Abstract
When classifying breeds of dogs, the accuracy of classification significantly affects breed identification and dog research. Using images to classify dog breeds can improve classification efficiency; however, it is increasingly challenging due to the diversities and similarities among dog breeds. Traditional image classification methods primarily rely on extracting simple geometric features, while current convolutional neural networks (CNNs) are capable of learning high-level semantic features. However, the diversity of dog breeds continues to pose a challenge to classification accuracy. To address this, we developed a model that integrates multiple CNNs with a machine learning method, significantly improving the accuracy of dog images classification. We used the Stanford Dog Dataset, combined image features from four CNN models, filtered the features using principal component analysis (PCA) and gray wolf optimization algorithm (GWO), and then classified the features with support vector machine (SVM). The classification accuracy rate reached 95.24% for 120 breeds and 99.34% for 76 selected breeds, respectively, demonstrating a significant improvement over existing methods using the same Stanford Dog Dataset. It is expected that our proposed method will further serve as a fundamental framework for the accurate classification of a wider range of species.
Keywords: dog breed classification, convolutional neural network, support vector machine, multi-network integration, feature selection, Stanford dog dataset
1. Introduction
Dogs were one of the earliest domestic animals and have a diversity of phenotypes. Currently, there are more than 400 dog breeds worldwide [1], and 283 breeds of them have been registered with the AKC (American Kennel Club; https://www.akc.org (accessed on 22 October 2024)). An accurate classification of dog breeds is crucial in various fields, including veterinary diagnosis, genetic disease research and pet care [2]. However, it is becoming increasingly difficult due to the diversities and similarities among dog breeds [3,4], which rely heavily on expert experiences. Therefore, identifying dog breeds easily, accurately and cost-effectively is a fascinating challenge for dog breeders, managers or fanciers.
To address the challenge of dog image identification, several methods have been proposed, which can be categorized into three main groups.
The first group consists of machine learning methods that focus on geometric features. These methods primarily involve training on geometric features extracted from dog face images and classifying these features using machine learning techniques such as principal component analysis (PCA) [5,6,7]. However, some of these methods were tested on a limited number of dog breeds (35 breeds) [5,7], while another used a larger number of dog breeds but achieved insufficient accuracy (67% for 133 breeds) [6]. PCA is a dimensionality reduction technique that aims to highlight patterns in data by emphasizing variance and capturing strong patterns in high-dimensional data. However, PCA’s effectiveness is predicated on the assumption that the data can be embedded in a globally linear or approximately linear low-dimensional space. Moreover, PCA focuses on the total variance in the explanatory variables, which does not fully reflect the amount of information, and the classification information in the original data is not fully utilized. The compressed data may even be detrimental to pattern classification.
The second group is based on convolutional neural networks (CNN) [8,9,10,11,12,13], which typically employ a single CNN model for the Stanford Dog Dataset, with most achieving an accuracy rate up to 80%. For instance, VGGNet increases network depth by using multiple 3 × 3 convolutional filters while reducing model parameters. The NIN model combines MLP and convolution, using more complex micro neural network structures in place of traditional convolutional layers. These models extract high-level semantic information to improve classification performance but may overlook contextual information around convolution and pooling kernels, leading to the loss of some feature information.
The third group combines CNN with machine learning, where several studies focusing on improving CNN models [14,15,16,17] have delivered better accuracy rates (up to 90%) than single CNN models. However, the classification accuracy is still hindered by the diversity of dog breeds. When applied to birds, cats and sheep, these models have achieved classification rates of 95% [18,19], 80% [20,21,22] and 85% [23,24,25], respectively. Notably, when using the combination of a CNN and Support Vector Machine (SVM) for flower identification, a satisfactory accuracy rate of 97% [26,27] was achieved. However, whether the accuracy of classification can be improved in dog image classification remains to be further explored.
To address this issue, we initiated this study to comprehensively research the integration of multiple CNNs and machine learning methods, with the aim of improving dog image classification accuracy. Additionally, the model was combined with dimension reducing and feature selection processing to optimize the exaction and fusion of image features. The experimental results demonstrate that our proposed model outperforms existing methods to achieve better identification efficiency.
The key contributions of this paper are given below: We propose a new method to combine CNN models and a machine learning model for dog image classification; we increased the accuracy of dog image classification to over 95%; and we used transfer learning for the CNN model training process which improved the efficiency and accuracy of this task. The rest of this paper is structured as follows: Section 2 introduces the materials and methods, including data sources, data preprocessing and model architecture; Section 3 introduce the experimental setup including parameter details; Section 4 analyzes the experimental results and discusses the selection of model combination; and finally, Section 5 discusses the findings and limitations of the study, concludes the research and explores its significance and potential applications in related fields.
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
We used the Stanford Dogs Dataset [28] to train and evaluate our model, which is named DataSet1 for convenience. DataSet1 includes 20,580 annotated dog images from 120 breeds, and each breed has about 180 images. Sample images are shown in Figure 1.
Figure 1.
Sample images of Stanford Dog Dataset. (A) Golden retriever; (B) Shih-Tzu; (C) Old English sheepdog; (D) American Eskimo dog.
