factors may compromise the ability to repair and maintain epithelial barrier function, thereby resulting in decreased absorptive and digestive functions. Thus, the insidious effects of HIV-1 infection in LTNP patients may eventually contribute to the loss of viral suppression, dysfunctional mucosal immune responses, and disease progression. Future investigations of the mucosal immune system in LTNP patients and individuals with primary HIV-1 infection and gene expression profiling of isolated mucosal cell types will provide unique opportunities to identify molecular targets for the development of effective vaccines and therapeutic strategies.
Supplementary Material
Acknowledgments
We thank patients for their participation, the nursing staff for their support, and the School of Medicine Microarray Core Facility and the Lucy Whittier Molecular Core Facility for technical support. This study was supported by National Institutes of Health Grant DK-61297, Northern California Center for AIDS Research Grant PHS-AI49366, and University-Wide AIDS Research Program Grant UARP D03-D-408).
Author contributions: S.S. and S.D. designed research; S.S., M.G., E.R., J.F., and T.P. performed research; S.S., M.G., M.D.G., J.F., T.P., and S.D. analyzed data; S.S., M.D.G., and S.D. wrote the paper; J.F. contributed to patient study design; and S.D. identified objectives and designed and directed study.
Abbreviations: LTNP, long-term nonprogressor; HVL, high viral load; ART, antiretroviral therapy; SIV, simian immunodeficiency virus; GALT, gut-associated lymphoid tissue.
References
- 1.Valdez, H., Carlson, N. L., Post, A. B., Asaad, R., Heeger, P. S., Lederman, M. M., Lehmann, P. V. & Anthony, D. D. (2002) AIDS 16, 1113–1118. [DOI] [PubMed] [Google Scholar]
- 2.Paroli, M., Propato, A., Accapezzato, D., Francavilla, V., Schiaffella, E. & Barnaba, V. (2001) Immunol. Lett. 79, 127–129. [DOI] [PubMed] [Google Scholar]
- 3.Guadalupe, M., Reay, E., Sankaran, S., Prindiville, T., Flamm, J., McNeil, A. & Dandekar, S. (2003) J. Virol. 77, 11708–11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley, J. M., Schacker, T. W., Ruff, L. E., Price, D. A., Taylor, J. H., Beilman, G. J., Nguyen, P. L., Khoruts, A., Larson, M., Haase, A. T. & Douek, D. C. (2004) J. Exp. Med. 200, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrovas, C., Mueller, Y. M. & Katsikis, P. D. (2004) Curr. HIV Res. 2, 153–162. [DOI] [PubMed] [Google Scholar]
- 6.Heise, C., Miller, C. J., Lackner, A. & Dandekar, S. (1994) J. Infect. Dis. 169, 1116–1120. [DOI] [PubMed] [Google Scholar]
- 7.Stone, J. D., Heise, C. C., Miller, C. J., Halsted, C. H. & Dandekar, S. (1994) AIDS 8, 1245–1256. [DOI] [PubMed] [Google Scholar]
- 8.George, M. D., Sankaran, S., Reay, E., Gelli, A. C. & Dandekar, S. (2003) Virology 312, 84–94. [DOI] [PubMed] [Google Scholar]
- 9.George, M. D., Reay, E., Sankaran, S. & Dandekar, S. (2005) J. Virol. 79, 2709–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heise, C., Vogel, P., Miller, C. J., Halsted, C. H. & Dandekar, S. (1993) Am. J. Pathol. 142, 1759–1771. [PMC free article] [PubMed] [Google Scholar]
- 11.Komanduri, K. V., Viswanathan, M. N., Wieder, E. D., Schmidt, D. K., Bredt, B. M., Jacobson, M. A. & McCune, J. M. (1998) Nat. Med. 4, 953–956. [DOI] [PubMed] [Google Scholar]
- 12.Zhong, S., Li, C. & Wong, W. H. (2003) Nucleic Acids Res. 31, 3483–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leutenegger, C. M., Mislin, C. N., Sigrist, B., Ehrengruber, M. U., Hofmann-Lehmann, R. & Lutz, H. (1999) Vet. Immunol. Immunopathol. 71, 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewar, R. L., Highbarger, H. C., Sarmiento, M. D., Todd, J. A., Vasudevachari, M. B., Davey, R. T., Jr., Kovacs, J. A., Salzman, N. P., Lane, H. C. & Urdea, M. S. (1994) J. Infect. Dis. 170, 1172–1179. [DOI] [PubMed] [Google Scholar]
- 16.Olsson, J., Poles, M., Spetz, A. L., Elliott, J., Hultin, L., Giorgi, J., Andersson, J. & Anton, P. (2000) J. Infect. Dis. 182, 1625–1635. [DOI] [PubMed] [Google Scholar]
- 17.Anton, P. A., Mitsuyasu, R. T., Deeks, S. G., Scadden, D. T., Wagner, B., Huang, C., Macken, C., Richman, D. D., Christopherson, C., Borellini, F., et al. (2003) AIDS 17, 53–63. [DOI] [PubMed] [Google Scholar]
- 18.Li, Q., Schacker, T., Carlis, J., Beilman, G., Nguyen, P. & Haase, A. T. (2004) J. Infect. Dis. 189, 572–582. [DOI] [PubMed] [Google Scholar]
- 19.Motomura, K., Toyoda, N., Oishi, K., Sato, H., Nagai, S., Hashimoto, S., Tugume, S. B., Enzama, R., Mugewa, R., Mutuluuza, C. K., et al. (2004) Int. Immunopharmacol. 4, 1829–1836. [DOI] [PubMed] [Google Scholar]
- 20.Geiss, G. K., Bumgarner, R. E., An, M. C., Agy, M. B., van't Wout, A. B., Hammersmark, E., Carter, V. S., Upchurch, D., Mullins, J. I. & Katze, M. G. (2000) Virology 266, 8–16. [DOI] [PubMed] [Google Scholar]
- 21.van't Wout, A. B., Lehrman, G. K., Mikheeva, S. A., O'Keeffe, G. C., Katze, M. G., Bumgarner, R. E., Geiss, G. K. & Mullins, J. I. (2003) J. Virol. 77, 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGowan, I., Elliott, J., Fuerst, M., Taing, P., Boscardin, J., Poles, M. & Anton, P. (2004) J. Acquir. Immune Defic. Syndr. 37, 1228–1236. [DOI] [PubMed] [Google Scholar]
- 23.Wacher, V. J., Silverman, J. A., Zhang, Y. & Benet, L. Z. (1998) J. Pharm. Sci. 87, 1322–1330. [DOI] [PubMed] [Google Scholar]
- 24.Ingelman-Sundberg, M. (2004) Naunyn Schmiedebergs Arch. Pharmacol. 369, 89–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.