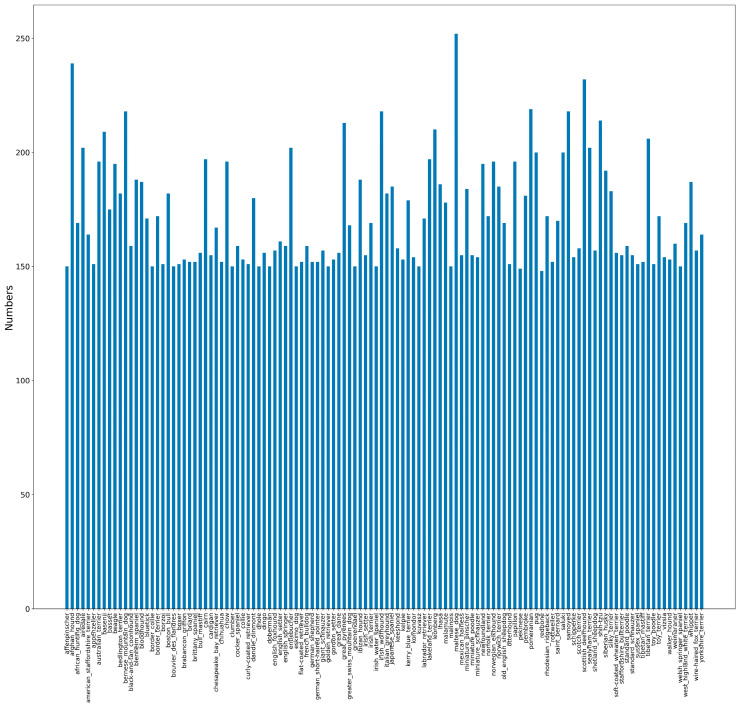
The distribution of the number of dogs for each breed is shown in Figure 2. Among these 120 breeds, Redbone has the lowest number of images with 148 and Maltese has the largest number of images with 252. To mitigate the effects of uneven data distribution on accuracy, 120 images were manually filtered for each breed according to image size, aspect ratio and background ratio to form a new dataset named DataSet2. In addition, preprocessing operations were performed on the images, including image size resetting, center cropping, and normalization. The reset image size was set according to requirements of different networks. Both datasets are divided into training and testing sets according to the ratio of 8:2 and trained separately.
Figure 2.
The distribution of images of each breed for Stanford Dog Dataset.
2.2. Proposed Architecture Details
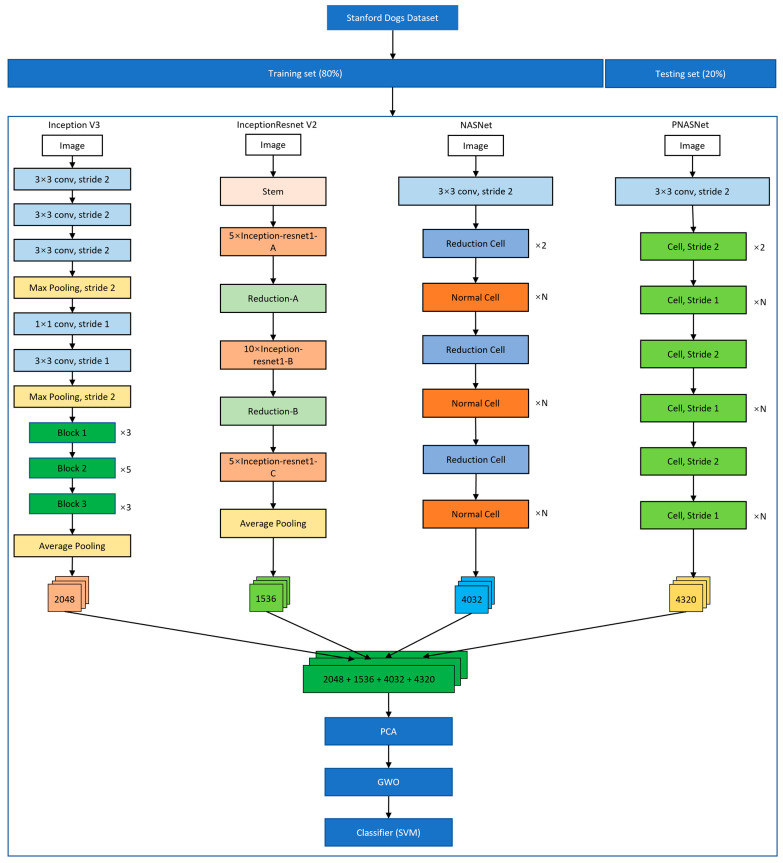
The model we propose is shown in Figure 3, which consists of three main steps as follows.
Figure 3.
The architecture of proposed model. Dataset1 and Dataset2 are divided into an 8:2 ratio separately, 80% of the data are used for training the model and 20% is used for testing its performance. Four CNN models including Inception V3, InceptionResNet V2, NASNet and PNASNet are fine-tuned using transfer learning approach and the features before fully connected layer are extracted, respectively. The numbers 2048, 1536, 4032 and 4320 represent the number of extracted features before fully connected layer of Inception V3, InceptionResNet V2, NASNet and PNASNet, respectively, and then these features are concatenated and flattened. PCA is used to reduce the feature size and GWO is used to select the specific features. Finally, SVM is used to perform the classification task.
2.2.1. Feature Extraction Using Four Known CNN Models
We used Inception V3, InceptionResNet V2, NASNet and PNASNet [29,30,31,32,33], which are independent fine-tuning models and have a high performance in ImageNet competitions to extract features before fully connected layers, respectively. Inception V3 is a part of GoogLeNet, which is a deep neural network model based on the Inception module launched by Google. Inception is used to assemble multiple convolution or pooling operations together into a network module [30]. InceptionResNet V2 is an improvement over Inception V3 by introducing residual connections, which reduces computational costs and speeds up network training [31]. NASNet is a new architecture proposed by Google in 2018, which combines features learned in ImageNet classification with the Faster-RCNN framework to exceed the best predictive performance of previously released COCO object detection task. The mean accuracy rate (mAP) of the model was 43.1%, which has 4% improvement over the best results published [33]. PNASNet is an improved model based on NASNet and proposes a search strategy using Sequential model-based optimization (SMBO). Compared with NASNet, PNASNet is 5 times more efficient, and significantly reduces the requirements of computation resources [32]. The size of each input image in this research was set to 299 × 299 pixels for Inception V3 and InceptionResNet V2, and 331 × 331 pixels for NASNet and PNASNet.
2.2.2. Feature Fusion and Feature Selection Methods
The features of the above four models extracted were combined together, and then filtered using feature selection methods including principal component analysis (PCA) and the gray wolf optimization (GWO) algorithm [34] to obtain specific features. Feature selection can help to improve the accuracy of classification [35]. PCA is a linear dimensionality reduction method, which can reduce the storage space required and improve the transmission efficiency. We achieved PCA using Python’s sklearn.decomposition module. GWO is a meta-heuristic algorithm inspired by gray wolves in nature, which mimics their leadership hierarchy and hunting process. The GWO algorithm designs four agents for simulating the leadership hierarchy, which, from high to low, are gray wolf , , and . Here, we set the population size to 30, the number of iterations to 5 and the search range from −1 to 1, all these considered as initial parameters of the GWO algorithm, and the number of rotations was set to 30. In this study, we first used PCA and then used the GWO algorithm for feature selection.
2.2.3. Classification for Dog Images
Support vector machine (SVM) was adopted to classify the above selected features, which was originally designed for binary classification problems. When dealing with multi-class problems, it is necessary to construct a suitable multi-class classifier. At present, there are two methods available: (1) The direct method, which can directly modify the objective function and combine the parameter solutions of multiple classification surfaces into an optimization problem. It is simple but difficult to implement because of its high computational complexity and is only suitable for small problems. (2) The indirect method, which mainly realizes multi-classification by combining multiple binary classifiers. Two common methods are supported, one-against-one and one-against-all [36]. In this study, we used the one-against-one strategy, which was implemented using Python’s sklearn.svm module, and we selected the RBF kernel function. In our experiments, the default parameters of the SVM were used without any hyperparameter tuning to ensure fair and reproducible results. The default parameters can perform consistently on multiple benchmark datasets and can also provide a reasonable baseline performance to focus on other aspects of the algorithm.
3. Experimental Setup
3.1. Training and Implementation Details
We integrated the three processes mentioned earlier to develop our proposed model, successfully combining CNNs with machine learning techniques. The code is implemented in Anaconda Python 3.7, and the deep learning framework is PyTorch (https://pytorch.org/ (accessed on 16 November 2024)). The whole leaning procedure is runnable on a Linux machine which is equipped with an Intel Xeon CPU with 24 GB memory and another Linux machine equipped with an NVIDIA GV100GL GPU with 32 GB memory.
All CNN models were trained using the transfer learning [37] approach in this study. The size of each input image was set to 299 × 299 pixels for Inception V3 and InceptionResNet V2, and 331 × 331 pixels for NASNet and PNASNet. These four proposed CNN models for dog breed classification were trained using a set of carefully selected hyperparameters. A batch size of 16 was chosen to balance computational efficiency and training stability. The initial learning rate was set to , and this was adjusted dynamically using a scheduler that reduced the rate by a factor of 0.1 upon detecting a plateau in validation performance. The Stochastic Gradient Descent (SGD) optimizer, known for its fast convergence and freedom from local optima, was utilized with the momentum of 0.9. The loss function we used is Cross Entropy for a multi-class classification task. The network was trained for 50 epochs. The epoch and batch size parameter values were chosen after our experimental results, and the momentum and learning rate parameter values were set according to reference [26,27]. The relevant parameters are shown in Table 1. After employing transfer learning, the features before fully connected layers of Inception V3, InceptionResNet V2, NASNet and PNASNet were extracted, with dimension of 2048, 1536, 4032 and 4320 respectively. These features were then used for feature fusion, selection and breed classification.
Table 1.
Parameter values of CNN architectures.
| Software Used | Model | Image Size | Optimizer | Momentum | Minibatch | Learning Rate |
|---|---|---|---|---|---|---|
| Anaconda Python 3.7 PyTorch v1 | Inception V3 | 299 × 299 | SGD | 0.9 | 16 | |
| InceptionResnet V2 | ||||||
| NASNet | 331 × 331 | |||||
| PNASNet |
Data augmentation techniques were employed to enhance model robustness, including random rotations, flips and intensity variations; here, we used random resized crop, center crop and random flip. We normalize the input images before training. During training, the dataset from Stanford Dog Dataset (http://vision.stanford.edu/aditya86/ImageNetDogs/ (accessed on 22 October 2024)) named Dataset1 and images after manual filtering named Dataset2 were split into training and testing sets in an 80–20 ratio, respectively. Input images were normalized so that they have a mean of zero and a unit variance.
3.2. Performance Metric
In this study, the evaluation metric used for the analysis of experiments was accuracy. In the following equation, TP represents True positives, TN represents true-negative, FP means false-positive and FN means false-negative [27]:
| (1) |
4. Results
To enhance the model’s performance and the accuracy of the classification of the dog breeds, we first tried four known CNN models with good classification performances for dog images. Then, we tried to extract the features before the fully connected layer with the known CNN models mentioned and implemented classification with SVM, we also compared the effect of using a single model to extract the features and multiple models to extract the features and then fusing them on the classification accuracy. After feature fusion, we also used two feature selection methods, PCA and GWO. For the PCA method, the effects of feature dimension reduction to 5000, 4000, 3000 and 2000 on the results were compared.
4.1. Comparative Analysis of Single CNN Models
The single CNN model can achieve better accuracy, but not the best. We used four transfer-learning CNNs to perform the classification, including Inception V3, InceptionResNet V2, NASNet and PNASNet. The results show that NASNet and PNASNet have better classification accuracies than Inception V3 and InceptionResNet V2 (93.03% and 89.64% vs. 84.33% and 85.97% in DataSet1, 93.96% and 89.27% vs. 86.25% and 84.13% in DataSet2), and NASNet has the highest accuracy for both datasets. The results from the two datasets are closely aligned, with DataSet2 (the manually filtered dataset) showing minimal significant benefits over the other. Table 2 summarizes the classification accuracy of each CNN model.
Table 2.
The performance of four different transfer learning CNN models.
| Models | Test Accuracy (Dataset1) | Test Accuracy (Dataset2) |
|---|---|---|
| Inception V3 | 84.33% | 86.25% |
| InceptionResNet V2 | 85.97% | 84.13% |
| PNASNet | 89.64% | 89.27% |
| NASNet | 93.03% | 93.96% |
4.2. The Evaluation of Fusion CNNs and SVM Classification
The combined multiple CNNs and SVM improved the classification accuracy compared with the single model shown above. In our study, fusing the the Inception V3, InceptionResnet V2, NASNet and PNASNet with SVM separately achieved a better effect in both datasets. Furthermore, fusing the merged four CNN models with SVM achieved a higher accuracy of 94.1% for 120 breeds in DataSet1 and the highest accuracy of 94.9% for 120 breeds in DataSet2. Meanwhile, we assembled any two of the CNNs mentioned above and fused them with SVM; for example, the PNASNet and NASNet (two higher accuracy models) with SVM has higher accuracy (94.2% in DataSet1 and 94.7% in DataSet2) than the combination of Inception V3 and InceptionResNet V2 (two lower-accuracy models, with 92.6% accuracy in DataSet1 and 93.5% in DataSet2). The results showed that a combination with an individual CNN model with a high accuracy overall has a better performance. Table 3 summarizes the classification accuracy of the different models.
Table 3.
Comparative result of CNN and SVM classification.
| Methods | Num of Features | Test Accuracy (Dataset1) | Test Accuracy (Dataset2) |
|---|---|---|---|
| Inception V3 + SVM | 2048 | 89.50% | 91.30% |
| InceptionResNet V2 + SVM | 1536 | 92.00% | 92.10% |
| PNASNet + SVM | 4320 | 93.70% | 94.30% |
| NASNet + SVM | 4032 | 93.30% | 94.50% |
| Inception V3 and InceptionResNet V2 + SVM | 3584 | 92.60% | 93.50% |
| NASNet and InceptionResNet V2 + SVM | 5568 | 93.20% | 94.60% |
| PNASNet and InceptionResNet V2 + SVM | 5856 | 93.80% | 94.20% |
| NASNet and Inception V3 + SVM | 6080 | 93.20% | 94.50% |
| Inception V3&PNASNet + SVM | 6368 | 93.80% | 94.20% |
| NASNet and PNASNet + SVM | 8352 | 94.20% | 94.70% |
| Inception V3 and NASNet and PNASNet and InceptionResNet V2 + SVM | 11,936 | 94.10% | 94.90% |
Bold means that in both datasets, four CNNs and SVM model have better accuracy than single CNN and SVM.
4.3. Ablation Analysis of Our Model
We conducted a multi-round feature selection experiment using various feature sizes, such as 5000, 4000, 3000, and 2000, to assess classification accuracy. The experiment first employed PCA, followed by a combination of PCA and the GWO algorithm. PCA together with the GWO feature selection algorithm improved the accuracy on both datasets, which reached 94.3% on DataSet1 and 95.24% on DataSet2, respectively. Based on the combined four CNN modules above, we used PCA to reduce the features to 5000, 4000, 3000 and 2000, respectively, then used GWO to select features. To evaluate the effective influence of PCA or GWO, we validated the combined patterns of different strategies, for example, without PCA or GWO, and the results indicate that GWO has a greater impact on accuracy than PCA (Table 4).
Table 4.
Ablation result of four CNN and SVM classification with feature selection.
| Methods | Number of Features After PCA | Number of Features After GWO | Test Accuracy (Dataset1) | Test Accuracy (Dataset2) | |
|---|---|---|---|---|---|
| Dataset1 | Dataset2 | ||||
| Inception V3 and NASNet and PNASNet and InceptionResNet V2 + SVM | 5000 | - | - | 94.00% | 94.40% |
| 4000 | - | - | 94.00% | 94.40% | |
| 3000 | - | - | 94.00% | 94.40% | |
| 2000 | - | - | 94.00% | 94.40% | |
| 5000 | 2508 | 2499 | 94.30% | 95.24% * | |
| 4000 | 2088 | 2045 | 94.45% | 95.00% | |
| 3000 | 1532 | 1483 | 94.23% | 95.07% | |
| 2000 | 1038 | 1004 | 94.35% | 94.97% | |
| - | 5991 | 5974 | 94.28% | 95.03% | |
* Using only PCA cannot improve the results, but after dimensionality reduction to 5000 using PCA and adding GWO algorithm, the accuracy was better in both datasets.
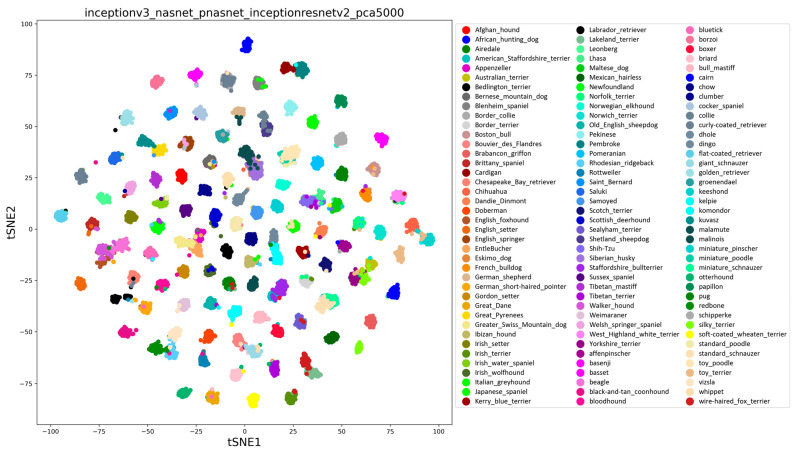
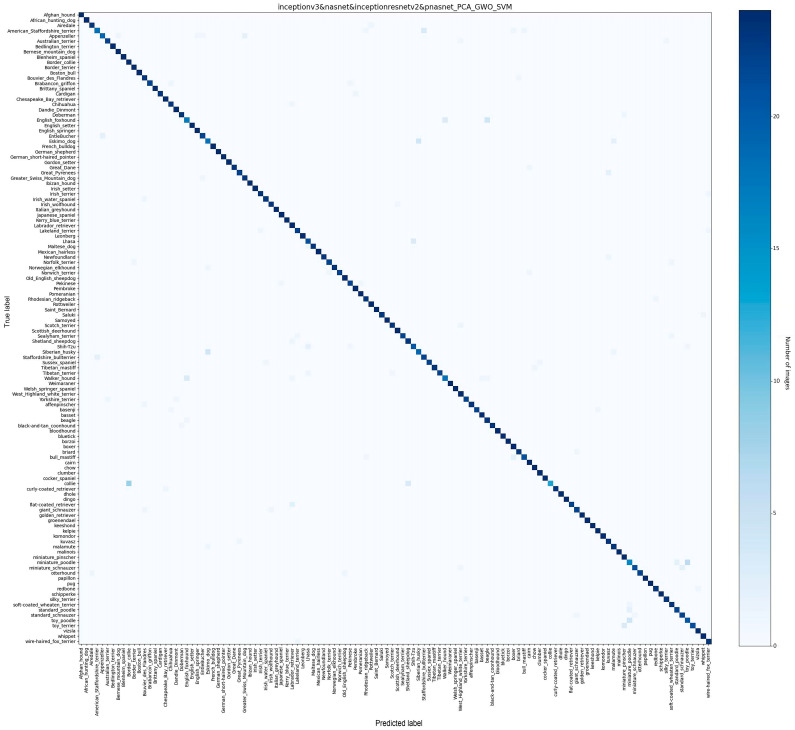
The accordingly extracted features of maximum classification accuracy (95.24%) visualized by the t-SNE method are shown in Figure 4, which clearly shows the different classifications of 120 breeds, and the corresponding confusion matrix (Figure 5) shows the same pattern.
Figure 4.
The visualized t-SNE map of extracted features from the training dataset of the 120 breeds from the Stanford Dog Dataset, which achieved the maximum classification accuracy of 95.24% on the testing dataset.
Figure 5.
The confusion matrix of maximum classification accuracy (95.24%) for the test data for 120 breeds from the Stanford Dog Database. The y axis shows the actual labels for the dog breeds, while the x axis shows the predicted labels.
4.4. Summary and Analysis of Results
In total, using the combined method mentioned above for the identification of 120 dog breeds, the average accuracy of each breed for the best model can be seen in Figure 2 and the accuracy statistics’ distribution can be seen in Table 5. There are 76 breeds with more than 95% average classification accuracy, of which 48 breeds can be distinguished perfectly with an average accuracy of 100%. Three breeds, including the English foxhound, Miniature poodle and Collie, have the lowest average accuracies, under 70%. We took a closer look at the English foxhound which has average accuracy of 67%. This breed looks similar to the Beagle and Walker hound, which are difficult to identify even by eye. In addition, the Stanford images of this breed are different in dog size, photo pose or distance. For the Miniature poodle which has average accuracy 66% and the Collie which has average accuracy 56%, the lower accuracy may be mainly caused by the vast inner differences in the breed such as coat color. We used the top 76 breeds (average accuracy above 95%) to evaluate our model, and the accuracy was up to 99.34%.
Table 5.
Statistics of accuracy distribution of each breed.
| Accuracy Range | Num of Breeds |
|---|---|
| 1 | 48 |
| 0.95~0.99 | 28 |
| 0.90~0.95 | 28 |
| 0.80~0.90 | 10 |
| 0.50~0.75 | 6 |
In addition, we developed an online dog image classification tool called DogVC. This tool integrates the mentioned CNN models, and was integrated into iDog [38] for NGDC [39], which is a database integrating genome, phenotype, disease and variation information of Canis lupus familiaris. DogVC is available at https://ngdc.cncb.ac.cn/dogvc/ (accessed on 22 October 2024); users can upload a dog image, and then the prediction result will be shown.
5. Discussion
The classification of dog breed images is a type of fine-grained image classification. The proposed model has the advantages of comprehensive feature coverage, high-quality feature output and complete automatic implementation. One of the most characteristic features of this study is that we proposed a comprehensive multi-CNNs model architecture and conducted experiments of the factors that affect accuracy including model combination, feature size and classification method. We achieved a 95.24% accuracy, which is better than other reported deep learning methods using the same Stanford Dog Dataset of 120 breeds regardless of various used hyper parameters (Table 6). Three improvements contribute to the result: Compared to the existing methods that only use a single CNN model as backbone model, we combined four CNN models and concatenated the extracted features. Our results show that the fusion of high-performance CNN models has a higher accuracy than single CNN model. (2) Existing methods directly use extracted features for classifying, while we use two feature selection methods, PCA and GWO, to obtain improved features. Our results show that PCA in combination with GWO shows better improvements in accuracy than PCA or GWO alone. (3) Most of the existing methods directly use the SoftMax function to classify dog breeds, while we use SVM. Our results show that the combination of CNN and SVM can improve accuracy. The whole set of results show the feasibility and effectiveness of our proposed model. Meanwhile, the number of images may have no direct effect on breed accuracy (Figure 2).
Table 6.
The results of existing studies using Stanford Dog Dataset.
| Study | Year | Model/Method | Backbone CNN | Hyperparameters | Feature Selection | Classification | Accuracy |
|---|---|---|---|---|---|---|---|
| [14] | 2016 | Fully Convolutional Attention Networks (FCANs) | ResNet-50 | Initial learning rate: 0.01, batch size: 512 | no | SoftMax | 88.90% |
| [15] | 2017 | Recurrent Attention Convolutional Neural Network (RA-CNN) | VGG-16 | - | no | SoftMax | 87.30% |
| [40] | 2018 | Fine-tuned CNN | ResNet-50 | - | no | SoftMax | 89.66% |
| [8] | 2018 | Fine-tuned CNN | Inception-Resnet V2 | Initial learning rate: 0.1, optimizer: Nesterov, batch size: 64 | no | SoftMax | 90.69% |
| [13] | 2021 | Fine-tuned CNN | ResNet-50 | learning rate:0.0001 | no | SoftMax | 90.12% |
| [16] | 2019 | Weakly Supervised Data Augmentation Network (WS-DAN) | Inception V3 | Initial learning rate: 0.001, optimization: SGD, momentum: 0.9, batch size: 16 | no | - | 92.20% |
| [17] | 2021 | Vision Transformers (ViT) | ViT-B/16 | - | no | MLP | 93.20% |
| Proposed Approach | 2022 | multi-CNNs and Feature selection and SVM | Inception V3, InceptionResnet V2, NASNet and PNASNet | learning rate: 0.001, optimization: SGD, momentum: 0.9, batch size: 16 | PCA and GWO | SVM | 95.24% * |
* Compared with other previous methods used on Stanford Dogs Dataset, our approach has better accuracy.
Historically, dog image classification has focused on dog facial geometry features such as dog face profile or facial local features such as ear shape. Due to complexity of localized features, classification tasks were only trained on limited dog breeds (35 breeds) and/or images (less than 1000 images) followed by classical machining learning methods such as PCA. Since 2012, the deep learning methods show best-in-class performance in several application tasks, which facilitate the application to dog image classification. By using the Stanford Dog Dataset, single CNN models can classify 120 breeds with state-of-the-art accuracy.
6. Conclusions
We started with the image data and manually filtered it based on background size and dog coat color so that there were the same number of images for each breed of dog in Dataset2; the model classification was slightly better for Dataset2 than for the raw data. The performance of multiple CNNs combined with SVM is superior to that of a single CNN combined with SVM. Two feature selection methods, PCA and GWO, also showed an improvement in dog breed classification accuracy. Selecting different CNN models for feature extraction also impacts the results.
Despite these positive outcomes from the study, there are also some limitations to consider. Firstly, the model’s accuracy may be somewhat compromised by the fact that some breeds have such subtle differences that even the human eye finds it challenging to distinguish among them, such as the English foxhound, Beagle and Walker. Secondly, the model’s accuracy is further affected by breeds with significant internal variations, like the Collie, which exhibits a diverse range of coat colors. Additionally, the complexity of the model needs to be addressed in future work.
Future research could be carried out in the following directions. Firstly, we will continue to monitor and update our proposed model in three aspects: We will incorporate the Tsinghua Dogs Dataset (https://cg.cs.tsinghua.edu.cn/ThuDogs/ (accessed on 22 October 2024)), which contains 70,428 images of 130 breeds, to increase the variety and quantity of image data, thereby enhancing the model’s generalization ability and robustness. (2) We will explore the use of modern architectures, such as autoencoders or large models in the computer vision field like Vision Transformers (ViT) [17], which can perform various vision tasks such as image classification, target detection, image segmentation, pose estimation, face recognition and so on by training on large-scale image data. (3) Extending our model to other animal images, such as cats, sheep and birds, will allow us to assess its scalability and versatility for classification tasks. Secondly, to facilitate the download and use of our model, we have developed and released all codes on BioCode (https://ngdc.cncb.ac.cn/biocode/tool/BT7319 (accessed on 22 October 2024)). And we plan to establish an email group accompanied by discussion workshops to garner feedback and suggestions. At present, our proposed model has been integrated into the iDog database (https://ngdc.cncb.ac.cn/dogvc/ (accessed on 22 October 2024)) for dog image classification, and further developing other applications such as a mobile APP will widely promote the usage of our model. And the model’s explanatory ability also be needed to improve the effectiveness of our applications. Moreover, we advocate for the collaboration of the global canine community research including researchers, veterinarians, dog owners, dog breed experts and data scientists to advance our proposed model. This would include having dog breed experts validate the image classification results using their domain knowledge, data scientists annotate and outline the images to improve classification accuracy and dog owners and veterinarians collect images to enlarge the image scale.
Acknowledgments
We thank Guo-Dong Wang and Ya-Ping Zhang from Kunming Institute of Zoology, Chinese Academy of Sciences, and Yi-Ming Bao, Zhang Zhang, Jing-Fa Xiao from Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation for their constructive suggestions.
Abbreviations
| English Abbreviations | Full English Title |
| CNN | Convolutional Neural Network |
| PCA | Principal Component Analysis |
| GWO | Gray Wolf Optimization |
| SVM | Support Vector Machine |
| AKC | American Kennel Club |
| RBF | Radial Basis Function |
| SGD | Stochastic Gradient Descent |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
| FCANs | Fully Convolutional Attention Networks |
| RA-CNN | Recurrent Attention Convolutional Neural Network |
| WS-DAN | Weakly Supervised Data Augmentation Network |
| ViT | Vision Transformers |
| t-SNE | t-distributed stochastic neighbor embedding |
Author Contributions
Conceptualization, B.T., W.Z. and Y.C.; methodology, B.T. and Y.C.; software, Y.C.; validation, Y.C. and B.T.; formal analysis, Y.C.; investigation, Y.C., G.W., L.L. and Z.D.; resources, B.T. and W.Z.; writing—original draft preparation, Y.C.; writing—review and editing, B.T., X.Z. and W.Z.; visualization, Y.C.; supervision, B.T. and W.Z.; project administration, B.T. and W.Z.; funding acquisition, W.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Stanford Dog Dataset can be downloaded at http://vision.stanford.edu/aditya86/ImageNetDogs/ (accessed on 22 October 2024). DataSet1 (the Stanford Dog Dataset after divided into training set and test set according to 8:2) can be downloaded at https://download.big.ac.cn/idog/dogvc/dataset_120_raw.zip (accessed on 22 October 2024). DataSet2 (the Stanford Dog Dataset after manually filtered) can be downloaded through https://download.big.ac.cn/idog/dogvc/dataset_120_10.zip (accessed on 22 October 2024). All codes can be found at https://download.big.ac.cn/idog/dogvc/code/ or https://ngdc.cncb.ac.cn/biocode/tool/BT7319 (accessed on 22 October 2024) (Here, we provide detailed usage manuals with a video). In addition, all results in this study can be available at https://download.big.ac.cn/idog/dogvc/run_result/ (accessed on 22 October 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences, grant number XDB38050300 and the National Natural Science Foundation of China, grant number 32100506 and 32170678.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ostrander E.A., Wayne R.K., Freedman A.H., Davis B.W. Demographic history, selection and functional diversity of the canine genome. Nat. Rev. Genet. 2017;18:705–720. doi: 10.1038/nrg.2017.67. [DOI] [PubMed] [Google Scholar]
- 2.Simpson R.J., Simpson K.J., VanKavage L. Rethinking dog breed identification in veterinary practice. J. Am. Vet. Med. Assoc. 2012;241:1163–1166. doi: 10.2460/javma.241.9.1163. [DOI] [PubMed] [Google Scholar]
- 3.Parkhi O.M., Vedaldi A., Zisserman A., Jawahar C. Cats and dogs; Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition; Providence, RI, USA. 16–21 June 2012; pp. 3498–3505. [Google Scholar]
- 4.Zou D.-N., Zhang S.-H., Mu T.-J., Zhang M. A new dataset of dog breed images and a benchmark for finegrained classification. Comput. Vis. Media. 2020;6:477–487. doi: 10.1007/s41095-020-0184-6. [DOI] [Google Scholar]
- 5.Chanvichitkul M., Kumhom P., Chamnongthai K. Face recognition based dog breed classification using coarse-to-fine concept and PCA; Proceedings of the 2007 Asia-Pacific Conference on Communications; Bangkok, Thailand. 18–20 October 2007; pp. 25–29. [Google Scholar]
- 6.Liu J., Kanazawa A., Jacobs D., Belhumeur P. Dog breed classification using part localization; Proceedings of the Computer Vision–ECCV 2012: 12th European Conference on Computer Vision; Florence, Italy. 7–13 October 2012; pp. 172–185. Proceedings, Part I 12. [Google Scholar]
- 7.Prasong P., Chamnongthai K. Face-Recognition-Based dog-Breed classification using size and position of each local part, and pca; Proceedings of the 2012 9th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology; Phetchaburi, Thailand. 16–18 May 2012; pp. 1–5. [Google Scholar]
- 8.Ráduly Z., Sulyok C., Vadászi Z., Zölde A. Dog breed identification using deep learning; Proceedings of the 2018 IEEE 16th International Symposium on Intelligent Systems and Informatics (SISY); Subotica, Serbia. 13–15 September 2018; pp. 000271–000276. [Google Scholar]
- 9.Sinnott R.O., Wu F., Chen W. A mobile application for dog breed detection and recognition based on deep learning; Proceedings of the 2018 IEEE/ACM 5th International Conference on Big Data Computing Applications and Technologies (BDCAT); Zurich, Switzerland. 17–20 December 2018; pp. 87–96. [Google Scholar]
- 10.Uno M., Han X.-H., Chen Y.-W. Comprehensive Study of Multiple CNNs Fusion for Fine-Grained Dog Breed Categorization; Proceedings of the 2018 IEEE International Symposium on Multimedia (ISM); Taichung, Taiwan. 10–12 December 2018; pp. 198–203. [Google Scholar]
- 11.Borwarnginn P., Thongkanchorn K., Kanchanapreechakorn S., Kusakunniran W. Breakthrough conventional based approach for dog breed classification using CNN with transfer learning; Proceedings of the 2019 11th International Conference on Information Technology and Electrical Engineering (ICITEE); Pattaya, Thailand. 10–11 October 2019; pp. 1–5. [Google Scholar]
- 12.Lai K., Tu X., Yanushkevich S. Dog identification using soft biometrics and neural networks; Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN); Budapest, Hungary. 14–19 July 2019; pp. 1–8. [Google Scholar]
- 13.Kamdar D. Implementation of pre-trained deep learning model for dog breed classification. Turk. J. Comput. Math. Educ. 2021;12:555–558. [Google Scholar]
- 14.Liu X., Xia T., Wang J., Yang Y., Zhou F., Lin Y. Fully convolutional attention networks for fine-grained recognition. arXiv. 20161603.0676 [Google Scholar]
- 15.Fu J., Zheng H., Mei T. Look closer to see better: Recurrent attention convolutional neural network for fine-grained image recognition; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Honolulu, HI, USA. 21–26 July 2017; pp. 4438–4446. [Google Scholar]
- 16.Hu T., Qi H., Huang Q., Lu Y. See better before looking closer: Weakly supervised data augmentation network for fine-grained visual classification. arXiv. 20191901.09891 [Google Scholar]
- 17.Conde M.V., Turgutlu K. Exploring vision transformers for fine-grained classification. arXiv. 20212106.10587 [Google Scholar]
- 18.Chen X., Wang G. Few-shot learning by integrating spatial and frequency representation; Proceedings of the 2021 18th Conference on Robots and Vision (CRV); Burnaby, BC, Canada. 26–28 May 2021; pp. 49–56. [Google Scholar]
- 19.Ermolov A., Mirvakhabova L., Khrulkov V., Sebe N., Oseledets I. Hyperbolic vision transformers: Combining improvements in metric learning; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; New Orleans, LA, USA. 18–24 June 2022; pp. 7409–7419. [Google Scholar]
- 20.Zhang X., Yang L., Sinnott R. A mobile application for cat detection and breed recognition based on deep learning; Proceedings of the 2019 IEEE 1st International Workshop on Artificial Intelligence for Mobile (AI4Mobile); Hangzhou, China. 24–24 February 2019; pp. 7–12. [Google Scholar]
- 21.Zhou J., Wang S., Chen Y., Sinnott R.O. A Web Application for Feral Cat Recognition through Deep Learning; Proceedings of the Big Data–BigData 2020: 9th International Conference, Held as Part of the Services Conference Federation, SCF 2020; Honolulu, HI, USA. 18–20 September 2020; pp. 85–100. Proceedings 9. [Google Scholar]
- 22.Zhang R. Classification and Identification of Domestic Catsbased on Deep Learning; Proceedings of the 2021 2nd International Conference on Artificial Intelligence and Computer Engineering (ICAICE); Hangzhou, China. 5–7 November 2021; pp. 106–110. [Google Scholar]
- 23.Jwade S.A., Guzzomi A., Mian A. On farm automatic sheep breed classification using deep learning. Comput. Electron. Agric. 2019;167:105055. doi: 10.1016/j.compag.2019.105055. [DOI] [Google Scholar]
- 24.Salama A., Hassanien A.E., Fahmy A. Sheep identification using a hybrid deep learning and bayesian optimization approach. IEEE Access. 2019;7:31681–31687. doi: 10.1109/ACCESS.2019.2902724. [DOI] [Google Scholar]
- 25.Meng X., Tao P., Han L., CaiRang D. Sheep identification with distance balance in two stages deep learning; Proceedings of the 2022 IEEE 6th Information Technology and Mechatronics Engineering Conference (ITOEC); Chongqing, China. 4–6 March 2022; pp. 1308–1313. [Google Scholar]
- 26.Ensari T., METE B.R. Flower Classification with Deep CNN and Machine Learning Algorithms; Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT); Ankara, Turkey. 11–13 October 2019; pp. 1–5. [Google Scholar]
- 27.Toğaçar M., Ergen B., Cömert Z. Classification of flower species by using features extracted from the intersection of feature selection methods in convolutional neural network models. Measurement. 2020;158:107703. doi: 10.1016/j.measurement.2020.107703. [DOI] [Google Scholar]
- 28.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. Imagenet: A large-scale hierarchical image database; Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition; Miami, FL, USA. 20–25 June 2009; pp. 248–255. [Google Scholar]
- 29.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 30.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Going deeper with convolutions; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Boston, MA, USA. 7–12 June 2015; pp. 1–9. [Google Scholar]
- 31.Szegedy C., Ioffe S., Vanhoucke V., Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning; Proceedings of the AAAI Conference on Artificial Intelligence; San Francisco, CA, USA. 4–9 February 2017. [Google Scholar]
- 32.Liu C., Zoph B., Neumann M., Shlens J., Hua W., Li L.-J., Fei-Fei L., Yuille A., Huang J., Murphy K. Progressive neural architecture search; Proceedings of the European Conference on Computer Vision (ECCV); Munich, Germany. 8–14 September 2018; pp. 19–34. [Google Scholar]
- 33.Zoph B., Vasudevan V., Shlens J., Le Q.V. Learning transferable architectures for scalable image recognition; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Salt Lake City, UT, USA. 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- 34.Basak H., Kundu R., Chakraborty S., Das N. Cervical cytology classification using PCA and GWO enhanced deep features selection. SN Comput. Sci. 2021;2:369. doi: 10.1007/s42979-021-00741-2. [DOI] [Google Scholar]
- 35.Chandrashekar G., Sahin F. A survey on feature selection methods. Comput. Electr. Eng. 2014;40:16–28. doi: 10.1016/j.compeleceng.2013.11.024. [DOI] [Google Scholar]
- 36.Weston J., Watkins C. Support vector machines for multi-class pattern recognition; Proceedings of the Esann; Bruges, Belgium. 21–23 April 1999; pp. 219–224. [Google Scholar]
- 37.Weiss K., Khoshgoftaar T.M., Wang D. A survey of transfer learning. J. Big Data. 2016;3:1–40. doi: 10.1186/s40537-016-0043-6. [DOI] [Google Scholar]
- 38.Tang B., Zhou Q., Dong L., Li W., Zhang X., Lan L., Zhai S., Xiao J., Zhang Z., Bao Y. iDog: An integrated resource for domestic dogs and wild canids. Nucleic Acids Res. 2019;47:D793–D800. doi: 10.1093/nar/gky1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Y., Bao Y., Zhang Z., Zhao W., Xiao J., He S., Zhang G., Li Y., Zhao G., Chen R., et al. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022;50:27–38. doi: 10.1093/nar/gkab951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayanzadeh A., Vahidnia S. Modified deep neural networks for dog breeds identification. Preprints. 2018:2018120232. doi: 10.20944/preprints201812.0232.v1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Stanford Dog Dataset can be downloaded at http://vision.stanford.edu/aditya86/ImageNetDogs/ (accessed on 22 October 2024). DataSet1 (the Stanford Dog Dataset after divided into training set and test set according to 8:2) can be downloaded at https://download.big.ac.cn/idog/dogvc/dataset_120_raw.zip (accessed on 22 October 2024). DataSet2 (the Stanford Dog Dataset after manually filtered) can be downloaded through https://download.big.ac.cn/idog/dogvc/dataset_120_10.zip (accessed on 22 October 2024). All codes can be found at https://download.big.ac.cn/idog/dogvc/code/ or https://ngdc.cncb.ac.cn/biocode/tool/BT7319 (accessed on 22 October 2024) (Here, we provide detailed usage manuals with a video). In addition, all results in this study can be available at https://download.big.ac.cn/idog/dogvc/run_result/ (accessed on 22 October 2024).